Abstract
Reactive oxygen species (ROS) are a family of molecules that are continuously produced from oxygen consumption in aerobic cells. Controlled generation of ROS in normal cells serves useful purposes to regulate important cellular processes such as cell proliferation, inflammation, and immune response, but overproduction of ROS causes oxidative stress that contributes to the development of cancer, chronic disease, and aging. These hugely different consequences of ROS exposure demand a carefully balanced control of ROS production and disposition, which is largely achieved through the body’s elaborate antioxidant system. The human antioxidant system consists of small antioxidants, antioxidant proteins, ROS-metabolizing enzymes, as well as many regulator proteins that mediate adaptive responses to oxidant stress. How such a complex system reacts with oxidants and achieves the required specificity and sensitivity for proper anti-oxidation is incompletely understood. In this respect, new advances in the understanding of the chemistry that determines the reaction of a given oxidant or antioxidant with a protein target provide considerable insights into these and related questions. The findings hold certain promise for new drug development for preventing and treating diseases associated with oxidant tissue damage.
Introduction: Oxidant Stress and Anti-oxidation Are Consequences of Oxygen Utilization
Eukaryotes are constantly exposed to reactive oxygen species (ROS) as a result of internal metabolism and external exposure (Balaban et al., 2005). During the course of evolution, molecular oxygen (O2) was selected as the terminal electron acceptor to generate biologically useful energy in the form of adenosine 5′-triphosphate (ATP) from carbon fuels, which reflects an adaptation of organismal life to the rising abundance of O2 in the atmosphere as well as the favorable thermodynamic properties of O2 (Berner et al., 2007). In eukaryotes, controlled oxidation of NADH and FADH2 generates an energy potential of protons across the mitochondrial inner membrane to drive the phosphorylation of ADP to ATP and the reduction of O2 to H2O. However, the electrons may leak from the respiratory chain to directly react with O2 producing superoxide anion (O2•−) and hydrogen peroxide (H2O2), two major forms of ROS in the cell. In fact, most, if not all, enzymes that are capable of metabolizing oxygen can generate ROS, either purposely or as by-products. In addition, numerous exogenous agents, such as infectious microbes and environmental toxicants, stimulate ROS production in the body by interfering with the internal metabolism and utilization of O2 (Ma, 2010).
ROS avidly react with cellular macromolecules such as nucleic acids, proteins, and membranous lipids. Uncontrolled production of ROS results in oxidative stress that potentially causes harm to the body and is implicated in aging, tumorigenesis, chronic inflammation, neurodegeneration, and chemical toxicity (Bossy-Wetzel et al., 2004; Finkel, 2005). On the other hand, ROS function as physiologic signaling molecules in normal cells regulating critical cellular processes such as cell division, differentiation, inflammation, immune function, and stress response (Dickinson and Chang, 2011; Finkel, 2011). Therefore, it has been increasingly believed that reactive oxidants play important roles in the development of many physiological and pathophysiological outcomes. Accordingly, the production and disposition of ROS are tightly controlled in tissue, cell type, and time-specific fashions, such that cells can utilize the beneficial effects but avoid the toxicity of reactive oxidants.
The homeostasis of ROS in the body is achieved largely through anti-oxidation via an intricate antioxidant system. Human antioxidants consist of low-molecular-weight antioxidants such as reduced glutathione (GSH) and vitamins C and E; noncatalytic antioxidant proteins such as thioredoxin (Trx) and glutaredoxin (Grx); and ROS-metabolizing enzymes such as superoxide dismutase (SOD), catalase, peroxiredoxin (Prx), and glutathione peroxidase (GPx). Importantly, the antioxidants are regulated by a web of regulator proteins that mediate adaptive responses to oxidant stress and antioxidant inducers, which is best exemplified by the activation of the nuclear factor erythroid 2 (NFE2)-related factor 2 (Nrf2) to induce the expression of antioxidant response element (ARE)-controlled cytoprotective proteins including ROS-metabolizing enzymes (Kensler et al., 2007; Ma, 2013). In addition, many plant-derived chemicals (phytochemicals and derivatives) exhibit antioxidant properties and protect cells from the damaging effects of reactive oxidants by means of chemical quenching and/or activation of adaptive responses. Anti-oxidation through exogenous antioxidants by way of dietary supplements, herbal medicine, and clinical drugs has been utilized as an effective and economic measure to prevent and treat cancer, chronic disease, and aging (Ma and He, 2012; Suzuki et al., 2013a; Talalay et al., 2003).
The molecular mechanisms of how ROS elicit a specific response and by which the homeostasis of oxidants is controlled in the body remain largely unclear. In this respect, recent data obtained from molecular characterization of oxidant signaling and antioxidant-induced transcriptional regulation of oxidant defense provided new mechanistic insights into mammalian oxidant signaling and anti-oxidation (Dickinson and Chang, 2011; Ma, 2013). An emerging recurrent theme from both oxidant signaling and antioxidant response is reactive cysteine thiol-based redox reaction. In the former case, ROS induced by a proliferation signal such as the ligation of a growth factor receptor (GFR) oxidizes the thiol group of critical cysteine residues of GFR to boost GFR activity and proliferation signaling (Paulsen et al., 2012). In the latter case, an antioxidant, such as the phytochemical sulforaphane, covalently binds to selective cysteine thiols of Keap1 (Kelch-like erythroid cell-derived protein with CNC homology-associated protein 1), a repressor of Nrf2, resulting in the activation of gene transcription through Nrf2 (Ma and He, 2012; Taguchi et al., 2011). In both scenarios, the versatile and reversible reactions of reactive cysteine thiols appear to serve as the chemical basis to determine the specificity and sensitivity of these responses.
