Systematic imaging studies and modeling reveal new details of the regulation of the Gq-coupled GPCR, protease-activated receptor 2, by phosphorylation and β-arrestin.
Abstract
Activated Gq protein–coupled receptors (GqPCRs) can be desensitized by phosphorylation and β-arrestin binding. The kinetics and individual contributions of these two mechanisms to receptor desensitization have not been fully distinguished. Here, we describe the shut off of protease-activated receptor 2 (PAR2). PAR2 activates Gq and phospholipase C (PLC) to hydrolyze phosphatidylinositol 4,5-bisphosphate (PIP2) into diacylglycerol and inositol trisphosphate (IP3). We used fluorescent protein–tagged optical probes to monitor several consequences of PAR2 signaling, including PIP2 depletion and β-arrestin translocation in real time. During continuous activation of PAR2, PIP2 was depleted transiently and then restored within a few minutes, indicating fast receptor activation followed by desensitization. Knockdown of β-arrestin 1 and 2 using siRNA diminished the desensitization, slowing PIP2 restoration significantly and even adding a delayed secondary phase of further PIP2 depletion. These effects of β-arrestin knockdown on PIP2 recovery were prevented when serine/threonine phosphatases that dephosphorylate GPCRs were inhibited. Thus, PAR2 may continuously regain its activity via dephosphorylation when there is insufficient β-arrestin to trap phosphorylated receptors. Similarly, blockers of protein kinase C (PKC) and G protein–coupled receptor kinase potentiated the PIP2 depletion. In contrast, an activator of PKC inhibited receptor activation, presumably by augmenting phosphorylation of PAR2. Our interpretations were strengthened by modeling. Simulations supported the conclusions that phosphorylation of PAR2 by protein kinases initiates receptor desensitization and that recruited β-arrestin traps the phosphorylated state of the receptor, protecting it from phosphatases. Speculative thinking suggested a sequestration of phosphatidylinositol 4-phosphate 5 kinase (PIP5K) to the plasma membrane by β-arrestin to explain why knockdown of β-arrestin led to secondary depletion of PIP2. Indeed, artificial recruitment of PIP5K removed the secondary loss of PIP2 completely. Altogether, our experimental and theoretical approaches demonstrate roles and dynamics of the protein kinases, β-arrestin, and PIP5K in the desensitization of PAR2.
INTRODUCTION
Here, we study the early steps of desensitization of a Gq protein–coupled receptor (GqPCR), protease-activated receptor 2 (PAR2). These steps turn off receptor signaling despite the presence of an activating ligand. Such desensitization can avert excessive responses and minimize potential cytotoxic effects of their intracellular messengers. However, the mechanisms responsible for GPCR desensitization are not fully described for many GPCRs and need not always be the same.
GPCRs are diverse membrane proteins that transfer environmental information to the cell interior. They recognize photons and extracellular ligands, including neurotransmitters and hormones to generate intracellular signals. Historically, rhodopsin has been a model to study receptor desensitization; it can be desensitized in 1 s after the absorption of a single photon through a cascade involving receptor phosphorylation by a G protein–coupled receptor kinase (GRK) followed by binding of visual arrestin to the phosphorylated receptor (Ranganathan and Stevens, 1995; Kirchberg et al., 2011). Desensitized rhodopsin does not become internalized. Another well-studied GPCR is the Gs-coupled β2-adrenergic receptor. It generates cAMP and regulates many cAMP-dependent pathways (Lefkowitz et al., 2000; Violin et al., 2008; Guellich et al., 2014). Ligand binding to β2-adrenergic receptors promotes homologous phosphorylation by GRK, which enhances binding of β-arrestin to the phosphorylated receptor. β-Arrestin binding to the ligand-occupied and phosphorylated receptor is a rate-limiting step for homologous desensitization (Dixon et al., 1986; Luttrell and Lefkowitz, 2002; Rasmussen et al., 2011). The arrestin complex promotes internalization of the receptor by endocytosis (Shenoy and Lefkowitz, 2011). For other less well-studied GPCRs, such as GqPCRs, it has been natural to assume that they may use the same mechanisms with β-arrestin binding to homologously phosphorylated receptors for desensitization and internalization, but for many receptors that hypothesis is not completely tested.
Desensitization of GqPCRs has been investigated previously (Soh et al., 2010; Heitzler et al., 2012; Ramachandran et al., 2012; Bojjireddy et al., 2015), but several fundamental questions remain. For example, the separate contributions of phosphorylation, β-arrestin binding, and recycling of the receptors to terminate the receptor activity are not well defined. The PAR family is an attractive model system to address these questions because of its unique mechanism of receptor activation. They are cellular sensors for extracellular proteolytic enzymes, including those released during inflammation. When the extracellular N terminus of the receptor is cleaved by proteases such as thrombin or trypsin, a tethered ligand sequence is revealed that binds to the receptor binding pocket for activation (Ramachandran et al., 2012). Thus, the receptors are irreversibly activated by this cleavage; they then need to be desensitized and internalized to minimize potential cellular damage caused by protracted hydrolysis of plasma membrane (PM) phosphatidylinositol 4,5-bisphosphate (PIP2) and to permit signaling from other GqPCRs that share the same cellular messengers. Previous work suggested that PAR2 is shut off by two mechanisms: desensitization and internalization (Böhm et al., 1996; Ricks and Trejo, 2009). The latter step retrieves the receptors via dynamin- and clathrin-dependent endocytosis for permanent shutdown of the signaling (Ricks and Trejo, 2009; McMahon and Boucrot, 2011). Again, the first step is the phosphorylation of receptors by a kinase. Because PAR2 signaling generates diacylglycerol (DAG) and activates PKC, the literature has emphasized PKC more than GRK as the desensitizing kinase. It is supposed that the phosphorylated receptor, after binding to β-arrestin, cannot interact with Gq anymore. However, it is still not clear whether phosphorylation desensitizes the receptors by itself or whether the binding of β-arrestin to receptor is necessary for the desensitization.
Here, we address these questions by using fluorescent probes to monitor several intracellular signals from PAR2 optically. These include monitoring PM PIP2, DAG, inositol trisphosphate (IP3), and Ca2+. In addition, we manipulate the level of β-arrestin with siRNA to determine the role of β-arrestin in PAR2 desensitization. Mathematical modeling is used to verify the key steps in a quantitative way. Our results support the view that the phosphorylation of PAR2 leads to receptor desensitization even without binding of β-arrestin, but subsequent binding of β-arrestin to the receptor is key to keeping the phosphorylated state from being dephosphorylated and reactivated by serine/threonine phosphatases.
MATERIALS AND METHODS
Cell culture
tsA201 cells are transformed HEK293 cells (Sigma-Aldrich). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin and subcultured every 3–4 d using 0.05% trypsin containing EDTA. Nontransformed pancreatic duct epithelial cells (PDECs), a cell line originally derived from the main pancreatic duct of a dog (Oda et al., 1996), were cultured on Transwell inserts (Corning) coated with collagen (Advanced BioMatrix Inc.) over a feeder layer of confluent human gallbladder myofibroblasts (Oda et al., 1996). For intracellular Ca2+ measurement, the single PDECs were plated on 5-mm small glass chips coated with Vitrogen. The tsA201 cells were plated on small chips coated with poly-l-ornithine for Ca2+ imaging and confocal experiments. All experiments were performed at room temperature (22–24°C).
Chemicals
PMA was purchased from Tocris Bioscience; bisindolylmaleimide I (BIS I), BIS V, and calyculin A from EMD Biosciences; rapamycin from LC Laboratories; PAR2 activating peptide (AP; N-SLIGKT-C) from United Biosystems Inc.; Compound101 (Lowe et al., 2015) from Hello Bio Limited; and Trypsin (T8802) from Sigma-Aldrich. The stocks of PMA (1 mM), compound101 (30 mM), BIS I (1 mM), BIS V (1 mM), rapamycin (5 mM), and calyculin A (1 mM) were dissolved in DMSO. AP (10 mM) and trypsin (500 µM) were reconstituted into distilled water.
Transfection of cDNA
tsA201 cells were transfected with 0.5 µg of several cDNA constructs except for 0.3 µg of the pleckstrin homology (PH)–red fluorescent protein (RFP) probe. 10 µl X-tremeGENE 9 (Roche) and cDNA were added to Opti-MEM solution (∼87 µl) sequentially. For transfection, cells at ∼75% confluency in a 35-mm culture dish were washed with Opti-MEM and supplemented with 0.5 ml Opti-MEM solution. Then the mixture of the transfection reagent and ∼100 µl cDNA was added into the culture dish. We incubated the cells for at least for 4 h. Finally, cells were detached with trypsin (0.05%), transferred onto small 5-mm glass chips coated with 1 mg/ml poly-l-ornithine, and incubated for 1 d before experiments. LIBRA version III (LIBRAvIII), PH-RFP, human PAR2-GFP, and C kinase activity reporter (CKAR) were provided by A. Tanimura (Health Sciences University of Hokkaido, Tobetsu, Japan), C.S. Kearn (University of Washington, Seattle, WA), N.W. Bunnett (Monash Institute of Pharmaceutical Sciences, Parkville, VIC, Australia), and J.D. Scott (University of Washington), respectively. Dark PAR2 and cyan fluorescent protein (CFP)–tagged PAR2 were subcloned into pcDNA3.1 vector (Invitrogen) and pcDNA3-CFP vector (Addgene), respectively, and the citrine-tagged C1 domain of PKCγ (C1-citrine) was generated from C1A(PKCγ)-EGFP provided by T. Meyer (Stanford University, Stanford, CA; Oancea et al., 1998). The PH-RFP (PIP2) probe that was used for most of experiments has specificity to PIP2 at the PM (van der Wal et al., 2001; Horowitz et al., 2005). LIBRAvIII has ∼500 nM affinity for IP3 (Tanimura et al., 2009; Dickson et al., 2013). DAG-binding C1-citrine has specificity for DAG and DAG analogues like phorbol esters (Oancea et al., 1998). The cytosolic CKAR probe monitors activity of PKC (Violin et al., 2003).
For the recruitable phosphatidylinositol 4-phosphate 5 kinase (PIP5K) experiment using rapamycin in Fig. 10, cells were transfected with the plasmids for cytosolic PIP5K-FKBP-CFP and PM anchoring Lyn11-FRB-CFP (Suh et al., 2006). Rapamycin induces heterodimerization between FK506-binding protein (FKBP) and FRB, translocating the PIP5K to the PM. β-Arrestin 2–yellow fluorescent protein (YFP) was obtained from M.J. Lohse (University of Würzburg, Würzburg, Germany).
Figure 10.
A role for PIP5K in the delayed PIP2 hump with β-arrestin knockdown cells. (A) Recruitment of PIP5K using the rapamycin system. Cells were transfected with β-arrestin 1/2 siRNA and then transfected with PIP5K-FKBP-CFP, Lyn11-FRB-CFP, PAR2-GFP, and PH-RFP. The kinase PIP5K-FKBP-CFP translocated from cytoplasm to PM during treatment with 5 µM rapamycin and AP (red symbols; n = 6). However, the kinase did not translocate during treatment with AP alone (black symbols; n = 6). (B) PIP5K translocation to the PM restores recovery of PIP2 in β-arrestin knockdown cells. Symbols indicate the effect of AP without (black) or with (red) PIP5K recruitment using rapamycin (for both cases, n = 6). Error bars are SEM.
