Abstract
Prostate cancer is the most frequently diagnosed and second most fatal non-skin cancer among men in the United States. African American men are two times more likely to develop and die of prostate cancer compared with men of other ancestries. Previous whole genome or exome tumor sequencing studies of prostate cancer have primarily focused on men of European ancestry. In this study, we sequenced and characterized somatic mutations in aggressive (Gleason ≥7, stage ≥T2b) prostate tumors from 24 African American patients. We describe the locations and prevalence of small somatic mutations (up to 50 bases in length), copy number aberrations, and structural rearrangements in the tumor genomes compared with patient-matched normal genomes. We observed several mutation patterns consistent with previous studies, such as large copy number aberrations in chromosome 8 and complex rearrangement chains. However, TMPRSS2-ERG gene fusions and PTEN losses occurred in only 21% and 8% of the African American patients, respectively, far less common than in patients of European ancestry. We also identified mutations that appeared specific to or more common in African American patients, including a novel CDC27-OAT gene fusion occurring in 17% of patients. The genomic aberrations reported in this study warrant further investigation of their biological significance in the incidence and clinical outcomes of prostate cancer in African Americans.
Keywords: prostate cancer, aggressive tumors, somatic mutations, sequencing, African Americans
INTRODUCTION
In the United States (US), prostate cancer is the most commonly diagnosed non-skin cancer, and the second most common cause of cancer-related deaths among men (American Cancer Society 2015). However, incidence and mortality rates are highly variable among different individuals and subpopulations within the US. African American (AA) men have the highest risk of developing and dying of prostate cancer (National Cancer Institute, 2015). Age-adjusted incidence and mortality rates are about twice as high in AA compared to European American (EA) men (SEER Cancer Statistics Review, 2007–2011). This discrepancy is likely due to a combination of social, environmental, and genetic factors (1).
Several recent studies have focused on the genetics and genomics of prostate cancer in AA men (2–5). However, no studies have used whole genome sequencing (WGS) to describe genome-wide somatic mutations in prostate tumors from AA patients. Previous WGS studies have focused primarily on men of European ancestry (6–10). However it is important to conduct these studies in diverse populations to help decipher the genetic and genomic contributions to the differences in incidence and mortality.
In this study, we characterize the somatic mutations found in aggressive prostate tumors (Gleason grades of seven or TNM Classification of Malignant Tumors stages of T2b or higher) in 24 AA patients through whole genome DNA sequencing of tumor and matched normal samples. We considered tumors at this stage and grade as aggressive because they put these patients at an increased risk of recurrence, metastases and mortality (11). In doing so, we hope to identify potential genomic risk factors and therapeutic targets for men with the most clinically relevant disease. We compare our findings to those of other prostate cancer patients using data publically available through various publications (6–10), The Cancer Genome Atlas (TCGA) Research Network (12), and the Catalog Of Somatic Mutations In Cancer (COSMIC) (13).
MATERIALS AND METHODS
Selection of patients and evaluation of ancestry/admixture
Twenty-four patients were selected for this study from three health care centers where they were initially diagnosed and treated for prostate cancer. Thirteen patients were selected at the Henry Ford Health System in Detroit, Michigan, seven at Northwestern University in Chicago, Illinois, and four at the University of California in San Francisco, California. Tumor samples were collected under Institutional Review Board approval at each health care center. Patients were eligible if they were diagnosed with an aggressive prostate tumor, defined as having a Gleason grade 7 or TNM stage of T2b or higher, and if radical prostatectomy was the first line of treatment. All patients identified themselves as AA and consented to the use of their biospecimens for genomic research. Prostate-specific antigen (PSA) levels were measured prior to prostatectomy surgery.
To determine whether our patients had ancestry profiles similar to self-identified AA samples in other studies (14, 15), we analyzed 128 ancestry informative markers identified by Kosoy et al. (16). Using the Structure software version 2.3 (17), we ran models assuming correlated allele frequencies for between two and eight populations, using likelihood statistics to determine the most appropriate number of underlying populations for the final model. We compared the predicted allele frequencies to actual allele frequencies in our samples to assess overall fit, and to actual allele frequencies from the AA and EA samples studied by Kidd et al. (14) and to AA individuals from the Midwestern and Western US (where our patients were recruited) studied by Zakharia et al. (15). We also estimated the proportions of African and European ancestry for each gene using the RFMix method (18) in order to determine whether these proportions were correlated with the frequency of gene mutations.
Tissue preparation and sequencing
Tumor samples submitted for sequencing were required to contain ≥80% of cells that were morphologically abnormal upon inspection by a pathologist. Normal specimens were obtained from whole blood samples for 21 of the cases prior to surgery. For the other three cases blood was not available, and normal prostate tissue was obtained from as far as possible (a minimum of 1cm) from abnormal prostate tissue and glands.
DNA was extracted from tumor and normal specimens at each health care center using QIAGEN’s EZ1® Advanced DNA Tissue Kit and Tissue Card or the DNAeasy Blood and Tissue Kit (www.qiagen.com), suspended in 10mM Tris/1mM EDTA at pH 8.0, frozen at −80°C, and shipped to Complete Genomics, Inc. (CGI). Quality was determined by using gel electrophoresis to ensure high molecular weight (>20kb) and double stranded genomic DNA. The Quant-iT™ PicoGreen® dsDNA Assay Kit and the Qubit® Flourometer Assays from Life Technologies (www.lifetechnologies.com) were used to ensure that DNA quantities and concentrations were adequate for sequencing. In addition, CGI confirmed that each tumor and normal sample pair came from the same male individual by genotyping a panel of 96 single nucleotide polymorphisms (SNPs).
