ABSTRACT
Succinic semialdehyde (SSA), an important metabolite of γ-aminobutyric acid (GABA), is a ligand of the repressor AttJ regulating the expression of the attJ-attKLM gene cluster in the plant pathogen Agrobacterium tumefaciens. While the response of A. tumefaciens to GABA and the function of attKLM have been extensively studied, genetic and physiological responses of A. tumefaciens to SSA remain unknown. In combination with microarray and genetic approaches, this study sets out to explore new roles of the SSA-AttJKLM regulatory mechanism during bacterial infection. The results showed that SSA plays a key role in regulation of several bacterial activities, including C4-dicarboxylate utilization, nitrate assimilation, and resistance to oxidative stress. Interestingly, while the SSA relies heavily on the functional AttKLM in mediating nitrate assimilation and oxidative stress resistance, the compound could regulate utilization of C4-dicarboxylates independent of AttJKLM. We further provide evidence that SSA controls C4-dicarboxylate utilization through induction of an SSA importer and that disruption of attKLM attenuates the tumorigenicity of A. tumefaciens. Taken together, these findings indicate that SSA could be a potent plant signal which, together with AttKLM, plays a vital role in promoting the bacterial prosurvival abilities during infection.
IMPORTANCE Agrobacterium tumefaciens is a plant pathogen causing crown gall diseases and has been well known as a powerful tool for plant genetic engineering. During the long history of microbe-host interaction, A. tumefaciens has evolved the capabilities of recognition and response to plant-derived chemical metabolites. Succinic semialdehyde (SSA) is one such metabolite. Previous results have demonstrated that SSA functions to activate a quorum-quenching mechanism and thus to decrease the level of quorum-sensing signals, thereby avoiding the elicitation of a plant defense. Here, we studied the effect of SSA on gene expression at a genome-wide level and reported that SSA also promotes bacterial survival during infection. These findings provide a new insight on the biological significance of chemical signaling between agrobacteria and plant hosts.
INTRODUCTION
During the long history of bacteria-plant interactions, Agrobacterium tumefaciens has evolved sophisticated signal cross talk mechanisms to ensure successful infection. Previous studies have shown that the bacterium can perceive a wide variety of plant-derived metabolites and transduce them to regulate a particular set of bacterial genes (1). Succinic semialdehyde (SSA), the metabolite of γ-aminobutyric acid (GABA), is one such metabolite. SSA has been implicated in the regulation of bacterial quorum-quenching activity, the starvation response, and GABA catabolism and consequently affects the severity of plant symptom development (2–5).
SSA exerts its function through the proteins encoded by the attJ-attKLM (also named bclR-bclABC) genes. In A. tumefaciens, attJ and attKLM are localized at the same locus in opposite orientations, where attKLM constitute a cotranscribed operon. In the attKLM operon, attK encodes a semialdehyde dehydrogenase, attL encodes an alcohol dehydrogenase, and attM encodes a lactonase. These enzymes convert gamma-butyrolactone (GBL) sequentially to gamma-hydroxybutyrate (GHB), SSA, and succinic acid (SA), the last of which is integrated into the tricarboxylic acid (TCA) cycle (3, 6, 7). These enzymes also enable A. tumefaciens to metabolize γ-butyrolactone (GABA) as a sole carbon source for bacterial growth. In addition, AttM efficiently degrades the quorum-sensing (QS) signal acylated homoserine lactone (AHL) and controls the replication and conjugation of Ti plasmid (8, 9). The transcription of attKLM is repressed by AttJ. AttJ, an IclR-type transcription factor, physically binds to the promoter region of attKLM and tightly represses its expression. The repression of AttJ is relieved in the presence of SSA (3). It has been demonstrated that SSA, serving as the cognate ligand, directly binds to AttJ and attenuates its association with the promoter of attKLM. Crystal structural analysis of AttJ further indicates that SSA regulates its DNA-binding activity through protein oligomerization (10). Thus, supplementation of SSA induces the expression of attKLM, while depletion of SSA suppresses attKLM transcription (2–4).
In addition to the role of SSA in activation of attKLM transcriptional expression, several lines of evidence suggest that SSA may also play other regulatory roles. First, the IclR-type transcriptional regulators are known to regulate a variety of bacterial activities ranging from carbon metabolism to virulence expression (11). For example, the regulator IclR represses the genes encoding acetate utilization in Escherichia coli, KdgR is involved in exoenzyme production in Erwinia chrysanthemi, and SggR is associated with sporulation development in Streptomyces coelicolor (12–16). Regulation of quorum quenching by AttJ in A. tumefaciens adds another new role of the IclR-type regulators in bacteria. In this regard, it is interesting that an in silico search found more than 16 IclR-type regulators in the genome of A. tumefaciens, suggesting that IclR-type regulators may regulate a wide range of biological activities in this bacterial species. It is not clear yet whether SSA may also influence the functionality of these AttJ homologues. Second, SSA is produced by both eukaryotes and prokaryotes and is implicated in various biological functions. In human, abnormal accumulation of SSA exerts a wide range of adverse effects within the nervous system (17, 18). In Arabidopsis, a defect in SSA degradation enhances cell death under stressful conditions (19, 20). In Saccharomyces cerevisiae, mutation of SSA dehydrogenase increases the susceptibility of the microbe to salinity and osmotic and oxidative stresses (21). In Ralstonia eutropha, disruption of SSA metabolism impairs the ability of the bacterium to metabolize 4-hydroxybutyric acid as the sole carbon source (22). Third, although current knowledge indicates that A. tumefaciens takes up the plant signaling molecule GABA through a specific importer and converts it to produce SSA (23), a recent study showed that the expression of attKLM could be induced in the presence of wounded plant tissues even if the A. tumefaciens importer of GABA is disrupted (4), suggesting that SSA could be a specific plant signal that can be recognized by the bacterial cells and is worthy of further investigations.
In this study, we conducted microarray analysis to test the impact of SSA on transcriptional expression of the genes in A. tumefaciens. Based on the microarray results, we further performed a series of genetic and biochemical analyses to explore the roles of SSA and regulatory mechanisms in modulation of bacterial physiology. Our results identified a few new functions regulated by SSA, including utilization of C4-dicarboxylates, nitrate assimilation, and reactive oxygen species (ROS) resistance. In addition, we showed that functional AttKLM are required for SSA-mediated nitrate assimilation, ROS resistance, and virulence, which strongly suggests that A. tumefaciens could recognize and utilize SSA as a potent plant signal to modulate its survival capability during pathogen-host interactions.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table S1 in the supplemental material. E. coli strains were grown at 37°C in LB medium. A. tumefaciens strains were grown at 28°C in LB or in BM medium (8). For the carbon and nitrogen utilization experiments, bacterial strains specified in the legend to Fig. 2B were freshly grown on LB agar plates. After 16 h, bacterial cells were directly scraped from the plates and suspended in BM medium without carbon or nitrogen sources (10 mM K2HPO4, 10 mM KH2PO4, 2.5 mM NaCl, 2 mM MgSO4 · 7H2O, 0.7 mM CaCl2, 9 μM FeSO4 · 7H2O, pH 7.2). d-Mannitol (0.2%) was the carbon source when KNO3 (0.2%) was used as the sole nitrogen source, and (NH4)2SO4 was the nitrogen source when malate (0.2%) was the sole carbon source. Chemicals used in this study were purchased from Sigma, and the final concentrations were 0.2 mM unless otherwise specified. The bacterial cells (10 ml of liquid culture) were grown in 50-ml Falcon tubes, and the initial inoculation was at an optical density at 600 nm (OD600) of 0.05, with shaking at 220 rpm at 28°C. The OD600 values were measured after 24 h of growth. The results are presented as means ± standard deviations (SD) based on three experimental repeats.
FIG 2.