Knockdown of β-arrestins using siRNA
To knock down β-arrestin 1 and 2, we used chemically synthesized double-stranded siRNAs with 19-nt duplex RNA and 2-nt 3′ dTdT overhangs purchased from GE Healthcare. The sense (target) sequences in β-arrestin 1 (NM_020251) and β-arrestin 2 (NM_004313) are 5′-AAAGCCUUCUGCGCGGAGAAG-3′ and 5′-AAGGACCGCAAAGUGUUUGUG-3′, respectively. For control siRNA, we used 5′-AAGUGGACCCUGUAGAUGGCG-3′ (Ahn et al., 2003). For the siRNA experiment, tsA201 cells were transfected with siRNA samples for 4–5 h and subsequently with the cDNAs, e.g., PH-probe for at least 4 h.
Western blot analysis
After transfection with siRNA, cells were cultured for 3 d, lysed in mammalian protein extraction reagent (Thermo Fisher Scientific) containing EDTA-free protease inhibitor mixture (Roche) and then centrifuged at 12,000 rpm for 10 min at room temperature. The supernatants were separated by 4–12% NuPAGE gel using a running buffer and electrotransferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked for 2 h in Tris-buffered saline with Tween 20 (TBST; 10 mM Tris-HCl and 0.1% Tween 20, pH 7.4) containing 5% nonfat dried milk and then incubated with rabbit anti–β-arrestin 1 and 2 (1:1,000 dilution; Cell Signaling Technology) or goat anti-actin (1:1,000 dilution; Santa Cruz Biotechnology, Inc.) antibodies at 4°C overnight. After a washing step with TBST, the membrane was incubated with horseradish peroxidase–conjugated goat anti–rabbit or rabbit anti–goat secondary antibody (1:5,000 dilution; Santa Cruz Biotechnology, Inc.) and visualized using enhanced chemiluminescence (ECL; GE Healthcare) followed by autoradiography. The intensity of the bands in the autoradiograms was calculated using ImageJ software (National Institutes of Health).
Confocal microscopy
Confocal images were taken every 7, 12, or 22 s depending on the experiment with a 710 confocal microscope (Carl Zeiss). The excitation wavelengths were 488-nm argon laser for GFP, 561-nm diode-pumped solid-state (DPSS) laser for RFP, and 633-nm helium-neon laser for Alexa Fluor 647 dye. Laser and emission light were delivered and collected by a 40× oil immersion lens (1.3 NA). Agonist and drugs were applied by pipette into the imaging chamber without perfusion. This solution addition was completed within 5 s.
Ca2+ imaging
Ratiometric Ca2+ measurements were performed with the Ca2+-sensitive fura-2 indicator (Jung et al., 2009). PDECs were loaded with 2 µM fura-2 AM for 30 min in the presence of 0.01% Pluronic F-127 and 100 µM sulfinpyrazone. For tsA201 cells, we reduced the amount of fura-2 AM to 1 µM and incubation time to 15 min without Pluronic F-127 and sulfinpyrazone. The fura-2 dye was excited at 340 and 380 nm using a Polychrome IV monochromator (Till photonics; FEI Life Sciences), and the emission signal was recorded at >470 nm every 2 s using an EMCCD camera (Photometrics) in an inverted TE2000 microscope (Nikon) equipped with a 20× objective lens. Background fluorescence measured in a cell-free area was subtracted.
Fluorescence resonance energy transfer (FRET) measurement
Previously, we described the method in detail (Jensen et al., 2009). In brief, we used epifluorescence photometry to measure the FRET between CFP and YFP in CKAR and LIBRAvIII probes or between PAR2-CFP and β-arrestin 2–YFP probes. Initially, CKAR and LIBRAvIII probes are localized in the cytosol and at the PM, respectively. We collected CFP or YFP emission via photomultipliers in photon-counting mode using an inverted Diaphot microscope (Nikon) equipped with a 40× 1.3 NA oil-immersion objective. CFP was excited at 440 nm, and emissions for CFP and YFP were collected at 480 nm and 535 nm, respectively. The bleed through from CFP to YFP channel was 17%, and the direct excitation of YFP by CFP excitation wavelength was negligible. FRET ratio (FRETr) was taken as the ratio of YFP emission divided by CFP emission after corrections. FRETr was normalized to remove cell to cell variation.
Single-molecule counting of PAR2-GFP
No. 1 coverslips of 25-mm diameter were cleaned with 100% ethanol for 1 d and washed with distilled water at least three times. Water drops on the coverslips were removed by suction. After complete drying, the coverslips were coated with 1 mg/ml poly-l-ornithine for 1–3 h. tsA201 cells transfected with a PAR2-GFP construct (0.05 µg) to produce PAR2 at a very low density were plated onto the coverslip. Most of the background signal from organic molecules bleached quickly after exposure to the laser light. Single-molecule TIRF images were taken on a custom-built microscope based on an Eclipse Ti microscope base (Nikon) with an APO TIRF 100× (1.49 NA) objective. A 488-nm laser was used for the illumination. Images were collected by EMCCD camera and a custom controller written in LabVIEW as reported previously (Sarangapani et al., 2014).
Modeling
The kinetic model of PAR2 desensitization was formulated as a compartmental model in the Virtual Cell simulation frame work (University of Connecticut Health Center): model “PAR2_Desensitization2015” is available at http://www.vcell.org under shared models/hillelab. Model species and reactions are shown in Fig. 6. We started with the basic model from our previous papers (Dickson et al., 2013; Falkenburger et al., 2013). In this study, we added steps for receptor desensitization. The kinetic parameters are listed in Tables 1 and 2. They were chosen manually based on the literature and on results from this paper without further optimization by statistical refinement.
Figure 6.
Schematic reaction diagram of the model including PAR2 signaling, desensitization, and internalization. Reactions in the dashed box were taken from the previous model for muscarinic receptor signaling (Falkenburger et al., 2013) except for the Ca2+-dependent PLC activation and β-arrestin–dependent PIP5K activity. After binding of ligand (L) to PAR2 (R), heterotrimeric Gαqβγ protein (Gq) binds to the receptor forming RLGq* (PAR2 bound to both ligand and Gqβγ). The soluble ligand can dissociate from the RLG state reversibly to yield RG. RLGq* generates active Gαq (Gq*), which in turn activates PLC to hydrolyze PIP2 into IP3 and DAG. The DAG-bound PKC is the activated form (active PKC) that catalyzes reaction 3. Phosphorylation of ligand-bound receptor (reaction 1) is mediated in parallel by two enzymes, GRK and active PKC. Based on literature, phosphorylated and ligand-bound receptors recruit two types of β-arrestins (β-arrestin 1 and 2; Ricks and Trejo, 2009). We assumed that translocation and binding of the arrestins to RLP can be described as a one-step reaction. To describe intracellular Ca2+ dynamics, the model considers only Ca2+ release from ER via IP3 receptors and uptake by SERCA pumps. Phosphatidylinositol 4-kinase (4K) and phosphatidylinositol 4-phosphate 5-kinase (5K) phosphorylate PI and PI(4)P, and 5-phosphatase (5P) and 4-phosphatase (4P) dephosphorylate PI(4,5)P2 (or PIP2) and PI(4)P (or PIP), respectively.
Table 1.
Model initial conditions and parameters
| Species/constants | Value | Rationale |
| R for PAR2 | 5,000 µm−2 | Receptor density from this paper (Fig. 5) with overexpression. If the receptors were able to dissolve in cytosol, it would be 5 µM. |
| G (G protein) | 40 µm−2 | Number of free G protein at the PM |
| PLC | 10 µm−2 | Number of free PLC at the PM |
| PI | 140,000 µm−2 | Number of free PI at the PM |
| PIP | 3,600 µm−2 | Number free PIP at the PM |
| PIP2 | 5,000 µm−2 | Number of free PIP2 at the PM |
| IP3 | 0.015 µM | IP3 at cytosol before receptor activation: Steady-state balance from PLC and IP3ase |
| DAG at PM | 23 µm−2 | Number of DAG at the PM before receptor activation |
| Arrestin (cytosol) = Arrestin1 + Arrestin2 | 15 µM | β-Arrestin 1 (Arrestin1) and β-arrestin 2 (Arrestin2) have 7.5 µM, respectively, to leave proper amount β-arrestins in the cytosol after their binding to the PAR2 at the PM. We assumed that β-arrestin 1 has higher affinity to PAR2 compared with the β-arrestin 2. |
| PKC_cyto (PKC Cytosol) | 1 µM | The concentration was chosen to make reasonable fitting of DAG-bound PKC |
| GRK | 600 | Fixed value to similar to the peak value of PKC_DAG |
| Weighting_factor_PKC_DAG or Weighting_factor_GRK | 0.5 | Active PKC (PKC_DAG) gives the same contribution to phosphorylation of ligand bound receptor as GRK |
| kf_PKC_DAG | 0.02 µM−1*s−1 | Forward rate constant for binding of PKC and DAG |
| Kr_PKC_DAG | 0.06 s−1 | Reverse rate constant for dissociation of DAG from PKC, giving Kd = 3 µM |
| Ca2+ (cytosol) | 0.13 µM | Typical intracellular Ca2+ level is 0.1–0.2 µM |
| fold PIP2 | 3 | Making the size of the of the total PIP2 (bound plus free) three times the free pool |
| k_4K (basal) | 0.00078 s−1 | Fold PIP2 * 0.00026 s−1 |
| k_5K (basal) | 0.06 s−1 | Fold PIP2 * 0.02 s−1 |
| k_4K (stimulated) | 6* k_4K (basal) | To fit the recovery kinetics of PIP2 probe in Fig. 2 A |
| k_5K (stimulated) | 3* k_5K (basal) | To fit the recovery kinetics of PIP2 probe in Fig. 2 A and to explain the PIP2 hump, we added β-arrestin–dependent PIP5K activity only in Fig. 9 C |
| k_4P (basal) | 0.03 s−1 | 0.03 s−1 * k4r_basal (k4r_basal = 1) |
| k_5P (basal) | 0.014 s−1 | 0.014 s−1 * k5r_basal (k5r_basal = 1) |
| k_4P (stimulated) | 0.19 s−1 | 0.03 s−1 * k4r_stim (k4r_stim = 6.25) |
| k_5P (stimulated) | 0.042 s−1 | 0.014 s−1 * k5r_stim (k5r_stim = 3) |
| kf_RL | 0.75 µm2*molecules−1*s−1 | Rate constant for phosphorylation of ligand-bound receptor-reaction 1 |
| kr_RLP | 0.0125 s−1 | Rate constant for dephosphorylation of ligand-bound phosphorylated receptor-reaction 1 |
| kf_R (basal) | 0.0001 s−1 | Basal phosphorylation rate constant of receptor-reaction 3 |
| kr_RP (basal) | 2 s−1 | Basal dephosphorylation rate constant of phosphorylated receptor-reaction 3 |
| kf_R (stimulated) | 10 s−1 | Stimulated phosphorylation rate constant of receptor |
| kr_RP (stimulated) | 4 s−1 | Stimulated dephosphorylation rate constant of receptor. We assumed that rate of dephosphorylation of RP doubles after ligand treatment-reaction 3 |
| kf_L2 | 0.09333 µM−1 * s−1 | Binding rate constant of ligand to RP, depending on the dissociation constant (K_L2) and dissociation rate constant (kr_L2) |
| kr_L2 | 5.6 s−1 | Dissociation rate constant of ligand from RP, assuming that it has same dissociation rate constant compared to the native receptor (R) |
| K_L2 | 60 µM | Dissociation constant of ligand bound to RP, which has slightly lower affinity compared with native receptor (R) based on the supplemental data (Fig. S5) |
| kf_RLP | 0.003 s−1 | Rate constant of β-arrestin 2 binding to phosphorylated ligand-bound receptor to make best fitting compared with Figs. 4 F and 8 F. For β-arrestin 1, we used 0.006 s−1-reaction 2 |
| kr_RLPA1 or kr_RLPA2 | 10−6 s−1 | Rate constant of β-arrestin dissociation from phosphorylated ligand-bound receptor. Same for Arrestins 1 and 2-reaction 2 |
The rate constants for the phosphorylation of receptors were chosen to fit experimental data. In our model, we did not consider spatial information, e.g., diffusion of molecules in space. The rate constant for the PI4K (k_4K during stimulation) = fold PIP2 * (k_4K_rest + (stim_4K − k_4K_rest + k_4K_basal) * scale_4K * (1 − exp(−t/tau_on))). Where, tau_on = 1 s, fold PIP2 = 3, scale_4K = 0.75, k_4K_rest = 0.00026 s−1, k_4K_basal = 0.0002353 s−1, and stim_4K = 0.00117 s−1. The rate constant for PIP5K (k_5K during stimulation) = fold PIP2 * (k_5K_rest + (stim_5K − k_5K_rest + k_5K_basal) * scale_5K * (1 − exp(−t/tau_on))), where tau_on = 1 s, fold PIP2 = 3, scale_5K = 1, k_5K_rest = 0.02 s−1, k_5K_basal = 0.0181 s−1, stim_5K = 5.75 * [unspecified contribution (0.5) + β-arrestin–dependent contribution (0.5) * exp(−t/tau_off)], tau_off = 40 * [β-arrestin]4, and β-arrestin = β-arrestin 1 + β-arrestin 2.
