Abstract
Craniorachischisis is a severe neural tube defect (NTD) resulting from failure to initiate closure, leaving the hindbrain and spinal neural tube entirely open. Clues to the genetic basis of this condition come from several mouse models, which harbour mutations in core members of the planar cell polarity (PCP) signalling pathway. Previous studies of humans with craniorachischisis failed to identify mutations in the core PCP genes VANGL1 and VANGL2. Here we analysed other key PCP genes: CELSR1, PRICKLE1, PTK7 and SCRIB, with the finding of eight potentially causative mutations in both CELSR1 and SCRIB. Functional effects of these unique or rare human variants were evaluated using known protein-protein interactions as well as subcellular protein localisation. While protein interactions were not affected, variants from 5 of the 36 patients exhibited a profound alteration in subcellular protein localisation, with diminution or abolition of trafficking to the plasma membrane. Comparable effects were seen in the crash and spin cycle mouse Celsr1 mutants, and the line-90 mouse Scrib mutant. We conclude that missense variants in CELSR1 and SCRIB may represent a cause of craniorachischisis in humans, as in mice, with defective PCP protein trafficking to the plasma membrane a likely pathogenic mechanism.
Keywords: Neural tube defects, craniorachischisis, planar cell polarity, CELSR1, SCRIB
Introduction
Neural tube closure is an event of early embryogenesis that is required for normal development of the brain and spinal cord. Primary neurulation in mammals begins with a de novo closure event at the boundary of the future hindbrain and cervical spine (so-called Closure 1) on day 22 post-fertilization in humans (O’Rahilly and Muller, 2002) and embryonic day 8.5 in mice (Copp, et al., 2003). From this site, the neural tube ‘zips up’ in a double wave of closure that spreads rostrally into the hindbrain region and caudally along the spine. Subsequently, closure in human embryos initiates separately at the rostral edge of the forebrain, generating a caudally directed wave of closure that meets the rostrally directed (hindbrain) wave to complete brain closure at the anterior neuropore (O’Rahilly and Muller, 2002). In mice, there is a slightly more complex sequence of cranial closure events, with a second initiation site at the forebrain-midbrain boundary (Closure 2) and then bidirectional closure between this site and the rostral edge of the forebrain, and also between this site and Closure 1 (Golden and Chernoff, 1993). In the spinal region of both humans and mice, closure is completed when zipping down the body axis reaches the upper sacral level, where the posterior (caudal) neuropore closes.
Defective closure during neurulation results in severe malformations of the central nervous system, termed neural tube defects (NTDs). Failure to complete low spinal closure causes spina bifida whereas incomplete cranial closure results in anencephaly. These are common birth defects, affecting 0.5-2 per 1000 pregnancies, world wide (Botto, et al., 1999). The most severe NTD, craniorachischisis (CRN), arises earlier in development as a failure of Closure 1, leaving the neural tube open from the midbrain or rostral hindbrain to the base of the spine (Copp, et al., 2003). CRN is considered rare, although estimates of prevalence vary from 1/100,000 in Atlanta (Johnson, et al., 2004) to 1/1000 in Northern China (Moore, et al., 1997).
Despite the high prevalence of NTDs, the genes responsible for their largely sporadic occurrence have proven elusive. This likely reflects a complex inheritance pattern and an important contribution of non-genetic factors. Indeed, many genes are known to be essential for neurulation in mice (Harris and Juriloff, 2010), with increasing evidence of phenotypic modulation through gene-gene and gene-environment interactions (Copp, et al., 2003). The first mouse gene recognised as a cause of CRN was Vangl2, which is mutated in the loop tail mouse (Kibar, et al., 2001b; Murdoch, et al., 2001a) and encodes a key component of a β-catenin-independent, Wnt/frizzled signalling cascade, called the planar cell polarity (PCP) pathway (Strutt, 2008). Subsequently, other PCP components were found to be essential for initiation of neural tube closure (closure 1) in mice, including Celsr1, Dsh1/2, Scrib, Fz3/6, Ptk7 and Sec2b (Greene, et al., 2009; Merte, et al., 2010). The developmental basis of this association is the requirement for convergent extension cell movements, which shape the early neural plate. Disturbance of PCP gene function in Xenopus (Wallingford and Harland, 2002) and Vangl2 mutant mice (Ybot-Gonzalez, et al., 2007) both abolish convergent extension, producing a short, wide neural plate in which the neural folds are spaced too far apart to initiate closure.
Although putative mutations in VANGL2 (MIM# 600533) and its closely related paralog VANGL1 (MIM# 610132) have been reported in patients with several other types of NTD (Kibar, et al., 2007; Lei, et al., 2010), no evidence has been presented to suggest a role in human CRN. Indeed, our preliminary investigation of CRN cases did not reveal mutations in either VANGL1 or VANGL2 (Doudney, et al., 2005). In the present study, we have increased the size of our cohort to 36 patients – to our knowledge, the only such cohort of this rare NTD anywhere in the world – and have used DNA from these patients to investigate four further genes in the PCP signalling pathway: the core PCP gene CELSR1 (MIM# 604523), and the PCP-associated genes SCRIB (SCRB1; MIM# 607733), PTK7 (MIM# 601890) and PRICKLE1 (MIM# 608500). CELSR1, the ortholog of flamingo in Drosophila, is mutated in crash and spin cycle mice in which homozygosity yields CRN (Curtin, et al., 2003). Of the three PCP-associated genes, SCRIB causes CRN in both the circle tail (Murdoch, et al., 2003) and line-90 (Zarbalis, et al., 2004) mouse mutants, and PTK7 causes CRN in mice homozygous for a gene trap mutation (Lu, et al., 2004) and in the ENU-induced chuzhoi mutant (Paudyal, et al., 2010). Neither of these genes has previously been implicated in human NTDs. In contrast, PRICKLE1, which is the ortholog of a Drosophila PCP gene, is required for regulation of apico-basal polarity in mouse epiblast (Tao, et al., 2009) and was recently found to be mutated in patients with epilepsy (Tao, et al., 2011). Interestingly, rare variants have been reported amongst patients with open or closed spina bifida, although CRN cases were not examined (Bosoi et al., 2011).
Like VANGL1 and VANGL2 (Doudney, et al., 2005), in the present study we did not detect putative mutations in patients with CRN in the genes for PRICKLE1 or PTK7. However, we do find putative heterozygous missense mutations in CELSR1 and SCRIB, prompting screening and identification of further sequence variants in a second series of cases. Comparative functional analysis with known pathogenic mouse mutations in Celsr1 and Scrib was then used to evaluate the likely effect of these human variants. We demonstrate, by in vitro assay of reporter constructs, a profound effect for several missense mutations of both genes on the subcellular localisation of these normally membrane-associated proteins, indicating a possible mechanism resulting in loss of function.
