Abstract
Inflammation is an important part of the innate immune response and is involved in the healing of many disease processes; however, chronic inflammation is a harmful component of many diseases. The regulatory mechanisms of inflammation are incompletely understood. One possible regulatory mechanism is the endocannabinoid system. Endocannabinoids such as 2-arachidonoylglycerol (2-AG) and anandamide (AEA) are generally anti-inflammatory via engagement of the cannabinoid receptor 2 (CB2) on innate cells; therefore, preventing the degradation of endocannabinoids by specific serine hydrolases such as fatty acid amide hydrolase (FAAH), monoacylglycerol lipase (MAGL), and carboxylesterases (CES) might decrease inflammation. We hypothesized that the activities of these catabolic enzymes would decrease with a subsequent increase in 2-AG and AEA in a model of inflammation. Mice were injected with lipopolysaccharide (LPS) for 6 or 24 hr, and inflammation was confirmed by an increase in interleukin-6 (il6) and il17 gene expression. Activity-based protein profiling (ABPP) of serine hydrolases showed no significant difference in various serine hydrolase activities in brain or liver, whereas a modest decrease in Ces activity in spleen after LPS administration was noted. 2-AG hydrolase activity in the spleen was also decreased at 6 hr post LPS, which was corroborated by LPS treatment of splenocytes ex vivo. ABPP-MudPIT proteomic analysis suggested that the decreased 2-AG hydrolysis in spleen was due to a reduction in Ces2g activity. These studies suggest that the endocannabinoid system could be activated via suppression of a 2-AG catabolic enzyme in response to inflammatory stimuli as one mechanism to limit inflammation.
Keywords: inflammation, endocannabinoid, carboxylesterase, 2-arachidonylglycerol, activity-based protein profiling
Introduction
Immune cells are recruited to tissues during acute inflammation to remove the inflammatory stimuli, clear cellular debris, and initiate healing. On the other hand, chronic inflammation is implicated as a contributor to several diseases, including autoimmune diseases, atherosclerosis, heart disease, metabolic disease, and cancer. Thus, regulatory mechanisms exist to control and/or resolve inflammation to prevent further tissue damage from pro-inflammatory mediators, such as cytokines, chemokines, reactive oxygen or nitrogen species, and proteolytic enzymes.
Cannabinoids are plant-derived compounds from Cannabis sativa that possess anti-inflammatory effects, in part via engagement of CB2 receptors on innate cells 1. Endocannabinoids, such as anandamide (AEA) and 2-arachidonlyglycerol (2-AG), also exhibit anti-inflammatory effects 2–7. This suggests that increasing endocannabinoid levels might be one mechanism by which inflammation is regulated in vivo. In fact, many studies have demonstrated that inhibition of endocannabinoid degradative enzymes, such as monoacylglycerol lipase (MAGL) or fatty acid amide hydrolase (FAAH), resulted in attenuated inflammatory responses 8–10. For instance, the MAGL inhibitor JZL184 attenuated acute lung injury in response to intranasal LPS 9, reduced carrageenan-induced inflammatory pain 10, and inhibited trinitrobenzene sulfonic acid-induced colitis 8.
Carboxylesterases are promiscuous hydrolytic enzymes of the serine hydrolase family that have been shown to hydrolyze the endocannabinoid 2-AG 11, 12. In fact, recombinant human CES1 has the same catalytic efficiency as human MAGL when it comes to the hydrolysis of 2-AG 12. On the basis of amino acid homology, the CES family of enzymes is classified into five groups in mammals (CES 1–5), with the majority of isoforms in the CES1 and CES2 groups 13, 14. There are eight Ces1 genes and eight Ces2 genes encoded in the murine genome compared to just one human CES1 gene and one human CES2 gene. Further, the individual murine Ces1 and Ces2 genes within their group are highly redundant in terms of sequence homology, due to multiple gene duplication events that have occurred throughout the evolutionary history of the mouse 13.
The goal of the current work was to determine whether the activities of endocannabinoid catabolic enzymes are attenuated in response to an inflammatory stimulus as one mechanism by which inflammation is limited and/or resolved. Specifically, we hypothesized that LPS would simultaneously induce pro-inflammatory mediators and suppress activity of endocannabinoid degradative enzymes in brain, liver, and spleen following a single intraperitoneal injection. Upon verification that pro-inflammatory cytokines were induced in spleen and liver, gel-based activity-based protein profiling (ABPP) of serine hydrolases was performed on all tissues at 6 and 24 hr post LPS injection. In addition, mass spectrometry-based ABPP of spleen serine hydrolases indicated that Ces2g activity was selectively down regulated and 2-AG hydrolytic activity in spleen was concomitantly decreased early following LPS injection. We then verified that Ces2g is a bona fide 2-AG hydrolase by overexpressing recombinant murine Ces2g in COS7 cells and assessing its ability to metabolize 2-AG. Together, these results demonstrate that LPS, in addition to inducing pro-inflammatory mediators, also inhibits endocannabinoid metabolizing enzyme activity, suggesting a possible negative feedback mechanism to control inflammation.
Materials and Methods
Chemicals and reagents
The activity-based serine hydrolase probe, fluorophosphonate-biotin (FP-biotin), was from Toronto Research Chemicals (North York, ON). β-mercaptoethanol, penicillin, streptomycin, p-Nitrophenyl valerate (pNPV), streptavidin–agarose beads, avidin–horseradish peroxidase and all buffer components were from Sigma (St. Louis, MO). Authentic standards of 2-AG, AEA, arachidonic acid (AA), and AA-d8 were from Cayman Chemical (Ann Arbor, MI). O,O′-Diethyl p-nitrophenyl phosphate (paraoxon, PO) was a gift from H. Chambers (Mississippi State University). High-performance liquid chromatography (HPLC) grade solvents were from Thermo-Fisher. Reagents for ABPP-MudPIT were purchased from sources described in Wang, et, al,.11
Mice
C57BL/6 female mice were obtained from Harlan Laboratories (Indianapolis, IN). Mice were randomly assigned to cages and housed at 5/cage. Mice were acclimated for two weeks prior to use in experiments. Animal rooms were kept at 20–25°C and 40–60% humidity with a 12-hr light cycle. All protocols were conducted in accordance with the Mississippi State University Institutional Animal Care and Use Committee in an AAALAC-approved facility.
