Abstract
Five isolates of non-pigmented, rapidly growing mycobacteria were isolated from three patients and, in an earlier study, from zebrafish. Phenotypic and molecular tests confirmed that these isolates belong to the Mycobacterium chelonae–Mycobacterium abscessus group, but they could not be confidently assigned to any known species of this group. Phenotypic analysis and biochemical tests were not helpful for distinguishing these isolates from other members of the M. chelonae–M. abscessus group. The isolates presented higher drug resistance in comparison with other members of the group, showing susceptibility only to clarithromycin. The five isolates showed a unique PCR restriction analysis pattern of the hsp65 gene, 100 % similarity in 16S rRNA gene and hsp65 sequences and 1–2 nt differences in rpoB and internal transcribed spacer (ITS) sequences. Phylogenetic analysis of a concatenated dataset including 16S rRNA gene, hsp65, and rpoB sequences from type strains of more closely related species placed the five isolates together, as a distinct lineage from previously described species, suggesting a sister relationship to a group consisting of M. chelonae, Mycobacterium salmoniphilum, Mycobacterium franklinii and Mycobacterium immunogenum. DNA–DNA hybridization values >70 % confirmed that the five isolates belong to the same species, while values < 70 % between one of the isolates and the type strains of M. chelonae and M. abscessus confirmed that the isolates belong to a distinct species. The polyphasic characterization of these isolates, supported by DNA–DNA hybridization results, demonstrated that they share characteristics with M. chelonae–M. abscessus members, but constitute a different species, for which the name Mycobacterium saopaulense sp. nov. is proposed. The type strain is EPM 10906T ( = CCUG 66554T = LMG 28586T = INCQS 0733T).
Nontuberculous mycobacteria are ubiquitous environmental organisms and several species can cause opportunistic infections in humans, in particular the members of the Mycobacterium chelonae–Mycobacterium abscessus group. This group comprises closely related, rapidly growing mycobacteria that can cause a broad spectrum of infections mainly affecting lung, skin and soft tissue (Simmon et al., 2011; Wallace et al., 1983). The ubiquitous distribution of these organisms facilitates the contamination of medical equipment and solutions that, associated with the growing number of therapeutic interventions, generate nosocomial infections and outbreaks representing a serious public health problem in some settings (Leão et al., 2010; Tortoli, 2009).
Until recently, the M. chelonae–M. abscessus group was composed of M. chelonae, M. abscessus (Kusunoki & Ezaki, 1992), Mycobacterium immunogenum (Wilson et al., 2001), Mycobacterium massiliense (Adékambi et al., 2004, 2006b), Mycobacterium bolletii (Adékambi et al., 2006a) and Mycobacterium salmoniphilum (Whipps et al., 2007). Taxonomic changes have been proposed and M. abscessus, M. massiliense and M. bolletii have been assigned to a single species (M. abscessus) with two subspecies, M. abscessus subsp. abscessus and M. abscessus subsp. bolletti, the latter including those isolates previously identified as M. massiliense and M. bolletii (Leão et al., 2011; Leão et al., 2009). Two novel members of this group have been recently described, Mycobacterium franklinii (Nogueira et al., 2015; Simmon et al., 2011) and ‘Mycobacterium fukienense’ (Zhang et al., 2013).
The aim of the present study was to define the taxonomic position of five mycobacterial isolates without conclusive species assignments (isolates EPM 10906T, EPM 10695, IAL 3785, JAN1 and JAN2), which share a PCR restriction analyses (PRA) profile of the hsp65 gene not present in the PRASITE database (http://app.chuv.ch/prasite/index.html). Our data indicate that these five isolates belong to a single taxon and represent a novel species of the M. chelonae–M. abscessus group.
The first two isolates (EPM 10906T and EPM 10695) were obtained in 1999 from corneal specimens of two patients with infectious crystalline keratopathy after LASIK surgery (laser-assisted in situ keratomileusis) performed in the same ophthalmological clinic, in São Paulo city (Brazil). These isolates were first misidentified as M. chelonae (Alvarenga et al., 2002) and subsequently as M. abscessus (Sampaio et al., 2006) on the basis of PRA of the hsp65 gene. Typing of these isolates by pulsed-field gel electrophoresis (PFGE) using a previously described protocol (Matsumoto et al., 2011), revealed that they share indistinguishable patterns and thus might belong to a single strain (Fig. 1). The third isolate (IAL 3785) was obtained in 2007 from a cervical abscess in the city of Ribeirão Preto, São Paulo (Brazil). The other two isolates (JAN1 and JAN2) were isolated from zebrafish (Danio rerio) (Kent et al., 2004) and initially categorized as M. chelonae. Using greater taxon sampling in a later phylogenetic analysis, these strains were recognized as phylogenetically distinct (Whipps et al., 2007), but they were not described as a novel species at the time. Isolates JAN1 and JAN2 share highly similar PFGE patterns, differing only in a single band, and were isolated from zebrafish at the same research facility about two months apart in 2003 (Kent et al., 2004); considering this, they could represent a single strain.
