Abstract
Chromosomal aneuploidies are observed in essentially all sporadic carcinomas. These aneuploidies result in tumor-specific patterns of genomic imbalances that are acquired early during tumorigenesis, continuously selected for and faithfully maintained in cancer cells. Although the paradigm of translocation induced oncogene activation in hematologic malignancies is firmly established, it is not known how genomic imbalances affect chromosome-specific gene expression patterns in particular and how chromosomal aneuploidy dysregulates the genetic equilibrium of cells in general. To model specific chromosomal aneuploidies in cancer cells and dissect the immediate consequences of genomic imbalances on the transcriptome, we generated artificial trisomies in a karyotypically stable diploid yet mismatch repair-deficient, colorectal cancer cell line and in telomerase immortalized, cytogenetically normal human breast epithelial cells using microcell-mediated chromosome transfer. The global consequences on gene expression levels were analyzed using cDNA arrays. Our results show that regardless of chromosome or cell type, chromosomal trisomies result in a significant increase in the average transcriptional activity of the trisomic chromosome. This increase affects the expression of numerous genes on other chromosomes as well. We therefore postulate that the genomic imbalances observed in cancer cells exert their effect through a complex pattern of transcriptional dysregulation.
INTRODUCTION
Aneuploidy is a consistent genetic alteration of the cancer genome (1–4). When the first quantitative measurements of the DNA content of cancer cells were performed, aneuploidy was defined as a variation in nuclear DNA content in the population of cancer cells within a tumor (5). With increased resolution of cytogenetic techniques, such as chromosome banding (6), comparative genomic hybridization (CGH; ref. 7), spectral karyotyping (SKY), and multiplex fluorescence in situ hybridization (8, 9), it has become clear that in addition to nuclear aneuploidy, specific nonrandom chromosomal imbalances (heretofore referred to as chromosomal aneuploidy) exist. Indeed, despite genetic instability in cancer genomes, cancer cell populations as a whole display a surprisingly conserved, tumor-specific pattern of genomic imbalances (4, 10, 11). At early steps in the sequence of malignant transformation during human tumorigenesis, e.g., in preinvasive dysplastic lesions, such chromosomal aneuploidies can be the first detectable genetic aberration (12–17). This suggests that there is both an initial requirement for the acquisition of specific chromosomal aneuploidies and a requirement for the maintenance of these imbalances despite genomic and chromosomal instability. This would be consistent with continuous selective pressure to retain a specific pattern of chromosomal copy number changes in the majority of tumor cells (4, 18–20). Additionally, in cell culture model systems in which cells are exposed to different carcinogens, chromosomal aneuploidy is the earliest detectable genomic aberration (21, 22).
The conservation of these tumor and tumor-stage–specific patterns of chromosomal aneuploidies suggests that they play a fundamental biological role in tumorigenesis. It remains, however, unresolved how such genomic imbalances affect global gene expression patterns. One could postulate that expression levels of all transcriptionally active genes on trisomic chromosomes would increase in accordance with the chromosome copy number. Alternatively, changing the expression level of only one or a few genes residing on that chromosome through tumor-specific chromosomal aneuploidies may be the selective advantage necessary for tumorigenesis. This would require the permanent transcriptional silencing of most of the resident genes. Another formal possibility that must be entertained is that chromosomal copy number changes are either neutral or inversely correlated with respect to gene expression levels. This would mean that gains or losses of chromosomes are a by product of specific gene mutation and may not offer any selective advantage. Methodology to analyze the consequences of chromosomal imbalances in tumor genomes has become available through the development of microarray based gene expression profiling, yet the few reports that attempt to specifically address this problem come to quite different conclusions (23–27).
Because of the many chromosomal aberrations usually found in cancer cells, it is difficult, if not impossible, to identify the consequences of specific trisomies, independent from other coexisting genomic imbalances, gene mutations, or epigenetic alterations (28). To develop a model system that allows direct correlation of acquired chromosome copy number alterations with transcriptional activity in genetically identical cells, we have used microcell-mediated chromosome transfer methodology. The introduction of three different chromosomes into karyotypically diploid, mismatch repair-deficient colorectal cancer cells and into immortalized normal breast epithelial cells allowed an assessment of the consequences of specific aneuploidies on global gene expression levels relative to their diploid parental cells.
MATERIALS AND METHODS
Microcell-Mediated Chromosome Transfer
Mouse/human hybrid cell lines were purchased from the Coriell Repository5 and the Japan Health Sciences Foundation6. All hybrids were cultured according to manufacturers’ recommendations. The diploid colorectal cancer cell line, DLD1, was purchased from American Type Culture Collection.7 The telomerase immortalized mammary epithelial cell line (hTERT-HME) was purchased from Clontech (Palo Alto, CA) and cultured in the recommended medium. Both recipient cell lines were first tested for the optimal concentration of G418 (Geneticin; Invitrogen, Carlsbad, CA). Microcell-mediated chromosome transfer methodology was performed as described previously (29, 30). Briefly, donor A9 cells were grown in six Nunclon T-25 flasks at 1 × 106 cells/flask in media containing 500 µg/mL Geneticin. Cells were incubated with 0.05 µg/mL Colcemid in media plus 20% serum for 48 hours to induce micronuclei formation. Cells were centrifuged in the presence of 10 µg/mL cytochalasin B at 8000 rpm for 1 hour at 34°C to isolate micronuclei. Micronuclei were purified by sequential filtration through sterile 8-, 5-, and 3-µm filters (Millipore, Billerica, MA). Purified micronuclei were incubated with the recipient cells for 15 to 20 minutes in phytohemagglutinin P containing medium (100 µg/mL). The medium was removed, and cells were coated with 1 mL of PEG 1500 (Roche, Indianapolis, IN) for 1 minute followed by three washes with serum-free medium and incubated overnight in serum containing medium. Cells were plated onto 100 mm2 plates at 1 × 106 cells/plate in medium plus Geneticin (200 µg/mL for DLD1 clones and 50 µg/mL for hTERT-HME clones) for 2 to 3 weeks, until clones appeared. Clones were expanded and tested for incorporation of neomycin-tagged chromosome by fluorescence in situ hybridization (FISH) using whole chromosome-specific paint probes and a neomycin-specific DNA probe. We generated four derivative cell lines: DLD1 containing an extra copy of chromosome 3 (DLD1 + 3); an extra copy of chromosome 7 (DLD1 + 7); and chromosome 13 (DLD1 + 13). In addition, chromosome 3 was introduced into the karyotypically normal immortalized mammary epithelial cell line (hTERT-HME + 3).
