ABSTRACT
Antimicrobial resistance in Staphylococcus aureus presents an increasing threat to human health. This resistance is often encoded on mobile plasmids, such as pSK41; however, the mechanism of transfer of these plasmids is not well understood. In this study, we first examine key protein-DNA interactions formed by the relaxase enzyme, NES, which initiates and terminates the transfer of the multidrug resistance plasmid pSK41. Two loops on the NES protein, hairpin loops 1 and 2, form extensive contacts with the DNA hairpin formed at the oriT region of pSK41, and here we establish that these contacts are essential for proper DNA cleavage and religation by the full 665-residue NES protein in vitro. Second, pSK156 and pCA347 are nonconjugative Staphylococcus aureus plasmids that contain sequences similar to the oriT region of pSK41 but differ in the sequence predicted to form a DNA hairpin. We show that pSK41-encoded NES is able to bind, cleave, and religate the oriT sequences of these nonconjugative plasmids in vitro. Although pSK41 could mobilize a coresident plasmid harboring its cognate oriT, it was unable to mobilize plasmids containing the pSK156 and pCA347 variant oriT mimics, suggesting that an accessory protein like that previously shown to confer specificity in the pWBG749 system may also be involved in transmission of plasmids containing a pSK41-like oriT. These data indicate that the conjugative relaxase in trans mechanism recently described for the pWBG749 family of plasmids also applies to the pSK41 family of plasmids, further heightening the potential significance of this mechanism in the horizontal transfer of staphylococcal plasmids.
IMPORTANCE Understanding the mechanism of antimicrobial resistance transfer in bacteria such as Staphylococcus aureus is an important step toward potentially slowing the spread of antimicrobial-resistant infections. This work establishes protein-DNA interactions essential for the transfer of the Staphylococcus aureus multiresistance plasmid pSK41 by its relaxase, NES. This enzyme also processed variant oriT-like sequences found on numerous plasmids previously considered nontransmissible, suggesting that in conjunction with an uncharacterized accessory protein, these plasmids may be transferred horizontally via a relaxase in trans mechanism. These findings have important implications for our understanding of staphylococcal resistance plasmid evolution.
INTRODUCTION
Antimicrobial-resistant strains of Staphylococcus aureus are a growing concern for hospital- and community-acquired infections. Most S. aureus bacteria examined clinically harbor at least one plasmid that encodes antimicrobial resistance, and many plasmids carry multiple antimicrobial resistance determinants. The pSK41 family of plasmids is made up of large, low-copy-number conjugative plasmids, for which pSK41 is used as a prototype for characterization (1–4). These plasmids carry a variety of antimicrobial resistance determinants, including those against aminoglycosides, penicillins, tetracycline, bleomycin, trimethoprim, macrolides, lincosamides, mupirocin, antiseptics, and disinfectants (2, 4–9). This family of plasmids also played a key role in the rise of vancomycin-resistant S. aureus (VRSA) (7, 8, 10). In addition to encoding antimicrobial resistance, they also carry transfer (tra) genes encoding the proteins necessary to conduct the conjugative plasmid transfer that spreads these plasmids among S. aureus strains and other Gram-positive bacteria (6, 7, 10, 11).
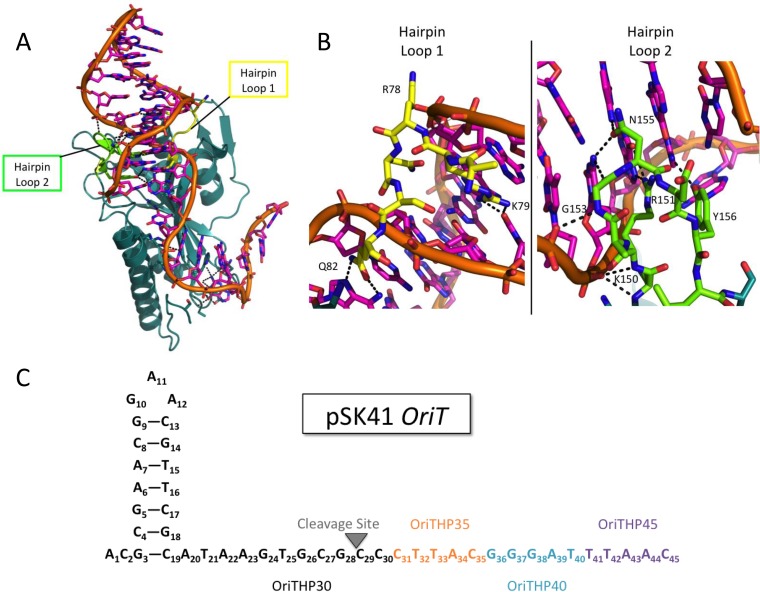
One of the proteins essential for conjugative plasmid transfer is the relaxase enzyme. A relaxase is responsible for initiation and completion of the transfer process, as it cleaves one strand of the double-stranded plasmid to begin transfer and then ligates the strands back together to complete transfer (8, 12–15). There are two classes of relaxases: multityrosine relaxases, which use a “thumb” motif to position the plasmid DNA for processing, and single-tyrosine relaxases, which lack this thumb motif (9, 16). The relaxase of pSK41 is termed the NES nicking enzyme in S. aureus, and it is a single-tyrosine relaxase (1–6, 8, 9). NES contains a relaxase N-terminal 220 residues and a C-terminal 350 residues necessary for in vivo function via an uncertain mechanism (5, 7, 8, 10). The crystal structure of the relaxase domain of NES was the first of a single-tyrosine relaxase bound to its target DNA, allowing for more detailed characterization of the protein-DNA interactions than was previously possible (6–8, 11). This structure revealed two sets of important protein-DNA interactions. The first is that the “thumb” used by multityrosine relaxases to position the DNA appears to be replaced by 12 protein-DNA contacts, including a buried nucleotide 3 bases upstream of the nic site that places the DNA in the correct position to be nicked by the single, catalytically active tyrosine. The second set of protein-DNA interactions unique to the NES-DNA complex is composed of two protein loops, termed hairpin loops 1 and 2, that surround the DNA hairpin formed upstream of the nic site (Fig. 1A and B). NES hairpin loop 1, shown in yellow in Fig. 1A and B, forms two base-specific contacts with the minor groove of the oriT DNA and one contact with the phosphate backbone. NES hairpin loop 2, shown in green in Fig. 1A and B, contacts the DNA more extensively, with six base-specific interactions and four phosphate contacts in the major groove of the DNA hairpin. Edwards et al. previously showed in vitro that these loops disrupt DNA cleavage by the relaxase domain alone and in vivo that a full-length NES protein lacking these loops was not able to facilitate plasmid transfer (7, 8, 10, 14). However, this important protein-DNA interaction had not been characterized in vitro in the context of the full-length 665-residue NES protein, and we set out to determine in which steps of conjugation this interaction plays a role.