This review discusses recent progress in the understanding of the molecular mechanism that governs the interaction between reactive oxidants and mammalian cells and, in light of the apparent therapeutic and preventive potentials of anti-oxidation, will focus on the emerging role and mechanism of action of Nrf2 in the control of inducible anti-oxidation pertinent to medicine.
Biologically Relevant Reactive Oxidants
ROS
ROS encompass a family of molecules that represent key reactive oxidants in aerobic cells (Dickinson and Chang, 2011). Major biologically relevant ROS include hydroxyl radical (•OH), singlet oxygen (1O2), lipid peroxides (ROOH), ozone (O3), and hypochlorous acid (HOCl), in addition to O2•− and H2O2 described earlier. ROS are frequently generated at specific subcellular locations and associated with defined physiologic functions. Besides the mitochondrial respiratory chain, ROS are produced in phagosomes to kill pathogens and in peroxisomes for energy metabolism. Protein folding via disulfide bond formation in the endoplasmic reticulum (ER) by the glycoprotein Erol and the thioredoxin protein disulfide isomerase results in the release of H2O2 that may contribute to ER stress. A group of NADPH oxidases (Nox) localize in the plasma membrane and produce ROS to mediate redox signaling that regulates cell proliferation and immune response, and phagocytic killing to ward off invading microbes. Determining the chemical identity, subcellular localization, and local concentration of ROS is critical for elucidating the chemistry and function of ROS at a molecular level, which remains a formidable challenge for most investigators due to the lack of sensitive, specific, and easy-to-use probes and technology to detect reactive oxidants at the present time.
RNS
Reactive nitrogen species (RNS), such as nitric oxide (•NO), are another group of reactive oxidants important in human biology (Pacher et al., 2007). Nitric oxide is produced by NO synthase (NOS) and serves as a gaseous signaling molecule (gasotransmitter) for certain physiologic functions, such as blood vessel relaxation and neurotransmitter function. RNS react with ROS to form stronger oxidants, such as the potent peroxynitrite (ONOO−) that can directly react with various biological molecules. Overproduction of RNS causes nitrosative stress that often accompanies oxidative stress and contributes to the overall oxidant stress-associated tissue damage. There is also a lack of specific markers to distinguish between nitrosative and oxidative stresses in a biological sample. For these reasons, nitrosative stress is frequently discussed together with oxidative stress in the literature.
The Chemical Basis of Oxidant Signaling
Reactive cysteine thiols
Reactive cysteine residues are a small set of protein cysteines that have low pKa values in the range of 4 to 5 as a result of the influence from their surrounding amino acid microenvironment; on the contrast, most protein cysteines have pKa values of about 8.5. At pH 7.4, reactive cysteine thiols exist as thiolate anions (-S−) that are more reactive than sulfhydryl groups (-SH) toward ROS, RNS, and electrophiles (Finkel, 2011).
ROS such as H2O2 easily oxidize reactive cysteine thiols to produce cysteine thiol oxidation products in the sequence of sulfenic acid (-SOH), sulfinic acid (-SO2H), and sulfonic acid (-SO3H) (Figure 1). Nitrosation of the cysteine thiols by •NO gives rise to S-nitrosothiol (-SNO) that is predisposed to further modifications. Sulfhydration of the cysteine thiols with hydrogen sulfide (H2S), another gasotransmitter, forms sulfhydrate (-SSH). Reaction of the thiols with a protein -SH group results in the formation of inter- and intra-protein disulfides, or S-glutathiolation if the other -SH group comes from glutathione.
Figure 1.

Reactive cysteine thiol reactions. Reactive cysteine thiols exist as thiolate anions and are more reactive toward ROS, RNS, and electrophiles than sulfhydryl groups. See text for details. Srx, sulfiredoxin.
Reactive cysteine thiols are also modified by a variety of chemicals through alkylation, thiol-ether and thiol-amide bond formation, and metal chelation. Recent evidence reveals that these modifications provide an important means of sensing and distinguishing various endogenous and environmental chemicals. Pertinent to this review, many antioxidants were found to covalently bind to selective cysteine thiols of target proteins to induce adaptive responses for anti-oxidation (Ma, 2013).
