Abstract
Mice carrying Collagen2a1-cre-mediated deletions of Lrp5 and/or Lrp6 were created and characterized. Mice lacking either gene alone were viable and fertile with normal knee morphology. Mice in which both Lrp5 and Lrp6 were conditionally ablated via Collagen2a1-cre-mediated deletion displayed severe defects in skeletal development during embryogenesis. In addition, adult mice carrying Collagen2a1-cre-mediated deletions of Lrp5 and/or Lrp6 displayed low bone mass suggesting that the Collagen2a1-cre transgene was active in cells that subsequently differentiated into osteoblasts. In both embryonic skeletal development and establishment of adult bone mass, Lrp5 and Lrp6 carry out redundant functions.
Introduction
The Wnt/β-catenin signaling pathway affects cell migration, proliferation, and differentiation and is a key regulator of skeletal development.1 This pathway is initiated when a Wnt ligand engages a receptor complex that includes a member of the Frizzled family of seven-transmembrane receptors and either low-density lipoprotein-related receptor 5 (Lrp5) or Lrp6.2 This results in the phosphorylation of the carboxyl terminus of Lrp5 or Lrp6, creating a binding site for Axin. Axin is a component of a multiprotein complex that also includes the Adenomatous polyposis coli protein and the serine/threonine protein kinase glycogen synthase kinase 3 (GSK3). In the absence of Wnt, this complex facilitates the GSK3-dependent phosphorylation of β-catenin, targeting it for ubiquitin-dependent proteolysis. Binding of Axin to the phosphorylated carboxyl terminus of Lrp5/6 inhibits GSK3 activity toward β-catenin, resulting in stabilization of β-catenin in the cytoplasm. β-catenin is then available to enter the nucleus where it complexes with members of the LEF/TCF family of DNA-binding proteins and induces the activation of target gene promoters.
Numerous studies have indicated that alterations in Wnt/β-catenin levels can influence chondrocyte differentiation and/or function.3–11 For example, ectopic expression of β-catenin in chondrocyte lineage cells inhibits chondrocyte differentiation. Increased levels of canonical Wnt/β-catenin signaling may inhibit Sox9 expression and activity thus reducing chondrocyte differentiation.12 In addition, both increased and decreased β-catenin signaling has been observed in degenerative osteoarthritis (OA) models.13–18 Lrp5 mRNA and protein is increased in cells from human OA patients and Lrp5 polymorphisms analyzed as a group were correlated with human OA.19–23
During the last decade, we and others have utilized genetically engineered mouse models to gain insight into the roles of components of the Wnt pathway in skeletal development (reviewed in ref. 1). Germline deletion of Lrp6 leads to neonatal lethality associated with defects in the axial skeletal and limbs.24 Tissue-specific and heterozygous mutations that inactivate Lrp6 early in development produce skeletal phenotypes that are typically more severe than those in mice with mutations in Lrp5.24–26 Gaining insight into how Wnt/β-catenin signaling influences the development and function of normal cartilage will provide insight into utilizing Lrp5/6 as therapeutic targets for OA.27,28
In this study, we examined the effect of loss of Lrp5 and Lrp6 function in Collagen2a1 expressing cells and their descendants in both developing and adult mouse skeletons. We used the cre-lox system in which cre was driven by the Col2a1 promoter,29 which is expressed by chondrocytes and osteoblast progenitors and has been used in a variety of animal models to achieve tissue-specific gene expression in chondrocytes.7,30 We generated mice with single conditional knockout mutations in either Lrp5 or Lrp6 and mice with combined mutations in Lrp5 and Lrp6.
Materials and methods
Mouse strains
Lrp5flox, Lrp6flox, Col2a1-cre, and mT/mG mice have been described previously.29,31–33 All experiments were done in compliance with the Guidelines for the Care and Use of Animals for Scientific Research.34 The Institutional Animal Care and Use Committee of the Van Andel Research Institute approved all experimental procedures.
PCR-based genotyping
Genomic DNA was extracted from tail biopsies using an AutoGenprep 960 automated DNA isolation system (AutoGen Inc., Holliston, MA, USA). PCR-based strategies were then used to genotype the mice (details available upon request).35 For allele-specific PCR, genomic DNA was extracted from various tissue types of mice using DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany).
Whole-Mount Skeletal Staining
Whole-mount skeletal staining of embryos was performed according to McLeod with modifications.36 Briefly, skin and viscera were removed and embryos were placed in 95% EtOH for 1–2 days. Embryos (E18.5) were then placed in acetone for 1 day and then stained with alizarin red (0.1% alizarin red S in 95% EtOH) and alcian blue (0.3% alcian blue 8GX in 70% EtOH) for 3 days at 37 °C. Embryos were then dipped in dH2O, cleared in 1% aqueous KOH until nearly all of the soft tissue was gone, cleared in 80% 1% KOH/20% glycerol, cleared in 50% 1% KOH/50% glycerol, cleared in 20% 1% KOH/80% glycerol, and finally transferred to 100% glycerol for storage.
Histology
To investigate the effect of Lrp5/6 loss of function driven by the Col2 promoter during embryonic development and in adult bone and cartilage, limbs from E16.5, E17.5, and E18.5 embryos Lrp5/6 mutant and WT mice and knees from 6-month-old male and female Col2-cre;Lrp5Fl/Fl, Col2-cre;Lrp6Fl/Fl, Col2-cre;Lrp5Fl/+;Lrp6Fl/+, Col2-cre;Lrp5Fl/FL;Lrp6Fl/+, and Col2-cre;Lrp5Fl/+;Lrp6Fl/FL mice and respective WT littermates were fixed in 10% neutral buffered formalin for 48 h and decalcified with Immunocal (Decal Chemical Corporation, Tallman, NY, USA) for 48 h. Samples were infiltrated with an alcohol series, cleared with xylene, and infiltrated with paraffin. Samples were embedded on edge into a paraffin mold using the Leica Embedding center, sectioned at 5 μm using a microtome, collected onto glass slides, deparaffinized, and hydrated in distilled water. Sagittal knee sections were stained with Safranin O (0.1% Safranin O in distilled water) and Fast Green (0.05% Fast Green FCF in distilled water), and counterstained with Hematoxylin. Sections from embryos were stained with pentachrome according to the Movat method. Sections were imaged with a Nikon Eclipse 55i microscope and Nikon Digital Sight camera (Nikon, Melville, NY, USA).
DEXA
Mice were anesthetized via inhalation of 2% isoflurane (TW Medical Veterinary Supply) with oxygen (1.0 L·min−1) for 10 min prior to imaging and during the procedure (⩽5 min). The mice were placed on a specimen tray in a PIXImus II bone densitometer (GE Lunar) for analysis. Bone mineral density (BMD) was calculated by the PIXImus software based on the active bone area in the subcranial region within the total body image and specifically in the femur, humerus, and axial skeleton.
