Cytokinetic abscission, the final stage of cell division, is mediated by the ESCRT machinery. Here, Christ et al. dissect the regulation of ESCRT-III recruitment and abscission timing and identify an intersection with abscission checkpoint signaling in cells with chromatin bridges.
Abstract
Cytokinetic abscission, the final stage of cell division where the two daughter cells are separated, is mediated by the endosomal sorting complex required for transport (ESCRT) machinery. The ESCRT-III subunit CHMP4B is a key effector in abscission, whereas its paralogue, CHMP4C, is a component in the abscission checkpoint that delays abscission until chromatin is cleared from the intercellular bridge. How recruitment of these components is mediated during cytokinesis remains poorly understood, although the ESCRT-binding protein ALIX has been implicated. Here, we show that ESCRT-II and the ESCRT-II–binding ESCRT-III subunit CHMP6 cooperate with ESCRT-I to recruit CHMP4B, with ALIX providing a parallel recruitment arm. In contrast to CHMP4B, we find that recruitment of CHMP4C relies predominantly on ALIX. Accordingly, ALIX depletion leads to furrow regression in cells with chromosome bridges, a phenotype associated with abscission checkpoint signaling failure. Collectively, our work reveals a two-pronged recruitment of ESCRT-III to the cytokinetic bridge and implicates ALIX in abscission checkpoint signaling.
Introduction
The endosomal sorting complex required for transport (ESCRT) machinery controls topologically similar membrane scission events during cytokinetic abscission (Carlton and Martin-Serrano, 2007; Morita et al., 2007), multivesicular endosome (MVE) formation (Katzmann et al., 2002), virus budding (Morita and Sundquist, 2004), neuron pruning (Loncle et al., 2015), plasma membrane repair (Jimenez et al., 2014), and nuclear envelope reassembly (Olmos et al., 2015; Vietri et al., 2015). The prototypical ESCRT function in the formation of intraluminal vesicles in MVEs is orchestrated by specific modules, such as ESCRT-0, ESCRT-I, and ESCRT-II, that nucleate assembly of cytosolic ESCRT-III monomers into membrane-associated filaments that cooperate with the AAA ATPase VPS4 to mediate membrane constriction and scission. ESCRT-III assemblies are composed of different charged multivesicular body proteins (CHMPs), of which CHMP4B is thought to be the main constituent. Additional cofactors include Bro1 domain proteins such as ALIX (ALG2-interacting protein X) and HD-PTP, which are recruited to sites of ESCRT function, where they are thought to provide a second mode of ESCRT-III recruitment by association with CHMP4 paralogs (McDonald and Martin-Serrano, 2009; Raiborg and Stenmark, 2009; Hurley, 2010; Hurley and Hanson, 2010; Caballe and Martin-Serrano, 2011; Henne et al., 2011, 2013; Peel et al., 2011; Guizetti and Gerlich, 2012; Jouvenet, 2012; Morita, 2012; Carlson and Hurley, 2012; McCullough et al., 2013; Mierzwa and Gerlich, 2014; Schuh and Audhya, 2014). Finally, ESCRT-III proteins recruit several effector proteins, most notably the AAA ATPase VPS4 that coordinates membrane constriction and scission by depolymerization of ESCRT-III filaments.
All ESCRT-III subunits consist of four α-helices forming a bundled core and a negatively charged C-terminal region containing α-helix 5 and a MIM element (microtubule interacting and transport [MIT] interacting motif) that mediates interaction with MIT containing effectors such as VPS4 or Spastin. Moreover, ESCRT-III subunits can cycle between an inactive closed conformation, where the acidic C terminus folds across the basic N-terminal core to autoinhibit the protein, and an active open confirmation, exposing the C-terminal motifs and enabling interaction with other ESCRT-III molecules (Zamborlini et al., 2006; Shim et al., 2007; Kieffer et al., 2008; Bajorek et al., 2009; Hurley and Hanson, 2010; Merrill and Hanson, 2010; Adell and Teis, 2011; McCullough et al., 2015; Tang et al., 2015). In vitro studies and experiments in yeast show that CHMP6/VPS20 recruitment by the ESCRT-II subunit EAP20/VPS25 during MVE formation generates a membrane curvature–sensing supercomplex. Activated CHMP6 can then bind and initiate oligomerization of CHMP4/SNF7 to form membrane-bound filaments that sequentially assemble CHMP3/VPS24, CHMP2/VPS2, CHMP1/DID2, and IST1 (Babst et al., 2002a,b; Teo et al., 2004; Yorikawa et al., 2005; Langelier et al., 2006; Teis et al., 2008, 2010; Im et al., 2009; Saksena et al., 2009; Wollert et al., 2009; Wollert and Hurley, 2010; Fyfe et al., 2011; Henne et al., 2012; Mageswaran et al., 2015; Lee et al., 2015).
During the final step of cell division, karyokinesis and cytokinesis are completed by abscission, cleavage of the thin bridge of membrane connecting the two daughter cells. The importance of correct cytokinetic abscission is most apparent in the presence of chromosome bridges traversing the intercellular bridge, whereby failure to coordinate abscission with removal of the physical impediment can lead to furrow regression and formation of tetraploid cells or premature scission in the presence of lagging chromosomes to induce DNA damage and aneuploidy, all phenomena associated with carcinogenesis (Lens et al., 2010; Chen et al., 2012; Fededa and Gerlich, 2012; Ganem and Pellman, 2012; Green et al., 2012; Holland and Cleveland, 2012; Agromayor and Martin-Serrano, 2013; Hayashi and Karlseder, 2013; Potapova et al., 2013).
Despite central roles during cytokinetic abscission and abscission checkpoint signaling, ESCRT-III recruitment during cytokinetic abscission remains poorly understood. The centrosomal protein of 55 kD (CEP55) directly recruits the ESCRT-I subunit TSG101 (tumor-susceptibility gene 101) and ALIX to the midbody, the electron-dense structure at the center of the cytokinetic bridge (Lee et al., 2008; Elia et al., 2011). Depletion of ALIX results in a dramatic increase in cytokinetic profiles and multinucleation, but whether this reflects defective CHMP4B recruitment or other functions remains to be elucidated (Cabezas et al., 2005; Carlton and Martin-Serrano, 2007; Morita et al., 2007; Chu et al., 2012). Furthermore, despite the important role for ESCRT-II and CHMP6 in bridging ESCRT-I and ESCRT-III as well as nucleating ESCRT-III filaments, depletion assays have led to the conclusion that they are dispensable for cytokinesis (Morita et al., 2007, 2010; Peel et al., 2011; Schuh and Audhya, 2014). Instead, TSG101 contribution has been proposed to be limited to stabilization of ALIX at the midbody, reflected by much weaker cytokinetic defects upon its depletion (Carlton and Martin-Serrano, 2007; Morita et al., 2007). Furthermore, it remains unresolved to what extent other CHMP4 paralogs, such as the Aurora B–dependent abscission checkpoint regulator CHMP4C, rely on similar recruitment mechanisms (Steigemann et al., 2009; Capalbo et al., 2012; Carlton et al., 2012).
Here, we set out to elucidate the regulation of CHMP4B recruitment to the midbody. We demonstrate a bona fide role for ESCRT-II and CHMP6 in cytokinetic abscission and identify two parallel “arms” for ESCRT-III recruitment to the cytokinetic bridge. One arm recruits CHMP4B via ESCRT-I, ESCRT-II, and CHMP6, with the other arm consisting of ALIX directly associating with the C terminus of CHMP4B. Codepletion experiments show a strong correlation between abscission timing and CHMP4B abundance at the midbody, arguing that CHMP4B recruitment is a limiting factor for cytokinetic abscission. Whereas CHMP4B recruitment overlaps between both arms, the abscission checkpoint CHMP4C isoform is predominantly recruited via ALIX. Our data suggest that impairment of CHMP4C recruitment accounts for the previously described furrow regression phenotype upon ALIX depletion, prominently in cells exhibiting DNA segregation defects. Collectively, our work resolves the molecular dependencies underlying CHMP4B recruitment during cytokinetic abscission and identifies ALIX as a novel abscission checkpoint signaling node.
Results
CHMP4B can be recruited to the midbody via ALIX or ESCRT-I
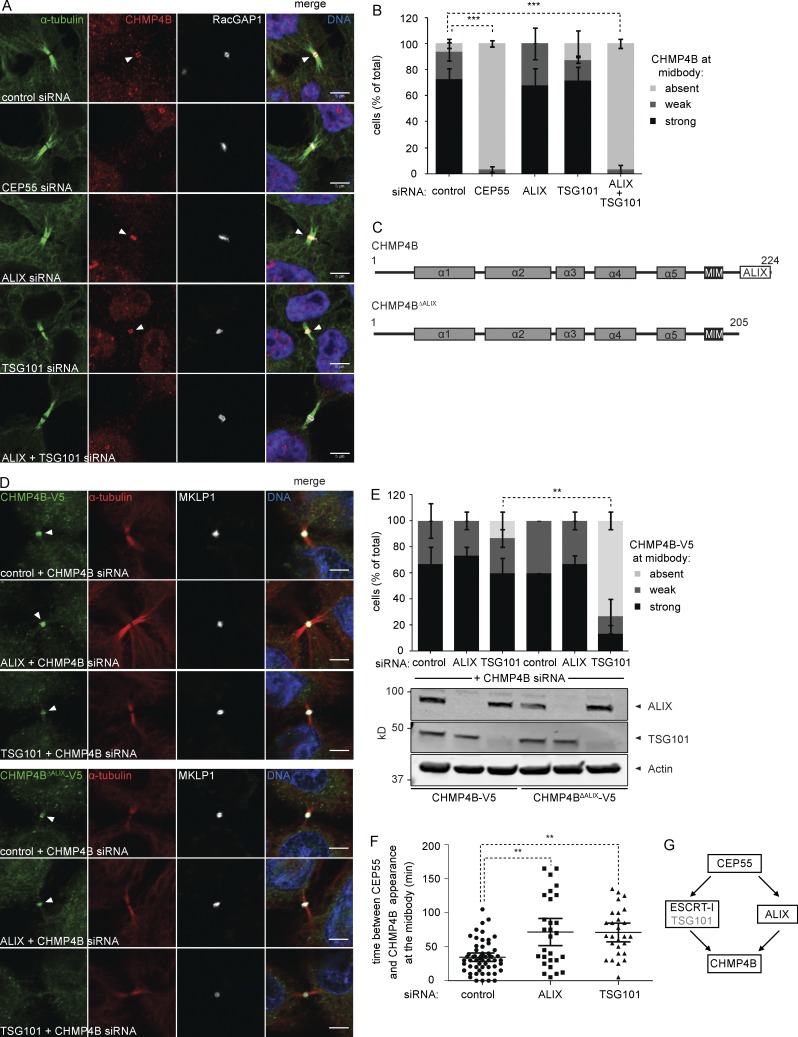
Considering the lacking understanding of CHMP4B recruitment to late cytokinetic bridges by upstream regulators, we addressed this major issue by analyzing localization of endogenous CHMP4B in cells depleted for ALIX, TSG101, or CEP55. As expected, depletion of the abscission master regulator CEP55 completely abrogated CHMP4B recruitment (Fig. 1, A and B; and Fig. S1). In contrast, efficient depletion of ALIX or TSG101 alone did not affect CHMP4B localization, with codepletion of ALIX and ESCRT-I subunits (TSG101 or VPS28) required to abrogate this recruitment (Fig. 1, A and B; and Figs. S1 and S2). These data indicated that ALIX and TSG101 could play redundant roles in CHMP4B recruitment. To address this in more detail, we generated stable cell lines expressing near-endogenous levels of siRNA-resistant full-length CHMP4B or a CHMP4B mutant lacking its C-terminal ALIX-interacting helix (CHMP4BΔALIX-V5; Fig. 1 C), fused at their C termini to the small V5 tag. Whereas depletion of ALIX or TSG101 (and endogenous CHMP4B) had no effect on midbody localization of CHMP4B-V5 (Fig. 1, D and E), TSG101 depletion was sufficient to completely abrogate midbody localization of CHMP4BΔALIX-V5. Together, these data indicated overlapping roles for ALIX and TSG101 in CHMP4B recruitment and extend TSG101 function beyond stabilization of ALIX (Carlton and Martin-Serrano, 2007; Morita et al., 2007).
