Summary
The prostate gland consists of basal and luminal cells arranged as pseudo-stratified epithelium. In tissue recombination models, only basal cells reconstitute a complete prostate gland, yet murine lineage-tracing experiments show that luminal cells generate basal cells. It has remained challenging to address the molecular details of these transitions and whether they apply to humans, due to the lack of culture conditions that recapitulate prostate gland architecture. Here we describe a 3D culture system that supports long-term expansion of primary mouse and human prostate organoids, composed of fully differentiated CK5+ basal and CK8+ luminal cells. Organoids are genetically stable, reconstitute prostate glands in recombination assays and can be experimentally manipulated. Single human luminal and basal cells give rise to organoids, yet luminal cell-derived organoids more closely resemble prostate glands. These data support a luminal multilineage progenitor cell model for prostate tissue and establish a robust, scalable system for mechanistic studies.
Introduction
The prostate is a male sex gland responsible for approximately 30% of all seminal fluid. Although prostate glands differ between species macroscopically prostatic acini are organized similarly at the cellular level. Prostatic ducts are lined by a pseudo-stratified epithelium. Three major cell types are identified within the epithelium: 1) secretory luminal cells marked by cytokeratin (CK) 8, CK18, Androgen receptor (AR) and secretory proteins like prostate specific antigen (PSA), 2) basal cells, identified by the expression of CK5, CK14 and p63, and 3) rare neuroendocrine cells (Shen and Abate-Shen, 2010). In the developing and adult prostate rare, intermediate cells expressing both luminal and basal markers are present (Hudson et al., 2001; Xue et al., 1998).
The identity of prostatic stem cells and how they give rise to these three cell types remains unclear. The classic urogenital sinus mesenchyme (UGSM) recombination model, where prostate epithelial cells are combined with mesenchymal cells derived from the UGS of murine embryos, are transplanted under the kidney capsule (Cunha, 1973; Xin et al., 2003) suggests that only basal cells are capable of generating glandular tissue(Goldstein et al., 2008). Other approaches to identify prostate stem cells involve in vitro culture methods of primary prostate epithelium(Garraway et al., 2010; Liu et al., 2012; Niranjan et al., 2013). In these, basal cells appear bipotent, i.e. capable of generating both luminal and basal lineages, indicating that basal cells have stem-like potential. However, none of these in vitro systems generate tissues that resemble the in vivo composition of the prostate gland or contain AR at physiological levels.
Recently, novel insights have been generated into the cellular hierarchy of the prostatic epithelium in mice through lineage tracing. Studies marking Ck5-expressing (Ck5+) basal cells and Ck8+ luminal cells suggest that basal and luminal lineages both harbor stem cell activity in the adult prostate (Choi et al., 2012; Ousset et al., 2012). However, in a separate study, rare multipotent basal cells reside in the adult prostate (Wang et al., 2013). While lineage tracing from Ck8+ and Ck18+ cells suggests unipotency in the luminal lineage (Choi et al., 2012; Ousset et al., 2012), a subset of luminal cells defined by Nkx3.1 expression post-castration can generate both lineages during regeneration of the prostate (Wang et al., 2009). Taken together, these studies suggest that in mice both luminal and basal cells sporadically are bipotent.
Although these studies provide important insights into prostate biology, translating these results to a human setting is difficult. One challenge is the expression pattern of the proposed stem cell markers c-kit, CD177 and CD133, which are exclusively expressed by basal cells in humans, but in mice are expressed by basal cells and a subset of luminal cells (Leong et al., 2008; Missol-Kolka et al., 2011). Translation to a human setting is also hampered by the lack of suitable human experimental systems.
We have previously described 3D culture conditions that allow long-term expansion of primary mouse and human epithelial organoids from small intestine (Sato et al., 2009), colon (Sato et al., 2011), stomach (Barker et al., 2010) and liver (Huch et al., 2013). These cultures can be initiated from single Lgr5+ stem cells and are based on the addition of the Lgr4/5 ligand R-spondin1, a potent Wnt pathway agonist (Binnerts et al., 2007; Carmon et al., 2011; de Lau et al., 2011). Organoids remain genetically and phenotypically stable in culture, exemplified by pathology-free transplantation of multiple mice with the organoid offspring of single Lgr5+ cells from colon (Yui et al., 2012) or liver (Huch et al., 2013).
Here we describe the development of an R-spondin1-based culture method that allows long-term propagation of murine and human prostate epithelium. Using this method, we show that both basal and luminal populations contain bipotent progenitor cells which retain full differentiation towards basal and luminal lineages in vitro and the UGSM transplantation model. Moreover, we show that organoid cultures can be used to study prostate cancer initiation.
Results
Establishment of primary murine prostate organoid cultures with basal and luminal epithelial layers
To establish murine prostate organoid cultures, we embedded dissociated cells of wildtype murine prostate epithelium in MatrigelR and added ‘generic’ organoid medium containing the growth factors EGF, Noggin, and R-spondin1 (ENR) (Sato et al., 2009). We also included the Alk3/4/5 inhibitor A83-01 to inhibit TGF-β pathway signaling to prevent a proliferative block in prostate cells (Ding et al., 2011; Qin et al., 2013). Because the murine prostate is composed of different lobes with some distinct properties(Marker et al., 2003), we separately cultured the anterior prostate (AP), dorsolateral prostate (DLP) and ventral prostate (VP) epithelium (Figure S1A). Prostate cells from each lobe yielded expanding organoids. These have been cultured at a split ratio of 1:3 for >1.5 years with stable, normal karyotypes (Figure S1B) and no changes in morphology.
Prostate epithelium formed cystic structures composed of a basal (outer) layer exclusively expressing typical basal prostate markers, such as p63 and Ck5 and a luminal (inner) layer exclusively expressing Ck8 (Figure 1A-B, Figure S1D), thus demonstrating that organoids retain an architecture that resembles prostate glands in vivo, with the exception that we did not detect neuroendocrine cells.
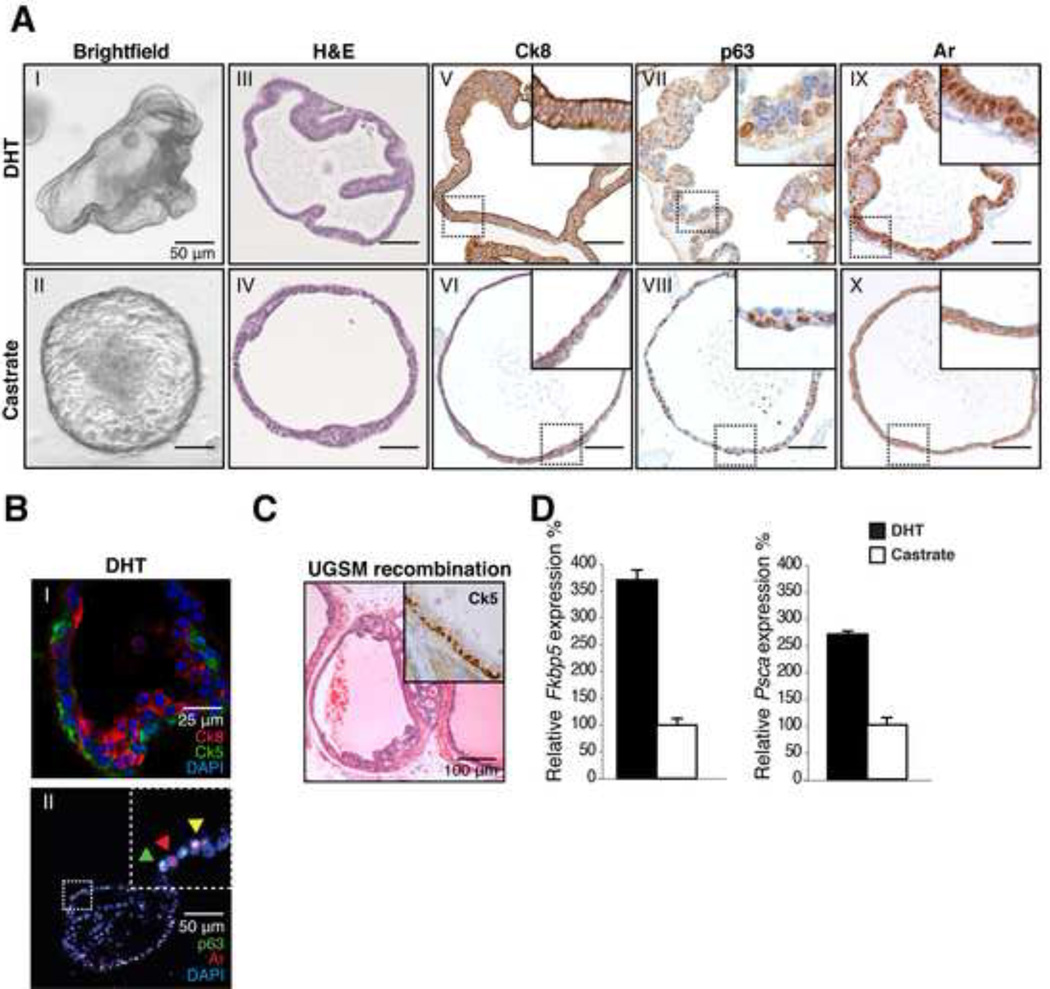
Figure 1. Establishment of murine prostate cultures.
