Abstract
The brain is thought to represent specific memories through the activity of sparse and distributed neural ensembles. In this review, we examine the use of immediate early genes (IEGs), genes that are induced by neural activity, to specifically identify and genetically modify neurons activated naturally by environmental experience. Recent studies using this approach have identified cellular and molecular changes specific to neurons activated during learning relative to their inactive neighbors. By using opto- and chemogenetic regulators of neural activity, the neurons naturally recruited during learning can be artificially reactivated to directly test their role in coding external information. In contextual fear conditioning, artificial reactivation of learning-induced neural ensembles in the hippocampus or neocortex can substitute for the context itself. That is, artificial stimulation of these neurons can apparently cause the animals to “think” they are in the context. This represents a powerful approach to testing the principles by which the brain codes for the external world and how these circuits are modified with learning.
Genetic tools in mice (involving the intermediate early genes) allow studies to focus on neurons that are activated by environmental stimuli. These studies provide insight into the molecular changes that underlie learning.
A central feature of nervous systems is that, to function properly, specific neurons must become active in response to specific stimuli. The nature of this selective activation and its modification with experience is the focus of much neuroscience research, ranging from studies of sensory processing in experimental animals to disorders of thought such as schizophrenia in humans. The central dogma of neuroscience is that perceptions, memories, thoughts, and higher mental functions arise from the pattern and timing of the activity in neural ensembles in specific parts of the brain at specific points in time. Until quite recently, the investigation of these “circuit”-based questions has primarily been limited to observational techniques, such as single unit recording, functional magnetic resonance imagery (fMRI), and calcium imaging, to document the patterns of neural activity evoked by sensory experience or even complex psychological contingencies in human fMRI studies. These techniques have been enormously successful and created a framework for understanding information processing in the brain. For example, recordings in the visual system have indicated that, in the primary visual cortex, neurons are tuned to the orientation of linear stimuli (Hubel and Wiesel 1962). In contrast, neurons in higher brain areas can respond to discrete items. The most striking example of this specificity comes from in vivo recording in the human medial temporal lobe in which single units have been identified that respond to photos of the actress Halle Berry as well as her written name (Quiroga et al. 2005). This highly selective tuning of neural activity is suggestive of function, but how can this be directly tested? What would be the effect of stimulating just this rare population of neurons, a memory of the actress, a sensory illusion of her image? How does this type of specific firing arise? Do these neurons differ from their nonresponsive neighbors in terms of biochemistry, cell biology, or connectivity? Do they undergo molecular alterations when new information is learned about this individual and are these changes required for the learning? These types of questions have recently become accessible to study in mice through the use of activity-based genetic manipulation, in which neurons that are activated by a specific sensory stimulus can be altered to express any gene of experimental interest. These studies and approaches will be the focus of this work.
IMMEDIATE EARLY GENES FOR ACTIVITY MAPPING
The first observation that the expression of certain genes was responsive to neural activity was made almost 30 years ago with the identification of increased expression of cfos in the brain following seizure (Morgan et al. 1987). cfos is one of a class of genes known as immediate early genes (IEGs) that are defined by their rapid induction by pre-existing transcription factors without the need for de novo protein synthesis (Greenberg et al. 1986). This allows rapid transcription initiation, within 1 min of electrical stimulation, supported by RNA polymerase II that is bound to the promoter region under resting conditions (Saha et al. 2011). On neuronal activation, the stalled RNA polymerase is released, thereby enabling the extremely rapid induction of IEG transcription. In addition to rapid induction, the cfos protein and messenger RNA (mRNA) have a relatively short half-life, such that following the end of active transcription the levels of expressed protein rapidly return to baseline (Rahmsdorf et al. 1987; Wellington et al. 1993). In the induction of cfos with seizure, the expression peaks at 1 h and returns to baseline by 3 h, allowing the expression of this gene to provide a snapshot view of brain activity with this 3-h time window (Morgan et al. 1987). Since the initial identification of cfos, genetic screens have identified a wide array of immediate early genes that show similar responses to neural activity (Nedivi et al. 1993; Qian et al. 1993; Yamagata et al. 1993). The most notable are arc and zif268/erg1, which, along with cfos, have been used extensively as a surrogate measure for neural ensemble activity in experimental animals (Worley et al. 1991; Lyford et al. 1995).
There is an extensive literature on the regulation of IEGs in cultured neurons and non-neuronal cells in which transcription can be triggered in response to multiple classes of stimuli, including growth factors and cAMP as well as strong depolarization (Ghosh et al. 1994). The degree of synaptic or intrinsic neural activity required to induce IEG expression is not straightforward and varies between genes and brain regions. For example, in vivo stimulation of dentate gyrus (DG) granule cells with high-frequency bursts that are sufficient to produce long-term potentiation (LTP) causes strong induction of zif268, but cfos is only induced in these neurons with more prolonged stimulation protocols. In both cases, the induction requires N-methyl-d-aspartate (NMDA) receptor function (French et al. 2001). In hippocampal CA1 neurons, similar stimulation protocols failed to induce any of the IEGs. In hippocampal slice, the cfos gene is induced with high fidelity by 30 or more action potentials delivered at 10 Hz or greater frequency (Schoenenberger et al. 2009). Although the precise nature of activity required to induce IEG expression is not clear, the expression of IEGs has been used extensively as a measure of neural activation in response to environmental stimulation and has given results that often track with activity determined by other techniques, such as electrophysiological recording or with known anatomical function and connectivity. For example, cfos is activated in a tonotopic pattern in the auditory brainstem with auditory stimulation (Saint Marie et al. 1999) and in a somatotopic pattern in somatosentory cortex following wisker stimulation (Wagener et al. 2010). In learning and memory paradigms, such as fear conditioning, there is cfos activation in an array of areas, such as hippocampus, amygdala, prefrontal cortex, and anterior cingulate cortex, that are known to be active and functionally necessary for the learning or retrieval of contextual fear memories (Milanovic et al. 1998; Radulovic et al. 1998; Bontempi et al. 1999; Frankland et al. 2004; Knapska and Maren 2009).
Until the development of Ca2+ imaging approaches (Jercog et al. 2015), detection of IEG expression was one of the only ways of assessing large ensembles of neural activity with cellular resolution. A critical question in learning and memory (and neural processing more generally) is how similar is neural circuit activation with learning and with retrieval or during multiple retrieval trials. One approach to this question that takes advantage of the rapid and transient nature of IEG expression is known as catFISH (for cellular compartment analysis of temporal activity by fluorescence in situ hybridization) (Guzowski et al. 1999). Because mRNA is initially produced in the nucleus in an unspliced form and is later transported to the cytoplasm following splicing, the use of probes targeting unspliced nuclear RNA (currently active neurons) and processed cytoplasmic RNA (previously active neurons) can be used to determine the activity of neurons at two different but closely spaced time points. This approach was used to examine the activity of CA1 hippocampal neurons following the exploration of two identical environments relative to the exploration of two distinct environments. The results showed that there was a greater overlap in arc expression in CA1 neurons following the two epochs exploring identical environments. This is consistent with the role of the hippocampus in place recognition and with in vivo electrophysiological studies of place cell activity and its role in encoding location. It also supports the idea that IEG expression reliably reports behaviorally relevant neural activity.