Whole genome DNA sequencing was performed on the tumor and blood or normal tissue samples by CGI, and the CGI Cancer Sequencing Pipeline Version 2.2 was used for mapping, assembly, and variant calling (19, 20). Variations in the normal sample were called relative to the GRCh37/hg19 reference genome or (where indicated below) to the 16 genomes sequenced by CGI from disease-free individuals of African ancestry (19). Somatic mutations in the tumor sample were called relative to the patient-matched normal genome.
Characterization of small somatic mutations
Small somatic mutations were defined as somatic insertions, deletions, or substitutions of up to 50 bases long. Prior to analysis, we filtered these mutations on several criteria using QIAGEN’s Ingenuity® Variant Analysis (IVA) software (www.qiagen.com/ingenuity) and data from the University of California, Santa Cruz Genome Browser (22). The latest Exome Variant Server (EVS-v.0.0.30, 23) available through the National Heart, Lung, and Blood Institute (NHLBI) was also used for annotations. We excluded mutations that matched germline SNPs occurring in ≥5% of African genomes in the EVS or in the 1,000 Genomes Project (24), or in the CGI reference samples (19). We also excluded mutations that fell in repetitive regions of the GRCh37/hg19 reference genome or in regions with ambiguous zygosity calls in either the tumor or normal genomes. Finally, we removed mutations that were supported by sequencing read depths below 20× or CGI Phred-like scores below 50 in the tumor or normal genomes. The latter four filters were validated for CGI data by Reumers et al. (25).
For internal validation purposes, we first assessed whether there were differences between transition/transversion (Ti/Tv) ratios among the 21 blood versus three prostate tissue reference samples. We also tested for differences in Ti/Tv ratios and individual transition and transversion events in our data compared to the prostate cancer data from TCGA. Finally, we ran an Illumina 1M SNP/CNV array (http://www.illumina.com) on a subset of five tumor and matched normal samples and calculated percent agreement between these genotypes and the WGS genotypes. We also used this Illumina SNP array to estimate the normal content in the tumor samples, in order to determine if this supported the pathology reports for this subset.
For the genes having the highest mutation frequency and rates in our cohort, we compared the frequency of mutations in the 227 TCGA prostate cancer patients (12) with the similar Gleason grades and stages as our patients and with self-identified European or AA ancestry, and to all others represented in the COSMIC database (13) (Supplementary Table S1). We also compared mutation frequencies in the genes (SPOP, FOXA1, and IDH1) that helped to identify subtypes of prostate cancer in TCGA (12), as well as the other most frequently mutated genes in TCGA (TP53, PTEN, and BRCA2) by race.
To assess the impact of ancestry on overall exonic single base substitutions (ESBS) in RefSeq genes (26), we calculated the average “rate” of these mutations (log10-transformed average number per megabase [Mb]) among tumors with one or more mutations. We directly compared the ESBS rates to the Berger et al. (6) and Baca et al. (7) studies using a generalized estimating equation (GEE) model (27) adjusting for patient age, pre-operative PSA and clinical Gleason grade, log10-transformed gene length and Guanine-Cytosine (GC) content, and proportion of European ancestry (18) for each gene (assuming 100% for EA patients).
Characterization of copy number aberrations
Copy number was determined by CGI using relative read depths. For normal samples, copy number was determined by comparing coverage levels to that of the 16 African reference genomes sequenced by CGI (19). For tumor samples, copy number was determined by comparing coverage levels to the patient-matched normal samples in contiguous non-overlapping segments of 100 kilobases (Kb) spanning the entire genome (autosomes only). Segments that had very high or low GC content in the normal genome and that had an average Phred-like quality score below 50 were removed. For segments that were diploid in the normal sample, we classified copy number aberrations (CNAs) in the corresponding tumor sample as losses if copy number was ≤1.5, and gains if copy number was ≥2.5. The diploid regions of the CGI-sequenced African reference genomes also had copy number levels well within these bounds (19). We then ranked the CNA segments by the number of tumors harboring them, followed by the magnitude of gain or loss in copy number, and determined which genes were contained within these segments. We then compared the frequency of CNAs in these genes to the TCGA and COSMIC databases.
To assess genome-wide patterns of tumor versus normal copy number (normalized log2 ratios) we performed unsupervised hierarchical clustering. In addition, we determined whether our cohort had more or less prevalent CNAs in regions where CNAs have previously been associated with clinical factors and outcomes. In particular, we checked for CNAs in the MYC gene family since these have previously been associated with Gleason grade (9), and in 38 loci called the Genomic Evaluators of Metastatic Prostate Cancer (GEMCaP) which have previously been associated with biochemical recurrence and metastases (5, 28).