A: IHC analysis of murine organoids in the presence of DHT (1 nM) and in castrate conditions. Brightfield image (I-II) H&E staining (III-IV) Ck8 (V-VI) p63 (VII-VIII) Ar (IX-X). Strong increase in cell size is observed upon DHT addition as well as nuclear localization of the Ar. Scale bars represent 50 microns.
B: IF staining (I) of basal Ck5 (Green) and luminal Ck8 (Red) in organoids. Distinct luminal and basal cell populations are present in organoids. Bottom (II) staining for p63 (green) and Ar (Red) showing single positive (Green & Red arrowhead) and double positive Ar/p63 cells (Yellow arrowhead).
C: H&E stain of UGSM recombination 8 weeks after transplantation of 50,000 organoid cells derived from wildtype mice. Inset; Ck5 staining, confirming presence of basal cells.
D: Quantitative RT-PCR of Ar targets Psca and Fkbp5 in the presence of DHT (1 nM) and in castrate conditions (24 hours). Psca and Fkbp5 transcripts are increased upon DHT addition. Expression was normalized to Hprt. Results are shown as mean ± SD.
See also Figure S1
Organoids also retained robust expression of the Ar (Figure 1A) and the prostate-specific transcription factor Nkx3.1 (Figure S1C)(Bhatia-Gaur et al., 1999). Ar signaling was intact, as demonstrated by nuclear translocation of Ar in organoids treated with dihydrotestosterone (DHT). Within 72 hours after DHT addition, Ck8+ luminal cells became strongly polarized with an apparent increase in size (Figure 1A). Moreover we found occasional double p63+ and Ar+ cells (Figure 1B), consistent with the differentiation-promoting function of Ar in normal prostate cells (Litvinov et al., 2006). Two classic Ar target genes Fkbp5 and Psca were robustly upregulated (Figure 1D). DHT also enhanced the rate at which organoids expanded, allowing weekly split ratios of 1:5 rather than 1:3. Organoids also retained DHT responsiveness after multiple cycles of DHT addition and withdrawal (Figure S1E), consistent with the response of the normal prostate gland to castration and DHT add-back in vivo.
As has been shown for primary murine prostate tissue, prostate organoids gave rise to reconstituted prostate glands when placed under the kidney capsule with urogenital mesenchymal cells (Figure 1C, Figure S1J), using a tissue recombination assay (Xin et al., 2003). Importantly, this reconstitution property was retained after 8 passages with organoids derived from the AP, DLP or VP, indicating long-term retention of prostate progenitor cells. In the grafts, we did not observe morphologies reminiscent of the respective lobes of the prostate, suggesting that this is imparted by local, non-epithelial cues in the prostate as has been shown by others (Hayashi et al., 1993; Timms et al., 1995).
We examined the relative requirement of each ingredient by quantifying the number of organoids formed after seeding 1000 single cells and tracking successive passages. Noggin increased the number of organoids formed during initial plating (Figure S1G) and resulted in faster expansion rates, but was not absolutely required for serial passaging (Figure S1H). This is in line with observations in the development of the prostate in Noggin knock-out mice, where reduced epithelial proliferation and reduced prostate budding is observed due to hyperactive BMP signaling (Cook et al., 2007). Similarly, the Wnt agonist R-spondin enhanced organoid formation and expansion, but was not absolutely required for expansion. R-spondins and Wnts are widely expressed in the urogenital sinus during prostate development (Mehta et al., 2011) and Wnt signaling is essential for prostate development (Francis et al., 2012). Moreover, the R-spondin receptor Lgr4 is essential for both prostate development as well as differentiation (Luo et al., 2013). We found that organoids express Lgr4 and Lgr5 (Figure S1I). DHT was not essential but significantly enhanced the efficiency of organoid formation (Figure S1G,H). EGF was essential for both establishment and passaging. TGF-β pathway inhibition was required for passaging of AP- and VP-derived organoids, but not for DLP-derived organoids (Figure S1 F-H).
Both luminal and basal cells can generate murine prostate organoids
The availability of a robust in vitro murine prostate organoid system with basal and luminal layers allowed us to determine whether isolated basal or luminal cells are capable of establishing organoids. We separated basal and luminal cells from primary mouse prostate tissue using FACS. Basal cells were isolated based on high CD49f expression (also called α6-integrin)(Goldstein et al., 2008), luminal cells were captured based on expression of CD24 (also called Heat Stable Antigen (HSA) (Figure 2A). CD24 was previously shown to specifically mark luminal cells in the human prostate (Liu et al., 2004) and CD24 expression has been used in conjunction with CD49f to isolate murine luminal cells (Lawson et al., 2007). We confirmed luminal-specific high CD24 expression in murine prostate tissue by IHC (Figure S2A). The purity of our sorted basal and luminal population was verified using qPCR for basal- and luminal-specific markers, Ck5 and Probasin (Pbsn) (Figure 2B) and co-staining for luminal cell-specific Ck8 and basal cell-specific Ck5 (Figure S2B).
