Abstract
The National Cancer Institute (NCI) Cancer Biomedical Informatics Grid® (caBIG®) program established standards and best practices for biorepository data management by creating an infrastructure to propagate biospecimen resource sharing while maintaining data integrity and security. caTissue suite, a biospecimen data management software tool, has evolved from this effort. More recently, the caTissue suite continues to evolve as an open source initiative known as OpenSpecimen. The essential functionality of OpenSpecimen includes the capture and representation of highly granular, hierarchically-structured data for biospecimen processing, quality assurance, tracking, and annotation. Ideal for multi-user and multi-site biorepository environments, OpenSpecimen permits role-based access to specific sets of data operations through a user-interface designed to accommodate varying workflows and unique user needs. The software is interoperable, both syntactically and semantically, with an array of other bioinformatics tools given its integration of standard vocabularies thus enabling research involving biospecimens. End-users are encouraged to share their day-to-day experiences in working with the application, thus providing to the community board insight into the needs and limitations which need be addressed. Users are also requested to review and validate new features through group testing environments and mock screens. Through this user interaction, application flexibility and interoperability have been recognized as necessary developmental focuses essential for accommodating diverse adoption scenarios and biobanking workflows to catalyze advances in biomedical research and operations. Given the diversity of biobanking practices and workforce roles, efforts have been made consistently to maintain robust data granularity while aiding user accessibility, data discoverability, and security within and across applications by providing a lower learning curve in using OpenSpecimen. Iterative development and testing cycles provide continuous maintenance and up-to-date capabilities for this freely available, open-access, web-based software application that is globally-adopted at over 25 institutions.
Keywords: biopecimen, tissue banks, specimen collection, biobanking, medical informatics, databases
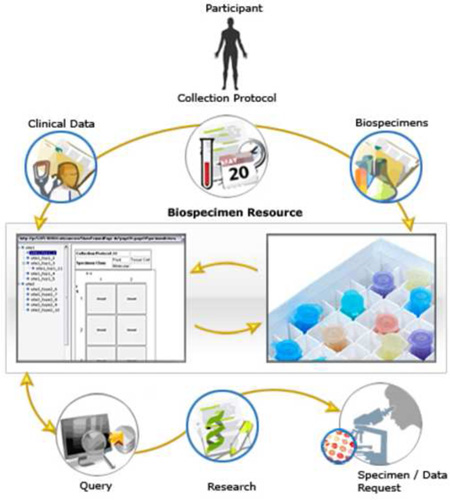
Graphical abstract
1. BACKGROUND AND SIGNIFICANCE
Resources for collecting, processing, storing, and tracking human biospecimens are vital to advances in molecular technologies, sophisticated clinical trial design, personalized medicine, and collaborative research.[1] Repositories at cancer and academic health centers collect and distribute diverse human biospecimens for molecular and genomic studies utilizing a wide variety of technology platforms[2]; thus to accommodate the evolving functional complexity of biospecimen resources, an informatics system is required.[3] However, many biospecimen resource centers have limitations, including: ad hoc, inadequate, or closed-source biospecimen banking/tracking informatics systems; large, sophisticated legacy systems that lack semantic and syntactic interoperability; and, non-modular informatics systems that cannot easily accommodate the expanding functionality of biospecimen resource centers.[4] Moreover, there is a need for different user classes (i.e. scientists, clinicians, biospecimen bank personnel) to access specific data types based upon their unique responsibilities. Though biospecimen resource centers have common functionality, each may have a unique workflow, levels of hierarchical data granularity, and end-user requirements.[1, 3]
Appreciating these deficits and the significance of biospecimens in research, the National Cancer Institute (NCI) formed standards and best practices for biorepositories creating a common infrastructure to propagate resource sharing while maintaining data integrity and security.[5] Toward this effort, the NCI constructed the Office of Biorepositories and Biospecimen Research (OBBR) in 2005 to: “i) Serve as the coordination and management center for biospecimen-related policies, practices, and other related issues across the NCI’s biorepositories; and, ii) Provide leadership for biobanking activities supporting all types of cancer research funded by the NCI.[6] While the OBBR developed the best practices directives, NCI also realized meeting the functional needs of the biospecimen resource community would require the development of informatics tools. For this, the NCI established the cancer Biomedical Informatics Grid (caBIG®) program, aimed at developing an open source, open access information technology infrastructure with a network of physicians, researchers, and patients within the cancer community.[7]
Guided by the Tissue Banks and Pathology Tools (TBPT) workspace, the OBBR guidelines, the caBIG® community, and the caGRID infrastructure,[8] researchers at seven institutions developed a database application for biospecimen management, caTissue Core. Two additional applications were built to meet natural language processing and clinical data annotation needs: i) The Cancer Text Information Extraction System (caTIES) automated coding, de-identifying, storing, and retrieving data from free-text pathology reports; and ii) The Clinical Annotation Engine (CAE) provided a web-based user-interface for standards-based manual annotation of biospecimens with clinical information. Since its initial conception, the caTissue project has evolved from this set of three complementary software applications into a comprehensive biospecimen information management system that has been adopted worldwide by cancer centers, academic and commercial entities and continues to be developed by the biospecimen informatics community. [9] The caTissue suite application has grown throughout multiple developmental releases based on NCI directives and the biospecimen user community input. Due to a shift from NCI-supported development to an open-source model of funding and development, caTissue underwent a community-elected name change to OpenSpecimen in September 2014.[10] The development efforts of caTissue suite and now with OpenSpecimen represent how accommodating the evolving functional complexity of biospecimen data and biobanking workflows can be effectively accomplished through an informatics system – one that leverages the varying needs of the biobanking community. To facilitate integration across other applications (e.g., clinical data management systems) and workflows, the informatics tool needs a capacity for interoperative flexibility. The caTissue/OpenSpecimen development trajectory described in this manuscript reveals how the support, effectiveness, and propagation such an application may offer the biospecimen community may be desirably enhanced when development efforts evolve with input from the community of users, is freely available, and customizable.
