Drp1-dependent mitochondrial fission through MiD49 and MiD51, but not through Mff, is essential for cristae remodeling to facilitate cytochrome c release during the early phase of intrinsic apoptosis.
Abstract
Mitochondrial fission facilitates cytochrome c release from the intracristae space into the cytoplasm during intrinsic apoptosis, although how the mitochondrial fission factor Drp1 and its mitochondrial receptors Mff, MiD49, and MiD51 are involved in this reaction remains elusive. Here, we analyzed the functional division of these receptors with their knockout (KO) cell lines. In marked contrast to Mff-KO cells, MiD49/MiD51-KO and Drp1-KO cells completely resisted cristae remodeling and cytochrome c release during apoptosis. This phenotype in MiD49/51-KO cells, but not Drp1-KO cells, was completely abolished by treatments disrupting cristae structure such as OPA1 depletion. Unexpectedly, OPA1 oligomers generally thought to resist cytochrome c release by stabilizing the cristae structure were similarly disassembled in Drp1-KO and MiD49/51-KO cells, indicating that disassembly of OPA1 oligomers is not directly linked to cristae remodeling for cytochrome c release. Together, these results indicate that Drp1-dependent mitochondrial fission through MiD49/MiD51 regulates cristae remodeling during intrinsic apoptosis.
Introduction
Cytochrome c release from the cristae into the cytoplasm constitutes the key step of intrinsic apoptosis (Frank et al., 2001; Detmer and Chan, 2007). A majority of total cytochrome c is encapsulated within the mitochondrial cristae folds that are connected to the intermembrane space (IMS) by relatively narrow structures named cristae junctions. At the early phase of intrinsic apoptosis, apoptotic signals induce cristae remodeling to redistribute cytochrome c into the IMS. Cytochrome c is then released into the cytoplasm through the mitochondrial outer membrane (MOM) pores generated by Bax and Bak, which are activated by BH3-only proteins such as proapoptotic truncated Bid (tBid), and initiates caspase cascade activation leading to cell death (Suen et al., 2008; Tait and Green, 2010).
The mitochondrial inner membrane (MIM) profusion GTPase OPA1 plays a key role in maintaining healthy cristae junctions to protect cells from apoptosis; its oligomer stabilizes cristae morphology and prevents cristae remodeling and cytochrome c release (Olichon et al., 2003; Frezza et al., 2006; Varanita et al., 2015). Therefore, OPA1 down-regulation not only causes mitochondrial fragmentation but also alters cristae morphology, rendering cells susceptible to apoptosis. A current model indicates that the MIM-bound long forms of OPA1 (L-OPA1) and the processed soluble short forms (S-OPA1) constitute high-molecular-weight OPA1 oligomers, and the L- to S-OPA1 balance is critical for maintaining cristae integrity; intrinsic apoptotic signals in vivo or incubation of isolated mitochondria with tBid induces the release of cytochrome c concomitant with stimulation of L-OPA1 processing to S-OPA1 and disassembly of OPA1 oligomers (Frezza et al., 2006; Jiang et al., 2014).
So far, three MIM proteins are involved in cristae morphogenesis through the regulation of OPA1 function: prohibitin-1 and -2 (PHB1 and PHB2) and reactive oxygen species modulator protein 1 (ROMO1; Mgr2 in yeast). Prohibitins form large oligomeric structures with a membrane scaffold function and regulate cristae morphogenesis through OPA1 regulation (Merkwirth et al., 2008). Loss of PHB2 in PHB2−/− cells (which also induces PHB1 degradation) leads to selective loss of L-OPA1 isoforms, resulting in aberrant cristae morphogenesis and increased susceptibility to apoptosis. Re-expression of a noncleavable L-OPA1 mutant in PHB2−/− cells restores normal cristae structures and growth phenotypes, demonstrating that L-OPA1 is crucial for maintaining healthy cristae structures (Merkwirth et al., 2008). ROMO1, the MIM redox-regulated protein, is required for maintaining cristae junctions through the regulation of OPA1 oligomerization (Norton et al., 2014).
The MOM proteins Fis1, Mff, MiD49/MIEF2, and MiD51/MIEF1 are reported to act as receptors of Drp1 in mammals, but recent studies revealed that Fis1 has little or no role in mitochondrial fission (Otera et al., 2010; Palmer et al., 2011; Zhao et al., 2011). During mitochondrial fission, ER tubules cross the mitochondria to constrict the membrane where the Drp1 receptor Mff accumulates to drive Drp1-dependent mitochondrial fission, although the contribution of MiD proteins to this process is not known (Friedman et al., 2011). Mff, MiD49, and MiD51 independently function as Drp1 receptors based on the detection of discretely assembled Drp1 foci on the MOM depending on their overexpression (Koirala et al., 2013; Losón et al., 2013; Palmer et al., 2013), but the morphological responses to overexpression are distinct between Mff and MiD proteins; Mff promotes mitochondrial fission, whereas MiD49/MIEF2 or MiD51/MIEF1 promotes mitochondrial fusion, probably because of the sequestration and inactivation of Drp1 on the MOM (Liu et al., 2013; Losón et al., 2013). However, recent observations with advanced imaging methods have raised a different possibility: MiD proteins and Mff colocalize within the same division foci at the mitochondria-associated ER membrane (MAM) and these proteins may cooperatively function as the same fission machinery (Elgass et al., 2015). Thus, the physiological and functional division of these proteins remains to be elucidated.
Mitochondrial fission is closely associated with the initial process of apoptosis; down-regulation of Drp1 or overexpression of a dominant-negative form of Drp1 delays the release of cytochrome c from within the cristae, but not other IMS-soluble apoptotic factors such as Smac/DIABLO, suggesting that mitochondrial fission, but not Bax/Bak-dependent MOM-pore formation, facilitates apoptotic cristae remodeling (Frank et al., 2001; Otera et al., 2010). Whether mitochondrial fission is essential for apoptosis and, if so, how Drp1 and its receptor proteins contribute to the initial process of cytochrome c release remains unclear (James and Martinou, 2008: Scorrano, 2009). Here, we generated Drp1-knockout (KO) and Drp1 receptor–KO HeLa cell lines and investigated their mitochondrial morphological and apoptotic responses. Our findings indicated that Drp1-dependent mitochondrial fission through MiD49 and MiD51, but not through Mff, is essential for cristae remodeling to facilitate cytochrome c release into the cytoplasm during the early phase of intrinsic apoptosis.
Results
Characterization of mitochondrial fission-related protein-KO HeLa cell lines
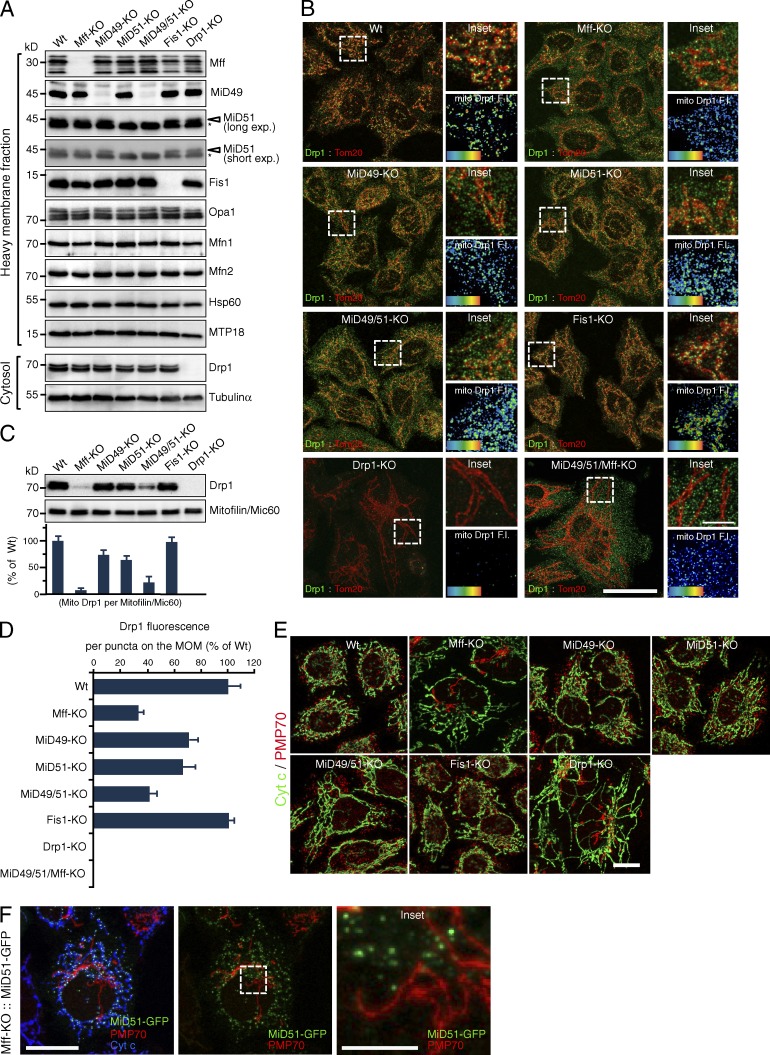
To elucidate the functional relations between Mff and MiD proteins in mitochondrial fission and apoptosis, we generated Drp1-KO, Mff-KO, MiD49-KO, MiD51-KO, MiD49/51-KO, and Fis1-KO HeLa cell lines using the CRISPR/Cas9 system and compared their mitochondrial morphological and apoptotic phenotypes. They were completely depleted of the target proteins, but the levels of other mitochondrial dynamics–related proteins were essentially unaffected (Fig. 1 A). Many Drp1 foci were detected on the mitochondrial tubules in wild-type cells (Fig. 1 B). Mitochondria in Mff-KO cells were elongated, and the number of Drp1 foci on the MOM was substantially decreased (Fig. 1, B and D). Like Mff-KO cells, mitochondria in MiD49-KO and MiD51-KO cells were elongated, but the number of Drp1 foci on the MOM was reduced more moderately than that in Mff-KO cells. Consistent with our previous observations using Fis1−/− HCT116 cells (Otera et al., 2010), neither the mitochondrial morphology nor the Drp1 foci on the MOM were affected by Fis1-KO. Supporting these observations, the Drp1 levels in mitochondrial fractions were substantially lower in Mff-KO cells than in wild-type and Fis1-KO cells (Fig. 1 C). Drp1 levels in the mitochondrial fraction were slightly lower in MiD49-KO or MiD51-KO cells than in wild-type cells, and MiD49/51-KO cells had substantially decreased levels of Drp1, suggesting that the phenotype of MiD51 loss was synergistically enhanced by removing MiD49. These mitochondrial phenotypes of Mff-KO and MiD49/51-KO cells were not as strong as that of Drp1-KO cells, but Mff-depletion in MiD49/51-KO cells induced almost a complete release of Drp1 foci into the cytoplasm (Fig. 1, B and D). Thus, Mff, MiD49, and MiD51 proteins redundantly function as Drp1 receptors, and the functional sum of these three proteins is responsible for almost all mitochondrial Drp1 recruitment activity; mitochondrial morphology and Drp1 recruitment to mitochondria in the Mff/MiD49/51 triple-depleted cells closely reflect those of Drp1-KO cells (Fig. 1, B and D; and Fig. S1 B).