Treatment
Mice (N = 5/group) received either saline or LPS (25 μg/mouse delivered in 0.5 ml saline i.p.). Mice were euthanized at 6 or 24 hr post saline or LPS injection and spleen, liver, and brain were harvested. The spleen was divided: half was flash frozen, and the other half was used for immune function and/or mRNA analysis. Liver was also divided: the left cranial liver lobe was isolated for mRNA analysis and the remaining lobes were flash-frozen. The brain was removed and dissected on a stainless steel metal plate placed on dry ice. The brain was dissected into left and right hemispheres (excluding the optic chiasm, cerebellum, and olfactory bulb), and each portion was flash-frozen in liquid nitrogen. All frozen tissue and mRNA samples were stored at −80°C until sample preparation.
Splenocyte culture
Splenocytes were prepared in a single cell suspension by mechanical disruption. Cells were seeded at 2 × 107 cells/well in complete media (1X RPMI containing 1% v/v bovine calf serum, 1% w/v penicillin-streptomycin and 50 μM 2-mercaptoethanol). Splenocytes were either not treated or treated with LPS (1 μg/ml) for 3.5 hr after which cells were lysed in ice-cold 50 mM Tris-HCl (pH 7.4) buffer by sonication. The lysates were assayed for 2-AG hydrolytic activity, as described below.
RNA Isolation
Spleen and liver samples were stored in TRI reagent (Sigma Aldrich, St. Louis, MO). Samples were thawed at room temperature (RT) and then vigorously mixed with 0.2 ml bromochloropropane (BCP). After a 15-min incubation at RT, liquid phases were separated by centrifugation at 12000g for 15 min at 4°C. The aqueous phase was transferred to a fresh tube and RNA was precipitated with 0.5 ml isopropanol. Samples containing precipitated RNA were mixed and incubated for 10 min before centrifugation at 12000g for 10 min at 4°C. The RNA pellet was then subjected to further purification and DNase treatment using the SV Total RNA Isolation System (Promega, Madison, WI) according to manufacturer’s instructions.
Real-time PCR
cDNA was prepared from total RNA using a High Capacity cDNA Archive Kit (Applied Biosystems/Life Technologies, Foster City, CA). cDNA was then used in a Taqman PCR Reaction with Universal 2X Master Mix and il6 or il17 primer/probes (Applied Biosystems/Life Technologies). Fold change was calculated using the ΔΔCt method 15 using the 6-hr saline control as the group to which the other groups were compared. Water controls were included during cDNA synthesis and Taqman PCR reaction.
Preparation of Native Tissue Proteomes
For liver, ~100 mg tissue was homogenized (20% w/v) in sucrose buffer (50 mM Tris-HCl, 0.32 M sucrose, pH 7.4) using a dounce homogenizer on ice. Homogenized samples were transferred to a 1.5-ml centrifuge tube and centrifuged at full speed (16,100g) for 30 min at 4°C. The supernatant was retained and stored at −80°C until utilized further. Brain was processed the same way as liver using the entire right hemisphere. Spleen halves were homogenized in 250 μl sucrose buffer using a micro tissue homogenizer. Each sample was centrifuged at 1,000g for 30 min at 4°C to remove debris, and the supernatant was retained and stored at −80°C.
Protein Concentration Determination
Samples from the supernatants were diluted 1:30 v/v in deionized water and incubated with BCA reagent (ThermoPierce) for 30 min at 37°C. Absorbance of solutions at 560 nm was measured using a plate reader. The absorbance values were compared against a bovine serum albumin standard to determine protein concentrations.
ABPP
The native tissue proteomes for liver and brain were diluted with sucrose buffer (50 mM Tris-HCl, 0.32 M sucrose, pH 7.4) to a final protein concentration of 2 mg/ml in a 100 μl reaction volume (spleen was diluted to 0.5 mg/ml due to limited sample). FP-biotin was added to each sample (final concentration, 8 μM; 1.9% v/v DMSO) and allowed to react at room temperature for 60 min. The reaction was stopped using 20 μl of 6X SDS-PAGE loading buffer (reducing) and samples heated at 90°C for 5 min. Samples were cooled and resolved on 10% SDS-PAGE gels. Electrophoresis proceeded at 100 V for 15 min and 120 V for approximately 75 min. Following gel electrophoresis, proteins were transferred to a polyvinylidene fluoride (PVDF) membrane. Each membrane was blocked in 5% (w/v) non-fat milk for 60 min then washed with tween buffer. The membranes were incubated with avidin-peroxidase (1:2000 v/v) in 1% (w/v) non-fat milk for at least 120 min before washing the membrane with tween buffer (3 × 10 min). The membranes were then incubated with chemiluminescent substrate for 5 min and images were obtained using X–ray film for various exposure times. Negative controls included proteomes that were heat denatured at 90°C for 5 min before adding FP-biotin. In some experiments, proteomes were pre-incubated with PO (1 and 10 μM), a broad-spectrum serine hydrolase inhibitor, for 15 min at 37°C before adding FP-biotin.
Ces Western Blots
Proteomes from naïve mouse spleen were resolved by 10% SDS-PAGE. Mouse Ces1 and Ces2 proteins were detected by incubation with rabbit monoclonal anti-human CES1 (1:25,000 v/v; Abcam) and goat polyclonal anti-mouse Ces2 (1:5,000 v/v; R&D Systems), respectively, followed by incubation with HRP-conjugated goat anti-rabbit or donkey anti-goat secondary antibody. The anti-human CES1 antibody can recognize mouse Ces1 protein. Equal protein loading of the gel was verified by detecting β-actin with anti-β-actin antibody.
