Abstract
Urine is a metabolite-rich biofluid that reflects the body’s effort to maintain chemical and osmotic homeostasis. Clinical diagnosis routinely relies on urine samples because the collection process is easy and noninvasive. Despite these advantages, urine is an under-investigated source of biomarkers for multiple sclerosis (MS). Nuclear magnetic resonance spectroscopy (NMR) has become a common approach for analyzing urinary metabolites for disease diagnosis and biomarker discovery. For illustration of the potential of urinary metabolites for diagnosing and treating MS patients, and for differentiating between MS and other illnesses, 38 urine samples were collected from healthy controls, MS patients, and neuromyelitis optica-spectrum disorder (NMO-SD) patients and analyzed with NMR, multivariate statistics, one-way ANOVA, and univariate statistics. Urine from MS patients exhibited a statistically distinct metabolic signature from healthy and NMO-SD controls. A total of 27 metabolites were differentially altered in the urine from MS and NMO-SD patients and were associated with synthesis and degradation of ketone bodies, amino acids, propionate and pyruvate metabolism, tricarboxylic acid cycle, and glycolysis. Metabolites altered in urine from MS patients were shown to be related to known pathogenic processes relevant to MS, including alterations in energy and fatty acid metabolism, mitochondrial activity, and the gut microbiota.
Keywords: urine biomarkers, multiple sclerosis, neuromyelitis optica-spectrum disorder, NMR, metabolomics, multivariate statistics
Graphical Abstract
Introduction
Multiple sclerosis (MS) is a chronic disease of the central nervous system (CNS) with both neurodegenerative and inflammatory demyelinating components.1 The disease has a heterogeneous clinical presentation and is characterized by clinical symptoms that involve different parts of the CNS. Especially in the early stages, MS shares features with other demyelinating diseases like neuromyelitis optica-spectrum disorders (NMO-SD).2 Despite recently updated classification criteria,3,4 differentiation between the two diseases can be difficult but nevertheless vital, because misclassification can lead to increased disease activity due to incorrect treatment.5 Thus, the identification of metabolite biomarkers for MS may help improve MS diagnostic protocols and help better understand the pathogenesis of the disease.
Metabolite biomarkers are small chemical entities (<1500 Da) found in biofluids where their presence or concentration has a correlation with either the prognosis, existence, or progression of a disease or the therapeutic response to a medication or treatment.6 Metabolites are the end products of enzymatic reactions or protein activity that are readily modulated by genetic alterations, environmental stress, toxins, or drugs.7 Thus, all phenotypic alteration caused by a disease or a medical treatment is expected to exhibit a unique metabolic profile or fingerprint.8 The ability to accurately and efficiently detect these metabolic alterations presents a potential avenue for personalized medicine and disease diagnosis through the identification of metabolite biomarkers.9 Unlike a single gene or protein routinely used as a medical biomarker, a metabolomics biomarker is commonly comprised of a dozen or more metabolites and provides a highly unique signature that may increase the likelihood of a correct diagnosis.10
The analysis of urine to obtain a metabolic profile has a number of well-known advantages that includes ready availability; a rapid, easy, inexpensive, and noninvasive sample collection procedure; the ability to collect multiple, large samples over a range of time-points; and well-established protocols for storing, handling, and examining urine samples.11 The investigation of urine metabolites using nuclear magnetic resonance spectroscopy (NMR) is experiencing a rapid growth of interest, where NMR metabolomics is routinely being used for drug and biomarker discovery.12 NMR is an attractive technique because it requires minimal sample preparations and is able to simultaneously detect and quantify a variety of compounds from a complex mixture without separation. NMR is commonly combined with multivariate statistics to efficiently identify and statistically validate the metabolomics profile.13
To date, investigations into MS metabolite biomarkers has primarily focused on the analysis of cerebrospinal fluid (CSF) and serum samples from MS patients.14,15 Little attention has been given to the analysis of urine16 despite the fact that metabolites excreted into urine are readily accessible and are easily detected compared to CSF or blood samples.16 We previously reported an NMR metabolomics analysis of urinary markers of MS using the animal model experimental autoimmune encephalomyelitis (EAE).17 The results of our prior EAE animal study demonstrated the potential of using urine as a source of metabolite biomarkers for MS. Herein, we report an NMR metabolomics analysis of human urine samples collected from healthy controls, MS patients, and NMO-SD patients. Our results demonstrate a statistically significant difference in the urinary metabolites observed between MS patients and healthy controls and between MS and NMO-SD patients.
Methods and Materials
Patient Information and Clinical Manifestation
To ensure as much homogeneity as possible within the groups, we selected definite aquaporin-4 (AQP4)-seropositive NMO-SD patients with high antibody titers in the serum directed against the water channel AQP4, and ensured that all were diagnosed according to the criteria proposed by Wingerchuk 2006.4 Seven of the patients had definite seropositive NMO, whereas 2 had seropositive NMO-SD in the form of optic neuritis (ON) and longitudinally extensive transverse myelitis (LETM) (Table 1 and Table S1). The mean age was 39.3 with a female predominance as expected,18 and all received immunosuppressive therapy in the form of azathioprine.
Table 1.
Demographic Data of Hungarian Cohort*
| HS | AQP4-NMO/NMO-SD | MS | |
|---|---|---|---|
| n = 7 | n = 9 | n = 8 | |
| Disease Subtype | |||
| NMO-SD | 0 | 7 | 0 |
| ON | 0 | 1 | 0 |
| LETM | 0 | 1 | 0 |
| RR-MS | 0 | 0 | 8 |
| Sex | |||
| female | 4 | 6 | 6 |
| male | 3 | 3 | 2 |
| mean age (range) | 26.7 (25–30) | 39.3 (22–55) | 44.6 (31–70) |
| Treatment | |||
| azathioprine | 0 | 9 | 0 |
| natalizumab | 0 | 0 | 1 |
| interferon β-1b | 0 | 0 | 3 |
| glatiramer acetate | 0 | 0 | 2 |
| none of the above | 7 | 0 | 2 |
HS, healthy subjects; AQP4, aquaporin 4; NMO, neuromyelitis optica; NMO-SD, neuromyelitis optica-spectrum disorder; MS, multiple sclerosis; ON, optic neuritis; LETM, longitudinally extensive transverse myelitis; RR-MS, relapsing-remitting multiple sclerosis
Similarly, all MS patients were diagnosed with relapsing-remitting MS (RR-MS) according to the McDonald’s 2010 criteria19 and accordingly received immunomodulatory therapy. Among this group, 5 patients received first-line therapy (glatiramer acetate and interferon-β), 1 patient received second-line therapy (natalizumab), and 2 patients received no treatment (Table 1 and Table S1). The mean age was 44.6 years with a female predominance. On average, the age at disease onset is 34 years; however, none of these patients were newly diagnosed. All healthy subjects were healthy volunteers with no known neurological or autoimmune diseases. Neither MS nor NMO/NMO-SD patients had experienced a relapse within 30 days of the sample collection. The study was conducted in accordance with both the Hungarian and Danish National Ethics Committee (38.93.316-12464/KK4/2010, 42341-2/2013/EKU, S-20120066).
