Abstract
We report on a patient conceived via in vitro fertilization (IVF) with a 22q11.2 deletion due to an unusual unbalanced translocation involving chromosomes 6 and 22 in a karyotype with 45 chromosomes. Cytogenomic studies showed that the patient has a 3.3-Mb deletion of chromosome 22q and a 0.4-Mb deletion of chromosome 6p, which resulted in haploinsufficiency of the genes responsible for the 22q11.2 deletion syndrome and also of the IRF4 gene, a member of the interferon regulatory factor family of transcription factors, which is expressed in the immune system cells. The rearrangement could be due to the manipulation of the embryo or as a sporadic event unrelated to IVF. Translocation involving chromosome 22 in a karyotype with 45 chromosomes is a rare event, with no previous reports involving chromosomes 6p and 22q.
Key Words: Deletion syndrome 22q11.2, In vitro fertilization, IRF-4 transcription factor
The 22q11.2 deletion syndrome (OMIM 188400; http://omim.org/) is caused by a submicroscopic deletion of the long arm of chromosome 22 and affects ∼1 in 4,000 births, making it the second most prevalent genetic syndrome after Down syndrome [Sedghi et al., 2012]. The high frequency is related to the susceptibility of the 22q11.2 region to rearrangements due to the presence of 8 chromosome-specific low-copy repeats (LCRs) within 22q11.2, named LCR-A to LCR-H [Shaik et al., 2007]. These LCRs contain a complex modular structure and a high degree of DNA sequence identity (>96%), which mediates nonallelic homologous recombination, resulting in chromosome 22 rearrangements. The most frequent (in ∼90% of the patients) are the 3-Mb deletions between LCR-A and D and the 1.5-Mb deletions (∼10-12% of the patients) between LCR-A and B. Rare, atypical 22q11.2 deletions can also affect distinct chromosome regions involving other LCRs [Shaikh et al., 2007; Nogueira et al., 2008].
The main characteristics observed in 22q11.2 deletion syndrome are dysmorphic facial features, cardiac anomalies, cleft palate, velopharyngeal and thymic insufficiency, hypoparathyroidism, immune deficiency, developmental delay, and psychiatric disorder [Basset et al., 2011; McDonald-McGinn and Sullivan, 2011; Baker and Vortsman, 2012]. However, there is a wide phenotypic variability in 22q11.2 deletion syndrome, even within family members [McDonald-McGinn and Sullivan, 2011].
Deletion 6p (OMIM 612582) is a rare event within the population [Mirza et al., 2004]. Patients with similar distal deletions involving the 6p25pter region have a recognizable malformation pattern including downslanting palpebral fissures, flat nasal bridge, small nose, hypertelorism, midface hypoplasia, low-set ears, hearing loss, high-arched palate, ophthalmological, anterior eye chamber and craniofacial abnormalities, Dandy-Walker malformation/variant, congenital heart defects, language impairment, and developmental delay [Lin et al., 2005; Nakane et al., 2013]. Impairment of the central nervous system and developmental delay of variable degrees are observed in all patients described [Martinet et al., 2008]. The clinical phenotype of these patients is probably caused by haploinsufficiency of one or more genes located in 6p25 [Cellini et al., 2012] and based on the patient described by Anderlid et al. [2003]; the critical region of the 6pter deletion syndrome was narrowed down to a 2.1-Mb interval.
Here, we present a 4-year-old girl conceived via in vitro fertilization (IVF) with characteristic features of a 22q11.2 deletion, who showed an unusual unbalanced translocation involving chromosomes 6 and 22 in a karyotype with 45 chromosomes.