IEG-Based Transgenics
Although the IEGs offer a temporally crude measure of neural activation, they do parallel other methods of assessing activity in many brain areas. In addition, genetic studies have suggested a role for cfos, zif268, and arc in synaptic plasticity and memory, suggesting that their activation may be particularly important in identifying circuits undergoing plastic changes (Fleischmann et al. 2003; Plath et al. 2006; Baumgartel et al. 2008). One advantage of IEGs over more direct and temporally precise measures of neural activity is that by providing a transcriptional response they offer a molecular genetic conduit into environmentally activated neural ensembles. This was first shown in a simple transgenic mouse in which a promoter element from the cfos gene was used to drive expression of a lacZ marker protein (Smeyne et al. 1992). The expression of the marker was strongly induced throughout the brain following seizure. In addition, the lacZ expression was responsive to environmental stimuli showing strong induction in the superchiasmatic nucleus, an area involved in circadian rhythm regulation, following a light pulse during the animal’s dark cycle. The transgene incorporated just 600 bp of promoter region containing four different transcriptional response elements, all of which were required for normal stimulus-induced gene expression (Robertson et al. 1995).
More recently, a number of transgenic mouse lines have been developed that drive green fluorescent protein (GFP) in an activity-dependent manner (see Table 1 for a list of published IEG-based transgenic lines) (Barth et al. 2004; Wang et al. 2006; Reijmers et al. 2007). The use of a fluorescent marker compatible with live tissue imaging allows the recording of neurons in brain slices or in anesthetized animals specifically from recently active cfos-positive cells. This approach was used to examine spontaneously active neurons in the somatosentory cortex, where approximately 15% of neurons in layer 2/3 are cfos-GFP positive in the absence of any specific environmental stimuli (Yassin et al. 2010). These cfos-positive neurons showed an increase in excitatory drive, a reduction in inhibitory drive, and a greater degree of interconnectivity than their cfos-negative neighbors. These results indicate that ensembles of neurons with increased activity are maintained for several hours (the timing between cfos expression and electrophysiological recording) and that there may be a stable structure and connectivity in these spontaneously active ensembles. The presence of such spontaneously active ensembles is consistent with electrophysiological recordings that have suggested the importance of posttraining replay of neural activity patterns evoked during learning in the consolidation of memory (Wilson and McNaughton 1994; Foster and Wilson 2006; Karlsson and Frank 2009).
Table 1.
Currently available immediate early genes (IEGs)-based transgenic mouse lines
| Gene | Type | References | JAX stock No. |
|---|---|---|---|
| Fos-lacZ | Transgenic | Smeyne et al. 1992 | - |
| Fos-taulacZ | Transgenic | Mehta et al. 2002 | - |
| Fos-GFP | Transgenic | Barth et al. 2004 | 014135 |
| Fos-GFP/tTA | Transgenic | Reijmers et al. 2007 | 018306 |
| Fos-CreERT2 | Knockin | Guenthner et al. 2013 | 021882 |
| Arc-CreERT2 | Knockin | Guenthner et al. 2013 | 021881 |
| Arc-CreERT2 | BAC transgenic | Denny et al. 2014 | 022357 |
| Arc-GFP | Knockin | Wang et al. 2006 | 007662 |
| ERG1-GFP | BAC transgenic | Xie et al. 2014 | 014709-UCD |
List of IEG-based transgenic and knockin mouse lines. Most are available from the Jackson Laboratory repository (www.jax.org).
IEG-linked GFP reporter mice have also been used as a convenient means of repeatedly measuring neural activation patterns in intact animals by two-photon imaging through cranial windows. To examine the role of arc in the orientation selectivity responses of neurons in the primary visual cortex, Wang et al. (2006) generated a mouse in which GFP replaced the functional arc protein to produce a knockout in which the activation of neural ensembles could be imaged through measurement of GFP expression levels. Heterozygous mice (carrying one functional arc allele and the activity-dependent arc-GFP) showed normal orientation tuning, whereas the homozygous, arc knockout mice showed impaired tuning of individual neurons measured electrophysiologically. Wang et al. then examined the activity of all neurons in the field using the GFP reporter and determined that the overall loss of tuning specificity was because of an increase in the number of weakly tuned cells rather than a loss of specificity in the highly tuned neurons. In another study, a transgenic mouse in which GFP was driven by the zif268 promoter was used to examine the formation of contextual representations in the neocortex following fear conditioning (Xie et al. 2014). These investigators found slowly developing and very sparse contextual representations that were stable for up to 2 months. Similar results were obtained using a cfos-GFP reporter to examine the formation of spatial representations in the retrosplenial cortex (Czajkowski et al. 2014).
Finally, IEG-linked GFP reporters have been used to detect neural activity on a brain-wide scale (Kim et al. 2015; Vousden et al. 2015). Serial two-photon tomography is a method for obtaining fluorescent images in an automated manner with the capacity to capture 260 50 μm serial sections throughout an entire mouse brain at 10× resolution within several hours (Ragan et al. 2012). This imaging approached was applied to cfos-GFP transgenic mice and combined with an analysis pipeline for automated cell counting and anatomical partitioning to obtain whole brain activity maps at cellular resolution in social behavior (Kim et al. 2015). The results were generally consistent with previous studies showing activation in appropriate areas of the hypothalamus and amygdala with social interaction and served as a proof of principle for automated whole brain imaging of activity using an IEG-based reporter.
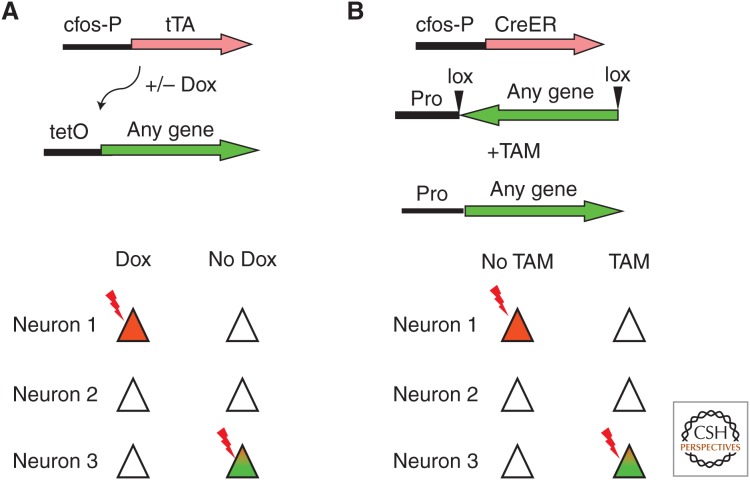
The use of activity-dependent promoters to drive expression of fluorescent markers allows for targeted recording, repeated imaging for prolonged time periods, and whole brain activity mapping at a single time point. To probe the function of active neural ensembles, several groups have developed IEG-based transgenic lines that allow the expression of effector molecules in neurons that are active at a specific time point. By using the cfos promoter in conjunction with the tetracycline system for control of gene expression, Reijmers et al. 2007 produced a transgenic mouse in which neurons that are naturally active in a given time window could be genetically modified to express essentially any gene, as shown in Figure 1A. The approach requires two transgenes introduced into the same animal. The first transgene consists of the cfos promoter driving expression of the tetracycline transactivator (tTA), which is a transcription factor that can be regulated with the antibiotic doxycycline (Dox) (Gossen and Bujard 1992). The second transgene carries a tetracycline-responsive element promoter (TRE) to drive expression of any gene of interest. In the presence of Dox delivered in the animal’s diet, the cfos promoter will drive expression of tTA in active neurons; however, transcription of the second TRE-linked gene will be blocked. When Dox is removed, a time window is open during which neurons that are sufficiently active to induce the cfos-tTA will express the TRE-linked transgene via tTA-driven transcription at the TRE promoter. The investigators used these mice to examine the neural ensembles in the amygdala that are activated with learning and retrieval in contextual fear conditioning. They used fos-tTA to drive expression of a long-lasting lacZ reporter protein in neurons activated during learning and compared the degree to which these neurons were reactivated following memory retrieval. They found that the amygdala neurons activated during learning are responsive primarily to shock, the unconditioned stimulus (US), and that after training these shock-US-responsive neurons were activated by the context, the conditioned stimulus (CS), alone and that the degree of reactivation was correlated with the strength of the expressed fear memory. These results suggest a model whereby pairing of CS and US during training produces plasticity within the circuit that allows the CS to now recruit US neurons, linking the two pathways during recall. These results are consistent with the literature on amygdala circuitry in fear conditioning (and also with fear conditioning in Aplysia; see Byrne and Hawkins 2015; Hawkins and Byrne 2015) and provide a validation of this genetic model, which has been used extensively in studies of the circuitry underlying memory as discussed in later sections.
