Abstract
The genus Acinetobacter encompasses multiple nosocomial opportunistic pathogens that are of increasing worldwide relevance because of their ability to survive exposure to various antimicrobial and sterilization agents. Among these, Acinetobacter baumannii, Acinetobacter nosocomialis, and Acinetobacter pittii are the most frequently isolated in hospitals around the world. Despite the growing incidence of multidrug-resistant Acinetobacter spp., little is known about the factors that contribute to pathogenesis. New strategies for treating and managing infections caused by multidrug-resistant Acinetobacter strains are urgently needed, and this requires a detailed understanding of the pathobiology of these organisms. In recent years, some virulence factors important for Acinetobacter colonization have started to emerge. In this review, we focus on several recently described virulence factors that act at the bacterial surface level, such as the capsule, O-linked protein glycosylation, and adhesins. Furthermore, we describe the current knowledge regarding the type II and type VI secretion systems present in these strains.
INTRODUCTION
Infections caused by pathogenic members of the genus Acinetobacter are emerging as a significant threat to human health. These Gram-negative bacteria are responsible for increasing numbers of infections encountered in hospitals, particularly among immunocompromised patients (1, 2), and community-acquired infections are also increasing in prevalence (3). A recently recognized population at particular risk for Acinetobacter infections is military service members who have suffered combat-related injuries, who often acquire these infections in field hospitals (4–6). Although Acinetobacter spp. primarily cause pneumonia, they are also frequent causes of wound and burn infections, meningitis, urinary tract infections, and sepsis (7). There is a growing trend for these isolates to display high levels of antibiotic resistance, with some being resistant to all clinically available antibiotics (8). Collectively, these attributes have led to a pressing need to elucidate the mechanisms used by pathogenic Acinetobacter to cause disease.
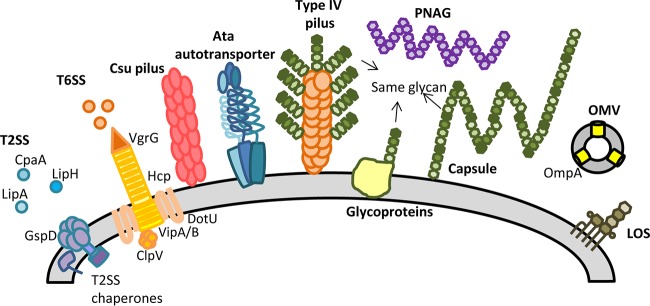
The bacterial cell surface plays essential roles in sensing of the environment, interactions with the host, and maintenance of cellular homeostasis (9). The molecular structures present on the cell surface, and those that extend beyond the surface, are of central importance for understanding the pathogenesis of an organism; these are often the key determinants that mediate bacterial virulence and thus represent important targets for novel antimicrobials and vaccines (10). This review highlights the recent research on pathogenic Acinetobacter spp., which has led to several key findings on the strategies used by these bacteria to elaborate cell surface and secretion components that are vital for causing disease (Fig. 1).
FIG 1.
Cell surface components and secretion systems identified in Acinetobacter spp.
CELL SURFACE
The cell surface of Gram-negative bacteria plays myriad roles in the physiology of these organisms, including transport of molecules into and out of the cell, interaction with and sensing of the extracellular environment, and protection from external stresses (9). While all bacterial cell surfaces are composed primarily of lipids, carbohydrates, and proteins, the diversity in the molecular composition and arrangement of these structures has vast implications for virulence in pathogenic bacteria. Recent experimental investigation into the arrangement and composition of these structures on Acinetobacter cell surfaces has provided important insights into their role in the pathobiology of these important human pathogens.
Lipooligosaccharide.
Lipopolysaccharide (LPS), the major component of the outer leaflet of the outer membrane (OM) of many Gram-negative bacteria, is an immunostimulatory molecule that plays an important role in bacterial resistance to external stresses (11). LPS is composed of the endotoxic lipid A, a core oligosaccharide, and a repeating sugar structure called the O antigen (11). During biosynthesis of LPS, the core oligosaccharide is built onto the lipid A moiety in the cytoplasm and flipped into the periplasmic space. The repeat subunit of the O antigen is synthesized separately onto an undecaprenyl phosphate (Und-P) carrier, which is then flipped to the periplasm and ligated to the lipid A core by the WaaL ligase enzyme (12); thus, the WaaL ligase is essential for the production of O antigen. Many pathogens, such as Escherichia coli and Salmonella spp., produce diverse O-antigen structures that form the basis for serotyping schemes (13–15). However, other important bacterial pathogens, such as Neisseria and Campylobacter, lack a WaaL ligase ortholog and do not produce O antigen, elaborating only lipooligosaccharide (LOS) (16).
Whether Acinetobacter spp. elaborate LPS or LOS on their cell surface has been a topic of considerable debate. Several reports have described the presence of LPS in Acinetobacter, many of which have used structural or antibody-based methods to detect O-antigen carbohydrate moieties (17–26). However, most Acinetobacter spp. do not show “typical” LPS laddering upon silver staining of isolated LPS (27, 28), leading to doubts regarding whether Acinetobacter spp. actually produce true O antigen. Intriguingly, Acinetobacter spp. possess either one or two genes, depending on the strain, that encode proteins with domains similar to those found in WaaL ligase orthologs (29–32); however, those domains are also found in PglL, the enzyme responsible for O-linked protein glycosylation (see below) (29). Bioinformatic analysis alone is not sufficient to distinguish between WaaL and PglL orthologs, and careful experimental analysis is required to determine the true function of the proteins possessing these domains. Recent work has now conclusively demonstrated that, in Acinetobacter spp. with a single “waaL-like” gene, the gene actually encodes a PglL enzyme with no O-antigen ligase activity (29). In Acinetobacter spp. with two waaL-like genes, the genes were initially suggested to encode one PglL enzyme and one WaaL enzyme (31), but experimental analysis has identified both enzymes as being exclusively involved in protein O-linked glycosylation, with no role in O-antigen biosynthesis (30). In light of these recent data, and in the absence of experimental data showing a protein possessing divergent O-antigen ligase activity, it seems most likely that Acinetobacter spp. produce LOS but not LPS. In any case, targeted mutagenesis and random mutagenesis of genes involved in synthesis of the LOS core oligosaccharide have shown that this component is a major contributor to Acinetobacter survival and virulence (33–35). The cluster of genes that synthesize the sugar component of LOS are extremely diverse among Acinetobacter spp., and a number of different structures have been determined or predicted (36). Modification of the LOS has also been shown to impart resistance to antimicrobials, similar to the findings for other bacterial species. Specifically, these modifications occur on the lipid A structure of Acinetobacter spp. and lead to decreased susceptibility to antibiotic and antimicrobial peptides and increased survival during desiccation (37–39). Acinetobacter spp. have also been shown to acquire mutations in the lipid A biosynthetic pathway when treated with colistin, resulting in resistance to the antibiotic (40–43). Interestingly, those studies found that mutations in certain lipid A genes resulted in complete loss of LOS.