Results
The coding sequence and exon-intron boundary regions of CELSR1, PRICKLE1, PTK7 and SCRIB were sequenced in twenty fetuses with CRN, identifying a variety of synonymous and non-synonymous coding sequence variants. Data reporting a lack of likely causative mutations in VANGL1 and VANGL2 in the same set of patients has previously been published (Doudney, et al., 2005). For PRICKLE1 and PTK7, two coding variants each (rs3747563 – F633F and rs3747562 – S634S; rs6905948 G617G and rs34764696 - A777V) were identified, all present in controls and dbSNP132 (http://www.ncbi.nlm.nih.gov/sites/entrez). Although there is a possibility of reduced penetrance for some mutations, we considered these alleles less likely to be causative and not investigated further in this study.
Missense changes identified in CELSR1 and SCRIB
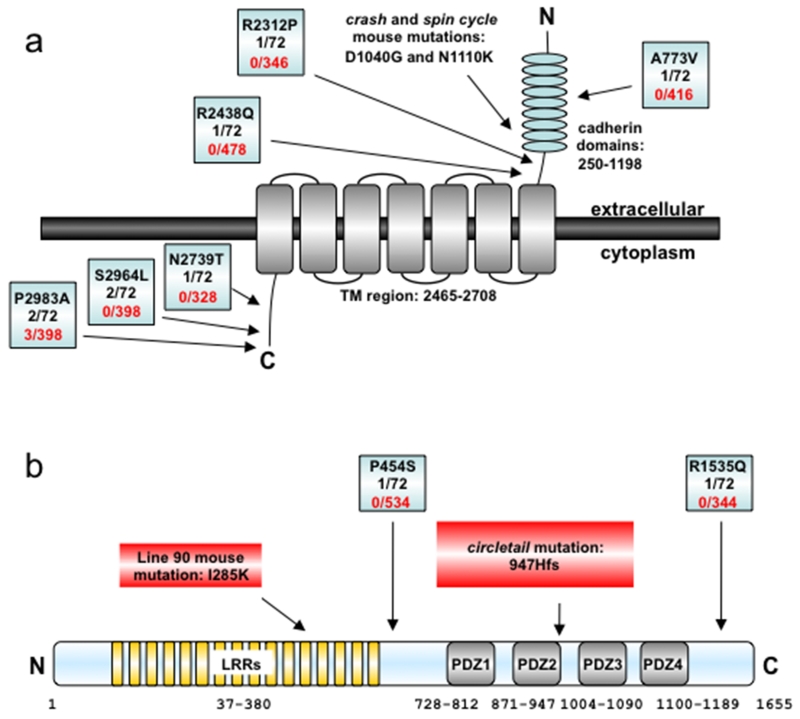
For CELSR1 and SCRIB, a variety of single nucleotide coding variants were detected. Non-synonymous variants were investigated for their presence in dbSNP and the 1000 Genomes Project (release 6) data (http://browser.1000genomes.org/index.html) and/or frequency in white European controls (Supp. Tables S1 and S2). In the initial patient set (n=20), two unique missense substitutions were identified in CELSR1: CRN3 - c.2318 C>T (A773V) and CRN19 - c.8216 A>C (N2739T) (Figure 1a and Supp. Figure S1). Three patients were each found to carry two missense changes: CRN8 (R2312P + S2964L), CRN3 (A773V + P2983A) and CRN6 (S2964L + P2983A). These include the changes c.6935 G>C (R2312P); C>T c.8891 (S2964L); and c.8947 C>G (P2983A) which are reported as rare alleles in dbSNP (rs7287089, rs6008777 and rs61741871 respectively). The c.8947 C>G (P2983A) change was also found to be heterozygous in 3 of 199 unaffected individuals, although none of the others were present in our control cohort. In addition, all of these variant residues were evolutionarily conserved in most mammalian species (except for the arginine at position R2312, which is a glutamine in mouse) but were poorly conserved with the invertebrate Drosophila (Supp. Table S1 and Figure S2). Screening the second set of patients (n=16) revealed overlap with the frequent variants but also revealed a third unique missense change located at a conserved CELSR1 residue, R2438Q (c.7313 G>A) (Supp. Table S1).
Figure 1. Schematic diagrams of CELSR1 and SCRIB proteins.
Location of missense changes and mouse mutations with frequency in patient and control chromosomes are shown for (a) CELSR1 and (b) SCRIB relative to their functional domains.
For SCRIB, two unique changes were identified in patients CRN5 and CRN20 respectively, comprising a C>T transition at c.1360 (P454S) and a G>A transition at c.4604 (R1535Q) (Figure 1b and Supp. Figure S1). The proline residue at P454 was not conserved between species (Supp. Table S2). In those cases where a parent or additional family member was available for screening, we found evidence for inheritance, indicating that if causal, the mutations have incomplete penetrance (Supp. Table S3).
Functional analysis of missense mutations: co-immunoprecipitation
Functional analysis of previously reported PCP missense variants has relied on two main strategies: perturbation of normal protein-protein interaction, as detected by co-immunoprecipitation, and mislocalisation of GFP-tagged proteins within polarised cells (Iliescu, et al., 2011; Kibar, et al., 2007; Lei, et al., 2010). In the present study, the first of these approaches was investigated for SCRIB only, since no protein-protein interactions had yet been described for CELSR1.