1.2. OBJECTIVES
The OpenSpecimen project provides a fundamental and extensible system design allowing researchers to comprehensively, electronically record and search for biospecimen data related to clinical and pathologic parameters across multiple biospecimen resources using a seamless and secure user-interface. The specific requirements for developing OpenSpecimen, guided by community users, include: using data element standards to develop an informatics system for biospecimen data storage, processing, quality assurance, tracking, and annotation; having the ability to store complex and hierarchical biospecimen tracking data with high granularity; creating a user-interface to accommodate varying workflows present at diverse biospecimen resource centers; allowing alternate functionality for specific user classes for rapid deployment at any biospecimen resource center; and the means to electronically interoperate with other informatics systems via an Application Program Interface (API). Most importantly, wider adoption and feedback with the biospecimen user community – through monthly calls, annual meetings, and on-line forums comprised of over 25 adopters across United States of America, Europe, Australia, Canada, Africa, and the Middle East – allow new enhancements to be developed and incorporated to create the latest release of OpenSpecimen. Thus, the product consumers drive the informatics tool development.
2. MATERIALS AND METHODS
2.1. Architecture
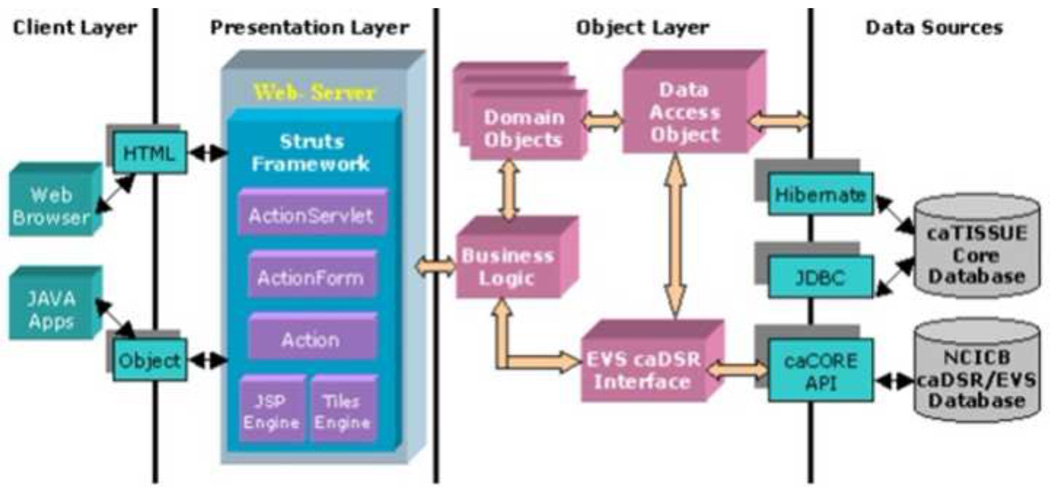
The architecture is that of a modern web-based application where each tier is optimized for speed, reliability and data integrity. This structure supports both application business and usability cases developed by the NCI and the greater biospecimen user community. OpenSpecimen comprises an n-tiered architecture: i) Client Layer consists of both a web-based UI and API containing HTML and JAVA applications passing data objects. These applications request operations on the server side. The Web browser and API are two client programs in OpenSpecimen application, ii) Presentation Layer provides a web interface, as well as support for the API. The Presentation Layer comprises a web server with a Struts Framework, ActionServlet, ActionForm, Action, JSP Engine and Tiles Engine, iii) Object Layer is the heart of the application and contains domain objects, model classes, and the data access layer. The Object Layer fulfills all tissue banking related business requirements. In the Object Layer, Domain Objects are passed to a Data Access Object.iv) Data Sources is the backend of the application and is a local database that stores all the tissue banking information. Hibernate and JDBC pass information to the database. Figure 1 depicts the n-tiered architecture described above
Figure 1.
Diagrammatic representation of OpenSpecimen architecture.
2.1.1. Technical Specifications
The software stack has been updated and can run on modern operating systems using proven, current technologies. The business logic is written in Java/J2EE, while the interaction with the database is performed using either Hibernate or JDBC based on expected performance requirements. Hibernate is used in cases where Java objects are to be inserted/updated in the database. JDBC is used in cases like the query interface where high throughput queries can be expected. The user interface is developed using JSP, JavaScript, Flex, and Ajax technologies. The Security infrastructure is built using the NCI CBIIT’s Common Security Module (CSM). The application program interface (API), to read and write data, is developed using the caCORE SDK toolkit with additional OpenSpecimen specific enhancements. Sharing of data on caGrid using caGrid Query Language (CQL) is also supported. OpenSpecimen installation is a command-line based process built using ANT scripts. The technology stack for latest OpenSpecimen is comprised of Apache 2.2, JBoss 5.1.0 GA, Ant 1.7.1, JDK 1.6, Windows or Linux server, and MySQL 5.1.× or Oracle 10g or Oracle 11g, and is compatible with Internet Explorer 9.0, Mozilla Firefox 8.0, and Safari 5.0. Hardware requirements include a least 4GB of RAM, 100 GB HDD, and Server class CPU. Additionally, OpenSpecimen is easily portable to other non-supported database (e.g., PostgreSQL or MS SQL), web server (e.g., Microsoft IIS) or browser with some minor coding adjustments.
2.1.2. Development
Two criteria are essential for enabling collaborators to share data: a transportation mechanism and understanding. To meet the desire for interoperability in biomedical research, the National Cancer Institute Center for Bioinformatics (NCICB) developed a cancer Common Ontologic Representation Environment (caCORE). Systems built using the caCORE paradigm, like OpenSpecimen, address two aspects of interoperability: the ability to access data (syntactic interoperability) and the ability to understand the data once retrieved (semantic interoperability) as described below. For further information concerning OpenSpecimen's development approach; testing methodology; programmatic access using caCORE, API, and caGrid; and security management refer to the resources listed in the bibliography and Appendix A.[11–13]
2.1.3. Data Model
OpenSpecimen is primarily modeled using the Unified Modeling Language (UML) with well-represented application semantics through a logical model in UML. The object model provides a description of object-oriented classes, attributes, associations, and eXtensible Markup Language (XML) tags. Due to adjustments in the OpenSpecimen database structure to preserve efficiency throughout iterative developments, a direct mapping between entity classes within the model to elements within database tables may present minor differences. However, the classes in the model conceptually represent database elements and such a mapping can be easily inferred. The OpenSpecimen data model follows the caCORE Software Development Kit (SDK) guidelines for naming classes, attributes, and associations. To facilitate semantic interoperability, OpenSpecimen data elements are annotated with standard vocabulary (NCI Metathesaurus) concepts and stored in the Cancer Data Standards Registry and Repository (caDSR).