Microcomputed tomography
Trabecular and cortical BMD and architecture were assessed after collecting samples at 6 months of age using standardized methods37 at the distal femoral metaphysis and femoral midshaft, respectively, using a desktop SkyScan 1172 microCT imaging system (SkyScan, Kontich, Germany). Scans were acquired using a 13.3-μm3 isotropic voxel size, with 130 CT slices evaluated at the distal femur and 230 CT slices at the femoral midshaft. For trabecular and cortical bone analyses, fixed thresholds of 85 and 113, respectively, were used to determine the mineralized bone fraction. These values were calculated by averaging together all individual threshold levels. Individual CT slices were reconstructed with SkyScan reconstruction software and data were analyzed with CTan(Comprehensive TeX Archive Network). The region of interest of trabecular bone was drawn manually a few voxels away from the endocortical surface. The SkyScan was calibrated daily to hydroxyapatite cores of known density.
Results
Generation of mice carrying Col2-cre-mediated deletions in Lrp5 and/or Lrp6
A detailed summary of the mouse strains and genetic backgrounds associated with this study is provided in Table 1. Mouse strains carrying floxed alleles of Lrp5 and Lrp6 have been previously described and have no deficit in Lrp5 or Lrp6 function in the absence of exposure to cre recombinase.31,35 These strains were crossed to mice expressing the cre recombinase under the control of the Collagen2A1 promoter (Col2-cre).29 This process generated mice with several potential genotypes. Col2-cre expressing mice homozygous for either Lrp5flox (Col2-cre;Lrp5Fl/Fl) or Lrp6flox (Col2-cre;Lrp6Fl/Fl) were viable and fertile. In addition, mice carrying several different combinations of Col2-cre-induced in Lrp5 and Lrp6 were also viable. Mice heterozygous for both Lrp5flox and Lrp6flox (Col2-cre;Lrp5Fl/+;Lrp6Fl/+) or heterozygous for one such allele and homozygous for the other (Col2-cre;Lrp5Fl/FL;Lrp6Fl/+ or Col2-cre;Lrp5Fl/+;Lrp6Fl/FL were all viable and fertile. In contrast, mice homozygous for Col2-mediated deletion of both Lrp5 and Lrp6 (Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL) died in utero or shortly after birth. Mice lacking the Col2-cre transgene are considered WT and used as controls for these studies as the Lrp5 and Lrp6 loci function normally in the absence of cre (Figure 1). A summary of the nomenclature used in the text and in the figures for the genotypic cohorts of mice in this study is provided in Table 2.
Table 1. Mouse strains used in this work.
| Nomenclature | Extended allele name | JAX database stock # | Genetic background | References |
|---|---|---|---|---|
| Col2-cre | Tg(Col2a1cre)1Bhr/J | 003554 | C57Bl/6 J; SJL | 29 |
| Lrp5flox | Lrp5tm1.1Vari/J | 026269 | 129/SvJ; C57Bl/6 J | 31 |
| Lrp6flox | Lrp6tm1.1Vari/J | 026267 | 129/SvJ; C57Bl/6 J | 31 |
| Mt/mG | Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | 007576 | 129X1/SvJ | 33 |
WT, wild type.
In each case, the nomenclature used in this study is listed, followed by the strain name and stock number for each strain at the Jackson Laboratories (JAX).
Figure 1.
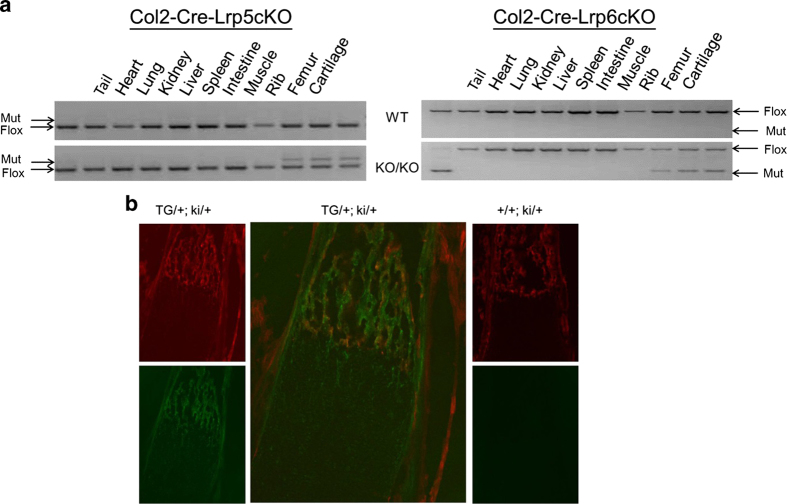
Evaluating Col2-cre activity and tissue-specific deletion of Lrp5 and Lrp6. (a) Allele-specific PCR was used to assess where cre-mediated activity resulted in an inactivated allele of Lrp5 or Lrp6 (mutant) and where the floxed allele remained intact (flox). The Col2-cre-Lrp5cKO mutant band was detected in the rib, femur, and cartilage, whereas the Col2-cre-Lrp6cKO mutant band was detected in the rib, femur, cartilage, and tail. (b) Col2-cre expressing mice were crossed to the mTmG reporter strain. The presence of Tomato protein (red fluoresence) marks cells that have not undergone cre-mediated recombination of the reporter gene, whereas the expression of GFP (green) is indicative of cells derived from Col2-cre expressing cells. The left panel demonstrates red (top) and green (top) fluoresence in sections from the femur of a Col2-cre;mtmG mouse (TG/+;ki/+), whereas the right panel shows the same staining for a femur from a control littermate (+/+;ki/+). The middle panel is a higher power view of a combination of the images in the left panels.
Table 2. Summary of nomenclature for genetically engineered mice characterized in this study.
| Nomenclature | Abbreviation in figures | Col2-cre genotype | Lrp5Floxgenotype | Lrp6Floxgenotype | Survival |
|---|---|---|---|---|---|
| Control | WT | +/+ | All | All | Viable |
| Col2-cre;Lrp5Fl/FL | 5cKO | TG/+ | Flox/Flox | +/+ | Viable |
| Col2-cre;Lrp6Fl/FL | 6cKO | TG/+ | +/+ | Flox/Flox | Viable |
| Col2-cre;Lrp5Fl/+;Lrp6Fl/+ | 5cHET6cHET | TG/+ | Flox/+ | Flox/+ | Viable |
| Col2-cre;Lrp5Fl/FL;Lrp6Fl/+ | 5cKO6cHET | TG/+ | Flox/Flox | Flox/+ | Viable |
| Col2-cre;Lrp5Fl/+;Lrp6Fl/FL | 5cHET6cKO | TG/+ | Flox/+ | Flox/Flox | Viable |
| Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL | 5cKO6cKO | TG/+ | Flox/Flox | Flox/Flox | Neonatal lethal |
WT, wild type.
A “+” is indicative of an unmodified (wild type) locus. A designation of “TG” denotes that a mouse is carrying the Col2-cre transgene while “Flox” indicates the presence of an allele of a gene that can be inactivated upon exposure to cre recombinase. The abbreviations used in the figures to refer to the different genotypes is also shown.