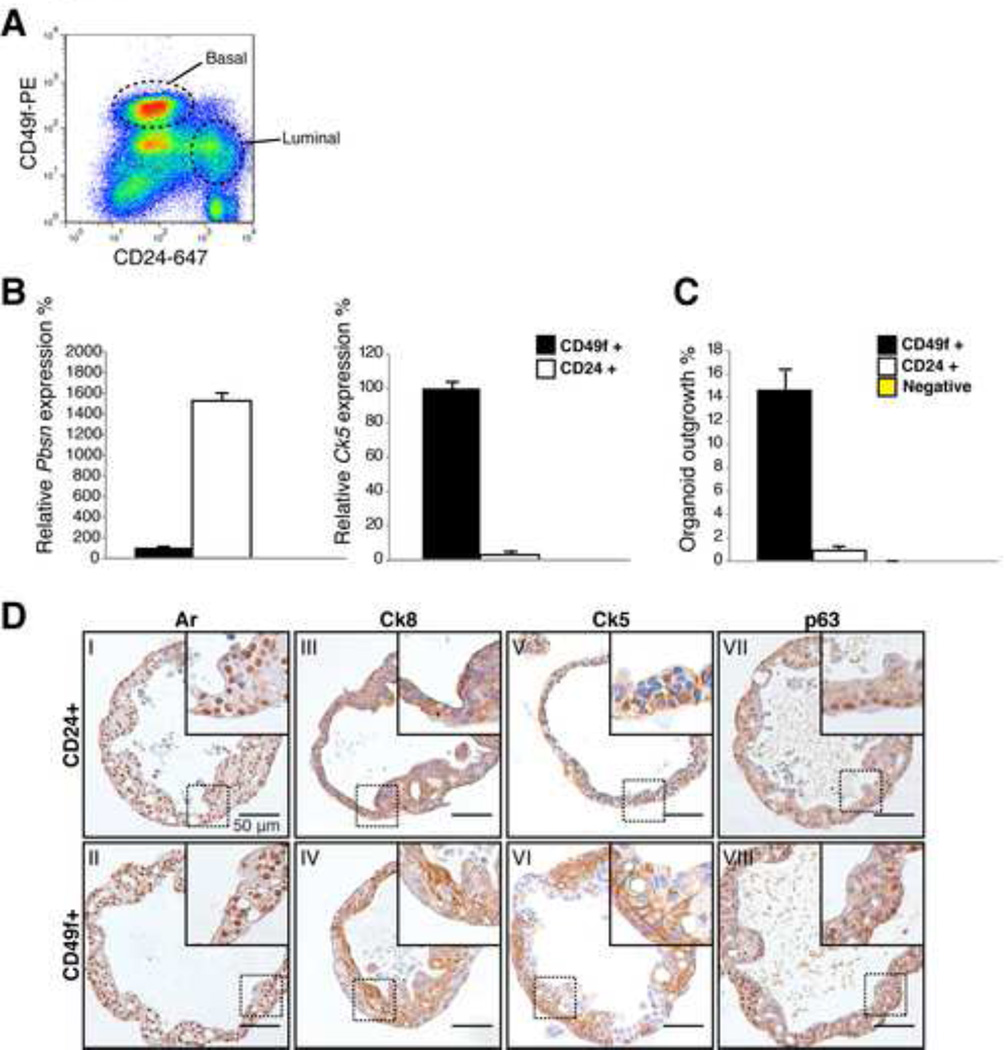
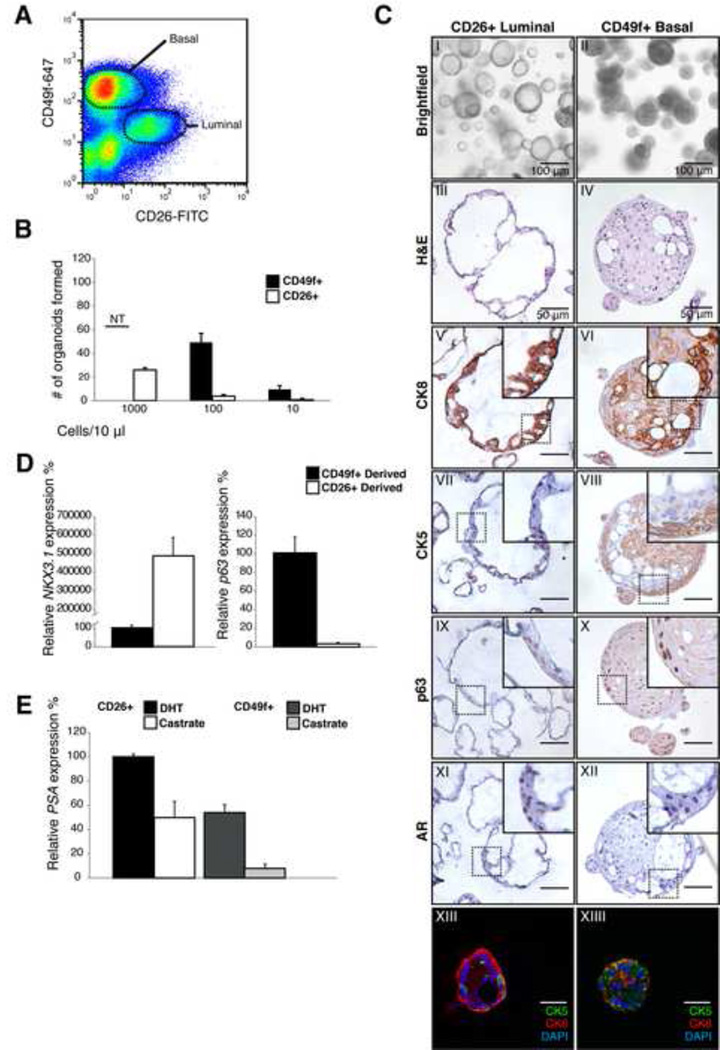
Figure 2. Basal and luminal cells give rise to organoids.
A: FACS plot of CD24 (luminal) CD49f (basal) stained murine prostate.
B: Quantitative RT-PCR expression analysis of basal (Ck5) and luminal (Probasin, Pbsn) marker expression in CD24+ luminal and CD49f+ basal cells. Pbsn is strongly expressed in luminal cells, Ck5 is strongly expressed in basal cells. Expression was normalized to Hprt. Results are shown as mean ± SD.
C: Percentage of organoids established by 200 single cells 14 days post seeding. Approximately 15% of basal cells generate an organoid. 1% of luminal cells generate an organoid. Results are shown as mean ± SD.
D: IHC stainings of single cell derived murine prostate organoids, showing similar staining patterns of Ar (I-II), Ck8 (III-IV), Ck5 (V-VI) and p63 (VII-VIII) and morphology, showing that basal cells and luminal cells are both capable of giving rise to both epithelial lineages.
Scale bars represent 50 microns.
See also Figure S2
We found a contamination of 0.04% of Ck5+ basal cells (~2 cells in 5000 cells, determined in 2 independent samples) in the CD24+ fraction.
To measure their organoid-forming capability, we seeded 200 single basal- or luminal-derived cells in culture, i.e. one cell per Matrigel drop. Approximately 15% of basal cells formed an organoid, compared to 1% of luminal cells (Figure 2C). Yet, the growth of luminal cells could not be explained by the presence of contaminating basal cells. Considering 15% of basal cells grow out, a basal cell contamination of >6% is required to explain an outgrowth efficiency of 1% of luminal cells, whereas we only found ~0.04% of contaminating basal cells. Finally, CD24/CD49f double-negative cells, which represent non-epithelial cells, did not grow (Figure 2C).
Despite the difference in cloning efficiency, we did not observe morphological differences in organoid development from single cells. Both luminal and basal cells initially formed a solid ball and subsequently developed into a sphere within 10 to 14 days (Figure S2C). IHC analysis showed that the organoids derived from either basal or luminal cells expressed both basal (p63, Ck5) and luminal (Ck8) markers and express Ar (Figure 2D). Furthermore, both luminal- and basal-derived murine organoids could be passaged long-term. These data are consistent with recent lineage tracing studies (Choi et al., 2012; Wang et al., 2009; Wang et al., 2013) and support the existence of prostate stem cells in both the basal and luminal compartments of the normal mouse prostate.
Murine prostate organoids as a tool for cancer biology
To explore the suitability of prostate organoid cultures for genetic studies, we asked if we could generate cancer phenotypes using known prostate cancer oncogenes or tumor suppressor genes that have been previously characterized in genetically engineered mouse models (GEMMs). We established organoids from mice with a floxed Pten allele (PtenloxP/loxP) (Di Cristofano et al., 1998), a TMPRSS2:ERG gene fusion (Rosa26 LSL-ERG) (Chen et al., 2013)or bigenic Pten/ERG mice and compared their phenotypes to organoids derived from wildtype mice. In these experiments, Cre-driven excision of loxP-flanked regions was mediated by the prostate-specific composite promoter ARR2PB (PBCre) (Figure S3A). Cultures carrying the prostate-specific Pten deletion initially formed normal-appearing organoids containing a lumen, but within a week the organoids had multiple layers of epithelium, eventually forming a nearly solid 3D structure (Figure 3A). Histologically, PBCre PtenloxP/loxP organoid resembled phenotypes observed in vivo (Carver et al., 2011), with features reminiscent of high-grade PIN. Organoids derived from PBCre Rosa26 LSL-ERG mice showed no signs of neoplasia (Figure 3A, Figure S3B), consistent with the in vivo phenotype (Carver et al., 2009; Chen et al., 2013). The combination of Pten deletion and ERG overexpression yielded a hyperplastic phenotype similar to that seen with Pten deletion alone (Figure 3A, Figure S3B), but with “fingers” protruding into the Matrigel (Figure 3A), suggesting invasive behavior. All organoids responded to DHT withdrawal with reduced Fkbp5 expression (Figure S3C). Furthermore, the histologic features attributable to Pten and ERG in vitro were confirmed in UGSM tissue recombination experiments of transplanted passage-8 organoids (Figure S3B).