Figure 1.
Characterization of mitochondrial fission-related protein KO HeLa cell lines. (A) Cytosol and mitochondrial fractions were analyzed by Western blotting. Asterisks, nonspecific bands. (B) Confocal images of the indicated cell lines; Drp1 (green) and Tom20 (red). Bar, 20 µm. Insets, magnified images of the boxed regions. Bar, 5 µm. Bottom right, fluorescence obtained for the Drp1 channel. Heat map, Drp1 fluorescence intensity (FI). (C) Western blotting of mitochondrial fractions for Drp1 and mitofilin. Histogram, quantification of three independent biological replicates. n = 3. Data represent mean ± SD. (D) Drp1 fluorescence per focus on the MOM. 200 foci were analyzed from randomly selected regions for each group in B. n = 200. In C and D, the data were normalized to the wild-type cells. Data represent mean ± SD. (E) Confocal images of the indicated cells; cytochrome c (green) and PMP70 (red; a peroxisomal marker). Bar, 10 µm. (F) Subcellular localization of MiD51-GFP. Mff-KO cells were transfected with MiD51-GFP and immunostained with the indicated antibodies. Bars, 10 µm. Magnified image of the boxed region is shown as an inset. Bar, 2.5 µm. Wt, wild type.
Mammalian peroxisomes and mitochondria share several fission-related components, including Drp1, Mff, Fis1, and GDAP1, but not MiD49 and MiD51 (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010; Huber et al., 2013). Consistently, almost all peroxisomes in both Drp1-KO and Mff-KO cells, but not in Fis1-KO cells, had an extremely elongated morphology (Fig. 1 E). In contrast, no significant peroxisomal morphological changes were observed in MiD49-KO, MiD51-KO, and MiD49/51-KO cells (Fig. 1 E). Consistent with this and previous observations (Palmer et al., 2013), MiD51 was not localized to peroxisomes in Mff-KO HeLa cells (Fig. 1 F). Thus, MiD49 and MiD51 function as mitochondria-specific receptors for Drp1.
Mff and MiD51 independently function as Drp1 receptors
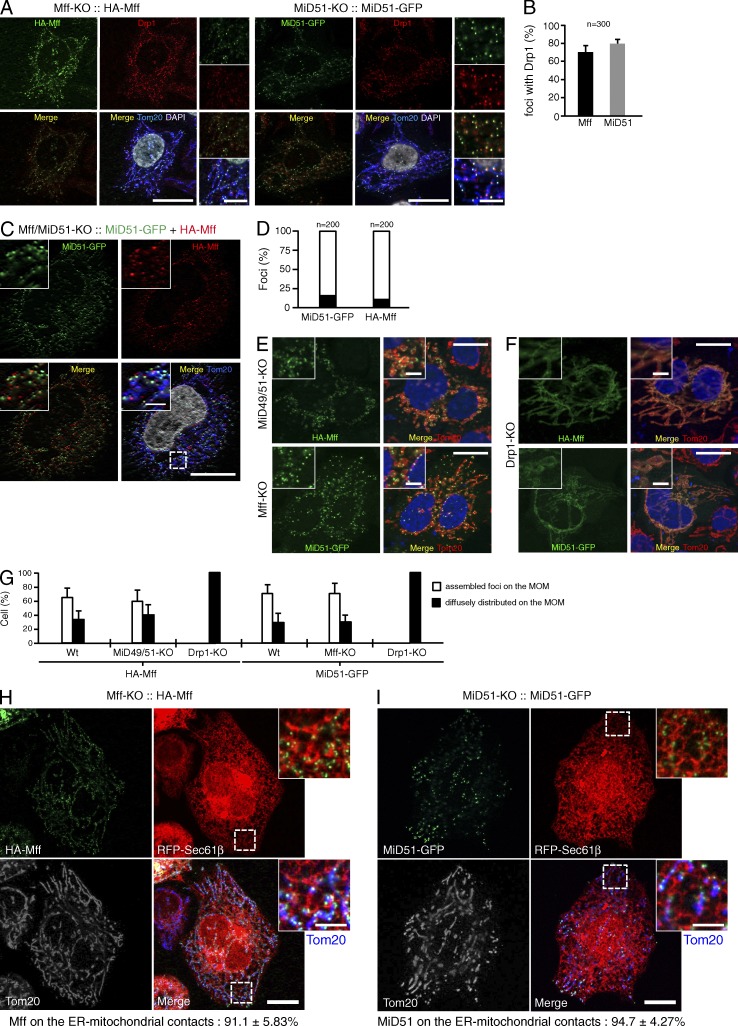
We next investigated the interrelation of Mff and MiD proteins in Drp1-dependent mitochondrial fission and found that (1) stably expressed HA-Mff (the splice isoform 8) and MiD51-GFP had discrete foci on the MOM in Mff-KO and MiD51-KO cells, respectively (Fig. 2 A); (2) double immunofluorescence staining with anti–Drp1-antibodies revealed that more than 70% of Mff and 80% of MiD51 foci were colocalized with Drp1 (Fig. 2 B); (3) stably expressed HA-Mff and MiD51-GFP in Mff/MiD51-KO cells had distinct foci on the MOM (85–90%; Fig. 2, C and D); (4) HA-Mff and MiD51-GFP separately expressed in MiD49/51-KO and Mff-KO cells, respectively, assembled into foci independently of each other (Fig. 2, E and G); (5) HA-Mff and MiD51-GFP failed to assemble into foci to distribute smoothly through the extended mitochondrial tubules in Drp1-KO cells (Fig. 2, F and G); (6) consistent with previous studies (Friedman et al., 2011; Elgass et al., 2015), 91% of HA-Mff (ncells = 4) and 94% of MiD51-GFP (ncells = 4) localized as foci on the MAM (Fig. 2, H and I); and (7) Mff functioned as a Drp1 recruiter on the MOM and mediated mitochondrial fission in the absence of MiD proteins (Fig. 3 A). Thus, Mff, MiD51, and probably MiD49 independently recruit Drp1 and mediate mitochondrial fission, which may proceed on the MAM, although interplay of these fission components cannot completely be ruled out (Elgass et al., 2015).
Figure 2.
Mff and MiD51 independently assemble into foci at ER–mitochondria contact sites in the presence of Drp1. (A) Confocal immunofluorescence microscopy. Left, Mff-KO cells stably expressing HA-Mff; right, MiD51-KO cells stably expressing MiD51-GFP. High magnification images are shown as insets. Bars: 10 µm; (magnification) 2 µm. (B) Percentage of HA-Mff and MiD51-GFP foci colocalized with Drp1. 300 foci were counted from randomly selected regions. n = 300. Data represent mean ± SD (C) Confocal images of Mff/MiD51-KO cells stably expressing MiD51-GFP and HA-Mff. Cells were immunostained with the indicated antibodies. High magnification images are shown as insets. Bars: 10 µm; (magnification) 2 µm. (D) Quantitative analysis of colocalizing (black) versus individual (white) foci on the MOM. 200 foci were counted from randomly selected regions. (E) MiD49/51-KO cells or Mff-KO cells were transfected as indicated and immunostained with the indicated antibodies. Magnified images are shown as insets. Bars: 10 µm; (magnification) 2 µm. (F) Drp1-KO cells were transfected as indicated and immunostained with the indicated antibodies. Magnified images are shown as insets. Bars: 10 µm; (magnification) 2 µm. Nuclei were stained with DAPI in A, C, D, and E. (G) Quantitative analysis of distribution of HA-Mff or MiD51-GFP on the MOM in the indicated cells. 200 cells each were counted in three independent experiments. n = 3. Data represent mean ± SD. Wt, wild type. (H) RFP-Sec61β (an ER marker) was transfected in Mff-KO cells stably expressing HA-Mff or (I) MiD51-KO cells stably expressing MiD51-GFP. Confocal images were obtained by immunostaining for HA-Mff and Tom20. Magnified images of the boxed regions are shown as insets. Bars: 10 µm; (magnification) 2 µm. The foci localizing on the ER–mitochondria contact sites in H and I were counted in eight ROIs from four cells for each group (n = ∼200).
Figure 3.