Table 2.
Model differential equations
| Reaction | Flux |
| PI to PI(4)P | k_4K * [PI] − k_4P * [PIP] |
| PI(4)P to PI(4,5)P2 | k_5K * [PIP] − k_5P * [PIP2] |
| PLC (basal) | [PIP2] * (PLC_basal + k_ PLC * fold PIP2 * Ga_GTP_PLC), where PLC_basal = 0.0001 s−1, k_PLC = 0.2 µm2molecule−1s−1 and fold PIP2 = 3 |
| PLC (stimulated) | [PIP2] * (PLC_basal + k_PLC * fold PIP2 * Ga_GTP_PLC) + [PIP2] * PLC_stim * Ga_GTP_PLC * [Ca_C/(Ca_C + Kd_PLC_Ca)], where PLC_basal = 0.0001 s−1, k_PLC = 0.2 µm2molecule−1s−1, fold PIP2 = 3, PLC_stim = 7 s−1, and Kd_PLC_Ca = 1 µM |
| RL to RLP | kf_RL * [RL] * (weighting_factor_PKC_DAG * [PKC_DAG] + weighting_factor_GRK * [GRK]) − kr_RLP * [RLP] |
| PKC to active PKC | kf_PKC_DAG * [PKC] * [DAG] − kr_PKC_DAG * [PKC_DAG] |
| R to RP | kf_R * [R] * [PKC_DAG] − kr_RP * [RP] |
| RP to RLP | kf_L2 * [RP] − kr_L2 * [RLP] |
| RLP to RLPA1 | kf_RLP * [RLP] * [Arrestin1] − kr_RLPA1 * [RLPA1] |
| RLP to RLPA2 | kf_RLP * [RLP] * [Arrestin2] − kr_RLPA2 * [RLPA2] |
Many additional reactions that have been already described previously in our paper are not in this table. The parameters are in previous papers (Dickson et al., 2013; Falkenburger et al., 2013). We added Ca2+-sensitive PLC activation in our model. In this table and calculation, the units of concentration and area are µM and µm2, respectively.
Statistical analysis
All numerical values and error bars in figures are given as mean ± SEM. Statistical significance was determined by Student’s t test, and P < 0.05 was considered significant.
Online supplemental material
Fig. S1 shows a lack of endogenous functional PAR2 in tsA201 cells. Fig. S2 shows a comparison of downstream signaling with nonfluorescent “dark” PAR2 and GFP-tagged PAR2. Fig. S3 shows monitoring of IP3 and PKC during the activation of PAR2. Fig. S4 shows a reduction of β-arrestin 1 and 2 proteins by knockdown. Fig. S5 shows the FRETr between CFP-tagged PAR2 and YFP-tagged β-arrestin 2. Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201511477/DC1.
RESULTS
Expression of PAR2 in a heterologous system
We chose to use tsA201 cells to study the termination of PAR2 signaling based on the following advantages. First, they do not express functional endogenous PAR2 as tested by the absence of a Ca2+ response to a PAR2 agonist (Fig. S1). Second, several engineered proteins including PAR2 can be expressed simultaneously in these cells. Third, we have already developed and refined a kinetic model for GPCRs coupled to Gq (the M1 muscarinic receptor) in this cell line (Horowitz et al., 2005; Jensen et al., 2009; Falkenburger et al., 2010a,b, 2013; Dickson et al., 2013). Because M1 and PAR2 receptors share common downstream signals, we could use the existing model parameters as a starting point for PAR2.
1 d after transfection, PAR2, labeled supravitally with anti-PAR2 Alexa Fluor 647 primary antibody or tagged with GFP, appears at the cell surface (e.g., see Fig. S2 A and Fig. 5 E). To confirm that PAR2-GFP overexpressed in tsA201 cells behaves like the endogenous PAR2 of other cells, we compared one of the endpoints, the intracellular Ca2+ signal between tsA201 cells and PDECs. Trypsin cleaves the N terminus of PAR2, and the exposed tethered ligand activates the receptors. Treatment of PDECs with 100 nM trypsin evoked a transient Ca2+ increase (Fig. 1 A). Then, even in the continuous presence of the stimulus, intracellular free Ca2+ (Ca2+) returned to a low steady-state level within minutes, reflecting in part receptor desensitization. Activation of endogenous PAR2 in PDECs with a specific AP that mimics the N-terminal ligand of cleaved receptor elicits similar Ca2+ signals (Kim et al., 2008). In tsA201 cells overexpressing PAR2, AP also evoked a transient Ca2+ rise with recovery kinetics almost identical to those in PDECs (Fig. 1 B), suggesting that the rate-limiting steps for receptor activation and desensitization are similar in the two systems. We could not test trypsin in tsA201 cells because it detached the cells from the substrate.
Figure 5.
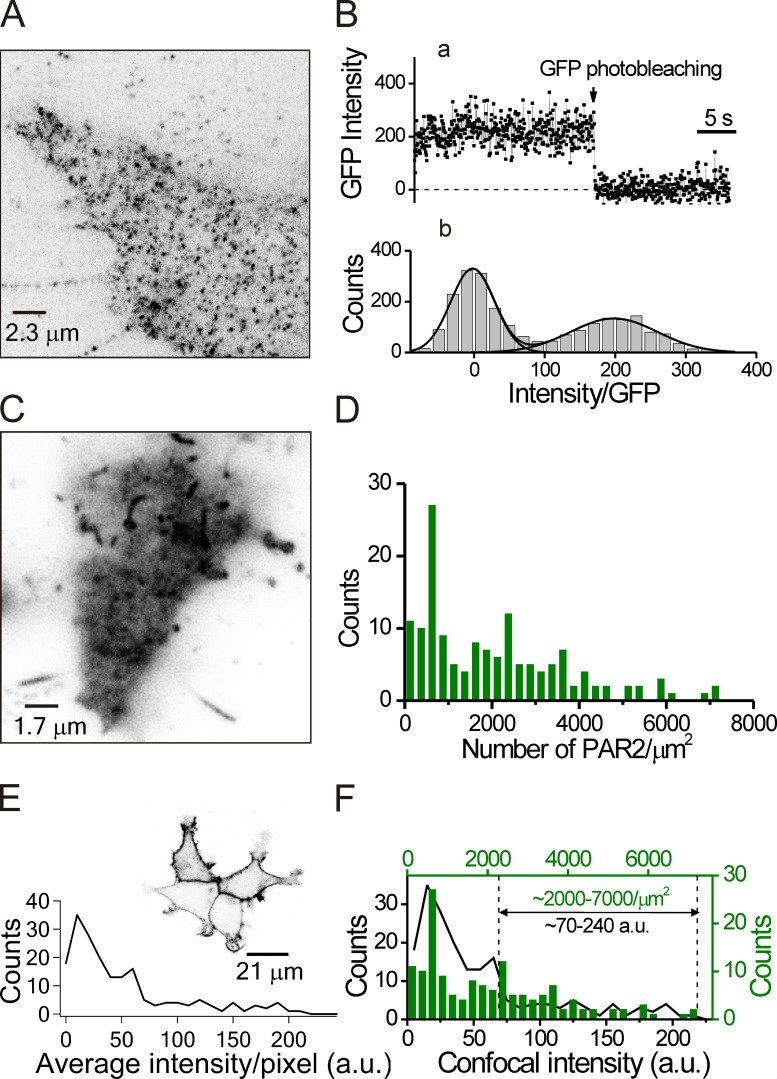
Counting PAR2 molecules. (A) TIRF microscopy of GFP-tagged PAR2 receptors expressed at a low density to observe single PAR2-GFP molecules. Although the cells were fixed with 4% paraformaldehyde, most PAR2-GFPs were still mobile. We chose immobilized molecules to analyze single GFP bleaching steps. (B) Photobleaching of single GFP molecules. (B, a) Time course of intensity during one photobleaching event. (b) Intensity histogram for the fluorescence of the basal and first levels for 21 GFP molecules in different regions of three cells. The single-step size was estimated by fitting two Gaussian curves, giving an intensity difference between the two peaks of 198 a.u. The background intensity after GFP photobleaching was subtracted. (C) TIRF image of a single cell overexpressing a typical amount of PAR2-GFP using the same microscope but with 20-fold lower gain (see section Counting single receptor molecules at the PM). (D) Frequency distribution of PAR2-GFP density in 147 overexpressing cells using TIRF microscopy from three independent experiments. (E) Frequency distribution of PAR2-GFP fluorescence in 187 overexpressing cells observed by conventional confocal microscopy. As seen in the insert image, most of the PAR2 was at the PM. (F) Comparison of the frequency distributions for confocal intensity (black line) and PAR2 molecular density (green bars) made by scaling the two curves for optimal register. Vertical dotted lines define the range of GFP fluorescence of the cells selected for our confocal experiments in Figs. 2, 3, 4, and 10.
Figure 1.
Intracellular Ca2+ concentration rises transiently during the activation of PAR2. (A and B) PDECs with endogenous PAR2 (A) and tsA201 cells with overexpressed PAR2-GFP (B) were preloaded with Ca2+-sensitive fura-2 AM dye and treated with 100 nM trypsin or 100 µM AP (sequence SLIGKT), respectively. Ca2+ is expressed as fluorescence excitation ratios (F340/F380) of the dye. The traces are means of seven PDECs and five tsA201 cells each, respectively. The dashed lines indicate the basal Ca2+ level before stimulation, and error bars (SEM) are gray.