FISH and Spectral Karyotyping
Chromosome-specific painting probes were hybridized to confirm the incorporation of a given chromosome in the derived clones and centromere specific probes (Vysis, Downers Grove, IL) were used for quantitation of chromosome incorporation rate. FISH was performed as described previously (31). Briefly, slides were pretreated with RNase, fixed, and denatured in 70% formamide/2× SSC for 1.5 minutes at 80°C. Centromere/telomere probes were denatured at 74°C for 5 minutes and placed on the denatured slides, coverslipped, and incubated at 37°C overnight (16 to 20 hours). Slides were washed, counterstained with 4′,6-diamidino-2-phenylindole and mounted with antifade solution. Images were acquired on a DMRXA epifluorescence microscope (Leica, Wetzlar, Germany) using Q-Fluoro software (Leica, Cambridge, United Kingdom). SKY was performed as described previously (8). Slides were pretreated with RNase, followed by pepsin to remove cytoplasm and denatured as described above. Slides were then hybridized with a SKY probe mixture for 72 hours at 37°C. Images were acquired and processed as described previously (8).
Microarray Analysis
RNA samples were prepared from multiple passages from each cell culture, and total RNA was extracted using TRIZOL (Invitrogen, Carlsbad, CA) followed by Qiagen RNeasy column purification (Qiagen, Valencia, CA) at least two separate times for each cell line. Each replicate RNA preparation was hybridized using a slightly modified protocol from ref. 32 on separate occasions. Extraction and hybridization protocols used can be viewed in detail online.8 In brief, 20 µg of total RNA were reverse transcribed using random primers and converted into cDNA using reverse transcriptase. After incorporation of aminoallyl-conjugated nucleotides, the RNA was indirectly labeled with Cy3 (cell line RNA) and Cy5 (reference RNA; Amersham, Piscataway, NJ). Each sample was hybridized against universal human reference RNA (Stratagene, La Jolla, CA) in a humid chamber (Arraylt Hybridization Cassette, TeleChem Intl., Sunnyvale, CA) for 16 hours at 42°C, washed, and scanned by the Axon GenePix 4000B Scanner (Axon Instruments, Union City, CA). Three replicates and a reverse fluorochrome labeling were performed for each sample (see Supplementary Table 1). We used customized arrays obtained from the National Cancer Institute’s Advanced Technology Center. All arrays were hybridized within two print batches and composed of 9128 cDNA denatured and immobilized on a poly-l-lysine–coated glass surface. The gene annotation file (GAL file) used for both print batches (Hs-UniGEM2-v2px-32Bx18Cx18R.gal) can be found at the Advanced Technology Center web site.9 GenePix software version 4.0.1.17 was used to apply the GAL file through an interactive gridding process (33).
Microarray Quality Assessment and Data Analysis
Log ratios for each spot were calculated as follows: in each channel, signal was calculated as foreground mean minus background median. If the signal was <100 in a single channel, the signal value in that channel was set to 100. If the signal was <100 in both channels, the spot was flagged as unreliable and not used in further analyses. Ratios for nonflagged spots were calculated by dividing the green (Cy3) signal by the red (Cy5) signal and then a log base 2 transformation was applied. Log ratios were normalized within each array by subtracting from each the median log ratio value across the spots on the array. These median-normalized log ratios (removing all flagged spots) were used in all subsequent analyses. Intensity-based normalization was considered; however, based on the diagnostic plots (Supplementary Fig. 1), a global normalization approach (with the truncation of signal at 100) was considered superior for this data set (34).
To determine whether the hybridizations were of sufficient quality to extract meaningful and statistically relevant conclusions, we examined the correlation among the four replicate arrays within each experimental condition. These pairwise correlations were calculated to assess the variability between hybridizations (technical repeats), cells at the time of harvest (biological repeats), and due to the physical properties of the individual fluorochromes (reverse-fluorochrome labeling). This was done for each of the six different experimental groups: DLD1, DLD1 + 3, DLD1 + 7, DLD1 + 13, hTERT-HME, and hTERT-HME +3. DLD1 was analyzed on two print batches (I and II). For the three purely technical replicates, the pairwise correlations were 0.83, 0.87, and 0.87 (see Supplementary Table 1). In subsequent analyses, all replicates within experimental conditions were treated equally, regardless of whether they were between-extraction, between-hybridization, or reverse fluorochrome replicates because the correlation analysis indicated that in this experimental system, between-extraction variability, or reverse fluorochrome variability (ratios inverted) contributed little additional variability over and above that due to hybridization. In Tables 1–5, columns labeled Ratio.0 for DLD1 represent the average value of eight replicate arrays from both print batches, Ratio.3 is normalized to the DLD1 Ratio.0 from print batch 2, and Ratio.7 and Ratio.13 are normalized to the DLD1 Ratio.0 from print batch 1.
The second phase of the analysis was to produce summary statistics of the expression levels within each group at the gene level, chromosome level, and chromosome arm level. All calculations were done on the log base 2 scale. To produce gene level summaries, the mean and SD of the log ratios for each gene across the four arrays within each group (missing values dropped) were calculated. Diagnostic plots (see Supplementary Fig. 1) suggested that the SD was constant as a function of mean log ratio across genes within a group, so an overall average SD was computed for each group as the median SD overall genes. For each array and each chromosome, chromosome level summaries were calculated by averaging log ratios from genes known to be located on that chromosome. When average expression ratios are reported, they have been obtained by taking antilog of the mean log ratios. Comparisons between experimental groups were conducted at each of the chromosome, chromosome arm, and gene levels (see Supplementary Materials and Methods for details).
RESULTS
Microcell-Mediated Chromosome Transfer
To analyze the immediate consequences of chromosomal copy number changes on gene expression profiles, we generated derivatives of the diploid colorectal cancer cell line, DLD1, and immortalized human mammary epithelial cell line (hTERT-HME) using microcell-mediated chromosome transfer to introduce extra copies of neomycin-tagged chromosomes 3, 7, and 13. We were successful in establishing clones for each of the derivative lines. The experiments conducted here were performed at low cell passage numbers and FISH with chromosome-specific probes confirmed the maintenance of extra copies of these chromosomes under neomycin selective cell culture conditions (Fig. 1A). A summary of the quantitative FISH data for each cell clone is given in Fig. 1B. We also performed SKY analysis to determine whether the chromosome transfer process induced secondary karyotypic changes. With the exception of loss of the Y chromosome in all DLD1 + 3 cells analyzed and in a small percentage of DLD1 + 7 cells, all four derivative lines maintained the diploid karyotype of the parental cell line and contained the additional copy of the introduced chromosome (Fig. 1, C and D). These results were also confirmed by CGH (the results can be viewed in detail online).10
Fig. 1.
Characterization of chromosome transfer clones. A, representative interphase/metaphase FISH experiment with 4′,6-diamidino-2-phenylindole counterstained DNA and centromere probes for chromosomes 3 (yellow) and 7 (red) in the DLD1 + 7 clone. B, population of cells within each clone containing two (yellow), three (red), or more than three (blue) copies of the introduced chromosome. C, representative karyotype from a SKY analysis of the parental cell line, DLD1. D, a spectral karyotype from the DLD1 + 3 clone clearly showing three copies of chromosome 3 and maintenance of the diploid DLD1 background.