Urine Collection
Using the NMO-SD and MS database of the University of Pecs in Hungary, we collected urine samples from 9 patients with NMOSD seropositive for antibodies against AQP4, 8 patients with RRMS, and 7 healthy subjects. Treatment, age, and gender of the study populations are shown in Table 1 and Table S1. Urine samples were collected as spot urine in the morning before breakfast and administration of drug treatment and processed within 2 h of collection. The time intervals between replicate urine sample collections were variable. All patients were on chronic treatment as indicated in Table 1 and Table S1. Azathioprine and glatiramer acetate were administered daily, interferon β-1b every other day, and natalizumab was administered once a month. In cases of interferon β-1b treatment, urine was collected the day after administration. Urine was collected the day before the monthly natalizumab infusion in the single patient treated with natalizumab. Samples were centrifuged at 20,000g for 20 min at room temperature (RT) to pellet cell debris, and the supernatants were stored at −80 °C until use. Except for one MS patient, two analytical replicate urine samples were obtained from each MS patient and each healthy subject. Conversely, only one urine sample was collected from each NMO-SD patient.
NMR Sample Preparation
The urine samples were thawed and then centrifuged at 13000 rpm for 5 min at RT to remove any precipitate. Then, 100 μL of each urine sample was transferred into a new Eppendorf tube and mixed with 500 μL of 50 mM phosphate buffer in 99.8% D2O (Isotec, St. Louis, MO) at pH 7.2 (uncorrected); 50 μM of 3-(trimethylsilyl) propionic acid-2,2,3,3-d4 (TMSP-d4) was added to each sample as a chemical shift reference. The urine samples were then transferred to a 5 mm NMR tube for NMR data acquisition.
NMR Data, Collection, and Processing and Multivariate Statistical Analysis
The one-dimensional (1D) 1H NMR experiments were collected and processed as described previously.17 The NMR data processing and multivariate statistical analysis were accomplished using our MVAPACK software suite (http://bionmr.unl.edu/mvapack.php).20 The 1D 1H NMR spectra were aligned with the icoshift algorithm when the full-resolution spectra were modeled using orthogonal projections to latent structure discriminant analysis (OPLS-DA). Alternatively, the 1D 1H NMR spectra were binned using an intelligent adaptive binning algorithm when S-plots and a shared and unique structure (SUSplot) plot were generated from the OPLS-DA model. The data were normalized with a probabilistic quotient normalization function and Pareto scaled prior to multivariate statistical analysis. Fractions of explained variation (R2 X and R2 Y) were computed during OPLS-DA model training. OPLS-DA models were internally cross-validated using 7-fold Monte Carlo cross validation to compute Q2 values, which were compared to a distribution of null model Q2 values in 1000 rounds of response permutation testing. Model results were further validated using CV-ANOVA significance testing.
Metabolite Identification
An SUS plot was generated from the OPLS-DA models using MVAPACK to compare the MS and NMO-SD group against healthy controls. The plot visualizes the correlation between predictive components of each model and was used to identify metabolite changes unique to either the MS or the NMO-SD group. The chemical shift information from the loadings and SUS plots were assigned to metabolites using Chenomx NMR suite 7.0 (Chenomx Inc., Edmonton, Alberta, Canada) and the Human Urine Metabolome database (http://www.urinemetabolome.ca/). A 1H chemical shift error of 0.08 ppm was used to match the experimental chemical shifts with database values. Metabolite pathway analysis was accomplished using the Metabolomics Pathway Analysis (MetPA) Web server (http://metpa.metabolomics.ca/MetPA).
One-Way ANOVA and Univariate Statistical Analysis
One-way ANOVA and univariate calculations were conducted using the R statistical package version 3.2.0.21 One-way ANOVA was used to determine the statistical significance of individual metabolite differences between healthy, MS, and NMO-SD patients. Metabolites with a p-value ≤ 0.05 were then subjected to Tukey’s multiple comparison of means test to identify the set of metabolites that are statistically different between the paired groups.22 A Student’s t test was also applied to determine the statistical significance of metabolite differences between the two groups. The p-values from the Student’s t test were further adjusted using the Benjamini–Hochberg multiple hypothesis method.23
Results
Urine Metabolomics Signature for MS Patients
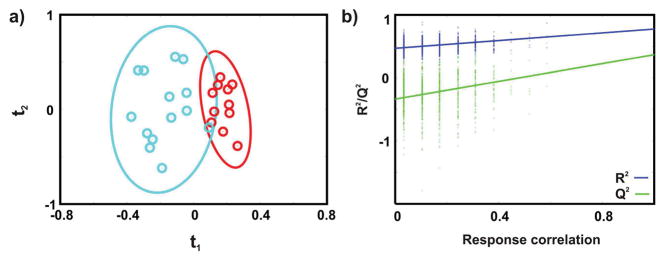
A 1D 1H NMR spectrum was acquired for each of the 29 urine samples collected from seven healthy individuals and eight patients previously diagnosed with MS. The 1D 1HNMR spectra capture a “snapshot” of the state of the urinary metabolome and provides a direct means of determining if the metabolic profiles differ between healthy and MS patients. The NMR data set was modeled by OPLS-DA, and the resulting scores plot (Figure 1a) shows a clear separation between the healthy controls and MS patients. Importantly, all of the biological replicates were assigned to the correct class in the OPLS-DA scores plot. The leave-n-out cross-validation metrics of R2Y = 0.77 and Q2 = 0.39 indicates an acceptable level of fit and predictive ability. A reliable model is also indicated by a p-value of 7.8 × 10−4 from the CV-ANOVA test and a p-value of 8.0 × 10−3 from the response permutation test (Figure 1b).