Figure 1.
Two systems for the genetic manipulation of active neural ensembles. (A) In this tetracycline (TET)-based system, two transgenes are required, a cfos promoter-driven tetracycline transactivator (tTA) and a tetracycline-responsive element (TRE) promoter-driven gene of interest. In the presence of doxycycline (Dox) the tTA is expressed in electrically active (cfos+) neurons but is prevented from activating expression of the gene of interest by the presence of Dox. In the absence of Dox, a window is opened during which active neurons that express tTA drive expression of the gene of interest from the TRE promoter. (B) This Cre-based system also uses two transgenes, a cfos promoter-driven CreERt2 and a gene of interest that is flanked by loxP sites and positioned in an inverted orientation to any neuronal promoter (Pr). The loxP sites are arranged such that Cre activity will lead to a single inversion event of the flanked DNA. In the absence of tamoxifin (TAM) the Cre recombinase is inactive so that no recombination takes place even in active neurons. On administration of TAM, any active (cfos+) neurons will express the Cre, which is now active, and inverts the orientation of the gene of interest. This gene is then constitutively and permanently expressed from the neurons specific promoter.
More recently, several groups have developed mouse lines in which IEG-promoters are used to drive expression of a tamoxifen-regulated form of Cre recombinase (CreERT2) (Guenthner et al. 2013; Denny et al. 2014). As shown in Figure 1B, these mice can be used to activate or delete genes from subsets of neurons that are active at a particular point in time. In the absence of tamoxifen, the cfos-linked CreERT2 will be expressed in active neurons but fail to induce loxP-mediated recombination because the protein is restricted to the cytoplasm by the estrogen receptor (ERT2) component of the molecule. Administration of tamoxifen opens a time window during which CreERT2 expressed in any cfos-positive neurons will be translocated to the nucleus to induce loxP-based recombination to activate or delete target genes. The use of Cre-based systems offers the advantage of permanent genetic tagging of active neural ensembles, the ability to delete endogenous genes, and the availability of a greater number of Cre-responsive genetic tools than are available for the tTA system.
IEG-Based Manipulation of Circuit Function
As discussed in the introductory paragraphs, there is an extensive body of literature characterizing the patterns of neuronal firing in response to sensory stimuli. For example, neurons in the medial temporal lobe of human patients have been identified that seem to respond only to a specific individual (Quiroga et al. 2005). The IEG-based genetic tools provide a mechanism for directly probing the function of these sparse and dispersed ensembles in the representation of sensory information and memories. By driving the expression of genetic regulators of neural activity into the neural ensembles activated naturally by a specific environmental stimulus, the activity of these distributed ensembles can be experimentally controlled to test their behavioral relevance. In this section, we will discuss several recent studies using this approach to the study of the representation of context in fear conditioning.
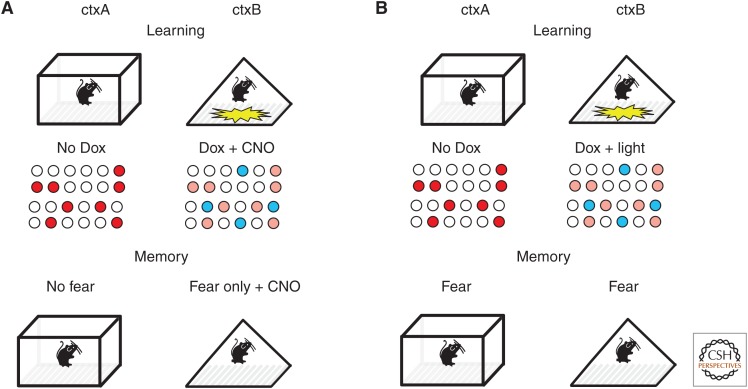
Contextual fear conditioning is a hippocampal-dependent task in which animals learn to fear a place (the context) where they receive aversive foot shocks. How is the context (characterized by specific visual, tactile, and olfactory cues) represented by dispersed neural activity? Would artificially stimulating neurons naturally activated by the context be sufficient to substitute for the actual sensory experience of the context during fear conditioning? One attempt to address this question used the cfos-tTA system to drive expression of the excitatory hM3Dq receptor into neural ensembles that were active during contextual learning (Fig. 2A) (Garner et al. 2012). hM3Dq is a human Gq coupled muscarinic receptor that has been mutated so that it no longer responds to acetylcholine but responds instead to the synthetic ligand clozapine-N-oxide (CNO) (Alexander et al. 2009). In neurons expressing hM3Dq, application of CNO causes an 8–10-mV depolarization of the membrane potential and a subsequent increase in action potential firing. In this study, mice carrying both the cfos-tTA and TRE-hM3Dq transgenes were allowed to explore a distinct context (ctxA) causing the cfos ensembles activated by this environmental stimulation to express the hM3Dq transgene. The animals were then fear conditioned in a separate context (ctxB), and CNO was delivered to depolarize the hM3Dq-expressing ctxA ensemble of neurons. Under these conditions, the animals fail to develop a fear response to either ctxA or ctxB, suggesting that the artificial activity produced in the ctxA neurons was not sufficient to substitute as a neural representation of ctxA but did interfere with the ability of the animals to represent the context in which they actually received foot shocks (ctxB). However, when the animals were placed in ctxB and received CNO to concurrently activate the ctxA neural ensemble, they showed a fear response that approached that of control animals. This suggests that the experimental animals formed an artificial contextual representation that incorporated both ctxA (artificial-internally generated) and ctxB (natural-sensory driven) neural ensembles.
Figure 2.
Manipulating contextual memory representations. This figure shows the arrangement of two sets of experiments to test the role of distributed neural ensembles in the coding of contextual memory. Both experiments use the cfos-tTA system discussed in Figure 1A to label neurons that are activated by exploration of a context (ctxA). In A, the active neurons express hM3Dq and in B, the neurons express ChR2. The animals are then fear conditioned in a distinct context (ctxB), whereas the ctxA neurons are activated with either clozapine N-oxide (CNO) or light, panels A and B, respectively. Memory retrieval is then tested in ctxA and ctxB. In panel A, the mice only show a fear response when the ctxA neurons are artificially activated while the animal is in ctxB, suggesting formation of a hybrid representation. In panel B, the animals show fear in ctxA even though they never received a shock in that context, suggesting that the artificial stimulation of the ChR2 positive neurons tagged in ctxA was able to substitute for (represent) that context. (Red circles) Neural ensembles expressing the genetic effector hM3Dq in A and ChR2 in B. (Blue circles) Neural ensembles naturally active by sensory input during training in ctxB.