Oxidant signaling through reactive cysteines
Many proteins undergo reversible cysteine thiol oxidation and the list is rapidly growing covering a wide range of biological functions. Oxidant signaling through reactive cysteine thiols is best illustrated by epidermal growth factor (EGF)-induced signaling through EGF receptor (EGFR) (Finkel, 2011; Paulsen et al., 2012). Ligand binding to EGFR induces the production of O2•− and H2O2 through membrane-bound Nox. In turn, H2O2 augments EGFR signaling by two mechanisms: (a) oxidization of the active site cysteine thiols of local protein tyrosine phosphatases (PTP) resulting in PTP inactivation and reduction of protein dephosphorylation; (b) oxidization of EGFR at Cys797 to increase the EGFR kinase activity. Both actions increase the local protein tyrosine phosphorylation near the plasma membrane and, consequently, enhance EGFR signaling.
In addition to cysteine residues, other amino acid residues, such as lysine, arginine, proline, and histidine can be oxidized at their side chains by ROS/RNS, some of which are reversible and potentially serve as signaling mechanisms for oxidants. However, examples of redox signaling via amino acids other than cysteine are currently scarce in the literature.
The Evolving Concepts of Antioxidant and Anti-oxidation
Plants and animals produce various antioxidants to cope with the damaging effects of ROS generated from oxygen consumption. The term antioxidant initially refers to a chemical that prevents the consumption of O2. The study on how vitamin E prevents lipid peroxidation led to the realization that antioxidants are reducing agents that prevent oxidative reactions by scavenging ROS. Further research revealed that many antioxidants also elicit adaptive responses such as the induction of enzymes that detoxify ROS and ROS-promoting chemicals besides directly quenching ROS. Thus, mammalian anti-oxidation is achieved through both direct (scavenging) and indirect (adaptive) modes of action of antioxidants.
The recognition that controlled production of oxidants in normal cells serves useful purposes to regulate critical cellular functions has also changed the concept of biological anti-oxidation in several ways.
First, it implies that the function of anti-oxidation in cells is not to remove ROS entirely, but instead to keep them at an optimum level. In this view, induction of adaptive anti-oxidation increases oxidant defense quickly in response to inducing signals, but induction subsides as the inducers are eliminated, avoiding sustained elevation of anti-oxidation once a homeostatic state of ROS is achieved. Therefore, adaptive (inducible) anti-oxidation is perhaps better fitted for the control of oxidant homeostasis in the body than non-inducible antioxidant mechanisms, especially when oxidant levels exceed the buffering capacity of existing antioxidants.
Second, many physiologic functions are accompanied with the production of ROS and/or RNS that, in turn, impact the functions either positively or negatively. Consistent with the notion, many antioxidants exhibit activities other than anti-oxidation such as anti-cancer, anti-viral, anti-inflammatory, and anti-immune functions, indicating that anti-oxidation generally does not occur alone. Therefore, anti-oxidation by an antioxidant should be analyzed in the context of the overall biological system and include other functions such as inflammation and immune response.
Third, anti-oxidation is boosted by many endogenous signals, which is likely through reactive cysteine thiols of target proteins. Physiologic adaptations, such as physical exercise and micronutrient control, enhance body’s anti-oxidation and, accordingly, the overall health and resistance to disease (del Valle and Hernandez, 2013; Nkenggack et al., 2013). In physical exercise, the stimulus for adaptive anti-oxidation is likely ROS themselves, because acute and chronic exercise is associated with increased production of ROS from increased muscular O2 consumption and activation of the endothelial xanthine oxidase that converts hypoxanthine and xanthine to uric acid and H2O2. In this scenario, continued presence of low concentrations of ROS induces the expression of antioxidant enzymes and other defense mechanisms. This phenomenon can be explained by the concept of hormesis in which the interaction between the stimulus and response follows a particular dose-response relationship: at high doses, ROS is inhibitory and harmful to the body, but at low doses, ROS stimulate anti-oxidation adaptation to induce beneficial effects.
The Nrf2/ARE Paradigm as a Model of Inducible Anti-oxidation
The interest on inducible anti-oxidation for human health has an origin in the observation that phenolic antioxidants, such as butylated hydroxyanisole (BHA), protected animals from benzo[a]pyrene (BaP)-induced tumor formation, which correlated with inhibition of the formation of BaP mutagenic metabolites and induction of GSTs and NQO1, drug-metabolizing enzymes that detoxify many carcinogens and toxicants (Talalay et al., 1978; 1988; Watternberg, 1985). These findings led to the concept of “chemoprevention” in which pre- or co-treatment with antioxidants prevents against chemical carcinogenesis. Induction of detoxifying enzymes has since become a mechanistic model for analyzing antioxidant biological effects. Subsequent molecular studies of the induction events revealed an inducible, xenobiotic-activated receptor (XAR)-mediated transcriptional response to small chemicals that consists of several major components (Figure 2) (Ma and He, 2012; Nguyen et al., 2003).
Figure 2.