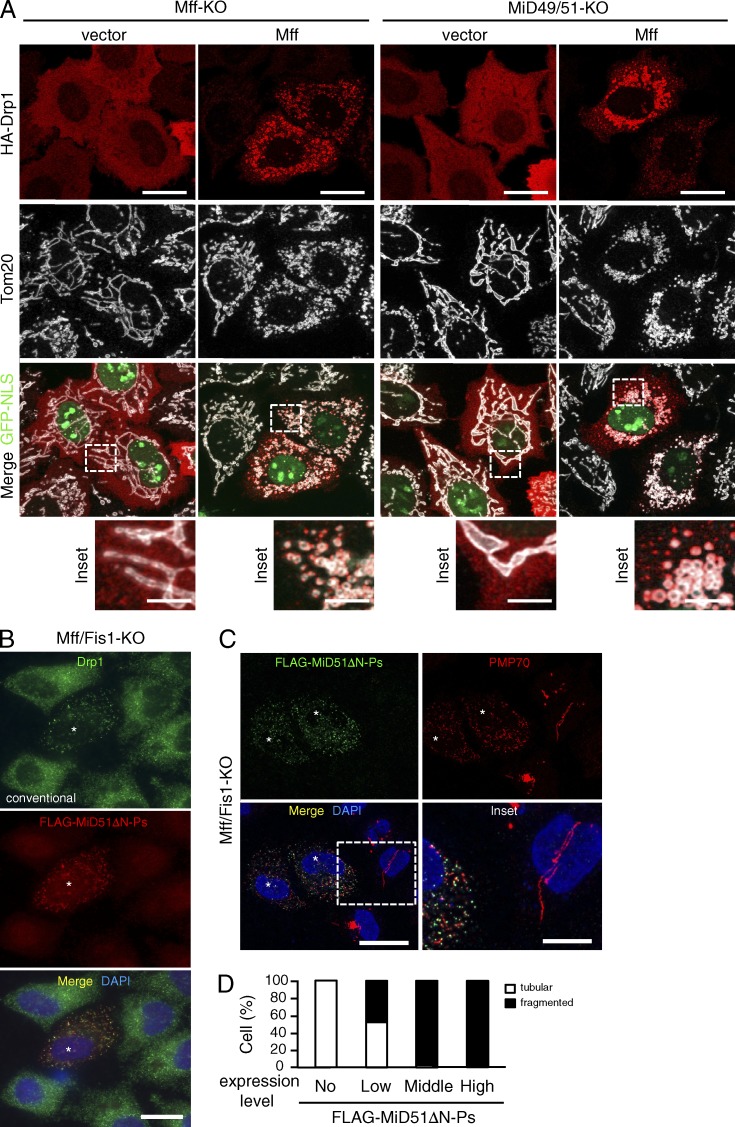
Mff and MiD51 independently mediate membrane fission. (A) Confocal images of the indicated cells cotransfected with HA-Drp1 and empty vector or pMff-IRES-GFP-NLS. Cells were immunostained with the indicated antibodies. Bottom, magnified images of the boxed regions. Bars: 10 µm; (magnification) 2.5 µm. (B–D) Mff/Fis1-KO cells were transfected with FLAG-MiD51ΔN-Ps and subjected to immunostaining with the indicated antibodies. Conventional (B) and confocal images (C) are shown. Asterisks, FLAG-MiD51ΔN-Ps expressing cells. Magnified image of the boxed region is shown as inset. Bars: 20 µm; (magnification) 10 µm. Nuclei were stained with DAPI in B and C. (D) Percentages of cells with indicated peroxisomal morphologies in cells (n = 100).
Of note, when a MiD51 mutant in which its N-terminal MOM-targeting signal anchor (N-SA; N-terminal 28–amino acid residue segment localizing in the IMS followed by an 18–amino acid residue transmembrane domain [TMD]; Palmer et al., 2011; Zhao et al., 2011) was deleted and an artificial peroxisome-targeted C-terminal signal anchor (modified C-SA of Omp25; Setoguchi et al., 2006; Yagita et al., 2013) was ligated to the C terminus (FLAG-MiD51ΔN-Ps) and expressed in Mff/Fis1-KO cells, it localized to peroxisomes, where it efficiently recruited Drp1 and induced their fission (Fig. 3, B–D). Together, these findings indicate that the cytoplasmic domain of MiD51 is sufficient to recruit Drp1 and induce Drp1-dependent peroxisomal fission when artificially targeted to peroxisomes.
Drp1-dependent mitochondrial fission through MiD49 and MiD51 regulates cytochrome c release downstream of Bax/Bak activation
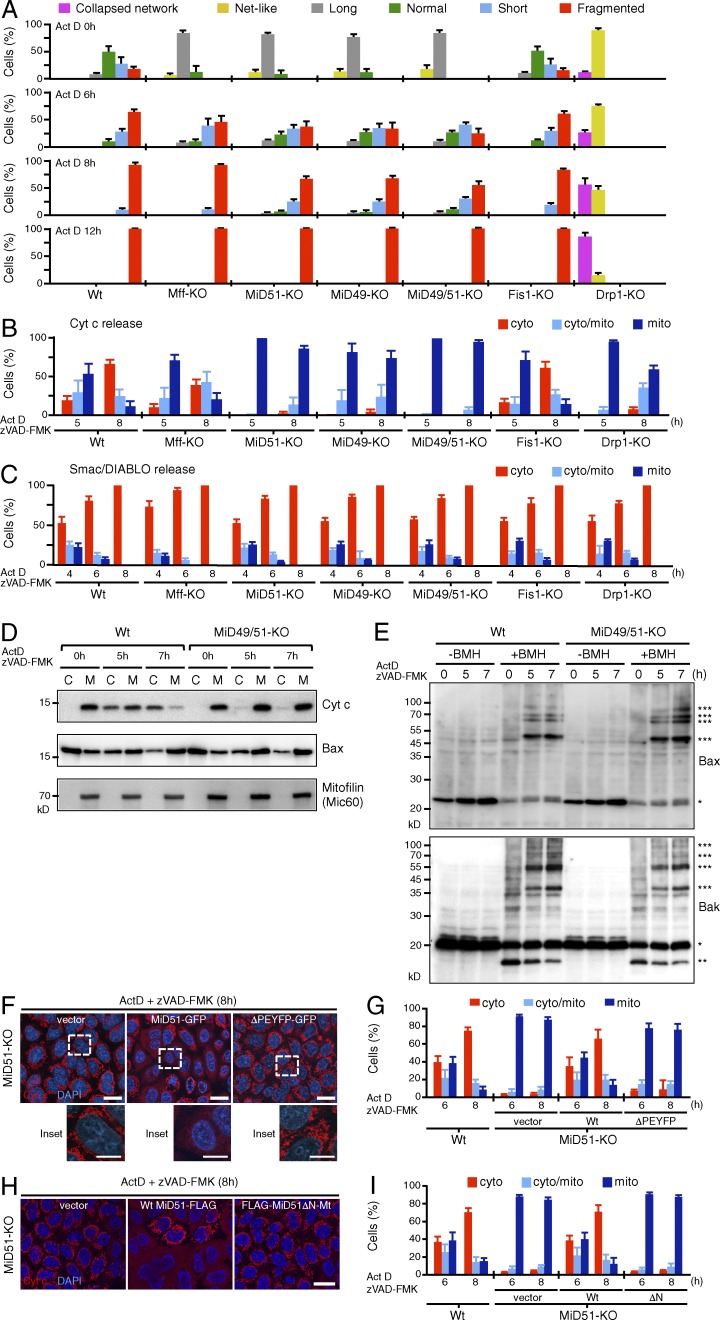
Because mitochondrial fission is induced by several physiological stimuli, we examined the contribution of Mff and MiD proteins to the cells’ response to detrimental stressors, including actinomycin D (Act D), carbonyl cyanide m-chlorophenylhydrozone (CCCP), antimycin A, valinomycin, and oligomycin. As a control, we also examined the mitochondrial response to the viral mitochondria-localized inhibitor of apoptosis (vMIA), which is a suppressor of apoptosis functioning in the lipid rafts of MAM and induces extensive mitochondrial fission (Bhuvanendran et al., 2014). As summarized in Fig. 4 A (for Act D) and Fig. S1 (for others), these treatments induced rapid mitochondrial fission in wild-type and Fis1-KO cells, confirming the minor role of Fis1 in mitochondrial fission. In contrast, fission rates were reduced in Mff-, MiD49-, MiD51-, and MiD49/51-KO cells, although their mitochondria eventually underwent extensive fission. In contrast, no such fission was detected in Drp1-KO cells. Mitochondria in Mff/MiD49/51-KO cells, however, completely resisted fission against these treatments (Fig. S1 B), confirming that cooperation of these three factors is responsible for almost all of the Drp1 recruitment activity (Fig. 1, B–D).
Figure 4.
Loss of MiD49/51 inhibits Drp1-dependent cytochrome c release downstream of Bax/Bak activation. The indicated cells were treated with Act D in the presence of zVAD-FMK for the indicated time points and analyzed by confocal immunofluorescence microscopy for Tom20, cytochrome c, and Smac/DIABLO. (A) Mitochondrial morphology, (B) cytochrome c release, and (C) Smac/DIABLO release were quantitated. (D) Wild-type and MiD49/51-KO cells were fractionated into cytosol (C) and membrane (M) fractions. They were subjected to SDS-PAGE, and endogenous cytochrome c, Bax, and mitofilin/Mic60 as a mitochondria marker were detected by Western blotting. (E) Membrane fractions isolated from wild-type and MiD49/51-KO cells treated with Act D for the indicated times were cross-linked with 2 mM 1,6-bismaleimidohexane (BMH). Cross-linked samples were analyzed by SDS-PAGE followed by Western blotting for Bax (top) or Bak (bottom). * and *** show Bax or Bak monomer and Bax or Bak complexes, respectively. Bak** is likely internally cross-linked Bak. (F) MiD51-KO cells stably transfected with the indicated plasmids were treated with Act D and immunostained with anti–cytochrome c antibodies. Magnified images of the boxed regions are shown as insets. Bars: 20 µm; (magnification) 10 µm. (G) Quantification of time-dependent release of cytochrome c as described in F. (H) MiD51-KO cells stably transfected with the indicated plasmids were treated as in F. Bar, 20 µm. (I) Time course of cytochrome c release as performed in H. The percentages in A, B, C, G, and I are means of three independent experiments with 300 cells per data point. n = 3. Nuclei were stained with DAPI (F and H). Data represent mean ± SD in A, B, C, G, and H. Wt, wild type.
Cytochrome c release from mitochondria is a crucial step in intrinsic apoptosis, associated with the disruption of mitochondrial tubular networks into small punctate structures, although whether Drp1-dependent mitochondrial fission is essential for cytochrome c release has been disputed (James and Martinou, 2008; Scorrano, 2009). To address this, we took advantage of fission component-KO cell lines and analyzed cytochrome c release in response to intrinsic apoptotic signals. Immunofluorescence microscopy and cell fractionation revealed that cytochrome c was efficiently released into the cytoplasm in wild-type cells and Fis1-KO cells upon Act D treatment, and the release was only weakly affected in Mff-KO cells (Fig. 4, B and D). In contrast, MiD49-, MiD51-, MiD49/51-, and Drp1-KO cells strongly resisted the cytochrome c release reaction (Fig. 4, B and D). Bax/Bak activation and MOM permeabilization, as examined by the release of the IMS-soluble protein Smac/DIABLO (Fig. 4 C), Bax/Bak recruitment to the MOM (Fig. 4 D and Fig. S2, B and C), and their oligomerization (Fig. 4 E) occurred in all of these cell lines with similar kinetics up to the maximum levels (Fig. 4, C–E; and Fig. S2). Thus, the Drp1-MiD49/51 fission system acts downstream of the Bax/Bak-activated mitochondrial outer membrane permeabilization (MOMP). A comparable cellular response was also observed in H2O2-induced apoptosis (unpublished data). Unexpectively, Bax activation occurred even in Drp1-KO cells (Fig. 4 C and Fig. S2, B and C) in contrast to the previous notion that Bax oligomerization proceeds through the Drp1-induced mitochondrial membrane hemifission/hemifusion intermediates during apoptosis (Montessuit et al., 2010), suggesting in our case that Bax activation can proceed in the absence of Drp1-induced mitochondrial membrane constrictions. However, we cannot strictly rule out the possibility that inefficient recruitment of activated Bax/Bak to the MiD49/51-dependent mitochondrial fission sites causes defects of cristae remodeling for cytochrome c release in MiD49/51-KO and Drp1-KO cells where MiD proteins failed to form foci and diffusely localized throughout the MOM.