Glycoproteins.
The posttranslational modification of proteins with glycans, which were once thought to be exclusive to eukaryotes, has been identified in all forms of life. In bacteria, carbohydrates can be attached via the amide of asparagine residues (N-linked) or the hydroxyl of serine/threonine residues (O-linked). In a series of steps analogous to O-antigen biosynthesis, glycans are assembled onto the Und-P lipid carrier and flipped to the periplasmic face of the inner membrane, where an O-oligosaccharyltransferase (O-OTase) enzyme catalyzes the transfer of the complete carbohydrate structure to a serine or threonine residue on the cognate acceptor protein (44).
Bioinformatic analysis identified a protein in Acinetobacter baumannii that showed homology to the O-OTase from Neisseria meningitidis, named PglL (29). PglL proteins often contain domains similar to those of WaaL ligases (Wzy_C domains), and bioinformatic identification of a PglL-like protein is not sufficient to differentiate it from WaaL ligases, thus necessitating experimental characterization. When the pglL-like gene from A. baumannii was deleted, loss of a carbohydrate-specific band was detected after SDS-PAGE analysis, with no change in the LOS profile (29). In-depth characterization of the A. baumannii pglL mutant by mass spectrometry determined that the pglL-deficient strain lacked a total of seven glycoproteins, which were glycosylated with a pentasaccharide in the wild-type strain. Loss of protein glycosylation in A. baumannii resulted in pleiotropic effects on several virulence-associated phenotypes, including biofilm formation and survival in a mouse model of systemic infection (29). Protein glycosylation has been shown to be a conserved modification present throughout the genus Acinetobacter, but the composition of the glycan moiety and the number and identity of the modified proteins vary among strains (45). Interestingly, the carbohydrate structure attached to glycoproteins is identical to the repeat units found in capsular polysaccharide (46). Although the phenotypes associated with complete loss of protein glycosylation have been studied, the contributions of individual glycoproteins to these phenotypes remain unknown.
The pglL gene in A. baumannii is located immediately downstream of the predicted major type IV pilin gene, pilA, which encodes a common target for glycosylation in several bacteria. In A. baumannii ATCC 17978, this protein was not found to be glycosylated under laboratory conditions but was glycosylated upon overexpression in the presence of PglL (29, 30). Intriguingly, most Acinetobacter spp. actually have two proteins with a domain from the Wzy_C superfamily that is common to PglL and WaaL orthologs. It was originally suggested that this second gene could be a WaaL ligase involved in O-antigen biosynthesis (31). Through mutagenesis and functional studies, however, it was determined that both genes actually encode O-OTases (30). The O-OTase encoded by the gene near the pilin gene was found to exclusively glycosylate the cognate pilin protein, similar to the PilO/TfpO protein from Pseudomonas aeruginosa (47), while the second gene encoded a general O-OTase that was responsible for the glycosylation of multiple proteins, similar to PglL from Neisseria (48, 49). Although the functional significance of having two glycosylation systems remains to be determined, this represents the first known case of multiple O-OTases present in a single bacterium.
Capsule.
Like many other pathogens, Acinetobacter spp. produce an extracellular capsule that provides a layer of protection from external threats such as complement-mediated killing (50). Capsule production and protein glycosylation are exquisitely linked in Acinetobacter, as the carbohydrate repeat unit found in the capsule is the same as the single repeat unit attached to proteins (46). The sugar subunits for capsule and protein glycosylation are derived from the same pathway, in which an initiating glycosyltransferase, PglC/ItrA, attaches the first carbohydrate to Und-P, followed by the addition of other sugar monomers by glycosyltransferase enzymes to complete the repeat unit (46). This repeat unit is then flipped to the periplasm, and the capsule and protein glycosylation pathways diverge at this point. In the case of protein glycosylation, this single repeat unit is attached to the target protein by the O-OTase (29). For capsule production, individual sugar repeat units are instead polymerized and exported to the cell surface. This bifurcated pathway represents a novel mechanism that illustrates the evolutionary connections between capsule and protein glycosylation, which may allow Acinetobacter to adapt rapidly to changing environments. How A. baumannii partitions a given carbohydrate repeat unit to the protein glycosylation pathway or capsule production remains to be determined. Although the carbohydrate structures produced by different strains are highly variable, functional studies have shown that capsule production is essential for Acinetobacter survival during infection and growth in serum (32, 46, 50, 51). It was recently reported that capsule production could be increased by the presence of subinhibitory levels of antibiotics, which increased resistance to complement-mediated killing and led to a hypervirulent phenotype in a mouse model of systemic infection (52). This capsule hyperproduction phenotype was shown to be controlled by the two-component BfmRS system, which regulates several other important virulence factors in Acinetobacter (53). Acinetobacter spp. also produce a surface-associated poly-β-1-6-N-acetylglucosamine (PNAG) polysaccharide, which is important for virulence and biofilm formation (54).
Pili.
Filamentous bacterial surface appendages, termed pili, mediate interactions between the producing organism and their environment. Acinetobacter pili have been studied since 1975, when Henrichsen and Blom observed that Acinetobacter calcoaceticus strains displaying surface fimbrial structures exhibited twitching motility (55, 56), a form of bacterial locomotion now known to be dependent on functioning type IV pili (57). Furthermore, it has been shown that the nonpathogenic model organism Acinetobacter baylyi ADP1 produces both thin and thick pili (58); however, the roles pili play in the biology and pathobiology of pathogenic Acinetobacter spp. have been only partially elucidated.
