Abstract
Mycoplasma hyopneumoniae is a genome-reduced, cell wall-less, bacterial pathogen with a predicted coding capacity of less than 700 proteins and is one of the smallest self-replicating pathogens. The cell surface of M. hyopneumoniae is extensively modified by processing events that target the P97 and P102 adhesin families. Here, we present analyses of the proteome of M. hyopneumoniae-type strain J using protein-centric approaches (one- and two-dimensional GeLC–MS/MS) that enabled us to focus on global processing events in this species. While these approaches only identified 52% of the predicted proteome (347 proteins), our analyses identified 35 surface-associated proteins with widely divergent functions that were targets of unusual endoproteolytic processing events, including cell adhesins, lipoproteins and proteins with canonical functions in the cytosol that moonlight on the cell surface. Affinity chromatography assays that separately used heparin, fibronectin, actin and host epithelial cell surface proteins as bait recovered cleavage products derived from these processed proteins, suggesting these fragments interact directly with the bait proteins and display previously unrecognized adhesive functions. We hypothesize that protein processing is underestimated as a post-translational modification in genome-reduced bacteria and prokaryotes more broadly, and represents an important mechanism for creating cell surface protein diversity.
Keywords: two-dimensional gels, adhesins, global proteome, processing, protein-centric
1. Background
Mycoplasma spp. are bacteria that evolved by a process of degenerative evolution from the low G + C Firmicutes. Mycoplasmas have lost genes for cell wall biosynthesis, and many anabolic processes (including a TCA cycle) are reliant on glycolysis for the production of cellular ATP [1,2]. Mycoplasmas typically have small genomes of less than 1000 kbp and are dependent on the host for the supply of cholesterol for membrane biosynthesis, amino acids, nucleotides and other macromolecular building blocks for cell growth [3]. As such, mycoplasmas are excellent model organisms to examine the complexity of post-translational modifications in prokaryotes.
Mycoplasma hyopneumoniae is an agriculturally significant swine respiratory pathogen that causes substantial economic losses, estimated in the billions of dollars per annum [4]. Complete genome sequences of four geographically distinct strains of M. hyopneumoniae are available [3,5,6], shedding light on the metabolic capacity, host specialization and evolutionary background of this minimal organism. Genomes range in size from 850 to 920 kb and encode approximately 700 open reading frames (ORFs). The M. hyopneumoniae strain 232 genome contains 691 known proteins and 728 annotated genes. A recent proteome analysis of strain 232 identified 8607 unique peptide sequences (false discovery rate of 0.53%) confirming the expression of 70% (483) of the 691 predicted ORFs during culture in Friis broth. This included 171 of the 328 predicted hypothetical proteins (52%), 80% of the lipoprotein genes, and all the P97/P102 adhesin gene families. In the same study, proteogenomic analysis of strain 232 uncovered previously unidentified genes and 5′ extensions to several genes [7]. Transcriptome studies indicate that 92% of predicted ORFs are transcribed in M. hyopneumoniae strain 7448 [8]. Seventy-eight non-coding RNAs were also identified in the analysis. Genes with the highest expression levels primarily encoded proteins involved in basal metabolism, as well as chaperones, adhesins, surface proteins, transporters and RNase P. A number of uncharacterized proteins were also identified. The M. hyopneumoniae gene encoding the P216 adhesin protein was also presented with a significant number of transcripts (RPKM, reads per kilobase of transcript per million mapped reads: 10 796.4) [8]. While these approaches have shed light on the protein coding capacity of M. hyopneumoniae, they do little to understand the extent by which it modifies its proteome post-translationally.
During the early, critical stages of infection, M. hyopneumoniae adheres specifically along the entire length of cilia of ciliated epithelial cells that line the trachea, bronchi and bronchioles in the upper respiratory tract of pigs. This association causes ciliostasis, loss of cilia and eventual epithelial cell death, which effectively perturbs mucociliary function. The P97 and P102 adhesin families are central to mediating attachment of M. hyopneumoniae to epithelial cilia [9–19]. Notably, all members of the P97 and P102 adhesin families are processed post-translationally to the extent that it is difficult to find evidence of adhesin pre-proteins [9–12,15,17,18,20–23]. Most members of the P97 and P102 families are processed via highly efficient cleavage events typically at S/T–X–F↓–X–D/E sites, but also within stretches of hydrophobic amino acids and by numerous, less efficient cleavage events often in a manner consistent with trypsin-like activity [20–22,24]. Consequently, the surface protein architecture of M. hyopneumoniae displays cleavage fragments derived via processing of the P97 and P102 adhesin families by several endopeptidases. What is unclear is how endoproteolysis alters the presentation of surface proteins not related to the P97 and P102 adhesin families, including members of the lipoprotein family.
The current trend in global proteomic analysis has been to use high-speed, ultra-sensitive mass spectrometers combined with orthogonal upfront chromatographic fractionation (i.e. two-dimensional LC–MS/MS) in a peptide-centric manner to characterize proteomes. These high-throughput protocols rely on all proteins in a sample being digested with an efficient protease (e.g. trypsin) into peptides for downstream analysis. Peptide-centric or ‘bottom-up’ approaches are used widely, because peptides are more readily solubilized for fractionation and are amenable to chromatographic separation, and mass spectrometry is more sensitive when analysing peptides, rather than intact proteins [25]. Conversely, protein-centric approaches aim to preserve intact proteins throughout fractionation steps, so that proteoform information may be retained [26], and then discrete proteins or fractions are digested to peptides and analysed individually by mass spectrometry. Protein-centric methods are thus not necessarily ‘top-down’ approaches that aim to analyse individual intact proteins by mass spectrometry [27]. Without selective enrichment, high-throughput peptide-centric approaches can fail to capture post-translational proteolytic modifications and can lead to an oversimplification of the complexity of the proteome. In this study, we applied protein-centric approaches that retain mass context with the aim of identifying proteins that are targets of processing events in M. hyopneumoniae-type strain J.
2. Experimental procedures
2.1. Preparation of Mycoplasma hyopneumoniae whole cell lysate
Mycoplasma hyopneumoniae (strain J) was grown in modified Friis broth [28] and harvested as described previously [29]. A 0.1 g pellet of M. hyopneumoniae cells was resuspended in 7 M urea, 2 M thiourea, 40 mM Tris–HCl pH 8.8, 1% w/v C7BzO and disrupted with four rounds of sonication at 50% power for 30 s bursts on ice. Proteins were reduced and alkylated with 5 mM tributylphosphine and 20 mM acrylamide monomers for 90 min. Insoluble material was pelleted by centrifugation at 16 000g for 10 min, and the remaining soluble protein was precipitated in five volumes of ice-cold acetone for 30 min and the pellet air-dried.
For one-dimensional SDS–PAGE, the pellet was resuspended in SDS sample buffer (0.25 M Tris–HCl pH 6.8; 0.25% w/v SDS; 10% glycerol and 0.0025% w/v bromophenol blue). For two-dimensional-PAGE, protein pellets were resuspended in 7 M urea, 2 M thiourea, 1% w/v C7BzO. If solution conductivity was measured to be greater than 200 µS cm−1, samples were desalted and buffer exchanged into 7 M urea, 2 M thiourea, 1% w/v C7BzO using a microBioSpin column (Bio-Rad) according to manufacturer's instructions.
2.2. TX-114 extraction
Mycoplasma hyopneumoniae cell pellets were resuspended in 1% Triton buffer (1% Triton X-114, 10 mM Tris–HCl pH 8.0, 150 mM sodium chloride, 1 mM EDTA) and extracted as previously described [11,30]. The detergent phase sample was mixed with SDS–sample buffer and separated as for GeLC–MS/MS.
2.3. Two-dimensional polyacrylamide gel electrophoresis
Two-dimensional gels were run using 250 µg of whole cell lysate with 0.2% pH 3–10 carrier ampholytes (Bio-Rad). Isoelectric focusing was performed using 11 cm pH 4–7 IPG strips (Bio-Rad) and 11 cm pH 6–11 immobiline drystrips (GE Healthcare). Focusing was carried out using a Protean IEF system (Bio-Rad) at a constant 20°C and 50 µA current limit per strip with a three-step programme: slow ramp to 4000 V for 4 h, linear ramp to 10 000 V for 4 h, then 10 000 V until 120 kVh was reached. Following IEF, the strips were equilibrated with 5 ml equilibration solution (2% SDS, 6 M urea, 250 mM Tris–HCl pH 8.5, 0.0025% (w/v) bromophenol blue) for 20 min before the second-dimension SDS–PAGE. The second-dimension gels were run using precast Bio-Rad TGX midi gels with TGS running buffer (Bio-Rad). Reference gels were stained with Coomassie blue G250 overnight and destained with 1% acetic acid to remove background. All visible spots (180 from the pH 4–7 gel and 160 from the pH 6–11 gel) were manually excised from the gel and subjected to in-gel trypsin digestion, before analysis by LC–MS/MS.
2.4. One-dimensional gel electrophoresis liquid chromatography tandem mass spectrometry
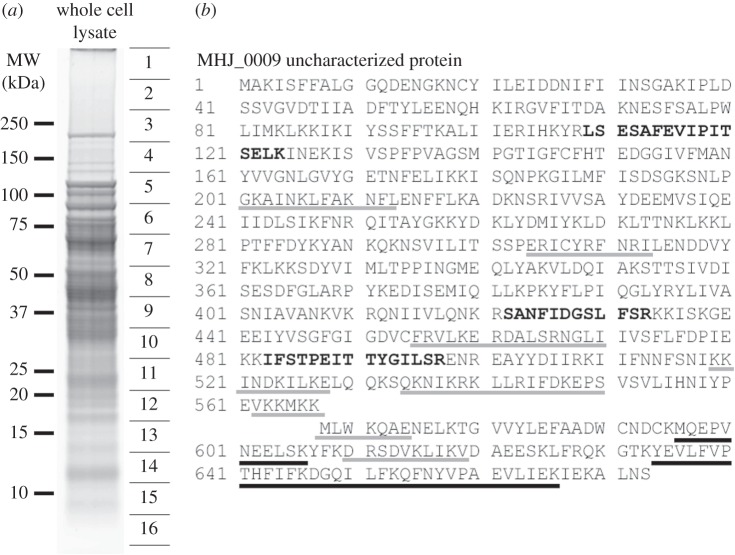
GeLC–MS/MS was performed on three biological replicates of M. hyopneumoniae whole cell lysates, with technical replicates analysed by ion trap MS/MS and Q-TOF MS/MS (representative lane shown in figure 4a). One-dimensional GeLC–MS/MS was also performed on a TX-114 detergent fraction and on a high-load lane of whole cell extract (where mass context was not reliably retained owing to macromolecular crowding effects), and these were also analysed by Q-TOF MS.
Figure 4.
GeLC–MS/MS analysis identified cleavage fragment of MHJ_0009 (Q4AAU0). (a) Representative one-dimensional gel of M. hyopneumoniae whole cell lysates. The gel lanes were cut into 16 slices (as shown), digested in-gel with trypsin and analysed by LC–MS/MS using ion trap and Q-TOF instruments, allowing protein mass context to be retained. (b) Identified peptides mapping to uncharacterized protein MHJ_0009 (Q4AAU0) in bold. Peptides in bold were identified from gel slice 6 at the approximate predicted intact mass (77 kDa). Peptides underlined in black were generated from proteins identified only from slices 13 and 14. Analysis of the C-terminal cleavage fragment spanning amino acids 568–664 with ProtParam indicated that it was 12.5 kDa with a predicted pI of 5.47 (see also figure 1). MHJ_0009 was also identified by GeLC–MS/MS from slices at approximately 12 kDa from low-affinity heparin chromatography elutions. Putative heparin binding motifs are underlined in grey.