2-AG and AEA Hydrolysis Activity in Spleen Proteomes
Spleen proteomes were adjusted with 50 mM Tris-HCl (pH 7.4) buffer to a final concentration of 0.5 mg/ml in a 100 μl reaction volume and preincubated for 5 min at 37°C. 2-AG or AEA was added (50 μM final concentration, 1.9% v/v ethanol) and the samples were incubated for 10 min at 37°C (a preliminary experiment showed that production of AA after 10 min was in the linear range of a reaction-time course). The reactions were terminated with 200 μl of cold acetonitrile (doped with AA-d8) and placed on ice for 15 min. The samples were centrifuged at full speed (16,100g) for 10 min at 4°C and supernatants transferred to HPLC vials with reducing inserts. The samples were stored at −20°C until LC-MS/MS analysis. To account for the level of endogenous AA in homogenates, “blank” spleen samples were prepared without either exogenous 2-AG or AEA. Non-tissue proteome controls (i.e., non-enzymatic controls) were also prepared in 50 mM Tris-HCl (pH 7.4) buffer with 50 μM 2-AG or AEA. In some experiments, either PO (1 μM final concentration) or ethanol was added to spleen proteomes and incubated for 30 min at 37°C prior to the addition of 2-AG. Analysis of AA by LC-MS/MS was done as described previously 11.
ABPP-MudPIT Serine Hydrolase Profiling
Spleen proteomes were adjusted to 1 mg/ml protein concentration in 50 mM Tris-HCl (pH 7.4) buffer in a total volume of 1 ml. The samples were treated with FP-biotin (8 μM) for 60 min. Reactions were terminated by adding SDS (0.5% w/v final concentration) and heating at 90°C for 10 min. The samples were allowed to cool on ice. Meanwhile, a 3-kDa MWCO Amicon Ultra centrifugal filter in cartridge (Millipore, Billerica, MA) was set-up in 2-ml centrifuge tubes for each sample to remove excess unreacted FP-biotin. The samples were transferred to the 3-kDa MWCO filter and centrifuged at 14,000 x g for 20 min, and the filtrate was discarded. The samples were washed three times with 500 μl of 50 mM Tris-HCl (pH 7.4) buffer and centrifuged each time. After the final wash, the filter was reversed into a new tube and centrifuged at 100 x g for 2 min. The filter was rinsed three times with 100 μl of 50 mM Tris buffer and each rinse transferred to sample tube. The samples were stored at 4°C. Streptavidin-agarose beads (200 μl; Thermo) were washed in PBS and proteome samples added to the washed beads and incubated at room temperature for 3.5 h on a rotator. This is done to attach the serine hydrolases that had reacted with the activity probe FP-biotin on the streptavidin beads. After attachment, the beads are pelleted by centrifugation at 1,400 x g for 3 min and the supernatant was discarded. The beads were then washed with 0.2% SDS in PBS (1 ml) once for 10 min before being centrifuged at 1,400 x g for 3 min. The supernatant was discarded. This wash process was repeated 3 times using PBS and 3 times using water. After washing the beads, the samples were reduced in 500 μl of 6 M urea in 50 mM Tris-HCl (pH 7.4) and 10 μl of 500 mM TCEP [Tris (2-carboxyethyl) phosphine hydrochloride; final concentration of 10 mM]. Samples were incubated at 65°C for 15 min then cooled to 37°C. Fresh 500 mM IAA (iodoacetamide; 20 μl) was added to alkylate free cysteines and the samples incubated at 37°C for 30 min in the dark. Following the alkylation reaction, 950 μl of 50 mM Tris-HCl (pH 7.4) was added. Samples were centrifuged at 1,400 x g for 2 min to pellet the beads, and the supernatant was discarded. Two-hundred μl of 2 M urea in 50 mM Tris-HCl (pH 7.4) and 4 μl of trypsin (Pierce) (0.5 mg/ml) were added to the beads. Trypsin was activated with 2 μl of 100 mM CaCl2 (1 mM final concentration) and the samples were incubated overnight at 37°C with gentle mixing. The samples were transferred to a spin-desalting column (Zeba 7-kDa MWCO, Thermo) to remove the beads and collect the filtrate containing the peptides. The beads in the column were washed two times with water and the filtrates were pooled together (total volume 300 μl). Three μl of 10% v/v formic acid in water was added to the filtrates containing peptides and the filtrates were desalted with a Sep-Pak C18 cartridge (Waters Associates, Milford, MA) following the manufacturer’s instructions. The peptides were eluted in 65% aqueous acetonitrile and the solvent was evaporated completely in a Speedvac concentrator. The residues were re-suspended in 20 μl of 0.1% v/v formic acid in water for proteomic LC-MS/MS analysis as described previously 11. In brief, tryptic peptides were injected on a 75 μm (inside diameter) × 15 cm reverse phase C18 column (Thermo) controlled by an Ultimate 3000 nanoflow HPLC system (Dionex) and eluted using a water/acetonitrile (containing 0.1% formic acid) gradient at a flow rate of 0.3 μl/min. The separated peptides were introduced into an LTQ OrbiTrap Velos mass spectrometer (ThermoFisher). Normalized spectral counts for each identified serine hydrolase were determined using the commercially available Scaffold version 4.0.7 11.
Overexpression of Ces2g and Assessment of 2-AG Hydrolytic Activity
Recombinant murine Ces2g was overexpressed in COS-7 cells transfected with an expression vector containing Ces2g cDNA (Origene), as described previously 11, 12. Control transfections of COS-7 cells included an expression vector without the Ces2g cDNA insert (mock transfection). Cells were harvested 24–48 hr after transfection and lysed in 50 mM Tris-HCl (pH 7.4) buffer by sonication. The esterase activity of the lysates was assessed using pNPV, as described previously 12. 2-AG hydrolytic activity of the lysates was determined by adding exogenous 2-AG (10 μM final concentration) and incubating for 10 min at 37°C. Reactions were quenched by addition of an equal volume of cold acetonitrile doped with internal standard (AA-d8). The amount of AA produced in the reaction was determined by LC-MS/MS as previously described 11.
