Abstract
Riemerella anatipestifer is an important pathogen of waterfowl, which causes septicemia anserum exsudativa in ducks. In this study, an AS87_03730 gene deletion R. anatipestifer mutant Yb2ΔAS87_03730 was constructed to investigate the role of AS87_03730 on R. anatipestifer virulence and gene regulation. By deleting a 708-bp fragment from AS87_03730, the mutant Yb2ΔAS87_03730 showed a significant decreased growth rate in TSB and invasion capacity in Vero cells, compared to wild-type strain Yb2. Moreover, the median lethal dose (LD50) of Yb2ΔAS87_03730 was 1.24 × 107 colony forming units (CFU), which is about 80-fold attenuated than that of Yb2 (LD50 = 1.53 × 105 CFU). Furthermore, RNA-Seq analysis and Real-time PCR indicated 19 up-regulated and two down-regulated genes in Yb2ΔAS87_03730. Functional analysis revealed that 12 up-regulated genes were related to “Translation, ribosomal structure and biogenesis”, two were classified into “Cell envelope biogenesis, outer membrane”, one was involved in “Amino acid transport and metabolism”, and the other four had unknown functions. Polymerase chain reaction and sequence analysis indicated that the AS87_03730 gene is highly conserved among R. anatipestifer strains, as the percent sequence identity was over 93.5%. This study presents evidence that AS87_03730 gene is involved in bacterial virulence and gene regulation of R. anatipestifer.
Riemerella anatipestifer is a significant pathogen of waterfowl, turkey and other birds1,2. It causes epizootic infectious polyserositis in ducks characterized by lethargy, diarrhea, respiratory and nervous symptoms, which led to high mortality and consequently to great economic losses3. In view of its veterinary importance, many investigations have been carried out on the virulence factors of R. anatipestifer, including CAMP cohemolysin, OmpA, nucleoside-diphosphate-sugar epimerase, and glycosyl transferase etc.4,5,6,7. Recently, 49 virulence associated genes were identified by random transponson mutagenesis8.
The luxE gene from a number of bioluminescent bacteria had been well defined by genetic and biochemical analysis, including Vibrio harveyi, Vibrio fischeri, Photobacterium phosphoreum, etc9,10,11. The luxE gene encodes acyl-protein synthetase (LuxE) which activates the fatty acid, results in the formation of a fatty acyl-AMP intermediate and functions as the second step in the bioluminescent fatty acid reduction system12. Up to date, most studies focus on the role of luxE in bacterial bioluminescence reaction, and the role of luxE in R. anatipestifer has remained unknown. In this study, a luxE homology gene deletion mutant strain Yb2ΔAS87_03730 was constructed by allelic exchange, and the roles of AS87_03730 gene on bacterial growth, adherence and invasion capability, as well as colonization and development during infection were investigated. Furthermore, the function of AS87_03730 on the gene regulation at genome level of R. anatipestifer was investigated using RNA-Seq, the differentially expressed genes between mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2 were analyzed. The distribution of AS87_03730 gene in 36 R. anatipestifer strains with different serotypes and the gene homology were also analyzed.
Results
Characterization of mutant strain Yb2ΔAS87_03730
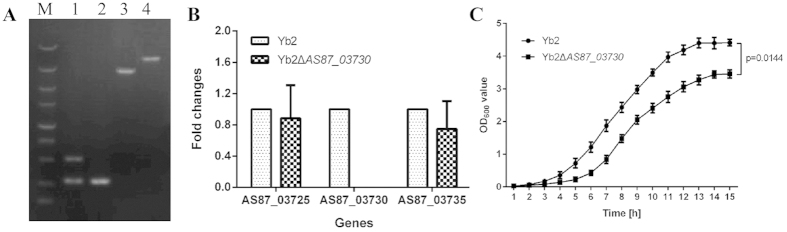
The AS87_03730 gene was deleted from the chromosome of R. anatipestifer Yb2 by alletic exchange, which was replaced by a SpecR cassette. The mutant strain was screened on TSA plates containing kanamycin (50 μg/ml) and spectinomycin (80 μg/ml), and confirmed by PCR amplification of the AS87_03730 and 16S rRNA fragments from transconjugants (Fig. 1A). Real-time PCR analysis further confirmed that AS87_03730 transcription was abolished in the mutant strain; however, inactivation of the AS87_03730 gene had no effect on the transcription of chromosomally upstream AS87_03725 and downstream AS87_03735 genes (Fig. 1B). The AS87_03730 deletion mutant strain was designated as Yb2ΔAS87_03730.
Figure 1. Characterization of mutant strain Yb2ΔAS87_03730.
(A) PCR amplification. Lane M: DM5000 DNA Marker (CWBIO, Beijing, China). Lane 1: Partial sequences of AS87_03730 gene and 16S rRNA were amplified from R. anatipestifer wild-type strain Yb2 using primers AS87_03730 in-F/AS87_03730 in-R and RA 16S rRNA-F/RA 16S rRNA-R, showing a 534-bp fragment of AS87_03730 or a 496-bp fragment of RA 16S rRNA. Lane 2: Partial sequence of R. anatipestifer 16S rRNA was amplified from mutant strain Yb2ΔAS87_03730 using primers RA 16S rRNA-F/RA 16S rRNA-R, showing a 496-bp fragment of RA 16S rRNA, no 534-bp fragment of AS87_03730 gene was amplified using primers AS87_03730 in-F/AS87_03730 in-R. Lane 3: A 3,681-bp flanking fragment of AS87_03730 gene was amplified from wild-type strain Yb2 using primers AS87_03730 out-F/AS87_03730 out-R. Lane 4: A 4,092-bp flanking fragment of AS87_03730 gene was amplified from mutant strain Yb2ΔAS87_03730 using primers AS87_03730 out-F/AS87_03730 out-R, indicating that a 708-bp fragment of AS87_03730 gene was replaced with the SpecR cassette. (B) Real-time PCR analysis. The transcription levels of upstream AS87_03725 gene and downstream AS87_03735 gene were measured. The changes of mRNAs were expressed as fold expression and calculated using the comparative CT (2−ΔΔCT) method. No transcription of AS87_03730 was detected from mutant strain Yb2ΔAS87_03730. Error bars represent SD from three replicates. (C) Bacterial growth curves. The mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2 were grown on TSB medium, and growth of each strain was monitored by measuring the OD600 values. The bacterial growth rate of mutant strain Yb2ΔAS87_03730 was significantly lower than that of wild-type strain Yb2 (p = 0.0144). The data was analyzed using Two-way ANOVA. Error bars represent the standard deviation of three independent experiments.