FIG 1.
Structure of NES relaxase domain and pSK41 oriT. (A) Structure of the NES relaxase domain in complex with DNA from the pSK41 origin of transfer (PDB entry 4HT4) (7). NES hairpin loop 1 is shown in yellow, and NES hairpin loop 2 is shown in green. (B) Hairpin loop 1 (yellow) of NES binds in the minor groove of the DNA hairpin formed by the pSK41 origin of transfer, while hairpin loop 2 (green) binds to the major groove. (C) Schematic of pSK41 oriT and the oligonucleotides used in these studies. When only the black portion is used, the oligonucleotide is referred to as OriTHP30. When the sequence is extended to include the orange portion, it is referred to as OriTHP35, to the teal is OriTHP40, and to the purple is OriTHP45.
pSK41-like conjugative plasmids have also been shown to mobilize several smaller coresident plasmids, such as pC221 and pSK639, which carry their own mob genes (9, 12, 13, 15, 16). Recently, O'Brien et al. showed that another staphylococcal conjugative plasmid, pWBG749, which is unrelated to pSK41, can facilitate the mobilization of other plasmids that lack mob genes (17). They demonstrated that this transfer is facilitated by origin-of-transfer sequences on the mobilizable plasmids that mimic the pWBG749 origin-of-transfer sequence, suggesting a conjugative relaxase in trans mechanism (18). We identified sequences similar to the pSK41 origin of transfer on numerous nonconjugative staphylococcal resistance plasmids (see Data Sets S1 and S2 in the supplemental material), raising the possibility that pSK41 family plasmids likewise facilitate mobilization of other plasmids by an analogous relaxase in trans mechanism mediated by NES. To investigate this possibility, we characterized the pSK41 oriT mimic sequences from two divergent nonconjugative plasmids. The first plasmid, pSK156, was isolated from a clinical strain in 1951 and is the earliest known multidrug efflux-encoding plasmid (19). The second plasmid, pCA347, was first sequenced in 2013 after isolation from a USA600 methicillin-resistant strain of S. aureus and encodes resistance to penicillin and heavy metals (20). Importantly, the variation in the origin-of-transfer sequences of pSK41 and the mimics of pSK156 and pCA347 is in the hairpin region of the DNA (see Fig. 3A). Based on these observations, we sought to explore the ability of NES to bind to and process putative oriT regions from pSK156 and pCA347 and the ability of pSK41 to facilitate transfer of plasmids containing these putative oriT regions in order to examine the potential for mobilization of plasmids containing pSK41 oriT mimics in staphylococci.
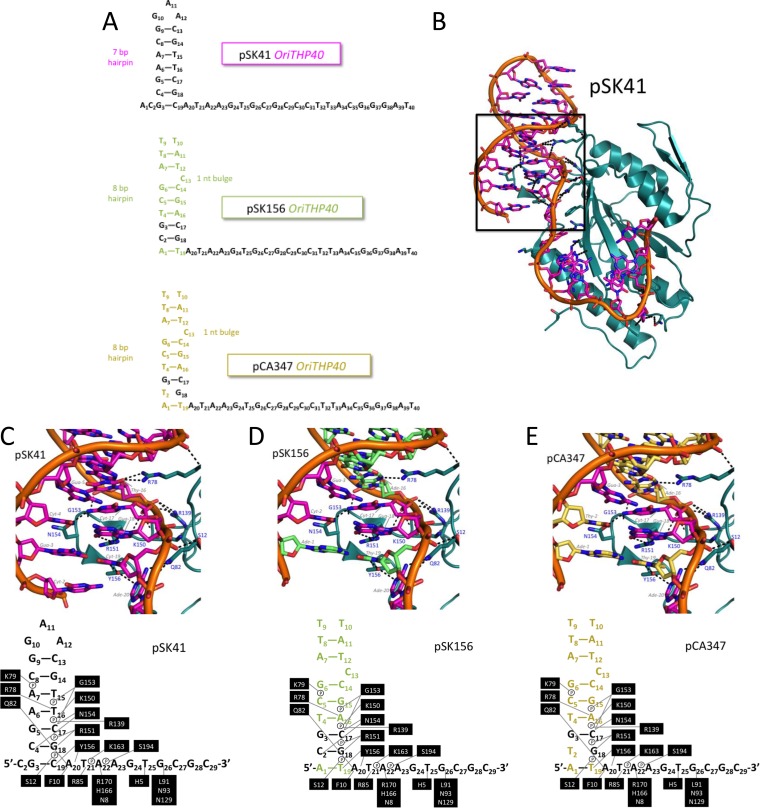
FIG 3.
Modeled structures of pSK41, pSK156, and pCA347 oriT regions. (A) Schematic of the pSK41, pSK156, and pCA347 oligonucleotides used in these studies. Colored nucleotides indicate differences in sequence from the pSK41 oriT. (B) Relaxase domain of NES in complex with pSK41 oriT. The box shows the region focused on for panels C to E. (C) Contacts between NES relaxase domain hairpin loop 1 and 2 amino acids and the pSK41 oriT nucleotide. (D) Contacts between NES relaxase domain hairpin loop 1 and 2 amino acids and the modeled pSK156 oriT nucleotide. Green nucleotides differ from the pSK41 oriT region. (E) Contacts between NES relaxase domain hairpin loop 1 and 2 amino acids and the modeled pCA347 oriT nucleotide. Gold nucleotides differ from the pSK41 oriT region.
MATERIALS AND METHODS
Cloning, expression, and purification of NES.