Chromosome-Specific Gene Expression Patterns
With the selection of these clones, we could now examine the consequence of chromosome copy number increases on the average transcriptional activity of the trisomic chromosome. Table 1 shows the average gene expression ratios for each of the introduced chromosomes. This was obtained by averaging the expression ratios of all of the genes on that chromosome. The expression level of genes in the parental cell lines DLD1 and hTERT-HME was compared with a common reference RNA (Stratagene, La Jolla, CA; Table 1, Ratio.0). Ratios from the four derivative cell lines compared with the same reference RNA were subsequently normalized to the respective parental cell line ratios (Table 1, Ratio.3 vs 0, Ratio.7 vs 0, Ratio.13 vs 0 for DLD1 and Ratio.3 vs 0 for hTERT-HME). Because of the large number of P values being examined, we only discuss here results when P < 0.001 for comparisons where no hypothesis was prespecified. The introduction of chromosome 3 resulted in a significant increase in average gene expression on chromosome 3 (Ratio.3 vs 0 = 1.17, P < 0.0001). The addition of chromosome 7 resulted in a similar increase in the average expression ratio for chromosome 7 genes (Ratio.7 vs 0 = 1.19, P < 0.0001) and trisomy of chromosome 13 resulted in an increase in the average expression ratio for genes on chromosome 13 (Ratio.13 vs 0 = 1.26, P < 0.0009).
Table 1.
Average gene expression profiles by chromosome
| DLD1 | hTERT-HME | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Ratio.0 | Ratio.7 vs 0 | P.7 vs 0 | Ratio.13 vs 0 | P.13 vs 0 | Ratio.3 vs 0 | P.3 vs 0 | Ratio.0 | Ratio.3 vs 0 | P.3 vs 0 |
| 1 | 0.96 | 1.03 | 0.0242 | 1.01 | 0.99 | 0.98 | 1.02 | |||
| 2 | 0.98 | 1.03 | 1.00 | 1.00 | 1.00 | 0.98 | ||||
| 3 | 0.92 | 0.98 | 1.00 | 1.17 | <0.0001 | 0.98 | 1.20 | <0.0001 | ||
| 4 | 1.04 | 0.98 | 0.98 | 0.98 | 1.11 | 0.96 | ||||
| 5 | 0.93 | 1.00 | 0.99 | 1.02 | 0.99 | 0.99 | ||||
| 6 | 0.93 | 1.05 | 0.0183 | 1.02 | 0.99 | 0.94 | 0.96 | |||
| 7 | 0.86 | 1.19 | <0.0001 | 0.98 | 0.99 | 0.88 | 1.02 | |||
| 8 | 0.93 | 1.02 | 1.02 | 1.01 | 1.00 | 0.98 | ||||
| 9 | 0.96 | 1.00 | 1.00 | 0.98 | 0.96 | 1.00 | ||||
| 10 | 1.01 | 1.01 | 0.99 | 1.00 | 1.03 | 1.01 | ||||
| 11 | 0.92 | 1.01 | 0.97 | 1.00 | 0.95 | 0.99 | ||||
| 12 | 0.92 | 1.05 | 0.0147 | 0.99 | 1.00 | 0.93 | 0.99 | |||
| 13 | 0.99 | 1.01 | 1.26 | 0.0009 | 1.04 | 1.11 | 0.99 | |||
| 14 | 0.92 | 1.02 | 1.01 | 1.02 | 0.93 | 1.00 | ||||
| 15 | 0.90 | 1.03 | 1.02 | 1.01 | 0.97 | 1.01 | ||||
| 16 | 0.92 | 1.05 | 1.01 | 0.99 | 0.87 | 1.00 | ||||
| 17 | 1.00 | 1.06 | 1.02 | 0.99 | 0.98 | 0.98 | ||||
| 18 | 1.03 | 1.01 | 1.04 | 1.01 | 1.19 | 0.94 | ||||
| 19 | 0.94 | 1.02 | 0.95 | 0.99 | 0.94 | 1.01 | ||||
| 20 | 0.87 | 1.01 | 1.01 | 0.99 | 0.93 | 1.00 | ||||
| 21 | 0.89 | 1.04 | 1.03 | 0.99 | 0.87 | 1.00 | ||||
| 22 | 0.94 | 1.04 | 0.98 | 0.97 | 0.91 | 1.00 | ||||
| X | 0.79 | 1.03 | 0.99 | 0.99 | 0.85 | 1.02 | ||||
| Y | 1.10 | 1.00 | 0.99 | 0.74 | 0.0002 | 0.99 | 0.99 | |||
NOTE. For each chromosome, the average expression ratio for all genes on that chromosome relative to the reference RNA pool is given for the parental line (Ratio.0). The average chromosome expression ratios relative to the same reference RNA pool for each chromosome transfer clone is then normalized to the parental ratio value (Ratio.3 vs 0, Ratio.7 vs 0, and Ratio.13 vs 0) and their respective P values from two-sample t tests. Ratio values having P < 0.001 were considered significant and are in bold. P values ≥ 0.05 are not reported.
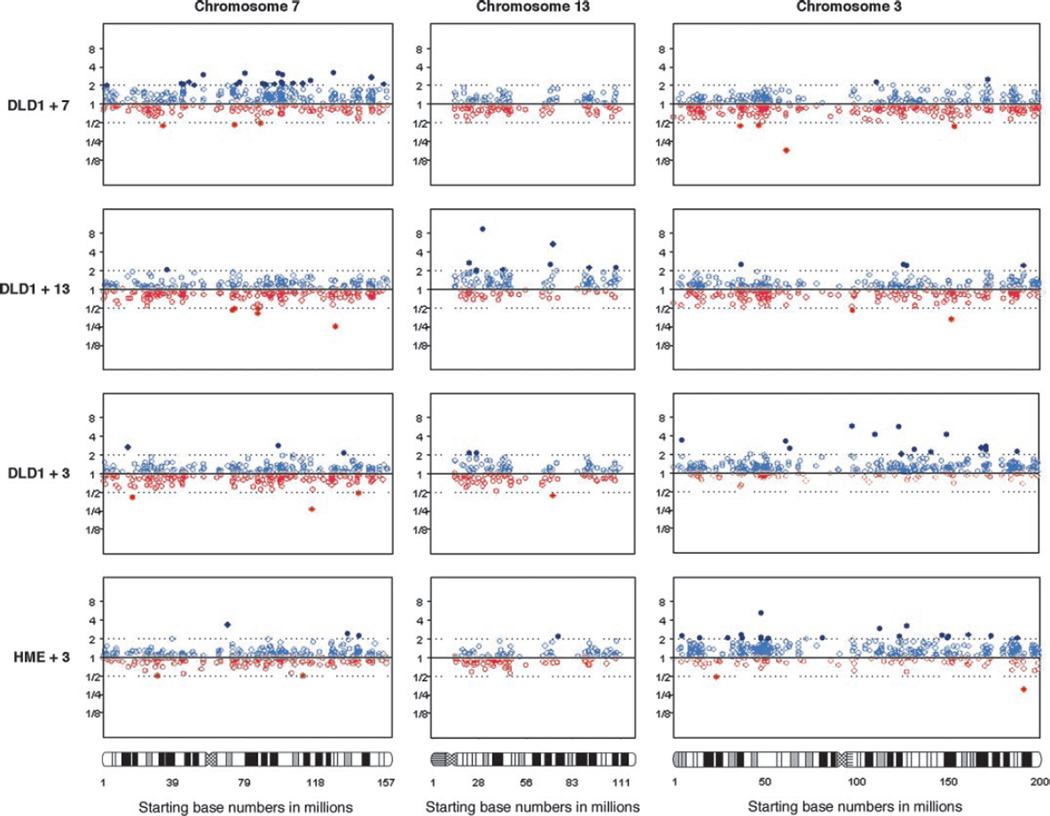
To determine whether this effect was specific for DLD1 per se or specific for the tissue of origin (colon), we analyzed the effect of an additional copy of chromosome 3 in a mammary epithelial cell line (hTERT-HME). The introduction of chromosome 3 into telomerase immortalized, karyotypically normal mammary epithelial cells resulted in an increase in the average gene expression of chromosome 3 genes as well (Ratio.3 vs 0 = 1.20, P < 0.0001). The results for the average gene expression increases are pictorially presented in Fig. 2. This figure clearly shows the average increase in expression of genes located on the trisomic chromosome compared with diploid chromosomes. When this analysis was repeated at the level of chromosome arms, no additional significant changes became apparent at the P < 0.001 level in any of the derivative cell lines (see Supplementary Table 2). Likewise, we did not observe any obvious local (i.e., chromosome band) clustering or region specific distribution of upregulated genes on the trisomic chromosome.