Mutant strain Yb2ΔAS87_03730 was viable when grown on TSB medium supplemented with kanamycin (50 μg/ml) and spectinomycin (80 μg/ml), however, the growth rate was significantly lower than that of wild-type strain Yb2 (Fig. 1C). When grown on TSA, mutant strain Yb2ΔAS87_03730 formed smooth, slightly raised and non-pigmented colony, which is similar to that of wild-type strain Yb2. Biochemical analysis demonstrated that both mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2 were unable to ferment saccharides (glucose, fructose, galactose, lactose, maltose, sucrose), but produced urease and liquefied gelatin (data not shown). The drug sensitivity of mutant strain Yb2ΔAS87_03730 was also determined, no differences were observed when compared to wild-type strain Yb2 (data not shown). These data suggest that the AS87_03730 gene has no influence on biochemical property and drug sensitivity of R. anatipestifer.
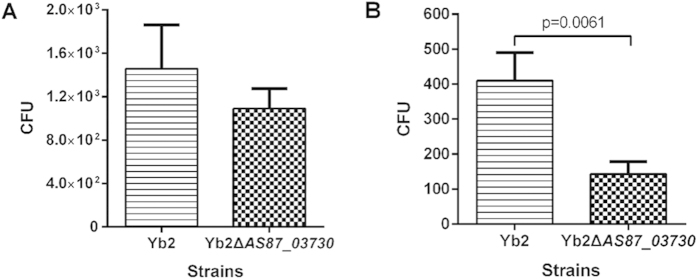
Deletion of AS87_03730 gene decreased bacterial adherence and invasion abilities
The adherence and invasion abilities of mutant strain Yb2ΔAS87_03730 to Vero cells were compared with wild-type strain Yb2 to investigate the role of AS87_03730 gene on the attachment and invasion of R. anatipestifer to host cells. When infection was performed at a multiplicity of infection (MOI) of 50, the adhered bacteria of mutant strain Yb2ΔAS87_03730 was 1,090 CFU/well, which was slightly decreased in comparison with that of wild-type strain Yb2 (1,456 CFU/well). After additional 1 h of incubation with 100 μg/ml gentamicin, the invaded bacterial counts of mutant strain Yb2ΔAS87_03730 was 140 CFU/well, about 3-fold lower than that of wild-type strain Yb2 (410 CFU/well) (p < 0.001). The results demonstrated that deletion of AS87_03730 gene has no influence on the bacterial adherence, but decreased bacterial invades significantly (Fig. 2).
Figure 2. Bacterial adherence and invasion assays.
The assays were performed on Vero cells. (A) Adherence assay. (B) Invasion assay. The Vero cells were infected with mutant strain Yb2ΔAS87_03730 or wild-type strain Yb2 at an MOI of 50, and incubated for 1.5 h to count the number of cell-adherent bacteria. For the invasion assay, the extra-cellular bacteria were killed by incubation of the monolayer with DMEM medium supplemented with 100 μg/ml gentamicin for an additional 1 h. Data were presented as mean ± standard deviations from three independent experiments. The data was analyzed using Student’s t-tests.
Determination of the bacterial virulence
Bacterial virulence was evaluated by the median lethal dose (LD50) using 18-day-old Cherry Valley ducks. The LD50 for mutant strain Yb2ΔAS87_03730 was 1.24 × 107 CFU, which was 80-fold attenuated virulence than that of wild-type strain Yb2 (1.53 × 105 CFU).
To further investigate the role of AS87_03730 gene on systemic infection in vivo, the bacterial loading in blood of infected ducks was quantified. The bacterial recovery of mutant strain Yb2ΔAS87_03730 was 1,255 CFU/ml, 1,845 CFU/ml, and 2,980 CFU/ml at 6, 12 and 24 h post infection (h.p.i.), respectively, whereas the bacterial recovery of wild-type strain Yb2 was 1,580 CFU/ml at 6 h.p.i., 2.36 × 104 CFU/ml at 12 h.p.i., and 3.80 × 104 CFU/ml at 24 h.p.i. respectively, indicating that mutant strain Yb2ΔAS87_03730 was drastically attenuated (Fig. 3).
Figure 3. Blood bacterial loadings in R. anatipestifer infected ducks.
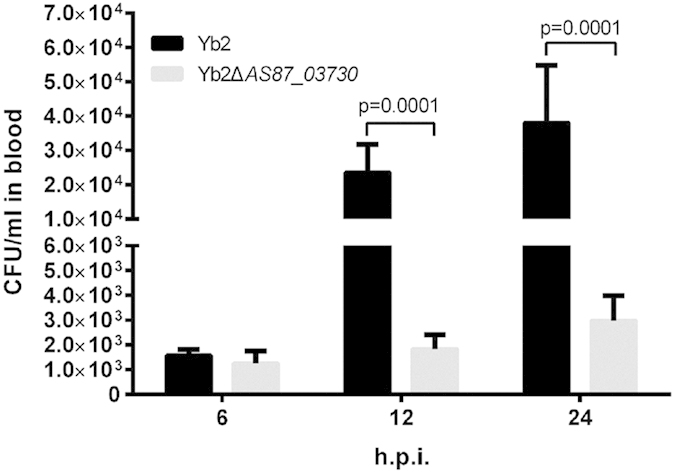
Six ducks were injected intramuscularly with 1 × 106 CFU of each bacterial strain. Blood samples were collected at 6, 12, and 24 h post infection and the bacterial CFU were counted. Data were presented as mean ± standard deviations from six infected ducks. The data was analyzed using Student’s t-tests.