About 150 µg of protein from any preparation was separated by SDS–PAGE, and fixed and stained with Coomassie blue G-250. Additionally, a high-load lane was run using 500 µg protein from whole cell lysates. Entire gel lanes were cut into 16 equal slices for whole cell lysates, 30 for the high-load lane or 15 for the TX-114 fraction. Gel slices were further diced into approximately 1 mm2 cubes, destained, washed and digested in-gel with trypsin for analysis. Identification of proteins was performed following clean-up of peptide fractions using OMIX C18 SPE pipette tips, using one of the LC–MS/MS methods described below.
2.5. Expression of recombinant proteins and creation of polyclonal antisera
Expression of recombinant P65 and creation of polyclonal antisera was carried out as described previously [9,14,31].
2.6. Blotting
Proteins separated on pH 6–11 two-dimensional gels were transferred to PVDF membranes as described previously [12]. Blots were blocked with 5% (w/v) skim milk powder in PBS with 0.1% Tween 20 (v/v) (PBS-T) at room temperature for 1 h. For detection of immunogenic proteins, membranes were probed with pooled convalescent sera collected from low-health-status M. hyopneumoniae-infected pigs described previously [9] diluted 1 : 100 in PBS-T for 1 h, followed by incubation with peroxidase-conjugated anti-pig antibodies diluted 1 : 3000 in PBS-T for 1 h. For detection of adhesin R1 cilium binding domains, membranes were probed with antisera raised against the F3 recombinant fragment that spans the R1 cilium binding domain of MHJ_0194 (F3P97); described previously [14] diluted 1 : 100 in PBS-T for 1 h, then peroxidase-conjugated anti-rabbit antibodies diluted 1 : 1500 in PBS-T for 1 h. For detection of P65 fragments, membranes were probed with antisera raised against recombinant P65 diluted 1 : 200 in PBS-T for 1 h, then peroxidase-conjugated anti-rabbit antibodies diluted 1 : 2000 in PBS-T for 1 h. Membranes were washed in three changes of PBS-T between incubations and were developed with SIGMAFAST 3,3′-diaminobenzidine tablets (Sigma-Aldrich) as per manufacturer's instructions.
2.7. Affinity chromatography for identification of protein interactions
Heparin affinity chromatography and avidin purification of fibronectin-binding proteins and PK15 cell surface protein interactors were performed as described previously [20–22].
Avidin purification of actin- and plasminogen-binding proteins was carried out as follows. Actin from bovine muscle (Sigma-Aldrich) was solubilized in 8 M urea, 20 mM triethylammonium bicarbonate, pH 8.0. Cysteine residues were reduced and alkylated with 5 mM tributylphosphine and 20 mM acrylamide monomers for 90 min at room temperature. Actin monomers were labelled in 20-fold molar excess Sulfo-NHS-LC-Biotin for 3 h at room temperature. Plasminogen from human serum (Sigma-Aldrich) was labelled in 20-fold molar excess Sulfo-NHS-LC-Biotin for 3 h at room temperature. Excess biotin was removed by buffer exchange into PBS using a PD-10 Desalting Column (GE Healthcare, Life Sciences). Biotinylated actin and plasminogen were incubated with avidin agarose (Thermo Scientific) on a rotating wheel for 5 h. The separate slurries were packed into columns and the flow-through collected from each. Unbound ligand was thoroughly washed with PBS. M. hyopneumoniae cells were pelleted by centrifugation at 10 000g for 20 min, washed with PBS, and gently lysed in 0.5% Triton X-100/PBS. Insoluble material was removed by centrifugation at 16 000g for 10 min, and the cleared lysate was incubated with biotinylated ligand–avidin agarose mixtures overnight on a rotating wheel at 4°C. The mixtures were packed into columns, and the unbound proteins were thoroughly washed and collected in PBS. Interacting proteins were eluted with 30% acetonitrile, 0.4% trifluoroacetic acid. The eluting proteins were concentrated using a 3000 Da cut-off filter and acetone precipitated before pelleting by centrifugation. Elutions were subsequently subjected to one-dimensional SDS–PAGE for transfer and detection by blotting or GeLC–MS/MS for protein identification.
Surface proteins were identified by enzymatic cell surface shaving using trypsin for 5 min at 37°C as previously described [12] and cell surface labelling using Sulfo-NHS-LC-Biotin for 30 s at 4°C as previously described [10].
2.8. One-dimensional liquid chromatography tandem mass spectrometry using Q-TOF
These methodologies were performed as described previously [21,22]. Briefly, samples were loaded using an Eksigent AS-1 autosampler connected to a Tempo nanoLC system (Eksigent, USA) at 20 µl min−1 onto a C8 trap column (Michrom, USA) before washing and elution at 300 nl min−1 onto a PicoFrit column (75 µm × 150 mm) packed with Magic C18AQ resin (Michrom, USA). Peptides were eluted and ionized into the source of a QSTAR Elite hybrid quadrupole time-of-flight mass spectrometer (AB Sciex) at 2300 V using the following programmes: 5−50% MS solvent B (98% acetonitrile + 0.2% formic acid) over 30 min for gel slices or 15 min for gel spots, 50−80% MS B over 5 min, 80% MS B for 2 min, 80−5% for 3 min. An intelligent data acquisition experiment was performed, with a mass range of 350−1500 Da scanned for peptides of charge state 2+ to 5+ with an intensity of more than 30 counts scan−1. Selected peptides were fragmented, and the product ion fragment masses were measured over a mass range of 50−1500 Da. The mass of the precursor peptide was then excluded for 120 s for gel slices or 15 s for gel spots.
2.9. One-dimensional liquid chromatography–mass spectrometry/mass spectrometry using ion trap
Peptide samples were analysed by nanoflow LC–MS/MS (nanoLC-MS/MS) using a LTQ-XL linear ion trap mass spectrometer (Thermo, San Jose, CA), using a fused silica capillary with an integrated electrospray tip (75 µm ID × 70 mm) packed with 100 Å, 5 µm Zorbax C18 resin (Agilent Technologies, CA, USA). An electrospray voltage of 1800 V was applied via a liquid junction upstream of the C18 column. Samples were injected onto the column using a Surveyor autosampler, which was followed by an initial wash step with buffer A (5% v/v acetonitrile, 0.1% v/v formic acid) for 10 min at 1 µl min−1. Peptides were eluted from the column with 0–50% buffer B (95% v/v acetonitrile, 0.1% v/v formic acid) for 58 min at 500 nl min−1. The column eluate was directed into a nanospray ionization source of the mass spectrometer. Spectra were scanned over the range of 400–1500 amu and, using Xcalibur software (version 2.06, Thermo), automated peak recognition, dynamic exclusion and MS/MS of the top six most intense precursor ions at 35% normalization collision energy were performed.
2.10. Mass spectrometry/mass spectrometry data analysis
The MS/MS data files were searched using Mascot (provided by the Australian Proteomics Computational Facility, hosted by the Walter and Eliza Hall Institute for Medical Research Systems Biology Mascot Server) against the LudwigNR database, comprising the UniProt, plasmoDB and Ensembl databases (vQ209. 8785680 sequences, 3087386706 residues), with the following parameter settings. Fixed modifications: none; variable modifications: propionamide, oxidized methionine, deamidated asparagine, n-terminal pyroglutamic acid and carbamoylmethylcysteine cyclization; enzyme: semitrypsin; number of allowed missed cleavages: 3; peptide mass tolerance: 100 ppm or 2.0 Da for data generated by Q-TOF or ion trap instruments, respectively. MS/MS mass tolerance: 0.2 Da or 0.4 Da for data generated by Q-TOF or ion trap instruments, respectively; charge state: 2+ and 3+.
Scaffold v. 3.00.02 (Proteome Software Inc., Portland) was used to validate and compare MS/MS-based peptide and protein identifications. Peptide identifications were accepted if their calculated probability was greater than 95.0% with a false discovery rate of 1.27%, and protein identifications were accepted if their calculated probability using the Peptide Prophet algorithm was greater than 80.0% with a false discovery rate of 2.4%. Protein probabilities were assigned by the Protein Prophet algorithm. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
The use of multiple techniques improved confidence in ‘one-hit wonders'; proteins identified by a single peptide in a single replicate. Adopting the approach of White et al. [32], if the same single peptide was identified in two or more replicates or experiments, the protein was considered to be present, rather than a ‘one-hit wonder’. Similarly, if a single peptide identified a protein in one replicate and a different single peptide identified the same protein in a separate replicate, then the protein was considered to be expressed. Single peptide hits were only retained in the dataset if, after being subjected to manual validation, the MS/MS spectra had a considerable sequence of b- and y-ions that were the dominant ions in the spectra. Six proteins were identified to be true one-hit wonders, with the identifying spectra and fragmentation data shown in electronic supplementary material, figure S4.
2.11. In silico analyses
Predicted MW and pI information for intact proteins and fragments was obtained using ProtParam via ExPASy bioinformatics resource portal (http://web.expasy.org/protparam/) [33]. Transmembrane domain predictions were made using TMPred (http://embnet.vital-it.ch/software/TMPRED_form.html) [34] with default minimum 17 and maximum 33 amino acid length of the hydrophobic portion of the transmembrane helix. The PONDR VSL2 algorithm was used to predict regions of protein disorder (http://pondr.com) [35]. All analyses were performed using sequence data obtained from UniProt (http://www.uniprot.org/) [36].
3. Results
3.1. Protein-centric approaches to mapping the Mycoplasma hyopneumoniae proteome
We applied a series of fractionation technologies that retain mass context to lysates of M. hyopneumoniae-type strain J, to determine the diversity of proteins that are targets of endoproteolytic processing. Members of two adhesin families related to P97 and P102 respectively are known to be extensively processed on the cell surface of M. hyopneumoniae, but the extent to which proteins on the cell surface are targets of endoproteolytic processing has not been explored. Three hundred and forty-seven unique M. hyopneumoniae strain J proteins, representing approximately 52% of the predicted proteome, were identified from the combined experiments following analysis by Scaffold (electronic supplementary material, table S1). Table 1 summarizes the identification of proteins expressed in M. hyopneumoniae as detected by each of these methods. Interestingly, two uncharacterized proteins were identified mapping only to strain 232: an 8.8 kDa protein, Q5ZZV3, identified by one peptide in two runs on both ion trap and Q-TOF; and an 11.3 kDa protein, Q5ZZV5, identified by two peptides in one run from ion trap data. A BLAST search of the UniProt database shows that these proteins are conserved among strains 232, 7448 and 168; however, they are not annotated to be present in strain J. Seventy-seven (22%) of the identified proteins are named in UniProt as ‘uncharacterized protein’, despite some sharing homology with proteins that are well characterized in the literature such as P97 and P102 paralogues, MHJ_0369 and MHJ_0368 (Q4A9W4 and Q4A9W5), homologues of Mhp385 and Mhp384 (Q600R9 and Q600S0) respectively, in M. hyopneumoniae strain 232 [12].
Table 1.
