Comparison of autophagy inhibition by deletion of Atg5, Atg16L1, Atg7, or Fip200 reveals a critical role for increased p62 in determining the fate of autophagy-deficient neural stem cells by controlling intracellular superoxide.
Abstract
Autophagy plays important roles in many biological processes, but our understanding of the mechanisms regulating stem cells by autophagy is limited. Interpretations of earlier studies of autophagy using knockouts of single genes are confounded by accumulating evidence for other functions of many autophagy genes. Here, we show that, in contrast to Fip200 deletion, inhibition of autophagy by deletion of Atg5, Atg16L1, or Atg7 does not impair the maintenance and differentiation of postnatal neural stem cells (NSCs). Only Fip200 deletion, but not Atg5, Atg16L1, or Atg7 deletion, caused p62/sequestome1 aggregates to accumulate in NSCs. Fip200 and p62 double conditional knockout mice demonstrated that p62 aggregate formation triggers aberrant superoxide increases by impairing superoxide dismutase functions. By comparing the inhibition of autophagy by deletion of Atg5, Atg16L1, or Atg7 with Fip200 deletion, we revealed a critical role of increased p62 in determining the fate of autophagy-deficient NSCs through intracellular superoxide control.
Introduction
Autophagy is a highly conserved process to degrade bulk cytoplasmic materials for cell homeostasis and in response to starvation. Dysfunctions in autophagy are implicated in cancer, neurodegenerative, and autoimmune diseases (He and Klionsky, 2009; Mizushima and Levine, 2010; Mizushima and Komatsu, 2011; Rubinsztein et al., 2011; White, 2012). Because many stem cells, including neural stem cells (NSCs) are long lived, quality-control mechanisms are crucial. The processes of stem cell self-renewal and differentiation also require strict control of cellular remodeling (Zeng and Zhou, 2008). Consistent with such expectations, we have reported that deletion of Fip200, an essential component of the complex required for the induction of autophagy in mammals (Hara et al., 2008), resulted in NSC depletion and defective differentiation by aberrant reactive oxygen species (ROS) accumulation (Wang et al., 2013).
Fip200 is the only autophagy gene analyzed in NSCs so far, but studies in other tissues and cancers suggest divergent outcomes by deletion of different autophagy genes (Kenific and Debnath, 2015). For example, beclin1 haplodeficiency promoted spontaneous malignancies in mouse models (lung and liver cancer, lymphomas; Liang et al., 1999; Qu et al., 2003; Yue et al., 2003), but deletion of other autophagy genes did not (Takamura et al., 2011). In contrast, recent studies showed that autophagy gene deletion, including Fip200, inhibited growth of breast, lung, and pancreatic cancers (Guo et al., 2011; Wei et al., 2011; Yang et al., 2011). Deletion of beclin1 led to early and Fip200 to late embryonic lethality (Yue et al., 2003; Gan et al., 2006), but Atg5- and Atg7-null mice survive until shortly after birth (Kuma et al., 2004; Komatsu et al., 2005). These effects may be caused by gene functions beyond canonical autophagy. Atg7 was recently shown to regulate cell-cycle and cell-death pathways independently of its autophagy-essential enzymatic activity (Lee et al., 2012). Beclin1 and Fip200 were shown to interact with other proteins to regulate cell functions independently of or in addition to autophagy (Gan and Guan, 2008; Mizushima and Levine, 2010; Liu et al., 2011; Boya et al., 2013). Thus, gaining insights into the mechanisms of NSC regulation by autophagy necessitates determining if autophagy genes other than Fip200 regulate NSCs.
The multifunctional adaptor protein p62/sequestome1 was the first selective autophagy receptor described (Bjørkøy et al., 2005; Pankiv et al., 2007). Knockout of Fip200 or other autophagy genes leads to abnormal p62 aggregate accumulation in several cell types including Fip200-null NSCs (Bjørkøy et al., 2005; Komatsu et al., 2007a; Liang et al., 2010; Mizushima and Komatsu, 2011). Previous studies suggested that p62 ablation could rescue liver injury, but not neuronal degeneration, induced by Atg7 conditional knockout (cKO; Komatsu et al., 2007a). The role and significance of p62 aggregates in Fip200-null NSCs defects remains to be characterized.
Prior studies showed that depletion of NSCs by autophagy inhibition was associated with aberrantly increased ROS. Aberrant ROS increases were also shown in Fip200-null NSCs (Wang et al., 2013). A major source of endogenous ROS is superoxide (O2•−) from mitochondrial respiration (Murphy, 2009). Superoxide dismutase (SOD) enzymes convert O2•− to hydrogen peroxide (H2O2), which is then converted to H2O by glutathione peroxidase (Rhee et al., 2005). This antioxidant defense system maintains cellular redox balance (Matés, 2000; Apel and Hirt, 2004). Cytoplasmic SOD1 and mitochondrial SOD2 eliminate intracellular O2•− (Zelko et al., 2002). Mitochondria accumulation and/or damage or SOD deficiency can lead to increased O2•−, causing oxidative stress and cell death (Dröge, 2002; Lombard et al., 2005; Naka et al., 2008).
Here, we show that deletion of Atg5, Atg16L1, and Atg7 in mice inhibited autophagy and led to mitochondria accumulation in postnatal NSCs. However, unlike Fip200 deletion, deletion of the other autophagy genes did not affect NSC maintenance and differentiation. Further, p62 aggregates accumulated in Fip200-null NSCs, but not in NSCs depleted of other autophagy genes. p62 deletion rescued Fip200-null NSCs. Reduced cytoplasmic levels of SOD1, but not SOD2 or increased mitochondria mass, correlated with increased levels of O2•− in Fip200-null NSCs. SOD mimetics also rescued the defective phenotype. These results show that p62 determines the outcome of autophagy inhibition by impeding O2•− elimination by SOD1 in postnatal NSCs.
Results
Atg5 and Atg16L1 deletion cause autophagy deficiency and mitochondria accumulation in postnatal NSCs
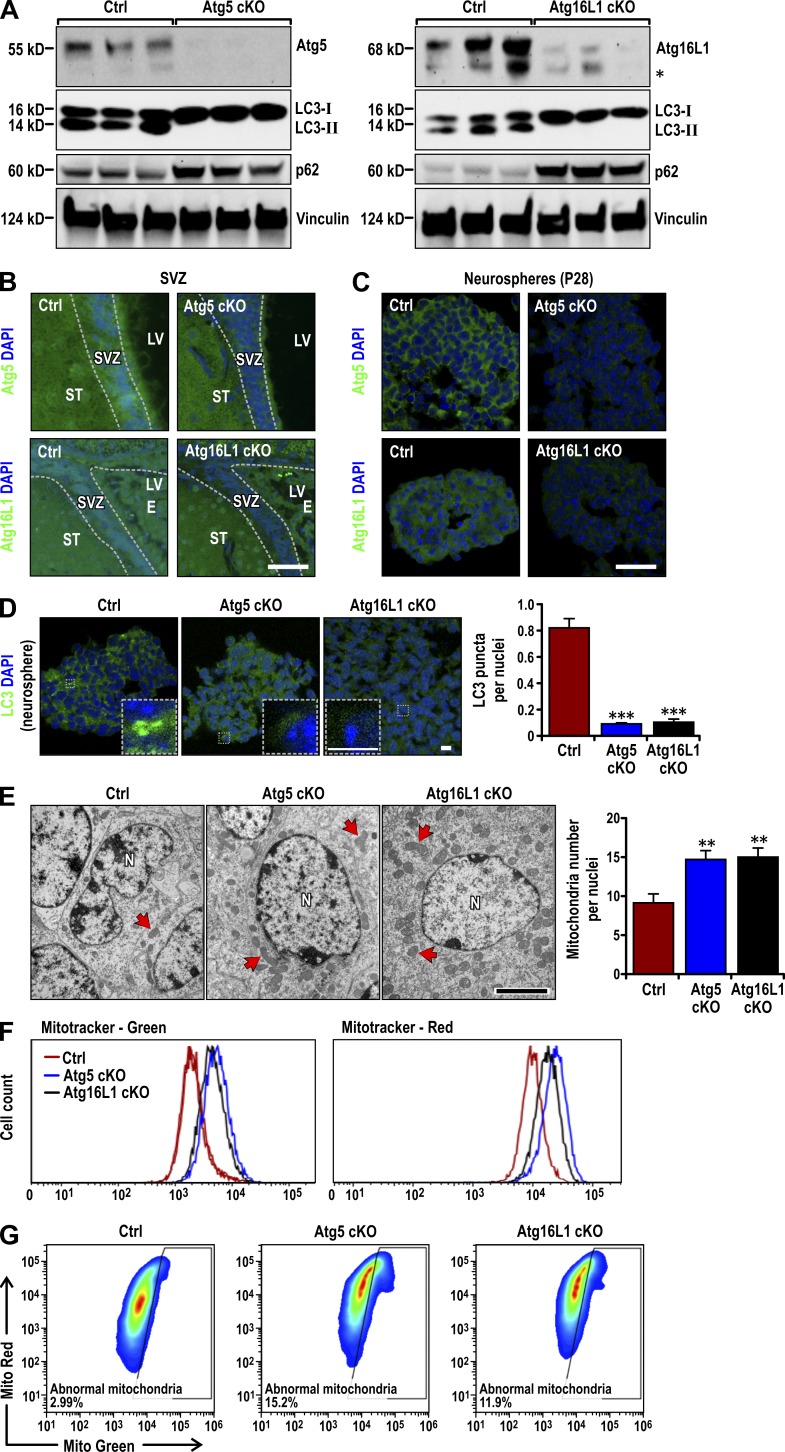
To explore if autophagy genes other than Fip200 regulate NSCs, we conditionally deleted the essential autophagy genes Atg5 and Atg16L1 using hGFAP-Cre transgenic mice expressing Cre in postnatal NSCs, as described previously (Zhu et al., 2005; Wang et al., 2013). We found that Atg5f/f;hGFAP-Cre (designated Atg5GFAP cKO) and Atg16L1f/f;hGFAP-Cre (designated Atg16L1GFAP cKO) mice were born at the expected Mendelian ratio and were indistinguishable from their control littermates (Atg5f/f, Atg5f/+;hGFAP-Cre, Atg16L1f/f, and Atg16L1f/+;hGFAP-Cre mice, all designated Ctrl mice). Subventricular zone (SVZ) tissues were microdissected from Ctrl and mutant mice at postnatal day 14 (P14). Mice were pretreated with chloroquine for 7 d to inhibit LC3-II degradation, as described previously (Wang et al., 2013). Immunoblots of lysates from the SVZs showed significantly decreased levels of Atg5 or Atg16L1 in cKO mice (Fig. 1 A). Immunofluorescent staining at P28 also showed reduced expression of Atg5 and Atg16L1 in cKO SVZ compared with controls (Fig. 1 B). Atg5 and Atg16L1 were also reduced in neurospheres derived from SVZ cells of P28 cKO mice (Fig. 1 C). Atg5 and Atg16L1 antibody specificity was verified by loss of staining in internal granular layer cells (targeted by hGFAP-Cre), but not Purkinje cells (not targeted by hGFAP-Cre), in the cerebellum of cKO mice (Fig. S1 A). These results showed that floxed Atg5 and Atg16L1 were efficiently deleted in NSCs in the SVZ of cKO mice.
Figure 1.
