SUMMARY
This article outlines the protein modules that target methylated lysine histone marks and 5mC DNA marks, and the molecular principles underlying recognition. The article focuses on the structural basis underlying readout of isolated marks by single reader molecules, as well as multivalent readout of multiple marks by linked reader cassettes at the histone tail and nucleosome level. Additional topics addressed include the role of histone mimics, cross talk between histone marks, technological developments at the genome-wide level, advances using chemical biology approaches, the linkage between histone and DNA methylation, the role for regulatory lncRNAs, and the promise of chromatin-based therapeutic modalities.
Various protein modules bind to modified histones and methylated DNA. These impact many cellular processes ranging from gene transcription to DNA replication, recombination, and repair.
OVERVIEW
This article focuses on the readout of histone and DNA methylation posttranslational modifications (PTMs) and their impact on chromatin structure and function. PTMs serve as docking sites for protein reader modules containing attached chromatin modifier and remodeling activities. The additional activities can alter noncovalent contacts within and between nucleosomes, thereby impacting on function. At a specific genomic site, there can be distinct combinations of methylation and other PTMs. The multivalent (more than a single mark) readout of these PTMs impacts on many DNA-templated processes ranging from gene transcription to DNA replication, recombination, and repair. Dysregulation of the readout due to mutated readers can result in aberrant gene expression patterns and/or genomic alterations, facilitating the onset of disease. A new generation of epigenetic drugs is being developed as a novel therapeutic approach to target these dysfunctions.
The article begins by introducing the landscape of histone and DNA methylation marks and then categorizes the various families of single and tandem reader modules that use an aromatic cage capture mechanism for readout of methyllysine (Kme) and methylarginine (Rme) marks. Next, the text highlights recent reader modules that target unmodified lysines and arginine marks, as well as reader cassettes involved as regulatory platforms for mediating functional output. The article also outlines the potential for cross talk between PTMs, whereby the binding of a reader module to a particular mark either sterically blocks an adjacent modification site or facilitates recruitment of additional modules to modify nearby residues. In addition, “histone mimics” are discussed as a distinct set of nonhistone proteins that are methylation targets, thereby expanding available methylated lysine recognition principles beyond the boundaries of direct chromatin regulation. The article next addresses DNA cytosine methylation (5mC) marks and their readout by 5mC-binding domains (MBDs) and zinc-finger-containing modules with the capacity to sequence specifically recognized 5mC-containing fully methylated CpG DNA sites. The article also highlights the contribution of 5mC-binding SRA (SET- and RING-associated) domains required for the establishment and/or maintenance of DNA methylation marks at hemimethyated CpG DNA sites in both mammals and plants.
The article ends by highlighting new initiatives and advances, as well as future challenges that promise to enhance our current mechanistic understanding of the readout of histone and DNA methylation marks. These include technological developments at the genome-wide level, chemical biology approaches to designer nucleosomes, and structural approaches to histone mark readout at the nucleosomal level. The article also outlines new developments related to readout of oxidative 5mC DNA adducts, the functional role for regulatory noncoding RNAs in epigenetic regulation, and the linkage between histone and DNA methylation. This article addresses the consequences of dysregulation of methylated lysine reader modules and long intergenic noncoding RNAs on epigenetic pathways resulting in the onset of disease states and outlines challenges toward identification and functional characterization of small molecules site-specifically targeted to aromatic-lined pockets involved in methyllysine readout.
1. INTRODUCTION
The nucleosome core particle is composed of almost two turns of a DNA superhelix amounting to 147 bp wrapped around a compact histone octamer core containing four subunits labeled H2A, H2B, H3, and H4 (Luger et al. 1997). Nuclesomes are packaged into progressively higher-order folds to ultimately form chromosomes. Projecting from the four histone cores are amino-terminal tails that are subject to covalent posttranslational modifications (PTMs) (Allfrey et al. 1964), depositing marks such as methylation, acetylation, phosphorylation, and ubiquitination. Methylation of cytosines on DNA is also possible. More recently, with the advent of advanced mass spectroscopic and antibody-based techniques, PTMs have also been identified within the carboxy-terminal end of histone tails and even within the globular central histone fold. In addition, new covalent modifications have recently been identified such as sumoylation, ADP-ribosylation, proline isomerization, citrullination, and glycosylation (see Zhao and Garcia 2014).
PTM marks are dynamic, being deposited and erased in the time frame of minutes. The recognition of a mark by a reader module that is part of a multidomain protein complex facilitates the recruitment and tethering of enzymatic activities intrinsic to other subunits to chromatin. Hence, histone and DNA covalent PTMs provide a scaffold for the assembly of activities that control the site- (e.g., lysine 4 of H3) and state-specific (e.g., mono-, di-, or trimethylated) readout of marks at the nucleosomal level. They also have the capacity to modulate higher-order chromatin structure and/or the ordered recruitment of nonhistone proteins and enzymes critical for DNA remodeling activities. Thus, PTMs serve as epigenetic information carriers that extend the message beyond that encoded in the DNA sequence. This capacity of chromatin to both store and transmit heritable information in the form of PTMs results in altered histone–DNA interactions. This leads to dynamic changes in chromatin-templated processes, including altered transcription rates reflecting the accessibility of particular segments of DNA.
The advent of histone mark-specific ChIP-chip (chromatin immunoprecipitation with DNA microarray analysis) and ChIP-seq (chromatin immunoprecipitation with next-generation sequence technology) has enabled the characterization of histone marks at the genome-wide level. This has provided unique insights into the distribution of marks to distinct segments of the genome. It has also identified global correlations of specific marks with downstream functional outcome. Genome-wide chromatin organization studies performed by the modENCODE consortium have, in addition, identified the linkage of PTMs with regulatory circuits at defined developmental stages in model organisms.
The methylation of certain histone residues and cytosine bases on the DNA are well-known and well-studied instances of PTMs. This article focuses on the structural aspects of how methylation mark readers interact with methylation marks, the functional consequence of these interactions, and how cross talk occurs with other histone PTMs.
1.1. Lysine and Arginine Methylation of Histone Tails
Lysine methylation is unique among known PTMs because of its relative stability, its multivalency (i.e., existence in the mono-, di-, and trivalent state), and its potential to engage in cross talk with other modifications. The hydrophobicity of lysine is increased on methylation with no net change in charge. This contrasts with the change in charge that occurs on lysine acetylation or serine phosphorylation.
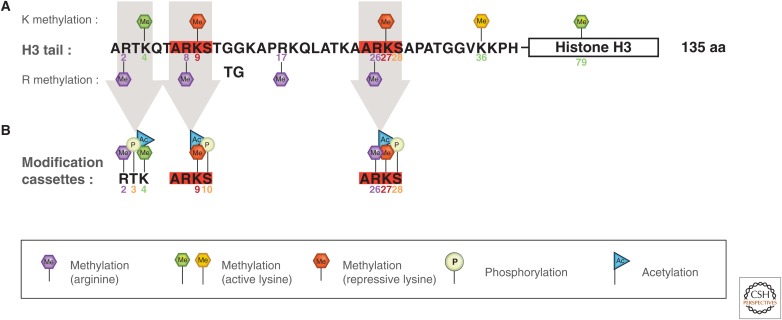
The major lysine (K) methylation sites on histone H3 are at positions K4, K9, K27, K36, and K79 (Fig. 1A), together with a single site on H4 at K20 and one on H1 at K26. Of these, H3K4, H3K36, and H3K79 methylation marks are linked to the activation of transcription, whereas H3K9, H3K27, and H4K20 methylation marks are linked to transcriptional repression (Fig. 1A). Some sites such as H3K9 and H3K27 are embedded within a common A-R-K-S sequence context (Fig. 1B). Each site-specific mark is written by a specific enzyme classed as a histone lysine (K) methyltransferase, or KMT, and erased by other enzymes called histone lysine demethylases or KDMs. The catalytic activity of each writer and eraser enzyme is substrate specific—that is, effective on an individual or set of methylation states, such as mono-, di-, or trimethylated lysines (reviewed in Black et al. 2012; Greer and Shi 2012). Enrichment of particular lysine marks can be found at specific regions along the genome.
Figure 1.
Sequence of H3 tail and distribution of PTMs. (A) Sequence of H3 tail and positioning of methyllysine (Kme) and methylarginine (Rme) marks. (B) Positioning of adjacent marks within the H3 tail at R2-T3-K4, A7-R8-K9-S10, and A25-R26-K27-S28 segments. Kme marks are partitioned between those that are activating and those that are repressive.
The major arginine (R) methylation sites are at R2, R8, R17, and R26 on histone H3 (Fig. 1A), at R3 on H4, and at R11 and R29 on H2A. The marks are written by protein arginine methyltransferase (PRMT) enzymes and erased by deiminases as a function of methylation state (mono-, symmetrical di-, and asymmetrical dimethylated arginines) context.
It has been proposed that multivalent readout of methylation together with other marks (e.g., acetylation, phosphorylation, ubiquitination) by reader modules regulates the transcriptional outcome. This occurs by directing the activity of chromatin-remodeling complexes (discussed in Becker and Workman 2013) in a precisely controlled manner according to the developmental stage and physiological state of cells, with transcription rates also susceptible to external stimuli (reviewed in Strahl and Allis 2000; Jenuwein and Allis 2001; Gardner et al. 2011). Meaningful variation in the chromatin polymer can also result from exchange of primary sequence histone variants such as H2A.Z in place of H2A or H3.3 instead of H3.1/2 (see Henikoff and Smith 2014).
The binding pockets of reader modules recognize histone methylation marks in a sequence- and state-specific manner. The architecture of these binding pockets and principles of molecular recognition are of fundamental importance to our understanding of chromatin function. A salient question is whether recognition can occur by processes beyond the first identified mode involving aromatic cage capture of the methylated lysine side chain discussed in Section 2.1 and reviewed in Taverna et al. (2007). There is also interest as to whether known methyllysine-binding pockets can be engineered to recognize and distinguish between distinct (mono-, di-, and tri-) lysine methylation states.
An increasing number of human diseases ranging from autoimmune disorders to cancer are associated with aberrant writers, readers, and erasers of histone methylation marks (reviewed in Chi et al. 2010; Dawson and Kouzarides 2012). Research has revealed that many mutations in these molecules have affected the function of these epigenetic regulators and often entire chromatin-remodeling complexes. This has led to research looking for viable epigenetic therapies, including the identification of small molecules that selectively target Kme-binding pockets on reader modules.
There have been several excellent reviews covering earlier structure–function research on the readout of histone marks (Kouzarides 2007; Kouzarides and Berger 2007; Ruthenburg et al. 2007a,b; Taverna et al. 2007; see also more recent reviews by Yap and Zhou 2010; Bannister and Kouzarides 2011; Khorasanizadeh 2011; Musselman et al. 2012b). In this article, we provide a comprehensive up-to-date (until the end of December 2012) structural overview on the readout of histone methylation marks and place these results in a functional context. In companion articles, a structure–function perspective is given on writers and erasers of histone lysine methylation marks (Cheng 2014) and on writers, readers, and erasers of histone lysine acetylation marks (Marmorstein and Zhou 2014; Seto and Yoshida 2014).
1.2. Cytosine Methylation in DNA
Methylation of cytosines at CpG sites along the mammalian genomes constitutes an ancient evolutionary epigenetic mark. It operates as an epigenetic regulatory mark predominantly in mammals and plants (the latter of which is discussed in Pikaard and Mittelsten Scheid 2014), although Neurospora and Drosophila contain this chromatin mark to a more limited extent. This epigenetic modification in mammals is associated with gene silencing and contributes to chromatin structure and genome stability (reviewed in Li and Bird 2007; Jones and Liang 2009; Law and Jacobsen 2010). The methyl mark is deposited at the 5-position of cytosines (5mC) in CpG steps by the de novo DNA methyltransferases, DNMT3A and DNMT3B, together with regulator DNMT3L, during embryonic development. The mark is faithfully maintained during multiple cell divisions by DNMT1, the maintenance DNA methyltransferase, thereby establishing a form of cellular memory through epigenetic marking of the genome (illustrated in Fig. 2 of Li and Zhang 2014). The importance of DNA methylation is highlighted by the fact that targeted disruption of DNA methyltransferases results in lethality. Mechanistically, gene expression profiles can be altered by DNA methylation as a consequence of influencing the binding affinities of transcription factors. Alternatively, transcriptional silencing results from 5mC binding proteins recruiting repressor complexes to methylated promoter segments. Thus, DNA methylation plays a critical role in the establishment and maintenance of tissue-specific gene expression patterns at distinct stages of development. Critical developmental processes such as X-inactivation, monoallelic expression of imprinted genes, and suppression of transposable elements and proviral genomes use DNA methylation as a component of complex regulatory networks (further discussed in Barlow and Bartolomei 2014; Brockdorff and Turner 2014; Pikaard and Mittelsten Scheid 2014).
Figure 2.
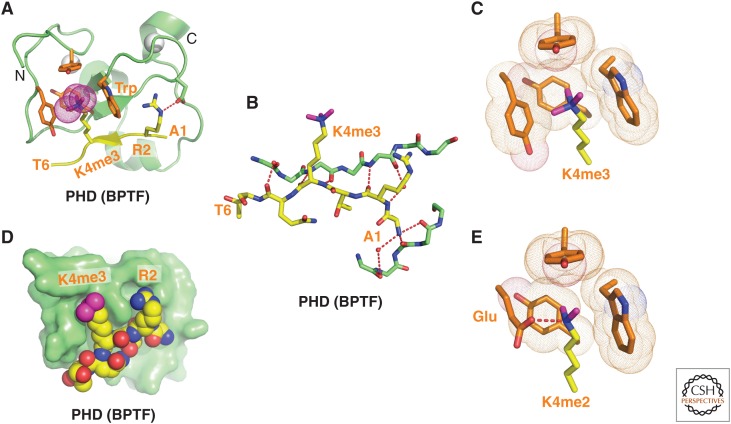
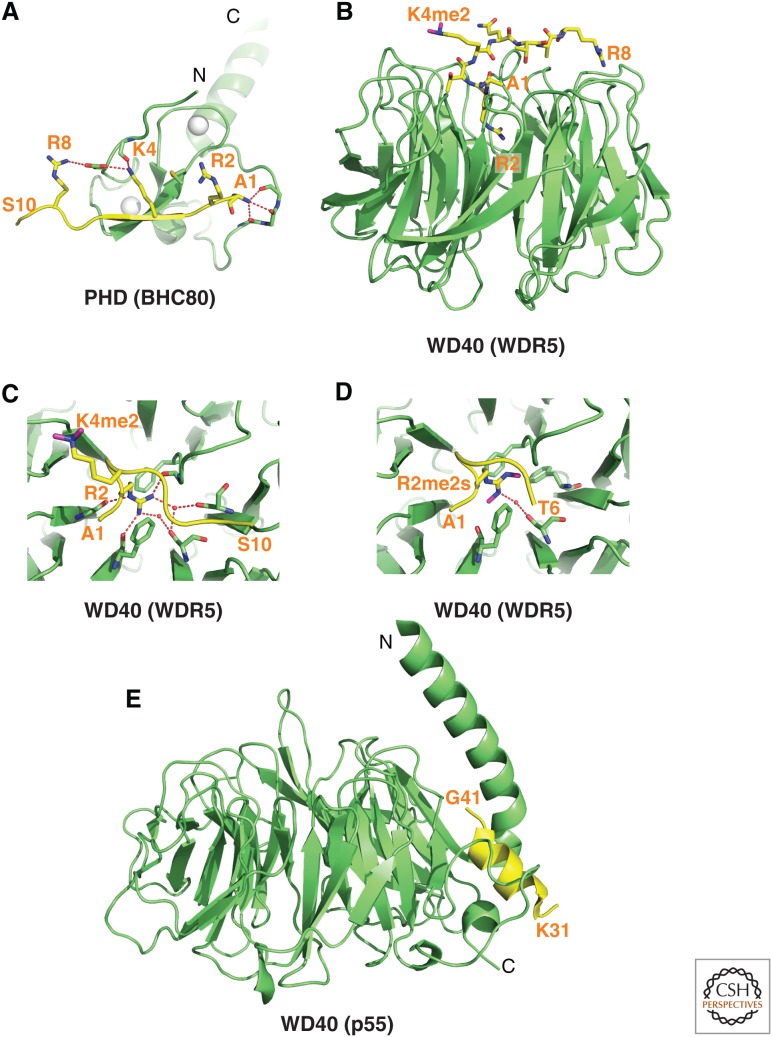
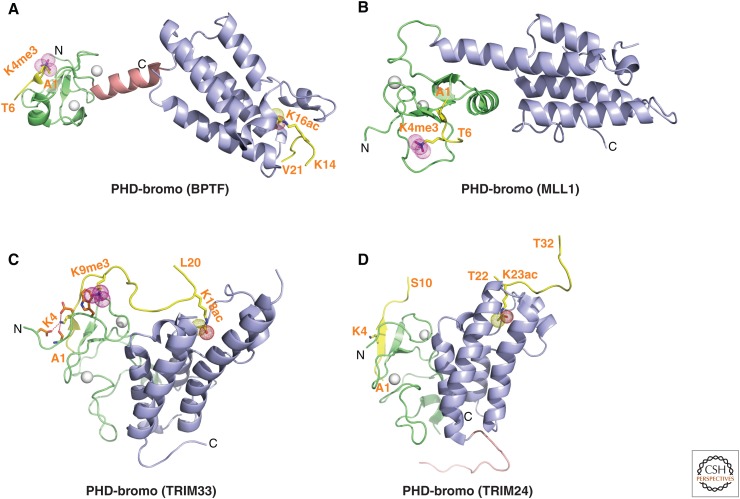
Structure of BPTF PHD finger bound to H3K4me3-containing peptide. (A) 2.0-Å crystal structure of the complex of the BPTF PHD finger bound to H3(1-15)K4me3 peptide (PDB: 2F6J). The PHD finger (as part of a PHD finger-bromo cassette) in a ribbon representation is in green, with two stabilizing bound Zn ions in silver balls. The bound peptide from A1 to T6 is shown in yellow with the trimethyl group of Kme3 shown by dotted balls in magenta. The residues forming the aromatic-lined cage are colored in orange. (B) Details showing the antiparallel alignment of the β-strands of the bound H3K4me3-containing peptide and PHD finger, resulting in formation of an antiparallel β-pleated sheet on complex formation. Note that the positively charged amino terminus is anchored in its own pocket. (C) Positioning of the K4me3 group within the aromatic-lined cage in the complex. (D) Positioning of R2 and K4me3 side chains in adjacent open surface pockets (surface groove mode), separated by the indole ring of an invariant Trp in the complex. The PHD finger and peptide are shown in surface- and space-filling representations, respectively. (E) Positioning of the K4me2 group into an engineered pocket, containing a Glu residue replacing the Tyr residue in C (PDB: 2RIJ).
Greater than 70% of CpG sites are methylated in the DNA of somatic tissues, but they are asymmetrically distributed along the genome within both CpG-rich and -poor regions. Pericentric heterochromatin, for example, is hypermethylated, yet overall CpG-poor. The global DNA methylation of such inaccessible heterochromatic regions requires the participation of SWI/SNF-like chromatin-remodeling proteins to allow DNA methyltransferase access. CpG islands, in contrast, are hypomethylated and immune to otherwise global DNA methylation. CpG islands are short CpG-rich sequences of ∼1 kb in length, making up <1% of genomic DNA, and mark the promoters and 5′ end of genes. Improved genome-scale mapping has provided more detailed insights into the distribution of DNA methylation patterns at transcription start sites, gene bodies, regulatory elements, and repeat sequences, with a relationship emerging between DNA methylation and transcription repression (see Li and Zhang 2014; also reviewed in Jones 2012). DNA methylation is a dynamic mark, which is mostly deposited, removed, and reestablished during early mammalian development.
A growing number of human diseases ranging from imprinting disorders (e.g., Beckwith–Wiedemann, Prader–Willi, and Angelman syndromes) to repeat-instability diseases (e.g., fragile X syndrome and facioscapulohumeral muscular dystrophy) and cancer are associated with aberrant DNA methylation (as discussed in Robertson 2005; Baylin and Jones 2011, 2014; Zoghbi and Beaudet 2014). This may involve the improper establishment or maintenance of methylation, resulting in the alteration of chromatin states and/or nucleosome positioning (reviewed in Baylin and Jones 2011). Another feature of DNA methylation patterns that may be occasionally causal to disease is the susceptibility of 5mC to spontaneously deaminate to thymine. This instability at CpG sites is reflected by the fact that a third of point mutations are C-to-T transitions at CpG sequences, and explains the fivefold underrepresentation of CpG steps in mammalian genomes generated over evolutionary time. Mutations in components of the DNA methylation machinery can also lead to disease, such as the documented mutations in DNMT3B leading to immune deficiency and blood cancers, whereas mutations in MeCP2, a 5mCpG-binding protein, cause Rett syndrome, a severe neurological disorder.
Li and Zhang (2014), as well as several excellent reviews, cover the topic of genomic DNA methylation, its establishment and maintenance (reviewed in Klose and Bird 2006; Jones and Liang 2009; Law and Jacobsen 2010). In this article, we provide a comprehensive up-to-date (until the end of December 2012) structural overview on the readout of 5mCpG marks, placing these results in a functional context.
2. READOUT OF Kme MARKS BY PHD FINGER AND BAH MODULES
We start our description of reader modules targeting methylated lysine marks by focusing on PHD (plant homeodomain) fingers and BAH (bromo-adjacent homology) domains. The PHD finger is a very common module found among chromatin remodelers and is often positioned adjacent to other reader modules. Although it was initially thought to partake in protein–protein and protein–lipid interactions, in 2006, structure–function studies showed that PHD fingers are readers of methylated lysine marks on histone tails. This promoted a more structure-based perspective in the field of epigenetic regulation. The PHD reader often participates in multivalent readout (i.e., readout of combinations of histone PTMs with other reader modules) at the nucleosome level as elaborated in Sections 9.1 and 10.1. The BAH domain was also initially thought to be a protein–protein interaction module until it, too, was shown to be a reader of methylated lysine marks on histone tails in 2012. Both PHD finger and BAH domains are of great interest given that reports on their dysfunction results in the onset of disease, such as Meier–Gorlin primordial dwarfism syndrome in the case of specific mutants in the BAH domain of the ORC1 (origin of replication 1 protein) protein.
2.1. PHD Finger Domains
Lysine methylation of histone tails constitutes a key component of an epigenetic indexing system, demarcating the transcriptional activity of chromatin domains. The H3K4me3 mark, in particular, is associated with nucleosomes near the promoters and the 5′-ends of highly transcribed genes (Santos-Rosa et al. 2002; Bernstein et al. 2006). BPTF is a protein named after its two reader modules: bromodomain and PHD domain transcription factor. Functional studies of the PHD finger established its role in nucleosome remodeling factor (NURF) complex-mediated ATP-dependent chromatin remodeling, directly coupling the complex to H3K4me3 so as to maintain HOX gene expression patterns during development (Wysocka et al. 2006).
The PHD finger (50–80 residues) (reviewed in Bienz 2006) has limited secondary structure (a two-stranded β-sheet and a short α-helix) wherein cross-brace topology is stabilized by two zinc ions coordinated to a Cys4-His-Cys3-containing segment (Pascual et al. 2000). Simultaneous structure–function studies on the PHD fingers of BPTF (Li et al. 2006; Wysocka et al. 2006), ING2 (inhibitor of growth 2) (Pena et al. 2006; Shi et al. 2006), and YNG1 (Taverna et al. 2006) bound to higher methylated lysine states of H3K4 revealed the principles of molecular recognition occurring in a sequence and methylation-state-specific manner (reviewed in Ruthenburg et al. 2007a; Taverna et al. 2007). We focus below on the BPTF system because this PHD finger is part of a PHD finger-bromodomain cassette discussed at greater length in Sections 9 and 10, and because structure–function studies were undertaken both at the peptide (reviewed in Ruthenburg et al. 2007a) and nucleosomal (Ruthenburg et al. 2011) levels.
Unbiased pull-down assays using nuclear extracts initially identified the second PHD finger of BPTF as a specific reader of the H3K4me3 mark (Wysocka et al. 2006). This PHD finger bound to higher methylation states of H3K4 that are linked to transcriptional activation with dissociation constants (Kd) of 2.7 µm for H3K4me3- and 5.0 µm for H3K4me2-containing peptides, while discriminating against monomethylated (me1) and unmodified (me0) counterparts. The observed intermediate range (i.e., μm; high range, nm; and low range, mm) of dissociation constants reflects the required balance between binding and dissociation of these marks by reader and eraser modules during epigenetic regulation. The structure of the H3(1-15)K4me3 peptide bound to the BPTF PHD finger solved by both X ray (Fig. 2A) and nuclear magnetic resonance (NMR) analysis showed that the histone peptide adopts a β-conformation paired with the β-sheet of the PHD finger to form a three-stranded, antiparallel β-sheet stabilized by backbone intermolecular hydrogen bonds (Fig. 2B) on the surface of the PHD finger on complex formation (Li et al. 2006). The sequence specificity for H3K4me3 involves recognition of the amino terminus, the R2 side chain, and the K4me3 mark in the complex. This distinguishes the H3 A1-R2-T3-K4me3 sequence segment from other Kme marks such as H3K9 and H3K27 embedded within an A-R-Kme3-S sequence context (Fig. 1B), and H3K36 and H4K20. The K4me3 is positioned in a preformed pocket composed of four aromatic residues, referred to as an “aromatic cage” and stabilized by electrostatic cation–π (Ma and Dougherty 1997) and hydrophobic interactions (Fig. 2A,C). The long side chains of R2 and K4me3 are positioned in adjacent preformed “surface groove” recognition pockets separated by the indole group of an invariant Trp residue (Fig. 2D), with this stapling role contributing to the specificity of H3K4me3 recognition. The guanidinium group of Arg2 is restrained by the formation of intermolecular hydrogen bonds between backbone carbonyls and an acidic side chain, whereas amino-terminal recognition (through hydrogen-bonding to a backbone carbonyl) is also important for complex formation (Fig. 2B), reflected by the discrimination against peptides with amino-terminal extensions. The binding affinity decreases dramatically on mutation of the aromatic cage residues, especially of the Trp residue separating R2 and K4me3, consistent with developmental defects associated with this mutation (Wysocka et al. 2006). The observed modest preference for H3K4me3 (Kd of 2.7 µm) over H3K4me2 (Kd of 5 µm) can be reversed following replacement of one of the cage-lining aromatic amino acids by a Glu, thereby facilitating formation of a hydrogen bond between the dimethylammonium proton of Kme2 and the carboxylate of the Glu side chain (Fig. 2E) (Li et al. 2007a). The above structural results provide an explanation for why global loss of H3K4me3 results in the loss of chromatin association of BPTF (Wysocka et al. 2006).