Wild-type full-length NES was previously cloned into the cysteine protease domain (CPD) fusion protein expression system developed by Shen et al. and optimized by our lab (7, 21). Loop deletion mutants were made through site-directed mutagenesis to remove hairpin loop 1 (residues 77 to 82) and hairpin loop 2 (residues 150 to 157) and replace each with a linker composed of one glycine and one serine. Cleavage-inactive mutants used in binding studies were made by replacing the tyrosine at amino acid position 25 with a phenylalanine. The resulting plasmids were transformed into Escherichia coli BL21(DE3) AI cells and grown in 1.5 liters of lysogeny broth (LB) in the presence of 0.1 mg/ml ampicillin at 37°C with shaking. At an optical density of 0.6 to 0.8, an l-arabinose solution was added to a final concentration of 0.2% (vol/vol), and the temperature was reduced to 18°C. After 30 min, protein expression was induced with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG), and cells were allowed to grow for 16 h. The cells were pelleted and stored at −80°C. Individual cell pellets were resuspended in buffer A (500 mM NaCl, 20 mM KH2PO4, pH 7.4, 25 mM imidazole, 0.02% [vol/vol] sodium azide) along with protease inhibitor tablets (Roche), DNase, and lysozyme. The mixture was sonicated and then clarified via centrifugation. The supernatant was filtered and loaded onto a HisTrap column (GE Healthcare). The CPD expression system contains a His6 tag in addition to the CPD tag, which has self-cleavage abilities in the presence of inositol hexakisphosphate (InsP6). Therefore, after the His-CPD-NES fusion protein was bound to the column via the His6 tag, the column was incubated with 2 mM InsP6 for 3 h at 4°C. The NES protein was then eluted off the column in buffer A, while the His6 and CPD tags remained bound to the column. The NES protein was then passed over a Superdex 200 column (GE Healthcare) preequilibrated in sizing buffer (25 mM HEPES, pH 7.4, 300 mM NaCl, 0.02% [vol/vol] sodium azide). The purity of each fraction was assessed by SDS-PAGE, and fractions containing >95% pure protein were combined and concentrated to approximately 1.2 mg/ml.
DNA binding studies.
DNA oligonucleotides 5′-end labeled with 6-carboxyfluorescein (FAM) were ordered from Integrated DNA Technologies and resuspended in annealing buffer (10 mM Tris, pH 7.5, 50 mM NaCl, 0.05 mM EDTA), and hairpins were formed by heating the oligonucleotides to 98°C for 1 min and then cooling the solution by 3°C/s. The dissociation constant of binding was calculated using fluorescence anisotropy as described by Edwards et al. (7). Briefly, protein was serially diluted in a buffer of 100 mM NaCl, 0.1 mg/ml bovine serum albumin (BSA), 5 mM magnesium acetate, and 25 mM Tris acetate, pH 7.5, to give 40-μl aliquots at final protein concentrations ranging from 0 to 0.5 μM. Assays were conducted in a 384-well black assay plate (Costar), allowing for 16 concentrations of protein. Ten microliters of the DNA probe was added to each 40-μl protein solution, resulting in a final concentration of DNA of 50 nM in a total volume of 50 μl in each well. Fluorescence anisotropy of the fluorescein-labeled DNA was observed via excitation at 485 nm and emission at 520 nm, using a PHERAstar plate reader (BMG Labtech). Measurements were made in triplicate, and reported values are the averages for three separate triplicate runs. Data were plotted as average fluorescence anisotropy values as a function of protein concentration by using Graphpad Prism v6.05. The following equation was employed to fit the data and to calculate the KD (equilibrium dissociation constant) for the substrate:
where f is the average fluorescence anisotropy signal, T is the total DNA concentration (set to 50 nM), x is the total protein concentration, K is the KD, “min” is the average fluorescence anisotropy signal of the no-protein control, and “max” is the average fluorescence anisotropy signal of the sample at a saturating concentration of protein. A single binding site was assumed, and the standard error is reported for each measurement. The reported values are averages for at least 5 independent experiments.
DNA cleavage assays.
The same 5′-end-FAM-labeled DNA oligonucleotides used for the DNA binding studies were used to measure equilibrium DNA cleavage in PAGE gels. Each 10-μl reaction mixture contained 1.52 μM NES protein, 1 μM DNA substrate, and electrophoretic mobility shift assay (EMSA) buffer (50 mM NaCl, 20 mM Tris, pH 7.4, 0.02% [vol/vol] sodium azide). The reaction mixture was incubated at 37°C for 1 h and quenched by the addition of 2× running buffer (0.01% xylene cyanol, 0.01% bromophenol blue, 85% formamide, 20 mM EDTA, 2× Tris-acetate-EDTA [TAE], 0.2% SDS). The resulting 20-μl reaction mixtures were run through a denaturing 16% polyacrylamide gel (35 ml of 16% acrylamide gel stock [8 M urea, 16% polyacrylamide-bisacrylamide, 1× Tris-borate-EDTA {TBE}], 300 μl of 10% ammonium persulfate [APS], and 33 μl of tetramethylethylenediamine [TEMED]) in 1× TBE running buffer to separate cleaved product DNA from the substrate. Using the fluorescein tag, oligonucleotides were visualized using a VersaDoc 4400 MP imaging system (Bio-Rad) and QuantityOne software (Bio-Rad). ImageJ 1.45s software was used to quantify band intensities, and the percent cleavage product formation was calculated by dividing the product band intensity by the product plus substrate band intensities. The averages for at least six individual cleavage experiments are presented, and individual data points are available in Fig. S2, S3, and S5 in the supplemental material.
DNA strand transfer assays.
DNA strand transfer assays were performed similarly to DNA cleavage assays, except that two pieces of DNA were used. The first piece of DNA was an unlabeled substrate with the same sequence and length as the oligonucleotides used in the DNA binding and cleavage studies (red DNA in Fig. 2B). The second piece of DNA was a 5′-end-FAM-labeled DNA oligonucleotide with the same sequence as the unlabeled substrate, but ending at the NES cleavage site (black DNA in Fig. 2B). Each 10-μl reaction mixture contained 1.52 μM NES protein, 1 μM unlabeled DNA substrate, 1 μM labeled DNA substrate, and EMSA buffer. The reaction mixture was incubated, run, and analyzed as in the DNA cleavage assays. Percent strand transfer was calculated by dividing the product band intensity by the product plus labeled substrate band intensities. The unlabeled DNA substrate was not visualized or quantified. The averages for at least six individual cleavage experiments are presented, and individual data points are available in Fig. S2, S4, and S5 in the supplemental material.