Atg5 and Atg16L1 deletion causes deficient autophagy and mitochondria accumulation in postnatal NSCs. (A) Lysates were extracted from the SVZ of P14 Ctrl, Atg5GFAP cKO, and Atg16L1GFAP cKO mice treated with chloroquine for 7 d and analyzed by immunoblot to detect Atg5 (left), Atg16L1 (right), LC3, p62, and Vinculin. The asterisk on the right marks a band detected by anti-Atg16L1 antibody either nonspecifically or as a Atg16L1 degradation product in some samples. (B and C) Atg5, Atg16L1, and DAPI immunofluorescence in the SVZ (B; n = 3 mice each) and neurospheres (C) from Ctrl, Atg5GFAP cKO, and Atg16L1GFAP cKO SVZ cells at P28. (D) LC3 and DAPI immunofluorescence in neurospheres from Ctrl, Atg5GFAP cKO, and Atg16L1GFAP cKO SVZ cells at P28. Boxed areas show more detail for LC3 puncta. LC3 puncta number per nuclei (mean ± SEM) are graphed on the right (n = 3 mice each, >300 cells from three mice). (E) TEM of mitochondria (examples marked by arrows) in Ctrl, Atg5GFAP cKO and Atg16L1GFAP cKO SVZ tissues at P28. Mitochondria number per nuclei (mean ± SEM; n = 2 mice each, >30 cells from each mouse). (F) Total (MitoTracker Green) and functional (MitoTracker Red) mitochondria by FACS analysis of dissociated neurospheres cells from Ctrl, Atg5GFAP cKO, and Atg16L1GFAP cKO SVZ at P28 (n = 3 mice each). (G) Dysfunctional mitochondria were evaluated by the percentage of cells with decreased ratios of MitoTracker Red to MitoTracker Green (n = 2 mice each). E, ependymal layer; LV, lateral ventricle; N, nuclei; ST, striatum. Bars: (B and C) 30 µm; (D and insets) 6 µm; (E) 1 µm. **, P < 0.01; ***, P < 0.001.
Consistent with their essential roles in LC3 lipidation and autophagosome formation (He and Klionsky, 2009; Mizushima and Komatsu, 2011), Atg5- and Atg16L1-deficient SVZ cells showed reduced LC3-I to LC3-II conversion and increased p62 accumulation compared with controls (Fig. 1 A). LC3 puncta number was also decreased in neurospheres from the mutant mice (Fig. 1 D). LC3 puncta number in neurospheres was low compared with other cells because of a low cytoplasmic-to-nuclear volume in NSCs in 3D culture. As autophagy is required for mitochondria clearance in various cells, including NSCs (Wang et al., 2013), we examined mitochondria in SVZ cells of cKO mice. Similar to Fip200hGFAP cKO mice, the number of mitochondria per nuclei at P28 by transmission electron microscopy (TEM) increased in cKO mice (15 ± 1 for both) compared with Ctrl mice (9 ± 1; Fig. 1 E). Dissociated neurosphere cell labeling with MitoTracker Green or MitoTracker Red followed by FACS showed that both total and functional mitochondria were increased in cells from Atg5GFAP and Atg16L1GFAP cKO mice (Fig. 1 F). We gated for cells with increased dysfunctional mitochondria (those with a low ratio of functional to total mitochondria) as described previously (Misawa et al., 2013; Wen et al., 2015). SVZ cells in both cKO mice showed increased dysfunctional mitochondria compared with controls (Fig. 1 G). These results demonstrated that, like Fip200 deletion, Atg5 or Atg16L1 deletion leads to defective autophagy and increased functional and dysfunctional mitochondria mass in postnatal NSCs.
Atg5, Atg16L1, or Atg7 loss does not affect NSC maintenance or differentiation
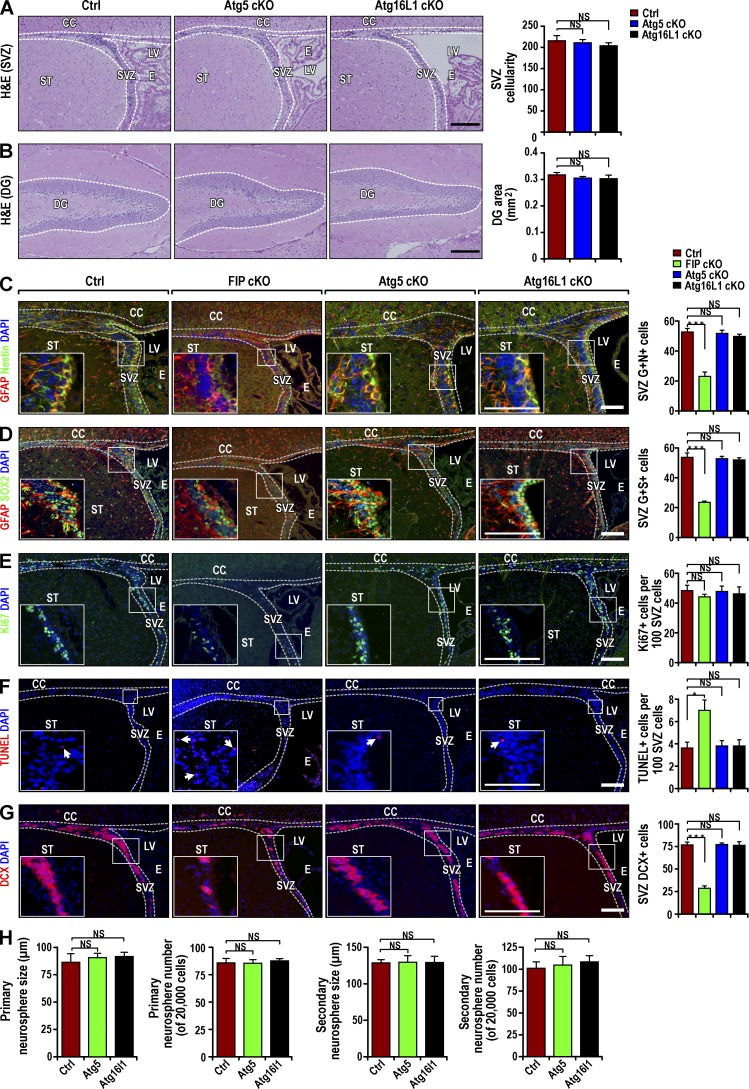
To determine if impaired autophagy affects postnatal NSCs, we examined the SVZ and dentate gyrus (DG) in Atg5GFAP cKO and Atg16L1GFAP cKO mice at P28. Surprisingly, histological analysis showed apparently normal SVZ and DG width and cellularity in the mutant mice (Fig. 2, A and B). This was in sharp contrast to the significant degeneration of these areas in Fip200GFAP cKO mice (Wang et al., 2013). Analysis with several lineage markers (GFAP+Nestin+, GFAP+SOX2+) showed both cKO mice contained similar numbers of NSCs as controls in the SVZ (Fig. 2, C and D) and DG (Fig. S1, B and C). Consistent with the normal NSC morphology and number, we did not observe changes in cell proliferation or apoptosis in the SVZ (Fig. 2, E and F) or DG (Fig. S1, D and E) of either cKO mice relative to Ctrls. Unlike Fip200GFAP cKO mice (Wang et al., 2013), GFAP+Nestin− or GFAP+SOX2− cells were not increased in the SVZ (Fig. 2, C and D) or DG (Fig. S1, B and C) of either cKO mice. Comparable levels of neuroblasts and mature neurons were detected in the SVZ (Fig. 2 G), rostral migratory stream (RMS), DG (Fig. S1, F and G), and olfactory bulb (OB; not depicted) of cKO and control mice. In vitro neurosphere formation also showed comparable numbers and size of primary and secondary neurospheres from cKO and control mice at P28 (Fig. 2 H).
Figure 2.
Postnatal NSC pool and neurogenesis are intact in Atg5GFAP cKO and Atg16L1GFAP cKO mice. (A and B) H&E staining of SVZ (A) and DG (B) from Ctrl, Atg5GFAP cKO, and Atg16L1GFAP cKO mice at P28. Dotted lines indicate the SVZ and DG boundaries. SVZ cellularity (mean ± SEM; A) and DG area (B) per section from different mice (n = 4 mice each). (C and D) Immunofluorescence for GFAP, Nestin (C), SOX2 (D), and DAPI in Ctrl, Fip200GFAP cKO, Atg5GFAP cKO, and Atg16L1GFAP cKO SVZ at P28. Mean ± SEM of GFAP+ and Nestin+ (C) and GFAP+ and Sox2+ (D) cell number per SVZ section (n = 3 mice each). (E and F) Immunofluorescence for Ki67 and DAPI (E) and TUNEL and DAPI (F) in Ctrl, Fip200GFAP cKO, Atg5GFAP cKO, and Atg16L1GFAP cKO SVZ and RMS at P28. Arrows mark examples of TUNEL+ cells in insets. Mean ± SEM of the Ki67+ (E) and TUNEL+ (F) cell number per SVZ section (n = 3 mice each). (G) Immunofluorescence for DCX and DAPI in Ctrl, Fip200GFAP cKO, Atg5GFAP cKO, and Atg16L1GFAP cKO SVZ and RMS at P28. Mean ± SEM of DCX+ cell number per SVZ section (n = 3 mice each). (H) Mean ± SEM of the number and size of primary and secondary neurospheres from Ctrl, Atg5GFAP cKO and Atg16L1GFAP cKO mice at P28 (n = 3 mice each). CC, corpus callosum; E, ependymal layer; LV, lateral ventricle; ST, striatum. Bars: (A and B) 100 µm; (C–G and insets) 40 µm. *, P < 0.05; ***, P < 0.001; NS, not significant.
As Fip200GFAP cKO mice showed gradual depletion of NSCs at P28 but apparently normal NSCs at P0, we wondered if prolonged blockage of autophagy in Atg5GFAP cKO or Atg16L1GFAP cKO mice could lead to NSC defects in older mice. To test this possibility, we examined Atg16L1GFAP cKO and control mice at P90. We did not find obvious differences in histology, the number of NSCs, neuroblasts, or mature neurons between Atg16L1GFAP cKO and control mice at P90 (Fig. S1, H–L). These results suggest that although Atg5, Atg16L1, and Fip200 are all required for autophagy, deletion of Atg5 or Atg16L1, unlike deletion of Fip200, does not affect NSC self-renewal or differentiation.
The differential role of Fip200 versus Atg5 and Atg16L1 appears to be specific to NSCs, given the previous findings of a similar cerebellar ataxia phenotype in nestin-Cre Atg5 and Fip200 cKO mice (Hara et al., 2006; Liang et al., 2010). Consistent with previous studies, we saw elevated apoptosis in the cerebellar granule neurons of Atg5GFAP cKO, Atg16L1GFAP cKO, and Fip200GFAP cKO mice (Fig. S1 M), indicating a requirement for all these autophagy genes to maintain neuron survival. We also analyzed the inducible deletion of Atg7 in NSCs in adult mice using Ubc-CreERT2/+;Atg7f/f mice (Karsli-Uzunbas et al., 2014). At 5 wk after Atg7 deletion induced by tamoxifen, the mice showed a cerebellar ataxia phenotype similar to nestin-Cre–induced neural-specific Atg7, Atg5, or Fip200 cKO mice reported previously (Hara et al., 2006; Komatsu et al., 2006; Liang et al., 2010; Karsli-Uzunbas et al., 2014). However, the mutant mice with Atg7 deletion did not show defects in SVZ, changes in NSC number, or neuronal progenies (Fig. S1, H–L). Our results suggest a distinct role of Fip200, but not Atg5, Atg16L1, or Atg7, in postnatal NSC maintenance and differentiation, despite the fact that all four genes are required for neuronal functions in the brain.
