Abstract
Head and neck squamous cell carcinoma remains a highly morbid and fatal disease. Importantly, genomic sequencing of head and neck cancers has identified frequent mutations in tumor suppressor genes. While targeted therapeutics increasingly are being investigated in head and neck cancer, the majority of these agents are against overactive/overexpressed oncogenes. Therapy to restore lost tumor suppressor gene function remains a key and under-addressed niche in trials for head and neck cancer. Recent advances in gene editing have captured the interest of both the scientific community and the public. As our technology for gene editing and gene expression modulation improves, addressing lost tumor suppressor gene function in head and neck cancers is becoming a reality. This review will summarize new techniques, challenges to implementation, future directions, and ethical ramifications of gene therapy in head and neck cancer.
Keywords: gene therapy, head and neck cancer, HNSCC, CRISPR, tumor suppressor
Introduction
As we progress into the genomics and personalized medicine era, targeted therapeutics increasingly are being investigated and utilized in head and neck squamous cell carcinoma (HNSCC). High mutation rates have been found in these tumors, with a majority of mutations occurring in tumor suppressor genes (Cancer Genome Atlas Network, 2015). Currently, multiple ongoing clinical trials are addressing targeted therapy in HNSCC (Birkeland and Brenner, 2015); however, the majority of agents being investigated are inhibitors of overexpressed or overactive oncogenes. Tumor suppressor genes have not been a primary focus of most current trials in HNSCC. Thus, despite recent advances in implementing targeted therapy paradigms towards personalized cancer care (Do et al., 2015), available therapeutic options have been limited by our ability to effectively restore lost tumor suppressor gene function.
Historically, gene therapy to correct disease has generated intense research interest. Conceptually, restoring mutated or deleted genes to a normal physiologic level has been appealing as a potential cure for a number of monogenic diseases. However, this field remains hotly debated, as it has been fraught with controversies in application for patient care. Excitingly, recent advances in genetic engineering and gene therapy have reinvigorated hope of application of gene therapy in human disease. Indeed, such therapy has captured renewed interest from both scientists and the public alike (Ledford, 2015; Reardon, 2015; Specter, 2015; Wallace-Wells, 2015). For HNSCC, this raises the possibility of restoring key tumor suppressor genes commonly mutated in head and neck cancers as a therapeutic option. Despite these recent significant advances in genetic engineering, however, many hurdles remain before we can further advance gene therapy in HNSCC. Key among these include overcoming technical challenges, optimizing patient selection, and addressing ethical dilemmas.
Gene Therapy in Human Disease
Gene therapy has been increasingly investigated in many diseases to restore lost function of key gene products. Historically, diseases that were investigated were established monogenic disorders with germline mutations, and included inherited enzymatic deficiencies (Wirth et al., 2013), blood dyscrasias (Cavazzana-Calvo et al., 2010), and immunodeficiencies (Cavazzana-Calvo et al., 2000; Aiuti et al., 2009). While some results from initial studies were encouraging, many concerning issues also arose. Specifically, iatrogenic harm to patients was noted in a few of the first reported gene therapy cases, including adverse fulminant reactions to the viral vector used in gene therapy, and off-target unintended gene alterations with resultant pre-malignant T cell proliferation (Marshall, 1999; Hacein-Bey-Abina et al., 2003).
Subsequently, gene therapy fell out of favor in some segments of the medical and scientific community. Nevertheless, the concept of correcting genetic defects through novel technology has remained alluring for scientists and clinicians alike. As a result, with significant scientific advances in recent years, there has been a renewed interest in gene therapy. In this past year, novel advances in gene editing have been successful in treating patients with leukemia (Reardon, 2015) and HIV (Tebas et al., 2014). In both instances, disease regression was achieved. Notably, in these instances, the modified cells were autologous T cells, which have the advantage over solid organ systems of being easily isolated, harvested and reintroduced into patients.
Gene therapy in HNSCC has been discussed and some early studies have been performed, but the field has yet to be robustly investigated. For HNSCC, gene therapy paradigms may be most applicable in restoring lost tumor suppressor gene function, particularly as an adjuvant to current standards of care. While many agents exist to inhibit activated or overexpressed oncogenes, currently therapies for restoring lost tumor suppressor genes are lacking.