Structure–function studies on additional PHD fingers have highlighted the plasticity (i.e., aromatic cages formed by both aromatic and nonaromatic amino acids) in their recognition of methylated lysine marks on histone tails (reviewed in Musselman and Kutateladze 2011; Sanchez and Zhou 2011). Thus, parallel structure–function studies on the PHD finger of ING2, a native subunit of the repressive mSin3a-HDAC1 histone deacetylase complex, explained why, in response to DNA damage, recognition of H3K4me3 by the PHD finger of ING2 stabilizes the mSin3a-HDAC1 complex at the promoters of proliferation genes (Pena et al. 2006; Shi et al. 2006). In the ING2 complex, the aromatic cage is composed of a Trp and a Tyr, with a Met side chain contributing to pocket formation. Disruption of binding interactions in the H3K4me3-ING2 PHD finger complex impacts on function, impairing the ability of ING2 to induce apoptosis in vivo (Pena et al. 2006).
In another important structure–function study, the PHD finger of RAG2, an essential component of the RAG1/2V(D)J recombinase, that mediates antigen-receptor gene assembly (explained in Sec. 4 of the Busslinger and Tarakhovsky 2014) couples H3K4me3 readout with V(D)J recombination (Matthews et al. 2007; Ramon-Maiques et al. 2007). The investigators solved the structure of the RAG2 PHD finger bound to H3K4me3, evaluated the impact of Arg2 methylation, and then showed that mutations that abrogate intermolecular recognition severely impact on V(D)J recombination in vivo, as does depletion of the H3K4me3 mark. Interestingly, the Trp residue that is bracketed by the side chains of Arg2 and K4me3 in the RAG2 PHD finger-H3K4me3 peptide complex was found to be mutated in patients with immunodeficiency syndromes, highlighting the concept that disruption of the readout of a Kme mark can be the causal factor underpinning an inherited human disease (Matthews et al. 2007).
Histone mark dysregulation by PHD reader modules can impact on human cancers as reflected by the impact of somatic mutations in ING (inhibitor of growth) PHD fingers on solid tumors (reviewed in Chi et al. 2010).
2.2. BAH Domains
BAH domains are protein folds associated with epigenetic inheritance and gene regulation processes, having been identified in mammalian ORC1, MTA1 (a subunit of NuRD, a histone deacetylase and nucleosome remodeling complex), Ash1 (a SET domain-containing H3K36 methyltransferase), and the Saccharomyces cerevisiae Sir3 protein (part of the Sir2-Sir3-Sir4 complex required for silencing at the yeast mating-type loci). Tandem BAH domain pairs have been found in DNMT1 (the mammalian maintenance DNA methyltransferase) and the budding yeast RSC (a chromatin-remodeling complex) (reviewed in Callebaut et al. 1999; Goodwin and Nicolas 2001; Yang and Xu 2012).
The BAH domain adopts a conserved β-sheet core from which emerge loop and short helical segments shown in the structural determination of yeast ORC1p (Fig. 3A) (Zhang et al. 2002). The BAH domain (approximately 130 residues) was initially thought of as solely a protein–protein interaction module of unknown function. The recent demonstration that BAH domains recognize methylated lysine as constituents of the mammalian ORC1 (Kuo et al. 2012) and Arabidopsis thaliana CMT3 (ZMET2 in maize) (Du et al. 2012) proteins at the peptide level, as well as recognition of unmodified mononucleosomes by yeast Sir3 (Armache et al. 2011), has now brought prominent attention to this reader module. The BAH domain is often flanked by reader modules of other histone marks, such as bromo and PHD domains, suggesting the potential for combinatorial readout (reviewed in Ruthenburg et al. 2007b).
Figure 3.
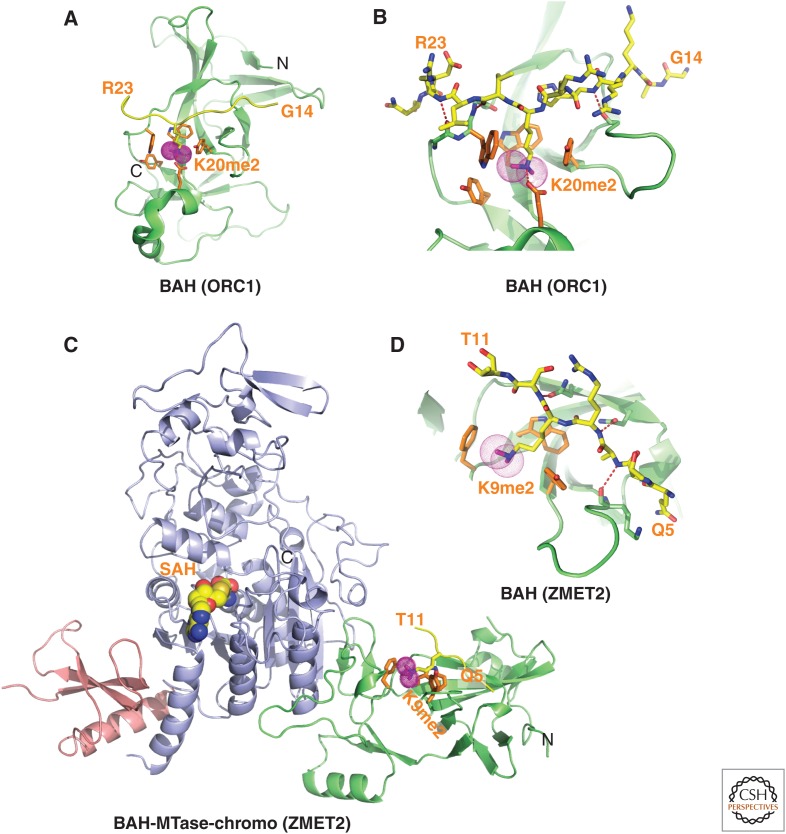
Structures of BAH domains of mammalian ORC1 and plant ZMET2 bound to methylated lysine histone peptides. (A) 1.95-Å crystal structure of the complex of mouse ORC1 BAH domain bound to H4(14-25)K20me2 peptide (PDB: 4DOW). The bound K20me2-containing H4 peptide can be traced from G14 to R23. (B) Enlargement of (A) showing details of the alignment of the K20me2-containing H4 peptide from G14 to R23 positioned on the mouse ORC1 BAH domain in the complex. The dimethylammonium group of H4K20 inserts into an aromatic-lined pocket in the BAH domain. (C) 2.7-Å crystal structure of the complex of maize ZMET2 BAH domain bound to H3(1-32)K9me2 peptide (PDB: 4FT4). The chromodomain, methyltransferase, and BAH domains are colored in pink, blue, and green, respectively. The bound K9me2-containing H3 peptide in yellow can be traced from Q5 to T11. (D) Enlargement showing details of the alignment of the K9me2-containing H3 peptide from Q5 to T11 positioned on the maize ZMET2 BAH domain in the complex. The dimethylammonium group of H3K9me2 inserts into an aromatic-lined pocket in the BAH domain.
A major advance in our understanding of the function of the BAH domain emerged following structure–function studies demonstrating that the mammalian ORC1 protein is a reader of the H4K20me2 mark, a property shared with diverse metazoans, but not the yeast ORC1 proteins (Kuo et al. 2012). Mammalian ORC1 constitutes the largest subunit of ORC (origin of replication complex) and it is the only ORC subunit of the six-subunit complex that contains a BAH domain. ORC1, which mediates pre-DNA replication licensing (Duncker et al. 2009), contributes both to ORC complex association and regulation of origin activity. The structure of the ORC1 BAH domain bound to the H4K20me2 peptide (Fig. 3A) showed that the side chain of K20me2 inserts into an aromatic-lined cage (surface-groove recognition) within the BAH domain of ORC1, with the dimethylammonium group hydrogen-bonded to a Glu side chain and stabilized by hydrophobic and cation–π interactions (Fig. 3B). The side chains of Trp and Glu pocket-forming residues undergo conformational changes, thereby generating a fully walled aromatic cage pocket on complex formation. Mutation of the ORC1 BAH domain aromatic cage residues impairs ORC1 occupancy at replication origins, ORC chromatin loading, and cell-cycle progression (Kuo et al. 2012). The Meier–Gorlin syndrome, a form of primordial dwarfism (Klingseisen and Jackson 2011), has been linked to mutations in the ORC1 BAH domain (Bicknell et al. 2011). In fact, phenotypic rescue of growth retardation in orc1-depleted zebrafish morphants using wild-type, but not H4K20me2-binding mutants confirmed the central role the ORC1 BAH domain in this syndrome (Kuo et al. 2012). The above study thus identified the first direct link between histone methylation and the metazoan DNA replication machinery and linked the canonical histone H4K20me2 mark recognized by ORC1 with a primordial dwarfism syndrome.
More recently, studies have shown that the mammalian ORC1 BAH domain has several protein–protein interacting surfaces. Thus, in addition to the aromatic cage segment involved in H4K20me2 interaction, the BAH domain interacts with the protein kinase Cyclin E-CDK2 to inhibit the kinase activity involved in centrosome duplication (Hossain and Stillman 2012). The Meier–Gorlin syndrome mutants located within the BAH domain of ORC1 attenuated this inhibition activity. A mechanistic understanding of the inhibition characteristics of the kinase activity could emerge following attempts at structure determination of the complex between interacting partners.
The yeast ORC1 BAH domains are involved in the transcriptional silencing of the HM mating-type loci in budding yeast, which is an epigenetically regulated process (see Fig. 5 of Grunstein and Gasser 2013). Structural studies of Sir1 (silencing information regulator 1) bound to the BAH domain of Orc1 revealed that the ORC1 BAH domain used a surface distinct from its aromatic cage-binding face to target Sir1 (Hou et al. 2005; Hsu et al. 2005), which contrasts with the mode of binding of higher eukaryotic ORC1 proteins.
Figure 5.
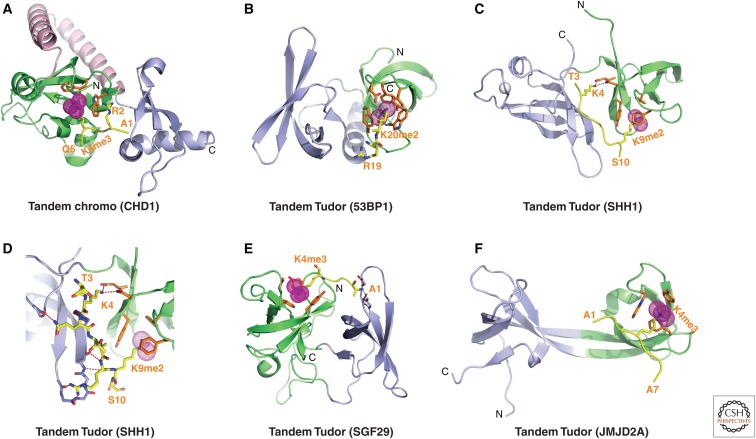
Structures of tandem Royal Family modules bound to methylated lysine histone peptides. (A) 2.4-Å crystal structure of the complex of the human CDH1 tandem chromodomains bound to H3(1-19)K4me3 peptide (PDB: 2B2W). Chromodomains 1 and 2 are colored in green and blue, respectively, with the connecting helix-turn-helix linker in pink. The bound K4me3-containing H3 peptide can be traced from A1 to Q5. (B) 1.7-Å crystal structure of the complex of 53BP1 tandem Tudor domains bound to H4(15-24)K20me2 peptide (PDB: 2IG0). Tudor domains 1 and 2 are colored in green and blue, respectively. The bound K20me2-containing H4 peptide can be traced for the R19-K20me2 step. (C) 2.7-Å crystal structure of the complex of tandem Tudor domains of A. thalania SHH1 protein bound to a H3(1-15)K9me2 peptide (PDB: 4IUT). A bound zinc ion is shown in a silver ball. Tudor domains 1 and 2 are colored in green and blue, respectively. The bound K9me2-containing H3 peptide can be traced from T3 to S10. (D) The enlargement shows details of the alignment of the K9me2-containing H3 peptide from T3 to S10 positioned on the A. thaliana SHH1 domain in the complex with intermolecular interactions formed with both Tudor domains. (E) 1.26-Å crystal structure of the complex of tandem Tudor domains of SGF29 bound to H3(1-11)K4me3 peptide (PDB: 3MEA). Tudor domains 1 and 2 are colored in blue and green, respectively. The bound K4me3-containing peptide can be traced from A1 to K4me3. (F) 2.1-Å crystal structure of the complex of tandem Tudor domains of JMJD2A bound to H3(1-10)K4me3 peptide (PDB: 2GFA). Individual Tudor domains are colored in green and blue, respectively. The bound K4me3-containing peptide can be traced from A1 to A7.
In a groundbreaking study, structural details of the complex between the yeast Sir3 BAH domain (containing hypermorphic D205N mutant) bound to a nucleosome core particle (NCP) have emerged (Armache et al. 2011). Sir3 is part of the distinct Sir2-3-4 silent chromatin-forming structure used in S. cerevisiae, which contrasts with the more widely used H3K9me-HP1 rich heterochromatin found in higher eukaryotes. The Sir3-NCP structure is discussed briefly so as to highlight key features of the recognition process. Two Sir3 molecules are bound on either side of the pseudosymmetric NCP surface, contacting all four core histones on complex formation (schematically illustrated in Fig. 5 of Grunstein and Gasser 2013). The complex is formed through a broad contiguous interaction between a surface on the NCP and a complementary surface on the BAH domain of Sir3 that is critical for transcriptional silencing. Complex formation results in the structural rearrangement of disordered segments on both the BAH domain and the nucleosome. The specific intermolecular contacts readily explain the numerous genetic mutations identified previously, as well as regulation of the silencing complex by modifications of H3K79 and H4K16 positions.
Tandem BAH domains in the structure of DNMT1 are separated by an α-helix, thereby ensuring a fixed separation and relative orientation between domains (Song et al. 2011). The DNMT1 BAH1 domain notably contains an aromatic cage along the same surface as mammalian ORC1. The BAH2 domain, however, lacks such an aromatic cage, but instead projects a very long loop, which interacts at its tip with the TRD (target recognition domain) of the methyltransferase domain, thereby holding the TRD in a retracted position away from the DNA in its complex with unmethylated DNA (illustrated in Fig. 6 of Cheng 2014) and facilitating the formation of an autoinhibitory conformation. Importantly, both BAH1 and -2 of DNMT1 show large accessible surfaces available for further recognition, potentially with histone tails of nucleosomes and/or other interacting protein partners.
Figure 6.
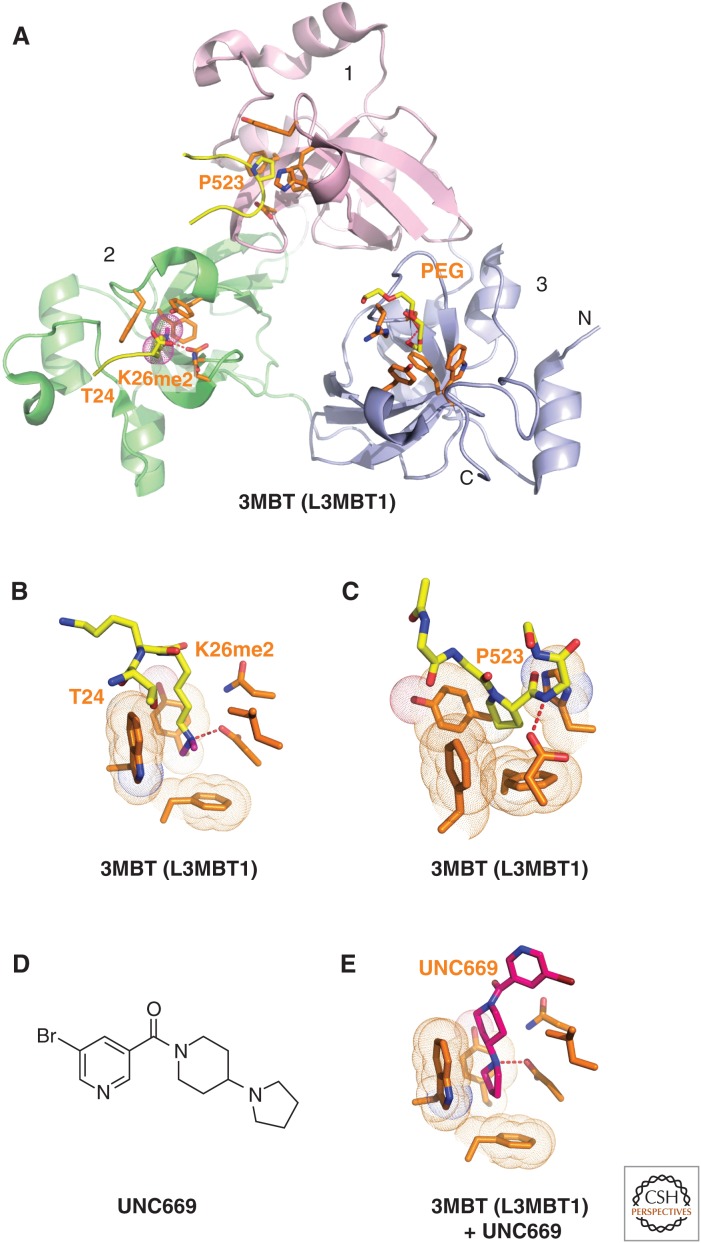
Structures of L3MBTL1 bound to methylated lysine histone peptides and an inhibitor. (A) 1.66-Å crystal structure of the complex-containing L3MBTL1 bound to H1(22-26)K26me2 peptide (PDB: 2RHI). A carboxy-terminal peptide from an adjacent L3MBTL1 in the crystal lattice inserts Pro523 into the aromatic-lined pocket of MBT domain 1 (in pink). The dimethylammonium group of bound K26me2 inserts into the aromatic-lined pocket of MBT domain 2 (in green) with the K26me2-containing H1 peptide traced from T24 to K26me2. A polyethylene glycol (PEG) molecule inserts into the aromatic-lined pocket of MBT domain 3 (in blue). (B) Details of how the H1K26me2 dimethylammonium group inserts into the aromatic-lined pocket of MBT domain 2. (C) Structural detail showing how the proline from an adjacent L3MBTL1 in the crystal lattice inserts into the aromatic-lined pocket of MBT domain 1. This pocket is shallower than the one shown in B. (D) Chemical formula of UNC669. (E) Details of how UNC669 inserts into the aromatic-lined pocket of MBT domain 2, based on the 2.55-Å crystal structure of L3MBTL1 bound to UNC669 (PDB: 3P8H).
A novel example of recognition of methylated lysine histone marks by dual domains was highlighted in structure–function studies of the plant CMT3 protein (chromomethylase 3; ZMET2 in maize) (Du et al. 2012). CMT3 is a plant-specific DNA methyltransferase that methylates CpHpG (H stands for C, T, or G) sites in an H3K9me2-dependent manner (discussed in Sec. 2.1 of Pikaard and Mittelsten Scheid 2014). CMT3 is composed of an amino-terminal BAH domain and a DNA methyltransferase domain, which has a chromodomain (for chromatin organization modifier) embedded within it. The structure of ZMET2 shows that the chromodomain and BAH domain are positioned at the corners of a triangular architecture, both binding H3K9me2-containing peptides through aromatic cage capture of the K9me2 side chain. The structure of the H3K9me2 peptide bound with directionality to the BAH domain of ZMET2 is shown in Fig. 3C, with details of the intermolecular interactions shown in Fig. 3D. Functional studies established a perfect correlation along the genome between methylated H3K9 and CMT3, and also showed that CMT3 is stably associated with H3K9me2-containing nucleosomes. Triple mutations of aromatic cage residues from the BAH domain or chromodomain disrupt CMT3 binding to nucleosomes and show a complete loss of CMT3 activity in vivo (Du et al. 2012). These studies definitively establish that DNA methylation in plants is directed by dual binding of the CMT3 BAH domain and chromodomain to H3K9me2-containing nucleosomes.
3. READOUT OF METHYLLYSINE MARKS BY SINGLE “ROYAL FAMILY” MODULES
The “Royal Family” of reader modules include the chromo, Tudor, PWWP (named after the conserved Pro-Trp-Trp-Pro motif), and MBT (malignant brain tumor) repeat domains (reviewed in Maurer-Stroh et al. 2003). Structural studies of these readers were among the first to elucidate the molecular principles behind methylated lysine recognition. In this section, we outline results on the structural principles by which single domains recognize lysine methylation marks and, in the next section, we look at tandem domains of the Royal Family.
3.1. Chromodomains
Structural studies of the chromodomains of HP1 (heterochromatin-associated protein 1) and Polycomb provided the first insights into how the aromatic cage pockets (illustrated in orange stick representation form in Fig. 4A) create a structural framework for methylated lysine recognition (reviewed in Yap and Zhou 2011). These proteins are known repressors that contribute to epigenetic silencing. More specifically, HP1 is a chromatin-associated protein first discovered in Drosophila and hallmark of condensed and highly repetitive heterochromatin (elaborated in Elgin and Reuter 2013). Polycomb is another protein originally characterized in Drosophila shown to direct heritable changes in chromatin organization (see Grossniklaus and Paro 2014).
Figure 4.
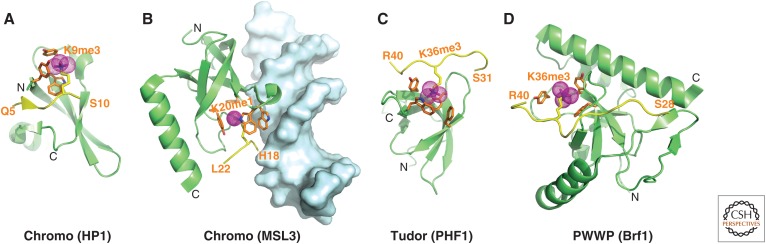
Structures of single Royal Family modules bound to methylated lysine histone peptides. (A) 2.4-Å crystal structure of the complex containing an HP1 chromodomain bound to H3(1-15)K9me3 peptide (PDB: 1KNE). The bound K9me3-containing H3 peptide can be traced from Q5 to S10. The HP1 residues in orange illustrate the aromatic cage that captures K9me3. (B) 2.35-Å crystal structure of the complex between the male-specific lethal (MSL)3 chromodomain bound to a H4(9-31)K20me1 peptide in the presence of duplex DNA (in surface representation) (PDB: 3OA6). The bound K20me1-containing H4 peptide can be traced from H18 to L22. (C) 1.85-Å crystal structure of the complex containing the PHF1 (a Polycomb-like protein) Tudor domain bound to H3(31-40)K36me3 peptide (PDB: 4HCZ). The bound K36me3-containing H3 peptide can be traced from S31 to R40. (D) 1.5-Å crystal structure of the complex of Brf1 PWWP domain bound to H3(22-42)K36me3 peptide complex (PDB: 2X4W). The bound K36me3-containing H3 peptide can be traced from S28 to R40.
Before structural studies, in situ immunofluorescence showed that H3K9me marks and HP1 colocalized to heterochromatic regions of Drosophila polytene chromosomes (illustrated in Figs. 3 and 4 of Elgin and Reuter 2013; also see Jacobs et al. 2001). Next, X-ray and NMR studies (Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002) independently corroborated the fact that the H3K9me3 mark was recognized by the HP1 chromodomain. We focus below on X-ray structural studies of the complex (Fig. 4A) that form with a Kd = 2.5 µm, illustrative of an intermediate affinity interaction. The H3 tail in an extended β-strand conformation (residues 5–10) binds to one face of the incomplete β-barrel architecture of HP1, where it aligns between β-strands of HP1 through an antiparallel-aligned-induced β-sandwich alignment. The K9me3 side chain inserts into a pocket lined by three conserved aromatic amino acids (i.e., the so-called aromatic cage pocket), where it is stabilized by cation–π interactions. The complex is stabilized by intermolecular contacts involving the ARKS (Ala-Arg-Lys-Ser) motif (Fig. 1), associated with two amino acids preceding and one following the K9me3 mark, providing a sequence context for the readout of the H3K9me3 mark. Mutation of the conserved aromatic amino acids lining the cage of HP1 results in a substantial loss of binding affinity (approximately 20-fold), as does mutation of Ala7 and Arg8 on the H3 peptide, with a smaller loss (threefold) observed on mutation of Ser10 (Jacobs and Khorasanizadeh 2002).
