Abstract
Accurate chromosome segregation during mitosis and meiosis is crucial for cellular and organismal viability. Kinetochores connect chromosomes with spindle microtubules and are essential for chromosome segregation. These large protein scaffolds emerge from the centromere, a specialized region of the chromosome enriched with the histone H3 variant CENP-A. In most eukaryotes, the kinetochore core consists of the centromere-proximal constitutive centromere-associated network (CCAN), which binds CENP-A and contains 16 subunits, and of the centromere-distal Knl1 complex, Mis12 complex, Ndc80 complex (KMN) network, which binds microtubules and contains 10 subunits. In the fruitfly, Drosophila melanogaster, the kinetochore underwent remarkable simplifications. All CCAN subunits, with the exception of centromeric protein C (CENP-C), and two KMN subunits, Dsn1 and Zwint, cannot be identified in this organism. In addition, two paralogues of the KMN subunit Nnf1 (Nnf1a and Nnf1b) are present. Finally, the Spc105R subunit, homologous to human Knl1/CASC5, underwent considerable sequence changes in comparison with other organisms. We combined biochemical reconstitution with biophysical and structural methods to investigate how these changes reflect on the organization of the Drosophila KMN network. We demonstrate that the Nnf1a and Nnf1b paralogues are subunits of distinct complexes, both of which interact directly with Spc105R and with CENP-C, for the latter of which we identify a binding site on the Mis12 subunit. Our studies shed light on the structural and functional organization of a highly divergent kinetochore particle.
Keywords: kinetochore, centromere, KMN network, Mis12, Spc105, Ndc80
1. Introduction
Accurate chromosome segregation in dividing cells is of utmost importance for the propagation of unicellular organisms, for organismal development and for sexual reproduction [1]. Perturbations of this process have been associated with congenital diseases, premature ageing and cellular transformation [2].
The mitotic spindle, a complex structure made of microtubules, microtubule-associated proteins and molecular motors, is devoted to chromosome capture and segregation [1]. Microtubules capture chromosomes at specialized and evolutionarily conserved structures named kinetochores [3,4]. Kinetochores are multi-protein assemblies that are built on a specialized chromatin domain called the centromere [5]. The crucial and universal feature that distinguishes the centromere from any other chromatin domain on chromosomes is the enrichment of a variant of histone H3 named CENP-A (CID in Drosophila melanogaster; figure 1a) [5]. In most organisms, this histone variant recruits the components of a constitutive centromere-associated network (CCAN), a group of approximately 16 proteins organized in different subcomplexes [8–12]. The CCAN, in turn, recruits the components of a 10-subunit complex named the KMN network (for Knl1 complex, Mis12 complex, Ndc80 complex, the three subcomplexes of which the KMN network is composed) [13]. Within the KMN network, the Ndc80 complex (Ndc80-C) has been implicated as the microtubule receptor at the kinetochore [14,15]. The Knl1 complex (Knl1-C), on the other hand, has been implicated in the coordination of the spindle assembly checkpoint, a signalling mechanism that prevents premature mitotic exit in the presence of unattached or incorrectly attached kinetochores [16]. Finally, the Mis12 complex (Mis12-C, also known as the MIND complex in Saccharomyces cerevisiae) acts as a ‘hub’ that interacts with all other KMN complexes and that also mediates the interaction with the inner kinetochore CCAN subunits [6,17–31]. Furthermore, the Mis12 complex may increase the binding affinity of the Ndc80 complex for microtubules, possibly through an allosteric mechanism [32].
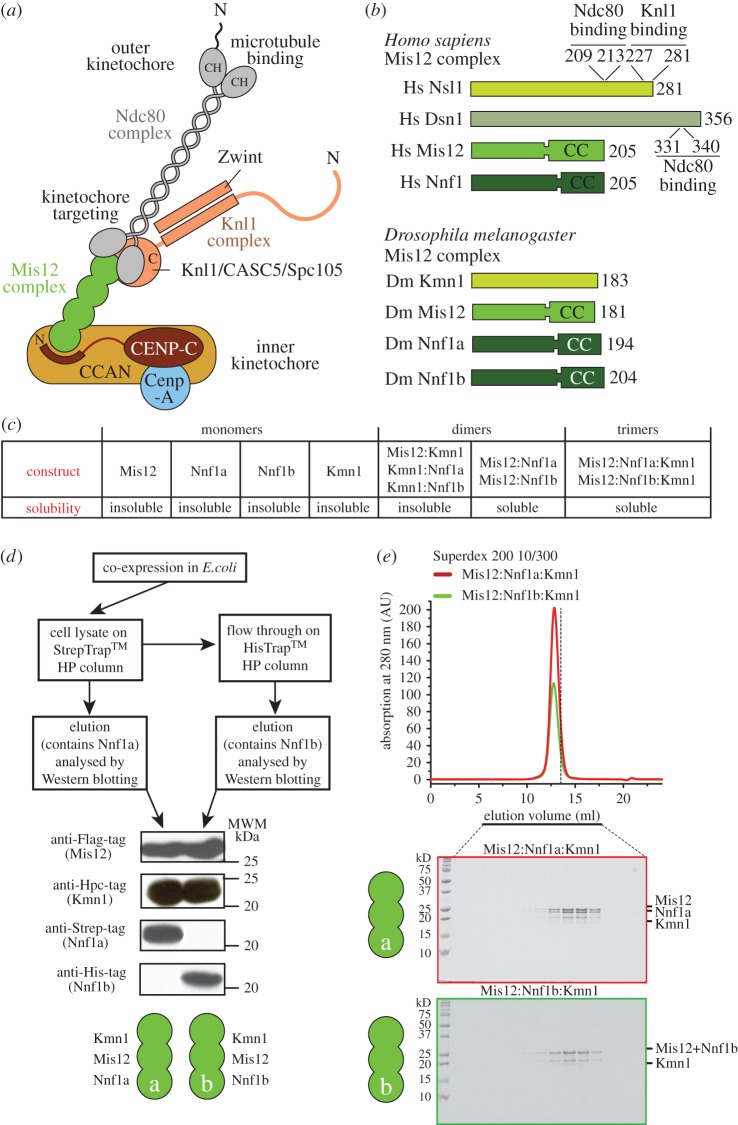
Figure 1.
Two Mis12 complexes in Drosophila melanogaster. (a) Schematic of the human kinetochore. Orthologues of the indicated subunits and complexes are generally conserved in evolution, and are for instance also identified in S. cerevisiae. (b) A presentation of the constitutive subunits of the Mis12 complex in humans and in Drosophila. Segments identified for their ability to interact with Knl1 or Ndc80 complex subunits [6,7] are indicated. (c) Summary of expression experiments. ‘Soluble’ or ‘insoluble’ indicates that the protein could/could not be identified in the bacterial cell lysate. (d) A strategy for determining whether Nnf1a and Nnf1b are part of the same or different complexes. (e) Size-exclusion chromatography (SEC) experiment on the DmMis12a and DmMis12b complex showing the two complexes elute in a single peak and appear monodisperse. The vertical dashed bar is a reference indicating the elution volume of the dimeric constructs shown in electronic supplementary material, figure S1A.