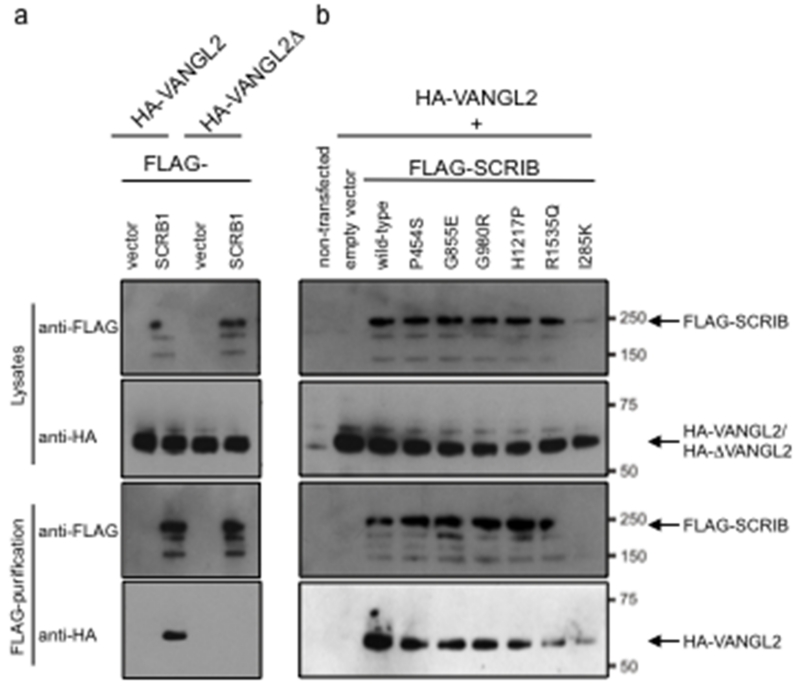
SCRIB is known to interact with both VANGL2 and LGL2 (Kallay, et al., 2006). We found that wild-type FLAG-tagged SCRIB could immunoprecipitate HA-tagged VANGL2 but not a similar construct lacking the NH3-terminal amino acids ETSV, which correspond to the PDZ binding domain of VANGL2 (Figure 2a). This strongly implicates binding through one or more of the four PDZ binding sites in SCRIB. A series of missense mutations were then introduced into the wild-type construct using in vitro mutagenesis. This included the pathogenic mutation found in the line-90 mouse (I285K), the putative human mutations P454S and R1535Q, and several variants that are also present in dbSNP (G855E, G980R and H1217P). None of the variants were found to ablate co-immunoprecipitation (Figure 2b).
Figure 2. Effect of missense variants on SCRIB-VANGL2 interactions by Co-Ip.
(a) Specific interaction of SCRIB with VANGL2 is through the PDZ binding domain. Western blotting of the lysate with anti-FLAG and anti-HA demonstrates the presence of FLAG-SCRIB and HA-VANGL2 proteins respectively. Following immunoprecipitation with anti-FLAG, full-length VANGL2 is pulled down but VANGL2Δ (lacking PDZ binding domain) is not. (b) A similar experiment was conducted with full-length HA-VANGL2 and FLAG-SCRIB constructs with various missense changes or wild type sequence. None of the variants, including the mouse pathogenic change I285K affect the ability of SCRIB to pull down VANGL2.
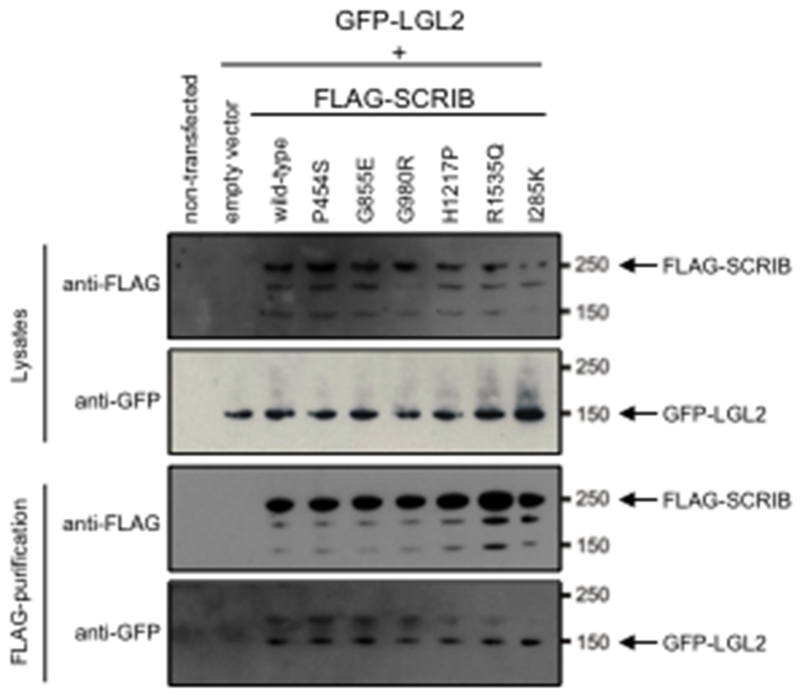
Next, we investigated the interaction between SCRIB and LGL2 (lethal giant larvae 2), previously shown to occur via the leucine rich repeat region of SCRIB (Kallay, et al., 2006). A similar co-immunoprecipitation experiment was performed as for HA-VANGL2, this time using GFP-tagged LGL2. Wild-type FLAG-SCRIB was able to pull down GFP-LGL2, as was the same set of SCRIB missense variants, including the line-90 mouse mutant (Figure 3). These assays confirm the specific binding of SCRIB to both VANGL2 and LGL2 and confirm that none of the missense changes in SCRIB affect these protein interactions. Notably, this includes the known pathogenic mouse mutation I285K (line-90), demonstrating that SCRIB pathogenicity, in terms of causing CRN, can occur in the absence of alteration to protein-protein interactions.
Figure 3. Effect of missense variants on SCRIB-LGL2 interactions by Co-Ip.
Western blotting of the lysate with anti-FLAG demonstrates the presence of wildtype FLAG-SCRIB protein and mutant proteins with different missense changes. Following FLAG immunoprecipitation, blotting with anti-GFP confirms interaction between SCRIB and LGL2 with all of the missense variants tested.
Functional analysis of missense mutations: subcellular localisation
To assess PCP protein function through a more general assay, we investigated whether subcellular localisation might be disrupted by the known pathogenic, and putative, mutations in the CELSR1 and SCRIB genes.
CELSR1 is a cadherin EGF LAG seven-pass G-type receptor transmembrane protein while the cytoplasmic scaffold protein SCRIB, although intracellular, localises tightly to the plasma membrane of polarised epithelial cells (Figure 1a). Therefore, as a means of assessing the effect of missense changes on protein trafficking, we used site-directed mutagenesis to introduce individual variants into either full-length mouse Celsr1 cDNA (NM_009886) expressed from a pCDNA3-Celsr1-GFP construct, or into full-length SCRIB cDNA (NM_015356) cloned downstream of the EGFP open reading frame in a pEGFP-C1 expression vector.
Clones were transfected into polarised MDCK epithelial cells (strain II) and subcellular localisation was studied at 24, 48 and 72 hour time points, with 48 hours chosen for quantitative analysis of CELSR1 and SCRIB variants (Supp. Figure S1). Cells were dual stained with anti-GFP and either anti-CELSR1 or anti-SCRIB. The subcellular localisation of GFP expression was classified for each cell into one of the following three different categories: (i) GFP exclusively at the membrane; (ii) GFP both at the membrane and in the cytoplasm; (iii) GFP exclusively in the cytoplasm.