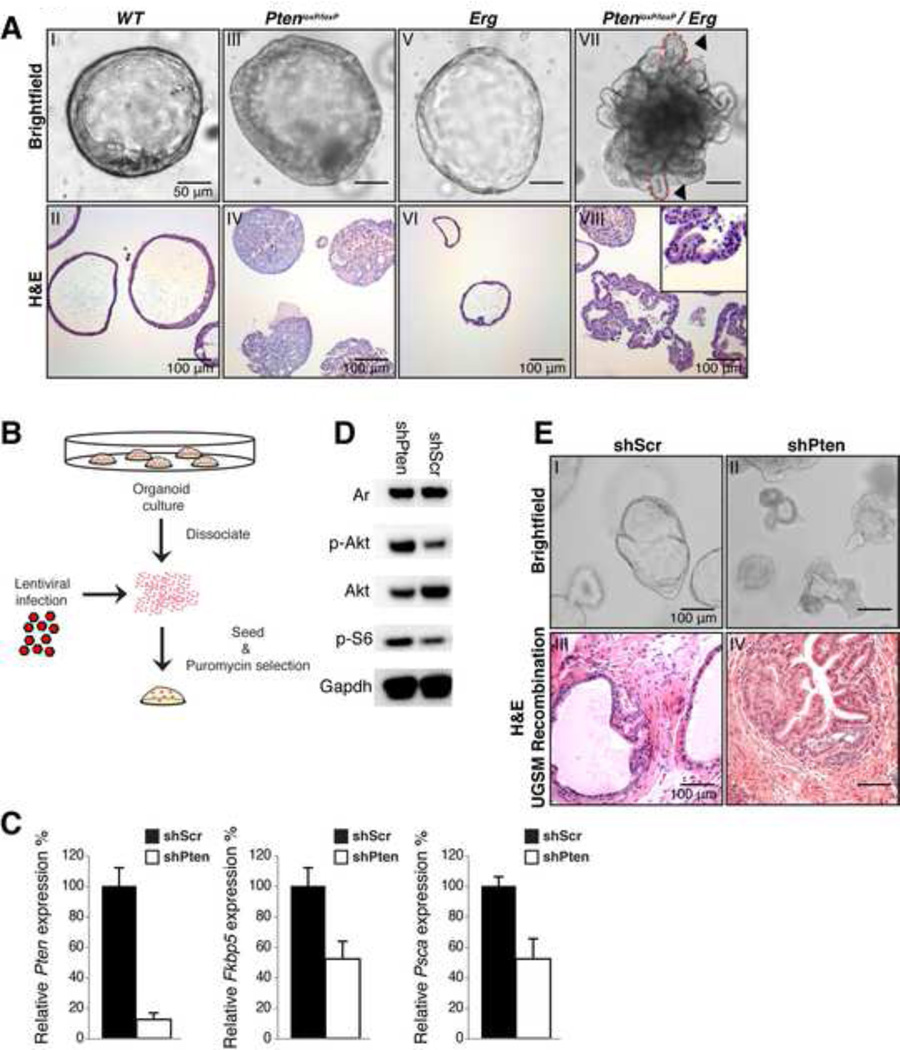
Figure 3. Genetically engineered mouse prostate cancer models are recapitulated in organoids.
A: Brightfield and H&E images of wildtype (WT) (I-II) PBCre PtenloxP/loxP (III-IV) PBCre Rosa LSL-ERG (V-VI) and PBCre PtenloxP/loxP Rosa LSL-ERG (VII-VIII). In Pten loxP/loxP organoids the lumen is filled with hyperplastic cells. PtenloxP/loxP /ERG organoids make fingerlike protrusions are made into the Matrigel (Arrowheads, dashed lines, inset VIII).
B: Schematic overview of retroviral infection.
C: Quantitative RT-PCR analysis of Pten, Fkbp5 and Psca expression in shPten and shScr-control infected PBCre Rosa26 LSL-ERG organoids. Upon Pten knockdown Ar target gene expression is diminished. Expression was normalized to Hprt. Results are shown as mean ± SD.
D: Western blot analysis of PI3K pathway activation in shPten and ShScr-control infected PBCre Rosa26 LSL-ERG organoids. p-Akt and p-S6 levels are increased upon Pten knockdown.
E: Brightfield image of shPten-RFP shScr–RFP infected PBCre Rosa26 LSL-ERG organoids (I-II) and corresponding H&E staining of shPten/shScr organoid-UGSM recombinations (III & IV) showing hyperplastic phenotype in organoids and in UGSM recombinations upon Pten knock down. Scale bars in A, B and E represent 50 microns.
See also Figure S3
To determine the suitability of prostate organoids for ex vivo genetic manipulation, we silenced Pten expression in PBCre Rosa26 LSL-ERG organoids by infection with lentivirus encoding a shRNA targeting Pten. Puromycin selection was applied to ensure that only infected organoids remained (Figure 3B). 7 days post infection, we observed an 80–90% decrease in Pten mRNA levels (Figure 3C; Figure S3D), accompanied by increased phosphorylation of AKT and ribosomal protein S6 (Figure 3D, Figure S3E), consistent with increased PI3K activation. Remarkably, the shPten-infected organoids displayed hyperplastic phenotypes (Figure 3E). Similarly, shPten-infected Rosa26 LSL-ERG organoids generated hyperplastic prostate glandular tissue in UGSM tissue recombination experiments, similar to that seen in the ERG; PtenloxP/loxP grafts, control shRNA-infected organoids showed no signs of hyperplasia (Figure 3E). Similar to earlier work in GEMM models showing that hyperactive PI3K signaling in Pten null prostate tissue results in reduced AR transcriptional activity due to negative feedback on Her kinase signaling(Carver et al., 2011), we observed reduced expression of the Ar target genes Psca and Fkbp5 in organoids with Pten knockdown (Figure 3C, Figure S3D). Collectively, these results show that in vitro prostate organoid phenotypes mimic in vivo phenotypes and that retroviral gene expression or gene knockdown in organoids can be used to identify and study genes involved in prostate cancer.
Establishment of human prostate organoids with basal and luminal cells
Based on our previous experience (Sato et al., 2011), we anticipated that human prostate organoid cultures might require additional growth factors. Fibroblast growth factor-10 (FGF10), FGF2, and prostaglandin E2 (PGE2) have been shown to support proliferation of prostate cell lines and/or human colon epithelium. Therefore, we added these to the 'mouse' medium (ENR+A83-01+DHT). We also included Nicotinamide and the p38 inhibitor SB202190, which have been previously identified as being essential for human small intestinal cultures. This combination of growth factors enabled us to establish and propagate human prostate epithelial organoids from 40 independent normal prostate specimens from patients undergoing radical prostatectomy. Human prostate organoids have been passaged for >12 months with no obvious phenotypic changes over time and with stable, normal karyotypes (Figure S4A-B). To determine the genetic stability of our organoid cultures at the nucleotide level, we performed whole exome sequencing on passage 2 and passage 7 organoids. To detect genetic changes that developed during in vitro expansion, we filtered single nucleotide variants (SNVs) and small insertions and deletions (indels) observed in the long-term culture for presence in the short-term culture (see Experimental Procedures). Subsequently, candidate changes that emerged using this approach were independently examined (Table S1). In addition, we also checked for cases of loss of heterozygosity in the passage 7 cultures compared to passage 2 cultures. We did not observe any genetic variants in the passage 7 culture that were not present in the passage 2 culture, implying that the organoid cultures remain genetically stable over time.
Initially, we observed solid spherical structures with multiple layers of epithelium, which progressed over 2–3 weeks to organoids with easily visible lumens. Areas of single-layered as well as double-layered epithelium were observed. The majority of cells were positive for the luminal epithelial markers CK8 and AR (Figure 4A-C). Typically, cells expressed either the basal marker CK5 or the luminal marker CK8. Cells positive for basal markers p63 and CK5 were also present and were always located in the outer layer of organoids (Figure 4A-C). Similar to mouse-derived prostate organoids, human organoids expressed prostate-specific genes such as PSA (Figure 4D-E) and NKX3.1 (Figure 4B,E). Additionally, human organoids retained intact AR signaling as measured by dose-dependent induction of PSA mRNA by DHT (Figure 4D) and reduced expression of NKX3.1 and PSA upon DHT withdrawal (Figure 4E). In addition, human organoids formed prostate glands in UGSM tissue recombination assays when transplanted after 8 passages (Figure 4F), as also seen with murine prostate organoids.