Assessment of Col2-cre activity
To confirm the pattern of Col2-cre activity, allele-specific PCR was performed. In Col2-cre;Lrp5Fl/FL mice, PCR products specifically derived from alleles that had undergone cre-mediated recombination were detected in the ribs, femur, and cartilage. In Col2-cre;Lrp6Fl/FL mice, mutant bands were detected in the tail, ribs, femur, and cartilage (Figure 1a). To further confirm activity, we crossed Col2-cre transgenic mice to a strain carrying the mT/mG cre reporter.33 In the mT/mG mice, loxP sites are present around the tdTomato (mT) expression cassette. In the absence of Cre-mediated recombination, the tomato protein is expressed in all tissues which, as a result, display red fluorescence. Upon exposure to cre, recombination excises the mT expression cassette, allowing expression of green fluorescent protein (GFP). GFP expression, indicative of Cre recombination, was detected in hypertrophic chondrocytes and both trabecular and cortical bone regions of Col2-cre-expressing mice (Figure 1b).
Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL mice have skeletal abnormalities during development
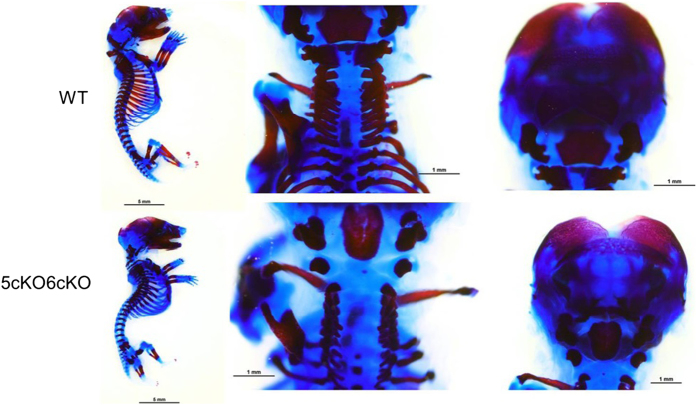
Once it was apparent that Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL mice did not survive beyond birth, we set up timed matings and collected embryos at e18.5. To assess differences in skeletal development, whole mount staining of embryos with Alizarin Red and Alcian Blue was performed. We did not observe any gross differences between control embryos and mice homozygous for conditional deletion of either Lrp5 (Col2-cre;Lrp5Fl/FL) or Lrp6 Col2-cre;Lrp6Fl/FL; data not shown). In contrast, gross differences in skeletal morphology were observed between Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL and WT littermates at this stage (Figure 2). In Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL embryos, the supraoccipital bone was not mineralized; and the dorsal arches of the C1 vertebrae and C2 vertebrae were underdeveloped (Figure 3a and 3b). These mutant embryos had smaller exoccipial bones, little cartilage ossification in the primordium body of the hyoid bone, and a gap within vertebrae (Figure 3a and 3b). A lack of bone growth and fusion was observed in the palate and reduced membranous ossification of the nasal bone was also observed in Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL embryos. A lack of nasal bone ossification and growth was apparent in addition to limited growth and ossification of the palatine bone, and underdevelopment and irregular shape of basisphenoid bone (Figure 3a and b). There were smaller projections of the vertebrae and reduced mineralization within the spine and ribs (Figure 3c) of Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL embryos.
Figure 2.
Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL (5cKO6cKO) embryos have significant defects in mineralization. E18.5 alizarin red and alcian blue stains of the entire skeleton (sagittal view) (left panels), top view of the spine and ribs (middle panel); and back of the cranium (right panels).
Figure 3.
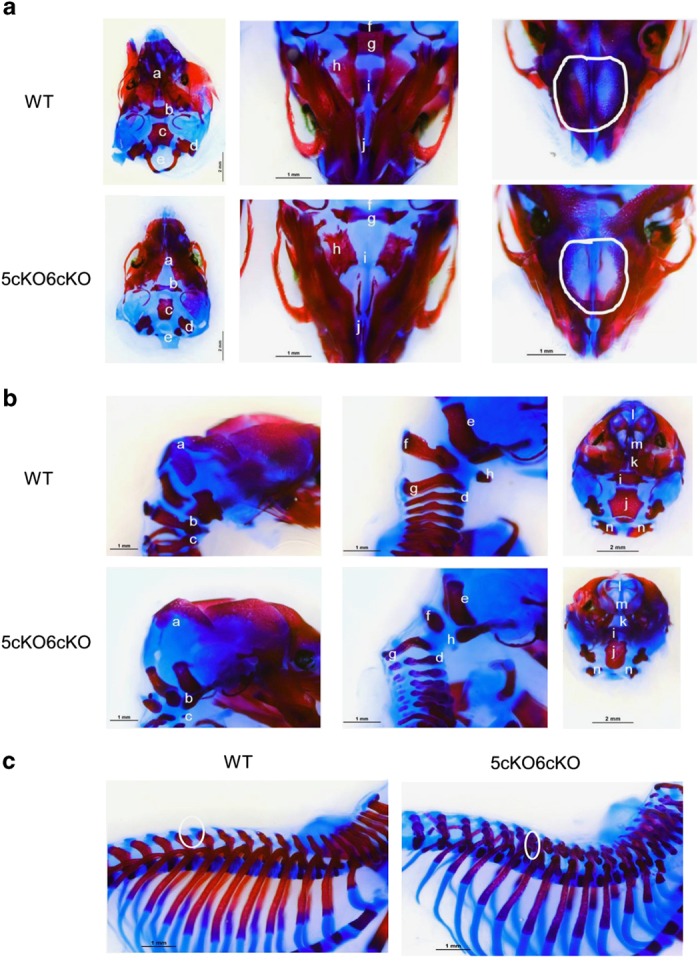
Higher magnification views of Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL (5cKO;6cKO) embryo mineralization defects. (a) E18.5 alizarin red and alcian blue stain of a top view of the skull with the supraoccipital bone removed; view from the underside of the skull; and top view of the skull looking down at the mouse nasal cavity. (b) E18.5 alizarin red and alcian blue stains of the skull and top cervical vertebra; base of the skull and cervical vertebra; and posterior view of the skull with the supraoccipital bone removed. Specific areas are noted in a and b by the following notations: a. Supraoccipital bone; b. Dorsal arch of the first cervical vertebrae (C1); c. C2 vertebra; d. Abnormal vertebral structure; e. Exocipital bone; f. Dorsal arch; g. C2 vertebrae; h. Cartilage primordium of body of hyoid bone. i. Basisphenoid bone; j. Basioccipital bone; k. Palatine bone; l. nasal bone; m. Putative extension of nasal bone; n. Dorsal arch of C1 vertebrae. (c) Representative example of alizarin red and alcian blue stains of the spine and ribs from a 5cKO6cKO E18.5 embryo and a WT littermate. WT, wild type.
Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL mice have altered cartilaginous zones during development
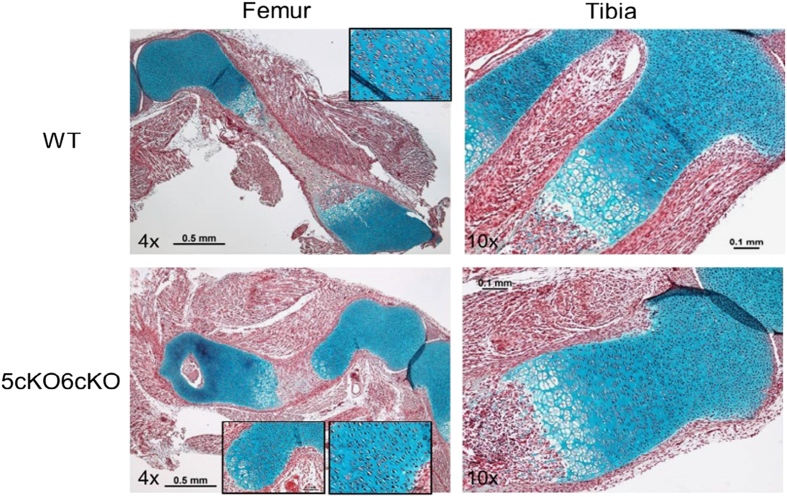
Limbs from E18.5 Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL embryos were bowed with the hypertrophic zone extending further from the growth plate. This was associated with increased alcian blue staining (Figure 4). This phenotype is similar to that seen in Dermo1-cre;Lrp5Fl/FL;Lrp6Fl/FL embryos.31
Figure 4.
Col2-cre;Lrp5Fl/FL;Lrp6Fl/FL (5cKO;6cKO) mice have altered cartilaginous zones during development. Cartilage (blue) in pentachrome stained femur (left panels) and tibia (right panels) sections from E18.5 5cKO6cKO (bottom panels) and WT littermates (top panels). Boxed regions are shown at higher magnification. WT, wild type.
Adult Col2-cre;Lrp5Fl/FL or Col2-cre;Lrp6Fl/FL mice and mice with combinatorial deletions of Lrp5 and Lrp6 have low bone mass
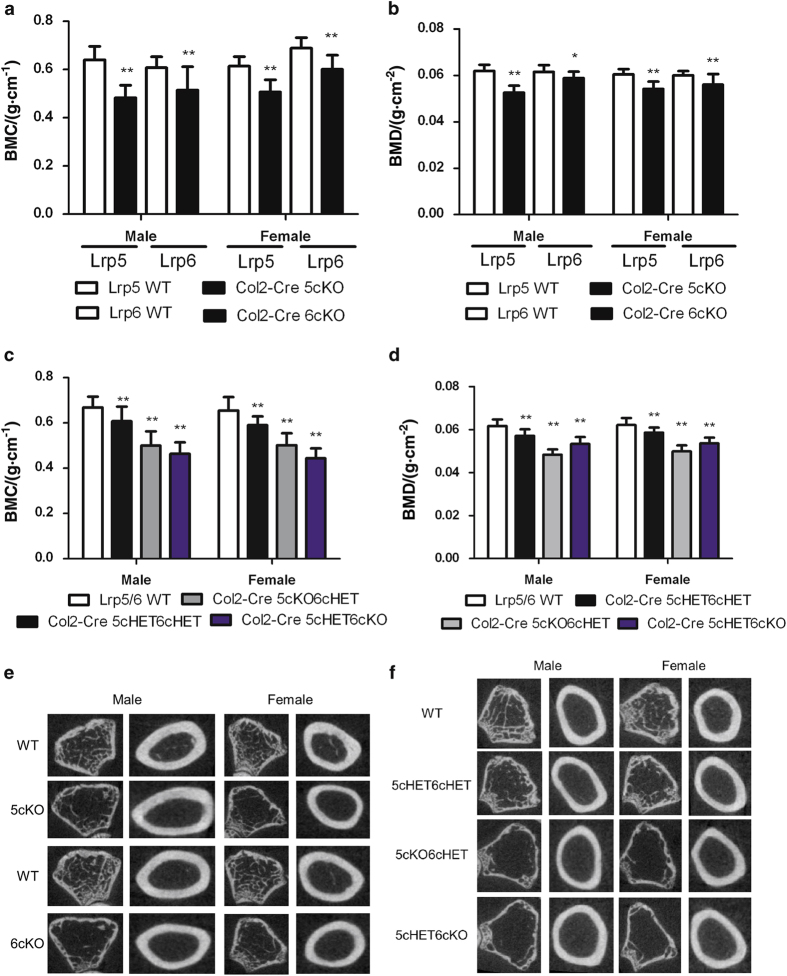
To assess the effect of Col2-cre-induced loss of Lrp5 or Lrp6 on adult bone at 6 months of age, dual X-ray absorbtiometry was carried out to measure total body BMD. This demonstrated that BMD was significantly lower for male and female Col2-cre;Lrp5Fl/FL and Col2-cre;Lrp6Fl/FL mice relative to control littermates (Figure 5). Total body BMD was also significantly reduced in male and female Col2-cre;Lrp5Fl/+;Lrp6Fl/+, Col2-cre;Lrp5Fl/FL;Lrp6Fl/+, and Col2-cre;Lrp5Fl/+;Lrp6Fl/FL mice compared with WT littermates (Figure 5).
Figure 5.
Adult Col2-cre;Lrp5Fl/FL (5cKO) or Col2-cre;Lrp6Fl/FL (6cKO) mice and mice with combinatorial deletions of Lrp5 and Lrp6 have low bone mass at six months of age. (a) Whole-body bone mineral content DEXA measurements (mean±s.d.) for male and female mice with single Lrp5 and single Lrp6 conditional loss of function mutations driven by Col2-cre and respective WT littermates; **P<0.01 compared to WT littermates. (b) Whole-body bone mineral density DEXA measurements (mean±s.d.) for male and female mice with single Lrp5 and single Lrp6 conditional loss of function mutations driven by Col2-cre and respective WT littermates; **P<0.01 and *P=0.01 compared to WT littermates. (c) Whole-body bone mineral content DEXA measurements (mean±s.d.) for male and female mice with double combinations of Lrp5 and Lrp6 conditional knockout or heterozygous loss of function mutation driven by Col2-cre and respective WT littermates; **P<0.01 compared with WT littermates. (d) Whole-body bone mineral density DEXA measurements (mean±s.d.) for male and female mice with combinations of Lrp5 and Lrp6 conditional knockout or heterozygous loss of function mutation driven by Col2-cre and respective WT littermates; **P<0.01 compared with WT littermates. (e) MicroCT cross sectional images of trabecular and cortical bone from male and female mice with single Lrp5 and single Lrp6 conditional knockout mutation driven by Col2-cre and of respective WT littermates. (f) MicroCT cross sectional images of trabecular and cortical bone from male and female mice with combinations of Lrp5 and Lrp6 conditional homozygous knockout or heterozygous loss of function mutations and of WT littermates. BMC, bone mineral content; BMD, bone mineral density.