A system of chaperone/usher pili, designated Csu pili, has been identified in all sequenced pathogenic Acinetobacter spp.; however, the Csu pili have been primarily studied in A. baumannii (59). The Acinetobacter Csu pili are required for biofilm formation and maintenance in A. baumannii ATCC 19606 but were found not to play a role in adherence to human epithelial cells (60). Another study found that the CsuA/B pilin subunit was the most abundant protein identified within the pellicle matrix of multiple A. baumannii strains (61), further strengthening the role of Csu pili in biofilm formation and maintenance. Previous reports also identified a single nucleotide insertion in the csuB gene of A. baumannii ATCC 17978, suggesting that this system may be nonfunctional in this strain (62, 63); however, recent resequencing of the A. baumannii ATCC 17978 genome (GenBank accession no. CP012004) did not find the same insertion event (64). Lastly, other chaperone/usher pilus-like systems have been bioinformatically identified in many A. baumannii strains, but none of those systems has been functionally characterized (61, 62). Mass spectrometric characterization of pellicle-associated proteins did find that non-Csu-pilin subunits were present, indicating that these systems may be functional.
Medically relevant Acinetobacter spp. have also been shown to produce type IV pili (Tfp), which are dynamic bacterial surface appendages known to mediate twitching motility, horizontal gene transfer, and biofilm formation (65). Although bioinformatic studies have identified genes predicted to encode proteins required for the biogenesis of Tfp in A. baumannii, only Acinetobacter nosocomialis strain M2 has been shown conclusively to produce functioning Tfp (66, 67), which is glycosylated by a TfpO-like oligosaccharyltransferase (30). Many A. baumannii isolates have been found to be naturally transformable and to exhibit twitching motility (63, 68–70), two classic Tfp-associated phenotypes, which strongly indicates their presence. Tfp-like structures were also identified on A. baumannii ATCC 17978 (71); furthermore, mutants in predicted Tfp biogenesis components of A. baumannii ATCC 17978 exhibited impaired biofilm formation (71) but the major pilin subunit, PilA, has not been shown to be surface exposed and/or associated with the pilin structures observed. Although Tfp, with roles in Acinetobacter motility and natural transformation, has emerged as a possible virulence factor, no studies have conclusively linked Tfp to the pathobiology of Acinetobacter, as is the case for Pseudomonas and Neisseria.
PROTEIN SECRETION
Extracellular export of proteins is a fundamental process of all forms of life. Protein secretion systems of Gram-negative bacteria are extremely diverse in function and composition and often are important mediators of virulence. Recent research has elucidated several of the mechanisms Acinetobacter uses to secrete proteins and the role they play in the biology of Acinetobacter. Here we describe the recent insights into macromolecular protein secretion systems present in Acinetobacter, with a focus on the systems that export proteins out of the cell and that have been characterized experimentally.
Type II secretion.
The most recently described secretion system is a functional type II secretion system (T2SS) identified in both A. nosocomialis strain M2 (119) and A. baumannii ATCC 17978 (72); moreover, it was shown that clinical isolates of Acinetobacter pittii, A. baumannii, A. calcoaceticus, and Acinetobacter junii all were able to secrete type II substrates, indicating that functioning type II secretion systems seem to be the rule and not the exception. With regard to the T2SS of A. nosocomialis strain M2, a two-dimensional differential gel electrophoresis approach identified multiple putative type II substrates; the LipA and LipH lipases and the CpaA metallopeptidase were validated as bona fide type II secretion substrates. Interestingly, both LipA and CpaA required specific membrane-associated chaperones for secretion, which indicates that T2SS chaperones are more widespread than previously recognized. Importantly, it was shown that an A. nosocomialis strain M2 gspD mutant lacking the outer membrane secretin of the T2SS was severely attenuated in both the Galleria mellonella and murine pulmonary infection models. Specifically, mice intranasally infected with the gspD mutant strain had ∼2-log lower bacterial burdens in both the lungs and the spleen after 36 h, compared to both the parental strain and the complemented mutant. In A. baumannii ATCC 17978, Johnson et al. identified a lipase, LipA, secreted in a T2SS-dependent manner that was required for growth on medium containing lipids as a sole carbon source (72). Mutants with mutations in lipA or the T2SS structural gene gspD were less competitive than the wild-type strain in a mixed-infection murine model of bacteremia. Collectively, these findings indicate that the Acinetobacter T2SS is a previously unrecognized virulence factor mediating pathogenesis in a relevant mammalian model. Interestingly, a recent study by Wang et al. utilized an A. baumannii ATCC 17978 gspN mutant for validation of their insertion sequencing murine pulmonary infection studies, and they subsequently found that the gspN mutant did not display any virulence defect in survival or competition models, compared to the parent strain (51). Although these data are in contrast to the newly defined role of type II secretion in Acinetobacter, it was demonstrated previously that gspN homologs were not required for a functioning T2SS in Klebsiella oxytoca (73); furthermore, gspN homologs are absent from numerous known T2SSs in other Gram-negative bacteria (74), indicating the dispensable nature of GspN in functioning T2SSs.
Autotransporters.
A type V autotransporter has been characterized in A. baumannii. The Acinetobacter trimeric autotransporter (Ata) was found to be crucial for the ability of certain A. baumannii strains to adhere to extracellular matrix components, including collagen I, III, IV, and V (75). Ata is also an important mediator of A. baumannii biofilm formation and maintenance, as an A. baumannii ATCC 17978 ata mutant had significantly diminished biofilm production and was less virulent in a murine intraperitoneal infection model, compared to the parental and complemented strains (75).
Type VI secretion.