Patient and Methods
Clinical Report
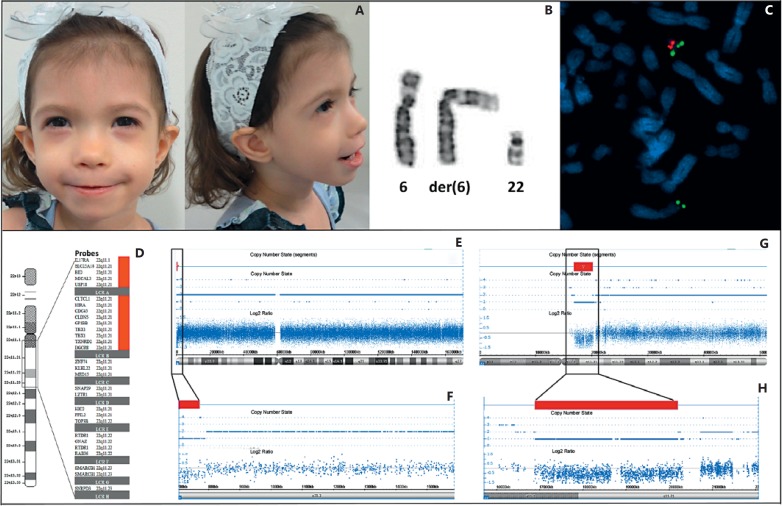
We report on a female patient born at 36 weeks in a triplet birth, conceived via IVF. It was the only pregnancy for the nonconsanguineous healthy couple. The Apgar score at birth was 7/8, and she had transient tachypnea of the newborn, requiring mechanically assisted ventilation. She remained in the intensive care unit for her first 4 months for treatment of congestive heart failure, when she was diagnosed with truncus arteriosus. During cardiac surgery, the thymus was not visualized. On physical examination at 2½ years (fig. 1A), she presented with generalized hypotonia, malar hypoplasia, flabby face, asymmetric ears, epicanthic folds, hypertelorism, small mouth, downslanted mouth commissures, long fingers, and swallowing disability. Cardiological evaluation revealed ventricular septal defects, pulmonary stenosis, aortic artery located on the right, persistent ductus arteriosus, and right ventricular hypertrophy. She also presented hypoparathyroidism, immunodeficiency and persistent hypocalcemia. Her weight was 9.4 kg (3rd centile), length 85.5 cm (3rd centile) and her head circumference was 45 cm (3rd centile). At 4½ years, she presented with hyperactivity, attention deficit disorder, hypernasal speech with a high-pitched voice, and mild conductive hearing loss. Her weight was 13.3 kg (3rd centile), length 104 cm (15-50th centile) and her head circumference was 47.5 cm (3rd-15th centile). Table 1 depicts the clinical features of the patient compared to other patients with 22q11.2 deletion syndrome evaluated in our institution.
Fig. 1.
A Frontal and lateral profile view of the patient at the age of 2½ years. B Partial karyotype showing the normal chromosome 6, the der(6) and the normal chromosome 22. C FISH with DiGeorge/VCFS TUPLE 1 probe showing one signal from the 22q11.21 probe in the normal chromosome 22 (red) and 2 signals from the 22q13.33 probe (green); one in chromosome 22, and another in the der(6). D Chromosome 22 diagram showing the 22q11 region between the LCR-A and LCR-H (grey bar) and the deleted region (red bar) shown by the SALSA MLPA P250 DiGeorge probemix kit probes. Genomic array showing the deletion in chromosome 6 highlighting the 6p25.3 region including the ∼0.4-Mb deleted segment (E, F) and in chromosome 22 highlighting the 22q11 region with the ∼3.3-Mb deleted segment (G, H). Red bars indicate deletions.
Table 1.
Clinical features of 21 patients with 22q11.2 deletion syndrome evaluated in our institution compared to the patient with t(6;22)
| Clinical features | Patients with deletiona (n = 21) | Our study |
|---|---|---|
| Congenital heart disease | 14/21 | + |
| Hypotonia | 7/19 | + |
| Asymetrie face | 5/21 | + |
| Ophtalmologic abnormalities | 2/17 | not evaluated |
| Downslanting palpebral fissures | 2/18 | + |
| Overfolded helices | 10/20 | – |
| Hearing loss | 8/18 | + |
| Prominent nasal bridge | 9/21 | – |
| Bulbous nasal tip | 9/21 | – |
| Hypoplastic nasal alae | 7/21 | – |
| Cleft palate | 5/21 | – |
| Velopharyngeal insufficiency | 11/21 | + |
| Hypernasal speech | 10/18 | + |
| Downturned oral commissures | 8/18 | + |
| Long and thin fingers | 15/21 | + |
| Hypocalcemia | 5/19 | + |
| Lymphopenia | 2/19 | – |
| Hypothyroidism | 3/18 | – |
| Thrombocytopenia | 0/18 | not evaluated |
| Thymus abnormalities | 1/4 | + |
| Lack of sociability | 11/18 | – |
| Learning disabilities | 15/19 | + |
| Psychiatric disorders | 3/18 | – |
Number of patients with the condition/number of patients evaluated.
