Abstract
Estrogenic endocrine disrupting chemicals are found in environmental and biological samples, commercial and consumer products, food, and numerous other sources. Given their ubiquitous nature and potential for adverse effects, there is a critical need for rapidly detecting these chemicals. We developed an estrogen-responsive recombinant human ovarian (BG1Luc4E2) cell line recently accepted by the USEPA and OECD as a bioanalytical method to detect estrogen receptor (ER) agonists/antagonists. Unfortunately, these cells appear to contain only one of the two known ER isoforms, ERα but not ERβ, and the differential ligand selectivity of these ERs indicates that the currently accepted screening method only detects a subset of total estrogenic chemicals. To improve the estrogen screening bioassay, BG1Luc4E2 cells were stably transfected with an ERβ expression plasmid and positive clones identified using ERβ-selective ligands (genistein and Br-ERβ-041). A highly responsive clone (BG1LucERβc9) was identified that exhibited greater sensitivity and responsiveness to ERβ-selective ligands than BG1Luc4E2 cells and qRT-PCR confirmed the presence of ERβ expression in these cells. Screening of pesticides and industrial chemicals identified chemicals that preferentially stimulated ERβ-dependent reporter gene expression. Together, these results not only demonstrate the utility of this dual ER recombinant cell line for detecting a broader range of estrogenic chemicals than the current BG1Luc4E2 cell line, but screening with both cell lines allows identification of ERα and ERβ-selective chemicals.
Keywords: Estrogen receptor bioassay, estrogenic chemicals, BG1Luc ER TA, environmental screening
INTRODUCTION
Many environmental contaminants can directly affect a multitude of hormonally regulated, physiological processes in humans and wildlife. Reports of alterations in reproduction and endocrine homeostasis, changes in behavior, reductions in sperm count, as well as dramatic increases in human breast and prostate cancers are responses which may result from chemicals that affect steroid hormone action [1]. Endocrine disrupting chemicals (EDCs) have been identified in a wide range of environmental samples (air, water, soil and sediment), commercial and consumer products and foods, and a significant number have been shown to exert their effects through mimicking and/or inhibiting the action of estrogen on estrogen receptors (ERs) [1, 2]. Accordingly, screening of extracts from diverse matrices for the presence of chemicals that can affect estrogen action using rapid ER-dependent mechanism-based bioassays is a relatively simple first step in determining the potential of chemicals, extracts, and products to affect wildlife and human health.
Numerous cell-based bioassays and bioanalytical methods have been described for the detection and relative quantitation of estrogenic/antiestrogenic activity of pure chemicals, mixtures and extracts of a broad range of matrices [3–8]. These bioassays are generally based on the ability of chemicals to stimulate or inhibit ER-dependent cell proliferation or expression of a native or transfected gene [6, 7, 9]. While the majority of these bioassays have widespread utility for screening purposes, very few have received official approval as an accepted bioassay for regulatory screening purposes. Only two ER-dependent gene expression (i.e. transactivation) cell bioassays have been included in the USEPA Endocrine Disruptor Screening Program (EDSP) as officially accepted methods [10, 11]. The first is the HeLa-9903 cell bioassay that uses a recombinant human cervical cancer (HeLa) cell line containing a stably transfected estrogen-/ER-responsive luciferase expression vector. Because these cells lack functional ER, they were also stably transfected with a human ERα expression vector. Curiously, the HeLa-9903 bioassay reportedly can only be utilized for detection of estrogen agonists and not antagonists [12]. The second accepted transactivation assay is the BG1Luc ER TA bioassay that uses a recombinant human ovarian carcinoma cell line (BG1Luc4E2) that contains a stably transfected estrogen-/ER-responsive luciferase reporter gene and endogenously expresses ERα [3]. In addition to the USEPA EDSP, the BG1Luc ER TA bioassay was also approved by the OECD as an officially accepted method (TG455/457) for detection of estrogenic/antiestrogenic chemicals [13, 14]. While the BG1Luc ER TA bioassay has some advantages over the HeLa cell bioassay method (such as the ability to detect estrogen antagonists) and has been used by many labs for screening purposes, there remain some limitations of this bioassay (and the HeLa bioassay) with regards to the spectrum of estrogenic chemicals that can be detected. This limitation derives from the fact that while BG1 cells, like most other ER-containing cancer cell lines, contain significant amounts of one ER subtype (ERα), they contain little or no ERβ, and thus may underestimate the overall estrogenic potency of a sample extract [3, 15, 16].
While ERα and ERβ are highly homologous in some functional domains, (i.e. the DNA binding domain), their ligand binding domain is distinctly different (~60% homology) [17]. Although both ER subtypes can bind and be activated/inhibited by many of the same compounds, numerous chemicals have been identified that can selectively bind and activate/inhibit ERs in a subtype-specific manner [18–21]. Given the critical role that ERβ plays in normal physiological/endocrinological processes and tissue health and in modulating the functional activity and levels of ERα [22, 23], detection by high throughput screening (HTS) assays of chemicals that also affect ERβ activity is necessary for more accurate assessment of the estrogenic/antiestrogenic activity of chemicals and sample extracts. Accordingly, here we describe the reengineering of the BG1Luc4E2 cell line so that it constitutively expresses both ERα and ERβ. The utility of this new recombinant cell line, BG1LucERβc9, was demonstrated through the use of ERβ-selective ligands and the identification of additional estrogenic chemicals present in a chemical library of common insecticides, herbicides, industrial and household chemicals that were not detected in BG1Luc4E2 cells containing only ERα.
MATERIALS AND METHODS
Materials
The ERβ-selective ligand Br-ERβ-041 was generously provided by Dr. John Katzenellenbogen (University of Illinois, Urbana-Champaign) and the chemical library of 176 compounds was obtained from Dr. Bruce Hammock (University of California, Davis). Fulvestrant (ICI-182,780) was from Santa Cruz Biotechnology (Santa Cruz, CA), genistein, 17β-estradiol (E2), tamoxifen (citrate salt) and 4-hydroxy tamoxifen (4-OHT) were purchased from Sigma Aldrich Chemical Company (St. Louis, MO), 4,4’,4”-(4-Proply-[1H]-pyrazole-1,3,5-triyl)trisphenol (PPT) from Tocris Biosciences (Ellisville, Missouri) and DMSO and EtOH from EMD Chemicals (San Diego, CA). Translation grade L-[35S]-methionine (>400 ci/mmol) was from MP Biomedical (Solon, OH). Primers for quantitative real-time polymerase chain reaction (qRT-PCR) were from Applied Biosystems (ABI, Grand Island, NY) and restriction enzymes from New England Biolabs (Ipswich, MA). Premium and charcoal-stripped fetal bovine serum (FBS) were obtained from Atlanta Biologicals (Lawrenceville, GA), Zeocin and alpha Minimal Essential Medium (α-MEM) were from Invitrogen (San Diego, CA), Dulbecco’s Modified Eagle’s Medium (DMEM) from Sigma (St. Louis, MO). FuGene6 transfection reagent, Cell Culture Lysis Buffer, and Passive Lysis Buffer were from Promega (Madison, WI), Lipofectamine 2000 transfection reagent from Invitrogen (San Diego, CA). Mini-Protean TGX gels (10%) and Protein Assay Dye Reagent were from Bio-Rad (Hercules, CA) and PageRuler prestained protein ladder from ThermoScientific (Rockford, IL). Human breast carcinoma (SKBR3) cells were obtained from American Tissue Culture Collection (Manassas, VA). Test chemicals and standards were dissolved in DMSO (with the exception of 4-OHT which was dissolved in EtOH) and stored at 4°C in borosilicate amber vials with PTFE-lined caps.
Construction of receptor expression vectors
To construct the human ERβ expression plasmid, ERβ/pcDNA3.1+Zeo pcDNA3.1+Zeo/ERβ, a 2367 bp fragment including the human ERβ sequence was excised from the ERβ/pcDNA3 plasmid (originally isolated from the ERβ/pCMV5 plasmid obtained from Dr. John Katzenellenbogen (University of Illinois, Urbana-Champaign)) and inserted into the HindIII/XbaI sites in the vector pCDNA3.1+Zeo (Invitrogen) [24]. The human ERα expression plasmid, pcDNA3.1+Zeo/ERα, was constructed by excision of a 2497 bp XhoI/BstEII fragment (human ERα cDNA) from the plasmid ERα/pcDNA3 and ligation into the same sites in pcDNA3.1+Zeo [24, 25]. The resulting plasmids were confirmed by sequencing. Plasmids ERβ/pcDNA3.1+Zeo and ERα/pcDNA3.1+Zeo were used as templates for in vitro expression using the Promega rabbit reticulocyte lysate TnT Quick coupled transcription/translation (with empty vector pcDNA3.1+Zeo and unprogrammed lysate [UPL] as controls) and the molecular weight of the resulting [35S]-radiolabeled expressed proteins was determined as previously described [25].