Figure 1.
(a) OPLS-DA scores resulting from modeling of the 1D 1H NMR data matrix from human urine samples collected from MS patients (cyan) and healthy controls (red). A statistically significant degree of separation is observed between the two experimental classes. The leave-n-out cross-validation metrics are R2Y = 0.77 and Q2 = 0.39, and the CV-ANOVA and a response permutation test p-values are 7.8 × 10−4 and 8.0 × 10−3, respectively. Ellipses enclose the 95% confidence intervals estimated by the sample means and covariances of each class. (b) Response permutation testing results for OPLS-DA scores after 1000 random permutations of the group membership information (Y). The model significance is inferred from the degree of vertical separation between the null distribution (leftmost) and the true R2Y and Q2 values (rightmost). The apparent discretization along the correlation axis is a result of using binary class labels in Y.
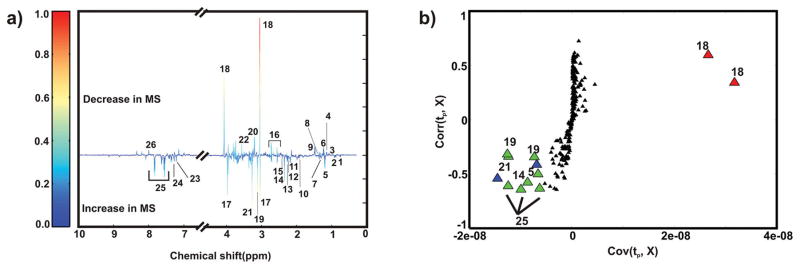
A back-scaled loadings plot was generated from the OPLS-DA model to identify the spectral regions (metabolites) that primarily contribute to the observed class separation in the scores plot (Figure 2a). Twenty-six metabolites are differentially altered in the urine samples collected from healthy individuals and MS patients. The identified metabolites are from metabolic pathways associated with energy metabolism, fatty acid synthesis, and gut microflora, which include amino acid derivatives and amino acid degradation products (Table S2). An S-plot was then used to identify the major contributors to these observed class differences in the OPLS-DA scores plot (Figures S1 and 2b). The S-plot identified creatinine, hippurate, 3-hydroxybutyrate, malonate, oxaloacetate, and trimethylamine N-oxide as having the highest covariance and correlation (from the 26 metabolites) with the OPLS-DA model from the healthy and MS data sets. A one-way ANOVA analysis was performed for each metabolite identified from the multivariate statistical analysis to determine if a statistically significant difference exists between the healthy and MS groups. Metabolites with a p-value ≤ 0.05 were selected (Table S3) and further subjected to Tukey’s multiple comparisons of means test.22 This analysis indicated that the set of metabolites, including creatinine (p-value = 4.2 × 10−4), 3-hydroxyisovalerae (p-value = 3.2 × 10−02), and oxaloacetate (p-value = 5.0 × 10−02) discriminate between healthy controls and MS patients (Figure S2). A Student’s t test followed by the Benjamini–Hochberg multiple hypothesis test (Figure S3) identified acetate and creatinine as statistically different between healthy controls and MS patients.23 The Benjamini–Hochberg adjusted p-value, one-way ANOVA, and multivariate analyses are fundamentally distinct techniques that emphasize different aspects of the data and have no expectations of producing identical results. Thus, a subset of the eight metabolites corresponding to acetate, creatinine, hippurate, 3-hydroxybutyrate, 3-hydroxyisovalerae, malonate, oxaloacetate, and trimethylamine N-oxide have the potential of differentiating between healthy controls and MS patients.
Figure 2.
(a) Back-scaled OPLS-DA loadings plot resulting from modeling of the 1D 1H NMR data matrix from human urine samples collected from MS patients and healthy controls. (b) S-plot from the OPLS-DA model generated from binned 1D 1H NMR spectra from the MS and healthy controls data sets (Figure S1). The x- and y-axis of the S-plot measures the covariance and correlation, respectively. The green and red triangles identify metabolites with a relative increase or decrease in concentration in urine samples from MS patients compared to healthy controls, respectively. The blue triangles correspond to unknown metabolites. The black triangles correspond to all other bins or metabolites. The metabolites are labeled as follows: 1, 2-hydroxyisovalerate; 2, isovalerate; 3, 3-hydroxyisobutyrate; 4, propylene glycol; 5, 3-hydroxybutyrate; 6, methylmalonate; 7, 3-hydroxyisovalerate; 8, lactate; 9, alanine; 10, acetate; 11, N-acetylglutamine; 12, acetone; 13, acetoacetate; 14, oxaloacetate; 15, succinate; 16, citrate; 17, creatine; 18, creatinine; 19, malonate; 20, choline-containing compounds; 21, trimethylamine N-oxide; 22, glycine; 23, phenylalanine; 24, phenylacetylglycine; 25, hippurate; and 26, xanthine.