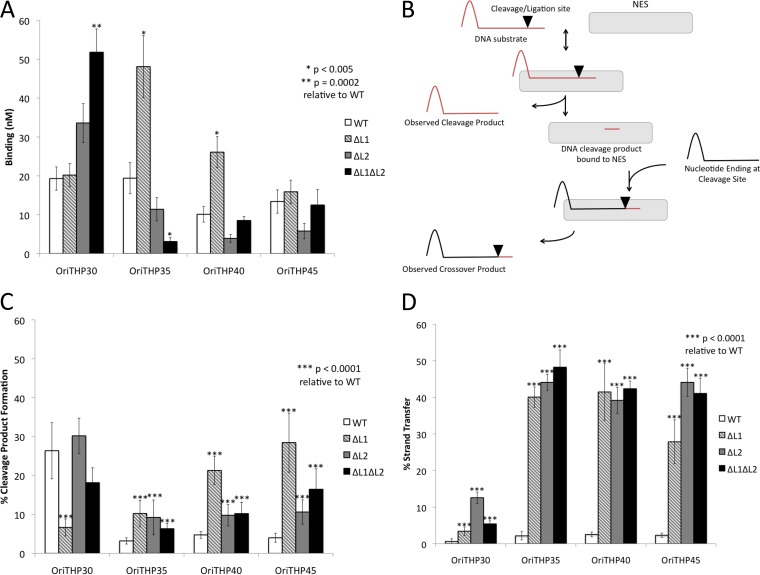
FIG 2.
Functional analysis of NES loop deletion mutants. (A) DNA binding (mean KD ± standard deviation) measured by fluorescence anisotropy for the indicated pSK41 oligonucleotides and NES mutants. (B) Schematic of DNA cleavage and strand transfer assays. DNA cleavage assays involved only the red DNA substrate, which was labeled with FAM at the 5′ end. DNA strand transfer assays involved both the red and black DNA substrates, with the red substrate being unlabeled and the black substrate being labeled with FAM at the 5′ end. (C) Functional analysis of cleavage activity of NES mutants on various pSK41 oligonucleotides. (D) Functional analysis of strand transfer activity of NES mutants on various pSK41 oligonucleotides to mimic religation during conjugative plasmid transfer.
Structure modeling.
The NES relaxase domain-DNA complex structure reported previously (RCSB PDB accession code 4HT4) was employed for Fig. 1A and B and 3B and C (7). For Fig. 3D and E, in which pSK156 and pCA347 were modeled in place of the original pSK41 DNA, Coot (22) was used to mutate each DNA residue, and the final figures were rendered in PyMol (version 1.5.0.5; Schrödinger, LLC).
Plasmid sequence analysis.
The plasmid database compiled and analyzed for pWBG749 family oriT sequences by O'Brien et al. was analyzed for oriT sequences similar to that of pSK41 (18). The online interface of BLASTN was used to search these plasmids for the sequence ATAAGTGCGCCCTTACGGGATTTAAC from the pSK41 oriT, and each sequence with a match was manually inspected for an adjacent DNA hairpin sequence (23). Plasmids were then grouped according to various sequences found in the DNA hairpin. Plasmids determined to carry potential pSK41 oriT mimics were then searched for the NES relaxase gene (accession number NC_005024.1; nucleotides 8115 to 10112) to determine if they were conjugative plasmids.
Bacterial strains, plasmids, and growth and assay conditions.
Strains and plasmids used are listed in Table S1 in the supplemental material. E. coli and S. aureus were cultured at 37°C on LB agar or in liquid LB medium with aeration (200 rpm). When required, the growth medium was supplemented with antibiotics at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 10 μg/ml; gentamicin (Gm), 20 μg/ml; novobiocin (Nb), 5 μg/ml; and streptomycin (Sm), 50 μg/ml.
DNA fragments encompassing oriT regions were synthesized as GeneArt Strings (Life Technologies) (see Fig. S6 in the supplemental material) and cloned into HindIII and/or BamHI sites of the pSK1-based S. aureus-E. coli shuttle vector pSK5632. The insert integrity was verified by sequencing. pSK5632 constructs were introduced into the restriction-deficient S. aureus strain RN4220 by electroporation. pSK41 was introduced into each resulting strain by conjugation with strain SK5428, and resulting Cmr Gmr transconjugants were used as donors in mobilization experiments. Mobilization assays were conducted in brain heart infusion (BHI) liquid medium (Sigma-Aldrich) containing 40% (final concentration) polyethylene glycol (PEG) as described previously (18). The WBG4515 strain was used as a recipient, and Sm and Nb were used to select against donors. Transconjugants were isolated on medium additionally carrying either Gm (for pSK41) or Cm (for pSK5632).
RESULTS
Characterization of NES hairpin loops 1 and 2.
The crystal structure of the relaxase domain of NES in complex with the pSK41 oriT DNA hairpin, reported previously by Edwards et al., revealed two features unique to this class of relaxase, namely, two protein loops, termed hairpin loops 1 and 2 (Fig. 1A and B), that clamp around the hairpin duplex of the oriT DNA (7). These contacts are unique to this class of relaxase compared to those observed with the longer, multityrosine relaxases, such as F-encoded TraI, and they have yet to be characterized in the context of the full-length protein. Hence, we sought to determine the impact that deleting these unique loops would have on NES functions in vitro. Hairpin loop 1 deletion (ΔL1), hairpin loop 2 deletion (ΔL2), and double-deletion (ΔL1ΔL2) forms of the full-length NES protein, in which the loops were replaced with Gly-Ser linkers, were created using site-directed mutagenesis. The proteins were expressed recombinantly in E. coli and purified to homogeneity. DNA binding, cleavage, and strand transfer assays were conducted using DNA oligonucleotides similar to that employed in the complex presented in the crystal structure and possessing the same sequence as the origin of transfer (oriT) of NES-conjugated plasmid pSK41 (Fig. 1C).