During her first 3 years of life, she presented recurrent respiratory infections, 2 of which were severe, requiring intensive care treatment. Immune system evaluation on different occasions during these first years showed normal IgG (range from 404 to 739 mg/dl) and IgA (range from <40 to 88 mg/dl) levels and a low IgM level (range from 27 to 60 mg/dl) according to age. In this same period, T and B lymphocytes were analyzed on 4 different occasions: T CD4+ ranged from 33 to 47% (824-2,545/ml); T CD8+ ranged from 4,8 to 8% (117-547/ml); CD19+ ranged from 13 to 23% (309-684/ml). At 1 year of age, naive T lymphocytes were normal (39 and 60% for CD4 and CD8, respectively), and she showed a predominant proportion of naive B-lymphocytes (92%). Natural killer cell numbers were normal at all evaluations. She was routinely immunized according to the Brazilian schedule and showed positive response for protein antigens and absence of response to 7 serotypes (4, 6B, 9V, 14, 18C, 19F, and 23F) of conjugate pneumococcus vaccine. Intravenous immunoglobulin was recommended when she was 3 years old, with an improvement of respiratory infections.
She was clinically diagnosed with a 22q11.2 deletion syndrome when she was 2 years old. Her 2 brothers had no major abnormalities; both had transient hypogammaglobulinemia of infancy, which they overcame.
Cytogenetic and Molecular Studies
Chromosome analysis was performed on 72-hour lymphocyte cultures from the patient, her parents and brothers, according to standard procedures. G-banding karyotype at the 550-band level revealed a translocation involving chromosomes 6 and 22 in a 45 chromosomes karyotype, with additional material in the short arm of chromosome 6 and one missing chromosome 22 (fig. 1B).
DNA was extracted with Gentra Puregene Blood Kit (Qiagen, Germantown, Md., USA). MLPA (MRC-Holland, Amsterdam, The Netherlands) was performed with SALSA MLPA P250-B2 DiGeorge kit and analyzed with GeneMarker Software (SoftGenetics, State College, Pa., USA). The analysis revealed a deletion of the probes corresponding to the proximal region of chromosome 22q up to the LCR-B (fig. 1D) showing the result: rsa 22q11.2(P250)×1. Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, Calif., USA), analyzed using Affymetrix Chromosome Analysis Suite Software (Affymetrix) showed a 0.4-Mb deletion of chromosome 6p and a 3.3-Mb deletion of chromosome 22q (fig. 1E-H), with the following result: arr[hg19] 6p25.3(366,332-771,504)×1,22q11.21(16,894,612-20,196,469)×1.
FISH performed in metaphase chromosomes using DiGeorge/VCFS TUPLE 1 probe (Cytocell) showed one copy of the 22q11.2 probe in the normal chromosome 22 and 2 copies of the 22q13.3 probe - one in the normal chromosome 22 and another in 6p (fig. 1C). The patient's parents and siblings presented normal cytogenetic and molecular technique results, indicating a de novo rearrangement. Thus, the karyotype of the patient was given as 45, XX, der(6)t(6;22)(p25.3;q11.21)dn,-22.
Discussion
Our patient was conceived via IVF in a triplet pregnancy. While several studies found a high degree of abnormalities in pregnancies conceived through infertility treatment [Hansen et al., 2002; Vanneste et al., 2009; Fortunato and Tosti, 2011], including the risk of imprinting disorders [Lazaraviciute et al., 2014], others were unable to find an association between assisted reproduction technologies and chromosomal abnormalities [Shevell et al., 2005; Martinez et al., 2010; Conway et al., 2011]. The genetic alterations in children conceived after assisted reproduction can be attributed to epigenetic errors from manipulation of the embryo culture that follows IVF [Cetin et al., 2003], to the effects of controlled ovarian hyperstimulation and ovulation induction, in vitro sperm preparation, or an inherent defect in the infertile couple possibly leading to both the infertility and the birth defect [Olson et al., 2005]. Thus, it is difficult to draw general conclusions about the overall levels of chromosome abnormality in embryos generated by IVF [Ioannou et al., 2012]. In our patient, the deletion could be either the result of manipulation or a sporadic event not directly related to IVF.