Figure 1.
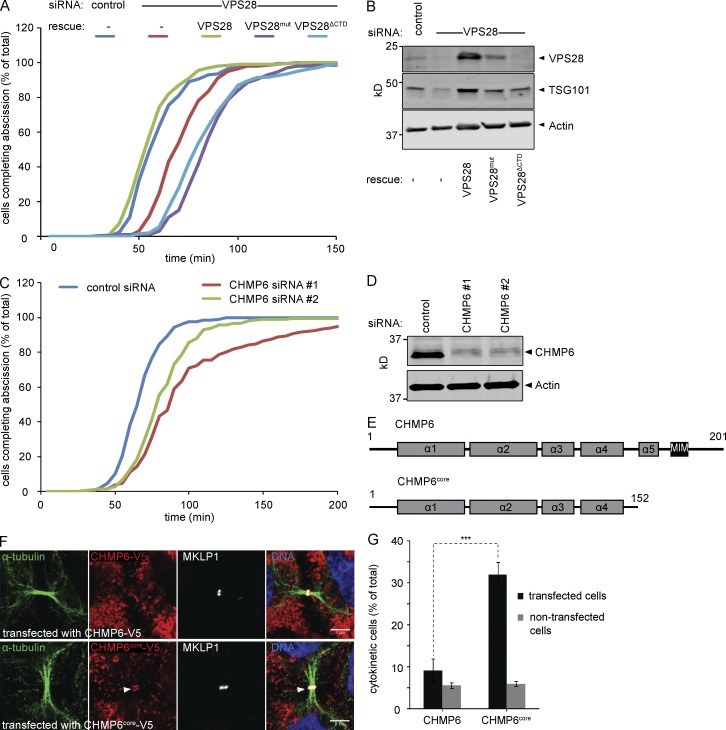
ALIX and ESCRT-I constitute parallel CHMP4B recruitment arms. (A) Confocal images showing CHMP4B at the cytokinetic bridge of HeLa cells stained for DNA (Hoechst), α-tubulin, CHMP4B, and the midbody marker RacGAP1 upon siRNA treatment as indicated. Bars, 5 µm. (B) Quantification of CHMP4B localization at the midbody (error bars indicate SEM; n > 30 cells from three independent experiments; unpaired t test; ***, P < 0.001). (C) Schematic view of the CHMP4B constructs used in D and E. (D) Confocal images of HeLa cells stably expressing CHMP4B-V5 or CHMP4BΔALIX-V5 stained for DNA (Hoechst), α-tubulin, V5, and MKLP1 upon siRNA treatment as indicated. Bars, 5 µm. (E) Quantification of the CHMP4B or CHMP4BΔALIX signal at the midbody in cells stably expressing CHMP4B-V5 or CHMP4BΔALIX-V5 (error bars indicate SEM; n = 15 cells from three independent experiments; unpaired t test; **, P < 0.01). Knockdown of TSG101 and ALIX is shown by Western blot. (F) Live imaging of CHMP4B enrichment at the midbody relative to CEP55 in HeLa cells stably expressing mCherry-CEP55 and CHMP4B-eGFP (error bars indicate mean with 95% confidence interval; n ≥ 27 cells from five independent experiments; control siRNA 35 ± 23.2 min, ALIX siRNA 72 ± 51.4 min, TSG101 siRNA 71 ± 34.7 min (± SD); mixed factor model; **, P ≤ 0.01). (G) Schematic view of cytokinetic CHMP4B recruitment.
To assess temporal effects on CHMP4B recruitment, we monitored CHMP4B recruitment in HeLa cells stably expressing mCherry-CEP55 and CHMP4B-eGFP, upon depletion of ALIX or TSG101. Relative to CEP55 recruitment, CHMP4B association with midbodies was significantly delayed (P ≤ 0.01) both upon ALIX and TSG101 knockdown to 72 ± 51.4 and 71 ± 34.7 min, respectively (± SD; Fig. 1 F), relative to control siRNA-treated cells (35 ± 23.2 min). Collectively, our data indicate that ALIX and TSG101 play overlapping and unique roles in controlling CHMP4B accumulation at the cytokinetic bridge (Fig. 1 G).
CHMP6 and ESCRT-II are required for CHMP4B localization to the midbody
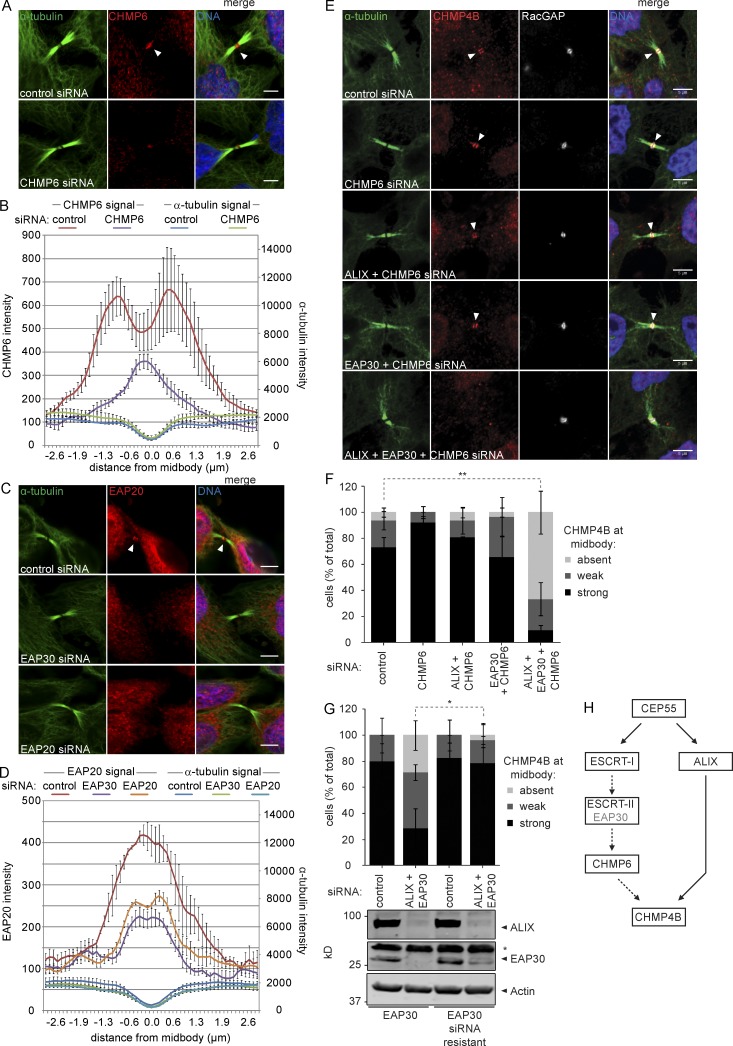
During MVE formation, ESCRT-I and CHMP4B are bridged by ESCRT-II and CHMP6, but these factors were considered dispensable during cytokinetic abscission (Morita et al., 2007; Carlton et al., 2008; McCullough et al., 2008, 2013; Schuh and Audhya, 2014). In light of our data (Fig. 1) and recent observations (Goliand et al., 2014), we decided to revisit the role for these factors during cytokinesis. To this end, affinity-purified antibodies specifically targeting CHMP6 and the ESCRT-II subunit EAP20 (Fig. S3) were used for confocal microscopy. Probing cytokinetic cells with the anti-CHMP6 antibody resulted in the strong midbody-flanking staining characteristic of ESCRT-III proteins (Fig. 2 A, top). This staining was highly sensitive to treatment with siRNAs targeting CHMP6 (Fig. 2, A [bottom] and B), indicating that the midbody signal represents bona fide CHMP6 localization. EAP20 similarly localized to the midbody, with staining sensitive to depletion of EAP20 or of the EAP30 subunit, that results in codegradation of EAP20 (Fig. 2, C and D; Malerød et al., 2007; Stieler and Prange, 2014). These datasets provide the first evidence that endogenous CHMP6 and ESCRT-II proteins localize to the midbody and suggest a role for these factors in cytokinetic abscission.
Figure 2.
CHMP6 and ESCRT-II localize to the cytokinetic bridge and contribute to CHMP4B recruitment. (A and C) Confocal images of HeLa cells stained for DNA (Hoechst), α-tubulin, CHMP6, and EAP20 upon siRNA treatment as indicated. Bars, 5 µm. (B and D) Intensity of CHMP6, EAP20, and α-tubulin along the intercellular bridge (error bars indicate SEM from three independent experiments; n = 30 cells; mean CHMP6 intensity in the bridge upon CHMP6 siRNA relative to control siRNA [set to 1, ± SEM] 0.47 ± 0.07 [P = 0.02], mean EAP20 intensity upon EAP30 siRNA relative to control siRNA 0.59 ± 0.05 [P = 0.01], mean EAP20 intensity upon EAP20 siRNA relative to control siRNA 0.69 ± 0.03 [P = 0.01]; P-values obtained using one-sample t test). (E) Confocal images showing CHMP4B at the cytokinetic bridge of HeLa cells stained for DNA (Hoechst), α-tubulin, CHMP4B, and the midbody marker RacGAP1 upon siRNA treatment as indicated. Bars, 5 µm. (F) Quantification of cells with or without CHMP4B at the midbody (error bars indicate SEM; n > 30 cells from four independent experiments; unpaired t test; **, P < 0.01) upon siRNA treatment as indicated. (G) Quantification of CHMP4B recruitment to the midbody in HeLa cells stably expressing siRNA sensitive or resistant EAP30 (error bars indicate SEM; n ≥ 24 cells from three independent experiments; unpaired t test; *, P < 0.05). Knockdown of ALIX and EAP30 is shown by Western blot (*, nonspecific immunoreactivity). (H) Schematic view of cytokinetic CHMP4B recruitment.