Characterization of somatic structural rearrangements
Rearrangement breakpoints were identified by locating adjacent paired reads on the tumor genome that mapped to non-adjacent positions on the reference genome. These must have been supported by a minimum of ten discordant reads between the tumor and reference genomes, a minimum of 70bp of high quality sequence data on each side, and local de novo assembly by CGI. Rearrangements were classified as deletions, duplications, inversions, translocations, or complex if the type was unclear. All fusions were verified and visualized using the iFuse software (29). We then ranked genes impacted by rearrangement breakpoints or fusions according to the number of tumors affected followed by the number of events per gene, and compared rearrangement frequencies for the most impacted genes to the COSMIC database.
We analyzed all rearrangements that could potentially cause gene fusions for evidence of chromoplexy, or chained rearrangements, using the ChainFinder software (7). Using Wilcoxon rank-sum tests, we then compared the number and percentage of breakpoints involved in any rearrangement chain, any inter-chromosomal chain, and the log10-transformed number of chromosomes per chain in tumors that were ETS fusion positive versus negative. Tumors were considered ETS-positive if they contained fusions involving any gene in the ELF, ELK, ETS or ETV family, EHF, ERG, FEV, or SPDEF. In addition, we compared these chromoplexy measures with the Baca et al. (7) data in separate linear regression models adjusted for age, PSA, Gleason grade at diagnosis, ETS fusion status, and proportion of European ancestry (assuming 100% for the EA patients). We also compared fusion frequencies in genes that helped to identify subtypes of prostate cancer in TCGA (ERG, ETV1, ETV4, and FLI1 fusions) (12).
RESULTS
Characteristics of the 24 patients and sequenced samples
Patient characteristics are described in Table 1. As expected since all patients identified themselves as African American, there were varying degrees of African and European ancestry as estimated using the Structure software (17). The best fitting Structure model according to maximum likelihood statistics indicated three ancestral populations. Allele frequencies estimated from this model (Supplementary Fig. S1) were nearly perfectly correlated with the actual allele frequencies in our cohort (Pearson correlation r>0.99). They were also highly correlated (r=0.98) with the 146 AA individuals but not well correlated (r=0.18) with the 204 EA individuals studied by Kidd et al. (14). Admixture proportions were also similar to those observed for AA individuals studied by Zakharia et al. (15) from the same regions of the US as our patients, which estimated 50% Yoruban, 30% other African, and 20% European ancestry. We found similar proportions in our cohort, with an estimated average of 50% from one population, 28% from another, and 22% from a third ancestral population (Supplementary Fig. S2). Estimated allele frequencies from the third population closely resembled those expected of European ancestry, further providing evidence that this population represented the European ancestry among our patients. These estimates of European ancestry ranged from 10% to 36% in our cohort (Supplementary Fig. S2).
Table 1.
Characteristics of the 24 African American patients included in the study
| Patient | Clinica | Normal tissue source |
Ageb | Pre-op PSAc |
Gleason grade |
TNM staged |
|---|---|---|---|---|---|---|
| 1 | Henry Ford | Blood | 66 | 16.4 | 9 (5+4) | T2b |
| 2 | Henry Ford | Blood | 59 | 4.9 | 8 (3+5) | T2b |
| 3 | Henry Ford | Blood | 54 | 11.5 | 7 (4+3) | T3a |
| 4 | Henry Ford | Blood | 50 | 6.7 | 7 (3+4) | T2b |
| 5 | Henry Ford | Blood | 68 | 26.5 | 9 (5+4) | T4 |
| 6 | Henry Ford | Blood | 52 | 4.9 | 8 (4+4) | T2c |
| 7 | Henry Ford | Blood | 60 | 5.6 | 7 (3+4) | T2c |
| 8 | UCSF | Prostate | 54 | 7.1 | 8 (3+5) | T2c |
| 9 | UCSF | Blood | 62 | 12.8 | 9 (4+5) | T3b |
| 10 | UCSF | Prostate | 61 | 4.1 | 7 (4+3) | T3a |
| 11 | UCSF | Prostate | 62 | 6.2 | 7 (3+4) | T2c |
| 12 | Henry Ford | Blood | 67 | 6.9 | 7 (4+3) | T2c |
| 13 | Henry Ford | Blood | 68 | 15.1 | 7 (4+3) | T3b |
| 14 | Henry Ford | Blood | 60 | 3.4 | 6 (3+3) | T2c |
| 15 | Henry Ford | Blood | 64 | 5.6 | 7 (3+4) | T3a |
| 16 | Henry Ford | Blood | 56 | 27.7 | 7 (3+4) | T2c |
| 17 | Henry Ford | Blood | 58 | 5.3 | 8 (3+5) | T2c |
| 18 | Northwestern | Blood | 44 | 11 | 7 (3+4) | T2c |
| 19 | Northwestern | Blood | 56 | 15 | 7 (3+4) | T3a |
| 20 | Northwestern | Blood | 58 | 5.3 | 7 (4+3) | N1 |
| 21 | Northwestern | Blood | 57 | 5.5 | 9 (4+5) | T2c |
| 22 | Northwestern | Blood | 54 | 15.3 | 9 (5+4) | T3b |
| 23 | Northwestern | Blood | 66 | 8.6 | 9 (4+5) | N1 |
| 24 | Northwestern | Blood | 56 | 13.8 | 7 (4+3) | T3b |
Clinic: Henry Ford=Henry Ford Health System, Detroit, Michigan; Northwestern=Northwestern University, Chicago, Illinois; UCSF=University of California, San Francisco
Age: patient age at the time of diagnosis
Pre-op PSA: prostate specific antigen levels measured prior to radical prostatectomy surgery
TNM stage: Classification of Malignant Tumors staging system
Tumor and normal sample sequencing statistics are presented in Table 2. On average, the tumor and matched normal samples had similar levels of sequence coverage genome-wide (105×), and over 98% of the genomes were covered by at least 20× read depths. The number of SNPs genotyped by sequencing was in the expected range for both tumor and normal genomes (approximately 4.1 million). Germline heterozygous to homozygous SNP and Ti/Tv ratios were also within expected ranges in all samples. Although sex chromosomes were included in all but the copy number analyses, most of the Y chromosome was removed from consideration since it is highly repetitive.