Optical imaging of intracellular signals generated by PAR2
To study receptor desensitization, we evaluated the ability of several intracellular probes to monitor the downstream activity of PAR2. The Ca2+ signals have been used in previous studies because they are convenient to measure. However, they are limited to a narrow range of receptor activation because even a slight increase of IP3 (to a few hundred nanomolar) after the activation of PLC suffices for full activation of IP3 receptors and for a maximal Ca2+ increase in these cells (Dickson et al., 2013). Considering that complete hydrolysis of PIP2 in the PM can generate 5–15 µM IP3 (Falkenburger et al., 2013), Ca2+ measurements will report only the foot of the activation curve (Dickson et al., 2013). Therefore, we tested two other signals generated by PAR2 activation: (1) depletion of PM PIP2 as reported by cytosolic PLCδ1-PH domain tagged with RFP (PH-RFP probe) and (2) production of DAG from PIP2 hydrolysis, as reported by citrine (yellow)-tagged PKCδ C1 domain (C1-citrine probe). The PH-RFP and C1-citrine probes expressed in the same cell can be discriminated optically (Fig. 2 A, insets). When the cells were exposed to AP to activate PAR2, the PH-RFP probe translocated toward the cytoplasm within 10 s as PIP2 became depleted by PLC. The fluorescence intensity measured in a cytoplasmic region of interest increased two- to threefold from the basal level (Fig. 2 A). Monitoring DAG also provides useful information about PIP2 hydrolysis because each molecule of PIP2 yields one DAG. 1 d after transfection, most of the C1-citrine was located in the cytoplasm and nucleus in confocal images (Fig. 2 A, bottom insets). Hydrolysis of PIP2 induced translocation of the C1-citrine probe from the cytoplasm toward the PM (Fig. 2 A). The activation and recovery kinetics reported by C1-citrine were nearly identical to those reported by PH-RFP (Fig. 2 B), and the estimated half-maximal agonist concentrations for PIP2 depletion (27 µM AP) and DAG generation (13 µM AP) were comparable (Fig. 2, C and D). This result is consistent with a previous dose–response curve of PAR2 as measured with Ca2+ signals (Déry et al., 1999). Based on a recent crystal structure of PAR1, the local concentration of tethered ligand has been estimated at equivalent to 400 µM (Zhang et al., 2012). Therefore, we used 100 µM AP for most of the following experiments.
Figure 2.
Monitoring of PIP2 and DAG during activation of overexpressed PAR2. (A) tsA201 cells were transfected with PH-RFP (PIP2 probe) for visualization of PIP2 depletion and C1-citrine (DAG probe) for estimation of DAG production during PAR2 activation. At rest, PIP2 and DAG probes are located at the PM and the cytoplasm, respectively (insets). After the addition of 100 µM AP, the two probes translocated toward the other compartment. Fluorescence intensity of both probes in a cytoplasmic region of interest was measured and normalized (Norm. cyt. intensity). (B) DAG and PIP2 probe translocation rates during receptor activation and desensitization. For A and B, n = 8 cells. (C and D) Concentration–response curves for PIP2 depletion (C) and DAG production (D). The peak values were measured at the indicated AP concentrations. Each point is the mean of three to nine cells. Error bars are SEM.
In various experiments, we used GFP-labeled or unlabeled (dark) forms of PAR2. Control experiments showed that PAR2-GFP and dark PAR2 elicited translocation and recovery of the PH-RFP probe with similar kinetics during AP treatment (Fig. S2 B), indicating that the GFP tag does not affect PAR2 activation and desensitization.
Next, we evaluated probes for IP3 and PKC. We measured IP3 using LIBRAvIII, a FRET probe engineered from the rat type-3 IP3 receptor with a membrane localization domain (Tanimura et al., 2009; Dickson et al., 2013). When IP3 binds to the probe, the normalized FRETr (YFP/CFP) decreases, indicating a separation of CFP from YFP (Fig. S3 A). As expected, during 100 µM AP treatment, the IP3 level increased (FRETr decreased) transiently and then the IP3 level decreased. Interestingly, the IP3 level became even lower than the original basal level, possibly because of the depletion of PIP2 and an overstimulation of IP3 clearance. Similarly, we measured phosphorylation by PKC using the CKAR FRET probe, which again has CFP and YFP (Dickson et al., 2013). When this probe is phosphorylated by PKC, the normalized FRETr falls, indicating greater separation between the CFP and YFP (Fig. S3 B). When cells were stimulated with AP, phosphorylation of the PKC probe increased (FRETr decreased) transiently and then phosphorylation fell, undershooting the basal level for several minutes, perhaps because of an acceleration of serine/threonine phosphatases. As would be anticipated, during treatment with okadaic acid, a blocker of the serine/threonine phosphatase PP2A, the AP agonist induced a greater accumulation of phosphorylated probe (drop of FRETr) with much less recovery. Thus, when active, the serine/threonine phosphatases limit the extent of phosphorylation by PKC after PAR2 activation. Therefore, we can consider phosphatase activity as a potential opponent of PAR2 desensitization (see section Contribution of β-arrestin binding to desensitization of PAR2).
Our comparison of several probes suggested that the best indicator of PAR2 activity is the level of PIP2. This measurement has the highest signal/noise ratio and the largest dynamic change compared with the IP3, PKC, and DAG measurements. It was used in many of the remaining experiments. For interpreting the experiments to be presented, note for practical reasons that we report the intensity of the PIP2-indicating PH-RFP probe signal in the cytoplasm as in Fig. 2 A. When this cytoplasmic signal goes up in Fig. 2 A, PIP2 at the PM is being depleted after AP addition. When it comes down again over the next 100 s, PIP2 is being resynthesized despite the presence of AP, meaning that PAR2 receptors have desensitized and have become uncoupled from signaling to Gq.
Contribution of phosphorylation to desensitization of PAR2
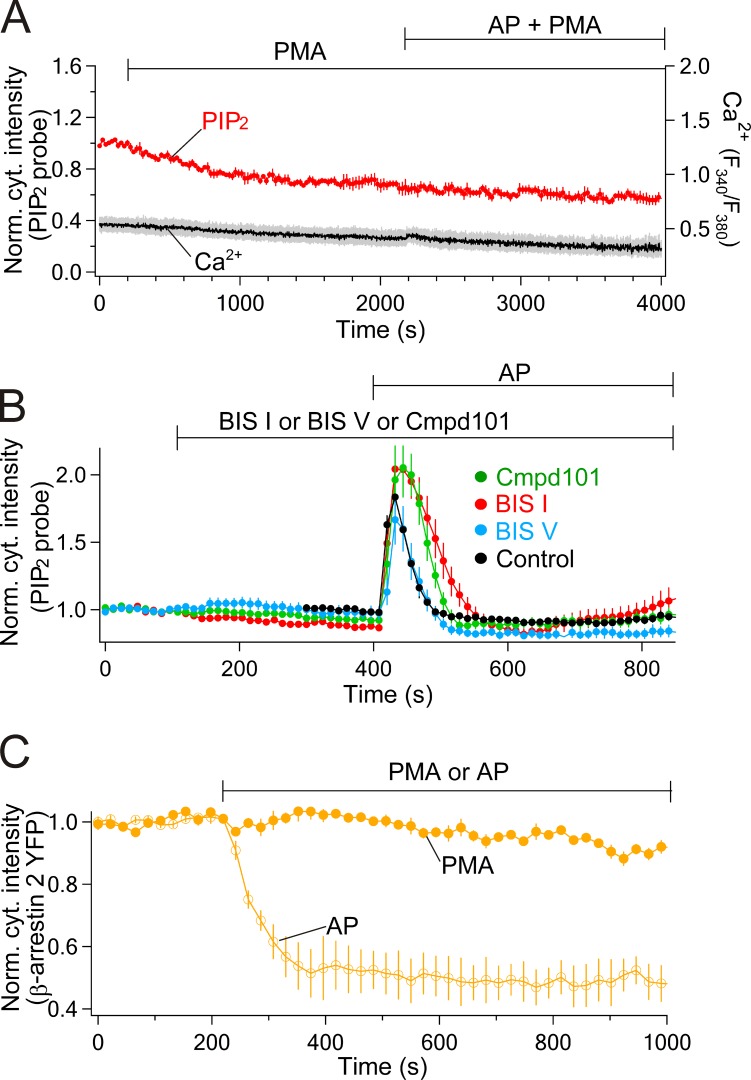
Now we address mechanisms of receptor desensitization beginning with kinases that may phosphorylate and turn down the activity of PAR2. Previous work suggested that pharmacological activation of PKC by phorbol esters will desensitize PAR2 (Böhm et al., 1996; DeFea et al., 2000). Accordingly, we treated tsA201 cells with PMA and tested the responses to subsequent application of AP. After 2,000 s of PMA treatment, AP could no longer elicit PIP2 depletion (Fig. 3 A, red line, PH-RFP) nor a rise of intracellular Ca2+ (gray line) compared with untreated cells (compare Fig. 2 A and with Fig. 1 B). In contrast, after 250 s in the PKC inhibitor BIS I, the response to AP was actually potentiated. There was more PIP2 depletion and a slower turn off (desensitization) of the response (Fig. 3 B, red) compared with control (Fig. 3 B, black). The inactive form of BIS I, BIS V, was no different from control. This suggests that PKC, which is activated by PAR2, can turn off receptor signaling completely when stimulated strongly and contributes to the dynamic desensitization during receptor stimulation and possibly also to a resting tonic desensitization. What about GRKs? Interestingly, Compound101, a blocker of GRK2/3, has nearly the same effects as BIS I, augmenting and prolonging the AP-stimulated PIP2 depletion. This suggests that during AP stimulation PKC and GRK2/3 participate in parallel in phosphorylating and desensitizing PAR2 (Fig. 3 B), and when either one of them is inhibited, the other will suffice to desensitize PAR2 fully, although much more slowly.
Figure 3.
Stimulation of PKC stops signaling from PAR2 without recruiting β-arrestin. Cells were treated with 100 nM PMA to activated PKC or 100 nM BIS I to block PKC. (A) PKC activator (PMA) stops PIP2 depletion and Ca2+ signaling from PAR2. Probes are PH-RFP for PIP2 (n = 4) and fura-2 for intracellular Ca2+ (n = 3). (B) PKC inhibitor (BIS I, n = 11) and GRK2/3 inhibitor (compound101 [Cmpd101]) potentiate the PIP2 depletion compared with 100 nM BIS V (n = 7) or control without the blockers (n = 17). AP was 10 µM. (C) 100 µM AP recruits β-arrestin (open circles, n = 8) and PMA does not (closed circles, n = 4). Error bars are SEM.