Subsequent structural studies showed that cage-capture of higher methylation groups (i.e., Kme3/2) was also observed for other chromodomains. Notably, this was shown for the chromodomain of Polycomb bound to an H3K27me3 peptide (Min et al. 2003), CHD1 bound to H3K4me2/3 peptides (Sims et al. 2005), MGR15 bound to an H3K36me2 peptide (Zhang et al. 2006), Eaf3 bound to an H3K36me3 peptide (Sun et al. 2008; Xu et al. 2008), and Chp1 bound to an H3K9me3 peptide (Schalch et al. 2009). This reinforces the principle that sequences flanking the Kme3/2 mark must determine the specificity of recognition by chromodomains (reviewed in Brehm et al. 2004; Yap and Zhou 2010).
A recent structural study has taken our understanding of chromodomains to a new level; the MSL3 chromodomain can target the lower lysine methylation mark H4K20me1 only in the presence of DNA (Kim et al. 2010). This unexpected corecognition was specific for GA-rich DNA, with binding greater by two orders of magnitude over RNA. MSL3 is a subunit of the male-specific lethal (MSL) complex, which is necessary for dosage compensation on Drosophila male X chromosomes (the topic of Lucchesi and Kuroda 2014). A preassembled complex contains the MSL3 chromodomain and DNA bound to H4K20me1 peptide with a Kd = 15 µm, with discrimination against unmodified and trimethylation states on H4K20. The crystal structure of this ternary complex is shown in Fig. 4B, with the chromodomain targeting the DNA minor groove and the K20me1 inserting into an adjacently positioned cage lined by four aromatic amino acids. Interestingly, the active H4K16ac mark antagonizes the DNA-mediated recognition of H4K20me1 by MSL3, suggesting that regulation of the MSL complex may be controlled by the readout of closely positioned marks. The investigators speculate that corecognition of the H4K20me1 mark and the DNA of two adjacent nucleosomes could contribute to the in vivo targeting of the MSL complex (Kim et al. 2010).
3.2. Tudor Domains
Polycomb group proteins are repressive chromatin modifiers essential for metazoan development, cellular differentiation, and the maintenance of cell fate. The Tudor fold, a domain within Polycomb, consists of a β-sheet core against which are packed one or more helical segments (Selenko et al. 2001). No fewer than four groups have recently undertaken functional studies of human Polycomb-like proteins PHF1 and PHF19, including the structural characterization of complex formation between its amino-terminal Tudor domain and H3K36me3-containing peptide (Ballare et al. 2012; Brien et al. 2012; Musselman et al. 2012a; Cai et al. 2013). X-ray and NMR studies of complex formation show that the H3K36me3 peptide in an extended conformation targets the five-stranded β-barrel of the Tudor domain (Fig. 4C). The trimethylammonium group of K36me3 inserts into an aromatic-lined pocket, whereas the flanking side chains of the peptide interact with a hydrophobic patch (bound by Pro38-His39 segment) and a shallow acidic groove (bound by Thr32-Gly33-Gly34 segment). Functionally, recognition of the H3K36me3 mark by polycomb-like proteins promotes the intrusion of Polycomb repressive complex 2 (PRC2) into active chromatin regions to then promote gene silencing, thereby impacting on the chromatin landscape during development.
3.3. PWWP Domains
The PWWP domain (containing a highly conserved Pro-Trp-Trp-Pro motif) along with chromodomain, MBT, and Tudor domains, belongs to the Royal Family of proteins that have the potential to recognize methylated lysine marks using an aromatic cage capture mechanism. It has been observed in many chromatin-associated proteins, often in combination with other domains such as SET modules, which are known writers of lysine methylation marks. PWWP-containing proteins are involved in diverse functions including transcriptional regulation, DNA repair, and DNA methylation (reviewed in Slater et al. 2003). The PWWP fold, as first determined by structural studies of the de novo methyltransferase DNMT3B, consists of a five-stranded β-barrel followed by an α-helical bundle with one of the α-helices packing against the β-barrel to generate a single structural motif (Qiu et al. 2002).
A recent structure of the PWWP domain of the Brf1 (bromodomain and plant homeodomain finger 1) protein bound to an H3K36me3 peptide (weak affinity of Kd = 2.7 mm) has validated the PWWP domain as a reader of methylated lysine marks (Fig. 4D) (Vezzoli et al. 2010). The peptide is positioned with directionality on a narrow surface groove with the K36me3 side chain positioned in an aromatic cage. Binding is abolished on mutation of any of the three conserved aromatic residues. The Brf1 protein associates specifically with the H3K36me3 mark and discriminates against other Kme3 marks on H3 and H4, in part, because of its ability to site-specifically accommodate Gly residues at positions 33 and 34. Functional studies in vivo indicate that Brf1 localizes to actively transcribed Hox genes, in which its enrichment parallels that of the H3K36me3 mark (Vezzoli et al. 2010). It should be noted that the binding of methylated lysine peptides to the chromodomain and the PWWP domain use different surfaces of the β-barrel for recognition.
4. READOUT OF METHYLLYSINE MARKS BY TANDEM ROYAL FAMILY MODULES
In this section, we outline how tandem reader modules of the Royal Family are involved in the recognition of Kme marks. These examples highlight the diversity of the recognition process between tandem chromo, tandem Tudor, and MBT repeats, and also within the tandem Tudor family.
4.1. Tandem Chromodomains
Proteins have been identified with tandem chromodomains. CHD (chromo-ATPase/helicase-DNA-binding) is one such protein that is involved in regulating ATP-dependent nucleosome assembly and mobilization at sites of transcription activity. It contains amino-terminal tandem chromodomains that target the H3K4me3 mark with a Kd = 5 µm. The structures of the human CHD1 tandem chromodomains have been solved both in the free state and bound to H3K4m3-containing peptides (Flanagan et al. 2005). Both chromodomains (1 and 2) adopt canonical chromodomain folds with a rigid helix-turn-helix motif connecting them such that the tandem chromodomains are juxtaposed together to form a continuous surface. One H3K4me3 peptide binds per CHD1 with the H3 peptide backbone positioned within an acidic surface between chromodomains 1 and 2 (Fig. 5A). The K4me3 side chain is positioned in a pocket composed of two Trp rings, with adjacent Arg2 also forming cation–π interactions with one of these Trp residues, thereby contributing to the specificity of recognition. Indeed, mutation of either Trp results in a substantial reduction in binding affinity. In addition, the binding affinity drops fourfold for complex formation with H3K4me3R2me2a, containing asymmetric dimethylation of Arg2 and a 25-fold reduction for complex formation with H3T3phK4me3-containing phosphorylation of adjacent Thr3 (Flanagan et al. 2005). It is important to note that chromodomains 1 and 2 of CHD1 do not use their canonical Kme-binding surfaces for peptide recognition partly because of sequence inserts in the CHD1 sequence that block canonical binding sites used by HP1 and Polycomb protein chromodomains.
4.2. Tandem Tudor Domains
Tandem Tudor domains have been identified as readers of methylated lysine marks and shown to use two distinct modes of recognition. The relative positioning of the pair of Tudor domains determines the binding mode; one class has a single linker that connects individually folded Tudor domains (examples include 53BP1, UHRF1, and SHH1) and another involves domain swapping and interdigitation of Tudor domains connected by a β-sheet whose β-strands are shared between domains (e.g., JMJD2A).
The tandem Tudor domain came into prominence following studies on 53BP1 (p53-binding protein). This study showed that histone lysine methylation marks facilitated the recruitment of 53BP1 and relocalized it to double-strand breaks on exposure to DNA-damaging agents. The recruitment element within 53BP1 was identified as a tandem Tudor domain (Huyen et al. 2004) and its mode of action emerged following the structural determination of its complex with H4K20me2 peptides (Kd = 19.7 µm) (Botuyan et al. 2006). The structure of the complex is shown in Fig. 5B with the peptide positioned between the Tudor domains, but primarily interacting with Tudor 1 (in green). The K20me2 side chain inserts into an aromatic cage positioned within Tudor 1 that is lined by four aromatic residues and an Asp that hydrogen bonds to the dimethylammonium proton, with the dimensions of the pocket preventing insertion of a K20me3 group (Kd = 1 mm) as a consequence of steric exclusion. The complex is also stabilized by cation–π interactions between the side chain of Arg19 of the peptide and a tyrosine ring of the protein. The importance of H4K20me2 recognition emerged following mutation of the aromatic residues lining the pocket, whereby binding was lost in vitro and also impacted on the targeting of 53BP1 to DNA double-strand breaks in vivo (Botuyan et al. 2006). A recent structural study has also reported that selective H3K4me3 recognition occurs by Spindlin1 Tudor domain 2 of this three-tandem Tudor domain-containing protein, with the Tudor domains aligned in a triangular-shaped architecture (Yang et al. 2012).
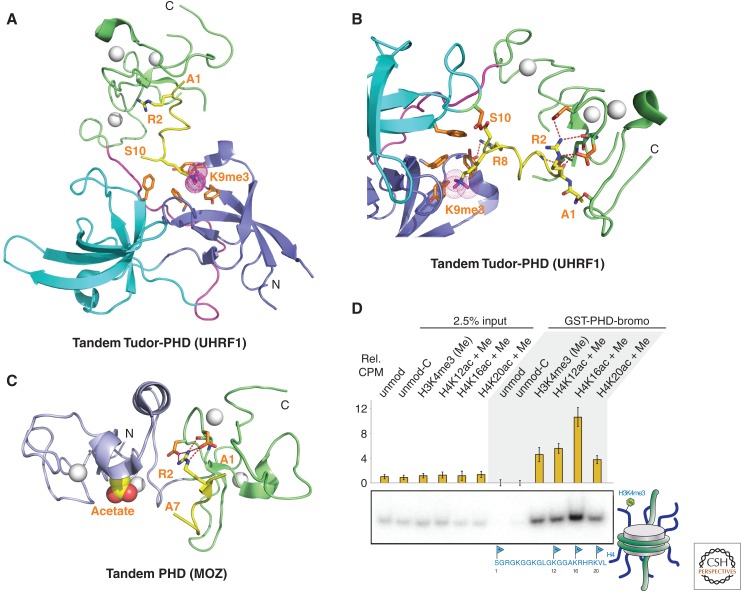
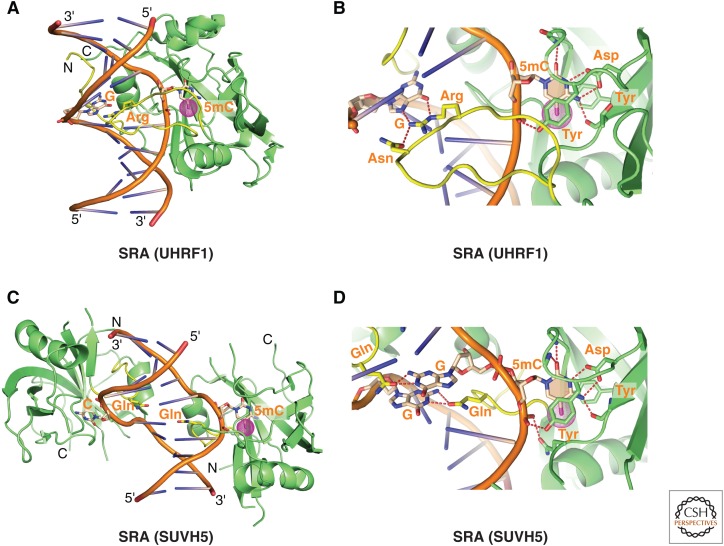
Structures have been solved for the tandem Tudor domains of UHRF1 (ubiquitin-like PHD and Ring finger 1) (Nady et al. 2011) and SHH1 (Sawadee homeodomain homolog 1) (Law et al. 2013) when bound to H3K9me2/3-containing peptides. These systems, like 53BP1, involve a single linker connecting individually folded Tudor domains, but unlike 53BP1, the bound H3 peptide interacts with both Tudor domains through positioning with directionality within a channel between domains. Importantly, the tandem Tudor domains within the UHRF1 and SHH1 proteins serve as dual lysine readers, probing for both unmethylated K4 (K4me0) and methylated K9 on the histone tail. Functional studies on the UHRF1 system show that Tudor domain mutants that can no longer bind H3K4me0K9me3 have reduced localization to heterochromatic chromocenters and fail to silence expression of the p16INK4A gene (Nady et al. 2011).
The structure of an H3(1-15)K9me2 peptide bound to the tandem Tudor domains of SHH1 (Kd = 1.9 µm) is shown in Fig. 5C with elements involved in the recognition of unmodified K4me0 and K9me2 shown in the expanded view in Fig. 5D. No conformational change was observed in the tandem Tudor fold when complexed with an H3K9me2 peptide (Law et al. 2013). Functional studies on the SHH1 system in plants show that the protein acts upstream of the RNA-dependent DNA methylation (RdDM) pathway (described in Sec. 3.5 of Pikaard and Mittelsten Scheid 2014) to enable small interfering RNA production from a large subset of the most active RdDM targets. SHH1 is required for polymerase-IV (pol-IV) occupancy at these same loci (Law et al. 2013). Further, key residues within both lysine-binding pockets of SHH1 are required to maintain DNA methylation in vivo, thereby providing the first insights into the mechanism by which SHH1 recruits RNA pol-IV to RdDM targets in plants.
SGF29, a component of the SAGA (Spt-Ada-Gcn5 acetyltransferase) complex, contains a pair of tandem Tudor domains that bind to H3K4me2/3 peptides. Binding occurs along one surface of the tightly packed face-to-face dimeric alignment (Fig. 5E) (Bian et al. 2011). The side chains of Ala1 insert into a pocket of the Tudor 1 domain (in blue) whereas the K4me2/3 inserts into an aromatic cage of the Tudor 2 domain (in green). Functionally, SGF29 targets active chromatin via H3K4me2/3, thereby recruiting the SAGA complex to mediate acetylation of H3 tails.
The jumonji histone KDM, JMJD2A, demethylates H3K9me2 via its jumonji domain (discussed in Sec. 1.5 of Cheng 2014). Its tandem Tudor domains adopt a domain-swapped interdigitated topology with a two-stranded β-sheet serving as a linker between the well-separated domains (Huang et al. 2006; Lee et al. 2008). The structure of the interdigitated tandem Tudor domains bound to an H3K4me3 peptide is shown in Fig. 5F; the side chain of K4me3 is positioned within a three-aromatic-residue-lined cage associated primarily with Tudor 2 (in green), yet binding affinity is also attributed to the intermolecular contacts it makes with side chains from Tudor 1 (in blue) within the interdigitated Tudor scaffold. No conformational change was observed in the bilobal interdigitated Tudor motif on proceeding from the free to the H3K4me3-bound state. In a related structural study (Lee et al. 2008), it was shown that the JMJD2A interdigitated Tudor domains bind with similar affinities to H3(1-10)K4me3 (Kd = 0.50 µm) and H4(16-25)K20me3 (Kd = 0.40 µm) peptides, although the two peptides share no amino acid sequence similarity other than the trimethylated lysine. Strikingly, the two peptides are aligned in opposite orientations despite the Kme3 side chains of both peptides inserting into the same aromatic-lined pocket in Tudor 2. Details of the intermolecular contacts led Lee et al. (2008) to identify single point mutants that inhibited recognition of H4K20me3, but not H3K4me3 or vice versa.
4.3. Tandem MBT Repeats
The MBT repeats are approximately 70 residues long and aligned in tandem. MBT acts as a transcription repressor with its repeats often perturbed in hematopoietic malignances (Koga et al. 1999). At the functional level, MBT proteins impact on diverse processes, ranging from regulation of mitosis and tumor suppression to the maintenance of cellular identity and body pattern during development (Bonasio et al. 2010).
The MBT fold was first identified following structure determination of the three MBT repeats from human L3MBTL1 (Wang et al. 2003) and the two MBT repeat-containing Drosophila SCML2 (Sex comb on midleg-like 2) protein (Sathyamurthy et al. 2003). The MBT unit is composed of a four-stranded β-barrel core followed by an extended arm of helices. Interdigitation occurs between the extended arms and cores of adjacent MBT subunits. In the case of L3MBTL1, this results in the formation of a three-leaved propeller-like architecture with each MBT unit containing an aromatic-lined pocket located on the same face of the triangular architecture (Fig. 6A) (Wang et al. 2003). Binding studies with histone peptides established that L3MBTL1 shows a preference for lower (mono- and di-) lysine methylation states. However, binding to a particular histone Kme1/2 mark is somewhat promiscuous and of relatively low to intermediate affinity (in the range of Kd = 5–40 µm). The Kme1/2 marks that L3MBTL1 binds to include H1.4K26me, H3K4me, H3K9me, H3K27me, H3K36me, and H4K20me, as measured at the peptide level by fluorescence polarization-based binding assays (Li et al. 2007a). Structural studies showed that the side chains of Kme1 and Kme2 insert deep into an aromatic cage pocket of the second MBT repeat of L3MBTL1 (Fig. 6B). Simultaneously, an adjacent L3MBTL1 protomer in the crystal lattice inserts its Pro ring from a carboxy-terminal tail Pro-Ser segment into a shallow aromatic cage pocket of the first MBT repeat (Fig. 5C). Pocket 2 is both deep and narrow and serves as a size-selective filter with the side chains that project from the gating and caging loops restricting access to the larger Kme3 group (Li et al. 2007a). Such a “cavity insertion” mode of methyllysine recognition (Fig. 6B) is distinct from the surface groove mode of methyllysine recognition (Fig. 2D) observed for other Royal Family members, PHD finger, and BAH domain reader complexes. A parallel structural study looking at the binding of the H4K20me2 peptide to L3MBTL1 also identified pocket 2 as a reader of lower lysine methylation marks (Min et al. 2007). In addition, it proposed an unanticipated mode of peptide-mediated dimerization leading to a model for chromatin compaction by L3MBTL1. More recent binding studies concluded that some MBT proteins showed sequence specificity, whereas others were promiscuous in the targeting of lower methylated lysine marks (Nady et al. 2012).
Reconstituted L3MBTL1-histone complexes analyzed by electron microscopy showed that L3MBTL1 does indeed compact nucleosomal arrays containing lower lysine methylation marks at H1K26 and H4K20. Thus, a combinatorial readout of methylated H1K26 and H4K20 by L3MBTL1 and methylated H3K9 by HP1γ results in chromatin condensation at Rb-regulated genes (Trojer et al. 2007; reviewed in Trojer and Reinberg 2008). Somewhat unexpectedly, recent functional studies indicate that the related protein L3MBTL2 can act in concert with PcG protein-mediated ubiquitination of H2A to establish a repressive chromatin structure without entailing the contribution of histone lysine methylation marks (Trojer et al. 2011).
Structural studies have also been undertaken on the four MBT repeats in human L3MBTL2 (Guo et al. 2009) and its Drosophila counterpart, dSfmbt (Grimm et al. 2009), bound to an H4K20me1 peptide. The four MBT repeats adopt an asymmetric rhomboid scaffold, with MBT repeats 2, 3, and 4 forming the triangular architecture and repeat 1 projecting off of it. The K20me1 side chain inserts into pocket 4 (corresponds to pocket 2 of L3MBTL1) with preferential recognition of lower methylation states. It was proposed that the observed lack of sequence specificity between bound histone peptides by MBT repeats could reflect the absence of distinct surface contours surrounding the methylated lysine-binding pocket (Guo et al. 2009). Functional experiments suggest that dSfmbt interacts with a related MBT repeat protein, Scm, and these two proteins together synergize in the repression of target genes crucial for Polycomb silencing (Grimm et al. 2009).
5. READOUT OF METHYLLYSINE MARKS BY EXPANDED AND PAIRED MODULES
We outline, below, two examples in which the readout of methylated lysine marks requires an expanded PHD finger module in one case and the modulation of PHD finger binding by a cofactor in the other case. The expanded PHD finger module example is most illuminating because the structural studies have identified a new principle for the recognition of Kme marks that is distinct from the surface groove (e.g., parts of PHD finger illustrated in Fig. 2D) and “insertion cavity” (e.g., Fig. 6B) modes of recognition already outlined in Sections 2.1 and 4.3. In addition, we outline recognition of Kme marks by ankyrin repeats.
5.1. GATA-1 and PHD Fingers of the ADD Domain
The ADD (ATRX-DNMT3A-DNMT3L) domain is found both in the ATRX protein, whose mutated form is associated with X-linked mental retardation (ATR-X) syndrome, and mammalian DNMT3A-DNMT3L required for de novo DNA methylation. ATRX is a large protein composed of adjacently positioned zinc-coordinated GATA and PHD fingers, referred to as the expanded PHD finger module. Half of the missense mutations in ATRX are associated with disease, many of them identified with pancreatic endocrine tumors. Further, about half are clustered toward the amino terminus within the Cys-rich ADD domain (Jiao et al. 2011); the other half cluster within the helicase/ATPase domain. The NMR solution structure of the ADD domain of ATRX in the free state showed that the GATA finger, PHD finger, and long carboxy-terminal α-helix form a single globular domain (Argentaro et al. 2007). Disease-causing mutations lie within this ADD domain dispersed between Zn-coordinating residues, those involved in packing, and others distributed along the outer surface.
The structure of H3K4me0-containing peptides bound to the ADD domain of DNMT3A has been solved (see Sec. 3 and Fig. 10 of Cheng 2014 and Otani et al. 2009). More recently, two groups have solved the structures of the ATRX ADD domain bound to H3K9me3-containing peptides (Eustermann et al. 2011; Iwase et al. 2011). The binding is promoted by H3K9me3, but inhibited by H3K4me3. A crystal structure of the complex reported at 1.0-Å resolution has provided details of intermolecular contacts and bridging water molecules at the highest resolution (Fig. 7A,B) (Iwase et al. 2011). The structure explains the requirement for unmodified H3K4; this side chain ammonium group is hydrogen bonded to the carboxyate groups of acidic amino acids within the PHD component of the ADD domain, leaving no room to accommodate methylation marks (Fig. 7B). Strikingly, the K9me3 mark is positioned within an “interfacial composite pocket” composed of residues from both GATA-1 and PHD fingers, with the dimensions of the pocket exquisitely sculpted to perfectly allow accommodation of the bulky trimethyllysine group (Fig. 7C). Unlike classical trimethyllysine readers, which usually consist of an aromatic-lined cage for higher methylation state-specific readout (reviewed in Taverna et al. 2007), the interfacial composite pocket of the ADD domain that binds K9me3 involves favorable van der Waals contacts associated with a high degree of surface complementarity (Fig. 7D), supplemented by a set of carbon–oxygen hydrogen bonds with the K9me3 group (Fig. 7C) (Iwase et al. 2011). The K4 and K9me3 binding pockets adopt a rigid mutual orientation (Fig. 7B), thereby contributing to the combinatorial readout of dual marks as reflected in the large increase in affinity (sevenfold on comparing H3K4me0K9me3, Kd = 0.5 µm, with H3K9me0K9me0, Kd = 3.6 µm) and enthalpy (twofold on comparing H3K4me0K9me3, ΔH = −12.2 Kcal mol−1, with H3K9me0K9me0, ΔH = −6.1 Kcal mol−1) for their joint readouts compared with their individual mark counterparts (Eustermann et al. 2011; Iwase et al. 2011). The structure of the complex containing the H3K9me3 peptide bound to the ATRX ADD domain highlights the role of integrated modules and the unanticipated composite reader pockets in methyllysine recognition. Functionally, ATRX localization at pericentric heterochromatin is lost for mutations in the H3K9me3 binding pocket and ones identified from ATRX syndrome patients (Iwase et al. 2011). In addition, the in vivo studies of Eustermann et al. (2011) showed that the readout of H3 with unmodified K4 and K9me3 by the ADD domain of ATRX was facilitated by recruitment of HP1, which independently recognizes H3K9me3. Such tripartite recognition could have the potential for spanning neighboring nucleosomes.
Figure 10.
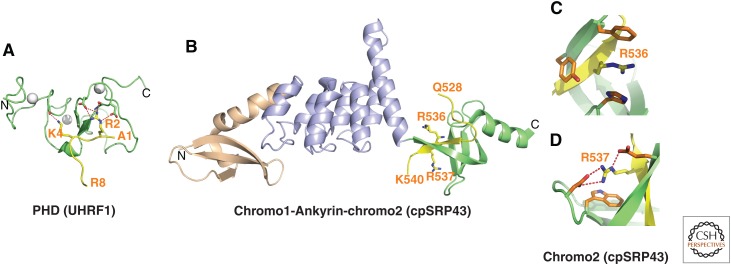
Structures of PHD finger and chromodomains bound to unmodified arginines. (A) 1.8-Å crystal structure of the complex of the PHD finger of UHRF1 bound to H3(1-9) peptide (PDB: 3SOU). The bound H3 peptide can be traced from A1 to R8. Zinc ions are shown by silver balls. (B) 3.18-Å crystal structure of the complex of the chromodomains and Ankyrin repeats of A. thaliana chloroplast signal recognition particle (cpSRP)43 bound to an RRKR (Arg-Arg-Lys-Arg)-containing peptide (yellow) (PDB: 3UI2). The side chains of R536 and R537 of the bound RRKR-containing peptide are positioned in adjacent pockets at the interface between the fourth Ankyrin repeat (purple) and the second chromodomain (green) in the complex. (C) Positioning of Arg536 of the RRKR-containing peptide within an aromatic-lined cage in the complex. (D) Positioning of Arg537 of the RRKR-containing peptide in a pocket lined by a Trp and two acidic side chains in the complex.
Figure 7.