Table 2.
Median (range) of per-genome sequencing measures for the 24 patients
| Tumor | Normal | ||
| Genome-wide average coverage depth | 105 (56–112) | 105 (56–113) | |
| Percent of genome with ≥ 10× | 99.2 (98.7–99.3) | 99.3 (98.6–99.4) | |
| Percent of genome with ≥ 20× | 98.1 (94.8–98.6) | 98.2 (93.9–98.6) | |
| Percent of genome with ≥ 40× | 91.9 (66.9–94.5) | 91.9 (67.6–94.7) | |
| Percent of genome fully called (all alleles) | 97.6 (97.1–97.8) | 97.6 (97.1–97.8) | |
| Single nucleotide polymorphisms (×1000) | 4,115 (3,975–4,183) | 4,110 (3,975–4,180) | |
| Heterozygous/homozygous ratio | 2.11 (1.96–2.23) | 2.11 (2.04–2.23) | |
| Transition/transversion ratio | 2.13 (2.13–2.14) | 2.13 (2.13–2.14) | |
| Tumor versus Normala | |||
| Small indels and substitutions (≤50 bases) | 14,537 (9,684–21,445) | ||
| Genicb | 6,474 (4,004–9,709) | ||
| Exonic | 77 (33–229) | ||
| Single base substitutions | 5,309 (3,323–11,603) | ||
| A ↔ G transitions | 1,566 (927–4,511) | ||
| C ↔ T transitions | 1,588 (907–4,374) | ||
| A ↔ C transversions | 535 (364–853) | ||
| A ↔ T transversions | 589 (474–854) | ||
| C ↔ G transversions | 448 (272–640) | ||
| G ↔ T transversions | 542 (379–864) | ||
| Copy number aberrationsc | 148 (1–3,557) | ||
| Losses | 41 (0–2,052) | ||
| Gains | 6 (1–1,820) | ||
| Rearrangements | 87 (29–309) | ||
| Deletions | 27 (9–106) | ||
| Duplications | 14 (7–82) | ||
| Inversions | 12 (0–92) | ||
| Translocations | 5 (0–22) | ||
| Complexd | 20 (11–58) | ||
| Chromoplexice | 7 (0–41) | ||
| Gene fusionsf | 4 (0–11) | ||
Refers to high-confidence somatic mutations (see Methods)
Genic mutations are defined as those occurring in exons, introns, promoters, 5’UTR, 3’UTR, and splice site regions of RefSeq genes.
Copy number aberrations are defined as tumor copy number of less than 1.5 (losses) or more than 2.5 (gains).
Complex rearrangements are those not easily classified as any one of the other categories
Chromoplexic rearrangements are those which are interdependent and adjacent to other rearrangements in the tumor genome (7).
Gene fusions can result from deletions, inversions, translocations, or complex rearrangements
Small somatic mutations
Small somatic mutation rates and Ti/Tv ratios did not differ when blood (21 patients) versus normal prostate tissue (three patients) was used as the reference (Mann-Whitney p=0.37 and p=0.96, respectively). In addition, individual somatic transition and transversion rates were similar to those seen in the TCGA prostate cancer data, and Ti/Tv ratios were not significantly different between them (Mann-Whitney p=0.86, Supplementary Figure S3). Genotypes based on Illumina 1M SNP arrays run on a subset of five patients were in significant agreement with sequence-based genotypes for both tumor and normal samples (Kappa statistic p<0.001 for all). The SNP arrays were also used to estimate ploidy in the five tumor samples, and confirmed the pathological reports indicating over 80% abnormal cell content.