Transient transfection
Transient transfection of SKBR3 cells was performed as we have described previously [25]. Briefly, SKBR3 cells were maintained in DMEM containing 10% premium FBS and 48 hours before transfection switched to phenol-red free DMEM (referred to as ‘estrogen-stripped medium’ [ESM]) containing 10% charcoal-stripped FBS. Twenty four hours before transfection, cells were seeded in 24-well plates at a density of 300,000 cells/mL. SKBR3 cells were transfected with 0.2 µg of ER-responsive reporter plasmid pGudLuc7.ERE [3] and 0.05 µg of ER expression plasmid (ERα/pcDNA3.1+Zeo or ERβ/pcDNA3.1Zeo) using Lipofectamine reagent (2 µL/well) according to the manufacturer’s recommendations (µg DNA/well was normalized to 0.8 µg/well with the addition of empty vector DNA [pcDNA3.1+Zeo]) and cells allowed to incubate for 24 h [3]. Transfected SKBR3 cells were then incubated for 24 h with the desired test chemical (triplicate wells per chemical or concentration), the cells harvested, and both protein concentration and luciferase activity determined as described [25, 26].
Stable transfection
To produce the stable BG1LucERβ cell line, human ovarian carcinoma (BG1Luc4E2) cells containing a stably transfected estrogen-responsive luciferase plasmid [3] were transfected with ERβ/pcDNA3.1+Zeo using FuGene6 transfection reagent according to the manufacturer’s recommendation. Following 24 h of incubation in regular medium, the transfected cells were split 1 to 15 and replated into selective medium containing the antibiotic Zeocin (600 mg/L) which was replaced every three days with fresh Zeocin-containing medium. After approximately 3 weeks of growth, 28 individual cell colonies were isolated, their ERβ responsiveness was determined, and those clones exhibiting the greatest induction by ERβ-selective ligands (Br-ERβ-041 (31.6 nM) and genistein (10 nM)), were selected for further evaluation.
RNA isolation and Real time PCR
Cells in 10 cm plates (four replicates per cell line) were grown in maintenance medium with the addition of Zeocin for the BG1LucERβc9 cells. Total RNA was isolated using RNeasy (Qiagen) according to the manufacturer’s instructions (β-mercaptoethanol was added to the lysis buffer immediately before use, and cells were homogenized using QIAshredder homogenizer). Complementary DNA (cDNA) was generated from 2 µg RNA using a High Capacity cDNA Reverse Transcription kit (ABI) with random primers followed by a 1:10 dilution with RNAse/DNAse-free water. Real-time quantitative PCR reactions (20 µL) were performed with TaqMan Fast Universal PCR Master Mix and TaqMan Gene Expression assays (ABI) in an ABI 7500 Fast Sequence Detection instrument. ERα and ERβ mRNA levels were quantitated using 7500 Software (v.2.0.6), normalized to β-actin (internal control), and results presented relative to levels in BG1Luc4E2 cells (set to value of 1.0). Primers/probes for human ERβ (ESR2, Hs01100353_m1), human ERα (ESR1, Hs00174860_m1) and human β-actin (ACTB, Hs999999031_m1) were obtained from ABI.
Chemical treatment and luciferase analysis
For the screening and characterization studies, BG1LucERβ cells were switched from maintenance medium (α-MEM, containing 10% premium FBS and 600 mg/L Zeocin) to ESM, containing 10% charcoal-stripped FBS and incubated three days prior to plating into white, clear-bottomed 96-well tissue culture plates at a density of 750,000 cells/mL. Cells were allowed to attach for 24 hours and were then incubated with carrier solvent DMSO (1% final solvent concentration) or the indicated concentration of chemical for 24 hours (unless otherwise indicated) at 37°C with triplicate wells per chemical and controls. After incubation cells were rinsed twice with PBS, lysed with Promega cell lysis buffer, and shaken for 20 min at room temperature to allow complete cell lysis. Luciferase activity in each well was measured using an Orion microplate luminometer as previously described [27].
Chemical compound library
The chemical library used for screening contained 176 different compounds in DMSO [28] and cells were incubated with each chemical at a final incubation concentration of 10 µM in triplicate. For comparative purposes, luciferase induction values were normalized to maximal luciferase induction obtained with 1 nM E2 in each plate (set at 100%). Values represented the mean ± SD of triplicate incubations in the single screening analysis of the entire chemical compound library. Accordingly, all positive chemicals from the initial library screen that induced luciferase activity in BG1LucERβc9 cells greater than 40% of maximum E2 activity and also induced luciferase activity significantly greater in BG1LucERβc9 cells than in BG1Luc4E2 cells were confirmed in subsequent induction analysis experiments (triplicate analysis in three separate experiments).
Statistical analysis
Significant differences between results were determined using One-way ANOVA (i.e. Student’s t-test, 2-tailed, type 2, p < 0.05). Luciferase activity of control (solvent treated) cells was subtracted from that of treated cells to obtain final induced activity (RLU). Final RLU values < 0 were set at 0 RLUs. Half-maximal concentrations induced (EC50) or repressed (and IC50) by chemical or extract were determined using SigmaPlot (v.12) by the concentration of chemical which induced exactly 50% of maximal E2-induced luciferase activity unless otherwise noted, in which case the values were determined as previously described [27].
RESULTS AND DISCUSSION
Subtype selectivity of ligands
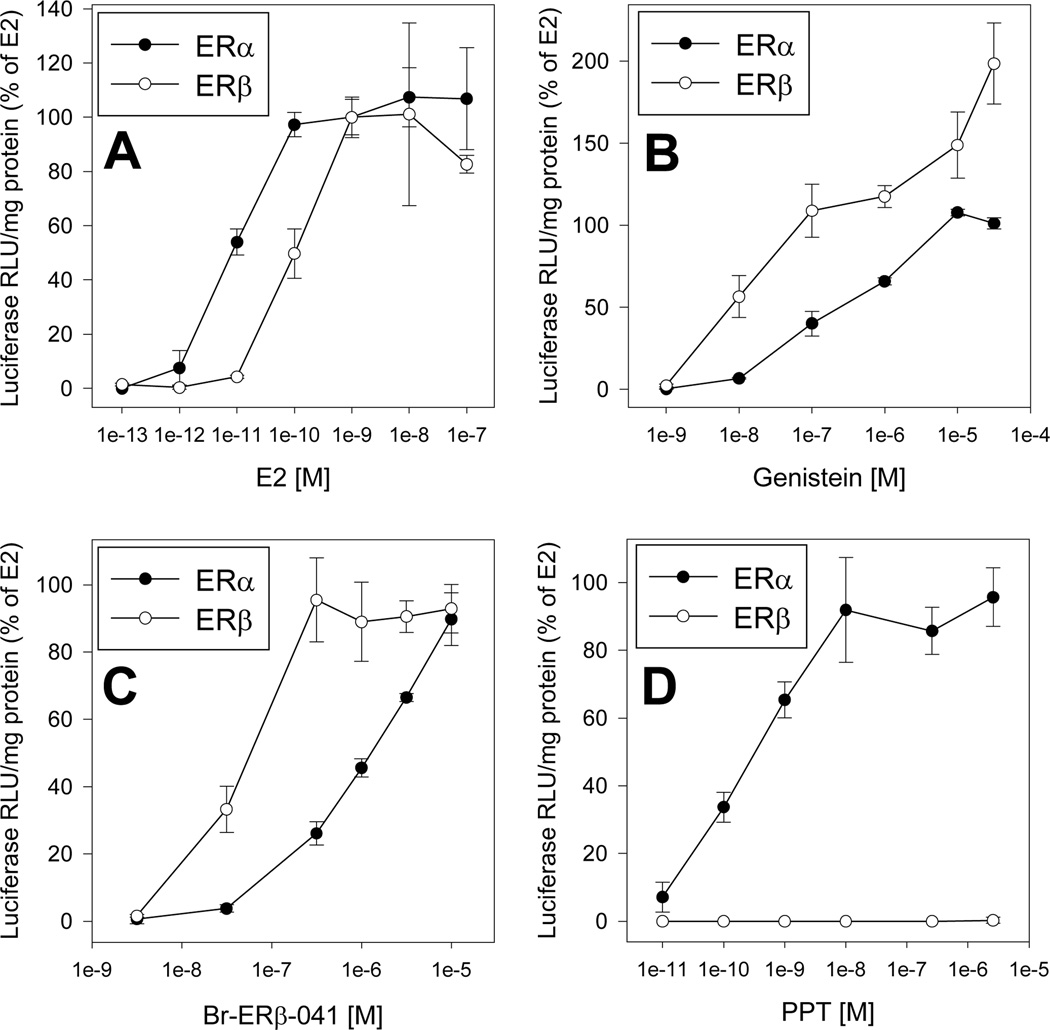