Overview of number of identifications by each method.
| method | protein IDs | peptide IDs | unique spectra | proteins unique to method |
|---|---|---|---|---|
| Ge ion trap | 331 | 2774 | 3832 | 7 |
| Ge Q-TOF | 297 | 1701 | 1961 | 2 |
| Ge high-load | 331 | 1748 | 2093 | 6 |
| Ge TX-114 | 206 | 846 | 897 | 5 |
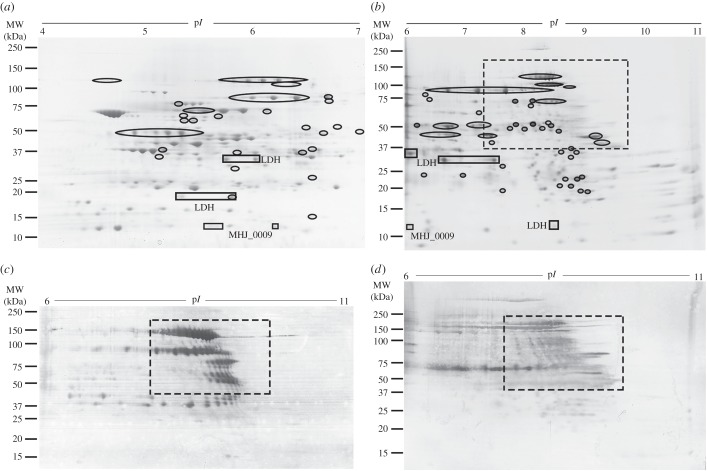
GeLC–MS/MS preserves the intact molecular weight of proteins and was a valuable strategy to identify cleavage events that affected the migration of members of the P97 and P102 adhesin families [10,20–22]. Much finer resolution of cleavage fragments was achieved using two-dimensional PAGE. pH 4–7 and 6–11 gels were run using whole cell extracts of M. hyopneumoniae (figure 1). Overall, 340 spots comprising 180 spots from a 4–7 isoelectric point gradient gel and 160 spots from a 6–11 isoelectric point gradient gel were resolved well enough to be excised and analysed by LC–MS/MS. Identifications were obtained for 302 spots (159 from pI 4–7 and 143 from pI 6–11; electronic supplementary material, figures S2 and S3). One hundred and thirty unique proteins were identified from these 302 spots, representing 19% of the predicted proteome (37% of the identifiable proteome).
Figure 1.
Two-dimensional gels and immunoblots. (a) Two-dimensional gel image (pH 4–7) with locations of relevant spots indicated. (b) Two-dimensional gel image (pH 6–11) with locations of relevant spots indicated and ‘cloud region’ boxed by a dashed line. Spots identified to contain protein cleavage fragments are circled. Full description of all cut spots and identifications from pH 4–7 and pH 6–11 two-dimensional gels can be found in electronic supplementary material, figures S2 and S3, respectively. (c) Two-dimensional blot probed with rabbit serum raised against the F3 recombinant fragment that spans the R1 cilium binding domain of MHJ_0194 (F3P97). Strongly staining protein fragments carrying regions of R1 or R1-like fragments of the cilium adhesin P97 and Mhp271 show that proline-rich repeats are highly antigenic. (d) A blot probed with pooled convalescent sera from sero-positive pigs. The ‘cloud regions' are also boxed, showing overlap between adhesin fragments and immunoreactive regions of the blots.
Eighty-seven proteins were identified from multiple spots. Not all of these, however, could be attributed to processing events, with a significant number of proteins appearing as ‘spot trains' at a specific molecular weight that track along the pI gradient. This is likely to be the result of other post-translational modifications that affect pI, such as deamidation, or phosphorylation, which has been previously documented in M. hyopneumoniae [18]. Of particular interest was the presence of ‘cloud regions' where numerous spots could be detected, but could not be individually resolved (figure 1, boxed). These cloud regions are significant, as similar patterns in the same region have been previously identified when M. hyopneumoniae proteins were separated over nonlinear pH 6–11 gels using a different gel system, carried out in a different laboratory and are thus unlikely to be an artefact of sample preparation or gel separation methods [23]. We postulated that these low-abundance cleavage fragments are generated by endoproteolysis of abundantly expressed members of the P97 and P102 adhesin families. A two-dimensional blot probed with rabbit anti-F3P97 serum [14] showed that P97, P66 and a range of lower abundance fragments of MHJ_0194 are recognized (figure 1c). Identical blots probed with a pool of convalescent sera sourced from pigs testing positive for infection with M. hyopneumoniae showed a strong reaction to the low-abundance P97 and P102 adhesin cleavage fragments (figure 1d). These observations are consistent with the highly immunoreactive nature of proteins carrying proline-rich repeats [18] such as those recognized by anti-F3P97 serum.
3.2. The extent of protein processing in Mycoplasma hyopneumoniae
Overall, 35 proteins (10% of the identified proteome) showed convincing evidence of proteolytic processing through identification from protein-centric experiments at molecular masses that were not in agreement with the predicted intact mass (table 2; also denoted by asterisks in electronic supplementary material, table S1). Ten of these belong to the P97/P102 adhesin families. Consistent with these data, endoproteolytic processing events have been characterized in P97/P102 adhesin families including MHJ_0194 (P97) [20,23], MHJ_0493 (P97 paralogue P216) [18,22], MHJ_0663 (P97 paralogue P146) [9], MHJ_0369 (P97 paralogue Mhp385) [12], MHJ_264 (P97 paralogue Mhp107) [16], MHJ_0195 (P102) [17,23], MHJ_0494 (P102 paralogue P159/P76/P110) [21,37], MHJ_0662 (P102 paralogue P135/Mhp683) [10], MHJ_0638 (P102 paralogue Mhp384) [12] and MHJ_263 (P102 paralogue P116/Mhp108) [15]. Other proteins showing evidence of cleavage include five uncharacterized proteins, three known surface antigens, two annotated proteases, multiple annotated cytosolic proteins and glycolytic enzymes, such as pyruvate dehydrogenase complex components A, B and D, and lactate dehydrogenase. It is important to note that this list is not exhaustive, as many other proteins were not identified with sufficient sequence coverage to be confirmed as cleavage fragments.
Table 2.
Cleaved proteins identified from protein-centric analyses. Intact MW shows the calculated mass of the predicted intact protein. Identified masses from one-dimensional GeLC–MS/MS experiments and two-dimensional gels also shown. Putative Fn binding and Heparin binding show mass ranges at which proteins were identified from fibronectin- and heparin-affinity chromatography GeLC–MS/MS experiments, respectively. Masses provided in kDa. D: identified from TX-114 detergent phase GeLC-MS/MS. (N) or (C): identified fragment mapping to N- or C-terminus of the protein. Shading indicates not detected in cell surface analyses. References are provided where proteins have been previously characterized.
| accession | putative cleaved protein ID | gene | intact MW | identified mass | mass from 4 to 7 two-dimensional gels | mass from 6–11 two-dimensional gels | putative Fn binding | heparin binding | |
|---|---|---|---|---|---|---|---|---|---|
| adhesins | Q4A925 | putative adhesin-like protein P146 [9] | MHJ_0663 | 147 | 120–70, 50–40, ∼37–25D | 94 | 35, 38, 45, 92 | intact | 95–21 |
| Q4A926 | uncharacterized protein [10] | MHJ_0662 | 135 | 50–45 | 52, 74 | 47, 53, 55, 57 | — | 57–27, 21–16 | |
| Q4A9J1 | putative P76 membrane protein (P159) [21] | MHJ_0494 | 161 | ∼110–20 | [21] | [21] | ∼20 | 153–27, 21–16 | |
| Q4A9J2 | putative P216 surface protein [22] | MHJ_0493 | 216 | ∼120, 85–20 | [22] | [22] | — | 153–27, 21–16 | |
| Q4A9W4 | uncharacterized protein [12] | MHJ_0369 | 114 | ∼30 | 25 | 23, 25 | — | 95–73, 21–16 | |
| Q4A9W5 | putative Lppt protein [12] | MHJ_0368 | 109 | ∼50, 30–20D | 57 | — | — | 57–44 | |
| Q4AA66 | putative P97-like protein [16] | MHJ_0264 | 120 | ∼25 | — | 100 | — | — | |
| Q4AA67 | putative P102-like protein [15] | MHJ_0263 | 116 | ∼25, 37, 20 | — | 19, 41 | — | — | |
| Q4AAD5 | uncharacterized protein (P102) [17,23] | MHJ_0195 | 102 | ∼60 (N), ∼42 (C) | — | 41–46 | 22–30, 37–70 | 73–21 | |
| Q4AAD6 | uncharacterized protein (P97) [20,23] | MHJ_0194 | 123 | ∼120–60, ∼37, 30–20 | [20] | [20] | [20] | 198–153, 122–57, 27–21, 16–12 | |
| surface Ags | P0C0J8 | 46 kDa surface antigen (p46) | p46 MHJ_0511 | 46 | 25–15, >10 | 44, 47 (multimers) | 23, 45 | intact | 44–27 |
| Q4A932 | putative prolipoprotein p65 | MHJ_0656 | 71 | 71–50 | 50, intact | — | intact, 23–37 | 73–57 | |
| Q4A981 | ABC transporter xylose-binding lipoprotein | xylF MHJ_0606 | 50 | ∼20D | ∼50 | — | intact | 57–44 | |
| annotated cytosolic proteins | Q4A9G1 | elongation factor Tu (EfTu) | tuf MHJ_0524 | 44 | 44, 21 | 44 | 21C | intact, ∼20 C | 95–57, 44–27, 21–16 |
| P0C0J3 | l-lactate dehydrogenase (l-LDH) (EC 1.1.1.27) (immunogenic protein p36) | ldh ictD MHJ_0133 | 34 | 34–10, ∼20 | 18, 32, 34 | 10, 17, 26, 28, 31, 35 | intact | 34–21, 16–12 | |
| Q4A9I0 | acetate kinase (EC 2.7.2.1) | ackA MHJ_0505 | 44 | ∼10 (N) | 44 | 43–45 | — | 44–34 | |
| Q4A9I1 | dihydrolipoamide dehydrogenase (EC 1.8.1.4) | pdhD MHJ_0504 | 66 | fragments as multimers | 76, 84 | — | intact | 95–44 | |
| Q4A9P9 | 3-hexulose-6-phosphate synthase (EC 4.1.2.-) | sgaH MHJ_0436 | 25 | ∼15 (C) | 23 | — | — | 44–34 | |
| Q4A9V3 | putative thioredoxin | MHJ_0380 | 13 | 10 (C) | 10 | 10, 14 | — | — | |
| Q4AAA3 | periplasmic sugar-binding protein | rbsB MHJ_0227 | 44 | 44–13D | 40–44 | 45 | intact | 44–34 | |
| Q4AAB1 | putative methylmalonate-semialdehyde dehydrogenase (EC 1.2.1.27) | MHJ_0219 | 54 | ∼25 (C) | — | 51, 53 | — | — | |
| Q4AAL7 | pyruvate dehydrogenase (EC 1.2.4.1) | pdhB MHJ_0112 | 37 | 37–15 | 35–40 | 31, 35 | intact | 34–27 | |
| Q4AAL8 | pyruvate dehydrogenase E1-alpha subunit (EC 1.2.4.1) | pdhA MHJ_0111 | 42 | 42–15+multimers | 40, 53, 97 | 45 | intact | 44–34 | |
| Q4AAL9 | adenine phosphoribosyltransferase (APRT) (EC 2.4.2.7) | apt MHJ_0110 | 19 | <10D+multimers | 21, 23, 38, 40 | 31, 35, 45 | intact | 16–12 | |
| Q4AAR4 | chaperone protein DnaK (HSP70) | dnaK MHJ_0063 | 66 | 66, <50 | 47, 68 | — | intact, ∼40 | 73–57, 44–34 | |
| Q4AAR8 | glyceraldehyde 3-phosphate dehydrogenase (EC 1.2.1.12) | gap MHJ_0031 | 37 | 37-∼10 | 35, 38 | 31, 35 | intact | — | |
| Q4AAV7 | ATP synthase subunit beta (EC 3.6.3.14) | atpD MHJ_0049 | 52 | 20–10D (N) | 52 | 26, 43 | — | — | |
| proteases | Q4AAC8 | ATP-dependent zinc metalloprotease FtsH (EC 3.4.24.-) | ftsH MHJ_0202 | 79 | 79–37 | — | — | intact | 73–57, 44–34 |
| Q4A9G3 | Oligoendopeptidase F (EC 3.4.24.-) | pepF MHJ_0522 | 71 | ∼25, ∼50 15D | — | — | — | 73–57 | |
| uncharacterized | Q4AA06 | uncharacterized protein | MHJ_0326 | 25 | 20-<10D | 21 | — | intact | 21–16 |
| Q4A974 | uncharacterized protein | MHJ_0613 | 95 | 20–15D (N) | — | 100 | — | — | |
| Q4A9G2 | uncharacterized protein | MHJ_0523 | 230 | 200–75D (C) | — | — | — | — | |
| Q4A9Q4 | uncharacterized protein | MHJ_0431 | 75 | ∼25 | — | — | — | — | |
| Q4AAB8 | uncharacterized protein | MHJ_0212 | 236 | 250, ∼100 | 191 | 100 104, 197 | — | 198–122 | |
| Q4AAU0 | uncharacterized protein | MHJ_0009 | 78 | intact, ∼12 | — | 10 (C) | — | 12–10 |
We selected lipoprotein P65, an uncharacterized protein of unknown function and the cytosolic protein lactate dehydrogenase, which we show are targets of endoproteolytic processing events, to provide an insight into the sequences that are targeted by the processing machinery and present some the putative functions of the cleavage fragments that are generated by these processing events.
