Abstract
During cytokinetic abscission, the endosomal sorting complex required for transport (ESCRT) proteins are recruited to the midbody and direct the severing of the intercellular bridge. In this issue, Christ et al. (2016. J. Cell Biol. http://dx.doi.org/10.1083/jcb.201507009) demonstrate that two separate but redundant pathways exist to recruit ESCRT-III proteins to the midbody.
Over the past 140 years, eukaryotic cell division has been extensively studied and is now understood to be an elaborate, tightly regulated set of events that culminates in the formation of two distinct daughter cells. The M phase of animal cells is characterized by a profound structural reorganization, regulated by a cohort of mitotic kinases and performed by mitosis-specific cytoskeletal structures, including the spindle apparatus and the cytokinetic midbody (Scholey et al., 2003). The completion of cytokinesis, called abscission, involves the severing of the intercellular bridge on both sides of the midbody. In 2007, two landmark studies demonstrated that several ESCRT proteins localize to the midbody and are required for the completion of cytokinesis (Carlton and Martin-Serrano, 2007; Morita et al., 2007). Aside from abscission, the ESCRTs participate in the formation of multivesicular endosomes (MVEs), function in plasma membrane repair, and participate in numerous other cellular processes (Katzmann et al., 2002; Morita and Sundquist, 2004; Hurley, 2015).
The canonical model for ESCRT function at MVEs involves the hierarchical recruitment of ESCRT proteins in four unique complexes: ESCRT-0 through ESCRT-III. The ESCRTs cluster cargos and deform membrane, and current models suggest that ESCRT-III subunits polymerize to form filaments that spiral down into the neck of a nascent intralumenal vesicle (Schuh and Audhya, 2014). With the assistance of the VPS4 AAA ATPase, ESCRT filaments are remodeled to facilitate vesicle fission (Shen et al., 2014). Though MVE maturation utilizes all four ESCRT complexes, cytokinetic abscission has been previously thought to require only ESCRT-I, ESCRT-III, and the ESCRT-associated ALG2-interacting factor ALIX (Morita et al., 2010). ALIX interacts with both the ESCRT-I protein TSG101 and all three ESCRT-III CHMP4 isoforms and has been postulated to act as an ESCRT-II bypass for linking ESCRT-I and ESCRT-III in abscission (Schuh and Audhya, 2014). However, the precise mechanism underlying the recruitment of the ESCRT-III complex to the midbody during cytokinesis has remained ambiguous.
In this issue, Christ et al. address how the ESCRT-III component CHMP4B (Vps32 in other metazoan systems) is recruited to the midbody and demonstrate the necessity of the ESCRT-II complex in this process. They observed that recruitment of CHMP4B to the midbody was abrogated when they codepleted ALIX and the ESCRT-I component TSG101 in cultured HeLa cells, but that CHMP4B did accumulate when only one of these components was depleted. These data indicate that CHMP4B can be recruited to the midbody via TSG101 or ALIX, but that the two proteins are unlikely to perform this function as a complex, suggesting that CHMP4B recruitment to the midbody involves two independent pathways.
After immunofluorescence staining of fixed cells, Christ et al. (2016) found that the endogenous ESCRT-III protein CHMP6 and the ESCRT-II protein EAP20 (VPS20 and VPS25 in other systems, respectively) localize to the midbody, consistent with a previous overexpression study (Thoresen et al., 2014). They additionally performed several depletion experiments to establish that ESCRT-II recruits CHMP6 without affecting TSG101 localization, demonstrating that CHMP6 acts downstream of ESCRT-I and ESCRT-II. This shows that ESCRT-I recruits ESCRT-III to the cytokinetic midbody the same way it does at the MVE.
Christ et al. (2016) also show that CHMP4B can still be recruited normally when the ESCRT-II component EAP30 (VPS22 in other systems) is depleted, but not when EAP30 is codepleted with ALIX, strongly suggesting that ALIX-dependent accumulation of CHMP4B does not involve CHMP6 and, more generally, that there are two pathways that can each recruit CHMP4B to the midbody: an ESCRT-I–ESCRT-II–CHMP6 pathway and an ALIX-dependent pathway. It will be important for future work to consider the partial redundancy between these two pathways when assaying the dispensability of early acting ESCRT complexes in cellular processes.