Although these results failed to show that artificial activation of a distributed pattern of sensory evoked neurons could function to represent that sensory experience, it provided a number of important insights. First, the neurons that were activated artificially in this study were widely distributed throughout the neocortex and hippocampus. Previous studies had shown that local stimulation of a particular brain region or fiber bundle could serve as a CS in a conditioning paradigm (e.g., Huber et al. 2008), but the current results suggest that widely distributed, even global, patterns of activity can be meaningfully incorporated into neural representations. In fact, the brain is not silent in the absence of experimenter-provided stimuli yet this ongoing and dispersed “spontaneous” activity is rarely taken into account. Studies in the hippocampus, but also in the neocortex, indicate that spontaneous activity may not be random but instead reflect the replay of patterned activity associated with recent experiences (Kenet et al. 2003; Foster and Wilson 2006; Ji and Wilson 2007). The study by Garner et al. 2012 suggests that the integration of internally activated ensembles of neurons (ctxA in these experiments) at the time of new sensory-evoked learning (ctxB in this case) may represent a mechanism for integrating new information with previous experience, an important component of all higher forms of learning.
A more direct test of the functional relevance of distributed neural ensembles used the cfos-based genetic tagging approach in conjunction with channelrhodopsin (ChR2) (Boyden et al. 2005) and local stimulation in the hippocampus (Liu et al. 2012). In this study, the cfos-tTA transgenic mouse was used to express a virally delivered TRE-ChR2 transgene in the DG region of the hippocampus (Fig. 2B). Animals received contextual fear conditioning to allow the expression of ChR2 in the learning-activated population of neurons. The animals were then moved to a neutral context that did not induce fear. Light-induced firing of the ChR2-labeled DG neurons in this neutral context was able to produce a fear response and this response was tightly linked to the light activation of the DG. Would stimulating any set of DG neurons produce a fear response, or do the neurons carry specific information about the context in which they were labeled? To address this question, active DG neurons were labeled with ChR2, while the animal explored one context (ctxA) but did not receive foot shocks. The animals were then fear conditioned in a distinct context (ctxB) while receiving Dox to prevent further labeling of active neurons. Stimulation of the ChR2-containing ctxA neurons did not produce a response in animals trained to fear only ctxB, indicating that the neurons carry context specific information. Taken together, the results suggest that the activation of a sparse (<5% of total neurons in the DG), distributed, population of neurons, originally activated during learning, was sufficient to produce an apparent context-specific memory retrieval event.
In a follow-up study using the same techniques (Ramirez et al. 2013), the neurons activated by exploration of a specific context (ctxA), in the absence of shock, were labeled with ChR2. The artificial stimulation of these ChR2-expressing neurons with light was then paired with foot shock in a different context (ctxB). When animals were placed back into ctxA, where they had never received the foot-shock US, they showed a fear response. This result suggests that the artificial stimulation of the distributed cfos-positive population of neurons labeled during the natural exploration of ctxA was sufficient to produce a neural representation of that complex environment. In essence, the artificial stimulation and the natural sensory experience were interchangeable, at least for the purposes of fear conditioning. The arrangement of this experiment is similar to that discussed previously in Garner et al. (2012) with the training taking place in ctxB, while a competing neural ensemble representing ctxA was stimulated. The results were similar; the animal's fear memory in the natural conditioning chamber (ctxB) was strongly reduced by the stimulation of ctxA neurons during training but recovered to normal levels during concurrent activation of the ctxA cells. Whether this reflects the integration of the A and B ensembles into a single complex representation, as suggested by Garner et al. (2012), or simply reflects the effects of the compound CS-A and CS-B each acquiring a lesser amount of associative strength is unclear.
Taken together these results indicate that the artificial activation of a sparse and distributed sensory-evoked population of neurons is sufficient to instantiate a neural representation of that sensory experience. The observation that artificial stimulation with ChR2, which will tend to activate all neurons simultaneously and was not synchronized to endogenous rhythms, is sufficient to produce this effect suggests that the temporal patterning seen in natural activity may not be critical, at least for this particular brain region and behavior. There are many remaining questions relating to this effect, such as in what other brain regions can it be produced, how many neurons need to be stimulated, what frequency parameters are required to produce these effects, what downstream circuits are required to produce the behavior, and how are they recruited? Perhaps the most interesting but difficult question to address is what is the animal experiencing when the neurons for ctxA are activated. Is it experiencing a memory of that context, a hallucinatory sensory experience, or a poorly defined sense of fear or anxiety independent of coherent contextual features?
Some of these questions are examined in another recent paper studying contextual representations in the retrosplenial cortex (Cowansage et al. 2014). The retrosplenial cortex is one of the output areas of the hippocampus and projects to a wide variety of other cortical areas. It is required for the encoding and retrieval of both recent and remote contextual fear memories (Keene and Bucci 2008a,b). Similar to what was seen in the DG studies, Cowansage et al. (2014) showed that stimulation of the ChR2 labeled neural ensembles naturally activated in the retrosplenial cortex at the time of contextual fear learning was sufficient to produce a fear response. The hippocampus has been the focus of intense study regarding its role in spatial and contextual learning; however, the cortical contribution has been less well characterized. This study shows that at the time of learning, a neural ensemble representing the context forms in the neocortex as well as the hippocampus. To investigate the circuit relationship and relative contribution of these two neural representations of context, the investigators inactivated the hippocampus pharmacologically and found that, although this produced the well-characterized amnesia for natural context recall (Kim and Fanselow 1992), this effect could be overcome by direct stimulation of the retrosplenial neural ensembles. This shows that the retrosplenial contextual representation is downstream from and functions independent of the hippocampal representation.
Early studies by Penfield (1968) showed that, in a small number of cases, direct brain stimulation resulted in apparent perceptions or memories. Although it is not clear whether these results represent normal or pathological responses or actual past experience, they do suggest that direct stimulation of the brain can, in some cases, produce a coherent psychological representation. Because mice are unable to report subjective experience, it is difficult to know the psychological effect of artificial stimulation. To provide some insight into this question, Cowansage et al. (2014) compared the activation of downstream circuit elements with both natural recall (exposure to the context in which animals were shocked) and artificial ChR2 stimulation-induced recall. Using the catFISH approach they found that stimulation of retrosplenial ctxA ensembles and natural memory retrieval in ctxA-activated overlapping subsets of neurons in the entorhinal cortex, as well as basolateral and central amygdala, suggesting that the two methods of producing memory retrieval are processed in a similar manner. This is perhaps the best insight that can be achieved in a nonverbal species; whatever is experienced with artificial stimulation in a mouse, we can say that it produces a state of brain activity that is similar to posttraining context exposure, which we define as natural memory retrieval.