Unbalanced translocations involving monosomy 22pterq11 in a 45 chromosome constitution have been described in association with 22q11.2 deletion [Back et al., 1980; Kelley et al., 1982; Faed et al., 1987; Jancar and Karki, 1989; Reddy et al., 1996; Jaquez et al., 1997; Carratalá et al., 1998; Damatova et al., 2009; Dundar al., 2009; McGoey and Lacassie, 2009; Shuib et al., 2009; Zrnová et al., 2012; Hu et al., 2014], none of these with a proven LCR 22q11.2 breakpoint. Our patient showed a breakpoint within the LCR-B, which has AT-rich palindromic sequences frequently involved in constitutional translocations, such as in the recurrent translocation t(11;22)(q23;q11.2) [Gotter et al., 2004; Kato et al., 2012]. Considering that similar repetitive regions cannot be observed in chromosome 6p25.3, the patient's translocation could be randomly formed by the nonhomologous end-joining mechanism.
Since we found one metaphase cell with 46 chromosomes including the der(6) and a small marker chromosome similar to what could have been a der(22), we can hypothesize that the zygote presented a balanced t(6;22) translocation, originating during meiosis or due to a germ cell mosaicism in one of her parents. The zygote may have subsequently lost the der(22), resulting in a 45 chromosome karyotype.
In the majority of patients with these translocations, the loss of the proximal 22q region usually results in 22q11.2 deletion syndrome, although the concomitant deletion of another autosome chromosome in some patients may result in both deletion syndrome phenotypes [Reddy et al., 1996]. There is only one case in the literature involving chromosome 6 in a 45 chromosome karyotype, although it resulted in a larger deletion of the long arm of chromosome 6 that caused a more severe phenotype than the 22q11.2 deletion [Jancar and Karki, 1989].
The 6p deletion in our patient is much smaller (0.4 Mb) when compared to other patients reported in the literature, who had deletions from 2.1 to 8.1 Mb, resulting in a higher number of hemyzigous genes and more severe phenotypes [Descipio, 2007; Martinet et al., 2008; Cellini et al., 2012; Nakane et al., 2013]. The 6p deletion in our patient encompassed only 3 genes: IRF4 (interferon regulatory factor 4) [OMIM 601900], EXOC2 (exocyst complex component 2) [OMIM 615329], HUS1B [HUS1 checkpoint homolog b (S. pombe)] [OMIM 609713] and a pseudogene LOC100421511 (MAP/microtubule affinity-regulating kinase 2 pseudogene). While EXOC2 and HUS1B appear to have no correlation with the patient's phenotype, IRF4, a member of the IRF family of transcription factors, is expressed exclusively in the immune system cells and is part of the biological processes involved with the immunological system. The IRF4 gene appears to play a critical role in the process of immunoglobulin class-switch recombination [De Silva et al., 2012]. During the generation of B cells in the bone marrow, IRF4 is largely redundant with the IRF8, which is part of the same family of genes [Lu et al., 2003]. They are more related to one another than to other genes of the IRF family [Taniguchi et al., 2001]. Considering that our patient shows the 3 classes of immunoglobulins, a single copy of the IRF4 gene, or the redundancy of function of IRF4 and IRF8 genes may maintain the operation of class-switch recombination. Despite the hemizigosity of the IRF4 gene, which appears to have an important role in the immunological system, the immunodeficiency showed by our patient appears to be a result of the 22q11.2 deletion, since the patients described with pure 6p deletion do not show immune defects [Descipio, 2007; Cellini et al., 2012]. Considering the restricted number of genes deleted in 6p25.3 and their functions, we can attribute the patient's phenotype to the 22q11.2 deletion.