To explore the roles for ESCRT-II and CHMP6, we monitored localization of endogenous CHMP4B upon depletion of these factors. Depletion of CHMP6 alone or together with EAP30 had no detectable effect on CHMP4B recruitment. However, CHMP4B recruitment was abrogated upon codepletion of ALIX, EAP30, and CHMP6 (Fig. 2, E and F). Inhibition of CHMP4B recruitment by codepletion of ALIX and EAP30 could be rescued by expression of siRNA-resistant EAP30 (Fig. 2 G). In contrast, codepletion of EAP30 and CHMP6 with TSG101 did not exacerbate the TSG101 knockdown phenotype (unpublished data), indicating that they function in the same recruitment pathway. Collectively, our data illustrate an important role for CHMP6 and ESCRT-II in recruitment of CHMP4B to the intercellular bridge (Fig. 2 H).
Establishing the ESCRT-I–ESCRT-II–CHMP6 pathway in cytokinetic abscission
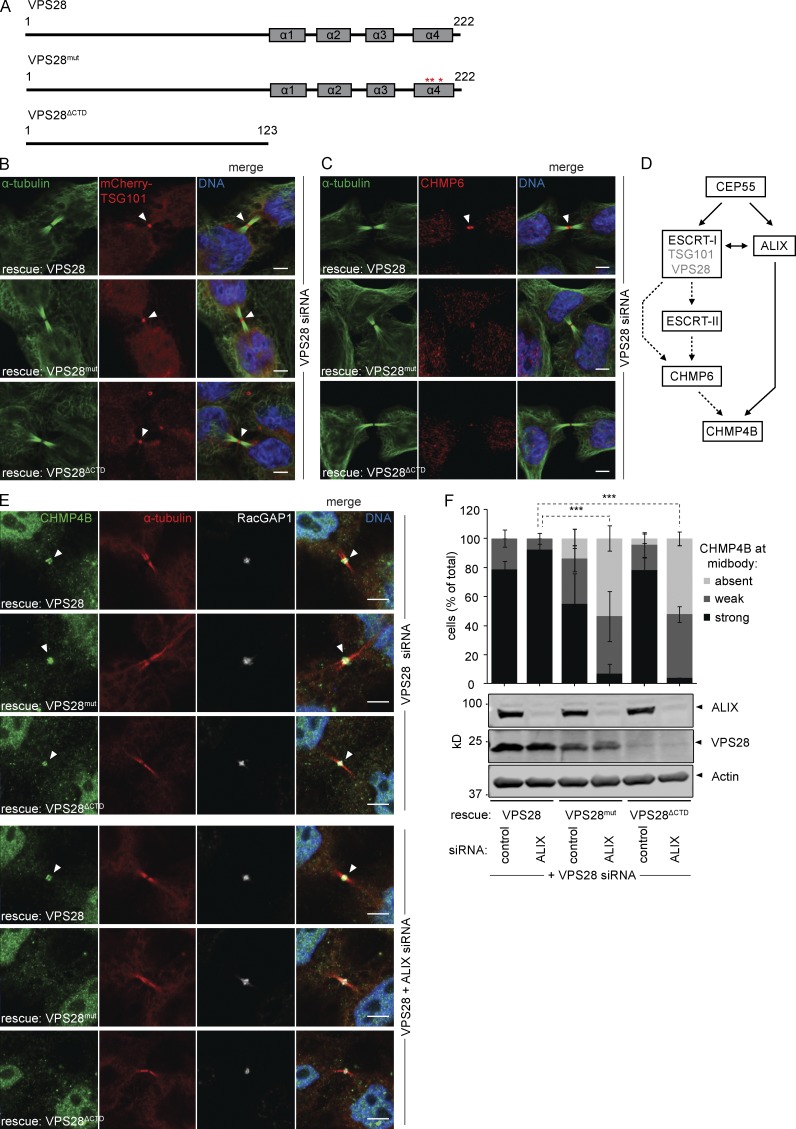
Previous work has highlighted the important role for the VPS28 C-terminal domain (CTD) in establishing interactions with ESCRT-II and CHMP6 (Pineda-Molina et al., 2006; Im and Hurley, 2008). To assess the conservation of such linearity, we generated stable cell lines expressing siRNA-resistant VPS28 mutants carrying mutations that abolish CHMP6 binding (VPS28mut; F206A D207S E209A) or that lack the CTD altogether, also abolishing ESCRT-II binding (VPS28ΔCTD; Fig. 3 A; Pineda-Molina et al., 2006; Im and Hurley, 2008). All VPS28 alleles were able to assemble into ESCRT-I complexes, as adjudged from midbody localization of mCherry-TSG101 (Fig. 3 B) upon knockdown of endogenous VPS28. In striking contrast, CHMP6 recruitment to the midbody was strongly impaired in cells expressing the VPS28mut and VPS28ΔCTD mutants depleted of endogenous VPS28 (Fig. 3 C and Fig. S4), indicating that CHMP6 functions in a linear path downstream of ESCRT-I (Fig. 3 D). Whereas VPS28 depletion did not affect CHMP4B recruitment in these cell lines, codepletion of ALIX abrogated CHMP4B recruitment in the VPS28mut or VPS28ΔCTD cell lines (Fig. 3, E and F). These data are consistent with our other experiments and further indicated that ALIX-dependent accumulation of CHMP4B did not involve the classical nucleation factor CHMP6.
Figure 3.
VPS28 binds ESCRT-II and CHMP6 to recruit CHMP4B. (A) Schematic view of the VPS28 constructs used. (B) Confocal images of HeLa cells stably expressing siRNA-resistant VPS28 alleles and mCherry-TSG101 upon siRNA treatment as indicated, stained for DNA (Hoechst), α-tubulin, and mCherry. Bars, 5 µm. (C) Confocal images of CHMP6 recruitment in HeLa cells stably expressing VPS28 alleles. Bars, 5 µm. (D) Schematic view of cytokinetic CHMP4B recruitment pathways. (E) Confocal images of CHMP4B recruitment in HeLa cells stably expressing VPS28 alleles. Bars, 5 µm. (F) Quantification of CHMP4B at the midbody (error bars indicate SEM; n ≥ 23 cells from four independent experiments; unpaired t test; ***, P < 0.001). Knockdown of ALIX and expression of siRNA-resistant VPS28 is shown by Western blot.
Collectively, our experiments highlight the existence of two parallel paths controlling CHMP4B recruitment to the midbody, namely an ESCRT-I–ESCRT-II–CHMP6 pathway and an ALIX-dependent pathway.
Relation between CHMP4B midbody recruitment and abscission timing
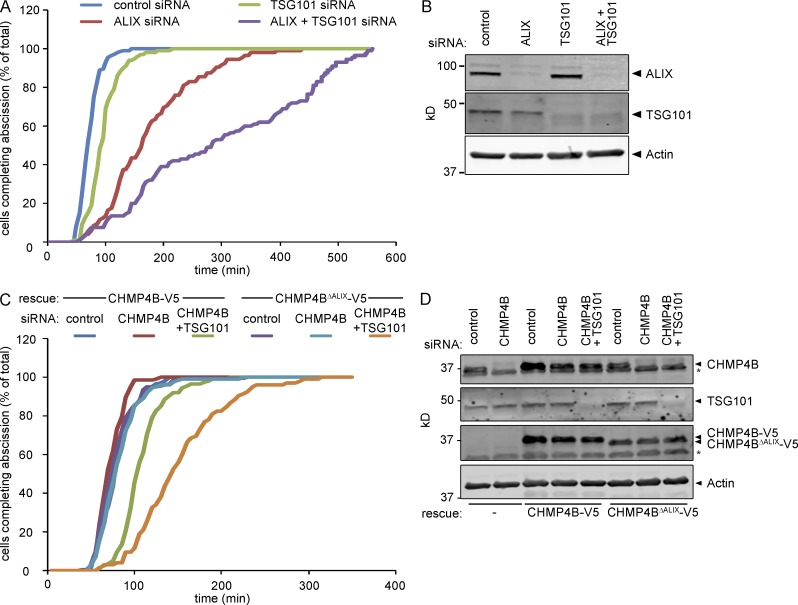
Based on cytokinetic defects, previous work has shown a dominant role for ALIX in regulation of abscission (Carlton and Martin-Serrano, 2007; Morita et al., 2007, 2010; Carlton et al., 2008). To understand the relationship between ALIX, TSG101, and CHMP4B recruitment in abscission timing, we performed live imaging of HeLa cells stably expressing Histone2B-mCherry and eGFP–α-tubulin upon treatment with siRNAs against ALIX and TSG101, using microtubuli severing at the secondary ingression as a proxy for abscission timing. Whereas the mean time between furrow ingression and abscission was 74 ± 17.3 min in control cells, TSG101-depleted cells display a significant abscission delay (98 ± 28.4 min [P < 0.05]; Fig. 4, A and B; and Video 1). ALIX knockdown delayed abscission more prominently, with cells requiring 177 ± 78.7 min (P < 0.001 compared with TSG101 alone) to traverse abscission (Fig. 4, A and B; and Video 1). Importantly, codepletion of ALIX and TSG101 synergistically delayed cytokinetic abscission to a mean of 338 ± 228.9 min (Fig. 4, A and B; and Video 1; P < 0.01 compared with ALIX alone). These data underscore the partially redundant roles for ALIX and TSG101 in cytokinetic abscission and are consistent with their overlapping roles in CHMP4B recruitment.
Figure 4.