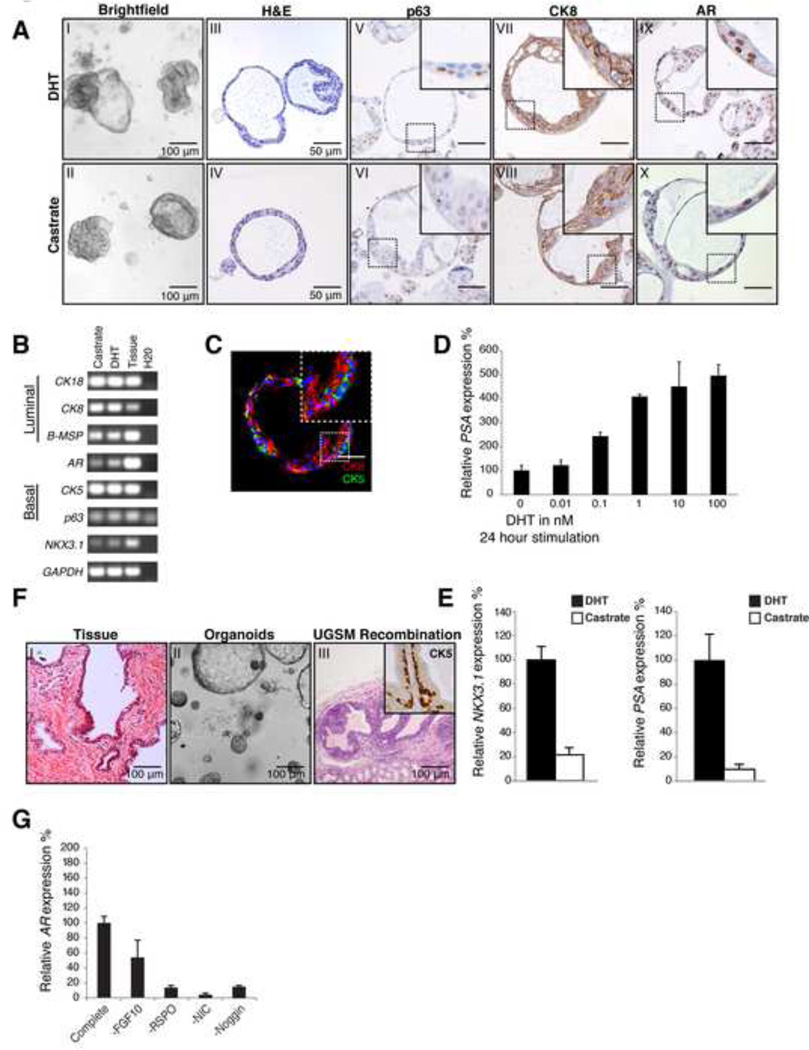
Figure 4. Establishment of human prostate organoid cultures.
A: IHC analysis of passage 6 (12 week) human organoids in the presence of DHT (1 nM) and in castrate conditions. Brightfield image (I-II) H&E staining (III-IV) p63 (V-VI) CK8 (VII-VIII) AR (IX-X). Upon DHT addition nuclear localization of the AR is observed. Scale bars represent 50 microns.
B: RT-PCR analysis of prostate organoids show that both luminal and basal markers are expressed in the absence and presence of DHT (1 nM).
C: IF staining of basal CK5 (Green) and luminal CK8 (Red) in organoids. Distinct luminal and basal cell populations are present in organoids.
D: Quantitative RT-PCR Analysis of PSA 24 hours after DHT stimulation (0 nM – 100 nM). Expression was normalized to GAPDH. Results are shown as mean ± SD (n=3)
E: Quantitative RT-PCR analysis of AR targets NKX3.1 and PSA in the presence of DHT (1 nM) and without DHT (castrate) (24 hours). Upon DHT addition a strong increase of PSA and NKX3.1 transcript is observed. Expression was normalized to GAPDH. Results are shown as mean ± SD (n=3).
F: H&E stain of UGSM recombination 8 weeks after transplantation of 50,000 human organoid cells derived. Inset; CK5 staining, confirming presence of basal cells.
Scale bars represent 50 microns.
G: Quantitative RT-PCR of AR mRNA levels in the absence of growth factors. Withdrawal of Noggin, R-spondin, FGF10 and Nicotinamide led to reduced AR expression. Expression was normalized to GAPDH. Results are shown as mean ± SD.
See also Figure S4
Having established conditions for human prostate organoid cultures, we examined the relative requirement for each growth factor in maintaining the cultures. EGF, FGF2, Nicotinamide, PGE2 and the TGF-β pathway inhibitor A83-01 were all essential to sustain prolonged organoid growth (Figure S4C, S4D). Removal of the p38 kinase inhibitor SB202190 did not lead to a decrease in organoid formation capacity but did result in histologic evidence of keratinization, perhaps due to increased stress signaling. Although passaging was not affected by the removal of FGF10, Noggin or R-spondin, the efficiency of organoid formation upon initial plating was reduced. Moreover, removal of any one of these factors, as well as of Nicotinamide, resulted in decreased AR expression levels. In particular, removal of R-spondin or Noggin led to a virtual disappearance of AR expression (Figure 4G, Figure S4E). Withdrawal of EGF resulted in a modest upregulation of AR mRNA and protein level together with a potent increase in expression of the AR target gene PSA (Figure S4F-H). These data are consistent with a negative feedback loop involving EGF and AR signaling and/or an inhibitory effect of high levels of EGF signaling on the generation or survival of luminal cells.
Human prostatic epithelium harbors both luminal and basal stem cells
Existing human prostate culture methods favor outgrowth of basal epithelial cells that, in UGSM tissue recombination assays, can generate prostate tissue with luminal cells, providing evidence for human prostate stem cells in the basal epithelium(Garraway et al., 2010; Goldstein et al., 2008).. However, experimental evidence for the existence of a human luminal stem cell is lacking. Our success in propagating human organoids with basal and luminal cells provided a unique opportunity to address this question.
As with our mouse studies, we separated basal and luminal cells from human prostate tissue by FACS. Basal cells were isolated on the basis of high CD49f expression, and luminal cells by the cell surface marker CD26 (also called DPPIV) (Liu et al., 2004)(Figure 5A). Staining of these two populations for CK5 and CK8 post-isolation verified their basal and luminal cellular identities (Figure S5A). Additionally, we stained for the luminal marker NKX3.1 and found that essentially all CD26-sorted cells express NKX3.1 (Figure S5B). Although CD26 and CD49f are also expressed on immune cells, we only found very low numbers of contaminating hematopoietic cells as measured by the pan-leukocyte marker CD45 (<0.1% in 10.000 cells, 3 independent samples). We detected very little contamination of CK5+ basal cells (<0.2%, 18±1.7 in 10.000 cells, 3 independent samples) in the FACS-purified luminal population.
Figure 5. Establishment of human prostate organoid cultures from luminal and basal cells.
A: FACS plot of human prostate cells stained with CD26 (luminal) and CD49f (basal) markers
B: Organoid outgrowth from CD49f+ basal and CD26+ luminal cells at varying densities. Results are shown as mean ± SD. *NT, Non Testable. At higher densities CD49f-derived organoids fuse, making counting of organoid number therefore unreliable.
C: Analysis of CD26-derived (luminal) and CD49f-derived (basal) organoids at passage 4. Brightfield picture (I-II) H&E staining (III-IV) CK8 (V-VI) CK5 (VII-VIII) p63 (IX-X) AR (XI-XII) IF stain of CK5 (green) and CK8 (red) showing distinct basal and luminal cells (XIII-XIV). Scale bars represent 50 microns.
D: Quantitative RT-PCR expression analysis of the luminal marker NKX3.1 and basal marker p63 in luminal and basal organoids at passage 4. Expression was normalized to GAPDH. Results are shown as mean ± SD.
E: Quantitative RT-PCR of the AR target PSA in the presence of DHT (1 nM) and in castrate conditions (24 hours). Increased PSA mRNA levels are observed in both luminal- and basal-derived organoids after DHT treatment. Expression was normalized to GAPDH. Results are shown as mean ± SD.