Statistical Analysis
The mean ± SE (or SD) was determined for each treatment group. Differences between means were determined with a two-way analysis of variance. When significant differences were detected, treatment groups within time points were compared to the 6 hr saline using Bonferroni’s test. For RT-PCR data, Grubb’s outlier test was performed for each treatment group using Delta Ct (Ct target gene − Ct18S). In addition, fold change values were transformed using natural log (fold change +1) prior to statistical analysis. Statistical analyses were performed using GraphPad Prism version 4.0a for Macintosh OSX, GraphPad software (San Diego, CA). Comparison of spectral count data from saline and LPS-treated samples following ABPP-MudPIT were assessed by Student’s t-test. Enzymatic activities of mock and Ces2g-overexpressed homogenates were assessed by Student’s t-test.
Results
LPS-induced inflammatory response
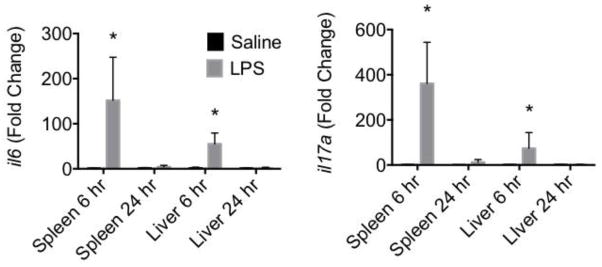
To verify that LPS induced inflammation in the mice, the levels of two pro-inflammatory cytokines, il6 and il17a, were assessed in spleen and liver (Fig. 1). Both il6 and il17a mRNA levels increased significantly in spleen and liver by 6 hr post LPS injection. By 24 hr, the mRNA levels of il6 and il17a had returned to baseline levels.
Figure 1. LPS induces inflammatory cytokine mRNA in liver and spleen.
Real time RT-PCR was performed on total RNA isolated from liver or spleen at various times following saline or LPS injection. Taqman RT-PCR was performed for il6 and il17. Fold change was calculated as compared with 6 or 24 hr saline. *p < 0.05 as compared to 6 hr saline in each tissue.
LPS-induced modulation of serine hydrolase activity
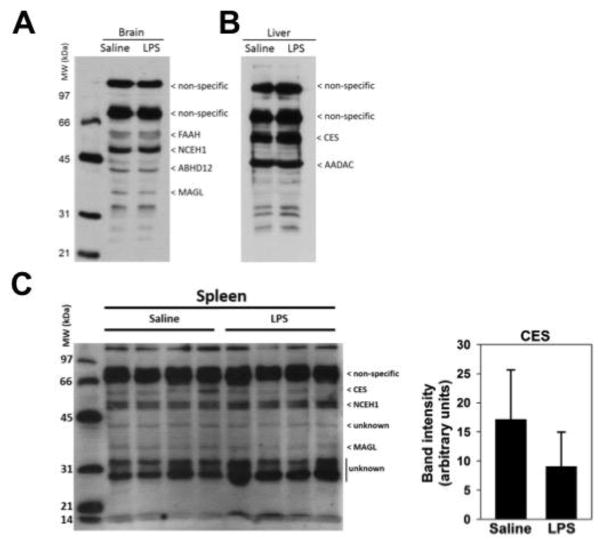
ABPP enables the activities of enzymes with conserved catalytic mechanisms, such as the serine hydrolases, to be evaluated in their native environments within tissues and cells 16. Serine hydrolase activities were assessed in brain, liver, and spleen proteomes in response to LPS using the activity probe FP-biotin and gel-based ABPP (Fig. 2). This probe reacts with most, but not all, active serine hydrolases. In brain, the serine hydrolases FAAH, NCEH1, ABHD12 and MAGL were detected, as we and others and others have previously shown 17, 18; however, none were altered by LPS treatment (Fig. 2A). In liver, Ces and AADAC activities were detected, but again, were not changed by LPS (Fig 2B). In spleen, there was a modest decrease in Ces activity in response to LPS at the 6 hr timepoint (Fig. 2C), although it did not reach statistical significance. This Ces band at 60 kDa in the gel-based ABPP is a composite of all Ces isoforms in the homogenate. No altered serine hydrolase activities were detected in any tissue at 24 hr post LPS (data not shown). Non-specific bands indicate those proteins in tissues that are known to be detected by avidin-peroxidase blotting in the absence of FP-biotin 17.
Figure 2. Profile of serine hydrolases in brain, liver, and spleen following treatment with saline or LPS.
Tissue homogenates were prepared from brain (A), liver (B) or spleen (C). Active serine hydrolases in the homogenates were detected following treatment with FP-biotin and subsequent resolution by SDS-PAGE. Probe-modified proteins were detected using streptavidin-conjugated horseradish peroxidase. Enzymes are identified based on molecular weight and previous publications 17, 18, 37. Saline- or LPS-treated tissues at the 6 hr timepoint are shown. Non-specific labels refer to proteins that are endogenously biotinylated and are detected in tissue homogenates that are not treated with FP-biotin (data not shown). Densitometry of CES bands in spleen samples are shown next to the blot (p = 0.17).
Characterization of Ces activity in spleen
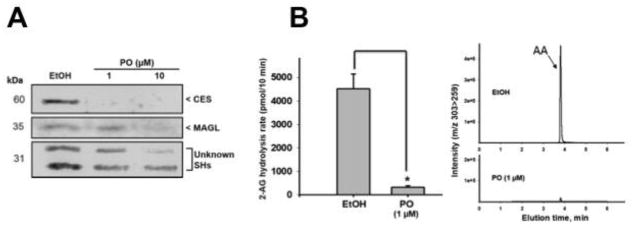
In order to verify that Ces and MAGL were active in the spleen, saline-treated spleen homogenates were incubated in the presence of PO, the bioactive metabolite of the insecticide parathion, which selectively inhibits Ces (at 1 μM) and at higher concentrations (≥ 10 μM) can also inhibit MAGL (PO is a more selective inhibitor of Ces relative to MAGL) 12, 19. As seen in Fig. 3A, pre-treatment of spleen proteomes with PO at a concentration of 1 μM resulted in the complete abrogation of Ces activity, as demonstrated by the lack of a visible band, indicating that the FP-biotin probe was unable to modify the PO-inactivated Ces proteins. On the other hand, the intensity of the band annotated as MAGL was unchanged relative to vehicle control; however, pre-treatment with a higher concentration of PO (10 μM) caused a decrease in its activity. Two serine hydrolases ~31 kDa were also inhibited by PO, particularly at 10 μM, although their identities are unknown. Moreover, using PO (1 μM) to inhibit all Ces activity, the hydrolysis of exogenous 2-AG in spleen homogenates was completely abolished (Fig. 3B), suggesting that Ces can hydrolyze 2-AG in the spleen.