Bacteria interact with each other in a multitude of ways; these interactions are often competitive in nature and play important roles in niche establishment (76). The bacterial type VI secretion system was first formally described for Vibrio cholerae and P. aeruginosa and was suggested to play a role against eukaryotic hosts (77, 78). While several T6SSs have been determined to secrete antieukaryotic toxins, it has recently been appreciated that many bacteria use their T6SSs to secrete antibacterial toxins to kill competing bacteria (79, 80). The T6SS is composed of approximately 15 conserved structural proteins and a variable number of accessory factors, which work in concert to secrete proteins extracellularly (81). Important components include Hcp, which forms a polymerized tubular structure that is secreted out of the cell and is essential for protein secretion, and VgrGs, which are present at the tip of this structure and can have effector activity or facilitate effector secretion (82). The T6SS bears striking similarity to bacteriophage, both structurally and functionally (83).
The presence of a T6SS was initially predicted bioinformatically for A. baylyi, and Hcp was subsequently detected in supernatants of A. baumannii ATCC 19606 (84, 85). The genetic organization and sequences of T6SS genes are remarkably well conserved across Acinetobacter spp. Based on homology with T6SS genes in other bacteria, the single Acinetobacter T6SS locus includes most of the genes required for apparatus assembly and function (86, 87). Notably, the main T6SS cluster does not contain vgrG genes, which are instead scattered throughout the genome. The VgrG proteins of Acinetobacter, which differ in number from strain to strain, do not seem to include effector domains (86, 88). Instead, bioinformatic analyses suggest that the proteins are most likely to mediate the secretion of adjacently encoded toxic effectors, with cognate immunity proteins being encoded nearby (88). However, no bona fide T6SS-dependent effectors have been experimentally characterized in Acinetobacter. In A. baylyi, mutation of three PAAR proteins, which interact with VgrG proteins, results in loss of Hcp secretion, and one of those PAAR proteins has been experimentally determined to be secreted, although it is not clear whether the proteins have any effector functions themselves (89). The primary function of the T6SS in Acinetobacter seems to be to kill competing bacteria, and Acinetobacter spp. with active T6SSs are able to kill a wide variety of other bacteria, including other strains of the same species (64, 87–89).
T6SS expression often is tightly controlled and is activated only under certain conditions; the molecular mechanisms used to achieve this regulation are extremely diverse and complex and differ from organism to organism and even between strains of a given species (82). Although little is known about T6SS regulation in Acinetobacter, recent studies have provided insight into some of the regulatory mechanisms used by these organisms. T6SS activities vary widely in different strains and species of Acinetobacter, with some strains showing robust T6SSs and bacterial killing and others seeming to have inactive systems under laboratory conditions (64, 86–88). However, the available data suggest that strains with T6SSs invariably express the main protein Hcp to at least some level, with variations in whether the protein is secreted (thus determining whether the system is active) (64, 86, 88). In A. baumannii ATCC 19606, Hcp is constitutively secreted in wild-type cells but is lost in mutants lacking lipid A, potentially due to membrane disruptions (85). In A. baumannii ATCC 17978, T6SS activity is controlled by a plasmid (see below), and a chromosomally encoded histone-like nucleoid-structuring (H-NS) protein may also regulate the T6SS (90). Several A. baumannii isolates harbor a resistance plasmid that encodes repressors of the T6SS (64). Upon spontaneous loss of this plasmid and subsequent loss of the repressors, the T6SS is activated and the resistance genes are lost. The functional significance of this remains to be elucidated but, considering the tremendous amount of energy required for T6SS activity (91) and the fitness defects often caused by harboring multiple antibiotic resistance genes (92), this may represent a mechanism to maintain both systems while avoiding potentially deleterious effects of having them be active at the same time. In the absence of antibiotic pressure, Acinetobacter strains do not require resistance genes but are more likely to encounter competitors, thus losing the resistance plasmid and activating the T6SS, which could provide a competitive advantage. Because the cells that lose the plasmid will lose resistance to antibiotics, this strategy could constitute an altruistic mechanism to ensure Acinetobacter population survival. It should be noted that several recent multidrug-resistant (MDR) A. baumannii strains seem to have permanently inactivated their T6SSs through chromosomal gene loss; it has been suggested that this may be a result of the antibiotic pressure being great enough to make it evolutionarily advantageous to completely lose the T6SS, rather than maintaining it in an inactive state (93). Although there are limited data, there appears to be a link between antibiotic resistance and T6SS status in Acinetobacter; strains that are multidrug resistant express but do not secrete Hcp, while those that are not multidrug resistant are more likely to have an active T6SS (64, 86, 88).
Outer membrane vesicles.
A special case of protein secretion is the production of outer membrane vesicles (OMVs), which are blebs of outer membrane (OM) released from the bacterial cell surface (94). There is significant debate regarding whether OMVs are produced by a directed process or simply represent cellular debris. Proteomic comparisons between the OM and OMVs of some bacteria have shown that the protein profiles differ between these two fractions, indicating that some OM proteins are excluded from OMV recruitment and suggesting that OMV formation is a directed process (94). However, many studies also detected cytoplasmic proteins in OMV preparations, indicating that cell lysis could also be a major contributor to OMV formation (94). OMVs have been implicated in numerous biological functions, with particular attention being devoted to their role in virulence (95). Several studies on OMVs in Acinetobacter have suggested that they have many functions, including roles in horizontal gene transfer, antibiotic resistance, and virulence. A wide variety of cargo types have been identified in OMVs from different Acinetobacter strains, including virulence proteins, antibiotic resistance determinants, and DNA (96–102). An important virulence factor of A. baumannii, OmpA, has also been found to be associated with OMVs, and OMVs have been suggested to act as a mechanism for delivery for this protein to host cells (103). Furthermore, OmpA has been suggested to be involved in the biogenesis of OMVs (104). OMVs from Acinetobacter may have an important role in the development of novel therapeutics, as they can stimulate a strong immune response and are protective when administered as a vaccine (105–107).