Next, we considered the recruitment of β-arrestin to the phosphorylated receptors. Cells were transfected with β-arrestin 2–YFP, which was normally localized in cytosol. As expected, the labeled arrestin translocated to the PM upon activation of receptors with AP (Fig. 3 C, open circles). Presumably it recognized phosphorylated receptors and the presence of agonist, as is known for β2-adrenergic receptors (Krasel et al., 2005; Vayttaden et al., 2010; Shukla et al., 2014). Unlike the other responses to agonist that we have seen so far (PIP2, DAG, PKC, IP3, and Ca2+), the translocation of β-arrestin developed slowly and was not transient. Presumably it is downstream from receptor. In contrast, β-arrestin did not translocate upon treatment of cells with PMA without agonist (Fig. 3 C, closed circles). We suggest that the receptors are phosphorylated by PKC that is turned on by the phorbol ester, but in the absence of an activating ligand, β-arrestin 2 does not bind–although the receptors are desensitized with respect to signaling (Böhm et al., 1996; Déry et al., 1999; Stalheim et al., 2005).
Contribution of β-arrestin binding to desensitization of PAR2
Phosphorylation of PAR2 and binding of β-arrestin have been implicated in PAR2 receptor desensitization (Böhm et al., 1996; Déry et al., 1999). However, it is difficult to extract the effect of phosphorylation alone because it may be followed quickly by a cascade of recruitment of other accessory proteins, and it is opposed by dephosphorylation (Ricks and Trejo, 2009). We chose to knock down β-arrestin using siRNA, which should slow the second step of desensitization and thus the return of PIP2 to resting levels. In the knockdown cells (Fig. 4, A and B), PIP2 recovery kinetics were significantly delayed. The rate constant for spontaneous recovery was slowed from 25 ± 3 × 10−3 s−1 (n = 16) with scrambled siRNA (Fig. 4 A, Scrambled) to 11 ± 3 × 10−3 s−1 with β-arrestin 1 and 2 siRNA (Fig. 4 A, β-arr 1/2; n = 5). In addition, the time course of PH-RFP intensity in the cytoplasm became biphasic with two peaks. We will refer to the biphasic kinetics as the “PIP2 hump.” These observations imply that the development of receptor desensitization was slowed by β-arrestin reduction and even became incomplete. We will be considering several mechanisms potentially contributing to the PIP2 hump both experimentally and by mathematical modeling. Western blot analysis revealed that β-arrestins were reduced by ∼40% in the whole dish of siRNA-treated cells compared with cells transfected with scrambled siRNA (Fig. 4 C and Fig. S4). Noting that only a fraction of the cells would have taken up the siRNA in the transfection, the β-arrestin reduction in those that did must have been considerably >40%, and the others may still have normal β-arrestin levels. Our assumption is that those cells that visibly overexpress PAR2-GFP will also have taken up siRNA.
Figure 4.
β-Arrestin locks in the desensitized state of PAR2. (A) PIP2 loss and recovery during maintained AP was monitored by the PIP2 probe in the cells transfected with scrambled siRNA (Scrmbld) or knocked down with β-arrestin 1/2 siRNA (β-arr 1/2). (B) Rate of translocation of the PIP2 probe measured in A. **, P < 0.01. (C) Amounts of β-arrestin in dishes of cells transfected with β-arrestin 1/2 siRNA (green) compared with cells transfected with scrambled siRNA. The amount of protein from Western blots was normalized to the value for cells transfected with scrambled siRNA. ***, P < 0.001. (D) Calyculin A restores PIP2 recovery in β-arrestin 1/2 knockdown cells (red symbols). For comparison, the data from A are plotted as dashed lines (scrambled, black; and β-arrestin 1/2 siRNA, green). (E) Rate recovery with (red) or without (green) calyculin A. The dotted line indicates the value for scrambled siRNA. (F) Overexpressing β-arrestin 2–YFP (yellow) does not speed up PIP2 recovery (red). (G and H) Two different AP concentrations were used, 10 and 100 µM. Rates of PIP2 translocation (G) and β-arrestin 2–YFP translocation (H). Error bars are SEM.
For the β2-adrenergic receptor, the phosphorylated receptor is said to have two possible fates: it may next recruit β-arrestin or it may become dephosphorylated (Tran et al., 2007). Thus, there is potential competition between the β-arrestin binding reaction and the dephosphorylation by phosphatases. For PAR2, we hypothesized that after knockdown of β-arrestin such competition would become biased so that phosphatases would act more effectively to restore receptors to their active state. This might generate a PIP2 hump. Conversely, to restore competitive equity, it should be possible to further block the activity of phosphatases pharmacologically. This might prevent formation of a PIP2 hump. To test these ideas, we used calyculin A, an inhibitor of the serine/threonine phosphatases PP1 and PP2A, as was done in studies of the β2-adrenergic receptor (Tran et al., 2007). We again knocked down β-arrestin, which slowed spontaneous PIP2 recovery and then found that addition of calyculin A restored the fast kinetics of PIP2 recovery (Fig. 4, D and E, red symbols) to control values (Fig. 4 D, black dashed line). Presumably, the phosphatase inhibitor allowed phosphorylated receptors to accumulate, thus reducing the active receptor pool despite knockdown of β-arrestin. We also tested an alternative hypothesis that accumulation of phosphorylation of the receptor by the phosphatase inhibitor speeds up the translocation of β-arrestin to PAR2 and accelerates PAR2 desensitization. To address this, we used another FRET assay between CFP-tagged PAR2 and YFP-tagged β-arrestin 2 to monitor direct interaction between PAR2 and β-arrestin. As shown in Fig. S5, the rate of translocation of β-arrestin 2 to PAR2 was not significantly affected by calyculin A treatment, suggesting that inhibiting receptor dephosphorylation does not facilitate the recruitment of β-arrestin. We speculate that in normal cells, β-arrestin binding to phosphorylated receptors locks the receptors in the phosphorylated state by blocking the access of serine/threonine phosphatases. Such a mechanism has been proposed for the β2-adrenergic receptor based on the effect of GRK on the receptor (Vayttaden et al., 2010).
Next, we tested whether overexpression of β-arrestin 2 might accelerate receptor turn off and the spontaneous recovery of PIP2. Initially, in these cells, β-arrestin 2 labeled with YFP was localized to the cytosol, and the PH-RFP (PIP2) probe was on the PM. Within seconds of the addition of AP, the PH-RFP probe entered the cytoplasm. Then, during continued AP treatment, both the labeled β-arrestin and the PH-RFP probe translocated toward the PM. The rate constants of these migrations were the same (24 ± 0.4 × 10−3 s−1 vs. 22 ± 2.4 × 10−3 s−1; n = 8; Fig. 4, F and G). Overexpression of β-arrestin 2–YFP did not speed the return of the PH-RFP probe to the membrane, as if receptor phosphorylation and not β-arrestin binding is normally the rate-limiting step of receptor desensitization.
Lowering the AP concentration from 100 to just 10 µM slowed β-arrestin 2–YFP translocation to the membrane sevenfold (Fig. 4, F–H). This suggests that β-arrestin binding depends on the ligand-bound PAR2, as is known for the β2-adrenergic receptor (see Fig. 8 [E and F] in this paper; Vayttaden et al., 2010; Ostermaier et al., 2014; Shukla et al., 2014).
Figure 8.
Simulated time courses of PIP2 depletion and β-arrestin recruitment. (A and B) Depletion of PM PIP2 by different concentrations of AP (A) replotted as a dose versus peak response curve (black squares; B) in simulation. For easier comparison with the experimental data, the y axes of simulated data in A and C were inverted. The peak response at various concentrations of AP was normalized with the value at 300 µM. Experimental results in B (Fig. 2 C), D (Fig. 2 C), F (Fig. 4 H), and H (Fig. 2 C) are overlaid for comparison (red symbols). (C and D) PM PIP2 levels during 100 µM AP treatment for different surface densities of PAR2 receptors. (E and F) β-Arrestin translocation time course and rates at different AP concentrations. (G and H) β-Arrestin translocation time course and rates at different PAR2 densities. Concentrations and densities used in the simulations are as follows: (A, B, E, and F) 0.1, 3, 10, 30, 100, 300, and 1,000 µM AP; (C and D) 5, 50, 500, 1,500, 5,000, and 10,000 µm−2; and (G and H) 5, 50, 500, 1,500, 5,000, and 10,000 µm−2.
Counting single receptor molecules at the PM
Based on these results, we hypothesized that (a) phosphorylation of PAR2 leads to receptor desensitization even without binding of β-arrestin and (b) subsequent binding of β-arrestin to the receptor is key to keeping the phosphorylated state from being dephosphorylated by serine/threonine phosphatases. As a test, we formulated a mathematical description of PAR2 signaling and its desensitization. To make a model, we required an estimate of the density of overexpressed PAR2 at the PM. This was calculated by dividing the total fluorescence intensity of PAR2-GFP by the fluorescence intensity of one molecule, both obtained from TIRF microscopy. To measure single molecules required cells that expressed individual PAR2-GFP molecules very sparsely. We reduced the amount of PAR2-GFP cDNA used for transfection 10-fold (0.05 µg) and then selected cells with only a low density of PAR2 at the membrane (<5 receptors/µm2; Fig. 5 A). Fluorescence of spots was measured during photobleaching steps using a 488-nm laser (Fig. 5 B, a). The trace shows a single down step of fluorescence terminating in complete photobleaching. The mean single-GFP intensity integrated over individual multi-pixel regions of interest was 198 arbitrary units (a.u.) in the TIRF microscope (Fig. 5 B, b).
For observing bulk PAR2-GFP overexpression (Fig. 5 C), the measurement sensitivity of the same microscope was set 20-fold lower than for the single-molecule measurement (5-fold lower laser power and 4-fold lower EMCCD gain), making the expected equivalent single-molecule fluorescence 9.9 a.u. (198/20). We plotted the distribution of mean receptor densities obtained from such TIRF images as receptors per square micrometer (Fig. 5 D) for comparison with the fluorescence intensity distribution obtained from our conventional confocal images (Fig. 5 E). Scaling these two distributions to match the minimum and maximum values, we could estimate the receptor density for our confocal experiments. We had been choosing cells with 70–240 a.u. of fluorescence in the confocal experiments. They would express 2,000–7,000 receptors/µm2 (Fig. 5 F) with a mean receptor density of 3,600 receptors/µm2. This value is consistent with previous results for overexpressed M1 muscarinic receptors in the same cells (Falkenburger et al., 2010a).
Mathematical modeling of PAR2 signaling
Rather than draw conclusions only by qualitative reasoning, it seemed prudent to cast all inferences as a mathematical model that could be checked against our real-time measurements of intermediates in PAR2 signaling. We built a kinetic description (Fig. 6) that derives from our model for muscarinic M1 receptor signaling in tsA201 cells (Dickson et al., 2013; Falkenburger et al., 2013) and adds a new focus on steps underlying receptor desensitization: receptor phosphorylation, receptor dephosphorylation, and β-arrestin binding to phosphorylated and ligand-bound receptor.
The dashed box of the schematic diagram (Fig. 6) encompasses the classical receptor activation and intracellular signals of Gq-coupled receptors. They include ligand (L) and Gq protein (Gq) binding to receptors (R), activation of PLC, and cleavage of PIP2 (see figure legend for details). The new features related to receptor shutdown are across the top, outside the box. PKC phosphorylates both empty receptor (R, reaction 3) and ligand-bound receptor (RL, reaction 1), giving RP and RLP. GRK phosphorylates only ligand-bound receptors (reaction 1). Phosphatases dephosphorylate these forms. A key assumption represented in the model is that phosphorylation suffices to turn off the activity of the receptor, and thus RLP is already a desensitized receptor that does not signal.