Fig. 1.
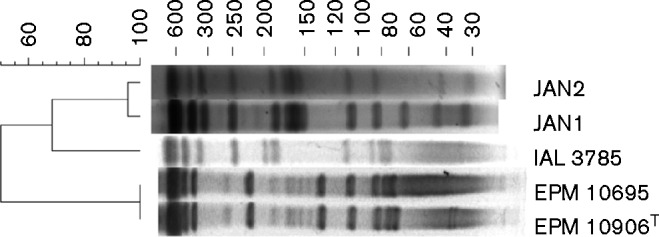
Pulsed-field gel electrophoresis (PFGE) patterns of DraI digested DNA of the five isolates studied in this work. PFGE images were analysed with the BioNumerics program v. 7.1 (Applied Maths). The band-based Dice unweighted-pair group method using average linkages was used to prepare a dendrogram of PFGE profiles, based on 1.5 % optimization and position tolerance.
We investigated the classification of these five isolates, comprising three different strains, using a polyphasic approach that included microscopic and macroscopic morphological examination, cultural and biochemical tests, drug susceptibility testing, HPLC analysis of cell-wall mycolic acids, PRA, sequencing of three housekeeping genes and DNA–DNA hybridization. The results were compared with those displayed by M. abscessus subsp. abscessus ATCC 19977T, M. abscessus subsp. bolletti CCUG 50184T, M. chelonae ATCC 35752T, M. immunogenum ATCC 700505T, M. salmoniphilum ATCC 13758T and M. franklinii DSM 45524T.
Cultures were grown on solid media [Löwenstein-Jensen (LJ), Middlebrook 7H10 supplemented with OACD (oleic acid, albumin, glucose and catalase) and Luria–Bertani agar], and in liquid Muller-Hinton medium or Luria–Bertani broth with 1 % Tween 80 at 28–30 °C for 5 days. Microscopic examination of colony smears by Ziehl–Neelsen staining confirmed that the isolates were acid-fast bacilli. Analysis of pigment production, single-source carbon utilization (mannitol, inositol and citrate), growth at 26 °C and 37 °C, and tolerance to 5 % NaCl, 0.2 % picric acid, para-nitrobenzoic acid (PNB) and nitrite were performed on 7H10-OADC and LJ. Nitrate reduction, Tween 80 hydrolysis and arylsulfatase production were also examined. All tests were performed as described in standard protocols for the biochemical identification of mycobacteria (Kent & Kubica, 1985; Leão et al., 2004; Tsukamura, 1984). The five isolates exhibited indistinguishable phenotypic and biochemical characteristics, which are listed in Table S1 (available in the online Supplementary Material). These cultural and biochemical tests were not helpful for distinguishing these isolates from other members of the M. chelonae–M. abscessus group (Table S1).
Antimicrobial drug-susceptibility testing was performed using a microdilution method in cation-supplemented Mueller–Hinton broth, according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI, 2011) for rapidly growing mycobacteria. Amikacin, cefoxitin, ciprofloxacin, clarithromycin, doxycycline, minocycline, moxifloxacin and tobramycin were tested. The five isolates were more drug resistant than the other members of the M. chelonae–M. abscessus group, showing susceptibility only to clarithromycin at 3 and 14 days incubation. The isolates were resistant to doxycycline, cefoxitin and tobramycin, and resistant or intermediate to amikacin, ciprofloxacin, minocycline and moxifloxacin (Table 1). These results are consistent with susceptibility testing previously conducted for strain JAN1 (Chang & Whipps, 2015). The drug resistance profile of the novel species described in this study highlights the importance of its correct identification for patient management.
Table 1. Antimicrobial susceptibility results for isolates and type strains included in this study.