FUTURE DIRECTIONS
The surge of infections caused by Acinetobacter spp. has led to increased interest from the scientific and medical communities and attempts to understand the disease-causing mechanisms of these organisms. Many recent studies have greatly increased our understanding of Acinetobacter infection mechanisms but have also illuminated what a formidable pathogen the health care community is facing. There is intense interest in discovering novel strategies to fight this pathogen, which is quickly becoming untreatable with our current antibiotic repertoire. Although the vast majority of patients who become infected with Acinetobacter spp. are immunocompromised, which complicates intervention strategies, vaccines have been proposed as an alternative method to fight MDR Acinetobacter (108, 109), and several promising candidates have been described (107, 110–114). Individuals for whom immunocompromise could potentially be predicted a priori, such as patients undergoing cancer treatments or surgery and military personnel entering conflicts, may benefit from a prophylactic vaccination strategy (115). Carbohydrate structures present in Acinetobacter, such as the capsule and glycoproteins, represent attractive antigenic targets for vaccine development; since the carbohydrate moieties are the same in both, targeting these structures may provide broad protection (46). Indeed, capsule-based vaccines have shown efficacy in soft tissue, pneumonia, and bacteremia rodent models (116, 117). A drawback to this approach, however, is that the strain-to-strain variations in carbohydrate structures are so great that a multivalent vaccine to target all pathogenic Acinetobacter strains is unrealistic. Extensive epidemiological data on the capsular/glycoprotein serotypes most prevalent in a given health care institution may allow for a more directed approach to vaccine design. Indeed, PCR-based schemes have been proposed to accomplish this, and advances in the speed and cost of genome sequencing may make this a feasible approach (118). Because the production of capsular polysaccharides and glycoproteins is essential for virulence, targeting common steps in the biosynthetic pathway of these structures may be more feasible. Given the genomic plasticity of Acinetobacter, it is likely that a “one size fits all” solution to the problem is not possible, and multiple strategies should be investigated in order to determine the most beneficial approach for a given health care setting.
Funding Statement
M.F.F. is a Washington University School of Medicine Faculty Diversity Scholar. B.S.W. is a recipient of the NSERC Postgraduate Scholarship-Doctoral award.
REFERENCES
- 1.Peleg AY, Hooper DC. 2010. Hospital-acquired infections due to Gram-negative bacteria. N Engl J Med 362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaynes R, Edwards JR. 2005. Overview of nosocomial infections caused by Gram-negative bacilli. Clin Infect Dis 41:848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 3.Dexter C, Murray GL, Paulsen IT, Peleg AY. 2015. Community-acquired Acinetobacter baumannii: clinical characteristics, epidemiology and pathogenesis. Expert Rev Anti Infect Ther 13:567–573. doi: 10.1586/14787210.2015.1025055. [DOI] [PubMed] [Google Scholar]
- 4.Scott P, Deye G, Srinivasan A, Murray C, Moran K, Hulten E, Fishbain J, Craft D, Riddell S, Lindler L, Mancuso J, Milstrey E, Bautista CT, Patel J, Ewell A, Hamilton T, Gaddy C, Tenney M, Christopher G, Petersen K, Endy T, Petruccelli B. 2007. An outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus complex infection in the US military health care system associated with military operations in Iraq. Clin Infect Dis 44:1577–1584. doi: 10.1086/518170. [DOI] [PubMed] [Google Scholar]
- 5.Turton JF, Kaufmann ME, Gill MJ, Pike R, Scott PT, Fishbain J, Craft D, Deye G, Riddell S, Lindler LE, Pitt TL. 2006. Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J Clin Microbiol 44:2630–2634. doi: 10.1128/JCM.00547-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sebeny PJ, Riddle MS, Petersen K. 2008. Acinetobacter baumannii skin and soft-tissue infection associated with war trauma. Clin Infect Dis 47:444–449. doi: 10.1086/590568. [DOI] [PubMed] [Google Scholar]
- 7.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 8.Valencia R, Arroyo LA, Conde M, Aldana JM, Torres MJ, Fernandez-Cuenca F, Garnacho-Montero J, Cisneros JM, Ortiz C, Pachon J, Aznar J. 2009. Nosocomial outbreak of infection with pan-drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect Control Hosp Epidemiol 30:257–263. doi: 10.1086/595977. [DOI] [PubMed] [Google Scholar]
- 9.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandi G. 2010. Bacterial surface proteins and vaccines. F1000 Biol Rep 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitfield C, Amor PA, Koplin R. 1997. Modulation of the surface architecture of Gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol Microbiol 23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 13.Stenutz R, Weintraub A, Widmalm G. 2006. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol Rev 30:382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Reeves PR, Wang L. 2014. Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol Rev 38:56–89. doi: 10.1111/1574-6976.12034. [DOI] [PubMed] [Google Scholar]
- 15.Orskov I, Orskov F, Jann B, Jann K. 1977. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev 41:667–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preston A, Mandrell RE, Gibson BW, Apicella MA. 1996. The lipooligosaccharides of pathogenic Gram-negative bacteria. Crit Rev Microbiol 22:139–180. doi: 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- 17.Pantophlet R, Brade L, Brade H. 1999. Identification of Acinetobacter baumannii strains with monoclonal antibodies against the O antigens of their lipopolysaccharides. Clin Diagn Lab Immunol 6:323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haseley SR, Traub WH, Wilkinson SG. 1997. Structures of polymeric products isolated from the lipopolysaccharides of reference strains for Acinetobacter baumannii O23 and O12. Eur J Biochem 244:147–154. doi: 10.1111/j.1432-1033.1997.00147.x. [DOI] [PubMed] [Google Scholar]
- 19.Traub WH. 1989. Acinetobacter baumannii serotyping for delineation of outbreaks of nosocomial cross-infection. J Clin Microbiol 27:2713–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haseley SR, Wilkinson SG. 1998. Structure of the O-7 antigen from Acinetobacter baumannii. Carbohydr Res 306:257–263. doi: 10.1016/S0008-6215(97)10039-8. [DOI] [PubMed] [Google Scholar]