ALIX and TSG101 cooperatively control abscission timing. (A) Live imaging of HeLa cells stably expressing Histone2B-mCherry and eGFP–α-tubulin upon siRNAs treatment as indicated. Cumulative frequency plot showing the time interval between furrow ingression and abscission (n > 100 cells from three independent experiments; control, 74 ± 17.3 min; ALIX siRNA, 177 ± 78.7 min; TSG101 siRNA, 98 ± 28.4 min; ALIX + TSG101 siRNA, 338.2 ± 228.9 min [± SD]; ALIX relative to control, P < 0.001; TSG101 relative to control, P = 0.04; ALIX+TSG101 relative to ALIX, P = 0.002; ALIX+TSG101 relative to all others, P < 0.001; P-values obtained using mixed factor model). (B) Knockdown efficiency of ALIX and TSG101. (C) Live imaging of HeLa cells stably expressing Histone2B-mCherry, eGFP–α-tubulin, and CHMP4B-V5 or CHMP4BΔALIX-V5 upon siRNA treatment as indicated. Cumulative frequency plot showing the time interval between furrow ingression and abscission (n > 100 cells from three independent experiments; CHMP4B: control, 82 ± 19.9 min; CHMP4B siRNA, 74 ± 14.5 min; CHMP4B+TSG101 siRNA, 110 ± 25.6 min; CHMP4BΔALIX control, 80 ± 24.0 min; CHMP4B siRNA, 84 ± 24.7 min; CHMP4B+TSG101 siRNA, 153 ± 50.4 min [± SD]; CHMP4B with control siRNA or CHMP4B siRNA relative to CHMP4B with CHMP4B+TSG101 siRNA, P < 0.05; CHMP4BΔALIX with CHMP4B+TSG101 siRNA relative to all others, P < 0.001; P-values obtained using mixed factor model). (D) Knockdown efficiency of CHMP4B and TSG101 as well as expression of CHMP4B-V5 or CHMP4BΔALIX-V5 (*, nonspecific immunoreactivity).
To relate cytokinetic abscission to CHMP4B recruitment more directly, we measured abscission timing in Histone2B-mCherry and eGFP–α-tubulin cells additionally expressing siRNA-resistant CHMP4B-V5 or CHMP4BΔALIX-V5 (also see Fig. 1 D). Codepletion of endogenous CHMP4B with TSG101 in CHMP4B-V5–expressing cells resulted in a mild abscission delay (110 ± 25.6 min) compared with controls (82 ± 19.9 min [P < 0.05]; Fig. 4, C and D) similar to our earlier observations (Fig. 4 A). In contrast, expression of an ALIX-binding–defective CHMP4B allele significantly aggravated the abscission delay upon codepletion of TSG101 and endogenous CHMP4B (153 ± 50.4 min [P < 0.001]; Fig. 4, C and D), providing a direct correlation between CHMP4B recruitment through ALIX and TSG101 and abscission timing. It should be noted that depletion of endogenous CHMP4B did not evoke any abscission timing phenotypes in cells expressing CHMP4BΔALIX-V5, in contrast to ALIX depletion (compare Fig. 4, A–C). This could be taken to mean that other CHMP4 paralogs contribute to regulation of abscission timing in an ALIX-dependent fashion.
Normal abscission timing requires a functional ESCRT-I–ESCRT-II–CHMP6 axis
To assess the function of the ESCRT-I–ESCRT-II–CHMP6 axis in abscission timing, we stably expressed siRNA-resistant VPS28, VPS28mut, or VPS28ΔCTD alleles in a Histone2B-mCherry and eGFP–α-tubulin background. To minimize artifacts caused by accumulation at class E–like structures, we limited VPS28 depletion to 30 h. Nevertheless, a significant abscission delay was observed upon VPS28 depletion in the parental cells (Fig. 5, A and B; 73 ± 15.0 min, P < 0.01 compared with control), which was fully suppressed by expression of siRNA-resistant VPS28 (58 ± 14.0 min). In contrast, neither VPS28 mutant was able to rescue this phenotype, but rather slightly aggravated it (88 ± 18.1 and 86 ± 23.3 min, P < 0.01 compared with control).
Figure 5.
Depletion of CHMP6 and overexpression of dominant-negative CHMP6 results in abscission delay. (A) Cumulative frequency plot showing the time interval between furrow ingression and abscission in HeLa cells stably expressing Histone2B-mCherry, eGFP–α-tubulin, and VPS28 alleles upon siRNA treatment as indicated (n ≥ 100 cells per treatment from four independent experiments; control, 61 ± 15.2 min; VPS28 siRNA, 73 ± 15.0 min; VPS28 with VPS28 siRNA, 58 ± 14.0 min; VPS28mut with VPS28 siRNA, 88 ± 18.1 min; VPS28ΔCTD with VPS28 siRNA, 86 ± 23.3 min [± SD]; control and VPS28 rescue relative to VPS28 siRNA, P < 0.01; control and VPS28 rescue relative to VPS28mut and VPS28ΔCTD, P < 0.01; VPS28 siRNA relative to VPS28mut and VPS28ΔCTD, P < 0.01; P-values obtained using mixed factor model). (B) Knockdown efficiency of VPS28 and codepletion of TSG101. (C) Cumulative frequency plot showing the time interval between furrow ingression and abscission in HeLa cells stably expressing Histone2B-mCherry and eGFP–α-tubulin upon siRNA treatment as indicated (n ≥ 200 cells per treatment from seven independent experiments; control, 69 ± 14.9 min; CHMP6 siRNA#1, 104 ± 53.7 min; CHMP6 siRNA#2, 85 ± 28.2 min [± SD]; CHMP6 1 relative to control, P < 0.001; CHMP6 2 relative to control, P < 0.001; P-values obtained using mixed factor model). (D) Knockdown efficiency of CHMP6. (E) Schematic view of the CHMP6 constructs used in F and G. (F) Confocal images of HeLa cells transfected with wild-type or truncated CHMP6 and stained for DNA (Hoechst), α-tubulin, and MKLP1. Bars, 5 µm. CHMP6core localizes to the midbody. (G) HeLa cells were transfected with wild-type or truncated CHMP6 and analyzed by high-throughput widefield microscopy. The mean of the total cell population is shown (error bars indicate SEM from six independent experiments; n > 1,000 cells per experiment; unpaired t test; ***, P < 0.001).
Extending these data, we observed that depletion of CHMP6 induces a significant delay in abscission using two independent siRNAs (P < 0.001; Fig. 5, C and D; 104 ± 53.7 min [siRNA 1] or 85 ± 28.2 min [siRNA 2]) compared with control siRNA–treated cells (69 ± 14.9 min). These results point toward a contribution of CHMP6 in regulation of cytokinetic abscission. Because the mild phenotypes observed could be because of our inability to fully deplete CHMP6 (with knockdown efficiency never surpassing 85%; Fig. 5 D), we explored alternative ways to perturb CHMP6 function. To this end, we generated a dominant-negative CHMP6core allele, consisting only of the four core α-helices (aa 1–152; Fig. 5 E) and lacking the autoinhibitory C terminus. Overexpressed CHMP6core accumulated at the midbody and dramatically arrested cells in cytokinesis, as illustrated by a strong increase in cytokinesis profiles among cells transiently overexpressing this allele (Fig. 5, F and G).
Together with results in Fig. 3, our data provide a strong correlation between the integrity of the ESCRT-I–ESCRT-II–CHMP6 axis and completion of cytokinetic abscission.
CHMP4B-independent furrow regression in ALIX-depleted cells
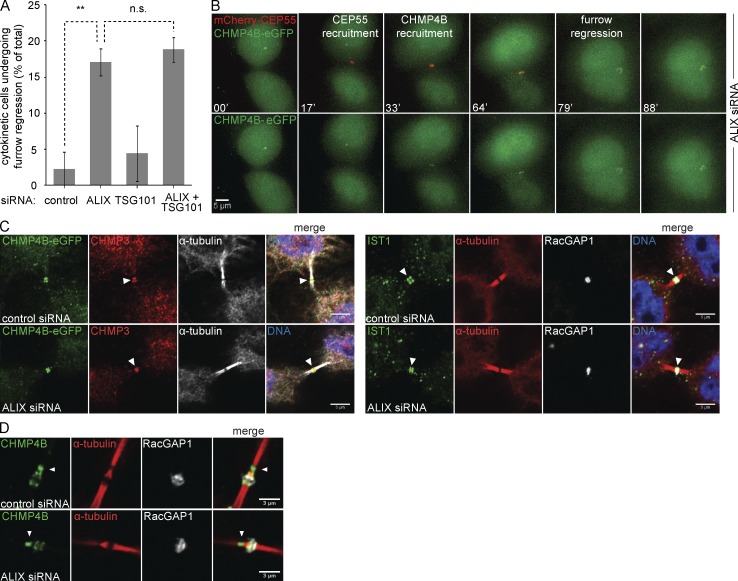
In line with previous studies (Morita et al., 2007), many ALIX-depleted cells, but not TSG101-depleted cells, underwent furrow regression and binucleation, after a brief or extended cytokinetic arrest (100–600 min after onset of furrow ingression; Fig. 6 A). In contrast to the redundancy of ALIX and TSG101 with regard to CHMP4B recruitment and abscission timing, this regression phenotype was not aggravated by codepletion of TSG101 with ALIX (Fig. 6 A), suggesting a unique role for ALIX in preventing binucleation. Even though CHMP4B recruitment was slightly delayed in ALIX-depleted cells (Fig. 1 F), it localized invariably to midbodies at the time of cytokinetic bridge regression (Fig. 6 B and Video 2). Furthermore, we observed no correlation between early or late CHMP4B recruitment and furrow regression. Because ALIX depletion was very efficient (up to 99%) and we clearly detected CHMP4B-eGFP at midbodies in the absence of ALIX (Fig. S5, A and B), our data argue that the furrow regression phenotype in ALIX-depleted cells is not directly caused by lack of CHMP4B recruitment.
Figure 6.
ALIX, but not TSG101, controls furrow regression. (A) Live imaging of HeLa cells stably expressing Histone2B-mCherry and eGFP–α-tubulin upon siRNA treatment as indicated. Percentage of cells undergoing furrow regression (error bars indicate SEM; n > 90 cells from three independent experiments; unpaired t test; **, P < 0.01; n.s., not significant). (B) Live imaging of HeLa cells stably expressing mCherry-CEP55 and CHMP4B-eGFP upon siRNA treatment as indicated. Selected frames of ALIX-depleted cells undergoing cytokinesis, showing enrichment of CHMP4B at the midbody at time of furrow regression (top, merged channels; bottom, CHMP4B-eGFP channel alone. Bar, 5 µm. (C) Confocal images of HeLa cells transfected with indicated siRNAs and stained for DNA (Hoechst), α-tubulin, RacGAP1, GFP, CHMP3, and IST1, as indicated. Bars, 5 µm. (left) HeLa cells stably expressing CHMP4B-eGFP. (D) Localization of CHMP4B to the secondary ingression of HeLa cells is unaffected by ALIX knockdown. Bars, 3 µm.
CHMP4B can recruit downstream factors and localizes to the secondary ingression in ALIX-depleted cells
Because ALIX is essential for successful cytokinetic abscission but not essential for CHMP4B recruitment, we asked whether ALIX knockdown affects CHMP4B-dependent processes downstream of its recruitment. We therefore stained ALIX-depleted cells for CHMP3, a direct interactor of CHMP4B, and IST1, a downstream ESCRT-III–like protein, and checked their presence at the midbody. Both CHMP3 and IST1 localized to the cytokinetic bridge in the absence of ALIX (Fig. 6 C), suggesting that CHMP4B is functional in the absence of ALIX. Similarly, CHMP4B was readily detectable at the secondary ingression upon ALIX depletion (Fig. 6 D), indicating that ALIX is dispensable for the formation of the secondary ingression and the recruitment of CHMP4B to the secondary ingression zone.