See also Figure S5
To determine their relative organoid-forming ability, we seeded single basal or luminal cells at varying densities. Human basal cells were highly efficient at establishing organoids, be it at single cell density (70%) or at high density (1000 cells/10 µl) (Figure 5B). Luminal cells also established organoids, albeit at much lower efficiency (1–2%), both at single cell and at high density seeding. Of note, this could not be explained by contaminating basal cells, as these were present at a >10-fold lower frequency (<0.2%), suggesting the presence of a rare luminal organoid-forming cell.
Within 7 days of plating, single basal cells formed solid spheres that, within 2–3 weeks, contained lumens resembling organoids grown from bulk tissue, when viewed by brightfield microscopy. However, unlike bulk-derived organoids, IHC analysis revealed that the majority of cells in basal-derived organoids expressed CK5, with CK8 cells surrounding a sporadic array of lumens within a single organoid (Figure 5C). AR expression was patchy, but tended to be near lumens (Figure 5C, Figure S5C). In contrast to basal cell-derived organoids, those generated from luminal cells immediately formed lumens and thus could be easily distinguished from basal-derived organoids by brightfield. IHC analysis of luminal-derived organoids revealed a majority of cells expressing CK8 and AR, consistent with a true luminal phenotype. Strikingly, CK5-positive, CK8-negative cells were also observed (Figure 5C), indicating that human luminal cells can generate basal cells. Consistent with their differing histology, the luminal markers NKX3.1 and PSA were strongly expressed in luminal organoids whereas the basal marker p63 was more highly expressed in basal organoids (Figure 5D, 5E). Both organoid types retained AR pathway signaling, as DHT withdrawal led to a significant decrease in PSA expression levels (Figure 5E) and a reduced nuclear localization of AR (Figure S5H).
Both basal- and luminal-derived organoids could be passaged for at least 4 months and maintained a stable karyotype (Figure S5D). A more detailed analysis of relative growth factor requirements revealed few differences compared to bulk human organoid cultures (Figure S5E,F), most notably that luminal organoids were more dependent on R-spondin for serial passaging. Expression analysis using RT-PCR of Lgr receptors revealed that LGR4 is expressed in both types of human organoids (Figure S5G). The relative dependence of luminal cells on R-spondin/Lgr signaling is also suggested by the reported failure of luminal differentiation by Lgr4−/− prostate progenitor cells (Luo et al., 2013). DHT drastically enhanced luminal organoid formation but, as observed with mouse, was not absolutely required for organoid maintenance (Figure S5E, F). The p38 inhibitor was not required to prevent keratinization. As seen with organoids derived from bulk tissue, we observed a loss of nuclear AR staining as well as a decrease in AR protein levels upon DHT withdrawal (Figure S5H). We also observed a strong increase in PSA expression upon EGF withdrawal in both luminal- and basal cell-derived organoids (Figure S5I).
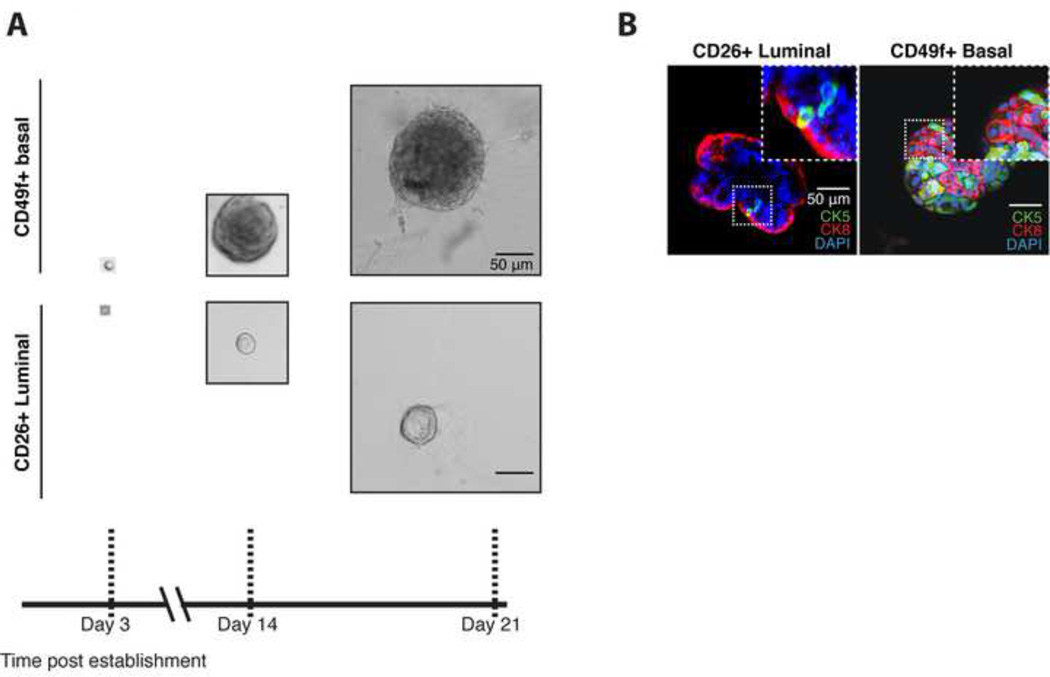
To formally demonstrate that single luminal cells can give rise to organoids containing both lineages, we repeated these experiments after plating single luminal cells. As observed in bulk, single basal cell-derived organoids proliferated much faster over time than single luminal cell-derived organoids. As observed in bulk, luminal cells quickly formed lumens, whereas basal cells first formed solid spheres (Figure 6A). IF staining of single cell-derived organoids showed that both basal and luminal cells generated organoids containing both lineages as demonstrated by the presence of CK8+ luminal and CK5+ basal cells (Figure 6B), providing conclusive evidence for a human multi-progenitor prostate cell in the luminal compartment.
Figure 6. Single human basal and luminal cells give rise to organoids.
A: Brightfield images of organoids grown from single CD26+ luminal and CD49f+ basal cells.
B: IF stain of CK5 (Green) and CK8 (Red) showing distinct basal and luminal cells in single luminal cell- and single basal cell-derived organoids (Passage 4). Scale bars represent 50 microns.
Discussion
Leveraging our previous experience establishing organoid cultures, we have devised a culture system that allows long-term expansion of murine and human prostatic epithelium. The resulting organoids retain both basal and luminal epithelial layers and preserve androgen-responsiveness in culture. Organoids differ significantly from earlier descriptions of prostate spheres, which consist primarily of CK5+ basal cells with luminal-like features in response to DHT treatment, but lack NKX3.1+ cells or secretions (Xin et al., 2007). In contrast, our culture conditions induce full luminal differentiation. These conditions were developed using growth factors originally optimized for intestinal epithelia, with some modifications. We conducted studies to address the requirement for each of these growth factors when removed from the complete cocktail, but have not exhaustively examined all possible perturbations.
Organoids proliferate at a much higher rate compared to normal prostate. We assume that organoid culture conditions mimic a regenerative response. In contrast to gastrointestinal organoid cultures, R-spondin is not essential for maintenance of prostate organoid cultures. However, organoid establishment and growth is enhanced by R-spondin. Importantly, expression of AR receptor is strongly dependent on R-spondin. Taken together, our data and previous studies suggest that R-spondin/Lgr signaling is important for luminal cell differentiation and luminal cell maintenance.
Several other factors, like FGF10 and Noggin, favor differentiation towards a luminal phenotype, by positively regulating AR expression. It is plausible that, in vivo, the stroma supplies most if not all of these factors. Work from others has demonstrated that the Rho/ROCK kinase inhibitor Y-27632 allows long-term culture of primary epithelial tissue cultures, including prostate, in conjunction with stroma-derived feeder cells(Liu et al., 2012). Possibly, the growth factor composition of our culture method mimics the factors secreted by the feeder cells in this method. In contrast to our method, AR responsiveness is not well defined in this system. Preliminary studies of Y-27632 in our system suggest a benefit in initiating single cell organoid cultures, but not in propagation of established organoids.