- 21.Haseley S, Wilkinson SG. 1997. Structure of the O18 antigen from Acinetobacter baumannii. Carbohydr Res 301:187–192. doi: 10.1016/S0008-6215(97)00095-5. [DOI] [PubMed] [Google Scholar]
- 22.Haseley SR, Wilkinson SG. 1994. Structure of the putative O10 antigen from Acinetobacter baumannii. Carbohydr Res 264:73–81. doi: 10.1016/0008-6215(94)00179-0. [DOI] [PubMed] [Google Scholar]
- 23.MacLean LL, Perry MB, Chen W, Vinogradov E. 2009. The structure of the polysaccharide O-chain of the LPS from Acinetobacter baumannii strain ATCC 17961. Carbohydr Res 344:474–478. doi: 10.1016/j.carres.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 24.Vinogradov EV, Brade L, Brade H, Holst O. 2003. Structural and serological characterisation of the O-antigenic polysaccharide of the lipopolysaccharide from Acinetobacter baumannii strain 24. Carbohydr Res 338:2751–2756. doi: 10.1016/j.carres.2003.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Vinogradov EV, Duus JO, Brade H, Holst O. 2002. The structure of the carbohydrate backbone of the lipopolysaccharide from Acinetobacter baumannii strain ATCC 19606. Eur J Biochem 269:422–430. doi: 10.1046/j.0014-2956.2001.02647.x. [DOI] [PubMed] [Google Scholar]
- 26.Vinogradov EV, Pantophlet R, Dijkshoorn L, Brade L, Holst O, Brade H. 1996. Structural and serological characterisation of two O-specific polysaccharides of Acinetobacter. Eur J Biochem 239:602–610. doi: 10.1111/j.1432-1033.1996.0602u.x. [DOI] [PubMed] [Google Scholar]
- 27.Pantophlet R. 2008. Lipopolysaccharides of Acinetobacter. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 28.Fregolino E, Gargiulo V, Lanzetta R, Parrilli M, Holst O, Castro CD. 2011. Identification and structural determination of the capsular polysaccharides from two Acinetobacter baumannii clinical isolates, MG1 and SMAL. Carbohydr Res 346:973–977. doi: 10.1016/j.carres.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 29.Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S, Feldman MF. 2012. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog 8:e1002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harding CM, Nasr MA, Kinsella RL, Scott NE, Foster LJ, Weber BS, Fiester SE, Actis LA, Tracy EN, Munson RS Jr, Feldman MF. 2015. Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol Microbiol 96:1023–1041. doi: 10.1111/mmi.12986. [DOI] [PubMed] [Google Scholar]
- 31.Schulz BL, Jen FE, Power PM, Jones CE, Fox KL, Ku SC, Blanchfield JT, Jennings MP. 2013. Identification of bacterial protein O-oligosaccharyltransferases and their glycoprotein substrates. PLoS One 8:e62768. doi: 10.1371/journal.pone.0062768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. doi: 10.1371/journal.pone.0062160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McQueary CN, Kirkup BC, Si Y, Barlow M, Actis LA, Craft DW, Zurawski DV. 2012. Extracellular stress and lipopolysaccharide modulate Acinetobacter baumannii surface-associated motility. J Microbiol 50:434–443. doi: 10.1007/s12275-012-1555-1. [DOI] [PubMed] [Google Scholar]
- 34.Luke NR, Sauberan SL, Russo TA, Beanan JM, Olson R, Loehfelm TW, Cox AD, St Michael F, Vinogradov EV, Campagnari AA. 2010. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect Immun 78:2017–2023. doi: 10.1128/IAI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin L, Tan B, Pantapalangkoor P, Ho T, Baquir B, Tomaras A, Montgomery JI, Reilly U, Barbacci EG, Hujer K, Bonomo RA, Fernandez L, Hancock RE, Adams MD, French SW, Buslon VS, Spellberg B. 2012. Inhibition of LpxC protects mice from resistant Acinetobacter baumannii by modulating inflammation and enhancing phagocytosis. mBio 3:e00312-12. doi: 10.1128/mBio.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenyon JJ, Nigro SJ, Hall RM. 2014. Variation in the OC locus of Acinetobacter baumannii genomes predicts extensive structural diversity in the lipooligosaccharide. PLoS One 9:e107833. doi: 10.1371/journal.pone.0107833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boll JM, Tucker AT, Klein DR, Beltran AM, Brodbelt JS, Davies BW, Trent MS. 2015. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. mBio 6(3):e00478-15. doi: 10.1128/mBio.00478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock RE. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin CY, Gregg KA, Napier BA, Ernst RK, Weiss DS. 2015. A PmrB-regulated deacetylase required for lipid A modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 59:7911–7914. doi: 10.1128/AAC.00515-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KR, Doi Y, Ernst RK. 2013. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 57:4831–4840. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hood MI, Becker KW, Roux CM, Dunman PM, Skaar EP. 2013. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect Immun 81:542–551. doi: 10.1128/IAI.00704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hug I, Feldman MF. 2011. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology 21:138–151. doi: 10.1093/glycob/cwq148. [DOI] [PubMed] [Google Scholar]
- 45.Scott NE, Kinsella RL, Edwards AV, Larsen MR, Dutta S, Saba J, Foster LJ, Feldman MF. 2014. Diversity within the O-linked protein glycosylation systems of Acinetobacter species. Mol Cell Proteomics 13:2354–2370. doi: 10.1074/mcp.M114.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lees-Miller RG, Iwashkiw JA, Scott NE, Seper A, Vinogradov E, Schild S, Feldman MF. 2013. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol Microbiol 89:816–830. doi: 10.1111/mmi.12300. [DOI] [PubMed] [Google Scholar]
- 47.Castric P. 1995. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 141:1247–1254. doi: 10.1099/13500872-141-5-1247. [DOI] [PubMed] [Google Scholar]
- 48.Faridmoayer A, Fentabil MA, Haurat MF, Yi W, Woodward R, Wang PG, Feldman MF. 2008. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J Biol Chem 283:34596–34604. doi: 10.1074/jbc.M807113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faridmoayer A, Fentabil MA, Mills DC, Klassen JS, Feldman MF. 2007. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J Bacteriol 189:8088–8098. doi: 10.1128/JB.01318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. 2010. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun 78:3993–4000. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5(3):e01163-14. doi: 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geisinger E, Isberg RR. 2015. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 11:e1004691. doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. 2008. Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 54.Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litran T. 2009. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol 191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henrichsen J. 1975. The occurrence of twitching motility among Gram-negative bacteria. Acta Pathol Microbiol Scand B 83:171–178. [DOI] [PubMed] [Google Scholar]