In certain organisms, including Drosophila melanogaster and Caenorhabditis elegans, most CCAN subunits cannot be identified, suggesting that these kinetochores underwent significant structural simplifications in the course of evolution [4,13,20,33–36]. To date, the only residual CCAN subunit to be clearly recognizable in these organisms is CENP-C [37–40]. CENP-C, which is the largest CCAN subunit, has been shown to act as a linker between CENP-A in the centromeric chromatin and the Mis12-C in the outer kinetochore [19–21,23,41–44]. In organisms that retained CCAN, CENP-C also contains binding sites for other CCAN subunits, including the CENP-HIKM and CENP-NL subcomplexes [45–49]. Finally, CENP-C has been shown to participate in the cell-cycle-dependent deposition of new CENP-A required to re-establish the CENP-A pool after its halving during chromosome replication [50–60].
Besides the loss of most CCAN subunits in the inner kinetochore, in D. melanogaster additional evolutionary changes affected the composition of the outer kinetochore, and in particular of the Mis12-C complex. These changes include the apparent loss of the Dsn1 subunit, the appearance of two paralogues of the Nnf1 subunit (Nnf1a and Nnf1b, also named Nnf1R-1 and Nnf1R-2), and the loss of the Zwint subunit in the Knl1-C, which therefore consists exclusively of the Spc105R subunit (Spc105-related, homologous to human Knl1/Blinkin/CASC5 and indicated here as Spc105RKnl1) [17,26,31,33,34,61–63]. How these changes affect the overall organization and stability of the outer kinetochore and of its interactions with CENP-C is currently unclear. Here, we used biochemical reconstitution and biophysical characterization as an entry point to characterize the outer kinetochore of D. melanogaster and its interaction with CENP-C. We report the main conclusions of our effort.
2. Results and discussion
2.1. Reconstitution of two related Mis12 complexes in Drosophila melanogaster
To gain insights into the organization of the DmMis12 complex, we expressed recombinant versions of its subunits (figure 1b) or their combination, as summarized in figure 1c. Mis12, Nnf1a, Nnf1b and Kmn1 (the latter indicated as Kmn1Nsl1 to remind readers that it is the Nsl1 orthologue) were all insoluble when expressed in isolation in Escherichia coli (not shown). Co-expression of different combinations of two subunits with the pST44 vector [64] resulted in soluble complexes of Mis12 with Nnf1a or Nnf1b, whereas binary combinations containing Kmn1Nsl1 were insoluble (figure 1c; electronic supplementary material, figure S1a; some data not shown). Overall, these results suggest that Mis12 and Nnf1 can form a stable pair within the Drosophila Mis12 complex, in line with previous observations [18,25,28,63,65]. Solubilization of Kmn1Nsl1 was only observed when it was co-expressed in combination with Mis12 and Nnf1a or Nnf1b (figure 1c).
The Nnf1a and Nnf1b paralogues have been previously shown to be functionally redundant, but their developmental expression patterns are not identical [34,63]. It is unclear if these proteins are incorporated in the same complex or in separate complexes. The question is particularly relevant in the specific case of the Drosophila Mis12-C, because no Dsn1 has been identified in this organism, suggesting that Mis12-C might have a different composition. To address this question, we co-expressed Mis12, Nnf1a, Nnf1b and Kmn1Nsl1, each fused to a distinct tag, in E. coli (figure 1d). Cleared cell lysates were incubated, in consecutive steps, with affinity resins designed to interact with the affinity tags of Nnf1a (Strep tag) and Nnf1b (polyhistidine), and after elution each bound fraction was analysed by Western blotting (figure 1d). This showed that Nnf1a and Nnf1b are both able to bind Mis12 and Kmn1Nsl1, but do not appear to interact with each other in the same complex.
We reconstituted the Mis12a and Mis12b complexes by bacterial co-expression and purified them to homogeneity (see Methods). Separation of these complexes by size-exclusion chromatography (SEC, which separates based on shape and molecular mass) demonstrated that both complexes are monodisperse and that they elute essentially identically, suggesting similar shape and overall mass (figure 1e). Overall, these data demonstrate that Nnf1a and Nnf1b form distinct and stoichiometric complexes with Kmn1Nsl1 and Mis12, which we define as the DmMis12a and DmMis12b complexes, respectively.
2.2. Characterization of the DmMis12a and DmMis12b complexes
By analytical ultracentrifugation (AUC) sedimentation velocity experiments, we observed molecular masses of the DmMis12a and DmMis12b complexes of 64.5 and 67.1 kDa, respectively (figure 2a and table 1). These values are in excellent agreement with the predicted molecular masses if each subunit was represented in a single copy (table 1). Frictional ratios (f/fo) of 1.7 indicate that both complexes are elongated.
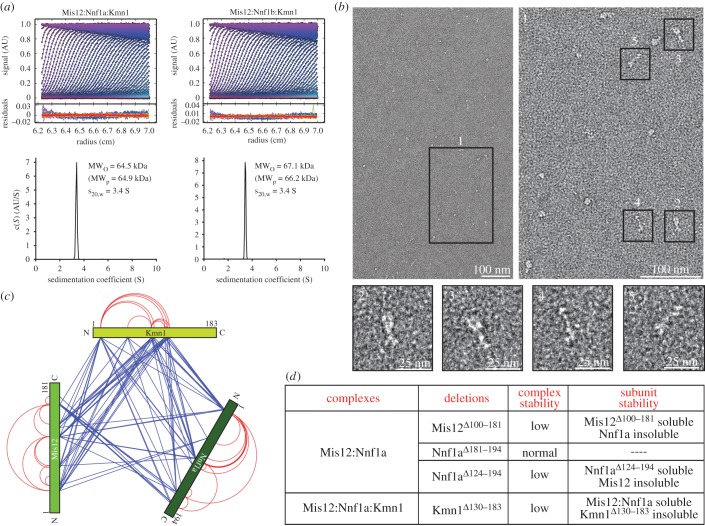
Figure 2.
Biophysical analysis of the DmMis12 complexes. (a) Sedimentation velocity absorbance profiles of the DmMis12a and DmMis12b complexes, with residuals of the fit showing the deviation of the c(S) model from the observed signals; the best-fit continuous-size c(S) distribution of the DmMis12a and DmMis12b complexes is shown in the bottom part of the panel. (b) Representative negative stain EM images of the DmMis12a complex. Scale bars are indicated. (c) Cross-linking-mass spectrometry (XL-MS) analysis of the DmMis12a complex. Blue and red lines indicate inter- and intramolecular cross-links, respectively. (d) Summary of expression experiments with different deletion mutants of the subunits of the DmMis12 complex.