The availability of a robust culture model that generates prostate organoids containing both basal and luminal cells allowed us to address the important question of whether basal or luminal cells (or both) are required to generate a complete prostate organoid. Prior efforts to explore the "prostate stem cell" question have primarily relied on UGSM tissue recombination assays. These experiments have shown that tissue-regenerating activity primarily resides in basal cell populations(Goldstein et al., 2008). However, murine lineage tracing studies demonstrate that both basal and luminal cells are self-renewing, including evidence that luminal cells can give rise to basal cells (Choi et al., 2012; Wang et al., 2013). Our culture method confirms that basal and luminal cells can each generate a complete multi-layer prostate organoid and shows repopulating potential of human prostate luminal cells which can generate both basal and luminal lineages. Although these luminal progenitor cells occur at lower frequency than their basal counterparts, they give rise to more normal appearing organoids and therefore could have greater physiologic relevance. Their frequency (~1% of all luminal cells) raises the question of whether these CD26+ and NKX3.1+ luminal cells may be the human equivalent of the murine castration-resistant Nkx3.1+ (CARN) (Wang et al., 2009).
This culture method, compared to other experimental procedures, uniquely allows for elucidation of the cellular identity of this human luminal progenitor. In the absence of evidence of neuroendocrine differentiation, we refer to these luminal and basal cells as multipotent progenitors rather than stem cells. Modifications to the culture conditions may reveal that these cells may al so have neuroendocrine cell repopulating potential.
In addition to their utility in defining prostate epithelial lineage relationships, the organoid model has the potential to greatly facilitate functional genetic studies, particularly in cancer genomics. Several large scale prostate cancer genome resequencing studies have now defined a plethora of novel genomic alterations in primary and metastatic cancer whose functions are currently unknown (Barbieri et al., 2012; Grasso et al., 2012; Taylor et al., 2010). Here we have shown that murine prostate organoids faithfully recapitulate in vivo phenotypes known from prostate cancer GEMMs. Moreover, the organoids are easily manipulated with inhibitors, retroviruses and CRISPR/Cas9 (Schwank et al., 2013), offering a tool to study the impact of these manipulations on growth, invasive behavior and drug-sensitivity. Organoid culture may also represent a straightforward, cheap and robust alternative to xenografting.
There is considerable interest in whether these same culture conditions can be used to generate organoids from human prostate cancers. If successful, this technology could help address a huge need in the prostate cancer biology field for a broad array of human prostate cancer cell lines, which has been forced to rely on the very few such cell lines that have been established to date. In a companion paper, Gao et al. show that this culture method does allow efficient and sustained growth of human prostate cancer organoids isolated from men with advanced disease that faithfully retain the genomic characteristics of the originating human tissue sample (Gao et al. this issue). We have conducted similar studies using primary human prostate cancer samples but failed to establish renewable primary cancer organoid lines, due to overgrowth by normal prostate epithelial cells present within each tumor sample. The notion that cells harboring oncogenic mutations grow slower than normal cells may be counterintuitive, yet we also observe this consistently for colorectal and pancreas cancers. The observed increased rates of apoptosis in cancer organoid cultures may reflect the high levels of genomic instability. The absence of normal prostate cells in metastatic samples may explain the high rate of success of our method in that context. Efforts to overcome this complication by purification of tumor cells from normal epithelium are ongoing. Taken together, prostate organoid technology offers a platform for mechanistic studies of prostate development, homeostasis and cancer.
Experimental Procedures
Isolation & culture of prostate epithelial cells
Murine prostates were divided into three lobe pairs; AP, DLP and VP. Lobes were enzymatically digested with collagenase type II (Gibco) and subsequently with TrypLE (Gibco). Cells were seeded in growth factor reduced Matrigel (Corning) and overlayed with medium containing the growth factors: EGF 5–50 ng/ml (Peprotech), R-spondin1 conditioned medium or 500 ng/ml recombinant R-spondin1 (Peprotech), Noggin conditioned medium or 100 ng/ml recombinant Noggin (Peprotech) and the 200 nM TGF-β/Alk inhibitor A83-01 (Tocris). Dihydrotestosterone (DHT) (Sigma) was added at 0.1–1 nM. Human prostate samples were obtained from patients undergoing radical prostatectomy according to guidelines from the UMC Utrecht. Prostate tissue was enzymatically digested with collagenase type II and subsequently with TrypLE. Cells were seeded in growth factor reduced Matrigel and cultured in medium containing growth factors as above, with the addition of 10 ng/ml FGF10 (Peprotech), 5 ng/ml FGF2 (Peprotech), 1 µM Prostaglandin E2 (Tocris), 10 µM SB202190 (Sigma-Aldrich), 10 mM Nicotinamide (Sigma-Aldrich) and DHT 0.1–1 nM. For detailed summary of culture medium composition see supplementary methods.
FACS
Single cell suspensions of human cells were stained using CD49f-alexa 647 (1:200, GoH3, BD Biosciences) and CD26-FITC (1:200, M-A261, eBioscience). Murine cells were stained using a CD49f-PE (1:200, GoH3, BD Biosciences) and CD24-alexa 647 (1:200, 30-F1, eBioscience).
UGSM essay
UGSM recombination essay was performed as described previously (Xin et al., 2003). All mouse work was performed in accordance with national guidelines and regulations of the Hubrecht institute or Memorial Sloan Kettering Cancer Center.
Genetic analysis
DNA was isolated from early (P2) and late (P7) passage organoids. Whole exome sequencing and genetic analysis were done as described previously(DePristo et al., 2011; Li and Durbin, 2009). Detailed discription is given in supplementary methods. The data for the whole exome sequencing were deposited to the EMBL European Nucleotide Archive, accession number: ERP006541.
Immunohistochemistry and Immunofluorescence
Organoids were processed and stained as described previously(Sato et al., 2009). Following antibodies were used for staining on murine and human prostate organoids AR (1:1000, N-20, Santa Cruz) CK5 (1:2000, AF-138, Covance), CK8 (1:50, C-51, Santa Cruz), p63 (1:800, 4A4, Millipore). Stainings were visualized with bright vision (Dako). For IF organoids were stained for AR (1:200) Ck5 (1:500), Ck8 (1:50), p63 (1:500) using the antibodies described above and NKX3.1 (Rabbit, 1:100, Kind gift of Michael Shen). Secondary antibodies were conjugated with alexa-fluor (-488, -568, -647). DNA was stained with DAPI.
Lentiviral infections
Lentiviral infections were performed as described previously (Koo et al., 2012) using pLKO.1-puro-UbC-TagFP635 (Sigma) targeting Pten (CGACTTAGACTTGACCTATAT) or control Scramble (CCTAAGGTTAAGTCGCCCTCG).
Western blot
Membranes were probed with antibodies directed against AR (1:000, N-20, Santa Cruz), GAPDH (1:2000 Abcam) AKT (1:1000 Cell Signaling #9272), phosphorylated-AKT (Ser473) (1:1000 Cell Signaling #4060) and Phosphorylated-S6 (Ser235/Ser236) (1:1000, Cell Signaling #2211). Signal was visualized with secondary HRP conjugated antibodies and ECL.
Supplementary Material
Highlights.
Prostate organoid culture method that recapitulates in vivo glandular morphology
Organoids recapitulate GEMM models and are amenable to experimental manipulation
Mouse and human prostate organoids are androgen-sensitive
Characterization of human prostate luminal progenitor cells
Acknowledgments
WK and HC conceived and oversaw project. WK and PI performed isolation of mouse organoids. PI and JW performed xenograft experiments. WK JD AG performed human organoid experiments. CD DG provided assistance with cell-isolation and xenografts. WK and HB performed IHC/IF. NS and RV gave essential input. YC provided GEMM models. WK, JD, CS and HC wrote manuscript. RB and EC performed sequencing experiments. RB and EC were supported by the Netherlands Genomics Initiative Zenith grant (935.12.003) JD was supported by NWO-ZonMw VENI (91614138). NS was supported by EU/232814-StemCellMark. CS and YC were supported by the PCF. HC and WK are inventors of several patents related to organoid culture.