Determination and functional categories of the differentially expressed genes
Differentially expressed genes between mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2 was investigated using strand-specific Illumina RNA-Seq analysis. In total, 31 genes were up-regulated and three genes were down-regulated in mutant strain Yb2ΔAS87_03730, compared to wild-type strain Yb2 (see Supplementary Table S1). Of them, real-time PCR further verified that 19 genes in Yb2ΔAS87_03730 were up-regulated by >2-fold at the transcriptional levels (Table 1). The proteins encoded by these genes were categorized on the basis of their putative functions. The products of up-regulated gene were mostly (63.16%, 12/19) related to “Translation, ribosomal structure and biogenesis”, and two proteins, encoded by AS87_03970 and AS87_06085, were classified into “Cell envelope biogenesis, outer membrane”. The protein encoded by AS87_02350 was involved in “Amino acid transport and metabolism”. There are only two down-regulated genes for hypothetical proteins, and one is involved in “Defense mechanisms”, the other has unknown function. Pathway analysis for the differentially expressed genes was performed according to the latest KEGG database (http://www.genome.jp/kegg/), and the differentially expressed genes were distributed in clusters. Furthermore, the up-regulated genes mostly (63.16%, 12/19) occurred within ribosome pathway (see Supplementary Figure S1), indicating AS87_03730 regulates the genes mainly responsible for translation process.
Table 1. Real-time PCR verification of differentially expressed genes in mutant strain Yb2ΔAS87_03730.
| Gene locusa | Description of genes | Subcellular locationb | Function group (COGs)c | 2—ΔΔCte |
|---|---|---|---|---|
| AS87_05650 | hypothetical protein | Cytoplasmic | –d | 4.31 |
| AS87_04530 | 50S ribosomal protein L10 | Unknown | COG0244 J | 4.30 |
| AS87_06160 | 50S ribosomal protein L28 | Cytoplasmic | COG0227 J | 4.25 |
| AS87_03970 | dTDP-4-dehydrorhamnose 3,5-epimerase | Cytoplasmic | COG1898 M | 4.19 |
| AS87_07745 | 50S ribosomal protein L23 | Cytoplasmic | COG0089 J | 3.88 |
| AS87_07845 | 50S ribosomal protein L36 | Unknown | – | 3.76 |
| AS87_06455 | 50S ribosomal protein L21 | Cytoplasmic | COG0261 J | 3.54 |
| AS87_06085 | dolichyl-phosphate beta-D-mannosyltransferase | Cytoplasmic | COG0463 M | 3.45 |
| AS87_07850 | 30S ribosomal protein S13 | Cytoplasmic | COG0099 J | 3.31 |
| AS87_07760 | 50S ribosomal protein L22 | Cytoplasmic | COG0091 J | 3.22 |
| AS87_02350 | cystathionine beta-synthase | Cytoplasmic | COG0031 E | 2.82 |
| AS87_07855 | 30S ribosomal protein S11 | Cytoplasmic | COG0100 J | 2.53 |
| AS87_07860 | 30S ribosomal protein S4 | Cytoplasmic | COG0522 J | 2.24 |
| AS87_07775 | 50S ribosomal protein L29 | Unknown | – | 2.13 |
| AS87_00900 | hypothetical protein | Cytoplasmic | – | 2.08 |
| AS87_07790 | 50S ribosomal protein L24 | Cytoplasmic | COG0198 J | 2.04 |
| AS87_07830 | 50S ribosomal protein L15 | Cytoplasmic | COG0200 J | 2.01 |
| AS87_07815 | 50S ribosomal protein L18 | Cytoplasmic | COG0256 J | 2.01 |
| AS87_07805 | 30S ribosomal protein S8 | Cytoplasmic | COG0096 J | 2.00 |
| AS87_05175 | 30S ribosomal protein S12 | Cytoplasmic | COG0048 J | 1.97 |
| AS87_04540 | 50S ribosomal protein L11 | Cytoplasmic | COG0080 J | 1.88 |
| AS87_05170 | 30S ribosomal protein S7 | Cytoplasmic | COG0049 J | 1.78 |
| AS87_02865 | hypothetical protein | Cytoplasmic | – | 1.75 |
| AS87_04500 | hypothetical protein | Unknown | – | 1.72 |
| AS87_07455 | membrane protein | Unknown | – | 1.65 |
| AS87_07825 | 50S ribosomal protein L30 | Cytoplasmic | COG1841 J | 1.65 |
| AS87_07870 | 50S ribosomal protein L17 | Cytoplasmic | COG0203 J | 1.57 |
| AS87_01795 | membrane-binding protein | Unknown | – | 1.55 |
| AS87_05160 | 30S ribosomal protein S10 | Cytoplasmic | COG0051 J | 1.55 |
| AS87_04930 | glycosyltransferase | Cytoplasmic | 1.29 | |
| AS87_02265 | S-adenosylmethionine tRNA ribosyltransferase | Cytoplasmic | COG0809 J | 1.09 |
| AS87_03335 | hypothetical protein | Unknown | – | 0.61 |
| AS87_05990 | hypothetical protein | Unknown | COG1002 V | 0.51 |
| AS87_03730 (mutated gene) | acyl-protein synthetase | Cytoplasmic | – | 0.01 |
Functional categories: (1) Information storage and processing: (J: Translation, ribosomal structure and biogenesis; K: Transcription; L: DNA replication, recombination and repair); (2) Cellular processes and signaling: (D: Cell division and chromosome partitioning; V: Defense mechanisms; O: Posttranslational modification, protein turnover, chaperones; M: Cell envelope biogenesis, outer membrane; N: Cell motility; T: Signal transduction mechanisms); (3) Metabolism: (C: Energy production and conversion; G: Carbohydrate transport and metabolism; H: Coenzyme transport and metabolism; P: Inorganic ion transport and metabolism; E: Amino acid transport and metabolism; F: Nucleotide transport and metabolism; Q: Secondary metabolites biosynthesis, transport and catabolism); (4) Poorly characterized: (R: General function prediction only; S: Function unknown).
aBased on R. anatipestifer Yb2 genome (accession number: CP007204).
bSubcellular locations were predicted by the PSORTb Version 3.0 server (http://www.psort.org/).
cFunctional characterization of the proteins was predicted by searching against eggNOG (Version 3) database using BLASTP.
dNo related COG.
eResults are presented as 2ΔΔCt. Figures = 1 indicated that the gene is expressed similarly in both mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2, figures >1 indicated that the gene is over expressed in mutant strain Yb2ΔAS87_03730, and figure <1 indicated that the gene is expressed less in mutant strain Yb2ΔAS87_03730.