A qualitative reduction in trabecular number and bone volume fraction, and an increase in trabecular spacing were observed in the distal femur of Col2-cre;Lrp5Fl/FL, Col2-cre;Lrp6Fl/FL, Col2-cre;Lrp5Fl/+;Lrp6Fl/+, Col2-cre;Lrp5Fl/FL;Lrp6Fl/+, and Col2-cre;Lrp5Fl/+;Lrp6Fl/FL mice compared with WT littermates (Figure 5). Compared with respective WT littermates cortical cross sectional area and cortical thickness was significantly lower for male and female Col2-cre;Lrp5Fl/FL and Col2-cre;Lrp6Fl/FL (Table 3). Cortical mineral density was significantly lower for female Col2-cre;Lrp5Fl/FL and Col2-cre;Lrp6Fl/FL mice compared with WT (Table 3). Cortical thickness and mineral density were significantly lower for male and female mice with mutations in both Lrp5 and Lrp6 (Table 3). Cortical cross sectional area was significantly lower for all female mice with mutations in both Lrp5 and Lrp6 and male Col2-cre;Lrp5Fl/FL;Lrp6Fl/+ mice compared with respective WT mice (Table 3). Compared with respective WT littermates, trabecular bone volume fraction and number were significantly lower and separation significantly higher for Col2-cre;Lrp5Fl/FL and Col2-cre;Lrp6Fl/FL male and female mice (Table 3). Trabecular thickness was significantly lower for male Col2-cre;Lrp5Fl/FL mice compared with WT (Table 3). Compared with respective WT littermates, trabecular bone volume fraction and number were significantly lower and separation higher for male and female Col2-cre;Lrp5Fl/FL;Lrp6Fl/+ and Col2-cre;Lrp5Fl/+;Lrp6Fl/FL mice (Table 3). Trabecular thickness was significantly lower in female Col2-cre;Lrp5Fl/FL;Lrp6Fl/+ and Col2-cre;Lrp5Fl/+;Lrp6Fl/FL mice and male Col2-cre;Lrp5Fl/FL;Lrp6Fl/+ mice compared with WT (Table 3).
Table 3. Average microCT cortical and trabecular bone geometrical measurements on samples from mice at 6 months of age.
| Lrp5 |
Lrp6 |
Lrp5Lrp6 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male |
Female |
Male |
Female |
Male |
Female |
|||||||||||
| Bone | WT 10 | cKO 10 | WT 10 | cKO 10 | WT 10 | cKO 10 | WT 10 | cKO 10 | WT 10 | cHETcHET 10 | cKOcHET 9 | cHETcKO 6 | WT 10 | cHETcHET 10 | cKOcHET 12 | cHETcKO 10 |
|
Trabecular bone
| ||||||||||||||||
| Percent bone volume/% | 20.114 | 6.447 | 13.676 | 8.212 | 15.602 | 7.024 | 9.561 | 5.506 | 12.762 | 10.362 | 2.206 | 3.945 | 12.050 | 7.049 | 4.385 | 4.880 |
| Standard deviation | 4.345 | 3.552 | 3.204 | 2.099 | 6.054 | 3.918 | 2.717 | 1.981 | 4.268 | 5.338 | 0.545 | 1.579 | 2.067 | 1.814 | 2.751 | 1.646 |
| P value | 1.1E−06** | 6.14E−04** | 0.032 | 0.004** | 0.002** | 0.307 | 3.56E−05** | 1.05E−04** | 3.72E−05** | 7.16E−07** | 2.71E−07** | |||||
| Trabecular thickness/mm | 0.033 | 0.026 | 0.029 | 0.027 | 0.032 | 0.028 | 0.030 | 0.029 | 0.031 | 0.032 | 0.026 | 0.030 | 0.034 | 0.032 | 0.027 | 0.028 |
| Standard deviation | 0.003 | 0.004 | 0.003 | 0.002 | 0.004 | 0.004 | 0.001 | 0.003 | 0.005 | 0.006 | 0.002 | 0.005 | 0.003 | 0.003 | 0.003 | 0.002 |
| P value | 2.54E−04** | 0.180 | 0.075 | 0.158 | 0.678 | 0.008** | 0.791 | 0.228 | 7.34E−05** | 9.12E−05** | ||||||
| Trabecular separation/mm | 0.136 | 0.452 | 0.192 | 0.326 | 0.190 | 0.448 | 0.312 | 0.552 | 0.236 | 0.342 | 1.197 | 0.834 | 0.254 | 0.452 | 0.986 | 0.601 |
| Standard deviation | 0.023 | 0.170 | 0.048 | 0.085 | 0.051 | 0.169 | 0.086 | 0.178 | 0.080 | 0.145 | 0.288 | 0.280 | 0.045 | 0.114 | 0.860 | 0.201 |
| P value | 3.29E−04** | 0.001** | 0.001** | 0.003** | 0.075 | 7.11E−06** | 0.004** | 4.30E−04** | 0.017* | 5.16E−04** | ||||||
| Trabecular number/mm−1 | 5.992 | 2.374 | 4.671 | 2.983 | 4.780 | 2.408 | 3.137 | 1.887 | 4.063 | 3.040 | 0.860 | 1.266 | 3.537 | 2.165 | 1.553 | 1.788 |
| Standard deviation | 0.782 | 0.919 | 0.735 | 0.670 | 1.182 | 0.940 | 0.885 | 0.582 | 1.110 | 1.023 | 0.183 | 0.357 | 0.437 | 0.461 | 0.894 | 0.675 |
| P value | 5.5E−08** | 7.83E−05** | 0.000** | 0.003** | 0.057 | 9.06E−06** | 1.61E−05** | 4.36E−06** | 6.34E−06** | 8.39E−06** | ||||||
|
Cortical bone
| ||||||||||||||||
| Mean total crossectional bone area/mm2 | 0.988 | 0.726 | 0.920 | 0.720 | 0.955 | 0.826 | 0.861 | 0.713 | 0.913 | 0.882 | 0.646 | 0.776 | 0.944 | 0.842 | 0.629 | 0.676 |
| Standard deviation | 0.076 | 0.121 | 0.057 | 0.071 | 0.066 | 0.080 | 0.052 | 0.083 | 0.085 | 0.112 | 0.077 | 0.141 | 0.060 | 0.054 | 0.080 | 0.041 |
| P value | 5.86E−05** | 4.33E−06** | 0.002** | 4.04E−04** | 0.512 | 3.24E−06** | 0.089 | 0.001** | 3.42E−09** | 7.92E−09** | ||||||
| Crossectional thickness/mm | 0.229 | 0.191 | 0.230 | 0.193 | 0.210 | 0.201 | 0.212 | 0.195 | 0.205 | 0.189 | 0.161 | 0.188 | 0.225 | 0.208 | 0.168 | 0.183 |
| Standard deviation | 0.009 | 0.017 | 0.007 | 0.010 | 0.010 | 0.009 | 0.007 | 0.012 | 0.010 | 0.009 | 0.010 | 0.015 | 0.012 | 0.008 | 0.017 | 0.007 |
| P value | 3.18E−05** | 5.27E−08** | 0.071 | 0.002** | 0.002** | 5.52E−08** | 0.050* | 0.003** | 3.36E−08** | 2.61E−07** | ||||||
| Mineral density/(g·cm−2) | 1.151 | 1.122 | 1.160 | 1.124 | 1.109 | 1.107 | 1.135 | 1.119 | 1.170 | 1.087 | 1.078 | 1.103 | 1.189 | 1.136 | 1.087 | 1.103 |
| Standard deviation | 0.038 | 0.037 | 0.021 | 0.020 | 0.030 | 0.027 | 0.013 | 0.018 | 0.040 | 0.034 | 0.029 | 0.043 | 0.045 | 0.020 | 0.065 | 0.015 |
| P value | 0.117 | 0.001** | 0.926 | 0.052* | 1.77E−04** | 4.48E−05** | 0.017* | 0.007** | 5.32E−04** | 2.24E−04** | ||||||
Abbreviation: WT, wild type.