Shore and collaborators have previously demonstrated with a dominant-negative Drp1 mutant and an inhibitor of mitochondrial permeability transition pore (MPTP; permeable to molecules <1,500 D) that ER-BIK initiates Drp1-regulated mitochondrial cristae remodeling and MPTP opening to apoptosis through ER Ca2+-influx to mitochondria (Germain et al., 2005). We therefore investigated the involvement of MPTP in the Drp1-MiD49/51-dependent cristae remodeling. Calcium ionophore ionomycin induced MPTP opening to stimulate the release of calcein trapped within mitochondria in both wild-type and MiD49/51-KO cells (Fig. S3, A and B). As expected, MPTP opening stimulated tBid-induced release of cytochrome c in wild-type cells (Fig. S3, C and D). However, it failed to resume cytochrome c release in tBiD-induced MiD49/51-KO cells (Fig. S3, C and D). Together, these data indicate that intact cristae structure is maintained in these KO cells despite Ca2+-dependent MPTP opening. Thus, the Drp1-MiD49/51 fission system acts downstream or independently of the Ca2+-dependent MPTP opening during intrinsic apoptosis, although whether Ca2+-dependent MPTP opening is involved in apoptotic cytochrome c release is controversial, because mice deficient in cyclophilin D, a component of MPTP, had a normal response to various stimuli including staurosporine or etoposide (Baines et al., 2005; Nakagawa et al., 2005). The role of MPTP opening in apoptotic cristae remodeling needs to be further determined.
Next, we addressed the structural requirements of MiD51 for the restoration of Act D–induced cytochrome c release in MiD51-KO cells. First, a MiD51-GFP fusion construct lacking the Drp1-recruiting segment MiD51ΔPEYFP (Losón et al., 2014; Richter et al., 2014) was stably expressed in MiD51-KO cells. Wild-type MID51-GFP assembled into foci on the MOM together with Drp1 and restored cytochrome c release (Fig. 4, F and G; and Fig. S4 A). In contrast, mitochondria-targeted MiD51ΔPEYFP did not assemble into foci with Drp1, distributed throughout mitochondrial tubules, and failed to restore cytochrome c release (Fig. 4, F and G; and Fig. S4 A). Second, we examined the requirement of MiD51’s N-SA for cytochrome c release. The N-SA was deleted, and the MOM-targeted C-SA of Omp25 (Yagita et al., 2013) was ligated to the C-terminal segment (FLAG-MiD51ΔN-Mt). When expressed in MiD51-KO cells, it was diffusely distributed throughout the MOM very much like GFP-Omp25 (unpublished data) and successfully recruited Drp1 (Fig. S4 B), but it failed to restore the Act D–dependent cytochrome c release (Fig. 4, H and I). Together, these results suggest that the N-SA of MiD51 is required for correct positioning of the Drp1-MiD51 complex in the vicinity of the cristae remodeling system in response to intrinsic apoptotic signals, regulating cristae junction disruption to release cytochrome c (see following paragraph).
Loss of MiD49 and MiD51 prevents cristae remodeling during intrinsic apoptosis
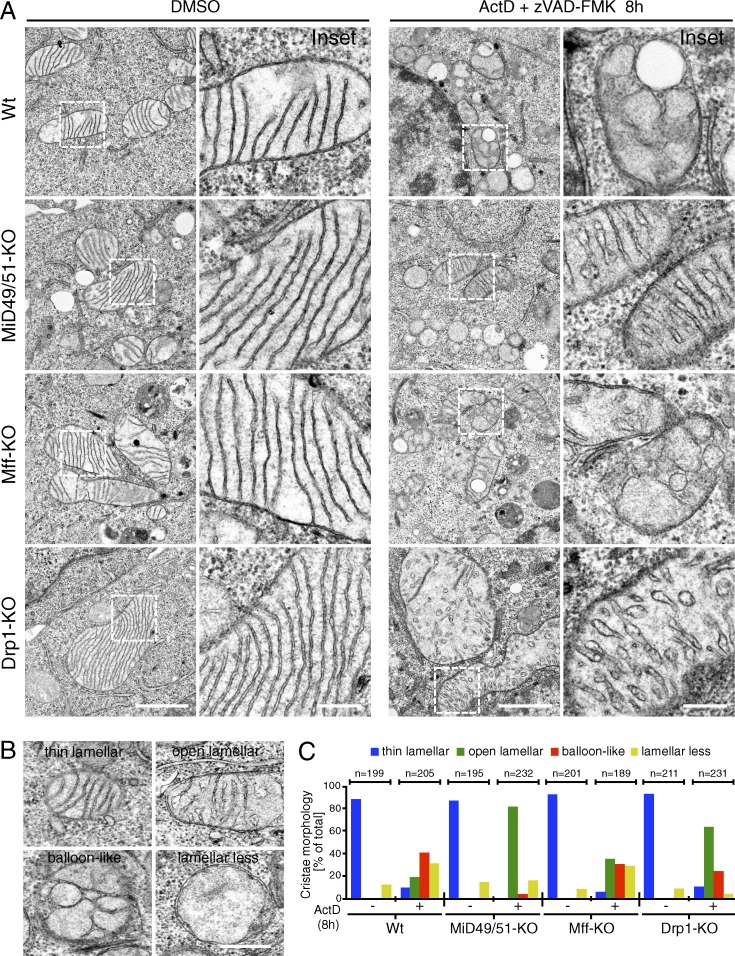
Intrinsic apoptotic signals induce mitochondrial cristae remodeling by disrupting oligomerized OPA1 to induce cytochrome c release from the cristae folds (Olichon et al., 2003; Frezza et al., 2006). Based on the results described in Fig. 4 B, we speculated that depletion of MiD proteins or Drp1 stabilized cristae structures against intrinsic apoptotic signals. To examine this, we first analyzed the mitochondrial structures in our cell lines by EM. Intact thin lamellar cristae structures connected by cristae junctions to the IMS were observed in MiD49/51-KO cells as well as in all other cell lines, and no significant structural differences were observed between these cells under nonstressed conditions (Fig. 5 A). In contrast, when the cells were treated with Act D for 8 h, both wild-type and Mff-KO cells displayed disorganized cristae structures in a large fraction of mitochondria, where cytochrome c release was observed in ∼50% of the cells (Figs. 4 B and 5 A). We evaluated the cristae morphology in these cells based on the categories shown in Fig. 5 B. Mitochondria with thin lamellar-shaped cristae disappeared and the number of aberrant mitochondria with balloon-like or lamella-less cristae dramatically increased in wild-type and Mff-KO cells (Fig. 5, A and C), although the morphological changes were rather moderate in Mff-KO cells. In marked contrast, a large fraction of mitochondria in the MiD49/51-KO and Drp1-KO cells still retained lamellar cristae structures upon apoptosis induction, although the thin lamellar cristae were slightly widened (open lamellae) after apoptosis induction compared with mock treatment (Fig. 5, A and C). These findings suggested that cristae disorganization to balloon-like or lamella-less and efficient cytochrome c release proceeded in parallel, whereas cristae stabilization evoked in MiD49/51-KO and Drp1-KO cells made the cells cytochrome c release resistant. Collectively, Drp1-MiD49/51–dependent fission pathways, but not the Mff-dependent pathway, are critical for cristae remodeling and complete cytochrome c release during apoptosis.
Figure 5.
Cristae remodeling during apoptosis is inhibited in MiD49/51-KO cells. (A) The indicated cells were treated with DMSO or Act D in the presence of zVAD-FMK for 8 h and analyzed by transmission EM. Magnified images in the boxed regions are shown as insets. Bars: 1 µm; (magnification) 200 nm. (B) Representative cristae morphology obtained by EM. Bar, 0.5 µm. (C) Quantification of cristae morphology shown in A. The indicated numbers of mitochondria were counted. Wt, wild type.