AS87_03730 gene is highly conserved in R. anatipestifer strains
The AS87_03730 gene was successfully amplified from all 36 R. anatipestifer strains tested. Eight of the PCR products were further subjected to sequence analysis, indicating that Yb2 AS87_03730 gene shares a sequence identity of more than 93.5% with other 7 strains (Fig. 4). In addition, BLAST analysis showed that Yb2 AS87_03730 gene exhibits 100%, 100%, 97%, 97% and 91% identity compared to RA-CH-2 (GenBank accession no. CP004020.1), DSM15868 (GenBank accession no. CP002346.1), CH3 (GenBank accession no. CP006649.1), RA-CH-1 (GenBank accession no. CP003787.1) and RA-GD (GenBank accession no. CP002562.1), respectively. These data indicated that AS87_03730 gene was highly conserved in R. anatipestifer. Moreover, AS87_03730 gene shares 71% similarity to the BD94_0721 gene encoding putative acyl protein synthase/acyl-CoA reductase of Elizabethkingia anopheles NUHP1 (GenBank accession no. CP007547.1). The predicted LuxE exhibits sequence identity to acyl transferase from other bacteria, including Chryseobacterium formosense (GenBank accession no. KFE99559.1; 72%), Epilithonimonas sp. FH1 (GenBank accession no. KFC20122.1; 71%), Elizabethkingia menigoseptica (GenBank accession no. WP_026149215.1; 69%) and Bergeyella zoohelcum (GenBank accession no. WP_002687639.1; 61%).
Figure 4. Identity and divergence analysis of AS87_03730 gene in R. anatipestifer strains.
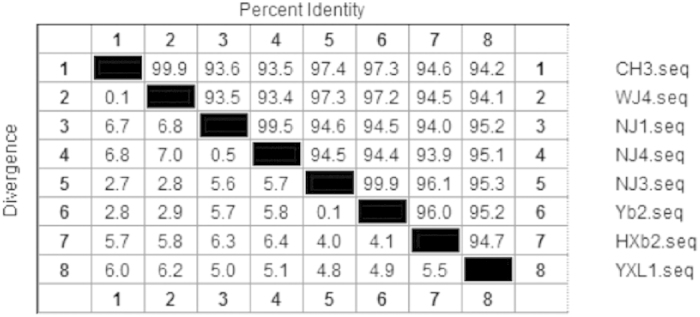
The nucleotide sequences of AS87_03730 from eight R. anatipestifer strains with different serotypes were sequenced and analyzed by DNASTAR software for percent identity and divergence. The percent sequence identity was >93.5%.
Discussion
LuxE is an acyl-protein synthetase found in bioluminescent bacteria, which catalyzes the formation of an acyl-protein thiolester from a fatty acid and protein. In addition to found in the bioluminescence system, a LuxE domain is also found in the Vibrio cholera RfbN protein, which is involved in the biosynthesis of the O-antigen component 3-deoxy-L-glycero-tetronic acid13. In R. anatipestifer strain Yb2, the homology gene of luxE is AS87_03730, the prevalence of the gene in R. anatipestifer that do not produce detectable light has led to the question of what role, if any, there is for the activity encoded by the AS87_03730 gene in non-luminescent bacteria. To better evaluate the role of AS87_03730 in R. anatipestifer, we constructed a mutant strain Yb2ΔAS87_03730, in which the AS87_03730 gene was inactivated by deleting a 708-bp fragment from the gene in wild-type strain Yb2. The mutant strain Yb2ΔAS87_03730 showed different growth characteristics in TSB, decreased capacity of invasion, and attenuated virulence in ducks.
R. anatipestifer infection is characterized as septicemia, hence, this species of bacteria have ability to colonize and develop in the tissues of their host. We found that the bacterial loadings in the blood of ducks infected with mutant strain Yb2ΔAS87_03730 was significantly decreased than that of Yb2 infected ducks. In addition, infection with mutant strain Yb2ΔAS87_03730 resulted in a marked delay in mortality, and more than 80-fold attenuated virulence. Our results indicated that the AS87_03730 gene is important for R. anatipestifer to establish a systemic infection. A previous study has revealed that the lux gene loci (luxA, luxI, and luxR) of Vibrio fischeri play an important role in colonization and development of host light organ14.
A report has suggested that the products of luxI and luxR control the expression of several non-lux loci, including one required for V. fischeri to remain competitive in mixed symbiotic infections15. To investigate whether AS87_03730 gene regulates the expression of other genes in the genome, RNA-Seq analysis was performed to identify differentially expressed genes in mutant strain Yb2ΔAS87_03730. As a result, 31 genes were up-regulated and three genes were down-regulated in mutant strain Yb2ΔAS87_03730. Real-time PCR verification further confirmed 19 genes were up-regulated by over 2-fold. Among them, 12 were classified into “Translation, ribosomal structure and biogenesis”. The ribosome is responsible and essential for translation process. The bacterial ribosome consists of three rRNA molecules and approximately 55 proteins16, components that are put together in an intricate and tightly regulated way. Regulation of gene expression on transcriptional level relies on signals transferred to the RNA polymerase (RNAP) that alter the enzyme’s activity or specificity17. Normally, rRNA transcription was affected by the stringent response and growth rate regulation. Stringent control leads to a reduction of stable RNA synthesis in response to amino acid starvation, while the growth rate control leads to adjustment of stable RNA synthesis in response to changes in the nutritional quality of growth medium18. In this study, the expressions of 12 rRNA proteins were up-regulated in mutant strain Yb2ΔAS87_03070, indicating that it is attempting to adapt to changing condition as a result of inactivation of AS87_03730 gene.