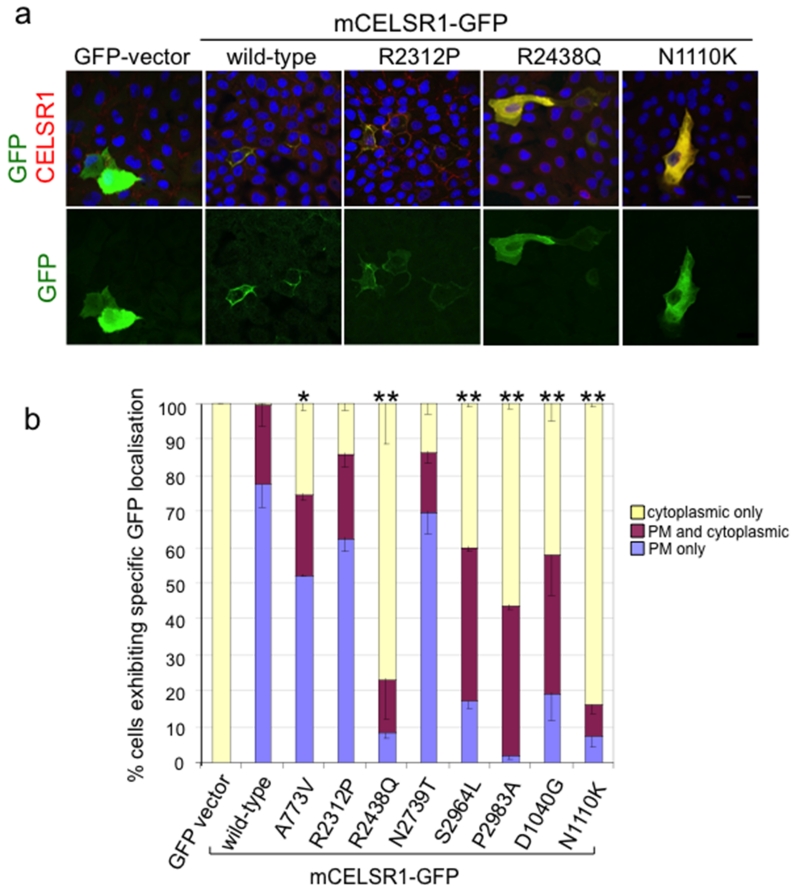
A characteristic pattern of localisation was observed at the plasma membrane for wild-type CELSR1, with very few cells showing diffuse signal in the cytosol (Figure 4a,b). CELSR1 missense variants S2964L, R2438Q and P2983A resulted in a significantly reduced frequency of membrane localisation, closely parallel to the effects of the crash (D1040G) and spin cycle (N1110K) mouse pathogenic mutants (Curtin, et al., 2003) (Figure 4a,b). The A773V variant showed a less dramatic, although statistically significant, reduction in membrane-localisation, whereas the reduction observed in the R2312P and N2739T variants did not reach statistical significance.
Figure 4. Effect of putative missense mutations in CELSR1 on subcellular localisation.
(a) Protein localisation in MDCK cells: Top - immunofluorescence with anti-GFP (green) and anti-CELSR1 (red); Bottom - anti-GFP alone. (b) Quantitative analysis of protein localisation for GFP +ve cells were scored as cytoplasmic only, plasma membrane (PM) and cytoplasmic combined, or PM only. ** Statistically significant differences of % cytoplasmic and % PM compared with wild-type (p < 0.001) were determined by two-way analysis of variance.
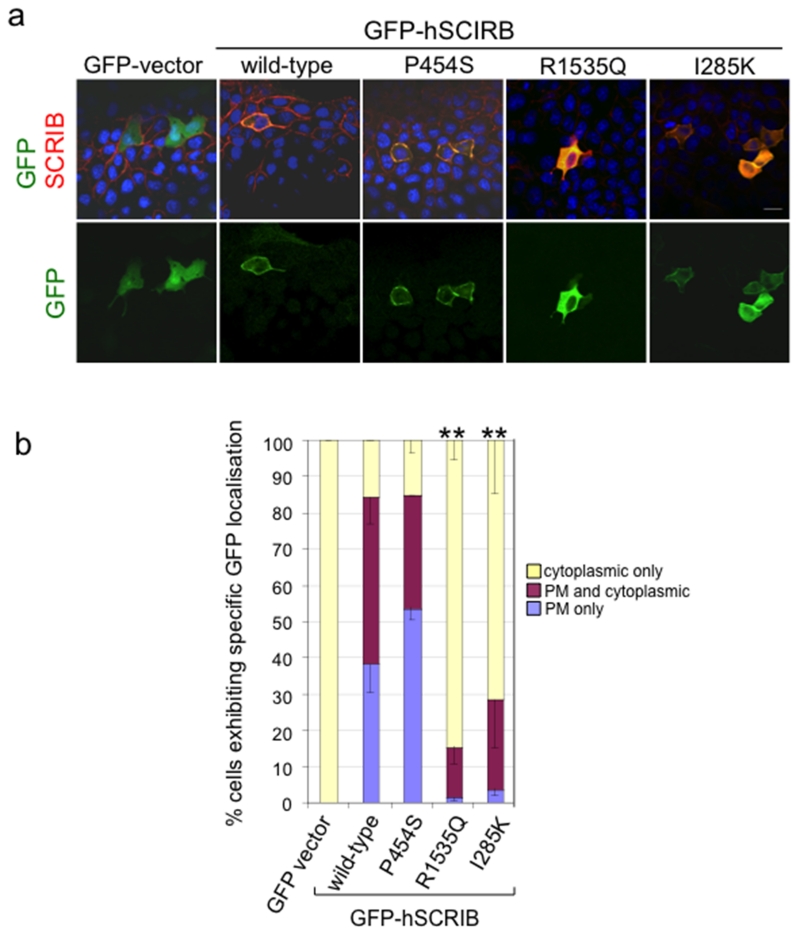
Both wild-type SCRIB, as expressed from the construct, and endogenous SCRIB exhibited strong plasma membrane localisation (Figure 5a,b). With the R1535Q variant, a significant increase was observed in the proportion of cells with cytosolic localisation; very few cells showed specific accumulation at the cell membrane. This was similar to findings with the mouse pathogenic line-90 mutant (I285K) (Zarbalis, et al., 2004) (Figure 5a,b). In contrast, distribution of the non-conserved P454S variant mimicked that of wild-type.
Figure 5. Effect of putative missense mutations in SCRIB on subcellular localisation.
(a) Protein localisation in MDCK cells: Top - immunofluorescence with anti-GFP (green) and anti-SCRIB (red); Bottom - anti-GFP alone. (b) Quantitative analysis of protein localisation for GFP +ve cells were scored scored as cytoplasmic only, plasma membrane (PM) and cytoplasmic combined, or PM only. ** Statistically significant differences of % cytoplasmic and % PM compared with wild-type (p < 0.001) were determined by two-way analysis of variance.