Disorganization of cristae by depletion of OPA1, PHB2, or ROMO1 restores cytochrome c release in MiD49/51-KO cells during apoptosis
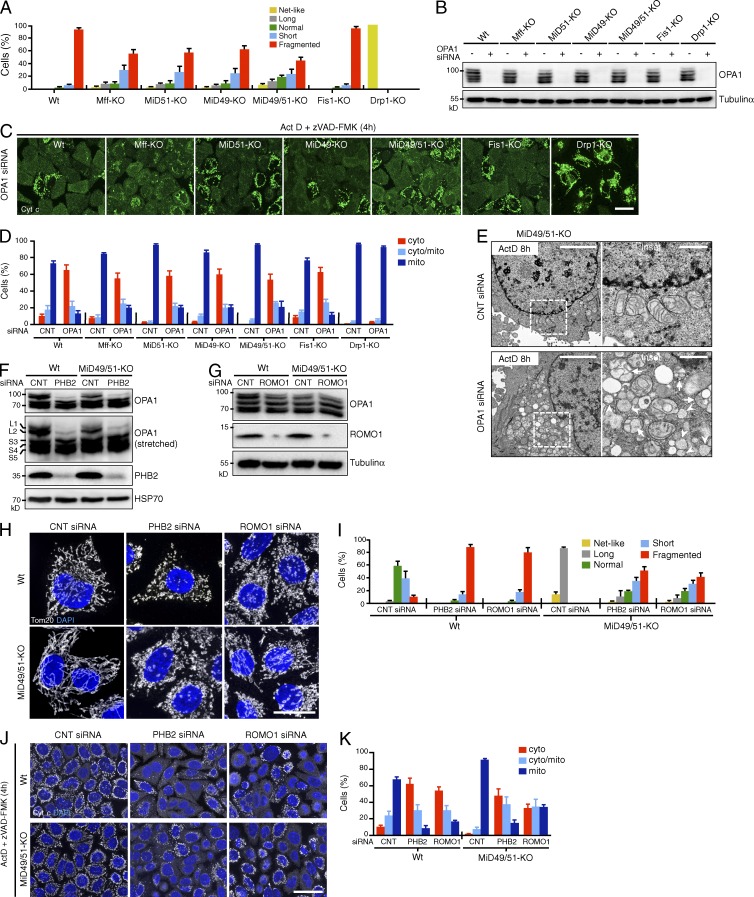
Depletion of OPA1 triggers cristae remodeling, leading to the complete release of cytochrome c upon the induction of apoptosis. OPA1 knockdown caused extensive mitochondrial fragmentation in all of our cell lines except Drp1-KO cells (Fig. 6 A); mitochondria in Drp1-KO cells completely resisted fragmentation against OPA1 depletion, indicating that Drp1 is involved in OPA1 knockdown–induced mitochondrial fragmentation (Fig. 6, A and B). Moreover, OPA1 down-regulation efficiently stimulated cytochrome c release upon Act D treatment (4 h) in all the cell lines except the Drp1-KO cells (Fig. 6, C and D). Of note, OPA1 down-regulation restored Act D–induced cytochrome c release in otherwise resistant MiD49/51-KO cells, and, accordingly, EM analysis revealed an increase in mitochondria with a balloon-like or cristae-less morphology in the cells (Fig. 6 E). Drp1-KO cells completely resisted Act D–induced cytochrome c release even after OPA1 depletion (Fig. 6, C and D), confirming our previous observations (Otera et al., 2010) and clearly indicating that Drp1-dependent mitochondrial fission is essential. These observations apparently correspond with previous observations in yeast that disruption of the cristae by mutation in Mgm1p (yeast homolog of OPA1) could be suppressed by mutations in Dnm1 (yeast homolog of Drp1; Sesaki et al., 2003). Together, these results clearly indicated that MOMP, cristae disorganization, and mitochondrial fission are essential as distinct steps of cytochrome c release.
Figure 6.
Down-regulation of OPA1, PHB2, or ROMO1 disrupts cristae morphology and restores the release of cytochrome c in MiD49/51-KO cells during apoptosis. (A–D) The indicated cells were transfected with control or OPA1 siRNA. (A) Quantified mitochondrial morphology. (B) Effect of OPA1 RNA interference as detected by Western blotting. (C) The cells in A were treated with Act D and immunostained with anti–cytochrome c antibodies. Bar, 20 µm. (D) Control and OPA1-siRNA cells were treated with Act D for 4 h and intracellular cytochrome c was quantified. (E) MiD49/51-KO cells were transfected as indicated. After 96 h, cells were treated with Act D in the presence of zVAD-FMK for 8 h and analyzed by transmission EM. Magnified images in the boxed regions are shown as insets. Bars: 4 µm; (magnification) 1 µm. Arrows, mitochondria with balloon-like structure; arrowheads, cristae-less mitochondria. (F and G) Effect of PHB2 (F) or ROMO1 (G) knockdown on OPA1 isoform patterns and the indicated proteins in the indicated cells as detected by Western blotting using the indicated antibodies. (H) Wild-type or MiD49/51-KO cells were transfected with control, PHB2, or ROMO1 siRNA. Confocal images were obtained by immunostaining for Tom20. Nuclei were stained with DAPI. Bar, 20 µm. (I) Percentages of the indicated mitochondrial morphologies of wild-type or MiD49/51-KO cells (n = 300) transfected with the indicated siRNA. (J) Wild-type or MiD49/51-KO cells were transfected as indicated. After 96 h, Act D–induced cytochrome c release was analyzed as in C. Nuclei were stained with DAPI. Bar, 30 µm. (K) Quantified cytochrome c release in J. Percentages in A, D, I, and K are means of three independent experiments with 300 cells per data point. n = 3. Data represent mean ± SD. Wt, wild type.
Because PHB1, PHB2, and ROMO1 also regulate cristae organization through OPA1 (Merkwirth et al., 2008; Norton et al., 2014), we examined whether knockdown of PHB2 or ROMO1 restores Act D–induced cytochrome c release in MiD49/51-KO cells. We first examined if these treatments affect the OPA1 processing pattern as processing of L-OPA1 to the S-isoform is thought to be tightly coupled with cristae remodeling, leading to cytochrome c release (Olichon et al., 2003; Jiang et al., 2014). In HeLa cells, five OPA1 bands were detected by Western blotting with anti-OPA1 antibodies (Fig. 6 F): two long membrane-bound isoforms, L1 and L2, and three short membrane-free isoforms, S3, S4, and S5 (Ishihara et al., 2006; Anand et al., 2014). As expected, knockdown of PHB2 stimulated the conversion of L1 and L2 to S-isoforms in wild-type and MiD49/51-KO cells concomitant with extensive mitochondrial fragmentation (Fig. 6, F, H, and I), confirming previous results in PHB2−/− MEFs (Merkwirth et al., 2008). In contrast, no such changes were induced by ROMO1 depletion in either wild-type or MiD49/51-KO cells (Fig. 6 G), although this manipulation induced extensive mitochondrial fragmentation (Fig. 6, H and I). Furthermore, depletion of PHB2 or ROMO1 accelerated cytochrome c release (Fig. 6, J and K) and overall cell death upon apoptosis induction (unpublished data) not only in wild-type cells but also in MiD49/51-KO cells, suggesting that the intramitochondrial L- to S-OPA1 balance is not always directly linked to cytochrome c release through cristae remodeling.
Interestingly, and contrary to previous studies, depletion of Mic60/Mitofilin (>90%), which is a central component of the mitochondria contact site (MICOS) complex and reported to be important for cristae morphology (John et al., 2005; Yang et al., 2012), neither induced mitochondrial fragmentation nor restored the Act D–induced cytochrome c release in MiD49/51-KO cells (Fig. S5, A–D). Supporting these findings, overall cell death and caspase-3 activation were also insensitive to Mic60/Mitofilin-depletion (Fig. S5, E and F). Considering that PHB2, OPA1, and ROMO1 are not found within the seven components of MICOS complex conserved in mammals (Guarani et al., 2015; note that ROMO1 associates with, but does not affect the integrity of, MICOS [Norton et al., 2014]), the components involved in the Drp1-dependent regulation of cristae remodeling might be functionally distinct from the MICOS complex.
Processing of L-OPA1 to S-OPA1 is insufficient for cristae remodeling and cytochrome c release at the early stage of apoptosis
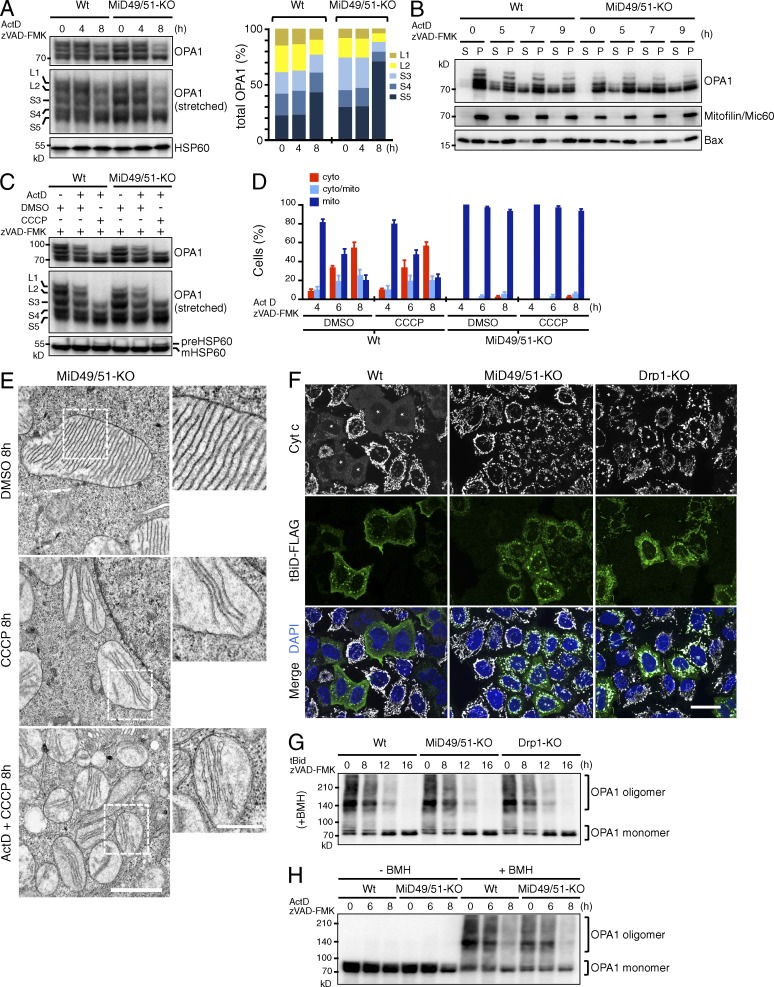
Because stimulation of processing of L-OPA1 to S-isoforms and extramitochondrial release of S-isoforms are thought to be linked to the feasibility of cytochrome c release by cristae remodeling, we examined whether activation of L-OPA1 processing and subsequent release of S-OPA1 occur in MiD49/51-KO as well as in wild-type cells at an early stage of apoptosis. When wild-type and MiD49/51-KO cells were treated with Act D, L-OPA1 (L1 and L2) processing in both cells occurred in similar kinetics (Fig. 7 A), suggesting that the increased processing of L-OPA1 to the S-isoforms is not directly related to cristae stabilization in MiD49/51-KO cells. We noted that loss of MiD49/51 led to an imbalance in OPA1 isoform abundance that favored the accumulation of isoform S3 and S5. Upon induction of apoptosis, MiD49/51-KO cells released substantial amounts of S-OPA1 into the cytosol with similar kinetics as wild-type cells (Fig. 7 B), also suggesting that cytosolic release of S-OPA1 alone does not explain the suppression of cytochrome c release in MiD49/51-KO cells.
Figure 7.