Figure 3. Ces hydrolyzes 2-AG in the spleen.
(A) Saline-treated spleen homogenates (6 hr) were preincubated with ethanol (EtOH) or PO (1–10 μM) for 15 min at 37°C, followed by treatment with FP-biotin for 1 h. Ces and MAGL activity was then detected using gel-based ABPP. (B) 2-AG hydrolytic activity was measured by quantifying AA production. Saline-treated spleen homogenates (6 hr) were preincubated with ethanol (EtOH) or PO (1–10 μM) for 15 min at 37°C, followed by the addition of 2-AG (50 μM). The amount of AA produced in 10 min was measured by LC-MS/MS.
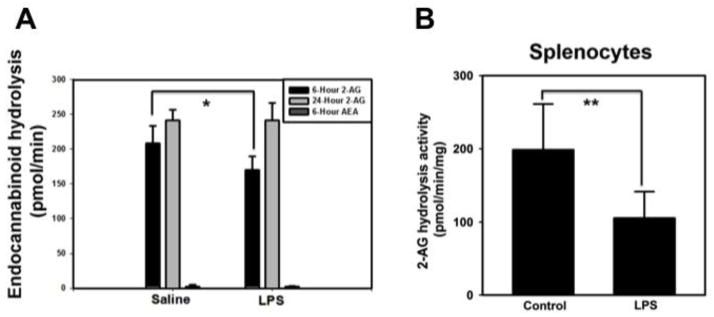
In response to LPS, there was a significant inhibition of 2-AG hydrolysis activity at 6 hr post LPS, with no effect on 2-AG hydrolysis at the 24 hr timepoint. It was also notable that the AEA hydrolytic activity in spleen was markedly lower than the 2-AG hydrolytic activity (Fig. 4A). In addition, there was no difference in AEA hydrolysis activity detected in the spleen at 6 h post LPS compared to saline control. The LPS-induced suppression of 2-AG hydrolytic activity was confirmed in vitro using splenocytes treated with LPS (Fig. 4B).
Figure 4. LPS suppresses 2-AG hydrolysis in the spleen.
(A) Saline- or LPS-treated spleen homogenates were incubated with 2-AG or AEA as substrates (50 μM) for 10 min. * p < 0.05 as compared to 6 hr saline-treated spleen. (B) Splenocytes (n=6) were treated in vitro with LPS (1 μg/ml) for 3.5 hr. 2-AG hydrolytic activity (50 μM, 10 min) of cell lysate was measured by quantifying AA production by LC-MS/MS. ** p < 0.01 as compared to control.
Identification of Ces isoforms in the spleen
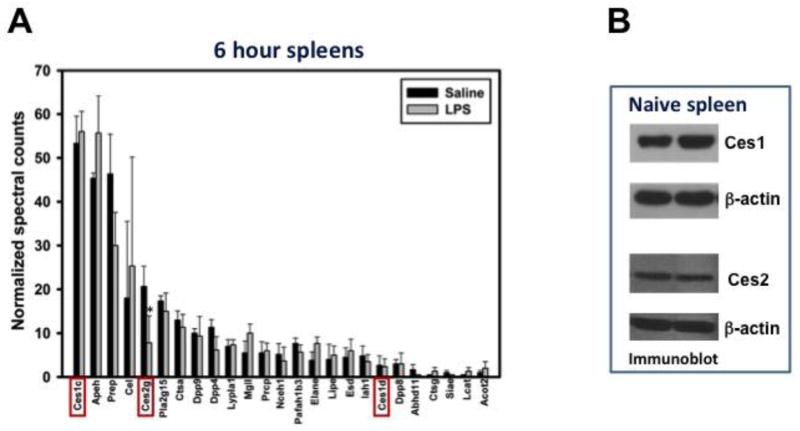
We next assessed the serine hydrolase activities in the 6 hr saline- and LPS-treated spleen samples using ABPP-MudPIT to specifically identify which serine hydrolase isoforms were modulated by LPS, because the gel-based ABPP results indicated that LPS-induced suppression of a Ces isoform was involved in the reduction in 2-AG hydrolysis by LPS (Fig. 2C). Indeed, Ces1c, Ces2g, and Ces1d were all identified in spleen with Ces1c being most abundant (Fig. 5A). Interestingly, of the three Ces isoforms, only the activity of Ces2g was suppressed by LPS (Fig. 5A), suggesting that the reduced 2-AG hydrolysis in spleen at 6 hr (Fig. 4A) was due to LPS-induced suppression of Ces2g activity specifically. It was notable that FAAH was not detected in the spleen by ABPP-MudPIT (Fig. 5A), which is consistent with the minimal AEA hydrolysis activity detected (Fig. 4A). Moreover, MAGL (Mgll) activity was not significantly altered by LPS when assessed by ABPP-MudPIT, again supporting Ces2g as the primary target of suppression by LPS contributing to reduced 2-AG hydrolysis. Immunoblots of control spleen homogenates verified that Ces1 and Ces2 isoforms were expressed (Fig. 5B), although specific subclasses of Ces isoforms, such as Ces1c and Ces1d, cannot be distinguished by immunoblotting due to the lack of specific antibodies.
Figure 5. LPS suppresses Ces2g activity in the spleen.
(A) Saline or LPS-treated spleen homogenates were subjected to ABPP-MudPIT proteomic analysis. The activity of the identified serine hydrolases was quantified at 6 hr following saline or LPS treatments. Ces isoforms are designated with red boxes. * p < 0.05 when compared to 6 hr saline-treated spleen. (B) Immunoblots of Ces1 and Ces2 in naïve spleens verified the expression of these Ces family members.