Isolates/strains: 1, EPM 10695; 2, EPM 10906T; 3, IAL 3785; 4, JAN1; 5, JAN2; 6, M. abscessus subsp. abscessus ATCC 19977T; 7, M. abscessus subsp. bolletti CCUG 50184T; 8, M. chelonae ATCC 35752T; 9, M. immunogenum ATCC 700505T; 10, M. salmoniphilum ATCC 13758T; 11, M. franklinii DSM 45524T. r, Resistant; i, intermediate; s, susceptible – criteria for rapidly growing mycobacteria established by CLSI (2011); nd, not determined.
| Drug | MIC (μg ml− 1) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| Amikacin | 32 i | 128 r | 32 i | 32 i | 32 i | 8 s | 16 s | 8 s | ≤ 4 s | 16 s | ≤ 4 s |
| Ciprofloxacin | 2 i | 2 i | 4 r | 4 r | 4 r | 4 r | 8 r | 0.5 s | 1 s | 1 s | 0.5 s |
| Clarithromycin | |||||||||||
| 3 days | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s |
| 14 days | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | ≤ 0.5 s | >64 r | >64 r | ≤ 0.5 s | ≤ 0.5 s | nd | ≤ 0.5 s |
| Doxycycline | >32 r | >32 r | >32 r | >32 r | >32 r | >32 r | >32 r | 4 i | >32 r | >32 r | ≤ 0.25 s |
| Cefoxitin | 512 r | >512 r | 512 r | >512 r | 512 r | 64 i | 32 i | 512 r | 256 r | 512 r | 16 s |
| Tobramycin | 16 r | 32 r | 16 r | 8 r | 16 r | 8 r | 16 r | 2 s | 4 i | 4 i | 2 s |
| Minocycline | 32 r | 16 r | 4 i | 16 r | 8 r | 8 r | 16 r | 2 i | 8 r | 2 i | ≤ 0.25 s |
| Moxifloxacin | 2 i | 8 r | 4 r | 4 r | 2 i | 8 r | 8 r | ≤ 0.25 s | 1 s | 2 i | 1 s |
For HPLC analysis of cell-wall mycolic acids, two strains of the panel characterized here were selected (the proposed type strain EPM 10906T and JAN1) and three reference strains belonging to the closely related M. chelonae–M. abscessus group (M. abscessus subsp. abscessus ATCC 19977T, Mycobacterium massiliense CCUG 48898T and M. abscessus subsp. bolletti CCUG 50184T). The cells of these strains, grown in culture on Middlebrook 7H10 agar, were saponified, extracted and derivatized as recommended by the Sherlock Mycobacteria Identification System (SMIS; MIDI) and separated using a gradient of methanol and 2-propanol. All the strains analysed produced nearly identical HPLC patterns characterized by two late emerging clusters of peaks (Fig. 2). The Sherlock software (version Myco 1.0) identified all the strains as M. chelonae–M. abscessus with very high similarity indexes (range 0.802–0.899). The low discriminatory power of HPLC analysis in differentiating most rapidly growing mycobacterial species (Tortoli, 2003) is therefore confirmed for the proposed novel species, with this approach being unsuitable to go further in the assignation to the M. chelonae–M. abscessus group.
Fig. 2.
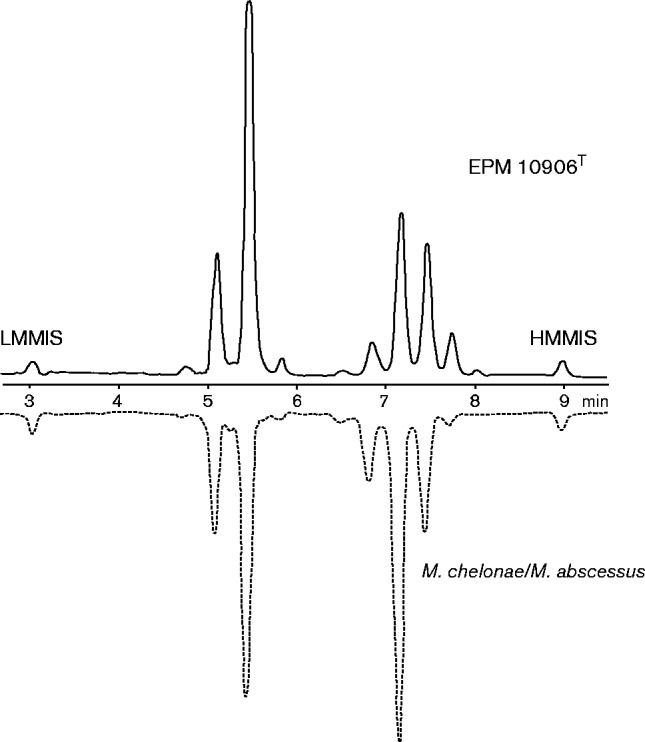
Representative mycolic acid pattern of isolate EPM 10906T paired with the reference profile of Mycobacterium chelonae–Mycobacterium abscessus (Sherlock database). LMMIS, Low molecular mass internal standard; HMMIS, high molecular mass internal standard.