P-values were calculated with respect to WT littermates; *P<0.05 and **P<0.01.
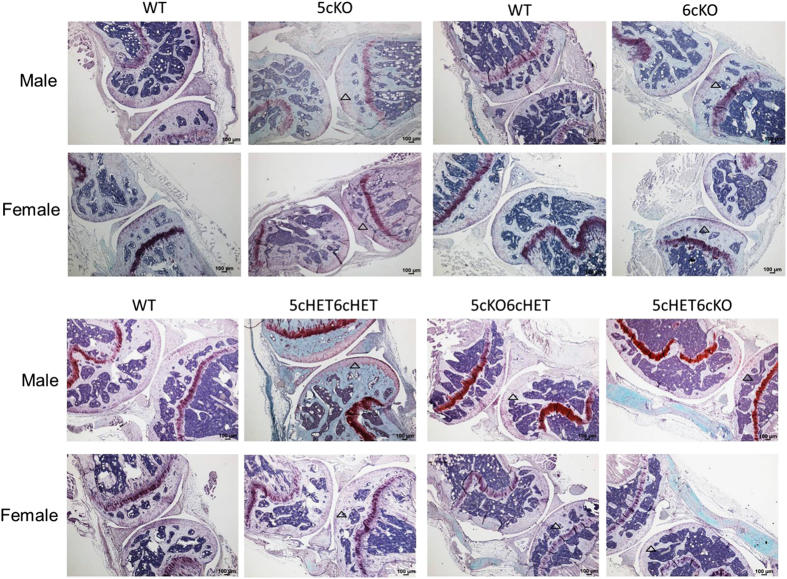
Adult mice with Col2-cre single Lrp5 or Lrp6 loss of function and double combinations of heterozygous and complete loss of Lrp5/6 function have normal articular cartilage and subchondral bone in knee joints
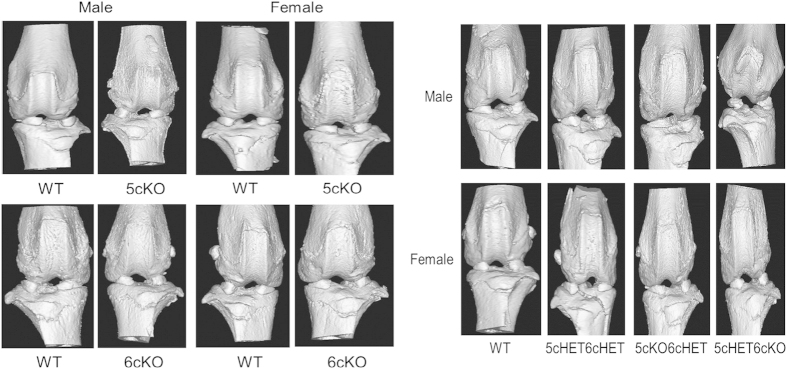
There were no gross morphological differences in knee joints between WT and mutant mice. Safranin O staining intensity was slightly reduced in articular cartilage from Col2-cre;Lrp5Fl/FL;Lrp6Fl/+ and Col2-cre;Lrp5Fl/+;Lrp6Fl/FL mice compared with littermates (Figure 6). The tidemark was well preserved in all mice and there was no obvious difference in cartilage thickness (Figure 6). Upon microcomputed tomography (microCT), knee joint subchondral bone morphology also appeared similar between mutant mice and WT littermates and there was no pathological evidence of degenerative joint diseases including subchondral bone erosion or osteophyte formation (Figure 7).
Figure 6.
Adult mice carrying Col2-cre-mediated deletions of Lrp5 and/or Lrp6 display normal knee joints Safranin O/fast green stained sagittal sections of the articular cartilage of knee joints from male and female Col2-cre5cKO, Col2-cre6cKO, Col2-cre5cHET6cHET, Col2-cre 5cKO6cHET, Col2-cre5cHET6cKO, and wild-type littermate mice at 6 months of age (4×).
Figure 7.
Adult mice carrying Col2-cre-mediated deletions of Lrp5 and/or Lrp6 have normal knee joints after evaluation by microCT. (Left) 3D reconstructed micro CT images of the knee joints from male and female Col2-cre5cKO, Col2-cre6cKO, and WT littermate mice. (Right) 3D reconstructed micro CT images of the knee joints from male and female Col2-cre5cHET6cHET, Col2-cre 5cKO6cHET, Col2-cre5cHET6cKO and WT littermate mice. WT, wild type.
Discussion
Studies have linked Wnt/β-catenin signaling to degenerative joint disease in humans and mice.19,23,38–43 The fact that Col2-cre-mediated deletion of Lrp5 results in normal appearing knee joints is not surprising given that global deletion of Lrp5 was previously shown to not alter skeletal morphogenesis.25,44–46 However, homozygosity for an inactivating allele of Lrp6 results in neonatal lethality,24 so it was previously not possible to evaluate the effects of loss of Lrp6 on the adult knee. Col2-cre-mediated deletion of Lrp6 did not result in any discernable knee phenotype. Furthermore, mice in which either Lrp5 or Lrp6 were homozygously deleted with Col2-cre, whereas the other gene was heterozygously deleted also displayed normal knee morphology. As homozygous deletion of both genes results in embryonic lethality,26 it is not possible to assess the effects of double deletion on the adult knee in mice carrying global deletions. We feel this is consistent with the idea that it is likely that retention of at least one allele of either Lrp5 or Lrp6 within the descendants of Col2-cre expressing cells is sufficient to mediate normal joint development. Alternatively, it is also possible that signaling from either Lrp5 or Lrp6 is not required for normal joint development. The use of chondrocyte-specific cre drivers that can be controlled temporally to induce simultaneous loss of both genes may be a way to fully discern between these two possibilities.