We conclude that all three known pathogenic mouse mutations, and some but not all of the human CELSR1 and SCRIB variants, abolish the membrane association, a feature that characterises wild-type proteins and is known to be required for PCP pathway function (Strutt, 2008).
Discussion
Our findings provide the first evidence for a causal association between human PCP mutations and CRN, a relationship that has been strongly established through studies of mouse mutants (Doudney and Stanier, 2005). Indeed, it is remarkable that, of the more than 150 different genes, which cause NTDs when mutated in mice, only genes of the PCP signalling pathway are associated with the CRN phenotype (Harris and Juriloff, 2010). Hence, the link between non-canonical Wnt signalling and failure of Closure 1 at the onset of mouse neurulation appears highly specific.
We did not detect any effects of the SCRIB pathogenic or putative mutations on the interaction of SCRIB protein with VANGL2 or LGL2 (Kallay, et al., 2006). A similar assay of the CELSR1 missense changes was not possible, owing to a lack of current knowledge of CELSR1’s binding partners. The lack of effect of missense genomic alterations on protein-protein interactions could reflect specificity for the particular site of protein contact, in this case SCRIB’s PDZ domain in the interaction with VANGL2, and its LRR domain in the interaction with LGL2 (Kallay, et al., 2006). The SCRIB missense changes we detected in CRN patients were all outside these functional domains, hence perhaps explaining the lack of effect on protein partner binding.
In contrast, use of a protein subcellular localisation assay in polarised MDCK cells, showed a clearly detrimental effect of many of the putative mutations in both CELSR1 and SCRIB. This suggests that one likely pathogenic mechanism of PCP mutation in mammals may be through impaired protein targeting to the plasma membrane. The disruption of membrane localisation by the CELSR1 and SCRIB mutations is similar to that identified for loss-of-function missense mutations in mouse Vangl2, including the Lp mouse mutant S464N (Gravel, et al., 2010) and two other mutant alleles, D255E and R259L (Iliescu, et al., 2011). Here, for the first time, we show that the mouse Celsr1 and Scrib mutations, found in the crash, spin cycle and line-90 alleles, all behave similarly in disrupting membrane localisation. This suggests a common mechanism for missense mutations in membrane associated PCP genes and provides a plausible mode of action to explain the effect of the missense changes found in our human CRN cohort.
Work in Drosophila has shown that the ‘core’ PCP proteins adopt distinctive asymmetric plasma membrane localisation, during establishment of planar polarity. The two transmembrane proteins, Flamingo (Celsr) and Frizzled, the latter receiving external Wnt signals, localise specifically to one side of cell-cell junctions. Intracellularly, these proteins associate with the cytoplasmic protein Dishevelled, which is essential for transduction of the PCP signal. On the opposite side of the cell junction, but in contact with Frizzled-Celsr via the extracellular space, is a complex of Vangl and Celsr membrane-localised proteins, in association with cytoplasmic Prickle. Such asymmetric complexes between cells may enable polarised cell–cell communication, during the establishment of long-range planar polarity within a tissue (Strutt, 2008). While there is accumulating evidence, as in the present study, for plasma membrane localisation of PCP proteins also in mammalian cells, there is little indication that asymmetric complexes are formed as in Drosophila. This may point to a difference in functioning of the PCP pathways between insects and mammals, or could merely reflect a limitation on subcellular protein imaging in mammalian cells.
Initiation of mammalian neural tube closure appears uniquely sensitive to loss of function in the PCP pathway, owing to the need for this pathway in shaping the embryo prior to neurulation (Colas and Schoenwolf, 2001). During the late stages of gastrulation, the elliptical gastrula is converted into the keyhole-shaped neurula largely as a result of the cell movements of convergent extension, which involve lateral to medial displacement of cells in the plane of the ectoderm, and in the underlying mesoderm (Keller, 2002). Cell intercalation in the midline leads to rostro-caudal extension and narrowing of the body axis, which ensures the neural folds are sufficiently closely spaced for closure (Wallingford and Harland, 2002; Ybot-Gonzalez, et al., 2007).
Heterozygous mutation of PCP genes diminishes pathway function in mice, as demonstrated by the predisposition of Vangl2Lp/+ embryos to develop CRN in the presence of concentrations of Rho kinase inhibitor that are too low to affect wild-type littermates (Ybot-Gonzalez, et al., 2007). Moreover, mice that are compound heterozygotes for two independent Vangl2 mutant alleles develop CRN in a manner indistinguishable from mice homozygous for a single mutation (Kibar, et al., 2001a). The present data suggest a similar dependence of neural tube closure initiation on PCP function in human embryogenesis, where 5/36 of our CRN cases carry putative mutations with deleterious effects on protein subcellular localisation of CELSR1 or SCRIB. It therefore seems likely, that diminution of PCP pathway function, perhaps in association with other (as yet unknown) genetic defects, leads to failure of closure initiation resulting in CRN.
In this study, we used a resequencing strategy to identify putative mutations in a targeted patient cohort, based on the assumption that causative alleles will be extremely rare or unique to a given family. These alleles are therefore not comparable with typical association studies since they cannot be part of a common haplotype shared with other unrelated patients who are affected with the same phenotype. Our model is essentially an autosomal dominant mode of inheritance with reduced penetrance, where penetrance is modified by the genetic background (i.e. a second compounding allele) or other non-specific environmental factors. Mouse studies demonstrate the potential for CRN inheritance to be strongly influenced by interactions at multiple loci. For example, all three pairwise combinations of Vangl2, Celsr1 and Scrib heterozygous mutations cause CRN (Iliescu, et al., 2011; Murdoch, et al., 2001b). Since convergent extension cell movements appear defective in homozygotes of all three of these genes, it seems likely that partial diminution of PCP signalling at different levels of the pathway can have additive effects, leading to overall loss of function. It is however, interesting to note that when Vangl2 interacts with the PCP gene Ptk7, or with the non-PCP genes Cthrc1, Grhl3 or Cobl, the resulting defect is spina bifida or anencephaly, but not CRN (Carroll, et al., 2003; Lu, et al., 2004; Stiefel, et al., 2007; Yamamoto, et al., 2008). Clearly, there is significant diversity of genetic interactions, and embryonic mechanisms, in determining the precise pattern of neural tube closure disruption (Copp and Greene, 2010). It will be interesting to determine whether CELSR1 and/or SCRIB mutations are found in non-CRN NTDs, as has previously been reported for VANGL1, VANGL2 and PRICKLE1 (Kibar, et al., 2009; Kibar, et al., 2007; Lei, et al., 2010); Bosoi et al., 2011).