3.3. Evidence that the P65 lipoprotein is processed on the surface of Mycoplasma hyopneumoniae
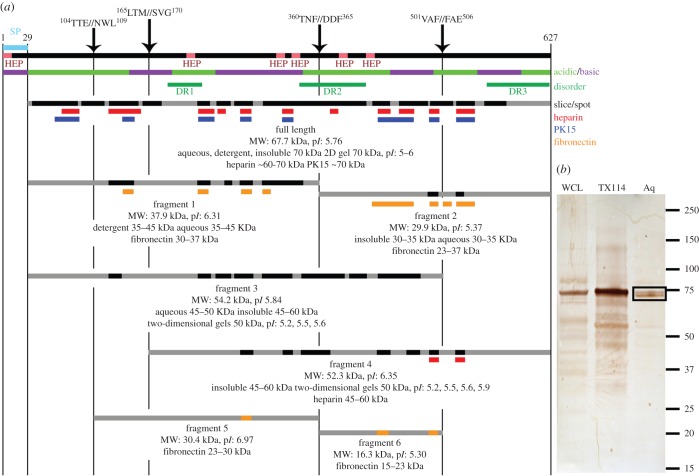
P65, MHJ_0656 (Q4A932), comprises 627 amino acids and encodes a 71 kDa lipolytic lipoprotein with preference for short-chain fatty acids [38]. The N-terminal 29 amino acids comprise the signal sequence and are expected to be removed followed by lipid modification of the cysteine residue at position 30, generating a mature lipoprotein with a mass of 68 kDa and a pI of 5.8. We identified P65 as a series of protein spots on a two-dimensional gel with a mass of approximately 68 kDa and a pI of 5.8 (figure 2, peptide coverage in black). This 68 kDa molecule was also identified in separate affinity-capture assays using heparin and biotinylated porcine epithelial-like surface proteins as bait (figure 2, peptide coverage in red and blue, respectively). P65 is predicted to display three regions of protein disorder from amino acids 189–228 (DR1), 340–418 (DR2) and 553–627 (DR3) according to the PONDR VSL2 algorithm. One of these, DR1, also overlaps with a coiled coil region (100% probability using the COILS algorithm) between amino acids 214–245, suggesting that this region may not be disordered [39]. Efficient cleavage events are known to occur in S/T–X–F↓X–D/E and related motifs that reside within acidic, disordered regions in the P97 and P102 adhesin families in M. hyopneumoniae [9,10,12,17,20–22]. We identified an S/T–X–F↓X–D/E motif in P65 with sequence 360T–N–F↓D–D364 that resides in DR2, and a cleavage site that cuts at phenylalanine with sequence 501V–A–F↓F–A505 that is not located within a region of disorder. Both motifs reside within acidic regions that display a pI of 5 or less (figure 2). Cleavage at 360T–N–F↓D–D364 is expected to generate an N-terminal fragment of 38 kDa and a C-terminal fragment of 30 kDa. Tryptic peptides that mapped to the N-terminal 38 kDa (amino acids 30–362) and to the C-terminal 30 kDa regions of P65 (amino acids 365–627) were identified when M. hyopneumoniae proteins were enriched by extraction with TX-114 and characterized by LC–MS/MS in gel slices representing proteins with masses between 35–45 and 30–35 kDa, respectively. Cleavage fragments with these masses were also identified by LC–MS/MS during affinity-capture experiments using fibronectin as bait (see fragments 1 and 2 in figure 2). No semi-tryptic peptides were identified to further validate this cleavage site, as lysine resides closely flank this region, making peptide identification following trypsin digestion unlikely. Affinity capture experiments using fibronectin as bait also provided evidence that the 30 kDa C-terminal fragment was cleaved at the 501V–A–F↓F–A505 site, where cleavage is expected to generate a fragment of 16 kDa. Consistent with this, several tryptic peptides that mapped between amino acids 363–501 were identified in a gel slice containing M. hyopneumoniae proteins with masses between 15 and 23 kDa (fragment 6 in figure 2). Protein spots migrating with a mass of approximately 50 kDa on two-dimensional gels produced tryptic peptides mapping to P65, consistent with a fragment that started at position 30 and ended at position 503 (fragment 3 in figure 2), and the semi-tryptic peptide F.504FAELNTDQEIK514 was identified from gel slices and gel spots by LC–MS/MS, providing further evidence that cleavage occurred at position 503 at the 501V–A–F↓F–A505 site (table 3; electronic supplementary material, figure S4).
Figure 2.
Cleavage map of P65 (MHJ_0656, Q4A932). (a) Major features of P65, including a putative signal peptide in light blue (SP), putative heparin binding motifs in light red (HEP), acidic/basic regions and disordered regions with four proposed cleavage sites. Peptides mapping to protein fragments identified from multiple analyses are indicated. Black regions indicate peptides obtained from gel spot or slice data. Peptides identified by affinity chromatography using heparin, PK15 surface proteins and fibronectin-coupled GeLC–MS/MS data are indicated in red, blue and orange, respectively. (b) Western blot of Mycoplasma hyopneumoniae proteins probed with antisera raised against recombinant P65. The lane labelled WCL contains M. hyopneumoniae whole cell lysate. Lanes labelled TX114 and Aq contain biotinylated surface proteins of M. hyopneumoniae strain J that partitioned to the detergent and aqueous phases, respectively. Biotinylated proteins were recovered from the Triton and aqueous phases by avidin chromatography prior to gel loading. Multiple cleavage fragments of P65 were detected at masses lower than the abundant intact form. The boxed proteins in the aqueous phase extract at approximately 70 kDa could be attributed to the loss of the lipid anchor in the N-terminus, explaining its abundance in the aqueous phase.
Table 3.
Semi-tryptic peptides denoting cleavage sites in P65. Site of cleavage that semi-tryptic peptide denotes in P65 according to figure 2, with peptide sequence showing amino acid positions and semi-tryptic terminus and modified amino acids underline. No indicates the number of times a peptide was identified by a given method. In the case where peptides were identified multiple times or in multiple runs, only the highest-scoring peptide is shown.
| site | peptide sequence | score | E-value | identified | no. |
|---|---|---|---|---|---|
| 1–1 | K.90NSLVSYDNLAISGTTTE106.N | 61 | 0.0011 | peptide-centrica | 5 |
| 1–2 | E.107NWLYLLNPTK116.Y | 50 | 0.011 | peptide-centric | 1 |
| 1–3 | E.107NWLYLLNPTKYPNGK121.M+deamidated (N) | 51 | 0.011 | peptide-centric | 1 |
| 2–1 | M.168SVGANDPFLAIFNEFK184.K | 74 | 4.4 × 10−6 | WCL ion trap | 2 |
| 2–2 | M.168SVGANDPFLAIFNEFK184.K | 50 | 0.0025 | TX114 Q-TOF | 1 |
| 2–3 | M.168SVGANDPFLAIFNEFK184.K | 82 | 0.011 | gel spots | 1 |
| 4–1 | F.504FAELNTDQEIK514.E | 56 | 0.0024 | WCL Q-TOF | 1 |
| 4–2 | F.504FAELNTDQEIK514.E | 90 | 0.0014 | gel spots | 6 |
| 4–3 | F.504FAELNTDQEIK514.E+deamidated (Q) | 108 | 2.84 × 10−5 | gel spots | 2 |
aPeptide-centric methods described in electronic supplementary material, figure S4.
Two additional putative cleavage sites were identified in the N-terminal third of P65, 104T–T–E↓N–W–L109 and 165L–T–M↓S–V–G170. These sites were identified by the identification of semi-tryptic peptides from GeLC–MS/MS and peptide-centric methods (table 3; electronic supplementary material, figure S4). Complementary C-terminal and N-terminal tryptic peptides identified the cleavage site 104T–T–E↓N–W–L109, whereas the N-terminal semi-tryptic peptide M.168SVGANDPFLAIFNEFK184 indicating cleavage at 165L–T–M↓S–V–G170 was identified from analysis by three different methods (table 3). Cleavage at position 167 is expected to generate two fragments spanning amino acids 30–167 (15.5 kDa; pI 4.87) and amino acids 168–627 (52.4 kDa; pI 6.35). A fragment with peptide coverage consistent with cleavage at this site was identified by LC–MS/MS in a series of protein spots with mass of approximately 50 kDa and with pIs ranging from 5.2 to 5.9. Peptides mapping to the same fragment were also identified from a gel slice containing Triton-X114 insoluble proteins with masses between 45 and 50 kDa, and again following heparin affinity purification, in a gel slice containing proteins with masses between 45 and 60 kDa (fragment 4 in figure 2). We were unable to find a 15.5 kDa protein spanning amino acids 30–167. Cleavage at amino acid position 106 is expected to generate an N-terminal 8.5 kDa protein (pI = 4.36) and a C-terminal 59.3 kDa protein (pI = 7.00). While neither of these cleavage fragments were identified in our studies, we did identify a single tryptic peptide in a gel slice spanning 30–35 kDa that contained M. hyopneumoniae proteins captured during affinity chromatography using fibronectin as bait (fragment 5 in figure 2). This fragment is consistent with cleavage at positions 106 and 362, generating a protein with a mass of 30.4 kDa with a pI of 6.97.