- 56.Henrichsen J, Blom J. 1975. Correlation between twitching motility and possession of polar fimbriae in Acinetobacter calcoaceticus. Acta Pathol Microbiol Scand B 83:103–115. [DOI] [PubMed] [Google Scholar]
- 57.Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 58.Gohl O, Friedrich A, Hoppert M, Averhoff B. 2006. The thin pili of Acinetobacter sp. strain BD413 mediate adhesion to biotic and abiotic surfaces. Appl Environ Microbiol 72:1394–1401. doi: 10.1128/AEM.72.2.1394-1401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 60.de Breij A, Gaddy J, van der Meer J, Koning R, Koster A, van den Broek P, Actis L, Nibbering P, Dijkshoorn L. 2009. CsuA/BABCDE-dependent pili are not involved in the adherence of Acinetobacter baumannii ATCC19606(T) to human airway epithelial cells and their inflammatory response. Res Microbiol 160:213–218. doi: 10.1016/j.resmic.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 61.Nait Chabane Y, Marti S, Rihouey C, Alexandre S, Hardouin J, Lesouhaitier O, Vila J, Kaplan JB, Jouenne T, De E. 2014. Characterisation of pellicles formed by Acinetobacter baumannii at the air-liquid interface. PLoS One 9:e111660. doi: 10.1371/journal.pone.0111660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. 2014. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. doi: 10.1186/1471-2164-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. 2011. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol Lett 323:44–51. doi: 10.1111/j.1574-6968.2011.02362.x. [DOI] [PubMed] [Google Scholar]
- 64.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112:9442–9447. doi: 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol 66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 66.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS Jr. 2013. Acinetobacter baumannii strain M2 produces type IV pili which play a role in natural transformation and twitching motility but not surface-associated motility. mBio 4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carruthers MD, Harding CM, Baker BD, Bonomo RA, Hujer KM, Rather PN, Munson RS Jr. 2013. Draft genome sequence of the clinical isolate Acinetobacter nosocomialis strain M2. Genome Announc 1:e00906-13. doi: 10.1128/genomeA.00906-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, Tolmasky ME. 2010. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol 48:1488–1490. doi: 10.1128/JCM.01264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilharm G, Piesker J, Laue M, Skiebe E. 2013. DNA uptake by the nosocomial pathogen Acinetobacter baumannii occurs during movement along wet surfaces. J Bacteriol 195:4146–4153. doi: 10.1128/JB.00754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoon EJ, Chabane YN, Goussard S, Snesrud E, Courvalin P, De E, Grillot-Courvalin C. 2015. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio 6:e00309-15. doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5:e01313-14. doi: 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Johnson TL, Waack U, Smith S, Mobley H, Sandkvist M. 14 December 2015. Acinetobacter baumannii is dependent on the type II secretion system and its substrate LipA for lipid utilization and in vivo fitness. J Bacteriol doi: 10.1128/JB.00622-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Possot OM, Vignon G, Bomchil N, Ebel F, Pugsley AP. 2000. Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J Bacteriol 182:2142–2152. doi: 10.1128/JB.182.8.2142-2152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Campos M, Cisneros DA, Nivaskumar M, Francetic O. 2013. The type II secretion system: a dynamic fiber assembly nanomachine. Res Microbiol 164:545–555. doi: 10.1016/j.resmic.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 75.Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litran T. 2012. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J Bacteriol 194:3950–3960. doi: 10.1128/JB.06769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Russell AB, Peterson SB, Mougous JD. 2014. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pukatzki S, McAuley SB, Miyata ST. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol 12:11–17. doi: 10.1016/j.mib.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 82.Silverman JM, Brunet YR, Cascales E, Mougous JD. 2012. Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brunet YR, Henin J, Celia H, Cascales E. 2014. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep 15:315–321. doi: 10.1002/embr.201337936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C, Durot M, Kreimeyer A, Le Fevre F, Schachter V, Pezo V, Doring V, Scarpelli C, Medigue C, Cohen GN, Marliere P, Salanoubat M, Weissenbach J. 2008. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol 4:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. 2012. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob Agents Chemother 56:59–69. doi: 10.1128/AAC.05191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, Feldman MF. 2013. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 8:e55142. doi: 10.1371/journal.pone.0055142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carruthers MD, Nicholson PA, Tracy EN, Munson RS Jr. 2013. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. doi: 10.1371/journal.pone.0059388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Repizo GD, Gagne S, Foucault-Grunenwald ML, Borges V, Charpentier X, Limansky AS, Gomes JP, Viale AM, Salcedo SP. 2015. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One 10:e0138265. doi: 10.1371/journal.pone.0138265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eijkelkamp BA, Stroeher UH, Hassan KA, Elbourne LD, Paulsen IT, Brown MH. 2013. H-NS plays a role in expression of Acinetobacter baumannii virulence features. Infect Immun 81:2574–2583. doi: 10.1128/IAI.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Basler M. 2015. Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci 370:20150021. doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. [DOI] [PubMed] [Google Scholar]
