Abstract
Background & Aims
Intestinal epithelial stem cells that express leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5) and/or B cell specific Moloney murine leukemia virus integration site 1 (Bmi1) continuously replicate and generate differentiated cells throughout life. Previously, Paneth cells were suggested to constitute an epithelium-intrinsic niche that regulates the behavior of these stem cells. However, ablating Paneth cells has no effect on the maintenance of functional stem cells. Here, we show definitively that a small subset of mesenchymal subepithelial cells expressing the winged-helix transcription factor forkhead box l1 (Foxl1) are a critical component of the intestinal stem cell niche.
Methods
We genetically ablated Foxl1+ mesenchymal cells in adult mice using 2 separate models by expressing either the human or simian diphtheria toxin receptor under Foxl1 promoter control.
Conclusions
Killing Foxl1+ cells by diphtheria toxin administration led to an abrupt cessation of proliferation of both epithelial stem- and transit-amplifying progenitor cell populations that was associated with a loss of active Wnt signaling to the intestinal epithelium. Therefore, Foxl1-expressing mesenchymal cells constitute the fundamental niche for intestinal stem cells.
Keywords: Intestinal Stem Cell Niche, Wnt, Mesenchyme
Abbreviations used in this paper: BAC, bacterial artificial chromosome; cDNA, complementary DNA; Foxl1, forkhead box l1; hDTR, human diphtheria toxin receptor; iDTR, inducible diphteria toxin receptor; mRNA, messenger RNA; Myh11, rabbit myosin heavy chain 11; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; YFP, yellow fluorescent protein
Summary.
Forkhead box l1+ cells are a small subset of mesenchymal subepithelial fibroblasts. These cells are a critical component of the intestinal stem cell niche as shown by using 2 separate models to ablate these cells.
Adult multipotent stem cells replenish the gut epithelium both during homeostasis and after injury throughout life. The mammalian intestinal epithelium undergoes complete regeneration every 3–5 days; this renewal is supported by multipotent intestinal stem cells.1, 2, 3, 4, 5, 6 Epithelial stem cell populations reside in 2 zones: at the crypt base and at the +4 position, 4–6 cell widths above the crypt base. Cells in both zones express markers associated with stem cell behavior, such as CD24 and Lrig1.4, 5 In addition, there are zone-specific markers. For the small intestine, the crypt base cells are marked by leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5)1 and Olfm4,7 and the +4 position cells express Bmi-1, HopX, and mTert.2, 3, 8 These epithelial stem cells divide to produce progenitor (or transit-amplifying) cells that proliferate rapidly and then differentiate into Paneth cells, which are located at the crypt base, or into goblet cells, enteroendocrine cells, M cells, and absorptive enterocytes that are located closer to the gut lumen.
Stem cell niches are defined as local microenvironments that provide physical support and/or molecular signals necessary for proper stem and progenitor cell replication and differentiation. Wnt signaling has been established as the major driver of intestinal stem and progenitor cell proliferation, as evidenced, for instance, by a rapid loss of proliferation when the secreted Wnt inhibitor Dickkopf-1 is overexpressed in the epithelium.9, 10 However, the cellular identity of the intestinal stem cell niche has remained controversial. Sato et al11 concluded that epithelial Paneth cells constitute the niche for Lgr5+ stem cells in intestinal crypts, based on the fact that Paneth cells elaborate important signaling molecules such as Wnt3 and EGF and on the observation that in vitro organoid formation by Lgr5+ stem cells was enhanced by co-culture with a Paneth cell–enriched population. However, complete and permanent absence of Paneth cells in mice deficient for the transcription factor Math1 (Atoh1) had no impact on intestinal stem cell maintenance and proliferation.12, 13 The latter studies support earlier work by Garabedian et al14 who used 2 independent methods to ablate mature Paneth cells and concluded that “stemness in the crypt is not defined by instructive interactions involving the Paneth cells.” Moreover, epithelial-specific deletion of Wnt3 had no effect on intestinal stem cells in mice, suggesting the presence of other Wnt source(s).15 These findings point to the existence of an extraepithelial source of Wnt and other signaling molecules necessary to maintain epithelial homeostasis.
Materials and Methods
Derivation of Forkhead Box l1–Human Diphtheria Toxin Receptor Mice
All protocols were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. The human diphtheria toxin receptor (hDTR) coding sequence (NM_001945.1) was introduced into the coding region of the mouse forkhead box l1 (Foxl1) gene in the bacterial artificial chromosome (BAC) RP23-446J14 by means of BAC recombineering as described previously.16 The targeting primers were as follows:
forward: GGGGCAAAGTCCTTAGGACTCCCCGGTGGAGCGGAGAGGCTGCTGTCGCCGAATTCGGCACGAGGGCTACGCGGG; reverse: AGGCCCCTCAGTGCACGACTTTGGCCGGCACGGGTACGCTGCTCCAAACC AGCTCCACCGCGGTGGCGGCCGCTC.