Urine Metabolomics Signature for NMO-SD Patients
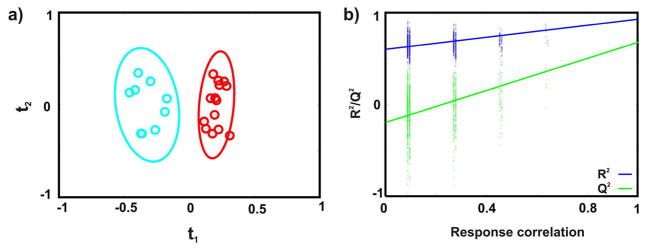
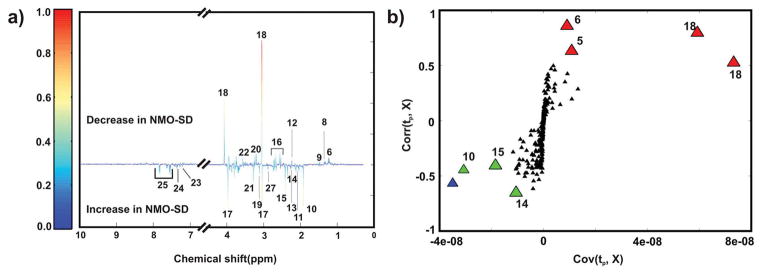
Because NMO-SD is an inflammatory demyelinating disease of the CNS similar to MS, but antibodies play a major role in the pathogenesis in contrast to the supposedly heterogeneous pathogenesis in MS, we considered NMO-SD as a valuable negative control for identifying biomarkers specific to MS.2 A 1D 1H NMR spectrum was acquired for each of the 23 urine samples collected from seven healthy individuals and nine patients previously diagnosed with NMO-SD. The NMR data set was modeled by OPLS-DA, and the resulting scores plot (Figure 3a) shows a clear separation between the healthy controls and NMOSD patients. All of the biological replicates were assigned to the correct class in the OPLS-DA scores plot. The leave-n-out cross-validation metrics of R2Y = 0.93 and Q2 = 0.68 indicates a reasonable level of fit and predictive ability. A reliable model is also indicated by a p-value of 7.3 × 10−3 from the CV-ANOVA test and a p-value of zero from the response permutation test (Figure 3b). A back-scaled loadings plot (Figure 4a) identified 20 metabolites differentially altered in the urine samples collected from healthy individuals and NMO-SD patients. The identified metabolites are amino acids and amino acid derivatives, tricarboxylic acid (TCA) cycle intermediates, choline-containing compounds, and metabolites from the gut microflora (Table S2). An S-plot was then used to identify the major contributors to this observed class differences in the OPLS-DA scores plot (Figures S4 and 4b). The S-plot identified acetate, creatinine, 3-hydroxybutyrate, methylmalonate, oxaloacetate, and succinate as having the highest covariance and correlation (from the 20 metabolites) with the OPLS-DA model from the healthy and NMO-SD data sets. A one-way ANOVA analysis was performed for each metabolite identified from the multivariate statistical analysis to determine if a statistically significant difference exists between the healthy and NMO-SD groups. Metabolites with a p value ≤ 0.05 were selected (Table S3) and further subjected to Tukey’s multiple comparisons of means test.22 This analysis indicated that the set of metabolites, including creatinine (p value 4.0 × 10−07), 3-hydroxybutyrate (p-value 2.9 × 10−02), oxaloacetate (p-value 2.7 × 10−02), and methylmalonate (p-value 2.6 × 10−06) discriminates between healthy controls and NMOSD patients (Figure S2). A Student’s t test followed by the Benjamini–Hochberg multiple hypothesis test (Figure S3) also identified the same set of metabolites as statistically different between healthy controls and NMO-SD patients. Similar to our comparison between healthy controls and MS patients, we identified a subset of the six metabolites corresponding to acetate, creatinine, 3-hydroxybutyrate, methylmalonate, oxaloacetate, and succinate that have the potential of differentiating between healthy controls and NMO-SD patients.
Figure 3.
(a) OPLS-DA scores resulting from modeling of the 1D 1H NMR data matrix from human urine samples collected from NMO-SD patients (cyan) and healthy controls (red). A statistically significant degree of separation is observed between the two experimental classes. The leave-n-out cross-validation metrics are R2Y = 0.93 and Q2 = 0.68, and the CV-ANOVA and a response permutation test p-values are 7.3 × 10−3 and 0, respectively. Ellipses enclose the 95% confidence intervals estimated by the sample means and covariances of each class. (b) Response permutation testing results for OPLSDA scores after 1000 random permutations of the group membership information (Y). The model significance is inferred from the degree of vertical separation between the null distribution (leftmost) and the true R2Y and Q2 values (rightmost). The apparent discretization along the correlation axis is a result of using binary class labels in Y.
Figure 4.
(a) Back-scaled OPLS-DA loadings plot resulting from modeling of the 1D 1H NMR data matrix from human urine samples collected from NMO-SD patients and healthy controls. (b) S-plot from the OPLS-DA model generated from binned 1D 1H NMR spectra from NMO-SD and healthy controls data sets (Figure S2). The x- and y-axis of the S-plot measures the covariance and correlation, respectively. The green and red triangles identify metabolites with a relative increase or decrease in concentration in urine samples from NMO-SD patients compared to healthy controls. The blue triangles correspond to unknown metabolites. The black triangles correspond to all other bins or metabolites. The metabolites are labeled as follows: 6, methylmalonate; 8, lactate; 9, alanine; 10, acetate; 11, N-acetylglutamine; 12, acetone; 13, acetoacetate; 14, oxaloacetate; 15, succinate; 16, citrate; 17, creatine; 18, creatinine; 19, malonate; 20, choline-containing compounds; 21, trimethylamine N-oxide; 22, glycine; 23, phenylalanine; 24, phenylacetylglycine; 25, hippurate; and 27, timethylamine.
Metabolite Pathway Analysis
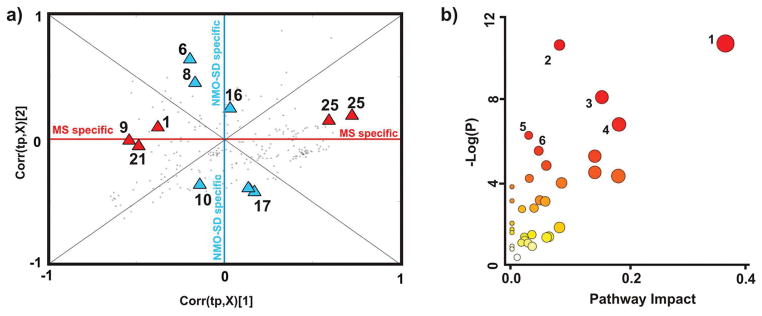
The complete list of 27 metabolites (Tables S2 and S4) differentially altered in urine samples collected from MS and NMO-SD patients relative to healthy controls were uploaded to the MetPA Web server. MetPA used the Homo sapiens pathway library, the hypergeometric test for the over-representation analysis, and the out-degree centrality for the pathway topology analysis. MetPA estimates a metabolite’s relative importance, provides a global overview of the metabolic changes, and assists in identifying important pathways associated with the disease phenotype. The synthesis and degradation of ketone bodies, amino acid metabolism, propionate metabolism, pyruvate metabolism, the TCA cycle, and glycolysis were identified as high impact pathways in MS and NMO-SD patients (Figure 5 and Table S5).