Fig. 2.
Global gene expression profiles. Each scatterplot displays all genes and their corresponding normalized expression ratio values along the length of each chromosome. Values in open light blue circles and open light orange circles represent ratio values between 0.5 and 2.0. Dark blue dots represent expression ratios ≥ 2.0 and dark orange dots are ratios ≤ 0.5. The X axis shows the starting bp location of each gene.
Global Gene Expression Patterns
In addition to analyzing average, chromosome-specific gene expression levels for the introduced chromosomes, we were interested in identifying additional, significantly dysregulated genes throughout the genome. The influence of chromosomal aneuploidy on the expression level of individual genes was examined by considering only those genes whose expression ratios were >2.0 (up-regulated) or <0.5 (down-regulated) when compared with the parental cell line. For clone DLD1 + 7, 155 genes were up-regulated and 47 down-regulated (23%) beyond these thresholds. Of those genes with known map locations, 18% (35 of 194) map to chromosome 7 and 32 of these were up-regulated (Table 2). Regarding chromosome 13, 92 genes were up-regulated and 72 were down-regulated (44%). Of those with known map locations, 6% (10 of 162) mapped to chromosome 13, of which all were up-regulated (Table 3). The introduction of chromosome 3 in DLD1 resulted in up-regulation of 81 genes and down-regulation of 67 genes (45%). Here, those with known map location, 12% (17/144) mapped to this chromosome and all were up-regulated (Table 4). Interestingly, this derivative cell line was the only one that demonstrated a highly significant decrease in the average expression of a chromosome other than that which was made trisomic (chromosome Y; Ratio.3 vs 0 = 0.74, P = 0.0002). The introduction of this same chromosome 3 into the hTERT-HME cells results in up-regulation of 91 genes and down-regulation of 49 genes (35%). Of those with known map location 17% (23 of 135) mapped to chromosome 3, 21 of which were up-regulated (Table 5). Strikingly, no genes were affected in common among any of the four cell lines. Table 6 displays the quantitative summary and respective chromosome designation of all genes whose expression was altered 2-fold in each cell line. The comprehensive gene lists of the above data are available in the Supplementary Tables 3–6. Five percent of all genes on the array mapped to each chromosome 7 and 3. For chromosome 13, the percentage was 1.7%. The observed percentages of up-regulated genes located on chromosomes 7 and 3 were each >20%, and the observed percentage of up-regulated genes residing on chromosome 13 was >10%. Thus, the percentages of up-regulated genes residing on the introduced chromosomes were substantially greater than would have been expected by chance if up-regulation occurred at random. The percentages of down-regulated genes residing on the introduced chromosomes were no more than expected by chance. Thus, we see not only a very specific increase in the overall average expression of genes on the trisomic chromosomes but also increases in a significant number of genes on those chromosomes, more than would be expected by chance due to the composition of the arrays. Additionally, a large number of genes on diploid chromosomes in these derivatives were also significantly increased, revealing a more complex global transcriptional dysregulation.
Table 2.
Genes significantly altered in expression in DLD1 + 7 and located on chromosome 7
| Ratio.7 vs 0 | Map | Gene | Description |
|---|---|---|---|
| Up-regulated genes | |||
| 12.40 | 7p21.2 | TWIST | Twist homologue (acrocephalosyndactyly 3; Saethre-Chotzen syndrome) (Drosophila) |
| 3.26 | 7q21.3-q22.1 | NPTX2 | Neuronal pentraxin II |
| 3.15 | 7q32.3 | SMO | Smoothened homologue (Drosophila) |
| 3.14 | 7q21 | GNAI1 | “Guanine nucleotide binding protein (G protein), α inhibiting activity polypeptide 1” |
| 3.13 | 7q21.3-q22.1 | NPTX2 | Neuronal pentraxin II |
| 2.98 | 7q22 | TRIP6 | Thyroid hormone receptor interactor 6 |
| 2.93 | 7p12 | GBAS | Glioblastoma amplified sequence |
| 2.69 | 7q36.1 | RARRES2 | Retinoic acid receptor responder (tazarotene induced) 2 |
| 2.56 | 7q22 | PCOLCE | Procollagen C-endopeptidase enhancer |
| 2.41 | 7q21.3 | ASNS | Asparagine synthetase |
| 2.40 | 7q21 | STEAP | Six transmembrane epithelial antigen of the prostate |
| 2.37 | 7q31.1 | CAV2 | Caveolin 2 |
| 2.27 | 7q11.23 | PMS2L9 | Postmeiotic segregation increased 2-like 9 |
| 2.26 | 7q11.23 | PTPN12 | “Protein tyrosine phosphatase, nonreceptor type 12” |
| 2.25 | 7p13-p12 | HUS1 | HUS1 checkpoint homologue (Schizosaccharomyces pombe) |
| 2.25 | 7q11.23 | BCL7B | B-cell CLL/lymphoma 7B |
| 2.17 | 7q31.1 | HBP1 | HMG-box containing protein 1 |
| 2.13 | 7q31.1 | ZNF277 | Zinc finger protein (C2H2 type) 277 |
| 2.13 | 7q21.3-q22 | SERPINE1 | “Serine (or cysteine) proteinase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1” |
| 2.12 | 7p14-cen | BLVRA | Biliverdin reductase A |
| 2.10 | 7q21 | STEAP2 | Six transmembrane epithelial antigen of prostate 2 |