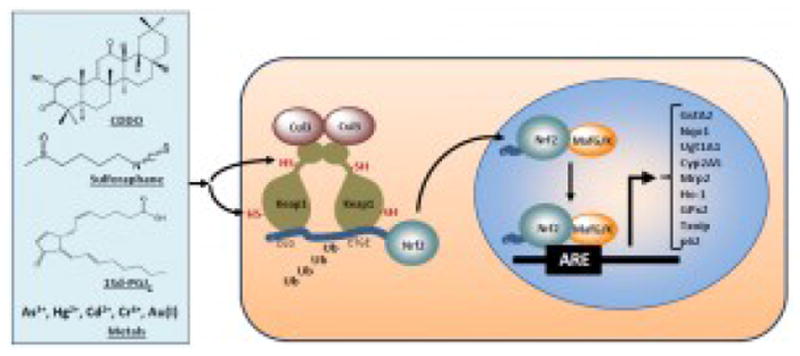
Model for activation of Nrf2 by ARE inducers. See text for details. CDDO, 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid.
Inducers
The chemicals that induce GSTs and NQO1 include phytochemicals and derivatives such as sulforaphane and triterpenoids; therapeutics such as oltipraz, auranofin, and dimethyl fumerate; environmental agents such as paraquat and arsenic; and endogenous inducers such as 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2), nitro-oleic acid, and 4-hydroxynonenal (4-HNE) (Kensler et al., 2007; Ma and He, 2012; Talalay et al., 1988). The inducers differ considerably in structure but share certain chemical property in that many inducers are electrophiles or redox-active compounds capable of modifying protein cysteine thiols by oxidation, reduction, alkylation, and metal chelation. For instance, sulforaphane and triterpenoids are Michael reaction acceptors that covalently bind to specific Keap1 cysteine thiols to activate Nrf2. Recent studies also identified a list of endogenous inducers, among which 15d-PGJ2, nitro-fatty acids, 4-HNE, acrolein, and nitric oxide directly modify the sensor motifs of Keap1. A few protein factors such as autophage cargo receptor protein p62 and cell cycle inhibitor p21 activate Nrf2 by interfering Nrf2-Keap1 binding through their structural motifs that bind Keap1.
Antioxidant response element (ARE) and ARE controlled genes
A core DNA sequence, 5′-TGACnnGC-3′ (n=any base) was identified from rat Gsta2 and later from mouse Gsta1 and rat and human NQO1 gene promoters as the antioxidant response element necessary for induction of the genes by phenolic antioxidants (Nguyen et al., 2003). The core ARE was later expanded into a 16-base-pair consensus sequence, 5′-TMAnnRTGAYnnGCR-3′ (M=A or C, R=A, Y=C or T, W=A or T) (Hayes et al., 2010). In recent years, genome-wide search for Nrf2 target genes has led to the identification of an array of ARE-regulated genes (Malhotra et al., 2010). Three notable groups of ARE-controlled genes are closely related to the cellular response to oxidants and electrophiles, including drug metabolizing enzymes and transporters such as UGT1A1, ALDH3A1, CYP2A5, and MRP2 besides GSTs and NQO1; antioxidant defense enzymes such as GPx2, Prx1, and TXNIP; and oxidant signaling molecules such as p62, DJ-1, and PTP1 (Ma, 2013). In addition, the Nrf2/ARE pathway regulates proteasomal protein degradation (Kwak et al., 2003), cell proliferation (Malhotra et al., 2010), and metabolic reprogramming (Mitsuishi et al., 2012). The presence of ARE in a broad range of genes provides a rational explanation for the multiple functions of Nrf2 and the involvement of anti-oxidation defense in various distinct physiologic and pathophysiologic processes.
Nrf2 and small Mafs
Nrf2 belongs to the cap ‘n’ collar (CNC) subfamily of basic leucine zipper transcription factors. Nrf2 was identified by way of its binding to the NFE2-binding motif, a cis-DNA sequence that controls β-globin expression critical for erythropoiesis and platelet development (Moi et al., 1994). Nrf2 does not seem to be essential for hematopoietic functions, but was instead identified as a transcription factor to mediate antioxidant induction of GSTs and NQO1 through ARE (Itoh et al., 1997). Nrf2 was later found to be involved in the cellular defense against a wide range of toxic and disease conditions that are characteristically associated with oxidative pathology (Kensler et al., 2007; Ma, 2013). Induction via Nrf2 increases the detoxification and elimination of many exogenous and some endogenous toxic chemicals including ROS. In this role, Nrf2 functions as an XAR to regulate the adaptive response to oxidants and electrophiles. Nrf2 hetero-dimerizes with a small Maf (musculoaponeurotic fibrosarcoma) protein (MafG, MafK) that has a bZip domain but lacks a transactivation domain in the nucleus (Motohashi et al., 1997). The Nrf2/small Maf dimer binds to ARE where Nrf2 binds to the ARE core and provides the transactivation activity, and MafG/K recognizes the GC dinucleotide of ARE core to increase binding affinity and specificity.
Keap1
Keap1 was identified as a cytoplasmic Nrf2-binding protein by using yeast two-hybrid screening with the Nrf2 N terminal inhibitory motif Neh2 as bait (Itoh et al., 1999). Keap1 contains two notable protein-protein interaction domains. The BTB (bric-a-brac, tramtrack, broad-complex) domain in the N terminal region mediates homodimerization of Keap1 and binding of Keap1 to Cullin (Cul) 3, a scaffold protein of the Nrf2 ubiquitin ligase (E3). The Kelch repeat (or double glycine repeat, DGR) domain in the C terminal region is responsible for binding to actin and Nrf2. An intervening region (IVR) or linker region (LR) locates between BTB and DGR and is rich in cysteine residues. Keap1 binds to the Neh2 domain of Nrf2 through its Kelch repeat to inhibit Nrf2 in the cytoplasm (Taguchi et al., 2011).