Two up-regulated genes (AS87_03970 and AS87_06085) were involved in “Cell envelope biogenesis, outer membrane”. The predicted product of AS87_03970 is a dTDP-4-dehydrorhamnose 3, 5-epimerase, which is involved in the biosynthesis of dTDP-1-rhamnose, an essential component of the bacterial cell wall19. More importantly, dTDP-1-rhamnose is the precursor of L-rhamnose, a saccharide required for the virulence of some pathogenic bacteria20. The AS87_06085 gene encodes a dolichyl-phosphate beta-D-mannosyltransferase, which transfers mannose from Dol-P-mannose to Ser or Thr residues on proteins21. In Saccharomyces cerevisiae, dolichyl-phosphate beta-D-mannosyltransferase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation22. In glycoprotein biosynthesis of Candida albicans, dolichol phosphate mannose synthease is activated by cAMP-mediated protein phosphorylation23. Similarly, our results showed that AS87_03730 gene defects in mutant strain Yb2ΔAS87_03730 up-regulated the expression of dolichyl-phosphate beta-D-mannosyltransferase.
One up-regulated gene AS87_02350 encoding cystathionine beta-synthase (CBS) was related to “Amino acid transport and metabolism”. CBS, primary found in eukaryotes, is the first enzyme in the transsulfuration pathway, catalyzing the conversion of serine and homocysteine to cystathionine and water24. The enzyme uses the cofactor pyridoxal-phosphate (PLP) and can be allosterically regulated by effectors such as the ubiquitous cofactor S-adenosyl-L-methionine25. In addition, CBS deficiency leads to changes in protein unfolding26. Due to AS87_03730 gene deletion, the biosynthesis of proteins involved in fatty acid metabolism was stalled in mutant strain Yb2ΔAS87_03730. Disturbance of fatty acid metabolism consequently resulted in up-regulated expression of AS87_02350.
In conclusion, for the first time, we demonstrated that the AS87_03730 gene is involved in bacterial virulence and gene regulation in R. anatipestifer. Further investigation is necessary to clarify the regulation mechanisms of the AS87_03730 gene in R. anatipestifer.
Methods
Bacterial strains, plasmids and culture conditions
The bacterial strains and plasmids used in this study, and their relevant characteristics, are described in Table 2. R. anatipestifer strains were grown on tryptic soy agar (TSA, Difco, USA) at 37 °C in 5% CO2 or tryptic soy broth (TSB, Difco). Escherichia coli strains were grown at 37 °C on Luria-Bertani (LB) plates or in LB broth. For selective growth of bacterial strains, antibiotics were added at following concentrations: ampicillin (100 μg/ml), kanamycin (50 μg/ml), spectinomycin (80 μg/ml) and chloramphenicol (30 μg/ml). R. anatipestifer serotype 2 strain Yb2 was selected to construct an AS87_03730 gene deletion mutant is because serotype 2 is one of the most prevalent serotypes in china27 and Yb2 is a kanamycin resistant (KanR) and spectinomycin sensitive (SpecS) strain which is good for the further mutant selection.
Table 2. Strains, plasmids and primers used in this study.
| Strains, plasmids or primers | Descriptions | Source or reference |
|---|---|---|
| Strains | ||
| Yb2 | Riemerella anatipestifer wild-type strain, serotype 2, virulent strain, KanR | 27 |
| Yb2ΔAS87_03730 | AS87_03730 gene deletion Yb2 mutant strain, SpecR | This study |
| CH3, NJ1, NJ4, WJ4, CQ1, CQ3, CQ4, CQ5, NN-2, NN-3, NN-4, YL4, YXb12, YXb14 | Riemerella anatipestifer wild-type strains, serotype 1 | 27 |
| NJ3, GD-3, GD-4, GD-5, GD-7, JY1, JY5, SC2, Th4, YXb1 | Riemerella anatipestifer wild-type strains, serotype 2 | 27 |
| HXb2, YXL1, YXb11 | Riemerella anatipestifer wild-type strains, serotype 10 | 27 |
| NN-6, NN-8 | Riemerella anatipestifer wild-type strains, serotype 15 | This study |
| NN-7, NN-9, NN-11, GD-1, GD-2, GD-6 | Riemerella anatipestifer wild-type strains, undefined serotype | This study |
| Plasmids | ||
| pGEM-T easy vector | TA cloning vector | Promega |
| pDS132 | sacB, mobRP4, oriR6K, CmR | 5 |
| pFW5 | E. coli.-streptococci shuttle vector | 5 |
| Primers | ||
| AS87_03730 orf-F | CCGCTCGAGATGCCTTCTATTTTTGATATTA (XhoI underlined) | This study |
| AS87_03730 orf-R | CATGCATGCCTAAGAAACCAAAAGGCTAC (SphI underlined) | This study |
| AS87_03730 Left-F | CATGCATGCATCCTGAGCTGGGAGAAACA (SphI underlined) | This study |
| AS87_03730 Left-R | AAACTGCAGCATCTGGGTAGTGCCTGAAC (PstI underlined) | This study |
| AS87_03730 Right-F | AAACTGCAGGACCATTCCGACATTAGAGG (PstI underlined) | This study |
| AS87_03730 Right-R | CGCGTCGACGTGATTTCAATAGCCGTTTT (SalI underlined) | This study |
| Spec-F | AAACTGCAGCGTTCGTGAATACATGTTAT (PstI underlined) | 5 |
| Spec-R | AAACTGCAGGCGCTTACCAATTAGAATGA (PstI underlined) | 5 |
| AS87_03730 in-F | ACAATTTATAGGCACTCCCG | This study |
| AS87_03730 in-R | TTAATAGCTCCTGTTCTCCC | This study |
| AS87_03730 out-F | GCGACCCACTCATAACCC | This study |
| AS87_03730 out-R | TTCCTAATGGCGACTTTG | This study |
| RA 16S rRNA-F | CAACCATGCAGCACCTTGAAAA | 6 |
| RA 16S rRNA-R | GACGAAAGCGTGGGGAGCGAAC | 6 |
RResistance.