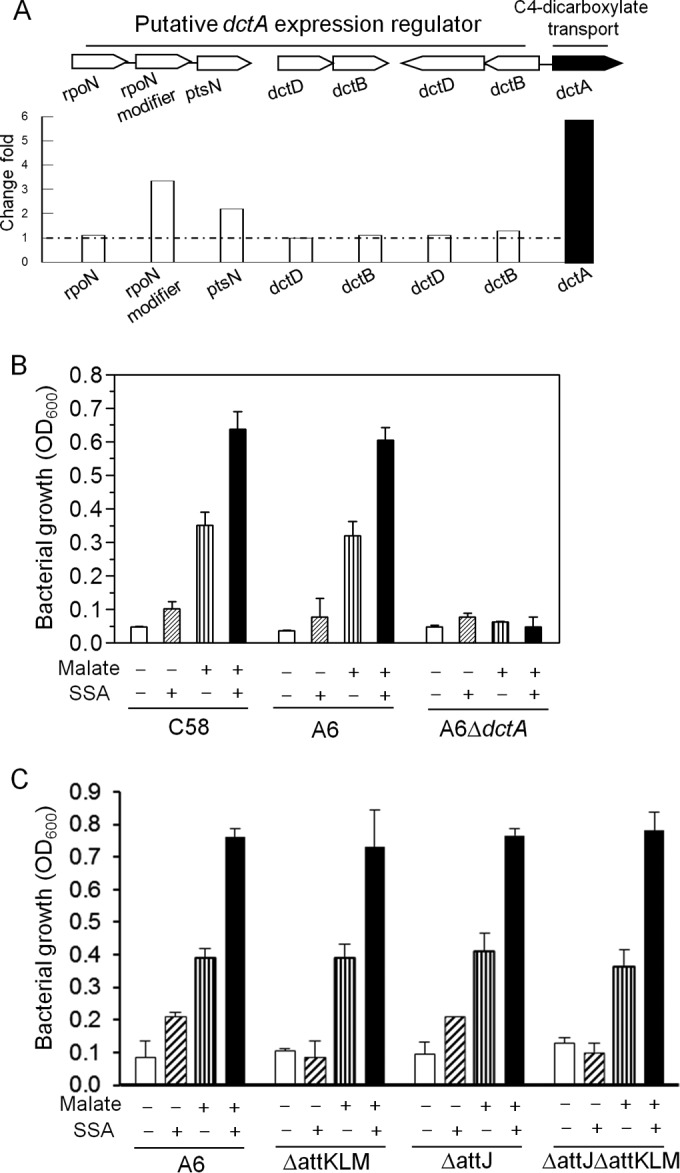
SSA promotes C4-dicarboxylate utilization in A. tumefaciens. (A) Organization of C4-dicarboxylate transporter gene clusters and their induction profiles in the microarray. The gene clusters are composed of a dctA transporter gene, three rpoN-related regulators, and two sets of dctB-dctD genes encoding two-component regulatory systems. The arrows indicate the direction of transcription. Fold change was calculated based on the average readings of two repeated microarrays. (B) Growth of A. tumefaciens C58, A6, and the A6 dctA deletion mutant (ΔdctA strain) using malate as the sole carbon source with or without SSA. (C) Growth of A. tumefaciens A6 and its mutant using malate as the sole carbon source with or without SSA. −, no chemical addition; +, chemical addition. Error bars denote standard deviations.
RNA preparation for RT-PCR and microarray hybridization.
A. tumefaciens strain C58 was used for microarray analysis in this study because of the availability of its genome sequence (24, 25). Bacterial cells were cultivated in BM medium at 28°C with shaking at 200 rpm and collected for RNA isolation when the OD600 reached ∼0.4. The total RNAs were isolated with an RNeasy minikit (Qiagen), and the residual DNAs were removed by RNase-free DNase I (Promega). The absence of residual DNAs was confirmed by the lack of PCR products after 35 cycles of PCR amplification with primers specific for the 16S RNA gene. RNA integrity was monitored by agarose gel electrophoresis, and RNA concentration was measured by a Nanodrop ND-1000 instrument (Nanodrop Technologies). For reverse transcription-PCR (RT-PCR), an aliquot of 0.2 μg of total RNA was serially diluted 10-fold and used as the template for one-step RT-PCR analysis (Qiagen). PCR primers used for RT-PCR analysis are listed in Table S1 in the supplemental material.
For microarray hybridization, cDNA was synthesized by using SuperScript II reverse transcriptase (Invitrogen) with 10 μg of total RNA and purified by phenol extraction following RNase digestion (NimbleGen Systems). The cDNA molecules were then fragmented into 50 to 200 nucleotides by partial digestion using DNase I (Promega) and labeled with biotin for microarray hybridization. The DNA microarray chips were synthesized by NimbleGen Systems (Madison, WI), and each chip included 4,661 open reading frames (ORFs) of the compiled genome sequence of A. tumefaciens strain C58 (University of Washington). Each ORF was represented by 20 unique and perfectly matched oligonucleotides (24-mer), which were in situ synthesized on the glass slide in duplicate. Therefore, each chip included two sets of 93,220 synthesized oligonucleotides, representing the 4,661 annotated ORFs. Labeled cDNA samples were individually hybridized to the A. tumefaciens C58-specific chips according to the NimbleGen standard procedures.
Microarray data analysis.
Following hybridization, the arrays were scanned, and the median signal intensity for each probe on the array was calculated by using NimbleGen's extraction software. For each probe pair, the difference between the perfect match (PM) and mismatch (MM) signal intensities was calculated together with the Tukey biweight mean from the 20 probe pairs for each ORF. The data were then further processed with the tools provided by Bioconductor (http://www.bioconductor.org). Gene calls were generated using the robust multiarray average (RMA) algorithm (26). The RMA value is the log to base 2, and ratios of SSA-treated to untreated cDNAs were calculated based on normalized data so that the ratio of signal from the SSA-induced sample to that of the untreated sample for a given ORF should represent the relative abundance of the transcripts of that ORF under the two conditions. Unless otherwise stated, the annotated ORFs were retrieved from the BioCyc Database Collection (http://biocyc.org/server.html).
Gene cloning and gene deletion.
DNA manipulation and transformation of E. coli were performed according to standard procedures. To generate the suicide plasmid construct pK18-ΔdctA, two DNA fragments were amplified from A. tumefaciens C58 with the primer pair 5′-GCTCTAGATGACAGAGGACTGCGTG-3′ and 5′-CGGGATCCCGGTGATGGTCTGCTCATG-3′ and the pair 5′-CGGGATCCTTCTGCTGCTCGTGG-3′ and 5′-GCTCTAGAAACAGACCGCGAAGACG-3′. After enzymatic digestion and ligation of T4 DNA ligase, the reaction mixture was aliquoted for another round of PCR amplification using the primer pair 5′-GCTCTAGATGACAGAGGACTGCGTG-3′ and 5′-GCTCTAGAAACAGACCGCGAAGACG-3′. DNA fragments with predicted sizes were then recovered from a 1% agarose gel, treated with XbaI, and linked into the vector pK18mobsacB that was also treated with XbaI. The resulting pK18-ΔdctA plasmid was screened by PCR and confirmed by DNA sequencing. Similarly, the pK18-ΔdctB1 plasmid was constructed by using the following PCR primers: the pair 5′-GCTCTAGAAATACGCATGCCAGACTTGC-3′ and5′-GCTCTAGAGCATAATGTTCGGACATGC-3′ and the pair 5′-GCTCTAGACTCGGTCTTGGCCTCGTC-3′ and 5′-GCTCTAGAACATCCACATCCGTATCGG-3′.
In-frame deletion of dctA and dctB1 in A. tumefaciens A6 was carried out according to a modification of a procedure described previously (27). Briefly, the transconjugants were screened on the BM medium containing kanamycin. The purified transconjugants were further selected on fresh BM agar plates containing 5% sucrose. The potential deletion mutants were then identified by loss of kanamycin resistance and PCR confirmation.
NADH/NAD and NADPH/NADP ratio measurement.
NADH/NAD and NADPH/NADP ratios were, respectively, measured by NAD/NADH and NADP/NADPH quantification kits (Sigma-Aldrich). Briefly, fresh bacteria were cultivated in minimal medium with or without SSA treatment. Ratios of free NADH/NAD and NADPH/NADP were determined with the method recommended by the manufacturer. The fold changes in the NADH/NAD and NADPH/NADP ratios were derived by dividing the NADH/NAD or NADPH/NADP ratio with SSA induction by the respective ratio without SSA induction.
H2O2 resistance assay.
A. tumefaciens strains C58 and A6 were grown in LB medium with or without SSA to an OD600 of ∼0.5 and used for subsequent experiments. For an H2O2 sensitivity assay, the cell cultures were added with H2O2 at a final concentration of 20 mM and grown at 28°C with shaking at 200 rpm for 25 min. A serial dilution of bacterial cells was immediately prepared and plated on LB agar plates. Colonies were enumerated after growth for 3 days, and the survival rate of each strain treated with H2O2 was calculated relative to that of the blank control.
Tumorigenicity assay.
Fresh bacterial cells grown in LB were collected and washed once and resuspended in phosphate-buffered saline (PBS) buffer at an OD600 of 1.0. The bacterial suspension was diluted as indicated (see Fig. 7A), and 5 μl of bacterial samples was used to infect leaves of Kalanchoe daigremontiana. The growth conditions and calculation of tumor size were described previously (4, 28).
FIG 7.