To confirm that the ERα and ERβ expression vectors were constructed appropriately, we first demonstrated that ER expressed in vitro from these vectors produced proteins of the expected size (66 and 59 kDa, respectively (Supplemental Figure S1)). To test the functionality of the two ER expression vectors, ER negative SKBR3 cells were transiently cotransfected with an estrogen-responsive luciferase reporter construct (pGudLuc7.ERE) and either the ERα expression vector (ERα/pcDNA3.1+Zeo) or the ERβ expression construct (ERβ/pcDNA3.1+Zeo) and their response to E2, genistein, Br-ERβ-041, or PPT determined (Figure 1). E2 induced luciferase activity only in SKBR3 cells cotransfected with either ER expression vector (Supplemental Figure S2), confirming both the absence of functional ERs in these cells [29] and the functionality of our ER expression vectors. E2 induced luciferase activity in a concentration-dependent manner in SKBR3 cells cotransfected with the ER-responsive luciferase reporter plasmid and either ERα or ERβ expression vectors (Figure 1A) with half-maximal induction of 8.5 ± 2.2 pM and 75 ± 29 pM E2, respectively (Figure 1A; Table 1). These EC50 values are comparable to previously published results in human osteosarcoma (U2OS) cells transfected with ERα or ERβ [30]. Concentration-dependent analysis revealed that the ERβ-selective ligands genistein or Br-ERβ-041 were more potent activators of ERβ-dependent luciferase reporter gene expression than that observed with ERα (Figures 1B and C), with EC50 induction values in ERβ- and ERα-transfected SKBR3 cells for genistein of 44 ± 19 nM and 260 ± 79 nM, and for Br-ERβ-041 54 ± 8.5 nM and 970 ± 130 nM, respectively (Table 1). Interestingly, although genistein resulted in superinduction of reporter gene expression only in ERβ-transfected cells (Figure 1B), no superinduction was observed with Br-ERβ-041 (Figure 1C). The mechanism for this ligand selective superinduction effect remains to be elucidated. Finally, as expected, incubation of ERα- and ERβ-transfected SKBR3 cells with PPT, an ERα-selective ligand, only stimulated ERα-dependent reporter gene expression (Figure 1D) with an EC50 of 250 ± 21 pM (Table 1) [18, 19, 31]. This is consistent with previously published studies demonstrating selective activation of ERα by PPT [18, 19]. Together, these results both confirm the functionality of our ERα and ERβ expression vectors and the selectivity of the ligands needed for identifying for ERβ-containing stable cell clones.
Figure 1.
ER subtype-specific induction of ER-dependent luciferase reporter gene expression in transiently transfected SKBR3 cells. Cells were cotransfected with the ER-responsive luciferase reporter plasmid pGudLuc7.ERE and the ERα or ERβ expression vector, incubated with the indicated concentration of (A) E2, (B) genistein, (C) Br-ERβ-041, or (D) PPT for 24 h, lysed and luciferase and total protein levels analyzed as described in Materials and Methods. Luciferase activity is expressed as a percent of maximum luciferase induction by 1 nM E2 and results represent the mean ± SD of triplicate determinations from each of three separate experiments after subtraction of the luciferase activity present in cells exposed to DMSO.
Table 1.
ER-subtype selectivity of genistein, Br-ERβ-041, and PPT in SKBR3 cells transiently co-transfected with a estrogen-responsive reporter gene plasmid (pGudLuc7.ERE) and either a ERα or ERβ expression plasmid.
| ERα | ERβ | |||
|---|---|---|---|---|
| Chemical | EC50 [M] | Maximal Inductiona | EC50 [M] | Maximal Inductiona |
| E2 | 8.5 ± 2.2 × 10−12 | 100 ± 8% | 7.5 ± 2.9 × 10−11 | 100 ± 7% |
| Genistein | 2.6 ± 0.8 × 10−7 | 101 ± 3% | 4.4 ± 1.9 × 10−8 | 199 ± 25%* |
| Br-ERβ-041 | 9.7 ± 1.3 × 10−7 | 90 ± 8% | 5.4 ± 0.9 × 10−8 | 93 ± 7% |
| PPT | 2.5 ± 0.2 × 10−10 | 92 ± 16% | No induction | No induction |
Luciferase induction is expressed as the mean ± SD of three individual experiments (with chemicals analyzed in triplicate) and values are presented as the percent of maximum luciferase induction obtained with E2 (1 nM). Maximum luciferase induction by genistein, Br-ERβ-041, and PPT was observed at concentration of 3.16 µM, 10 µM, and 10 nM, respectively.
Significantly different from that of 1 nM E2 at p < 0.05 as determined by Student’s t-test.
Selection of stably transfected BG1LucERβ clones
To prepare the desired ERα- and ERβ-containing stably transfected cell line, BG1Luc4E2 cells, which contain endogenously expressed ERα and a stably integrated ER-responsive luciferase reporter gene [3], were stably transfected with ERβ/pcDNA3.1+Zeo, and Zeocin-resistant clones were isolated and screened for the presence of functional ERβ. This was accomplished by evaluating the ability of the cell clones to respond to low concentrations of the ERβ-selective ligands Br-ERβ-041 (31.6 nM) and genistein (10 nM) with the induction of luciferase activity. Preliminary studies demonstrated that while these concentrations of Br-ERβ-041 and genistein failed to induce ERα-dependent reporter gene induction in BG1Luc4E2 cells (data not shown), they are sufficient to simulate ERβ-dependent reporter gene expression at least in transfected SKBR3 cells (Figure 1B, C). The response of BG1Luc4E2 cells and selected stably transfected BG1LucERβ clones to Br-ERβ-041, genistein, and E2 is shown in Supplemental Figure 3. While initial screening revealed several cell clones that responded to genistein and Br-ERβ-041 with the induction of luciferase activity, BG1LucERβ clones 2 and 9 exhibited the greatest response to these ERβ selective ligands (Supplemental Figure 3); little induction was observed in the parental BG1Luc4E2 cell line. Given that the absolute level of luciferase induction was significantly greater in clone 9 than in clone 2 (data not shown), BG1LucERβ clone 9 (BG1LucERβc9) was selected as the ERβ containing cell line for chemical analysis.
Characterization of the BG1LucERβc9 cell line
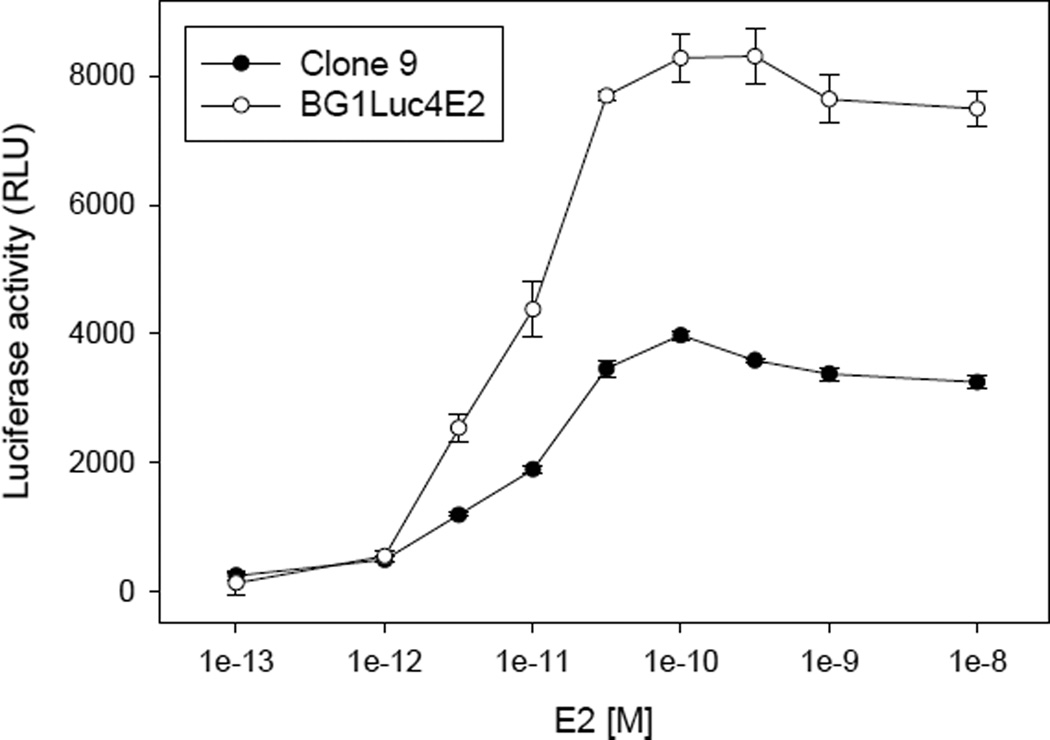
To examine the characteristics of the BG1LucERβc9 cell line, we first compared its ability to respond to E2 in a concentration-dependent manner to that induced in the parental cell line BG1Luc4E2 (Figure 2). Interestingly, these results revealed that clone 9 cells exhibited a significantly reduced response to E2, when compared to the parental BG1Luc4E2 cell line. While the mechanism responsible for this reduction remains to be determined, it may be related to the previously documented ability of ERβ to decrease ERα levels and functionality [22, 32–36].