Molecular Mechanism of Chemical Sensing by Nrf2/Keap1
The mechanism by which Nrf2 is activated to mediate gene induction is two-fold: suppression of Nrf2 under a basal condition and activation of Nrf2 by an inducer.
Ubiquitination/proteasomal degradation of cytoplasmic Nrf2 through Keap1
Nrf2 mRNA is expressed broadly and independently of inducers; on the other hand, inhibition of protein synthesis completely blocks the basal and induced expression of ARE genes, suggesting a post-transcriptional regulation through protein turnover as a major mechanism of Nrf2 regulation (Ma and Kinneer De-Fede, 2001). Indeed, in unstimulated cells, Nrf2 is rapidly ubiquitinated and degraded with a half-life of ~20 minutes (He et al., 2006). Ubiquitination is mediated through the Keap1/Cul3 ubiquitin ligase complex in which Cul3 serves as a scaffold protein, Keap1 as an adaptor to bring Nrf2 into the E3 by binding to Cul3 via its BTB domain, and RING box protein 1 as a recruiter for ubiquitin-conjugating enzyme (E2) that catalyzes the polyubiquitination of Nrf2 to signal proteasomal degradation.
Structural studies reveal that Keap1 forms a homo-dimer that resembles a cherry-bob (Figure 2). Each monomer forms a large globular sphere (representing Keap1 IVR, Kelch repeats, and the C terminal end) that is linked to the other monomer through short linker arms to the sides of a small forked-stem structure (representing the dimerization interphase of two BTB domains) (Li et al., 2004; Ogura et al., 2010). The Kelch repeats fold into a drum-shaped, six-bladed β-propeller structure with an inner cavity buried in the central core and open to the top and bottom of the sphere. The overall structure of Nrf2 is not available at the present time, but the Neh2 domain was found to be intrinsically disordered with certain secondary structures. A α-helix locates at the center of rod-shaped Neh2 and contains seven lysine residues. Juxtaposed to the carboxyl end of the α-helix is a mini anti-parallel β-sheet with a conserved ETGE sequence between the two sheets. A β-hairpin structure containing a conserved DLG sequence flanks the α-helix at its amino side. Nrf2 binds to Keap1 in a 1:2 ratio through its Neh2 domain where the ETGE and DLG motifs each binds to one globular sphere of Keap1 at the binding cleft near the bottom opening of the central tunnel. The α-helix between DLG and ETGE is exposed to E2 with six lysine residues facing the enzyme for ubiquitination. The ETGE-Keap1 binding is two orders of magnitude stronger than the DLG-Keap1 binding, suggesting a hinge-and-latch-like model of binding where the Nrf2-Keap1 binding occurs first through ETGE, followed by binding at DLG.
Activation of Nrf2 through inducer-cysteine thiol binding
Many ARE inducers are electrophiles suggesting an inducer-cysteine thiol interaction as the initial chemical reaction for Nrf2 activation (Dinkova-Kostova et al., 2004). Evidence for binding of inducers to Keap1 cysteine thiols was provided using labelled inducers, stoichiometry analysis, mass spectrometry, and mutational studies (Dinkova-Kostova et al., 2001; Eggler et al., 2005; He and Ma, 2010; Hong et al., 2005; Zhang et al., 2004). Notably, the Keap1 IVR domain is rich in cysteine residues, among which Cys273, Cys288, and Cys297 were frequently labeled by inducers. Reconstitution in Keap1 knockout (KO) mice revealed that Cys151 is required for activation of Nrf2 by electrophiles, and Cys273 and Cys288 for suppression of Nrf2 under a basal condition (Yamamoto et al., 2008). Cumulative evidence indicates that inducers bind to Keap1 and/or Nrf2 through distinctive sets of cysteine residues, coined “cysteine codes,” to activate Nrf2. For instance, Keap1 Cys288 is a sensor for alkenals such as 4-HNE, and Cys151 together with a cluster of neighboring basic amino acids, namely His129, Lys131, Arg135, Lys150, and His154, forms an NO sensor (McMahon et al., 2010). In contrast to Keap1 that has 25 or more cysteine residues, Nrf2 has only 6 (human) or 7 (mouse and rat) cysteine residues that are highly conserved and were shown to have multiple effects on Nrf2 activation and function (He and Ma, 2009). The structural basis for the selectivity of inducer-cysteine code interaction remains to be elucidated for most inducers.