The artificial stimulation of small ensembles of neurons in the DG region of the hippocampus allows the retrieval of contextual memories, presumably by activating more distributed ensembles of neurons in downstream hippocampal and cortical regions. Given the highly parallel nature of neural connectivity and processing, it is possible that although these ensembles may be sufficient for retrieval, they may not be necessary because other pathways could compensate. The question of necessity was addressed in two recent studies using a light-gated proton pump from archaebacteria (ArchT) (Chow et al. 2010) to hyperpolarize and silence cfos-tagged neural ensembles in experiments analogous to those described above with ChR2 (Denny et al. 2014; Tanaka et al. 2014). Both papers also used contextual fear conditioning and examined the requirement for ensembles in three different hippocampal regions, the DG, CA3, and CA1. Expression of ArchT was driven into neurons that were active during learning in context fear conditioning and these ensembles were subsequently silenced in either the DG, CA3, or CA1 during memory retrieval. In each case, the silencing impaired the retrieval of the contextual fear memory. These results show that in each of the three major hippocampal subregions, the reactivation of the ensemble of neurons active during initial learning is necessary for the subsequent retrieval of the memory. This is consistent with the view that retrieval involves the recruitment of patterns of brain activity produced during initial learning.
In associative conditioning tasks, the unconditioned stimulus (US) generally has an emotional valence; for example, food is generally positive or rewarding, whereas shock, the US in fear conditioning, is aversive. Two recent studies examined the circuits that encode emotional valence in the hippocampus and amygdala using the cfos-tagging approach. The first study extended on the finding of a neural representation for context fear in the DG (Redondo et al. 2014). The investigators genetically tagged DG neurons active during fear conditioning with channelrhodopsin (ChR2), and when stimulated with light these neurons produced a fear response. They next asked whether the emotional valence of this fear memory could be reversed by pairing light stimulation of the DG ensembles with a positive valence US. Following the pairing, animals that previously avoided the context in which they received a shock now sought out the context, suggesting that there had been a switch in emotional valence associated with the context. This result is consistent with the view that the DG neural ensemble encodes a contextual representation (i.e., substitutes for the conditioning box), so that when the artificial activation of these ensembles is paired with a positive or negative stimulus, the box itself acquires this emotional valence as occurs in natural associative conditioning. Interestingly, when the same experiment was conducted using stimulation of cfos-labeled amygdala neural ensembles, the switch in valence was not observed. This suggests that the amygdala ensembles activated during learning are hard wired for a particular emotional valence.
The amygdala circuitry for emotional valence was explored in another recent paper (Gore et al. 2015). The study genetically introduced ChR2 into neurons activated by either a positive valence US (nicotine), or a negative valence US (shock). The investigators found two sparse (approximately 3%) and distinct populations of basolateral amygdala neurons were labeled by the stimuli, with little overlap in the cells activated by positive and negative valence stimuli. They went on to show that the artificial stimulation of these neurons could serve as a US (positive or negative valence depending on the cell population stimulated) in an array of associative and instrumental conditioning tasks. To examine the effects of conditioning on the US neurons, animals were conditioned by pairing a CS with artificial stimulation of the US neurons to produce an associative memory. After this training, the CS alone was now able to activate the US neurons of the paired emotional valence, similar to what was seen in previous studies with fear conditioning (Reijmers et al. 2007). Taken together, this study presents a view in which the amygdala contains distinct populations of neurons that confer positive or negative valence to a stimulus, presumably because of their output targets. During associative conditioning, the CS develops the ability to activate the subpopulation of neurons that were activated by the US to which it was paired and those neurons output the appropriate emotional valence to guide behavior.
Biochemical and Cellular Studies in Active Circuits
The formation of memories presumably involves cellular and molecular changes at specific points within the processing stream of incoming sensory information such that it is altered to produce new behavioral outputs following learning. For example, in our fear-conditioning model, when exposed to a new environment (context), mice show exploratory behavior. However, if the animal received an aversive foot shock while in this context, their subsequent behavior to the same sensory cues of the context will be immobility or freezing. The physical instantiation of this context memory, the engram, is presumably reflected in cellular and molecular changes at specific points in processing of the contextual cues, which diverts or enhances activity to the fear-output circuits in the amygdala. Where these changes occur, whether they are distributed or localized, their cellular and molecular nature and how they alter the processing of information from sensory input to motor output are unknown but required for a deep understanding of the engram in this behavioral model. The IEG-based genetic technology enables the targeting of cellular and biochemical studies specifically to active neurons as opposed to their inactive neighbors and has been used in several recent studies to look for specific changes that may be relevant to the underlying cellular mechanisms of memory.
Forms of activity-dependent synaptic plasticity, such as LTP and long-term depression (LTD), represent attractive candidates for a cellular mechanism of learning. The idea remains controversial and a detailed discussion of the issue can be found in Angelakos and Abel (2015) and Basu and Siegelbaum (2015) and in several recent reviews (Morris 2013; Kandel et al. 2014; Mayford 2014). Perhaps the best support for LTP and LTD as a synaptic mechanism of memory comes from a recent study in which pairing optogenetic stimulation of inputs to the amygdala with foot shock produced a fear memory in which the optogenetic stimulation served as the CS. When LTD and LTP were subsequently produced by the appropriate stimulation of the ChR2 positive fibers carrying the CS, the behavioral fear response showed a corresponding decrease and increase, respectively, providing a direct link between these forms of plasticity and behavior, albeit in a nonnatural memory. The molecular signaling involved in the production of LTP is complex but may converge on at least one common mechanism, the insertion of new 2-amino-3-5-methyl-3-oxo-1,2-oxazol-4-yl propanoic acid (AMPA)-type glutamate receptors into dendritic spines that have undergone plasticity leading to an increased postsynaptic response (Shi et al. 1999; Rumpel et al. 2005; Kessels and Malinow 2009).
To examine glutamate receptor trafficking specifically in circuits activated during learning, Matsuo et al. (2008) used the cfos-tTA-based genetic tagging approach to drive expression of a GFP-linked GluR1 receptor subunit. Animals received either standard contextual fear conditioning or an unpaired protocol in which they were exposed to the conditioning chamber and later received immediate foot shocks in a separate context. Novel context exposure induced GFP-GluR1 expression in approximately 25% of hippocampal CA1 neurons and this was not altered by the shock US, consistent with a role for the hippocampus in encoding contextual information. Twenty-four hours after conditioning, the investigators examined the distribution of GFP-tagged glutamate receptors on dendritic spines and found that the paired training caused an increase in receptor trafficking specifically to mushroom-type spines. Because the expression of the GFP-GluR1 transgene was induced by the training itself and the protein did not reach the dendrites for several hours, the results were consistent with the idea that the training produced a molecular change or tag at some synapses allowing them to capture the newly synthesized receptors when they arrived. The notion of synaptic tagging was developed in studies of LTP to explain how newly expressed genes, required for long-term maintenance of the plasticity and of memories, could influence only those synapses that underwent the initial short-term plasticity (Frey and Morris 1997, 1998; see also Martin et al. 1997 in Aplysia and Si and Kandel 2015). Although these results are purely correlative, they are consistent with a subtle associative change in hippocampal synaptic function following fear conditioning. They also serve to indicate some of the difficulties likely to be encountered in trying to identify and functionally test the role of molecular changes with learning in mammalian systems that are likely to occur in a limited group of neurons and on a limited number or type of synapse on those neurons.
One of the weakest links in the LTP-memory connection is the paucity of studies that directly observe synaptic potentiation with learning. This may be attributable to the presumed paucity of neurons and synapses that contribute to any single learning event. To overcome this problem, Whitlock et al. (2006) used an array of recording electrodes implanted in the apical dendrites of the hippocampal CA1 region to measure synaptic responses before and after learning in an inhibitory avoidance task. They found a modest increase in synaptic response on 14 of 44 electrodes with the conditioning, indicating a sparse but localized potentiation. Several studies have reported LTP-like synaptic potentiation in the lateral amygdala with auditory fear conditioning (McKernan and Shinnick-Gallagher 1997; Rogan et al. 1997; Rumpel et al. 2005). The ability to detect plasticity in this region may reflect the recruitment of a large number of neurons in this particular behavioral task. In one study, it was estimated that one-third of the lateral amygdala neurons showed plasticity following conditioning (Rumpel et al. 2005). The identification of plastic changes in more sparsely recruited networks may be more difficult.