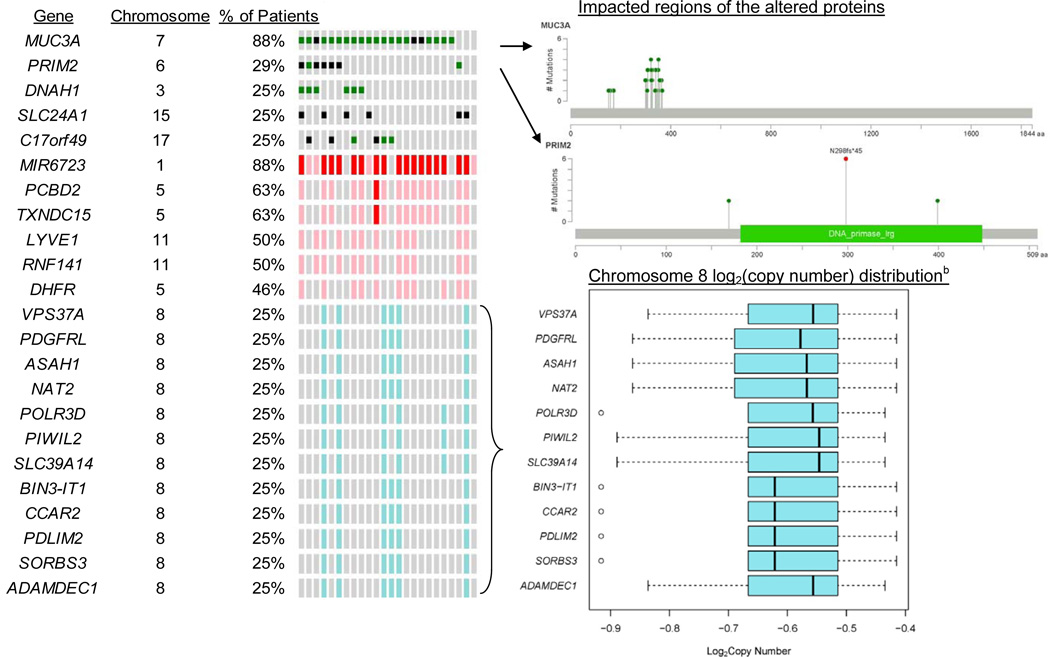
Among all 24 tumor samples, 10,883 of all known RefSeq genes harbored one or more small mutations. Sixteen percent (1,722) of the genes were mutated in only one tumor, and 2% (177) genes were mutated in all 24 tumors. Of the genes mutated in all patients, PRIM2 had the highest average mutation rate. However, MUC3A had the highest frequency and rate of ESBS and affected protein changes in most (88%) of the patients (Figure 1). PRIM2 mutations were the second most impactful on protein changes, occurring in 29% of patients. PRIM2 mutations appeared in the TCGA and COSMIC databases but only for 1.4% of samples tested, and MUC3A did not appear in TCGA or COSMIC (Supplemental Table S1). However, for the three individual genes (SPOP, FOXA1, and IDH1) with small mutations identifying subtypes in the TCGA cohort (12), frequencies of FOXA1 differed significantly by race (Figure 2). So did two of the other most frequently mutated individual genes in TCGA (TP53 and PTEN).
Figure 1.
Figure 2.
Comparisons of overall ESBS rates in EA patients from Berger et al. (6) and Baca et al. (7) to our AA patients showed no significant differences by race (p=0.35) or ancestry (p=0.64) after adjusting for age, pre-op PSA and Gleason grade, gene length and GC content.
Copy number aberrations
There was a median of 26,652 high-quality 100Kb copy number segments in the tumor genomes, representing 82% of the whole genome. CNAs occurred in 148 of these segments, affecting 2,551 genes. Of these, MIR6723 was the most frequently impacted gene with amplifications occurring in 21 (88%) of tumors, followed by PCBD2 and TXNDC15 with gains in 63% (Figure 1). These genes were far less frequently affected by CNAs in TCGA and COSMIC (5% or less). However, EBF2 and all other genes that were frequently affected by losses on chromosome 8 in our cohort were similarly affected in TCGA and COSMIC (Supplementary Figure S1).
From genome-wide hierarchical clustering analysis of log2 copy number ratios in tumor versus normal samples, we found some distinct overall patterns. For example, the short arm of chromosome 8 contained most of the copy number losses and the long arm contained many of the gains (Supplementary Figure S4). Our cohort had a lower frequency of CNAs in regions that were previously associated with Gleason grade in the MYC gene family (9) as well as in 38 loci that have previously been associated with biochemical recurrence and metastases (5, 28) (Supplementary Table S2).
Structural rearrangements
Several rearrangements occurred in multiple tumors, most of which occurred on chromosomes 17 and Y. On chromosome 17, 18 tumors (75%) had rearrangements in q21.31 and/or q21.32. The most frequently impacted gene in this region was KIAA1267 (or KANSL1), which contained rearrangement breakpoints in 12 tumors. Following KIAA1267 in rearrangement frequencies were TMPRSS2, CDC27, and ERG (Supplementary Table S1). Eleven tumors had duplications of nearly 1Mb on chromosome 17 which entirely covered eight other genes. On the Y chromosome, 16 tumors (67%) had rearrangements, all having breakpoints in q11.21. In half of these tumors, all genes on the long arm of chromosome Y were deleted. Of the 20 genes most frequently impacted by rearrangement breakpoints in our data, only four (TMPRSS2, ERG, EXT1, and DECR1) were previously reported in COSMIC or TCGA (Supplementary Table S1).