Extrapolating from the mouse studies, it seems possible that human CRN cases may involve combined heterozygous mutation of two or more genes each affecting PCP signalling, while spina bifida and anencephaly are more likely to involve a combination of heterozygous PCP and non-PCP mutations (or entirely non-PCP gene defects). Use of high throughput genomic or exomic deep sequencing will, enable testing of this hypothesis, by identifying combinatorial hits in interacting PCP and non-PCP genes in individuals with varying NTD phenotypes.
Materials and Methods
Human Subjects
Ethical approval for the collection and analysis of the human DNA samples used in this study was granted by the Hammersmith and Queen Charlotte’s and Chelsea Hospitals and the Institute of Child Health/Great Ormond Street Hospital Research Ethics Committees. The study included 36 fetuses with CRN of which 17 were collected at the Greenwood Genetic Center, South Carolina and three were from the Hammersmith and Queen Charlotte’s and Chelsea Hospitals, London as previously described (Doudney, et al., 2005). A further 16 fetal samples affected with CRN were obtained from Groupe Hôpitalier Necker-Enfants Malades, Paris. Control DNAs were randomly selected from a total of 350 consecutive consenting normal pregnancies of white European ancestry recruited at Queen Charlotte’s and Chelsea Hospital (Apostolidou, et al., 2007). Details of a variety of parameters such as sex and birth weight were collected for these controls, and none of the patients or close relatives had an NTD.
DNA sequencing
Coding exons and flanking exon-intron boundaries were amplified by PCR from genomic DNA, for CELSR1 (NM_014246), SCRIB (NM_182706), PTK7 (NC_000006.11) and PRICKLE1 (NG_012965.1). Primer sequences are available on request. The PCR products were subjected to exonuclease I/shrimp alkaline phosphatase (Exo/SAP) treatment before sequencing with BigDye terminator v3.1 cycle sequencing kit (ABI) and loading onto the ABI Prism 3100 Genetic Analyzer. Sequence analysis was performed using Sequencher software (V4.8, GeneCodes Corporation) and putative variants were confirmed by re-sequencing both DNA strands. Sequence variants were investigated for population frequency in control DNAs (described above) as well as dbSNP132 (http://www.ncbi.nlm.nih.gov/snp) and (release 6) of the 1000 genomes project (http://browser.1000genomes.org/). Nucleotide numbering of the coding sequence variants described (designated c.) are based on cDNA sequences with +1 corresponding to A of the ATG translation initiation codon of the respective reference sequences (CELSR1: NM_014246.1 and SCRB1: NM_015356). Peptide sequence conservation was investigated using the web-based programs BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and CLUSTAL X (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Expression constructs
The full-length hSCRIB cDNA cloned into pEGFP-C1 expression plasmid was obtained from Dr. Patrick Humbert (Peter MacCallum Cancer Centre, Melbourne, Australia). FLAG-SCRIB fusion proteins were generated from the same full-length clone. Constructs expressing the putative SCRIB interacting proteins HA-VANGL2, HA-VANGL2Δ4 and GFP-LGL2 were obtained from Dr. Lelita Braiterman (Johns Hopkins University School of Medicine, Baltimore, USA). The pCDNA3-Celsr1-GFP construct expressing full-length mouse Celsr1 cDNA (NM_009886) was kindly provided by Dr. Caroline Formstone (MRC Centre for Developmental Neurobiology, Kings College, London). SCRIB and CELSR1 missense changes were introduced to the respective constructs by PCR-mediated site-directed mutagenesis using PfuTurbo DNA polymerase (Stratagene). For each point mutation a pair of complementary mutagenesis oligonucleotide primers was designed (sequences available on request). In addition to DNA sequencing to confirm the introduction of a unique and correct change, constructs were tested for expression following cell transfection by western blotting.
Cell transfection
MDCK (strain II) cells were cultured in minimum essential medium (MEM) (Invitrogen) supplemented with 5% (v/v) fetal bovine serum (FBS) and 2 mM glutamine, as recommended by the supplier (Health Protection Agency Culture Collections; HPACC). Between 8 and 12 hours before transfection, cells were seeded in 24-well TC plates (0.5 × 105 cells/well). Transfection was performed using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions. Twenty hours prior to localisation studies, transfected MDCK cells were re-plated onto poly-L-lysine (10 μg/mL) coated glass coverslips (1.5×105cells/coverslip). Cells were examined for localisation studies at 24, 48 and 72 hours post-transfection.
Protein extracts, co-purifications and western blotting
Protein extraction was performed at 4°C. Cells harvested 24 hours post-transfection were washed in phosphate-buffered saline (PBS) and lysed in 1% (v/v) Nonidet P-40 buffer (150 μM NaCl, 10 μM Tris-HCl, pH 8, 1 × complete protease inhibitor cocktail, in PBS) for 1 hour. Cell extracts were cleared by centrifugation at 3,000 × g for 20 min. For co-purification of FLAG fusion proteins and associated proteins, lysates were incubated with anti-FLAG agarose (Sigma) for 2 hours. The bound agarose was washed 3 times with 0.1% (v/v) Nonidet P-40 wash buffer (150 μM NaCl, 10 μM Tris-HCl; pH 7.8) before eluting the bound FLAG protein complex by the addition of FLAG peptide (Sigma). Co-purification or total lysate samples were resolved by 10% SDS-PAGE and subjected to western blot analysis with mouse monoclonal anti-FLAG antibody (M2; Sigma), rabbit polyclonal anti-GFP antibody (Invitrogen) or rat monoclonal anti-HA antibody (3F10; Roche). Bands were visualized by enhanced chemiluminescence (Amersham Biosciences) (see Supp. Figures S3, S4).