| 2.10 | 7q36.3 | KIAA0010 | Ubiquitin-protein isopeptide ligase (E3) |
| 2.09 | 7p12.3 | LOC285958 | Hypothetical protein LOC285958 |
| 2.08 | 7q11.23 | MK-STYX | MAP kinase phosphatase-like protein MK-STYX |
| 2.08 | 7q21.3-q22.1 | DSS1 | Deleted in split-hand/split-foot 1 region |
| 2.06 | 7q36 | INSIG1 | Insulin-induced gene 1 |
| 2.05 | 7q32 | IRF5 | IFN regulatory factor 5 |
| 2.04 | 7q21-q22 | AKAP9 | A kinase (PRKA) anchor protein (yotiao) 9 |
| 2.03 | 7p22.2 | LOC221955 | KCCR13L |
| 2.02 | 7p22 | LFNG | Lunatic fringe homologue (Drosophila) |
| 2.02 | 7p12.1 | COBL | KIAA0633 protein |
| 2.00 | 7p22.3 | EIF3S9 | “Eukaryotic translation initiation factor 3, subunit 9 η, 116kDa” |
| Down-regulated genes | |||
| 0.44 | 7p14 | B1 | PTH-responsive osteosarcoma B1 protein |
| 0.46 | 7q11.23 | NCF1 | “Neutrophil cytosolic factor 1 (47 kDa, chronic granulomatous disease, autosomal 1)” |
| 0.49 | 7q21.12 | MGC26647 | Hypothetical protein MGC26647 |
NOTE. For all of the genes in this table, the P value associated with the approximate z-test of the chromosome transfer clone expression ratio compared to the parental clone expression ratio was <0.0001 with the exception of the following genes: NCF1, ′neutrophil cytosolic factor 1 (47 kDa, chronic granulomatous disease, autosomal 1)′ (P = 0.0155); and MGC26647, hypothetical protein MGC26647 (P = 0.0016). This table shows the summary of genes up- or down regulated (>2.0 and <0.5, respectively) in the DLD1 + 7 cell line that reside on chromosome 7. The ratio value, chromosome map location, name, and description are given for each gene.
Table 3.
Genes significantly altered in expression in DLD1 + 13 and located on chromosome 13
| Ratio.13 vs 0 | Map | Gene | Description |
|---|---|---|---|
| Up-regulated genes | |||
| 20.94 | 13q13 | MAB21L1 | Mab-21-like 1 (Caenorhabditis elegans) |
| 9.27 | 13q13.1 | SPG20 | “Spastic paraplegia 20, spartin (Troyer syndrome)” |
| 5.24 | 13q22 | SCEL | Sciellin |
| 3.25 | 13q32 | ZIC2 | “Zic family member 2 (odd-paired homologue, Drosophila)” |
| 2.64 | 13 | “ESTs, weakly similar to cytokine receptor-like factor 2; cytokine receptor CRL2 precursor (Homo sapiens)” | |
| 2.48 | 13q21.33 | LMO7 | LIM domain only 7 |
| 2.21 | 13q31.2-q32.3 | STK24 | “Serine/threonine kinase 24 (STE20 homologue, yeast)” |
| 2.20 | 13q34 | C13orf11 | Chromosome 13 open reading frame 11 |
| 2.03 | 13q14.3 | ITM2B | Integral membrane protein 2B |
| 2.01 | 13 | “Human BRCA2 region, mRNA sequence CG011” |
NOTE. For all of the genes in this table, the P value associated with the approximate z-test of the chromosome transfer clone expression ratio compared to the parental clone expression ratio was <0.0001 with the exception of the following genes: OTC, ornithine carbamoyltransferase (P = 0.0001); C14orf2, chromosome 14 open reading frame 2 (P = 0.0017); and NCF1, ′neutrophil cytosolic factor 1 (47 kDa, chronic granulomatous disease, autosomal 1)′ (P = 0.0027). This table shows the summary of genes up- or down-regulated (>2.0 and <0.5, respectively) in the DLD1 + 13 cell line that reside on chromosome 13. The ratio value, chromosome map location, name, and description are given for each gene.
Table 4.
Genes significantly altered in expression in DLD1 + 3 and located on chromosome 3
| Ratio.3 vs 0 | Map | Gene | Description |
|---|---|---|---|
| Up-regulated genes | |||
| 5.71 | 3q12.1 | ESDN | Endothelial and smooth muscle cell-derived neuropilin-like protein |
| 5.55 | 3q21 | MYLK | “Myosin, light polypeptide kinase” |
| 4.43 | 3p22-p21.3 | CCK | Cholecystokinin |
| 4.21 | 3q25.1-q26.1 | SMARCA3 | “SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 3” |
| 4.18 | 3q13 | PVRL3 | Poliovirus receptor-related 3 |
| 3.38 | 3p26 | BHLHB2 | “Basic helix-loop-helix domain containing, class B, 2” |
| 3.23 | 3p21-p14 | PTPRG | “Protein tyrosine phosphatase, receptor type, G” |
| 3.17 | 3p21 | SCN5A | “Sodium channel, voltage-gated, type V, α polypeptide [long (electrocardiographic) QT syndrome 3]” |
| 3.10 | 3q23 | RBP1 | “Retinol binding protein 1, cellular” |
| 2.65 | 3q26.2-q26.3 | CLDN11 | Claudin 11 (oligodendrocyte transmembrane protein) |
| 2.50 | 3q26.2 | SERPINI1 | “Serine (or cysteine) proteinase inhibitor, clade I (neuroserpin), member 1” |
| 2.44 | 3p14.3-p14.2 | ADAMTS9 | “A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 9” |
| 2.38 | 3q21-q23 | MRPL3 | Mitochondrial ribosomal protein L3 |
| 2.33 | 3q26 | SKIL | SKI-like |
| 2.18 | 3q27-q28 | SIAT1 | “Sialyltransferase 1 (β-galactoside α-2,6-sialyltransferase)” |
| 2.17 | 3q23-q24 | CLSTN2 | Calsyntenin 2 |
| 2.00 | 3q21.2 | KIAA1237 | KIAA1237 protein |
NOTE. For all of the genes in this table, the P value associated with the approximate z-test of the chromosome transfer clone expression ratio compared to the parental clone expression ratio was <0.0001. This table shows the summary of genes up- or down-regulated (>2.0 and <0.5, respectively) in the DLD1 + 3 cell line that reside on chromosome 3. The ratio value, chromosome map location, name, and description are given for each gene.
Table 5.