Table 1.
Summary of sedimentation velocity experiments. All predicted molecular masses assumed each subunit was present in a single copy.
| complexes | frictional ratio | observed molecular mass (kDa) | predicted molecular mass (kDa) | S (20,w) |
|---|---|---|---|---|
| Mis12:Nnf1a:Kmn1 | 1.7 | 64.5 | 64.9 | 3.4 |
| Mis12:Nnf1b:Kmn1 | 1.7 | 67.1 | 66.2 | 3.4 |
| Mis12:Nnf1a:Kmn1:Cenp-C1–105 | 1.9 | 76.4 | 76.8 | 3.4 |
| Mis12:Nnf1b:Kmn1:Cenp-C1–105 | 1.9 | 75.6 | 78.0 | 3.4 |
| Mis12:Nnf1a:Kmn1:Spc105R1707–1960 | 1.7 | 93.3 | 95.2 | 4.3 |
| Mis12:Nnf1b:Kmn1:Spc105R1707–1960 | 1.6 | 95.7 | 96.4 | 4.8 |
| Mis12:Nnf1b:Kmn1-Spc105R1707–1960-Cenp-C1–105 | 1.8 | 106.5 | 109.3 | 4.4 |
This was confirmed by negative-stain electron microscopy (EM) experiments on the DmMis12a complex (figure 2b). In each field of view, the majority of single particles appeared elongated, with a thicker end and an overall length of approximately 20 nm. The appearance of the DmMis12a complex is largely comparable to that of the previously observed human and budding yeast complexes [6,24,25,65]. Thus, loss of Dsn1 does not dramatically alter the structure of the DmMis12 complex. However, despite high purity, compositional homogeneity and monodispersity of the Mis12 emerging from SEC experiments (figure 1e), we observed more structural heterogeneity of the complex by negative stain EM (figure 2b) than previously observed with the human complex [6,24], complicating the calculation of class averages. In summary, the EM and AUC analyses indicated that the DmMis12 complex has an elongated appearance, a feature previously observed with the human and yeast complexes [6,19,24,25,65].
To gain additional insights into the organization of the DmMis12a and DmMis12b complexes, we resorted to chemical cross-linking with the bi-functional reagent BS2G (bis[sulfo-succinimidyl]glutarate), which cross-links the primary amines of lysine side chains within a distance compatible with the length of the cross-linker (7.7 Å) (electronic supplementary material, figure S1b), followed by protease digestion and mass spectrometry (XL-MS) [66]. The analysis revealed a very extensive network of interactions between the Mis12 and Nnf1a or Nnf1b subunits, extending all along their sequences (figure 2c; electronic supplementary material, figure S1c). Both subunits also become extensively cross-linked to the N-terminal region of Kmn1Nsl1, extending approximately to residue 120. However, residues 130–183 in the C-terminal region were required for a stable interaction of Kmn1Nsl1 with the rest of the DmMis12a complex, because their deletion (Kmn1Δ130–183) generated an unstable mutant that failed to be incorporated in a complex with Nnf1a and Mis12 (figure 2d). Large C-terminal deletions of Mis12 and Nnf1a also strongly reduced the stability of the binary Mis12:Nnf1a complex (figure 2d; some data not shown).
2.3. DmMis12-C interacts directly with CENP-C
CENP-C, a subunit of the CCAN complex, interacts directly with the specialized CENP-A nucleosome in the centromere chromatin underlying the kinetochore (see Introduction). Comparison of the overall organization of CENP-C in Drosophila melanogaster and in other metazoans reveals remarkable differences (figure 3a). For instance, DmCENP-C is approximately 500 residues longer than HsCENP-C [38]. Within its N-terminal half, DmCENP-C sequence contains two regions, the arginine-rich (R-rich) domain and the drosophilids CENP-C homology (DH) domain [38], that cannot be detected in the human sequence. It also contains two predicted AT-hooks domain (AT1 and AT2), which may mediate interactions with DNA [38]. The function of all these domains unique to the Drosophila sequence is currently unclear. In humans, a region of CENP-C also located in the N-terminal half of the protein has been recently implicated in binding to CCAN subunits such as CENP-H, CENP-I and others (figure 3a) [46,49]. Thus, divergence in the N-terminal region of CENP-C may reflect the specific evolutionary history of Drosophila that led to the loss of other CCAN subunits. On the other hand, the C-terminal region of DmCENP-C, containing a CENP-C motif implicated in CENP-A binding and a dimerization domain [38,67], is related to metazoans' (figure 3a).
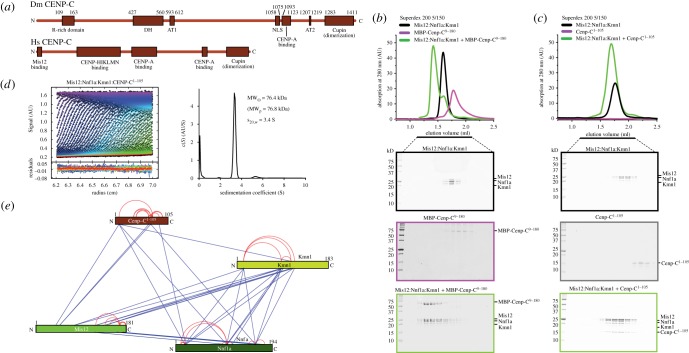
Figure 3.
The DmMis12 complex binds the N-terminal region of CENP-C. (a) Schematic comparison of the domain structure of CENP-C in Drosophila and humans. Domains in the Drosophila sequence are as follow: R-rich, arginine-rich; DH, drosophilid Cenp-C homologues; AT1 and AT2, AT hooks; NLS, nuclear localization signal; CENP-A binding motif, also known as the CENP-C motif; Cupin, a dimerization domain near the C-terminal region (C-term). For more detail, see [38]. Human CENP-C contains an N-terminal Mis12 binding domain [19,20,28], a domain for binding to the CENP-HIKM and CENP-NL complexes [46,49], and domains related to those present in Drosophila. (b) Analytical size-exclusion chromatography shows that the DmMis12a complex binds directly to MBP-CENP-C9–180. (c) Analytical size-exclusion chromatography shows that the DmMis12a complex binds directly to CENP-C1–105. (d) Sedimentation velocity absorbance profiles of the DmMis12a and DmMis12b complexes, with residuals of the fit showing the deviation of the c(S) model from the observed signals; the best-fit continuous-size c(S) distribution of the DmMis12a and DmMis12b complexes is shown on the right-hand side of the panel. (e) Cross-linking-mass spectrometry (XL-MS) analysis of the DmMis12a complex. Blue and red lines indicate inter- and intramolecular cross-links, respectively.