Figure 5.
(a) SUS plot generated from the OPLS-DA models compare the MS and NMO-SD groups against healthy controls. NMR bins within the MS specific or NMO-SD specific regions of the SUS plot are labeled with red or blue triangles, respectively. The resulting chemical shift information from the SUS plot was assigned to metabolites using ChenomxNMRsuite 7.0. The metabolites are numbered accordingly: 1, 2-hydroxyisovalerate/isovalerate; 6, methylmalonate; 8, lactate; 9, alanine; 10, acetate; 16, citrate; 17, creatine; 21, trimethylamine N-oxide; and 25, hippurate. (b) Overview of the pathway topology analysis produced by MetPa (http://metpa.metabolomics.ca/MetPA). The highest ranked pathways are numbered accordingly: 1, synthesis and degradation of ketone bodies; 2, propionate metabolism; 3, pyruvate metabolism; 4, TCA cycle; 5, alanine, aspartate, and glutamate metabolism; and 6, glycolysis or gluconeogenesis.
Urine NMR Metabolomics Signatures That Differentiate MS and NMO-SD Patients
The two back-scaled loadings plots and the two S-plots were directly compared to identify metabolites that were distinctly altered in the urine samples from either MS or NMO-SD patients relative to healthy controls (Figures 2 and 4 and Figure S5). Eight metabolites, alanine, hippurate, 2-hydroxyisovalerate, 3-hydroxybutyrate, isovalerate, malonate, oxaloacetate, and trimethylamine N-oxide were identified as being uniquely altered in urine samples from MS patients relative to NMO-SD patients. Eight other metabolites, acetate, acetone, citrate, creatine, creatinine, lactate, methylmalonate, and succinate were identified as being uniquely altered in urine samples from NMO-SD patients relative to MS patients.
For further refinement of a metabolic signature unique to MS patients, an SUS plot (Figure 5a) was generated from the MS vs healthy and the NMO-SD vs healthy OPLS-DA models using binned NMR data (Figures S1 and S4). The two sets of metabolites identified from the overlaid back-scaled loading plots were then assigned to appropriate bins in the SUS plot. In this manner, only metabolites that clearly fell within the MS specific or NMO-SD specific regions of the SUS plot were identified as statistically significant and retained. This analysis reduced the number of urine metabolites specific to MS (2-hydroxyisovalerate, alanine, hippurate, isovalerate, trimethylamine N-oxide) or to NMO-SD (acetate, creatine, lactate, methylmalonate, and citrate) to five each.
The results of the multivariate statistical analysis was further supported by a follow-up univariate analysis.24 A one-way ANOVA analysis was performed for each metabolite identified from the multivariate statistical analysis to determine if a statistically significance difference exists between the healthy, MS, and NMO-SD groups. Metabolites with a p-value ≤ 0.05 were selected (Table S3) and further subjected to Tukey’s multiple comparisons of means test.22 This analysis indicated that the set of metabolites, including creatinine (p-value = 1.5 × 10−02), 3-hydroxybutyrate (p-value = 8.0 × 10−3), 3-hydroxyisovalerate (p-value = 8.4 × 10−05), and methylmalonate (p-value = 1.1 × 10−4), discriminate between MS and NMO-SD patients (Figure S2). An identical set of metabolites was also obtained using the Student’s t test followed by the Benjamini–Hochberg multiple hypothesis test (Figure S–3).23 It is important to note that, in addition to being identified by the univariate analysis, 3-hydroxybutyrate was also identified in both S-plots but was absent in the SUS plot. 3-Hydroxybutyrate was likely missing in the SUS plot due to a serendipitous cancellation because it was located in opposite regions in the two S-plots (Figures 2 and 4).
Discussion
A wide range of chemicals from food, medication, environmental contaminants, normal biological processes, and disease conditions are routinely excreted into the urine to maintain chemical homeostasis. Consequently, urinalysis has been used to diagnose diseases and evaluate a patient’s well-being for years. More recently, there has been a growing interest in identifying urinary metabolite biomarkers to monitor the prognosis, existence, or progression of various cancers, cardiovascular diseases, and neurological diseases.25 Despite the high-rate of misdiagnosis, little attention has been paid toward the analysis of urinary metabolites for diagnosing MS.26 To address this oversight, we previously demonstrated that urinary metabolites can differentiate between EAE mice (prototypic disease model for MS) from healthy and fingolimod (MS drug)-treated EAE mice.17 We extended this initial animal study by using NMR, multivariate statistics, one-way ANOVA, and univariate statistics to analyze changes in urine samples collected from MS and NMO-SD patients. Although the sample size was limited, we were still able to observe a potentially unique metabolic signature in urine samples collected from MS patients. Importantly, the MS metabolic signature was likely to be distinct from the urinary metabolites identified for NMO-SD patients. Our results are also consistent with a recent study by Moussallieh et al.15 that showed an increase in serum acetate levels in NMO-SD patients relative to MS patients. However, the limited sample size requires us to describe our statistical model and the set of potential urinary metabolite biomarkers as only a working hypothesis. A major revision may occur as the number of patients per group increases significantly. Nevertheless, we are encouraged by the fact that we were able to link all of the metabolites and metabolic pathways identified from the urine of MS patients to known pathologic processes associated with MS.