Gene Engineering and Restoration Tools
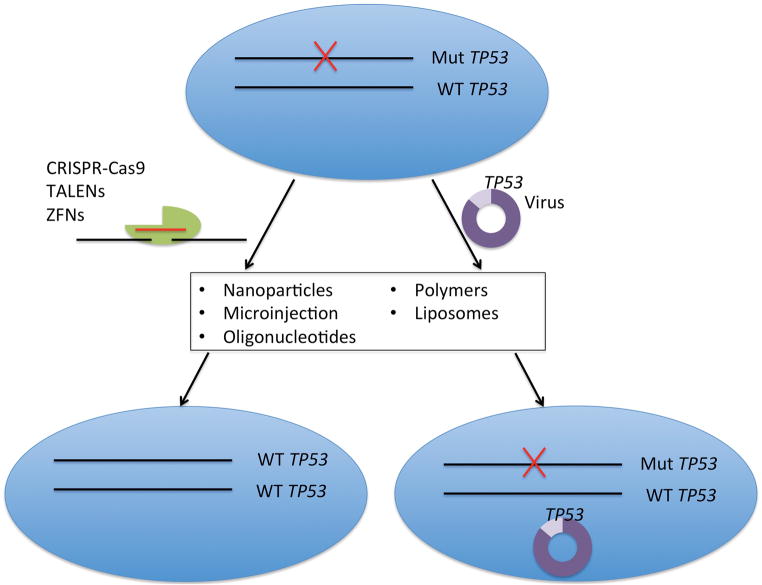
Two key components of gene therapy are the gene restoration or editing tool and the cellular targeting/delivery mechanism for the tool (Figure 1). Various iterations of these tools and delivery systems have been investigated, and this field continues to evolve.
Figure 1. Options for Gene Therapy in HNSCC.
Potential options for gene restoration in HNSCC, in this example for TP53. Gene editing of mutant TP53 or viral-mediated expression of WT TP53 are both potential options. An ideal gene therapy tool would incorporate 100% specificity, no footprint, no off target effects, selective tropism to HNSCC cells, and 100% efficacy.
Viral Gene Therapy and Gene Editing Therapy
Historically, recombinant gene products, driven by viral promoters, were the primary methodology for gene therapy. In these instances, gene function is restored by expression of a wild-type copy of the gene product, leading to constitutive expression of a functional gene product (Figure 1).
More recently, novel gene editing techniques have been utilized in translational research, with significant implications for future patient care. The most studied amongst these include zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat (CRISPR) associated Cas9 nucleases. Each system involves an efficient DNA nuclease, and a specific guide system (zinc fingers for ZFNs, transcription activator-like effectors for TALENs, and guide RNAs for CRISPR/Cas9) to orient the nuclease to a specific DNA region. In all cases, the nucleases cleave DNA, causing double-strand breaks. The DNA is then repaired either by random non-homologous end joining, which frequently causes disruptive insertion/deletion mutations, or by homologous recombination with a DNA template, which can result in corrective gene changes. These systems can be engineered to target specific segments within genes with high fidelity (Figure 1). Each of these nucleases has their benefits and limitations, discussed elsewhere in detail (LaFountaine et al., 2015), but overall each has garnered strong consideration in application for corrective editing in human diseases. ZFNs and TALENs have already been used to edit T cells in humans to provide successful therapy for HIV and leukemia, as described above (Reardon, 2015; Tebas et al., 2014). More recently, the CRISPR/Cas9 system has been investigated for specific gene editing (Jinek et al., 2013; Ran et al., 2013; Yin et al., 2014), and has been implemented into correcting hematopoietic stem cell mutations (Genovese et al., 2014). These systems have the potential for restoration of deleterious genetic mutations in tumor suppressor genes in HNSCC.
Gene Delivery Methods
Most frequently, the recombinant gene product or editing tool is delivered via a viral vector. A key distinction in viral vectors is whether the vector integrates into the human genome. A variety of viruses have been investigated, with the most common currently being adenoviruses. Some viruses exist episomally without DNA integration, whereas others will insert randomly in the human genome, thus introducing the possibility of a deleterious insertion (Thomas et al., 2003).
Novel techniques under investigation include direct microinjection (Cottle et al., 2015), nanoparticles and nanocarriers (Kemp et al., 2015), liposomes, and cationic polymers, among others (Mitra et al., 2015). It is important to note that these systems are utilized in in vitro experiments, with limited data suggesting their feasibility in humans (LaFountaine et al., 2015). Additionally, some techniques may not be optimal for large gene editing or gene therapy vectors. Moreover, these delivery systems do not address the necessary tissue specificity required for specifically targeting tumor cells. Specifying viral vectors or other delivery techniques to cancer cells, specifically in vivo, remains a challenging prospect. While some advances have been made in targeting specific cellular receptors overexpressed on cancer cells (Dosio et al., 2015), much more work is needed in this area before targeted gene deliver to cancer cells can become a reality.