Structures of expanded and paired modules bound to methylated lysine histone marks. (A) 1.6-Å crystal structure of the complex containing the ADD domain of ATRX bound to the H3(1-15)K9me3 peptide (PDB: 3QLA). The ADD GATA-1 and PHD fingers are colored in blue and green, respectively. Bound Zn ions are shown as silver balls. The H3 peptide containing K9me3 is traced from A1 to S10. (B) Enlargement of A showing intermolecular contacts between the K9me3-containing H3 peptide, traced from A1 to S10, complexed with the ADD domain. The me3 are shown as magenta spheres. (C) Ribbon and stick representation of K9me3 positioned to interact with the GATA-1 and PHD finger domains in the complex. (D) Surface and space-filling representation of surface complementarity between K9me3 and the walls of the pocket lined by the GATA-1 and PHD finger domains in the complex. (E) 1.7-Å crystal structure of the ternary complex of the Pygo PHD finger (in green) bound to H3(1-7)K4me2 peptide (in yellow) in the presence of the HD1 domain of BCL9 (in pink) (PDB: 2VPE). (F) 2.99-Å crystal structure of the complex consisting of the G9a ankyrin repeats (green) bound to an H3(1-15)K9me2 peptide traced from A7 to G13 in yellow (PDB: 3B95). The K9me2 aromatic-lined binding pocket is positioned between the fourth and fifth ankyrin repeats of G9a.
5.2. Pygo PHD Finger and Its Cofactor BCL9
The Pygopus (Pygo) protein contains a PHD domain, which, together with its cofactor BCL9, operates during development via the Wnt signaling pathway by regulating β-catenin-mediated transcription through an interaction with methylated H3K4 marks. This regulatory function relies on the interaction between the carboxy-terminal PHD domain of Pygo and the homology domain 1 (HD1) of BCL9. Structural studies on the Pygo PHD finger (Nakamura et al. 2007), its binary complex with BCL9 HD1 (Fiedler et al. 2008), and its ternary complex with added H3K4me2 (Fig. 7E) (Fiedler et al. 2008; Miller et al. 2010) identified the principles underlying K4me2 mark recognition. The H3K4me2 mark and HD1 are positioned on opposite faces of the Pygo PHD finger, with efficient mark recognition requiring association of the Pygo PHD finger with HD1. HD1 binding to the PHD finger, in fact, triggers an allosteric transition, thereby facilitating optimal recognition of the H3K4me2 mark. The K4me2-binding pocket is composed of four aromatic residues and an Asp with a twofold preference for the Kme2 over the Kme3 state.
5.3. Ankyrin Repeats
G9a and GLP (G9a-like) are euchromatin-associated KMTs composed of amino-terminal ankyrin repeats and a carboxy-terminal methyltransferase SET domain. These enzymes repress transcription following SET domain-catalyzed writing of H3K9me1 and H3K9me2 marks. It was subsequent to identifying the enzymatic H3K9me writing activity that G9a and GLP were also shown to be readers of H3K9me marks, using the ankyrin repeats within the protein (Collins et al. 2008). Ankyrin repeats involve helix-turn-helix-β-turn modules, aligned through the stacking of helices, with the β-turns projecting outward at right angles. The H3K9me1 and H3K9me2 peptides bind to the ankyrin repeats of G9a with intermediate binding affinity (Kd = 14 µm and 6 µm, respectively), with a crystal structure solved for the complex of ankyrin repeats with the H3K9me2 peptide. The bound H3 peptide is sandwiched between β-turns and helices of the fourth and fifth ankyrin repeats, with the dimethylammonium group of K9me2 inserted into an aromatic pocket lined by three Trp residues and a Glu (Fig. 7F). Intermolecular recognition involves peptide residues 9 to 11, which includes K9me2. Mutation of these peptide or aromatic cage residues adversely impacts on complex formation (Collins et al. 2008). Perturbation of methyllysine recognition by the ankyrin repeats, however, has no effect on G9a’s SET domain-mediated methyltransferase activity, indicating that the reading and writing domains function independently.
6. READOUT OF METHYLARGININE MARKS BY TUDOR MODULES
Tudor modules, as well as recognizing lysine methylation (Kme) marks, recognize methylated arginine (Rme) marks, and the latter can be subdivided into two classes: the canonical and the expanded Tudor reader modules. Tudor proteins play key roles in a range of cellular processes ranging from germ cell development to RNA metabolism, processing, and silencing, as well as DNA damage response and chromatin-remodeling (reviewed in Bedford and Clarke 2009; Siomi et al. 2010; Chen et al. 2011). The structural studies described in this section are relevant to chromatin biology not so much for directly reading arginine methylation marks contained on the histone proteins, but on proteins such as PIWI, which are part of the machinery involved in chromatin-templated processes such as RNA silencing. Although we describe the details of the binding structures of Tudor domains with nonchromatin targets, the details become relevant to epigenetic regulation in that they affect Piwi, a component of RNA interference (RNAi) pathways.
6.1. Canonical Tudor Domains
The first insights into the canonical Tudor domain fold emerged from structural studies of the SMN (survival of motor neuron) protein. When mutated, SMN causes spinal muscular atrophy, a degenerative motor neuron disease. The SMN protein contains a single highly conserved canonical Tudor domain required for uridine-rich small nuclear ribonucleoprotein complex assembly. The Tudor domain facilitates binding to the seven-membered hetero-oligomeric ring-shaped Sm proteins. The SMN Tudor domain forms a strongly bent five-stranded antiparallel β-sheet barrel-like fold (Selenko et al. 2001), with a cluster of aromatic amino acids forming a cage on one face of the Tudor domain (Sprangers et al. 2003). This targets methylated arginines positioned within the carboxy-terminal Arg-Gly-rich tails of Sm proteins (Brahms et al. 2001). The Tudor-containing protein, TDRD3, is also a reader of methylated arginine marks on histone tails (H3R17me2a and H4R3me2a), acting as a transcriptional coactivator (Yang et al. 2010).
NMR-based solution structures have been solved for the Tudor domains of SMN and SFP30 bound to short symmetrical dimethylated arginine peptides corresponding to the carboxy-terminal Arg-Gly-rich tails of Sm proteins (Tripsianes et al. 2011). The binding specificity decreases in the order symmetrical dimethylated arginine (Rme2s) > asymmetrical dimethylated arginine (Rme2a) > monomethylated arginine (Rme1). Binding specificity for Rme2s marks was independent of residues within the peptide on either side of the mark. The dimethyl guanidinium moiety of the Rme2s mark, in an anti–anti alignment (anti orientation of both N-CH3 bonds in relation to the side chain C-Nε bond), inserts into a cage formed by four aromatic residues and an Asn residue (Fig. 8A), in which it is stabilized by cation–π interactions. The E134K mutation associated with spinal muscular atrophy impairs a hydrogen bond between the Glu134 side chain and hydroxyl group of a Tyr residue lining the aromatic cage, highlighting the role of aromatic amino acids lining the cage for optimally orienting the dimethyl guanidinium group of the Rme2s, ensuring recognition.
Figure 8.
Structures of expanded and paired modules bound to methylated arginine histone marks. (A) NMR solution structure of the complex containing the SMN Tudor domain (green ribbon representation) bound to a symmetrical Rme2s-containing peptide (yellow) (PDB: 4A4E). The methyl groups are illustrated with magenta spheres and the aromatic-lined cage in orange. (B) 2.8-Å crystal structure of the complex containing the SND1 extended Tudor module bound to the amino-terminal PIWI peptide, traced from R10 to R17, with the R14me2s modification (PDB: 3NTI). The core fold of the Tudor domain is shown in green, whereas the extensions are shown in blue. (C) Enlargement of B showing the positioning of R14me2s in the aromatic-lined cage of the SND1 Tudor domain in the complex.
6.2. Expanded Tudor Domains
Certain proteins contain expanded multiple tandem repeats as a 180-residue module, which itself consists of a 60-residue canonical Tudor core domain and flanking amino- and carboxy-terminal conserved elements. Many of these Tudor proteins are germline-specific and exert their regulatory effect via the recognition of arginine methylation at the amino-terminal end of PIWI family proteins. PIWI proteins are themselves important regulators of the germline via the RNA silencing pathway. This pathway functions predominantly in silencing transposable elements in early development (reviewed in Siomi et al. 2010; Chen et al. 2011).
Several structure–function studies have been performed on complexes containing the expanded Tudor domains bound to methylated arginine peptides from the Arg-Gly/Arg-Ala-rich amino terminus of PIWI-family proteins. These studies have shed light on the principles underlying recognition of the symmetrical dimethylated arginine mark (Rme2s) (Liu et al. 2010a; Liu et al. 2010b; Mathioudakis et al. 2012). The structures of complexes containing other Tudor group proteins led to similar conclusions about the nature of expanded Tudor domain interactions with methylated arginines (Liu et al. 2010a; Liu et al. 2010b; Mathioudakis et al. 2012). The results of the complex between the SND1 (staphylococcal nuclease domain-containing 1) extended Tudor module and the R14me2s-containing amino-terminal PIWI peptide is discussed here (Liu et al. 2010b).
The SN-like domain of the SND1 protein is split into two segments by the canonical Tudor domain with an α-helical linker connecting the Tudor (in green) and SN-like (in blue) domains (Fig. 8B). This so-called extended Tudor module forms an OB (oligonucleotide and oligosaccharide-binding) fold. The R14me2s-containing amino-terminal PIWI peptide binds with directionality within a wide and negatively charged groove of the SND1 Tudor domain (Fig. 8B), in the process inserting the planar dimethyl-guanidinium group of R14me2s in an anti–syn orientation into an aromatic cage lined by four aromatic amino acids and an Asn residue (Fig. 8C). SND1 can also form a complex with the PIWI R4me2s-containing peptide and its structure has also been reported (Liu et al. 2010a).
Unlike canonical Tudor domains, binding affinity on complex formation is affected by proximal residues flanking the Rme2s mark. And both the canonical Tudor domain and flanking amino- and carboxy-terminal extensions are equally required for binding because deletion of either of the extensions resulted in loss of binding. Mutation of either individual aromatic amino acids or the Asn (involved in hydrogen bonding to the dimethyl guanidinium group) lining the pocket resulted in 11- to 22-fold loss in binding affinity. The SND1 extended Tudor module shows specificity for the R4me2s mark (Kd = 10 µm), with reduction in binding by fourfold for the R4me2a mark, twofold for the R4me1 mark, and a larger ninefold for the unmodified R4 mark. It is likely that monomethylation would reduce hydrophobic interactions with the aromatic pocket, and asymmetrical dimethylation could disrupt the intermolecular hydrogen bond with the Asn, accounting for the modest reduction in binding affinity. Interestingly, the R4me2s-containing amino-terminal PIWI peptide binds to the SND1 extended Tudor in the opposite orientation to the R14me2s peptide yet retains insertion of R4me2s into the aromatic cage pocket, thereby suggesting plasticity in the recognition process. Functional studies have identified an intricate interplay between writers and readers of the Rme2s mark and the mark itself and in regulating transposon silencing and germ cell development (reviewed in Chen et al. 2011). A remaining challenge in the field will be to identify potential readers of Rme2a and Rme1 marks and the principles underlying optimal selectivity as a function of methylation state of arginine.
7. READOUT OF UNMODIFIED LYSINE MARKS
Although the focus of this article is largely on reader modules that target methylated lysine marks, it should be noted that, as introduced in Sections 4.2 and 5.1, certain reader modules target unmodified lysines on histone tails. Such recognition events can then be blocked or weakened by methylation with functional consequences.
7.1. PHD Fingers
The first indication that unmethylated lysines contribute to molecular recognition emerged from the structure of the BHC80 PHD finger bound to the histone H3(1-10) peptide (Kd = 30 µm) (Lan et al. 2007). A high-resolution (1.4-Å) crystal structure study readily revealed details of the intermolecular contacts; the ammonium group of K4 hydrogen bonded to both an acidic side chain and backbone carbonyl group (Fig. 9A). There was no room to accommodate a methylated K4 in this complex, thereby accounting for the specificity of unmodified lysine recognition. The intermolecular interactions are stabilized by interdigitation of side chains involving the H3 peptide (R2 and K4) with the PHD finger (Met and Asp); binding is lost on mutation of these Met and Asp residues (Lan et al. 2007). RNAi knockdown of BHC80 results in derepression of LSD1 target genes, whereas ChIP studies show that BHC80 and LSD1 mutually depend on each other to associate with chromatin. These results point to BHC80 function being coupled to that of LSD1, which is consistent with BHC80 having a role in LSD1-mediated gene repression via the recognition of unmodified H3K4 (Lan et al. 2007).
Figure 9.
Structures of reader modules bound to lysines and arginines. (A) 1.43-Å crystal structure of the complex of the PHD finger of BHC80 bound to H3(1-10) peptide (in yellow) (PDB: 2PUY). The bound H3 peptide can be traced from A1 to S10. Zinc ions are shown as silver balls. (B) 1.5-Å crystal structure of the complex of the WD40 motif of WDR5 bound to H3(1-9)K4me2 peptide. The bound K9me2-containing peptide can be traced from A1 to R8 (PDB: 2H6N). (C) Insertion of R2 into the central channel of the WD40 motif in the H3(1-9)K9me2-WDR5 complex. (D) Insertion of R2me2s into the central channel of the WD40 motif in the H3(1-15)R2me2sK9me2-WDR5 complex solved at 1.9 Å (PDB: 4A7J). (E) 3.2-Å crystal structure of the complex of the WD40 motif of p55 bound to H4(15-41) peptide (PDB: 3C9C). The bound H4 peptide can be traced from K31 to G41.
Other examples of reader modules that recognize unmodified lysines at H3K4 include DNMT3l (Ooi et al. 2007), the ADD domains of DNMT3A and ATRX (discussed in Sec. 5.1), PHD finger domains of TRIM24 and TRIM33 (to be discussed in Sec. 9), and tandem Tudor domains of UHRF1 and SSH1 (discussed in Sec. 4.2). Recognition of unmodified H3K4 and discrimination against methylation at this position is achieved by intermolecular hydrogen bonding between the ammonium group of lysine and acidic side chains, or the backbone carbonyl groups, or a combination of both.
8. READOUT OF UNMODIFIED ARGININE MARKS
The readout of unmodified lysines can solely be achieved by PHD fingers, as outlined in Section 7. In contrast, unmodified arginines can be read by several distinct reader modules including WD40 motifs, PHD fingers, and chromodomains. We outline below the distinct recognition principles by which each of these reader modules binds with unmodified arginine.
8.1. WD40 Motifs
WDR5 is a member of the WD-40 (for 40 amino acids) repeat family. Individual WD40 repeats form a toroidal B-propeller fold, much like the blades of a propeller. WDR5 is a common component of the SET1-family of KMT complexes that play an essential role in Hox gene activation and vertebrate development. Several groups simultaneously solved the structure of the H3K4me2 peptide-WDR5 complex (Couture et al. 2006; Han et al. 2006; Ruthenburg et al. 2006; Schuetz et al. 2006) with the anticipation that the structure would identify the principles underlying H3K4me2 recognition, given that WDR5 associates with and is essential for H3K4 methylation and vertebrate development (Wysocka et al. 2005). The H3K4me2 peptide binds on the surface above the central channel of the toroid-shaped fold of WDR5 (Fig. 9B), where it is stabilized by a set of extensive intermolecular contacts between the bound H3 peptide and WDR5. Unexpectedly, it is the side chain of unmodified Arg2 rather than K4me2 that inserts into the narrow central channel of the toroidal β-propeller fold (cavity insertion mode), in which it is stabilized by stacking with staggered Phe side chains and oriented through direct and water-mediated hydrogen bonds (Fig. 9C).
A further surprise came from a recent structure–function study demonstrating that the central channel of the WDR5 WD40 fold also accommodates the symmetrically dimethylated H3R2me2s mark (Fig. 9D), but not its asymmetrical H3R2me2a counterpart (Migliori et al. 2012). Comparison of the H3R2 and H3R2me2s complexes indicated that the guanidinium group forms direct and water-mediated hydrogen bonds involving two anchored water molecules in the H3R2 complex (Fig. 9C), with one of these waters displaced in the H3R2me2s complex, together with protrusion of the dimethylated side chain toward the Phe ring (Fig. 9D) to form enhanced hydrophobic interactions. Functionally, the H3R2me2s mark retains euchromatic genes in a poised state ready for transcriptional activation following transit from the cell cycle and differentiation.
Another study has investigated the structure of WRD5 complexed with a peptide from the KMT writer enzyme, MLL1 (mixed lineage leukemia 1) (Patel et al. 2008; Song and Kingston 2008). Unexpectedly, the histone H3-like MLL1 peptide bound in an almost identical orientation as an H3 peptide and, in addition, similarly inserted the Arg side chain into the central channel of the WD40 scaffold. It was also shown that H3K4me-containing peptides compete with the MLL1 peptide for WDR5 because disruption of WDR5-MLL1 binding was more effective by H3K4me1/me2 peptides over H3K4me0/me3 peptides. One interpretation of these observations is that there is a delicate interplay between components of the MLL1 complex, namely WDR5 (the reader), the SET domain of MLL1 (the writer), and the histone H3 tail (the substrate) (Song and Kingston 2008).
In another structural study, the WD40 motif of p55, a common component of several chromatin-modifying complexes, bound the first A-helix of H4 within a surface binding channel located on the side of the β-propeller scaffold (Fig. 9E) (Song et al. 2008). Because the first helix of H4 is buried within the canonical fold of the nucleosome, a substantial alteration of the conformation of this H4 segment was observed on complex formation with p55. Further, it was shown that the histone H4-binding pocket was important for the functional activities of p55-containing complexes involved in chromatin assembly (CAF1), remodeling (NURF), and deacetylation (NuRD) (Song et al. 2008).
8.2. PHD Fingers
The PHD finger is a reader module that can recognize unmodified arginines. A recent example includes the recognition of unmodified R2 on the H3 tail by the PHD finger of UHRF1, an important regulator of CpG methylation (discussed in Sec. 2 of Cheng 2014; Rajakumara et al. 2011b; see also Hu et al. 2011; Wang et al. 2011; Xie et al. 2012). In this complex, the guanidinium group of R2 participates in an extensive intermolecular hydrogen bond network (Fig. 10A), and methylation of H3R2, but not H3K4 or H3K9, disrupts complex formation. Functionally, the ability of UHRF1 to suppress the expression of its target genes is dependent on the PHD finger of UHRF1 binding to unmodified H3R2, thereby linking this recognition event involving UHRF1 to regulation of euchromatic gene expression (Rajakumara et al. 2011b).
Another example of a PHD finger that recognizes the Rme0 mark is the MOZ (monocytic leukemia zinc-finger protein) PHD tandem cassette, detailed in Section 9.3.
8.3. Chromodomains
Recent research reported an unexpected interaction in the cytosol between cpSRP43 (composed of chromodomains and Ankyrin repeats) and a peptide containing the RRKR motif from cpSRP54. The latter is associated with the chloroplast signal recognition particle in A. thaliana (Holdermann et al. 2012). The structure of cpSRP43ΔCD3 bound to an RRKR-containing peptide of cpSRP54 is shown in Fig. 10B, with the peptide binding at the interface between the fourth Ankyrin repeat and the second chromodomain (CD2) (Kd = 6.4 µm). In the complex, the RRKR-containing peptide adopts an extended conformation except for a type-II β-turn at its amino terminus, with recognition occurring through a β-completion mode within the hydrophobic binding groove of CD2. The specificity of molecular recognition resides in two adjacent arginines in the peptide (Arg536 and Arg537), which are positioned in adjacent pockets of cpSRP43, one of which is composed of a three-aromatic residue-lined cage (Arg536; Fig. 10C), whereas the other is composed of an aromatic amino acid and two acidic residues (Arg537; Fig. 10D), with replacement of either Arg on the peptide or Tyr and Trp residues lining the pockets, abolishing complex formation and its functional readout (targeting of Ankyrin repeats to light-harvesting chlorophyll a,b binding proteins). The investigators also propose that a similar recognition mechanism likely exists for the interaction between cpSRP43 and the carboxy-terminal tail of the membrane insertase Alb3 involved in recruiting cpSRP to the thylakoid membrane (Falk et al. 2010). This insightful contribution introduces the novel concept of readout of adjacent unmodified arginines by a twinned aromatic cage (Holdermann et al. 2012) to generate the cpSRP complex (Goforth et al. 2004) and defines an unexpected nonnuclear function for chromodomains in posttranslational targeting.
9. MULTIVALENT READOUT BY LINKED BINDING MODULES AT THE PEPTIDE LEVEL
Given the low affinity in the readout of single marks by their reader modules, it should not be surprising that the affinity of binding can be increased following multivalent readout of two or more marks by linked binding modules (reviewed in Ruthenburg et al. 2007b; Wang and Patel 2011). In this section, we outline recent results on histone combinatorial PTM readout at the peptide level and, in the following section, discuss results at the nucleosomal level.
9.1. PHD Finger-Bromo Cassettes
The adjacent positioning of PHD finger and bromodomain to form the PHD finger-bromodomain (designated PHD-bromo) cassette is the most frequently observed combination of dual reader modules impacting on epigenetic regulation (Ruthenburg et al. 2007b). The PHD finger and bromodomain read Kme and Kac marks, respectively, and different proteins with PHD-bromo cassettes show the potential for reading different combinations of these two marks. To date, structure–function studies have been undertaken on the PHD-bromo cassettes of BPTF (Li et al. 2006; Ruthenburg et al. 2011; Wysocka et al. 2006), MLL1 (Wang et al. 2010b), TRIM24 (Tsai et al. 2010b), TRIM33 (Xi et al. 2011), and KAP1 (Ivanov et al. 2007; Zeng et al. 2008). There are similarities and differences in structure–function aspects associated with these PHD-bromo cassettes at the peptide level. We outline their distinct roles in epigenetic regulation below.
The PHD-bromo cassette of the BPTF component of the NURF chromatin remodeler introduced in Section 2.1 provided the first insights into the structure and function of this cassette. The structure of the BPTF PHD-bromo cassette showed that the two domains are separated by an α-helical linker (Fig. 11A), resulting in a fixed distance between domains, with their histone mark-binding pockets defined by a fixed relative orientation (Li et al. 2006). The PHD finger binds the H3K4me3 mark (Li et al. 2006; Wysocka et al. 2006), whereas the bromodomain binds a range of Kac marks with a preference for H4K16ac (Ruthenburg et al. 2011). Thus, the BPTF PHD-bromo cassette shows capabilities for targeting distinct marks on different histone tails (Ruthenburg et al. 2011).
Figure 11.
Structures of PHD-bromo cassettes involved in multivalent readout. (A) 2.0-Å crystal structure of the PHD-bromo cassette of BPTF bound to H3(1-15)K4me3 peptide (PDB: 2F6Z). A separate 1.8-Å crystal structure of the BPTF bromodomain bound to H4(12-21)K16ac peptide was also solved (PDB: 3QZS) and that information was superpositioned on the structure shown in this panel. The bound H3(1-15)K4me3-containing peptide can be traced from A1 to T6, whereas the bound H4(12-21)K16ac-containing peptide can be traced from K14 to V21 in the complexes. (B) 1.9-Å crystal structure of the MLL1 PHD-bromo cassette bound to H3(1-9)K4me3 peptide (PDB: 3LQJ). The bound H3(1-9)K4me3-containing peptide can be traced from A1 to T6 in the complex. (C) 2.7-Å crystal structure of the TRIM33 PHD-bromo cassette bound to H3(1-22)K9me3K18ac peptide (PDB: 3U5O). The bound H3(1-22)K9me3K18ac peptide can be traced from A1 to L20 in the complex. (D) The crystal structures of the TRIM24 PHD-bromo cassette bound to H3(1-10) peptide (2.0 Å) (PDB: 3O37) and bound to H3(13-32)K23ac peptide (1.9 Å) (PDB: 3O37). The structures were superpositioned to generate the composite structure shown in this panel. The bound H3(13-32)K23ac peptide can only be traced from T22 to T32 in the complex.
The leukemia-inducing MLL1 protein targets its SET domain methyltransferase activity to HOX gene promoters during hematopoiesis (Milne et al. 2002). This protein contains adjacently positioned PHD finger (PHD3) and bromodomain, which adopt the characteristic individuals folds of these motifs, with the connecting linker adopting a turn segment that leads into the extended αZ-helix of the bromodomain (Fig. 11B) (Wang et al. 2010b). This alignment results in interactions between the PHD and bromodomains that manifests as an increase in binding affinity for PHD finger recognition of the H3K4me3 mark as compared with the isolated PHD finger. The K4me3 side chain positions into an aromatic-lined pocket (Fig. 11B), and a conformational change closes an additional face of the aromatic-lined cage on complex formation. The bromodomain part of the cassette has lost its ability to target Kac-containing histone peptides. ChIP measurements established colocalization of MLL1 and H3K4me3 marks on HOX genes, suggesting that the binding of MLL1 PHD3 to the H3K4me3 mark contributes to MLL1 localization at target genes (Milne et al. 2010; Wang et al. 2010b).
Structure–function studies on the carboxy-terminal PHD-bromo cassettes of TRIM24 (Tsai et al. 2010b) and TRIM33 (Xi et al. 2011) provide further insights into combinatorial recognition of dual marks. For both TRIM24 and TRIM33, the adjacently positioned PHD finger and bromodomain folds interact extensively with each other to generate a single folded unit (Fig. 11C,D). Histone peptide binding studies on TRIM33 elucidated that the PHD finger recognized a combination of unmodified K4 (me0) and K9me3 on the same H3 tail, whereas the bromodomain recognized K18ac. Further, the binding affinity for H3(1-28)K9me3K18ac (Kd = 0.06 µm) dual mark was greater than the sum of binding to individual H3(1-28)K9me3 (Kd = 0.20 µm) and H3(1-28)K18ac (Kd = 0.21 µm) marks, consistent with combinatorial recognition occurring, even at the peptide level.