3.4. Processing events identified in atypical cell surface proteins of Mycoplasma hyopneumoniae
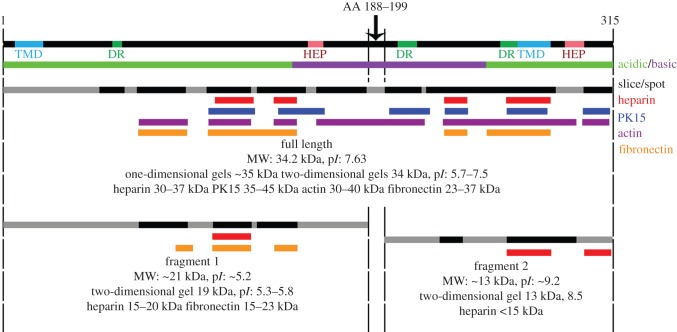
Metabolic proteins such as elongation factor Tu, pyruvate dehydrogenase complex components A, B and D, glyceraldehyde-3-phosphate dehydrogenase and l-lactate dehydrogenase (LDH) showed evidence of post-translational processing and were also identified in cell surface analyses (table 2). Evidence that LDH is processed is presented in figure 3. LDH was identified at its predicted mass of 35 kDa and at multiple pI between 5.7–7.5 on pH 4–7 and 6–11 two-dimensional gels (figure 3, peptide matches in black). Peptides mapping to LDH were also identified from gel spots at apparent molecular mass of 19 kDa and pI 5.3–5.8 on pH 4–7 gels and at 13 kDa and pI 8.5 on pH 6–11 gels. The full-length LDH protein was identified in separate affinity-capture assays using heparin, biotinylated fibronectin, actin and porcine epithelial-like surface proteins as bait (figure 3, peptide coverage in red, orange, purple and blue, respectively). While further studies are needed to confirm biologically meaningful interactions between LDH and these host molecules, affinity-capture assays provide independent evidence that regions within LDH bind host molecules and enrich for cleavage fragments.
Figure 3.
Cleavage map of l-lactate dehydrogenase (LDH; MHJ_0133, P0C0J3). The major features of l-lactate dehydrogenase are shown, including putative transmembrane domains (TMD), putative heparin binding motifs (HEP), putative disordered regions (DR) and acidic/basic regions. A single proposed cleavage site is shown between amino acids 188 and 199, based on peptide coverage. Peptides mapping to protein fragments identified from multiple analyses are indicated. Black regions indicate peptides obtained from gel spot (figure 1) or slice data. Peptides identified by affinity chromatography using heparin, PK15 surface proteins, actin and fibronectin-coupled GeLC–MS/MS data are indicated in red, blue, purple and orange, respectively, at masses as indicated.
A single cleavage event between amino acids 188–199 would result in a theoretical N-terminal fragment of approximately 21 kDa with a pI of 5.2 and C-terminal fragment of approximately 13 kDa and a pI of 9.2, which is similar to the fragments of LDH identified from two-dimensional gels. The shift in pI may be attributed to deamidation of asparagine residues at position 121 in the N-terminal fragment and positions 269 and 279 in the C-terminal fragment as detected in peptides identified by GeLC–MS/MS. Neither fragment of LDH was detected from actin or PK15 cell surface binding pulldowns; however, peptides mapping to LDH were identified at masses between 40 and 200 kDa in actin pulldowns, possibly indicating incomplete disassociation from multimeric complexes prior to SDS–PAGE. Peptides mapping to the N-terminal fragment were identified from heparin and fibronectin affinity GeLC–MS/MS experiments in slices at masses 15–20 and 15–23 kDa respectively, whereas peptides mapping to the C-terminal fragment were identified only from heparin affinity GeLC–MS/MS experiments in a slice encompassing masses less than 15 kDa. Although no heparin-binding motif was identified in the C-terminal fragment, the lysine-rich sequence 290DKEKEKFAKS300 could facilitate interaction with heparin. Further work is needed to determine if 290DKEKEKFAKS300 binds heparin.
3.5. Processing in uncharacterized proteins
MHJ_0009 encoding a 77.5 kDa uncharacterized protein (Q4AAU0) was identified consistently in slices 6 (approx. 70–90 kDa), 13 (approx. 13–16 kDa) and 14 (approx. 10–13 kDa) of GeLC–MS/MS using ion trap and Q-TOF analyses from whole cell lysates (figure 4). Peptides identified from replicates of slice 6 mapped to the N-terminus and middle regions of the protein at approximately the predicted mass of the intact protein. Slices 13 and 14, however, are taken from regions of the gel with mass 10–16 kDa, and peptides identified from replicates of these slices mapped only to the C-terminal region of the protein. This C-terminal fragment may represent the product of post-translational proteolytic cleavage. Predicting the true N-terminus of the C-terminal fragment at amino acid position 567 (M) would generate a protein with a mass of 12.5 kDa and pI of 5.47 as predicted by ProtParam. We identified the C-terminal fragment from two-dimensional gels at the same approximate molecular mass, with pI ranging from approximately 5.5 to 6.2 (figure 1). MHJ_0009 was also identified from GeLC–MS/MS of samples following heparin affinity chromatography from a slice at molecular mass of approximately 10–12 kDa, in elutions carrying proteins with low heparin binding affinity (elution in 150–600 mM NaCl). This is consistent with the presence of putative heparin binding motifs within the C-terminus. Eight putative heparin-binding motifs were identified within MHJ_0009 similar to those described previously [21,22] in both the N- and C-terminal fragments, as denoted by grey underlined regions in figure 4. The protein was identified by the same two C-terminal peptides identified from low molecular mass slices in GeLC–MS/MS (underlined in black in figure 4). The C-terminal fragment of MHJ_0009 contains a thioredoxin-like domain, and a BLAST search of this fragment gives approximately 60% identity to thioredoxin from other Mycoplasma species (M. bovoculi: E-value: 2 × 10−43, score: 375, identity: 62%). Further work is needed to confirm if the C-terminal cleavage fragment displays oxidoreductase activity.
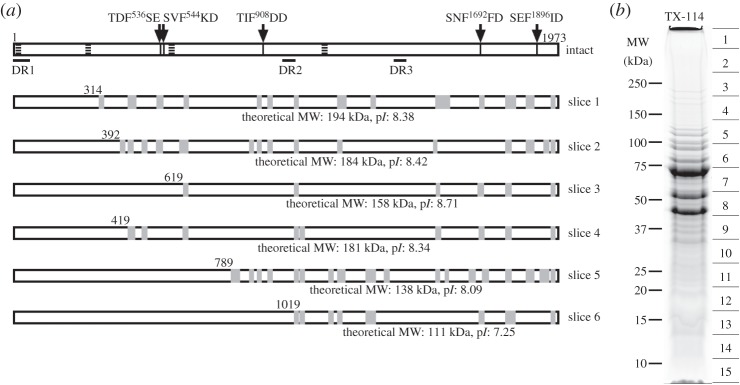
Only one of the cleaved proteins listed in table 2, the uncharacterized protein MHJ_0523, has not also been identified in surfaceome studies using enzymatic shaving and/or cell surface biotinylation [40]. MHJ_0523 encodes a 230 kDa putative lipoprotein and is predicted to possess a transmembrane domain at the N-terminus (TMPred score 1612) and three other putative transmembrane domains (figure 5), which would suggest that the protein is likely to traverse the cell membrane. Extraction of M. hyopneumoniae with TX-114 is likely to have concentrated MHJ_0523 into the detergent-soluble fraction, indicating that it may be surface-exposed but expressed at low levels, rendering it undetectable by our shaving/biotin labelling methods. Detection of MHJ_0523 in slice 1 indicates that the molecule is poorly soluble during SDS–PAGE or that it forms large mass multimeric structures. Fragments identified were from the C-terminus ranging from masses upwards of 75 kDa on the TX114 gel, with no coverage of the first 314 amino acids. Five putative S/T–X–F–X–D/E cleavage motifs were identified along the length of the ORF, but we were unable to confirm if processing does occur at these sites.
Figure 5.
MHJ_0523 cleavage map. (a) General features and peptides mapping to MHJ_0523 identified from (b) GeLC-MS/MS of a M. hyopneumoniae Triton X-114 detergent phase enrichment. Transmembrane domains are indicated by horizontally striped regions and three disordered regions spanning more than 40 amino acids were detected (DR1-3). Five putative S/T-X-F-X-D/E cleavage motifs are indicated by arrows. Peptide coverage identified from individual slices is indicated in grey, and theoretical molecular weight and isoelectric points of protein fragments are shown (according to peptide coverage).
3.6. Proteases identified in Mycoplasma hyopneumoniae
Eighteen ORFs have been annotated in the UniProt database (GO annotation) to have putative protease activity, 11 of which have been identified in our study (table 4). Identified proteins with annotated endoprotease activity include MHJ_0522 (Q4A9G3) oligoendopeptidase F, MHJ_0525 (Q4A9G0) Lon protease, MHJ_0636 (Q4A952) tsaD, MHJ_0202 (Q4AAC8) ftsH and MHJ_0568 (Q4A9B9), an uncharacterized protein. These identifications are consistent with those identified in strain 232 [7]. These proteases are likely to carry out the major proteolytic actions that give rise to adhesin fragments, as well as potentially processing other proteins. Lon proteases are bioinformatically predicted to cleave at hydrophobic residues, including phenylalanine (F), and so may play a role in processing at the dominant cleavage motif S/T–X–F↓–X–D/E [9,10]. Additionally, uncharacterized protein MHJ_0568 is predicted to possess a trypsin-like domain, which may be responsible for trypsin-like cleavage events at lysine (K) and arginine (R) residues. Efforts are currently under way to confirm these bioinformatically predicted results.
Table 4.
Identified proteases of M. hyopneumoniae.
| accession and locus | identified proteases | surface | gene ontology (GO) |
|---|---|---|---|
| Q4AAC8 MHJ_0202 | ATP-dependent zinc metalloprotease FtsH (EC 3.4.24.-) | Y | cell division; integral component of membrane; metalloendopeptidase activity; zinc ion binding |
| Q4A9G0 MHJ_0525 | Lon protease (EC 3.4.21.53) (ATP-dependent protease La) | Y | cellular response to stress; cytoplasm; serine-type endopeptidase activity |
| Q4AAK4 MHJ_0125 | putative aminopeptidase | Y | aminopeptidase activity |
| Q4A9G3 MHJ_0522 | oligoendopeptidase F (EC 3.4.24.-) | Y | metalloendopeptidase activity; zinc ion binding |
| Q4A9M4 MHJ_0461 | leucyl aminopeptidase (EC 3.4.11.1) | Y | aminopeptidase activity; manganese ion binding; metalloexopeptidase activity |
| Q4A929 MHJ_0659 | XAA-PRO aminopeptidase (EC 3.4.11.9) | Y | aminopeptidase activity; metalloexopeptidase activity |
| Q4AAM9 MHJ_0098 | ATP-dependent protease binding protein | N | ATP binding; nucleoside-triphosphatase activity; peptidase activity |
| Q4A952 MHJ_0636 | tRNA N6-adenosine threonylcar-bamoyltransferase (EC 2.6.99.4) | N | cytoplasm; iron ion binding; metalloendopeptidase activity |
| Q4AAG1 MHJ_0169 | methionine aminopeptidase (MAP) (MetAP) (EC 3.4.11.18) | N | metal ion binding; metalloaminopeptidase activity; protein initiator methionine removal |
| Q4AAS7a MHJ_0022 | signal peptidase I (EC 3.4.21.89) | N | integral component of membrane; serine-type peptidase activity |
| Q4A9B9 MHJ_0568 | uncharacterized protein | N | serine-type endopeptidase activity |
aAlthough MHJ_0022 has a signal peptidase I signature motif, existing biochemical data from amino-terminal sequence analysis of amino-terminal cleavage products indicates that this species lacks SPase I activity. ‘Surface’ indicates proteins were (Y) or were not (N) identified in cell surface shaving or biotinylation experiments.