Gene Therapy and Gene Editing in HNSCC
Investigations into gene therapy in HNSCC have been limited. Conceptually, restoration of lost tumor suppressor genetic checkpoints is alluring, as these are key for carcinogenesis (Hanahan and Weinberg, 2011). From genomic sequencing data, we have identified that the majority of HNSCCs have distinct deleterious mutations in one of a few key tumor suppressor genes (Cancer Genome Atlas Network, 2015; Birkeland et al., 2015a). As a result, checks on cell cycling (TP53, CDKN2A), squamous differentiation (NOTCH1) and apoptosis (CASP8) are lost (Table I). Despite this fact, while a number of targeted therapies are currently being advanced in early clinical trials for HNSCC (Birkeland and Brenner, 2015), these agents almost universally target oncogenes.
Table I. Commonly Mutated Tumor Suppressors in HNSCC.
HPV- HNSCC patients with mutated or deleted tumor suppressor genes from The Cancer Genome Atlas.
| Gene | Pathway | % Mutated or Deleted |
|---|---|---|
| TP53 | Cell Cycle | 84% |
| CDKN2A | Cell Cycle | 57% |
| NOTCH1 | Squamous Differentiation | 21% |
| CASP8 | Apoptosis | 10% |
| PTEN | Cell Cycle | 2% |
TP53, by far the most commonly mutated tumor suppressor gene, has been the focus of some prior investigations into gene therapy in HNSCC. It has made the most clinical progress in China, where a recombinant p53 protein delivered via an adenoviral vector (Gendicine) is in phase II/III clinical trials (Liu et al., 2013) as an adjuvant therapy to standard of care (surgery, chemotherapy and radiation). While early studies from China suggested a potential role for virally delivered p53, notably, there have been concerns about the reliability of these results (Xin, 2006). In the United States, investigations into adenoviral-delivered p53 remain in early clinical phases (Yoo et al., 2009), with few recent advances. Investigation into restoration of other tumor suppressor genes in HNSCC is lacking.
Challenges to and Future of Gene Therapy
While great advances have recently been made in gene therapy, a number of significant hurdles remain in its applicability for treatment for HNSCC.
Optimizing Gene Therapy Techniques
Current gene editing techniques remain imperfect. Algorithms and methodologies to enhance ZFN, TALEN, or CRISPR/Cas9 nuclease efficiency are currently being investigated, but in vitro efficiency can be quite low (Shi et al., 2015). Thus, repeated transfections or constitutive expression of the nucleases may be necessary in order to achieve a modified gene product over time. Additionally, these gene editing tools are currently best suited for point mutations. Larger insertion/deletion mutations and gene copy number deletions currently are hard to address with these gene-editing systems. Moreover, each gene editing technique has specific limitations in its targetable genetic segments. For instance, CRISPR/Cas9 needs a specific motif in the DNA that is recognized by its guide RNA, so if a patient’s mutation is not near such a motif, this tool may not be usable.
For viral gene delivery, dose titration remains imperfect. Increased gene activity above intended endogenous levels could lead to unexpected and unwanted effects in some instances. This may be of particular importance as some genes may act as both tumor suppressors and oncogenes in different contexts. The best example of this is NOTCH1, which frequently acts as a tumor suppressor in HNSCC, but may play an oncogenic role in subsets of HNSCCs (Rettig et al., 2015).
Enhancing Gene Therapy Delivery Systems
One of the most immediately challenging aspects of gene therapy is delivering the intended modulations into the intended cells. Ideally, gene therapy will involve editing the intended cancer gene without leaving a genomic footprint. While advances are being made in modulating viral vectors to have more specific tropism for cancer cells, further research into increasing specificity for delivering gene therapy specifically to cancer cells while avoiding surrounding normal tissue is an area in need of important advances. Furthermore, gene therapy would ideally be deliverable to all cells within a tumor. Thus, not only would an ideal vector target only cancer cells, but it would be able to deliver it’s gene editing/restoring tool with 100% efficiency. Such tools still elude us.