In addition, Christ et al. (2016) observed that ALIX depletion led to furrow regression and binucleation in dividing cells with chromatin spanning the intercellular bridge, the same phenotype observed in cells expressing a CHMP4C construct lacking the ALIX interaction domain. Further, they showed that CHMP4C localization to the midbody is abrogated after ALIX depletion but is unaffected by TSG101 knockdowns, strongly implicating ALIX in CHMP4C recruitment independently of ESCRT-I.
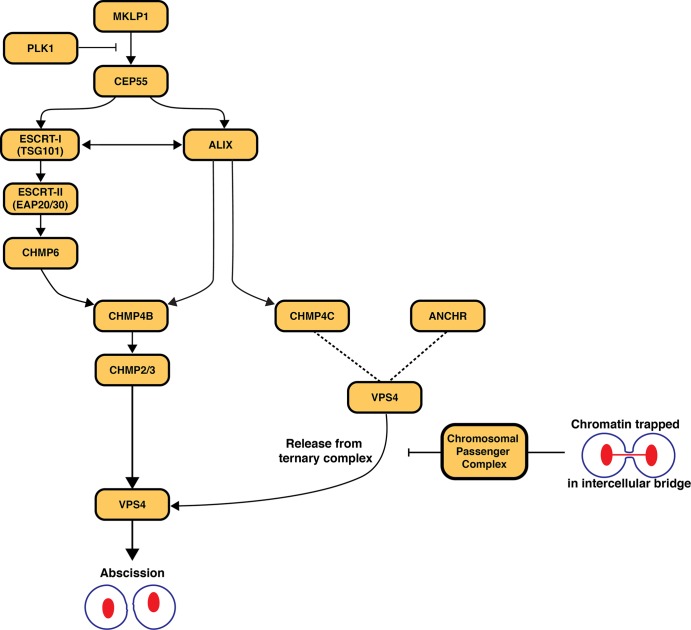
Our overall understanding of the regulation of abscission still remains elementary (Fig. 1). In addition to the roles of the ESCRT machinery, the chromosomal passenger complex (CPC) regulates the timing of cytokinesis and abscission via interactions with the Polo-like kinase PLK1, the mitotic kinesin-like protein MKLP1, and CEP55, a key component of the midbody that associates directly with both ESCRT-I and ALIX (Schuh and Audhya, 2014). One current model is that the CPC promotes the formation of a ternary complex consisting of CHMP4C, ANCHR, and VPS4 and prevents premature action by VPS4 in response to chromatin trapped in the midbody (Thoresen et al., 2014). It has also been suggested that CHMP4C phosphorylation by the enzymatic core of the CPC, the Aurora B kinase, directs CHMP4C localization to the midbody and its retention of VPS4 (Carlton et al., 2012). With the new findings by Christ et al. (2016), the relationship between Aurora B–mediated phosphorylation of CHMP4C and its ability to bind ALIX must now be further explored. Additionally, because ALIX appears to be the primary factor that recruits CHMP4C to the midbody, it may represent a novel therapeutic target for activation or bypass of the NoCut abscission checkpoint.
Figure 1.
Model for the recruitment of CHMP4B and CHMP4C to the midbody and their roles in regulating the timing of abscission. PLK-1 phosphorylation of CEP55 inhibits its binding to MKLP1. At the end of anaphase, PLK1 is degraded and MKLP1 recruits CEP55 to the midbody. CEP55 recruits TSG101 and ALIX to the midbody, and Christ et al. (2016) demonstrate that there are two pathways that lead to the subsequent recruitment of CHMP4B: one through ESCRT-I–ESCRT-II–CHMP6 and the second directly through ALIX. ALIX also recruits CHMP4C, which, upon phosphorylation by the CPC, is hypothesized to form a ternary complex with ANCHR and VPS4. Formation of this complex prevents VPS4 from facilitating the completion of abscission until all chromatin is cleared from the intercellular bridge.