For DNA binding studies, these variant proteins contained an active site Tyr25Phe mutation. Previous evidence suggested that NES relaxase activity is dependent on oligonucleotide length (see Fig. S1 and S2 in the supplemental material; see also reference 7), so various lengths of oriT were used to verify this trend and named as shown in Fig. 1C. Longer oligonucleotides should better mimic in vivo plasmid transfer. As shown in Fig. 2A, the ΔL1 form of full-length NES exhibited increased DNA binding (P < 0.005) compared to wild-type NES on the OriTHP35 and OriTHP40 oligonucleotides. ΔL2 NES did not demonstrate significantly different DNA binding on any oligonucleotide. In contrast, ΔL1ΔL2 NES showed significantly increased DNA binding (P = 0.0002) on the shortest oligonucleotide tested, OriTHP30, but decreased binding (P < 0.005) on OriTHP35. For the longest oligonucleotide tested, OriTHP45, no difference in binding was observed for any variant proteins compared to wild-type NES. Thus, we concluded that eliminating hairpin loop 1 or 2 from full-length NES can alter DNA binding in vitro in an oligonucleotide length-dependent manner.
DNA cleavage and strand transfer assays were conducted as described in Fig. 2B. The cleavage assay mimics the cleavage of the plasmid oriT to produce the single strand transferred during conjugation. On OriTHP30, only ΔL1 NES exhibited a significant (P < 0.0001) difference in DNA cleavage, in this case a reduction, relative to that by wild-type NES (Fig. 2C). On the longer OriTHP35, OriTHP40, and OriTHP45 oligonucleotides, all three variant proteins (ΔL1, ΔL2, and ΔL1ΔL2) demonstrated statistically significant (P < 0.0001) increases in DNA cleavage levels relative to those with wild-type NES. For these longer oligonucleotides, the wild-type cleavage rate was ∼4%, while the variant proteins exhibited 2- to 7-fold increases in cleavage. We concluded that eliminating the DNA hairpin-associated loops from NES increases DNA cleavage by the enzyme.
A more dramatic effect was observed in examining DNA strand transfer by the variant full-length forms of NES. The strand transfer assay measured the ligation of a portion of DNA covalently linked to NES following cleavage to a new piece of DNA containing the hairpin characteristic of oriT (Fig. 2B). This mimics the ligation step of conjugation that ends plasmid transfer. For OriTHP30, all NES variants (ΔL1, ΔL2, and ΔL1ΔL2) showed 5- to 15-fold increases in DNA strand transfer relative to wild-type NES (Fig. 2D). For OriTHP35, -40, and -45, the increases were even larger: 25% to nearly 50% of the substrate oligonucleotides provided to the NES variants were processed to strand transfer, while ∼5% of the oligonucleotides formed strand transfer products with wild-type NES. Thus, eliminating the DNA hairpin-contacting loops of NES produces significant and dramatic increases in the level of DNA strand transfer in vitro compared to that with wild-type NES. It can be concluded that the hairpin loop 1 and loop 2 regions of NES play an important role, particularly on longer DNA substrates that are more relevant to transfer in vivo, in limiting the level of DNA religation during conjugation.
Modeling of NES bound to pSK156 and pCA347.
We next sought to determine if related DNA sequences from other plasmids might serve as substrates for pSK41 NES. We examined the S. aureus plasmids of known sequence and selected two with sequences identical to the pSK41 oriT cleavage site. These two plasmids, pSK156 and pCA347, exhibited the same sequence as pSK41 in the 20-nucleotide region from the predicted hairpin through the nic site but deviated somewhat from the DNA hairpin region of the pSK41 oriT (Fig. 3A). We predicted based on modeling that pSK156 and pCA347 might each form 8-bp DNA hairpins with a one-nucleotide bulge; in contrast, pSK41 is known from its crystal structure to form a 7-bp DNA hairpin with no bulge (Fig. 3A and B). Interestingly, within the predicted DNA hairpins of pSK156 and pCA347, nucleotides at the base of the DNA hairpin (G3, C17, and G18) are conserved with the sequence of pSK41 (Fig. 3A and C to E). Furthermore, we noted that the 8-bp hairpins predicted for pSK156 and pCA347 are nearly identical in sequence (Fig. 3A, D, and E).
We next modeled the pSK156 and pCA347 DNA sequences into the pSK41 NES relaxase domain-DNA hairpin complex crystal structure. For reference, Fig. 3B shows the NES relaxase domain in complex with the pSK41 DNA hairpin, highlighting the interactions between the protein and DNA; the boxed region contains all the protein contacts with the DNA hairpin and will remain the focus of the pSK156 and pCA347 models. As shown in Fig. 3C, NES makes base-specific contacts with the pSK41 DNA hairpin at C4 via N154, T16 via R78, C17 via G153 and N154, G18 via R151, and C19 via Y156. All but one contact with pSK156 and two contacts with pCA347, along with six phosphate contacts, are maintained in the models despite the changes in DNA sequence between these plasmids and pSK41 (Fig. 3D and E). Because C17 and G18 are conserved in both pSK156 and pCA347, the contacts via G153, N154, and R151 are maintained. The cytosine at position 19 in pSK41 is replaced by a thymine in pSK156 and pCA347; however, the para-oxygen of thymine appears to be capable of receiving a hydrogen bond from Y156 of NES. Position 16 of pSK156 and pCA347 contains an adenine rather than the thymine found in pSK41. In our models, the ring nitrogen of adenine appears to be capable of receiving the same hydrogen bond from R78 as the oxygen of thymine; however, while the thymine oxygen can form two hydrogen bonds, the adenine nitrogen can form only one. Thus, despite sequence differences between pSK41 and these other two S. aureus plasmids, contacts between NES and the predicted oriT regions of all three plasmids are largely maintained.
An additional contact is predicted to be lost between NES and pCA347. While a cytosine is conserved in the same positions in pSK41 (position −4) and pSK156 (position −2), it is a thymine in pCA347 (T2) (Fig. 3E). In pSK41 and pSK156, the amine group of C4 donates a hydrogen bond to the oxygen of N154; however, the para-oxygen of thymine cannot form the same interaction. However, it is possible that the asparagine side chain could rotate to allow the thymine oxygen to receive a hydrogen bond from the N154 side chain amine. In doing so, though, this side chain would lose an interaction with C17. Despite this potential change, five base-specific contacts and six phosphate contacts are maintained in our models between NES and the sequences of plasmids pSK156 and pCA347 in this region. Thus, we hypothesize that NES is capable of binding to and utilizing these potential oriT regions of pSK156 and pCA347 as substrates.