Off-Target Effects
As mentioned above, a number of off-target effects are real concerns for those interested in gene therapy. Viral vectors themselves can have unintended consequences: many of the viral vectors used for gene delivery currently will randomly insert into the human genome. While in many cases no effect may be evidenced from this insertion, in some instances important genes may be disrupted. Indeed, previous reports of adverse off-target events (Hacein-Bey-Abina et al., 2003; Kim and Kim, 2014) and fulminant systemic responses (Marshall, 1999) with gene therapy tempered the findings of initial trials. Additionally, gene editing enzymes currently do not have 100% fidelity, and off-target genomic alteration is a real concern (Kim and Kim, 2014), as a risk for off-target deleterious alterations of unintended genes exists.
Challenges in Selecting Gene Targets
Restoration of single gene function likely will not be sufficient in HNSCC. With the average HNSCC containing over 140 somatic mutations (Cancer Genome Atlas Network, 2015), it is not feasible to restore normative function to all mutated genes in all instances. Identifying and targeting the few key genes for gene therapy will need to be carefully considered. Whether the same genes, or a certain combination of genes, and in what personalized contexts remains an important question.
Paradigms for Gene Therapy Application
As gene therapy improves, discrete protocols will need to be introduced in order to guide who may be best served by gene therapy and in what contexts. Clinical trials incorporating gene therapy in conjunction with standard of care, either as adjuvant or neoadjuvant or palliative treatment, may be the most readily applicable paradigm. Consideration of additional treatment algorithms, including prevention of second primary malignancies, application to premalignant lesions, and implementation for embryos of patients with heritable cancer syndromes may alleviate significant morbidity and disease-burden. Certainly, however, these latter paradigms are fraught with significant ethical dilemmas.
Gene Therapy as Adjuvant HNSCC Therapy
Current clinical trials with adenovirus-mediated TP53 restoration employ this gene therapy as an adjuvant to current standard of care for HNSCC (Liu et al., 2013; Yoo et al., 2009). Likely, future investigations into gene therapy will follow a similar model. While it is unlikely that restoration of individual tumor suppressor genes will be sufficient for cancer therapy (given the large number of mutations in each tumor, and the multiple hits needed for carcinogenesis), it may be useful as an adjuvant treatment. In particular, restoration of tumor suppressor genes may result in chemosensitizing or radiosensitizing agent in conjunction with standardized therapy by restoring cell cycle checkpoint or apoptosis functions.
Editing multiple genes at once may be of importance for HNSCC as these tumors will frequently carry multiple lost tumor suppressor genes. Combinations to restore both TP53 and CDKN2A may be a useful initial universal step in gene therapy for HPV- HNSCCs given their exceedingly high rates of mutation in these tumors (Table I). Notably, gene-editing technologies can be used to create knockout mutations in oncogene pathways as well. Thus, one could conceivably deliver gene-editing technologies to individual cells to simultaneously restore lost tumor suppressor gene function (e.g. TP53) and inhibit oncogene activity (e.g. PIK3CA). Potentially, personalized combination cocktails could be delivered to patients to help restore some wild-type genetic function to tumor cells, which could affect tumor behavior and response to standard therapies.
Additional considerations may be made into application of gene therapy into premalignant lesions. As these tissues carry a lower overall mutational burden, and have acquired only some of the mutations in tumor suppressors necessary for carcinogenesis (Hanahan and Weinberg, 2011), these lesions may be the ideal targets for specific restoration of lost tumor suppressor gene function. In these lesions, the goal would be to halt the progression from dysplasia to frank carcinoma.
HPV+ HNSCC
HPV+ HNSCC, an increasingly prevalent subset of HNSCCs, carries a different mutational signature from traditional smoking and drinking-associated, HPV- HNSCCs (Cancer Genome Atlas Network, 2015). In these tumors, HPV viral genes E6 and E7 inhibit TP53 and RB1, respectively. As opposed to HPV- tumors, HPV+ HNSCCs frequently have wild-type copies of TP53 and CDKN2A. Thus, rather than attempting to edit these tumor suppressor genes, using gene editing to knock out the HPV E6 and E7 proteins and subsequently restore TP53 and RB function is a conceivable adjuvant treatment modality for these patients (Kennedy et al., 2014). Additionally, as there may be a strong immunogenic response component in these tumors, engineering of immune cells may prove to be a more attractive option.
Heritable HNSCC Syndromes
Heritable genetic diseases are being actively investigated for corrective gene editing in other frameworks, as mentioned above (e.g. monogenic immunodeficiency syndromes). Notably, a number of monogenic genetic syndromes exist (e.g. Lynch syndrome, Fanconi anemia) that predispose patients to HNSCC (Birkeland et al., 2015b). Potentially, these patients could undergo gene therapy in tissues at high risk (e.g. upper aerodigestive tract mucosa in Fanconi anemia), or in existing premalignant lesions. Additionally, there could potentially be a future role for germline or embryonic editing for offspring of these patients to avoid propagation of these genetic diseases, although this is currently an intensely debated topic, as discussed below.