Genes significantly altered in expression in hTERT-HME +3 and located on chromosome 3
| Ratio.3 vs 0 | Map | Gene | Description |
|---|---|---|---|
| Up-regulated genes | |||
| 5.21 | 3p21-p22 | PFKFB4 | “6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4” |
| 3.97 | 3p11-q11.2 | PROS1 | Protein S (α) |
| 3.17 | 3q21 | MCM2 | “MCM2 minichromosome maintenance deficient 2, mitotin (Saccharomyces cerevisiae)” |
| 2.92 | 3q13.2 | LOC152185 | Hypothetical protein AY099107 |
| 2.32 | 3q26.1 | SMC4L1 | SMC4 structural maintenance of chromosomes 4-like 1 (yeast) |
| 2.27 | 3p22-p21.3 | PLCD1 | “Phospholipase C, δ 1” |
| 2.24 | 3q23-q24 | PLOD2 | “Procollagen-lysine, 2-oxoglutarate 5-dioxygenase (lysine hydroxylase) 2” |
| 2.21 | 3q26.1-q26.2 | ECT2 | Epithelial cell-transforming sequence 2 oncogene |
| 2.20 | 3p26 | BHLHB2 | “Basic helix-loop-helix domain containing, class B, 2” |
| 2.17 | 3q21 | MYLK | “Myosin, light polypeptide kinase” |
| 2.15 | 3q23-q24 | TAZ | Transcriptional co-activator with PDZ-binding motif (TAZ) |
| 2.10 | 3 | Homo sapiens clone 24627 mRNA sequence | |
| 2.08 | 3p24.1-p21.2 | CELSR3 | “Cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homologue, Drosophila)” |
| 2.06 | 3p22 | TGFBR2 | “Transforming growth factor, β receptor II (70/80 kDa)” |
| 2.04 | 3p25-p24 | SLC6A6 | “Solute carrier family 6 (neurotransmitter transporter, taurine), member 6” |
| 2.04 | 3q27 | RFC4 | “Replication factor C (activator 1) 4, 37 kDa” |
| 2.03 | 3p22-p21.3 | XYLB | Xylulokinase homologue (Haemophilus influenzae) |
| 2.02 | 3q25.1-q26.1 | SMARCA3 | “SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 3” |
| 2.02 | 3p12.3 | GBE1 | “Glucan (1,4-α-), branching enzyme 1 (glycogen branching enzyme, andersen disease, glycogen storage disease type IV)” |
| 2.01 | 3p21.3 | SLC26A6 | “Solute carrier family 26, member 6” |
| 2.01 | 3p21.31 | STAB1 | Stabilin 1 |
| Down-regulated genes | |||
| 0.30 | 3q28-q29 | CLDN1 | Claudin 1 |
| 0.48 | 3p24.1 | NR1D2 | “Nuclear receptor subfamily 1, group D, member 2” |
NOTE. For all of the genes in this table, the P value associated with the approximate z-test of the chromosome transfer clone expression ratio compared with the parental clone expression ratio was <0.0001. This table shows the summary of genes up- or down-regulated (>2.0 and <0.5, respectively) in the hTERT-HME +3 cell line that reside on chromosome 3. The ratio value, chromosome map location, name, and description are given for each gene.
Table 6.
Summary of 2-fold altered gene lists
| DLD1 + 7 | DLD1 + 13 | DLD1 + 3 | HME +3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of genes two-fold altered | 202 | 164 | 148 | 140 | ||||||||
| No. of up-regulated genes | 155 | 92 | 81 | 91 | ||||||||
| Map on chromosome | 32 | 10 | 17 | 21 | ||||||||
| Map off chromosome | 117 | 82 | 64 | 66 | ||||||||
| Map unknown | 6 | 0 | 0 | 4 | ||||||||
| No. of down-regulated genes | 47 | 72 | 67 | 49 | ||||||||
| Map on chromosome | 3 | 0 | 0 | 2 | ||||||||
| Map off chromosome | 42 | 70 | 63 | 46 | ||||||||
| Map unknown | 2 | 2 | 4 | 1 |
NOTE. Genes up or down regulated (normalized ratio > 2.0 and < 0.5, respectively).
DISCUSSION
The application of cytogenetic techniques, in particular SKY and CGH, to the analysis of chromosomal aberrations in carcinomas has shown that recurrent patterns of genomic imbalances and chromosomal aneuploidies are common themes in tumors of epithelial origin (4, 35, 36). These imbalances are specific for different tumors and, to a certain extent, also tumor-stage specific and are maintained even after prolonged propagation under cell culture conditions (37). Genomic and chromosomal instability therefore do not seem to affect the distribution of these acquired genetic alterations. In general, imbalances of entire chromosomes, also referred to as chromosomal aneuploidies, occur at early stages of tumorigenesis, whereas regional amplifications typically occur in more advanced tumors. For instance, one of the earliest genetic abnormalities observed in the development of sporadic colorectal tumors is trisomy of chromosome 7 (19). Usually, once acquired, these specific imbalances are maintained despite ongoing chromosomal instability (38). It is therefore reasonable to assume that continuous selective pressure for the maintenance of established genomic imbalances exists in cancer genomes.
With the advent of technologies for the parallel monitoring of global gene expression patterns, it has become possible to analyze the consequences of such tumor-specific chromosomal aneuploidies on the transcriptome. One can hypothesize that chromosomal aneuploidies influence gene expression levels in several distinct ways: firstly, the expression levels of all genes on an aneuploid chromosome are affected by genomic copy number changes such that monosomy or trisomy of a chromosome results in an overall reduction or increase of absolute expression levels, respectively, of all or most genes; alternatively, chromosomal copy number changes affect only a few relevant target genes on the aneuploid chromosome, and chromosome missegregation and resulting aneuploidy are merely convenient mechanisms for simultaneously increasing and maintaining increased dosage of a limited number of chromosomally linked genes; thirdly, aneuploidies do not have any appreciable affect (or even an inverse affect) on gene expression levels.
Several groups have therefore engaged to address these hypotheses. Phillips et al. (26) used a model system for prostate tumorigenesis and found that relatively few genes on trisomic chromosomes were up-regulated beyond a certain threshold; however, the average expression level of all genes on that particular chromosome increased significantly. Similarly, Platzer et al. (25) described silencing of genes on chromosomes that are gained or amplified in primary liver metastases of colorectal carcinomas. Quite a different picture emerges from four other reports in which amplicons in primary breast cancers and breast cancer-derived cell lines were analyzed by CGH and expression profiling. These studies suggest a more direct impact of genomic copy number changes on gene expression levels (23, 24, 27, 39). Although the results of these six studies appear to be at odds with one another, a direct comparison of the results is not easy. To begin with, four different tumor types, including nonepithelial hematologic cells, were analyzed. Although one could presuppose a general principle by which gene copy number predictably affects gene expression patterns, there is no compelling reason to assume this is the case. Therefore, we must allow for the possibility that distinct tumors with dissimilar chromosomal aneuploidies may in fact behave differently. Additionally, the chromosome aberrations in the studies by both Phillips et al. (26) and Platzer et al. (25) primarily involved whole chromosomes or chromosome arms, whereas the breast cancers tended to have more regional copy number changes (0.2 to 12 Mb; ref. 23). Finally, the methodologies used were often different (e.g., chromosome CGH versus array CGH and different platforms for expression analyses), making it even more difficult to determine the source of the disparity.