4. Discussion
4.1. Protein-centric approaches to mapping the Mycoplasma hyopneumoniae proteome
While our protein-centric separation approaches identified only 347 proteins, representing 52% of the M. hyopneumoniae proteome [32,41–44], they enabled us to characterize endoproteolytic processing events in 35 functionally diverse, surface-associated proteins. The unidentified portion of the proteome consisted of 198 uncharacterized ORFs, which may be of low abundance or have a high rate of turnover, or may not be transcribed under the growth conditions used in our analyses. Additionally, some ORF sequences that remain unidentified contain too many (or rarely too few) lysine and/or arginine residues, making the tryptic peptides generated by digestion undetectable by the methods used. Instrument sensitivity only partially explains why we did not identify a greater proportion of the proteome, given that we were only able to identify 70% (483) of the 691 predicted ORFs in strain 232 during culture in Friis broth [7]. Our approach is consistent with our primary goal to preserve mass-context prior to mass spectrometry as a means to identify the gamut of proteins targeted by processing mechanisms. Two-dimensional PAGE was able to resolve individual proteins and isoforms, providing information about post-translational modifications, whereas one-dimensional GeLC–MS/MS methods are higher-throughput, making them better suited for global proteome identification. The protein-centric approaches used in our studies provided insights into the extent of protein processing in M. hyopneumoniae. Table 2 lists the proteins cleaved in M. hyopneumoniae. Notably, almost all of the proteins in table 2 were identified in a comprehensive surfaceome analysis conducted using cell shaving and surface biotinylation methodologies (J.L.T., B.B.A.R. & S.P.D. 2012 unpublished data). Our data suggest that protein processing is a post-translational modification that occurs with greater frequency than is currently recognized and occurs in a wide range of functionally diverse cell surface proteins. This is consistent with the processing machinery being associated with the cell surface or with the general secretory pathway. On a cautionary note, it remains to be determined what mechanisms are needed to export proteins with canonical functions in the cytosol onto the cell surface [45–47]. Nonetheless, we provide strong evidence that numerous proteins with functions in the cytosol are bound on the surface of M. hyopneumoniae where they are targets of endoproteolytic processing. We detected cleavage at S/T–X–F↓–X–D/E sites consistent with the hypothesis that the same enzyme that cleavages the P97 and P102 families is also targeting other surface accessible proteins.
Enrichment procedures such as TX-114 fractionation and affinity-capture chromatography techniques were useful for enriching the low-abundance proteome, delineating regions of proteins that bind host molecules and enriching for cleavage fragments, all of which provided clues to protein function. TX-114 extraction enriches for hydrophobic membrane proteins, which partition to the detergent phase [48]. As M. hyopneumoniae lacks a cell wall, the cell membrane is the mediator of contact between the bacteria and extracellular environment; hence, membrane-bound proteins are potentially valuable targets for vaccine and therapeutic development. While the TX-114 GeLC–MS/MS protocol detected the fewest protein identifications, at 206, it contributed five unique proteins to the overall analysis, all of which were uncharacterized proteins described as lipoproteins and/or predicted to contain transmembrane domains using TMpred. Overall, 26 of 50 M. hyopneumoniae lipoproteins were identified by all methods, and LC–MS/MS analysis of TX-114 solubilized proteins identified 22 of the 26.
While the precise functions of bacterial lipoproteins remain poorly understood, there is mounting evidence to suggest they are pathogen-associated molecular pattern (PAMP) molecules on the surface of Gram-positive bacteria. PAMPs are recognized by Toll-like receptors that trigger innate immune responses [49–52]. Most mycoplasma lipoproteins are surface-exposed with acyl groups anchoring these proteins in the cell membrane, where they are thought to function as cytadhesins, transport proteins or virulence factors with immunomodulatory capabilities [53]. P65 is an abundantly expressed, immunoreactive and lipolytic lipoprotein that selectively partitions to the detergent phase during extraction with TX-114 [38,54]. Schmidt et al. showed that anti-P65 antibodies inhibit the lipolytic activity of P65 and growth of M. hyopneumoniae, indicating that P65 performs a primary function on the external membrane surface by providing a source of essential lipids for growth [38]. It has also been suggested that P65 may alter surfactant properties in the lungs of pigs in vivo [38]. In our studies, P65 was recovered during affinity capture protocols using different host molecules as bait. Although these are preliminary data that require quantitative studies to confirm a direct role for P65 in these interactions, this suggests that P65 displays motifs that facilitate binding to a diverse range of host molecules. Consistent with these preliminary observations, we show here for the first time that P65 is a target of several processing events that generate cleavage fragments which are selectively retained during affinity chromatography using porcine epithelial cell surface proteins, fibronectin or porcine heparin as bait. The ability of the cleavage fragments of P65 to bind the same bait proteins as P65 lends weight to the hypothesis that the interactions with host molecules are direct and biologically relevant. Cleavage occurred at a number of sites in the P65 protein sequence, including at a phenylalanine residue within a S/T–X–F↓X–D/E motif; a known processing site in the P97 and P102 adhesin families [9–12,15,18,20–23]. An immunoblot of biotinylated cell surface M. hyopneumoniae strain J proteins fractionated using TX-114 that was probed with anti-P65 polyclonal antibodies identified a 65 kDa protein and numerous smaller mass fragments of P65 consistent with cleavage at several sites within the molecule. These data show that P65 and cleavage fragments of P65 reside on the surface of M. hyopneumoniae. Notably, there is clear evidence of a doublet at approximately 65 kDa (boxed in figure 2b) in the lane containing M. hyopneumoniae aqueous phase proteins. Previous studies have shown that P65 may undergo clipping at the N-terminus and be a target of further post-translational processing events [54]. Our data suggest that the doublet may represent forms of P65 that have lost the lipid anchor because they partitioned to the aqueous phase. If correct, these data suggest that a small lipopeptide similar to the macrophage-activating lipopeptide 2 (MALP-2) of Mycoplasma fermentans may be produced from P65.
Lipoproteins of mycoplasmal origin are known targets of post-translational processing events. The first 14 amino acids of MALP-404, a 41 kDa lipoprotein in M. fermentans, are removed by a post-translational cleavage event generating a 2 kDa MALP-2 lipopeptide. The C-terminal 39 kDa cleavage fragment (known as RF) that results from this cleavage event has been isolated from culture supernatants, but its function remains unknown [55]. Unlike RF, both MALP-2 and MALP-404 are lipid-modified and remain associated with the membrane of M. fermentans. MALP-2 is a potent immunomodulatory molecule that engages Toll-like receptor 2 [50]. Like the MALP-2 lipopeptide, the N-terminus of P65 may play a similar immunomodulatory role in M. hyopneumoniae; however, further studies are required to confirm this. Similarly, MGA0674 is an 82 kDa lipoprotein in Mycoplasma gallisepticum whose expression is elevated in virulent strain Rlow compared with the attenuated vaccine strain F, suggesting that it may play a role in pathogenesis. MGA0674 is a target of a processing event at position 225 that releases a C-terminal 57 kDa fragment from the anchored N-terminal 22 kDa lipoprotein [56]. There are other reports of processing events that target lipoproteins in Mycoplasma pneumoniae but their functions have remained poorly characterized [57].
4.2. The extent of proteolytic processing in Mycoplasma hyopneumoniae
A significant number of the 35 proteins identified to be targets of post-translational processing were glycolytic enzymes and other metabolic proteins. Glycolytic enzymes are increasingly being identified as multitasking or moonlighting proteins in a wide range of organisms, including parasites [58], yeasts and fungi [59], mammalian cells [60], plants [61] and bacteria [62], and this is reflected in the range of entries seen in MultitaskProtDB [63]. In other members of the Mollicutes, proteins with canonical functions in the cytosol have also been found to be surface-exposed and interact with host components. For example, in Mycoplasma pneumoniae, elongation factor Tu (EfTu) and pyruvate dehydrogenase (PdhB) were identified as surface-exposed moonlighting proteins, through screening for fibronectin binding proteins by ligand blotting of whole cell lysates and fibronectin-coupled affinity chromatography, and their surface localization was confirmed by immunogold labelling and electron microscopy [64]. Further investigation of EfTu revealed specifically that the carboxyl-terminus was surface-exposed by immunogold labelling and responsible for fibronectin binding [65]. In Mycoplasma genitalium, glyceraldehyde-3-phosphate dehydrogenase was identified to be surface-exposed and bind mucin, probably functioning as an adhesin [66]. These proteins were identified here to be cleaved. Many processing events are likely to alter canonical (enzymatic) function and profoundly influence how cleavage fragments interact with the mycoplasma membrane and host molecules [67].
LDH is a highly immunogenic cytoplasmic protein involved in the glycolytic process of M. hyopneumoniae [68,69]. Here, we have identified LDH to be present at the cell surface both as a full-length molecule and as cleavage fragments. We identified a single putative cleavage site between amino acids 188–199 and the cleaved form of LDH is unlikely to carry out its primary function owing to significant structural alteration. In eukaryotic organisms, LDH has been recognized as a moonlighting protein, along with other glycolytic enzymes such as hexokinase, glyceraldehyde dehydrogenase and enolase, playing a role in transcriptional regulation [70]. LDH has also been identified as a single-stranded DNA-binding protein in eukaryotic cells [71,72]. In eukaryotic cells, this switch in function is likely to be due to translocation to the nucleus where these functions take place, possibly through post-translational modifications such as tyrosine phosphorylation [70,73,74]. It is possible that, as in eukaryotic cells, post-translational modifications may also affect localization and function of LDH in M. hyopneumoniae, or direct a subset towards processing. Intact LDH was identified from spots on two-dimensional gels between pI of 5.7 and 7.5. With a theoretical pI of 7.63, this indicates an acidic shift is likely to be caused by a variable degree of post-translational modification such as deamidation affecting a proportion of LDH [75]. LDH has also been identified from extracellular supernatants of various Lactobacillus and Bifidobacterium species from the honeybee Apis mellifera [76]. These lactic acid bacteria belong to the Firmicutes, and are genetically similar to the low G + C content mycoplasma species. It was hypothesized that LDH, once localized to the surface, could evolve alternative functions as a moonlighting protein, functioning as an auxiliary adhesin [76]. Indeed, we have also previously identified a glutamyl aminopeptidase from M. hyopneumoniae, MHJ_0125, which moonlights as a multifunctional adhesin at the cell surface [77], and a leucyl aminopeptidase, MHJ_0461, which functions as a multi-substrate peptidase and binds heparin, plasminogen and foreign DNA [47]. The cleavage fragments of LDH identified here bound to heparin, used as a structural mimic for glycosaminoglycans in the respiratory tract, and the N-terminal fragment also bound to fibronectin, an extracellular matrix component, indicating fragments may also have adhesin functions.