Alternative mechanisms of Nrf2 activation
Activation of Nrf2 by some inducers involves mechanisms other than inducer-cysteine thiol binding. Some modifications serve as part of the signaling mechanism triggered by inducer-cysteine binding. For example, human Nrf2 is phosphorylated by casein kinase 2 in response to ARE inducers and the phosphorylation is necessary for nuclear translocation of Nrf2 (Apopa et al., 2008). Several protein factors including p62, p21, PGAM5, and DJ-1 can compete with Keap1-Nrf2 binding through their structural motifs that resemble the Nrf2 ETGE or DLG motif and, thereby, persistently activate Nrf2 under certain physiological and disease conditions (Chen et al., 2009; Clements et al., 2006; Komatsu et al., 2010; Lo and Hannink, 2008). The Nrf2 Neh6 region contains several serine residues that can be phosphorylated by GSK-3 to form a recognition motif for β-TrCP (β-transducin repeats-containing proteins) leading to Nrf2 ubiquitination through the β-TrCP/Cul1 E3 complex and proteasomal degradation (Rada et al., 2011).
Translational Aspects of the Nrf2/Keap1/ARE Pathway
The mechanistic elucidation of the Nrf2/Keap1 pathway for induction of ARE-controlled genes has invigorated the enthusiasm toward application of inducible anti-oxidation in human health. In supporting the notion, cumulative evidence has shown that activation of Nrf2 by either pharmacological means (treatment with inducers) or genetic manipulation (knockout of Keap1) protects animals from damages associated with oxidant stress and pathology, whereas knockout of Nrf2 markedly reduces the expression of cellular defense genes leading to increased sensitivity to a range of disease pathology including cancer, chronic obstructive pulmonary disease (COPD), and neuro-degenerative disease, as well as chemical toxicity such as acetaminophen hepatotoxicity (drug-induced liver injury, DILI) and alcohol-induced liver disease (AILD) (Kensler et al., 2007; Ma and He, 2012). For example, Nrf2 KO mice had higher incidence of BaP-induced gastric tumor and BaP-DNA adducts than wild type, whereas Nrf2 inducers (oltipraz, sulforaphane) protected mice from BaP-induced gastric tumor formation in a Nrf2-dependent manner (Fahey et al., 2002; Ramos-Gomez et al., 2003; 2001). In humans, a single nucleotide polymorphism in the NRF2 upstream promoter region (rs6721961) was associated with reduced expression of Nrf2 and increased risk of lung cancer (Suzuki et al., 2013b). On the other hand, persistently elevated expression of ARE genes in tumors was found to be advantageous for tumor cells to proliferate and resist to drug therapy (Kensler and Wakabayashi, 2010). These findings suggest that translation of mechanistic and animal studies to medicine require careful weighing of the pros and cons of Nrf2 activation.
Nrf2 in cancer
Somatic mutations in the genes of the KEAP1 gene were found in human tumors; the mutations are mostly located at the DC region followed by IVR and BTB, suggesting that the mutations affect Keap1-Nrf2 binding and/or Nrf2 ubiquitination (Takahashi et al., 2010). Similarly, mutations were found in the ETGE and DLG motifs of Nrf2 in certain cancers that disrupt Keap1-Nrf2 binding leading to stabilization of Nrf2 in tumor cells. Some tumors have increased methylation in the KEAP1 gene that suppresses Keap1 expression and activates Nrf2 (Wang et al., 2008). In certain renal tumors, fumarate hydrotase inactivation leads to accumulation of fumarate that modifies Keap1 cysteine thiols to activate Nrf2 (Adam et al., 2011). In these cases, the tumors have elevated Nrf2 levels and increased expression of ARE genes. Elevated Nrf2 function is advantageous for tumor cells in at least three ways: (a) boosting tumor resistance to anti-cancer drugs and endogenous tumor killing chemicals; (b) enhancing proliferation by reducing ROS associated with cell proliferation and by promoting metabolic reprogramming of tumor cells; and (c) increasing Notch1 expression.
Targeting Nrf2 for drug development
Because Nrf2 is implicated in an increasing list of disease processes, there are substantial interests in developing Nrf2 activators as therapeutic drugs. A series of triterpenoids were derived from oleanolic acid, a phytoantioxidant, and were shown to be among the most potent inducers of Nrf2 genes (Liby et al., 2007). For comparison, the induction potency for NQO1 expression is 2 nM for CDDO-Im (an imidazolide of triterpenoid), 100 nM for sulforaphane, 10 mM for oltipraz, and 45 mM for butylated hydroxytoluene. Triterpenoid inducers have been shown effective in protection against cancer and various chronic diseases. Bardoxolone methyl (CDDO-Me) was effective for treating chronic kidney disease associated with type 2 diabetes, but was withdrawn from a phase III clinical trials due to adverse events (Pergola et al., 2011). In another example, dimethyl fumarate, a Michael reaction acceptor and ARE inducer, is safe and highly efficacious for the treatment of multiple sclerosis. BF-12 (Tecfidera), an oral drug containing dimethyl fumarate, was recently approved by FDA for treatment for multiple sclerosis. These examples illustrate the increasing demand for, in addition to high efficacy, a good safety profile for new Nrf2 activators. Because tumor cells can hijack the protective functions of Nrf2 by persistently activating Nrf2 genes, there is also an interest in developing Nrf2 inhibitors for the treatment of cancers that exhibit elevated Nrf2 functions. Specific and efficacious Nrf2 inhibitors for drug development are currently lacking.