Figure 2.
Concentration-dependent induction of luciferase in BG1LucERβ clone 9 and the parent cell line BG1Luc4E2 by estrogen. Cells were incubated with E2 (24 h), lysed, and luciferase levels measured and analyzed as described in Materials and Methods. Luciferase activity is expressed as relative light units (RLUs) and values represent the mean ± SD of triplicate determinations after subtraction of the luciferase activity obtained in cells exposed to DMSO and are representative of more than 15 different replicate experiments.
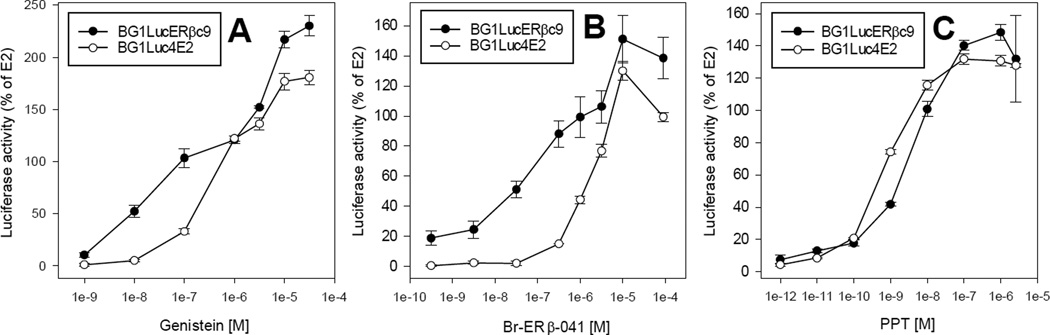
To further characterize the responsiveness of the stably transfected BG1LucERβc9 cell line, we examined the concentration-dependent ability of E2, the ERβ-selective ligands Br-ERβ-041 and genistein and the ERα-selective ligand PPT to induce luciferase reporter gene activity and compared the induction responses to those obtained using the ERα-containing parental cell line BG1Luc4E2. Induction in these cells was examined at 24 h of incubation, the earliest time resulting in maximal luciferase induction by all tested ligands (Supplemental Figure S4) and induction responses in each cell line was normalized to maximal induction observed with E2. The results of these analyses revealed the ability of the ERβ-selective ligands to induce luciferase reporter gene activity in the BG1LucERβc9 cells at a significantly lower concentration than in BG1Luc4E2 cells (Figure 3A, B). As expected, the relative induction by the ERα-selective ligand PPT was comparable in both cell lines (Figure 3C) when activity was normalized to the response to E2 since they both contain endogenously expressed ERα. Quantitative analysis of concentration-response experiments for each compound in BG1Luc4E2 and BG1LucERβc9 cell lines are presented in Table 2. These results not only reveal comparable EC50 and minimal detection limit (MDL) values for induction of reporter gene activity by E2, but they highlight the overall reduction in both the background luciferase activity of the BG1LucERβc9 cells compared to the BG1Luc4E2 cells and the maximal induction levels by all ER ligands. The reduction in the overall ER-dependent reporter gene induction response in the new BG1LucERβc9 cell line is consistent with several other reports demonstrating an ERβ-dependent down regulation/repression of ERα signaling in a variety of different transfected cell types [22, 32, 33]. Since ERα can result in dramatically greater levels of reporter gene activity than observed in the presence of ERβ (Supplemental Figure S2), the significant reduction in inducible gene expression in the ERβ cell line is not surprising since the level of ERα would be the primary driver of the gene expression response.
Figure 3.
Concentration-dependent induction of luciferase in BG1LucERβc9 and BG1Luc4E2 cells by ERα and ERβ selective ligands. BG1LucERβc9 cells (closed circles) or BG1Luc4E2 cells (open circles) were incubated with the ERβ-selective ligands genistein (A) or Br-ERβ-041 (B) or with the ERα-selective ligand PPT (C) for 24 h, cells lysed, and luciferase levels measured and analyzed as described in Materials and Methods. Luciferase activity is expressed as a percent of maximum induction by E2 (0.1 nM) in each cell line and results represent the mean ± SD of triplicate determinations after subtraction of the background luciferase activity obtained in cells exposed to DMSO and values are representative of more than 8 replicate experiments.
Table 2.
Half-maximal induction values (EC50) and minimal detection limits (MDL) of BG1Luc4E2 and BG1LucERβc9 cells incubated with genistein, Br-ERβ-041, E2 or PPT.
| Cell line | Compound | EC50a,b, [M] | MDL, [M] | DMSO RLU | Maximum Induction (RLU)c |
|---|---|---|---|---|---|
| BG1Luc4E2 | Genistein | 1.7 ± 0.8 × 10−7 | 1 × 10−7 | 2,773 ± 252 | 17,623 ± 575 |
| Br-ERβ-041 | 1.3 ± 0.7 × 10−6 | 3 × 10−7 | 3,147 ± 257 | 18,017 ± 737 | |
| E2 | 1.6 ± 0.6 × 10−11 | 3 × 10−12 | 2,807 ± 182 | 10,594 ± 250 | |
| PPT | 6.7 ± 3.8 × 10−10 | 1 × 10−10 | 2,482 ± 444 | 11,422 ± 356 | |
| BG1LucERβc9 | Genistein | 2.0 ± 0.6 × 10−8 | 1 × 10−8 | 1,529 ± 61 | 8,804 ± 306 |
| Br-ERβ-041 | 6.0 ± 2.0 × 10−8 | 3 × 10−8 | 1,724 ± 224 | 9,468 ± 111 | |
| E2 | 1.3 ± 1.0 × 10−11 | 1 × 10−12 | 1,259 ± 60 | 5,236 ± 71 | |
| PPT | 3.2 ± 0.8 × 10−9 | 1 × 10−10 | 1,116 ± 136 | 7,017 ± 188 | |
Values represent the mean ± SD of 8–16 individual concentration response experiments, each with 7–9 concentrations of the indicated chemical analyzed in triplicate in each experiment.
EC50 values for genistein and Br-ERβ-041 were determined from values at 50% when normalized to maximum E2 activity.
Maximum induction was obtained in both cell lines at the following chemical concentrations: 30 µM Genistein, 10 µM Br-ERβ-041, 0.1nME2, and 1 µM PPT.
The EC50 values for the ERβ-selective ligands, genistein and BR-ERβ-041, were 9- and 22-fold lower, respectively in the BG1LucERβc9 cells compared to the parental BG1Luc4E2 cells and the MDL for each was 10-fold lower (Table 2). Additionally, although BG1LucERβc9 cells display a small but significant increase in the relative potency of the ERα-selective ligand PPT compared to the parental cell line (3.2 nM compared to 0.67 nM, respectively), no differences in the relative potency of E2 were observed. This slight reduction in the potency of PPT may result from the presence of ERβ interfering with ERα functionality. In fact, a previous study demonstrated that while ERα:ERα homodimers bind PPT with high affinity, ERα:ERβ heterodimers and ERβ:ERβ homodimers bind PPT inefficiently and both receptors must bind and be activated by PPT in order to transactivate the induction response [31].
In addition to examining the response of BG1LucERβc9 cells to ER agonists, we demonstrated the ability of the ER antagonists fulvestrant (ICI 182780) and 4-hydroxytamoxifen (4-OHT) to inhibit E2-inducible reporter gene expression in these cells (Supplemental Figure S5). To examine the differential effect of the ER antagonist fulvestrant against ERα and ERβ, we determined the ability of fulvestrant to antagonize luciferase induction by both E2 and Br-ERβ-041 in BG1LucERβc9 and BG1Luc4E2 cells. While concentration-dependent antagonism of E2-induced luciferase activity by fulvestrant was comparable in the BG1LucERβc9 and BG1Luc4E2 cells, it was a significantly more potent antagonist of Br-ERβ-041 induced activity (p < 0.05 as determined by Student’s t-test) in BG1Luc4E2 cells (Supplemental Figure S6; Supplemental Table S1). Together, these results demonstrate that the ERβ-containing BG1LucERβc9 recombinant cell line responds appropriately to ER antagonists and can be used to detect both ER agonists and antagonists.