GenoType Mycobacterium (Hain Lifescience), a commercial DNA strip assay for mycobacteria identification, was performed according to the manufacturer's instructions. Using the GenoType CM strip, the isolates showed the M. chelonae profile (hybridization with probes 5 and 10). GenoType AS only identified the isolates at the level of Mycobacterium species.
For molecular identification, PRA of the hsp65 gene and of the ITS, and partial sequencing of the small subunit (16S) rRNA gene, rpoB, hsp65 and ITS were performed. The DNA was prepared by boiling one loop full of bacteria for 10 min in 300 μl TET (10 mM Tris, 1 mM EDTA, 1 % Triton X-100; pH 8.0) followed by centrifugation at 14 000 g for 2 min. For PRA-hsp65, a 441 bp fragment of the hsp65 gene was amplified using primers Tb11 and Tb12 (Table S2), and the amplicon was digested in two separate tubes with BstEII and HaeIII restriction enzymes (Telenti et al., 1993). For PRA-ITS, amplicons generated with primers Sp1 and Sp2 (Table S2) were digested with TaqI restriction enzyme (Roth et al., 2000). PRA-hsp65 and PRA-ITS digestion products were visualized in 3 % agarose gels stained with ethidium bromide after electrophoresis, using the 50 bp ladder as the molecular size standard. The restriction fragment sizes were estimated using the BioNumerics program version 7.1 (Applied Maths) and compared to the patterns included in the PRASITE database for PRA-hsp65 and published by Roth et al. (2000) for PRA-ITS. The five isolates showed identical PRA-hsp65 patterns – BstEII [bp] (235, 210) and HaeIII [bp] (145, 60, 50), which differs from patterns of other members of the M. chelonae–M. abscessus group. This profile was not registered in the PRASITE database. The isolates showed the same PRA-ITS pattern – TaqI [bp] (225, 30), which is common to M. abscessus and M. franklinii. Therefore, only PRA-hsp65 was useful for differentiation of this novel species.
Primers used for PCR amplification and partial sequencing of the 16S rRNA gene, hsp65, rpoB and ITS sequences are listed in Table S2. PCR products were purified using a QIAquick PCR purification kit (Qiagen). Dideoxy sequencing was performed using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) and an ABI PRISM 3100 DNA Analyser (Applied Biosystems). Using clustal w multiple alignment (BioEdit version 7.2.5), the obtained sequences of isolates EPM 10906T, EPM 10695, IAL 3785, JAN1 and JAN2 were aligned and the percentages of sequence similarity were calculated after alignment. The five isolates shared 100 % similarity in the partial hsp65 and 16S rRNA gene sequences and had 12 nt differences in the partial rpoB and ITS sequences. The high16S rRNA gene sequence similarity to M. chelonae had been previously recognized by Kent et al. (2004) who assigned the two isolates investigated (JAN1 and JAN2) to such species. Greater differences in other genes (rpoB, hsp65, ITS) and in the PRA-hsp65 pattern subsequently revealed that these isolates are distinct (Sampaio et al., 2006; Whipps et al., 2007), and this is supported by the data presented here.