Ethics Statement
One-day-old Cherry Valley ducks were purchased from Zhuanghang duck farm (Shanghai, China) and kept under a controlled temperature (28 to 30 °C). The ducks were housed in cages with a 12-h light/dark cycle and free access to food and water during this study. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Institutional Animal Care and Use Committee (IACUC) guidelines set by Shanghai Veterinary Research Institute, the Chinese Academy of Agricultural Sciences (CAAS). The protocol was approved by the Committee on the Ethics of Animal Experiments of Shanghai Veterinary Research Institute, CAAS (Permit Number: 13–11). All surgeries were performed under sodium pentobarbital anesthesia, and all efforts were made to minimize suffering.
Construction of mutant strain Yb2ΔAS87_03730
An AS87_03730 gene deletion mutant was constructed by allelic exchange through the recombinant suicide plasmid pDS132 as described previously5. Briefly, a SpecR cassette (1,119 bp) was amplified from plasmid pFW5 using primers Spec-F and Spec-R. A 1,076 bp left flanking region of AS87_03730 gene was amplified from R. anatipestifer Yb2 genomic DNA using primers AS87_03730 Left-F and AS87_03730 Left-R, and cloned into pGEM®-T easy vector (Promega, Madison, WI, USA). A 1,132 bp of right flanking region of AS87_03730 gene was amplified using primers AS87_03730 Right-F and AS87_03730 Right-R, and cloned into the above-mentioned plasmid; and then a SpecR cassette was inserted at the PstI site. The recombinant DNA was moved to SphI- and SalI-digested suicide plasmid pDS132 to produce pDS132-AS87_03730-LSR (SpecR). Subsequently, the recombinant plasmid pDS132-AS87_03730-LSR was transformed into R. anatipestifer strain Yb2 by conjugation. The transconjugants were selected by plating on TSA plates containing 50 μg/ml kanamycin and 80 μg/ml spectinomycin. Deletion of AS87_03730 gene in the mutant was confirmed by PCR analysis and resulting mutant strain was designated as Yb2ΔAS87_03730.
Phenotype characteristics of Yb2ΔAS87_03730
To determine whether AS87_03730 gene deletion would influence the bacterial growth, the growth curves of mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2 were measured as previously described28. Briefly, equal amount of each bacterial culture in mid-exponential phase was transferred into fresh TSB medium at a ratio of 1:100(v/v) for growing at 37 °C with shaking at 200 rpm. Optical density at 600 nm (OD600) was measured at 1 h intervals for 15 h using a spectrophotometer (BIO-RAD, USA). Bacterial growth rate was expressed as OD600 values.
The biochemical test for mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2 was carried out using bacterial biochemical tube (Hangwei, Hangzhou, China) following the manufacturer’s instruction. Both strains were also tested against ten commonly used antibiotic disc (ampicillin, amoxicillin, gentamicin, tetracycline, erythromycin, chloramphenicol, ciprofloracin, rifampicin, polymyxin B and trimethoprim-sulfamethoxazde). Inoculums from the tested bacteria were prepared depending on Kirby-Bauer antibiotic testing29.
Adhesion and invasion assays
Adhesion and invasion assays were performed on Vero cells (ATCC CCL-81) in 24-well plates (Corning, NY, USA). The mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2 were grown to mid-logarithmic phase and washed three times with phosphate-buffered saline (PBS) prior to use. Vero cells (approximately 2.5 × 105 cells) were inoculated with treated bacteria at a multiplicity of infection (MOI) of 50, and incubated at 37 °C with 5% CO2 for 1.5 h. The infected cells were rinsed three times with PBS to remove unbound bacteria, and then incubated with 0.1% trypsin (100 μl/well) to lyse the eukaryotic cells. The number of cell-adherent bacteria was counted after dilution and plating. For the invasion assay, the infected cells were further incubated for an additional 1 h with DMEM medium supplemented with 100 μg/ml gentamicin to kill extracellular bacteria. After washed three times with PBS, the infected cells were lysed and the amount of intracellular bacteria was determined. All samples were tested in triplicate, and experiments were repeated three times.
Determination of the bacterial virulence and survival in vivo
To determine whether AS87_03730 gene deletion had an influence on virulence of R. anatipestifer, the bacterial median lethal dose (LD50) of mutant Yb2ΔAS87_03730 was determined using 18-day-old Cherry Valley ducks as described and compared with that of wild-type strain Yb230. A total of 64 ducks were divided randomly into eight groups (8 ducks per group) and injected intramuscularly with respective bacterial strain at a dose of 104–108 CFU. Ducks in groups 1–4 were injected with 104, 105, 106 or 107 CFU of Yb2 bacteria, and ducks in groups 5–8 were injected with 105, 106, 107 or 108 CFU of Yb2ΔAS87_03730 bacteria, respectively. The infected ducks which appeared clinical signs (clinical signs include lethargy, diarrhea, rough hair coat, frequent seizure activity, paralysis and no eating or drinking, etc.) were sacrificed humanely with an intravenous injection of sodium pentobarbital at a dose of 120 mg/kg and counted as dead. The infected ducks were clinically observed three times at 8-h intervals daily for a period of 7 days post-challenge. The LD50 value was calculated by improved Karber’s method31.
The bacterial loadings in the blood of infected ducks were counted to evaluate bacterial survival in vivo32. Twelve 18-day-old Cherry Valley ducks were divided randomly into two groups, and infected intramuscularly with mutant strain Yb2ΔAS87_03730 or wild-type strain Yb2 at a dose of 106 CFU in 0.5 ml PBS. The blood samples were collected at 6 h, 12 h and 24 h post infection, diluted appropriately and plated on TSA for bacterial counting.
Bacterial RNA isolation
The mutant strain Yb2ΔAS87_03730 and wild-type strain Yb2 were grown to mid-logarithmic phase, harvested by centrifugation at 10,000 × g for 1 min, and then washed once with PBS for RNA extraction. Total RNA was extracted from bacteria (2.5 × 109 CFU) using Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. All RNA samples were treated with TURBO DNA-freeTM kit (Ambion, Grand Island, NY, USA) to remove DNA contamination. Quantification and quality assessment of RNA samples were performed with NanoDrop ND-100 instrument (NanoDrop Technologies, Inc., Wilmington, DE, USA). RNA integrity was checked using 1% agarose gel electrophoresis.