We represent phosphorylation as a single step to reduce unknown parameters but realize that there are 17 candidate serine/threonine residues in the C terminus of PAR2, several of which need to become phosphorylated (Ricks and Trejo, 2009). Although two different protein kinases are involved, we do not know which serines and threonines are targeted by each. Tentatively, the model makes no distinction and assumes that phosphorylated receptors are equivalent independent of the kinase or kinases involved.
Subsequently, β-arrestin 1 and 2 bind to RLP with similar binding affinity (reaction 2), giving RLPA1 and RLPA2, inactive forms of the receptor that are now sequestered from phosphatases. We did not include any binding of β-arrestin to RP, the phosphorylated receptor without ligand, because when we activated PKC by PMA, it did not induce translocation of β-arrestin to the receptor (Fig. 3 B). We included a step where the receptor can be phosphorylated first (giving RP) and then ligand can be bound to the phosphorylated receptor (giving RLP). Dephosphorylation by serine/threonine phosphatases counteracting phosphorylation was added because the recovery of PIP2 after knockdown of β-arrestins was significantly restored by a protein phosphatase inhibitor (Fig. 4 D). β-Arrestin and phosphatases are in competition to control the number of phosphorylated receptors. The schematic diagram also shows clathrin- and dynamin-dependent steps of internalization of PAR2 (Ricks and Trejo, 2009), but those steps are not included in the equations of the present model because endocytosis of PAR2 was not significantly detectable within the first 10 min (not depicted). The internalization takes many tens of minutes in our experimental condition.
Fig. 7 shows the simulated time courses of cellular signals and of several states of PAR2 before and during AP application in the new model. Changes of PM PIP2 and DAG, intracellular IP3 and free Ca2+, active PKC, and several states of the receptor (RP, RLP, and RLPA2) are shown. The measured time courses of PM PIP2, DAG, IP3, Ca2+, and active PKC are well described. The RLPA2 state can be estimated indirectly from the translocation of β-arrestin 2 (Fig. 4), but otherwise, the distribution among receptor states could be deduced only from the simulations. Adjusting the model to the data informed us that the rates of the PI4K and PIP5K enzymes had to be accelerated transiently during the application of agonist as we also found in our study of M1 muscarinic receptor signaling (Falkenburger et al., 2010a, 2013). Here, the rate constants of these enzymes were speeded after AP application by approximately six- and threefold, respectively, to account for fast recovery of PM PIP2 during the desensitization of PAR2.
Figure 7.
Simulation showing the predicted time courses of several intracellular signals triggered by AP and the distribution of PAR2 among different states. For PIP2 recovery at the PM (PM PIP2), the model accelerated PI4 and PIP5 kinases, with the maximal rate constant for PIP5K being regulated by the amount of β-arrestin. The PM PIP2 scale was inverted for easier comparison with the PH-domain signals from this study. The simulated concentration of AP was 100 µM. Parameters and equations for the underlying reactions are listed in Tables 1 and 2.
Next, we used the model to simulate agonist dose–response relations and the effects of receptor density. When the receptor density was set at 5,000 receptors/µm2, the simulated AP dose–response relation for PM PIP2 depletion (Fig. 8 A) was similar to our experimental data for overexpressed receptors (Fig. 8 B). Here, the PAR2 receptor was assigned the same kinetic signaling properties as the M1 muscarinic receptor in our earlier models. Simulations with lower receptor density (Fig. 8, C and D) suggested that PM PIP2 can be depleted strongly by 100 µM AP at PAR2 densities down to 3,000/µm2. Below that, depletion fell off in a graded manner. At receptor densities of only 5–50/µm2, a realistic density for endogenous receptors, there still was partial PM PIP2 depletion during simulated 100 µM AP treatment (Fig. 8, C and D), reminiscent of previous studies with endogenous purinergic receptors in tsA201 cells (Dickson et al., 2013) and with endogenous PAR2 in PDECs (Kim et al., 2008). The experimental and simulated translocation rates of β-arrestin 2 were dependent on the ligand concentration (Fig. 8, E and F), but in contrast to PM PIP2 depletion, the simulated rates were independent of the receptor density (Fig. 8, G and H). Thus, ligand binding, but not the total number of receptors, is important for β-arrestin recruitment to the receptors at the PM. Bound β-arrestin marks the receptors that are cleaved by trypsin, phosphorylated, and ready to be internalized. The simulations nicely explain the observed slower translocation rate of β-arrestin 2 with lower agonist concentration in Fig. 4 (G and H; see red symbols in Fig. 8 F).
To mimic the effect of β-arrestin knockdown, we reduced the concentration of β-arrestin in the simulation by 70% (Fig. 9). As explained before, this is more than the mean reduction in a whole dish of cells because many of those cells will not have incorporated the siRNA or the receptor plasmid at all. With this reduction, the predicted desensitization became incomplete, and PIP2 no longer recovered completely during agonist application (Fig. 9 A, green line). To check that dephosphorylation of PAR2 by phosphatases would oppose receptor desensitization and PIP2 recovery as we supposed from Fig. 4 D, we then also inhibited phosphatases in the model by setting their rate constants to zero (30% β-arrestin + calyculin A). As anticipated, after PAR2 activation, the PIP2 level initially fell but recovered completely, compensating for the β-arrestin knockdown (Fig. 9 B). As inferred from experiments, our simulations show that β-arrestin would normally protect the phosphorylated receptor from serine/threonine phosphatases, but after reduction of β-arrestins, a significant effect of the phosphatases is unmasked and receptors are able to return to their active state. This strengthens our proposal that phosphorylation itself suffices to terminate further activation of G proteins by the PAR2 receptor.
Figure 9.
Simulated effects of serine/threonine phosphatases in β-arrestin knockdown cells. AP was set at 100 µM. (A) PM PIP2 time course with 100% or only 30% of control β-arrestin. (B) PM PIP2 time course with 30% β-arrestin with and without inhibition of phosphatase. Calyculin A was simulated by setting the reverse rate constants in reactions 1 and 3 to zero (30% β-arrestin + calyculin A). Compare with experiments in Fig. 4 D. (C) Simulated PM PIP2 time course with 30% β-arrestin and with β-arrestin–dependent activity of PIP5K. The blue line indicates the PIP2 hump generated by reduced PIP5K activity. The black dashed line indicates control without β-arrestin knockdown.
Possible role for lipid kinase for maintaining PM PIP2 during PAR2 desensitization
Now we consider how β-arrestin knockdown might create a hump in the time course of PIP2 recovery (Fig. 4 A). Our concept is that the hump reflects dynamic changes in the activity of the lipid kinases PI4K and PIP5K. This idea was tested empirically using the mathematical model. The kinase PI4K phosphorylates PI to give PIP. This rate-limiting slow first step for PIP2 recovery in tsA201 cells is followed by a faster PIP5K, which makes PIP2 (Falkenburger et al., 2013). In the Falkenburger model for muscarinic receptors and in our model for PAR2, both kinases had to be accelerated empirically above their basal rates after receptor activation to account for the observed rates of PIP2 recovery and of IP3 production.
What factors might underlie such acceleration? We have not proven a mechanism, but mention a speculation that may suggest one factor. Both lipid kinases have been shown to interact with β-arrestin, which possibly provides a scaffold for them (Nelson et al., 2008; Kríz et al., 2014). We tried ad hoc modifications of the model to include a β-arrestin–dependent stabilization of an accelerated state of the either PIP5K or PI4K. The results with either assumption were similar. The concept is that agonist-accelerated lipid kinase activity is stable when free β-arrestin levels are high, but when β-arrestin is already knocked down and then depletes further as it binds to sensitized receptors, the free level falls so low that the lipid kinase activity turns off. For this calculation only, the PIP5K activity during PAR2 activation was taken as the sum two components, the fixed basal rate B and an additional accelerated rate. In the version used only for Fig. 9 C, the rate of PIP5K was given by
where tau_on is the time constant (1 s) for quick exponential approach to full accelerated kinase activity after receptor activation and tau_off is an arrestin-dependent time constant for fall of the accelerated kinase activity. This tau_off is proportional to [β-arrestin]4 (Table 1). The fall (tau_off) is very slow (>104 s) when β-arrestin is normal. However, it becomes fast as cytosolic free β-arrestin is strongly lowered. If we assumed that the knocked down β-arrestins have been lowered to 30%, the model generated an appropriate PIP2 hump (Fig. 9 C, blue line) compared with 100% β-arrestins (Fig. 9 C, black dashed line).
What might this calculation imply? One plausible scenario is that β-arrestin translocated to PAR2 may recruit the PIP5K to the PM where PIP5K generates PIP2 more effectively compared with the diffusible PIP5K in cytosol. We mimicked this idea experimentally. If the hump is caused by a curtailed PIP5K acceleration at the PM when β-arrestin is low, we would expect to be able to restore fuller recovery of PIP2 by recruiting extra PIP5K to the PM. In experiments in Fig. 10 (A and B), we expressed a PM-anchored FRB domain (Lyn11-FRB-CFP) and a CFP-tagged PIP5K coupled to an FKBP domain (PIP5K-FKBP-CFP; see Methods and Materials and legend for details). This pair of proteins can be chemically dimerized by adding rapamycin (Suh et al., 2006). When such cells with β-arrestin siRNA were treated with rapamycin plus AP, exogenous PIP5K-FKBP-CFP translocated to the anchor at the PM (Fig. 10 A). As predicted, with this forced enrichment of PIP5K at the PM, the hump was eliminated (Fig. 10 B). AP alone did not clearly translocate PIP5K-FKBP-CFP. To make such speculation more plausible would require an additional direct test and a matching more complete mechanistic model of the steps.
DISCUSSION
Our experiments suggest that PAR2 phosphorylation suffices to desensitize receptors and that β-arrestin secures this phosphorylated state for long-term deactivation of receptors. Using these ideas, we developed the first mathematical description of PAR2 desensitization. It fits the experiments and predicts additional parameters we could not measure directly.
Phosphorylation and PAR2 desensitization
Rhodopsin can be desensitized by phosphorylation at multiple sites (Doan et al., 2006) followed by visual arrestin binding, and conversely, visual arrestin binding to the receptor affects the kinetics of receptor phosphorylation (Mendez et al., 2000; Doan et al., 2009). Similarly, Gs-coupled receptors such as β2-adrenergic receptors use phosphorylation of activated receptors by GRKs followed by recruitment of β-arrestin to the phosphorylated and still ligand-bound receptors for desensitization (Carman et al., 2000; Penn et al., 2000; Violin et al., 2008; Vayttaden et al., 2010). Because this mechanism will desensitize only those receptors that are bound to agonist, it is called homologous desensitization. In contrast, receptor desensitization mediated by protein kinases such as PKA or PKC that may not care about whether the receptor is or ever was activated is called heterologous desensitization. What about PAR2? Numerous lines of evidence support desensitization by phosphorylation mediated by parallel actions of GRKs and PKC. (a) Agonist-induced desensitization is slowed both by an inhibitor of GRKs and by an inhibitor of PKC (Fig. 3 B). (b) Activated PAR2 recruits GRK2 (Zhao et al., 2015). (c) β-Arrestins 1 and 2 translocate to activated PAR2 (Fig. 3 C and Fig. S5; Ricks and Trejo, 2009; Zhao et al., 2015) but not after mutation of phosphorylation sites in the C terminus (Ricks and Trejo, 2009). (d) β-Arrestin knockdown slows desensitization, but desensitization is restored to normal simply by inhibiting serine/threonine phosphatases (Fig. 4, A–E). (e) Activating PAR2 suffices to produce DAG and activate PKC (Fig. 2 and Fig. S3; Kim et al., 2008). (f) Pharmacological activation of PKC desensitizes PAR2 (Fig. 3 A; Böhm et al., 1996; DeFea et al., 2000), although it does not recruit β-arrestin (Fig. 3 C). In sum, activation of PAR2 is likely to stimulate phosphorylation by both GRKs and by PKC. This phosphorylation seems to be enough to turn off signaling by the receptors, and subsequent binding of arrestins protects the phosphorylation from protein phosphatases that would otherwise reverse the desensitization.