2.1.4. Database Access and Audit Trail
Database access is achieved via the Data Access Object (DAO) layer. The DAO layer separates the business logic with the database interactions. The DAO layer also performs an audit of every operation including storing the user’s identifier, Identity Protocol address, timestamp, old value and new value of every data change.
3. RESULTS
Since the inception of caTissue in 2005 to its current status as OpenSpecimen, it has been designed as a generic, standardized, open-source system providing comprehensive functionality to track and annotate specimens and to search for biospecimens of interest for correlative science studies. As biobanking needs progressed, necessary enhancements to caTissue were identified and developed, and this practice continues in OpenSpecimen. Key features and functionalities; integration with other systems; and recent enhancements in OpenSpecimen are described below.
3.1. Features and Functionalities
Similar to many software development projects, there is a balance between meeting all end-user needs and making the software user-friendly. For electronic systems that support repository operational workflows, in general, we have learned it is of upmost significance that features around administration, privileges and security, data entry, and query be maximized for performance and usability. Systems that blend interoperability with ease of use will be accepted over ones that tend to favor either.
The features and functionalities described in this section are seen as essential to the biospecimen user community for facilitated workflow and research. Entry, interoperative distribution, tracking, and procurement of biospecimen data throughout OpenSpecimen is streamlined and secured by integrating unique capabilities within the administrative functions, data annotation, and access privileges.
3.1.1. Administrative Features and Functions
Managing biospecimen data robustly requires diverse features and functionalities, including:
3.1.1.2. Core data elements
The fundamental workflow involves tracking multiple biospecimens from the same patient or participant, creating and tracking refined materials (RNA, DNA, etc.) used for molecular analysis, and distributing biospecimens. The core data elements involved in managing the workflow are defined in Table 1.
Table 1.
Data elements used in OpenSpecimen.
| Data Element | Description |
|---|---|
| Participant | Participant is not necessarily a patient. Participant is an individual from whom biospecimens are collected for research purposes. |
| Participant Registration | Participants registered to a research study (collection Protocol) for collection of clinical and biospecimen information. |
| Specimen Collection Group | A group of one or more biospecimens collected at a single point in time from a Participant registered to a research study. Typically referred to as a case or accession in anatomic pathology and as an encounter event in a clinical study or trial. |
| Specimen | A circumscribed piece of tissue or body fluid collected from Participant at a particular point in time. |
| Specimen Events/Procedures | Any action performed on a biospecimen is referred to as an event/procedure. (e.g. transfer, review of the biospecimen, freeze, thaw, fixed, disposal, spun). |
| Specimen Array | A collection of biospecimens arranged in an ordered pattern (e.g. a 96 well PCR plate). |
| Storage Container | A physically discrete container used to store a biospecimen (e.g. freezer, shelf, rack, and box). |
| Distribution | The act of distributing, spreading, or apportioning based on a set of written procedures describing what, how many, and how much of a biospecimen or biospecimens will be utilized for research. Biospecimens may be collected under one collection protocol and then later utilized by single or multiple studies. |
3.1.1.3. Storage Containers
A storage container represents a physically discrete object used to store biospecimens, specimen arrays, or other containers. OpenSpecimen utilizes flexible and hierarchical storage representation to display biospecimen locations. Moreover, users can employ a storage type template during the creation of a new storage container, thus maintaining conformity among similar containers. Guidelines for storage containers make the storage system more organized, particularly when banking samples divided and stored by biospecimen type. For example, a storage container can be restricted to hold samples of a certain class (e.g. tissue, fluid, cell, or molecular) or type (e.g. fixed tissue, serum, cell pellet, DNA) and biospecimens procured under certain collection protocols.
3.1.1.4. Collection Protocol Creation
A collection protocol (CP) contains a set of guidelines delineating what, how, and when biospecimens are to be collected; hence, a CP contains the biospecimen collection schedule for all registered participants. Creating a CP provides the ability to define consent questions, time points, and biospecimen collected per time point, in addition to defining future derivatives and aliquots created from the collected biospecimens. The OpenSpecimen system uses this information to create anticipated biospecimens for each participant registration and builds a study calendar based on the anticipated biospecimens. This information can be used to predict workload for a specified future time period and generate missing biospecimen reports by querying for biospecimens past their anticipated collection date. Additionally, there is support for complex protocols such as studies with arms and phases.
3.1.1.5. Biospecimen Data Entry
Data captured are categorized in three ways: a) clinical data, b) pathology data and reports, and, c) quality assurance and availability data. Clinical data are represented as participants, collection protocols, and participant protocol identifier (PPI). The participant protocol identifier (PPI) is the study number assigned to each participant, uniquely identifying each person and their biospecimens in each study.
3.1.1.6. Data Entry Workflows
Biorepositories operate in diverse ways; thus, designing a single data management system is challenging. To account for multiple manual data entry scenarios, workflows are generalized so adopters can modify biorepository processes to create the most befitting environment. Biorepository staff can process biospecimens individually or in bulk, both methods requiring user action (e.g. add biospecimen events, transfer, distribute). The Bulk Operations functionality allows multiple biospecimen data to be loaded in aggregate rather than via a single record within the traditional data entry interface, thereby significantly decreasing data entry time. This function is akin to using a comma delimited file to load large amounts of data directly to a database. Users can download common ‘data templates’ that are available within the application. Data can then be pasted into the template and subsequently uploaded into OpenSpecimen. This feature can also be used for one-time loading of legacy data from simple spreadsheets or other data sources.
3.1.1.7. Specimen collection group (SCG)
Specimen collection groups (or time points) are regimented checkpoints throughout a clinical study during which specified biospecimen(s) are collected from a participant. Within the SCG are the biospecimens themselves (e.g. paraffin blocks, serum, plasma, frozen tissue) from which other biospecimens can be derived (e.g. whole blood to plasma) or aliquoted (e.g. four vials of plasma).
3.1.1.8. Distribution Protocol (DP)
The research study itself is a distribution protocol. Items registered in the DP include IRB number or exempt status, the principal investigator (PI), study title, and other standard study information. While the primary function of the DP allows the biospecimen resource staff to associate biospecimens with studies analyzing those specimens, the DP information is searchable by the research scientist. One can query data associated with the participant, pathology data associated with the SCG, and quality assurance and availability data associated with individual biospecimen.
3.1.1.9. Label/Barcode Generation and Printing
OpenSpecimen allows customized generation of labels and barcode for biospecimens. The contents of the label can be configured at the collection protocol level. Multiple barcode printers can be configured and OpenSpecimen can be configured to print on different types of labels (caps, slides, tubes), thus, enabling the essential feature of biospecimen tracking.