Lack of accumulation and aggregation of p62 in NSCs of Atg5GFAP cKO and Atg16L1GFAP cKO mice
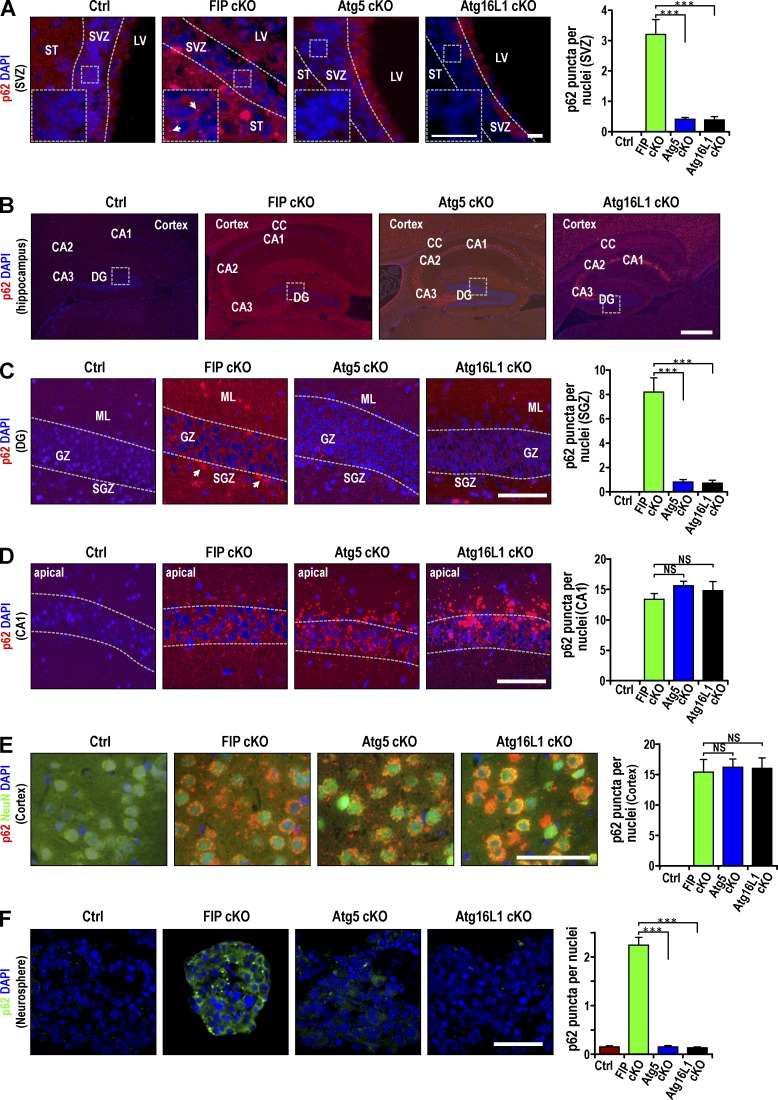
Abnormal p62 aggregate accumulation follows autophagy inhibition in different cell types (Komatsu et al., 2007a; Mathew et al., 2009; Wei et al., 2011). To study the mechanisms of the different effects of autophagy gene deletion in NSCs, we examined p62 aggregates in NSCs and other brain regions of our cKO mice to understand the different effects of autophagy gene deletion in NSCs. Many p62 aggregates were found in Fip200GFAP cKO mice (Fig. 3 A), as described previously (Wang et al., 2013). Surprisingly, few p62 aggregates were detected in the SVZ of Atg5GFAP cKO or Atg16L1GFAP cKO mice, although Atg5 and Atg16L1 are required for autophagy. Ubiquitin-positive protein aggregates were only detected in Fip200-deficient, but not Atg5- or Atg16L1-deficient, SVZ cells (Fig. S2 A). p62 staining was present throughout the hippocampus in Atg5GFAP cKO, Atg16L1GFAP cKO, and Fip200GFAP cKO mice (Fig. 3 B). In the DG, fewer p62 aggregates were found in Atg5GFAP cKO and Atg16L1GFAP cKO mice in both the granular zone (GZ) and subgranular zone (SGZ) compared with Fip200GFAP cKO mice (Fig. 3 C). However, p62 aggregates were detected for all three mutant mice in cornu ammonis regions 1–3 of the hippocampus, with larger p62 aggregates in the pyramidal neurons in these region of Atg5GFAP cKO and Atg16L1GFAP cKO mice than in Fip200GFAP cKO mice (Fig. 3 D). p62 aggregation number and size were comparable among Fip200GFAP cKO, Atg5GFAP cKO, and Atg16L1GFAP cKO mice in other neuron-enriched regions such as cerebral cortex (Fig. 3 E) and IGL of the cerebellum (Fig. S2 B). Double staining for p62 and NeuN verified that the majority of p62 aggregates were localized in NeuN+ neurons in the cerebral cortex (Fig. 3 E). These results suggest that unlike brain regions that are enriched with mature neurons, the SVZ and DG, where NSCs reside, contain significantly more p62 aggregates in Fip200GFAP cKO mice than in Atg5GFAP cKO or Atg16L1GFAP cKO mice.
Figure 3.
Differential p62 aggregation in postnatal NSCs of Fip200GFAP cKO mice, but not Atg5GFAP cKO and Atg16L1GFAP cKO mice. Immunofluorescence of p62 and DAPI in brain regions of Ctrl, Fip200GFAP cKO, Atg5GFAP cKO, and Atg16L1GFAP cKO mice at P28 (A–E) and in neurospheres from SVZ cells of these mice (F). E includes staining for NeuN. Dotted lines indicate the boundaries of the SVZ (A), GZ (C), and CA1 (D). Boxed areas in A show more detail, and p62+ aggregates are marked by arrows. Boxed areas in B show more detail in C. p62 aggregate number per cell (mean ± SEM; n = 3 mice each, >150 cells per mouse counted for graphs on the right). CA1-CA3, cornu ammonis regions 1–3; LV, lateral ventricle; ML, molecular layer; ST, striatum. Bars: (A and insets) 15 µm, (B) 100 µm; (C–F) 50 µm. ***, P < 0.001; NS, not significant.
We then analyzed neurospheres derived from SVZ cells of mutant mice. Consistent with our observations in vivo, we detected increased p62 aggregates only in neurospheres derived from SVZ cells of Fip200GFAP cKO mice, but not from Atg5GFAP cKO or Atg16L1GFAP cKO mice (Fig. 3 F). Similar levels of aggregate accumulation were found for two other autophagy adaptors, NBR1 (Kirkin et al., 2009) and TAX1BP1 (Newman et al., 2012), in neurospheres derived from all three mutant mice (Fig. S2, C and D). We also found p62 and Nbr1 aggregates colocalized in Fip200 cKO neurospheres (Fig. S2 E). These results suggest that p62 accumulation may contribute to the defective NSC phenotypes in Fip200GFAP cKO mice.
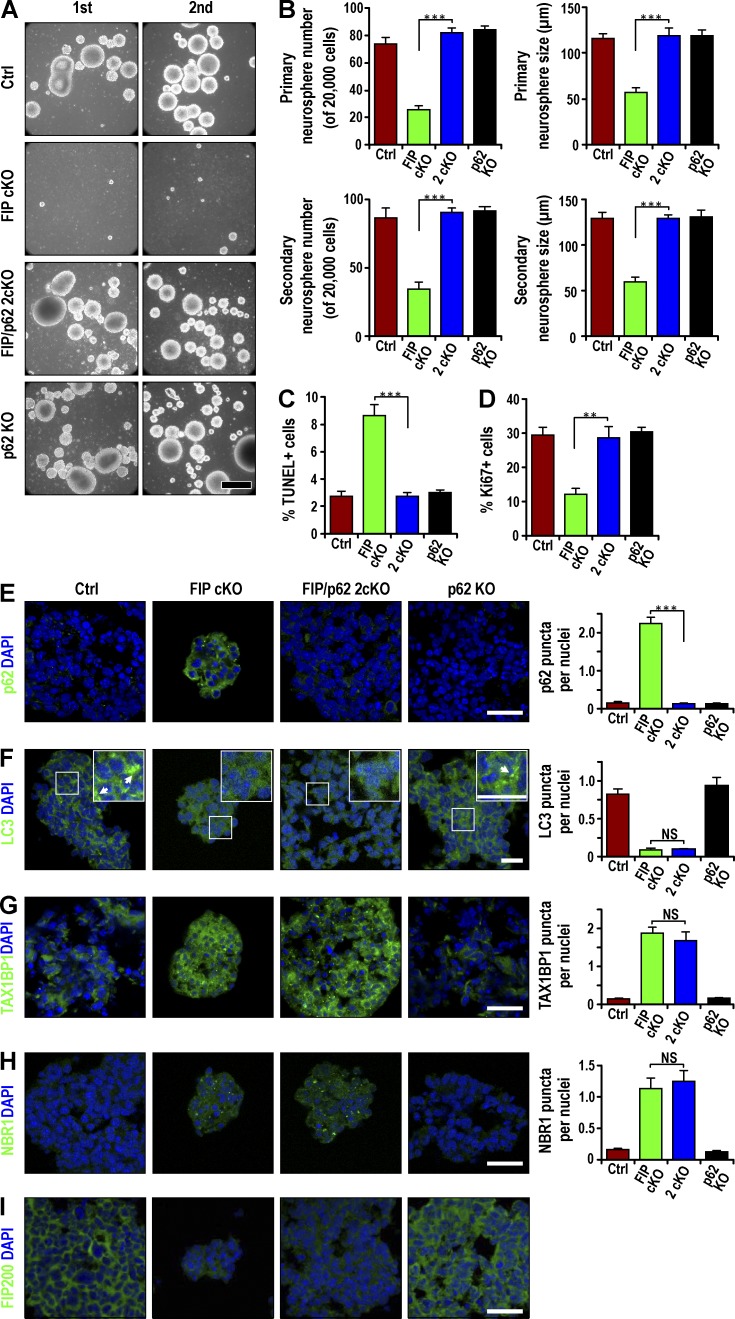
p62 deletion rescues NSC defects in Fip200GFAP cKO mice
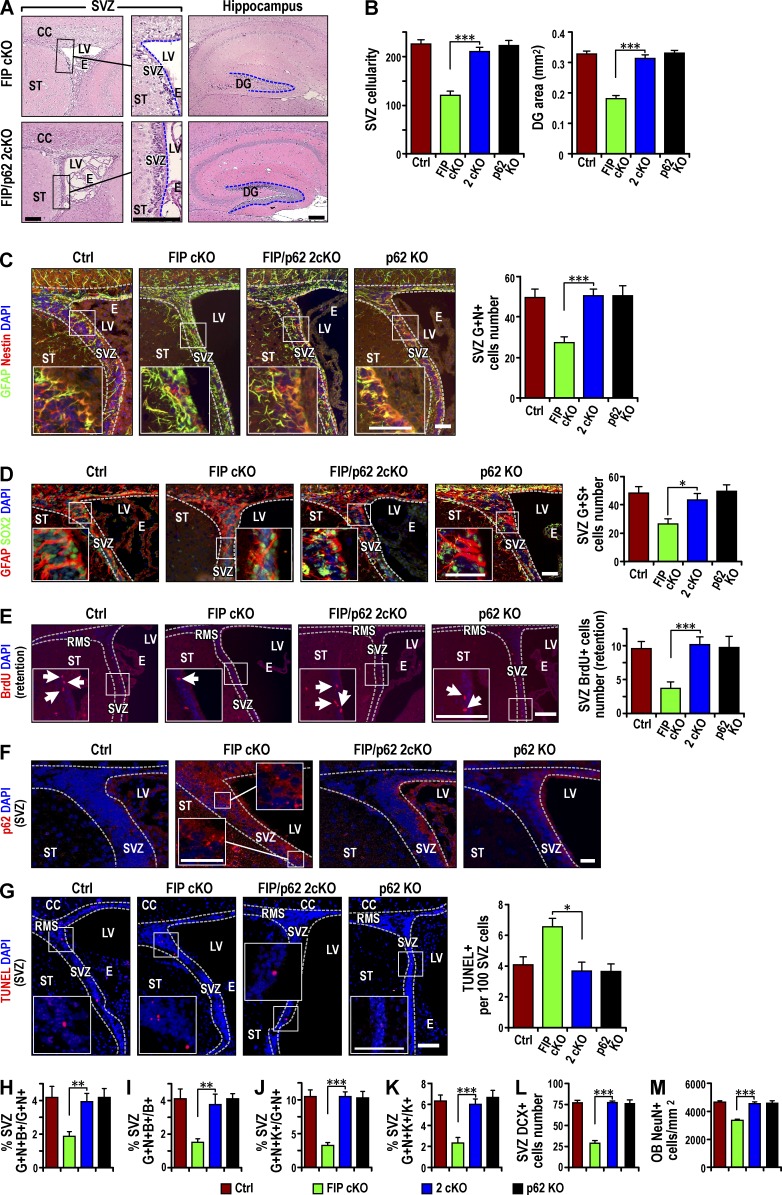
To directly assess the role of p62 aggregates in NSC defects of Fip200GFAP cKO mice, we generated double conditional KO mice with deletion of Fip200 and p62 (Fip200f/f;p62−/−;hGFAP-Cre; designated 2cKO) by crossing Fip200GFAP cKO and p62−/− mice (designated p62 KO; Komatsu et al., 2007a). Histological analysis showed that p62 KO mice had normal SVZ and DG regions compared with control mice. The low SVZ cellularity and DG area of Fip200GFAP cKO mice was rescued in 2cKO mice (Fig. 4, A and B). The decreased pool of NSCs in the SVZ (Fig. 4, C and D) and DG (Fig. S3, A and B) of Fip200GFAP cKO mice was restored in 2cKO mice to levels comparable to Ctrl mice (Fig. 4, C and D; and Fig. S3, A and B). The NSC increase was confirmed by BrdU retention in the SVZ (Fig. 4 E) and DG (Fig. S3 C) of 2cKO mice. p62 immunostaining verified that elevated p62 levels in the SVZ (Fig. 4 F) and DG (Fig. S3 D) of Fip200GFAP cKO mice was abolished in the 2cKO mice.