Footnotes
Author Contributions
The other authors declare no conflict of interest
REFERENCES
- Barbieri C, Baca S, Lawrence M, Demichelis F, Blattner M, Theurillat J-P, White T, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Huch M, Kujala P, van de Wetering M, Snippert H, van Es J, Sato T, Stange D, Begthel H, van den Born M, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell stem cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Bhatia-Gaur R, Donjacour A, Sciavolino P, Kim M, Desai N, Young P, Norton C, Gridley T, Cardiff R, Cunha G, et al. Roles for Nkx3.1 in prostate development and cancer. Genes & development. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnerts M, Kim K-A, Bright J, Patel S, Tran K, Zhou M, Leung J, Liu Y, Lomas W, Dixon M, et al. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14700–14705. doi: 10.1073/pnas.0702305104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmon K, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver B, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino P, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nature genetics. 2009;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrop J, van Wichen D, Koomans-Bitter M, van de Wetering M, de Weger R, van Dongen J, Clevers H. The human TCF-1 gene encodes a nuclear DNA-binding protein uniquely expressed in normal and neoplastic T-lineage lymphocytes. Blood. 1995;86:3050–3059. [PubMed] [Google Scholar]
- Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nature medicine. 2013;19:1023–1029. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C, Vezina C, Allgeier S, Shaw A, Yu M, Peterson R, Bushman W. Noggin is required for normal lobe patterning and ductal budding in the mouse prostate. Developmental biology. 2007;312:217–230. doi: 10.1016/j.ydbio.2007.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha G. The role of androgens in the epithelio-mesenchymal interactions involved in prostatic morphogenesis in embryonic mice. The Anatomical record. 1973;175:87–96. doi: 10.1002/ar.1091750108. [DOI] [PubMed] [Google Scholar]
- de Lau W, Barker N, Low T, Koo B-K, Li V, Teunissen H, Kujala P, Haegebarth A, Peters P, van de Wetering M, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nature genetics. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Ding Z, Wu C-J, Chu G, Xiao Y, Ho D, Zhang J, Perry S, Labrot E, Wu X, Lis R, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis JC, Thomsen MK, Taketo MM, Swain A. β-catenin is required for prostate development and cooperates with Pten loss to drive invasive carcinoma. PLoS genetics. 2012;9:e1003180. doi: 10.1371/journal.pgen.1003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway I, Sun W, Tran C, Perner S, Zhang B, Goldstein A, Hahm S, Haider M, Head C, Reiter R, et al. Human prostate sphere-forming cells represent a subset of basal epithelial cells capable of glandular regeneration in vivo. The Prostate. 2010;70:491–501. doi: 10.1002/pros.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso C, Wu Y-M, Robinson D, Cao X, Dhanasekaran S, Khan A, Quist M, Jing X, Lonigro R, Brenner J, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi N, Cunha GR, Parker M. Permissive and instructive induction of adult rodent prostatic epithelium by heterotypic urogenital sinus mesenchyme. Epithelial cell biology. 1993;2:66–78. [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj S, van Es J, Li V, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D, Guy A, Fry P, O'Hare M, Watt F, Masters J. Epithelial cell differentiation pathways in the human prostate: identification of intermediate phenotypes by keratin expression. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2001;49:271–278. doi: 10.1177/002215540104900214. [DOI] [PubMed] [Google Scholar]
- Jung P, Sato T, Merlos-Suárez A, Barriga F, Iglesias M, Rossell D, Auer H, Gallardo M, Blasco M, Sancho E, et al. Isolation and in vitro expansion of human colonic stem cells. Nature medicine. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- Koo B-K, Stange D, Sato T, Karthaus W, Farin H, Huch M, van Es J, Clevers H. Controlled gene expression in primary Lgr5 organoid cultures. Nature methods. 2012;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- Lawson D, Xin L, Lukacs R, Cheng D, Witte O. Isolation and functional characterization of murine prostate stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong K, Wang B-E, Johnson L, Gao W-Q. Generation of a prostate from a single adult stem cell. Nature. 2008;456:804–808. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov I, Vander Griend D, Xu Y, Antony L, Dalrymple S, Isaacs J. Low-calcium serum-free defined medium selects for growth of normal prostatic epithelial stem cells. Cancer research. 2006;66:8598–8607. doi: 10.1158/0008-5472.CAN-06-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Roudier M, True L. Heterogeneity in primary and metastatic prostate cancer as defined by cell surface CD profile. The American journal of pathology. 2004;165:1543–1556. doi: 10.1016/S0002-9440(10)63412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva O, Nealon C, Dakic A, Simic V, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. The American journal of pathology. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Rodriguez M, Valdez J, Zhu X, Tan K, Li D, Siwko S, Xin L, Liu M. Lgr4 is a key regulator of prostate development and prostate stem cell differentiation. Stem cells (Dayton, Ohio) 2013;11:492–505. doi: 10.1002/stem.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marker PC, Donjacour AA, Dahiya R, Cunha GR. Hormonal, cellular, and molecular control of prostatic development. Developmental Biology. 2003;253 doi: 10.1016/s0012-1606(02)00031-3. [DOI] [PubMed] [Google Scholar]
- Mehta V, Abler L, Keil K, Schmitz C, Joshi P, Vezina C. Atlas of Wnt and R-spondin gene expression in the developing male mouse lower urogenital tract. Developmental dynamics : an official publication of the American Association of Anatomists. 2011;240:2548–2560. doi: 10.1002/dvdy.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memarzadeh S, Xin L, Mulholland D, Mansukhani A, Wu H, Teitell M, Witte O. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer cell. 2007;12:572–585. doi: 10.1016/j.ccr.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missol-Kolka E, Karbanová J, Janich P, Haase M, Fargeas C, Huttner W, Corbeil D. Prominin-1 (CD133) is not restricted to stem cells located in the basal compartment of murine and human prostate. The Prostate. 2011;71:254–267. doi: 10.1002/pros.21239. [DOI] [PubMed] [Google Scholar]
- Niranjan B, Lawrence M, Papargiris M, Richards M, Hussain S, Frydenberg M, Pedersen J, Taylor R, Risbridger G. Primary culture and propagation of human prostate epithelial cells. Methods in molecular biology (Clifton, NJ) 2013;945:365–382. doi: 10.1007/978-1-62703-125-7_22. [DOI] [PubMed] [Google Scholar]
- Ousset M, Van Keymeulen A, Bouvencourt G, Sharma N, Achouri Y, Simons B, Blanpain C. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nature cell biology. 2012;14:1131–1138. doi: 10.1038/ncb2600. [DOI] [PubMed] [Google Scholar]
- Qin J, Wu S-P, Creighton C, Dai F, Xie X, Cheng C-M, Frolov A, Ayala G, Lin X, Feng X-H, et al. COUP-TFII inhibits TGF-β-induced growth barrier to promote prostate tumorigenesis. Nature. 2013;493:236–240. doi: 10.1038/nature11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange D, Ferrante M, Vries R, Van Es J, Van den Brink S, Van Houdt W, Pronk A, Van Gorp J, Siersema P, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries R, Snippert H, van de Wetering M, Barker N, Stange D, van Es J, Abo A, Kujala P, Peters P, et al. Single Lgr5 stem cells build cryptvillus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schwank G, Koo B-K, Sasselli V, Dekkers J, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent C, et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell stem cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Shen M, Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes & development. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver B, Arora V, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms BG, Lee CW, Aumüller G, Seitz J. Instructive induction of prostate growth and differentiation by a defined urogenital sinus mesenchyme. Microscopy research and technique. 1995;30:319–332. doi: 10.1002/jemt.1070300407. [DOI] [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides K, Walker D, Yu H, Halili M, Hu Y-P, Price S, Abate-Shen C, Shen M. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Mitrofanova A, Bergren S, Abate-Shen C, Cardiff R, Califano A, Shen M. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nature cell biology. 2013;15:274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Ide H, Kim Y, Dubey P, Witte O. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proceedings of the National Academy of Sciences of the United States of America 100 Suppl. 2003;1:11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Lukacs R, Lawson D, Cheng D, Witte O. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem cells (Dayton, Ohio) 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- Xue Y, Smedts F, Debruyne F, de la Rosette J, Schalken J. Identification of intermediate cell types by keratin expression in the developing human prostate. The Prostate. 1998;34:292–301. doi: 10.1002/(sici)1097-0045(19980301)34:4<292::aid-pros7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nature medicine. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.