Disassembly of OPA1 oligomers does not limit cytochrome c release through cristae remodeling during apoptosis in MiD49/51-KO cells. (A) Cells were treated with Act D for the indicated periods and endogenous OPA1 patterns were analyzed by Western blotting. Quantitation of isoform band intensity is shown in right figure. (B) Cells treated with Act D as indicated were fractionated into cytosolic (S) and membrane (P) fractions. They were subjected to SDS-PAGE; and endogenous OPA1, Bax as an apoptotic MOM marker, and mitofilin were detected by Western blotting. (C) The indicated cells were treated with Act D for 8 h with or without CCCP in the indicated combinations and OPA1 patterns were analyzed by Western blotting. (D) The cells were treated under the indicated conditions, and intracellular localization of cytochrome c was analyzed. The percentage is means of three independent experiments with 300 cells per data point. n = 3. Data represent as mean ± SD. (E) MiD49/51-KO cells were treated as indicated and mitochondrial morphology was analyzed by transmission EM. Magnified images in the boxed regions are shown as insets. Bars: 1 µm; (magnification) 500 nm. (F) The cells were transfected with FLAG-tagged tBid in the presence of zVAD-FMK. After 16 h, they were subjected to confocal immunofluorescence microscopy with anti–cytochrome c and anti-FLAG antibodies. Nuclei were stained with DAPI. Asterisks, FLAG-tBid–expressing cells. Bar, 20 µm. (G and H) The cells were transfected with tBid-FLAG as in F (G) or treated with Act D (H) for the indicated time points, and OPA1 oligomers were analyzed by Western blotting after treatment with the cross-linker 1,6-bismaleimidohexane. Wt, wild type.
Because CCCP treatment of cells stimulates rapid processing of L-OPA1 to the S-isoforms (Ishihara et al., 2006), we examined if enforced processing of L-OPA1 by CCCP would restore the Act D–induced cytochrome c release in MiD49/51-KO cells. Act D and CCCP treatment induced complete conversion of L-OPA1 to S-isoforms (Fig. 7 C). This treatment, however, neither stimulated cytochrome c release in wild-type cells nor restored the cytochrome c release in MiD49/51-KO cells (Fig. 7 D). Thus, simple conversion of L-OPA1 to the S-OPA1 isoforms and cytosolic release of S-OPA1 isoforms did not induce cristae remodeling to induce cytochrome c release in MiD49/51-KO cells. Consistent with these findings, EM observations revealed that treatment of MiD49/51-KO cells with CCCP or with Act D and CCCP did not generate aberrant mitochondria with a balloon-like or cristae-less morphology (Fig. 7 E), but they still maintained thin lamellar cristae similar to those in untreated cells, although they frequently lost cristae junctions and were released into the matrix as thin lamellar membrane stacks (Fig. 7 E). We concluded that Drp1-dependent mitochondrial fission through MiD49 and MiD51 regulates cristae remodeling, but a simple stimulation of L-OPA1 processing does not or only weakly contributes to cytochrome c release under conditions in which other cristae organizing components such as prohibitins are maintained at normal levels; conditions distinct from those reported in PHB2−/− cells (Merkwirth et al., 2008).
Because tBid induces rapid cristae remodeling through the disassembly of OPA1 oligomers and subsequent cytochrome c release both in wild-type cells and in vitro (Frezza et al., 2006; Jiang et al., 2014), we analyzed whether tBid expression induces cytochrome c release in MiD49/51-KO cells and Drp1-KO cells. In wild-type cells, tBid-FLAG was targeted to the mitochondria and induced cytochrome c release (Fig. 7 F). In striking contrast, in MiD49/51-KO and Drp1-KO cells, tBid-FLAG failed to release cytochrome c, although it was correctly targeted to the mitochondria (Fig. 7 F, asterisk). We then analyzed the oligomeric states of OPA1 in these cells using a cell-permeable cross-linker, 1, 6-bismaleimidohexane (BMH), that is frequently used to detect OPA1 oligomer (Frezza et al., 2006; Jiang et al., 2014; Patten et al., 2014). Surprisingly, however, disassembly of OPA1 oligomers proceeded with similar kinetics in all these cell lines (Fig. 7 G). Similar OPA1 responses were also observed for wild-type and MiD49/51-KO cells during Act D–induced apoptosis (Fig. 7 H). This is in contrast to the current knowledge that disassembly of OPA1 oligomers is directly linked to cristae remodeling for cytochrome c release through the Bax/Bak-generated MOMP in wild-type cells. Altogether, Drp1-KO and MiD49/51-KO cells enabled detection, for the first time, of distinct steps of the cristae remodeling–coupled cytochrome c release reaction during intrinsic apoptosis.
Discussion
Taking advantage of mitochondrial fission components KO cell lines generated from the same genetic background, we could demonstrate for the first time that Drp1-dependent mitochondrial fission through MiD49 and MiD51, but not through Mff, plays an essential role in the progression of intrinsic apoptosis by regulating cristae remodeling. EM observations also revealed that MiD49/51-KO cells maintained tight cristae structures during the induction of intrinsic apoptosis and thus appeared to acquire resistance to cristae remodeling to preclude the release of cytochrome c. Most importantly, Drp1-KO cells also maintained tight cristae structures even after the induction of apoptosis. Together, these results indicated that the Drp1-dependent fission through MiD49/51 facilitates cristae remodeling required for cytochrome c release. The consensus has been that OPA1 oligomers have an important role in the regulation of cristae remodeling to release cytochrome c. In the present study, however, apoptosis induction or tBid expression induced disassembly of OPA1 oligomers in otherwise cristae remodeling– and cytochrome c release–resistant Drp1-KO and MiD49/51-KO cells. These cells allowed us to detect disassembly of OPA1 oligomers and cristae remodeling as separate events and further to identify Drp1-MiD49/51–dependent fission as the rate-limiting step of cristae remodeling during overall cytochrome c release.
We demonstrated that when FLAG-MiD51ΔN-Mt was expressed in MiD51-KO cells, it was correctly targeted to mitochondria and recruited Drp1 but failed to restore cytochrome c release upon Act D treatment. Similarly, when an MiD51 mutant in which the IMS-localized N-terminal 14–amino acid residues were deleted was expressed in MiD51-KO cells, it was targeted to the mitochondria but localized uniformly on the MOM and failed to restore cytochrome c release (unpublished data), suggesting that the N-SA of MiD51 may interact with the cristae remodeling system. Furthermore, the Drp1-binding site on MiD51 is also essential for the recovery of cytochrome c release. Thus, the N-SA of MiD51 is essential for propagating the Drp1 signal to the cristae remodeling system during intrinsic apoptosis; the N-SA of MiD51 might function as a spatiotemporal coupler to the cristae remodeling machinery in the presence of Drp1. The same is probably true for MiD49. Interestingly, in this relation, Langer’s group reported that S-OPA1 promotes mitochondrial fission and the GTPase domain mutant S-OPA1K301A assembles into punctate structures partially colocalizing with the MAM, mitochondrial constrictions, and foci containing both Drp1 and MiD49 (Anand et al., 2014). Thus, Drp1-dependent mitochondrial fission at the MiD49/51-containing foci in the vicinity of cristae junctions may couple with its remodeling system and release cytochrome c upon the induction of intrinsic apoptosis.
Our findings revealed that simple conversion of L-OPA1 to S-isoforms is not sufficient for cristae remodeling at the early stage of apoptosis. In this relation, however, Merkwirth et al. (2008) demonstrated that depletion of PHB1/2 induced complete conversion of L-OPA1 to the S-isoforms concomitant with cristae disorganization, and these defective phenotypes were rescued by the expression of an uncleavable mutant of L-OPA1. This apparent discrepancy could be reconciled considering that the substrate amounts of L-OPA1 containing oligomers could substitute for the loss of PHB1/2 complexes with a scaffolding function, and vice versa, our present experiments were performed under conditions in which the normal levels of PHB1/2 complexes were maintained. Another possibility might be that the remaining amounts of S-isoforms containing oligomers in our system maintained the cristae structures in cooperation with PHB1/2 complexes. In any case, the long-term absence of L-OPA1 in PHB2−/− cells might destabilize OPA1 oligomers that eventually lead to the loss of cristae structures.
It is known that efficient activation of Bax/Bak require mitochondrial sphingolipids (Chipuk et al., 2012) or cardiolipin (Lucken-Ardjomande et al., 2008; Montessuit et al., 2010). Because these events normally proceeded in MiD49/51-KO cell lines, the functional ER–mitochondria contacts required for these metabolism should be maintained. Similarly, Bax/Bak activation proceeded normally in Drp1-KO cells, although MiD proteins failed to form foci to diffuse throughout the MOM, also suggesting that MiD proteins might not be involved in maintaining the MAM integrity for the lipid translocation. In this context, stabilization of the ER–mitochondrial contacts by Bax/Bak-promoted Drp1 sumoylation has been reported (Wasiak et al., 2007; Prudent et al., 2015). The functional significance and mechanism of MAM localization of the Drp1-MiD49/51 fission system in intrinsic apoptosis remain to be solved.
Materials and methods
Antibodies and reagents
Commercial antibodies used in this study were rabbit anti-Mff (Proteintech), rabbit anti-MiD49 (Proteintech), rabbit anti-MiD51 (Proteintech), rabbit anti-Fis1 (Atlas antibodies), mouse anti-OPA1 (BD Biosciences), mouse anti-Mfn2 (XX-1; Santa Cruz Biotechnology), mouse anti-Mfn1 (Santa Cruz Biotechnology), mouse anti-HSP60 (Enzo Life Sciences), mouse anti-HSP70 (Thermo Fisher Scientific), rabbit anti-mitofilin/Mic60 (Origene), rabbit anti-MTP18 (Proteintech), mouse anti-Drp1 (BD Biosciences), rabbit anti-PMP70 (Invitrogen), rabbit anti-Tubulinα (Invitrogen), rat anti-HA (clone 3F10; Roche), rabbit anti-Tom20 (sc-11415; Santa Cruz Biotechnology), rabbit anti-FLAG (Sigma-Aldrich), mouse anti–cytochrome c (BD Biosciences), rabbit anti Bax (Upstate), mouse anti-Bax 6A7 (Enzo Life Sciences), mouse anti-Bak Ab-1 (Oncogene), rabbit anti-Bak NT (Merck Millipore), rabbit anti-ROMO1 (Sigma-Aldrich), rabbit anti-PHB2 (Proteintech), rabbit anti-cleaved caspase-3 (Cell Signaling), mouse anti-Fas (MBL), and rabbit anti-Smac/DIABLO (ProSci). Act D (Sigma-Aldrich) was used at a final concentration of 10 µM. zVAD-FMK (Peptide Institute) was used at a final concentration of 100 µM. CCCP (Sigma-Aldrich) was used at a final concentration of 20 µM. Antimycin A (Sigma-Aldrich) was used at a final concentration of 50 µg/ml. Valinomycin was used at a final concentration of 20 µM. Oligomycin was used at a final concentration of 10 µM. MitoTracker Red CMXRos (Molecular Probes) was used at a final concentration of 20 nM. BMH cross-linker was obtained from Thermo Fisher Scientific.