Figure 4.
p62 KO rescues the maintenance and differentiation defects of Fip200-null NSCs in vivo. (A) H&E staining of SVZ and hippocampus from FIPGFAP cKO and 2cKO mice at P28. Dotted lines indicate boundaries of SVZ and DG (n = 4 mice each). (B) Mean ± SEM of SVZ cellularity (left) and DG area (right) per section from Ctrl, FIPGFAP cKO, 2cKO, and p62 KO mice at P28. (C and D) Immunofluorescence for GFAP, Nestin (C), SOX2 (D), and DAPI in SVZ from Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO mice at P28. Mean ± SEM of GFAP+ and Nestin+ (C) and GFAP+ and Sox2+ (D) cell number per SVZ section (n = 4 mice each). (E) Immunofluorescence of long-term retained BrdU and DAPI in Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO SVZ at P28. Mean ± SEM of BrdU+ cell number per SVZ section (n = 4 mice each). (F and G) Immunofluorescence for p62 and DAPI (F; n = 3 mice each) and TUNEL and DAPI (G) in SVZ of Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO mice at P28. Mean ± SEM of TUNEL+ cells per 100 SVZ cells (G, n = 4 mice each, >1,000 cells per mouse). (H and I) Immunofluorescence of Nestin, GFAP, and short-term labeled BrdU in Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO SVZ at P28. Mean ± SEM of the percentage of GFAP+Nestin+BrdU+ to total GFAP+Nestin+ cells (H) or total BrdU+ cells (I) per section (n = 5 mice each). (J and K) Immunofluorescence for Nestin, GFAP, and Ki67 in Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO SVZ at P28. Mean ± SEM of the percentage of GFAP+Nestin+Ki67+ to total GFAP+Nestin+ cells (J) or total Ki67+ cells (K) per section (n = 5 mice each). (L and M) Immunofluorescence for DCX in SVZ (L) and NeuN in olfactory bulb (M) in Ctrl, Fip200GFAP cKO, 2cKO and p62 KO mice at P28. Mean ± SEM of DCX+ cells per section (L) and NeuN+ cells per square millimeter (M; n = 4 mice each). Dotted lines indicate the boundaries of SVZ (C–G). CC, corpus callosum; E, ependymal layer; LV, lateral ventricle; OB, olfactory bulb; RMS, rostral migratory stream; ST, striatum. Bars: (A, E, and G and insets) 50 µm; (C, D, and insets) 25 µm; (F and insets) 15 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Our previous studies showed that NSC gradual depletion was caused by both increased apoptosis and reduced proliferation in Fip200GFAP cKO mice (Wang et al., 2013). Increased apoptotic cells in the SVZ (Fig. 4 G) and DG (Fig. S3 E) of Fip200GFAP cKO mice was restored to levels comparable to controls in 2cKO mice. The reduced proliferation of NSCs was also rescued in the SVZ (Fig. 4, H and I) and DG (Fig. S3, F and G) in 2cKO mice. Similar results were obtained when proliferation was measured by Ki67 labeling in both the SVZ (Fig. 4, J and K) and DG (Fig. S3, H and I). In addition to the gradual loss of NSCs, Fip200GFAP cKO mice exhibited defective neurogenesis. This phenotype was not rescued by p53 ablation that restored NSC number by rescue of the aberrant proliferation and apoptosis phenotypes (Wang et al., 2013). It was rescued by p62 deletion of defective neurogenesis, measured by both the neuroblast number in the SVZ (Fig. 4 L) and DG (Fig. S3 J) and the number of mature neurons in the OB (Fig. 4 M) and DG (Fig. S3 K). These results suggest that accumulated p62 aggregate removal by p62 ablation rescued defective NSC maintenance and neurogenesis in Fip200GFAP cKO mice.
We next examined SVZ cells from mutant mice for neurosphere formation in vitro. Consistent with previous findings (Wang et al., 2013), SVZ cells from P28 Fip200GFAP cKO mice exhibited significantly decreased neurosphere formation in both primary and secondary cultures (Fig. 5, A and B). p62 deletion alone did not affect SVZ cell neurosphere formation; the number and size of neurospheres formed by SVZ cells of 2cKO mice were comparable to control mice, supporting the idea that p62 deletion rescued the Fip200-null NSCs self-renewal defect. Increased apoptosis in neurospheres from Fip200GFAP cKO mice was abolished in neurospheres from 2cKO mice (Fig. 5 C). The Fip200-null NSC reduced proliferation was also restored in 2cKO mice, as measured by Ki67 staining (Fig. 5 D) or BrdU incorporation assay (unpublished data). The accumulated p62 aggregates in Fip200-null NSCs were eliminated in neurospheres derived from 2cKO mice (Fig. 5 E). However, defective autophagy in these cells was not affected. LC3 puncta were found in neurospheres from both Fip200GFAP cKO and 2cKO mice at lower levels compared with Ctrl or p62 KO mice (Fig. 5 F). The increased accumulation of autophagy adaptors TAX1BP1 and NBR1 was not reversed in neurospheres from 2cKO mice relative to Fip200GFAP cKO mice (Fig. 5, G and H). Deletion of p62 alone or in Fip200-null NSCs did not affect expression of Fip200 (Fig. 5 I). These results support the idea that accumulated p62 aggregates (but not other adaptors TAX1BP1 or NBR1) are responsible for the defective NSC phenotypes in Fip200GFAP cKO mice. These results also suggest that increased p62 aggregates act downstream of Fip200 loss–induced autophagy deficiency to cause the defective NSC phenotypes.
Figure 5.
Defective self-renewal of Fip200-deficient NSCs in vitro restored by p62 KO. (A) Primary (first) and secondary (second) neurosphere formation from Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO mice at P28. Representative phase contrast images. (B) Mean ± SEM of the number and size of primary and secondary neurospheres (n = 3 mice each). (C and D) Mean ± SEM of the percentage of TUNEL+ (C) and Ki67+ (D) cells in neurospheres from Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO mice at P28 by immunofluorescence (n = 3 mice each, >500 cells per mouse). (E–H) Immunofluorescence of p62 (E), LC3 (F), TAX1BP1 (G), NBR1 (H), Fip200 (I), and DAPI in neurospheres from Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO mice at P28. Mean ± SEM of p62 aggregates (E), LC3 puncta (F), TAX1BP1 aggregates (G), and NBR1 aggregates (H) per neurosphere cell. Boxed areas in F show more detail for LC3 puncta, marked by arrows in insets (n = 3 mice each, >200 cells from Ctrl, 2cKO, and p62 KO mice; 87 cells for Fip200GFAP cKO mouse). Bars: (A) 100 µm; (E–I and F and insets) 20 µm. **, P < 0.01; ***, P < 0.001; NS, not significant.
Increased O2•−, but not abnormal mitochondria accumulation alone, leads to defective NSCs in Fip200GFAP cKO mice
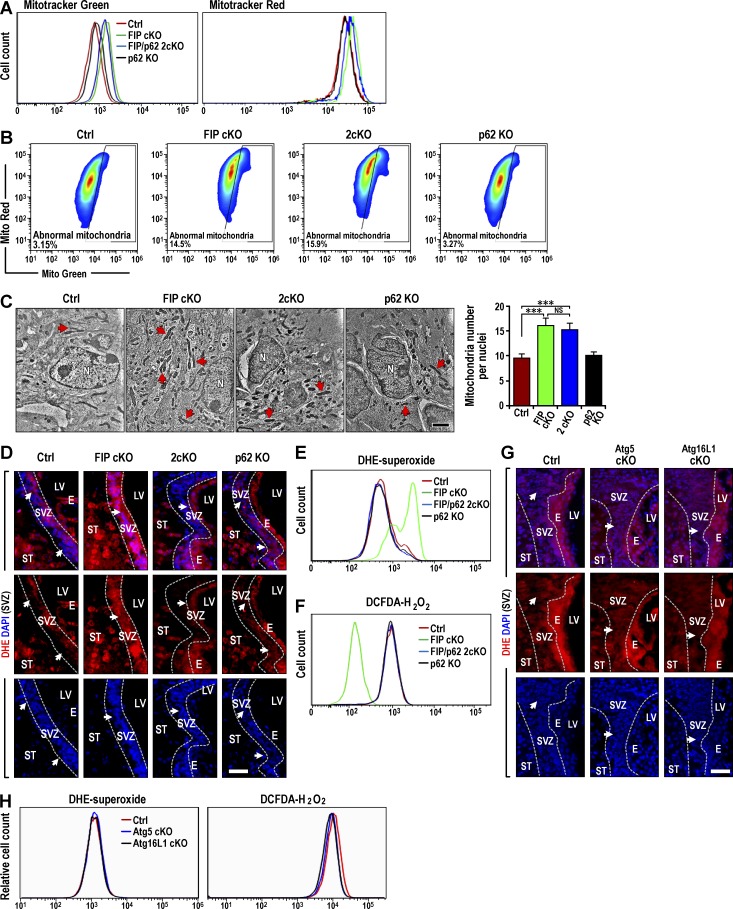
In addition to accumulated p62 aggregates, increased mitochondria mass observed in Fip200GFAP cKO mice could contribute to the aberrant ROS increase responsible for defective NSC maintenance and differentiation in these mice (Wang et al., 2013). We examined mitochondria content in neurospheres derived from mutant mice SVZ cells. Consistent with previous observations by TEM (Wang et al., 2013), we found elevated total and functional mitochondria in Fip200GFAP cKO SVZ cells compared with control mice using MitoTracker Green and MitoTracker Red, respectively (Fig. 6 A). Total and functional mitochondria in 2cKO SVZ cells of were also elevated to a high level, like Fip200GFAP cKO SVZ cells. p62 deletion alone did not affect mitochondria relative to Ctrl NSCs. Fip200 cKO and 2cKO SVZ cells also showed similar increased dysfunctional mitochondria compared with Ctrl and p62 KO cells (Fig. 6 B). We then directly examined mitochondria in SVZ tissues by TEM. Similar to our findings in vitro, p62 deletion did not reverse the increased number of mitochondria caused by Fip200 deletion. Both 2cKO and Fip200GFAP cKO mice displayed comparable increases in mitochondria compared with p62 KO and Ctrl mice (Fig. 6 C). These results suggest that p62 accumulation of aggregates and increase in mitochondria mass were independent events downstream of Fip200 deletion. The results also indicate that abnormal mitochondria accumulation alone in 2cKO, Atg5GFAP cKO, and Atg16L1GFAP cKO mice was insufficient to cause NSC defects (Fig. 1, E–G).
Figure 6.
p62 aggregates increase O2•− accumulation in Fip200GFAP cKO NSCs. (A) FACS analysis of total (MitoTracker Green) and functional (MitoTracker Red) mitochondria in dissociated cells from neurospheres from Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO SVZ of mice at P28. (B) Dysfunctional mitochondria were evaluated by the percentage of cells with decreased ratios of MitoTracker Red to MitoTracker Green (n = 2 mice each). (C) TEM of mitochondria from Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO in SVZ at P28. Arrows indicate mitochondria. Mean ± SEM of the mitochondria number per nuclei (n = 2 mice each, >30 cells per mouse). (D) Fluorescence of DHE and DAPI in Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO SVZ at P28. Arrows indicate SVZ cells (n = 3 mice each). (E and F) FACS of DHE (E) and DCFDA (F) in dissociated neurosphere cells of Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO mice at P28. (G) Fluorescence of DHE and DAPI in Ctrl, Atg5GFAP cKO, and Atg16L1GFAP cKO SVZ at P28. Arrows indicate SVZ cells (n = 3 mice each). (H) FACS of DHE (left) and DCFDA (right) in dissociated neurosphere cells of Ctrl, Atg5GFAP cKO, and Atg16L1GFAP cKO mice at P28. E, ependymal layer; LV, lateral ventricle; N, nucleus; ST, striatum. Bars: (C) 1 µm; (D and G) 50 µm. ***, P < 0.001; NS: not significant.