In previous studies, we and others demonstrated that Mis12-C binds directly to CENP-C in Drosophila, budding yeast and humans [19,20,28]. In humans, as little as approximately 20 residues at the N-terminus of CENP-C are sufficient to generate a relatively tight binding interaction with Mis12-C, whereas longer CENP-C segments bind more tightly [19]. Similar conclusions emerged from studies in S. cerevisiae [28]. An alignment of the N-terminal region of CENP-C in drosophilids, yeasts and vertebrates failed to reveal strictly conserved features, although a possible fuzzy pattern consisting of a stretch of positive charges followed by hydrophobic stretches might be envisioned (electronic supplementary material, figure S2).
Because the domain of DmCENP-C interacting with the Mis12 complex has not been mapped in detail, we tested binding of the DmMis12a complex to a fusion protein of maltose binding protein (MBP) with residues 9–180 of CENP-C (CENP-C9–180) in an SEC experiment (figure 3b). A clear shift in the elution pattern of both species was indicative of a tight interaction. Essentially identical results were obtained with DmMis12b complex (electronic supplementary material, figure S3a). Residues 1–8 of DmCENP-C are not conserved in other drosophilids, but conservation increases significantly in regions immediately C-terminal to this non-conserved region (electronic supplementary material, figure S2). Indeed, larger N-terminal deletions (DmCENP-C36–180) prevented an interaction with both the Mis12a and Mis12b complexes (electronic supplementary material, figures S3b,c), indicating that residues 9–35 contain essential interaction determinants.
We then tested the effects of C-terminal deletions from the DmCENP-C N-terminal region. A construct corresponding to DmCENP-C1–105 (devoid of affinity tags) interacted with the DmMis12a and DmMis12b complexes stoichiometrically (figure 3c; electronic supplementary material, figure S3d), and so did an even shorter deletion mutant, DmCENP-C9–71 (also devoid of tags; electronic supplementary material, figure S3e,f). Collectively, these results demonstrate that the Mis12 complex binds directly to the N-terminal region of CENP-C in Drosophila, similarly to what was previously observed in humans and yeast [19,28], and despite the very modest sequence identity in the CENP-C N-terminal region across species. The DmMis12a:DmCENP-C1–105 complex was monodisperse in SEC runs and sedimented essentially as a single peak in sedimentation velocity experiments (figure 3d), with a predicted molecular mass of 76.4 (table 1), indicating that the Mis12a complex and CENP-C interact with 1 : 1 stoichiometry. Essentially identical results were obtained with the Mis12b:CENP-C complex (table 1 and electronic supplementary material, figure S4a). XL–MS experiments confirmed an interaction of CENP-C1–105 with the Mis12 subunit, but also identified additional potential contacts with Nnf1 and Kmn1Nsl1 (figure 3e and electronic supplementary material, figure S4b).
2.4. A CENP-C binding site on the Mis12 subunit of the Mis12 complex
The determinants of the Mis12 complex required to interact with CENP-C are unknown, although a requirement for the Nnf1 subunit in vitro has been described [20]. In our attempts (until now unsuccessful; data not shown) to crystallize the D. melanogaster Mis12 complex, we generated a version of the Mis12a complex in which the first 15 residues of the Mis12 subunit had been deleted. Unexpectedly, the deletion mutant failed to bind CENP-C1–105 (electronic supplementary material, figure S5a).
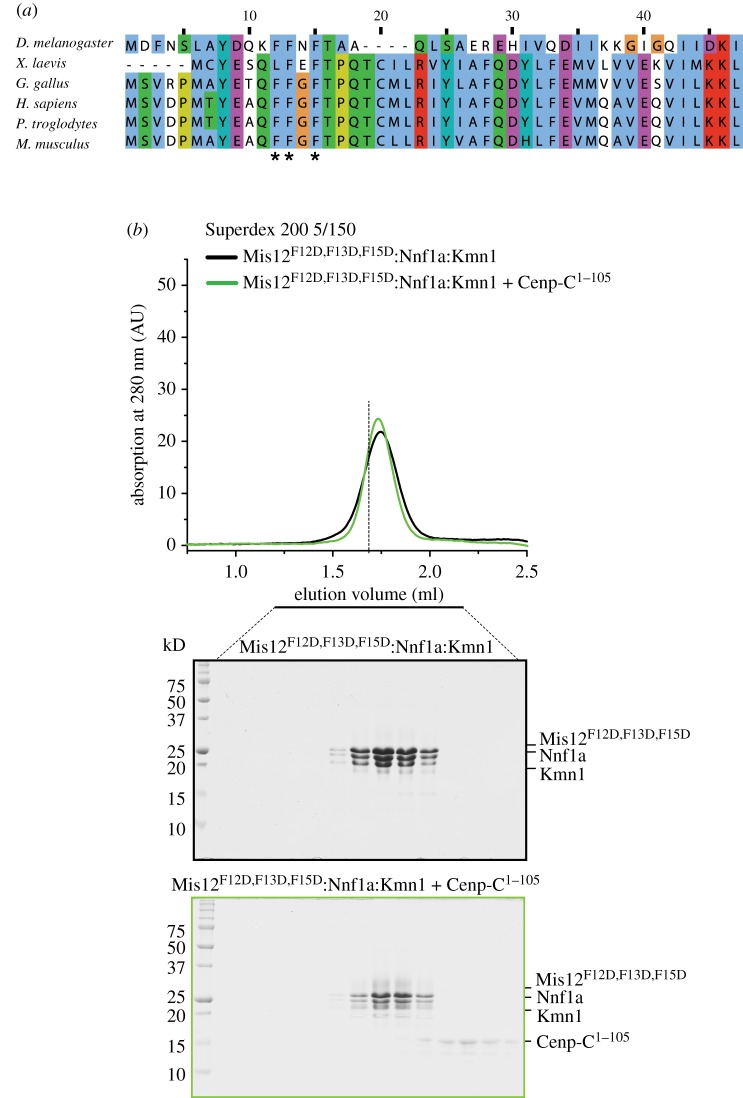
The sequence of the first 15 residues of the Mis12 subunit is evolutionarily conserved (figure 4a). Because removal of this region does not appear to be grossly detrimental to the stability of the Mis12 complex, we tested the role of three conserved phenylalanine (F) residues, F12, F13 and F15, in the interaction with CENP-C1–105. A DmMis12a complex containing mutations F12D, F13D and F15D in the Mis12 subunit was monodisperse, as judged by its SEC elution profile (figure 4b). In agreement with a role of the N-terminal region of Mis12 in CENP-C binding, the mutant was unable to interact with CENP-C1–105 in a SEC co-elution experiment, indicating that the mutations disrupt the interaction of Mis12 with CENP-C (figure 4b). Essentially identical results were obtained with a DmMis12b complex expressing mutations F12D, F13D and F15D (electronic supplementary material, figure S5b). Thus, our results implicate the N-terminal region of the Mis12 subunit as a necessary determinant of the interaction of the Mis12 complex with CENP-C.