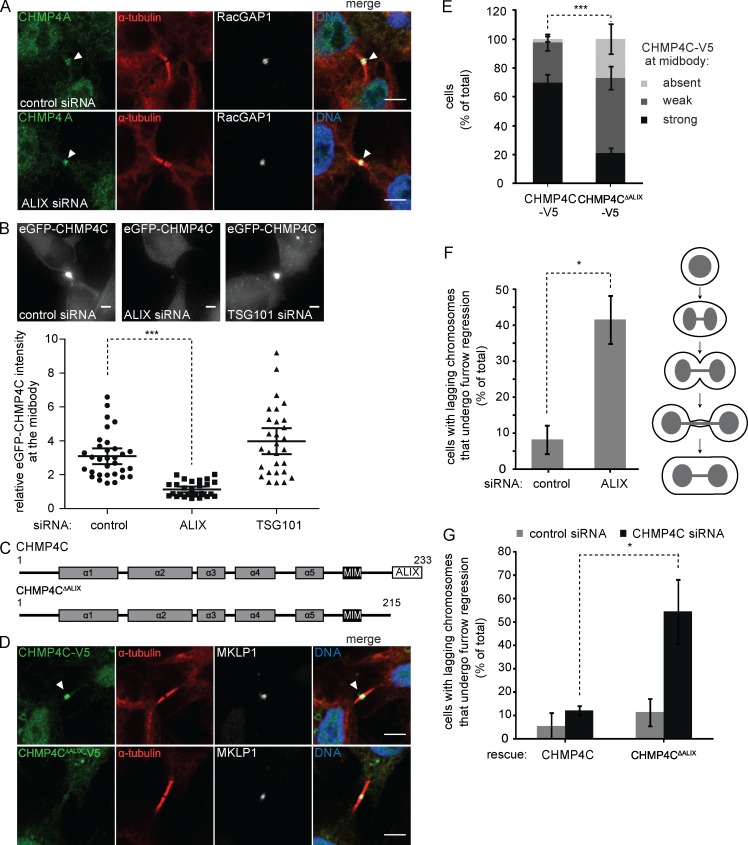
Previously, the lacking recruitment of ALIX in CEP55-depleted cells or of CHMP4 paralogs in ALIX-depleted cells was suggested to cause the formation of aberrant midbodies with continuous α-tubulin staining (Zhao et al., 2006; Carlton and Martin-Serrano, 2007; Carlton et al., 2008). We also noted the presence of such aberrant midbodies but observed no difference in CHMP4B recruitment patterns when comparing the two bridge morphologies (Fig. S5, B and C). Hence, the absence of the protein-dense gap in ALIX-depleted intercellular bridges does not appear to be a direct consequence of defective CHMP4B recruitment.
CHMP4C recruitment to the cytokinetic bridge is dependent on ALIX
Our experiments suggest that CHMP4B localization is maintained in the absence of ALIX, because of the ESCRT-I–ESCRT-II–CHMP6 recruitment arm, and indicate that the prominent furrow regression phenotype observed upon knockdown of ALIX is independent of CHMP4B recruitment. Because it had previously been shown that ALIX mutants unable to bind CHMP4 paralogs (F199D or I212D) could not rescue furrow regression phenotypes (Morita et al., 2007; Carlton et al., 2008), we decided to explore the recruitment of the CHMP4A and CHMP4C isoforms upon down-regulation of ALIX. Like CHMP4B, CHMP4A localization was largely unperturbed in the absence of ALIX (Fig. 7 A). In the absence of antibodies recognizing endogenous CHMP4C, we studied CHMP4C localization dynamics by live imaging using HeLa cell lines expressing inducible eGFP-CHMP4C. Strikingly, CHMP4C recruitment to the midbody was abrogated upon depletion of ALIX alone but unaffected by the depletion of TSG101 (Fig. 7 B and Video 3), suggesting that CHMP4C recruitment is dependent on ALIX. We confirmed this dependence with cell lines expressing full-length or truncated CHMP4C-V5 constructs lacking the ALIX interaction domain (aa 1–215; Fig. 7, C–E). CHMP4C-V5 was readily detectable at the midbody, whereas only a weak or no signal was observable for CHMP4CΔALIX-V5 (Fig. 7, D and E). These results point toward ALIX as dominant CHMP4C recruiter with little redundancy from the ESCRT-I–ESCRT-II–CHMP6 arm.
Figure 7.
CHMP4C recruitment to the midbody relies on ALIX. (A) CHMP4A recruitment is maintained in the absence of ALIX. Confocal images of HeLa cells upon siRNA treatment as indicated, stained for DNA (Hoechst), CHMP4A, α-tubulin, and RacGAP1. Bars, 5 µm. (B) CHMP4C recruitment to the midbody is perturbed in the absence of ALIX. Live imaging of HeLa cells stably expressing inducible eGFP-CHMP4C upon siRNA treatment as indicated. Images and quantification of the relative intensity of eGFP-CHMP4C at the midbody are shown (bars indicate mean with 95% confidence interval; n ≥ 30 cells from four independent experiments; mixed factor model; ***, P < 0.001). Bars, 5 µm. (C) Schematic view of the CHMP4C constructs used in D–F. (D) Confocal images of HeLa cells stably expressing CHMP4C-V5 or CHMP4CΔALIX-V5, stained for DNA (Hoechst), V5, α-tubulin, and MKLP1. Bars, 5 µm. (E) Quantification of full-length or truncated CHMP4C-V5 at the midbody (error bars indicate SEM; n > 50 cells from six independent experiments; unpaired t test; ***, P < 0.001). (F) Depletion of ALIX increases frequency of binucleation in the presence of lagging chromosomes. Live imaging of HeLa cells stably expressing Histone2B-mCherry and eGFP-α-tubulin upon siRNA treatment as indicated. Quantification of furrow regression in cells with lagging chromosomes as depicted in the cartoon (error bars indicate SEM; n = 24 cells from three independent experiments; unpaired t test; *, P < 0.05). (G) Live imaging of HeLa cells stably expressing Histone2B-mCherry, eGFP-α-tubulin, and siRNA-resistant CHMP4C or CHMP4CΔALIX upon CHMP4C siRNA treatment. Quantification of furrow regression in cells with lagging chromosomes (error bars indicate SEM; n ≥ 15 cells from three independent experiments; unpaired t test; *, P < 0.05).
ALIX represents a novel abscission checkpoint signaling node
CHMP4C is a key regulator of the Aurora B–dependent abscission checkpoint, and CHMP4C depletion has been shown to perturb persistent cytokinetic arrest in the presence of chromosomal impediments, culminating in furrow regression and binucleation (Steigemann et al., 2009; Capalbo et al., 2012; Carlton et al., 2012; Thoresen et al., 2014). Considering the essential role for ALIX in CHMP4C recruitment, we asked whether ALIX contributed to abscission checkpoint signaling. To address this, we tracked control and ALIX-depleted HeLa cells stably expressing Histone2B-mCherry and eGFP–α-tubulin and scored the cells with chromosome segregation defects that underwent furrow regression. Importantly, depletion of ALIX led to a dramatic increase in furrow regression in cells with chromatin bridges (Fig. 7 F and Video 4). To establish a direct connection between CHMP4C recruitment via ALIX and abscission checkpoint signaling, we generated stable cell lines expressing siRNA-resistant CHMP4C-V5 or CHMP4CΔALIX-V5 and monitored furrow regression in cells with chromatin bridges upon depletion of endogenous CHMP4C. Remarkably, CHMP4CΔALIX-expressing cells showed a significantly higher tendency to undergo regression (Fig. 7 G). Collectively, these results underscore the significance of the ALIX-dependent recruitment of CHMP4C and indicate a role for ALIX in abscission checkpoint signaling.
Discussion
During MVE formation, sequential assembly of ESCRT-0, ESCRT-I, and ESCRT-II recruits CHMP6 to nucleate the formation of CHMP4B filaments (Teis et al., 2008, 2010; Im et al., 2009; Saksena et al., 2009; Fyfe et al., 2011; Henne et al., 2012). However, based on depletion experiments (Morita et al., 2010), it has become a prevailing opinion that ESCRT-II and CHMP6 are dispensable for cytokinetic abscission, raising questions about how and whether ESCRT-I contributes to this process beyond interacting with ALIX. Furthermore, the mechanism of recruitment of CHMP4B by ALIX or ESCRT-I during cytokinetic abscission has not been established.
To resolve these issues, we set out to map the relative contribution of ESCRT-I and ALIX to CHMP4B recruitment and abscission timing. First, we show that ALIX and TSG101 functionally overlap to control CHMP4B accumulation at the midbody and use CHMP4B mutants to show that ALIX directly recruits CHMP4B to the midbody. Moreover, we show the conservation of an ESCRT-I–ESCRT-II–CHMP6 axis that recruits CHMP4B to the midbody, extending the function of TSG101 beyond ALIX binding. Previous studies have localized ESCRT-II and CHMP6 to the intercellular bridge and demonstrated perturbation of cytokinesis using mutant CHMP6 alleles. However, the use of high overexpression in these experiments warranted caution with respect to their interpretation (Goliand et al., 2014). Here, we provide the first evidence that endogenous CHMP6 and ESCRT-II subunits localize to the midbody. Furthermore, using functional rescue experiments with VPS28 mutants defective in CHMP6 and ESCRT-II recruitment we show that recruitment of CHMP6 and ESCRT-II by ESCRT-I is critical for controlling CHMP4B accumulation. Collectively, our data argue for the existence of two independent CHMP4B recruitment pathways (Fig. 8). We propose that the classical ESCRT-I–ESCRT-II–CHMP6 axis provides a membrane curvature-sensing supercomplex that targets CHMP4B to membrane necks (Im et al., 2009; Fyfe et al., 2011; Lee et al., 2015) and greatly enhances nucleation of CHMP4B filaments. Because ALIX has no reported CHMP4B filament nucleation capability, it could contribute by creating a high local CHMP4B concentration that bypasses the need for CHMP6, by stabilizing spontaneous nucleation events, or preventing disassembly of CHMP4B filaments (Wemmer et al., 2011; Chiaruttini et al., 2015).