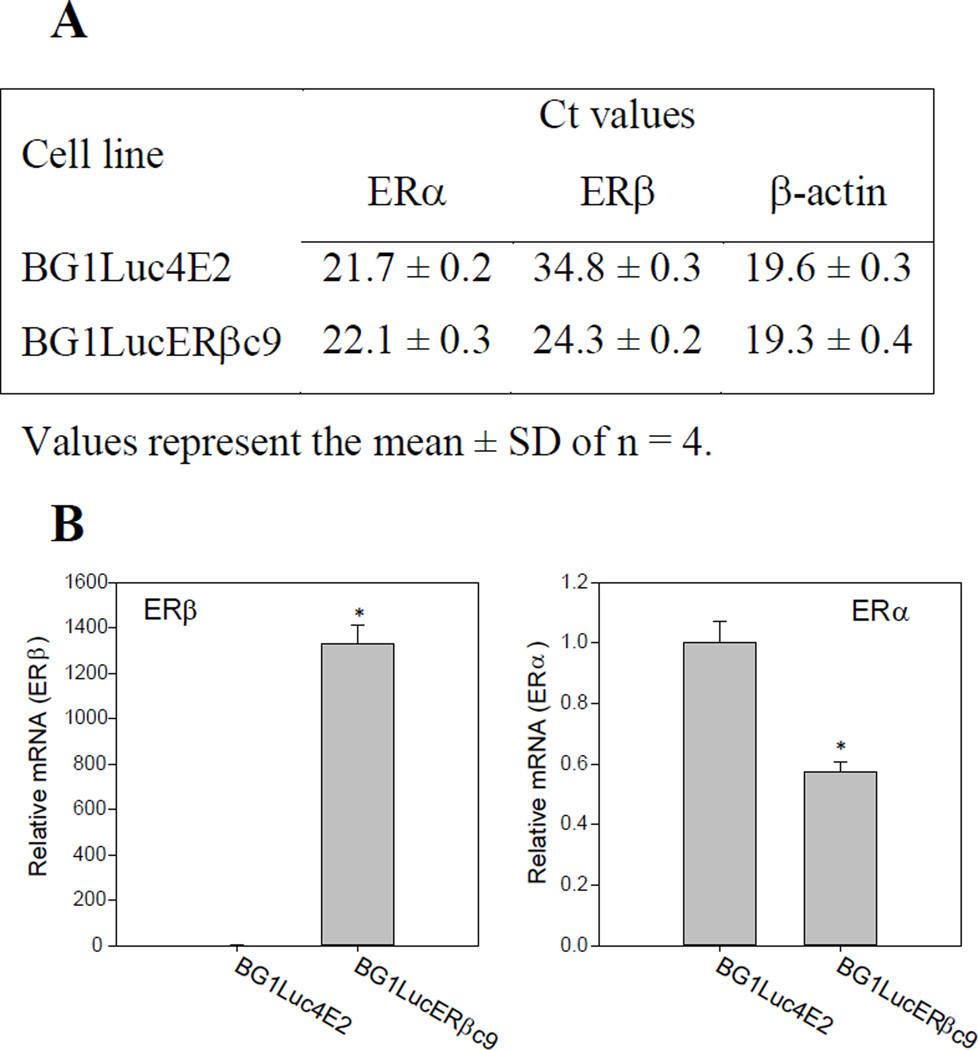
Finally, to confirm that the increased responsiveness of BG1LucERβc9 cells to ERβ-selective ligands was correlated with the increased ERβ expression levels in these cells, we compared ERβ mRNA levels in BG1LucERβc9 cells to those of the parental BG1Luc4E2 cells using quantitative rtPCR. The average threshold cycle (Ct) value for ERβ mRNA levels in the parental BG1Luc4E2 cells was 34.8 ± 0.3 indicating very little expression of this gene (Figure 4A), comparable to low levels of ERβ mRNA expression previously reported in wild type BG1 cells [32, 37]. In contrast, the average Ct value for ERβ mRNA in BG1LucERβc9 cells was 24.3 ± 0.2, approximately 1,100-fold more ERβ mRNA than that in BG1Luc4E2 cells (Figure 4A, B). We also examined whether ERα mRNA levels differed between BG1LucERβc9 cells and BG1Luc4E2 cells, especially given the significant reduction in overall ER-inducible gene expression and the decrease in potency of the ERα-specific ligand PPT observed in BG1LucERβc9 cells (Table 2). These analyses revealed that the level of ERα mRNA expression in BG1LucERβc9 cells was only 64 ± 6% of that found in the parental BG1Luc4E2 cells (Figure 4B). The significant reduction of ERα mRNA in BG1LucERβc9 cells not only agrees well with the apparent repressive effect of ERβ expression on ERα mRNA levels (Figure 4) and ER-dependent reporter gene activation (Figure 2; Table 2) observed in the BG1 cells, but also that previously reported by other investigators using a variety of other cell lines [22, 32, 33,31].
Figure 4.
Comparison of ERβ and ERα mRNA levels in BG1Luc4E2 and BG1LucERβc9 cells. (A) Threshold cycle values for ERα, ERβ, and β-actin mRNA levels in BG1Luc4E2 and BG1LucERβc9 cells. (B) Expression of ERβ and ERα mRNA in BG1LucERβc9 cells. Results represent the mean ± SD of four determinations, and ER mRNA levels were normalized to that of β-actin and expressed relative to levels in BG1Luc4E2 cells as described in Materials and Methods. Significant difference (p < 0.05, determined by Student’s t-test) in relative ERβ or ERα mRNA levels in BG1LucERβc9 cells from that of levels in BG1Luc4E2 cells is indicated by an asterisk *.
Application of the BG1LucERβc9 cell line for chemical screening
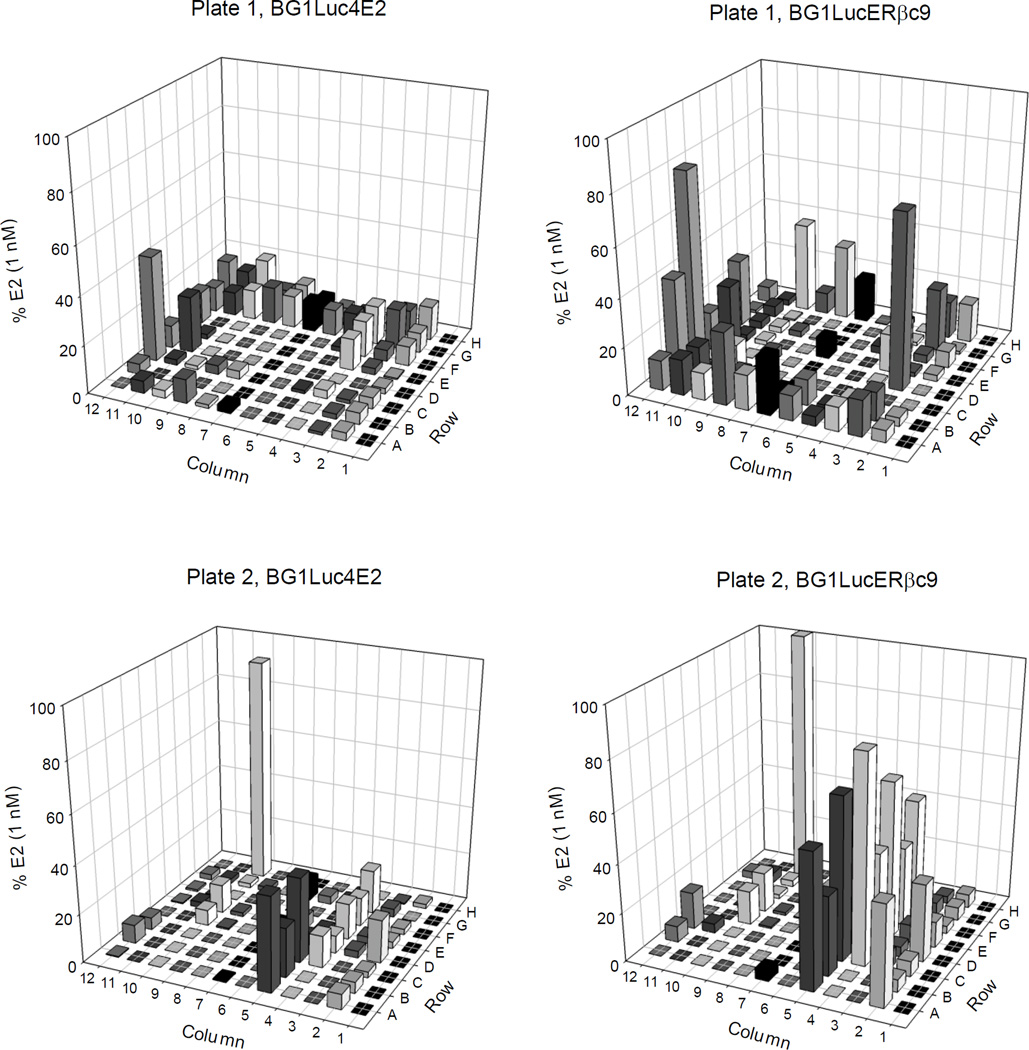
Given that the goal of generating the stably transfected BG1LucERβc9 cells was to develop a cell bioassay containing both ERα and ERβ that would be useful for more comprehensive chemical screening, the BG1LucERβc9 cell line was used to detect both ERα- and ERβ-selective agonists in a chemical library consisting of 176 pesticides and industrial chemicals (in two 96-well plates) [28]. The BG1Luc4E2 cells were also included in the screen for comparative purposes, with the resulting data allowing identification of chemicals that can activate ERα. Differences in response between the two BG1Luc4E2 and BG1LucERβc9 cell lines can be used to identify potential ERβ-selective agonists. Representative plates of luciferase induction data from these screens are shown in Figure 5 and Supplemental Table S2 and results normalized to the maximal response obtained with E2. While the induction results in BG1Luc4E2 cells identified compounds that induce reporter gene activity in an ERα-dependent manner, the induction observed in BG1LucERβc9 cells can result from activation of ERα and/or ERβ. These comparative results identified numerous chemicals in the library that increased reporter gene activity to a greater degree in BG1LucERβc9 cells compared to that in BG1Luc4E2 cells, consistent with a role of ERβ in the induction response by these chemicals.