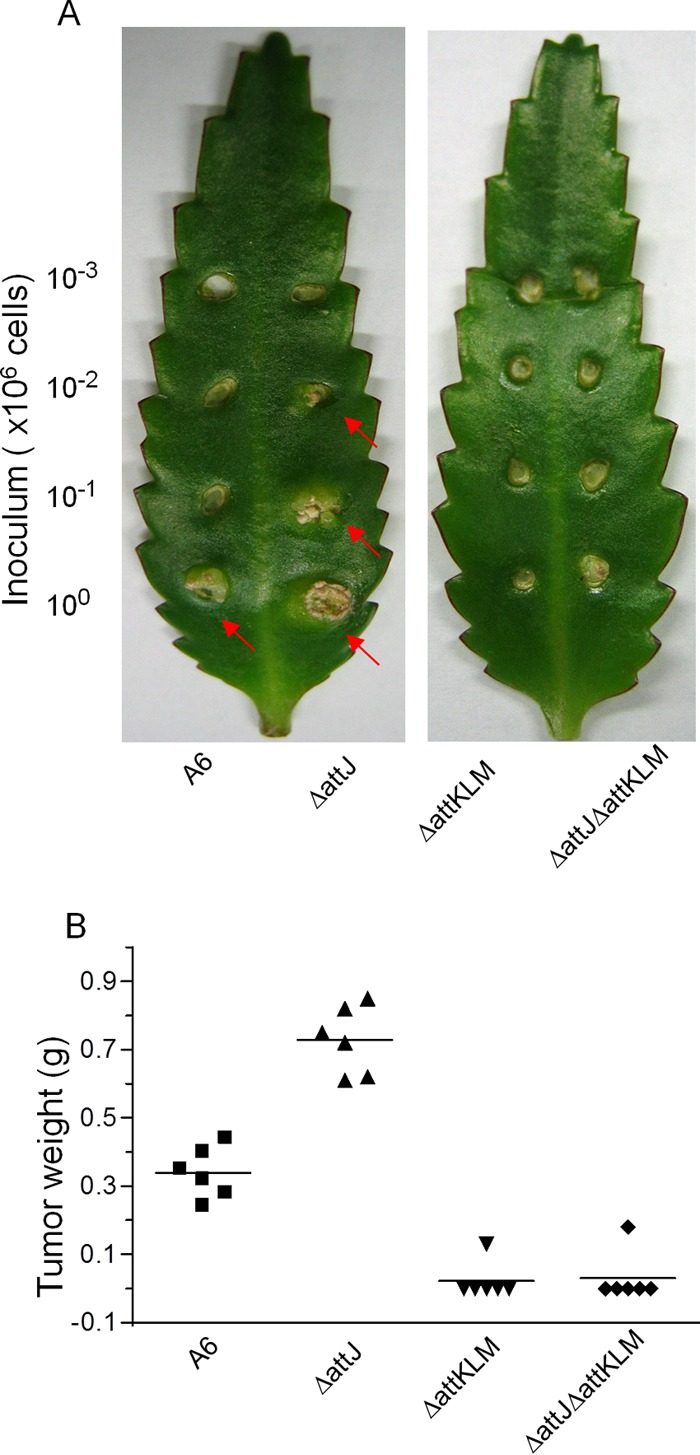
AttJ-AttKLM regulate the tumorigenicity of A. tumefaciens. (A) A representative photo for the tumorigenicity assay of A. tumefaciens A6 and its mutants. Bacteria were serially diluted 10-fold, and 5 μl of diluted samples was individually used to infect the leaves of K. daigremontiana. Arrows indicate visible tumor growth. (B) Tumor weights induced by A. tumefaciens A6 and its mutants.
RESULTS
Experimental design and microarray measurement.
For identification of the genes modulated by SSA on a genome-wide scale, a DNA microarray was performed on A. tumefaciens C58 in the presence or absence of SSA. Given that the bacterial intracellular SSA could transiently accumulate upon entry into stationary phase (3), we treated the cell culture with SSA at the early stage (OD600 of 0.5), and RNA samples were prepared from exponentially growing cells to minimize the interference of endogenous SSA. Thus, the results obtained from this study may miss those genes whose regulation requires a specific factor(s) present only in the late growth phase.
The microarray analysis showed that the transcriptional levels of 325 genes, falling into 14 functional categories (Fig. 1), were increased by more than 2-fold upon SSA treatment (see Table S2 in the supplemental material). Among these genes, 20% are the ORFs encoding hypothetical proteins. The remaining are the genes associated with protein synthesis, transportation, and energy metabolism, representing 17%, 12%, and 13%, respectively, of the total. In contrast, only 20 genes showed a >1.5-fold decrease (Fig. 1; see also Table S3 in the supplemental material). These results suggest that SSA may primarily activate rather than suppress gene expression under our experimental conditions.
FIG 1.
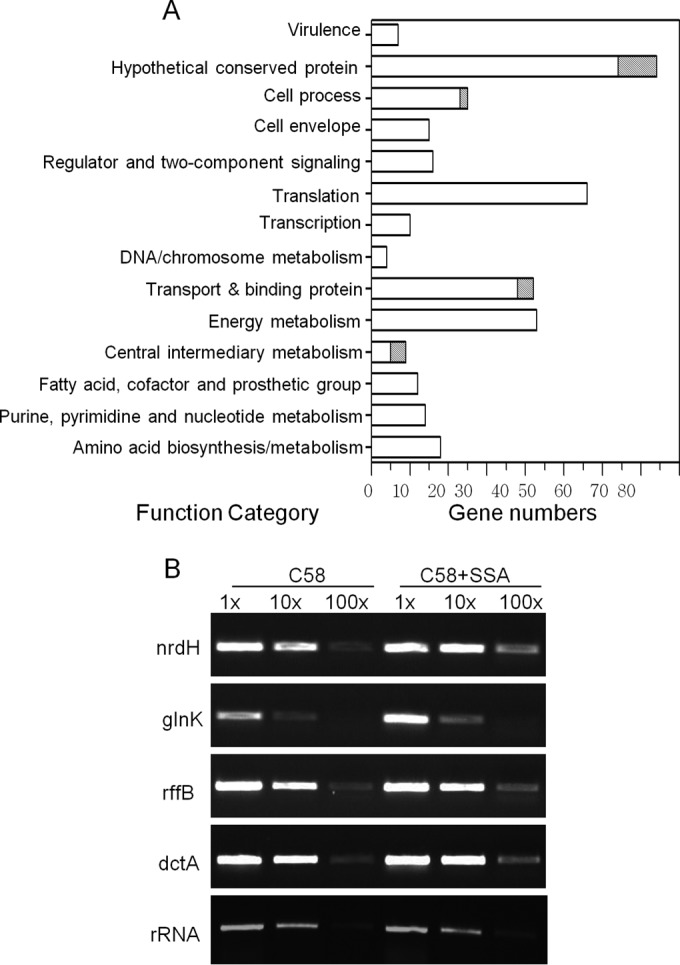
Microarray analyses of A. tumefaciens A6 with or without SSA treatment. (A) Functional groups and the numbers of genes whose expression levels were upregulated (open bars) or repressed (shaded bars) by external addition of SSA. Genes are categorized into functional classes based on the annotation available from the A. tumefaciens genome sequences (24, 25). (B) Validation of microarray results by RT-PCR.
The microarray data showed that the genes most strongly induced by SSA were attM, attL, and attK, with induction ratios of 21-, 14-, and 9-fold, respectively (see Table S2 in the supplemental material), which is highly consistent with the previous results that SSA is the inducer of the attMLK operon (3). In addition to attKLM, the transcriptional expression of attJ was also increased by about 3.7-fold (see Table S2), suggesting an SSA-dependent autoregulatory mechanism that governs AttJ transcription.
SSA promotes C4-dicarboxylate utilization.
The metabolism of C4-dicarboxylates, such as succinate and malate, has received considerable attention in rhizobia because of its important role in promoting bacterial survival in plant hosts (29). Rhizobial species take up these compounds via a high-affinity C4-dicarboxylate transporter encoded by the dctA gene (30). In Sinorhizobium meliloti, transcription of dctA is induced by C4-dicarboxylates via the DctB/DctD two-component system (31). Additionally, the sigma factor σ54 is also implicated in the regulation of dctA (32). Analysis of the A. tumefaciens genome sequences revealed not only dctA but also two copies of the σ54 gene rpoN and two sets of the dctB-dctD paralogs (Fig. 2A). Our microarray data showed that dctA transcription was induced up to about 5.8 times by SSA treatment, which was further verified by semiquantitative RT-quantitative PCR (RT-qPCR) analyses (Fig. 1B). In contrast, the mRNA levels of dctB-dctD and the nitrogen-limiting sigma factor σ54 remained unchanged (Fig. 2A; see also Table S2 in the supplemental material).
Given that DctA is essential for the growth of A. tumefaciens C58 with C4-dicarboxylates as the sole carbon source (30), induction of dctA by SSA suggests that SSA may promote C4-dicarboxylate utilization in this bacterial species. Therefore, we monitored the bacterial growth with malate as the sole carbon source. Results shown in Fig. 2B indicate that the A. tumefaciens strains C58 and A6 grew poorly when malate was used as the sole carbon source, with the OD600 reaching about 0.3 after 24 h of growth. In the presence of SSA, however, the growth of the two bacterial strains was substantially boosted, with the OD600 reaching approximately 0.65. Given that SSA alone could hardly support bacterial growth, these results indicate that SSA is able to promote C4-dicarboxylate utilization in A. tumefaciens. When the dctA gene is deleted in the octopine-type A6 strain, the bacterial cells failed to grow with either SSA or malate as the only carbon source (Fig. 2B). When mannitol or fumarate was used as a carbon source, however, no significant change was recorded in the growth rates between the dctA deletion mutant and the wild-type strain, and both grew well. Taken together, these results indicate that SSA might promote malate utilization by increasing the expression level of its transporter DctA although it could not be ruled out at this stage that SSA may also function through other unknown pathways.