5. Conclusion
We identified 347 (52%) of the 672 putative ORFs predicted from the genome sequence of M. hyopneumoniae strain J. The proteome coverage from well-resolved two-dimensional gels, while low, is unsurprising. The limitations of two-dimensional gels are well documented, particularly considering the nature of sample preparation required, which limits the ability to retain and resolve very basic, acidic, small, large or hydrophobic proteins [43]. However, protein-centric, gel-based separations provide a technique complimentary to high-throughput two-dimensional LC–MS/MS protocols by maintaining mass and pI context, allowing the identification of cleavage products and the extent of proteolytic processing. We show for the first time that proteins with canonical functions in the cytosol that moonlight on the cell surface are also targets of endoproteolytic events. This describes a new dimension to protein moonlighting and suggests that much more biological information is inherent in proteins. While we cannot yet determine the exact nature of cleavage events as they occur in vivo, the analysis presented here is an important first step in determining physiologically relevant cleavage events. Cleavage events will undoubtedly complicate efforts to correlate the transcriptome with the proteome in future studies [78,79], and the protein-centric approaches presented here will provide a solid foundation for further investigation of post-translational processing in proteins involved in pathogenesis of M. hyopneumoniae, and will assist with delineating functionally important binding motifs.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
An internal UTS grant (no. 2012001182) to S.P.D. assisted in supporting this research. J.L.T., B.B.A.R. and I.J.B. are recipients of UTS Doctoral Scholarships. M.W. and L.K.W. are recipients of Australian Postgraduate Awards.
Authors' contributions
J.L.T. performed proteomics and heparin-binding experiments, analysed data and wrote the manuscript; B.B.A.R. performed TX114 biotinylation experiments, actin-binding and PK15 interaction analyses; P.A.H. carried out ion trap mass spectrometry analyses; I.J.B. assisted in data analysis for identification of cleavage sites; M.W. performed fibronectin-binding experiments; D.R.B. expressed and purified P65 and generated polyclonal antisera; L.K.W. and C.J. provided convalescent porcine sera and contributed to review of the manuscript, F.C.M. provided the construct for P65 expression and purification; M.P.P. carried out QTOF mass spectrometry and assisted data analysis and validation; S.P.D. conceived and coordinated the study and co-wrote the manuscript.
Competing interests
The authors have declared no competing interests.
Funding
We received no funding for this study.
References
- 1.Razin S, Yogev D, Naot Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62, 1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woese CR. 1987. Bacterial evolution. Microbiol. Rev. 51, 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minion FC, Lefkowitz EJ, Madsen ML, Cleary BJ, Swartzell SM, Mahairas GG. 2004. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J. Bacteriol. 186, 7123–7133. (doi:10.1128/jb.186.21.7123-7133.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark LK, Armstrong CH, Freeman MJ, Scheidt AB, Sands-Freeman L, Knox K. 1991. Investigating the transmission of Mycoplasma hyopneumoniae in a swine herd with enzootic pneumonia. Vet. Med. 86, 543–550. [Google Scholar]
- 5.Liu W, et al. 2011. Complete genome sequence of Mycoplasma hyopneumoniae strain 168. J. Bacteriol. 193, 1016–1017. (doi:10.1128/JB.01305-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasconcelos AT, et al. 2005. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 187, 5568–5577. (doi:10.1128/JB.187.16.5568-5577.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pendarvis K, Padula MP, Tacchi JL, Petersen AC, Djordjevic SP, Burgess SC, Minion FC. 2014. Proteogenomic mapping of Mycoplasma hyopneumoniae virulent strain 232. BMC Genomics 15, 576 (doi:10.1186/1471-2164-15-576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siqueira FM, Gerber AL, Guedes RL, Almeida LG, Schrank IS, Vasconcelos AT, Zaha A. 2014. Unravelling the transcriptome profile of the swine respiratory tract mycoplasmas. PLoS ONE 9, e110327 (doi:10.1371/journal.pone.0110327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogema DR, et al. 2012. Characterization of cleavage events in the multifunctional cilium adhesin Mhp684 (P146) reveals a mechanism by which Mycoplasma hyopneumoniae regulates surface topography. mBio 3 (doi:10.1128/mBio.00282-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogema DR, et al. 2011. Sequence TTKF↓QE defines the site of proteolytic cleavage in Mhp683 protein, a novel glycosaminoglycan and cilium adhesin of Mycoplasma hyopneumoniae. J. Biol. Chem. 286, 41 217–41 229. (doi:10.1074/jbc.M111.226084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutscher AT, Jenkins C, Minion FC, Seymour LM, Padula MP, Dixon NE, Walker MJ, Djordjevic SP. 2010. Repeat regions R1 and R2 in the P97 paralogue Mhp271 of Mycoplasma hyopneumoniae bind heparin, fibronectin and porcine cilia. Mol. Microbiol. 78, 444–458. (doi:10.1111/j.1365-2958.2010.07345.x) [DOI] [PubMed] [Google Scholar]
- 12.Deutscher AT, et al. 2012. Mycoplasma hyopneumoniae Surface proteins Mhp385 and Mhp384 bind host cilia and glycosaminoglycans and are endoproteolytically processed by proteases that recognize different cleavage motifs. J. Proteome Res. 11, 1924–1936. (doi:10.1021/pr201115v) [DOI] [PubMed] [Google Scholar]
- 13.Hsu T, Artiushin S, Minion FC. 1997. Cloning and functional analysis of the P97 swine cilium adhesin gene of Mycoplasma hyopneumoniae. J. Bacteriol. 179, 1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins C, Wilton JL, Minion FC, Falconer L, Walker MJ, Djordjevic SP. 2006. Two domains within the Mycoplasma hyopneumoniae cilium adhesin bind heparin. Infect. Immun. 74, 481–487. (doi:10.1128/iai.74.1.481-487.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seymour LM, et al. 2010. A processed multidomain Mycoplasma hyopneumoniae adhesin binds fibronectin, plasminogen, and swine respiratory cilia. J. Biol. Chem. 285, 33 971–33 978. (doi:10.1074/jbc.M110.104463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seymour LM, Falconer L, Deutscher AT, Minion FC, Padula MP, Dixon NE, Djordjevic SP, Walker MJ. 2011. Mhp107 is a member of the multifunctional adhesin family of Mycoplasma hyopneumoniae. J. Biol. Chem. 286, 10 097–10 104. (doi:10.1074/jbc.M110.208140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seymour LM, et al. 2012. Mhp182 (P102) binds fibronectin and contributes to the recruitment of plasmin(ogen) to the Mycoplasma hyopneumoniae cell surface. Cell Microbiol. 14, 81–94. (doi:10.1111/j.1462-5822.2011.01702.x) [DOI] [PubMed] [Google Scholar]
- 18.Wilton J, et al. 2009. Mhp493 (P216) is a proteolytically processed, cilium and heparin binding protein of Mycoplasma hyopneumoniae. Mol. Microbiol. 71, 566–582. (doi:10.1111/j.1365-2958.2008.06546.x) [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Young TF, Ross RF. 1994. Microtiter plate adherence assay and receptor analogs for Mycoplasma hyopneumoniae. Infect. Immun. 62, 1616–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raymond BB, et al. 2014. Proteolytic processing of the cilium adhesin MHJ_0194 (P123) in Mycoplasma hyopneumoniae generates a functionally diverse array of cleavage fragments that bind multiple host molecules. Cell. Microbiol. 11, 1924–1936. (doi:10.1111/cmi.12377) [DOI] [PubMed] [Google Scholar]
- 21.Raymond BB, Tacchi JL, Jarocki VM, Minion FC, Padula MP, Djordjevic SP. 2013. P159 from Mycoplasma hyopneumoniae binds porcine cilia and heparin and is cleaved in a manner akin to ectodomain shedding. J. Proteome Res. 12, 5891–5903. (doi:10.1021/pr400903s) [DOI] [PubMed] [Google Scholar]
- 22.Tacchi JL, Raymond BB, Jarocki VM, Berry IJ, Padula MP, Djordjevic SP. 2014. Cilium adhesin P216 (MHJ_0493) is a target of ectodomain shedding and aminopeptidase activity on the surface of Mycoplasma hyopneumoniae. J. Proteome Res. 13, 2920–2930. (doi:10.1021/pr500087c) [DOI] [PubMed] [Google Scholar]
- 23.Djordjevic SP, Cordwell SJ, Djordjevic MA, Wilton J, Minion FC. 2004. Proteolytic processing of the Mycoplasma hyopneumoniae cilium adhesin. Infect. Immun. 72, 2791–2802. (doi:10.1128/iai.72.5.2791-2802.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moitinho-Silva L, et al. 2013. Mycoplasma hyopneumoniae in vitro peptidase activities: identification and cleavage of kallikrein-kinin system-like substrates. Vet. Microbiol. 163, 264–273. (doi:10.1016/j.vetmic.2013.01.011) [DOI] [PubMed] [Google Scholar]
- 25.Mann M, Kulak NA, Nagaraj N, Cox J. 2013. The coming age of complete, accurate, and ubiquitous proteomes. Mol. Cell. 49, 583–590. (doi:10.1016/j.molcel.2013.01.029) [DOI] [PubMed] [Google Scholar]
- 26.Smith LM, Kelleher NL, Consortium for Top Down Proteomics. 2013. Proteoform: a single term describing protein complexity. Nat. Methods 10, 186–187. (doi:10.1038/nmeth.2369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelleher NL. 2004. Top-down proteomics. Anal. Chem. 76, 197A–203A. (doi:10.1021/ac0415657) [PubMed] [Google Scholar]
- 28.Friis NF. 1975. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare a survey. Nord Vet. Med. 27, 337–339. [PubMed] [Google Scholar]
- 29.Scarman AL, Chin JC, Eamens GJ, Delaney SF, Djordjevic SP. 1997. Identification of novel species-specific antigens of Mycoplasma hyopneumoniae by preparative SDS-PAGE ELISA profiling. Microbiology 143, 663–673. (doi:10.1099/00221287-143-2-663) [DOI] [PubMed] [Google Scholar]
- 30.Jenkins C, Samudrala R, Geary SJ, Djordjevic SP. 2008. Structural and functional characterization of an organic hydroperoxide resistance protein from Mycoplasma gallisepticum. J. Bacteriol. 190, 2206–2216. (doi:10.1128/JB.01685-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minion FC, Menon SA, Mahairas GG, Wannemuehler MJ. 2003. Enhanced murine antigen-specific gamma interferon and immunoglobulin G2a responses by using mycobacterial ESAT-6 sequences in DNA vaccines. Infect. Immun. 71, 2239–2243. (doi:10.1128/IAI.71.4.2239-2243.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White MY, Brown DA, Sheng S, Cole RN, O'Rourke B, Van Eyk JE. 2011. Parallel proteomics to improve coverage and confidence in the partially annotated Oryctolagus cuniculus mitochondrial proteome. Mol. Cell Proteomics 10, M110004291 (doi:10.1074/mcp.M110.004291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. 1999. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann K, Stoffel W. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374, 166. [Google Scholar]
- 35.Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. 2005. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 61(Suppl 7), 176–182. (doi:10.1002/prot.20735) [DOI] [PubMed] [Google Scholar]
- 36.UniProt C. 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212. (doi:10.1093/nar/gku989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burnett TA, et al. 2006. P159 is a proteolytically processed, surface adhesin of Mycoplasma hyopneumoniae: defined domains of P159 bind heparin and promote adherence to eukaryote cells. Mol. Microbiol. 60, 669–686. (doi:10.1111/j.1365-2958.2006.05139.x) [DOI] [PubMed] [Google Scholar]
- 38.Schmidt JA, Browning GF, Markham PF. 2004. Mycoplasma hyopneumoniae p65 surface lipoprotein is a lipolytic enzyme with a preference for shorter-chain fatty acids. J. Bacteriol. 186, 5790–5798. (doi:10.1128/JB.186.17.5790-5798.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferron F, Longhi S, Canard B, Karlin D. 2006. A practical overview of protein disorder prediction methods. Proteins 65, 1–14. (doi:10.1002/prot.21075) [DOI] [PubMed] [Google Scholar]
- 40.Tacchi JL, Raymond BBA, Djordjevic SP. Characterisation of proteins on the surface of Mycoplasma hyopneumoniae. In preparation.