Figure 5.
Screening results of a chemical library of common pesticides and industrial chemicals in BG1Luc4E2 and BG1LucERβc9 cells. Chemicals in the library were screened once in triplicate for ER agonist activity in BG1Luc4E2 or BG1LucERβc9 cells at a final concentration of 10 µM. Cells were incubated for 24 h, lysed, and luciferase levels measured and analyzed as described in Materials and Methods. Luciferase activity is expressed as a percent of maximal luciferase induction obtained with 1 nM E2 in each cell line. Each bar represents the mean of triplicate determinations.
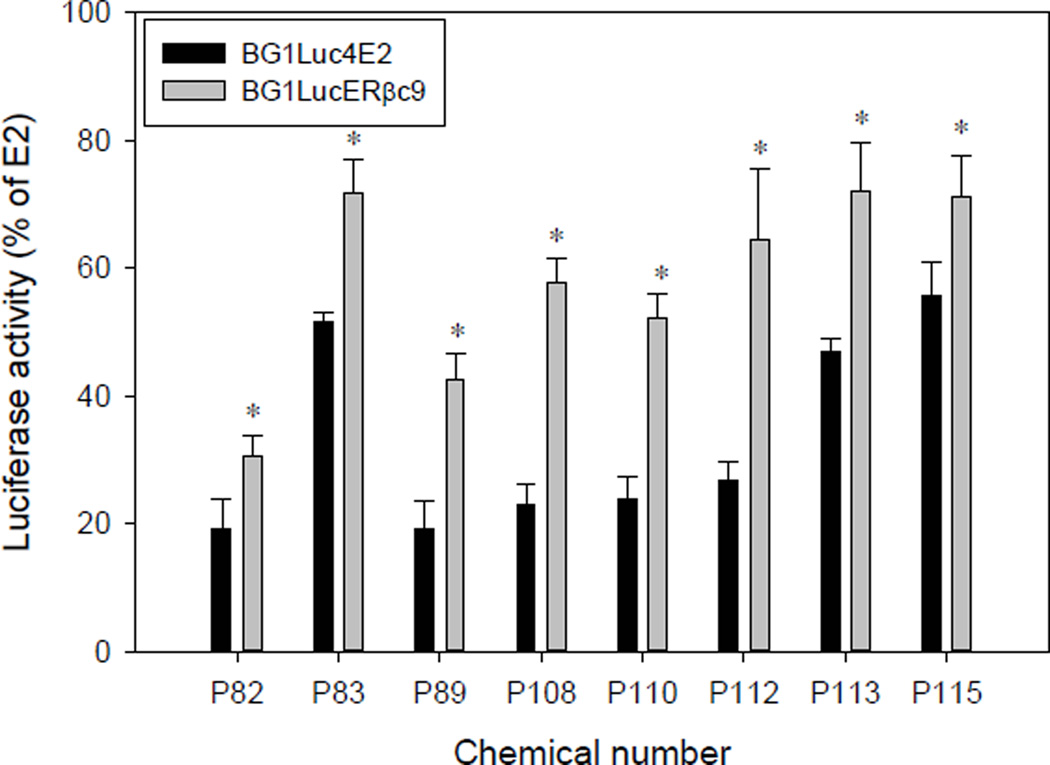
To confirm the ERβ-selectivity of the positive chemicals in these library screens, those chemicals that induced luciferase activity >40% of that of maximal induction by E2, and which exhibited a significantly greater induction response in BG1LucERβc9 cells (compared to that in the parental BG1Luc4E2 cells) in the initial screen were reanalyzed for their ability to induce luciferase in both cell lines. Based on these criteria, we selected chemicals P82, P83, P89, P108, P110, P112, P113, and P115 for subsequent reanalysis to confirm their ability to induce luciferase activity in both cell lines. Chemical P159 (bisphenol A, position H10 on plate 2), produced high activity in both cell lines in the initial screen and was selected as a second positive control (in addition to E2) for confirmatory analysis. Interestingly, the majority of the most potent estrogenic chemicals which met the criteria for inclusion in the confirmatory screen were organochlorine insecticides (Table 3; Supplemental Figure S7). In the initial screening analysis, chemical P12 (cyanazine, position 3D on plate 1) induced significant luciferase activity in BG1LucERβc9 cells (72% of maximum induction by E2 (Figure 5)) while not inducing significant luciferase activity above background in BG1Luc4E2 cells. However, in the confirmatory screening assays cyanazine failed to induce luciferase activity above background in either BG1LucERβc9 or BG1Luc4E2 cells. The reason for the observed activity of cyanazine in the initial screen only, is unclear, but these results clearly demonstrate the necessity for confirmatory screening analysis. The chemical which exhibited the greatest ERβ-selective induction of luciferase activity in BG1LucERβc9 cells was chemical P112 (Tedion, also known as tetradifon) inducing luciferase to 64 ± 11% of the maximum activity induced by E2 (0.1 nM); Tedion induced only to 27 ± 3% in BG1Luc4E2 cells (Figure 6, Table 3). While the endocrine disrupting activity of Tedion has been previously examined, no studies examined the ER-subtype selectivity of Tedion and the differential ability of Tedion to affect ovaries and bone remodeling and development remains to be examined [38, 39]. Lindane (P108), which resulted in the second largest ERβ-selective difference in induction (Figure 6, Table 3), was previously reported to inhibit E2-mediated activation of ERK1/2 and Akt (kinases known to stimulate cell proliferation) in ERα/ERβ-containing cerebellar granular cells [40]. Thiodan (P113, also known as endosulfan) is a known ER agonist and stimulated both ERβ- and ERβ-dependent gene expression (Figure 6, Table 3) [40, 41].
Table 3.
Identity of library chemicals subjected to confirmatory analysis for estrogenic activity.
| Chemical number |
Plate, Location |
Chemical name | Usage | Activity in BG1Luc4E2 cellsa |
Activity in BG1LucERβc9 cellsa |
|---|---|---|---|---|---|
| P82 | 1, B12 | p,p-DDT | Insecticide | 19 ± 5 | 30 ± 3 |
| P83 | 1, C12 | o,p’-DDD | Insecticide | 52 ± 1 | 72 ± 5 |
| P89 | 2, A2 | Heptachlor | Insecticide | 19 ± 5 | 43 ± 4 |
| P108 | 2, E4 | Lindane | Insecticide | 23 ± 3 | 58 ± 4 |
| P110 | 2, G4 | Endrin | Insecticide | 24 ± 3 | 52 ± 4 |
| P112 | 2, C4 | Tedion | Insecticide | 27 ± 3 | 64 ± 11 |
| P113 | 2, A5 | Thiodan | Insecticide | 47 ± 2 | 72 ± 8 |
| P115 | 2, C5 | Methoxychlor | Insecticide | 56 ± 5 | 71 ± 6 |
Activity represents the mean ± SD of results obtained in three separate experiments with each sample run in triplicate and values expressed as a percent ofthe maximal activity induced by E2 (0.1 nM).
Figure 6.