To examine whether attJ-attKLM are involved in the SSA-mediated C4-dicarboxylate utilization, we used the corresponding mutants to conduct growth tests. As shown in Fig. 2C, the wild-type and ΔattJ strains grew poorly when SSA was added under these experimental conditions, whereas no growth was recorded for the ΔattKLM and ΔattJΔattKLM strains. These results suggest that attKLM are essential for SSA-supported bacterial growth. When malate was added alone as the sole carbon source, however, all the tested mutants exhibited growth rates similar to the growth rate of the wild type, indicating that neither attJ nor attKLM is involved in malate metabolism. Furthermore, when SSA and malate were added together, all the tested mutants grew well, with growth at a rate comparable to that of the wild type. Taken together, these results suggest that SSA-promoted malate utilization is dependent on the DctA transporter but independent of the functions encoded by the attJ-attKLM gene cluster.
SSA promotes nitrate assimilation.
In bacteria, the nitrate assimilation pathway generally involves four steps (33). Nitrate is first reduced by a nitrate reductase into nitrite and then ammonia, which is further converted into glutamate by glutamine synthase and glutamate synthase (Fig. 3A). Microarray results showed that the genes encoding nitrate reductase, ammonia transportation, and glutamine synthase were significantly induced by SSA treatment (Fig. 3B), suggesting that SSA may be involved in nitrate assimilation. With validation by RT-PCR (Fig. 1B), we examined the growth of A. tumefaciens by using potassium nitrate (KNO3) as the sole nitrogen source and mannitol as the carbon source. As shown in Fig. 3C, both the nopaline strain C58 and the octopine strain A6, similar to other rhizobial species, could use KNO3 as the sole nitrogen source for growth (34). Addition of SSA to the same medium significantly promoted the growth of both strain C58 and strain A6. As a control, SSA alone, as expected, could not support bacterial growth in the absence of a nitrogen source. Moreover, when the nitrogen source was changed to (NH4)2SO4, SSA failed to promote bacterial growth (see Fig. S1 in the supplemental material). Cumulatively, the above findings established the specific role of SSA in promoting nitrate assimilation in A. tumefaciens through transcriptional upregulation of the related genes.
FIG 3.
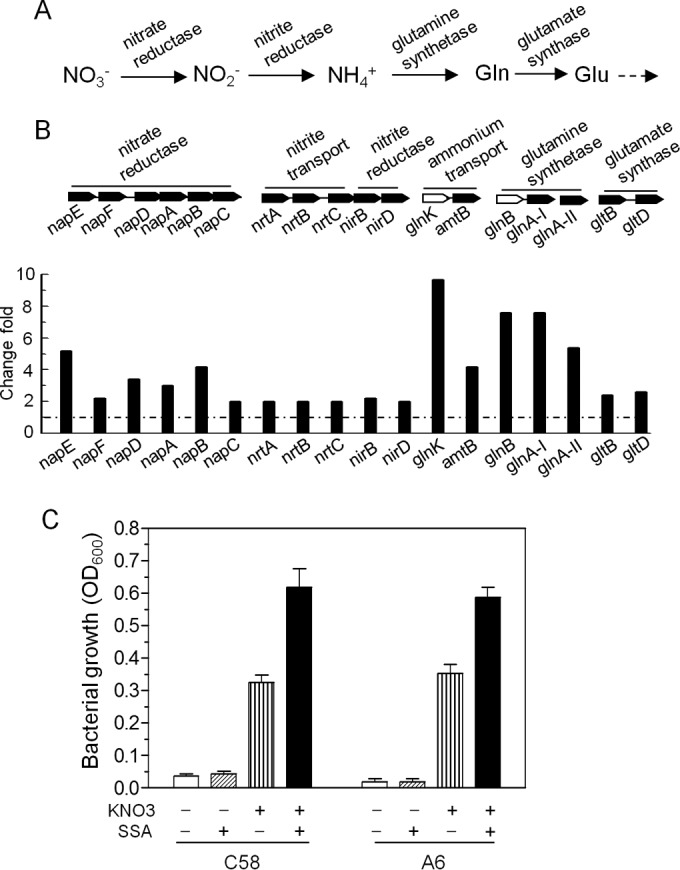
SSA promotes nitrate assimilation in A. tumefaciens. (A) The metabolic pathway for nitrate assimilation in A. tumefaciens. Glu, glutamate; Gln, glutamine. (B) Arrangement of nitrate assimilatory gene clusters and their induction profiles in microarray. The clusters are composed of the genes encoding putative nitrate reductase, nitrite reductase, glutamine synthetase, glutamate synthase, and related transporters and regulators (open columns). The arrows indicate the transcriptional direction. (C) Growth of A. tumefaciens using KNO3 as the sole nitrogen source with or without SSA treatment. The fresh bacteria from LB agar were resuspended in PBS and then inoculated at a ratio of 1% in a defined medium. −, no chemical addition; +, chemical addition. Error bars denote standard deviations.
AttK and AttL, but not AttM, are responsible for the SSA-mediated nitrate assimilation.
To examine whether AttJ-AttKLM are involved in the SSA-mediated nitrate assimilation, we used the attJ-attKLM mutants to investigate bacterial growth using nitrate as the sole nitrogen source. Results showed that the wild-type strain and its mutants grew at similar rates without SSA. In contrast, addition of SSA significantly boosted the growth of the wild-type strain and the ΔattJ mutant, whereas the ΔattKLM and ΔattKLM ΔattJ mutants failed to respond to SSA and had growth rates similar to those in the absence of SSA (Fig. 4A). To determine which gene in attKLM is responsible for the SSA-promoted nitrate assimilation, we constitutively expressed the corresponding individual genes in the ΔattKLM mutant. Results showed that expression of attK or attL could partially restore the bacterial response to the level of that with SSA treatment (Fig. 4B). In contrast, overexpression of attM displayed no significant effect as the ΔattKLM (attM) strain grew similarly with or without SSA (Fig. 4B). We also expressed attK, attL, and attM separately in the ΔattKLM ΔattJ strain in which attJ was further mutated in the background of ΔattKLM. Similar to the results for the ΔattKLM strain, overexpression of attK or attL but not of attM restored the bacterial response to SSA (Fig. 4C). Sequence analyses indicate that both attK and attL encode homologues of dehydrogenases, but it is not yet clear how these two dehydrogenases are involved in nitrate utilization.
FIG 4.
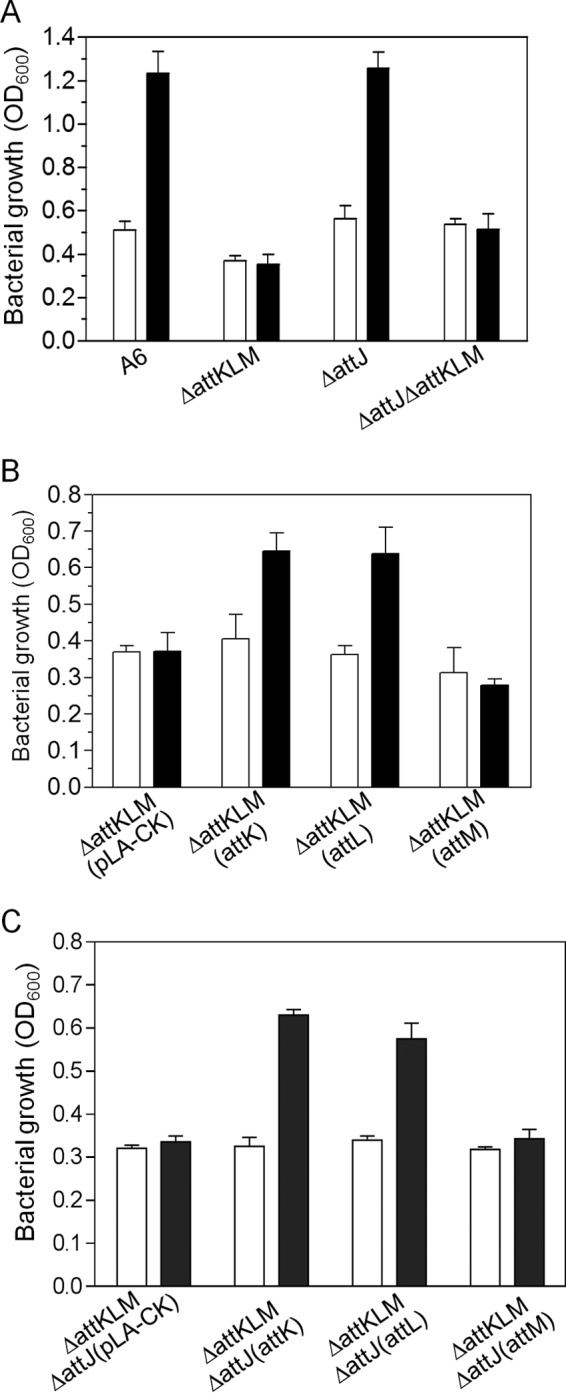
AttKL, but not AttJ and AttM, was required for nitrate utilization in A. tumefaciens. Bacterial growth of A. tumefaciens A6 and its mutants was determined using KNO3 as the sole nitrogen source. OD600 values were recorded for cells cultured with shaking at 220 rpm at 28°C for 24 h. Open bars, no SSA added; filled bars, SSA added. Error bars denote standard deviations.