The resulting Foxl1–hDTR BAC was linearized and microinjected into the pronucleus of C57Bl/6 mice. The positive transgenic founders were identified by genomic polymerase chain reaction (PCR) and crossed to C57Bl/6 mice for at least 5 generations. Animals were euthanized at 2–6 months of age for subsequent experiments.
Generation of Foxl1–Cre;Rosa–iDTR/YFP, Foxl1–Cre;Rosa–YFP, and Foxl1–Cre;Rosa–mT/mG Mice
Foxl1–Cre mice were generated and characterized previously.16 Foxl1–Cre mice were crossed to Rosa–inducible diphteria toxin receptor (iDTR)/yellow fluorescent protein (YFP) mice (Jackson Laboratories, Bar Harbor, ME) to obtain Foxl1–Cre;Rosa–iDTR/YFP mice. Foxl1–Cre;Rosa–membrane-targeted dimer tomato protein (mT)/membrane-targeted green fluorescent protein (mG) mice, Foxl1–Cre mice resulted from crossing to Rosa–mT/mG mice (Jackson Laboratories). Animals were killed at 2–6 months of age for subsequent experiments.
Diphtheria Toxin Treatment
For Foxl1–hDTR mice, diphtheria toxin (Sigma-Aldrich, St. Louis, MO) dissolved in 0.9% sodium chloride was administered intraperitoneally at 20 ng/g body weight. Mice were injected on days 0 and 2 and killed on day 3. Foxl1–Cre;Rosa–iDTR/YFP mice and their control littermate (Rosa–iDTR/YFP) mice were administered diphtheria toxin at 22 ng/g body weight, twice daily from days 0–3, and killed on day 4.
Anti-Foxl1 Antibody Production
A Foxl1–pUC57 codon-optimized plasmid containing amino acids 150–255 of mouse Foxl1 (avoiding regions of similarity with other Fox proteins, especially focusing on non–winged-helix domains with good antigenicity prediction and likely subdomain folding) with flanking BamHI/HindIII sites was generated (GenScript USA, Inc, Santa Clara, CA). The BamHI/HindIII fragment was inserted into pGexKG,17 confirmed by sequencing, and used to produce soluble GST-Foxl1 fusion protein from bacteria. GST-Foxl1 was dialyzed against phosphate-buffered saline (PBS) and used as an antigen in goat and guinea pig (SDIX, LLC) or chicken (Aves Labs). Raw sera/antibodies were used to purify Foxl1-specific antibodies via GST/Escherichia coli–extract depletion and GST-Foxl1 matrix-based affinity purification. GST-Foxl1 protein production and antibody purification were performed as described previously.18
Fluorescent Activated Cell Sorting, RNA Isolation, and Sequencing Library Preparation
Isolation of Foxl1+ cells using fluorescent-activated cell sorting was performed using Foxl1–Cre;Rosa–YFP mice. Small intestines were dissected and washed thoroughly with PBS. Intestinal villi were scraped using a coverslip and the remaining tissue was incubated in 30 mmol/L EDTA plus 1.5 mmol/L dithiothreitol and Hank's balanced salt solution on ice for 20 minutes and subsequently incubated in 30 mmol/L EDTA at 37° for 8 minutes to completely remove the epithelium. After vigorous washes, the remaining mesenchymal fraction was collected and cut into small pieces. The mesenchymal tissue was collected by centrifugation and resuspended in 7 mg/mL Dispase II/0.05% trypsin solution (Sigma-Aldrich, St. Louis, MO) at 37° until the solution became cloudy and the mesenchyme was dissociated. A single-cell suspension was obtained by collecting the supernatant and washing with Hank's balanced salt solution before cell sorting using a BD influx instrument (BD Biosciences, San Jose, CA). For RNA isolation, YFP+ cells were lysed and total RNA was isolated by column purification (Agilent Technologies). Messenger RNA (mRNA) was isolated using Poly(A) mRNA isolation magnetic beads and an mRNA sequencing library prepared using the NEBNext RNA library prep kit (New England BioLabs, Inc, Ipswich, MA). RNA sequencing was performed on an Illumina HiSeq instrument.
RNA Isolation and Library Formation From Crypt Cells
To isolate RNA from intestinal crypts after diphtheria toxin injection, Foxl1–hDTR and control mice were injected with diphtheria toxin at 20 ng/g body weight on day 0 and euthanized on day 3. Dissected intestines were washed in PBS to remove the luminal content, incubated in 5 mmol/L EDTA in PBS for 10 minutes, and scraped with a coverslip gently to remove villus cells. The remaining tissue was cut into 5-mm pieces and incubated in 5 mmol/L EDTA for 20 minutes with vigorous pipetting every 2–3 minutes. The tissue was washed and pipetted vigorously, and floating crypts were collected. Crypt RNA was isolated using TRIzol reagent (Life Technologies, Carlsbad, CA) and subjected to Poly-A selection using magnetic isolation (New England BioLabs, Inc). Libraries were prepared with the RNA Library Prep Kit (New England BioLabs, Inc) and sequenced using an Illumina HiSeq instrument.