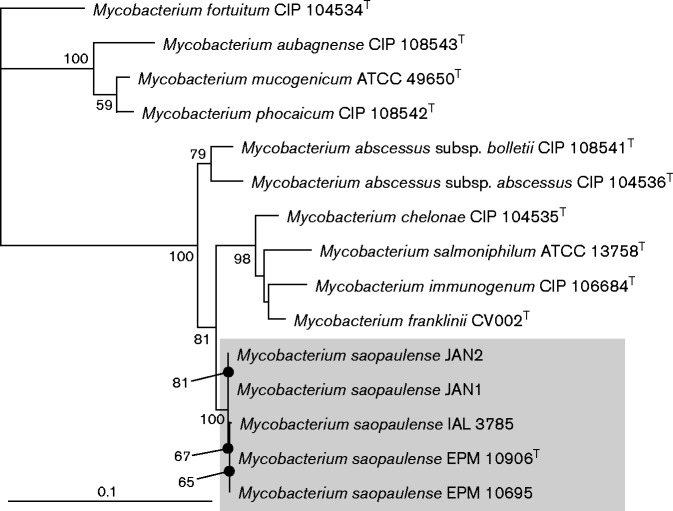
Previous phylogenetic analyses of individual genes (Whipps et al., 2007) had shown sister relationships of JAN1 and JAN2 isolates to different species. In the tree from hsp65 sequences, these isolates were sister to M. abscessus. In the tree from rpoB sequences, they showed a sister relationship to the entire M. chelonae–M. abscessus group. In the tree from ITS sequences, a sister relationship between the novel species and M. abscessus emerged. Analysis of 16S rRNA gene sequences alone revealed that sequences from the five isolates, M. abscessus and M. chelonae were intermixed (Fig. S1). Thus, the 16S rRNA gene alone was not sufficient to resolve these relationships and other genes yielded conflicting results. For this study, we chose to analyse a concatenated dataset using 16S rRNA gene, hsp65 and rpoB sequences from type strains of related species of the genus Mycobacterium (Table S3). Sequences were aligned with clustal x version 1.8 (Thompson et al., 1997) with default parameters (penalty for gap opening = 10, gap extension = 0.2). A maximum-likelihood analysis was performed using PhyML (Guindon & Gascuel, 2003) and bootstrap confidence values were calculated with 100 replicates. The resulting tree was edited and annotated in Adobe Illustrator (Adobe Systems). The ITS region was excluded from the final analysis because equivalent sites were difficult to confidently identify from outgroup taxa; however, when ITS data are included (unalignable data treated as missing), the tree topology was unaltered (data not shown). In either analysis, the five isolates showed a sister relationship to a clade made up of four species (M. chelonae, M. salmoniphilum, M. immunogenum and M. franklinii) (Fig. 3). These species, plus the five isolates, are sister to M. abscessus. These concatenated data present the phylogenetic hypothesis that the five isolates are sister to M. chelonae versus M. abscessus as suggested by individual gene trees from hsp65 and ITS sequences (Whipps et al., 2007). Regardless of the precise sister relationship, all analyses support inclusion in the M. chelonae–M. abscessus group, and a phylogenetic position for the five isolates that is distinct from all other nominal species.
Fig. 3.
Estimate of phylogeny of Mycobacterium saopaulense sp. nov. and other closely related rapid growers based on maximum-likelihood analysis of concatenated dataset of 16S rRNA gene, hsp65 and rpoB sequences. Bootstrap support values >50 % are shown at nodes. GenBank accession numbers of the 16S rRNA gene, hsp65 and rpoB sequences are shown in Table S3.
For DNA–DNA hybridization (DDH), high-molecular-mass DNA was prepared from 2 g cell mass using the protocol described by Pitcher et al. (1989) with modifications. Bacterial cells were centrifuged, inactivated at 90 °C for 30 min and resuspended in 3 ml lysis buffer containing ml− 1: 200 μg RNase, 25 mg fresh lysozyme and 100U mutanolysine. The suspensions were incubated overnight at 37 °C and the DNA was extracted with chloroform/isoamyl alcohol, treated with RNase and precipitated with ethanol as described by Marmur (1961). DDH was performed as described by Ezaki et al. (1989), using photobiotin-labelled probes in microplate wells. Fluorescence was measured in a HTS7000 Bio Assay Reader (Perkin-Elmer). DNA–DNA hybridization values are presented as means of reciprocal experiments, performed in quadruplicate hybridization reactions. The DNA G+C content, estimated as described by Mesbah & Whitman (1989), was 64.6 mol% (EPM 10906T), 64.8 mol% (EPM 10695), 64.7 mol% (IAL 3785), 64.5 mol% (JAN1) and 64.7 mol% (JAN2). These values were used for calculation of the 50 °C hybridization temperature, and are consistent with the DNA G+C contents of the genus Mycobacterium, between 59 mol% and 66 mol% (Devulder et al., 2005). DDH experiments performed with the five isolates yielded hybridization values above 70 % (data not shown), confirming that they belong to the same species. Isolate EPM 10906T was selected to perform DDH reciprocal experiments with the type strains of the M. chelonae–M. abscessus group. All values were below 70 %, confirming that EPM 10906T and the other four isolates belong to a distinct species of the M. chelonae–M. abscessus group (Table 2).