In addition to AttK, AldH was also demonstrated to convert SSA to SA, and inactivation of aldH in the ΔattK strain led to accumulation of SSA in A. tumefaciens (3, 7). To test the potential involvement of AldH in assimilation of C4-dicarboxylates and nitrates, the aldH gene was deleted from the mutant ΔattKLM strain, and bacterial growth was examined accordingly. With nitrate as the sole nitrogen source, the growth of the mutant lacking both attK and aldH was indistinguishable from that of the mutant lacking only attK (see Fig. S2 in the supplemental material). However, no growth of the ΔattKLM ΔaldH strain was observed with exogenous addition of SSA, even at a concentration as low as 20 μM (see Fig. S2). These results suggest the attK aldH double deletion mutant was highly sensitive to the toxicity of SSA. Further experiments are required to study the biological significance of AldH in A. tumefaciens physiology.
SA promotes nitrate utilization in an attJ-attKLM-independent manner.
It has been proposed that attK functions as a dehydrogenase which converts SSA into SA for the tricarboxylic acid (TCA) cycle. The requirement of attKLM for SSA-promoted nitrate assimilation tempted us to test whether SA, the hydrolyzing product of SSA, could also induce bacteria to use nitrate as the sole nitrogen source. The results showed that, similar to SSA, SA promoted the bacterial growth of the wild-type strain and the attJ mutant to an extent that was statistically significant, but growth was less than that with SSA (P < 0.005, t test). However, SA also promoted bacterial growth of ΔattKLM and ΔattKLM ΔattJ strains, which was different from the results with SSA (Fig. 5A). As a comparison, we further investigated whether other carbon sources could also promote bacterial growth with nitrate as the sole nitrogen source. The results showed that only SSA and its product SA could promote nitrate assimilation by the wild-type strain A6, whereas other C4-dicarboxylates, including malate and α-ketoglutaric acid, and common carbon sources, such as mannitol, glucose, and fructose, failed to promote nitrate assimilation (Fig. 5B). Altogether, these results suggest that SA, like its precursor SSA, is able to induce bacteria to assimilate nitrate for growth, and this promotion effect of SA is independent of the attJ-attKLM operon.
FIG 5.
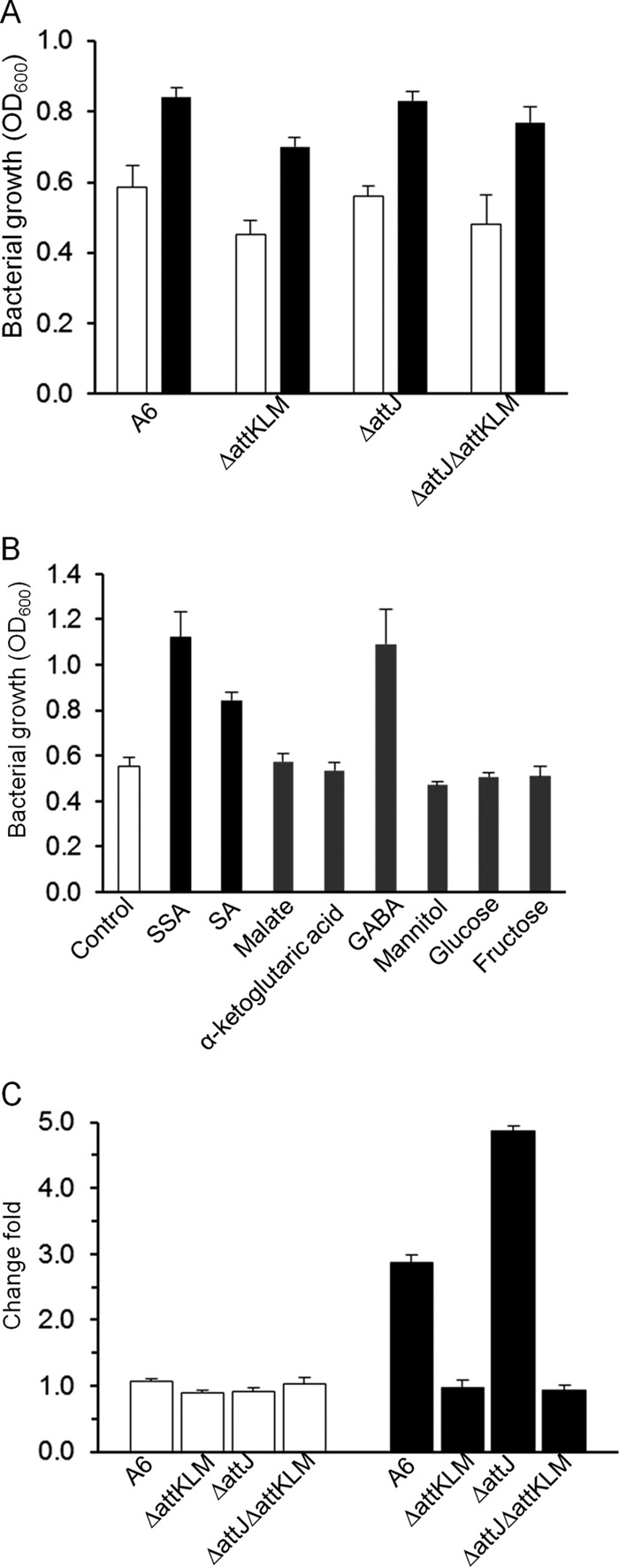
SA promotes nitrate utilization independent of attJ-attKLM. (A) Bacterial growth of A6 and its mutants using KNO3 as the sole nitrogen source with (filled bars) or without (open bars) SA. (B) Bacterial growth of A6 using KNO3 as the sole nitrogen source supplied by the given compounds. (C) Changes in the NADH/NAD (open bars) and NADPH/NADP (filled bars) ratios in A. tumefaciens A6 and its mutants upon SSA treatment.
In A. tumefaciens, AttK is an enzyme supposed to convert SSA into SA, and this conversion is accompanied by the oxidation of NAD(P)H. Results showing that SA promoted bacterial growth encouraged us to examine the changes in abundance of NAD(P)H in bacterial cells in the presence or absence of SSA. First, we treated bacterial cells with SSA and determined the changes in the NADH/NAD and NADPH/NADP ratios. As shown in Fig. 5C, no significant change was recorded for the NADH/NAD ratio in wild-type strain A6 and its attJ-attKLM mutant with and without SSA treatment. For the NADPH/NADP ratio, however, addition of SSA led to a 3-fold increase in the wild-type strain over the level of the blank control without SSA. In the ΔattJ mutant, the SSA-induced increase in the NADPH/NADP ratio appeared even higher than that of the wild type, with about a 4.5-fold increase above the level of the control without SSA. When attKLM was mutated, however, this SSA-stimulated increase vanished, as shown by results for the ΔattKLM and ΔattKLM ΔattJ mutant strains (Fig. 5C). Although purified AttK has been found to prefer NADH over NADPH as a cofactor in vitro (3, 7), our results demonstrate that SSA treatment significantly affected the intracellular ratio of NADPH/NADP but not of NADH/NAD, and the SSA-induced change in the NADPH/NADP ratio is heavily dependent on the functions encoded by attKLM.
SSA adapts bacteria to hydrogen peroxide protection.
In various bacteria, nitrate assimilation and changes in the NADPH/NADP ratio are closely associated with the generation of reactive oxygen species (ROS) (35). ROS, such as a superoxide anion radical (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (HO−), are unavoidably generated during normal respiratory processes, and they can damage diverse cellular molecules (36). To protect against ROS-induced damage, bacteria have evolved sophisticated molecular mechanisms to sense ROS levels and synthesize enzymes such as catalase and superoxide dismutase (SOD), small proteins like thioredoxin (Trx) and glutaredoxin (Grx), and oligomeric proteins such as bacterioferritin (Bfr) (37–39). SOD catalyzes O2− to H2O2 and H2O, and catalase converts H2O2 to O2 and H2O2, both of which directly reduce the toxicity of H2O2 and prevent the formation of hydroxyl radicals via the Fenton reaction (40). Trx and Grx are the protein disulfide reductases which serve as electron donors for enzymes such as peroxiredoxins (Prx) and sulfoxide reductases and therefore protect against oxidative damage (38, 41, 42). Bfr, on the other hand, oxidizes excess ferrous ions and protects bacterial cells against the oxidative stress associated with ferrous ions (43).