Illumina sequencing for RNA-Seq
Total RNA quantification and quality were assessed by spectrophotometer, and illumina RNA-Seq libraries were generated as following steps: 1) RNA fragmentation; 2) reverse transcription; 3) double-strand cDNA synthesis with dUTP; 4) cDNA fragmentation and end-repair; 5) 3′end “dA” base addition and illumina adapter ligation; 6) cleavage of the uridine containing strand with Uracial DNA Glycosylase; 7) PCR amplification; 8) size selection and purification33. The complete libraries were then quantified with Agilent 2100 Bioanalyzer using DNA 1000 Kit. The DNA fragments in these libraries were denatured with 0.1M NaOH to generate single-strand DNA molecules, loaded onto channels of Illumina flow cell at 8 pM concentration, amplified in situ using TruSeq SR Cluster Kit v3-cBot-HS (Illumina), and subsequently sequenced for 100 cycles on Illumina HiSeq 2000 according to the manufacturer’s instruction.
Differential expression analysis
Image analysis and base calling was performed using Solexa pipeline Version 1.8 (Off-Line Base Caller software, Version 1.8)34. Trimmed reads were aligned to R. anatipestifer Yb2 genome using TopHat2 software (Version 2.0.9)35. Transcript levels were calculated as FPKM (fragments per kilobase cDNA per million fragments mapped). Differential expressed genes were analyzed using Cufflinks software (Version 2.1.1) with fold change (cutoff = 2.0)36, and considered statistically significant if the fold change was >2.0 and the p-value was <0.05.
Real-time PCR analysis
Transcriptional levels of differently expressed genes obtained in the transcriptome analysis were further confirmed by real-time PCR. Primers were designed from R. anatipestifer Yb2 using primer3 online software Version.0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) and described in Supplementary Table S2. RNA samples were extracted and purified as mentioned above. cDNA was synthesized using PrimeScript RT Master Mix (Takara) according to the manufacturer’s protocol, and the resulting cDNAs were diluted 3-fold served as templates. Real-time PCR reaction was carried out in Go Taq qPCR® Master Mix (Promega, Fitchburg, WI, USA), and subjected to the Mastercycler ep realplex4 apparatus (Eppendorf, Germany). For each gene, reactions were performed for three RNA samples and each reaction was performed in triplicate. Quantification of transcriptional level was calculated according to the 2−ΔΔCt method using RA 16S rRNA gene for normalization37.
Distribution and sequence analyses of AS87_03730 gene in R. anatipestifer strains
AS87_03730 gene in R. anatipestifer strains was amplified using PCR. Genomic DNAs of 36 R. anatipestifer strains with different serotypes were extracted using TIANamp Bacteria DNA kit (Tiangen, Beijing, China). The primers AS87_03730 orf-F and AS87_03730 orf-R were used to amplify the AS87_03730 sequence using Premix LA Taq® (loading dye mix) (Takara, Dalian, China) according to the following cycle parameters: 94 °C for 4 min, 30 cycles of 94 °C for 40 s, 52 °C for 40 s and 72 °C for 1 min, followed by one cycle of 72 °C for 10 min. To investigate AS87_03730 homology among different R. anatipestifer strains, the PCR products from 8 strains were cloned into pGEM®-T easy vector (Promega, Madison, WI, USA), respectively. DNA sequencing was performed on an Applied Biosystems DNA sequencer (ABI 3730XL) by Thermo Fisher Scientific Inc. (Thermo Fisher Scientific, Shanghai, China). The AS87_03730 gene sequences from these R. anatipestifer strains have been submitted to the GenBank database under the accession numbers KM676074 ∼KM076081. Homology of the AS87_03730 gene sequences was analyzed with DNASTAR software (DNASTAR Inc., Madison, WI, USA).
Statistical analysis
The statistical significance of the data was determined by the Student’s t-test in the SPSS Version 17.0 software (SPSS Inc., Chicago, IL, USA). A p value of <0.05 was considered to be statistically significant.
Nucleotide sequence accession numbers
The accession number for R. anatipestifer strain Yb2 in the GenBank is CP007204.
Additional Information
How to cite this article: Wang, X. et al. Deletion of AS87_03730 gene changed the bacterial virulence and gene expression of Riemerella anatipestifer. Sci. Rep. 6, 22438; doi: 10.1038/srep22438 (2016).
Supplementary Material
Acknowledgments
This study was supported by the National Science Foundation of China (31272591, http://www.nsfc.gov.cn/).
Footnotes
Author Contributions Conceived and designed the experiments: S.Y. and X.W. Performed the experiments: X.W., J.Y. and S.W. Analyzed the data: X.W., J.Y., S.Y. and C.D. Contributed reagents/materials/analysis tools: X.W., B.L. and M.T. Wrote the paper: X.W. and S.Y. All authors received the manuscript.