- 93.Wright MS, Haft DH, Harkins DM, Perez F, Hujer KM, Bajaksouzian S, Benard MF, Jacobs MR, Bonomo RA, Adams MD. 2014. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio 5:e00963-13. doi: 10.1128/mBio.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haurat MF, Elhenawy W, Feldman MF. 2015. Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biol Chem 396:95–109. [DOI] [PubMed] [Google Scholar]
- 95.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liao YT, Kuo SC, Chiang MH, Lee YT, Sung WC, Chen YH, Chen TL, Fung CP. 2015. Acinetobacter baumannii extracellular OXA-58 is primarily and selectively released via outer membrane vesicles after Sec-dependent periplasmic translocation. Antimicrob Agents Chemother 59:7346–7354. doi: 10.1128/AAC.01343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li ZT, Zhang RL, Bi XG, Xu L, Fan M, Xie D, Xian Y, Wang Y, Li XJ, Wu ZD, Zhang KX. 2015. Outer membrane vesicles isolated from two clinical Acinetobacter baumannii strains exhibit different toxicity and proteome characteristics. Microb Pathog 81:46–52. doi: 10.1016/j.micpath.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 99.Fulsundar S, Kulkarni HM, Jagannadham MV, Nair R, Keerthi S, Sant P, Pardesi K, Bellare J, Chopade BA. 2015. Molecular characterization of outer membrane vesicles released from Acinetobacter radioresistens and their potential roles in pathogenesis. Microb Pathog 83–84:12–22. [DOI] [PubMed] [Google Scholar]
- 100.Fulsundar S, Harms K, Flaten GE, Johnsen PJ, Chopade BA, Nielsen KM. 2014. Gene transfer potential of outer membrane vesicles of Acinetobacter baylyi and effects of stress on vesiculation. Appl Environ Microbiol 80:3469–3483. doi: 10.1128/AEM.04248-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dallo SF, Zhang B, Denno J, Hong S, Tsai A, Haskins W, Ye JY, Weitao T. 2012. Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. ScientificWorldJournal 2012:128705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kwon SO, Gho YS, Lee JC, Kim SI. 2009. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett 297:150–156. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- 103.Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. 2011. Acinetobacter baumannii secretes cytotoxic outer membrane protein A via outer membrane vesicles. PLoS One 6:e17027. doi: 10.1371/journal.pone.0017027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moon DC, Choi CH, Lee JH, Choi CW, Kim HY, Park JS, Kim SI, Lee JC. 2012. Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J Microbiol 50:155–160. doi: 10.1007/s12275-012-1589-4. [DOI] [PubMed] [Google Scholar]
- 105.Nho JS, Jun SH, Oh MH, Park TI, Choi CW, Kim SI, Choi CH, Lee JC. 2015. Acinetobacter nosocomialis secretes outer membrane vesicles that induce epithelial cell death and host inflammatory responses. Microb Pathog 81:39–45. doi: 10.1016/j.micpath.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 106.Jun SH, Lee JH, Kim BR, Kim SI, Park TI, Lee JC, Lee YC. 2013. Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins. PLoS One 8:e71751. doi: 10.1371/journal.pone.0071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McConnell MJ, Rumbo C, Bou G, Pachon J. 2011. Outer membrane vesicles as an acellular vaccine against Acinetobacter baumannii. Vaccine 29:5705–5710. doi: 10.1016/j.vaccine.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 108.Garcia-Quintanilla M, Pulido MR, McConnell MJ. 2013. First steps towards a vaccine against Acinetobacter baumannii. Curr Pharm Biotechnol 14:897–902. [DOI] [PubMed] [Google Scholar]
- 109.Pachon J, McConnell MJ. 2014. Considerations for the development of a prophylactic vaccine for Acinetobacter baumannii. Vaccine 32:2534–2536. doi: 10.1016/j.vaccine.2013.10.064. [DOI] [PubMed] [Google Scholar]
- 110.Badmasti F, Ajdary S, Bouzari S, Fooladi AA, Shahcheraghi F, Siadat SD. 2015. Immunological evaluation of OMV(PagL)+Bap(1-487aa) and AbOmpA(8-346aa)+Bap(1-487aa) as vaccine candidates against Acinetobacter baumannii sepsis infection. Mol Immunol 67:552–558. doi: 10.1016/j.molimm.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 111.Lin L, Tan B, Pantapalangkoor P, Ho T, Hujer AM, Taracila MA, Bonomo RA, Spellberg B. 2013. Acinetobacter baumannii rOmpA vaccine dose alters immune polarization and immunodominant epitopes. Vaccine 31:313–318. doi: 10.1016/j.vaccine.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bentancor LV, Routray A, Bozkurt-Guzel C, Camacho-Peiro A, Pier GB, Maira-Litran T. 2012. Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect Immun 80:3381–3388. doi: 10.1128/IAI.06096-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McConnell MJ, Pachon J. 2010. Active and passive immunization against Acinetobacter baumannii using an inactivated whole cell vaccine. Vaccine 29:1–5. doi: 10.1016/j.vaccine.2010.10.052. [DOI] [PubMed] [Google Scholar]
- 114.Huang W, Wang S, Yao Y, Xia Y, Yang X, Long Q, Sun W, Liu C, Li Y, Ma Y. 2015. OmpW is a potential target for eliciting protective immunity against Acinetobacter baumannii infections. Vaccine 33:4479–4485. doi: 10.1016/j.vaccine.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 115.Rubin LG, Levin MJ, Ljungman P, Davies EG, Avery R, Tomblyn M, Bousvaros A, Dhanireddy S, Sung L, Keyserling H, Kang I, Infectious Diseases Society of America . 2014. 2013 IDSA clinical practice guideline for vaccination of the immunocompromised host. Clin Infect Dis 58:309–318. doi: 10.1093/cid/cit816. [DOI] [PubMed] [Google Scholar]
- 116.Russo TA, Beanan JM, Olson R, MacDonald U, Cox AD, St Michael F, Vinogradov EV, Spellberg B, Luke-Marshall NR, Campagnari AA. 2013. The K1 capsular polysaccharide from Acinetobacter baumannii is a potential therapeutic target via passive immunization. Infect Immun 81:915–922. doi: 10.1128/IAI.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bentancor LV, O'Malley JM, Bozkurt-Guzel C, Pier GB, Maira-Litran T. 2012. Poly-N-acetyl-β-(1-6)-glucosamine is a target for protective immunity against Acinetobacter baumannii infections. Infect Immun 80:651–656. doi: 10.1128/IAI.05653-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu D, Liu B, Dijkshoorn L, Wang L, Reeves PR. 2013. Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS One 8:e70329. doi: 10.1371/journal.pone.0070329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harding CM, Kinsella RL, Palmer LD, Skaar EP, Feldman MF. 2016. Medically relevant Acinetobacter species require a type II secretion system and specific membrane-associated chaperones for the export of multiple substrates and full virulence. PLoS Pathog 12:e1005391. doi: 10.1371/journal.ppat.1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]