A. tumefaciens contains two catalases (KatA and CatE), three SODs (SodB, SodF, and Atu4583), two Trxs (Atu0022 and Atu3698), two Grxs (Atu0068 and Atu3511), and three Prxs (Atu1480, Atu0779, and Atu2399) (44–46). Our microarray results showed that addition of SSA did not affect transcriptional expression of the genes encoding catalases and the regulator OxyR (47), indicating that direct detoxification of H2O2 is unlikely modulated by SSA in A. tumefaciens. However, the transcript levels of sodF, the Trx gene atu0022, the Grx gene atu0068, the Prx gene atu0779, and the Bfr gene atu2771 were significantly elevated with SSA treatment (Fig. 6A), suggesting that SSA may be involved in regulation of oxidative protection. To test this possibility, we carried out an H2O2 killing assay and found that the SSA pretreatment could significantly protect bacterial cells against the oxidative reagent. As shown in Fig. 6B, preexposure of the nopaline strain C58 to SSA conferred a greater than 10-fold increase in resistance to the lethal dosage of H2O2 over levels in the untreated cells. Similar results were also observed for the octopine strain A6 (Fig. 6B). Mutation of attJ did not affect the SSA-induced bacterial resistance to H2O2. However, deletion of attKLM entirely abolished this SSA-mediated resistance (Fig. 6B), suggesting a key role of attKLM and SSA in modulation of the bacterial defense against oxidative stresses.
FIG 6.
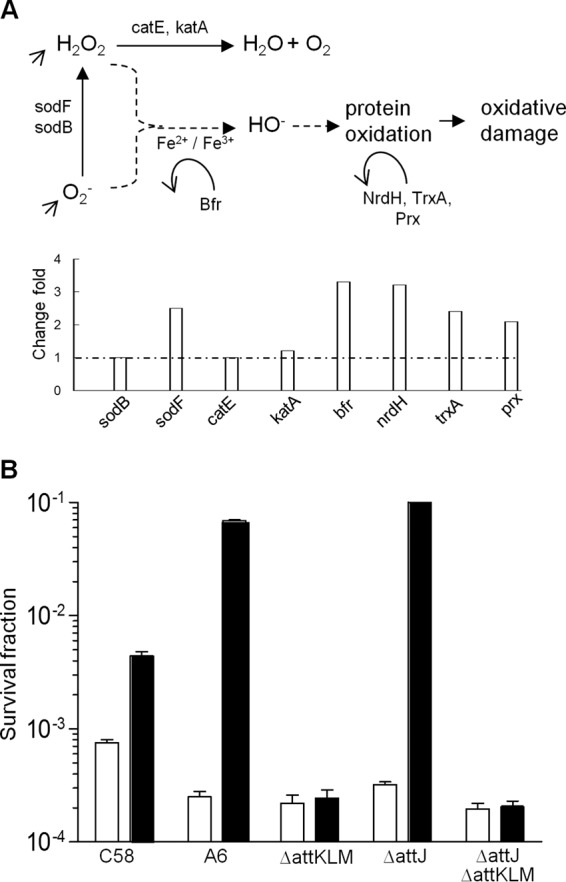
SSA induces oxidative stress resistance in A. tumefaciens. (A) Putative defense systems against oxidative stresses in A. tumefaciens and the expression profiles of related genes in microarray analysis. (B) SSA enhanced bacterial resistance against H2O2. The experiments were performed by adding 20 mM H2O2 to the fresh cell cultures with or without SSA induction. The cultures were then grown for an additional 30 min before aliquots of cells were removed, washed once prior to preparation of appropriate dilutions, and plated on LB agar. Colonies were counted after 48 h of incubation at 28°C. Surviving fractions were defined as the number of CFU recovered after the treatment divided by the number of CFU without treatment. Open bars, no SSA treatment; filled bars, SSA treatment. Error bars denote standard deviations.
attJ-attKLM regulate the tumorigenicity of A. tumefaciens.
To further study the physiological role of attJ-attKLM, we analyzed bacterial virulence by infecting the plant host Kalanchoe daigremontiana. We grew the wild-type and mutant strains in LB medium. After cells were washed with PBS, different amounts of bacterial cells were inoculated into the wound sites of K. daigremontiana. We found that both the wild-type and ΔattJ mutant of octopine-type A6 readily incited plant tumors with an inoculum of 106 cells, while the ΔattKLM and ΔattKLMΔattJ mutant strains hardly developed any plant tumors. With an inoculum of 105 cells, however, the wild type could not induce tumors, consistent with the previous results showing that the A6 strain is less virulent than the C58 strain. In contrast, the ΔattJ strain continued to induce tumors even with an inoculum of 104 cells, indicating a stronger virulence than that of the wild-type and the attKLM mutant strains (Fig. 7A). Analyses of tumor weights also confirmed that overexpression of attKLM by inactivation of the repressor gene attJ enhanced bacterial virulence and that mutation of attKLM impaired the development of plant tumors (Fig. 7B). These results appeared different from the findings of a previous study, in which the nopaline strain C58 was used to infect tobacco plants, and the results showed that the virulence of the attM mutant was similar to or even greater than that of the wild-type strain (4, 48). Strain specificity or a difference in host plants may explain this discrepancy, given that different plant hosts react differently to infections by different A. tumefaciens strains (49).
DISCUSSION
Previous results suggest that SSA, a metabolic intermediate of GABA, functions as a ligand of AttJ and induces the expression of attKLM, which leads to disruption of the QS signaling of A. tumefaciens during plant infections (4). However, a recent finding that attKLM does not affect Ti plasmid conjugation in plants argues the biological significance of SSA in regulation of bacterial virulence (50). In this study, we used the combined application of microarray and genetic analyses to understand the roles of SSA in regulation of bacterial physiology and virulence, and our results revealed three new biological functions influenced by SSA, i.e., promoting C4-dicarboxylate utilization, nitrate assimilation, and ROS resistance in A. tumefaciens. Unlike its related metabolites GABA and SA, SSA could not support bacterial growth when supplied as the sole carbon source (Fig. 2), which seems to exclude the possibility that SSA promotes the utilization of malate and nitrate to support bacterial growth through its downstream metabolite SA.
In addition to modulation of quorum quenching and GABA metabolism, our results demonstrate that SSA plays additional roles through two different types of mechanisms to promote the bacterial survival capability (Fig. 8). First, SSA, independent of the attJ-attKLM genes, induces the expression of the C4-dicarboxylate importer dctA and hence promotes the utilization of TCA intermediates (Fig. 2). It has been known that C4-dicarboxylates are abundant in wound sites and plant tumors, and the transporter dctA is regulated by the two-component system DctB/DctD in bacteria (31). In its genome, A. tumefaciens carries two sets of the DctB/DctD pair, with one located on the cell membrane and one in the cytosol (see Fig. S3 in the supplemental material). Our results showed that SSA induced dctA transcription independent of the attJ-attKLM genes, suggesting that SSA may bind to the membrane-bound DctB/DctD and consequently induce the expression of DctA, which imports the plant-produced C4-dicarboxylates, including SSA, leading to attKLM induction and utilization of C4-dicarboxylates. Consistent with this notion, inactivation of dctB1 reduced the bacterial growth rate when malate was supplied as the sole carbon source, and supplementation of SSA had no effect on the ΔdctB1 strain (see Fig. S5 in the supplemental material). While these results demonstrate the importance of DctB1 in the SSA-mediated promotion of C4-dicarboxylate utilization and nitrate assimilation, further characterization of DctB1 and DctB2 is required to provide a clear understanding of their physiological roles.
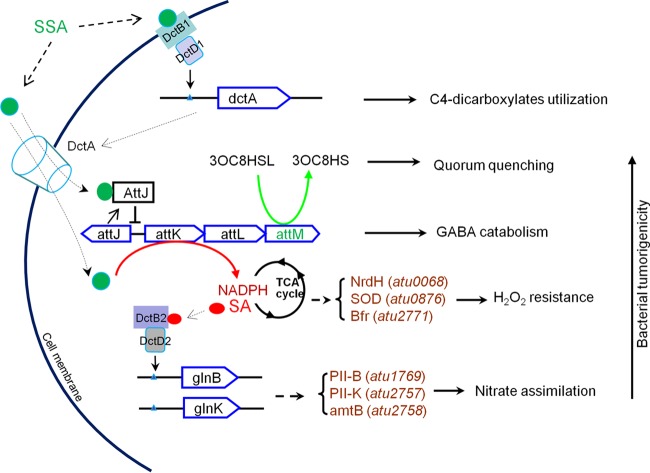
FIG 8.
Schematic presentation of the genetic and physiological response to SSA in A. tumefaciens. Solid lines indicate the processes supported by experimental evidence, while dashed lines indicate speculative processes that await further experimental validation. 3OC8HS(L), 3-oxo-octanoyl-homoserine (lactone); PII, nitrogen regulatory protein.