Ethical Ramifications of Gene Therapy
As with any new and investigational technology, particularly those aimed at affecting the human genome, a full assessment of the ethical ramifications is important. Foremost for physicians, nonmaleficience, or “doing no harm” must be fully considered. In cases of gene editing, soon the question may not be “can we do this?”, but rather “should we do this?”. Already, there has been a call for a moratorium on using CRISPR technology for germ-line gene editing (Wade, 2015). A key point in this debate is between somatic (non-heritable) and germline (or heritable) alterations.
The majority of HNSCC patients do not have germline mutations or cancer syndromes, but rather gain somatic mutations. Ethical issues for these patients are numerous. For instance, we may not appreciate long-term deleterious effects of gene therapy treatments in patients for quite some time. As described above, gene editing enzymes have imperfect fidelity, and if permanently expressed in a cell, could result in an increasing number of unwanted or deleterious mutations. Historical examples of iatrogenic harm to patients from viral vector-mediated gene therapy also highlight the need for further optimization of our techniques and careful follow-up of patients who have received such treatments. Finally, inherent in providing gene therapy to patients is a full characterization of their genome. With such characterization comes incidental findings, which lead to a variety of issues regarding disclosure, right to know, and management of such findings (Birkeland et al., 2015b).
A small, but important, subset of patients may have heritable cancer syndromes. While the notion of providing a genetic cure to such syndromes certainly seems beneficial, this opens up a gray area in consideration of other heritable diseases and traits. This can quickly become more ethically dubious, and may blend into genetic engineering for preferred or unwanted traits (e.g. hair color). Additionally, as our tools for gene editing remain imperfect, again concerns about iatrogenic harm from unintended effects of our investigational technologies remains.
Conclusion
In sum, gene therapy for HNSCC remains an alluring, potentially viable, yet underdeveloped arm for therapeutic treatments. Specific technological hurdles currently remain, but as significant advances are rapidly being achieved, an era of genetic engineering in HNSCC is rapidly approaching. New challenges for this era will include optimizing gene therapeutic techniques, designing paradigms for employing personalized gene therapy, and setting guidelines for ethical issues inherent to genomics and gene therapy.
Footnotes
Disclosure: The authors have no conflicts of interest to disclose.
References
- Aiuti A, Cattaneo F, Galimberti S, Benninghoff U, Cassani B, Callegaro L, Scaramuzza S, Andolfi G, Mirolo M, Brigida I, Tabucchi A, Carlucci F, Eibl M, Aker M, Slavin S, Al-Mousa H, Al Ghonaium A, Ferster A, Duppenthaler A, Notarangelo L, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360(5):447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- Birkeland AC, Brenner JC. Personalizing medicine in head and neck squamous cell carcinoma: The rationale for combination therapies. Med Res Arch. 2015;3 doi: 10.18103/mra.v0i3.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland AC, Ludwig ML, Meraj TS, Brenner JC, Prince ME. The tip of the iceberg: Clinical implications of genomic sequencing projects in head and neck cancer. Cancers (Basel) 2015a;7(4):2094–2109. doi: 10.3390/cancers7040879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland AC, Uhlmann WR, Brenner JC, Shuman AG. Getting personal: Head and neck cancer management in the era of genomic medicine. Head Neck. 2015b doi: 10.1002/hed.24132. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, Bousso P, Deist FL, Fischer A. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288(5466):669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, Cavallesco R, Gillet-Legrand B, Caccavelli L, Sgarra R, Maouche-Chrétien L, Bernaudin F, Girot R, Dorazio R, Mulder GJ, Polack A, et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassemia. Nature. 2010;467(7313):318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottle RN, Lee CM, Archer D, Bao G. Controlled delivery of β-globin-targeting TALENs and CRISPR/Cas9 into mammalian cells for genome editing using microinjection. Sci Rep. 2015;5:16031. doi: 10.1038/srep16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do K, O’Sullivan Coyne G, Chen AP. An overview of the NCI precision medicine trials-NCI MATCH and MPACT. Chin Clin Oncol. 2015;4(3):31. doi: 10.3978/j.issn.2304-3865.2015.08.01. [DOI] [PubMed] [Google Scholar]