Glycolysis and the synthesis and degradation of ketone bodies were identified as two potentially high-impact pathways from our NMR metabolomics analysis of urine samples from MS patients. Specifically, we observed significant concentration changes in the glycolysis intermediates lactate and acetate and the ketone bodies 3-hydroxybutyrate and acetoacetate. This observation is consistent with basic brain chemistry that is expected to be altered by MS. The brain has a high energy need and uses 25% of the total available glucose. Interestingly, the brain also uses ketone bodies as an alternative energy source.27 Thus, alteration in glycolysis and the synthesis and degradation of ketone bodies is consistent with an alteration in energy generation. Besides energy generation, ketone bodies are also associated with fatty acid metabolism. This is pertinent because prior analysis of CSF and serum samples from MS patients indicated altered energy and fatty acid metabolism.28
The observed alterations in energy generation have led to the speculation that MS might be associated with mitochondrial defects.29 In fact, Witte et al. demonstrated an occurrence of severe mitochondrial defects in MS lesions.30 Because the mitochondria are the primary source of energy in axons, the observed defects would be expected to alter energy generation. In the mitochondria, ATP is produced via the TCA cycle, the electron transport chain (respiratory chain), and oxidative phosphorylation. From our NMR metabolomics analysis, we identified the TCA cycle as another possible high-impact pathway that was altered in the urine samples from MS patients. Intermediates of the TCA cycle, such as citrate, oxaloacetate, and succinate were altered in the urine from MS patients. Of particular note, pyruvate and amino acid metabolism (alanine, aspartate, and glutamate) pathways, which are directly coupled to the TCA cycle, were also identified as potential high-impact pathways. Defects in the amino acid metabolism pathway are known to cause a range of neurological issues.31
We also observed an increase in creatine and a decrease in creatinine in the urine from MS patients. The creatine/phosphocreatine/creatine kinase system is critical for maintaining energy levels in the brain and as a high-energy phosphate shuttle from the mitochondria to the cytoplasm.32
Our NMR metabolomics analysis also identified propionate metabolism as another likely high-impact pathway altered in the urine of MS patients. Propionate (short-chain fatty acid) is primarily derived from the catabolism of lipids (fatty acid metabolism) or proteins, where its accumulation is toxic and inhibits TCA cycle enzymes and cell growth.33 Propionate and propionyl-CoA are detoxified by the mitochondria through the methyl-malonyl-CoA pathway.34 Genetic flaws in the propionate metabolism pathway cause faulty amino acid and fatty acid metabolism that lead to various neurological problems.34 Thus, the observed alteration in propionate metabolism appears to be consistent with a recent view that MS may also be associated with dysfunction in lipid metabolism.35
Gut microbiota are known to establish a symbiotic relationship that provides essential health benefits to the host.36 A similar correlation between gut microbiota and MS has been previously observed.36 In fact, Cantarel et al. described a difference in specific operational taxonomic units of gut microbiota in MS patients. Their finding also indicated that immunomodulatory medications cause alterations in the gut microbiota of MS patients.37 Herein, we observed hippurate, a mammalian microbial cometabolite that was altered in the urine of MS patients. Propionate (described above) is also a metabolite of the gut microbiota. Thus, differences in urinary hippurate and propionate levels may be a result of a change in the gut microbiota or alterations in the relevant metabolic pathway.38 Taken together, these findings suggest a potential role of gut microbiota in the pathogenesis and treatment of MS.
Conclusions
Although CNS and serum metabolites have been previously considered as a source of MS and NMO-SD biomarkers, we have demonstrated that the urine metabolome shows significant promise for investigating and diagnosing MS and NMO-SD. We observed a set of eight potential urinary metabolites (acetate, creatinine, hippurate, 3-hydroxybutyrate, 3-hydroxyisovalerae, malonate, oxaloacetate, and trimethylamine N-oxide) associated with MS that may be prospective biomarkers. Similarly, we observed a set of eight potential urinary metabolites (acetate, creatinine, 3-hydroxybutyrate, 3-hydroxyisovalerae, methylmalonate, oxaloacetate, and succinate) associated with NMO-SD. Critically, we observed a distinct set of urinary metabolites (creatinine, 3-hydroxybutyrate, 3-hydroxyisovalerate, methylmalonate) that possibly differentiates MS from NMO-SD patients. This is despite the fact that NMO-SD is also an inflammatory immune-mediated disease of the CNS that was once considered a variant of MS. The observation that all of the metabolites and metabolic pathways identified from the urine of MS patients are linked to known pathologic processes associated with MS supports the potential reliability of our study, although the sample size was small. Specifically, metabolite changes are associated with alterations in energy and fatty acid metabolism, mitochondrial activity, and the gut microbiota. Although our results highlight the promise of urinary biomarkers as a tool to diagnose MS and NMO-SD, the limited sample size necessitates interpreting our results as only a proof-of-principle that requires further validation. It is possible that an increase in the number of patients per group may lead to a change in the observed set of urinary metabolites that differentiates between MS patients, NMO-SD patients, and healthy controls. Nevertheless, as evident by a number of other recent MS and NMO-SD studies where practical considerations limit sample size,39–43 a conservative interpretation does not negate the inherent scientific value of our observations. Instead, given the ease and ready access of urine samples from MS and NMO-SD patients, our results establish the urine metabolome as a potentially valuable resource for investigating the pathology of MS and NMO-SD for obtaining a rapid and reliable diagnosis and for monitoring a patient’s response to treatment.
Supplementary Material
Table S1, individual demographic data of Hungarian cohort;
Table S2, list of metabolites assigned from the analysis of NMR spectral data;
Table S3, p-values calculated from one-way ANOVA and Tukey’s test of significance;
Table S4, mapping of metabolite nomenclature to MetPA database;
Table S5, result from pathway analysis using MetPA;
Figure S1, OPLS-DA model generated from binned spectra of MS and healthy controls data set;
Figure S2, box-plot for each metabolite with a univariate statistical significance;
Figure S3, Benjamini–Hochberg adjusted p-values;
Figure S4, OPLS-DA model generated from binned spectra of NMO-SD and healthy controls data set;
Figure S5, back-scaled loadings plot generated from the OPLS-DA models comparing MS patients and healthy controls and NMO-SD patients and healthy controls (PDF)
Acknowledgments
This manuscript was supported in part by funds from the National Institutes of Health (NIH), USA (P30 GM103335, R.P., J.R.; HL114669, J.R.), NIH National Center for Research Resources (P20 RR-17675, J.R.), University of Nebraska Research Council (J.R., R.P.), Scleroseforeningen (A-19412, Denmark, Z.I.), Lundbeckfonden (R118-A11472, Denmark, Z.I.), and a grant from Odense University Hospital (Z.I.). The research was performed in facilities renovated with support from the National Institutes of Health (RR015468-01).