3.1.2. Data Annotation Features
Rich data annotation capabilities for informed consent status, clinical data (e.g. treatment, follow-up events), pathology data (based on College of American Pathologists organ site checklists),[14] and textual pathology reports for the study participant and biospecimens allow for robust biospecimen data collection. Default clinical annotations were deemed necessary at three levels: Participant, Specimen Collection Group, and Specimen (Table 2). Standard vocabularies (e.g., ICD9, LOINC, RxNorm) are used for enumerated data elements, modeled in UML and imported in the software.
Table 2.
Clinical annotations in OpenSpecimen.
| Participant | Specimen Collection Group | Biospecimen |
|---|---|---|
| Laboratory Tests | Solid Tissue Pathology | Solid Tissue Pathology |
| Family History | Hematology Pathology | Colorectal Specimen Pathology |
| Treatment/Treatment Regimen | Pancreas Pathology | Pancreas Specimen Pathology |
| Radiation Therapy | Prostate Needle Biopsy Pathology | Melanoma Specimen Pathology |
| Chemotherapy | CNS Pathology | CNS Specimen Pathology |
| Environmental Exposures | Transurethral Prostate Resection Pathology | Prostate Specimen Pathology |
| Alcohol Use | Melanoma Pathology | Kidney Specimen Pathology |
| Tobacco Use | Lung Biopsy Pathology | Lung Specimen Pathology |
| Health Examination | Lung Resection Pathology | Breast Specimen Pathology |
| New Diagnosis | Kidney Biopsy Pathology | |
| Recurrence – local/distant | Breast Pathology | |
| Radical Prostatectomy Pathology | ||
| Retropubic Enucleation | ||
| Pathology | ||
| Kidney Nephrectomy Pathology | ||
| Colorectal Local Excision | ||
| Pathology | ||
| Colorectal Excisional Biopsy | ||
| Pathology | ||
| Colorectal Resection Pathology |
3.1.2.1. Local Extensions: Dynamic Extensions/Clinical Annotations
The ability to extend the application, known as Dynamic Extension/Clinical Annotations, allows users to modify OpenSpecimen to meet study-specific requirements. This functionality permits data entry forms to be created on-the-fly to support annotating biospecimens or capturing additional clinical data. This allows different clinical domains (e.g. cancer vs. cardiovascular disease) to have tailored data collection within OpenSpecimen without specialized programming. For example, with the correct permissions, a user could electronically mimic a study case report form or create a new form to capture specimen biomarker testing results. With the OpenSpecimen Graphical User-interface (GUI), users can create forms by adding controls (e.g. text box, drop down), data validation rules, and other attributes for custom data. The forms can then be added to annotate a participant, specimen collection group, or specimen as appropriate.