Second, our results revealed that SSA could promote nitrate assimilation in an attJKLM-dependent manner. Once inside bacterial cells, SSA acts as a signal directly binding to AttJ to activate the expression of attKLM (3), and their products AttK/AttL/AttM work together to metabolize GABA to support bacterial growth. Additionally, AttM functions as an AHL-lactonase to degrade QS signal and terminate or postpone the QS-dependent replication and conjugation of Ti plasmid (4, 8, 9, 48, 51). We showed in this study that conversion of SSA to SA could also influence the bacterial redox status, as reflected by the increased NADPH/NADP ratio, which was accompanied by enhanced nitrate assimilation (Fig. 3 to 6). Under normal conditions, nitrate is not a preferred nitrogen source for A. tumefaciens, but it is a major nitrogen source for plant growth and functions as a critical signal molecule regulating the whole plant development process (52–54). Nitrate is mainly taken up from the soil by plant roots, stored in vacuoles, and accumulated at the wound sites for healing (55). It has been reported that nitrate is capable of increasing the number of tumors on inoculated plant hosts by 200% (56). During infection, A. tumefaciens inevitably encounters nutrient starvation, and it is conceivable that this bacterium has evolved the SSA-AttJKLM system to take advantage of the nitrate compounds. At present, the precise mechanism by which SSA promotes nitrate assimilation is not clear. In A. tumefaciens, genes for nitrate assimilation are mainly located in two clusters (see Fig. S2 in the supplemental material). Our microarray results show that the majority of these genes are upregulated by SSA. In various bacteria, nitrate-assimilating genes are regulated by the nitrogen-limiting sigma factor RpoN (57). Unlike other sigma factors such as RpoS and RpoH, RpoN requires an extra coactivator for gene activation (58). In the case of the nitrogen-assimilating genes, this coactivator has been identified as the response regulator of the DctB/DctD two-component system. Interestingly, sequence analyses revealed that glnK and glnB, like the dctA gene, contain typical DNA motifs for DctD binding in their promoter regions (see Fig. S4 in the supplemental material). Similar to dctA, SSA may activate the nitrate-assimilating genes by activating the DctB/DctD two-component system. However, unlike the expression of DctA, SSA-mediated nitrate assimilation is dependent on the function of attKLM, and the SSA-hydrolyzing product SA can also promote bacterial nitrate assimilation, suggesting that the intracellular SA but not SSA itself is the direct signal for this phenomenon. Given that two sets of the DctB/DctD system are present with different localizations in A. tumefaciens, it is most likely that one DctB/DctD pair specifically senses extracellular SSA on the cell membrane and that the other specifically senses intracellular SA in the bacterial cytosol. Intracellular SA may directly bind to DctB/DctD and activate the expression of glnB and glnK, thus inducing the nitrate response. It is also possible that SA, which was converted from SSA, revamps the central metabolism of A. tumefaciens and alters the level of other metabolic regulators such as glutamate and acetate, which are responsible for the nitrate response.
In addition to modulation of C4-dicarboxylates and nitrate utilization, our results showed that SSA also contributes to ROS resistance and bacterial tumorigenicity (Fig. 6). For plants, a ROS burst is an early response to bacterial infection, where huge amounts of ROS are rapidly produced at the wound sites to directly kill the invading bacterial pathogens at the early stage of infection (59). Our results indicate that SSA could protect bacteria from killing by ROS. This finding results from the observation that conversion of SSA to SA is associated with the change in the NADPH/NADP ratio. NADPH usually serves as an electron donor for the respiratory chain; its oxidation inevitably produces ROS. At a sublethal concentration, ROS might trigger the protective mechanism of A. tumefaciens against subsequent oxidative stresses (60). It is not yet clear how SSA exactly activates this protective response. Given that the SSA-induced protection from ROS stress is dependent on the presence of attKLM, it is likely that SSA may need to be converted into succinate, which may increase the NADPH/NAD ratio and the ROS level and subsequently induce the expression of protective systems.
During a long history of coevolution with plants, A. tumefaciens has evolved complicated signaling and regulatory networks for establishing a successful infection. The SSA-regulated attJ-attKLM signaling pathway is one such example. The findings from this study allow us to propose a signaling and regulatory working model for SSA (Fig. 8). During A. tumefaciens infection, the plant-derived SSA is sensed by the membrane-bound sensor DctB1 of the bacterial pathogen, which leads to activation of the response regulator DctD1 and, consequently, induces the expression of dctA. The transporter DctA imports SSA into bacterial cells and induces the expression of attKLM by binding to the transcriptional repressor AttJ. While AttK/AttL/AttM work together to metabolize GBL to support bacterial growth as nitrogen and carbon sources, AttM also works alone as an AHL-lactonase to quench the QS system and provide fine control of the bacterial energy-consuming processes, such as Ti plasmid replication and conjugation. In addition, expressed AttK converts SSA into SA, which not only detoxifies the high concentration of SSA but also influences the NADPH/NADP ratio. Functioning as an intracellular signal, SA is sensed by the cytosolic sensor DctB2 and activates the corresponding regulator DctD2, which subsequently expresses the nitrate-assimilating genes and promotes bacterial resistance to oxidative stresses. While the detailed molecular mechanisms of this working model await further experimental validation, it is clear from the findings of this study that SSA could be a potent plant signal which modulates bacterial physiology and contributes to the survival capability and tumorigenicity of A. tumefaciens during infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by the Singapore Biomedical Research Council (A*STAR), the National Basic Research Program of China (973 Program, number 2015CB150600), and the Singapore National Research Foundation (NRF-RF2009-RF001-267).
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00373-15.
REFERENCES
- 1.Subramoni S, Nathoo N, Klimov E, Yuan ZC. 2014. Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front Plant Sci 5:322. doi: 10.3389/fpls.2014.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang HB, Wang C, Zhang LH. 2004. The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p)ppGpp. Mol Microbiol 52:1389–1401. doi: 10.1111/j.1365-2958.2004.04061.x. [DOI] [PubMed] [Google Scholar]
- 3.Wang C, Zhang HB, Wang LH, Zhang LH. 2006. Succinic semialdehyde couples stress response to quorum-sensing signal decay in Agrobacterium tumefaciens. Mol Microbiol 62:45–56. doi: 10.1111/j.1365-2958.2006.05351.x. [DOI] [PubMed] [Google Scholar]
- 4.Chevrot R, Rosen R, Haudecoeur E, Cirou A, Shelp BJ, Ron E, Faure D. 2006. GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 103:7460–7464. doi: 10.1073/pnas.0600313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan ZC, Haudecoeur E, Faure D, Kerr KF, Nester EW. 2008. Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and gamma-amino butyric acid reveals signalling cross-talk and Agrobacterium-plant co-evolution. Cell Microbiol 10:2339–2354. doi: 10.1111/j.1462-5822.2008.01215.x. [DOI] [PubMed] [Google Scholar]
- 6.Carlier A, Chevrot R, Dessaux Y, Faure D. 2004. The assimilation of gamma-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol Plant Microbe Interact 17:951–957. doi: 10.1094/MPMI.2004.17.9.951. [DOI] [PubMed] [Google Scholar]
- 7.Chai Y, Tsai CS, Cho H, Winans SC. 2007. Reconstitution of the biochemical activities of the AttJ repressor and the AttK, AttL, and AttM catabolic enzymes of Agrobacterium tumefaciens. J Bacteriol 189:3674–3679. doi: 10.1128/JB.01274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang HB, Wang LH, Zhang LH. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 99:4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pappas KM. 2008. Cell-cell signaling and the Agrobacterium tumefaciens Ti plasmid copy number fluctuations. Plasmid 60:89–107. doi: 10.1016/j.plasmid.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Pan Y, Fiscus V, Meng W, Zheng Z, Zhang LH, Fuqua C, Chen L. 2011. The Agrobacterium tumefaciens transcription factor BlcR is regulated via oligomerization. J Biol Chem 286:20431–20440. doi: 10.1074/jbc.M110.196154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina-Henares AJ, Krell T, Eugenia Guazzaroni M, Segura A, Ramos JL. 2006. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol Rev 30:157–186. doi: 10.1111/j.1574-6976.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Jiang G, Cui Y, Mukherjee A, Ma WL, Chatterjee AK. 1999. kdgREcc negatively regulates genes for pectinases, cellulase, protease, HarpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora. J Bacteriol 181:2411–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maloy SR, Nunn WD. 1982. Genetic regulation of the glyoxylate shunt in Escherichia coli K-12. J Bacteriol 149:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nasser W, Reverchon S, Robert-Baudouy J. 1992. Purification and functional characterization of the KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi. Mol Microbiol 6:257–265. doi: 10.1111/j.1365-2958.1992.tb02007.x. [DOI] [PubMed] [Google Scholar]