Figure 4.
A CENP-C binding region in the Mis12 subunit. (a) Sequence alignment of the N-terminal region of the Mis12 subunit of the Mis12 complex. The positions of three phenylalanine (F) residues that were mutated to Asp are indicated by asterisks. (b) Analytical size-exclusion chromatography shows that the CENP-C1–105 is unable to interact with the mutant Mis12 complex. The vertical dashed bar is a reference indicating the peak elution volume of the tetrameric MIs12:Nnf1a:Kmn1:CENP-C1–105 complex whose elution is shown in figure 3c.
2.5. The interaction of the Mis12 complex with Spc105R
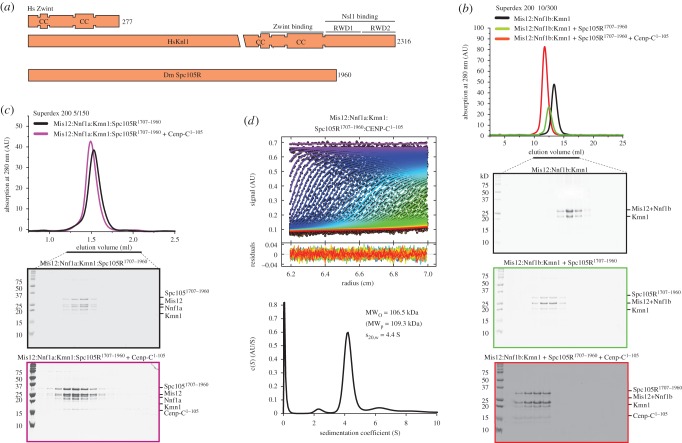
Another interesting difference between the KMN network in D. melanogaster and other eukaryotes lies in the Knl1 complex. One of the two subunits of the complex, Zwint, has not been identified in D. melanogaster (figure 5a). Conversely, DmSpc105RKnl1, related to the Knl1/CASC5 subunit, is shorter than in humans. Previously, it has been shown that the C-terminal region of human Knl1 contains two consecutive RWD (RING finger, WD repeat, DEAD-like helicases) domains preceded by a coiled-coil region. The latter mediates the interaction with Zwint, which is also a coiled-coil protein, whereas the former mediate binding to the C-terminal region of the Nsl1 subunit of the human Mis12 complex, homologous to Kmn1 in D. melanogaster [6,24,27] (figure 5a).
Figure 5.
Interaction of the Mis12 complex with the C-terminal region of DmSpc105. (a) Schematic comparison of the domain structure of Spc105R1Knl1 in Drosophila and of its human homologue Knl1/CASC5. The C-terminal region of Knl1/CASC contains a coiled-coil domain that has been implicated in a direct interaction with Zwint, a coiled-coil protein that has not been identified in Drosophila. It also contains two consecutive RWD domains implicated in a direct interaction with the Nsl1 subunit of the human Mis12 complex [24]. (b) Size-exclusion chromatography analysis of the indicated complexes demonstrates that Spc105R1707–1960 and CENP-C1–105 enter a single complex with the Mis12b complex. (c) The Mis12a:Spc105R1707–1960 complex binds CENP-C1–105. (d) AUC sedimentation velocity analysis of Mis12a:Spc105R1707–1960:CENP-C1–105 complex.
None of these features is evident in DmSpc105RKnl1. First, program COILS [68] only identifies a short sequence (residues 1850–1890) with (limited) potential for forming a coiled-coil, instead of the approximately 200-residue coiled-coil domain identified in the human protein. Second, there is no evidence that the C-terminal region of DmSpc105RKnl1 might contain RWD domains like the human counterpart Knl1/CASC5. For instance, BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches with the last approximately 200 residues of DmSpc105RKnl1 fail to detect homologous proteins beyond drosophilids (not shown). Additionally, three-dimensional modelling with the Phyre2 server [69] failed to identify structural homology of the C-terminal region of DmSpc105RKnl1 with structures deposited in the protein data bank, which include several structures of RWD domains, including those present in Knl1/CASC5 [24] (not shown). Nevertheless, secondary structure prediction servers, including JPRED4 (http://www.compbio.dundee.ac.uk/jpred/index_up.html) and PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) [70,71], identify a succession of secondary structure elements for residues 1850–1960 of DmSpc105RKnl1 that is, in principle, compatible with the presence of an RWD domain (data not shown). Thus, the detailed structural organization of the C-terminal region of DmSpc105RKnl1 remains uncertain. Despite possible evolutionary changes, however, previous evidence demonstrated that an approximately 600-residue construct containing the C-terminal region of DmSpc105RKnl1 can interact with Kmn1Nsl1 in a yeast two-hybrid (Y2H) experiment [17]. This suggests that the C-terminal regions of the human and fly sequences are, if not evolutionary conserved, at least functionally related.
To shed light on the interaction of DmSpc105RKnl1 with the Mis12 complex, we co-expressed several recombinant segments encompassing the C-terminal region of Spc105RKnl1 with the Mis12a or Mis12b complexes. Constructs approximately encompassing the predicted coiled-coil region (comprised in the segment 1852–1889), including Spc105R1707–1882 and Spc105R1707–1890, were insoluble. Constructs containing the C-terminal region downstream from the predicted coiled-coil, including Spc105R1887–1960, Spc105R1875–1960 and Spc105R1890–1960, were insoluble. Finally, constructs containing the predicted coiled-coil and the C-terminal region, including Spc105R1847–1960 and Spc105R1810–1960, were also insoluble. The only segment of Spc105RKnl1 that could be co-expressed in a soluble form with the Mis12a and Mis12b complexes was Spc105R1707–1960. In both cases, an apparently monodisperse and stoichiometric complex formed (figure 5b,c), whose behaviour in AUC sedimentation velocity experiments predicted a Mis12 complex:Spc105R1707–1890 stoichiometry of 1 : 1 (table 1; electronic supplementary material, figure S6a,b). Both the Mis12a:Spc105R1707–1890 and the Mis12b:Spc105R1707–1890 complexes further interacted with CENP-C1–105 in single monodisperse complexes (figure 5b,c). In agreement with this observation, AUC sedimentation velocity analysis of the Mis12a:Spc1051707–1960:CENP-C1–105 complex revealed a stable 1 : 1:1 assembly (table 1 and figure 5d).
2.6. Conclusion
Owing to the considerable array of interactions it mediates, the Mis12 complex is viewed as a ‘hub’ of kinetochore assembly and function. Biochemical reconstitution of the yeast and human Mis12 complexes has shed considerable light on their organization, revealing a conserved set of intra- and intercomplex interactions [6,7,24,28,65]. A detailed, high-resolution structural characterization of the Mis12 complex, however, has been missing, possibly because of the inherent flexibility of some of its domains.