3.1.3. Role-based Access Privileges
OpenSpecimen serves two key user groups: i) biorepository resource staff tracking and storing data about their biospecimen collection, inventory, and distribution, and ii) research scientists searching for biospecimens and associated data for translational research projects and requesting qualifying biospecimens for further study.
3.1.3.1. Roles
Users are assigned a ‘built-in’ role such as an administrator, supervisor, technician, or scientist, designating distinct permissions for data access (Table 3). The roles are customizable by adding or removing privileges based on the required activity.
Table 3.
User roles in OpenSpecimen.
| Role | Description |
|---|---|
| Super Administrator | A Super Administrator can perform any operation within the application. |
| Administrator | An Administrator registers new users, containers, and collection protocols and distribution protocols into the system, typically within a single physical biorepository. |
| Supervisor | A Supervisor adds participants and registers them to collection protocols. |
| Technician | A Technician adds biospecimens into the system. |
| Scientist | A Scientist performs a query and orders biospecimens and biospecimen arrays of interest. |
| Custom | The Custom role allows selected privileges to be assigned to a user. |
Access is controlled with individual user-group assigned privileges further controlled at the object, record, and attribute levels and defined within independent biospecimen repository sites. Moreover, role-based access allows individual users to have assigned privileges based upon their affiliation with an individual biorepository or their involvement in a particular collection protocol (study), independent of actual physical location. Hence, users at different institutions involved in the same study can have the same privileges assigned.
3.1.3.2. Acquiring and Sharing Data and Biospecimen
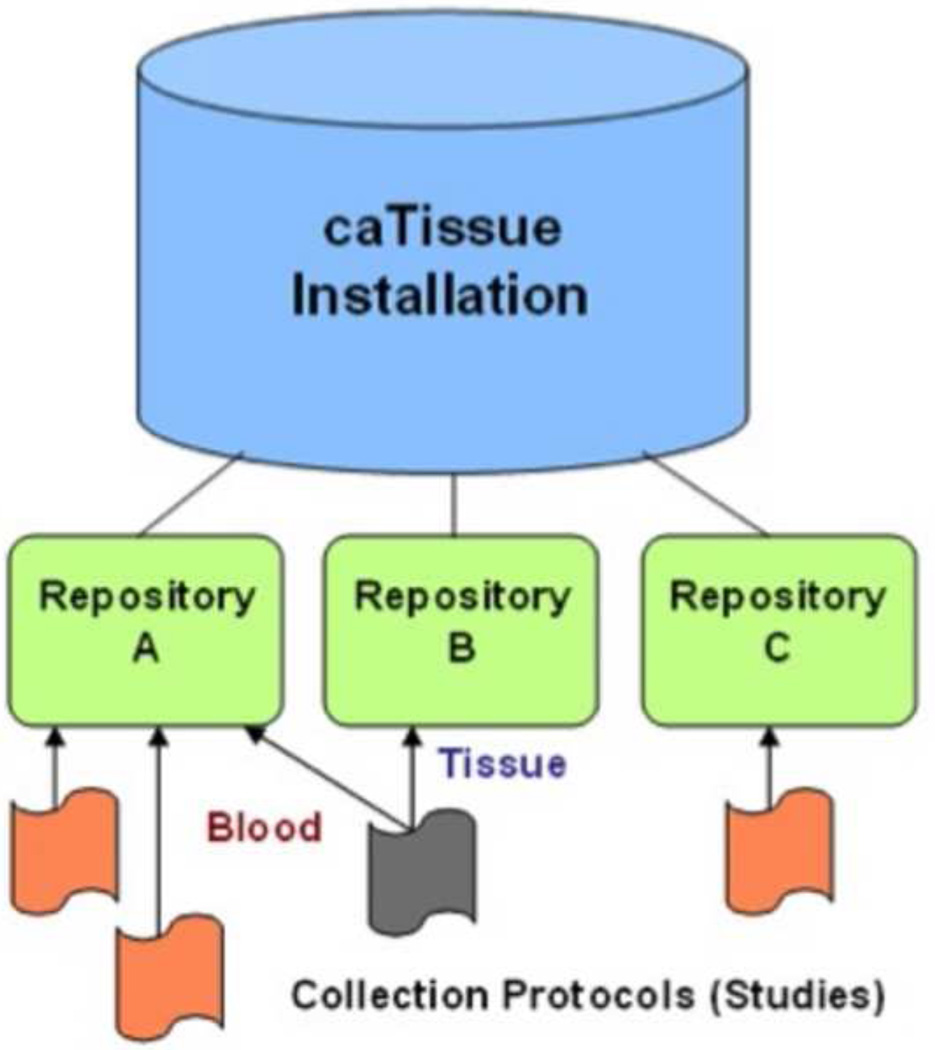
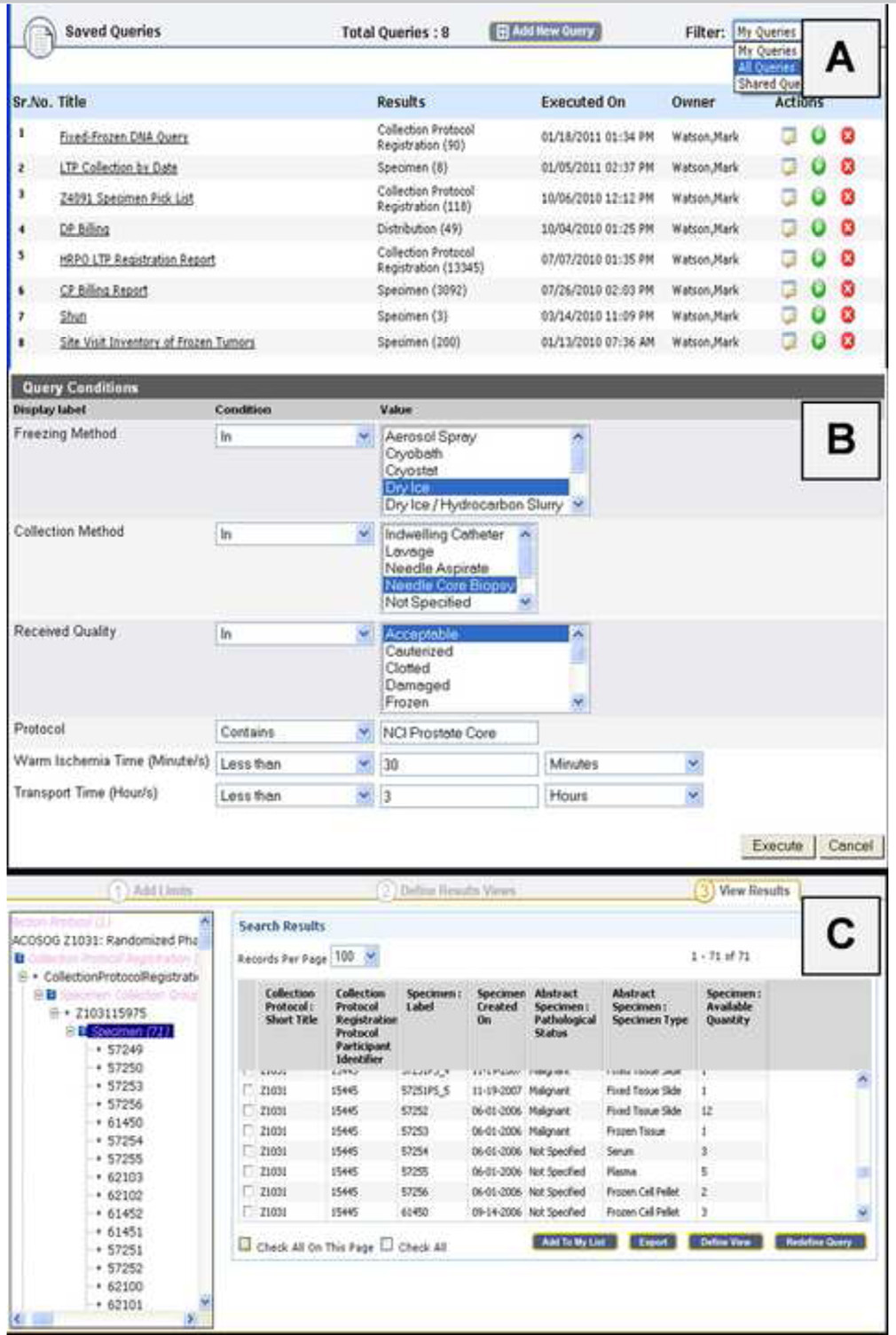
Physical biospecimen repositories are represented as logically separated entities generating optimized user experiences based on the assigned role (Figure 2). Users can create and save custom parameterized queries to view and export specific sets of data filtered for participant privacy in compliance with HIPAA guidelines and based on user privileges (Figure 3 (A–C)).
Figure 2.
Biospecimen repositories are logically separate entities.
Figure 3.
Users create and save parameterized queries (A); hence, queries can be reused by altering selected attributes (B). Results are presented and available to export (C) based on participant consent and user privileges.