The lack of rescue of the increased mitochondria mass was surprising, as this was proposed as the cause of increased ROS and defective Fip200GFAP cKO NSCs (Wang et al., 2013). We directly examined the mutant mice SVZ and DG for ROS levels by dihydroethidium (DHE) staining. We found that the ROS aberrant increase in Fip200GFAP cKO SVZ (Fig. 6 D) and DG (Fig. S3 L) was reversed in 2cKO mice, although the increased mitochondria mass was not reversed. These results were consistent with our previous studies demonstrating a critical contribution of increased ROS to the defective maintenance and differentiation of Fip200-null NSCs (Wang et al., 2013). However, the results also raised questions about the cause of increased ROS. We then analyzed dissociated cells from neurospheres for ROS by labeling with DHE (O2•−) and DCFDA (H2O2), followed by FACS analysis. Consistent with SVZ tissues, O2•− was increased in Fip200GFAP cKO neurosphere cells and rescued in 2cKO neurosphere cells to levels comparable to Ctrl and p62 KO neurospheres (Fig. 6 E). Contrary to our expectation, a significant decrease in H2O2 was seen in Fip200GFAP cKO neurosphere cells compared with control and p62 KO (Fig. 6 F). The decreased H2O2 content was recovered in 2cKO neurospheres cells. No differences in the levels of O2•− and H2O2 for Atg5 and Atg16L1 cKO mice compared with Ctrl mice was seen (Fig. 6, G and H). These results suggested that elevated O2•− was responsible for defective NSC maintenance and differentiation in Fip200GFAP cKO mice. They also suggested that aberrantly increased O2•− may be caused by a failure of O2•− conversion into H2O2.
Restored cytoplasmic activity of SOD1 eliminates elevated O2•− and rescues defective Fip200GFAP cKO NSCs
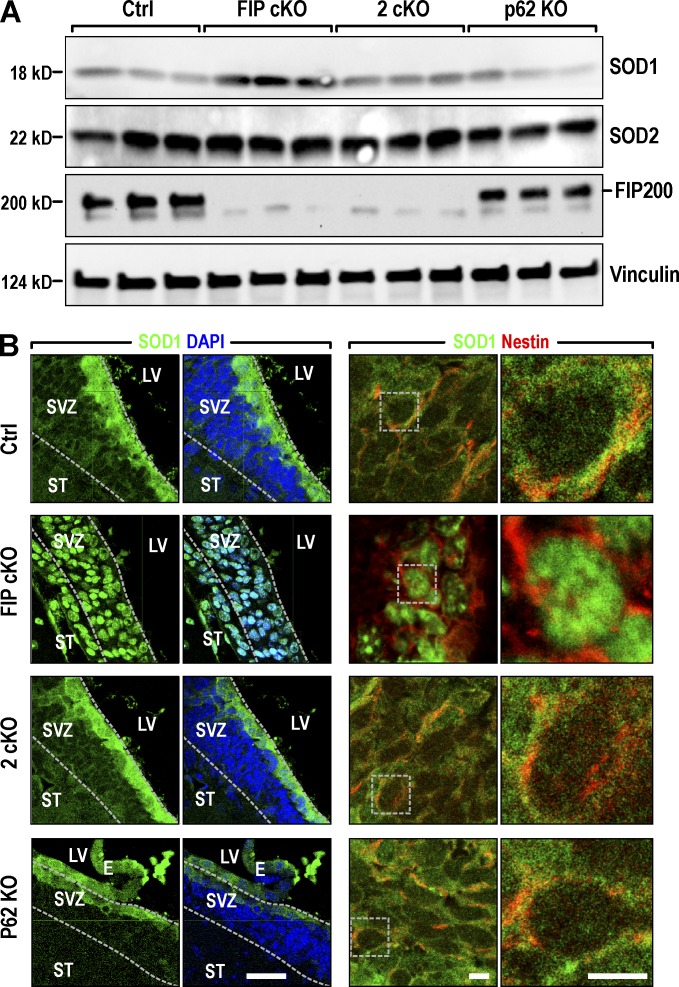
To explore the mechanisms of decreased conversion from O2•− to H2O2, we examined the expression of cytoplasmic SOD1 and nuclear SOD2 in mitochondria (Matés, 2000) in SVZ of mutant mice. Immunoblot analysis verified that Fip200 was completely deleted in Fip200GFAP cKO and 2cKO SVZ (Fig. 7 A). SOD1 expression was increased in Fip200GFAP cKO compared with Ctrls, slightly increased in 2cKO, and unchanged in p62 KO mice. No significant change in the expression of SOD2 was found in any mutant mice. Because NSCs are mainly localized in the boundaries of the SVZ with the lateral ventricle (LV) but the lysates in the analysis were from whole SVZ, we examined SOD expression in the SVZ directly by immunofluorescence. We found SOD1 highly expressed in cells lining the lateral wall of the SVZ in Ctrl mice, and mainly in cytoplasm (Fig. 7 B, top row). In Fip200GFAP cKO mice, SOD1 was detected both in this region and in the striatum of the SVZ (Fig. 7 B, row 2) and was almost exclusively localized in nuclei. Both the preferred expression of SOD1 in SVZ cells adjacent to the LV (the location of NSCs for neurospheres) and the cytoplasmic location of SOD1 were restored in 2cKO SVZ (Fig. 7 B, row 3). SOD1 in p62 KO showed similar distribution patterns as Ctrl mice (Fig. 7 B, row 4). No differences in the expression or subcellular localization of SOD2 were found in mutant mice relative to Ctrl mice (Fig. S3 M). These results suggest that Fip200 deletion and p62 aggregate accumulation kept cytoplasmic SOD1 in NSCs from translocation to the nucleus. This may be responsible for the lack of O2•− conversion to H2O2 and the defective phenotypes in Fip200GFAP cKO mice.
Figure 7.
p62 aggregates alter expression pattern of SOD1 localization in the cytoplasm of NSCs of Fip200GFAP cKO mice. (A) Lysates were extracted from SVZ of P28 Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO mice and analyzed by immunoblot. (B) Confocal images for Nestin, SOD1, and DAPI in SVZ from Ctrl, Fip200GFAP cKO, 2cKO, and p62 KO mice at P28. Boxed areas in the third column show detail for SOD1 and Nestin in the right column. Bars: (left two columns) 50 µm; (right two columns) 5 µm.
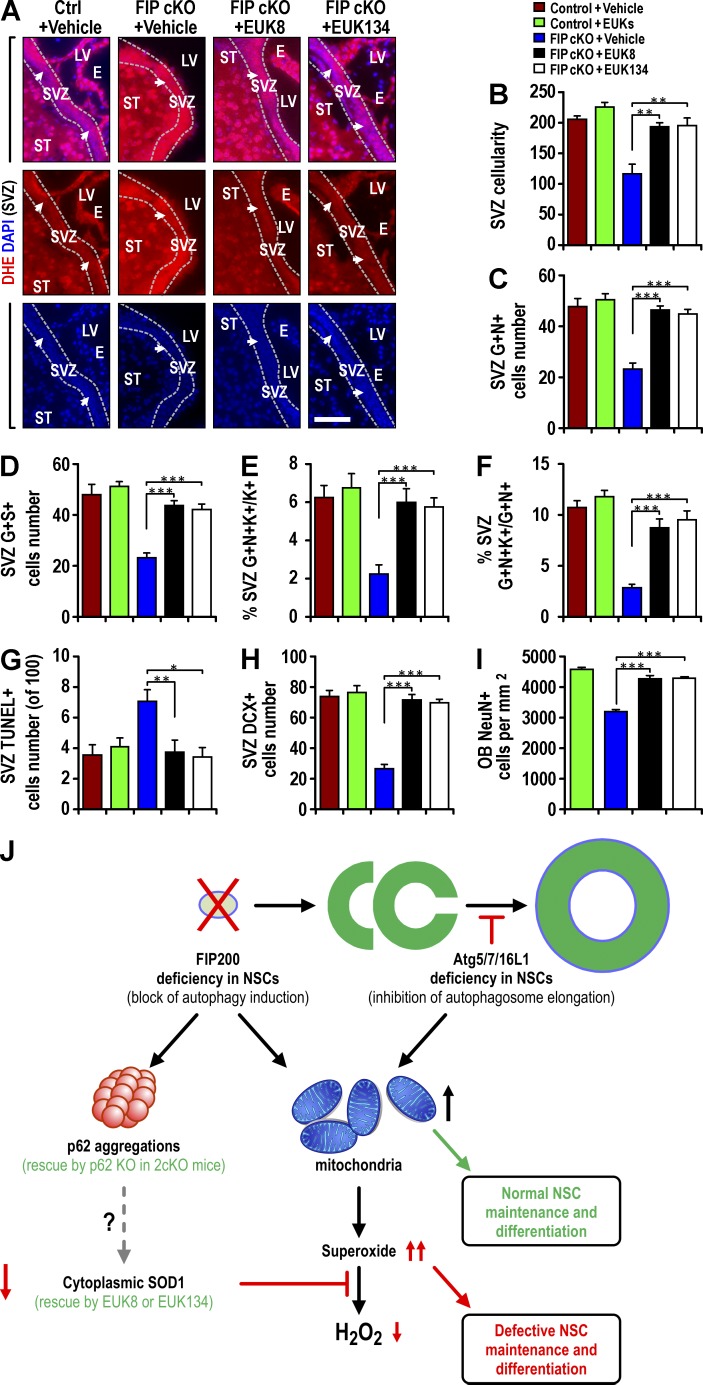
To test this hypothesis directly, we used two SOD mimetics, EUK-8 and EUK-134, which have been used extensively to rescue CNS phenotypes caused by increased O2•− (Jung et al., 2001; Melov et al., 2001). After daily treatment with EUK-8 or EUK-134 for 21 d, starting at P7, elevated O2•− in Fip200GFAP cKO SVZ (Fig. 8 A) and DG (Fig. S4 A) was restored to Ctrl levels. Treatment of Ctrl mice with the same doses of EUK-8 or EUK-134 did not affect O2•− levels in these regions (unpublished data). The elimination of aberrant O2•− with EUK-8 or EUK-134 returned decreased SVZ cellularity (Fig. 8 B) and DG area (Fig. S4 B) of Fip200GFAP cKO mice to Ctrl-mice levels. It also rescued the pool of NSCs as measured by double positivity for GFAP+Nestin+ or GFAP+SOX2+ (Fig. 8, C and D), the number of proliferative NSCs as either a fraction of total Ki67+ or total GFAP+Nestin+ cells (Fig. 8, E and F) and reverted the increased Fip200GFAP cKO SVZ apoptotic cells (Fig. 8 G). Similar results were obtained in the DG (Fig. S4, C–G). EUK-8 or EUK-134 treatment rescued Fip200GFAP cKO defective neurogenesis in SVZ (Fig. 8, H and I) and DG (Fig. S4, H and I). Similar treatment of Ctrl mice with EUK-8 or EUK-134 did not affect SVZ cellularity, DG area, NSC pool, or any NSC activity (Fig. 8, B–I; and Fig. S4, B–I). These results show that deficient cytoplasmic SOD1 activity linked to p62 aggregates upon Fip200 deletion is responsible for the aberrant accumulation of O2•− and consequent defects of NSC maintenance and differentiation in Fip200GFAP cKO mice.
Figure 8.
SOD mimetics EUK-8 and EUK-134 restore normal O2•− levels and rescue defective NSCs in Fip200GFAP cKO mice. (A–I) Ctrl and Fip200GFAP cKO mice at P28 were treated with vehicle, EUK-8, or EUK-134. (A) DHE and DAPI fluorescence in SVZ. Arrows indicate SVZ cells. Dotted lines indicate the boundaries of the SVZ (n = 4 mice each). (B) Mean ± SEM of SVZ cellularity per section (n = 3 mice each). (C and D) Mean ± SEM of GFAP+Nestin+ (C) and GFAP+SOX2+ (D) cells per section (n = 4 mice each). (E and F) Mean ± SEM of the percentage of GFAP+Nestin+Ki67+ to total Ki67+ cells (E) or total GFAP+Nestin+ cells (F) in SVZ (n = 4 mice each). (G–I) Mean ± SEM of TUNEL+ (G) or DCX+ (H) cells per SVZ section and NeuN+ cells in the OB per square millimeter (I) of mice (n = 4 mice each). E, ependymal layer; LV, lateral ventricle; ST, striatum. Bars, 50 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (J) A working model of differential p62 aggregate formation and O2•− accumulation by deletion of Fip200, but not Atg5, Atg16L1, or Atg7, leading to defective neural stem cell maintenance and differentiation.