Cell localisation studies
For localisation of wild-type and mutant SCRIB and CELSR1 proteins, cells plated on poly-L-lysine coated coverslips were fixed with 4% (w/v) paraformaldehyde on ice and permeabilized with ice-cold 100% methanol before being rehydrated in phosphate buffered saline (PBS). Samples were blocked in 1% (w/v) bovine serum albumin (BSA) in PBS, incubated with primary antibody for 1 hour, rinsed in PBS and incubated with secondary antibody for 1 hour. Samples were washed with DAPI (4′,6-diamidino-2-phenylindole) and mounted in Mowiol 4-88 mounting medium (Sigma; prepared with glycerol and 0.2 M Tris pH 6.8). Primary and secondary antibodies were diluted in 1% (w/v) BSA in PBS and all incubations were performed at room temperature. Primary antibodies used in this study were against SCRIB (goat, 1:300, sc-11049, Santa Cruz), Celsr1 (rabbit, 1:800, a kind gift from Dr. Caroline Formstone), GFP (rabbit, conjugated with Alexa Fluor 488, 1:1000, A21311, Invitrogen; chicken, 1:300, A10262, Invitrogen) and goat immunoglobulins (rabbit, biotinylated, 1:250, E0466; DAKO). Secondary antibodies used were Alexa Fluor 568 goat anti-rabbit IgG (A21069, Invitrogen), rabbit anti-chicken FITC conjugated (613111; Invitrogen), and fluorescent streptavidin (conjugated with Alexa Fluor 546, S11225, Invitrogen). Labelled cells were examined and quantified by epifluorescence on an inverted LSM710 confocal system mounted on an AxioObserver Z1 microscope (Carl Zeiss Ltd, United Kingdom). The images were acquired using 63× oil immersion objective. The thickness of optical sections was set at 0.2-0.9 µm thickness. The AlexaFluor 488 dye was excited by a 488 nm line of an Argon laser and AlexaFluor 546 and AlexaFluor 568 by a 561 nm diode laser. Z-projections of confocal stacks were created in ImageJ. Images were further processed in Photoshop CS3 (Adobe).
Cells over-expressing SCRIB and CELSR1 were initially examined for localization studies at 24, 48 and 72 hours post-transfection (Supp. Figure S4). The 48 hours time point was found to be optimal for investigating membrane localization and was therefore used in all localization experiments detailed in this study. The subcellular localisation of GFP expression was classified for each cell into one of the following three different categories: (i) GFP exclusively at the membrane; (ii) GFP exclusively in the cytoplasm; (iii) GFP at the membrane and in the cytoplasm. For each transfection, at least 200 cells were counted and each transfection was performed in triplicate (i.e. at least 600 cells were counted in total for each treatment). Quantitative data were analysed by two-way analysis of variance to determine the statistical significance of comparisons between treatments (wild-type and the various mutant proteins) (Sigmastat v 3.5).
Supplementary Material
Acknowledgements
We thank Gudrun Moore for control DNA samples, Cindy Skinner and Sayeda Abu-Amero for assistance with sample preparation and sequencing and Bertrand Vernay for technical support with confocal microscopy. The work was supported by grants from SPARKS, the EMB Charitable Trust, the Wellcome Trust and the Medical Research Council. PS is supported by Great Ormond Street Hospital Children’s Charity.
Grant Sponsors: SPARKS (06ICH06); Wellcome Trust (087525); Medical Research Council (G0801124).
References
- Apostolidou S, Abu-Amero S, O’Donoghue K, Frost J, Olafsdottir O, Chavele KM, Whittaker JC, Loughna P, Stanier P, Moore GE. Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. Journal of molecular medicine. 2007;85(4):379–87. doi: 10.1007/s00109-006-0131-8. [DOI] [PubMed] [Google Scholar]
- Botto LD, Moore CA, Khoury MJ, Erickson JD. Neural-tube defects. The New England journal of medicine. 1999;341(20):1509–19. doi: 10.1056/NEJM199911113412006. [DOI] [PubMed] [Google Scholar]
- Carroll EA, Gerrelli D, Gasca S, Berg E, Beier DR, Copp AJ, Klingensmith J. Cordon-bleu is a conserved gene involved in neural tube formation. Developmental biology. 2003;262(1):16–31. doi: 10.1016/s0012-1606(03)00323-3. [DOI] [PubMed] [Google Scholar]
- Colas JF, Schoenwolf GC. Towards a cellular and molecular understanding of neurulation. Developmental dynamics : an official publication of the American Association of Anatomists. 2001;221(2):117–45. doi: 10.1002/dvdy.1144. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Greene ND. Genetics and development of neural tube defects. The Journal of pathology. 2010;220(2):217–30. doi: 10.1002/path.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nature reviews. Genetics. 2003;4(10):784–93. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Quint E, Tsipouri V, Arkell RM, Cattanach B, Copp AJ, Henderson DJ, Spurr N, Stanier P, Fisher EM. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Current biology : CB. 2003;13(13):1129–33. doi: 10.1016/s0960-9822(03)00374-9. others. [DOI] [PubMed] [Google Scholar]
- Doudney K, Stanier P. Epithelial cell polarity genes are required for neural tube closure. American journal of medical genetics. Part C, Seminars in medical genetics. 2005;135C(1):42–7. doi: 10.1002/ajmg.c.30052. [DOI] [PubMed] [Google Scholar]
- Doudney K, Ybot-Gonzalez P, Paternotte C, Stevenson RE, Greene ND, Moore GE, Copp AJ, Stanier P. Analysis of the planar cell polarity gene Vangl2 and its co-expressed paralogue Vangl1 in neural tube defect patients. American journal of medical genetics. Part A. 2005;136(1):90–2. doi: 10.1002/ajmg.a.30766. [DOI] [PubMed] [Google Scholar]
- Golden JA, Chernoff GF. Intermittent pattern of neural tube closure in two strains of mice. Teratology. 1993;47(1):73–80. doi: 10.1002/tera.1420470112. [DOI] [PubMed] [Google Scholar]
- Gravel M, Iliescu A, Horth C, Apuzzo S, Gros P. Molecular and cellular mechanisms underlying neural tube defects in the loop-tail mutant mouse. Biochemistry. 2010;49(16):3445–55. doi: 10.1021/bi902180m. [DOI] [PubMed] [Google Scholar]
- Greene ND, Stanier P, Copp AJ. Genetics of human neural tube defects. Human molecular genetics. 2009;18(R2):R113–29. doi: 10.1093/hmg/ddp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. An update to the list of mouse mutants with neural tube closure defects and advances toward a complete genetic perspective of neural tube closure. Birth defects research. Part A, Clinical and molecular teratology. 2010;88(8):653–69. doi: 10.1002/bdra.20676. [DOI] [PubMed] [Google Scholar]