Characterization of NES processing of pSK156 and pCA347.
We next analyzed the ability of pSK41 NES to process the potential oriT regions of pSK156 and pCA347 by measuring the protein's ability to employ these DNAs for binding, cleavage, and strand transfer. For DNA binding studies, wild-type full-length NES with an active site Y25F mutation was employed along with OriTHP40-like forms of pSK41, pSK156, and pCA347 (Fig. 3A). The OriTHP40-like form alone was analyzed because longer oligonucleotides have been shown to be important for the regulatory function of the C-terminal domain (see Fig. S1 and S2 in the supplemental material; see also reference 7), but significant differences between OriTHP40 and -45 were not seen in assays with the NES loop deletion protein mutants. NES bound the oriT mimic regions of pSK156 and pCA347, but less well than its binding of the pSK41 oriT region (Fig. 4A). The KD of NES binding to pSK41 is 19.3 ± 3 nM; in contrast, NES binds to pSK156 and pCA347 3- and 9-fold less strongly, with KDs of 55.8 ± 9 nM and 175 ± 30 nM, respectively. While the loss of one or two hydrogen bonds is not sufficient to explain this decrease in binding affinity, it is interesting that the changes in binding affinity reflect the degrees of change in sequence and interactions seen in our models.
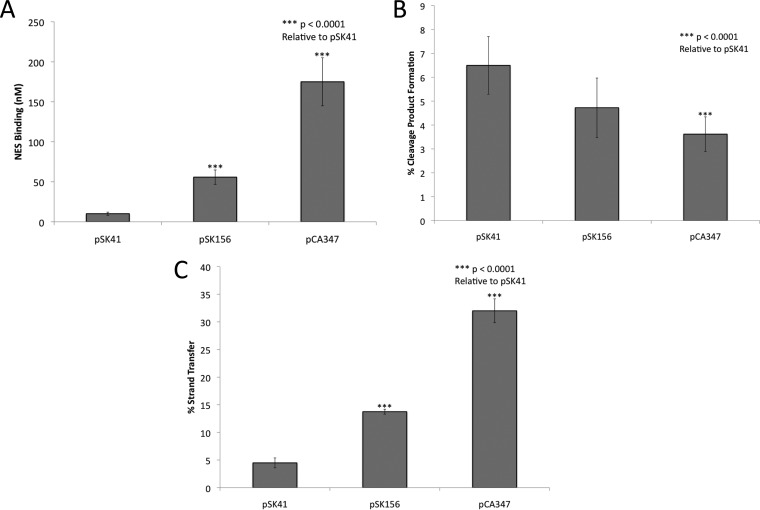
FIG 4.
NES processing of pSK41, pSK156, and pCA347 oriT oligonucleotides. (A) DNA binding (mean KD ± standard deviation) measured by fluorescence anisotropy for the pSK41, pSK156, and pCA347 oriT regions. The pSK41 value is the wild-type NES value as presented in Fig. 2A. (B) Cleavage activities of the pSK41-encoded NES protein on the pSK41, pSK156, and pCA347 oriT regions. (C) Strand transfer activities of the pSK41-encoded NES protein on the pSK41, pSK156, and pCA347 oriT regions.
DNA cleavage and strand transfer assays with full-length wild-type pSK41 NES and the OriTHP40-like regions of pSK156 and pCA347 were conducted as described in Fig. 2B. For DNA cleavage, pSK156 exhibited the same level of activity as pSK41, while pCA347 showed significantly decreased cleavage by NES (P < 0.0001) (Fig. 4B). However, because the cleavage process is dependent on NES first binding the DNA, this reduction in cleavage may have resulted from the decrease in binding seen in Fig. 4A for pCA347. Both pSK156 and pCA347 showed significantly increased DNA strand transfer, with 3- and 7-fold increases, respectively, relative to that with pSK41 (Fig. 4C). Taken together, these DNA binding, cleavage, and strand transfer data reveal that NES is capable of processing pSK156 and pCA347 oriT-like sites but does so at a lower efficiency than that at its cognate site.
Relaxase in trans mobilization by pSK41 in vivo.
To investigate the ability of pSK41 to facilitate relaxase in trans mobilization, the oriT-like sites corresponding to pSK156 and pCA347 and the pSK41 oriT sequence itself were synthesized and cloned into the nonmobilizable shuttle vector pSK5632 (24) to generate the plasmids pSK6881, pSK6879, and pSK6877, respectively; the DNA fragments cloned are shown in Fig. S6 in the supplemental material. These new plasmid constructs and pSK5632 were electroporated into S. aureus strain RN4220, and pSK41 was subsequently introduced via conjugation. These strains were then used as donors in mobilization assays with the recipient strain S. aureus WBG4515. As shown in Table 1, pSK41 was found to mobilize pSK6877, containing the pSK41 oriT sequence, at a frequency of 2.9 × 10−5, which is approximately 5-fold lower than that for pSK41 itself transferred in the same assay (1.4 × 10−4). However, despite repeated efforts, mobilization of the plasmids containing the pSK156 or pCA347 oriT mimic was never detected. These results demonstrate that pSK41-encoded NES can mediate trans-mobilization of a plasmid containing a copy of its own oriT site but suggest that its activity on the variant oriT-like sites from pSK156 and pCA347 is inadequate to facilitate plasmid transfer in vivo. As discussed below, an accessory protein may be required to complete relaxase-mediated transfer in vivo.
TABLE 1.
Relaxase in trans mobilization of plasmids containing oriT sites
| Plasmid | oriT site | Transfer frequencya |
|
|---|---|---|---|
| pSK41 | pSK5632 derivative | ||
| pSK5632 | None | 9.9 × 10−5 | Not detected |
| pSK6877 | pSK41 | 1.4 × 10−4 | 2.9 × 10−5 |
| pSK6879 | pCA347 | 6.6 × 10−5 | Not detected |
| pSK6881 | pSK156 | 8.7 × 10−5 | Not detected |
Transfer frequencies are presented as per-donor frequencies and are the averages for three experiments.