Induction of luciferase activity by selected library chemicals in BG1Luc4E2 and BG1LucERβc9 cells. Cells were incubated with the indicated chemicals (10 µM) for 24 h, lysed, and luciferase levels measured and analyzed as described in Materials and Methods. Luciferase activity is expressed as a percent of maximum induction by E2 (0.1 nM) in each cell line. Results represent the mean ± SD of three individual experiments (with triplicate wells analyzed per chemical per experiment) after subtraction of the luciferase activity obtained in cells exposed to DMSO. Significant differences (p < 0.05, determined by Student’s t-test) in luciferase induction in BG1LucERβc9 cells above that of the BG1Luc4E2 cell line from chemical treatment is indicated by an asterisk (*).
Interestingly, all the compounds which induced significantly greater luciferase activity in the BG1LucERβc9 cells compared to BG1Luc4E2 cells, were organochlorine insectides, some of which are known to be estrogenic (although ER subtype selectivity is not known), and/or they have structures which group them into known classes of ER ligands [42]. However, several of these compounds (Heptachlor (P89), Lindane (P108), Endrin (P110), and Thiodan (P113)) have structures which deviate significantly from established structure activity relationship (SAR) characteristics reported for estrogenic chemicals [42–44]. Although quantitative SAR models have been previously developed for both ERα and ERβ binding, it is unlikely that SAR approaches alone can anticipate binding of a novel compound to ERα/β, especially if the compound structure does not fall under the established classes of known estrogenic chemicals or tested compounds [45, 46]. Whether these novel ER activators directly bind to the ER ligand binding pocket and activate the ER and ER-dependent gene expression by the classical mechanism or whether they utilize a more novel mechanism (i.e. allosteroic modulation or stimulation of an ER activating kinase activity) remains to be determined. However the results presented here not only demonstrate the utility of this new bioassay to identify ERβ active chemicals, but these approaches can identify novel compounds whose chemical structure deviates from known QSAR predictions, providing new results that can be used to improve on existing QSAR models for inclusion of unexpected ERα/β activators.
Taken together, we have generated an improved BG1 ER-dependent screening bioassay that now can be used to identify a broader and more comprehensive spectrum of ER-dependent ligands (ERα/β agonists and antagonists), compared to our previous cell line (BG1Luc4E2) which lacks functional ERβ. The combined use of both the BG1LucERβc9 and BG1Luc4E2 cell lines for screening will also allow identification of ERα and/or ERβ-selectivity of a given chemical or chemical class. As such, the new BG1LucERβc9 has significant advantages over the parental cell line BG1Luc4E2 which is currently accepted by the OECD and the USEPA for detection of estrogenic chemicals. The ERβ-selective results presented here clearly demonstrate that the BG1Luc4E2 cell line may underestimate the overall potency and ability of chemicals and mixtures to activate ER-dependent gene expression. Additionally, the BG1LucERβc9 and parental cell lines provide a useful model system to examine both the molecular mechanism responsible for ERβ-mediated suppression of ERα expression and signaling and its impact on other signaling events in these cells.
Supplementary Material
Acknowledgments
Special thanks to Drs. Majorie Philips and Robert Rice for providing assistance with qRT-PCR and providing the β-actin primer/probe and to Dr. Jing Zhao for advice on stable transfection and the proliferation assay. Also, thanks to Dr. John Katzenellenbogen for advice on ERβ-selective ligands and for providing Br-ERβ-041 and the ERβ/pCMV5 plasmid, and to Dr. Bruce Hammock for the chemical compound library. This work was supported by the National Institutes of Environmental Health Sciences [R01-ES007685 and P42-ES004699 (MSD) and T32 ES007058-33 (JCB)], a UCDavis Jastro-Shields Research Award (JCB) and the California Agricultural Experiment Station.
Footnotes
The authors declare that they have no conflict or competing interests with regards to this manuscript.
REFERENCES
- 1.Bergman k, Heindel JJ, Jobling S, Kidd KA, Zoeller RT World Health Organization., United Nations Environment Programme. State of the science of endocrine disrupting chemicals - 2012: an assessment of the state of the science of endocrine disruptors prepared by a group of experts for the United Nations Environment Programme (UNEP) and WHO. Geneva, Switzerland: UNEP: WHO; 2013. [Google Scholar]
- 2.Tyler CR, Jobling S, Sumpter JP. Endocrine disruption in wildlife: a critical review of the evidence. Crit Rev Toxicol. 1998;28:319–361. doi: 10.1080/10408449891344236. [DOI] [PubMed] [Google Scholar]
- 3.Rogers JM, Denison MS. Recombinant cell bioassays for endocrine disruptors: Development of a stably transfected human ovarian cell line for the detection of estrogenic and anti-estrogenic chemicals. In Vitro Mol Toxicol. 2000;13:67–82. [PubMed] [Google Scholar]
- 4.Klein KO. Is there a role for estrogen activity assays? Recombinant cell bioassay for estrogen: Development and applications. Steroids. 2014;9:108–112. doi: 10.1016/j.steroids.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Van der Linden SC, Heringa MB, Man HY, Sonneveld E, Puijker LM, Brouwer A, Van der Burg B. Detection of multiple hormonal activities in wastewater effluents and surface water, using a panel of steroid receptor CALUX bioassays. Environ Sci Technol. 2008;42:5814–5820. doi: 10.1021/es702897y. [DOI] [PubMed] [Google Scholar]
- 6.Gutendorf B, Westendorf J. Comparison of an array of in vitro assays for the assessment of the estrogenic potential of natural and synthetic estrogens, phytoestrogens and xenoestrogens. Toxicol. 2001;166:79–89. doi: 10.1016/s0300-483x(01)00437-1. [DOI] [PubMed] [Google Scholar]
- 7.Bovee TFH, Helsdingen RJR, Rietjens I, Keijer J, Hoogenboom R. Rapid yeast estrogen bioassays stably expressing human estrogen receptors alpha and beta, and green fluorescent protein: a comparison of different compounds with both receptor types. J Steroid Biochem Mol Biol. 2004;91:99–109. doi: 10.1016/j.jsbmb.2004.03.118. [DOI] [PubMed] [Google Scholar]
- 8.Wilson VS, Bobseine K, Gray LE. Development and characterization of a cell line that stably expresses an estrogen-responsive luciferase reporter for the detection of estrogen receptor agonist and antagonists. Toxicol Sci. 2004;81:69–77. doi: 10.1093/toxsci/kfh180. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Rijk JC, Besselink HT, Houtman R, Peijnenburg AA, Brouwer A, Rietjens IM, Bovee TF. Extending an in vitro panel for estrogenicity testing: the added value of bioassays for measuring antiandrogenic activities and effects on steroidogenesis. Toxicol Sci. 2014;141:78–89. doi: 10.1093/toxsci/kfu103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.EPA. Endocrine Disruptor Screening Program (EDSP); Announcing the availability of the Tier 1 Screening Battery and related test guidelines. Federal Register. 2009;74:54415–54422. [Google Scholar]
- 11.Borgert CJ, Mihaich EM, Quill TF, Marty MS, Levine SL, Becker RA. Evaluation of EPA's Tier 1 Endocrine Screening Battery and recommendations for improving the interpretation of screening results. Regul Toxicol Pharmacol. 2011;59:397–411. doi: 10.1016/j.yrtph.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 12.EPA. Endocrine Disruptor Screening Program test guideline OPPTS 890.1300: Estrogen receptor transcriptional activation (human cell line-HeLa-9903), Standard Evaluation Procedure (SEP) Washington, DC: Environmental Protection Agency; 2009. pp. 1–23. [Google Scholar]
- 13.OECD. OECD Guidelines for the Testing of Chemicals. OECD Publishing; 2012. Test No. 457: BG1Luc Estrogen Receptor Transactivation Test Method for Identifying Estrogen Receptor Agonists and Antagonists. [Google Scholar]
- 14.LeBlanc G, Norris D, Kloas W, Kullman S, Baldwin W, Greally J. Detailed review paper state of the science on novel in vitro and in vivo screening and testing methods and endpoints for evaluating endocrine disruptors. Series on testing and assessment No. 178. 2012:1–213. [Google Scholar]