The crystal structure of the TRIM33 PHD finger-bromo cassette bound to H3(1-22)K9me3K18ac peptide has been solved (Fig. 11C) and provides a molecular explanation for the results obtained from binding studies. Key intermolecular contacts contributing to the specificity of recognition include interaction between the TRIM 33 amino terminus and H3 peptide backbone, between the unmodified K4 and three acidic side chains, as well as stacking between K9me3 and a Trp side chain, all involving the PHD finger. Additional specificity is associated with positioning of K18ac in an atypical binding pocket on the bromodomain (Xi et al. 2011). Functionally, nodal signaling-induced formation of TRIM33-Smad2/3 and Smad4-Smad2/3 complexes triggered the interaction between TRIM33-Smad2/3 and the H3K9me3 mark, thereby displacing the chromatin-compacting factor HP1γ, and making nodal response elements accessible to Smad4-Smad2/3 for RNA polymerase II recruitment. In essence, nodal TGF-β signals use the poised H3K9me3 mark as a platform to switch master regulators of stem cell differentiation from the poised to the active state (Xi et al. 2011).
The TRIM24 bromodomain, which is part of the PHD-bromo cassette, targets the H3K23ac mark (Tsai et al. 2010b). The difference in acetyllysine residue recognition between TRIM24 (H3K23ac recognition) and TRIM33 (H3K18ac recognition) is a reflection of distinct contributions of sequences that flank the Kac mark, translating into different intermolecular recognition capabilities of the two complexes. Crystal structures of two TRIM24 PHD finger-bromo cassette complexes, one bound to H3K9me3-, the other to H3K18ac-containing peptides, are superimposed and shown in Fig. 11D. The spatial alignment and extensive interaction between the paired reader domains generates a continuous binding surface, enhancing the combinatorial readout capacity (Tsai et al. 2010b). TRIM24 binds chromatin with H3K4me0 and H3K23ac marks and also to the estrogen receptor protein, which itself binds to estrogen response elements when signaled by estrogen to activate estrogen-dependent genes associated with cellular proliferation and tumor development. Thus, it is not surprising that the overexpression of TRIM24 frequently observed in breast cancer patients correlates with poor survival. The coexistence of estrogen response elements and distinct dual-marked H3 chromatin, thus, provides a distinct system conducive to regulating estrogen-dependent target genes (Tsai et al. 2010b).
9.2. The Tandem Tudor-PHD Finger Cassette
A striking pair of studies shows the multivalent readout (i.e., of multiple PTMs) on H3 tails by the tandem Tudor-PHD cassette, a linked binding module of UHRF1 (Arita et al. 2012; Cheng et al. 2013). The X-ray, NMR, small-angle X-ray scattering, and binding studies by Arita et al. (2012) show that a UHRF1-H3 peptide complex forms with a 1:1 stoichiometry and Kd = 0.37 µm. This strong binding affinity is suggestive of combinatorial recognition given that the tandem Tudor domain or PHD finger alone bind with affinities of Kd = 1.75 µm and Kd = 1.47 µm, respectively. The crystal structure of the complex is shown in Figure 12A, with the 17-residue linker between the tandem Tudor (in cyan and blue) and PHD finger (in green) domains essentially packed between the Tudor domains, with minimal contacts between the Tudor domains and the PHD finger. There is a central hole in the overall architecture of the protein scaffold, which accommodates the compactly folded bound H3K9me3-containing peptide in the complex (expanded view in Fig. 12B). On the H3K9me3-containing peptide side, residues 1 to 4 participate in intermolecular β-sheet formation, with the guanidine group of unmodified Arg2 involved in a network of intermolecular hydrogen bonds such that methylation at this position results in reduced binding affinity (see Sec. 8.2). Peptide residues 5 to 8 unexpectedly adopt an α-helical conformation stabilized by a N-capping hydrogen bond, but otherwise do not form specific intermolecular contacts in the complex. The 9-10 peptide segment forms contacts with the first Tudor domain (in blue), thereby positioning the trimethylammonium group of K9me3 within an aromatic cage. The phosphorylation state of the adjacent Ser10 (and also Thr3) residue of the bound peptide significantly impacts on binding affinity. On the UHRF1 protein side, the importance of the linker element connecting the tandem Tudor and PHD finger was highlighted by the loss of binding following dual Arg to Ala mutations of adjacent Arg residues within the linker segment. In addition, phosphorylation of a Ser (target of protein kinase A) within the linker segment resulted in a 30-fold loss in binding affinity. It is conceivable that modifications (such as phosphorylation) within the linker segment could act as a switch between potential regulatory pathways used by UHRF1 during its functional role in the maintenance of DNA methylation and transcription repression (detailed below in Sec. 12.1; Arita et al. 2012).
Figure 12.
Structures of linked binding modules involved in multivalent readout at the peptide and nucleosomal levels. (A) 2.9-Å crystal structure of the UHRF1 tandem Tudor-PHD finger cassette bound to H3(1-13)K9me3 peptide (PDB: 3ASK). The tandem Tudor domains are shown in cyan and purple, whereas the PHD finger is shown in green. Zinc ions are shown as silver balls. The bound H3(1-13)K9me3-containing peptide can be traced from A1 to S10. (B) Enlargement of A, showing details of the intermolecular contacts between H3(1-13)K9me3-containing peptide (A1 to S10) and the tandem Tudor-PHD finger cassette of UHRF1. (C) 1.47-Å crystal structure of the complex of the tandem PHD finger cassette of MOZ bound to H3(1-18)K14ac peptide (PDB: 3V43). There is a bound acetate (in space-filling representation) from buffer bound in a pocket in the amino-terminal PHD finger (in blue). The bound H3(1-18)K14ac-containing peptide can be traced from A1 to A7, in which it is bound to the carboxy-terminal PHD finger (in green). (D) glutathione S-transferase (GST) pull-down of modified nucleosomes with semisynthetic histones produced by expressed protein ligation. Nucleosomes containing dual marks involving H4K12ac, H4K16ac, or H4K20ac in combination with H3K4me3 are pulled down with resin bound GST-BPTF PHD-bromo cassette and detected by autoradiography after native gel electrophoresis. (D, Reprinted from Ruthenburg et al. 2011.)
9.3. Tandem PHD Finger Cassettes
The histone acetyltransferase (HAT), MOZ, which contains a pair of tandem PHD fingers, is important for the expression of HOX genes and contributes to embryonic and postnatal development. Two groups have now shown that the tandem PHD fingers of MOZ target H3 peptides containing unmodified R2 and K14ac marks (Ali et al. 2012; Qiu et al. 2012). In the crystal structure of the complex bound to an H3(1-18)K14ac peptide (Fig. 12C), the R2 side chain forms a network of hydrogen bonds to the side chains of Asp residues on the second PHD finger (in green), whereas the binding site for K14ac on the first PHD finger (in blue) (by analogy with the related structure of the tandem PHD fingers of DPF3b bound to H3K14ac peptide; Zeng et al. 2010) was occluded by bound acetate from buffer and crystal packing interactions (Qiu et al. 2012). NMR-binding studies by both groups showed that methylation of either R2 or K4 significantly impacted on complex formation. Fluorescence microscopy studies showed that both PHD fingers are necessary for binding to H3K14ac in vivo and localization to chromatin (Ali et al. 2012). Further, ChIP studies showed that the tandem PHD finger facilitated the localization of MOZ to the promoter locus of the HOXA9 gene, upregulating HOXA9 messenger RNA (mRNA) levels through promotion of H3 acetylation (Qiu et al. 2012).
Recently, it has been shown that the tandem PHD fingers of CHD4, a catalytic subunit of the NuRD (nucleosome remodeling and deacetylase) complex, are required for NuRD complex transcriptional repression. The tandem PHD fingers concurrently bind both H3 tails within the same nucleosome, displacing HP1γ from pericentric sites (Musselman et al. 2012c). This interaction is modulated by PTMs on the H3 tail, with binding enhanced by H3K9me and H3K9ac marks, and abolished by H3K4me mark.
10. MULTIVALENT READOUT BY LINKED BINDING MODULES AT THE NUCLEOSOMAL LEVEL
Although the emphasis of structure–function studies investigating the readout of Kme marks has been perfomed at the peptide level, the major challenge has been to extend these studies to the nucleosomal level. Recent progress in this area centers on the BPTF PHD-bromo cassette described in Section 9.1 at the H3 peptide level, now outlined below at the nucleosomal level.
10.1. PHD Finger-Bromo Cassette
In a seminal contribution, Ruthenburg et al. (2011) extended studies of the readout of dual histone marks by the BPTF PHD-bromo cassette from the peptide to the nucleosomal level, thereby experimentally validating aspects of multivalent engagement of chromatin marks by linked binding modules (Ruthenburg et al. 2007b). Contrary to peptide level analysis, they showed that the bromodomain of BPTF displayed discrimination among different acetylated lysines on H4 (K12ac, K16ac, K20ac) with marked selectivity for H4K16ac in combination with H3K4me3 at the mononucleosomal level (Fig. 12D). This is corroborated by the observation that a significant pool of nucleosomes in vivo are doubly modified for H3K4me3 and H4K16ac and the demonstration that the BPTF PHD-bromo cassette colocalizes with the H3K4me3 and H4K16ac marks in the genome (Ruthenburg et al. 2011). Intriguingly, the writers of these marks—MLL1 for H3K4me3 and MOF for H4K16ac—interact with each other (Dou et al. 2005). Structure-based modeling suggests that the BPTF PHD-bromo cassette can potentially be snugly docked on a mononucleosome, with bound PHD finger and bromodomains simultaneously recognizing adjacently positioned H3 and H4 tails (Ruthenburg et al. 2007b).
11. ALTERNATE FUNCTIONAL ROLES FOR PHD-BROMO CASSETTES
Recent structure–function studies have identified new non-chromatin-associated roles for the PHD-bromo cassettes beyond their identities as multivalent readers of Kme and Kac marks at the peptide (Sec. 9.1; Li et al. 2006) and nucleosomal level (Sec. 10.1; Ruthenburg et al. 2011). Two examples are described here, outlining alternate mechanisms of functional synergy between the PHD finger and bromodomain in the PHD-bromo cassette.
11.1. PHD Finger as an E3 SUMO Ligase of Adjacent Bromodomain
The PHD-bromo cassette of the KAP1 (KRAB-associated protein 1) corepressor binds neither the expected Kme nor Kac marks under conditions in which both reader domains interact extensively with each other (Ivanov et al. 2007; Zeng et al. 2008). Rather, the PHD finger unexpectedly functions as an intramolecular E3 small ubiquitin-related modifier (SUMO) ligase, which through interaction with the E2 enzyme facilitates SUMOylation of the linked bromodomain. This recruits the NuRD complex via SUMO-interacting motifs to facilitate the establishment and stabilization of silent chromatin. This silencing occurs through the NuRD-catalyzed deacetylation of nucleosomes, which stimulates SETDB1 (an H3K9 KMT) to methylate H3K9. This mark is, in turn, targeted by HP1γ to induce the silenced state (reviewed in Peng and Wysocka 2008). It should be noted that site-specific phosphorylation of a Ser located near the bromodomain of KAP1 appears to inhibit KAP1 SUMOylation (Li et al. 2007b), implying a role for PTM-mediated cross talk in the regulation of KAP1 function.
11.2. PHD-Bromo Cassette as a Regulatory Platform
The MLL1 gene is essential for embryonic development and hematopoiesis, as well as a frequent target for recurrent chromosomal translocations, resulting in transformation of hematopoietic precursors into leukemia stem cells. MLL1 has been shown to maintain Hoxa9 expression in stem and progenitor cells, yet is also required for Hoxa9 silencing during blood cell maturation. Failure to silence results in the expansion of self-renewing progenitor cells and the onset of leukemia (reviewed in Grow and Wysocka 2010). Indeed, induction of the epigenetically regulated switch during haematopoiesis depends on the PHD finger 3 of MLL1 (Xia et al. 2003), given its loss results in the immortalization of hematopoietic stem cells (Chen et al. 2008). The MLL1 PHD3-bromo cassette is targeted by the cyclophilin CyP33, composed of PPIase (peptidyl prolyl isomerase) and RRM (RNA recognition motif) domains. Functional studies have established that the MLL1-CyP33 interaction is required for the histone deacetylase-mediated repression of HOX target genes during blood cell maturation in vivo (Fair et al. 2001; Xia et al. 2003). Structural and biochemical studies show that the PPIase domain of CyP33 regulates the conformation of the MLL1 PHD-bromo cassette (Fig. 13A). It does this through cis-trans isomerization of a linker proline, thereby disrupting the PHD3-bromo interface and facilitating access of an otherwise occluded MLL1 PHD3 finger in the cassette to the Cyp33 RRM domain (Fig. 13B) (Wang et al. 2010b). This perspective is supported by an NMR-based structure of a segment of the MLL1 PHD finger bound to the Cyp33 RRM domain (Fig. 13C), which implies that the H3K4me3 mark and RRM domain target distinct surfaces of PHD3 and, further, can integrate distinct regulatory inputs by coexisting as a ternary complex. Collectively these results highlight the role of the MLL1 PHD3-bromo cassette both as a regulatory platform and a switch; the cis-trans proline isomerization within the linker segment connects H3K4me3 readout (see Fig. 11B) to CyP33 and HDAC-mediated repression (Wang et al. 2010b). Given that cellular RNAs can compete for the RRM motif in CyP33, unanswered questions remain as to the potential role of RNAs in the targeting, stabilization, and/or release of CyP33 from MLL1. Two additional studies on this system have focused solely on the binding of CyP33 to the MLL1 PHD3 finger in efforts to provide insights into regulation by CyP33 of MLL1-mediated activation and repression (Hom et al. 2010; Park et al. 2010).
Figure 13.
Structures of BPTF PHD-bromo cassette with cis and trans linker prolines. (A) 1.72-Å crystal structure of the MLL1 PHD-bromo cassette with a cis linker proline (circled in red) in the free state (PDB: 3LQH). The PHD finger and bromodomain are colored in green and blue, respectively. (B) Model of the MLL1 PHD-bromo cassette with a trans linker proline (circled in red) with this alignment stabilized by a bound RRM domain of CyP33 (magenta). (C) NMR solution structure of the complex containing the MLL1 PHD3 fragment (1603–1619, in green) and RRM domain of CyP33 (2-82, in magenta) (PDB: 2KU7).
12. CROSS TALK BETWEEN HISTONE MARKS
A notable feature of histone tails is their extreme density (adjacent or closely spaced) and variety of marks (see Fig. 6 of Allis et al. 2014). In some cases, a single amino acid such as lysine can be labeled by more than one kind of mark, including methylation, acetylation, or ubiquitination (e.g., H3K9). Such features have led to the concept of dynamic “binary switches” in which the readout of one mark is modulated by an adjacent/nearby second mark, thereby impacting on processes ranging from gene transcription, DNA replication, repair, and recombination (reviewed in Fischle et al. 2003a,b; Latham and Dent 2007; Garske et al. 2010; Oliver and Denu 2011). In this regard, the concept of “modification cassettes” was proposed (Fischle et al. 2003a), in which the segment of a histone tail consists of adjacent positions that are subject to different modifications, such as the R2(me)-T3(ph)-K4(me) and R26(me)-K27(me/ac)-S28(ph) segments on H3 (Fig. 1B). Thus, the readout of lysine methylation marks can be impacted by nearby serine/threonine phosphorylation, arginine methyation, lysine acetylation, and lysine ubiquitination marks. These can occur in cis within the same histone (intrahistone), in trans between histone pairs (interhistone, e.g., between H3 and H4), or even within (intranucleosomal) or across (internucleosomal) nucleosomes (see Fig. 10 of Allis et al. 2014). The binding of a reader module to a mark has the potential of sterically blocking an adjacent modification site or, conversely, may recruit an additional module to modify neighboring residues. Several examples of such cross talk are outlined below, with a complete listing of cross-regulation of histone marks found in comprehensive reviews (Latham and Dent 2007; Oliver and Denu 2011).
Genome-wide location analysis has uncovered the combinatorial patterns of histone marks and chromatin regulators along the genome in human cells (Ram et al. 2011). This study established that specific combination of chromatin marks and regulators colocalize in characteristic patterns with distinct genomic regions and chromatin environments. Interestingly, the chromatin regulators retain their modular and combinatorial associations even during redistribution to different loci as a function of cell type.
12.1. Kme-Sph Cross Talk
An examination of histone tail sequences indicates that lysines and serine/threonines are often proximally positioned in sequence. Examples on H3 include Thr3-Lys4, Lys9-Ser10, Thr22-Lys23, Lys27-Ser28, and Lys79-Thr80 (Fig. 1A). To take an example, Lys9 and Ser10 occupy adjacent positions on the histone H3 tail, with methylation of lysine a more stable mark than phosphorylation of serine. The H3K9me3 mark (written by the Suv39h KMT) serves as a recruitment site for HP1, involved in heterochromatin formation. Phosphorylation of adjacent Ser10 is written by the mitotic kinase Aurora B during mitosis, resulting in the ejection of HP1 from the adjacent H3K9me3 mark (Fischle et al. 2005; Hirota et al. 2005). Subsequent dephosphorylation of Ser10 at the end of mitosis reestablishes the association of HP1 with the H3K9me3 mark. These studies thus point to a “methyl/phospho switch” that dynamically controls the H3K9me3-HP1 interaction, and whose impairment impacts on chromosome alignment and segregation, spindle assembly, and cytokinesis.
The proposed methyl/phospho switch also plays a role in the recognition of the H3K4me3 mark by the PHD finger domains of BPTF and RAG2 (discussed in Sec. 2.1), and the tandem chromodomains of CHD1 (Sec. 4.1). Through the use of peptide microarrays containing a library of combinatorially modified histone peptides, the recognition of H3K4me3 by reader modules was found to be either blocked or attenuated when Thr3 or Thr6 were phosphorylated (Fuchs et al. 2011). The same conclusions were obtained by a parallel investigation, which also showed the structural basis for a 20-fold decrease in ING2 PHD finger binding affinity to H3K4me3, when Thr6 is phosphorylated and no longer able to be accommodated into the shallow pocket that targets the side chain hydroxyl of Thr6 (Garske et al. 2010).
The association of UHRF1 with the H3K9me3 mark through its tandem Tudor domain is insensitive to phosphorylation at the adjacent S10 site (Rothbart et al. 2012), unlike the cell-cycle restricted HP1 binding to H3K9me3. It appears that interactions between the side chains on the H3K9me3 reader modules and S10ph determine whether the reader module bound to H3K9me3 is retained or released on phosphorylation of S10. Thus, replacement of an Asn in the tandem Tudor domain of UHRF1 by Glu/Asp makes readout sensitive to phosphorylation of Ser10. Thus, given that the UHRF1 tandem Tudor domain is the only H3K9me3 reader that is insensitive to the K9me-S10ph switch, its function may be important in tethering DNMT1 to chromatin through mitosis in heterochromatic regions that require considerable maintenance of DNA methylation.
In another noteworthy study, the nonreceptor tyrosine kinase JAK2 was shown to phosphorylate Tyr41 on histone H3 in the nucleus of haematopoietic cells, thereby excluding HP1α from targeting an adjacent presumably lysine methylation site using its chromoshadow domain (Dawson et al. 2009). There are also examples of methyl/phospho switches in nonhistone proteins such as the S. cerevisiae Dam1 protein (Zhang et al. 2005).
12.2. Kme-Kac Cross Talk
A number of histone mark combinations have been shown to be synergistic or antagonistic. A few examples are described below.
The ING4 association with chromatin provides an example of synergistic Kme3-Kac cross talk. The PHD finger of ING4 (inhibitor of growth 4) targets the H3K4me3 mark. ING4 is a subunit of the HBO1 HAT complex, and acetylation activity augments the recognition of H3K4me3 on H3 tails in cis. This effectively activates ING4 target promoters and attenuates cellular transformation through its tumor-suppressor activity (Hung et al. 2009).
An antagonistic cross talk in cis (intrahistone) is observed between H4K20me3 and acetylation of lysines toward the amino terminus of H4; hyperacetylation of H4 antagonizes H4K20me3, and the reverse also holds, in which H4K20 methylation appears to inhibit acetylation of H4 (Sarg et al. 2004).
Another synergistic cross talk example occurs in trans (interhistone) in human cells; the H3K4 methylating MLL1 associates with the H4K16 acetylating MOF, thereby potentially linking these two marks during transcription (Dou et al. 2005).
12.3. Kme-Rme Cross Talk
The amino-terminal A1-R2-T3-K4 sequence of histone H3 has methylation sites on both R2 and K4 (Fig. 1A,B). The arginine methyltransferase PRMT6 deposits the asymmetrical R2me2a mark on H3 and this mark is distributed within the body and the 3′-ends of human genes (Guccione et al. 2007; Hyllus et al. 2007; Kirmizis et al. 2007). Antagonistic cross talk between the R2me2a and H3K4me3 mark seems to occur; that is, the R2me2a mark is prevented by the H3K4me3 mark and, conversely, the H3R3me2a mark prevents trimethylation of H3K4 by the MLL1 KMT and its associated factors (ASH2 and WDR5). It is conceivable that this mutual antagonism reflects the inability of WDR5 to recognize the H3R2me2a mark and, hence, is unable to recruit MLL1 necessary for H3K4 trimethylation (Guccione et al. 2007). Certainly, in S. cerevisiae, the H3R2me2a mark appears to occlude binding by the Set1 KMT via the Spp1 subunit PHD finger, necessary for H3K4 trimethylation (Kirmizis et al. 2007).
In another example, binding studies showed that recognition of unmodified H3K4 (me0) by the AIRE (autoimmune regulator) PHD finger is abrogated by H3R2me2 in vitro; this results in the reduction of AIRE target gene activation (Chignola et al. 2009).
Section 2.1 introduced the RAG2 PHD finger as being able to bind to the dual H3R2me2sK4me3 mark in vitro. In a follow-up study, high levels of the H3R2me2sK4me3 dual mark were found at antigen receptor gene segments poised for rearrangement (Yuan et al. 2012). Strikingly, this dual mark colocalizes at active promoters throughout the genome, with the implication that readout of the K4me3 mark on H3 can be modulated by the R2me2s mark.
12.4. Kme-Kub Cross Talk
Early studies established that ubiquitination (Ub) of histone H2B regulates H3 methylation and gene silencing (Ng et al. 2002; Sun and Allis 2002; Lee et al. 2007; reviewed in Shilatifard 2006). This has been elegantly investigated following the generation of designer mono- and dinucleosomes incorporating a site-specific ubiquitination mark on H2B at Lys120 (McGinty et al. 2008). Biochemical studies established that histone H2BK120-monoubiquitination (H2BK120ub1) stimulated intranucleosomal methylation of H3K79 (illustrated in Fig. 12 of Allis et al. 2014). This observed cross talk is mediated by the catalytic domain of hDot1, potentially through an allosteric mechanism (McGinty et al. 2008).
These Kme-Kub cross talk studies have been recently extended to the Drosophila MSL complex system, which regulates dosage compensation (the topic of Lucchesi and Kuroda 2014); H2B ubiquitination by MSL1/2 directly regulates H3K4 and H3K79 methylation through intranucleosomal trans-tail cross talk both in vitro and in vivo (Wu et al. 2011b). Given that MSL1/2 activity contributes to transcriptional activation at HOXA1 and MEIS1 loci, the MSL complex shows two distinct chromatin-modifying activities: MSL1/2-mediated H2BK34 ubiquitination and MOF-mediated H4K16 acetylation (see Fig. 3 of Lucchesi and Kuroda 2014). These studies provide insights into the underlying intricate network of interactions whereby chromatin-modifying enzymes function coordinately to program gene activation.
13. HISTONE MIMICS
In the above sections, we have largely outlined structure–function studies on the readout of methylation marks on histone tails by reader modules. This has raised the question of whether nonhistone chromatin proteins also use similar modification-recognition systems. In this regard, recent advances have highlighted the identification of “histone mimics” as a distinct set of nonhistone proteins that can have PTMs (Kme, Kac, Yph) written and read in a similar fashion to histone proteins. In this article, the focus will be on studies of histone mimics that are methylated, thereby expanding our insights of methylated lysine recognition principles beyond the boundaries of histones. We outline, below, four examples of histone mimicry involving methylated lysine marks on nonhistone proteins functioning in chromatin-templated processes, including KMTs, tumor suppressors, RNA polymerases, and influenza viral proteins.
13.1. G9a Methyltransferase
Our first insights into histone mimics emerged from structure–function studies on a member of the Suvar(3-9) family of SET domain lysine methyltransferases called G9a. This KMT writes methylation marks on H3K9. In a striking result, G9a was shown to be a self-methylating KMT, trimethylating itself at K165 and di- and trimethylating K94 (Sampath et al. 2007). These Kme sites in G9a have marked sequence similarity to H3K9 sites and are in vivo binding targets for the chromodomain protein HP1. Recognition of the G9aK165me3 mark by HP1 can be reversed by concomitant phosphorylation of adjacent Thr166. Thus, G9a contains a lysine methylation cassette that mimics features of sequence context, reader module recognition, and regulation by phosphorylation (Sampath et al. 2007), which are characteristic features of histone lysine methylation.