Table 2. DNA–DNA hybridization values of isolate EPM 10906T against the type strains of members of the M. chelonae–M. abscessus group.
Reciprocal experiments were performed in quadruplicate hybridization reactions.
| Strain | Mean DNA–DNA relatedness ± sd (%) | Reciprocal values |
|---|---|---|
| M. abscessus subsp. abscessus ATCC 19977T | 41.5 ± 2.5 | 39;44 |
| M. abscessus subsp. bolletti CCUG 50184T | 40 ± 1 | 39;41 |
| M. chelonae ATCC 35752T | 47 ± 5 | 42;52 |
| M. immunogenum ATCC 700505T | 40 ± 7 | 33;47 |
| M. salmoniphilum ATCC 13758T | 44.5 ± 1.5 | 43;46 |
| M. franklinii DSM 45524T | 51 ± 4 | 47;55 |
In conclusion, phenotypic and genotypic tests indicated that isolates EPM 10906T, EPM 10695, IAL 3785, JAN1 and JAN2 belong to the M. chelonae–M. abscessus group. In addition, several results clearly indicated that these isolates form a uniform group separated from the other members of M. chelonae–M. abscessus group. Therefore, we propose to classify these isolates as a novel species of the genus Mycobacterium in the M. chelonae–M. abscessus group, with the name Mycobacterium saopaulense sp. nov.
Description of Mycobacterium saopaulense sp. nov.
Mycobacterium saopaulense (sa.o.paul.en′se. N.L. neut. adj. saopaulense of or pertaining to the Brazilian state of São Paulo, where the first strains were isolated).
Cells are acid-fast bacilli and visible growth on solid media requires 3–5 days at 28 °C. Colonies are non-pigmented and smooth. After some days, the medium often acquires a brown colour. Growth occurs in the presence of 5 % NaCl, picric acid, para-nitrobenzoic acid (PNB) and nitrite. Growth is observed in the presence of citrate as a single source of carbon, but not in the presence of mannitol or inositol. Negative reactions are observed for nitrate reduction and Tween 80 hydrolysis. Conventional biochemical testing cannot distinguish this species from other members of the M. chelonae–M. abscessus group. The antimicrobial pattern is characterized by susceptibility to clarithromycin and resistance to doxycycline, tobramycin and cefoxitin. Variable results, intermediate or resistant, were obtained with amikacin, ciprofloxacin, minocycline and moxifloxacin. The mycolic acid profile is similar to that of M. chelonae–M. abscessus by HPLC analysis. Genotype CM shares the same profile as M. chelonae. The partial sequencing of rpoB and hsp65 genes can distinguish Mycobacterium saopaulense from other members of M. chelonae–M. abscessus group. The PRA-hsp65 pattern that characterizes this species is BstEII [bp] (235, 210) and HaeIII [bp] (145, 60, 50).
The type strain is EPM 10906T ( = CCUG 66554T = LMG 28586T = INCQS 0733T). The DNA G+C content of the type strain is 64.6 mol%.