This issue was recently addressed in a study using the cfos-tagging approach to look for physiological changes in DG neurons activated with contextual fear conditioning, the same neurons that can produce an apparent contextual representation when artificially stimulated (Ryan et al. 2015). Whole-cell recording of the cfos-positive DG neurons from fear-conditioned animals showed an increase in excitatory postsynaptic current (EPSC) amplitude and in the ratio of AMPA/NMDA current relative to their negative neighbors. The cfos-positive neurons also showed an increase in dendritic spine density. Both of these changes were blocked by immediate posttraining administration of the protein synthesis inhibitor anisomycin, which also blocked the contextual fear memory. Taken together, these results suggest that the active ensemble of neurons undergo a learning-related increase in synaptic strength and connectivity. However, a similar change was also found in neurons from the animals that received only context exposure. This could reflect plasticity resulting from the encoding of a novel context, which might be the expected function of hippocampal ensembles. Alternately, it could reflect differences in a constitutively active subpopulation of neurons as the results are quite similar to what has been described previously in cfos-positive cortical neurons from animals not receiving any environmental stimuli (Yassin et al. 2010).
Memory Allocation
Are the neurons that are recruited to encode a specific representation or memory determined exclusively by their preexisting connectivity and synaptic responses within the circuit processing the relevant sensory cues or is there a greater flexibility in the precise set of neurons recruited in any particular circumstance? A number of recent studies have been put forward to support a memory allocation model, which posits that in a given brain area many neurons have the potential to be recruited for the encoding of a particular stimulus and that the specific neurons are chosen for this task based on their preexisting state of excitability.
The initial idea for this model came from a study attempting to rescue memory deficits in a cyclic AMP response element-binding protein (CREB)-deficient mouse line (Han et al. 2007). Viral vectors carrying a wild-type copy of CREB were injected into the lateral amygdala of CREB-deficient animals and resulted in a complete rescue of the impairments in auditory fear conditioning, even though the virus infected only about 20% of the neurons. How could such a small number of restored cells support normal memory encoding? The viral construct carried a GFP marker to allow identification of the infected cells. When the investigators examined the neurons activated during a memory retrieval trial, using staining for the IEG arc, they found that almost all of the arc was expressed in GFP-positive cells, indicating that the infected neurons were selectively recruited to the fear memory representation. This was not an effect that was specific to the CREB mutant mouse line as viral elevation CREB levels in wild-type animals produce the same effect. In subsequent studies, it was shown that the specific silencing or lesioning of the CREB-elevated neurons impaired the fear memory showing that these specific neural ensembles contributed to expression of the memory (Han et al. 2009; Zhou et al. 2009).
How could CREB, a transcription factor implicated in the consolidation of memory, alter the recruitment of neurons during learning? The first clue came from the observation that CREB-transfected neurons showed a significant increase in excitability (Zhou et al. 2009). This suggests that it was not necessarily CREB itself that was responsible but simply that neurons with elevated excitability at the time of learning are those more likely to be recruited to the representation of that event. This idea was tested by increasing neuronal excitability directly using expression of a dominant negative mutant of the potassium channel KCNQ2 or the hM3Dq receptor in viral transfection experiments analogous to those described above for CREB (Yiu et al. 2014). In both cases, increased excitability was sufficient to recruit the transfected neurons to the fear memory representation. Using the regulated hM3Dq system, it was further shown that the increase in excitability was effective only immediately before training but not after learning had occurred. Although these studies were restricted to the lateral amygdala and fear conditioning, similar effects have recently been described in the insular cortex with conditioned taste aversion, suggesting that memory allocation may be a more general phenomena (Sano et al. 2014). The memory allocation model is attractive as a mechanism for linking memories for events that are closely spaced in time (Silva et al. 2009). According to this view, neural activity and the formation of long-term memory evokes CREB-dependent transcription, which in turn opens a time window of increased excitability in those active neurons. When new information is acquired during this time window, it will be more likely to recruit neurons that participated in the previous learning event creating a link at the circuit level between events that are closely spaced in time.
CONCLUSIONS
One of the critical difficulties in studying information processing and its modification with learning in the mammalian brain is identifying and manipulating the neurons involved in representing any specific environmental stimulus or memory. A particular complex item, a place, or an individual, is likely represented in higher brain areas in the activity pattern of a relatively small and anatomically dispersed group of neurons. In this review, we have discussed the use of genetic tools in the mouse that allow the molecular identification and genetic modification of neurons that are electrically active (because of natural environmental stimuli) at a specific point in time. This allows experiments to focus on just those neurons that were active in response to a given stimulus. This has been used to identify cellular and molecular changes that are specific to active neurons relative to their inactive neighbors. Active neural ensembles in the DG and somatosensory cortex have been shown to have increased excitability, interconnectivity, and dendritic spine density. In the hippocampus, learning induced an increase in glutamate receptor trafficking specifically in neurons activated during context learning. Although the functional significance of these cellular changes is still unclear, the approach is revealing that active neural ensembles show underlying molecular differences compared to apparently identical neurons in the same brain region that are not activated by behaviorally relevant stimuli.
In a second line of research, effectors of neural activity were introduced into naturally active ensembles to explore the nature of neural representations. Previous experiments were limited to recording the sensory- or learning-evoked activity of neurons and inferring a role in producing the psychological or perceptual manifestation of that sensory experience or memory. By introducing ChR2 into cfos-active neural ensembles during learning of a specific location in context fear conditioning, the activity of naturally activated neurons could actually be replayed artificially to directly test the relevance of that specific pattern of neural activity in encoding that specific context. The results suggest that even with the relatively crude IEG-based labeling of active ensembles, their reactivation via CHR2 produced a coherent representation/perception of the context. This result has been found in both the hippocampus and retrosplenial cortex with complementary studies in the hippocampus showing that suppression of these ensembles is sufficient to impair context recall. This is the first direct link between dispersed patterns of sensory-evoked neural activity and a coherent perceptual representation. In relation to learning and memory, these results should be viewed as an important first step in identifying the relevant principles of information processing rather than identification of the elusive “engram.” The studies show that the stimulation of the correct pattern of neurons in one brain area is able to substitute for, or represent, a complex environmental stimulus (the context box), not that these neurons undergo the physical changes necessary to link that context to the fear-evoking shock. For example, if one could stimulate retinal neurons in precisely the same pattern as when an animal explored a specific context, then the animal would presumably perceive that they were in the box, and freeze if fear conditioned, but we would not suggest that the engram for that memory lies in the retina.
The mammalian brain is an immensely complex information-processing system that transforms an ongoing stream of sensory information into evolutionarily advantageous behavioral outputs. Learning is the process by which specific environmental contingencies (e.g., paring a specific location with an aversive stimulus) alter this processing to produce altered behavioral outputs. The IEG-based genetic tagging approach provides the ability to focus studies specifically on those neurons that are activated by any given set of environmental contingencies. It should prove useful in future studies to identify and genetically test specific models for molecular changes that may underlie learning, such as Hebbian forms of plasticity. A deep understanding of learning and memory will require not just identifying these molecular mechanisms and their location(s) in the brain, but also identifying how they alter the processing of information to evoke the learned responses. The new approaches for recording and manipulating active neural ensembles described in this and previous work offers an experimental framework in which this type of question can begin to be addressed.