Construction of plasmids
MiD51 cDNA was obtained with RT-PCR using total RNA isolated from HeLa cells. The PCR product was cloned into the AflIII and NotI sites of pIRESneo3 vector (Clontech) and sequenced to role out mutations introduced by PCR. A FLAG-tag sequence was inserted by PCR at the 3′ end of the sequence. MiD51-GFP and MiD51ΔPEYFP-GFP were PCR amplified and cloned into the AflIII and NotI sites of pIRESneo3 vector. HA-Mff was PCR amplified and cloned into the AflIII and EcoRI sites of pIRESneo3 vector.
Construction of guide RNA (gRNA) expression vectors
Bicistronic expression vectors expressing Cas9 and gRNA were obtained from System Biosciences. A pair of gRNA oligonucleotides for each targeting site was annealed, ligated to linearized vector, and transformed into DH5α. The sequences of the gRNAs were verified by sequencing analysis.
gRNA target sequences
To generate KO cell lines, the following sequences were used as the target gRNAs in this study: MiD51, 5′-ACTTTGTGCTCTCCAATGCCCGG-3′; MiD49, 5′-TGTGCTGGGCATTGCCACCCTGG-3′; Mff, 5′-AGCACCGCCAAACGCTGACCTGG-3′; Drp1, 5′-AAATCAGAGAGCTCATTCTTCGG-3′; and Fis1, 5′-GCACGCAGTTTGAGTACGCCTGG-3′. These sequences are found in all predicted splice variants of each gene.
Cell culture and construction of knockout HeLa cell lines
HeLa cells were cultured in DMEM supplemented with 10% FBS under 5% CO2, 95% air. To generate the clonal KO cell line, HeLa cells were transfected with bicistronic expression vectors expressing Cas9 and gRNA for MiD51, MiD49, Mff, Drp1, and Fis1. Single clones were generated by limited dilution after transfection. Clonal populations were expanded and screened by SDS-PAGE and immunoblotting with specific antibodies. MiD49/51-KO cells and Mff/Fis1-KO cells were generated from MiD51-KO cells and Mff-KO cells, respectively. DNA transfection was performed using FuGENE HD (Roche) according to the manufacturer’s instructions. All stable cell lines were isolated by selection in the presence of 500 µg/ml G418. Single clones were generated by limited dilution.
RNAi
Oligonucleotide for siRNA for OPA1 was made by QIAGEN. The pair of OPA1 siRNA was based on the sequence 5′-AACACGUUUUAACCUUGAAAC-3′. For knockdown of PHB2 (M-018703-00-0005; sequences 5′-GCACGGCCCUGAAGCUGUU-3′, 5′-CAUCAAACUUCGCAAGAUU-3′, 5′-CAGAAUAUCUCCAAGACGA-3′, and 5′-CCACAUCACAGAAUCGUAU-3′), ROMO1 (M-015268-02-0005; sequences 5′-GGGAAAACCAUGAUGCAGA-3′, 5′-GCACAUUCAUGGCCAUUGG-3′, 5′-CUGCUUCGACCGUGUCAAA-3′, and 5′-CAGCCCAUGUACUAAUAAA-3′), mitofilin/Mic60 (M-019832-01-0005; 5′-GCACAUCGUCGUAUUGAUC-3′, 5′-GGCCAAGCCUCAUAUAACU-3′, 5′-GGGAUGACUUUAAACGAGA-3′, and 5′-GCGAGAUGUCCUUAGGGUA-3′), and control (D-001206-13-05; sequences 5′-AUGAACGUGAAUUGCUCAA-3′, 5′-UAAGGCUAUGAAGAGAUAC-3′, 5′-AUGUAUUGGCCUGUAUUAG-3′, and 5′-UAGCGACUAAACACAUCAA-3′), specific siRNAs were purchased from Dharmacon. Cells were transfected with siRNA duplexes at a concentration of 75 pM by Lipofectamine 2000 (Invitrogen). At 36 h after the initial transfection, a second siRNA transfection was performed was performed, and cells were grown for 60 h. At 96 h after initial transfection, cells were subjected to assays.
Immunofluorescence microscopy
HeLa cells grown on glass coverslips were fixed at room temperature for 15 min with 4% paraformaldehyde, permeabilized by incubation with 0.1% Triton X-100 for 5 min, and incubated with primary antibodies for 2 h. After washing with PBS, cells were incubated with Alexa-conjugated secondary antibodies (Invitrogen) for 1 h and washed with PBS. Cell nuclei were counterstained and mounted with a mounting medium with DAPI (Vectashield; Vector Laboratories). Confocal images were collected at room temperature using an inverted structured illumination microscope (ApoTome with Axio Observer.Z1; Carl Zeiss) with a 63×/1.4 NA oil differential interference contrast Plan-Apochromat objective, a digital CCD camera (AxioCam MRm; Carl Zeiss), and Zeiss ZEN 2010 acquisition software. Brightness and contrast of images were adjusted in Photoshop CS6 (Adobe). All images for a given experiment were collected and adjusted in an identical manner. The fluorescence intensity was measured using ImageJ software.
Immunoblotting
Protein samples were separated by SDS-PAGE and electrotransferred to a polyvinylidene fluoride membrane (Bio-Rad Laboratories). After blocking in PBS containing 0.3% nonfat dry milk and 0.1% Tween 20, blots were probed with appropriate primary and AP-conjugated secondary antibodies, developed with Immun-Star AP Western blotting detection reagents (Bio-Rad Laboratories). Blots were visualized using a LAS-1000 (FUJIFILM). The intensity of protein bands was analyzed using the Image Gauge Ver. 3.3 (FUJIFILM).
Cell fractionation
Cells were washed with PBS and collected by centrifugation at 800 g for 5 min. The cells were washed once with homogenization buffer (10 mM Hepes-KOH, pH 7.5, and 0.25 M sucrose), homogenized in 1 ml of homogenization buffer by passing through a 27G needle 10 times, and then centrifuged at 1,000 g for 10 min to obtain a postnuclear supernatant. The postnuclear supernatant was centrifuged at 8,000 g for 10 min to obtain a mitochondria-rich heavy membrane fraction. The resultant supernatant was further centrifuged at 100,000 g for 15 min to obtain the cytosol fraction.
Transmission EM
Cells were fixed with 2% PFA and 2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, for 30 min at 4°C. Thereafter, cells were fixed with glutaraldehyde in 0.1 M cacodylate buffer at 4°C overnight. The samples were additionally fixed with 0.5% tannic acid in 0.1 M cacodylate buffer, pH 7.4, at 4°C for 1 h. After the fixation, the samples were washed four times with 0.1 M cacodylate buffer for 30 min each and postfixed with 2% osmium tetroxide in 0.1 M cacodylate buffer at 4°C for 1 h. Fixed samples were dehydrated, embedded in Questol-812 (Nisshin EM), and polymerized at 60°C for 48 h. The blocks were ultrathin-sectioned at 70 nm with a diamond knife using an ultramicrotome (Leica). Sections were placed on copper grids and stained with 2% uranyl acetate at room temperature for 15 min and then rinsed with distilled water, followed by secondary staining with lead stain solution (Sigma-Aldrich) at room temperature for 3 min. The grids were observed under a JEM-1400Plus (JEOL) transmission electron microscope at an acceleration voltage of 80 kV.
Chemical cross-linking
OPA1 oligomers were analyzed by SDS-PAGE after cross-linking. In brief, cells were treated with 50 µM 1,6-bismaleimideohexane (BMH) at 30°C for 30 min. Cells were washed with phosphate-buffered saline containing 5 mM dithiothreitol, lysed, and subjected to SDS-PAGE followed by Western blotting for OPA1. Membrane fraction isolated from cells treated with Act D were treated with 2 mM BMH at 30°C for 30 min. Samples were lysed and subjected to SDS-PAGE followed by Western blotting for Bax or Bak.
Calcein-AM to monitor MPTP
HeLa cells were incubated with 2 µM calcein-AM (Dojindo) for 60 min at 37°C. Plasma membrane of calcein-AM–loaded HeLa cells were permeabilized with 25 µg/ml digitonin diluted in buffer A (20 mM Hepes-KOH, pH 7.4, 25 mM KCl, 2.5 mM Mg(OAc)2, 0.25 M sucrose, and 1 µM Taxol) to allow for leakage of the cytosolic calcein signal.
Cytochrome c and Smac/DIABLO release in semi-intact cells
The preparation of semi-intact cells was performed essentially as described previously (Setoguchi et al., 2006). In brief, HeLa cells on glass coverslips were incubated at 30°C for 5 min with buffer A containing 25 µg/ml digitonin and gently washed three times with the same buffer. The semi-intact cells were then incubated for 60 min at 30°C with 75 nM recombinant tBid (R&D Systems) with or without 20 µM ionomycin (Wako) in buffer A to induce cytochrome c and Smac/DIABLO release. The semi-intact cells were fixed with 4% PFA, completely permeabilized by incubation with 0.1% Triton X-100 for 5 min, and subjected to immunofluorescence analysis with antibodies to cytochrome c or Smac/DIABLO.
Trypan blue stain
Cells were harvested and resuspended in DMEM medium at a concentration 105 cells/ml. Cell death was assessed using trypan blue staining. 10 µl 0.05% trypan blue (Invitrogen) was mixed with 10 µl cell suspension, spread onto a Neubauer chamber, and covered with a coverslip. Although viable cells exclude the dye and appear translucent, nonviable cells appear blue-stained. Viability and mortality of three independent experiments were quantified.
Statistical analysis
The results of all quantitative experiments are reported as mean ± SD of three independent experiments.