13.2. p53 Tumor Suppressor
The transcriptional activity of the tumor suppressor p53 is modulated by multiple PTMs. Three lysine residues within its regulatory carboxy-terminal domain (CTD) undergo methylation by KMTs at Lys370, Lys372, and Lys382. The DNA repair factor 53BP1 (p53-binding protein 1) uses its tandem Tudor domain to recognize p53 during DNA damage via the K382me2 mark. Structural studies on the complex established that the p53K382me2 mark inserts into the aromatic-lined pocket of the tandem Tudor domain of 53BP1 in a similar manner to H4K20me2 (Roy et al. 2010). The adjacent His380 and Lys381, in a HKKme2 sequence context, contribute to the sequence specificity of molecular recognition (Kd = 0.9 µm). ChIP and DNA repair assays suggest that recognition of the p53K382me2 mark by the tandem Tudor domains of p53BP1 may facilitate accumulation of p53 at DNA damage sites and promote repair.
p53K382me1 is generated by the SET8 KMT and recognized by the MBT repeats of L3MBTL1 (whose structure is illustrated in complex with Kme2-containing histone peptides in Fig. 6) (West et al. 2010). Functionally, the activation of p53 by DNA damage is coupled with a decrease in p53K382me1 levels and, hence, an abrogation of the p53-L3MBTL1 interaction, which causes the dissociation of L3MBTL1 from p53 target promoters. This study provides a mechanistic explanation for the basis by which the MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at Lys382 to regulate p53 activity (West et al. 2010).
13.3. Carboxyl Terminus of RNA Polymerase II
The RNA polymerase II CTD can be modified by site-specific methylation of Arg1810. This modification, along with Ser2 and Ser5 phosphorylation, is essential for transcriptional initiation and elongation (Sims, III et al. 2011). Given that RNA polymerase II participates in chromatin-templated processes, the R1810me mark within the CTD mimics Arg methylation observed in histone tails.
Interestingly, the Tudor domain–containing protein TDRD3, but not SMN and SPF30, binds to the R1810me2a-containing CTD peptide, but not to its mono- or symmetrically dimethylated counterparts. This is in addition to its binding to histone tail sites discussed in Section 6.1. Mutation of the aromatic cage residues of the TDRD3 reader results in a loss of binding to the R1810me2a-containing CTD peptide.
13.4. Carboxyl Terminus of Influenza Virus NS1 Protein
In a landmark study, Marazzi et al. (2012) describe the suppression of the antiviral response by an influenza histone mimic residing within the carboxyl terminus of the viral NS1 (nonstructural protein 1) protein. The NS1 protein of the H3N2 influenza virus subtype contains an ARSK (Ala-Arg-Ser-Lys) sequence that is similar to the amino-terminal ARKS sequence of histone H3 (Fig. 1A). The SET1 KMT, which methylates H3K4, an activation mark, also methylates the lysine in the ARSK sequence of NS1, supporting the notion of histone mimicry by NS1. Normally, hPAF1C (PAF1 transcription elongation complex) function is potentiated for the transcriptional elongation of inducible antiviral gene sets when methylated H3K4 tails bind to it. The binding of the methylated NS1 mimic to hPAF1C sequesters it and the methylating Set1 enzyme from acting at the hosts normal genomic targets, thus interfering with the gene transcription program of the host cell. Therefore, histone mimicry provides a selective advantage to viruses by inducing specific suppression of host antiviral gene transcription (also discussed in Sec. 7 and Fig. 12 of Busslinger and Tarakhovsky 2014).
14. READOUT OF FULLY METHYLATED 5mCpG SITES ON DNA
DNA methylation may exert its repressive effect on transcription as a consequence of 5mC marks serving as docking sites for 5mC-binding proteins, which, in turn, recruit corepressors that can modify chromatin. In this section, we discuss structural studies involving 5mCpG-binding (MBDs) proteins and their zinc-finger counterparts, including Kaiso, bound to 5mC sites on duplex DNA.
14.1. Methylcytosine-Binding Proteins
Two MBD protein structural complexes are described below: first, the MBD1 protein and then MeCP2, bound to symmetrical 5mCpG/5mCpG sites. These structures illustrate the different principles by which recognition of 5mC marks occurs on duplex DNA.
MBD1 is a transcriptional regulator containing an amino-terminal 5mCpG-binding domain and a carboxy-terminal transcription repression domain (illustrated in Fig. 8 of Li and Zhang 2014). The MBD domain targets methylated CpG islands of tumor suppressors and imprinted genes, thereby inhibiting their promoter activities in cooperation with the transcription repressor domain. An NMR-based solution structure has been solved for the complex of MBD1 with a DNA duplex containing a fully methylated 5mCpG/5mCpG step (1 protein bound per DNA duplex). In this structure, loops project from the protein core (an α/β sandwich composed of four β-strands and an α-helix) and interact with the major groove of the DNA duplex (Fig. 14A) (Ohki et al. 2001). Loop L1 adopts a defined hairpin-like structure on complex formation and interacts with one of the DNA strands in the major groove. Loop L2 and a segment of α-helix toward the amino terminus interact with the other DNA strand in the major groove, with the loop forming base-specific contacts, whereas the helical segment forms contacts with the sugar-phosphate backbone (schematically shown in Fig. 14B). Five protein residues form a continuous hydrophobic patch that interacts with the methyl groups of the 5mC residues (indicated by magenta arrows in Fig. 13B), with distinct interactions observed for the 5mC residues on partner strands. In addition, a pair of arginines and a tyrosine interact with guanines of the 5mCpG sites (indicated by blue arrows), with binding affinity lost following mutation of the Arg residues. The small observed protein–DNA interface that is essentially restricted to the 5mCpG/5mCpG binding site suggests that MBD1 should be able to access such sites in the major groove of nucleosomes most likely without encountering steric interference.
Figure 14.
Structures of methylcytosine-binding proteins bound to fully methylated 5mCpG DNA. (A) NMR solution structure of MBD1 protein bound to fully methylated 5mCpG-containing DNA duplex (PDB: 1IG4). Two loops L1 and L2 are colored in yellow. The methyl groups of 5mC’s are marked by magenta dotted circles. (B) Schematic of intermolecular contacts centered about the 5mCpG/5mCpG site involving loops L1 and L2, and the amino-terminal α-helix adjacent to L2. Methyl group of 5mC is represented by the magenta circle. Hydrophobic interactions between the 5mC residues and side chains of MBD1 are indicated by magenta colored arrows. (C) 2.5-Å crystal structure of MeCP2 protein bound to fully methylated 5mCpG-containing DNA duplex (PDB: 3C2I). The methyl groups of 5mCs are marked by magenta dotted circles. (D) Intermolecular contacts between hydrophilic amino acids of MeCP2 (green stick representation) and the 5mC groups (magenta dotted circles) in the major groove of the duplex, including C—H••O hydrogen bonds to tightly bound water molecules.
A second 5mC-binding protein, MeCP2, is of interest because of its role in the maintenance of neuronal function; mutations in the MECP2 gene are responsible for the majority of cases of Rett syndrome, a late-onset autism spectrum disorder (discussed more extensively in Li and Zhang 2014 and Zoghbi and Beaudet 2014; see also Amir et al. 1999). The crystal structure of MeCP2 bound to a 5mCpG/5mCpG-containing DNA duplex (Fig. 14C) established that the methyl groups of 5mC make contacts predominantly with a hydrophilic surface along the major groove, including C—H••O hydrogen bonds to tightly bound water molecules (Fig. 14D) (Ho et al. 2008). In addition, Arg residues are involved in 5mC–Arg-G interactions, whereby the Arg is positioned in the plane of the guanine base and locked in place by a salt bridge to the carboxylate of an Asp, with the guanidinium group of the Arg positioned directly above the methyl group of the 5mC. The most frequently mutated residue in Rett syndrome involves a Thr residue, and it was shown that this Thr plays an important role in maintaining a structural motif within the MeCP2 fold (Ho et al. 2008).
14.2. Methylcytosine-Binding Zn-Finger Proteins
A subset of human zinc-finger-containing proteins have the capacity to specifically recognize 5mC-containing DNA (reviewed in Sasai et al. 2010). We outline the results obtained from structural studies of the Kaiso and Zpf57 zinc-finger proteins bound to symmetrical 5mCpG/5mCpG sites on duplex DNA.
Kaiso is a methylated DNA-binding factor involved in noncanonical Wnt signaling. It contains an amino-terminal BTB/POZ (BR-C, ttk and bab/Pox virus and Zn finger) domain involved in protein–protein interactions and three mCG-DNA-binding carboxy-terminal zinc fingers (Cys2His2 coordination) that repress transcription by recruiting chromatin-remodeling compression machinery to target genes (reviewed in Clouaire and Stancheva 2008). Recently, structural studies of the zinc-finger domains of Kaiso bound to a pair of sequential symmetrically methylated 5mCpG/5mCpG DNA sites from the E-cadherin promoter region have elucidated the details underlying the recognition process, in which 1 Kaiso molecule is bound per DNA duplex (Buck-Koehntop et al. 2012). The side chains of the first two zinc fingers target the major groove through base-specific recognition mediated by classical and C—H••O hydrogen bonds, as well as phosphate backbone contacts. The carboxy-terminal extension following the third zinc finger (disordered in the free structure) targets the minor groove and contributes to high-affinity binding (Fig. 15A). The amino-terminal regions of the first two zinc fingers provide hydrophobic environments that accommodate the methyl groups of 5mC in the complex (Fig. 15B). Unexpectedly, the three zinc fingers of Kaiso span a total of 4–5 base pairs, contacting both the major and minor grooves, in contrast to most other triple zinc-finger proteins that solely target the major groove and span a total of 9–10 base pairs (Wolfe et al. 2000). Functionally, Kaiso DNA-binding sites are limited to target genes regulated by the Wnt signaling pathway that play key roles in early development and tumor progression. Kaiso targets and silences aberrantly methylated DNA repair and tumor-suppressor genes, acting oncogenically to contribute to tumor progression (Lopes et al. 2008).
Figure 15.
Structures of methylcytosine-binding zinc-finger proteins bound to fully methylated 5mCpG DNA. (A) 2.8-Å crystal structure of three zinc fingers of Kaiso protein bound to a pair of fully methylated 5mCpG-containing DNA duplex (PDB: 4F6N). The first, second, and third zinc fingers are colored in green, blue, and pink, respectively. Note that although the majority of the intermolecular contacts are with the major groove, involving zing fingers 1 (green) and 2 (blue), there are also contacts with the minor groove, involving zinc finger 3 and the carboxy-terminal extension of Kaiso. Zinc ions are shown as silver balls. The methyl groups of 5mC are marked by magenta dotted circles. (B) Intermolecular contacts between amino acids of the first zinc finger (in green) of Kaiso and 5mC groups in the major groove of the duplex. (C) 0.99-Å crystal structure of two zinc fingers of Zfp57 protein bound to a fully methylated 5mCpG-containing DNA duplex (PDB: 4GZN). Zinc ions are shown as silver balls. The methyl groups of 5mC are marked by magenta dotted circles. (D) One of the 5mC groups in the Zfp57-DNA complex is involved in hydrophobic interactions through positioning between an Arg side chain and a neighboring guanine. (E) The second 5mC in the Zfp57-DNA complex interacts with a layer of ordered water molecules (red circles).
The structure of tandem zinc fingers (C2H2 coordination) of the Zfp57 transcription factor and a DNA duplex containing a fully methylated 5mCpG/5mCpG-step within the T-G-C-5mC-G-C sequence element (1 Zfp57 bound per DNA duplex) has been solved at high resolution (Liu et al. 2012). Zfp57 is expressed during very early embryogenesis, and is responsible for the maintenance of paternal and maternal imprinted loci (the topic of Barlow and Bartolomei 2014). Both zinc fingers target the DNA major groove on complex formation and together span 6 base pairs without perturbing the B-DNA conformation (Fig. 15C). The first zinc finger (depicted in green in the figure) contacts the 5′-T-G-C segment, whereas the second zinc finger (in blue) contacts the 5mC-G-C segment. The 5mC base is recognized asymmetrically: One case involves hydrophobic interactions with an Arg side chain (whose mutation results in loss in binding affinity) and a neighboring 3′-guanine (so called 5mC–Arg-G interaction) (Fig. 15D), whereas the other case is defined by a layer of ordered water molecules (Fig. 15E). The binding affinity of Zpf57 for its DNA target sites is reduced on formation of ten eleven translocation (Tet)-catalyzed oxidative products of 5mC such as 5-hydroxymethylcytosines (5hmC) (explained below in Sec. 16.4). Interestingly, in patients with transient neonatal diabetes, DNA-binding activity is abolished for two Zfp57 point mutations.
Future studies will have to elucidate the complex network of downstream protein–protein interactions through which 5mC-binding proteins regulate gene expression impacting on development and oncogenesis.
15. READOUT OF HEMIMETHYLATED 5mCpG SITES ON DNA
Proteins containing SRA 5mC-binding domains are required for the establishment and/or maintenance of DNA methylation in both mammals and plants. As we show below, the SRA domains of mammals and plants use both common and distinct recognition principles for targeting their DNA duplex sites. The SRA-containing mammalian UHRF1 protein plays a critical role in epigenetic inheritance of 5mC marks in a 5mCpG context. The SUVH (SU(VAR)3-9 homolog 5) family of proteins in plants, which contain the 5mC-binding SRA domains, have no obvious mammalian counterparts, and are more versatile, given that they target 5mCpG, 5mCpHpG, and 5mCpHpH sites.
15.1. Mammalian SRA Domains
Functional experiments initially showed that the protein UHRF1 plays a critical role in maintaining DNA methylation in mammalian cells by mediating epigenetic inheritance patterns through recruitment of DNMT1 to hemimethylated DNA at replication forks (Bostick et al. 2007; Sharif et al. 2007; reviewed in Cedar and Bergman 2009; Hashimoto et al. 2009). UHRF1 is composed of tandem Tudor, PHD-finger, and SRA domains, with the SRA domain involved in site-specific recognition of hemimethylated 5mCpG/CpG sites on duplex DNA (Fig. 6 of Cheng 2014). The challenge has been to understand the molecular basis underlying the ability of mammalian SRA domains to target hemimethylated 5mCpG/CpG-containing DNA and discriminate against both fully methylated 5mCpG/5mCpG- and unmethylated CpG/CpG-containing DNA.
Three groups simultaneously solved the structure of mammalian (human or mouse) SRA domains of UHRF1 in the free state and when bound to 12-mer DNAs containing a central hemimethylated 5mCpG/CpG site (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008). The SRA domain uses two loops (designated NKR [Asn-Lys-Arg]-containing loop and thumb) projecting from its highly conserved concave face to penetrate both major and minor grooves centered about the hemimethylated 5mCpG/CpG site (Fig. 16A), resulting in a 1:1 UHRF1 SRA:DNA duplex complex. Complex formation results in flipping out of the 5mC, thereby positioning it in a conserved binding pocket within the SRA domain, in which it is anchored through hydrogen-bonding along its Watson–Crick edge, as well as van der Waals and planar stacking interactions (Fig. 16B). The nature of the intermolecular contacts involving the flipped-out 5meC will discriminate against replacement by thymine, with specificity for 5mC over C conferred by the methyl group fitting precisely within an available hemispheric-like space. The orphan guanine remains stacked within the helix, with the DNA being both straight and adopting a B-DNA conformation, despite the flipping out of the 5mC on complex formation. An Arg side chain from the NKR-containing loop inserts from the major groove side into the cavity vacated by the flipped-out 5mC, in which it hydrogen bonds with both the orphan guanine and its adjacent cytosine of the CpG step on the unmodified partner strand. An Asn side chain interacts with and buttresses this inserted Arg, with both its main and side chain atoms of Asn positioned to clash with the C of the CG step, were it methylated. These results nicely explain why mammalian SRA domains bind exclusively to 5mCpG/CpG steps by recognizing both the flipped-out 5mC on the parental and opposing CpG step on the partner strand (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008). Direct contacts involving protein and DNA are limited to the 5mCpG/CpG segment, indicative of binding and recognition restricted to this site and independent of flanking sequence.
Figure 16.
Structures of SRA domain-containing proteins bound to hemimethylated 5mCpG DNA. (A) 1.6-Å crystal structure of SRA domain of UHRF1 bound to a hemimethylated 5mCpG-containing DNA duplex (PDB: 2ZKD). The stoichiometry of the complex is one SRA domain per DNA duplex. The methyl groups of 5mC are marked by magenta dotted circles. (B) Alignment of the flipped-out 5mC (ring is shaded for clarity) in a pocket within the SRA domain of UHRF1. (C) 2.37-Å crystal structure of SRA domain of plant SUVH5 bound to a hemimethylated 5mCpG-containing DNA duplex (PDB: 3Q0D). The stoichiometry of the complex is two SRA domains per DNA duplex. 5mC’s methyl groups are marked by magenta dotted circles. (D) Alignment of the flipped-out 5mC (ring is shaded for clarity) in a pocket within the SRA domain of plant SUVH5.
15.2. Plant SRA Domains
The 5mC-binding capacity of several plant SRA domains has been characterized, and each has a preference for DNA methylation in different sequence contexts (Johnson et al. 2007). Binding studies established that the SRA domain of SUVH5 differs from other SRA domains in that it binds DNA to similar extents in all sequence contexts.
Unexpectedly, the structures of complexes containing the SUVH5 SRA domain bound to 5mC-containing DNA in either hemimethylated or fully methylated 5mCpG contexts, or a methylated 5mCpHpH context, show an SUVH5 SRA:DNA duplex stoichiometry of 2:1. These complexes revealed a dual flip-out mechanism in which both the 5mC from the parental strand and a base (5mC in 5mCpG/5mCpG context, C in 5mCpG/CpG, or G in 5mCpHpH/HpHpG context) from the partner strand are simultaneously extruded from the DNA duplex and positioned within individual binding pockets of SRA domains (Rajakumara et al. 2011a). The structure of the SUVH5 SRA domain bound to hemimethylated 5mCpG/CpG DNA duplex is shown in Fig. 16C. The 5mC and C bases on partner strands are simultaneously flipped out and positioned in conserved pockets on the symmetry-related SRA domains with the resulting gaps filled by Gln side chains projecting from the thumb loops into the minor groove (Fig. 16D). Interestingly, a second example of a dual flip out involving 5mC was recently reported for the recognition of this mark by the Escherichia coli methyl-specific McrBC (modified cytosine restriction BC) endonuclease (Sukackaite et al. 2012). Thus, mammalian and plant SRAs use different amino acids originating from different loops while inserting into the duplex from different grooves of the DNA (compare Figs. 16B and 16D). Complementary functional studies established that a functional SUVH5 SRA domain is required for both DNA methylation and accumulation of the H3K9me2 modification in vivo, suggesting a role for the SRA domain in recruitment of SUVH5 to genomic loci (Rajakumara et al. 2011a).
16. PERSPECTIVES AND FUTURE CHALLENGES
In this section, we outline new initiatives and advances, as well as future challenges, with the promise of enhancing our current mechanistic understanding of the readout of histone and DNA methylation marks. On the technology front, PTMs and their distribution have been identified and characterized at the genome-wide level using high-throughput and mass spectroscopic approaches. Chemical biological approaches have been developed to generate designer nucleosomes containing site-specifically incorporated histone lysine mark mimics, nonnatural and modified amino acids, and PTMs. The availability of designer nucleosomes should permit attempts at structural characterization of multivalent PTM readout at the nucleosomal level.
The contribution of DNA methylation in epigenetic regulation has been greatly expanded by the identification of oxidative adducts of 5mC (reviewed in Kriaucionis and Tahiliani 2014), with current efforts aimed at understanding the role of these new PTMs as novel epigenetic states and/or intermediates in DNA demethylation. The potential role of long noncoding RNAs (lncRNAs) in mediating epigenetic regulation events has received increasing attention, but the available information from functional studies has yet to be matched at the structural level, heightening the need to understand the molecular basis for protein–RNA recognition facilitated by noncoding RNAs. Given the extensive information on both histone and DNA PTMs, attention has turned to the cross talk between these marks and the potential of structural and functional approaches, which can elucidate the interplay between these marks, preferably at the nucleosomal level.
Finally, dysregulation of chromatin is associated with autoimmune, neurological, and age-related disorders, as well as cancer. We consider the consequences caused by dysregulation of methylated lysine reader modules on epigenetic pathways and outline progress toward the identification of small molecules targeted to aromatic pockets involved in methyllysine readout. Each of these topics is addressed below in some detail so as to outline ongoing challenges and the future promise for further progress in the field.
16.1. Technological Developments at Genome-Wide Level
There have been several advances in microarray and next-generation sequencing technology that have enhanced approaches that precisely and comprehensively monitor the emerging patterns of PTM landscapes on a genome-wide basis. Such approaches have included PTM-modified peptide microarray platforms for proteome-wide identification of readers of histone marks (specific, or combinations thereof, discussed in Sec. 12.1) and the use of SILAC (stable isotope labeling by amino acids in cell culture)-based technology coupled with mass spectroscopy to identify the nucleosome-interacting proteins that read one or more PTMs incorporated into designer nucleosomes.
Modified histone peptide microarray platforms have been developed for high-throughput identification of chromatin reader modules, as well as assessing the epitope specificity of antibodies (Bua et al. 2009). The peptide arrays were built, for example, of 20-amino-acid-long H3 peptides that were unmodified or had one or a variety of PTMs. This approach led to the discovery that the BAH domain of mammalian ORC1 is a reader of the H4K20me2 mark, and was followed by the structural and functional characterization of this interaction and its implications for a dwarfism syndrome (Kuo et al. 2012). In another study, a large PTM-modified randomized combinatorial peptide library consisting only of amino-terminal H3 peptides was developed to investigate how the specificity of chromatin binding modules that read the H3K4me3 mark were affected by other PTMs (Garske et al. 2010). This approach showed that recognition of K4me3 was modulated by methylation of Arg2, as well as by phosphorylation of Thr3 and Thr6 on the H3 tail segment, discussed in Sections 12.1 and 12.3.
An alternate approach for monitoring the Kme3 mark involved a histone peptide pulldown approach coupled with quantitative SILAC proteomics technology to profile this mark on a genome-wide scale and identify readers of the Kme3 mark (Vermeulen et al. 2010). In an extension of this approach, SILAC nucleosome affinity purification technology based on designer nucleosomes labeled with specific modification states was used to identify nucleosome-interacting proteins regulated by DNA and histone methylation (Bartke et al. 2010). This study identified components of the ORC to be a methylation-sensitive nucleosome interactor based on cooperative recruitment by histone and DNA methylation. In another example, the recruitment of the jumonji-containing lysine demethylase KDM2A by methylated lysines on histone tails at the nucleosomal level was disrupted by DNA methylation.
More recently, an interspecies comparative epigenomics approach (using human, mouse, and pig pluripotent stem cells) has been undertaken to look at DNA and histone modifications as an approach for annotating the regulatory genome (Xiao et al. 2012). This approach identified conserved colocalization of different epigenetic marks with distinct regulatory functions during differentiation of embryoninc stem cells into mesendoderm cells.
These high throughput advances toward charting histone PTM modifications and the functional organization of mammalian genomes (reviewed in Zhou et al. 2011) have led to the development of a website titled the “structural genomics of histone tail recognition” (Wang et al. 2010a). Further advances are likely to involve both miniaturization and sensitivity enhancement of the assays so as to target individual cells in the future. In the longer term, the coupling of structural biology with these technological advances in the detection and distribution of PTMs should provide an improved molecular perspective of the multivalent readout of PTMs at the nucleosomal level.
16.2. Chemical Biology Approaches to Designer Nucleosomes
An emerging multidisciplinary challenge has been to harness the tools of chemical biology to address specific structure/function problems in chromatin biology, achievable through the preparation and manipulation of chemically defined chromatin (reviewed in Allis and Muir 2011; Voigt and Reinberg 2011; Fierz and Muir 2012). This type of research could result in an improved understanding of the underlying basic mechanisms that dictate the folding and inheritance of chromatin, the multivalent readout of chromatin marks at the nucleosomal level, as well as the principles underlying the spreading of PTMs within defined chromatin regions.
One chemical biology approach has been to use recombinant histones with lysines site-specifically replaced by cysteines. These sites can then be conjugated to introduce site-specific modifications of the cysteine that acts as a lysine methylation mimic within histone tails, even at the nucleosome level. Thus, mimics of methylated lysines have been introduced by S-alkylation through generation of N-methylated aminoethylcysteine residues (Simon et al. 2007) and this cysteine-based chemistry has also been applied to introduce ubiquitin modifications (Chatterjee et al. 2010) and acetyllysine analogs (Huang et al. 2010). Such approaches are limited by the standard constraints associated with mimics not being able to fully replicate their natural counterparts. Nevertheless, the simplicity of the cysteine conjugation approach has made it accessible to the nonchemist and could benefit from further efforts to mimic additional PTMs, as well as their incorporation in a multiple mark context.
In an alternate approach, a genetic code expansion methodology has been developed to introduce into recombinant proteins methylation, acetylation, and ubiquitination marks on Lys and phosphorylation marks on Ser/Tyr (reviewed in Davis and Chin 2012). The methodology incorporates nonnatural and modified amino acids site-specifically into recombinant proteins through the application of orthogonal aminoacyl-transfer RNA (tRNA) synthetase-tRNA pairs that direct incorporation in response to introduced amber stop codon placed in the gene of interest (reviewed in Liu and Schultz 2010; Davis and Chin 2012). Improvements in the methodology have resulted in the incorporation of a wider range of multiple unnatural amino acids (Neumann et al. 2010) and should be extendable to their multiple PTM counterparts.