Since the deletion of chromosome 22 in our patient includes the region between LCRs A and B, where the candidate genes for the syndrome are located, she presents some of the expected characteristics. The TBX1 (T-box transcription factor) gene is considered the major candidate for 22q11.2 deletion syndrome [Gao et al., 2015], being associated with cardiovascular defects [Lindsay et al., 2001; Jerome and Papaioannou, 2001], middle and inner ear defects, resulting in sensorineural hearing loss [Funke et al., 2001], and with craniofacial and dental development delay [Gao et al., 2015]; features we found in our patient.
Most patients with 22q11.2 deletion syndrome have diminished T-cell numbers in peripheral blood with severity ranging from absent thymic tissue with no circulating T cells, to completely normal T-cell counts [Sullivan, 2008; McDonald-McGinn and Sullivan, 2011]. Patients with 22q11.2 deletion syndrome and microscopic rests of thymic epithelial cells producing circulating T cells have also been reported [Bale and Sotelo-Avila, 1993]. In these patients, because of the limited organ space, the peripheral blood has a diminished supply of T cells [McDonald-McGinn and Sullivan, 2011]. Although the patient's thymus was not visualized during cardiac surgery, our patient must have remnant thymic tissue because of the T cells found in peripheral blood. Regarding B cells, it has already been demonstrated that patients with a 22q11.2 deletion have a higher frequency of naive B cells compared to controls [Finocchi et al., 2006] and also have an inability to mount an efficacious response to polysaccharide antigens [Schubert and Moss, 1992] as was observed in our patient.
The 22q11.2 deletion in our patient also includes the proximal cat eye syndrome critical region near the centromere. When triplicated, this region results in cat eye syndrome, and when duplicated in association with duplication of 11q, it results in Emanuel syndrome [Choudhary et al., 2013]. Interestingly, the loss of the same region may not cause clinically important features, such as in the family reported by Kriek et al. [2006] which has 5 family members carrying the deletion without phenotypic alteration, suggesting that haploinsufficiency of the cat eye syndrome region may have no clinical relevance.
In this report, the first of a 22q11.2 deletion associated to IVF, the use of analytical methods such as cytogenetic and molecular techniques improved the analysis of an unusual chromosomal rearrangement, enabling the correlation between the genomic imbalances found in our patient and her 22q11.2 deletion syndrome-associated phenotype.
Statement of Ethics
Informed consent for clinical and genetic analyses was obtained from the patient's parents in compliance with the ethics committee of our institution.
Disclosure Statement
The authors have no conflicts of interest to declare.
Acknowledgements
We would like to thank the family for their permission to publish and the São Paulo Research Foundation (FAPESP) and the Coordination for the Improvement of Higher Education Personnel (CAPES) for their financial support.
References
- 1.Anderlid BM, Schoumans J, Hallqvist A, Ståhl Y, Wallin A, et al. Cryptic subtelomeric 6p deletion in a girl with congenital malformations and severe language impairment. Eur J Hum Genet. 2003;11:89–92. doi: 10.1038/sj.ejhg.5200907. [DOI] [PubMed] [Google Scholar]
- 2.Back E, Stier R, Böhm N, Adlung A, Hameister H. Partial monosomy 22pter leads to q11 in a newborn with the clinical features of trisomy 13 syndrome. Ann Genet. 1980;23:244–248. [PubMed] [Google Scholar]