Our work on the Drosophila Mis12 complex was motivated by its considerable simplification in comparison with its counterparts in other organisms, with one of the four subunits, Dsn1, having apparently disappeared from the Drosophila genome. Furthermore, because CCAN subunits are absent in Drosophila (with the exception of CENP-C), it may be surmised that the Drosophila Mis12 complex does not require stabilization through additional protein–protein interactions at the kinetochore. By way of example, the yeast Mis12/MIND complex was found to interact with the COMA complex, consisting of the Ctf19, Okp1, Mcm21 and Ame1 subunits (and homologous to CCAN subunits CENP-O, CENP-P, CENP-Q and CENP-U of humans) [28], none of which is identified in Drosophila. Similarly, the human Mis12 complex has been proposed to interact with the CCAN subunit CENP-T [22,23,42]. The latter additionally interacts with the Ndc80 complex, contributing to its recruitment and to a general stabilization of the outer kinetochore [7,21,42,72–76].
We identify two distinct Drosophila Mis12 complexes, containing either the Nnf1a or the Nnf1b subunit. Our extensive biochemical and biophysical analyses failed to reveal significant differences in the behaviour of these complexes. In each of the complexes, the Mis12 and Nnf1 subunits (a or b) form a tight dimer and create the binding site for Kmn1Nsl1, which in turn creates a binding site for Spc105RKnl1. Furthermore, both complexes interact tightly with the N-terminal region of CENP-C and with the C-terminal region of Spc105RKnl1, suggesting that they have similar or indistinguishable interaction potentials. However, previous studies demonstrated different developmental expression patterns for Nnf1a and Nnf1b, suggesting the possibility of functional specialization of the two complexes [34,63].
Despite considerable sequence divergence of the DmCENP-C and DmSpc105Knl1 binding regions, the interactions they entertain with the Mis12 complex engage topologically equivalent regions of their primary structure (near the N-terminus of CENP-C and the C-terminus of Spc105RKnl1). Our mutational analysis identifies the N-terminal region of the Mis12 subunit as a primary determinant of CENP-C binding. An overall conclusion emerging from these studies, therefore, is that kinetochores display considerable evolutionary and structural plasticity. How this plasticity can be accommodated in the structure of the Mis12 complex remains unclear, and our future work will aim to address this urgent question by direct structural analysis.
3. Methods
3.1. cDNAs and DNA constructs
The cDNA for DmSpc1051707–1960 was amplified by the polymerase chain reaction (PCR) from the pOT2 vector containing the full-length DmSpc105RKnl1 sequence (isoform A; a generous gift of Christian Lehner's Lab in University of Zurich) and subcloned into the fourth cassette of pST44 [64]. Optimized (for E. coli) coding sequences for DmMis12, DmNnf1a, DmNnf1b, DmKmn1 and full-length DmCenp-C were obtained from GeneArt. DmCenp-C fragments were amplified by PCR and subcloned into the pETDuet-MBP8His, a modified version of pETDuet vector (Novagen) generated in house. Sequences encoding variant versions of the DmMis12 complexes were generated in the pST44 system using standard restriction enzyme-based cloning procedures. The QuikChange mutagenesis kit (Agilent Technologies) was used to generate all mutant versions of the plasmids.
3.2. Protein expression and purification Escherichia coli
BL21(DE3) Rosetta cells were used to express all recombinant proteins. Cells were grown in Terrific broth at 37°C to an OD600 of about 0.8. Protein expression was induced by addition of 0.1 mM IPTG at 20°C, and cells were incubated overnight. Cell pellets were resuspended in binding buffer (20 mM Tris/HCl pH 8.0, 300 mM NaCl, 5% (v/v) glycerol, 1 mM EDTA, 1 mM TCEP), lysed by sonication and cleared by centrifugation at 10 000g for 30 min. The cleared lysate was purified through a succession of His-Trap HP, HP ResourceQ and Superdex 200 10/300 columns (GE Healthcare).
3.3. Analytical size-exclusion chromatography
Analytical size-exclusion chromatography experiments were performed on calibrated Superdex200 5/150 column (GE Healthcare). All samples were eluted under isocratic conditions at 4°C in size-exclusion chromatography buffer (20 mM Tris, 150 mM NaCl, 1 mM TCEP) at a flow rate of 0.2 ml min−1. Elution of proteins was monitored at 280 nm. The loading volume for each injection was 50 µl. In order to detect complex formation, proteins were mixed at 1 : 1 (molar ratio) and incubated for 2 h on ice. SDS–PAGE, followed by Coomassie staining, was used to detect proteins.
3.4. Negative-stain electron microscopy
The Mis12 complex was diluted to 15 nM for EM grid preparation. About 4 µl of protein sample was adsorbed onto glow-discharged carbon-coated grids for 1 min at 25°C, prior to negative staining with 0.07% uranyl formate (SPI supplies/Structure Probe). Samples were imaged with a JEOL1400 microscope equipped with a LaB6 cathode operating at 120 kV. Images were recorded at low-dose conditions at a magnification of 67 200 on a 4 × 4 k charge-coupled device (CCD) camera (TVIPS GmbH).
3.5. Sedimentation velocity analytical ultracentrifugation
Sedimentation velocity experiments were performed in an Optima XL-A analytical ultracentrifuge (Beckman Coulter, Palo Alto, CA) with Epon charcoal-filled double-sector quartz cells and an An-60 Ti rotor (Beckman Coulter). Samples were dialysed against buffer (20 mM Tris pH 8, 0.15 M NaCl and 1 mM TCEP) that was used as a reference. Samples were centrifuged at 42 000 rpm at 20°C, and 500 radial absorbance scans at 280 nm were collected with a time interval of 1 min. The data were analysed using the SEDFIT software [77] in terms of continuous distribution function of sedimentation coefficients (c(S)). The protein partial specific volume was estimated from the amino acid sequence using the program SEDNTERP. Data were plotted using the program GUSSI.
3.6. Cross-linking/mass spectrometry
About 0.8 mg ml−1 DmMis12a was mixed with 0.6 mM BS2G-H6/D6 (Creative Molecules, www.creativemolecules.com) in a final volume of 50 µl. After incubation for 30 min at 37°C, the reaction was quenched by adding 100 mM ammonium bicarbonate and incubating 15 min at 37°C. Cross-linked proteins were digested, and the cross-linked peptides were enriched and analysed by liquid chromatography coupled to tandem mass spectrometry using a hybrid LTQ-Orbitrap Elite instrument (Thermo Fisher Scientific, Waltham, MA) [66]. Cross-links were identified by the dedicated software xQuest [78]. False discovery rates (FDRs) were estimated using xProphet [78], and results were filtered according to the following parameters: FDR < 0.05, min delta score = 0.90, MS1 tolerance window of −4 to 4 ppm, Id-score > 22. Cross-links were visualized using the xVis web server [79].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Marcin Przewloka and David Glover for communicating results prior to publication, Christian Lehner for sharing reagents, Thomas Vaccari and the Musacchio laboratory and especially Dongqing Pan, Marion Pesenti and Siva Jeganathan, for helpful discussions and comments. The authors declare no competing financial interests.