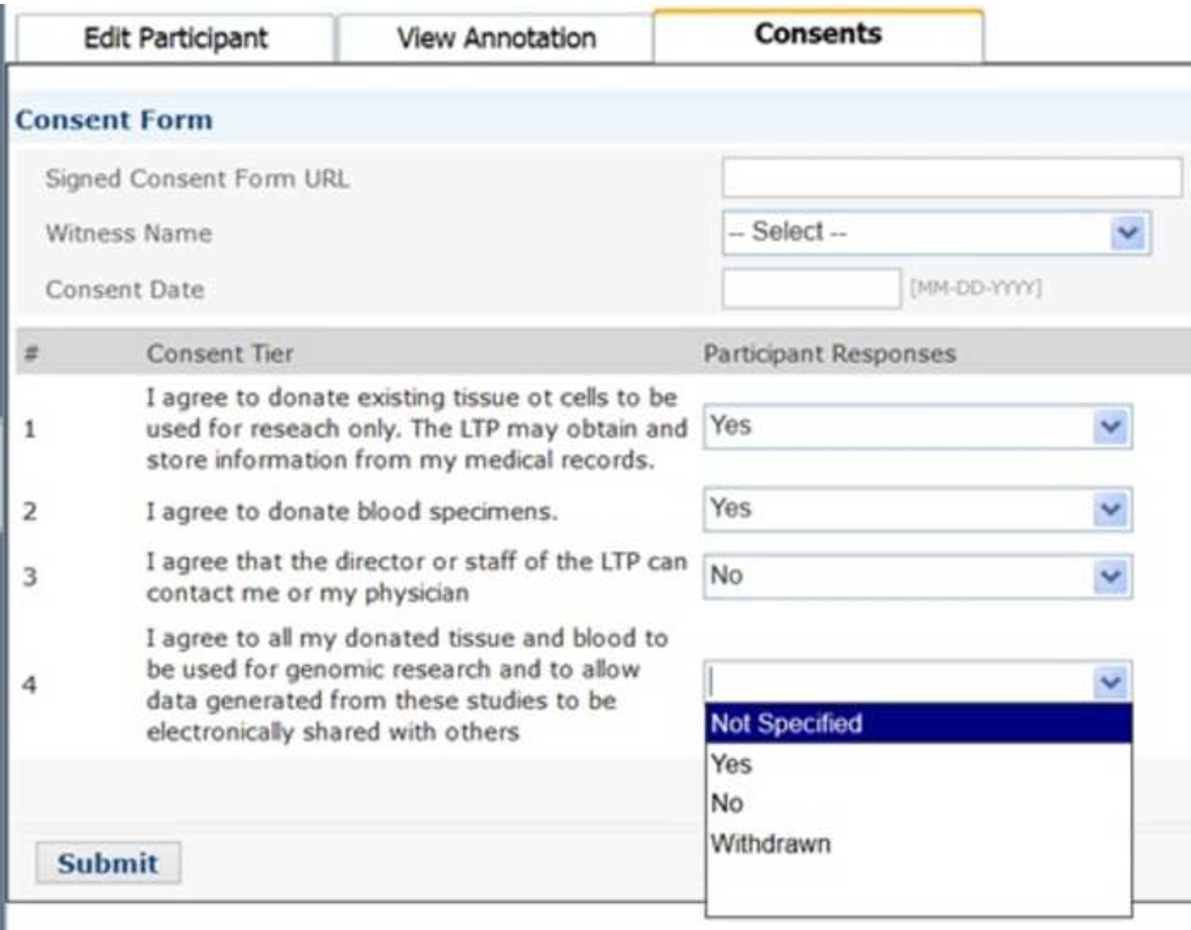
OpenSpecimen permits a scientist to query for biospecimens of interest and, when needed, place orders for those biospecimens or biospecimen arrays. Biorepository staff can view the order list, process the distribution, and generate a report, which can be exported in a comma-delimited format to use for analyses and integration with billing or other informatics tools. Participant consent responses, if recorded in the application, are displayed for the user to confirm prior to distribution (Figure 4). Users can then send and receive biospecimens from one site to another via the built-in shipping and tracking functionality. The biospecimens are placed in an electronic shipment, with a unique shipment identifier and barcode. The system tracks the shipments’ movement between the sites and sends email notifications to appropriate personnel during the processing cycle (e.g. order, in-transit, receipt).
Figure 4.
Consent Tiers tracks the specific participant permissions for sharing of the biospecimen in current and future research.
3.2. Integration with Other Systems
OpenSpecimen’s capability to integrate with other systems facilitates biorepository operations such as billing or regulatory compliance (e.g. audit or QA data) in addition to methods in biospecimen research (e.g., phenotype-genotype studies). Interoperability with other systems is achieved through the OpenSpecimen API. As previously referenced, caCORE compliant systems achieve syntactic and semantic interoperability, while non-caCORE compliant external systems can write and read data from OpenSpecimen but may lose semantics. Surgical pathology reports can be imported, and the system can create or programmatically match to participants in OpenSpecimen using the Mirth Connect, which queues HL7 messages to load the reports.[15]
To facilitate data interoperability, a Clinical Data Management System (CDMS) storing study participant clinical data can be used to populate participant demographics and extract biospecimen details. For example, ClinPortal, a custom CDMS developed by Washington University, is integrated with OpenSpecimen in both the application presentation and data layers. Via this integration, users may seamlessly traverse both applications with a single account, entering and accessing participant specific clinical study data and biospecimen data.
Research patient data warehouses can likewise extract data from OpenSpecimen to give the research-scientist access to inpatient and outpatient data along with corresponding biospecimens thus facilitating rapid application of advances in science to improvements in patient care.
The OpenClinica and EPIC applications support participant registration to clinical studies within OpenSpecimen. Both of these applications are separate from, yet interoperable with OpenSpecimen. To facilitate integration across clinical trial and biorepository systems and workflows, a participant registered in OpenClinica should automatically be registered in the corresponding Collection Protocol in OpenSpecimen. In the EPIC integration, subjects are registered to studies through the EPIC web interface, and EPIC broadcasts a registration notification including key identifiers of the participant. For clinical trials containing a biospecimen collection component, studies in OpenClinica link to collection protocols in OpenSpecimen through the SHMi hub (Secure HL7 Mirth Interface Interface), a toolkit specifically developed to integrate various applications with OpenSpecimen via HL7 messaging. SHMi is our messaging Hub that receives and sends messages between OpenClinica, EPIC, and OpenSpecimen.[16, 17] When a subject is registered or updated in EPIC or OpenClinica, the registration or notification of a change to the patient’s data broadcasts through SHMi hub to OpenSpecimen and this participant will be registered to the associated collection protocol. When a patient is registered to a study, the message conveys the consent, signature date, and registration date. OpenSpecimen will interoperate with both EPIC and OpenClinica in a number of different scenarios including: becoming aware of certain initiated events; obtaining data related to the aforementioned events; and executing its own workflow on the data obtained.