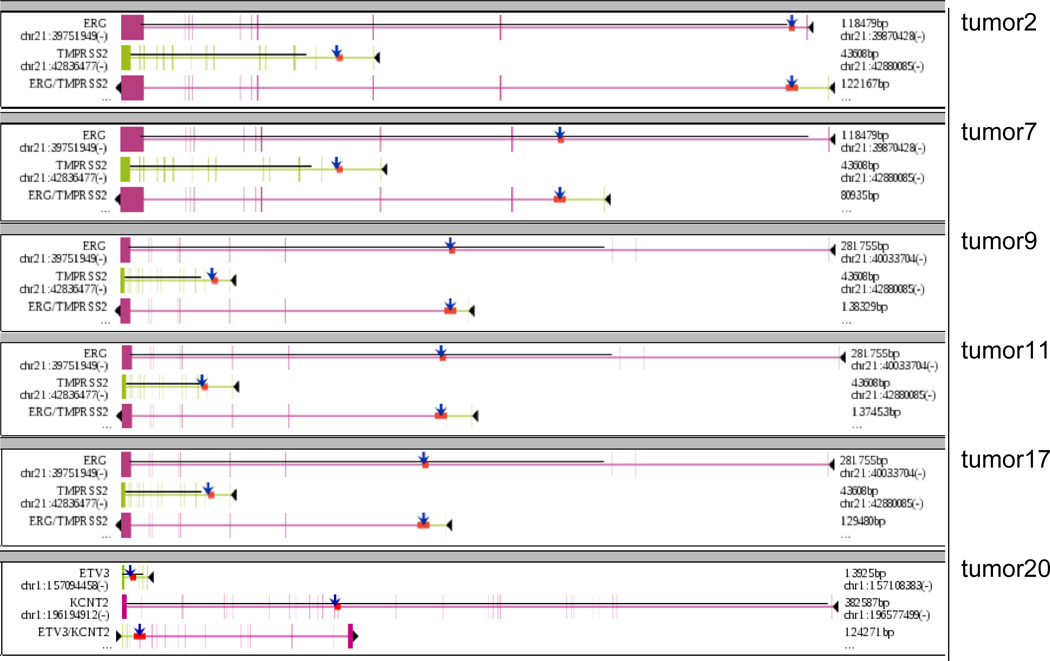
All 24 tumors had rearrangements that were caused by breakpoints falling in gene regions, resulting in a total of 99 gene fusions (Supplemental Table S3). Six tumors harbored ETS-related gene fusions (Figure 3), five of which were TMPRSS2-ERG fusions (all caused by deletions). In four patients, breakpoints in the CDC27 gene resulted in a fusion with the OAT gene on chromosome 10, making this the second most common fusion in our tumors after TMPRSS2-ERG. Fusions involving CDC27 and OAT have not yet been recorded in COSMIC or TCGA (Supplementary Table S1).
Figure 3.
To look for evidence of chromoplexy using the ChainFinder software (7), we analyzed a median of 128 (range 42–504) breakpoints per tumor. Seven percent (range 0–31%) of these breakpoints were involved in rearrangement chains. A median of 7% (range 0–50%) of the rearrangement chains were inter-chromosomal, involving up to four chromosomes per chain. The number of breakpoints per tumor, the percent of breakpoints involved in chains and inter-chromosomal chains, and the number of chromosomes involved per inter-chromosomal chain did not differ by ETS fusion status in our data. Circos plots (31) depict all rearrangements color-coded by chain in each tumor (Supplementary Fig. S4).
The 57 EA patients studied by Baca et al. (7) exhibited a similar number of breakpoints per tumor (median=124, unadjusted p=0.57, adjusted for patient and tumor characteristics p=0.59) and percentage of inter-chromosomal chains (median=13%, unadjusted p=0.61, adjusted p=0.42). There was a significantly higher percentage of breakpoints involved in chains overall in the EA compared with AA patient tumors; (median=50%, p<0.001), but this difference was not significant after adjusting for percent European ancestry (p=0.60) or for age, PSA, Gleason, and ETS fusion status (p=0.54). There was also a significantly higher number of chromosomes per chain in the EA patients (up to ten chromosomes per chain, unadjusted p<0.001), but this also was not significant after adjusting for percent European ancestry (p=0.77) and other characteristics (p=0.82).
The gene most commonly harboring breakpoints leading to rearrangement chains in our data was TMPRSS2 occurring in three tumors and chains, followed by ELK4 occurring in two tumors and three chains. Eleven of the top 20 genes in our data were also involved in rearrangement chains in the Baca et al. (7) data (Supplemental Table S1). For the genes involved in fusions (ERG, ETV1, ETV4, and FLI1) that helped to identify subtypes in the TCGA cohort (12), ERG was significantly less common in AA patients (Figure 2).
DISCUSSION
This study is the first to characterize the genomic alterations found through whole-genome DNA sequencing of aggressive tumors from African American (AA) prostate cancer patients. We analyzed the tumor genomes from 24 patients in reference to matched normal samples and identified the genes most frequently impacted by somatic mutations in three major categories: 1) small insertions, deletions, or substitutions of ≤50 bp in length; 2) large copy number aberrations (CNAs); 3) complex rearrangements. We also compared the frequency of mutations in each of these categories to those of previous WGS studies of prostate cancer patients with primarily European ancestry (6–10) and to the TCGA and COSMIC public databases (12, 13).