The PIP2 hump is consistent with possible interaction between PIP kinases and β-arrestin
When cytosolic β-arrestins were reduced by siRNA, the AP agonist evoked both two phases of PIP2 depletion: a first rapid phase of loss of PIP2 followed by the secondary hump (Fig. 4 A). One possible interpretation, crudely corroborated by the model, is that the second phase reflects a reduction of PIP2 synthesis when β-arrestin is reduced. According to recent findings in Wnt/β-catenin signaling, β-arrestin interacts with PI4K and PIP5K at the PM to augment PIP2 pools in human embryonic kidney (HEK) cells (Kríz et al., 2014), the cell line from which tsA201 cells were derived. If this concept also applies during PAR2 signaling, a knockdown of β-arrestin would reduce PM PI4K and PIP5K activity and attenuate PIP2 regeneration, favoring the secondary loss of PIP2. Indeed, adding β-arrestin dependence of PIP5K activity to our model predicted a hump in β-arrestin–depleted cells. A down-regulation of PI4K in our model had a similar effect.
Up-regulation of PI4K or PIP5K during PAR2 activation
Previously, to fit experimental data for M1 muscarinic receptor activation in tsA201 cells, we had postulated that PI4K and PIP5K are transiently accelerated by 10-fold by the muscarinic agonist (Falkenburger et al., 2013). Again, in the PAR2 model, both rate constants needed to be accelerated, sixfold for PI4K and threefold for PIP5K, to simulate the recovery of PIP2 during PAR2 desensitization. As demonstrated in Fig. 10 B, artificial recruitment of PIP5K to the PM reduced the PIP2 hump generated by β-arrestin knockdown. However, the concept of lipid kinase recruitment and activation by β-arrestins has to be regarded as just hypothetical, and we advance no direct experimental evidence for it. It is just a candidate idea to be considered in the future. Other candidate signals for lipid kinase acceleration need to be considered as well, such as the rise of intracellular Ca2+ reported to activate PI4K via neuronal Ca2+ sensor 1 (Gamper et al., 2004; Delmas et al., 2005; de Barry et al., 2006) and Rho kinase, which regulates PIP5K activity (Mao and Yin, 2007).
Concluding remarks
Significant roles for PAR2 are reported in many cell types (Gieseler et al., 2013; Yau et al., 2013). For example, it stimulates ion and mucin secretion in PDECs (Nguyen et al., 1999; Jung et al., 2006, 2009, 2010; Kim et al., 2008), it protects acinar and pancreatic duct cells from acute pancreatitis (Namkung et al., 2004), and it promotes inflammation in airway epithelial cells (Soh et al., 2010; Ramachandran et al., 2012; Yau et al., 2013). As this receptor is activated irreversibly by proteolytic cleavage, understanding its desensitization has broad relevance. In addition, desensitization of GqPCRs in general, of which PAR2 is a member, is still not deeply explored. As a first step, we focused on the effect of phosphorylation. Key conclusions were that phosphorylation alone suffices to desensitize the receptor and that arrestin and protein serine/threonine phosphatases compete dynamically for phosphorylated receptors. Our results and model imply that β-arrestin may have at least two roles: (1) stabilizing phosphorylated ligand-bound receptors and (2) interacting with PI4K and PIP5K to regulate the amount of PM PIP2. It will be interesting to extend such studies to other members of the family of GqPCRs.
Supplementary Material
Acknowledgments
We thank Drs. Fred Rieke, Eamonn J. Dickson, Martin Kruse, Oscar Vivas, Gucan Dai, and Haijie Yu for reading the manuscript, Lea M. Miller for technical assistance, and Dr. Jill B. Jensen for help with the FRET measurements.
The Virtual Cell is supported by National Institutes of Health (NIH) grant number P41 GM103313 from the National Institute for General Medical Sciences. Our work was supported by NIH grant numbers R01-080840 (to D.-S. Koh), R01-GM083913 and R37-NS08174 (to B. Hille), and R01-GM099373 and 1S10RR026406 (to C.L. Asbury).
The authors declare no competing financial interests.
Angus C. Nairn served as editor.
Footnotes
Abbreviations used in this paper:
- AP
- activating peptide
- a.u.
- arbitrary units
- CFP
- cyan fluorescent protein
- CKAR
- C kinase activity reporter
- DAG
- diacylglycerol
- FKBP
- FK506-binding protein
- FRET
- fluorescence resonance energy transfer
- FRETr
- FRET ratio
- GqPCR
- Gq protein–coupled receptor
- GRK
- G protein–coupled receptor kinase
- IP3
- inositol trisphosphate
- PDEC
- pancreatic duct epithelial cell
- PH
- pleckstrin homology
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PIP5K
- phosphatidylinositol 4-phosphate 5 kinase
- PM
- plasma membrane
- RFP
- red fluorescent protein
- YFP
- yellow fluorescent protein
References
- Ahn S., Nelson C.D., Garrison T.R., Miller W.E., and Lefkowitz R.J.. 2003. Desensitization, internalization, and signaling functions of β-arrestins demonstrated by RNA interference. Proc. Natl. Acad. Sci. USA. 100:1740–1744. 10.1073/pnas.262789099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm S.K., Khitin L.M., Grady E.F., Aponte G., Payan D.G., and Bunnett N.W.. 1996. Mechanisms of desensitization and resensitization of proteinase-activated receptor-2. J. Biol. Chem. 271:22003–22016. 10.1074/jbc.271.36.22003 [DOI] [PubMed] [Google Scholar]
- Bojjireddy N., Guzman-Hernandez M.L., Reinhard N.R., Jovic M., and Balla T.. 2015. EFR3s are palmitoylated plasma membrane proteins that control responsiveness to G-protein-coupled receptors. J. Cell Sci. 128:118–128. 10.1242/jcs.157495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman C.V., Barak L.S., Chen C., Liu-Chen L.Y., Onorato J.J., Kennedy S.P., Caron M.G., and Benovic J.L.. 2000. Mutational analysis of Gβγ and phospholipid interaction with G protein-coupled receptor kinase 2. J. Biol. Chem. 275:10443–10452. 10.1074/jbc.275.14.10443 [DOI] [PubMed] [Google Scholar]
- de Barry J., Janoshazi A., Dupont J.L., Procksch O., Chasserot-Golaz S., Jeromin A., and Vitale N.. 2006. Functional implication of neuronal calcium sensor-1 and phosphoinositol 4-kinase-β interaction in regulated exocytosis of PC12 cells. J. Biol. Chem. 281:18098–18111. 10.1074/jbc.M509842200 [DOI] [PubMed] [Google Scholar]
- DeFea K.A., Zalevsky J., Thoma M.S., Déry O., Mullins R.D., and Bunnett N.W.. 2000. β-arrestin–dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell Biol. 148:1267–1282. 10.1083/jcb.148.6.1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P., Coste B., Gamper N., and Shapiro M.S.. 2005. Phosphoinositide lipid second messengers: new paradigms for calcium channel modulation. Neuron. 47:179–182. 10.1016/j.neuron.2005.07.001 [DOI] [PubMed] [Google Scholar]
- Déry O., Thoma M.S., Wong H., Grady E.F., and Bunnett N.W.. 1999. Trafficking of proteinase-activated receptor-2 and β-arrestin-1 tagged with green fluorescent protein. β-Arrestin-dependent endocytosis of a proteinase receptor. J. Biol. Chem. 274:18524–18535. 10.1074/jbc.274.26.18524 [DOI] [PubMed] [Google Scholar]
- Dickson E.J., Falkenburger B.H., and Hille B.. 2013. Quantitative properties and receptor reserve of the IP3 and calcium branch of Gq-coupled receptor signaling. J. Gen. Physiol. 141:521–535. 10.1085/jgp.201210886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A., Kobilka B.K., Strader D.J., Benovic J.L., Dohlman H.G., Frielle T., Bolanowski M.A., Bennett C.D., Rands E., Diehl R.E., et al. 1986. Cloning of the gene and cDNA for mammalian β-adrenergic receptor and homology with rhodopsin. Nature. 321:75–79. 10.1038/321075a0 [DOI] [PubMed] [Google Scholar]
- Doan T., Mendez A., Detwiler P.B., Chen J., and Rieke F.. 2006. Multiple phosphorylation sites confer reproducibility of the rod’s single-photon responses. Science. 313:530–533. 10.1126/science.1126612 [DOI] [PubMed] [Google Scholar]
- Doan T., Azevedo A.W., Hurley J.B., and Rieke F.. 2009. Arrestin competition influences the kinetics and variability of the single-photon responses of mammalian rod photoreceptors. J. Neurosci. 29:11867–11879. 10.1523/JNEUROSCI.0819-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburger B.H., Jensen J.B., and Hille B.. 2010a. Kinetics of M1 muscarinic receptor and G protein signaling to phospholipase C in living cells. J. Gen. Physiol. 135:81–97. 10.1085/jgp.200910344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburger B.H., Jensen J.B., and Hille B.. 2010b. Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 135:99–114. 10.1085/jgp.200910345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburger B.H., Dickson E.J., and Hille B.. 2013. Quantitative properties and receptor reserve of the DAG and PKC branch of Gq-coupled receptor signaling. J. Gen. Physiol. 141:537–555. 10.1085/jgp.201210887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper N., Reznikov V., Yamada Y., Yang J., and Shapiro M.S.. 2004. Phosphatidylinositol 4,5-bisphosphate signals underlie receptor-specific Gq/11-mediated modulation of N-type Ca2+ channels. J. Neurosci. 24:10980–10992. 10.1523/JNEUROSCI.3869-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseler F., Ungefroren H., Settmacher U., Hollenberg M.D., and Kaufmann R.. 2013. Proteinase-activated receptors (PARs) - focus on receptor-receptor-interactions and their physiological and pathophysiological impact. Cell Commun. Signal. 11:86 10.1186/1478-811X-11-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guellich A., Mehel H., and Fischmeister R.. 2014. Cyclic AMP synthesis and hydrolysis in the normal and failing heart. Pflugers Arch. 466:1163–1175. 10.1007/s00424-014-1515-1 [DOI] [PubMed] [Google Scholar]