Footnotes
Editors: Eric R. Kandel, Yadin Dudai, and Mark R. Mayford
Additional Perspectives on Learning and Memory available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alexander GM, Rogan SC, Abbas AI, Armbruster BN, Pei Y, Allen JA, Nonneman RJ, Hartmann J, Moy SS, Nicolelis MA, et al. 2009. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron 63: 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Angelakos CC, Abel T. 2015. Molecular genetic strategies in the study of corticohippocampal circuits. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL, Gerkin RC, Dean KL. 2004. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J Neurosci 24: 6466–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Basu J, Siegelbaum SA. 2015. The corticohippocampal circuit, synaptic plasticity, and memory. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartel K, Genoux D, Welzl H, Tweedie-Cullen RY, Koshibu K, Livingstone-Zatchej M, Mamie C, Mansuy IM. 2008. Control of the establishment of aversive memory by calcineurin and Zif268. Nat Neurosci 11: 572–578. [DOI] [PubMed] [Google Scholar]
- Bontempi B, Laurent-Demir C, Destrade C, Jaffard R. 1999. Time-dependent reorganization of brain circuitry underlying long-term memory storage. Nature 400: 671–675. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. 2005. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8: 1263–1268. [DOI] [PubMed] [Google Scholar]
- *.Byrne JH, Hawkins RD. 2015. Nonassociative learning in invertebrates. Cold Spring Harb Perspect Biol 7: a021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, et al. 2010. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowansage KK, Shuman T, Dillingham BC, Chang A, Golshani P, Mayford M. 2014. Direct reactivation of a coherent neocortical memory of context. Neuron 84: 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowski R, Jayaprakash B, Wiltgen B, Rogerson T, Guzman-Karlsson MC, Barth AL, Trachtenberg JT, Silva AJ. 2014. Encoding and storage of spatial information in the retrosplenial cortex. Proc Natl Acad Sci 111: 8661–8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, Tomm NK, Turi GF, Losonczy A, Hen R. 2014. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron 83: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, Reschke M, Spanagel R, Sprengel R, et al. 2003. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci 23: 9116–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster DJ, Wilson MA. 2006. Reverse replay of behavioural sequences in hippocampal place cells during the awake state. Nature 440: 680–683. [DOI] [PubMed] [Google Scholar]
- Frankland PW, Bontempi B, Talton LE, Kaczmarek L, Silva AJ. 2004. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science 304: 881–883. [DOI] [PubMed] [Google Scholar]
- French PJ, O'Connor V, Jones MW, Davis S, Errington ML, Voss K, Truchet B, Wotjak C, Stean T, Doyere V, et al. 2001. Subfield-specific immediate early gene expression associated with hippocampal long-term potentiation in vivo. Eur J Neurosci 13: 968–976. [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RG. 1997. Synaptic tagging and long-term potentiation. Nature 385: 533–536. [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RG. 1998. Synaptic tagging: Implications for late maintenance of hippocampal long-term potentiation. Trends Neurosci 21: 181–188. [DOI] [PubMed] [Google Scholar]
- Garner AR, Rowland DC, Hwang SY, Baumgaertel K, Roth BL, Kentros C, Mayford M. 2012. Generation of a synthetic memory trace. Science 335: 1513–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Ginty DD, Bading H, Greenberg ME. 1994. Calcium regulation of gene expression in neuronal cells. J Neurobiol 25: 294–303. [DOI] [PubMed] [Google Scholar]
- Gore F, Schwartz EC, Brangers BC, Aladi S, Stujenske JM, Likhtik E, Russo MJ, Gordon JA, Salzman CD, Axel R. 2015. Neural representations of unconditioned stimuli in basolateral amygdala mediate innate and learned responses. Cell 162: 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci 89: 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Hermanowski AL, Ziff EB. 1986. Effect of protein synthesis inhibitors on growth factor activation of c-fos, c-myc, and actin gene transcription. Mol Cell Biol 6: 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. 2013. Permanent genetic access to transiently active neurons via TRAP: Targeted recombination in active populations. Neuron 78: 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. 1999. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci 2: 1120–1124. [DOI] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, Josselyn SA. 2007. Neuronal competition and selection during memory formation. Science 316: 457–460. [DOI] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, Bontempi B, Neve RL, Frankland PW, Josselyn SA. 2009. Selective erasure of a fear memory. Science 323: 1492–1496. [DOI] [PubMed] [Google Scholar]
- *.Hawkins RD, Byrne JH. 2015. Associative learning in invertebrates. Cold Spring Harb Perspect Biol 7: a021709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubel DH, Wiesel TN. 1962. Receptive fields, binocular interaction and functional architecture in the cat's visual cortex. J Physiol 160: 106–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D, Petreanu L, Ghitani N, Ranade S, Hromadka T, Mainen Z, Svoboda K. 2008. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature 451: 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji D, Wilson MA. 2007. Coordinated memory replay in the visual cortex and hippocampus during sleep. Nat Neurosci 10: 100–107. [DOI] [PubMed] [Google Scholar]
- *.Jercog P, Rogerson T, Schnitzer MJ. 2015. Large-scale fluorescence calcium-imaging methods for studies of long-term memory in behaving mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Dudai Y, Mayford MR. 2014. The molecular and systems biology of memory. Cell 157: 163–186. [DOI] [PubMed] [Google Scholar]
- Karlsson MP, Frank LM. 2009. Awake replay of remote experiences in the hippocampus. Nat Neurosci 12: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene CS, Bucci DJ. 2008a. Contributions of the retrosplenial and posterior parietal cortices to cue-specific and contextual fear conditioning. Behav Neurosci 122: 89–97. [DOI] [PubMed] [Google Scholar]
- Keene CS, Bucci DJ. 2008b. Neurotoxic lesions of retrosplenial cortex disrupt signaled and unsignaled contextual fear conditioning. Behav Neurosci 122: 1070–1077. [DOI] [PubMed] [Google Scholar]
- Kenet T, Bibitchkov D, Tsodyks M, Grinvald A, Arieli A. 2003. Spontaneously emerging cortical representations of visual attributes. Nature 425: 954–956. [DOI] [PubMed] [Google Scholar]
- Kessels HW, Malinow R. 2009. Synaptic AMPA receptor plasticity and behavior. Neuron 61: 340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Fanselow MS. 1992. Modality-specific retrograde amnesia of fear. Science 256: 675–677. [DOI] [PubMed] [Google Scholar]
- Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. 2015. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep 10: 292–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapska E, Maren S. 2009. Reciprocal patterns of c-Fos expression in the medial prefrontal cortex and amygdala after extinction and renewal of conditioned fear. Learn Mem 16: 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. 2012. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. 1995. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 14: 433–445. [DOI] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. 1997. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: A function for local protein synthesis in memory storage. Cell 91: 927–938. [DOI] [PubMed] [Google Scholar]