Acknowledgements
This study received financial support from Fundação de Amparo à Pesquisa do Estado de São Paulo (www.fapesp.br) (FAPESP) (grant 2011/18326-4). C. L. N. and C. K. M. received fellowships from FAPESP (2012/13763-0 and 2013/16018-6). This work has been partially supported by International Cooperation UAM-Banco Santander and Latin America (CEAL-UAM). Contributions to this work by C. M. W. were funded in part by the Office of Research Infrastructure Programs of the National Institutes of Health (NIH) under award number R24OD010998. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Supplementary Data
Supplementary Data
Abbreviations:
- DDH
DNA–DNA hybridization
- ITS
internal transcribed spacer
- PFGE
pulsed-field gel electrophoresis
- PRA
PCR restriction analyses
References
- Adékambi T., Reynaud-Gaubert M., Greub G., Gevaudan M. J., La Scola B., Raoult D., Drancourt M. (2004). Amoebal coculture of Mycobacterium massiliense sp. nov. from the sputum of a patient with hemoptoic pneumonia J Clin Microbiol 42 5493–5501 10.1128/JCM.42.12.5493-5501.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adékambi T., Berger P., Raoult D., Drancourt M. (2006a). rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov Int J Syst Evol Microbiol 56 133–143 10.1099/ijs.0.63969-0 . [DOI] [PubMed] [Google Scholar]
- Adékambi J., Reynaud-Gaubert M., Greub G., Gevaudan M. J., La Scola B., Raoult D., Drancourt M. (2006b). Mycobacterium massiliense sp. nov. In List of New Names and New Combinations Previously Effectively, but not Validly, Published, Validation List 111 Int J Syst Evol Microbiol 56 2025–2027 10.1099/ijs.0.64643-0 . [DOI] [PubMed] [Google Scholar]
- Alvarenga L., Freitas D., Hofling-Lima A. L., Belfort R., Jr, Sampaio J., Sousa L., Yu M., Mannis M. (2002). Infectious post-LASIK crystalline keratopathy caused by nontuberculous mycobacteria Cornea 21 426–429 10.1097/00003226-200205000-00020 . [DOI] [PubMed] [Google Scholar]
- Chang C. T., Whipps C. M. (2015). Activity of antibiotics against Mycobacterium species commonly found in laboratory zebrafish J Aquat Anim Health 27 88–95 10.1080/08997659.2015.1007176 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2011). Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes; Approved Standard, CLSI document M24-A2 2nd edn Wayne, PA: Clinical and Laboratory Standards Institute. [PubMed] [Google Scholar]
- Devulder G., Pérouse de Montclos M., Flandrois J. P. (2005). A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model Int J Syst Evol Microbiol 55 293–302 10.1099/ijs.0.63222-0 . [DOI] [PubMed] [Google Scholar]
- Ezaki T., Hashimoto Y., Yabuuchi E. (1989). Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains Int J Syst Bacteriol 39 224–229 10.1099/00207713-39-3-224. [DOI] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood Syst Biol 52 696–704 10.1080/10635150390235520 . [DOI] [PubMed] [Google Scholar]
- Kent P. T., Kubica G. P. (1985). Public Health Mycobacteriology. A Guide for the Level III Laboratory Atlanta: Centers for Disease Control. [Google Scholar]
- Kent M. L., Whipps C. M., Matthews J. L., Florio D., Watral V., Bishop-Stewart J. K., Poort M., Bermudez L. (2004). Mycobacteriosis in zebrafish (Danio rerio) research facilities Comp Biochem Physiol C: Toxicol Pharmacol 138 383–390 10.1016/j.cca.2004.08.005 . [DOI] [PubMed] [Google Scholar]
- Kusunoki S., Ezaki T. (1992). Proposal of Mycobacterium peregrinum sp. Nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov Int J Syst Bacteriol 42 240–245 10.1099/00207713-42-2-240 . [DOI] [PubMed] [Google Scholar]
- Leão S. C., Martin A., Mejia G. I., Palomino J. C., Robledo J., Telles M. A. S., Portaels F. (2004). Practical Handbook for the Phenotypic and Genotypic Identification of Mycobacteria Brugges: Vanden BROELLE. [Google Scholar]
- Leão S. C., Tortoli E., Viana-Niero C., Ueki S. Y., Lima K. V., Lopes M. L., Yubero J., Menendez M. C., Garcia M. J. (2009). Characterization of mycobacteria from a major Brazilian outbreak suggests that revision of the taxonomic status of members of the Mycobacterium chelonae-M. abscessus group is needed J Clin Microbiol 47 2691–2698 10.1128/JCM.00808-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leão S. C., Viana-Niero C., Matsumoto C. K., Lima K. V., Lopes M. L., Palaci M., Hadad D. J., Vinhas S., Duarte R. S., other authors (2010). Epidemic of surgical-site infections by a single clone of rapidly growing mycobacteria in Brazil Future Microbiol 5 971–980 10.2217/fmb.10.49 . [DOI] [PubMed] [Google Scholar]