Although we did not detect global differences in small mutation rates, copy number patterns, or rearrangement frequencies between the tumors from our cohort compared to others, we did find some differences in mutations for specific genes. In the small mutation category for example, we found that MUC3A and PRIM2 had the most frequent mutations with impacts on proteins in our cohort. Mutations in these genes were absent or far less common in the primarily European patients previously studied (e.g. those represented in TCGA and COSMIC). However, both of these genes could play a role in prostate cancer, possibly for AA patients especially. MUC3A is part of the MUC gene family that encodes mucin proteins lining various tissues including those of the reproductive system. These proteins are involved in cell signaling, growth and survival. Mutations in MUC genes have previously been associated with carcinogenesis (RefSeq 2015), and four other MUC genes were among the ten most frequently mutated genes in COSMIC for prostate cancer. PRIM2 encodes a subunit of DNA primase that participates in synthesizing Okazaki fragments during DNA replication (RefSeq 2015) and has been associated with carcinogenesis as well (32). Both of these genes deserve further attention in studies involving AA patients in particular, given the high prevalence of mutations we observed in them. Another gene that deserves further attention is FOXA1, since this gene identified a subtype of prostate cancer in TCGA (12) and had a significantly higher rate of mutation in patients with African ancestry (in our cohort and the TCGA cohort combined) compared to TCGA patients with European ancestry. TP53 and PTEN, also containing high rates of small mutations in TCGA (12), had significantly lower mutation rates in patients with African ancestry.
With a few notable exceptions, we found that overall copy number patterns in our cohort were similar to those observed in previous prostate cancer studies. For example, there were distinct copy number changes in chromosome 8, with losses primarily in the short arm and gains in the long arm. These patterns are commonly observed in prostate cancer (e.g. 33), and some have been associated with systemic tumor recurrence and mortality (34). However, like with small mutations, we observed some differences in rates of CNAs in specific genes. Only two of our patients (8%) had heterozygous losses of PTEN. In EA patients, heterozygous and homozygous PTEN losses are far more common (reported in up to 40% of patients) and are typically associated with poor outcomes (35). MYC gene CNAs that have been reported to be associated with Gleason grade (9) were not as prevalent or associated with grade in our cohort. CNAs in 38 different loci that have previously been associated with recurrence and metastases were also less prevalent in our cohort compared to EA patients previously studied (5, 28). Larger sample sizes and finer resolution of copy number data (using segments smaller than 100Kb) in tumor samples will be important for future studies in order to determine if these CNA patterns are observed in other AA patients.
The TMPRSS2, CDC27, ERG, and OAT genes were the most commonly involved in gene fusions. However, the prevalence of TMPRSS2-ERG fusions in our cohort was much lower (21%) than previously observed in patients of European ancestry (estimates range from 40% to 80%, 6–10, 37, 38). There is other evidence of lower prevalence rates for TMPRSS2-ERG fusions in patients with African ancestry (39–41). Furthermore, we found that the frequency of ERG fusions in general were significantly less common among AA patients within the TCGA cohort. The biological significance of these differences warrants additional investigation. As do inter-chromosomal fusions between CDC27 and OAT, which were observed in 17% of our tumor samples, since these have not previously been associated with prostate cancer to our knowledge, and since OAT is a target of the androgen receptor and regulated by testosterone (42, 43). The differences in chromoplexy/rearrangement chain frequencies in our tumors compared to those of previously studied EA patients (7) will also be important to investigate in future studies.
In this study, we have described our findings from the first WGS study focused on African American prostate cancer patients. Additional WGS studies with larger sample sizes and more clinical and follow-up data will be needed to further validate and confirm the biological and clinical significance of our findings. However, the patterns of somatic mutations that we have described in this study could help provide a basis for understanding the genomic contributions to aggressive tumors and associated outcomes in this understudied population.
Supplementary Material
Acknowledgments
Financial Support:
R25T CA112355, U01CA127298, and R01CA088164 (KJL, PLP, TJH, NJC, RK, JAM, JPS, VN, JSW); R01ES011126 (AML, DC, BAR); P50CA090386, U01CA089600, The Urological Research Foundation (BTH, WJC), and the UCSF Goldberg-Benioff Program in Cancer Translational Biology (JSW).
Footnotes
Conflicts of Interest: The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Witte JS. Prostate cancer genomics: towards a new understanding. Nat Rev Genet. 2009;10(2):77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haiman CA, Han Y, Feng Y, Xia L, Hsu C, Sheng X, et al. Genome-wide testing of putative functional exonic variants in relationship with breast and prostate cancer risk in a multiethnic population. PLoS Genet. 2013;9(3):e1003419. doi: 10.1371/journal.pgen.1003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46(10):1103–1109. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koochekpour S, Willard SS, Shourideh M, Ali S, Liu C, Azabdaftari G, et al. Establishment and characterization of a highly tumorigenic African American prostate cancer cell line, E006AA-hT. Int J Biol Sci. 2014;10(8):834–845. doi: 10.7150/ijbs.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin AM, Lindquist KJ, Avila A, Witte JS, Paris PL, Rybicki BA. Performance of the Genomic Evaluators of Metastatic Prostate Cancer (GEMCaP) tumor biomarker for identifying recurrent disease in African American patients. Cancer Epidemiol Biomarkers Prev. 2014;23(8):1677–1682. doi: 10.1158/1055-9965.EPI-13-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47(7):736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 10.Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet. 2015;47(4):367–372. doi: 10.1038/ng.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedland SJ, Humphreys EB, Mangold LA, Eisenberger M, Dorey FJ, Walsh PC, et al. Risk of prostate cancer-specific mortality following biochemical recurrence after radical prostatectomy. JAMA. 2005;294(4):433–439. doi: 10.1001/jama.294.4.433. [DOI] [PubMed] [Google Scholar]