3.3. Enhancements to Address Lessons Learned
Throughout the development of caTissue and OpenSpecimen, efforts have been made consistently to maintain robust granularity within the structure of the data while easing the access to the information. A key example of a lesson learned pertains to difficulties in query construction. Originally, query development needed a vast understanding of the data model, so this was addressed by adding the capability of storing queries for later use. This feature allows an individual who is knowledgeable of caTissue's object model and the application's functionality to create a "canned" query for use by others. Furthermore, the ability to specify query parameters was also added. To satisfy the need for researchers to easily discover and access biospecimens without advanced understanding of the biobanking process or database structure, which they were still unable to achieve, enhancements have been made to the query functionality within the latest release of OpenSpecimen.[18, 19]
These changes were not only cosmetic but also in line with the ‘bigger picture’ of aiding user accessibility and discoverability within and across applications by providing a lower learning curve. These enhancements in application approachability aid in exporting data from the application more readily to expedite research and clinical efforts in addition to collaborative use of the data across institutions. Such alterations to the application were in line with an overall shift in development perspective and aim of improving the user community’s ability to enter, access, track, and share both their data and biospecimens more readily within the application. This shift in development altered the focus of OpenSpecimen as an application to store and track biospecimen data to an application that can aid researchers in flexibly viewing and exporting data across applications to thus offer a more robust and malleable data resource.
Attaining such interoperability and bolstering of robust data sources through integration across systems represents a further developmental shift away from creating an exclusively unified application aimed to be adopted as a single closed package. Instead, aims are shifting towards an open application providing flexible interoperability with diverse systems and plugins, a more amenable interface and workflow, and freely available source-code to spark adoption, further innovation, and expansion within the biobanking community.
The inclusion of dynamic form extensions within caTissue Suite has evolved and expanded into the ability to generate custom forms within OpenSpecimen.[20] Through feedback from community adopters, the ability to expand default specimen events and collection procedures – such as freeze, spun, thaw – to adapt for the inclusion of unique events for emerging research. Users may create new dynamic events using the custom form builder and make it available and queryable for all data entry for specimens of all collection events. Thus, a research initiative need not sacrifice accuracy and complexity of documentation within collected biospecimen data to match default form values. Additional adaptations to lessons learned and concerns reported by the community include expansions of structured vocabulary to support SNOMED coding of tissue anatomic site, in addition to enhancements to encoding familial relationships and a streamlining of the shipping/distribution workflow interface.
As we transition from an NCI supported initiative to an open-source community driven initiative, we have also learned that one needs an initial driver and form of governance to direct the trajectory of the application’s development. We currently operate under the direction of a beneficent oligarchy wherein a community board comprised of research technicians, biorepository managers, application developers, research specialists, and data scientists review the application enhancements proposed by users within the End User Committee. End users are free to share their day-to-day experiences in working with the application, thus providing to the community board insight into the needs and limitations which need to be addressed. Users are also requested to review and validate new features through group testing environments and mock screens. End users are further engaged through monthly webinars, application demonstrations, and yearly face-to-face meetings during which they can report questions, concerns, or suggestions. Proposed enhancements and expansions are evaluated for feasibility, overall efficacy, and the expected scale of community adoption during monthly phone calls between the community board members. Approved enhancements are then put into the development queue for build and testing.
4. DISCUSSION
OpenSpecimen is a freely available, open-access, web-based software application allowing for the storage, inventory tracking, annotation, and retrieval of biospecimen data from a relational database. It permits biorepository resource staff to track the collection, storage, and distribution of biospecimens; monitor data and biospecimens for quality assurance; as well as derive and aliquot new biospecimens from existing ones (e.g. for nucleic acid analysis). It may be used by biorepository facilities regardless of the nature of biospecimen transactions occurring or the type of biospecimens involved in the transaction. Furthermore, research scientists can search for and request biospecimens to be used in translational studies.
As compared with other systems,[21–24] OpenSpecimen offers: developed, documented, and tested software using principles of commercial software development while remaining freely available for a variety of biorepository environments and workflows; open and documented API leveraged for interoperation with other systems; freely modifiable code base with documented instructions; and, a large and growing academic and commercial user community.
OpenSpecimen is developed and improved to satisfy continuous feedback from its growing global user community. Currently, nineteen repositories are represented in OpenSpecimen within a single database at Washington University School of Medicine, representing research in cancer, Alzheimer's, gastro-intestinal (GI), renal, cardiac and other disease groups. OpenSpecimen has been adopted by more than forty academic and industry institutions in the US, Australia, Europe, China and India including Washington University, Indiana University, Thomas Jefferson University, Yale University, Emory University, Louisiana Cancer Research Consortium (LCRC), Oregon Health & Science University, University of New South Wales, Vanderbilt University, and Cancer Research Center of Hawaii, and Ohio University. Technical and end-user documentation, training modules, discussion forums, FAQs, and application updates for OpenSpecimen have been supported through the Krishagni OpenSpecimen Support Center and community forum.[25–27] More support information can be found on-line in Appendix B.
5. CONCLUSION
Use of the software in daily biorepository operations by several NCI Cancer Centers and other biospecimen resource groups is providing a rapid and facilitated path toward standardizing biospecimen informatics and promoting biospecimen data sharing both nationally and globally. OpenSpecimen is sufficiently scalable and configurable for broad deployment across biorepositories of varying size and function.
OpenSpecimen is a freely available, supported, open-access software application for biobanking management systems, thus providing the only known free and open-source biospecimen informatics tool available. This has moved the biospecimen informatics community forward through growing from an NCI directed initiative to a community-based initiative to enhance biospecimen software. The user community can work to clarify enhancement requests, verify bugs, and prioritize fixes for inclusion in open source development cycles. Community code contributions are available for download and provide a way for developers to share locally developed code with the broader community of users. As we move forward with the Open Source Development Initiative the biopecimen community will be more empowered to set direction and goals for future enhancements and other application development.
Supplementary Material
Highlights.
Flexibly-tiered application accommodates diverse adoption and user requirements
High-throughput entry and display of robust, hierarchical biospecimen data
Focused development to meet workflow and research needs reported by user feedback
Scalable for multi-site collection, processing, tracking, and quality assurance.
OpenSpecimen evolved from NCI-developed biobanking standards and best practices.
Acknowledgments
OpenSpecimen and this manuscript have benefited from the work and insight of Amy Brink, Vicky Holtschlag, and the development teams at Persistent Systems Limited and Semantic Bits. Thomas Jefferson, Emory, and Yale universities were instrumental in the application development process as was Ian Fore, program manager at the National Cancer Institute (NCI). We acknowledge the entire OpenSpecimen user community for their input on OpenSpecimen requirements and functionality. We additionally wish to thank and acknowledge Srikanth Adiga for his assistance in preparation of this manuscript and for his continued effort in collaboration with Poornima Govindrao and the entire team of Krishagni Solutions Pvt Ltd to maintain adoption and developmental support of OpenSpecimen.