- Leão S. C., Tortoli E., Euzéby J. P., Garcia M. J. (2011). Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus Int J Syst Evol Microbiol 61 2311–2313 10.1099/ijs.0.023770-0 . [DOI] [PubMed] [Google Scholar]
- Marmur J. (1961). A procedure the for isolation of deoxyribonucleic acid from micro-organisms J Mol Biol 3 208–218 10.1016/S0022-2836(61)80047-8. [DOI] [Google Scholar]
- Matsumoto C. K., Chimara E., Bombarda S., Duarte R. S., Leão S. C. (2011). Diversity of pulsed-field gel electrophoresis patterns of Mycobacterium abscessus type 2 clinical isolates J Clin Microbiol 49 62–68 10.1128/JCM.01665-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah M., Whitman W. B. (1989). Measurement of deoxyguanosine/thymidine ratios in complex mixtures by high-performance liquid chromatography for determination of the mole percentage guanine + cytosine of DNA J Chromatogr A 479 297–306 10.1016/S0021-9673(01)83344-6 . [DOI] [PubMed] [Google Scholar]
- Nogueira C. L., Simmon K. E., Chimara E., Cnockaert M., Palomino J. C., Martin A., Vandamme P., Brown-Elliott B. A., Wallace R. J., Jr, other authors (2015). Mycobacterium franklinii sp. nov., a species closely related to members of the Mycobacterium chelonae-Mycobacterium abscessus group Int J Syst Evol Microbiol 65 2148–2153 10.1099/ijs.0.000234. [DOI] [PubMed] [Google Scholar]
- Pitcher D. G., Saunders N. A., Owen R. J. (1989). Rapid extraction of bacterial genomic DNA with guanidium thiocyanate Lett Appl Microbiol 8 151–156 10.1111/j.1472-765X.1989.tb00262.x. [DOI] [Google Scholar]
- Roth A., Reischl U., Streubel A., Naumann L., Kroppenstedt R. M., Habicht M., Fischer M., Mauch H. (2000). Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases J Clin Microbiol 38 1094–1104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio J. L., Viana-Niero C., de Freitas D., Höfling-Lima A. L., Leão S. C. (2006). Enterobacterial repetitive intergenic consensus PCR is a useful tool for typing Mycobacterium chelonae and Mycobacterium abscessus isolates Diagn Microbiol Infect Dis 55 107–118 10.1016/j.diagmicrobio.2006.01.006 . [DOI] [PubMed] [Google Scholar]
- Simmon K. E., Brown-Elliott B. A., Ridge P. G., Durtschi J. D., Mann L. B., Slechta E. S., Steigerwalt A. G., Moser B. D., Whitney A. M., other authors (2011). Mycobacterium chelonae-abscessus complex associated with sinopulmonary disease, Northeastern USA Emerg Infect Dis 17 1692–1700 10.3201/eid1709.101667 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti A., Marchesi F., Balz M., Bally F., Böttger E. C., Bodmer T. (1993). Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis In J Clin Microbiol 31 175–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools Nucleic Acids Res 25 4876–4882 10.1093/nar/25.24.4876 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E. (2003). Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s Clin Microbiol Rev 16 319–354 10.1128/CMR.16.2.319-354.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortoli E. (2009). Clinical manifestations of nontuberculous mycobacteria infections Clin Microbiol Infect 15 906–910 10.1111/j.1469-0691.2009.03014.x . [DOI] [PubMed] [Google Scholar]
- Tsukamura M. (1984). Identification of Mycobacteria Aichi: Mycobacteriosis Research Laboratory of the National Chubu Hospital. [Google Scholar]
- Wallace R. J., Jr, Swenson J. M., Silcox V. A., Good R. C., Tschen J. A., Stone M. S. (1983). Spectrum of disease due to rapidly growing mycobacteria Rev Infect Dis 5 657–679 10.1093/clinids/5.4.657 . [DOI] [PubMed] [Google Scholar]
- Whipps C. M., Butler W. R., Pourahmad F., Watral V. G., Kent M. L. (2007). Molecular systematics support the revival of Mycobacterium salmoniphilum (ex Ross 1960) sp. nov., nom. rev., a species closely related to Mycobacterium chelonae Int J Syst Evol Microbiol 57 2525–2531 10.1099/ijs.0.64841-0 . [DOI] [PubMed] [Google Scholar]
- Wilson R. W., Steingrube V. A., Böttger E. C., Springer B., Brown-Elliott B. A., Vincent V., Jost K. C., Jr, Zhang Y., Garcia M. J., other authors (2001). Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy Int J Syst Evol Microbiol 51 1751–1764 10.1099/00207713-51-5-1751 . [DOI] [PubMed] [Google Scholar]
- Zhang Y. Y., Li Y. B., Huang M. X., Zhao X. Q., Zhang L. S., Liu W. E., Wan K. L. (2013). Novel species including Mycobacterium fukienense sp. is found from tuberculosis patients in Fujian Province, China, using phylogenetic analysis of Mycobacterium chelonae/abscessus complex Biomed Environ Sci 26 894–901 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data