To address the question of how acquired chromosomal aneuploidies affect gene expression levels in a controlled setting, we generated a unique experimental model system in which the only genetic alteration between parental and derived cell lines is an extra copy of a single chromosome. In all instances, the average transcriptional activity of the trisomic chromosomes was significantly increased. Thus, several important conclusions regarding the impact of chromosomal aneuploidy on cellular transcription levels can be drawn from our analysis. First, alterations in the copy number of whole chromosomes results in an increase in average gene expression for that chromosome. The average increase in gene expression (1.21), however, was lower than the average increase of genomic copy number (1.44). These results were consistent with results from similar analyses of aneuploid colorectal, pancreatic and renal cancer derived cell lines in which we observed a trend, indicating that indeed chromosomal aneuploidies correlate with global transcription levels, at least for the majority of commonly aneuploid chromosomes.11 Second, chromosomes not observed to be aneuploid in particular tumor types (i.e., chromosome 3 in colorectal tumors) also have increased transcriptional activity when placed into that cellular environment. Thus, their presence is not neutral with respect to the transcriptome. In fact, the number of genes that increase when chromosome 3 is introduced in either colorectal or mammary gland cells is similar (however, only a few genes are the same). Additionally, long-term culture of the DLD1 chromosome transfer clones indicate that chromosome 3 is rather quickly eliminated from DLD1 cells (this differs from observations in DLD + 7 and DLD + 13 cell lines). One could therefore hypothesize that increased expression of certain genes residing on chromosome 3 results in selective pressure against maintaining extra copies of this chromosome in DLD1 cells. This is not a novel concept and is in fact supported by previous studies identifying tumor suppressor genes through reversion of the tumor phenotype upon microcell-mediated chromosome transfer (40, 41). Third, aneuploidy not only affects gene expression levels on the chromosomes present in increased copy numbers but a substantial number of genes residing on other chromosomes significantly increased or decreased, apparently in a stochastic manner. This is of course consistent with known mechanisms of gene regulation (e.g., activator and suppressor proteins, signaling pathways, and so on) and the fact that genes residing in a given pathway are for the most part distributed throughout the genome on different chromosomes.
Three groups have analyzed the consequences of constitutional chromosomal trisomies on transcriptional activity in noncancerous, fetal cells and in a mouse model of human trisomy 21 and reach similar conclusions as we do (42–44). These studies concluded that the average gene expression of trisomic chromosomes is clearly increased, although this was not due to high-level up-regulation of only a few specific genes. However, expression levels of multiple genes throughout the genome were dysregulated. Normal cells with constitutional chromosomal aneuploidies (or segmental duplications) cannot tolerate trisomy of >4.3% of the genome (45, 46). However, this limit is not merely a reflection of the DNA content because multiple copies of an inactivated X-chromosome can be tolerated. Therefore, this limit is more likely to be imposed by global disturbance of the transcriptome as a consequence of genomic imbalances. This hypothesis is supported by the recent identification of differential average expression levels of specific chromosomes. For instance, the average gene expression levels and gene density of chromosomes 13, 18, and 21 are lower than those of smaller chromosomes (46), yet trisomy of only these chromosomes is compatible with life in noncancerous cells. It is interesting to speculate that one of the specific features of emerging cancer cells, which would differ from nontransformed cells that carry constitutional trisomies, would be the ability to exceed this transcriptional threshold during the multiple steps required for tumorigenesis. This global dysregulation of the transcriptome of cancers of epithelial origin may also reflect on our ability for therapeutic intervention: although the consequences of a simple chromosomal translocation, such as the BCR/ABL fusion in chronic myelogenic leukemia, can be successfully targeted with an inhibitor of the resulting tyrosine kinase activity such as Gleevec (47), the normalization of the complex dysregulation of transcriptional activity in carcinomas requires a more general, less specific, and hence more complex interference.
Supplementary Material
Acknowledgments
We thank Buddy Chen and Joseph Cheng for information technological support and editorial assistance, Dr. Anitha Clarence for assistance with molecular cytogenetic analyses, and Dr. Simone Difilippantonio for technical assistance with chromosome transfer methodology.
Footnotes
Note: Supplementary data for this article can be found at Cancer Research Online (http://cancerres.aacrjournals.org).
Internet address: http://locus.umdnj.edu/nigms/.
Internet address: http://cellbank.nihs.go.jp/.
Internet address: http://www.atcc.org.
Internet address: http://www.riedlab.nci.nih.gov.
Internet address: http://nciarray.nci.nih.gov.
Internet address: http://www.ncbi.nlm.nih.gov/sky/skyweb.cgi.
M. J. Difilippantonio et al., personal communication.
References
- 1.Heim S, Mitelman F. Cancer cytogenetics. New York: Alan R. Liss; 1987. [Google Scholar]
- 2.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature (Lond.) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 4.Ried T, Heselmeyer-Haddad K, Blegen H, Schrock E, Auer G. Genomic changes defining the genesis, progression, and malignancy potential in solid human tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999;25:195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 5.Caspersson TO. Quantitative tumor cytochemistry: G.H.A. Clowes Memorial Lecture. Cancer Res. 1979;39:2341–2345. [PubMed] [Google Scholar]
- 6.Caspersson T, Zech L, Johansson C, Modest EJ. Identification of human chromosomes by DNA-binding fluorescent agents. Chromosoma. 1970;30:215–227. doi: 10.1007/BF00282002. [DOI] [PubMed] [Google Scholar]
- 7.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science (Wash. DC) 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 8.Schröck E, du Manoir S, Veldman T, et al. Multicolor spectral karyotyping of human chromosomes. Science (Wash. DC) 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 9.Speicher MR, Ballard SG, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet. 1996;12:368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- 10.Knuutila S, Björkqvist A-M, Autio K, et al. DNA copy number amplifications in human neoplasms. Am J Pathol. 1998;152:1107–1123. [PMC free article] [PubMed] [Google Scholar]