DISCUSSION
Transfer of conjugative and mobilizable plasmids is a major route by which antimicrobial resistance propagates, but the lack of details about the mechanism of this process impedes efforts to slow or prevent the spread of such resistance. We focused on the mechanism of action of the NES relaxase enzyme, encoded by pSK41 and related plasmids from staphylococci. Formation of a DNA hairpin in the pSK41 oriT and the importance of the associated NES hairpin loops 1 and 2 had been suggested previously (7). We demonstrate here that NES hairpin loops 1 and 2 are important for proper DNA cleavage and strand transfer (Fig. 2C and D) but not for DNA binding (Fig. 2A). The large increase in DNA strand transfer causing DNA to be ligated before transfer is complete is likely the biggest contributor to the large reduction of plasmid transfer seen when either hairpin loop 1 or loop 2 is eliminated from the encoded NES enzyme (7). It is also likely that accessory proteins in the pSK41 relaxosome complex with NES through interactions with NES hairpin loops 1 and 2, amplifying the effect of loss of these protein features.
Because relaxases are essential for transfer, share many common features, and are unique to the conjugative plasmid system, they represent a novel therapeutic target for decreasing the spread of antimicrobial resistance to allow current antimicrobial compounds to maintain efficacy. As explored previously, there are two potential sites of disruption common to relaxases: the metal binding site and specific protein-DNA interactions (7, 25). These results validate the NES hairpin loop 1 and 2 DNA interactions as target sites for such therapeutics. By disrupting the specific protein-DNA interactions in the NES hairpin loops, a molecule such as a sequence-specific polyamide could specifically disrupt cleavage and religation during pSK41 conjugation (7). As there seems to be some sequence conservation at the base of the DNA hairpin, this inhibitor molecule could target mobilizable plasmids in addition to the conjugative plasmid. Interestingly, there is a biological example of relaxase interference from Staphylococcus epidermidis strains carrying a CRISPR spacer that matches the nes gene of pSK41 and limits conjugative transfer (26). Targeted disruption of conjugation after initiation of the process and formation of the mating pore could cause cell death specifically in conjugative plasmid-containing bacteria. This targeted approach to bactericidal compounds is desirable as we learn more about the importance of the human skin microbiome (27).
Despite the importance of the protein-DNA interactions at the DNA hairpin, we show that there is some flexibility in the DNA hairpin sequence allowing sequences from pSK156 and pCA347 to be processed by NES. The oriT mimic sequences of pSK156 and pCA347 maintain all but one and two protein-DNA contacts, respectively, and are able to be bound, cleaved, and ligated by NES, though with an altered efficiency. We were therefore somewhat surprised to find that plasmid constructs containing the pCA347 or pSK156 oriT mimic could not be mobilized from cells harboring a pSK41 coresident, in contrast to a pSK41 oriT construct. However, the analogous relaxase in trans mobilization phenomenon recently described for the distinct pWBG749-like conjugative plasmids provides a precedent that likely explains this apparent paradox. Namely, pWBG749 oriT-like sequences exist as subtypes differentiated by sequence divergence in an inverted repeat (IR2) located adjacent to the nic site-containing core sequence (18). This results in specificity between various mobilizable plasmids and particular pWBG749-like conjugative plasmids. Thus, pWBG749 can mobilize plasmids with a pWBG749-like oriT of subtype OT49 but not those carrying an OT45 subtype, which instead can be mobilized by pWBG749-like conjugative plasmids that possess a cognate OT45 subtype oriT (18). Despite this, pWBG749 was able to stimulate recombination between OT49- and OT45-type oriT sequences carried on the same mobilizable plasmid, indicating that the pWBG749 relaxosome could recognize the OT45-type oriT even though it cannot mediate transfer of that subtype (18). By analogy, it seems plausible that the pSK156 and pCA347 oriT mimics examined here represent subtypes of pSK41-like oriT regions that can be recognized by NES but cannot be mobilized by the pSK41 relaxosome. In the case of the pWBG749 system, it has been shown that specificity for IR2 subtypes is dictated by a small putative DNA-binding accessory protein, SmpO, encoded adjacent to oriT on pWBG749, rather than by the relaxase SmpP (18). The involvement of accessory proteins in the pSK41 relaxosome has yet to be established.
Importantly, the scenario proposed above implies the existence of pSK41-like conjugative plasmids with variant oriT sequences that are capable of mobilizing plasmids such as pSK156, pCA347, and other plasmids listed in Data Set S2 in the supplemental material. Although no such variant pSK41-like plasmids have been detected to date, the presence of variant pSK41-like oriT mimic sequences on one-fifth of all sequenced staphylococcal plasmids (not including pSK41-like plasmids themselves) makes the whereabouts of such a reservoir an important question, since it is clearly influencing the evolution of plasmids in clinical staphylococci.
Interestingly, the oriT mimic sequence found in pCA347 is identical to a sequence found in pWBG757, a plasmid that could not be mobilized with pWBG749 in studies by O'Brien and colleagues (17). Comparative data such as these may allow us to classify mobilizable plasmids into families related to the relaxase(s) used for transmobilization. The oriT mimic sequence of pSK156 is also found in plasmid pWBG747, which could be mobilized by pWBG749, suggesting that pWBG747 harbors two distinct origin-of-transfer sequences to maximize its ability to be transferred (17, 18).
We searched the sequenced staphylococcal plasmids for other pSK41 oriT-like sequences and found 85 sequences from 83 different plasmids, including 14 pSK41 family conjugative plasmids (see Data Set S1 in the supplemental material). This represents 23.6% of Staphylococcus plasmids, which is significantly lower than the 53% of plasmids O'Brien et al. identified as harboring the pWBG749 oriT-like sequence (18). However, the set identified here includes 26 plasmids that do not have a pWBG749 oriT sequence, suggesting that NES is an important actor in the conjugative relaxase in trans mechanism.