Ces2g is a 2-AG hydrolytic enzyme
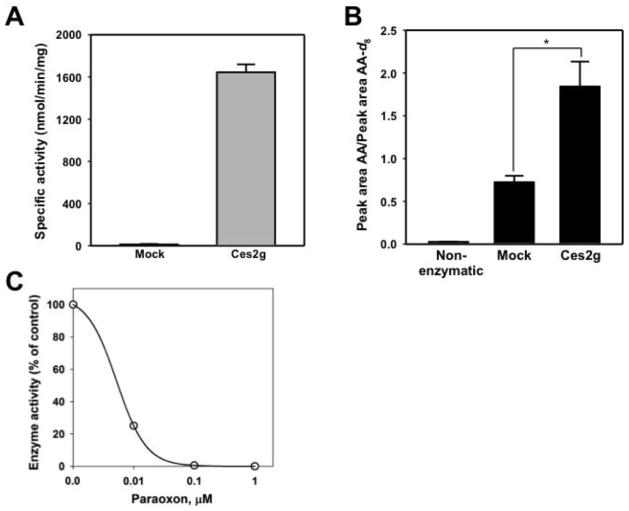
To verify that murine Ces2g is a 2-AG hydrolytic enzyme, we overexpressed it in COS7 cells following transient transfection (Fig. 6A) and showed that it could hydrolyze 2-AG to AA (Fig. 6B). Thus, Ces2g appears to be a bona fide 2-AG hydrolytic enzyme. Finally, we also verified that Ces2g is highly sensitive to the inhibitory effects of PO (Fig. 6C), as was expected for a Ces isoform 19.
Figure 6. Ces2g is a 2-AG hydrolytic enzyme.
COS-7 cells were transiently transfected with mouse Ces2g cDNA. The ability of the Ces2g-expressing COS-7 homogenates to hydrolyze p-NPV (A) and 2-AG (B) was assessed. Endocannabinoid hydrolytic activity was measured by quantifying AA production by LC-MS/MS. * p < 0.05 when compared to mock-transfected cell lysate. (C) Inhibition of Ces2g activity by PO. The enzyme was pre-incubated with the indicated concentrations of PO for 10 min (37°C) before assaying the activity of the enzyme using the substrate p-NPV. The estimated IC50 for PO is 5.2 nM. Error bars are depicted but fall within the symbol.
Discussion
The mechanisms involved in inflammation initiation are well-studied as compared to those involved in resolution. The inflammatory response we observed, as measured by il6 and il17a gene expression, in the spleen and liver in response to LPS demonstrates the classical, transient response with high expression early followed by almost complete resolution within 24 hr. The resolution of inflammation must involve more than simply clearance of LPS because in one study in which cecal ligation and puncture was utilized as a model of systemic endotoxemia, plasma IL-6 levels returned toward baseline even when plasma endotoxin levels remained high 20.
In this work, we examined the possibility that LPS not only induced inflammation but also inhibited endocannabinoid metabolizing enzymes as one mechanism to raise endocannabinoid levels as a potential negative feedback system to limit inflammation. Specifically in the spleen, Ces2g activity was suppressed by LPS. Because we also demonstrated that Ces2g was a 2-AG hydrolytic enzyme, these results show that LPS suppresses 2-AG hydrolysis in part through inhibition of Ces2g. This result is similar to reports in which FAAH activity was inhibited by LPS 21–23. Interestingly, FAAH activity was not detected in the spleen proteome by ABPP-MudPIT, nor was there any AEA hydrolysis activity detected in spleen homogenates. Thus, 2-AG is the predominant endocannabinoid in the spleen, consistent with other reports in which 2-AG was found in nmol/g range, whereas AEA was detected in pmol/g range in the spleen 24.
Ces1c was the most abundant serine hydrolase detected by ABPP-MudPIT; however, Ces1c mRNA was not detected in mouse spleen (unpublished observation and 25). On the other hand, Ces2g mRNA was detected in spleen 25, which is consistent with our ABPP-MudPIT result. The high levels of active Ces1c detected in spleen by ABPP-MudPIT likely stems from contaminating Ces1c, which is present in abundance in mouse blood 26. Spleen is a richly perfused tissue and spleen proteomes would be prone to Ces1c contamination via the blood. Ces2g, however, is not found in mouse blood.
The results demonstrate that 2-AG hydrolysis in the spleen is inhibited early following LPS administration. The LPS-induced suppression of 2-AG hydrolysis was also shown in vitro in splenocytes, suggesting that the mediator induced by LPS to suppress CES2g activity in the spleen could be found in the cells. One such mediator for inhibition of CES2g activity is IL-6. It was demonstrated in a human HepG2 liver cell line that IL-6 directly inhibited the CES2 promoter region 27. Studies are currently ongoing to determine if IL-6 is part of the mechanism by which LPS suppressed CES2g in splenocytes leading to inhibition of 2-AG hydrolysis.
The mechanism by which elevated 2-AG exhibits anti-inflammatory actions could be engagement of CB1 or CB2 on innate cells 5, 6, 28. However, it is also possible that Ces2g inhibition leading to suppression of 2-AG hydrolysis causes the elevated 2-AG to be shunted to an alternative metabolism pathway. For example, 2-AG can be metabolized by cyclooxygenase-2 (COX-2) to prostaglandin J2 glycerol esters, which are also anti-inflammatory through their interaction with PPAR-γ 29–32. Other anti-inflammatory eicosanoids can be produced following metabolism of 2-AG by COX-2, and 2-AG can also be metabolized by lipoxygenase and cytochrome P450 enzymes, which could also yield anti-inflammatory molecules 33–36.
The present studies demonstrate that inflammagens such as LPS possess pro-inflammatory actions and activate mechanisms to temper inflammatory responses simultaneously. Identification of the mediator(s) that participate in a negative feedback system to control inflammation would help guide therapy development; for instance, if IL-6 is an effective suppressor of endocannabinoid metabolizing enzymes, perhaps it is not a beneficial target of suppression for inflammatory diseases.
Highlights.
LPS inhibited Ces activity and 2-AG hydrolysis in the spleen.
Ces2g specifically was identified as one target of LPS suppression.
Ces2g is a bona fide 2-AG hydrolytic enzyme.