- 3.Baker K, Vorstman JA. Is there a core neuropsychiatric phenotype in 22q11.2 deletion syndrome? Curr Opin Neurol. 2012;25:131–137. doi: 10.1097/WCO.0b013e328352dd58. [DOI] [PubMed] [Google Scholar]
- 4.Bale PM, Sotelo-Avila C. Maldescent of the thymus: 34 necropsy and 10 surgical cases, including 7 thymuses medial to the mandible. Pediatr Pathol. 1993;13:181–190. doi: 10.3109/15513819309048205. [DOI] [PubMed] [Google Scholar]
- 5.Bassett AS, McDonald-McGinn DM, Devriendt K, Digilio MC, Goldenberg P, et al. Practical guidelines for managing patients with 22q11.2 deletion syndrome. J Pediatr. 2011;159:332–339. doi: 10.1016/j.jpeds.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carratalá F, Galán F, Moya M, Estivill X, Pritchard MA, et al. A patient with autistic disorder and a 20/22 chromosomal translocation. Dev Med Child Neurol. 1998;40(7):492–495. doi: 10.1111/j.1469-8749.1998.tb15400.x. [DOI] [PubMed] [Google Scholar]
- 7.Cellini E, Disciglio V, Novara F, Barkovich JA, Mencarelli MA, et al. Periventricular heterotopia with white matter abnormalities associated with 6p25 deletion. Am J Med Genet A. 2012;158A:1793–1797. doi: 10.1002/ajmg.a.35416. [DOI] [PubMed] [Google Scholar]
- 8.Cetin I, Cozzi V, Antonazzo P. Fetal development after assisted reproduction-a review. Placenta. 2003;24(Suppl B):S104–S113. doi: 10.1016/s0143-4004(03)00185-1. [DOI] [PubMed] [Google Scholar]
- 9.Choudhary MG, Babaji P, Sharma N, Dhamankar D, Naregal G, Reddy VS. Derivative 11;22 (Emanuel) syndrome: a case report and a review. Case Rep Pediatr. 2013;2013:237935. doi: 10.1155/2013/237935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conway DA, Patel SS, Liem J, Fan KJ, Jalian R, et al. The risk of cytogenetic abnormalities in the late first trimester of pregnancies conceived through assisted reproduction. Fertil Steril. 2011;95:503–506. doi: 10.1016/j.fertnstert.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Damatova N, Beyer V, Galetzka D, Schneider E, Napiontek U, et al. Haploinsufficiency of 16.4 Mb from chromosome 22pter-q11.21 in a girl with unilateral conductive hearing loss. Cytogenet Genome Res. 2009;125:241–247. doi: 10.1159/000230008. [DOI] [PubMed] [Google Scholar]
- 12.DeScipio C. The 6p subtelomere deletion syndrome. Am J Med Genet C Semin Med Genet. 2007;145C:377–382. doi: 10.1002/ajmg.c.30156. [DOI] [PubMed] [Google Scholar]
- 13.De Silva NS, Simonetti G, Heise N, Klein U. The diverse roles of IRF4 in late germinal center B-cell differentiation. Immunol Rev. 2012;247:73–92. doi: 10.1111/j.1600-065X.2012.01113.x. [DOI] [PubMed] [Google Scholar]
- 14.Dundar M, Kiraz A, Tasdemir S, Akalin H, Kurtoglu S, et al. Unbalanced 3;22 translocation with 22q11 and 3p deletion syndrome. Am J Med Genet A. 2010;152A:2791–2795. doi: 10.1002/ajmg.a.33249. [DOI] [PubMed] [Google Scholar]
- 15.Faed MJ, Robertson J, Beck JS, Cater JI, Bose B, Madlom MM. Features of di George syndrome in a child with 45,XX,-3,-22, +der(3),t(3;22)(p25;q11) J Med Genet. 1987;24:225–227. doi: 10.1136/jmg.24.4.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finocchi A, Di Cesare S, Romiti ML, Capponi C, Rossi P, et al. Humoral immune responses and CD27+ B cells in children with DiGeorge syndrome (22q11.2 deletion syndrome) Pediatr Allergy Immunol. 2006;17:382–388. doi: 10.1111/j.1399-3038.2006.00409.x. [DOI] [PubMed] [Google Scholar]
- 17.Fortunato A, Tosti E. The impact of in vitro fertilization on health of the children: an update. Eur J Obstet Gynecol Reprod Biol. 2011;154:125–129. doi: 10.1016/j.ejogrb.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Funke B, Epstein JA, Kochilas LK, Lu MM, Pandita RK, et al. Mice overexpressing genes from the 22q11 region deleted in velo-cardio-facial syndrome/DiGeorge syndrome have middle and inner ear defects. Hum Mol Genet. 2001;10:2549–2556. doi: 10.1093/hmg/10.22.2549. [DOI] [PubMed] [Google Scholar]