Analysis of all 85 sequences showed that oriT sequences identical to that of pSK41 are evident only on plasmids which encode their own NES protein. On other plasmids, the pCA347 and pSK156 hairpin sequences, with their one-nucleotide difference, are by far the most common oriT mimics, representing 73% of the sequences (see Data Set S2 in the supplemental material). The other two major pSK41 oriT mimic types are significantly different in sequence but are still predicted to form a DNA hairpin which will allow for most of the NES protein-DNA interactions seen with the pSK41 oriT to be maintained, again suggesting that these specific protein-DNA interactions may be a potential therapeutic target. It is likely that NES proteins are capable of acting on a wide range of nonconjugative staphylococcal plasmids that contain an oriT mimic sequence, ranging from the oldest known multidrug resistance plasmid, pSK156, to prevalent contemporary plasmids, such as pMW2 and pUSA300HOUMR. The results described here imply that the recently described relaxase in trans mechanism of mobilization extends beyond pWBG749-like conjugative plasmids to the clinically more prevalent pSK41-like plasmids, thereby further increasing the proportion of staphylococcal plasmids that are potentially mobilizable. These observations lend further weight to the recent proposal that relaxase in trans mobilization represents a significant driver of horizontal transfer in staphylococci (18).
Supplementary Material
Funding Statement
Funding sources were not involved in the study design, data collection and interpretation, or decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00832-15.
REFERENCES
- 1.Liu MA, Kwong SM, Jensen SO, Brzoska AJ, Firth N. 2013. Biology of the staphylococcal conjugative multiresistance plasmid pSK41. Plasmid 70:42–51. doi: 10.1016/j.plasmid.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Pérez-Roth E, Kwong SM, Alcoba-Florez J, Firth N, Méndez-Alvarez S. 2010. Complete nucleotide sequence and comparative analysis of pPR9, a 41.7-kilobase conjugative staphylococcal multiresistance plasmid conferring high-level mupirocin resistance. Antimicrob Agents Chemother 54:2252–2257. doi: 10.1128/AAC.01074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Climo MW, Sharma VK, Archer GL. 1996. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J Bacteriol 178:4975–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDougal LK, Fosheim GE, Nicholson A, Bulens SN, Limbago BM, Shearer JES, Summers AO, Patel JB. 2010. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob Agents Chemother 54:3804–3811. doi: 10.1128/AAC.00351-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shearer JES, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, Lamarre J, Lindsay JA, Bayles K, Nicholson A, O'Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. 2011. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 1:581–591. doi: 10.1534/g3.111.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg T, Firth N, Apisiridej S, Hettiaratchi A, Leelaporn A, Skurray RA. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J Bacteriol 180:4350–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards JS, Betts L, Frazier ML, Pollet RM, Kwong SM, Walton WG, Ballentine WK, Huang JJ, Habibi S, Del Campo M, Meier JL, Dervan PB, Firth N, Redinbo MR. 2013. Molecular basis of antibiotic multiresistance transfer in Staphylococcus aureus. Proc Natl Acad Sci U S A 110:2804–2809. doi: 10.1073/pnas.1219701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 9.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhu W, Clark N, Patel JB. 2013. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob Agents Chemother 57:212–219. doi: 10.1128/AAC.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grohmann E, Muth G, Espinosa M. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev 67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Projan SJ, Archer GL. 1989. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol 171:1841–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apisiridej S, Leelaporn A, Scaramuzzi CD, Skurray RA, Firth N. 1997. Molecular analysis of a mobilizable theta-mode trimethoprim resistance plasmid from coagulase-negative staphylococci. Plasmid 38:13–24. doi: 10.1006/plas.1997.1292. [DOI] [PubMed] [Google Scholar]
- 14.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caryl JA, Thomas CD. 2006. Investigating the basis of substrate recognition in the pC221 relaxosome. Mol Microbiol 60:1302–1318. doi: 10.1111/j.1365-2958.2006.05188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong JJW, Lu J, Glover JNM. 2012. Relaxosome function and conjugation regulation in F-like plasmids—a structural biology perspective. Mol Microbiol 85:602–617. doi: 10.1111/j.1365-2958.2012.08131.x. [DOI] [PubMed] [Google Scholar]
- 17.O'Brien FG, Ramsay JP, Monecke S, Coombs GW, Robinson OJ, Htet Z, Alshaikh FAM, Grubb WB. 2015. Staphylococcus aureus plasmids without mobilization genes are mobilized by a novel conjugative plasmid from community isolates. J Antimicrob Chemother 70:649–652. doi: 10.1093/jac/dku454. [DOI] [PubMed] [Google Scholar]
- 18.O'Brien FG, Eto KY, Murphy RJT, Fairhurst HM, Coombs GW, Grubb WB, Ramsay JP. 2015. Origin-of-transfer sequences facilitate mobilisation of non-conjugative antimicrobial-resistance plasmids in Staphylococcus aureus. Nucleic Acids Res 43:7971–7983. doi: 10.1093/nar/gkv755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulsen IT, Brown MH, Skurray RA. 1998. Characterization of the earliest known Staphylococcus aureus plasmid encoding a multidrug efflux system. J Bacteriol 180:3477–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stegger M, Driebe EM, Roe C, Lemmer D, Bowers JR, Engelthaler DM, Keim P, Andersen PS. 2013. Genome sequence of Staphylococcus aureus strain CA-347, a USA600 methicillin-resistant isolate. Genome Announc 1:e00517-13. doi: 10.1128/genomeA.00517-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen A, Lupardus PJ, Morell M, Ponder EL, Sadaghiani AM, Garcia KC, Bogyo M. 2009. Simplified, enhanced protein purification using an inducible, autoprocessing enzyme tag. PLoS One 4:e8119. doi: 10.1371/journal.pone.0008119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grkovic S, Brown MH, Hardie KM, Firth N, Skurray RA. 2003. Stable low-copy-number Staphylococcus aureus shuttle vectors. Microbiology (Reading, Engl) 149:785–794. doi: 10.1099/mic.0.25951-0. [DOI] [PubMed] [Google Scholar]
- 25.Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR. 2007. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc Natl Acad Sci U S A 104:12282–12287. doi: 10.1073/pnas.0702760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schommer NN, Gallo RL. 2013. Structure and function of the human skin microbiome. Trends Microbiol 21:660–668. doi: 10.1016/j.tim.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.