Discussion
Autophagy requires a set of core genes to produce the ULK1-Atg13-Fip200-Atg101 and Beclin1-Atg14L1-Vps34 protein complexes for the induction and initiation of autophagosomes, and two ubiquitin-like conjugation systems, Atg12-Atg5-Atg16L1 and Atg8, for autophagosome elongation (He and Klionsky, 2009; Mizushima and Levine, 2010; Kenific and Debnath, 2015). Many studies show each of these genes is essential for efficient autophagy, but accumulating evidence suggests additional nonoverlapping functions for them in vivo (Mizushima and Levine, 2010; Boya et al., 2013). The challenge in autophagy research is to determine whether changes in biological processes upon deletion of autophagy genes are caused by disruption of autophagy, autophagy blockage at different stages, or the loss of other nonautophagy functions of these genes (Boya et al., 2013). In this study, we generated cKO mice to study autophagy genes with autophagy induction (Fip200) and autophagosome elongation functions (Atg5, Atg7, and Atg16L1) in postnatal NSCs. We found that, unlike Fip200, ablation of Atg5, Atg16L1, or Atg7 blocked autophagy as expected but did not impair NSC maintenance and differentiation. Taking advantage of the differential outcomes in these mouse models, we demonstrated that the formation of p62 aggregates and the consequent accumulation of O2•− by decreased cytoplasmic SOD1, not mitochondria mass increase, mediated defective NSC maintenance and differentiation upon Fip200 deletion, as summarized in our working model (Fig. 8 J).
One interpretation of our observations is that nonautophagic functions of Fip200 are crucial to the regulation of p62 protein levels, maintenance, and differentiation of NSCs. Alternatively, autophagy induction or initiation (Fip200) and autophagosome elongation (Atg5, Atg16L1, and Atg7) play different roles in the regulation of NSCs, as outlined in the working model (Fig. 8 J). Consistent with the latter hypothesis, we showed that Atg13 knockdown decreased neurosphere formation to the same extent as Fip200 knockdown in postnatal NSCs (Fig. S4, J–L). Our previous study showed Spautin-1, an autophagy inhibitor that acts by decreasing the stability of Vps34 (Liu et al., 2011), inhibited neurospheres in vitro (Wang et al., 2013), consistent with autophagy initiation, and may be important for postnatal NSCs. Similarly, embryonic olfactory bulb stem cells (eOBSCs) from Ambra1 (a positive regulator of the Beclin1–Atg14L1–Vps34 complex) gene trapped (Ambra1gt/gt) mice could not form neurospheres, and haploinsufficient (Ambra1gt/+) Ambra1 cells formed small neurospheres (Vázquez et al., 2012). A recent study of Beclin1+/− mice also showed more apoptosis and less proliferation in the SVZ and decreased neurosphere formation (Yazdankhah et al., 2014). Ambra1 haploinsufficiency inhibited eOBSC neuronal differentiation (Vázquez et al., 2012). Although eOBSCs from Atg5 KO mice showed different neuronal defects in vitro (Vázquez et al., 2012), and neurogenesis defects in cerebral cortex after Atg5 knockdown during early brain development (Lv et al., 2014), we did not observe impaired neuronal differentiation in postnatal DG (Fig. S1, F and G) and OB (Fig. S1 L) of mice with Atg5, Atg16L1, or Atg7 deletion. It may be that the loss of both canonical (shared by deletion of Atg5, Atg16L1, or Atg7) and noncanonical (unique to Fip200 inactivation, independent of blocking its role in autophagy induction) functions trigger increased p62 protein levels, leading to defective NSC maintenance and differentiation. Future studies using similar cKO mice for Atg13, components of the Beclin1–Atg14L1–Vps34 complex, and reagents that specifically disrupt these protein complexes could directly ascertain mechanisms.
Previous studies showed that Fip200, Atg5, Atg16L1, and Atg7 are important for p62 degradation in many cell types, including neurons (Mizushima and Komatsu, 2011). However, unlike other regions of the brain, p62 aggregates were detected in the SVZ and SGZ of Fip200GFAP cKO mice, but not in mice with Atg5, Atg16L1, or Atg7 deletion. Our results demonstrated a role for p62 aggregate accumulation in defective NSCs with Fip200 deletion. p62 mRNA levels are decreased in Fip200-null neurospheres compared with Ctrls (Fig. S4 M), suggesting that p62 aggregate accumulation is caused by increased protein levels. However, eliminating p62 aggregates in 2cKO mice completely rescued the defective Fip200-null NSCs.
One explanation for increased p62 aggregates in Fip200-null NSCs may be that Atg5, Atg16L1, and Atg7 defective NSCs have turned on compensatory pathways to clear proteins. However, measurements of proteasome activities in neurospheres from Ctrl, Fip200 cKO, Atg5 cKO, and Atg16L1 cKO SVZ cells using various substrates (Liang et al., 2010) showed comparable proteasome activities in Ctrl and cKO NSCs, with the minor exception that Atg16L1-deficient NSCs showed reduced activity when Boc-LRR aminomethylcoumarin (AMC) was used as a substrate (Fig. S5, A–E). A compensatory increase in proteasome activity is an unlikely mechanism for the lack of p62 accumulation in Atg5 or Atg16L defective NSCs. An alternative possibility is that Fip200 may regulate p62 homeostasis through noncanonical autophagy pathways. A recent study of Caenorhabditis elegans embryogenesis showed that epg-7 (counterpart for Fip200) functions as a scaffold to facilitate degradation of aggregates containing SQST-1 (counterpart for p62; Lin et al., 2013). This supports the idea that Fip200 may have a role controlling p62 aggregate formation besides its “classic” autophagy function in NSCs. A previous study reported that Atg5/7-independent autophagy still required the classical autophagy components ULK1, Fip200, and beclin1 (Nishida et al., 2009). However, close examination of TEM images in other studies suggested that the Atg12- and LC3-conjugation systems are not essential for elongation of the phagophore membranes (Fujita et al., 2008; Sou et al., 2008; Noda et al., 2009; Kishi-Itakura et al., 2014). The observations by Nishida et al. may have been classic autophagy that requires Atg5 and Atg7 to complete (i.e., formation of closed autophagosomes). Blockage of autophagy induction or initiation led to p62 aggregate formation and ferritin clusters, whereas inhibition of autophagosome elongation by Atg5 deletion caused less accumulation of p62 in MEFs (Kishi-Itakura et al., 2014). The partial autophagosomes formed in Atg5- or Atg7-null (but not Fip200-null) MEFs (or NSCs) might still mediate p62 binding and degradation by mechanisms yet to be determined (i.e., not through fusion of the intact autophagosomes with lysosomes). This is consistent with the idea that autophagy induction or initiation, but not autophagosome elongation, plays a differential role in the regulation of NSCs by p62.
Our comparative analysis of Fip200GFAP cKO mice versus Atg5GFAP cKO and Atg16L1GFAP cKO mice, adult mice with Atg7 deletion, and 2cKO mice yielded additional mechanistic insights into the regulation of NSCs by modulating intracellular ROS levels. Although the p62–Keap1–Nrf2 axis regulating oxidative stress was well characterized in autophagy-deficient liver (Komatsu et al., 2007b), we did not find activation of Nrf2 in Fip200-deficient SVZ cells (Fig. S5, F–J). We cannot completely exclude the involvement of the p62–Keap1–Nrf2 axis, but our studies strongly suggest a critical role for SOD1 in the regulation of oxidative stress responses in Fip200-null NSCs. Under normal conditions, ROS is balanced by mitochondrial and cytoplasmic SODs, GPx, and catalase (Zelko et al., 2002). Several lines of evidence showed that deficient SODs, rather than mitochondria accumulation, are responsible for increased O2•− leading to Fip200GFAP cKO defective NSCs. Increased O2•− was observed in the SVZ and SGZ of Fip200GFAP cKO mice, but not Atg5GFAP cKO or Atg16L1GFAP cKO mice without NSC defects, although all mutant mice showed increased mitochondria mass. Decreased H2O2 levels were found in isolated Fip200GFAP cKO SVZ cells, consistent with reduced SOD activity. Increased O2•− and decreased H2O2, but not mitochondria accumulation, were rescued by p62 ablation concomitant with the restoration of NSC maintenance and differentiation in 2cKO mice. Treatment of Fip200GFAP cKO mice with SOD mimetics EUK-8 or EUK-134 also rescued the defective NSCs in these mice.
Previous studies showed that SOD1 KO mice are viable without overt abnormalities, but SOD2 KO mice exhibit severe mitochondrial damage, metabolic abnormalities, and die in mid-gestation or shortly after birth depending on their genetic background (Li et al., 1995; Huang et al., 2001, 2006). Our results implicating a role for SOD1, but not SOD2, in mediating Fip200GFAP cKO NSC defects are in contrast to SOD roles in development. A lack of SOD2 involvement is consistent with our observation that mitochondria accumulation after deletion of Fip200 and other autophagy genes, not rescued in 2cKO mice, did not cause defective NSCs. On the other hand, increased O2•− and defective NSCs correlated with decreased cytoplasmic SOD1 in Fip200GFAP cKO mice. Decreased cytoplasmic SOD1, increased O2•−, and defective NSCs were rescued by p62 ablation, suggesting that p62 aggregate accumulation in Fip200-null NSCs blocked the conversion of O2•− to H2O2 by decreasing cytoplasmic SOD1 and/or sequestering SOD1 in the nuclei. Given that the majority of p62 aggregates are cytoplasmic, it is unlikely that p62 directly sequesters SOD1 in the nucleus of Fip200-null NSCs. However, we cannot exclude the possibility that cytoplasmic p62 increases SOD1 nuclear localization by displacing the interaction of another protein that maintains SOD1 in the cytoplasm. This would be similar to the reported role of p62 promoting Nrf2 nuclear localization by displacing Keap1 binding to Nrf2 (Komatsu et al., 2010). Alternatively, a study showed that phosphorylation of SOD1 at S60 and S99 by the Mec1/ATM effector Dun1/Cds1 kinase leads to SOD1 localization in the nucleus under oxidative stress conditions (Tsang et al., 2014). p62 may regulate SOD1 nuclear translocation by modulating SOD1 phosphorylation at these sites. Future studies to examine SOD1 phosphorylation and/or identify interactions of p62 with SOD1 will help clarify mechanisms.
In summary, our studies revealed differential functions for an autophagy induction gene, as opposed to autophagy genes for autophagosome elongation, in the regulation of postnatal NSCs. Our studies identified the role of p62 aggregate accumulation in Fip200-null NSCs as a key determinant of NSC regulation through O2•− accumulation caused by decreased cytoplasmic SOD1 levels.
Materials and methods
Mice
Fip200GFAP cKO mice with Fip200 conditionally deleted in postnatal NSCs (Wang et al., 2013), hGFAP-Cre transgenic mice expressing Cre in postnatal NSCs (Zhu et al., 2005; Wang et al., 2013), Atg5f/f mice bearing two Atg5flox alleles with exon 3 flanked by two loxP sequences (Hara et al., 2006), Atg16L1f/f mice bearing two Atg16L1flox alleles with exon 3 flanked by two loxP sequences (gift of T. Saitoh and S. Akira, Osaka University, Osaka, Japan; Saitoh et al., 2008; Hwang et al., 2012; Conway et al., 2013), Ubc-CreERT2 transgenic mice expressing the TAM-regulated Cre-recombinase fusion protein under the control of the ubiquitously expressed ubiquitin C (Ubc) promoter (Karsli-Uzunbas et al., 2014), Atg7f/f bearing two Atg7flox alleles with exon 14 flanked by two loxP sequences (gift of M. Komatsu, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan; Komatsu et al., 2007a), and p62−/− mice with deletion of exons 1–4 of the gene (gift of T. Ishii, University of Tsukuba, Tsukuba, Japan; Okada et al., 2009) were as described previously. All mice were maintained on a mixed C57BL/6 (50%) and FVB (50%) backgrounds. Age- and littermate-matched control and mutant mice were used for analysis to minimize the impact of modifier genes. Mice were housed and handled according to local, state, and federal regulations. All experimental procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee of the University of Michigan and the University of Cincinnati. Genotyping for Fip200, Atg5, Atg16L1, p62, and Cre alleles was performed by PCR analysis of tail DNA, essentially as described previously (Zhu et al., 2005; Hara et al., 2006; Komatsu et al., 2007a; Hwang et al., 2012; Conway et al., 2013; Wang et al., 2013; Karsli-Uzunbas et al., 2014).