- 41.Lopez JL. 2007. Two-dimensional electrophoresis in proteome expression analysis. J. Chromatogr. B 849, 190–202. (doi:10.1016/j.jchromb.2006.11.049) [DOI] [PubMed] [Google Scholar]
- 42.Ly L, Wasinger VC. 2011. Protein and peptide fractionation, enrichment and depletion: tools for the complex proteome. Proteomics 11, 513–534. (doi:10.1002/pmic.201000394) [DOI] [PubMed] [Google Scholar]
- 43.Maillet I, Berndt P, Malo C, Rodriguez S, Brunisholz RA, Pragai Z, Arnold S, Langen H, Wyss M. 2007. From the genome sequence to the proteome and back: evaluation of E. coli genome annotation with a 2-D gel-based proteomics approach. Proteomics 7, 1097–1106. (doi:10.1002/pmic.200600599) [DOI] [PubMed] [Google Scholar]
- 44.Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. 2006. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5, 1543–1554. (doi:10.1021/pr050477f) [DOI] [PubMed] [Google Scholar]
- 45.Bendtsen JD, Kiemer L, Fausboll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 5, 58 (doi:10.1186/1471-2180-5-58) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79, 3476–3491. (doi:10.1128/IAI.00179-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jarocki VM, Santos J, Tacchi JL, Raymond BBA, Deutscher AT, Jenkins C, Padula MP, Djordjevic SP. 2015. MHJ_0461 is a multifunctional leucine aminopeptidase on the surface of Mycoplasma hyopneumoniae. Open Biol. 5, 140175 (doi:10.1098/rsob.140175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wise KS, Kim MF. 1987. Major membrane surface proteins of Mycoplasma hyopneumoniae selectively modified by covalently bound lipid. J. Bacteriol. 169, 5546–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashimoto M, Tawaratsumida K, Kariya H, Aoyama K, Tamura T, Suda Y. 2006. Lipoprotein is a predominant Toll-like receptor 2 ligand in Staphylococcus aureus cell wall components. Int. Immunol. 18, 355–362. (doi:10.1093/intimm/dxh374) [DOI] [PubMed] [Google Scholar]
- 50.Muhlradt PF, Kiess M, Meyer H, Sussmuth R, Jung G. 1997. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J. Exp. Med. 185, 1951–1958. (doi:10.1084/jem.185.11.1951) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc. Natl Acad. Sci. USA 97, 13 766–13 771. (doi:10.1073/pnas.250476497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. 2001. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13, 933–940. (doi:10.1093/intimm/13.7.933) [DOI] [PubMed] [Google Scholar]
- 53.Browning GF, Marenda MS, Noormohammadi AH, Markham PF. 2011. The central role of lipoproteins in the pathogenesis of mycoplasmoses. Vet. Microbiol. 153, 44–50. (doi:10.1016/j.vetmic.2011.05.031) [DOI] [PubMed] [Google Scholar]
- 54.Kim MF, Heidari MB, Stull SJ, McIntosh MA, Wise KS. 1990. Identification and mapping of an immunogenic region of Mycoplasma hyopneumoniae p65 surface lipoprotein expressed in Escherichia coli from a cloned genomic fragment. Infect. Immun. 58, 2637–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis KL, Wise KS. 2002. Site-specific proteolysis of the MALP-404 lipoprotein determines the release of a soluble selective lipoprotein-associated motif-containing fragment and alteration of the surface phenotype of Mycoplasma fermentans. Infect. Immun. 70, 1129–1135. (doi:10.1128/IAI.70.3.1129-1135.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Szczepanek SM, Frasca S Jr, Schumacher VL, Liao X, Padula M, Djordjevic SP, Geary SJ. 2010. Identification of lipoprotein MslA as a neoteric virulence factor of Mycoplasma gallisepticum. Infect. Immun. 78, 3475–3483. (doi:10.1128/IAI.00154-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Regula JT, Ueberle B, Boguth G, Gorg A, Schnolzer M, Herrmann R, Frank R. 2000. Towards a two-dimensional proteome map of Mycoplasma pneumoniae. Electrophoresis 21, 3765–3780. (doi:10.1002/1522-2683(200011)21:17<3765::AID-ELPS3765>3.0.CO;2-6) [DOI] [PubMed] [Google Scholar]
- 58.Gomez-Arreaza A, Acosta H, Quinones W, Concepcion JL, Michels PAM, Avilan L. 2014. Extracellular functions of glycolytic enzymes of parasites: Unpredicted use of ancient proteins. Mol. Biochem. Parasitol. 193, 75–81. (doi:10.1016/j.molbiopara.2014.02.005) [DOI] [PubMed] [Google Scholar]
- 59.Ikeda R, Ichikawa T. 2014. Interaction of surface molecules on Cryptococcus neoformans with plasminogen. FEMS Yeast Res. 14, 445–450. (doi:10.1111/1567-1364.12131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petit FM, Serres C, Bourgeon F, Pineau C, Auer J. 2013. Identification of sperm head proteins involved in zona pellucida binding. Hum. Reprod. 28, 852–865. (doi:10.1093/humrep/des452) [DOI] [PubMed] [Google Scholar]
- 61.Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P. 2013. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front. Plant Sci. 4, 450 (doi:10.3389/Fpls.2013.00450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang GQ, Xia Y, Cui J, Gu ZN, Song YD, Chen YQ, Chen HQ, Zhang H, Chen W. 2014. The roles of moonlighting proteins in bacteria. Curr. Issues Mol. Biol. 16, 15–22. [PubMed] [Google Scholar]
- 63.Hernandez S, Ferragut G, Amela I, Perez-Pons J, Pinol J, Mozo-Villarias A, Cedano J, Querol E. 2014. MultitaskProtDB: a database of multitasking proteins. Nucleic Acids Res. 42, D517–D520. (doi:10.1093/Nar/Gkt1153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dallo SF, Kannan TR, Blaylock MW, Baseman JB. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46, 1041–1051. (doi:10.1046/j.1365-2958.2002.03207.x) [DOI] [PubMed] [Google Scholar]
- 65.Balasubramanian S, Kannan TR, Baseman JB. 2008. The surface-exposed carboxyl region of Mycoplasma pneumoniae elongation factor Tu interacts with fibronectin. Infect. Immun. 76, 3116–3123. (doi:10.1128/IAI.00173-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alvarez RA, Blaylock MW, Baseman JB. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalium binds mucin. Mol. Microbiol. 48, 1417–1425. (doi:10.1046/j.1365-2958.2003.03518.x) [DOI] [PubMed] [Google Scholar]
- 67.Sueyoshi N, Nimura T, Onouchi T, Baba H, Takenaka S, Ishida A, Kameshita I. 2012. Functional processing of nuclear Ca2+/calmodulin-dependent protein kinase phosphatase (CaMKP-N): evidence for a critical role of proteolytic processing in the regulation of its catalytic activity, subcellular localization and substrate targeting in vivo. Arch. Biochem. Biophys. 517, 43–52. (doi:10.1016/j.abb.2011.10.017) [DOI] [PubMed] [Google Scholar]
- 68.Frey J, Haldimann A, Kobisch M, Nicolet J. 1994. Immune response against the l-lactate dehydrogenase of Mycoplasma hyopneumoniae in enzootic pneumonia of swine. Microb. Pathog. 17, 313–322. (doi:10.1006/mpat.1994.1077) [DOI] [PubMed] [Google Scholar]
- 69.Haldimann A, Nicolet J, Frey J. 1993. DNA sequence determination and biochemical analysis of the immunogenic protein P36, the lactate dehydrogenase (LDH) of Mycoplasma hyopneumoniae. J. Gen. Microbiol. 139, 317–323. (doi:10.1099/00221287-139-2-317) [DOI] [PubMed] [Google Scholar]
- 70.Kim JW, Dang CV. 2005. Multifaceted roles of glycolytic enzymes. Trends Biochem. Sci. 30, 142–150. (doi:10.1016/j.tibs.2005.01.005) [DOI] [PubMed] [Google Scholar]
- 71.Cattaneo A, Biocca S, Corvaja N, Calissano P. 1985. Nuclear localization of a lactic dehydrogenase with single-stranded DNA-binding properties. Exp. Cell Res. 161, 130–140. (doi:10.1016/0014-4827(85)90497-5) [DOI] [PubMed] [Google Scholar]
- 72.Grosse F, Nasheuer HP, Scholtissek S, Schomburg U. 1986. Lactate dehydrogenase and glyceraldehyde-phosphate dehydrogenase are single-stranded DNA-binding proteins that affect the DNA-polymerase-alpha-primase complex. Eur. J. Biochem. 160, 459–467. (doi:10.1111/j.1432-1033.1986.tb10062.x) [DOI] [PubMed] [Google Scholar]
- 73.Zhong XH, Howard BD. 1990. Phosphotyrosine-containing lactate dehydrogenase is restricted to the nuclei of PC12 pheochromocytoma cells. Mol. Cell Biol. 10, 770–776. (doi:10.1128/MCB.10.2.770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cooper JA, Esch FS, Taylor SS, Hunter T. 1984. Phosphorylation sites in enolase and lactate dehydrogenase utilized by tyrosine protein kinases in vivo and in vitro. J. Biol. Chem. 259, 7835–7841. [PubMed] [Google Scholar]
- 75.Sarioglu H, Lottspeich F, Walk T, Jung G, Eckerskorn C. 2000. Deamidation as a widespread phenomenon in two-dimensional polyacrylamide gel electrophoresis of human blood plasma proteins. Electrophoresis 21, 2209–2218. (doi:10.1002/1522-2683(20000601)21:11<2209::AID-ELPS2209>3.0.CO;2-T) [DOI] [PubMed] [Google Scholar]
- 76.Butler E, Alsterfjord M, Olofsson TC, Karlsson C, Malmstrom J, Vasquez A. 2013. Proteins of novel lactic acid bacteria from Apis mellifera mellifera: an insight into the production of known extra-cellular proteins during microbial stress. BMC Microbiol. 13, 235 (doi:10.1186/1471-2180-13-235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson MW, et al. 2013. MHJ_0125 is an M42 glutamyl aminopeptidase that moonlights as a multifunctional adhesin on the surface of Mycoplasma hyopneumoniae. Open Biol. 3, 130017 (doi:10.1098/rsob.130017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gunawardana Y, Niranjan M. 2013. Bridging the gap between transcriptome and proteome measurements identifies post-translationally regulated genes. Bioinformatics 29, 3060–3066. (doi:10.1093/bioinformatics/btt537) [DOI] [PubMed] [Google Scholar]
- 79.Olivares-Hernandez R, Bordel S, Nielsen J. 2011. Codon usage variability determines the correlation between proteome and transcriptome fold changes. BMC Syst. Biol. 5, 33 (doi:10.1186/1752-0509-5-33) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.