Potential drug targets associated with Nrf2 in chronic diseases
Nrf2 was associated with chronic disease through molecules that are likely important for disease, providing new therapeutic targets for treating chronic disease. For example, a recent study implicates Nrf2 in the resistance to severe malaria among carriers of the Sickle cell anemia mutation who are infected with malaria (Ferreira et al., 2011). In this scenario, the carriers have elevated levels of free heme in the blood. Free heme is converted by heme oxygenase-1 (HO-1) to iron that binds to ferritin and the antioxidant molecules carbon monoxide (CO) and biliverdin. CO and biliverdin prevent oxidative tissue damage and inhibit pathogenic CD8+ T cell immune responses, both of which are associated with severe malaria. Because Nrf2 is a major regulator of HO-1 induction, modulating Nrf2 function potentially impacts the resistance to malaria through induction of HO-1 expression. As discussed earlier, some protein factors such as p62 in autophage, p21 for cell cycle inhibition, and DJ-1 associated with Parkinson’s disease activate Nrf2 by interfering with Keap1-Nrf2 binding, whereas the ER stress protein PERK activates Nrf2 by phosphorylating Nrf2 at Thr80. The levels of these proteins are elevated in certain chronic diseases or disease processes. Therefore, it is tempting to speculate that modulating the levels of these proteins and/or the Nrf2/ARE pathway may influence the development of the disease.
Conclusion
There is growing appreciation of biological anti-oxidation as it controls the homeostasis of reactive oxidants in normal cells and copes with oxidant stress associated with cancer and chronic disease. The molecular mechanism of mammalian anti-oxidation is not well understood. Some recent studies provided new insights into the interaction between an oxidant or an inducing signal and a protein target, as demonstrated for oxidant signaling and adaptive anti-oxidation. In both scenarios, the reaction of the oxidant/inducer with a reactive cysteine thiol of the target protein triggers the molecular cascade for downstream functions. Inducible anti-oxidation is best exemplified by antioxidant activation of the Nrf2/Keap1 pathway for induction of ARE-controlled cytoprotective genes. Mechanistically, Nrf2 is suppressed under basal conditions through Keap1-controlled ubiquitination-proteasomal degradation, and is activated by oxidants and electrophilic inducers via modification of the critical cysteine thiols of Keap1 and Nrf2 to induce gene expression for anti-oxidation. The potentials of the Nrf2 pathway for drug development and medicine are intriguing as animal studies have provided a large body of evidence supporting that activation of Nrf2 protects against a variety of diseases. On the other hand, sustained activation of Nrf2 has been found to link to increased tumor growth and resistance to chemotherapy. Certain potent Nrf2 activators exhibited multiple effects, some of which induce undesirable events. A careful balance of both the protective and deleterious effects of Nrf2 activation is warranted in future translation of the Nrf2 pathway for human health.
Footnotes
Disclosure
The author reports no conflicts of interest. The findings and conclusions presented in this review are those of the author and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
References
- Adam J, Hatipoglu E, O’flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, Stevens M, Fischer R, Carmeliet P, Maxwell PH, Pugh CW, Frizzell N, Soga T, Kessler BM, El-Bahrawy M, Ratcliffe PJ, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer cell. 2011;20(4):524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apopa PL, He X, Ma Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J Biochem Mol Toxicol. 2008;22(1):63–76. doi: 10.1002/jbt.20212. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120(4):483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Berner RA, Vandenbrooks JM, Ward PD. Evolution. Oxygen and evolution. Science. 2007;316(5824):557–558. doi: 10.1126/science.1140273. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat Med. 2004;10(Suppl):S2–S9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- Chen W, Sun Z, Wang XJ, Jiang T, Huang Z, Fang D, Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34(6):663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements CM, Mcnally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Valle LG, Hernandez RG. Physical activity as antioxidant and palliative beneficial option in human immunodeficiency virus infection. Oxid Antioxid Med Sci. 2013;2(4):231–243. [Google Scholar]
- Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol. 2011;7(8):504–511. doi: 10.1038/nchembio.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Fahey JW, Talalay P. Chemical structures of inducers of nicotinamide quinone oxidoreductase 1 (NQO1) Methods Enzymol. 2004;382:423–448. doi: 10.1016/S0076-6879(04)82023-8. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P. Potency of Michael reaction acceptors as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc Natl Acad Sci U S A. 2001;98(6):3404–3409. doi: 10.1073/pnas.051632198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggler AL, Liu G, Pezzuto JM, Van Breemen RB, Mesecar AD. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc Natl Acad Sci U S A. 2005;102(29):10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci U S A. 2002;99(11):7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A, Marguti I, Bechmann I, Jeney V, Chora A, Palha NR, Rebelo S, Henri A, Beuzard Y, Soares MP. Sickle hemoglobin confers tolerance to Plasmodium infection. Cell. 2011;145(3):398–409. doi: 10.1016/j.cell.2011.03.049. [DOI] [PubMed] [Google Scholar]
- Finkel T. Radical medicine: treating ageing to cure disease. Nature reviews. Mol Cell Biol. 2005;6(12):971–976. doi: 10.1038/nrm1763. [DOI] [PubMed] [Google Scholar]
- Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194(1):7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Mcmahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal. 2010;13(11):1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- He X, Chen MG, Lin GX, Ma Q. Arsenic induces NAD(P)H-quinone oxidoreductase I by disrupting the Nrf2-Keap1-Cul3 complex and recruiting Nrf2-Maf to the antioxidant response element enhancer. J Biol Chem. 2006;281(33):23620–23631. doi: 10.1074/jbc.M604120200. [DOI] [PubMed] [Google Scholar]
- He X, Ma Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol Pharmacol. 2009;76(6):1265–1278. doi: 10.1124/mol.109.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Ma Q. Critical cysteine residues of Kelch-like ECH-associated protein 1 in arsenic sensing and suppression of nuclear factor erythroid 2-related factor 2. J Pharmacol Exp Therapeut. 2010;332(1):66–75. doi: 10.1124/jpet.109.160465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280(36):31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236(2):313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Develop. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31(1):90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278(10):8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279(52):54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat Rev Cancer. 2007;7(5):357–369. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res. 2008;314(8):1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Transcriptional responses to oxidative stress: pathological and toxicological implications. Pharmacol Ther. 2010;125(3):376–393. doi: 10.1016/j.pharmthera.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, He X. Molecular basis of electrophilic and oxidative defense: promises and perils of Nrf2. Pharmacol Rev. 2012 doi: 10.1124/pr.110.004333. sumitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Kinneer De-Fede K. A labile factor regulates induction of NQOR by TCDD and phenolic antioxidants. Toxicologist. 2001;60(1):364. [Google Scholar]
- Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38(17):5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci U S A. 2010;107(44):18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22(1):66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic Acids Res. 1997;25(15):2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu Rev Pharmacol Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- Nkenggack GN, Ngogang J, Englert H. Effects of “5 a day” fruit and vegetable intake on micronutrient level and oxidative stress markers in HIV positive patients: a cluster randomized trial. Oxid Antioxid Med Sci. 2013;2(4):275–284. [Google Scholar]
- Ogura T, Tong KI, Mio K, Maruyama Y, Kurokawa H, Sato C, Yamamoto M. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci U S A. 2010;107(7):2842–2847. doi: 10.1073/pnas.0914036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87(1):315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2012;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergola PE, Raskin P, Toto RD, Meyer CJ, Huff JW, Grossman EB, Krauth M, Ruiz S, Audhya P, Christ-Schmidt H, Wittes J, Warnock DG, Investigators BS. Bardoxolone methyl and kidney function in CKD with type 2 diabetes. N Engl J Med. 2011;365(4):327–336. doi: 10.1056/NEJMoa1105351. [DOI] [PubMed] [Google Scholar]
- Rada P, Rojo AI, Chowdhry S, Mcmahon M, Hayes JD, Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol Cell Biol. 2011;31(6):1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomez M, Dolan PM, Itoh K, Yamamoto M, Kensler TW. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24(3):461–467. doi: 10.1093/carcin/24.3.461. [DOI] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98(6):3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Motohashi H, Yamamoto M. Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol Sci. 2013a;34(6):340–346. doi: 10.1016/j.tips.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Shibata T, Takaya K, Shiraishi K, Kohno T, Kunitoh H, Tsuta K, Furuta K, Goto K, Hosoda F, Sakamoto H, Motohashi H, Yamamoto M. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Mol Cell Biol. 2013b;33(12):2402–2412. doi: 10.1128/MCB.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Sonobe M, Menju T, Nakayama E, Mino N, Iwakiri S, Nagai S, Sato K, Miyahara R, Okubo K, Hirata T, Date H, Wada H. Mutations in Keap1 are a potential prognostic factor in resected non-small cell lung cancer. J Surg Oncol. 2010;101(6):500–506. doi: 10.1002/jso.21520. [DOI] [PubMed] [Google Scholar]
- Talalay P, Batzinger RP, Benson AM, Bueding E, Cha YN. Biochemical studies of glutathione S-transferases in respnose to anticarcinogenic antioxidants, Cloning and measurement of messenger RNA. J Biol Chem. 1978;258:2052–2062. [PubMed] [Google Scholar]
- Talalay P, De Long MJ, Prochaska HJ. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc Natl Acad Sci U S A. 1988;85(21):8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talalay P, Dinkova-Kostova AT, Holtzclaw WD. Importance of phase 2 gene regulation in protection against electrophile and reactive oxygen toxicity and carcinogenesis. Adv Enzyme Regul. 2003;43:121–134. doi: 10.1016/s0065-2571(02)00038-9. [DOI] [PubMed] [Google Scholar]
- Wang R, An J, Ji F, Jiao H, Sun H, Zhou D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem Biophys Res Commun. 2008;373(1):151–154. doi: 10.1016/j.bbrc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Watternberg LW. Chemoprevention of cancer. Cancer Res. 1985;45:1–8. [PubMed] [Google Scholar]
- Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28(8):2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24(24):10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