Online supplemental material
Fig. S1 shows morphological response of mitochondrial fission-related protein-KO HeLa cell lines to various detrimental stressors. Fig. S2 shows apoptotic response of mitochondrial fission-related protein-KO HeLa cell lines to Act D. Fig. S3 shows that MiD49/51-KO cells resist tBiD-induced cytochrome c release even after MPTP opening. Fig. S4 shows subcellular localization and Drp1-recruiting activity of MiD51 mutants. Fig. S5 shows that down-regulation of mitofilin/Mic60 does not affect mitochondrial morphology, cytochrome c release, and apoptosis in MiD49/51-KO cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201508099/DC1.
Supplementary Material
Acknowledgments
We are grateful to Drs. M.T. Ryan, N. Ishihara, T. Rapoport, and A.M. Colberg-Poley for the gift of MiD51-GFP∆PEYFP cDNA, gRNA expression plasmid for Drp1-KO, RFP-Sec61β expression plasmid, and vMIA-GFP expression plasmid, respectively.
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants-in-aid for Scientific Research 25440088 (H. Otera), 25291046 (K. Mihara), and 22370071 (K. Mihara) and Challenging Exploratory Research 24657138 (K. Mihara); and a grant from the Takeda Science Foundation (H. Otera), the Inamori Foundation (H. Otera), the Uehara Memorial Foundation (H. Otera), and the Kanae Foundation for the Promotion of Medical Science (H. Otera).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Act D
- actinomycin D
- BMH
- 1,6-bismaleimidohexane
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- CHX
- cycloheximide
- C-SA
- C-terminal signal anchor
- IMS
- intermembrane space
- KO
- knockout
- MAM
- mitochondria-associated endoplasmic reticulum membrane
- MICOS
- mitochondria contact site
- MIM
- mitochondrial inner membrane
- MOM
- mitochondrial outer membrane
- MOMP
- mitochondrial outer membrane permeabilization
- MPTP
- mitochondrial permeability transition pore
- N-SA
- N-terminal signal anchor
References
- Anand R., Wai T., Baker M.J., Kladt N., Schauss A.C., Rugarli E., and Langer T.. 2014. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204:919–929. 10.1083/jcb.201308006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines C.P., Kaiser R.A., Purcell N.H., Blair N.S., Osinska H., Hambleton M.A., Brunskill E.W., Sayen M.R., Gottlieb R.A., Dorn G.W., et al. 2005. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 434:658–662. 10.1038/nature03434 [DOI] [PubMed] [Google Scholar]
- Bhuvanendran S., Salka K., Rainey K., Sreetama S.C., Williams E., Leeker M., Prasad V., Boyd J., Patterson G.H., Jaiswal J.K., and Colberg-Poley A.M.. 2014. Superresolution imaging of human cytomegalovirus vMIA localization in sub-mitochondrial compartments. Viruses. 6:1612–1636. 10.3390/v6041612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk J.E., McStay G.P., Bharti A., Kuwana T., Clarke C.J., Siskind L.J., Obeid L.M., and Green D.R.. 2012. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell. 148:988–1000. 10.1016/j.cell.2012.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer S.A., and Chan D.C.. 2007. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8:870–879. 10.1038/nrm2275 [DOI] [PubMed] [Google Scholar]
- Elgass K.D., Smith E.A., LeGros M.A., Larabell C.A., and Ryan M.T.. 2015. Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J. Cell Sci. 128:2795–2804. 10.1242/jcs.169136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L., and Youle R.J.. 2001. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 1:515–525. 10.1016/S1534-5807(01)00055-7 [DOI] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., and Scorrano L.. 2006. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 126:177–189. 10.1016/j.cell.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S., and van der Bliek A.M.. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19:2402–2412. 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M., Mathai J.P., McBride H.M., and Shore G.C.. 2005. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. EMBO J. 24:1546–1556. 10.1038/sj.emboj.7600592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarani V., McNeill E.M., Paulo J.A., Huttlin E.L., Fröhlich F., Gygi S.P., Van Vactor D., and Harper J.W.. 2015. QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. eLife. 4:1–23. 10.7554/eLife.06265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber N., Guimaraes S., Schrader M., Suter U., and Niemann A.. 2013. Charcot-Marie-Tooth disease-associated mutants of GDAP1 dissociate its roles in peroxisomal and mitochondrial fission. EMBO Rep. 14:545–552. 10.1038/embor.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., and Mihara K.. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25:2966–2977. 10.1038/sj.emboj.7601184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D.I., and Martinou J.C.. 2008. Mitochondrial dynamics and apoptosis: a painful separation. Dev. Cell. 15:341–343. 10.1016/j.devcel.2008.08.011 [DOI] [PubMed] [Google Scholar]
- Jiang X., Jiang H., Shen Z., and Wang X.. 2014. Activation of mitochondrial protease OMA1 by Bax and Bak promotes cytochrome c release during apoptosis. Proc. Natl. Acad. Sci. USA. 111:14782–14787. 10.1073/pnas.1417253111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John G.B., Shang Y., Li L., Renken C., Mannella C.A., Selker J.M., Rangell L., Bennett M.J., and Zha J.. 2005. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell. 16:1543–1554. 10.1091/mbc.E04-08-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S., Guo Q., Kalia R., Bui H.T., Eckert D.M., Frost A., and Shaw J.M.. 2013. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. USA. 110:E1342–E1351. 10.1073/pnas.1300855110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Yu R., Jin S.B., Han L., Lendahl U., Zhao J., and Nistér M.. 2013. The mitochondrial elongation factors MIEF1 and MIEF2 exert partially distinct functions in mitochondrial dynamics. Exp. Cell Res. 319:2893–2904. 10.1016/j.yexcr.2013.07.010 [DOI] [PubMed] [Google Scholar]
- Losón O.C., Song Z., Chen H., and Chan D.C.. 2013. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 24:659–667. 10.1091/mbc.E12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losón O.C., Liu R., Rome M.E., Meng S., Kaiser J.T., Shan S.O., and Chan D.C.. 2014. The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure. 22:367–377. 10.1016/j.str.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucken-Ardjomande S., Montessuit S., and Martinou J.C.. 2008. Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 15:929–937. 10.1038/cdd.2008.9 [DOI] [PubMed] [Google Scholar]
- Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F.T., von Kleist-Retzow J.C., Waisman A., Westermann B., and Langer T.. 2008. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 22:476–488. 10.1101/gad.460708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montessuit S., Somasekharan S.P., Terrones O., Lucken-Ardjomande S., Herzig S., Schwarzenbacher R., Manstein D.J., Bossy-Wetzel E., Basañez G., Meda P., and Martinou J.-C.. 2010. Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell. 142:889–901. 10.1016/j.cell.2010.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Shimizu S., Watanabe T., Yamaguchi O., Otsu K., Yamagata H., Inohara H., Kubo T., and Tsujimoto Y.. 2005. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 434:652–658. 10.1038/nature03317 [DOI] [PubMed] [Google Scholar]
- Norton M., Ng A.C., Baird S., Dumoulin A., Shutt T., Mah N., Andrade-Navarro M.A., McBride H.M., and Screaton R.A.. 2014. ROMO1 is an essential redox-dependent regulator of mitochondrial dynamics. Sci. Signal. 7:ra10 10.1126/scisignal.2004374 [DOI] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., and Lenaers G.. 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 278:7743–7746. 10.1074/jbc.C200677200 [DOI] [PubMed] [Google Scholar]
- Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., and Mihara K.. 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191:1141–1158. 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Osellame L.D., Laine D., Koutsopoulos O.S., Frazier A.E., and Ryan M.T.. 2011. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12:565–573. 10.1038/embor.2011.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Elgass K.D., Parton R.G., Osellame L.D., Stojanovski D., and Ryan M.T.. 2013. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J. Biol. Chem. 288:27584–27593. 10.1074/jbc.M113.479873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten D.A., Wong J., Khacho M., Soubannier V., Mailloux R.J., Pilon-Larose K., MacLaurin J.G., Park D.S., McBride H.M., Trinkle-Mulcahy L., et al. 2014. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 33:2676–2691. 10.15252/embj.201488349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent J., Zunino R., Sugiura A., Mattie S., Shore G.C., and McBride H.M.. 2015. MAPL SUMOylation of Drp1 Stabilizes an ER/Mitochondrial Platform Required for Cell Death. Mol. Cell. 59:941–955. 10.1016/j.molcel.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Richter V., Palmer C.S., Osellame L.D., Singh A.P., Elgass K., Stroud D.A., Sesaki H., Kvansakul M., and Ryan M.T.. 2014. Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. J. Cell Biol. 204:477–486. 10.1083/jcb.201311014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L. 2009. Opening the doors to cytochrome c: changes in mitochondrial shape and apoptosis. Int. J. Biochem. Cell Biol. 41:1875–1883. 10.1016/j.biocel.2009.04.016 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S.M., Yaffe M.P., and Jensen R.E.. 2003. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell. 14:2342–2356. 10.1091/mbc.E02-12-0788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi K., Otera H., and Mihara K.. 2006. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 25:5635–5647. 10.1038/sj.emboj.7601438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen D.F., Norris K.L., and Youle R.J.. 2008. Mitochondrial dynamics and apoptosis. Genes Dev. 22:1577–1590. 10.1101/gad.1658508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait S.W., and Green D.R.. 2010. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11:621–632. 10.1038/nrm2952 [DOI] [PubMed] [Google Scholar]
- Varanita T., Soriano M.E., Romanello V., Zaglia T., Quintana-Cabrera R., Semenzato M., Menabò R., Costa V., Civiletto G., Pesce P., et al. 2015. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab. 21:834–844. 10.1016/j.cmet.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak S., Zunino R., and McBride H.M.. 2007. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 177:439–450. 10.1083/jcb.200610042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita Y., Hiromasa T., and Fujiki Y.. 2013. Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J. Cell Biol. 200:651–666. 10.1083/jcb.201211077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R.F., Zhao G.W., Liang S.T., Zhang Y., Sun L.H., Chen H.Z., and Liu D.P.. 2012. Mitofilin regulates cytochrome c release during apoptosis by controlling mitochondrial cristae remodeling. Biochem. Biophys. Res. Commun. 428:93–98. 10.1016/j.bbrc.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlén P., Tomilin N., Shupliakov O., Lendahl U., and Nistér M.. 2011. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30:2762–2778. 10.1038/emboj.2011.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.