The most promising approach for site-specifically incorporating PTMs at the nucleosomal level has been to use chemical ligation strategies, generating so-called “designer nucleosomes” containing the marks of interest. It is based on native chemical ligation chemistry in which a carboxy-terminal α-thioester-containing peptide reacts with an amino-terminal cysteine-containing peptide to generate a normal peptide linkage (reviewed in Dawson and Kent 2000). The corresponding expressed protein ligation (EPL) version can use a recombinant histone amino-terminal peptide with an α-thioester building block, which can be chemically modified to mimic a particular PTM (e.g., Kme1), ligated to the remainder of the unmodified core histone. This approach, when used with yet to be determined DNA templates that allow more uniform positioning of core histones on the DNA, has the potential for being able to incorporate standard PTMs positioned at multiple sites on histone tails into mononucleosomes in good yields, thereby generating designer chromatin for structural and functional studies (reviewed in Allis and Muir 2011; Frederiks et al. 2011; Voigt and Reinberg 2011; Fierz and Muir 2012). The potential exists for extending this EPL technology to PTM probes site-specifically incorporated into dinucleosomes and nucleosomal arrays by ligating the DNA of preformed nucleosomes. This approach has the potential to provide more useful and biologically relevant biochemical/biophysical probes. Another challenge that chemical approaches are trying to tackle is the generation of asymmetric nucleosomes, in which, for instance, the nucleosome contains a copy of a canonical H3.1 and variant H3.3. Such approaches could set the stage for a better understanding of the structural consequences of nucleosome asymmetry. The availability of designer nucleosomes could also assist toward elucidating a structure-based understanding of whether PTMs have an impact on nucleosome positioning, occupancy, and dynamics and, if so, the consequence of DNA damage on these processes.
Looking to the future, one anticipates the emergence of high-throughput biochemical approaches that couple libraries of PTM designer chromatin with “lab-on-a-chip” microfabrication methodology facilitated by microfluidic devices to screen reader modules at the nano scale (see review by Fierz and Muir 2012). Such studies could identify new systems for conducting structural studies of PTMs at the nucleosomal level.
16.3. Histone Mark Readout at the Nucleosomal Level
The field is moving toward understanding histone PTM readout at the nucleosomal and chromatin fiber level. It is very likely that histone modifications may alter structure at various levels. Not only might it affect the stability and dynamics of individual nucleosomes, but also the organization and compactness of nucleosome arrays involving 10-nm (primary chromatin structure) and 30-nm (secondary chromatin structure) fiber formation. Furthermore, it might affect fiber–fiber interactions leading to higher-order compaction (tertiary chromatin structure) observed in condensed chromatin (reviewed in Luger et al. 2012). That is to say, histone PTMs may impact on histone–DNA interactions at the mononucleosomal level, as well as on short-range and long-range internucleosomal contacts and the ability of readers targeted to these marks to stabilize particular chromatin conformations. The potential linkage of histone PTM patterns with chromatin accessibility, combined with information on nucleosomal positioning, transcription factor binding, and the role of noncoding RNAs, should thus provide us with a more complete understanding of chromatin structure and function.
Currently, there are several examples of proteins that bind to the unmodified nucleosome core particle, ranging from the Kaposi sarcoma herpes virus peptide LANA (Barbera et al. 2006) to the chromosome factor RCC1 (Makde et al. 2010) and BAH domain of Sir3 (Armache et al. 2011). These examples have provided insights into the interactions and common principles (i.e., that all proteins target the acidic patch on H2A/H2B) associated with protein–nucleosome recognition (reviewed in Tan and Davey 2011). Nevertheless, an important emerging challenge is the structural characterization of complexes between reader modules and their PTM targets at the nucleosomal, and in the longer term, nucleosomal array level. Potential candidate reader modules include isolated domains (PHD fingers and BAH domains) and cassettes (PHD-bromo dual domains). In addition, the chemical biology approaches mentioned in the previous section, capable of generating the required appropriate PTM-containing designer nucleosomes, should provide ideal substrates for such structural studies. It should also be possible to gain insights into the spreading of histone marks at a molecular level by structurally characterizing dinucleosome complexes in which the reader component of a dual reader/writer protein site-specifically reads a histone mark on one nucleosome and is positioned to write the same site-specific mark on an adjacent nucleosome. Such studies could determine whether there is a directional component to spreading and its molecular basis, as well as the role of antagonistic PTMs to spreading, potentially providing molecular insights into the role of boundary elements.
An even greater and longer term challenge is to structurally characterize PTM-modified designer nucleosomes bound to ATP-dependent chromatin-remodeling complexes (containing reader modules) that either facilitate movement of nucleosomes to different positions on the DNA (SNF2H family) or those that transiently unravel nucleosomes (SWI/SNF family) (reviewed in Clapier and Cairns 2009; also the topic of Becker and Workman 2013). There has been promising initial progress toward this goal from X-ray studies on the chromatin-remodeling factor ISW1a bound to DNA (although the construct lacks the ATPase domain) and nucleosome-containing complexes of this system determined by cryo-electron microscopy (Yamada et al. 2011). Nevertheless, more needs to be performed toward the structural characterization of the complete system, including the use of PTM-modified nucleosomes.
Currently, there is an as yet unresolved controversy surrounding the structure of the compact 30-nm fiber: Does it consist of a one-start solenoid or two-start zig-zag model (reviewed in van Holde and Zlatanova 2007; Luger et al. 2012)? It is likely that the 30-nm structure and dynamics depend on several factors including the type and distribution of PTMs, linker length, the presence/absence of linker histones, and concentration of divalent cations. Thus, it has been shown that the charge change associated with the deposition of the H4K16ac mark disrupts formation of the 30-nm fiber and higher-order chromatin structures (Shogren-Knaak et al. 2006). This most likely reflects the disruption of internucleosomal interactions that promote chromatin folding via the interaction of H4K16 with an acidic patch on H2A on an adjacent nucleosome. The availability of designer nucleosomes with site-specifically incorporated fluorescence resonance energy transfer probes (see review by Allis and Muir 2011) could provide handles for investigating aspects of the architecture and dynamics of the 30-nm fiber.
Even less is known about higher-order tertiary folded states of chromatin, and the role of nuclear lamins and matrix proteins in the anchoring of looped-out chromatin domains to the nuclear periphery. Much work, however, has been performed to ascertain the likely genomic regions that interact with each other (see Dekker and Misteli 2014). It should be possible to expand on earlier experiments involving cross-linking agents added in trans to probe higher-order tertiary chromatin structure (Kan et al. 2009) through involvement of designer nucleosomes containing site-specifically incorporated chemical cross-linkers (see review by Allis and Muir 2011). Given that H3 tails have been shown to contribute to interarray interactions, the next step would be to probe the effect of PTMs on higher-order tertiary folded states of chromatin, which could potentially be elucidated using small optical probes and fluorescence microscopy-based biophysical approaches.
16.4. Readout of Oxidative 5mC DNA Adducts
The field of DNA methylation received an unexpected boost with the discovery of 5hmC, a new PTM found on cytosines validated in a biological context (described in Kriaucionis and Tahiliani 2014). One group focused on the enzymatic activity of Tet proteins, a common fusion partner of the MLL1 gene in acute leukemia; they showed Tet1 to be a 2-oxogulaterate and Fe2+-dependent hydroxygenase, which catalyzes the conversion of 5mC to 5hmC, both in vitro and in cultured cells (Tahiliani et al. 2009). The other group identified 5hmC in neurons and the brain, constituting 0.6% of total nucleotides in Purkinje cells and 0.2% in granule cells (Kriaucionis and Heintz 2009). Both groups predicted a role for 5hmC in epigenetic regulation with a potential to influence chromatin structure and local transcription activity. The significance of these results was reinforced by the demonstration that Tet1 plays a role in embryonic stem (ES) cell self-renewal and inner cell mass specification (Ito et al. 2011; discussed in Reik and Surani 2014).
Subsequent to identifying 5hmC, it was shown that 5hmC is converted by Tet to 5-formylcytosine (5fC) (Ito et al. 2011) and 5-carboxylcytosine (5caC) (He et al. 2011; Ito et al. 2011) in an enzymatic activity-dependent manner under in vitro and cell culture conditions. Further, 5hmC, 5fC, and 5caC can be detected in genomic DNA of mouse ES cells and organs with relative levels governed by the activity of the Tet proteins. Importantly, 5caC can be specifically recognized and excised by the enzyme TDG (thymine-DNA glycosylase); this provided the link for explaining how the conversion of 5mC to 5caC by Tet, when followed by the conversion of 5caC to C by TDG as part of the base-excision repair pathway, could constitute an active DNA demethylation pathway (He et al. 2011).
The identification of oxidative adducts of 5mC has led to the development of quantitative methods for 5mC and 5hmC detection. An oxidative bisulfite sequencing approach (oxidative conversion of 5hmC to 5fC, followed by bisulfite conversion of 5fC to uracil) has been developed to measure these adducts at single-base resolution (Booth et al. 2012). This research has identified high 5hmC levels at CpG islands associated with transcriptional regulators and in long interspersed nuclear elements, hinting that these regions may play a role in the epigenetic reprogramming of ES cells. Genome-wide studies of 5hmC and Tet1 have established that Tet1 controls 5mC and 5hmC levels in mouse ES cells by binding to CpG-rich regions thereby preventing unwanted DNA methyltransferase activity (Xu et al. 2011).
Functional studies have also identified a dual role of Tet1 in the transcriptional regulation of mouse ES cells, whereby Tet1 promotes transcription of pluripotency factors, as well as participates in the repression of Polycomb-targeted developmental regulators (Wu et al. 2011a). A regulatory link between histone PTMs and Tet has recently been established; maternal chromatin containing the H3K9me2 mark binds the maternal factor PGC7 (also known as STELLA), protecting 5mC from Tet3-mediated conversion to 5hmC (Nakamura et al. 2012; further explained in Sec. 3.2 of Reik and Surani 2014).
The above studies suggest that 5hmC, 5fC, and 5caC may either represent new epigenetic states in genomic DNA and/or key intermediates in DNA demethylation pathways mediated by Tet and glycosylases/deaminases (reviewed in Bhutani et al. 2011; Wu and Zhang 2011; Branco et al. 2012). Also, it appears that Tet proteins are important regulators of cellular identity given that Tet2 is frequently mutated in haematopoietic tumors. It may do this by playing a key role in DNA methylation fidelity, with perturbation of Tet levels contributing to DNA hypermethylation phenotypes associated with certain cancers (reviewed in Williams et al. 2012; see Baylin and Jones 2014).
At the molecular level, a structure has been solved for a 5caC adduct-containing DNA duplex bound to a human thymine DNA glycosylase (hTDG) (Zhang et al. 2012). In this complex, 5caC is specifically recognized through extrusion from the duplex and repositioning in the catalytic pocket of the enzyme, in which the flipped-out base is locked through polar interactions with pocket residues. This result is supportive of proposals that hTDG (together with Tet) plays a key role in mammalian 5-methylcytosine demethylation. A related study has reported the structure of the carboxy-terminal glycosylase domain of MBD4 (methyl-binding domain 4) bound to an extruded 5-hydroxylmethyl uridine, a deamination product of 5hmC (Hashimoto et al. 2012). Future challenges from the molecular perspective should focus on the identification and structural characterization of readers of oxidative 5mC marks, and whether such readers can selectively target and discriminate between 5mC, 5hmC, 5fC, and 5caC adducts, and whether this requires a base-flipping mechanism for adduct recognition (as illustrated in Fig. 6D of Cheng 2014).
16.5. A Functional Role for Noncoding Regulatory RNAs
The vast majority of the human genome is transcribed, which means there is a much larger repertoire of RNAs beyond just protein-coding mRNAs (summarized in Fig. 6 of Allis et al. 2014). lncRNAs are generally greater than 200 nucleotide in length and, in many cases, are an integral component of chromatin. They serve a regulatory role by interacting with chromatin remodelers and modifiers, thereby changing the epigenetic status of target genes (reviewed in Guil and Esteller 2012; Guttman and Rinn 2012; Kugel and Goodrich 2012). It is believed that lncRNAs function at the interface of histone-modifying enzymatic complexes and the genome, thereby regulating chromatin states and epigenetic inheritance. The regulation can occur in cis or trans with the former controlling transcription at or in close proximity to the segment of the genome from which the lncRNA is transcribed, whereas the latter controls transcription at genomic sites removed from its site of transcription. From a different perspective, small RNAs in the 20- to 30-nucleotide range also play a role in directing effector complexes to target nascent chromatin-bound lncRNAs, thereby impacting on the recruitment of chromatin-modifying complexes to specific chromosomal regions (reviewed in Moazed 2009; see Martienssen and Moazed 2014).
Long intergenic noncoding RNAs (lincRNAs) have been shown to play critical roles in imprinting, dosage compensation, and homeotic gene expression. One of the earliest identified lincRNAs acting in cis was XIST, expressed exclusively from the X chromosome and required for X inactivation in mammalian systems (the topic of Brown et al. 1991; Penny et al. 1996; Brockdorff and Turner 2014). XIST RNA interacts directly with the Polycomb complex, facilitating condensation of chromatin and initiating transcriptional repression of the entire X chromosome (reviewed in Lee 2012; Brockdorff and Turner 2014). From the perspective of this article, ∼30% of lincRNAs are known to associate with chromatin-regulatory complexes that include writers, readers, and erasers of histone marks. It has been proposed that lincRNAs contribute regulatory specificity through localization of chromatin regulatory complexes to genomic DNA targets. Some examples of trans-acting lincRNAs that control gene transcription by mediating changes in chromatin structure are HOTAIR (HOX antisense intergenic RNA), ANRIL (antisense noncoding RNA in the INK4 locus), and HOTTIP (HOXA transcript at the distal tip). We focus below on one such lincRNA, HOTAIR, whose discovery is described in Rinn (2014).
HOTAIR is one of the earliest and best-characterized lincRNAs (2.2 kb in length), expressed from the HOXC locus on chromosome 12 in human distal fibroblasts (Rinn et al. 2007) and metastatic breast tumors (Gupta et al. 2010). The 5′-domain of HOTAIR interacts with PRC2, whereas its 3′-counterpart interacts with the LSD1/CoREST/REST complex, thereby coordinating targeting of PRC2 and LSD1 to chromatin for coupled H3K27 methylation and H3K4me demethylation. Based on these observations, it has been proposed that HOTAIR and related lincRNAs could serve as scaffolds, providing binding surfaces for the assembly of select histone-modifying enzymes that can specify the pattern of histone modifications on target genes (Tsai et al. 2010a).
At the structural level, information is totally lacking both on the three-dimensional architecture of lincRNAs as well as the intermolecular contacts at protein–RNA interfaces, accounting for the specificity of the recognition process. It is likely that the inherent flexibility of lincRNAs will hinder attempts toward defining their structures in the free state. Nevertheless, this should be a tractable problem when considered at the complex level provided the lincRNAs adopt compact scaffolds in the presence of bound protein targets; such successful structural studies have been conducted on the riboswitch sensing domains bound to small cognate metabolites (reviewed in Serganov and Patel 2012). It is conceivable that lincRNAs may contain highly conserved surface patches involved in protein (and RNA and DNA) recognition and, hence, a tractable approach would require trimming of the RNA constructs down to a minimal functional size, preferably in the 200-nucleotide range (similar in size to the larger riboswitch sensing domains), as well as working with minimal protein constructs, so as to facilitate successful crystallization and structure determination of manageable complexes. Such an effort, premised on the functional modularity of lincRNAs, if successful, could provide insights into the RNA secondary structural elements involved in the recognition process and the diversity of such intermolecular interactions.
16.6. Linking Histone and DNA Methylation
There has been considerable interest in the potential cross talk linking histone and DNA methylation in mammals and the mechanisms underlying the relationships between these marks (reviewed in Cedar and Bergman 2009), given that DNA methylation correlates with histone methylation patterns in genome-scale DNA methylation maps of pluripotent and differentiated cells (Meissner et al. 2008). In this regard, DNA methylation is inversely correlated with H3K4 methylation and positively correlated with H3K9 methylation. Relevant questions relate to whether histone methylation serves as a template for directing DNA methylation and/or the converse. Other questions include how the relationship can be addressed on a mechanistic basis from a combined structural and functional perspective.
A notable example of cross talk is shown by the association of UHRF1 with H3K9me3 directing the maintenance of DNA methylation (Rothbart et al. 2012). UHRF1 is a multidomain protein composed of Ubl, tandem Tudor, PHD finger, SRA, and RING domains (see Fig. 6 of Cheng 2014). DNMT1 is also a multidomain protein composed of RFD, CXXC, BAH1/2, and methyltransferase domains. Structures are available for the UHRF1 tandem Tudor domain bound to H3K9me2/3 (Nady et al. 2011), UHRF1 PHD finger bound to unmodified H3K4 (Rajakumara et al. 2011b), and UHRF1 SRA domain bound to hemimethylated CpG DNA (Arita et al. 2008; Avvakumov et al. 2008; Hashimoto et al. 2008), as well as for truncated DNMT1 constructs bound to unmodified CpG (Song et al. 2011) and hemimethylated 5mCpG (Song et al. 2012) sites on DNA. Recent functional studies have shown that UHRF1 association with the H3K9me2/3 histone mark via its tandem Tudor domain is required for the maintenance of DNA methylation, and this association is insensitive to phosphorylation at adjacent Ser10 (Rothbart et al. 2012). It was further established that mitotic binding of UHRF1 to methylated H3K9-containing chromatin stabilizes DNMT1 and contributes to maintenance DNA methylation through the cell cycle. It is not clear at this time whether UHRF1 and DNMT1 can simultaneously bind and be adjacently positioned on hemimethylated 5mCpG DNA given that UHRF1 targets unmodified H3K4 (through its PHD finger), H3K9me2/3 (tandem Tudor domain), and 5mCpG (SRA domain), whereas DNMT1 targets 5mCpG (through its methyltransferase domain) and potentially other histone PTMs (through its BAH1 and/or BAH2 domains). A future challenge will be to attempt making complexes of UHRF1 and DNMT1 on designer nucleosomes containing H3K9me2/3 marks and, if structurally tractable, to define the relative alignments and interactions between components of the complex.
Another example of cross talk between histone and DNA methylation has been noted for the maintenance of CpHpG DNA methylation in plants (reviewed in Law and Jacobsen 2010; Pikaard and Mittelsten Scheid 2014). In this regard, KRYPTONITE (KYP) is a plant histone KMT that facilitates deposition of K9me2 marks on nucleosomal H3 tails (Jackson et al. 2002), whereas chromomethylase CMT3 is a plant DNA methyltransferase that facilitates deposition of 5mC methylation marks at CpHpG sites on nucleosomal DNA (Du et al. 2012). Genome-wide profiling of DNA and H3K9 methylation has established a high correlation between H3K9me2 and CpHpG methylation. In addition, loss of CMT3 or KYP results in a dramatic decrease in DNA methylation. It has been proposed that these two enzymes establish a feedback loop whereby methylated CpHpG DNA recruits KYP to maintain methylation at H3K9, whereas the H3K9me2 mark recruits CMT3 to methylate DNA, so completing the feedback loop. Structural information is available for the complex of ZMET2 (maize counterpart of A. thaliana CMT3) bound to H3K9me2 through its chromodomain and BAH domain (Fig. 2C; Du et al. 2012), but as yet no structural information is available for KYP (also designated SUVH4), which contains SRA, pre-SET, SET, and post-SET domains, either in the free state or when bound either to H3K9me2-containing peptide and/or methylated CpHpG-containing DNA (represented schematically in Fig. 10B of Cheng 2014). There is also the additional challenge of elucidating whether direct interactions occur between histone (KYP) and DNA (CMT3) methyltransferases in this system and, if so, to elucidate the principles underlying recognition.
The take-home message is that a combined structure–function approach, as shown for the two examples above, should shed further light on mechanistic aspects of cross talk linking histone and DNA methylation, which can mediate epigenetic regulation in diverse systems (also discussed in Cheng 2014).
16.7. Chromatin-Based Therapeutic Modalities
There is considerable interest in trying to determine the consequences of abnormal chromatin regulator expression and genome alterations and their impact in facilitating the onset of disease states. The types of diseases being looked at range from autoimmune to neurological disorders, and developmental abnormalities to cancer. A natural follow-up from such studies is research into the development of chromatin-based therapeutics, which holds promise given the potential for reversibility of epigenetic-based mutations (reviewed in Chi et al. 2010; Dawson and Kouzarides 2012).
For example, structure–function studies have highlighted that the dysregulation of a chromatin-binding PHD finger can give rise to hematological malignancies (Wang et al. 2009). This study examined the consequence of fusing the carboxy-terminal H3K4me3-binding PHD finger of the jumonji domain lysine demethylase JARID1A to nucleoporin-98 (NUP98), a common fusion partner, thereby generating potent oncoproteins that arrested haematopoietic differentiation and induced acute myeloid leukemia in murine models. The fused PHD finger-NUP98 cassette dominated over polycomb-mediated gene silencing to lock developmentally critical loci into a perpetually active chromatin state, essential for leukaemogenesis. Interestingly, the aromatic cage in the JARID1A PHD finger, which binds to H3K4me3 peptide, was composed of two orthogonally aligned Trp residues. The mutation of either Trp abrogated H3K4me3 binding, consequently abolishing leukaemic transformation.
Related studies have evaluated the consequences of somatic mutations in the ING PHD fingers on solid tumors (reviewed in Chi et al. 2010). Also, the impact of INK4/ARF pathway dysregulation has been investigated in aging and cancer (reviewed in Kim and Sharpless 2006).
There is very limited literature on inhibitors targeting the Kme-binding pockets of reader modules, in contrast to Kac-binding pockets (discussed in Sec. 6.3 of Busslinger and Tarakhovsky 2014; Sec. 3.5 of Marmorstein and Zhou 2014; Qi 2014; Schaefer 2014; and reviewed in Arrowsmith et al. 2012). In earlier sections, we have outlined three general modes of molecular recognition of methylated lysine marks by reader modules. First, higher methylation state Kme3/2 marks are predominantly recognized within aromatic-lined cage-type pockets that involve a surface groove mode of recognition (Fig. 2D) (reviewed in Taverna et al. 2007). These are open and shallow pockets and, hence, are difficult to target, and as yet no inhibitors have been identified for such pockets.
Second, lower methylation state Kme1/2 marks can be positioned within the aromatic cage of MBT pocket 2 of L3MBTL1 (Li et al. 2007a; Min et al. 2007) that involve a cavity insertion mode of recognition (Fig. 6B) (reviewed in Taverna et al. 2007). Such pockets are both narrow and deep and, hence, a promising target for inhibitors. Indeed, a ligand and structure-guided design approach (Kireev et al. 2010) has identified UNC669 (Fig. 6D), a pyrrolidine-containing small molecule that targets L3MBTL1 (Kd = 5 µm) and shows fivefold increased affinity compared with cognate peptide (H4K20me1) binding, as well as selectivity against close homologs (L3MBTL3 and L3MBTL4) (Herold et al. 2011b). An X-ray structure of L3MBTL1 bound to UNC669 established that the ligand inserts its pyrrolidine ring system into the aromatic-lined pocket of MBT domain 2, with good shape complementarity between the inserted ligand and the walls of the pocket (Herold et al. 2011b) (Fig. 6E).
Third, Kme marks can be recognized by the recently identified interfacial composite pocket composed of residues from adjacently positioned reader domains, with recognition involving favorable van der Waals contacts associated with a high degree of surface complementarity, supplemented by a set of carbon–oxygen hydrogen bonds (Fig. 7C,D) (Iwase et al. 2011). It remains to be seen whether inhibitors can be designed to target such pockets.
An alternate approach would be to design linked small molecule inhibitors that can target closely positioned pockets such as, for instance, those for unmodified K4 and K9me3 on the same histone tail. Progress on development of small molecule inhibitors targeted to Kme reader modules is still in its infancy (reviewed in Herold et al. 2011a) and new approaches will need to be developed for further progress. In this regard, current drugs have rather broad specificity profiles, requiring the next-generation epigenetic drugs to target dysregulated processes with increased specificity.
Recently, it has been shown that HOTAIR lincRNA can reprogram chromatin states to promote cancer metastasis (Gupta et al. 2010). HOTAIR expression is increased in primary breast tumors, with expression levels in primary tumors a diagnostic predicator for eventual metastasis and death. In this regard, enforced expression of HOTAIR in epithelial cancer cells resulted in genome-wide retargeting of the PRC2 complex leading to altered H3K27 methylation, together with increased cancer invasiveness and metastasis. These results implicate an active role for HOTAIR in modulating the cancer epigenome, implying that lincRNAs could serve as potential targets for the diagnosis and therapy of cancer.
Because several protein–lincRNA complexes are involved in disease states, detailed structural information of intermolecular contacts could lead to directed functional studies toward identification and implementation of chromatin-based therapeutic modalities.
ACKNOWLEDGMENTS
I am grateful to Dr. Zhanxin Wang for his assistance in the preparation of this review.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, Danny Reinberg, and Monika Lachner
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this subject collection.