- 12.The Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidd JR, Friedlaender FR, Speed WC, Pakstis AJ, De La Vega FM, Kidd KK. Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples. Investig Genet. 2011;2(1):1. doi: 10.1186/2041-2223-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zakharia F, Basu A, Absher D, Assimes TL, Go AS, Hlatky MA, et al. Characterizing the admixed African ancestry of African Americans. Genome Biol. 2009;10(12):R141. doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30(1):69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am J Hum Genet. 2013;93(2):278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327(5961):78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 20.Carnevali P, Baccash J, Halpern AL, Nazarenko I, Nilsen GB, Pant KP, et al. Computational techniques for human genome resequencing using mated gapped reads. J Comput Biol. 2012;19(3):279–292. doi: 10.1089/cmb.2011.0201. [DOI] [PubMed] [Google Scholar]
- 21.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: [accessed June, 2015]. (URL: http://evs.gs.washington.edu/EVS/) [Google Scholar]
- 23.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reumers J, De Rijk P, Zhao H, Liekens A, Smeets D, Cleary J, et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat Biotechnol. 2012;30(1):61–68. doi: 10.1038/nbt.2053. [DOI] [PubMed] [Google Scholar]
- 25.Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42(Database issue):D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang KY, Zeger S. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73(1):13–22. [Google Scholar]
- 27.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paris PL, Andaya A, Fridlyand J, Jain AN, Weinberg V, Kowbel D, et al. Whole genome scanning identifies genotypes associated with recurrence and metastasis in prostate tumors. Hum Mol Genet. 2004;13(13):1303–1313. doi: 10.1093/hmg/ddh155. [DOI] [PubMed] [Google Scholar]
- 29.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiltemann S, McClellan EA, van Nijnatten J, Horsman S, Palli I, Teles Alves I, et al. iFUSE: integrated fusion gene explorer. Bioinformatics. 2013;29(13):1700–1701. doi: 10.1093/bioinformatics/btt252. [DOI] [PubMed] [Google Scholar]
- 31.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yatsula B, Galvao C, McCrann M, Perkins AS. Assessment of F-MuLV-induced tumorigenesis reveals new candidate tumor genes including Pecam1, St7, and Prim2. Leukemia. 2006;20(1):162–165. doi: 10.1038/sj.leu.2404034. [DOI] [PubMed] [Google Scholar]
- 33.Beheshti B, Park PC, Sweet JM, Trachtenberg J, Jewett MA, Squire JA. Evidence of chromosomal instability in prostate cancer determined by spectral karyotyping (SKY) and interphase fish analysis. Neoplasia. 2001;3(1):62–69. doi: 10.1038/sj.neo.7900125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst. 1999;91(18):1574–1580. doi: 10.1093/jnci/91.18.1574. [DOI] [PubMed] [Google Scholar]
- 35.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97(5):678–685. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perinchery G, Sasaki M, Angan A, Kumar V, Carroll P, Dahiya R. Deletion of Y-chromosome specific genes in human prostate cancer. J Urol. 2000;163(4):1339–1342. [PubMed] [Google Scholar]
- 37.Clark J, Merson S, Jhavar S, Flohr P, Edwards S, Foster CS, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26(18):2667–2673. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 38.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8(7):497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15(14):4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magi-Galluzzi C, Tsusuki T, Elson P, Simmerman K, LaFargue C, Esgueva R, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71(5):489–497. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 41.Khani F, Mosquera JM, Park K, Blattner M, O'Reilly C, MacDonald TY, et al. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin Cancer Res. 2014;20(18):4925–4934. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levillain O, Diaz JJ, Blanchard O, Dechaud H. Testosterone down-regulates ornithine aminotransferase gene and up-regulates arginase II and ornithine decarboxylase genes for polyamines synthesis in the murine kidney. Endocrinology. 2005;146(2):950–959. doi: 10.1210/en.2004-1199. [DOI] [PubMed] [Google Scholar]
- 43.Jariwala U, Prescott J, Jia L, Barski A, Pregizer S, Cogan JP, et al. Identification of novel androgen receptor target genes in prostate cancer. Mol Cancer. 2007;6:39. doi: 10.1186/1476-4598-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarado C, Beitel LK, Sircar K, Aprikian A, Trifiro M, Gottlieb B. Somatic mosaicism and cancer: a micro-genetic examination into the role of the androgen receptor gene in prostate cancer. Cancer Res. 2005;65(18):8514–8518. doi: 10.1158/0008-5472.CAN-05-0399. [DOI] [PubMed] [Google Scholar]
- 45.Boyd LK, Mao X, Lu YJ. The complexity of prostate cancer: genomic alterations and heterogeneity. Nat Rev Urol. 2012;9(11):652–664. doi: 10.1038/nrurol.2012.185. [DOI] [PubMed] [Google Scholar]
- 46.Wyatt AW, Mo F, Wang K, McConeghy B, Brahmbhatt S, Jong L, et al. Heterogeneity in the inter-tumor transcriptome of high risk prostate cancer. Genome Biol. 2014;15(8):426. doi: 10.1186/s13059-014-0426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McFarland CD, Korolev KS, Kryukov GV, Sunyaev SR, Mirny LA. Impact of deleterious passenger mutations on cancer progression. Proc Natl Acad Sci U S A. 2013;110(8):2910–2915. doi: 10.1073/pnas.1213968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156(6):1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 49.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012 May;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.