Footnotes
The authors declare no competing financial interest.
References
- 1.Sospedra M, Martin R. Immunology of multiple sclerosis. Annu Rev Immunol. 2005;23:683–747. doi: 10.1146/annurev.immunol.23.021704.115707. [DOI] [PubMed] [Google Scholar]
- 2.Trebst C, Jarius S, Berthele A, Paul F, Schippling S, Wildemann B, Borisow N, Kleiter I, Aktas O, Kumpfel T. Update on the diagnosis and treatment of neuromyelitis optica: recommendations of the Neuromyelitis Optica Study Group (NEMOS) J Neurol. 2014;261(1):1–16. doi: 10.1007/s00415-013-7169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wingerchuk DM, Banwell B, Bennett JL, Cabre P, Carroll W, Chitnis T, de Seze J, Fujihara K, Greenberg B, Jacob A, Jarius S, Lana-Peixoto M, Levy M, Simon JH, Tenembaum S, Traboulsee AL, Waters P, Wellik KE, Weinshenker BG. International consensus diagnostic criteria for neuromyelitis optica spectrum disorders. Neurology. 2015;85(2):177–89. doi: 10.1212/WNL.0000000000001729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingerchuk DM, Lennon VA, Pittock SJ, Lucchinetti CF, Weinshenker BG. Revised diagnostic criteria for neuromyelitis optica. Neurology. 2006;66(10):1485–9. doi: 10.1212/01.wnl.0000216139.44259.74. [DOI] [PubMed] [Google Scholar]
- 5.Carroll WM, Saida T, Kim HJ, Kira J, Kermode AG, Tsai CP, Fujihara K, Kusunoki S, Tanaka M, Kim KK, Bates D. A guide to facilitate the early treatment of patients with idiopathic demyelinating disease (multiple sclerosis and neuromyelitis optica) Mult Scler. 2013;19(10):1371–80. doi: 10.1177/1352458512471092. [DOI] [PubMed] [Google Scholar]
- 6.Schnackenberg LK, Beger RD. Metabolomic biomarkers: their role in the critical path. Drug Discovery Today: Technol. 2007;4(1):13–16. doi: 10.1016/j.ddtec.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Fiehn O. Metabolomics–the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1–2):155–71. [PubMed] [Google Scholar]
- 8.Kruger NJ, Troncoso-Ponce MA, Ratcliffe RG. 1H NMR metabolite fingerprinting and metabolomic analysis of perchloric acid extracts from plant tissues. Nat Protoc. 2008;3(6):1001–12. doi: 10.1038/nprot.2008.64. [DOI] [PubMed] [Google Scholar]
- 9.Gebregiworgis T, Powers R. Application of NMR metabolomics to search for human disease biomarkers. Comb Chem High Throughput Screening. 2012;15(8):595–610. doi: 10.2174/138620712802650522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray P, Manach YL, Riou B, Houle TT. Statistical evaluation of a biomarker. Anesthesiology. 2010;112(4):1023–40. doi: 10.1097/ALN.0b013e3181d47604. [DOI] [PubMed] [Google Scholar]
- 11.Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5(10):1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Powers R. The current state of drug discovery and a potential role for NMR metabolomics. J Med Chem. 2014;57(14):5860–70. doi: 10.1021/jm401803b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worley B, Powers R. Multivariate analysis in metabolomics. Curr Metabolomics. 2013;1(1):92–107. doi: 10.2174/2213235X11301010092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinke SN, Broadhurst DL, Sykes BD, Baker GB, Catz I, Warren KG, Power C. Metabolomic profiling in multiple sclerosis: insights into biomarkers and pathogenesis. Mult Scler. 2014;20(10):1396–400. doi: 10.1177/1352458513516528. [DOI] [PubMed] [Google Scholar]
- 15.Moussallieh FM, Elbayed K, Chanson JB, Rudolf G, Piotto M, De Seze J, Namer IJ. Serum analysis by 1H nuclear magnetic resonance spectroscopy: a new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult Scler. 2014;20(5):558–65. doi: 10.1177/1352458513504638. [DOI] [PubMed] [Google Scholar]
- 16.Dobson R. Urine: An under-studied source of biomarkers in multiple sclerosis? Mult Scler Relat Disord. 2012;1(2):76–80. doi: 10.1016/j.msard.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Gebregiworgis T, Massilamany C, Gangaplara A, Thulasingam S, Kolli V, Werth MT, Dodds ED, Steffen D, Reddy J, Powers R. Potential of urinary metabolites for diagnosing multiple sclerosis. ACS Chem Biol. 2013;8(4):684–90. doi: 10.1021/cb300673e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandit L, Asgari N, Apiwattanakul M, Palace J, Leite MI, Paul F, Kleiter I, Chitnis T. Demographic and clinical features of neuromyelitis optica: A review. Mult Scler. 2015;21(7):845–53. doi: 10.1177/1352458515572406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polman CH, Reingold SC, Banwell B, Clanet M, Cohen JA, Filippi M, Fujihara K, Havrdova E, Hutchinson M, Kappos L, Lublin FD, Montalban X, O’Connor P, Sandberg-Wollheim M, Thompson AJ, Waubant E, Weinshenker B, Wolinsky JS. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Worley B, Powers R. MVAPACK: a complete data handling package for NMR metabolomics. ACS Chem Biol. 2014;9(5):1138–44. doi: 10.1021/cb4008937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.RCoreTeam. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 22.Miller RGJ. Simultaneous Statistical Inference. Springer-Verlag; New York: 1981. p. 299. [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 24.Saccenti E, Hoefsloot HCJ, Smilde AK, Westerhuis JA, Hendriks MMWB. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics. 2014;10(3):361–374. [Google Scholar]
- 25.Hecht SS. Human urinary carcinogen metabolites: biomarkers for investigating tobacco and cancer. Carcinogenesis. 2002;23(6):907–922. doi: 10.1093/carcin/23.6.907. [DOI] [PubMed] [Google Scholar]