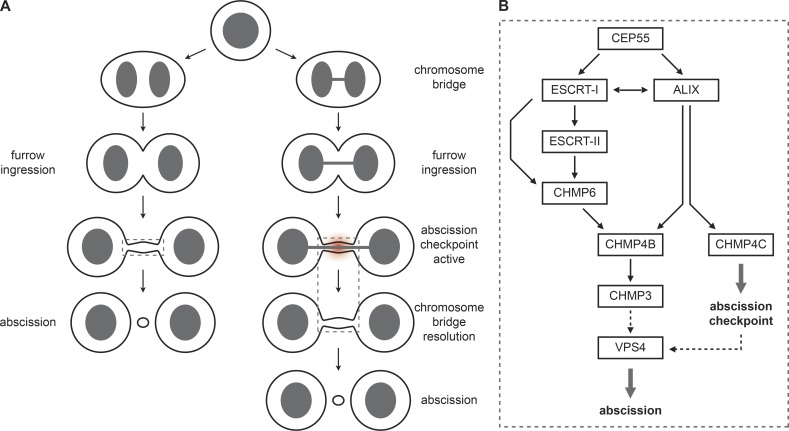
Figure 8.
ESCRT-III recruitment model. (A) Cartoon illustrating cytokinesis in the absence or presence of chromosome segregation defects. (B) Model for ESCRT-III recruitment during cytokinetic abscission and checkpoint signaling. CHMP4B, the main component of the ESCRT-III filaments, is recruited via two parallel arms, namely CEP55–ALIX or CEP55–ESCRT-I–ESCRT-II–CHMP6. This recruitment is essential for timing of cytokinetic abscission. In contrast, recruitment of the abscission checkpoint regulator CHMP4C relies on ALIX, indicating that ALIX represent a dual functionality as abscission regulator and checkpoint signaling node.
By combining depletion experiments with live- and fixed-cell imaging, we show that disruption of individual recruitment arms that only lead to minor delays in CHMP4B recruitment are associated with mild or moderate effects on cytokinetic abscission. We provide the first evidence of reproducible abscission delay in cells depleted for CHMP6 or in cells expressing VPS28 alleles defective in CHMP6 recruitment. Even though the abscission delay was small when compared with overexpression of dominant-negative CHMP6core (aa 1–152), it was highly significant and very similar to delays we observed upon depletion of the established cytokinetic regulator and ESCRT-I subunit TSG101. In contrast to CHMP4B recruitment (Fig. 2 G), expression of siRNA-resistant EAP30 did not rescue abscission timing, indicative of off-targets effects possibly related to mitotic defects observed in EAP30 knockdown cells (not depicted).
The mild ESCRT-I or CHMP6 depletion abscission phenotypes are likely caused by compensatory mechanisms via ALIX, as supported by our observation of prominent redundancies between ALIX and ESCRT-I–ESCRT-II–CHMP6 in CHMP4B recruitment. Indeed, simultaneous disruption of both recruitment arms completely abrogates CHMP4B recruitment and synergistically delays cytokinetic abscission. The difference in abscission timing defects between ALIX depletion and functional rescues using CHMP4BΔALIX alleles is likely explained by the role for CHMP4A in cytokinetic abscission, although CHMP4-independent roles for ALIX should not be excluded. Collectively, the strong correlation between effects on CHMP4B recruitment and abscission timing points toward CHMP4B recruitment as a key parameter in ALIX- and ESCRT-I axis dependent control of cytokinetic abscission.
In accordance with others (Carlton and Martin-Serrano, 2007; Morita et al., 2007, 2010; Carlton et al., 2008), we observed frequent furrow regression upon depletion of ALIX. However, in stark contrast to delayed cytokinetic abscission, no correlation between CHMP4B recruitment and severity of this phenotype was observed, with regression commonly occurring in the presence of CHMP4B at the midbody (Fig. 6). Furthermore, whereas codepletion of TSG101 with ALIX aggravated the abscission delay and abrogated CHMP4B recruitment, it had no discernible effect on the frequency of furrow regression. These results point toward a unique role for ALIX in maintaining intercellular bridge integrity beyond regulation of CHMP4B recruitment. Previously, it has been shown that ALIX mutants that could not bind CHMP4 isoforms (F199D or I212D) were unable to rescue the furrow regression induced by ALIX depletion (Morita et al., 2007; Carlton et al., 2008), but localization of CHMP4B or its paralogs CHMP4A and CHMP4C was not addressed in these studies. In contrast to CHMP4B, CHMP4C was predominantly reliant on ALIX for its localization at the midbody. Such recruitment selectivity between closely related paralogs is supported by previous yeast two-hybrid experiments indicating that CHMP4B has higher affinity for CHMP6 (von Schwedler et al., 2003; Tsang et al., 2006). It will be of interest to explore whether the differences between CHMP4B and CHMP4C are caused by intrinsic sequence differences (e.g., at the α1–α2 or α3–α4 junctions) or reflect differential cofactor binding. In this context, it is interesting to note that the chromosome passenger complex subunit Borealin preferentially binds CHMP4C compared with CHMP4A and CHMP4B (Carlton et al., 2012).
Because CHMP4C has been identified as a central regulator of the abscission checkpoint and CHMP4C depletion has been shown to induce furrow regression in the presence of chromatin bridges (Thoresen et al., 2014), the furrow regression phenotype observed upon ALIX depletion could be explained by defective CHMP4C recruitment. We tested this hypothesis by monitoring cytokinetic progression in cells displaying chromatin bridges and found that ALIX depletion dramatically increases the frequency of furrow regression in such cells. Complementary, rescue experiments showed that a CHMP4C allele defective for ALIX-binding was unable to rescue cytokinetic abscission in cells with chromatin bridges upon depletion of endogenous CHMP4C. Collectively, our findings point toward a novel role for ALIX as an abscission checkpoint signaling node. This is of special interest when considering the Aurora B–dependent checkpoint phosphorylation of CHMP4C juxtapositioned to its ALIX-binding motif (Capalbo et al., 2012; Carlton et al., 2012) and the recent ALIX-binding motif mutation in CHMP4C found in ovarian cancer (Pharoah et al., 2013). We note that ALIX depletion also increases furrow regression frequency in cells lacking apparent chromatin bridges (∼10% of cell divisions), indicating that ALIX either controls bridge stability through other means (Cabezas et al., 2005; Chu et al., 2012) or that abscission checkpoint senses additional cytokinetic stresses (Caballe et al., 2015).
In conclusion, the work presented here firmly implicates ESCRT-II and CHMP6 as participants in a canonical ESCRT recruitment cascade during cytokinetic abscission and reveals two parallel pathways for CHMP4B recruitment and cytokinetic abscission: via ALIX or via ESCRT-I–ESCRT-II–CHMP6. Importantly, we provide evidence for a dual role for ALIX in cytokinesis as a CHMP4B recruiter and abscission checkpoint regulator. It will therefore be important to further dissect the molecular interplay between these ALIX functionalities, CHMP4 paralogs, and ANCHR as well as their regulation by Aurora B and ULK3 (Caballe et al., 2015) in the abscission checkpoint signaling so as to understand their contributions to cancer development.
Materials and methods
Plasmid constructs
In short, eGFP, mCherry, or V5 fusions of transgenes were generated by PCR, synthetic DNA fragments (Integrated DNA Technologies), or direct subcloning into Gateway ENTRY plasmids. Subsequently, vectors used for transient expression (Gateway-modified pcDNA3.1(+)) or lentiviral vectors for stable transgenesis (Addgene vectors 41393, 19067, 19068, and derivatives thereof, as well as vectors based on pCDH-EF1α-MCS-IRES-PURO; SystemBiosciences) were generated by recombination (Campeau et al., 2009). Plasmids used in this study are described in Table S1. The original CEP55 construct was obtained from K. Kutsche (Institut für Humangenetik, Universitätsklinikum Hamburg-Eppendorf, Hamburg, Germany).
Cell culture and stable cell lines
HeLa “Kyoto” and HeLa “Paris” cells were maintained in DMEM (Gibco) supplemented with 10% FBS, 5 U/ml penicillin, and 50 µg/ml streptomycin at 37°C under 5% CO2. Stable cell lines expressing eGFP–α-tubulin and H2B-mCherry were made as previously described (Thoresen et al., 2014). Stable cell lines expressing CHMP4B-eGFP were obtained from A.A. Hyman (Max Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany; Poser et al., 2008). All other cell lines were lentivirus-generated pools, using plasmids described in Table S1 (Thoresen et al., 2014). Inducible eGFP-CHMP4C cell lines were imaged 5 h or more after induction with 2 µg/ml doxycycline (Sigma-Aldrich).
siRNA and plasmid transfections
siRNAs against CEP55 (5′-GGAGAAGAAUGCUUAUCAA-3′), ALIX (5′-CCUGGAUAAUGAUGAAGGA-3′), EAP20 (5′-CGAUCCAGAUUGUAUUAGA-3′), EAP30 (5′-CUUGCAGAGGCCAAGUAUA-3′), CHMP6 (#1, 5′-CUUGCAGAGGCCAAGUAUA-3′; #2, 5′-GGAAAUGAGUGUCUGAACA-3′), CHMP4B (5′-CAUCGAGUUCCAGCGGGAG-3′), TSG101 (5′-CCUCCAGUCUUCUCUCGUC-3′), VPS28 (5′-GGCUCAGAAAUCAGCUCUA-3′), CHMP4C (5′-AAUCGAAUCCAGAGAGAAA-3′), and nontargeting control siRNA (predesigned, catalog number 4390844) were purchased from Ambion. Cells at 50% confluency were transfected with 20–50 nM final siRNA concentration using Lipofectamine RNAiMax transfection reagent (Life Technologies) according to the manufacturer’s instructions, washed the next day, and harvested after 30, 48, or 72 h.
Cells were transfected with cDNA using jetPrime transfection reagent (PolyPlus) following the manufacturer’s instructions and collected the next day.
Antibodies
Antiserum against a peptide corresponding to amino acids 25–39 of human CHMP6 was raised in two rabbits and affinity purified on Affi-Gel beads containing immobilized peptide. Rabbit anti-CHMP3, rabbit anti-ALIX, rabbit anti-EAP30, rabbit anti-CHMP4B, and rabbit anti-EAP20 were described previously (Sharma et al., 2004; Cabezas et al., 2005; Bache et al., 2006; Malerød et al., 2007; Sagona et al., 2010). Mouse anti–α-tubulin (Sigma-Aldrich), sheep anti–α-tubulin (Cytoskeleton), mouse anti–β-actin (Sigma-Aldrich), goat anti-RacGAP1 (Abcam), rabbit anti-MKLP1 (Santa Cruz Biotechnology, Inc.), goat anti-V5 (Abcam), mouse anti-GFP (Roche), mouse anti-TSG101 (BD Transduction Laboratories), rabbit anti-CHMP6 (Santa Cruz Biotechnology, Inc.), rabbit anti-CEP55 (Abnova), rabbit anti-VPS28 (Santa Cruz Biotechnology, Inc.), rabbit anti-IST1 (Proteintech), rabbit anti-CHMP4A (Santa Cruz Biotechnology, Inc.), and rabbit anti-mCherry (Acris) were used as primary antibodies. Secondary antibodies included anti-mouse, anti-rabbit, and anti-goat Alexa Fluor 488 (Jackson ImmunoResearch), Alexa Fluor 555 (Molecular Probes), Alexa Fluor 568 (Molecular Probes), Alexa Fluor 647 (Jackson ImmunoResearch), and DyLight649 (Jackson ImmunoResearch).