- Matsuo N, Reijmers L, Mayford M. 2008. Spine-type-specific recruitment of newly synthesized AMPA receptors with learning. Science 319: 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M. 2014. The search for a hippocampal engram. Philos Trans R Soc Lond B Biol Sci 369: 20130161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan MG, Shinnick-Gallagher P. 1997. Fear conditioning induces a lasting potentiation of synaptic currents in vitro. Nature 390: 607–611. [DOI] [PubMed] [Google Scholar]
- Mehta MR, Lee AK, Wilson MA. 2002. Role of experience and oscillations in transforming a rate code into a temporal code. Nature 417: 741–746. [DOI] [PubMed] [Google Scholar]
- Milanovic S, Radulovic J, Laban O, Stiedl O, Henn F, Spiess J. 1998. Production of the Fos protein after contextual fear conditioning of C57BL/6N mice. Brain Res 784: 37–47. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T. 1987. Mapping patterns of c-fos expression in the central nervous system after seizure. Science 237: 192–197. [DOI] [PubMed] [Google Scholar]
- Morris RG. 2013. NMDA receptors and memory encoding. Neuropharmacology 74: 32–40. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. 1993. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature 363: 718–722. [DOI] [PubMed] [Google Scholar]
- Penfield W. 1968. Engrams in the human brain. Mechanisms of memory. Proc R Soc Med 61: 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, et al. 2006. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 52: 437–444. [DOI] [PubMed] [Google Scholar]
- Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. 1993. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature 361: 453–457. [DOI] [PubMed] [Google Scholar]
- Quiroga RQ, Reddy L, Kreiman G, Koch C, Fried I. 2005. Invariant visual representation by single neurons in the human brain. Nature 435: 1102–1107. [DOI] [PubMed] [Google Scholar]
- Radulovic J, Kammermeier J, Spiess J. 1998. Relationship between fos production and classical fear conditioning: Effects of novelty, latent inhibition, and unconditioned stimulus preexposure. J Neurosci 18: 7452–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. 2012. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods 9: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmsdorf HJ, Schonthal A, Angel P, Litfin M, Ruther U, Herrlich P. 1987. Posttranscriptional regulation of c-fos mRNA expression. Nucleic Acids Res 15: 1643–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez S, Liu X, Lin PA, Suh J, Pignatelli M, Redondo RL, Ryan TJ, Tonegawa S. 2013. Creating a false memory in the hippocampus. Science 341: 387–391. [DOI] [PubMed] [Google Scholar]
- Redondo RL, Kim J, Arons AL, Ramirez S, Liu X, Tonegawa S. 2014. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature 513: 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, Mayford M. 2007. Localization of a stable neural correlate of associative memory. Science 317: 1230–1233. [DOI] [PubMed] [Google Scholar]
- Robertson LM, Kerppola TK, Vendrell M, Luk D, Smeyne RJ, Bocchiaro C, Morgan JI, Curran T. 1995. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron 14: 241–252. [DOI] [PubMed] [Google Scholar]
- Rogan MT, Staubli UV, LeDoux JE. 1997. Fear conditioning induces associative long-term potentiation in the amygdala. Nature 390: 604–607. [DOI] [PubMed] [Google Scholar]
- Rumpel S, LeDoux J, Zador A, Malinow R. 2005. Postsynaptic receptor trafficking underlying a form of associative learning. Science 308: 83–88. [DOI] [PubMed] [Google Scholar]
- Ryan TJ, Roy DS, Pignatelli M, Arons A, Tonegawa S. 2015. Engram cells retain memory under retrograde amnesia. Science 348: 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM. 2011. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci 14: 848–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint Marie RL, Luo L, Ryan AF. 1999. Spatial representation of frequency in the rat dorsal nucleus of the lateral lemniscus as revealed by acoustically induced c-fos mRNA expression. Hearing Res 128: 70–74. [DOI] [PubMed] [Google Scholar]
- Sano Y, Shobe JL, Zhou M, Huang S, Shuman T, Cai DJ, Golshani P, Kamata M, Silva AJ. 2014. CREB regulates memory allocation in the insular cortex. Curr Biol 24: 2833–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenenberger P, Gerosa D, Oertner TG. 2009. Temporal control of immediate early gene induction by light. PLoS ONE 4: e8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SH, Hayashi Y, Petralia RS, Zaman SH, Wenthold RJ, Svoboda K, Malinow R. 1999. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science 284: 1811–1816. [DOI] [PubMed] [Google Scholar]
- *.Si K, Kandel ER. 2015. The role of functional prion-like proteins in the persistence of memory. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Zhou Y, Rogerson T, Shobe J, Balaji J. 2009. Molecular and cellular approaches to memory allocation in neural circuits. Science 326: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeyne RJ, Schilling K, Robertson L, Luk D, Oberdick J, Curran T, Morgan JI. 1992. fos-lacZ transgenic mice: Mapping sites of gene induction in the central nervous system. Neuron 8: 13–23. [DOI] [PubMed] [Google Scholar]
- Tanaka KZ, Pevzner A, Hamidi AB, Nakazawa Y, Graham J, Wiltgen BJ. 2014. Cortical representations are reinstated by the hippocampus during memory retrieval. Neuron 84: 347–354. [DOI] [PubMed] [Google Scholar]
- Vousden DA, Epp J, Okuno H, Nieman BJ, van Eede M, Dazai J, Ragan T, Bito H, Frankland PW, Lerch JP, et al. 2015. Whole-brain mapping of behaviourally induced neural activation in mice. Brain Struct Function 220: 2043–2057. [DOI] [PubMed] [Google Scholar]
- Wagener RJ, David C, Zhao S, Haas CA, Staiger JF. 2010. The somatosensory cortex of reeler mutant mice shows absent layering but intact formation and behavioral activation of columnar somatotopic maps. J Neurosci 30: 15700–15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. 2006. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell 126: 389–402. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Greenberg ME, Belasco JG. 1993. The destabilizing elements in the coding region of c-fos mRNA are recognized as RNA. Mol Cell Biol 13: 5034–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock JR, Heynen AJ, Shuler MG, Bear MF. 2006. Learning induces long-term potentiation in the hippocampus. Science 313: 1093–1097. [DOI] [PubMed] [Google Scholar]
- Wilson MA, McNaughton BL. 1994. Reactivation of hippocampal ensemble memories during sleep. Science 265: 676–679. [DOI] [PubMed] [Google Scholar]
- Worley PF, Christy BA, Nakabeppu Y, Bhat RV, Cole AJ, Baraban JM. 1991. Constitutive expression of zif268 in neocortex is regulated by synaptic activity. Proc Natl Acad Sci 88: 5106–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Liu Y, Zhu Y, Ding X, Yang Y, Guan JS. 2014. In vivo imaging of immediate early gene expression reveals layer-specific memory traces in the mammalian brain. Proc Natl Acad Sci 111: 2788–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Andreasson KI, Kaufmann WE, Barnes CA, Worley PF. 1993. Expression of a mitogen-inducible cyclooxygenase in brain neurons: Regulation by synaptic activity and glucocorticoids. Neuron 11: 371–386. [DOI] [PubMed] [Google Scholar]
- Yassin L, Benedetti BL, Jouhanneau JS, Wen JA, Poulet JF, Barth AL. 2010. An embedded subnetwork of highly active neurons in the neocortex. Neuron 68: 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu AP, Mercaldo V, Yan C, Richards B, Rashid AJ, Hsiang HL, Pressey J, Mahadevan V, Tran MM, Kushner SA, et al. 2014. Neurons are recruited to a memory trace based on relative neuronal excitability immediately before training. Neuron 83: 722–735. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Won J, Karlsson MG, Zhou M, Rogerson T, Balaji J, Neve R, Poirazi P, Silva AJ. 2009. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci 12: 1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]