- 11.Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends Genet. 1997;13:405–409. doi: 10.1016/s0168-9525(97)01244-4. [DOI] [PubMed] [Google Scholar]
- 12.Hittelman WN. Genetic instability in epithelial tissues at risk for cancer. Ann NY Acad Sci. 2001;952:1–12. doi: 10.1111/j.1749-6632.2001.tb02723.x. [DOI] [PubMed] [Google Scholar]
- 13.Hopman AHN, Ramaeker FCS, Raap AK, et al. In situ hybridization as a tool to study numerical chromosome aberrations in solid bladder tumors. Histochemistry. 1988;89:307–316. doi: 10.1007/BF00500631. [DOI] [PubMed] [Google Scholar]
- 14.Heim S, Mitelman F. Cancer cytogenetics. 2nd. New York: Wiley-Liss; 1995. [Google Scholar]
- 15.Ried T, Knutzen R, Steinbeck R, et al. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Solinas-Toldo S, Wallrapp C, Müller-Pillasch F, Bentz M, Gress T, Lichter P. Mapping of chromosomal imbalances in pancreatic carcinoma by comparative genomic hybridization. Cancer Res. 1996;56:3803–3807. [PubMed] [Google Scholar]
- 17.Heselmeyer K, Schrock E, du Manoir S, et al. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowak MA, Komarova NL, Sengupta A, et al. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci USA. 2002;99:16226–16231. doi: 10.1073/pnas.202617399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bomme L, Bardi G, Pandis N, Fenger C, Kronborg O, Heim S. Clonal karyotypic abnormalities in colorectal adenomas: clues to the early genetic events in the adenoma-carcinoma sequence. Genes Chromosomes Cancer. 1994;10:190–196. doi: 10.1002/gcc.2870100307. [DOI] [PubMed] [Google Scholar]
- 20.Desper R, Jiang F, Kallioniemi OP, Moch H, Papadimitriou CH, Schaffer AA. Distance-based reconstruction of tree models for oncogenesis. J Comput Biol. 2000;7:789–803. doi: 10.1089/10665270050514936. [DOI] [PubMed] [Google Scholar]
- 21.Oshimura M, Barrett JC. Chemically induced aneuploidy in mammalian cells: mechanisms and biological significance in cancer. Environ Mutagen. 1986;8:129–159. doi: 10.1002/em.2860080112. [DOI] [PubMed] [Google Scholar]
- 22.Barrett JC, Oshimura M, Tanaka N, Tsutsui T. Role of aneuploidy in early and late stages of neoplastic progression of Syrian hamster embryo cells in culture. Basic Life Sci. 1985;36:523–538. doi: 10.1007/978-1-4613-2127-9_36. [DOI] [PubMed] [Google Scholar]
- 23.Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- 24.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Platzer P, Upender MB, Wilson K, et al. Silence of chromosomal amplifications in colon cancer. Cancer Res. 2002;62:1134–1138. [PubMed] [Google Scholar]
- 26.Phillips JL, Hayward SW, Wang Y, et al. The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Res. 2001;61:8143–8149. [PubMed] [Google Scholar]
- 27.Virtaneva K, Wright FA, Tanner SM, et al. Expression profiling reveals fundamental biological differences in acute myeloid leukemia with isolated trisomy 8 and normal cytogenetics. Proc Natl Acad Sci USA. 2001;98:1124–1129. doi: 10.1073/pnas.98.3.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matzke MA, Mette MF, Kanno T, Matzke AJ. Does the intrinsic instability of aneuploid genomes have a causal role in cancer? Trends Genet. 2003;19:253–256. doi: 10.1016/s0168-9525(03)00057-x. [DOI] [PubMed] [Google Scholar]
- 29.Fournier RE, Frelinger JA. Construction of microcell hybrid clones containing specific mouse chromosomes: application to autosomes 8 and 17. Mol Cell Biol. 1982;2:526–534. doi: 10.1128/mcb.2.5.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxon PJ, Stanbridge EJ. Transfer and selective retention of single specific human chromosomes via microcell-mediated chromosome transfer. Methods Enzymol. 1987;151:313–325. doi: 10.1016/s0076-6879(87)51026-6. [DOI] [PubMed] [Google Scholar]
- 31.Ried T, Lengauer C, Cremer T, et al. Specific metaphase and interphase detection of the breakpoint region in 8q24 of Burkitt lymphoma cells by triple-color fluorescence in situ hybridization. Genes Chromosome Cancer. 1992;4:69–74. doi: 10.1002/gcc.2870040109. [DOI] [PubMed] [Google Scholar]
- 32.Hegde P, Qi R, Abernathy K, et al. A concise guide to cDNA microarray analysis. Biotechniques. 2000;29:548–550. 552–554. doi: 10.2144/00293bi01. 556 passim. [DOI] [PubMed] [Google Scholar]
- 33.Korn EL, Habermann J, Upender M, Ried T, McShane LM. An objective method of comparing DNA microarray image analysis systems. Biotechniques. 2004;36:960–967. doi: 10.2144/04366BI01. [DOI] [PubMed] [Google Scholar]
- 34.Simon RM, Korn EL, McShane LM, Radmacher MD, Wright GW, Zhao Y. Design and analysis of DNA microarray investigations. 1st. New York: Springer Verlag; 2003. [Google Scholar]
- 35.Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- 36.Knuutila S, Armengol G, Björkqvist A-M, et al. Comparative genomic hybridization study on pooled DNAs from tumors of one clinical-pathological entity. Cancer Genet Cytogenet. 1998;100:25–30. doi: 10.1016/s0165-4608(97)00001-0. [DOI] [PubMed] [Google Scholar]
- 37.Ghadimi BM, Sackett DL, Difilippantonio MJ, et al. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- 38.Roschke AV, Stover K, Tonon G, Schaffer AA, Kirsch IR. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia. 2002;4:19–31. doi: 10.1038/sj.neo.7900197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monni O, Barlund M, Mousses S, et al. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X, Tahin Q, Hu YF, et al. Functional roles of chromosomes 11 and 17 in the transformation of human breast epithelial cells in vitro. Int J Oncol. 1999;15:629–638. doi: 10.3892/ijo.15.4.629. [DOI] [PubMed] [Google Scholar]
- 41.Wan M, Sun T, Vyas R, Zheng J, Granada E, Dubeau L. Suppression of tumorigenicity in human ovarian cancer cell lines is controlled by a 2 cM fragment in chromosomal region 6q24–q25. Oncogene. 1999;18:1545–1551. doi: 10.1038/sj.onc.1202476. [DOI] [PubMed] [Google Scholar]
- 42.Saran NG, Pletcher MT, Natale JE, Cheng Y, Reeves RH. Global disruption of the cerebellar transcriptome in a Down syndrome mouse model. Hum Mol Genet. 2003;12:2013–2019. doi: 10.1093/hmg/ddg217. [DOI] [PubMed] [Google Scholar]
- 43.Mao R, Zielke CL, Zielke HR, Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81:457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 44.FitzPatrick DR, Ramsay J, McGill NI, Shade M, Carothers AD, Hastie ND. Transcriptome analysis of human autosomal trisomy. Hum Mol Genet. 2002;11:3249–3256. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 45.Brewer C, Holloway S, Zawalnyski P, Schinzel A, FitzPatrick D. A chromosomal duplication map of malformations: regions of suspected haplo- and triplolethality—and tolerance of segmental aneuploidy—in humans. Am J Hum Genet. 1999;64:1702–1708. doi: 10.1086/302410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caron H, van Schaik B, van der Mee M, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science (Wash. DC) 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 47.Druker BJ. Imatinib alone and in combination for chronic myeloid leukemia. Semin Hematol. 2003;40:50–58. doi: 10.1053/shem.2003.50000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.