In contrast to the necessity of ALIX during cytokinetic abscission, its role during MVE formation and ubiquitin-dependent cargo degradation remains debatable. Depletion studies suggest that ALIX is dispensable for the lysosomal sorting of several cargoes (Bowers et al., 2006). However, ALIX is capable of targeting to late endosomal membranes through its interaction with lysobisphosphatidic acid, and some data suggest that ALIX can promote ESCRT-III filament assembly at MVEs (Matsuo et al., 2004; Pires et al., 2009; Bissig and Gruenberg, 2014). In the future, it will be essential to elucidate the mechanisms by which ALIX and CHMP6 direct the nucleation of CHMP4B/ESCRT-III spiral filaments and to determine whether the membrane landscapes of the MVE and the cytokinetic bridge differ in a manner that promotes one pathway over the other. As cryoelectron microscopy–based approaches in cells and reconstituted systems advance, the answer to these questions may become more accessible.
Acknowledgments
The authors declare no competing financial interests.
References
- Bissig C., and Gruenberg J.. 2014. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 24:19–25. 10.1016/j.tcb.2013.10.009 [DOI] [PubMed] [Google Scholar]
- Bowers K., Piper S.C., Edeling M.A., Gray S.R., Owen D.J., Lehner P.J., and Luzio J.P.. 2006. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J. Biol. Chem. 281:5094–5105. 10.1074/jbc.M508632200 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., and Martin-Serrano J.. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 316:1908–1912. 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Caballe A., Agromayor M., Kloc M., and Martin-Serrano J.. 2012. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 336:220–225. 10.1126/science.1217180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L., Wenzel E.M., Liestøl K., Raiborg C., Campsteijn C., and Stenmark H.. 2016. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J. Cell Biol. 10.1083/jcb.201507009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H. 2015. ESCRTs are everywhere. EMBO J. 34:2398–2407. 10.15252/embj.201592484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D.J., Odorizzi G., and Emr S.D.. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893–905. 10.1038/nrm973 [DOI] [PubMed] [Google Scholar]
- Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N.S., Matile S., Dubochet J., Sadoul R., et al. 2004. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 303:531–534. 10.1126/science.1092425 [DOI] [PubMed] [Google Scholar]
- Morita E., and Sundquist W.I.. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425. 10.1146/annurev.cellbio.20.010403.102350 [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.-Y., Morham S.G., Gygi S.P., Rodesch C.K., and Sundquist W.I.. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227. 10.1038/sj.emboj.7601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E., Colf L.A., Karren M.A., Sandrin V., Rodesch C.K., and Sundquist W.I.. 2010. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. USA. 107:12889–12894. 10.1073/pnas.1005938107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires R., Hartlieb B., Signor L., Schoehn G., Lata S., Roessle M., Moriscot C., Popov S., Hinz A., Jamin M., et al. 2009. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 17:843–856. 10.1016/j.str.2009.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J.M., Brust-Mascher I., and Mogilner A.. 2003. Cell division. Nature. 422:746–752. 10.1038/nature01599 [DOI] [PubMed] [Google Scholar]
- Schuh A.L., and Audhya A.. 2014. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 49:242–261. 10.3109/10409238.2014.881777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.T., Schuh A.L., Zheng Y., Quinney K., Wang L., Hanna M., Mitchell J.C., Otegui M.S., Ahlquist P., Cui Q., and Audhya A.. 2014. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J. Cell Biol. 206:763–777. 10.1083/jcb.201403108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen S.B., Campsteijn C., Vietri M., Schink K.O., Liestøl K., Andersen J.S., Raiborg C., and Stenmark H.. 2014. ANCHR mediates Aurora-B-dependent abscission checkpoint control through retention of VPS4. Nat. Cell Biol. 16:550–560. 10.1038/ncb2959 [DOI] [PubMed] [Google Scholar]