References
- Glünder G. & Hinz K. H. Isolation of Moraxella anatipestifer from embryonated goose eggs. Avian Pathology 18, 351–355 (1989). [DOI] [PubMed] [Google Scholar]
- Sandhu T. S. & Rimler R. Riemerella anatipestifer infection. Diseases of Poultry 161–166 (1997). [Google Scholar]
- Sandhu T. in Duck production: science and world practice (ed Farrell D. J. & Stapleton P.) 111–134 (1986). [Google Scholar]
- Crasta K. C. et al. Identification and characterization of CAMP cohemolysin as a potential virulence factor of Riemerella anatipestifer. J. Bacteriol 184, 1932–1939 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. et al. OmpA is a virulence factor of Riemerella anatipestifer. Vet. Microbiol. 150, 278–283 (2011). [DOI] [PubMed] [Google Scholar]
- Wang X. et al. The AS87_04050 gene is involved in bacterial lipopolysaccharide biosynthesis and pathogenicity of Riemerella anatipestifer. PloS one 9, e109962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J. et al. Characterization and cross-protection evaluation of M949_1603 gene deletion Riemerella anatipestifer mutant RA-M1. Appl Microbiol Biotechnol. 99, 10107–10116 (2015). [DOI] [PubMed] [Google Scholar]
- Wang X., Ding C., Wang S., Han X. & Yu S. Whole-genome sequence analysis and genome-wide virulence gene identification of Riemerella anatipestifer strain Yb2. Appl Environ Microbiol. 81, 5093–5102 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston T. C., Hruska K. S. & Adams L. F. The nucleotide sequence of the luxE gene of Vibrio harveyi and a comparison of the amino acid sequences of the acyl-protein synthetases from V. harveyi and V. fischeri. Biochem. Biophys. Res. Commun. 163, 93–101 (1989). [DOI] [PubMed] [Google Scholar]
- Shadel G., Devine J. & Baldvvin T. Control of the lux regulon of Vibrio fischeri. J. Biolumin. Chemilumin. 5, 99–106 (1990). [DOI] [PubMed] [Google Scholar]
- Riendeau D., Rodriguez A. & Meighen E. Resolution of the fatty acid reductase from Photobacterium phosphoreum into acyl protein synthetase and acyl-CoA reductase activities. Evidence for an enzyme complex. J. Biol. Chem. 257, 6908–6915 (1982). [PubMed] [Google Scholar]
- Lin J. W., Chao Y. F. & Weng S. F. Nucleotide Sequence and Functional Analysis of the luxE Gene Encoding Acyl-Protein Synthetase of the lux Operon from Photobacterium leiognathi. Biochem. Biophys. Res. Commun. 228, 764–773 (1996). [DOI] [PubMed] [Google Scholar]
- Morona R., Stroeher U. H., Karageorgo L. E., Brown M. H. & Manning P. A. A putative pathway for biosynthesis of the O-antigen component, 3-deoxy-L-glycero-tetronic acid, based on the sequence of the Vibrio cholerae O1 rfb region. Gene 166, 19–31 (1995). [DOI] [PubMed] [Google Scholar]
- Visick K. L., Foster J., Doino J., McFall-Ngai M. & Ruby E. G. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182, 4578–4586 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan S. M. & Dunlap P. V. LuxR-and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 182, 2811–2822 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth B. S. et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 (2005). [DOI] [PubMed] [Google Scholar]
- Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annual Reviews in Microbiology 54, 499–518 (2000). [DOI] [PubMed] [Google Scholar]
- Dennis P. P., Ehrenberg M. & Bremer H. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol. Mol. Biol. Rev. 68, 639–668 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler R. W. & Gabriel O. Biological mechanisms involved in the formation of deoxy sugars VII. Biosynthesis of 6-deoxy -L-talose. J. Biol. Chem. 248, 6041–6049 (1973). [PubMed] [Google Scholar]
- Giraud M. F., Leonard G. A., Field R. A., Berlind C. & Naismith J. H. RmlC, the third enzyme of dTDP-L-rhamnose pathway, is a new class of epimerase. Nat. Struct. Biol. 7, 398–402 (2000). [DOI] [PubMed] [Google Scholar]
- Tanaka N. et al. Characterization of O-mannosyltransferase family in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 330, 813–820 (2005). [DOI] [PubMed] [Google Scholar]
- Orlean P. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 5796–5805 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo-Flores B. L., Calvo-Méndez C., Flores-Carreón A. & López-Romero E. Biosynthesis of glycoproteins in the pathogenic fungus Candida albicans: Activation of dolichol phosphate mannose synthase by cAMP-mediated protein phosphorylation. FEMS Immunol. Med. Microbiol. 45, 429–434 (2005). [DOI] [PubMed] [Google Scholar]
- Jhee K. H. & Kruger W. D. The role of cystathionine β-synthase in homocysteine metabolism. Antioxid Redox Sign 7, 813–822 (2005). [DOI] [PubMed] [Google Scholar]
- Banerjee R., Evande R., Kabil Ö., Ojha S. & Taoka S. Reaction mechanism and regulation of cystathionine β-synthase. BBA-Proteins Proteom 1647, 30–35 (2003). [DOI] [PubMed] [Google Scholar]
- Hnízda A., Jurga V., Raková K. & Kožich V. Cystathionine beta-synthase mutants exhibit changes in protein unfolding: conformational analysis of misfolded variants in crude cell extracts. J. Inherit. Metab. Dis. 35, 469–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. et al. Epidemiology investigation and study on the Riemerella anatipestifer infection in Jiangsu and Anhui provinces. Chinese Journal of Veterinary Science and Technology 31, 12–13 (2001). [Google Scholar]
- Peñuelas-Urquides K., Villarreal-Treviño L., Silva-Ramírez B., Rivadeneyra-Espinoza L. & Said-Fernández S. Measuring of Mycobacterium tuberculosis growth. A correlation of the optical measurements with colony forming units. Braz J Microbiol 44, 287–290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle V. J., Fancher M. E. & Ross R. W. Rapid, modified Kirby-Bauer susceptibility test with single, high-concentration antimicrobial disks. Antimicrob. Agents Chemother. 3, 418–424 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. et al. Characterization of biofilm formation by Riemerella anatipestifer. Vet. Microbiol. 144, 429–436 (2010). [DOI] [PubMed] [Google Scholar]
- Gu B., Li Y., Yu R. & Wang X. Summary of median lethal dose and its calculation methods. China Occupational Medicine 36, 507–508, 511 (2009). [Google Scholar]
- Wang X., Hu Q., Han X., Ding C. & Yu S. Screening of serotype 2 Riemerella anatipestifer candidate strains for the production of inactivated oil-emulsion vaccine. Chinese Journal of Animal Infectious Diseases 20, 5 (2012). [Google Scholar]
- Zhong S. et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011, pdb. prot5652 (2011). [DOI] [PubMed] [Google Scholar]
- Whiteford N. et al. Swift: primary data analysis for the Illumina Solexa sequencing platform. Bioinformatics 25, 2194–2199 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S. & Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6, 279–284 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.