Funding: Funding was provided through the Department of Health and Human Services (DHHS), grant HHSN261200800001E, with contracts from the NCI contracts 29XS203ST and 28XS205, and Science Applications International Corporation (SAIC) contract 29XS203.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests: All authors from Washington University School of Medicine declare no competing interests with this work.
BIBLIOGRAPHY
- 1.Vaught J, Rogers J, Myers K, et al. An NCI perspective on creating sustainable biospecimen resources. Journal of the National Cancer Institute Monographs. 2011;2011(42):1–7. doi: 10.1093/jncimonographs/lgr006. [DOI] [PubMed] [Google Scholar]
- 2.Hewitt RE. Biobanking: the foundation of personalized medicine. Current opinion in oncology. 2011 Jan;23(1):112–119. doi: 10.1097/CCO.0b013e32834161b8. [DOI] [PubMed] [Google Scholar]
- 3.Blow N. Biobanking: freezer burn. Nature methods. 2009;6(2):173–178. [Google Scholar]
- 4.Eder JDC, Schicho M, Stark K. Transactions on Large-Scale Data-And Knowledge-Centered Systems I. Berlin, Heidelberg: Springer Berlin Heidelberg; 2009. Information System for Federated Biobanks; pp. 156–190. [Google Scholar]
- 5.U.S. Department of Health and Human Services NIoH, Office of Biorepositories and Biospecimen Research, National Cancer Institute. NCI Best Practices for Biospecimen Resources. 2007 [Google Scholar]
- 6.Office of Biorepositories and Biospecimen Research (OBBR) [08/14/2013];2007 Available from: http://cabig.cancer.gov/action/nci/obbr/
- 7.caBIG Mission, Goals, and Principles. [Jan 1, 2013]; Available from: https://cabig.nci.nih.gov/overview.
- 8.CaGrid 1.4 Deprecation Plan: Oster caGrid 1.4. [Jan 01, 2013]; Available from: http://cagrid.org/display/caGrid14/Deprecation+Plan. [Google Scholar]
- 9.Institue NC. caTissue Suite 1.0. [12/02/2013 [04/25/2014]];2013 Available from: https://wiki.nci.nih.gov/display/caTissue/caTissue+Suite+1.0#caTissueSuite10-WhatsNew.
- 10.Solutions K. OpenSpecimen: Revolutionary Biobanking Informatics. [03/02/2015]; Available from: http://www.openspecimen.org/ [Google Scholar]
- 11.Overview for Technical Administration Guide of caTissue Suite v2.0. [08/14/2013]; Available from: https://wiki.nci.nih.gov/display/caTissuedoc/1+-+Overview+for+Technical+Administration+Guide+of+caTissue+Suite+v2.0#1-OverviewforTechnicalAdministrationGuideofcaTissueSuitev2.0-caTissueSuiteArchitecture.
- 12.Programmatic access using caCORE,API, and caGrid. [08/14/2013]; Available from: https://wiki.nci.nih.gov/display/caTissuedoc/3+-+API+Access+v2.0.
- 13.Security Management v2.0. [08/14/2013]; Available from: https://wiki.nci.nih.gov/display/caTissuedoc/2+-+Security+Management+v2.0.
- 14.CAP. Cancer Protocols and Checklists. [08/13/2015]; Available from: http://www.cap.org/apps/cap.portal?_nfpb=true&cntvwrPtlt_actionOverride=%2Fportlets%2FcontentViewer%2Fshow&_windowLabel=cntvwrPtlt&cntvwrPtlt{actionForm.contentReference}=committees%2Fcancer%2Fcancer_protocols%2Fprotocols_index.html&_state=maximize d&_pageLabel=cntvwr. [Google Scholar]
- 15.Ltd KSP. Index of Releases. [03/02/2015];2014 Available from: https://catissueplus.atlassian.net/wiki/display/CAT/OpenSpecimen+-+SPR+Integration+via+HL7. [Google Scholar]
- 16.Ltd KSP. OpenSpecimen - EPIC Integration. [03/02/2015]; Available from: https://catissueplus.atlassian.net/wiki/display/CAT/OpenSpecimen+-+EPIC+Integration. [Google Scholar]
- 17.Ltd KSP. OpenSpecimen - OpenClinica Integration. [08/13/2015]; Available from: https://catissueplus.atlassian.net/wiki/display/CAT/OpenSpecimen+-+OpenClinica+Integration. [Google Scholar]
- 18.Ltd KSP. What's New in OpenSpecimen 1.0-Advanced Query Construction. [03/02/2015]; Available from: https://catissueplus.atlassian.net/wiki/display/CAT/OpenSpecimen#OpenSpecimen-AdvanceQueryEnhancements. [Google Scholar]
- 19.Jack W, London PDC., Phd Using the semantically interoperable biospecimen repository application, caTissue end user deployment lessons learned. [03/02/2015];2012 Available from: https://wiki.nci.nih.gov/display/TBPTKC/Thomas+Jefferson+University.
- 20.Ltd KSP. What's New in OpenSpecimen - Custom Forms. [03/04/2015]; Available from: https://catissueplus.atlassian.net/wiki/display/CAT/OpenSpecimen#OpenSpecimen-CustomForms. [Google Scholar]
- 21.OnCore Medical Solutions. date accessed; Available from: http://www.oncorems.com/markets/medical.php. [Google Scholar]
- 22.LabVantage Laboratory Knowledge. [08/14/2013]; Delivered. Available from: http://www.labvantage.com/services/laboratory-strategy.aspx. [Google Scholar]
- 23.Freezerworks Sample Management Software. [08/13/2015]; Available from: http://www.freezerworks.com/ [Google Scholar]
- 24.Cryotrack Inventory Management System features. [08/14/2013]; Available from: http://www.cryotrack.com/ [Google Scholar]
- 25.Ltd KSP. OpenSpecimen Community Forums. [03/02/2015]; Available from: http://forums.openspecimen.org/ [Google Scholar]
- 26.Ltd KSP. Community Center. [03/02/2015]; Available from: https://catissueplus.atlassian.net/wiki/display/CAT/Community+Center. [Google Scholar]
- 27.Ltd KSP. Krishagni's OpenSpecimen Support Center: Everything you wanted to know about OpenSpecimen and more. [08/13/2015]; Available from: https://catissueplus.atlassian.net/wiki/display/CAT/Home. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.