- Heitzler D., Durand G., Gallay N., Rizk A., Ahn S., Kim J., Violin J.D., Dupuy L., Gauthier C., Piketty V., et al. 2012. Competing G protein-coupled receptor kinases balance G protein and β-arrestin signaling. Mol. Syst. Biol. 8:590 10.1038/msb.2012.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz L.F., Hirdes W., Suh B.C., Hilgemann D.W., Mackie K., and Hille B.. 2005. Phospholipase C in living cells: activation, inhibition, Ca2+ requirement, and regulation of M current. J. Gen. Physiol. 126:243–262. 10.1085/jgp.200509309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J.B., Lyssand J.S., Hague C., and Hille B.. 2009. Fluorescence changes reveal kinetic steps of muscarinic receptor-mediated modulation of phosphoinositides and Kv7.2/7.3 K+ channels. J. Gen. Physiol. 133:347–359. 10.1085/jgp.200810075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.R., Kim K., Hille B., Nguyen T.D., and Koh D.S.. 2006. Pattern of Ca2+ increase determines the type of secretory mechanism activated in dog pancreatic duct epithelial cells. J. Physiol. 576:163–178. 10.1113/jphysiol.2006.114876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.R., Kim M.H., Hille B., and Koh D.S.. 2009. Control of granule mobility and exocytosis by Ca2+-dependent formation of F-actin in pancreatic duct epithelial cells. Traffic. 10:392–410. 10.1111/j.1600-0854.2009.00884.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.R., Hille B., Nguyen T.D., and Koh D.S.. 2010. Cyclic AMP potentiates Ca2+-dependent exocytosis in pancreatic duct epithelial cells. J. Gen. Physiol. 135:527–543. 10.1085/jgp.200910355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Choi B.H., Jung S.R., Sernka T.J., Kim S., Kim K.T., Hille B., Nguyen T.D., and Koh D.S.. 2008. Protease-activated receptor-2 increases exocytosis via multiple signal transduction pathways in pancreatic duct epithelial cells. J. Biol. Chem. 283:18711–18720. 10.1074/jbc.M801655200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberg K., Kim T.Y., Möller M., Skegro D., Dasara Raju G., Granzin J., Büldt G., Schlesinger R., and Alexiev U.. 2011. Conformational dynamics of helix 8 in the GPCR rhodopsin controls arrestin activation in the desensitization process. Proc. Natl. Acad. Sci. USA. 108:18690–18695. 10.1073/pnas.1015461108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasel C., Bünemann M., Lorenz K., and Lohse M.J.. 2005. β-Arrestin binding to the β2-adrenergic receptor requires both receptor phosphorylation and receptor activation. J. Biol. Chem. 280:9528–9535. 10.1074/jbc.M413078200 [DOI] [PubMed] [Google Scholar]
- Kríz V., Pospíchalová V., Masek J., Kilander M.B., Slavík J., Tanneberger K., Schulte G., Machala M., Kozubík A., Behrens J., and Bryja V.. 2014. β-arrestin promotes Wnt-induced low density lipoprotein receptor-related protein 6 (Lrp6) phosphorylation via increased membrane recruitment of Amer1 protein. J. Biol. Chem. 289:1128–1141. 10.1074/jbc.M113.498444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz R.J., Rockman H.A., and Koch W.J.. 2000. Catecholamines, cardiac β-adrenergic receptors, and heart failure. Circulation. 101:1634–1637. 10.1161/01.CIR.101.14.1634 [DOI] [PubMed] [Google Scholar]
- Lowe J.D., Sanderson H.S., Cooke A.E., Ostovar M., Tsisanova E., Withey S.L., Chavkin C., Husbands S.M., Kelly E., Henderson G., and Bailey C.P.. 2015. Role of G protein–coupled receptor kinases 2 and 3 in µ-opioid receptor desensitization and internalization. Mol. Pharmacol. 88:347–356. 10.1124/mol.115.098293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell L.M., and Lefkowitz R.J.. 2002. The role of β-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell Sci. 115:455–465. [DOI] [PubMed] [Google Scholar]
- Mao Y.S., and Yin H.L.. 2007. Regulation of the actin cytoskeleton by phosphatidylinositol 4-phosphate 5 kinases. Pflugers Arch. 455:5–18. 10.1007/s00424-007-0286-3 [DOI] [PubMed] [Google Scholar]
- McMahon H.T., and Boucrot E.. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12:517–533. 10.1038/nrm3151 [DOI] [PubMed] [Google Scholar]
- Mendez A., Burns M.E., Roca A., Lem J., Wu L.W., Simon M.I., Baylor D.A., and Chen J.. 2000. Rapid and reproducible deactivation of rhodopsin requires multiple phosphorylation sites. Neuron. 28:153–164. 10.1016/S0896-6273(00)00093-3 [DOI] [PubMed] [Google Scholar]
- Namkung W., Han W., Luo X., Muallem S., Cho K.H., Kim K.H., and Lee M.G.. 2004. Protease-activated receptor 2 exerts local protection and mediates some systemic complications in acute pancreatitis. Gastroenterology. 126:1844–1859. 10.1053/j.gastro.2004.03.019 [DOI] [PubMed] [Google Scholar]
- Nelson C.D., Kovacs J.J., Nobles K.N., Whalen E.J., and Lefkowitz R.J.. 2008. β-arrestin scaffolding of phosphatidylinositol 4-phosphate 5-kinase Iα promotes agonist-stimulated sequestration of the β2-adrenergic receptor. J. Biol. Chem. 283:21093–21101. 10.1074/jbc.M800431200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.D., Moody M.W., Steinhoff M., Okolo C., Koh D.S., and Bunnett N.W.. 1999. Trypsin activates pancreatic duct epithelial cell ion channels through proteinase-activated receptor-2. J. Clin. Invest. 103:261–269. 10.1172/JCI2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oancea E., Teruel M.N., Quest A.F., and Meyer T.. 1998. Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells. J. Cell Biol. 140:485–498. 10.1083/jcb.140.3.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda D., Savard C.E., Nguyen T.D., Eng L., Swenson E.R., and Lee S.P.. 1996. Dog pancreatic duct epithelial cells: long-term culture and characterization. Am. J. Pathol. 148:977–985. [PMC free article] [PubMed] [Google Scholar]
- Ostermaier M.K., Schertler G.F., and Standfuss J.. 2014. Molecular mechanism of phosphorylation-dependent arrestin activation. Curr. Opin. Struct. Biol. 29:143–151. 10.1016/j.sbi.2014.07.006 [DOI] [PubMed] [Google Scholar]
- Penn R.B., Pronin A.N., and Benovic J.L.. 2000. Regulation of G protein-coupled receptor kinases. Trends Cardiovasc. Med. 10:81–89. 10.1016/S1050-1738(00)00053-0 [DOI] [PubMed] [Google Scholar]
- Ramachandran R., Noorbakhsh F., Defea K., and Hollenberg M.D.. 2012. Targeting proteinase-activated receptors: therapeutic potential and challenges. Nat. Rev. Drug Discov. 11:69–86. 10.1038/nrd3615 [DOI] [PubMed] [Google Scholar]
- Ranganathan R., and Stevens C.F.. 1995. Arrestin binding determines the rate of inactivation of the G protein-coupled receptor rhodopsin in vivo. Cell. 81:841–848. 10.1016/0092-8674(95)90004-7 [DOI] [PubMed] [Google Scholar]
- Rasmussen S.G., DeVree B.T., Zou Y., Kruse A.C., Chung K.Y., Kobilka T.S., Thian F.S., Chae P.S., Pardon E., Calinski D., et al. 2011. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 477:549–555. 10.1038/nature10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricks T.K., and Trejo J.. 2009. Phosphorylation of protease-activated receptor-2 differentially regulates desensitization and internalization. J. Biol. Chem. 284:34444–34457. 10.1074/jbc.M109.048942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarangapani K.K., Duro E., Deng Y., Alves F.L., Ye Q., Opoku K.N., Ceto S., Rappsilber J., Corbett K.D., Biggins S., et al. 2014. Sister kinetochores are mechanically fused during meiosis I in yeast. Science. 346:248–251. 10.1126/science.1256729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy S.K., and Lefkowitz R.J.. 2011. β-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol. Sci. 32:521–533. 10.1016/j.tips.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A.K., Westfield G.H., Xiao K., Reis R.I., Huang L.Y., Tripathi-Shukla P., Qian J., Li S., Blanc A., Oleskie A.N., et al. 2014. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature. 512:218–222. 10.1038/nature13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh U.J., Dores M.R., Chen B., and Trejo J.. 2010. Signal transduction by protease-activated receptors. Br. J. Pharmacol. 160:191–203. 10.1111/j.1476-5381.2010.00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalheim L., Ding Y., Gullapalli A., Paing M.M., Wolfe B.L., Morris D.R., and Trejo J.. 2005. Multiple independent functions of arrestins in the regulation of protease-activated receptor-2 signaling and trafficking. Mol. Pharmacol. 67:78–87. 10.1124/mol.104.006072 [DOI] [PubMed] [Google Scholar]
- Suh B.C., Inoue T., Meyer T., and Hille B.. 2006. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 314:1454–1457. 10.1126/science.1131163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura A., Morita T., Nezu A., Shitara A., Hashimoto N., and Tojyo Y.. 2009. Use of fluorescence resonance energy transfer-based biosensors for the quantitative analysis of inositol 1,4,5-triphosphate dynamics in calcium oscillations. J. Biol. Chem. 284:8910–8917. 10.1074/jbc.M805865200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.M., Friedman J., Baameur F., Knoll B.J., Moore R.H., and Clark R.B.. 2007. Characterization of β2-adrenergic receptor dephosphorylation: Comparison with the rate of resensitization. Mol. Pharmacol. 71:47–60. 10.1124/mol.106.028456 [DOI] [PubMed] [Google Scholar]
- van der Wal J., Habets R., Várnai P., Balla T., and Jalink K.. 2001. Monitoring agonist-induced phospholipase C activation in live cells by fluorescence resonance energy transfer. J. Biol. Chem. 276:15337–15344. 10.1074/jbc.M007194200 [DOI] [PubMed] [Google Scholar]
- Vayttaden S.J., Friedman J., Tran T.M., Rich T.C., Dessauer C.W., and Clark R.B.. 2010. Quantitative modeling of GRK-mediated β2AR regulation. PLOS Comput. Biol. 6:e1000647 10.1371/journal.pcbi.1000647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin J.D., Zhang J., Tsien R.Y., and Newton A.C.. 2003. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol. 161:899–909. 10.1083/jcb.200302125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin J.D., DiPilato L.M., Yildirim N., Elston T.C., Zhang J., and Lefkowitz R.J.. 2008. β2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J. Biol. Chem. 283:2949–2961. 10.1074/jbc.M707009200 [DOI] [PubMed] [Google Scholar]
- Yau M.K., Liu L., and Fairlie D.P.. 2013. Toward drugs for protease-activated receptor 2 (PAR2). J. Med. Chem. 56:7477–7497. 10.1021/jm400638v [DOI] [PubMed] [Google Scholar]
- Zhang C., Srinivasan Y., Arlow D.H., Fung J.J., Palmer D., Zheng Y., Green H.F., Pandey A., Dror R.O., Shaw D.E., et al. 2012. High-resolution crystal structure of human protease-activated receptor 1. Nature. 492:387–392. 10.1038/nature11701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Lieu T., Barlow N., Sostegni S., Haerteis S., Korbmacher C., Liedtke W., Jimenez-Vargas N.N., Vanner S.J., and Bunnett N.W.. 2015. Neutrophil elastase activates protease-activated receptor-2 (PAR2) and transient receptor potential vanilloid 4 (TRPV4) to cause inflammation and pain. J. Biol. Chem. 290:13875–13887. 10.1074/jbc.M115.642736 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.