Antibodies and reagents
Primary antibodies used were: mouse anti-BrdU (G3G4; DSHB), mouse anti-GFAP (Thermo Fisher Scientific), rhodamine-conjugated mouse anti-BrdU (Millipore), mouse anti-nestin (Rat-401), mouse anti-NeuN (Millipore), mouse anti–Nqo-1 (Santa Cruz Biotechnology), mouse anti-p62 (Abcam), mouse anti ubiquitin (Santa Cruz Biotechnology), mouse antivinculin (Sigma-Aldrich), rabbit anti-Atg5 (Novus Biologicals), rabbit anti-Atg16L1 (Abgent), rabbit anti-GFAP (Dako), rabbit anti-Ki67 (Spring Bioscience), rabbit anti-LC3 (Cell Signaling), rabbit anti-NBR1 (Cell Signaling), rabbit anti-Nrf2 (Abcam), rabbit anti-p62 (Enzo), rabbit anti-Sox2 (Millipore), rabbit anti-TAX1BP1 (Abgent), rabbit anti-Fip200 (ProteinTech), rabbit anti-olig2 (Millipore), rat anti-Ki67 (BioLegend), and guinea pig antidoublecortin (anti-DCX; EMD Millipore). Secondary antibodies were goat anti–rabbit IgG-FITC, goat anti–rabbit IgG–Texas red, goat anti–mouse IgG-FITC, goat anti–mouse IgG–Texas red, goat anti–mouse IgG-HRP, and goat anti–rabbit IgG-HRP (Jackson Immunology).
DHE and EUK-8 were purchased from Sigma-Aldrich. EUK-134 was purchased from Cayman Chemical. DCFDA, MitoTracker Red, and MitoTracker Green were purchased from Invitrogen.
Protein extraction, SDS-PAGE, and immunoblotting
The SVZ tissues from mice were microdissected from 300-µm-thick sagittal sections of P14 and P28 brains and used for protein extraction by homogenization in modified radioimmune precipitation assay buffer (50 mM Tris-HCl, pH 7.4, 1% Triton X-100, 0.2% sodium deoxycholate, 0.2% SDS, and 1 mM sodium EDTA) supplemented with protease inhibitors (5 µg/ml leupeptin, 5 µg/ml aprotinin, and 1 mm phenylmethylsulfonyl fluoride). In some experiments, sections containing SVZ tissues of Ctrl, Fip200hGFAP cKO, Fip200/p62 2cKO, and p62 KO mice at P28 were lysed with 1% SDS. After removing tissue and cell debris by centrifugation at 13,000 rpm for 10 min at 4°C, protein concentration was determined using a Bio-Rad protein assay reagent. Lysates were boiled for 5 min in 1× SDS sample buffer (50 mM Tris-HCl, pH 6.8, 12.5% glycerol, 1% SDS, and 0.01% bromphenol blue) containing 5% β-mercaptoethanol and analyzed by SDS-PAGE followed by immunoblotting using various antibodies, as described previously (Wang et al., 2013).
Neurosphere culture
Neurospheres from SVZ cells of mice at P28 were cultured essentially as described in our previous study (Wang et al., 2013). In brief, the SVZ tissues were isolated under a dissection microscope and cut into ∼1-mm3 cubes. The tissues were digested in 0.2% trypsin to obtain single-cell suspensions. The cells were cultured in neurobasal medium supplemented with B27 (Invitrogen), 10 ng/ml bFGF, and 20 ng/ml EGF (Invitrogen) in Ultra-Low Attachment dishes (Corning). Neurospheres with diameters larger than 50 µm were counted 9 d after culturing. For their passage, primary neurospheres were collected and then incubated in 0.2% trypsin at 37°C for 10 min with pipetting ∼50 times. After addition of trypsin inhibitor supplemented with DNase I (Worthington) to stop digestion, dispersed cells were collected by centrifugation at 400 g for 10 min, counted and cultured for the secondary neurosphere formation, and quantified the same way as the primary neurosphere formation described above.
Proteasomal activity assay
Proteasomal activity was analyzed essentially as described previously (Liang et al., 2010) using the synthetic peptide substrates linked to the fluorometric receptor and AMC (Proteasome Substrate Pack; Biomol). Neurospheres were lysed by proteolysis buffer (0.5 mm DTT, 5 mm ATP, and 5 mm MgCl2). The 250-µl aliquots containing equal amounts of protein were incubated for 30 min at 37°C with 2.5 µl Ac-Gly-Pro-Leu-Asp-AMC (5 mm), Z-Leu-Leu-Glu-AMC (5 mm), Suc-Leu-Leu-Val-Tyr-AMC (5 mm), Ac-Arg-Leu-Arg-AMC (5 mm), or Boc-Leu-Arg-Arg-AMC (5 mm) for caspase-like, chymotrypsin-like, or trypsin-like activity, respectively. The reaction was stopped by adding 252.5 µl of ice-cold ethanol (96%). Proteasomal activity was determined by measuring the fluorescence from AMC hydrolysis (350 nm excitation and 440 nm emission).
Histology and immunofluorescence (IF)
Mice were euthanized using CO2, and a complete tissue set was harvested during necropsy. Fixation was performed for 16 h at 4°C using 4% (wt/vol) freshly made, prechilled PBS-buffered PFA. Brain tissues were sagittal sectioned, embedded in paraffin, and sectioned at 5 µm with a Leica 2125 Microtome (Leica). Slides from histologically comparable positions (triangular lateral ventricle with intact RMS) were stained with hematoxylin and eosin (H&E) for routine histological examination or left unstained for IF. H&E-stained sections were examined under a BX41 light microscope (Olympus) equipped with a 20× 0.5 NA UPlanFl air objective lens, and images were captured at room temperature with a digital camera (model DP70; Olympus) using DP Controller software (Version 1.2.1.10 8). For IF, unstained tissues were first deparaffinized in three washes of xylene (3 min each) and then rehydrated in graded ethanol solutions (100%, 95%, 70%, 50%, and 30%, 1 min each). After heat-activated antigen retrieval (Retriever 2000; PickCell Laboratories) according to the manufacturer's specifications, sections were treated with Protein Block Serum Free (Dako) at room temperature for 10 min. Sections were then incubated with primary antibodies at 4°C for 16 h in a humidified chamber, washed in PBS three times (5 min each), and incubated with 1:200 FITC or Texas red–conjugated secondary antibodies for 1 h at room temperature. After incubation with secondary antibodies and washing three times (5 min each) in PBS, nuclei were stained with DAPI and mounted with Vectashield mounting medium (Vector Laboratories). Digital photography was performed as described in the previous paragraph.
Laser scanning confocal microscope imaging
Confocal image acquiring was performed at room temperature using an Axiovert 710 Meta laser scanning confocal microscope (Carl Zeiss) equipped with a 63×/1.2 water-corrected oil-immersion lens. Images were integrated and processed using Zen 2010 software. Raw data were used to reconstitute z-stacks and orthogonal sections, and all data were processed identically between samples. Orthogonal projections were created from z-stacks containing 20 images acquired at intervals of 0.4 µm using Zen 2010 software. Figures were reassembled in Photoshop (Adobe).
TEM
TEM studies of the SVZ samples were essentially performed as described previously (Wang et al., 2013). In brief, Ctrl, Atg5hGFAP cKO, Atg16L1hGFAP cKO, Fip200hGFAP cKO, Fip200/p62 2cKO, and p62 KO mice were perfused through the heart with a mixture of 2% PFA and 1.25% glutaraldehyde in 0.1 M phosphate buffer. Brains were removed and postfixed for 2 h with the same fixative before sectioned as 200-µm slices with a vibratome (Leica). After postfixation for 1 h with 1% osmium tetroxide, slices were washed in 0.1 M cacodylate buffer, pH 7.2. The slices were dehydrated through a series of ethanol solutions, infiltrated with epoxy resins, and flat embedded. Slices containing the lateral ventricle and the SVZ were trimmed and mounted on blocks and sectioned with an ultramicrotome. Ultrathin sections were counterstained with uranyl acetate and lead citrate and analyzed with a JEM-1230 transmission electron microscope (JEOL).
FACS
Dissociated cells from neurospheres were stained with MitoTracker Green and MitoTracker Red at 20 nM for 15 min at 37°C, or by 2′-7′-dichlorofluorscein diacetate at 5 µM and DHE at 3.2 µM for 15 min at 37°C, according to manufacturer's instructions. Cells were resuspended in 2 µg/ml DAPI to distinguish live from dead cells. Flow cytometry was performed on a FACSVantage SE-dual laser, three-line flow cytometer or an FACSCanto (BD Biosciences). Dysfunctional mitochondria was gated with decreased ratio of respiring mitochondria to total mitochondrial mass (MitoTracker Red/MitoTracker Green) in dissociated NSCs.
DHE, EUK-8, and EUK-134 administration
DHE was delivered intraperitoneally into P28 mice essentially as previously described (Wang et al., 2013). EUK-8 and EUK-134 were injected intraperitoneally into P7 mice every day at a concentration of 30 mg/kg for 21 d as described previously (Melov et al., 2001).
BrdU incorporation assay and TUNEL assay
For in vivo studies, BrdU was administrated intraperitoneally at 100 µg/g three times at 2-h intervals. Mice were either euthanized 2 h after the last injection (short-term incorporation) or 3 wk later (long-term retention), and tissues were processed as described in the Histology and immunofluorescence section. For BrdU detection, tissue samples were treated with 2M HCL at room temperature for 20 min to denature the nucleotides and then neutralized with 0.1 M sodium borate at room temperature for another 20 min. After three washes in PBS (5 min each), slides were incubated with mouse anti-BrdU antibody and secondary antibodies as described for immunofluorescent staining. Nuclei were stained with DAPI and mounted with Vectashield mounting medium (Vector Laboratories). For IF and detection of BrdU in the same tissue, IF was performed first and samples were postfixed with 4% PFA at room temperature for 20 min before nucleotide denaturing with HCL. Histological examination and digital photography were performed as described in the Histology and immunofluorescence section.
Apoptotic cells were detected by TUNEL according to the protocol provided by the manufacturer in the In Situ Cell Death Detection kit-TMR Red (Roche).
Fip200 and Atg13 shRNAs for lentiviral Infection of NSCs
Mouse Fip200 and Atg13 lentiviral shRNA constructs were purchased from Open Biosystems. Control and target shRNA constructs were cotransfected with psPAX2 and pMD2G package vectors into 293T cells. Supernatants containing lentivirus were collected and concentrated according to the manufacturer’s instructions. Viruses were used to infect isolated SVZ cells by spin infection at 2,000 rpm for 30 min. Infected cells were collected by FACS and cultured in the same conditions for neurosphere formation assay.
Statistical analysis
Lengths, areas, and the number of cells from comparable sections were quantified using ImageJ software. Statistical significance was evaluated by Student’s t test, with P < 0.05 indicative of statistical significance using Graph Pad Prism (version 5.0). The number of animals used for quantification is indicated in the figure legends.
Online supplemental material
Fig. S1 shows that deletion of Atg5, Atg16L1, or Atg7 does not affect postnatal neural stem cells in the SGZ. Fig. S2 shows analyses of ubiquitin, p62, TAX1BP1, and NBR1 aggregates in Fip200GFAP cKO, Atg5GFAP cKO, and Atg16L1GFAP cKO mice. Fig. S3 shows that p62 KO rescues degenerative neural stem cells in the SGZ of Fip200GFAP cKO mice. Fig. S4 shows that SOD mimetics restore normal superoxide levels and rescue defective neural stem cells in the DG of Fip200GFAP cKO mice, Atg13 knockdown decreases neurosphere formation of SVZ cells, and decreased p62 mRNA in Fip200-null neurospheres. Fig. S5 shows analysis of proteasome activity and activation of p62–Keap1–Nrf2 axis in neural stem cells of Ctrl, Fip200GFAP cKO, Atg5GFAP cKO, and Atg16L1GFAP cKO mice. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507023/DC1.