The presence of decreased bone mass in mice carrying deletions in Lrp5 and Lrp6 is consistent with previous observations that mutations in one of both of these genes within the osteochondral lineage cause reductions in bone mass.1,25,31,35,45,47 We did not carry out detailed histomorphometry in the context of these studies, but previously work showed that mice carrying OCN-cre-mediated deletions of either Lrp5 or Lrp6 developed osteopenia35 and therefore speculate that this could be the underlying mechanism for the reduced bone mass observed.
An alternative model in which Lrp5 functions to control bone mass via regulating the production of serotonin from enterchromaffin cells of the cells of the duodenum has also been proposed.48–50 However, this latter model has been debated.51–54 In any event, reductions in bone mass caused by loss of Lrp5 and/or Lrp6 in the descendants of Col2-cre-expressing cells could be either owing to widespread leakiness of the Col2-cre transgene within osteoblasts or reflect significant contribution of Col2-cre expressing cells to the osteoblast lineage found in the mature skeleton.
Although a detailed assessment of body composition was not part of this study, we did not observe differences in body weight that might be of interest for future studies. Assessment of the mice at the time of DEXA analysis demonstrated that both male (12% decrease) and female (9% decrease) Col2-cre;Lrp5Fl/FL mice weighed significantly less than their WT counterparts. Col2-cre;Lrp6Fl/FL female mice also weighed significantly less than controls (17% reduction), whereas male mice displayed a downward trend (4% decrease) that did not reach statistical significance. Whether this is related to relative changes in lean body mass and/or energy expenditure55 could be assessed in future studies.
The observation that Lrp5 and Lrp6 act in an overlapping and/or redundant manner within the descendants of Col2-cre expressing cells is consistent with observations in a wide variety of other cell types and tissues. These include mice carrying global deletions in the two genes as well as mice in which both genes are deleted in cells derived from the following cre drivers: Osteocalcin-cre (mature osteoblasts and osteocytes),35 Dermo1-cre (osteochondral progenitors),31 and Albumin-cre (hepatocytes).56 Future work will evaluate how Lrp5 and Lrp6 coordinately regulate downstream signaling pathways from Wnt ligands32,57,58 that are necessary for normal development in skeletal tissues.
Acknowledgments
We thank other members of the Williams Laboratory for helpful suggestions and assistance with genotyping. In addition, we thank the staffs of the VARI Vivarium and the VARI Pathology and Biorepository Core. This work was supported by the Van Andel Research Institute and by NIH/NIAMS R01 grant AR053293 to BOW.
References
- Maupin KA , Droscha CJ , Williams BO . A comprehensive overview of skeletal phenotypes associated with alterations in Wnt/beta-catenin signaling in humans and mice. Bone Res 2013; 1: 27–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner DM , Ke J , Zhong Z et al. LRP5 and LRP6 in development and disease. Trends Endocrinol Metab 2013; 24: 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X , Liu H , Huang H et al. The key role of canonical Wnt/beta-catenin signaling in cartilage chondrocytes. Curr Drug Targets 2015; 17: 475–485. [DOI] [PubMed] [Google Scholar]
- Shin Y , Huh YH , Kim K et al. Low-density lipoprotein receptor-related protein 5 governs Wnt-mediated osteoarthritic cartilage destruction. Arthritis Res Ther 2014; 16: R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T , Jiang M , Hsu W . Gpr177, a novel locus for bone mineral density and osteoporosis, regulates osteogenesis and chondrogenesis in skeletal development. J Bone Miner Res 2013; 28: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao DY , Jonason JH , Zhang Y et al. Cartilage-specific beta-catenin signaling regulates chondrocyte maturation, generation of ossification centers, and perichondrial bone formation during skeletal development. J Bone Miner Res 2012; 27: 1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ , McMahon AP . Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 2006; 133: 3231–3244. [DOI] [PubMed] [Google Scholar]
- Spater D , Hill TP , O'Sullivan RJ et al. Wnt9a signaling is required for joint integrity and regulation of Ihh during chondrogenesis. Development 2006; 133: 3039–3049. [DOI] [PubMed] [Google Scholar]
- Akiyama H , Lyons JP , Mori-Akiyama Y et al. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev 2004; 18: 1072–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagaki J , Iwamoto M , Liu JG et al. Activation of beta-catenin-LEF/TCF signal pathway in chondrocytes stimulates ectopic endochondral ossification. Osteoarthritis Cartilage 2003; 11: 36–43. [DOI] [PubMed] [Google Scholar]
- Hartmann C , Tabin CJ . Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development 2000; 127: 3141–3159. [DOI] [PubMed] [Google Scholar]
- Miclea RL , Karperien M , Bosch CA et al. Adenomatous polyposis coli-mediated control of beta-catenin is essential for both chondrogenic and osteogenic differentiation of skeletal precursors. BMC. Dev Biol 2009; 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodewyckx L , Luyten FP , Lories RJ . Genetic deletion of low-density lipoprotein receptor-related protein 5 increases cartilage degradation in instability-induced osteoarthritis. Rheumatology (Oxford) 2012; 51: 1973–1978. [DOI] [PubMed] [Google Scholar]
- Joiner DM , Less KD , Van Wieren EM et al. Heterozygosity for an inactivating mutation in low-density lipoprotein-related receptor 6 (Lrp6) increases osteoarthritis severity in mice after ligament and meniscus injury. Osteoarthritis Cartilage 2013; 21: 1576–1585. [DOI] [PubMed] [Google Scholar]
- Funck-Brentano T , Bouaziz W , Marty C et al. Dkk-1-mediated inhibition of Wnt signaling in bone ameliorates osteoarthritis in mice. Arthritis Rheumatol 2014; 66: 3028–3039. [DOI] [PubMed] [Google Scholar]
- Blom AB , Brockbank SM , van Lent PL et al. Involvement of the Wnt signaling pathway in experimental and human osteoarthritis: prominent role of Wnt-induced signaling protein 1. Arthritis Rheum 2009; 60: 501–512. [DOI] [PubMed] [Google Scholar]
- Zhu M , Tang D , Wu Q et al. Activation of beta-catenin signaling in articular chondrocytes leads to osteoarthritis-like phenotype in adult beta-catenin conditional activation mice. J Bone Miner Res 2009; 24: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M , Chen M , Zuscik M et al. Inhibition of beta-catenin signaling in articular chondrocytes results in articular cartilage destruction. Arthritis Rheum 2008; 58: 2053–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papathanasiou I , Malizos KN , Tsezou A . Low-density lipoprotein receptor-related protein 5 (LRP5) expression in human osteoarthritic chondrocytes. J Orthop Res 2010; 28: 348–353. [DOI] [PubMed] [Google Scholar]
- Velasco J , Zarrabeitia MT , Prieto JR et al. Wnt pathway genes in osteoporosis and osteoarthritis: differential expression and genetic association study. Osteoporos Int 2010; 21: 109–118. [DOI] [PubMed] [Google Scholar]