Acknowledgments
The authors would like to thank Dr. Mariola Edelman at Mississippi State University for help with proteomic data analysis. Drs. Russell Carr and Saphala Dhital helped with tissue harvest. Funding was provided by Mississippi State University College of Veterinary Medicine, NIH 5T35OD010432, and NIH 1R15ES015348-02 (to M.K.R.).
Abbreviations
- 2-AG
2-arachidonylclycerol
- AA
arachidonic acid
- ABPP
activity-based protein profiling
- AEA
arachidonylethanolamide (anandamide)
- Ces
carboxylesterase
- FAAH
fatty acid amide hydrolase
- IL
interleukin
- LPS
lipopolysaccharide
- MAGL
monoacylglycerol lipase
- PCR
polymerase chain reaction
- PO
paraoxon
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
Contributions
BLFK and MKR conceived the experimental design; BS, AB, JWL, MKR and BLFK performed experiments; BS, MKR and BLFK wrote the manuscript. All authors approved the final manuscript.
Submission declaration
The work has not been published previously (except in the form of an abstract), it is not under consideration for publication elsewhere, and it will not be published elsewhere in the event it is accepted.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- 1.Rom S, Persidsky Y. Cannabinoid receptor 2: potential role in immunomodulation and neuroinflammation. J Neuroimmune Pharmacol. 2013;8(3):608–20. doi: 10.1007/s11481-013-9445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Facchinetti F, Del Giudice E, Furegato S, Passarotto M, Leon A. Cannabinoids ablate release of TNFalpha in rat microglial cells stimulated with lypopolysaccharide. Glia. 2003;41(2):161–8. doi: 10.1002/glia.10177. [DOI] [PubMed] [Google Scholar]
- 3.Gallily R, Breuer A, Mechoulam R. 2-Arachidonylglycerol, an endogenous cannabinoid, inhibits tumor necrosis factor-alpha production in murine macrophages, and in mice. Eur J Pharmacol. 2000;406(1):R5–7. doi: 10.1016/s0014-2999(00)00653-1. [DOI] [PubMed] [Google Scholar]
- 4.Krishnan G, Chatterjee N. Endocannabinoids alleviate proinflammatory conditions by modulating innate immune response in muller glia during inflammation. Glia. 2012;60(11):1629–45. doi: 10.1002/glia.22380. [DOI] [PubMed] [Google Scholar]
- 5.Lourbopoulos A, Grigoriadis N, Lagoudaki R, et al. Administration of 2-arachidonoylglycerol ameliorates both acute and chronic experimental autoimmune encephalomyelitis. Brain Res. 2011;1390:126–41. doi: 10.1016/j.brainres.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Peng F, Dong M, Yang H. Endocannabinoid 2-arachidonylglycerol protects primary cultured neurons against LPS-induced impairments in rat caudate nucleus. J Mol Neurosci. 2014;54(1):49–58. doi: 10.1007/s12031-014-0246-2. [DOI] [PubMed] [Google Scholar]
- 7.Panikashvili D, Shein NA, Mechoulam R, et al. The endocannabinoid 2-AG protects the blood-brain barrier after closed head injury and inhibits mRNA expression of proinflammatory cytokines. Neurobiol Dis. 2006;22(2):257–64. doi: 10.1016/j.nbd.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Alhouayek M, Lambert DM, Delzenne NM, Cani PD, Muccioli GG. Increasing endogenous 2-arachidonoylglycerol levels counteracts colitis and related systemic inflammation. FASEB J. 2011;25(8):2711–21. doi: 10.1096/fj.10-176602. [DOI] [PubMed] [Google Scholar]
- 9.Costola-de-Souza C, Ribeiro A, Ferraz-de-Paula V, et al. Monoacylglycerol lipase (MAGL) inhibition attenuates acute lung injury in mice. PLoS One. 2013;8(10):e77706. doi: 10.1371/journal.pone.0077706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S, Wise LE, Chen Y, et al. The monoacylglycerol lipase inhibitor JZL184 suppresses inflammatory pain in the mouse carrageenan model. Life Sci. 2013;92(8–9):498–505. doi: 10.1016/j.lfs.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang R, Borazjani A, Matthews AT, Mangum LC, Edelmann MJ, Ross MK. Identification of palmitoyl protein thioesterase 1 in human THP1 monocytes and macrophages and characterization of unique biochemical activities for this enzyme. Biochemistry. 2013;52(43):7559–74. doi: 10.1021/bi401138s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie S, Borazjani A, Hatfield MJ, Edwards CC, Potter PM, Ross MK. Inactivation of lipid glyceryl ester metabolism in human THP1 monocytes/macrophages by activated organophosphorus insecticides: role of carboxylesterases 1 and 2. Chem Res Toxicol. 2010;23(12):1890–904. doi: 10.1021/tx1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes RS, Wright MW, Laulederkind SJ, et al. Recommended nomenclature for five mammalian carboxylesterase gene families: human, mouse, and rat genes and proteins. Mamm Genome. 2010;21(9–10):427–41. doi: 10.1007/s00335-010-9284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu Rev Pharmacol Toxicol. 1998;38:257–88. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Niphakis MJ, Cravatt BF. Enzyme inhibitor discovery by activity-based protein profiling. Annu Rev Biochem. 2014;83:341–77. doi: 10.1146/annurev-biochem-060713-035708. [DOI] [PubMed] [Google Scholar]