- 15.Sunnarborg A, Klumpp D, Chung T, LaPorte DC. 1990. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J Bacteriol 172:2642–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Wezel GP, van der Meulen J, Kawamoto S, Luiten RG, Koerten HK, Kraal B. 2000. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J Bacteriol 182:5653–5662. doi: 10.1128/JB.182.20.5653-5662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta M, Hogema BM, Grompe M, Bottiglieri TG, Concas A, Biggio G, Sogliano C, Rigamonti AE, Pearl PL, Snead OC III, Jakobs C, Gibson KM. 2003. Murine succinate semialdehyde dehydrogenase deficiency. Ann Neurol 54(Suppl 6):S81–S90. doi: 10.1002/ana.10625. [DOI] [PubMed] [Google Scholar]
- 18.Yogeeswari P, Sriram D, Vaigundaragavendran J. 2005. The GABA shunt: an attractive and potential therapeutic target in the treatment of epileptic disorders. Curr Drug Metab 6:127–139. doi: 10.2174/1389200053586073. [DOI] [PubMed] [Google Scholar]
- 19.Bouche N, Fait A, Bouchez D, Moller SG, Fromm H. 2003. Mitochondrial succinic-semialdehyde dehydrogenase of the gamma-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proc Natl Acad Sci U S A 100:6843–6848. doi: 10.1073/pnas.1037532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fait A, Yellin A, Fromm H. 2005. GABA shunt deficiencies and accumulation of reactive oxygen intermediates: insight from Arabidopsis mutants. FEBS Lett 579:415–420. doi: 10.1016/j.febslet.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 21.Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS. 2001. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J Biol Chem 276:244–250. doi: 10.1074/jbc.M007103200. [DOI] [PubMed] [Google Scholar]
- 22.Lutke-Eversloh T, Steinbuchel A. 1999. Biochemical and molecular characterization of a succinate semialdehyde dehydrogenase involved in the catabolism of 4-hydroxybutyric acid in Ralstonia eutropha. FEMS Microbiol Lett 181:63–71. doi: 10.1111/j.1574-6968.1999.tb08827.x. [DOI] [PubMed] [Google Scholar]
- 23.Wilms I, Voss B, Hess WR, Leichert LI, Narberhaus F. 2011. Small RNA-mediated control of the Agrobacterium tumefaciens GABA binding protein. Mol Microbiol 80:492–506. doi: 10.1111/j.1365-2958.2011.07589.x. [DOI] [PubMed] [Google Scholar]
- 24.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 25.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF Jr, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D Sr, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 27.Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 28.Garfinkel DJ, Nester EW. 1980. Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J Bacteriol 144:732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spaink HP. 1995. The molecular basis of infection and nodulation by rhizobia: the ins and outs of sympathogenesis. Annu Rev Phytopathol 33:345–368. doi: 10.1146/annurev.py.33.090195.002021. [DOI] [PubMed] [Google Scholar]
- 30.Wu ZL, Charles TC, Wang H, Nester EW. 1992. The ntrA gene of Agrobacterium tumefaciens: identification, cloning, and phenotype of a site-directed mutant. J Bacteriol 174:2720–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yurgel SN, Kahn ML. 2004. Dicarboxylate transport by rhizobia. FEMS Microbiol Rev 28:489–501. doi: 10.1016/j.femsre.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Ronson CW, Nixon BT, Albright LM, Ausubel FM. 1987. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J Bacteriol 169:2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin JT, Stewart V. 1998. Nitrate assimilation by bacteria. Adv Microb Physiol 39:1–30, 379. [DOI] [PubMed] [Google Scholar]
- 34.Spaink HP, Kondorosi A, Hooykaas PJJ. 1998. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Kluwer Academic Publishers, Boston, MA. [Google Scholar]
- 35.Shimizu K. 2013. Metabolic regulation of a bacterial cell system with emphasis on Escherichia coli metabolism. ISRN Biochem 2013:645983. doi: 10.1155/2013/645983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison J, Jamet A, Muglia CI, Van de Sype G, Aguilar OM, Puppo A, Frendo P. 2005. Glutathione plays a fundamental role in growth and symbiotic capacity of Sinorhizobium meliloti. J Bacteriol 187:168–174. doi: 10.1128/JB.187.1.168-174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofmann B, Hecht HJ, Flohe L. 2002. Peroxiredoxins. Biol Chem 383:347–364. [DOI] [PubMed] [Google Scholar]
- 39.Steinman HM, Fareed F, Weinstein L. 1997. Catalase-peroxidase of Caulobacter crescentus: function and role in stationary-phase survival. J Bacteriol 179:6831–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCord JM, Fridovich I. 1969. The utility of superoxide dismutase in studying free radical reactions. I. Radicals generated by the interaction of sulfite, dimethyl sulfoxide, and oxygen. J Biol Chem 244:6056–6063. [PubMed] [Google Scholar]
- 41.Arner ES, Holmgren A. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Manzaneque MT, Ros J, Cabiscol E, Sorribas A, Herrero E. 1999. Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol Cell Biol 19:8180–8190. doi: 10.1128/MCB.19.12.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JL. 2004. The physiological role of ferritin-like compounds in bacteria. Crit Rev Microbiol 30:173–185. doi: 10.1080/10408410490435151. [DOI] [PubMed] [Google Scholar]
- 44.Prapagdee B, Eiamphungporn W, Saenkham P, Mongkolsuk S, Vattanaviboon P. 2004. Analysis of growth phase regulated KatA and CatE and their physiological roles in determining hydrogen peroxide resistance in Agrobacterium tumefaciens. FEMS Microbiol Lett 237:219–226. doi: 10.1111/j.1574-6968.2004.tb09699.x. [DOI] [PubMed] [Google Scholar]
- 45.Xu XQ, Pan SQ. 2000. An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol Microbiol 35:407–414. doi: 10.1046/j.1365-2958.2000.01709.x. [DOI] [PubMed] [Google Scholar]
- 46.Eiamphungporn W, Nakjarung K, Prapagdee B, Vattanaviboon P, Mongkolsuk S. 2003. Oxidant-inducible resistance to hydrogen peroxide killing in Agrobacterium tumefaciens requires the global peroxide sensor-regulator OxyR and KatA. FEMS Microbiol Lett 225:167–172. doi: 10.1016/S0378-1097(03)00511-1. [DOI] [PubMed] [Google Scholar]
- 47.Nakjarung K, Mongkolsuk S, Vattanaviboon P. 2003. The oxyR from Agrobacterium tumefaciens: evaluation of its role in the regulation of catalase and peroxide responses. Biochem Biophys Res Commun 304:41–47. doi: 10.1016/S0006-291X(03)00535-7. [DOI] [PubMed] [Google Scholar]
- 48.Haudecoeur E, Planamente S, Cirou A, Tannieres M, Shelp BJ, Morera S, Faure D. 2009. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc Natl Acad Sci U S A 106:14587–14592. doi: 10.1073/pnas.0808005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ooms G, Klapwijk PM, Poulis JA, Schilperoort RA. 1980. Characterization of Tn904 insertions in octopine Ti plasmid mutants of Agrobacterium tumefaciens. J Bacteriol 144:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan SR, Farrand SK. 2009. The BlcC (AttM) lactonase of Agrobacterium tumefaciens does not quench the quorum-sensing system that regulates Ti plasmid conjugative transfer. J Bacteriol 191:1320–1329. doi: 10.1128/JB.01304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haudecoeur E, Faure D. 2010. A fine control of quorum-sensing communication in Agrobacterium tumefaciens. Commun Integr Biol 3:84–88. doi: 10.4161/cib.3.2.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang YY, Tsay YF. 2011. Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23:1945–1957. doi: 10.1105/tpc.111.083618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gojon A, Krouk G, Perrine-Walker F, Laugier E. 2011. Nitrate transceptor(s) in plants. J Exp Bot 62:2299–2308. doi: 10.1093/jxb/erq419. [DOI] [PubMed] [Google Scholar]
- 54.Vidal EA, Gutierrez RA. 2008. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Curr Opin Plant Biol 11:521–529. doi: 10.1016/j.pbi.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 55.Song J, Ding X, Feng G, Zhang F. 2006. Nutritional and osmotic roles of nitrate in a euhalophyte and a xerophyte in saline conditions. New Phytol 171:357–366. doi: 10.1111/j.1469-8137.2006.01748.x. [DOI] [PubMed] [Google Scholar]
- 56.Khalfa MDE, Lippincott JA. 1968. Promotion of crown-gall tumor initiation on primary pinto bean leaves by certain inorganic salts. Plant Cell Physiol 9:217–225. [Google Scholar]
- 57.Fischer HM. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev 58:352–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dombrecht B, Marchal K, Vanderleyden J, Michiels J. 2002. Prediction and overview of the RpoN-regulon in closely related species of the Rhizobiales. Genome Biol 3:RESEARCH0076. doi: 10.1186/gb-2002-3-12-research0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wojtaszek P. 1997. Oxidative burst: an early plant response to pathogen infection. Biochem J 322:681–692. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu XQ, Li LP, Pan SQ. 2001. Feedback regulation of an Agrobacterium catalase gene katA involved in Agrobacterium-plant interaction. Mol Microbiol 42:645–657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.