- 19.Gao S, Moreno M, Eliason S, Cao H, Li X, et al. TBX1 protein interactions and microRNA-96-5p regulation controls cell proliferation during craniofacial and dental development: implications for 22q11.2 deletion syndrome. Hum Mol Genet. 2015;24:2330–2348. doi: 10.1093/hmg/ddu750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gotter AL, Shaikh TH, Budarf ML, Rhodes CH, Emanuel BS. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum Mol Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen M, Kurinczuk JJ, Bower C, Webb S. The risk of major birth defects after intracytoplasmic sperm injection and in vitro fertilization. N Engl J Med. 2002;346:725–730. doi: 10.1056/NEJMoa010035. [DOI] [PubMed] [Google Scholar]
- 22.Hu H, Yao H, Dong Y, Long Y, Xu L, et al. Distinct karyotypes in two offspring of a man with jumping translocation karyotype 45, XY, der(16)t(16;22)(q24;q11.2),-22[59]/ 45, XY, der(1)t(1;22)(p36;q11.2),-22[11]/ 45, XY, der(22)t(22;22)(p13;q11.2),-22[10] Am J Med Genet A. 2014;164A:2048–2053. doi: 10.1002/ajmg.a.36560. [DOI] [PubMed] [Google Scholar]
- 23.Ioannou D, Fonseka KG, Meershoek EJ, Thornhill AR, Abogrein A, et al. Twenty-four chromosome FISH in human IVF embryos reveals patterns of post-zygotic chromosome segregation and nuclear organisation. Chromosome Res. 2012;20:447–460. doi: 10.1007/s10577-012-9294-z. [DOI] [PubMed] [Google Scholar]
- 24.Jancar J, Karki CB. Unbalanced form of translocation deletion between chromosomes 6 and 22 in a mentally handicapped female [45,XX,-6,-22, +der(6), t(6;22)(q25.1:q11.2)] J Ment Defic Res. 1989;33:255–259. doi: 10.1111/j.1365-2788.1989.tb01473.x. [DOI] [PubMed] [Google Scholar]
- 25.Jaquez M, Driscoll DA, Li M, Emanuel BS, Hernandez I, et al. Unbalanced 15;22 translocation in a patient with manifestations of DiGeorge and velocardiofacial syndrome. Am J Med Genet. 1997;70:6–10. doi: 10.1002/(sici)1096-8628(19970502)70:1<6::aid-ajmg2>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 26.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 27.Kato T, Kurahashi H, Emanuel BS. Chromosomal translocations and palindromic AT-rich repeats. Curr Opin Genet Dev. 2012;22:221–228. doi: 10.1016/j.gde.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley RI, Zackai EH, Emanuel BS, Kistenmacher M, Greenberg F, Punnett HH. The association of the DiGeorge anomalad with partial monosomy of chromosome 22. J Pediatr. 1982;101:197–200. doi: 10.1016/s0022-3476(82)80116-9. [DOI] [PubMed] [Google Scholar]
- 29.Kriek M, Szuhai K, Kant SG, White SJ, Dauwerse H, et al. A complex rearrangement on chromosome 22 affecting both homologues; haplo-insufficiency of the Cat eye syndrome region may have no clinical relevance. Hum Genet. 2006;120:77–84. doi: 10.1007/s00439-006-0185-2. [DOI] [PubMed] [Google Scholar]
- 30.Lazaraviciute G, Kauser M, Bhattacharya S, Haggarty P, Bhattacharya S. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum Reprod Update. 2014;20:840–852. doi: 10.1093/humupd/dmu033. [DOI] [PubMed] [Google Scholar]
- 31.Lin RJ, Cherry AM, Chen KC, Lyons M, Hoyme HE, Hudgins L. Terminal deletion of 6p results in a recognizable phenotype. Am J Med Genet A. 2005;136:162–168. doi: 10.1002/ajmg.a.30784. [DOI] [PubMed] [Google Scholar]
- 32.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, et al. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 33.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17:1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinet D, Filges I, Besuchet Schmutz N, Morris MA, Gaide AC, et al. Subtelomeric 6p deletion: clinical and array-CGH characterization in two patients. Am J Med Genet A. 2008;146A:2094–2102. doi: 10.1002/ajmg.a.32414. [DOI] [PubMed] [Google Scholar]