- 17.Carr RL, Borazjani A, Ross MK. Effect of developmental chlorpyrifos exposure, on endocannabinoid metabolizing enzymes, in the brain of juvenile rats. Toxicol Sci. 2011;122(1):112–20. doi: 10.1093/toxsci/kfr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long JZ, Nomura DK, Cravatt BF. Characterization of monoacylglycerol lipase inhibition reveals differences in central and peripheral endocannabinoid metabolism. Chem Biol. 2009;16(7):744–53. doi: 10.1016/j.chembiol.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crow JA, Bittles V, Herring KL, Borazjani A, Potter PM, Ross MK. Inhibition of recombinant human carboxylesterase 1 and 2 and monoacylglycerol lipase by chlorpyrifos oxon, paraoxon and methyl paraoxon. Toxicol Appl Pharmacol. 2012;258(1):145–50. doi: 10.1016/j.taap.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng M, Scott MJ, Loughran P, et al. Lipopolysaccharide clearance, bacterial clearance, and systemic inflammatory responses are regulated by cell type-specific functions of TLR4 during sepsis. J Immunol. 2013;190(10):5152–60. doi: 10.4049/jimmunol.1300496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maccarrone M, De Petrocellis L, Bari M, et al. Lipopolysaccharide downregulates fatty acid amide hydrolase expression and increases anandamide levels in human peripheral lymphocytes. Arch Biochem Biophys. 2001;393(2):321–8. doi: 10.1006/abbi.2001.2500. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Batkai S, Pacher P, et al. Lipopolysaccharide induces anandamide synthesis in macrophages via CD14/MAPK/phosphoinositide 3-kinase/NF-kappaB independently of platelet-activating factor. J Biol Chem. 2003;278(45):45034–9. doi: 10.1074/jbc.M306062200. [DOI] [PubMed] [Google Scholar]
- 23.Wolfson ML, Aisemberg J, Salazar AI, Dominguez Rubio AP, Vercelli CA, Franchi AM. Progesterone reverts LPS-reduced FAAH activity in murine peripheral blood mononuclear cells by a receptor-mediated fashion. Mol Cell Endocrinol. 2013;381(1–2):97–105. doi: 10.1016/j.mce.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 24.Pasquarelli N, Porazik C, Hanselmann J, Weydt P, Ferger B, Witting A. Comparative biochemical characterization of the monoacylglycerol lipase inhibitor KML29 in brain, spinal cord, liver, spleen, fat and muscle tissue. Neuropharmacology. 2015;91:148–56. doi: 10.1016/j.neuropharm.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Jones RD, Taylor AM, Tong EY, Repa JJ. Carboxylesterases are uniquely expressed among tissues and regulated by nuclear hormone receptors in the mouse. Drug Metab Dispos. 2013;41(1):40–9. doi: 10.1124/dmd.112.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Sedlacek M, Manoharan I, et al. Butyrylcholinesterase, paraoxonase, and albumin esterase, but not carboxylesterase, are present in human plasma. Biochem Pharmacol. 2005;70(11):1673–84. doi: 10.1016/j.bcp.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Shi D, Yang D, Song X, Yan B. Interleukin-6 alters the cellular responsiveness to clopidogrel, irinotecan, and oseltamivir by suppressing the expression of carboxylesterases HCE1 and HCE2. Mol Pharmacol. 2007;72(3):686–94. doi: 10.1124/mol.107.036889. [DOI] [PubMed] [Google Scholar]
- 28.Dong M, Lu Y, Zha Y, Yang H. Endocannabinoid 2-arachidonylglycerol protects primary cultured neurons against homocysteine-induced impairments in rat caudate nucleus through CB1 receptor. J Mol Neurosci. 2015;55(2):500–8. doi: 10.1007/s12031-014-0371-y. [DOI] [PubMed] [Google Scholar]
- 29.Raman P, Kaplan BL, Kaminski NE. 15-Deoxy-Delta(1)(2),(1)(4)-prostaglandin J(2)-glycerol, a putative metabolite of 2-arachidonyl glycerol and a peroxisome proliferator-activated receptor gamma ligand, modulates nuclear factor of activated T cells. J Pharmacol Exp Ther. 2012;342(3):816–26. doi: 10.1124/jpet.112.193003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raman P, Kaplan BL, Thompson JT, Vanden Heuvel JP, Kaminski NE. 15-Deoxy-delta12,14-prostaglandin J2-glycerol ester, a putative metabolite of 2-arachidonyl glycerol, activates peroxisome proliferator activated receptor gamma. Mol Pharmacol. 2011;80(1):201–9. doi: 10.1124/mol.110.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rockwell CE, Raman P, Kaplan BL, Kaminski NE. A COX-2 metabolite of the endogenous cannabinoid, 2-arachidonyl glycerol, mediates suppression of IL-2 secretion in activated Jurkat T cells. Biochem Pharmacol. 2008;76(3):353–61. doi: 10.1016/j.bcp.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Rockwell CE, Snider NT, Thompson JT, Vanden Heuvel JP, Kaminski NE. Interleukin-2 suppression by 2-arachidonyl glycerol is mediated through peroxisome proliferator-activated receptor gamma independently of cannabinoid receptors 1 and 2. Mol Pharmacol. 2006;70(1):101–11. doi: 10.1124/mol.105.019117. [DOI] [PubMed] [Google Scholar]
- 33.Rouzer CA, Marnett LJ. Endocannabinoid oxygenation by cyclooxygenases, lipoxygenases, and cytochromes P450: cross-talk between the eicosanoid and endocannabinoid signaling pathways. Chem Rev. 2011;111(10):5899–921. doi: 10.1021/cr2002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urquhart P, Nicolaou A, Woodward DF. Endocannabinoids and their oxygenation by cyclo-oxygenases, lipoxygenases and other oxygenases. Biochim Biophys Acta. 2015;1851(4):366–76. doi: 10.1016/j.bbalip.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Alhouayek M, Masquelier J, Cani PD, Lambert DM, Muccioli GG. Implication of the anti-inflammatory bioactive lipid prostaglandin D2-glycerol ester in the control of macrophage activation and inflammation by ABHD6. Proc Natl Acad Sci U S A. 2013;110(43):17558–63. doi: 10.1073/pnas.1314017110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turcotte C, Chouinard F, Lefebvre JS, Flamand N. Regulation of inflammation by cannabinoids, the endocannabinoids 2-arachidonoyl-glycerol and arachidonoyl-ethanolamide, and their metabolites. J Leukoc Biol. 2015;97(6):1049–70. doi: 10.1189/jlb.3RU0115-021R. [DOI] [PubMed] [Google Scholar]
- 37.Ross MK, Borazjani A, Wang R, Crow JA, Xie S. Examination of the carboxylesterase phenotype in human liver. Arch Biochem Biophys. 2012;522(1):44–56. doi: 10.1016/j.abb.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]