- 15.Bardin A, Boulle N, Lazennec G, Vignon F, Pujol P. Loss of ER beta expression as a common step in estrogen-dependent tumor progression. Endocr-Related Canc. 2004;11:537–551. doi: 10.1677/erc.1.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandenberger AW, Tee MK, Jaffe RB. Estrogen receptor alpha (ER-alpha) and beta (ER-beta) mRNAs in normal ovary, ovarian serous cystadenocarcinoma and ovarian cancer cell lines: Down-regulation of ER-beta in neoplastic tissues. J Clin Endocrinol Metab. 1998;83:1025–1028. doi: 10.1210/jcem.83.3.4788. [DOI] [PubMed] [Google Scholar]
- 17.Leitman DC, Paruthiyil S, Vivar OI, Saunier EF, Herber CB, Cohen I, Tagliaferri M, Speed TP. Regulation of specific target genes and biological responses by estrogen receptor subtype agonists. Curr Opin Pharmacol. 2010;10:629–636. doi: 10.1016/j.coph.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington WR, Sheng SB, Barnett DH, Petz LN, Katzenellenbogen JA, Katzenellenbogen BS. Activities of estrogen receptor alpha- and beta-selective ligands at diverse estrogen responsive gene sites mediating transactivation or transrepression. Mol Cell Endocrinol. 2003;206:13–22. doi: 10.1016/s0303-7207(03)00255-7. [DOI] [PubMed] [Google Scholar]
- 1.Escande A, Pillon A, Servant N, Cravedi JP, Larrea F, Muhn P, Nicolas JC, Cavailles V, Balaguer P. Evaluation of ligand selectivity using reporter cell lines stably expressing estrogen receptor alpha or beta. Biochem Pharmacol. 2006;71:1459–1469. doi: 10.1016/j.bcp.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Kuiper G, Carlsson B, Grandien K, Enmark E, Haggblad J, Nilsson S, Gustafsson JA. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors alpha and beta. Endocrinol. 1997;138:863–870. doi: 10.1210/endo.138.3.4979. [DOI] [PubMed] [Google Scholar]
- 21.Harris DM, Besselink E, Henning SM, Go VLW, Heber D. Phytoestrogens induce differential estrogen receptor alpha- or beta-mediated responses in transfected breast cancer cells. Exp Biol Med. 2005;230:558–568. doi: 10.1177/153537020523000807. [DOI] [PubMed] [Google Scholar]
- 22.Liu MM, Albanese C, Anderson CM, Hilty K, Webb P, Uht RM, Price RH, Pestell RG, Kushner PJ. Opposing action of estrogen receptors alpha and beta on cyclin D1 gene expression. J Biol Chem. 2002;277:24353–24360. doi: 10.1074/jbc.M201829200. [DOI] [PubMed] [Google Scholar]
- 23.Hall JM, McDonnell DP. The estrogen receptor beta-isoform (ERbeta) of the human estrogen receptor modulates ERalpha transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinol. 1999;140:5566–5578. doi: 10.1210/endo.140.12.7179. [DOI] [PubMed] [Google Scholar]
- 24.Soshilov A, Denison MS. Role of the Per/Arnt/Sim domains in ligand-dependent transformation of the aryl hydrocarbon receptor. J Biol Chem. 2008;283:32995–33005. doi: 10.1074/jbc.M802414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan JC, Denison MS, Holstege DM, Magiatis P, Dallas JL, Gutierrez EG, Soshilov AA, Millam JR. 2,3-cis-2R, 3R-(−)- epiafzelechin-3-O-p-coumarate, a novel flavan-3-ol isolated from Fallopia convolvulus seed, is an estrogen receptor agonist in human cell lines. BMC Comp Alt Med. 2013;13:14. doi: 10.1186/1472-6882-13-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Baston DS, Denison MS. Considerations for potency equivalent calculations in the Ah receptor-based CALUX bioassay: normalization of superinduction results for improved sample potency estimation. Talanta. 2011;83:1415–1421. doi: 10.1016/j.talanta.2010.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morisseau C, Merzlikin O, Lin A, He GC, Feng W, Padilla I, Denison MS, Pessah IN, Hammock BD. Toxicology in the fast Lane: application of high-throughput bioassays to detect modulation of key enzymes and receptors. Environ Health Perspect. 2009;117:1867–1872. doi: 10.1289/ehp.0900834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hevir N, Trošt N, Debeljak N, Rižner TL. Expression of estrogen and progesterone receptors and estrogen metabolizing enzymes in different breast cancer cell lines. Chem Biol Interact. 2011;191:206–216. doi: 10.1016/j.cbi.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 30.Evers NM, van den Berg JH, Wang S, Melchers D, Houtman R, de Haan LH, Ederveen AG, Groten JP, Rietjens IM. Cell proliferation and modulation of interaction of estrogen receptors with coregulators induced by ERα and ERβ agonists. J Steroid Biochem Mol Biol. 2014;143:376–385. doi: 10.1016/j.jsbmb.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Kim SH, Tamrazi A, Carlson KE, Katzenellenbogen JA. A proteomic microarray approach for exploring ligand-initiated nuclear hormone receptor pharmacology, receptor selectivity, and heterodimer functionality. Mol Cell Proteomics. 2005;4:267–277. doi: 10.1074/mcp.M400192-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Bossard C, Busson M, Vindrieux D, Gaudin F, Machelon V, Brigitte M, Jacquard C, Pillon A, Balaguer P, Balabanian K, Lazennec G. Potential role of estrogen receptor beta as a tumor suppressor of epithelial ovarian cancer. PLoS One. 2012;7:e44787. doi: 10.1371/journal.pone.0044787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodges-Gallagher L, Valentine CD, El Bader S, Kushner PJ. Estrogen receptor beta increases the efficacy of antiestrogens by effects on apoptosis and cell cycling in breast cancer cells. Breast Can Res Treat. 2008;109:241–250. doi: 10.1007/s10549-007-9640-6. [DOI] [PubMed] [Google Scholar]
- 34.Strom A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA. Estrogen receptor beta inhibits 17 beta-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Nat Acad Sci USA. 2004;101:1566–1571. doi: 10.1073/pnas.0308319100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: How do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- 36.Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA. A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene. 2008;27:1019–1032. doi: 10.1038/sj.onc.1210712. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Arao Y, Hall JM, Burkett S, Liu L, Gerrish K, Cavailles V, Korach KS. Research resource: STR DNA profile and gene expression comparisons of human BG-1 cells and a BG-1/MCF-7 clonal variant. Mol Endocrinol. 2014 doi: 10.1210/me.2014-1229. me20141229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badraoui R, Abdelmoula NB, Sahnoun Z, Fakhfakh Z, Rebai T. Effect of subchronic exposure to tetradifon on bone remodelling and metabolism in female rat. Comptes Rendus Biologies. 2007;330:897–904. doi: 10.1016/j.crvi.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 39.Badraoui R, Abdelmoula NB, Feki N, Ben Nasr H, Rebai T. Endocrine disruption and ovarian morphometric responses in rats following exposure to tetradifon. Gen Comp Endocrinol. 2010;166:268–272. doi: 10.1016/j.ygcen.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 40.Briz V, Molina-Molina JM, Sanchez-Redondo S, Fernandez MF, Grimalt JO, Olea N, Rodriguez-Farre E, Sunol C. Differential estrogenic effects of the persistent organochlorine pesticides dieldrin, endosulfan, and lindane in primary neuronal cultures. Toxicol Sci. 2011;120:413–427. doi: 10.1093/toxsci/kfr019. [DOI] [PubMed] [Google Scholar]
- 41.Grunfeld HT, Bonefeld-Jorgensen EC. Effect of in vitro estrogenic pesticides on human oestrogen receptor alpha and beta mRNA levels. Toxicol Lett. 2004;151:467–480. doi: 10.1016/j.toxlet.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 42.Fang H, Tong WD, Shi LM, Blair R, Perkins R, Branham W, Hass BS, Xie Q, Dial SL, Moland CL, Sheehan DM. Structure-activity relationships for a large diverse set of natural, synthetic, and environmental estrogens. Chem Res Toxicol. 2001;14:280–294. doi: 10.1021/tx000208y. [DOI] [PubMed] [Google Scholar]
- 43.Gao H, Williams C, Labute P, Bajorath J. Binary quantitative structure-activity relationship (QSAR) analysis of estrogen receptor ligands. J Chem Inform Comp Sci. 1999;39:164–168. doi: 10.1021/ci980140g. [DOI] [PubMed] [Google Scholar]
- 44.Gao H, Katzenellenbogen JA, Garg R, Hansch C. Comparative QSAR analysis of estrogen receptor ligands. Chem Rev. 1999;99:723–744. doi: 10.1021/cr980018g. [DOI] [PubMed] [Google Scholar]
- 45.Chesworth R, Wessel MD, Heyden L, Mangano FM, Zawistoski M, Gegnas L, Galluzzo D, Lefker B, Cameron KO, Tickner J, Lu BH, Castleberry TA, Petersen DN, Brault A, Perry P, Ng O, Owen TA, Pan L, Ke HZ, Brown TA, Thompson DD, DaSilva-Jardine P. Estrogen receptor beta selective ligands: Discovery and SAR of novel heterocyclic ligands. Bioorg Med Chem Lett. 2005;15:5562–5566. doi: 10.1016/j.bmcl.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Deng CL, Chen XX, Lu HY, Yang X, Luan F, Cordeiro M. Prediction of the estrogen receptor binding affinity for both hER(alpha) and hER(beta) by QSAR approaches. Lett Drug Design Discov. 2014;11:265–278. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.