Competing interests
We declare we have no competing interests.
Funding
F.H. is supported by the European Research Council (MolStruKT StG no. 638218) and by the DFG (GRK 1721). A.M. gratefully acknowledges funding by the European Research Council (ERC) Advanced Investigator award RECEPIANCE (GA number 669686) and the DFG's Collaborative Research Centre (CRC) 1093.
References
- 1.Foley EA, Kapoor TM. 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14, 25–37. (doi:10.1038/nrm3494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16, 473–485. (doi:10.1038/nrm4025) [DOI] [PubMed] [Google Scholar]
- 3.Cheeseman IM. 2014. The kinetochore. Cold Spring Harb. Perspect. Biol. 6, a015826 (doi:10.1101/cshperspect.a015826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westermann S, Schleiffer A. 2013. Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans. Trends Cell Biol. 23, 260–269. (doi:10.1016/j.tcb.2013.01.010) [DOI] [PubMed] [Google Scholar]
- 5.Fukagawa T, Earnshaw WC. 2014. The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508. (doi:10.1016/j.devcel.2014.08.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrovic A, et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190, 835–852. (doi:10.1083/jcb.201002070) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malvezzi F, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S. 2013. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32, 409–423. (doi:10.1038/emboj.2012.356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR III, Cleveland DW. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8, 458–469. (doi:10.1038/ncb1397) [DOI] [PubMed] [Google Scholar]
- 9.Izuta H, et al. 2006. Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11, 673–684. (doi:10.1111/j.1365-2443.2006.00969.x) [DOI] [PubMed] [Google Scholar]
- 10.Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. 2004. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells 9, 105–120. (doi:10.1111/j.1365-2443.2004.00705.x) [DOI] [PubMed] [Google Scholar]
- 11.Hori T, et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135, 1039–1052. (doi:10.1016/j.cell.2008.10.019) [DOI] [PubMed] [Google Scholar]
- 12.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR III, Desai A, Fukagawa T. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8, 446–457. (doi:10.1038/ncb1396) [DOI] [PubMed] [Google Scholar]
- 13.Cheeseman IM, Desai A. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46. (doi:10.1038/nrm2310) [DOI] [PubMed] [Google Scholar]
- 14.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127, 969–982. (doi:10.1016/j.cell.2006.09.047) [DOI] [PubMed] [Google Scholar]
- 15.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127, 983–997. (doi:10.1016/j.cell.2006.09.039) [DOI] [PubMed] [Google Scholar]
- 16.Musacchio A. 2015. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25, R1002–R1018. (doi:10.1016/j.cub.2015.08.051) [DOI] [PubMed] [Google Scholar]
- 17.Schittenhelm RB, Chaleckis R, Lehner CF. 2009. Intrakinetochore localization and essential functional domains of Drosophila Spc105. EMBO J. 28, 2374–2386. (doi:10.1038/emboj.2009.188) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiyomitsu T, Obuse C, Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13, 663–676. (doi:10.1016/j.devcel.2007.09.005) [DOI] [PubMed] [Google Scholar]
- 19.Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21, 391–398. (doi:10.1016/j.cub.2010.12.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. 2011. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 21, 399–405. (doi:10.1016/j.cub.2011.02.005) [DOI] [PubMed] [Google Scholar]
- 21.Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145, 410–422. (doi:10.1016/j.cell.2011.03.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rago F, Gascoigne KE, Cheeseman IM. 2015. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr. Biol. 25, 671–677. (doi:10.1016/j.cub.2015.01.059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Yu H. 2015. Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J. Cell Biol. 208, 181–196. (doi:10.1083/jcb.201407074) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrovic A, et al. 2014. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol. Cell 53, 591–605. (doi:10.1016/j.molcel.2014.01.019) [DOI] [PubMed] [Google Scholar]
- 25.Hornung P, Maier M, Alushin GM, Lander GC, Nogales E, Westermann S. 2011. Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J. Mol. Biol. 405, 548–559. (doi:10.1016/j.jmb.2010.11.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkei Z, Przewloka MR, Ladak Y, Albadri S, Sossick A, Juhasz G, Novak B, Glover DM. 2012. Spatiotemporal dynamics of Spc105 regulates the assembly of the Drosophila kinetochore. Open Biol. 2, 110032 (doi:10.1098/rsob.110032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiyomitsu T, Iwasaki O, Obuse C, Yanagida M. 2010. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J. Cell Biol. 188, 791–807. (doi:10.1083/jcb.200908096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hornung P, et al. 2014. A cooperative mechanism drives budding yeast kinetochore assembly downstream of CENP-A. J. Cell Biol. 206, 509–524. (doi:10.1083/jcb.201403081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan X, et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell 137, 672–684. (doi:10.1016/j.cell.2009.03.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joglekar AP, Bloom K, Salmon ED. 2009. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr. Biol. 19, 694–699. (doi:10.1016/j.cub.2009.02.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feijao T, Afonso O, Maia AF, Sunkel CE. 2013. Stability of kinetochore-microtubule attachment and the role of different KMN network components in Drosophila. Cytoskeleton (Hoboken) 70, 661–675. (doi:10.1002/cm.21131) [DOI] [PubMed] [Google Scholar]
- 32.Kudalkar EM, et al. 2015. Regulation of outer kinetochore Ndc80 complex-based microtubule attachments by the central kinetochore Mis12/MIND complex. Proc. Natl Acad. Sci. USA 112, E5583–E5589. (doi:10.1073/pnas.1513882112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7, R23 (doi:10.1186/gb-2006-7-3-r23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Przewloka MR, Zhang W, Costa P, Archambault V, D'Avino PP, Lilley KS, Laue ED, McAinsh AD, Glover DM. 2007. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE 2, e478 (doi:10.1371/journal.pone.0000478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barth TK, Schade GO, Schmidt A, Vetter I, Wirth M, Heun P, Thomae AW, Imhof A. 2014. Identification of novel Drosophila centromere-associated proteins. Proteomics 14, 2167–2178. (doi:10.1002/pmic.201400052) [DOI] [PubMed] [Google Scholar]
- 36.Drinnenberg IA, deYoung D, Henikoff S, Malik HS. 2014. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. Elife 3, 2104 (doi:10.7554/eLife.03676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitoh H, Tomkiel J, Cooke CA, Ratrie H III, Maurer M, Rothfield NF, Earnshaw WC. 1992. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell 70, 115–125. (doi:10.1016/0092-8674(92)90538-N) [DOI] [PubMed] [Google Scholar]