Supplementary Material
Acknowledgments
We are grateful to Drs. T. Saitoh and S. Akira of Osaka University for Atg16L1f/f mice, Dr. M. Komatsu of Tokyo Metropolitan Institute of Medical Science for Atg7f/f mice, and Dr. T. Ishii at University of Tsukuba for p62−/− mice. We thank our colleagues Drs. Czyzyk-Krzeska, Du, Lu, Plas, and Zheng and members of the Guan laboratory for critical reading and helpful comments on the manuscript. We thank Dr. B. Peace for professional editing and G. Doerman for figure preparation. We also thank V. Sui, Z.-C. Bian, A. Serna, and M. Haas for their assistance.
This research was supported by National Institutes of Health grants R01CA150926, R01CA163493, and R01HL073394 to J.-L. Guan, Japan Society for the Promotion of Science KAKENHI Grants-in-Aid for Scientific Research on Innovative Areas (grant 25111005) to N. Mizushima, and National Institutes of Health grant U19AI109725 to H.W. Virgin.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AMC
- aminomethylcoumarin
- cKO
- conditional knockout
- DCX
- doublecortin
- DG
- dentate gyrus
- DHE
- dihydroethidium
- eOBSC
- embryonic olfactory bulb stem cell
- GZ
- granular zone
- H&E
- hematoxylin and eosin
- IF
- immunofluorescence
- NSC
- neural stem cell
- OB
- olfactory bulb
- P
- postnatal day
- RMS
- rostral migratory stream
- ROS
- reactive oxygen species
- SGZ
- subgranular zone
- SOD
- superoxide dismutase
- SVZ
- subventricular zone
- TEM
- transmission electron microscopy
References
- Apel K., and Hirt H.. 2004. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55:373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., and Johansen T.. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614. 10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P., Reggiori F., and Codogno P.. 2013. Emerging regulation and functions of autophagy. Nat. Cell Biol. 15:713–720. 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway K.L., Kuballa P., Song J.H., Patel K.K., Castoreno A.B., Yilmaz O.H., Jijon H.B., Zhang M., Aldrich L.N., Villablanca E.J., et al. 2013. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology. 145:1347–1357. 10.1053/j.gastro.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge W. 2002. Free radicals in the physiological control of cell function. Physiol. Rev. 82:47–95. 10.1152/physrev.00018.2001 [DOI] [PubMed] [Google Scholar]
- Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., and Yoshimori T.. 2008. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell. 19:4651–4659. 10.1091/mbc.E08-03-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B., and Guan J.L.. 2008. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell. Signal. 20:787–794. 10.1016/j.cellsig.2007.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan B., Peng X., Nagy T., Alcaraz A., Gu H., and Guan J.L.. 2006. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 175:121–133. 10.1083/jcb.200604129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.Y., Chen H.Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G., Kamphorst J.J., Chen G., Lemons J.M., Karantza V., et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25:460–470. 10.1101/gad.2016311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N.. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J.L., and Mizushima N.. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181:497–510. 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., and Klionsky D.J.. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43:67–93. 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.T., Carlson E.J., Kozy H.M., Mantha S., Goodman S.I., Ursell P.C., and Epstein C.J.. 2001. Genetic modification of prenatal lethality and dilated cardiomyopathy in Mn superoxide dismutase mutant mice. Free Radic. Biol. Med. 31:1101–1110. 10.1016/S0891-5849(01)00694-3 [DOI] [PubMed] [Google Scholar]
- Huang T.T., Naeemuddin M., Elchuri S., Yamaguchi M., Kozy H.M., Carlson E.J., and Epstein C.J.. 2006. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum. Mol. Genet. 15:1187–1194. 10.1093/hmg/ddl034 [DOI] [PubMed] [Google Scholar]
- Hwang S., Maloney N.S., Bruinsma M.W., Goel G., Duan E., Zhang L., Shrestha B., Diamond M.S., Dani A., Sosnovtsev S.V., et al. 2012. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 11:397–409. 10.1016/j.chom.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C., Rong Y., Doctrow S., Baudry M., Malfroy B., and Xu Z.. 2001. Synthetic superoxide dismutase/catalase mimetics reduce oxidative stress and prolong survival in a mouse amyotrophic lateral sclerosis model. Neurosci. Lett. 304:157–160. 10.1016/S0304-3940(01)01784-0 [DOI] [PubMed] [Google Scholar]
- Karsli-Uzunbas G., Guo J.Y., Price S., Teng X., Laddha S.V., Khor S., Kalaany N.Y., Jacks T., Chan C.S., Rabinowitz J.D., and White E.. 2014. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 4:914–927. 10.1158/2159-8290.CD-14-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenific C.M., and Debnath J.. 2015. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 25:37–45. 10.1016/j.tcb.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516. 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Kishi-Itakura C., Koyama-Honda I., Itakura E., and Mizushima N.. 2014. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 127:4089–4102. 10.1242/jcs.156034 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., and Tanaka K.. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. 2007a Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 131:1149–1163. 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Wang Q.J., Holstein G.R., Friedrich V.L. Jr., Iwata J., Kominami E., Chait B.T., Tanaka K., and Yue Z.. 2007b Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. USA. 104:14489–14494. 10.1073/pnas.0701311104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12:213–223. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., and Mizushima N.. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036. 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- Lee I.H., Kawai Y., Fergusson M.M., Rovira I.I., Bishop A.J., Motoyama N., Cao L., and Finkel T.. 2012. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 336:225–228. 10.1126/science.1218395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang T.T., Carlson E.J., Melov S., Ursell P.C., Olson J.L., Noble L.J., Yoshimura M.P., Berger C., Chan P.H., et al. 1995. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 11:376–381. 10.1038/ng1295-376 [DOI] [PubMed] [Google Scholar]
- Liang C.C., Wang C., Peng X., Gan B., and Guan J.L.. 2010. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 285:3499–3509. 10.1074/jbc.M109.072389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., and Levine B.. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- Lin L., Yang P., Huang X., Zhang H., Lu Q., and Zhang H.. 2013. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J. Cell Biol. 201:113–129. 10.1083/jcb.201209098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Xia H., Kim M., Xu L., Li Y., Zhang L., Cai Y., Norberg H.V., Zhang T., Furuya T., et al. 2011. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 147:223–234. 10.1016/j.cell.2011.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard D.B., Chua K.F., Mostoslavsky R., Franco S., Gostissa M., and Alt F.W.. 2005. DNA repair, genome stability, and aging. Cell. 120:497–512. 10.1016/j.cell.2005.01.028 [DOI] [PubMed] [Google Scholar]
- Lv X., Jiang H., Li B., Liang Q., Wang S., Zhao Q., and Jiao J.. 2014. The crucial role of Atg5 in cortical neurogenesis during early brain development. Sci. Rep. 4:6010 10.1038/srep06010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matés J.M. 2000. Effects of antioxidant enzymes in the molecular control of reactive oxygen species toxicology. Toxicology. 153:83–104. 10.1016/S0300-483X(00)00306-1 [DOI] [PubMed] [Google Scholar]
- Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.Y., Bray K., Reddy A., Bhanot G., Gelinas C., et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 137:1062–1075. 10.1016/j.cell.2009.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S., Doctrow S.R., Schneider J.A., Haberson J., Patel M., Coskun P.E., Huffman K., Wallace D.C., and Malfroy B.. 2001. Lifespan extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J. Neurosci. 21:8348–8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa T., Takahama M., Kozaki T., Lee H., Zou J., Saitoh T., and Akira S.. 2013. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14:454–460. 10.1038/ni.2550 [DOI] [PubMed] [Google Scholar]
- Mizushima N., and Komatsu M.. 2011. Autophagy: renovation of cells and tissues. Cell. 147:728–741. 10.1016/j.cell.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Mizushima N., and Levine B.. 2010. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 12:823–830. 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.P. 2009. How mitochondria produce reactive oxygen species. Biochem. J. 417:1–13. 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka K., Muraguchi T., Hoshii T., and Hirao A.. 2008. Regulation of reactive oxygen species and genomic stability in hematopoietic stem cells. Antioxid. Redox Signal. 10:1883–1894. 10.1089/ars.2008.2114 [DOI] [PubMed] [Google Scholar]
- Newman A.C., Scholefield C.L., Kemp A.J., Newman M., McIver E.G., Kamal A., and Wilkinson S.. 2012. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS One. 7:e50672 10.1371/journal.pone.0050672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., and Shimizu S.. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 461:654–658. 10.1038/nature08455 [DOI] [PubMed] [Google Scholar]
- Noda T., Fujita N., and Yoshimori T.. 2009. The late stages of autophagy: how does the end begin? Cell Death Differ. 16:984–990. 10.1038/cdd.2009.54 [DOI] [PubMed] [Google Scholar]
- Okada K., Yanagawa T., Warabi E., Yamastu K., Uwayama J., Takeda K., Utsunomiya H., Yoshida H., Shoda J., and Ishii T.. 2009. The alpha-glucosidase inhibitor acarbose prevents obesity and simple steatosis in sequestosome 1/A170/p62 deficient mice. Hepatol. Res. 39:490–500. 10.1111/j.1872-034X.2008.00478.x [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112:1809–1820. 10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S.G., Yang K.S., Kang S.W., Woo H.A., and Chang T.S.. 2005. Controlled elimination of intracellular H2O2: regulation of peroxiredoxin, catalase, and glutathione peroxidase via post-translational modification. Antioxid. Redox Signal. 7:619–626. 10.1089/ars.2005.7.619 [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., Mariño G., and Kroemer G.. 2011. Autophagy and aging. Cell. 146:682–695. 10.1016/j.cell.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., et al. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 456:264–268. 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- Sou Y.S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 19:4762–4775. 10.1091/mbc.E08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., and Mizushima N.. 2011. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25:795–800. 10.1101/gad.2016211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang C.K., Liu Y., Thomas J., Zhang Y., and Zheng X.F.. 2014. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 5:3446 10.1038/ncomms4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez P., Arroba A.I., Cecconi F., de la Rosa E.J., Boya P., and de Pablo F.. 2012. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy. 8:187–199. 10.4161/auto.8.2.18535 [DOI] [PubMed] [Google Scholar]
- Wang C., Liang C.C., Bian Z.C., Zhu Y., and Guan J.L.. 2013. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat. Neurosci. 16:532–542. 10.1038/nn.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Wei S., Gan B., Peng X., Zou W., and Guan J.L.. 2011. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 25:1510–1527. 10.1101/gad.2051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Yeo S., Wang C., Chen S., Sun S., Haas M.A., Tu W., Jin F., and Guan J.L.. 2015. Autophagy inhibition re-sensitizes pulse stimulation-selected paclitaxel-resistant triple negative breast cancer cells to chemotherapy-induced apoptosis. Breast Cancer Res. Treat. 149:619–629. 10.1007/s10549-015-3283-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. 2012. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer. 12:401–410. 10.1038/nrc3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J.M., Dell’antonio G., et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25:717–729. 10.1101/gad.2016111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdankhah M., Farioli-Vecchioli S., Tonchev A.B., Stoykova A., and Cecconi F.. 2014. The autophagy regulators Ambra1 and Beclin 1 are required for adult neurogenesis in the brain subventricular zone. Cell Death Dis. 5:e1403 10.1038/cddis.2014.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z., Jin S., Yang C., Levine A.J., and Heintz N.. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 100:15077–15082. 10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelko I.N., Mariani T.J., and Folz R.J.. 2002. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33:337–349. 10.1016/S0891-5849(02)00905-X [DOI] [PubMed] [Google Scholar]
- Zeng M., and Zhou J.N.. 2008. Roles of autophagy and mTOR signaling in neuronal differentiation of mouse neuroblastoma cells. Cell. Signal. 20:659–665. 10.1016/j.cellsig.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Guignard F., Zhao D., Liu L., Burns D.K., Mason R.P., Messing A., and Parada L.F.. 2005. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 8:119–130. 10.1016/j.ccr.2005.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.