- Iliescu A, Gravel M, Horth C, Kibar Z, Gros P. Loss of membrane targeting of Vangl proteins causes neural tube defects. Biochemistry. 2011;50(5):795–804. doi: 10.1021/bi101286d. [DOI] [PubMed] [Google Scholar]
- Johnson KM, Suarez L, Felkner MM, Hendricks K. Prevalence of craniorachischisis in a Texas-Mexico border population. Birth defects research. Part A, Clinical and molecular teratology. 2004;70(2):92–4. doi: 10.1002/bdra.10143. [DOI] [PubMed] [Google Scholar]
- Kallay LM, McNickle A, Brennwald PJ, Hubbard AL, Braiterman LT. Scribble associates with two polarity proteins, Lgl2 and Vangl2, via distinct molecular domains. Journal of cellular biochemistry. 2006;99(2):647–64. doi: 10.1002/jcb.20992. [DOI] [PubMed] [Google Scholar]
- Keller R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 2002;298(5600):1950–4. doi: 10.1126/science.1079478. [DOI] [PubMed] [Google Scholar]
- Kibar Z, Bosoi CM, Kooistra M, Salem S, Finnell RH, De Marco P, Merello E, Bassuk AG, Capra V, Gros P. Novel mutations in VANGL1 in neural tube defects. Human mutation. 2009;30(7):E706–15. doi: 10.1002/humu.21026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibar Z, Torban E, McDearmid JR, Reynolds A, Berghout J, Mathieu M, Kirillova I, De Marco P, Merello E, Hayes JM. Mutations in VANGL1 associated with neural-tube defects. The New England journal of medicine. 2007;356(14):1432–7. doi: 10.1056/NEJMoa060651. others. [DOI] [PubMed] [Google Scholar]
- Kibar Z, Underhill DA, Canonne-Hergaux F, Gauthier S, Justice MJ, Gros P. Identification of a new chemically induced allele (Lp(m1Jus)) at the loop-tail locus: morphology, histology, and genetic mapping. Genomics. 2001a;72(3):331–7. doi: 10.1006/geno.2000.6493. [DOI] [PubMed] [Google Scholar]
- Kibar Z, Vogan KJ, Groulx N, Justice MJ, Underhill DA, Gros P. Ltap, a mammalian homolog of Drosophila Strabismus/Van Gogh, is altered in the mouse neural tube mutant Loop-tail. Nature genetics. 2001b;28(3):251–5. doi: 10.1038/90081. [DOI] [PubMed] [Google Scholar]
- Lei YP, Zhang T, Li H, Wu BL, Jin L, Wang HY. VANGL2 mutations in human cranial neural-tube defects. The New England journal of medicine. 2010;362(23):2232–5. doi: 10.1056/NEJMc0910820. [DOI] [PubMed] [Google Scholar]
- Lu X, Borchers AG, Jolicoeur C, Rayburn H, Baker JC, Tessier-Lavigne M. PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature. 2004;430(6995):93–8. doi: 10.1038/nature02677. [DOI] [PubMed] [Google Scholar]
- Merte J, Jensen D, Wright K, Sarsfield S, Wang Y, Schekman R, Ginty DD. Sec24b selectively sorts Vangl2 to regulate planar cell polarity during neural tube closure. Nature cell biology. 2010;12(1):41–6. doi: 10.1038/ncb2002. sup pp 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CA, Li S, Li Z, Hong SX, Gu HQ, Berry RJ, Mulinare J, Erickson JD. Elevated rates of severe neural tube defects in a high-prevalence area in northern China. American journal of medical genetics. 1997;73(2):113–8. [PubMed] [Google Scholar]
- Murdoch JN, Doudney K, Paternotte C, Copp AJ, Stanier P. Severe neural tube defects in the loop-tail mouse result from mutation of Lpp1, a novel gene involved in floor plate specification. Human molecular genetics. 2001a;10(22):2593–601. doi: 10.1093/hmg/10.22.2593. [DOI] [PubMed] [Google Scholar]
- Murdoch JN, Henderson DJ, Doudney K, Gaston-Massuet C, Phillips HM, Paternotte C, Arkell R, Stanier P, Copp AJ. Disruption of scribble (Scrb1) causes severe neural tube defects in the circletail mouse. Human molecular genetics. 2003;12(2):87–98. doi: 10.1093/hmg/ddg014. [DOI] [PubMed] [Google Scholar]
- Murdoch JN, Rachel RA, Shah S, Beermann F, Stanier P, Mason CA, Copp AJ. Circletail, a new mouse mutant with severe neural tube defects: chromosomal localization and interaction with the loop-tail mutation. Genomics. 2001b;78(1-2):55–63. doi: 10.1006/geno.2001.6638. [DOI] [PubMed] [Google Scholar]
- O’Rahilly R, Muller F. The two sites of fusion of the neural folds and the two neuropores in the human embryo. Teratology. 2002;65(4):162–70. doi: 10.1002/tera.10007. [DOI] [PubMed] [Google Scholar]
- Paudyal A, Damrau C, Patterson VL, Ermakov A, Formstone C, Lalanne Z, Wells S, Lu X, Norris DP, Dean CH. The novel mouse mutant, chuzhoi, has disruption of Ptk7 protein and exhibits defects in neural tube, heart and lung development and abnormal planar cell polarity in the ear. BMC developmental biology. 2010;10:87. doi: 10.1186/1471-213X-10-87. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiefel D, Copp AJ, Meuli M. Fetal spina bifida in a mouse model: loss of neural function in utero. Journal of neurosurgery. 2007;106(3 Suppl):213–21. doi: 10.3171/ped.2007.106.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt D. The planar polarity pathway. Current biology : CB. 2008;18(19):R898–902. doi: 10.1016/j.cub.2008.07.055. [DOI] [PubMed] [Google Scholar]
- Tao H, Manak JR, Sowers L, Mei X, Kiyonari H, Abe T, Dahdaleh NS, Yang T, Wu S, Chen S. Mutations in prickle orthologs cause seizures in flies, mice, and humans. American journal of human genetics. 2011;88(2):138–49. doi: 10.1016/j.ajhg.2010.12.012. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Suzuki M, Kiyonari H, Abe T, Sasaoka T, Ueno N. Mouse prickle1, the homolog of a PCP gene, is essential for epiblast apical-basal polarity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(34):14426–31. doi: 10.1073/pnas.0901332106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford JB, Harland RM. Neural tube closure requires Dishevelled-dependent convergent extension of the midline. Development. 2002;129(24):5815–25. doi: 10.1242/dev.00123. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Nishimura O, Misaki K, Nishita M, Minami Y, Yonemura S, Tarui H, Sasaki H. Cthrc1 selectively activates the planar cell polarity pathway of Wnt signaling by stabilizing the Wnt-receptor complex. Developmental cell. 2008;15(1):23–36. doi: 10.1016/j.devcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Ybot-Gonzalez P, Gaston-Massuet C, Girdler G, Klingensmith J, Arkell R, Greene ND, Copp AJ. Neural plate morphogenesis during mouse neurulation is regulated by antagonism of Bmp signalling. Development. 2007;134(17):3203–11. doi: 10.1242/dev.008177. [DOI] [PubMed] [Google Scholar]
- Zarbalis K, May SR, Shen Y, Ekker M, Rubenstein JL, Peterson AS. A focused and efficient genetic screening strategy in the mouse: identification of mutations that disrupt cortical development. PLoS biology. 2004;2(8):E219. doi: 10.1371/journal.pbio.0020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.