- 38.Heeger S, Leismann O, Schittenhelm R, Schraidt O, Heidmann S, Lehner CF. 2005. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 19, 2041–2053. (doi:10.1101/gad.347805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore LL, Roth MB. 2001. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 153, 1199–1208. (doi:10.1083/jcb.153.6.1199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153, 1209–1226. (doi:10.1083/jcb.153.6.1209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. 2013. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340, 1110–1113. (doi:10.1126/science.1235532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hori T, Shang WH, Takeuchi K, Fukagawa T. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200, 45–60. (doi:10.1083/jcb.201210106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Falk SJ, et al. 2015. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science 348, 699–703. (doi:10.1126/science.1259308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR III, Oegema K, Desai A. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18, 2255–2268. (doi:10.1101/gad.1234104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basilico F, et al. 2014. The pseudo GTPase CENP-M drives human kinetochore assembly. Elife 3, e02978 (doi:10.7554/eLife.02978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A. 2015. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol. 210, 11–22. (doi:10.1083/jcb.201412028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hinshaw SM, Harrison SC. 2013. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep. 5, 29–36. (doi:10.1016/j.celrep.2013.08.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka K, Chang HL, Kagami A, Watanabe Y. 2009. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev. Cell 17, 334–343. (doi:10.1016/j.devcel.2009.08.004) [DOI] [PubMed] [Google Scholar]
- 49.Nagpal H, Hori T, Furukawa A, Sugase K, Osakabe A, Kurumizaka H, Fukagawa T. 2015. Dynamic changes in the CCAN organization through CENP-C during cell-cycle progression. Mol. Biol. Cell 26, 3768–3776. (doi:10.1091/mbc.E15-07-0531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carroll CW, Milks KJ, Straight AF. 2010. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol. 189, 1143–1155. (doi:10.1083/jcb.201001013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milks KJ, Moree B, Straight AF. 2009. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol. Biol. Cell 20, 4246–4255. (doi:10.1091/mbc.E09-05-0378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moree B, Meyer CB, Fuller CJ, Straight AF. 2011. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol. 194, 855–871. (doi:10.1083/jcb.201106079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westhorpe FG, Fuller CJ, Straight AF. 2015. A cell-free CENP-A assembly system defines the chromatin requirements for centromere maintenance. J. Cell Biol. 209, 789–801. (doi:10.1083/jcb.201503132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen CC, Dechassa ML, Bettini E, Ledoux MB, Belisario C, Heun P, Luger K, Mellone BG. 2014. CAL1 is the Drosophila CENP-A assembly factor. J. Cell Biol. 204, 313–329. (doi:10.1083/jcb.201305036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jansen LE, Black BE, Foltz DR, Cleveland DW. 2007. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805. (doi:10.1083/jcb.200701066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunleavy EM, Beier NL, Gorgescu W, Tang J, Costes SV, Karpen GH. 2012. The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol. 10, e1001460 (doi:10.1371/journal.pbio.1001460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Unhavaithaya Y, Orr-Weaver TL. 2013. Centromere proteins CENP-C and CAL1 functionally interact in meiosis for centromere clustering, pairing, and chromosome segregation. Proc. Natl Acad. Sci. USA 110, 19 878–19 883. (doi:10.1073/pnas.1320074110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fachinetti D, et al. 2013. A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056–1066. (doi:10.1038/ncb2805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orr B, Sunkel CE. 2011. Drosophila CENP-C is essential for centromere identity. Chromosoma 120, 83–96. (doi:10.1007/s00412-010-0293-6) [DOI] [PubMed] [Google Scholar]
- 60.Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. 2008. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 183, 805–818. (doi:10.1083/jcb.200806038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venkei Z, Przewloka MR, Glover DM. 2011. Drosophila Mis12 complex acts as a single functional unit essential for anaphase chromosome movement and a robust spindle assembly checkpoint. Genetics 187, 131–140. (doi:10.1534/genetics.110.119628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Przewloka MR, Venkei Z, Glover DM. 2009. Searching for Drosophila Dsn1 kinetochore protein. Cell Cycle 8, 1292–1293. (doi:10.4161/cc.8.8.8159) [DOI] [PubMed] [Google Scholar]
- 63.Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner CF. 2007. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116, 385–402. (doi:10.1007/s00412-007-0103-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan S, Kern RC, Selleck W. 2005. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40, 385–395. (doi:10.1016/j.pep.2004.12.002) [DOI] [PubMed] [Google Scholar]
- 65.Maskell DP, Hu XW, Singleton MR. 2010. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 190, 823–834. (doi:10.1083/jcb.201002059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herzog F, et al. 2012. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352. (doi:10.1126/science.1221483) [DOI] [PubMed] [Google Scholar]
- 67.Sugimoto K, Kuriyama K, Shibata A, Himeno M. 1997. Characterization of internal DNA-binding and C-terminal dimerization domains of human centromere/kinetochore autoantigen CENP-C in vitro: role of DNA-binding and self-associating activities in kinetochore organization. Chromosome Res. 5, 132–141. (doi:10.1023/A:1018422325569) [DOI] [PubMed] [Google Scholar]
- 68.Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252, 1162–1164. (doi:10.1126/science.252.5009.1162) [DOI] [PubMed] [Google Scholar]
- 69.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. (doi:10.1038/nprot.2015.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394. (doi:10.1093/nar/gkv332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. (doi:10.1006/jmbi.1999.3091) [DOI] [PubMed] [Google Scholar]
- 72.Bock LJ, et al. 2012. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat. Cell Biol. 14, 614–624. (doi:10.1038/ncb2495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki A, Badger BL, Salmon ED. 2015. A quantitative description of Ndc80 complex linkage to human kinetochores. Nat. Commun. 6, 8161 (doi:10.1038/ncomms9161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krizaic I, Williams SJ, Sanchez P, Rodriguez-Corsino M, Stukenberg PT, Losada A. 2015. The distinct functions of CENP-C and CENP-T/W in centromere propagation and function in Xenopus egg extracts. Nucleus 6, 133–143. (doi:10.1080/19491034.2014.1003509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14, 604–613. (doi:10.1038/ncb2493) [DOI] [PubMed] [Google Scholar]
- 76.Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32, 424–436. (doi:10.1038/emboj.2012.348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619. (doi:10.1016/S0006-3495(00)76713-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walzthoeni T, Claassen M, Leitner A, Herzog F, Bohn S, Forster F, Beck M, Aebersold R. 2012. False discovery rate estimation for cross-linked peptides identified by mass spectrometry. Nat. Methods 9, 901–903. (doi:10.1038/nmeth.2103) [DOI] [PubMed] [Google Scholar]
- 79.Grimm M, Zimniak T, Kahraman A, Herzog F. 2015. xVis: a web server for the schematic visualization and interpretation of crosslink-derived spatial restraints. Nucleic Acids Res. 43, W362–W369. (doi:10.1093/nar/gkv463) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.