- Ali M, Yan K, Lalonde ME, Degerny C, Rothbart SB, Strahl BD, Cote J, Yang XJ, Kutateladze TG. 2012. Tandem PHD fingers of MORF/MOZ acetyltransferases display selectivity for acetylated histone H3 and are required for the association with chromatin. J Mol Biol 424: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci 51: 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Muir TW. 2011. Spreading chromatin into chemical biology. Chembiochem 12: 264–279. [DOI] [PubMed] [Google Scholar]
- *.Allis CD, Jeuwein T, Reinberg D. 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739. [DOI] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23: 185–188. [DOI] [PubMed] [Google Scholar]
- Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, Higgs DR, Neuhaus D, Rhodes D. 2007. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci 104: 11939–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. 2008. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455: 818–821. [DOI] [PubMed] [Google Scholar]
- Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, et al. 2012. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci 109: 12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. 2011. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334: 977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. 2012. Epigenetic protein families: A new frontier for drug discovery. Nat Rev Drug Discov 11: 384–400. [DOI] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. 2008. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455: 822–825. [DOI] [PubMed] [Google Scholar]
- Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, et al. 2012. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19: 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. 2006. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science 311: 856–861. [DOI] [PubMed] [Google Scholar]
- *.Barlow DP, Bartolomei MS. 2014. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 6: a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome—Biological and translational implications. Nat Rev Cancer 11: 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Baylin SB, Jones PA. 2014. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Becker PB, Workman JL. 2013. Nucleosome remodeling and epigenetics. Cold Spring Harb Perspect Biol 5: a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. 2009. Protein arginine methylation in mammals: Who, what, and why. Mol Cell 33: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. 2011. DNA demethylation dynamics. Cell 146: 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W, Barsyte D, Li J, Wu M, Zhou BO, et al. 2011. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J 30: 2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin CA, Yeyati P, Al Sanna N, Bober M, et al. 2011. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier–Gorlin syndrome. Nat Genet 43: 350–355. [DOI] [PubMed] [Google Scholar]
- Bienz M. 2006. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31: 35–40. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. 2012. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol Cell 48: 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Lecona E, Reinberg D. 2010. MBT domain proteins in development and disease. Semin Cell Dev Biol 21: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317: 1760–1764. [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7: 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. 2012. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet 13: 7–13. [DOI] [PubMed] [Google Scholar]
- Brehm A, Tufteland KR, Aasland R, Becker PB. 2004. The many colours of chromodomains. Bioessays 26: 133–140. [DOI] [PubMed] [Google Scholar]
- Brien GL, Gambero G, O’Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et al. 2012. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol 19: 1273–1281. [DOI] [PubMed] [Google Scholar]
- *.Brockdorff N, Turner BM. 2014. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44. [DOI] [PubMed] [Google Scholar]
- Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, et al. 2009. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One 4: e6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck-Koehntop BA, Stanfield RL, Ekiert DC, Martinez-Yamout MA, Dyson HJ, Wilson IA, Wright PE. 2012. Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso. Proc Natl Acad Sci 109: 15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Busslinger M, Tarakhovsky A. 2014. Epigenetic control of immunity. Cold Spring Harb Perspect Biol 6: a019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Rothbart SB, Lu R, Xu B, Chen WY, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, Strahl BD, et al. 2013. An H3K36 methylation-engaging Tudor motif of Polycomb-like proteins mediates PRC2 complex targeting. Mol Cell 49: 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Courvalin JC, Mornon JP. 1999. The BAH (bromo-adjacent homology) domain: A link between DNA methylation, replication and transcriptional regulation. FEBS Lett 446: 189–193. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet 10: 295–304. [DOI] [PubMed] [Google Scholar]
- Chatterjee C, McGinty RK, Fierz B, Muir TW. 2010. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol 6: 267–269. [DOI] [PubMed] [Google Scholar]
- Chen J, Santillan DA, Koonce M, Wei W, Luo R, Thirman MJ, Zeleznik-Le NJ, Diaz MO. 2008. Loss of MLL PHD finger 3 is necessary for MLL-ENL-induced hematopoietic stem cell immortalization. Cancer Res 68: 6199–6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Nott TJ, Jin J, Pawson T. 2011. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol 12: 629–642. [DOI] [PubMed] [Google Scholar]
- *.Cheng X. 2014. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 6: a018747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Yang Y, Fang J, Xiao J, Zhu T, Chen F, Wang P, Li Z, Yang H, Xu Y. 2013. Structural insight into coordinated recognition of H3K9me3 by the plant homeodomain (PHD) and tandem tudor domain (TTD) of the UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem 288: 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. 2010. Covalent histone modifications—Miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chignola F, Gaetani M, Rebane A, Org T, Mollica L, Zucchelli C, Spitaleri A, Mannella V, Peterson P, Musco G. 2009. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res 37: 2951–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Clouaire T, Stancheva I. 2008. Methyl-CpG binding proteins: Specialized transcriptional repressors or structural components of chromatin? Cell Mol Life Sci 65: 1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X. 2008. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol 15: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Trievel RC. 2006. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol 13: 698–703. [DOI] [PubMed] [Google Scholar]
- Davis L, Chin JW. 2012. Designer proteins: Applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol 13: 168–182. [DOI] [PubMed] [Google Scholar]
- Dawson PE, Kent SB. 2000. Synthesis of native proteins by chemical ligation. Annu Rev Biochem 69: 923–960. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. 2012. Cancer epigenetics: From mechanism to therapy. Cell 150: 12–27. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. 2009. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature 461: 819–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dekker J, Misteli T. 2014. Long-range chromatin interactions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121: 873–885. [DOI] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. 2012. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Chesnokov IN, McConkey BJ. 2009. The origin recognition complex protein family. Genome Biol 10: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Elgin SC, Reuter G. 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, Garrick D, Clynes D, Gibbons RJ, Rhodes D, et al. 2011. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol 18: 777–782. [DOI] [PubMed] [Google Scholar]
- Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz MO. 2001. Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol Cell Biol 21: 3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S, Ravaud S, Koch J, Sinning I. 2010. The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem 285: 5954–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M, Sanchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Muller J, Evans P, Bienz M. 2008. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol Cell 30: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B, Muir TW. 2012. Chromatin as an expansive canvas for chemical biology. Nat Chem Biol 8: 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. 2003a. Binary switches and modification cassettes in histone biology and beyond. Nature 425: 475–479. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. 2003b. Histone and chromatin cross-talk. Curr Opin Cell Biol 15: 172–183. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438: 1181–1185. [DOI] [PubMed] [Google Scholar]
- Frederiks F, Stulemeijer IJ, Ovaa H, van Leeuwen F. 2011. A modified epigenetics toolbox to study histone modifications on the nucleosome core. Chembiochem 12: 308–313. [DOI] [PubMed] [Google Scholar]
- Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. 2011. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol 21: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner KE, Allis CD, Strahl BD. 2011. Operating on chromatin, a colorful language where context matters. J Mol Biol 409: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, Garcia BA, Kutateladze TG, Denu JM. 2010. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat Chem Biol 6: 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goforth RL, Peterson EC, Yuan J, Moore MJ, Kight AD, Lohse MB, Sakon J, Henry RL. 2004. Regulation of the GTPase cycle in post-translational signal recognition particle-based protein targeting involves cpSRP43. J Biol Chem 279: 43077–43084. [DOI] [PubMed] [Google Scholar]
- Goodwin GH, Nicolas RH. 2001. The BAH domain, polybromo and the RSC chromatin remodelling complex. Gene 268: 1–7. [DOI] [PubMed] [Google Scholar]
- Greer EL, Shi Y. 2012. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet 13: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Muller J, Muller CW. 2009. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J 28: 1965–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Grossniklaus U, Paro R. 2014. Transcriptional silencing by Polycomb-group proteins. Cold Spring Harb Perspect Biol 6: a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow EJ, Wysocka J. 2010. Flipping MLL1’s switch one proline at a time. Cell 141: 1108–1110. [DOI] [PubMed] [Google Scholar]
- *.Grunstein M, Gasser SM. 2013. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol 5: a017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. 2007. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449: 933–937. [DOI] [PubMed] [Google Scholar]
- Guil S, Esteller M. 2012. Cis-acting noncoding RNAs: Friends and foes. Nat Struct Mol Biol 19: 1068–1075. [DOI] [PubMed] [Google Scholar]
- Guo Y, Nady N, Qi C, Allali-Hassani A, Zhu H, Pan P, Adams-Cioaba MA, Amaya MF, Dong A, Vedadi M, et al. 2009. Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2. Nucleic Acids Res 37: 2204–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J. 2006. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell 22: 137–144. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. 2008. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455: 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Cheng X. 2009. UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications. Epigenetics 4: 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Zhang X, Cheng X. 2012. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: Structural basis and implications for active DNA demethylation. Nucleic Acids Res 40: 8276–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Henikoff S, Smith MM. 2014. Histone variants and epigenetics. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold JM, Ingerman LA, Gao C, Frye SV. 2011a. Drug discovery toward antagonists of methyl-lysine binding proteins. Curr Chem Genomics 5: 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold JM, Wigle TJ, Norris JL, Lam R, Korboukh VK, Gao C, Ingerman LA, Kireev DB, Senisterra G, Vedadi M, et al. 2011b. Small-molecule ligands of methyl-lysine binding proteins. J Med Chem 54: 2504–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180. [DOI] [PubMed] [Google Scholar]
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. 2008. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell 29: 525–531. [DOI] [PubMed] [Google Scholar]
- Holdermann I, Meyer NH, Round A, Wild K, Sattler M, Sinning I. 2012. Chromodomains read the arginine code of post-translational targeting. Nat Struct Mol Biol 19: 260–263. [DOI] [PubMed] [Google Scholar]
- Hom RA, Chang PY, Roy S, Musselman CA, Glass KC, Selezneva AI, Gozani O, Ismagilov RF, Cleary ML, Kutateladze TG. 2010. Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain. J Mol Biol 400: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Stillman B. 2012. Meier–Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev 26: 1797–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Bernstein DA, Fox CA, Keck JL. 2005. Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing. Proc Natl Acad Sci 102: 8489–8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HC, Stillman B, Xu RM. 2005. Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing. Proc Natl Acad Sci 102: 8519–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Wang P, Lin Y, Xu Y. 2011. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res 21: 1374–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. 2006. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312: 748–751. [DOI] [PubMed] [Google Scholar]
- Huang R, Holbert MA, Tarrant MK, Curtet S, Colquhoun DR, Dancy BM, Dancy BC, Hwang Y, Tang Y, Meeth K, et al. 2010. Site-specific introduction of an acetyl-lysine mimic into peptides and proteins by cysteine alkylation. J Am Chem Soc 132: 9986–9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. 2009. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell 33: 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr., Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411. [DOI] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. 2007. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev 21: 3369–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. 2007. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28: 823–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, Allis CD, Picketts DJ, Patel DJ, Li H, et al. 2011. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol 18: 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295: 2080–2083. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J 20: 5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331: 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. 2007. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. 2009. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 10: 805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan PY, Caterino TL, Hayes JJ. 2009. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol 29: 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorasanizadeh S. 2011. Recognition of methylated histones: New twists and variations. Curr Opin Struct Biol 21: 744–749. [DOI] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. 2006. The regulation of INK4/ARF in cancer and aging. Cell 127: 265–275. [DOI] [PubMed] [Google Scholar]
- Kim D, Blus BJ, Chandra V, Huang P, Rastinejad F, Khorasanizadeh S. 2010. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat Struct Mol Biol 17: 1027–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireev D, Wigle TJ, Norris-Drouin J, Herold JM, Janzen WP, Frye SV. 2010. Identification of non-peptide malignant brain tumor (MBT) repeat antagonists by virtual screening of commercially available compounds. J Med Chem 53: 7625–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. 2007. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449: 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingseisen A, Jackson AP. 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev 25: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. 2006. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 31: 89–97. [DOI] [PubMed] [Google Scholar]
- Koga H, Matsui S, Hirota T, Takebayashi S, Okumura K, Saya H. 1999. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene 18: 3799–3809. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Kouzarides T, Berger SL. 2007. Chromatin modifications and their mechanism. In Epigenetics (ed. Allis CD, Jenuwen T, Reinberg D), pp. 191–209. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Kriaucionis S, Heintz N. 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kriaucionis S, Tahiliani M. 2014. Expanding the epigenetic landscape: Novel modifications of cytosine in genomic DNA. Cold Spring Harb Perspect Biol 6: a018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel JF, Goodrich JA. 2012. Non-coding RNAs: Key regulators of mammalian transcription. Trends Biochem Sci 37: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. 2012. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome. Nature 484: 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. 2007. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448: 718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham JA, Dent SY. 2007. Cross-regulation of histone modifications. Nat Struct Mol Biol 14: 1017–1024. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Du J, Hale CJ, Feng S, Krajewski K, Palanca AM, Strahl BD, Patel DJ, Jacobsen SE. 2013. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. 2012. Epigenetic regulation by long noncoding RNAs. Science 338: 1435–1439. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. 2007. Histone cross talk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096. [DOI] [PubMed] [Google Scholar]
- Lee J, Thompson JR, Botuyan MV, Mer G. 2008. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol 15: 109–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bird A. 2007. DNA methylation in mammals. In Epigenetics (ed. Allis CD, Jenuwen T, Reinberg D), pp. 341–356. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- *.Li E, Zhang Y. 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 6: a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fischle W, Wang W, Duncan EM, Liang L, Murakami-Ishibe S, Allis CD, Patel DJ. 2007a. Structural basis for lower lysine methylation state-specific readout by MBT repeats of L3MBTL1 and an engineered PHD finger. Mol Cell 28: 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, Ann DK. 2007b. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem 282: 36177–36189. [DOI] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. 2010. Adding new chemistries to the genetic code. Annu Rev Biochem 79: 413–444. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM. 2010a. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev 24: 1876–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Chen C, Guo Y, Lam R, Bian C, Xu C, Zhao DY, Jin J, MacKenzie F, Pawson T, et al. 2010b. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci 107: 18398–18403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. 2012. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev 26: 2374–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes EC, Valls E, Figueroa ME, Mazur A, Meng FG, Chiosis G, Laird PW, Schreiber-Agus N, Greally JM, Prokhortchouk E, et al. 2008. Kaiso contributes to DNA methylation-dependent silencing of tumor suppressor genes in colon cancer cell lines. Cancer Res 68: 7258–7263. [DOI] [PubMed] [Google Scholar]
- *.Lucchesi JC, Kuroda MI. 2014. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol 6: a019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ. 2012. New insights into nucleosome and chromatin structure: An ordered state or a disordered affair? Nat Rev Mol Cell Biol 13: 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JC, Dougherty DA. 1997. The Cation–π interaction. Chem Rev 97: 1303–1324. [DOI] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. 2010. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467: 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, et al. 2012. Suppression of the antiviral response by an influenza histone mimic. Nature 483: 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Marmorstein R, Zhou M-M. 2014. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol 6: a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Martienssen R, Moazed D. 2014. RNAi and heterochromatin assembly. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathioudakis N, Palencia A, Kadlec J, Round A, Tripsianes K, Sattler M, Pillai RS, Cusack S. 2012. The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors. RNA 18: 2056–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450: 1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. 2003. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci 28: 69–74. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. 2008. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453: 812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliori V, Muller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, et al. 2012. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol 19: 136–144. [DOI] [PubMed] [Google Scholar]
- Miller TC, Rutherford TJ, Johnson CM, Fiedler M, Bienz M. 2010. Allosteric remodelling of the histone H3 binding pocket in the Pygo2 PHD finger triggered by its binding to the B9L/BCL9 co-factor. J Mol Biol 401: 969–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117. [DOI] [PubMed] [Google Scholar]
- Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, Wang Z, Ruthenburg AJ, Elenitoba-Johnson KS, Roeder RG, et al. 2010. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell 38: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. 2007. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol 14: 1229–1230. [DOI] [PubMed] [Google Scholar]
- Moazed D. 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Kutateladze TG. 2011. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res 39: 9061–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nunez JK, Nickoloff J, et al. 2012a. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19: 1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. 2012b. Perceiving the epigenetic landscape through histone readers. Nat Struct Mol Biol 19: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Ramirez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, Mackay JP, Wade PA, Hagman J, Kutateladze TG. 2012c. Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc Natl Acad Sci 109: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, et al. 2011. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem 286: 24300–24311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nady N, Krichevsky L, Zhong N, Duan S, Tempel W, Amaya MF, Ravichandran M, Arrowsmith CH. 2012. Histone recognition by human malignant brain tumor domains. J Mol Biol 423: 702–718. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Umehara T, Hamana H, Hayashizaki Y, Inoue M, Kigawa T, Shirouzu M, Terada T, Tanaka A, Padmanabhan B, et al. 2007. Crystal structure analysis of the PHD domain of the transcription co-activator Pygopus. J Mol Biol 370: 80–92. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. 2012. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486: 415–419. [DOI] [PubMed] [Google Scholar]
- Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. 2010. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464: 441–444. [DOI] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277: 34655–34657. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416: 103–107. [DOI] [PubMed] [Google Scholar]
- Ohki I, Shimotake N, Fujita N, Jee J, Ikegami T, Nakao M, Shirakawa M. 2001. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell 105: 487–497. [DOI] [PubMed] [Google Scholar]
- Oliver SS, Denu JM. 2011. Dynamic interplay between histone H3 modifications and protein interpreters: Emerging evidence for a ‘histone language’. Chembiochem 12: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. 2009. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep 10: 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Osmers U, Raman G, Schwantes RH, Diaz MO, Bushweller JH. 2010. The PHD3 domain of MLL acts as a CYP33-regulated switch between MLL-mediated activation and repression. Biochemistry 49: 6576–6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. 2000. Structure of the PHD zinc finger from human Williams–Beuren syndrome transcription factor. J Mol Biol 304: 723–729. [DOI] [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, Cosgrove MS. 2008. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem 283: 32158–32161. [DOI] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. 2006. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442: 100–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Wysocka J. 2008. It takes a PHD to SUMO. Trends Biochem Sci 33: 191–194. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. [DOI] [PubMed] [Google Scholar]
- *.Pikaard CS, Mittelsten Scheid O. 2014. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Qi J. 2014. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: Chemical modulation of chromatin structure. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Sawada K, Zhang X, Cheng X. 2002. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat Struct Biol 9: 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Liu L, Zhao C, Han C, Li F, Zhang J, Wang Y, Li G, Mei Y, Wu M, et al. 2012. Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription. Genes Dev 26: 1376–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE. 2011a. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev 25: 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, et al. 2011b. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell 43: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. 2011. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147: 1628–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, Gozani O, Yang W. 2007. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci 104: 18993–18998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Reik W, Surani MA. 2014. Germline and pluripotent stem cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rinn JL. 2014. lncRNAs: Linking RNA to chromatin. Cold Spring Harb Perspect Biol 6: a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. 2005. DNA methylation and human disease. Nat Rev Genet 6: 597–610. [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. 2012. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 19: 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Musselman CA, Kachirskaia I, Hayashi R, Glass KC, Nix JC, Gozani O, Appella E, Kutateladze TG. 2010. Structural insight into p53 recognition by the 53BP1 tandem Tudor domain. J Mol Biol 398: 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. 2006. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol 13: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. 2007a. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. 2007b. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 8: 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, et al. 2011. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145: 692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. 2007. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell 27: 596–608. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. 2011. The PHD finger: A versatile epigenome reader. Trends Biochem Sci 36: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411. [DOI] [PubMed] [Google Scholar]
- Sarg B, Helliger W, Talasz H, Koutzamani E, Lindner HH. 2004. Histone H4 hyperacetylation precludes histone H4 lysine 20 trimethylation. J Biol Chem 279: 53458–53464. [DOI] [PubMed] [Google Scholar]
- Sasai N, Nakao M, Defossez PA. 2010. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res 38: 5015–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyamurthy A, Allen MD, Murzin AG, Bycroft M. 2003. Crystal structure of the malignant brain tumor (MBT) repeats in Sex Comb on Midleg-like 2 (SCML2). J Biol Chem 278: 46968–46973. [DOI] [PubMed] [Google Scholar]
- *.Schaefer U. 2014. Pharmacological inhibition of bromodomain-containing proteins in inflammation. Cold Spring Harb Perspect Biol 6: a018671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Noffsinger VJ, Shanker S, Kuscu C, Joshua-Tor L, Partridge JF. 2009. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell 34: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J. 2006. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J 25: 4245–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. 2001. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol 8: 27–31. [DOI] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. 2012. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys 41: 343–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Seto E, Yoshida M. 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6: a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. 2006. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. 2006. Chromatin modifications by methylation and ubiquitination: Implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311: 844–847. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. 2007. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128: 1003–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem 280: 41789–41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ 3rd, Eick D, Reinberg D. 2011. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 332: 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Mannen T, Siomi H. 2010. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev 24: 636–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater LM, Allen MD, Bycroft M. 2003. Structural variation in PWWP domains. J Mol Biol 330: 571–576. [DOI] [PubMed] [Google Scholar]
- Song JJ, Kingston RE. 2008. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem 283: 35258–35264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Garlick JD, Kingston RE. 2008. Structural basis of histone H4 recognition by p55 Genes Dev 22: 1313–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Rechkoblit O, Bestor TH, Patel DJ. 2011. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science 331: 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Teplova M, Ishibe-Murakami S, Patel DJ. 2012. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science 335: 709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M. 2003. High-resolution X-ray and NMR structures of the SMN Tudor domain: Conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol 327: 507–520. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Sukackaite R, Grazulis S, Tamulaitis G, Siksnys V. 2012. The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res 40: 7552–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108. [DOI] [PubMed] [Google Scholar]
- Sun B, Hong J, Zhang P, Dong X, Shen X, Lin D, Ding J. 2008. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J Biol Chem 283: 36504–36512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Davey CA. 2011. Nucleosome structural studies. Curr Opin Struct Biol 21: 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, et al. 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 24: 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. 2007. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat Struct Mol Biol 14: 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. 2011. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol 18: 1414–1420. [DOI] [PubMed] [Google Scholar]
- Trojer P, Reinberg D. 2008. Beyond histone methyl-lysine binding: How malignant brain tumor (MBT) protein L3MBTL1 impacts chromatin structure. Cell Cycle 7: 578–585. [DOI] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. 2007. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129: 915–928. [DOI] [PubMed] [Google Scholar]
- Trojer P, Cao AR, Gao Z, Li Y, Zhang J, Xu X, Li G, Losson R, Erdjument-Bromage H, Tempst P, et al. 2011. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol Cell 42: 438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010a. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. 2010b. TRIM24 links a non-canonical histone signature to breast cancer. Nature 468: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K, Zlatanova J. 2007. Chromatin fiber structure: Where is the problem now? Semin Cell Dev Biol 18: 651–658. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980. [DOI] [PubMed] [Google Scholar]
- Vezzoli A, Bonadies N, Allen MD, Freund SM, Santiveri CM, Kvinlaug BT, Huntly BJ, Gottgens B, Bycroft M. 2010. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol 17: 617–619. [DOI] [PubMed] [Google Scholar]
- Voigt P, Reinberg D. 2011. Histone tails: Ideal motifs for probing epigenetics through chemical biology approaches. Chembiochem 12: 236–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Patel DJ. 2011. Combinatorial readout of dual histone modifications by paired chromatin-associated modules. J Biol Chem 286: 18363–18368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WK, Tereshko V, Boccuni P, MacGrogan D, Nimer SD, Patel DJ. 2003. Malignant brain tumor repeats: A three-leaved propeller architecture with ligand/peptide binding pockets. Structure 11: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. 2009. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Mok MW, Harper H, Lee WH, Min J, Knapp S, Oppermann U, Marsden B, Schapira M. 2010a. Structural genomics of histone tail recognition. Bioinformatics 26: 2629–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. 2010b. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell 141: 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shen J, Yang Z, Chen P, Zhao B, Hu W, Lan W, Tong X, Wu H, Li G, et al. 2011. Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger. Cell Res 21: 1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, Kutateladze TG, Gozani O. 2010. The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. J Biol Chem 285: 37725–37732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. 2012. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep 13: 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29: 183–212. [DOI] [PubMed] [Google Scholar]
- Wu H, Zhang Y. 2011. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25: 2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. 2011a. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zee BM, Wang Y, Garcia BA, Dou Y. 2011b. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell 43: 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90. [DOI] [PubMed] [Google Scholar]
- Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. 2011. A poised chromatin platform for TGF-β access to master regulators. Cell 147: 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZB, Anderson M, Diaz MO, Zeleznik-Le NJ. 2003. MLL repression domain interacts with histone deacetylases, the polycomb group proteins HPC2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci 100: 8342–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Xie D, Cao X, Yu P, Xing X, Chen CC, Musselman M, Xie M, West FD, Lewin HA, et al. 2012. Comparative epigenomic annotation of regulatory DNA. Cell 149: 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Jakoncic J, Qian C. 2012. UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J Mol Biol 415: 318–328. [DOI] [PubMed] [Google Scholar]
- Xu C, Cui G, Botuyan MV, Mer G. 2008. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure 16: 1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. 2011. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell 42: 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ. 2011. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature 472: 448–453. [DOI] [PubMed] [Google Scholar]
- Yang N, Xu RM. 2012. Structure and function of the BAH domain in chromatin biology. Crit Rev Biochem Mol Biol 48: 211–221. [DOI] [PubMed] [Google Scholar]
- Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. 2010. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell 40: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Wang W, Wang Y, Wang M, Zhao Q, Rao Z, Zhu B, Xu RM. 2012. Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1. Proc Natl Acad Sci 109: 17954–17959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Zhou MM. 2010. Keeping it in the family: Diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol 45: 488–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Zhou MM. 2011. Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry 50: 1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CC, Matthews AG, Jin Y, Chen CF, Chapman BA, Ohsumi TK, Glass KC, Kutateladze TG, Borowsky ML, Struhl K, et al. 2012. Histone H3R2 symmetric dimethylation and histone H3K4 trimethylation are tightly correlated in eukaryotic genomes. Cell Rep 1: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, Rauscher FJ 3rd, Zhou MM. 2008. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol 15: 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou MM. 2010. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466: 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hayashi MK, Merkel O, Stillman B, Xu RM. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J 21: 4600–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, Chan C, Tatchell K, Hawke DH, Kobayashi R, Dent SY. 2005. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell 122: 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, Huang Q, Liu Q, Hao Q, Ding J. 2006. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res 34: 6621–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. 2012. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol 8: 328–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zhao Y, Garcia BA. 2014. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18. [DOI] [PubMed] [Google Scholar]
- *.Zoghbi HY, Beaudet AL. 2014. Epigenetics and human disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019497. [DOI] [PMC free article] [PubMed] [Google Scholar]