- 26.Dobson R. Urine: an under-studied source of biomarkers in multiple sclerosis? Mult Scler Relat Disord. 2012;1(2):76–80. doi: 10.1016/j.msard.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Mathur D, Lopez-Rodas G, Casanova B, Marti MB. Perturbed glucose metabolism: insights into multiple sclerosis pathogenesis. Front Neurol. 2014;5:250. doi: 10.3389/fneur.2014.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tavazzi B, Batocchi AP, Amorini AM, Nociti V, D’Urso S, Longo S, Gullotta S, Picardi M, Lazzarino G. Serum metabolic profile in multiple sclerosis patients. Mult Scler Int. 2011;2011:167156. doi: 10.1155/2011/167156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao P, Reddy PH. Is multiple sclerosis a mitochondrial disease? Biochim Biophys Acta, Mol Basis Dis. 2010;1802(1):66–79. doi: 10.1016/j.bbadis.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witte ME, Bo L, Rodenburg RJ, Belien JA, Musters R, Hazes T, Wintjes LT, Smeitink JA, Geurts JJ, De Vries HE, van der Valk P, van Horssen J. Enhanced number and activity of mitochondria in multiple sclerosis lesions. J Pathol. 2009;219(2):193–204. doi: 10.1002/path.2582. [DOI] [PubMed] [Google Scholar]
- 31.Yoon HJ, Kim JH, Jeon TY, Yoo S-Y, Eo H. Devastating metabolic brain disorders of newborns and young infants. Radiographics. 2014;34(5):1257–72. doi: 10.1148/rg.345130095. [DOI] [PubMed] [Google Scholar]
- 32.Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000;80(3):1107–213. doi: 10.1152/physrev.2000.80.3.1107. [DOI] [PubMed] [Google Scholar]
- 33.Matsuishi T, Stumpf DA, Seliem M, Eguren LA, Chrislip K. Propionate mitochondrial toxicity in liver and skeletal muscle: acyl CoA levels. Biochem Med Metab Biol. 1991;45(2):244–53. doi: 10.1016/0885-4505(91)90027-i. [DOI] [PubMed] [Google Scholar]
- 34.Fenton WA, Gravel RA, Rosenblatt DS. Disorders of Propionate and Methylmalonate Metabolism. In: Valle D, editor. OMMBID-The Online Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 2165–2193. [Google Scholar]
- 35.Corthals AP. Multiple sclerosis is not a disease of the immune system. Q Rev Biol. 2011;86(4):287–321. doi: 10.1086/662453. [DOI] [PubMed] [Google Scholar]
- 36.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10(11):735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 37.Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J, Venkatesan A, Fraser CM, Mowry EM. Gut Microbiota in Multiple Sclerosis: Possible Influence of Immunomodulators. J Invest Med. 2015;63(5):729–734. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams HR, Cox IJ, Walker DG, Cobbold JF, Taylor-Robinson SD, Marshall SE, Orchard TR. Differences in gut microbial metabolism are responsible for reduced hippurate synthesis in Crohn’s disease. BMC Gastroenterol. 2010;10:108. doi: 10.1186/1471-230X-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh V, Stingl C, Stoop MP, Zeneyedpour L, Neuteboom RF, Smitt PS, Hintzen RQ, Luider TM. Proteomics Urine Analysis of Pregnant Women Suffering from Multiple Sclerosis. J Proteome Res. 2015;14(5):2065–2073. doi: 10.1021/pr501162w. [DOI] [PubMed] [Google Scholar]
- 40.Vafaeyan H, Alavijeh SK, Madadi A, Rad HS, Faeghi F, Ebrahimzadeh SA, Rahimian N, Harirchian MH. Quantification of diagnostic biomarkers to detect multiple sclerosis lesions employing (1)H-MRSI at 3T. Australas Phys Eng Sci Med. 2015;38:611. doi: 10.1007/s13246-015-0390-1. [DOI] [PubMed] [Google Scholar]
- 41.Kroksveen AC, Jaffe JD, Aasebo E, Barsnes H, Bjorlykke Y, Franciotta D, Keshishian H, Myhr K-M, Opsahl JA, van Pesch V, Teunissen CE, Torkildsen O, Ulvik RJ, Vethe H, Carr SA, Berven FS. Quantitative proteomics suggests decrease in the secretogranin-1 cerebrospinal fluid levels during the disease course of multiple sclerosis. Proteomics. 2015;15(19):3361–3369. doi: 10.1002/pmic.201400142. [DOI] [PubMed] [Google Scholar]
- 42.Jurynczyk M, Leite I, Palace J, Selmaj K, Weinshenker B, Akman-Demir G, Asgari N, Barnes D, Wren D, Boggild M, Chaudhuri A, D’Hooghe M, Evangelou N, Tanasescu R, Geraldes R, Illes Z, Jacob A, Kim HJ, Kleiter I, Levy M, Marignier R, Vukusic S, McGuigan C, Murray K, Nakashima I, Pandit L, Paul F, Pittock S, de Seze J, Siva A, Wingerchuk D. Status of diagnostic approaches to AQP4-IgG seronegative NMO and NMO/MS overlap syndromes. J Neurol. 2016;263:140. doi: 10.1007/s00415-015-7952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tremlett H, Dai DLY, Hollander Z, Kapanen A, Aziz T, Wilson-McManus JE, Tebbutt SJ, Borchers CH, Oger J, Cohen Freue GV. Serum proteomics in multiple sclerosis disease progression. J Proteomics. 2015;118:2–11. doi: 10.1016/j.jprot.2015.02.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, individual demographic data of Hungarian cohort;
Table S2, list of metabolites assigned from the analysis of NMR spectral data;
Table S3, p-values calculated from one-way ANOVA and Tukey’s test of significance;
Table S4, mapping of metabolite nomenclature to MetPA database;
Table S5, result from pathway analysis using MetPA;
Figure S1, OPLS-DA model generated from binned spectra of MS and healthy controls data set;
Figure S2, box-plot for each metabolite with a univariate statistical significance;
Figure S3, Benjamini–Hochberg adjusted p-values;
Figure S4, OPLS-DA model generated from binned spectra of NMO-SD and healthy controls data set;
Figure S5, back-scaled loadings plot generated from the OPLS-DA models comparing MS patients and healthy controls and NMO-SD patients and healthy controls (PDF)