Immunoblotting
Cells were lysed in 2× sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT, and 0.004% bromophenol blue). Whole-cell lysates were subjected to SDS-PAGE on 10% or 4–20% gradient gels (mini-PROTEAN TGX; Bio-Rad). Proteins were transferred to Immobilon-P membranes (Millipore) followed by blocking and antibody incubation in 5% fat-free milk powder in Tris-buffered saline with 0.05% Tween 20. Membranes incubated with fluorescent secondary antibodies (IRDye680 and IRDye800; LI-COR) were developed with an Odyssey infrared scanner (LI-COR), whereas those incubated with horseradish peroxidase–conjugated antibodies were developed using Clarity Western ECL substrate solutions (Bio-Rad) with a ChemiDoc XRS+ imaging system (Bio-Rad).
High-throughput widefield microscopy
For the analysis of cytokinesis profiles, cells were seeded on coverslips, fixed with 3% paraformaldehyde (Sigma-Aldrich), permeabilized with 0.05% saponin in PBS, stained with antibodies, and imaged using an Olympus ScanR automated microscope equipped with an ULSAPO 40× objective. 64 fields from each coverslip were then scored manually.
Confocal fluorescence microscopy
For localization studies, cells were seeded on coverslips, fixed with 3% PFA (Sigma-Aldrich) or methanol, permeabilized with 0.05% saponin in PBS and stained with antibodies. In CHMP6 overexpression experiments, cells were permeabilized with PEM buffer (80 mM K-Pipes, pH 6.8, 5 mM EGTA, and 1 mM MgCl2) before fixation.
Stained coverslips were viewed with an LSM 710 or 780 confocal microscope (Carl Zeiss) equipped with an Ar-laser multiline (458/488/514 nm), a DPSS-561 10 (561 nm), a laser diode 405–30 CW (405 nm), and an HeNe laser (633 nm). The objective used was a Plan-Apochromat 63×/1.40 oil DIC M27 (Carl Zeiss). Image processing was performed with basic software ZEN 2009 (Carl Zeiss) and ImageJ software (National Institutes of Health). CHMP4B and CHMP4C intensities at the intercellular bridge were scored manually using constant settings for the relevant channels. For CHMP6, EAP20 and α-tubulin intensity assessments values of gray intensities from 16-bit images were measured in a line along the intercellular bridge and plotted relative to the distance from the midbody identified by lack of tubulin staining. The mean intensity along the line (control set to 1) was compared between different treatments. The SEM is calculated between individual experiments.
Live microscopy
Cells seeded in Lab-Tek chamber slides with coverslip bottom (Nunc) were imaged on a Deltavision microscope (Applied Precision) equipped with Elite TruLight Illumination System, a CoolSNAP HQ2 camera, and a 40× or 60× Plan-Apochromat (1.42 NA) lens. For temperature control during live observation, the microscope stage was kept at 37°C under 5% CO2 by an incubation chamber. Time-lapse images (15–16 z-sections 0.8–1 µm apart) were acquired every 5–8 min over a total time period of up to 24 h and deconvolved and z-projected using the softWoRx software (Applied Precision). To ensure that the statistical results did not depend on specific assumptions, tests were performed both by ordinary analysis of variance (using averages from each experiment) and using nested mixed factor models. Results from the mixed factor models are given, but results were always similar. To take multiple comparisons into account, Tukey’s HSD was used. For relative eGFP-CHMP4C intensity value measurements at the midbody, the ratio between mean midbody and mean cytoplasmic fluorescence intensities was calculated using ImageJ. The SD is calculated based on all cells from all experiments.
Online supplemental material
Fig. S1 shows the knockdown efficiency of siRNAs used. Fig. S2 demonstrates that depletion of VPS28 decreases TSG101 levels and phenocopies TSG101 depletion. Fig. S3 shows the characterization of CHMP6 and EAP20 antibodies used. Fig. S4 shows the intensity of CHMP6 and α-tubulin along the intercellular bridge of cells depleted of endogenous VPS28 and stably expressing different siRNA-resistant VPS28 alleles. Fig. S5 shows CHMP4B localization to the midbody in the absence of ALIX, including to intercellular bridges with continuous α-tubulin staining. Video 1 shows the abscission time of a cell depleted of TSG101 and ALIX. Video 2 shows CHMP4B recruitment to the midbody of an ALIX-depleted cell undergoing furrow regression. Video 3 shows CHMP4C recruitment in ALIX- and TSG101-depleted cells. Video 4 shows an ALIX depleted cell with chromosome segregation defects that undergoes furrow regression. Table S1 describes all plasmids used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507009/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201507009.dv.
Supplementary Material
Acknowledgments
We thank Trine Håve and Torill Høiby for technical support, Anne Engen for help with cell culture, and Chema Bassols for IT assistance. We are also grateful to Kerstin Kutsche for kindly providing the original CEP55 construct and Anthony A. Hyman and Ina Poser for the CHMP4B-eGFP BAC HeLa cells. The Advanced Light Microscopy core facility at Oslo University Hospital is acknowledged for providing access to microscopes. We also thank Marina Vietri, Kay O. Schink, Kia Wee Tan, and Sigrid B. Thoresen for helpful discussions. E.M. Wenzel is a senior research fellow of the Helse Sør-Øst RHF (grant 2015014). C. Campsteijn is a postdoctoral fellow and C. Raiborg a senior research fellow of the Norwegian Cancer Society (grant 4541899).
H. Stenmark was supported by a grant from the Norwegian Cancer Society (grant 605009) and an Advanced Grant from the European Research Council (grant 233146). This work was partly supported by the Norges Forskningsråd through its Centres of Excellence funding scheme (project number 179571).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CHMP
- charged multivesicular body protein
- CTD
- C-terminal domain
- ESCRT
- endosomal sorting complex required for transport
- MIT
- microtubule interacting and transport
- MVE
- multivesicular endosome
References
- Adell M.A., and Teis D.. 2011. Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett. 585:3191–3196. 10.1016/j.febslet.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agromayor M., and Martin-Serrano J.. 2013. Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends Cell Biol. 23:433–441. 10.1016/j.tcb.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Estepa-Sabal E.J., Meerloo T., and Emr S.D.. 2002a Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 3:271–282. 10.1016/S1534-5807(02)00220-4 [DOI] [PubMed] [Google Scholar]
- Babst M., Katzmann D.J., Snyder W.B., Wendland B., and Emr S.D.. 2002b Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289. 10.1016/S1534-5807(02)00219-8 [DOI] [PubMed] [Google Scholar]
- Bache K.G., Stuffers S., Malerød L., Slagsvold T., Raiborg C., Lechardeur D., Wälchli S., Lukacs G.L., Brech A., and Stenmark H.. 2006. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol. Biol. Cell. 17:2513–2523. 10.1091/mbc.E05-10-0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M., Schubert H.L., McCullough J., Langelier C., Eckert D.M., Stubblefield W.M., Uter N.T., Myszka D.G., Hill C.P., and Sundquist W.I.. 2009. Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 16:754–762. 10.1038/nsmb.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballe A., and Martin-Serrano J.. 2011. ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic. 12:1318–1326. 10.1111/j.1600-0854.2011.01244.x [DOI] [PubMed] [Google Scholar]
- Caballe A., Wenzel D.M., Agromayor M., Alam S.L., Skalicky J.J., Kloc M., Carlton J.G., Labrador L., Sundquist W.I., and Martin-Serrano J.. 2015. ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins. eLife. 4:e06547 10.7554/eLife.06547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas A., Bache K.G., Brech A., and Stenmark H.. 2005. Alix regulates cortical actin and the spatial distribution of endosomes. J. Cell Sci. 118:2625–2635. 10.1242/jcs.02382 [DOI] [PubMed] [Google Scholar]
- Campeau E., Ruhl V.E., Rodier F., Smith C.L., Rahmberg B.L., Fuss J.O., Campisi J., Yaswen P., Cooper P.K., and Kaufman P.D.. 2009. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 4:e6529 10.1371/journal.pone.0006529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capalbo L., Montembault E., Takeda T., Bassi Z.I., Glover D.M., and D’Avino P.P.. 2012. The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis. Open Biol. 2:120070 10.1098/rsob.120070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson L.-A., and Hurley J.H.. 2012. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc. Natl. Acad. Sci. USA. 109:16928–16933. 10.1073/pnas.1211759109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.G., and Martin-Serrano J.. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 316:1908–1912. 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Agromayor M., and Martin-Serrano J.. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA. 105:10541–10546. 10.1073/pnas.0802008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.G., Caballe A., Agromayor M., Kloc M., and Martin-Serrano J.. 2012. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 336:220–225. 10.1126/science.1217180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.T., Hehnly H., and Doxsey S.J.. 2012. Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission. Nat. Rev. Mol. Cell Biol. 13:483–488. 10.1038/nrm3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N., Redondo-Morata L., Colom A., Humbert F., Lenz M., Scheuring S., and Roux A.. 2015. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell. 163:866–879. 10.1016/j.cell.2015.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D., Pan H., Wan P., Wu J., Luo J., Zhu H., and Chen J.. 2012. AIP1 acts with cofilin to control actin dynamics during epithelial morphogenesis. Development. 139:3561–3571. 10.1242/dev.079491 [DOI] [PubMed] [Google Scholar]
- Elia N., Sougrat R., Spurlin T.A., Hurley J.H., and Lippincott-Schwartz J.. 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA. 108:4846–4851. 10.1073/pnas.1102714108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fededa J.P., and Gerlich D.W.. 2012. Molecular control of animal cell cytokinesis. Nat. Cell Biol. 14:440–447. 10.1038/ncb2482 [DOI] [PubMed] [Google Scholar]
- Fyfe I., Schuh A.L., Edwardson J.M., and Audhya A.. 2011. Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J. Biol. Chem. 286:34262–34270. 10.1074/jbc.M111.266411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., and Pellman D.. 2012. Linking abnormal mitosis to the acquisition of DNA damage. J. Cell Biol. 199:871–881. 10.1083/jcb.201210040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliand I., Nachmias D., Gershony O., and Elia N.. 2014. Inhibition of ESCRT-II-CHMP6 interactions impedes cytokinetic abscission and leads to cell death. Mol. Biol. Cell. 25:3740–3748. 10.1091/mbc.E14-08-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.A., Paluch E., and Oegema K.. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58. 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Guizetti J., and Gerlich D.W.. 2012. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 22:133–140. 10.1016/j.tcb.2011.11.007 [DOI] [PubMed] [Google Scholar]
- Hayashi M.T., and Karlseder J.. 2013. DNA damage associated with mitosis and cytokinesis failure. Oncogene. 32:4593–4601. 10.1038/onc.2012.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., and Emr S.D.. 2011. The ESCRT pathway. Dev. Cell. 21:77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., Zhao Y., and Emr S.D.. 2012. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 151:356–371. 10.1016/j.cell.2012.08.039 [DOI] [PubMed] [Google Scholar]