- Kerkhof JM , Uitterlinden AG , Valdes AM et al. Radiographic osteoarthritis at three joint sites and FRZB, LRP5, and LRP6 polymorphisms in two population-based cohorts. Osteoarthritis Cartilage 2008; 16: 1141–1149. [DOI] [PubMed] [Google Scholar]
- Urano T , Shiraki M , Narusawa K et al. Q89R polymorphism in the LDL receptor-related protein 5 gene is associated with spinal osteoarthritis in postmenopausal Japanese women. Spine (Phila Pa 1976) 2007; 32: 25–29. [DOI] [PubMed] [Google Scholar]
- Smith AJ , Gidley J , Sandy JR et al. Haplotypes of the low-density lipoprotein receptor-related protein 5 (LRP5) gene: are they a risk factor in osteoarthritis? Osteoarthritis Cartilage 2005; 13: 608–613. [DOI] [PubMed] [Google Scholar]
- Pinson KI , Brennan J , Monkley S et al. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 2000; 407: 535–538. [DOI] [PubMed] [Google Scholar]
- Holmen SL , Giambernardi TA , Zylstra CR et al. Decreased BMD and limb deformities in mice carrying mutations in both Lrp5 and Lrp6. J Bone Miner Res 2004; 19: 2033–2040. [DOI] [PubMed] [Google Scholar]
- Kelly OG , Pinson KI , Skarnes WC . The Wnt co-receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development 2004; 131: 2803–2815. [DOI] [PubMed] [Google Scholar]
- Bondeson J . Are we moving in the right direction with osteoarthritis drug discovery? Expert Opin Ther Targets 2011; 15: 1355–1368. [DOI] [PubMed] [Google Scholar]
- Schett G , Zwerina J , David JP . The role of Wnt proteins in arthritis. Nat Clin Pract Rheumatol 2008; 4: 473–480. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov DA , Deng JM , Ogunrinu G et al. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 2000; 26: 145–146. [PubMed] [Google Scholar]
- Wu Q , Zhu M , Rosier RN et al. Beta-catenin, cartilage, and osteoarthritis. Ann N Y Acad Sci 2010; 1192: 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeng KS , Schumacher CA , Zylstra-Diegel CR et al. Lrp5 and Lrp6 redundantly control skeletal development in the mouse embryo. Dev Biol 2011; 359: 222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z , Zylstra-Diegel CR , Schumacher CA et al. Wntless functions in mature osteoblasts to regulate bone mass. Proc Natl Acad Sci USA 2012; 109: E2197–E2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD , Tasic B , Miyamichi K et al. A global double-fluorescent Cre reporter mouse. Genesis 2007; 45: 593–605. [DOI] [PubMed] [Google Scholar]
- Research NACfLA. Guidelines on the Care and Use of Animals for Scientific Purposes. Available at http://wwwavagovsg/NR/rdonlyres/C64255C0-3933-4EBC-B869-84621A9BF682/13557/Attach3AnimalsforScientificPurposesPDFE2204 (serial on the Internet) 2004.
- Riddle RC , Diegel CR , Leslie JM et al. Lrp5 and Lrp6 exert overlapping functions in osteoblasts during postnatal bone acquisition. PLoS ONE 2013; 8: e63323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod MJ . Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology 1980; 22: 299–301. [DOI] [PubMed] [Google Scholar]
- Bouxsein ML , Boyd SK , Christiansen BA et al. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 2010; 25: 1468–1486. [DOI] [PubMed] [Google Scholar]
- Zhu M , Tang D , Wu Q et al. Activation of -catenin signaling in articular chondrocytes leads to osteoarthritis-like phenotype in adult -catenin conditional activation mice. Journal Bone Miner Res 2009; 24: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M , Chen M , Zuscik M et al. Inhibition of catenin signaling in articular chondrocytes results in articular cartilage destruction. Arthritis Rheum 2008; 58: 2053–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi H . Regulation of osteoarthritis development by Wnt– catenin signaling through the endochondral ossification process. Journal Bone Miner Res 2009; 24: 8–11. [DOI] [PubMed] [Google Scholar]
- Weng LH , Wang CJ , Ko JY et al. Control of Dkk-1 ameliorates chondrocyte apoptosis, cartilage destruction, and subchondral bone deterioration in osteoarthritic knees. Arthritis Rheum 2010; 62: 1393–1402. [DOI] [PubMed] [Google Scholar]
- Wang M , Tang D , Shu B et al. Conditional activation of beta-catenin signaling in mice leads to severe defects in intervertebral disc tissue. Arthritis Rheum 2012; 64: 2611–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco J , Zarrabeitia M , Prieto J et al. Wnt pathway genes in osteoporosis and osteoarthritis: differential expression and genetic association study. Osteoporos Int 21: 109–118. [DOI] [PubMed] [Google Scholar]
- Iwaniec UT , Wronski TJ , Liu J et al. PTH stimulates bone formation in mice deficient in Lrp5. J Bone Miner Res 2007; 22: 394–402. [DOI] [PubMed] [Google Scholar]
- Kato M , Patel MS , Levasseur R et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol 2002; 157: 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T , Asaba H , Kang MJ et al. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci USA 2003; 100: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C , Williams BO , Cao X et al. LRP6 in mesenchymal stem cells is required for bone formation during bone growth and bone remodeling. Bone Res 2014; 2: 14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode A , Obri A , Paone R et al. Lrp5 regulation of bone mass and serotonin synthesis in the gut. Nat Med 2014; 20: 1228–1229. [DOI] [PubMed] [Google Scholar]
- Yadav VK , Ryu JH , Suda N et al. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell 2008; 135: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BO , Insogna KL . Where Wnts went: the exploding field of Lrp5 and Lrp6 signaling in bone. J Bone Miner Res 2009; 24: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y , Niziolek PJ , MacDonald BT et al. Reply to Lrp5 regulation of bone mass and gut serotonin synthesis. Nat Med 2014; 20: 1229–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y , Niziolek PJ , MacDonald BT et al. Lrp5 functions in bone to regulate bone mass. Nat Med 2011; 17: 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brommage R . Genetic approaches to identifying novel osteoporosis drug targets. J Cell Biochem 2015; 116: 2139–2145. [DOI] [PubMed] [Google Scholar]
- Lee GS , Simpson C , Sun BH et al. Measurement of plasma, serum, and platelet serotonin in individuals with high bone mass and mutations in LRP5. J Bone Miner Res 2014; 29: 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey JL , Li Z , Ellis JM et al. Wnt-Lrp5 signaling regulates fatty acid metabolism in the osteoblast. Mol Cell Biol 2015; 35: 1979–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J , Mowry LE , Nejak-Bowen KN et al. beta-catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation!. Hepatology 2014; 60: 964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong ZA , Zahatnansky J , Snider J et al. Wntless spatially regulates bone development through beta-catenin-dependent and independent mechanisms. Dev Dyn 2015; 244: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J , Ivy Yu HM , Maruyama T et al. Gpr177/mouse Wntless is essential for Wnt-mediated craniofacial and brain development. Dev Dyn 2011; 240: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]