- 35.Martinez MC, Méndez C, Ferro J, Nicolás M, Serra V, Landeras J. Cytogenetic analysis of early nonviable pregnancies after assisted reproduction treatment. Fertil Steril. 2010;93:289–292. doi: 10.1016/j.fertnstert.2009.07.989. [DOI] [PubMed] [Google Scholar]
- 36.McDonald-McGinn DM, Sullivan KE. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Medicine (Baltimore) 2011;90:1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- 37.McGoey RR, Lacassie Y. Paternal balanced reciprocal translocation t(9;22)(q34.3;q11.2) resulting in an infant with features of the 9q subtelomere and the 22q11 deletion syndromes due to 3:1 meiotic segregation and tertiary monosomy. Am J Med Genet A. 2009;149A:2538–2542. doi: 10.1002/ajmg.a.33078. [DOI] [PubMed] [Google Scholar]
- 38.Mirza G, Williams RR, Mohammed S, Clark R, Newbury-Ecob R, et al. Refined genotype-phenotype correlations in cases of chromosome 6p deletion syndromes. Eur J Hum Genet. 2004;12:718–728. doi: 10.1038/sj.ejhg.5201194. [DOI] [PubMed] [Google Scholar]
- 39.Nakane T, Kousuke N, Sonoko H, Yuko K, Sato H, et al. 6p subtelomere deletion with congenital glaucoma, severe mental retardation, and growth impairment. Pediatr Int. 2013;55:376–381. doi: 10.1111/j.1442-200X.2012.03729.x. [DOI] [PubMed] [Google Scholar]
- 40.Nogueira SI, Hacker AM, Bellucco FT, Christofolini DM, Kulikowski LD, et al. Atypical 22q11.2 deletion in a patient with DGS/VCFS spectrum. Eur J Med Genet. 2008;51:226–230. doi: 10.1016/j.ejmg.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson CK, Keppler-Noreuil KM, Romitti PA, Budelier WT, Ryan G, et al. In vitro fertilization is associated with an increase in major birth defects. Fertil Steril. 2005;84:1308–1315. doi: 10.1016/j.fertnstert.2005.03.086. [DOI] [PubMed] [Google Scholar]
- 42.Reddy KS, Sulcova V, Siassi B. Two sibs with Wolf-Hirschhorn and DiGeorge deletions resulting from an unbalanced chromosome rearrangement, 45, XX/XY, der(4)t(4;22)(p16.3;q11.2)mat,-22. J Med Genet. 1996;33:852–855. doi: 10.1136/jmg.33.10.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubert MS, Moss RB. Selective polysaccharide antibody deficiency in familial DiGeorge syndrome. Ann Allergy. 1992;69:231–238. [PubMed] [Google Scholar]
- 44.Sedghi M, Nouri N, Abdali H, Memarzadeh M, Nouri N. A case report of 22q11 deletion syndrome confirmed by array-CGH method. J Res Med Sci. 2012;17:310–312. [PMC free article] [PubMed] [Google Scholar]
- 45.Shaikh TH, O'Connor RJ, Pierpont ME, McGrath J, Hacker AM, et al. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–491. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shevell T, Malone FD, Vidaver J, Porter TF, Luthy DA, et al. Assisted reproductive technology and pregnancy outcome. Obstet Gynecol. 2005;106:1039–1045. doi: 10.1097/01.AOG.0000183593.24583.7c. [DOI] [PubMed] [Google Scholar]
- 47.Shuib S, Abdul Latif Z, Abidin NZ, Akmal SN, Zakaria Z. Inherited t(9;22) as the cause of DiGeorge syndrome: a case report. Malays J Pathol. 2009;31:133–136. [PubMed] [Google Scholar]
- 48.Sullivan KE. Chromosome 22q11.2 deletion syndrome: DiGeorge syndrome/velocardiofacial Syndrome. Immunol Allergy Clin North Am. 2008;28:353–366. doi: 10.1016/j.iac.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 50.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 51.Zrnová E, Vranová V, Soukalová J, Slámová I, Vilémová M, et al. Unique combination of 22q11 and 14qter microdeletion syndromes detected using oligonucleotide array-CGH. Mol Syndromol. 2012;2:88–93. doi: 10.1159/000335334. [DOI] [PMC free article] [PubMed] [Google Scholar]