- Henne W.M., Stenmark H., and Emr S.D.. 2013. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 5:5 10.1101/cshperspect.a016766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland A.J., and Cleveland D.W.. 2012. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 13:501–514. 10.1038/embor.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H. 2010. The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 45:463–487. 10.3109/10409238.2010.502516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., and Hanson P.I.. 2010. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat. Rev. Mol. Cell Biol. 11:556–566. 10.1038/nrm2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Y.J., and Hurley J.H.. 2008. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev. Cell. 14:902–913. 10.1016/j.devcel.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im Y.J., Wollert T., Boura E., and Hurley J.H.. 2009. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev. Cell. 17:234–243. 10.1016/j.devcel.2009.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A.J., Maiuri P., Lafaurie-Janvore J., Divoux S., Piel M., and Perez F.. 2014. ESCRT machinery is required for plasma membrane repair. Science. 343:1247136 10.1126/science.1247136 [DOI] [PubMed] [Google Scholar]
- Jouvenet N. 2012. Dynamics of ESCRT proteins. Cell. Mol. Life Sci. 69:4121–4133. 10.1007/s00018-012-1035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D.J., Odorizzi G., and Emr S.D.. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893–905. 10.1038/nrm973 [DOI] [PubMed] [Google Scholar]
- Kieffer C., Skalicky J.J., Morita E., De Domenico I., Ward D.M., Kaplan J., and Sundquist W.I.. 2008. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 15:62–73. 10.1016/j.devcel.2008.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Langelier C., von Schwedler U.K., Fisher R.D., De Domenico I., White P.L., Hill C.P., Kaplan J., Ward D., and Sundquist W.I.. 2006. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol. 80:9465–9480. 10.1128/JVI.01049-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.H., Elia N., Ghirlando R., Lippincott-Schwartz J., and Hurley J.H.. 2008. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 322:576–580. 10.1126/science.1162042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.H., Kai H., Carlson L.A., Groves J.T., and Hurley J.H.. 2015. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc. Natl. Acad. Sci. USA. 112:15892–15897. 10.1073/pnas.1518765113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens S.M., Voest E.E., and Medema R.H.. 2010. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat. Rev. Cancer. 10:825–841. 10.1038/nrc2964 [DOI] [PubMed] [Google Scholar]
- Loncle N., Agromayor M., Martin-Serrano J., and Williams D.W.. 2015. An ESCRT module is required for neuron pruning. Sci. Rep. 5:8461 10.1038/srep08461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mageswaran S.K., Johnson N.K., Odorizzi G., and Babst M.. 2015. Constitutively active ESCRT-II suppresses the MVB-sorting phenotype of ESCRT-0 and ESCRT-I mutants. Mol. Biol. Cell. 26:554–568. 10.1091/mbc.E14-10-1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerød L., Stuffers S., Brech A., and Stenmark H.. 2007. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 8:1617–1629. 10.1111/j.1600-0854.2007.00630.x [DOI] [PubMed] [Google Scholar]
- McCullough J., Fisher R.D., Whitby F.G., Sundquist W.I., and Hill C.P.. 2008. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA. 105:7687–7691. 10.1073/pnas.0801567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Colf L.A., and Sundquist W.I.. 2013. Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82:663–692. 10.1146/annurev-biochem-072909-101058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J., Clippinger A.K., Talledge N., Skowyra M.L., Saunders M.G., Naismith T.V., Colf L.A., Afonine P., Arthur C., Sundquist W.I., et al. 2015. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 350:1548–1551. 10.1126/science.aad8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B., and Martin-Serrano J.. 2009. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J. Cell Sci. 122:2167–2177. 10.1242/jcs.028308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill S.A., and Hanson P.I.. 2010. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem. 285:35428–35438. 10.1074/jbc.M110.126318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa B., and Gerlich D.W.. 2014. Cytokinetic abscission: molecular mechanisms and temporal control. Dev. Cell. 31:525–538. 10.1016/j.devcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Morita E. 2012. Differential requirements of mammalian ESCRTs in multivesicular body formation, virus budding and cell division. FEBS J. 279:1399–1406. 10.1111/j.1742-4658.2012.08534.x [DOI] [PubMed] [Google Scholar]
- Morita E., and Sundquist W.I.. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425. 10.1146/annurev.cellbio.20.010403.102350 [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.Y., Morham S.G., Gygi S.P., Rodesch C.K., and Sundquist W.I.. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227. 10.1038/sj.emboj.7601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Colf L.A., Karren M.A., Sandrin V., Rodesch C.K., and Sundquist W.I.. 2010. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. USA. 107:12889–12894. 10.1073/pnas.1005938107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y., Hodgson L., Mantell J., Verkade P., and Carlton J.G.. 2015. ESCRT-III controls nuclear envelope reformation. Nature. 522:236–239. 10.1038/nature14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel S., Macheboeuf P., Martinelli N., and Weissenhorn W.. 2011. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem. Sci. 36:199–210. 10.1016/j.tibs.2010.09.004 [DOI] [PubMed] [Google Scholar]
- Pharoah P.D., Tsai Y.Y., Ramus S.J., Phelan C.M., Goode E.L., Lawrenson K., Buckley M., Fridley B.L., Tyrer J.P., Shen H., et al. 2013. GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 45:362–370: e1–e2. 10.1038/ng.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda-Molina E., Belrhali H., Piefer A.J., Akula I., Bates P., and Weissenhorn W.. 2006. The crystal structure of the C-terminal domain of Vps28 reveals a conserved surface required for Vps20 recruitment. Traffic. 7:1007–1016. 10.1111/j.1600-0854.2006.00440.x [DOI] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J.R., Hériché J.K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A.W., et al. 2008. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 5:409–415. 10.1038/nmeth.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova T.A., Zhu J., and Li R.. 2013. Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev. 32:377–389. 10.1007/s10555-013-9436-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., and Stenmark H.. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 458:445–452. 10.1038/nature07961 [DOI] [PubMed] [Google Scholar]
- Sagona A.P., Nezis I.P., Pedersen N.M., Liestøl K., Poulton J., Rusten T.E., Skotheim R.I., Raiborg C., and Stenmark H.. 2010. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat. Cell Biol. 12:362–371. 10.1038/ncb2036 [DOI] [PubMed] [Google Scholar]
- Saksena S., Wahlman J., Teis D., Johnson A.E., and Emr S.D.. 2009. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 136:97–109. 10.1016/j.cell.2008.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh A.L., and Audhya A.. 2014. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 49:242–261. 10.3109/10409238.2014.881777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Pampinella F., Nemes C., Benharouga M., So J., Du K., Bache K.G., Papsin B., Zerangue N., Stenmark H., and Lukacs G.L.. 2004. Misfolding diverts CFTR from recycling to degradation: quality control at early endosomes. J. Cell Biol. 164:923–933. 10.1083/jcb.200312018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S., Kimpler L.A., and Hanson P.I.. 2007. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 8:1068–1079. 10.1111/j.1600-0854.2007.00584.x [DOI] [PubMed] [Google Scholar]
- Steigemann P., Wurzenberger C., Schmitz M.H., Held M., Guizetti J., Maar S., and Gerlich D.W.. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 136:473–484. 10.1016/j.cell.2008.12.020 [DOI] [PubMed] [Google Scholar]
- Stieler J.T., and Prange R.. 2014. Involvement of ESCRT-II in hepatitis B virus morphogenesis. PLoS One. 9:e91279 10.1371/journal.pone.0091279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Henne W.M., Borbat P.P., Buchkovich N.J., Freed J.H., Mao Y., Fromme J.C., and Emr S.D.. 2015. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. eLife. 4:4 10.7554/eLife.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D., Saksena S., and Emr S.D.. 2008. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell. 15:578–589. 10.1016/j.devcel.2008.08.013 [DOI] [PubMed] [Google Scholar]
- Teis D., Saksena S., Judson B.L., and Emr S.D.. 2010. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 29:871–883. 10.1038/emboj.2009.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H., Perisic O., González B., and Williams R.L.. 2004. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev. Cell. 7:559–569. 10.1016/j.devcel.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Thoresen S.B., Campsteijn C., Vietri M., Schink K.O., Liestøl K., Andersen J.S., Raiborg C., and Stenmark H.. 2014. ANCHR mediates Aurora-B-dependent abscission checkpoint control through retention of VPS4. Nat. Cell Biol. 16:550–560. 10.1038/ncb2959 [DOI] [PubMed] [Google Scholar]
- Tsang H.T., Connell J.W., Brown S.E., Thompson A., Reid E., and Sanderson C.M.. 2006. A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 88:333–346. 10.1016/j.ygeno.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Vietri M., Schink K.O., Campsteijn C., Wegner C.S., Schultz S.W., Christ L., Thoresen S.B., Brech A., Raiborg C., and Stenmark H.. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 522:231–235. 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- von Schwedler U.K., Stuchell M., Müller B., Ward D.M., Chung H.Y., Morita E., Wang H.E., Davis T., He G.P., Cimbora D.M., et al. 2003. The protein network of HIV budding. Cell. 114:701–713. 10.1016/S0092-8674(03)00714-1 [DOI] [PubMed] [Google Scholar]
- Wemmer M., Azmi I., West M., Davies B., Katzmann D., and Odorizzi G.. 2011. Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J. Cell Biol. 192:295–306. 10.1083/jcb.201007018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., and Hurley J.H.. 2010. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 464:864–869. 10.1038/nature08849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., and Hurley J.H.. 2009. Membrane scission by the ESCRT-III complex. Nature. 458:172–177. 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorikawa C., Shibata H., Waguri S., Hatta K., Horii M., Katoh K., Kobayashi T., Uchiyama Y., and Maki M.. 2005. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem. J. 387:17–26. 10.1042/BJ20041227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A., Usami Y., Radoshitzky S.R., Popova E., Palu G., and Göttlinger H.. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA. 103:19140–19145. 10.1073/pnas.0603788103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.M., Seki A., and Fang G.. 2006. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell. 17:3881–3896. 10.1091/mbc.E06-01-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.