- Dosio F, Arpicco S, Stella B, Fattal E. Hyaluronic acid for anticancer drug and nucleic acid delivery. Adv Drug Deliv Rev. 2015 doi: 10.1016/j.addr.2015.11.011. epub. [DOI] [PubMed] [Google Scholar]
- Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, Moi D, Mazzieri R, Bonini C, Holmes MC, Gregory PD, van der Burg M, Gentner B, Montini E, Lombardo A, Naldini L. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510(7504):235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302(5644):415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp JA, Shim MS, Heo CY, Kwon YJ. “Combo” nanomedicine: Co-delivery of multi-modal therapeutics for efficient, targeted, and safe cancer therapy. Adv Drug Deliv Rev. 2015 doi: 10.1016/j.addr.2015.10.019. epub. [DOI] [PubMed] [Google Scholar]
- Kennedy EM, Kornepati AV, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88(20):11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15(5):321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- LaFountaine JS, Fathe K, Smyth HD. Delivery and therapeutic applications of gene editing technologies ZFNs, TALENs, and CRISPR/Cas9. Int J Pharm. 2015;494(1):180–194. doi: 10.1016/j.ijpharm.2015.08.029. [DOI] [PubMed] [Google Scholar]
- Ledford H. Mini enzyme moves gene editing closer to the clinic. Nature. 2015;520(7545):18. doi: 10.1038/520018a. [DOI] [PubMed] [Google Scholar]
- Liu S, Chen P, Hu M, Tao Y, Chen L, Liu H, Wang J, Luo J, Gao G. Randomized, controlled phase II study of post-surgery radiotherapy combined with recombinant adenoviral human p53 gene therapy in treatment of oral cancer. Cancer Gene Ther. 2013;20(6):375–378. doi: 10.1038/cgt.2013.30. [DOI] [PubMed] [Google Scholar]
- Marshall E. Gene therapy death prompts review of adenovirus vector. Science. 1999;286(5448):2244–2245. doi: 10.1126/science.286.5448.2244. [DOI] [PubMed] [Google Scholar]
- Mitra AK, Agrahari V, Mandal A, Cholkar K, Natarajan C, Shah S, Joseph M, Trinh HM, Vaishya R, Yang X, Hao Y, Khurana V, Pal D. Novel delivery approaches for cancer therapeutics. J Control Release. 2015 doi: 10.1016/j.jconrel.2015.09.067. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guide CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon S. Leukaemia success heralds wave of gene-editing therapies. Nature. 2015;527(7577):146–147. doi: 10.1038/nature.2015.18737. [DOI] [PubMed] [Google Scholar]
- Rettig EM, Chung CH, Bishop JA, Howard JD, Sharma R, Li RJ, Douville C, Karchin R, Izumchenko E, Sidransky D, Koch W, Califano J, Agrawal N, Fakhry C. Cleaved NOTCH1 expression pattern in head and neck squamous cell carcinoma is associated with NOTCH1 mutation, HPV status, and high-risk features. Cancer Prev Res (Phila) 2015;8(4):287–295. doi: 10.1158/1940-6207.CAPR-14-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33(6):661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specter M. The Gene Hackers. New Yorker. 2015 Nov 16; NewYorker.com. Web. 8 Dec. 2015.
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370(10):901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Wade N. Scientists seek moratorium on edits to human genome that could be inherited. New York Times. 2015 Dec 3; nytimes.com. Web. 8 Dec. 2015.
- Wallace-Wells D. What Will Happen When Gene Editing Is As Easy As Cut-and-Paste? New York Magazine. 2015 Dec 4; nymag.com. Web. 8 Dec. 2015.
- Wirth T, Parker N, Yia-Herttuala S. History of gene therapy. Gene. 2013;525(2):162–169. doi: 10.1016/j.gene.2013.03.137. [DOI] [PubMed] [Google Scholar]
- Xin H. Chinese gene therapy. Gendicine’s efficacy: hard to translate. Science. 2006;314(5803):1233. doi: 10.1126/science.314.5803.1233. [DOI] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo GH, Moon J, Leblanc M, Lonardo F, Urba S, Kim H, Hanna E, Tsue T, Valentino J, Ensley J, Wolf G. A phase 2 trial of surgery with perioperative INGN 201 (Ad5CMV-p53) gene therapy followed by chemoradiotherapy for advanced, resectable squamous cell carcinoma of the oral cavity, oropharynx, hypopharynx, and larynx: report of the Southwest Oncology Group. Arch Otolaryngol Head Neck Surg. 2009;135(9):869–874. doi: 10.1001/archoto.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]