Histology
For H&E staining, paraffin tissue sections were rehydrated, incubated in hematoxylin for 2.5 minutes, rinsed in water, dipped quickly in 0.5% acid alcohol, and washed in water. Tissues then were immersed in 0.2% NaHCO3, rinsed in water, dipped in eosin for 15 seconds, and briefly rinsed in water before dehydration and mounting.
Immunofluorescence and Immunohistochemistry
At time of death, mouse intestines were rinsed in PBS and fixed with 4% paraformaldehyde overnight, rinsed in PBS, and either dehydrated for paraffin embedding or immersed in 30% sucrose for 4 hours at 4°C, embedded in optimum cutting temperature compound (OCT), and frozen for cryosectioning. Antigen retrieval was achieved using citrate buffer, pH 6.0, with a pressure cooker (PickCell Laboratories, Agoura Hills, CA). Foxl1 staining required the use of cryosections, antigen retrieval, and amplification of signal using tyramide (TSA systems; PerkinElmer, Waltham, MA). Antibodies used were as follows: Ki67 (1:500; BD Pharmingen, Franklin Lakes, NJ), lysozyme (1:1000; Dako, Carpinteria, CA), Sox9 (1:300; Millipore, Temecula, CA), mucin 2, HBEGF (1:50; Santa Cruz Biotechnology, Dallas, TX), guinea pig Foxl1 (1:1500), goat green fluorescence protein (1:200; Abcam, Cambridge, MA), mouse E-cadherin (1:250; BD Transduction, Franklin Lakes, NJ), mouse β-catenin (1:200; BD Transduction), rabbit EpCAM (1:100; Abcam), rabbit α-smooth muscle actin (1:100; Abcam), and rabbit myosin heavy chain 11 (Myh11) (1:100; Abcam). Cy2-, Cy3-, and Cy5-conjugated donkey secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc. For immunohistochemistry, horseradish-peroxidase–conjugated secondary antibodies were incubated at room temperature for 2 hours. The Vectastain ABC kit (Vector Laboratories, Burlingame, CA) then was used to detect the signal, and the slides were washed with PBS. For signal development, the 3,3′-diaminobenzidine tetra hydrochloride substrate was used.
In Situ Hybridization
Olfm4 RNA in situ hybridization was performed as previously described using ornithine carbamyl transferase–embedded frozen tissues.7, 19 For other in situ hybridization experiments, double digoxigenin-labeled locked nucleic acid probes were designed (Exiqon, Vedbaek, Denmark). Paraffin sections were rehydrated, fixed, and pretreated as described by Silahtaroglu20 with the following modifications: in addition to acetylation, slides were pretreated with 10 ng/mg proteinase K for 10 minutes at 37°, and 80 nmol/L of probe mixture was used for hybridization. 4′,6-Diamidino-2-phenylindole was used to mark the nuclei (Vector Laboratories).
Western Blot
Small and large intestines were dissected from C57Bl/6 control and Foxl1-null7 mice. Isolated tissues were washed with PBS and scraped with a coverslip to remove the villus epithelium. Intestinal epithelial cells were separated from mesenchymal cells as described in fluorescent-activated cell sorting and RNA isolation. Tissues then were subjected to multiple rounds of sonication to lyse the cells in buffer containing 50 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Triton X-100, 0.5% deoxycholic acid, and 1% sodium dodecyl sulfate, supplemented with protease inhibitor cocktail (Roche, Indianapolis, IN) and phosphatase inhibitor cocktails 1 and 2 (Sigma-Aldrich). Cell lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes with the iBlot system (Life Technologies). Membranes were blocked in blocking buffer consisting of 5% milk and 0.1% Tween-20 in PBS for 2 hours and incubated with antibodies at 4° overnight in blocking buffer. Antibodies used were as follows: chicken Foxl1 (1:500), goat Foxl1 (1:500), mouse α-smooth muscle actin (1:10,000; Sigma), mouse E-cadherin (1:5000; BD Transduction), rabbit β-actin (1:10,000; Cell Signaling, Danvers, MA). Membranes then were washed and incubated with horseradish-peroxidase–conjugated goat-anti-rabbit IgG.
Fractional Separation of Mesenchyme and Reverse-Transcription PCR
Intestines from adult Foxm1–hDTR mice were cut open and washed in PBS. Intestinal epithelial cells were separated from mesenchymal cells as described in fluorescent-activated cell sorting and RNA isolation. The crypt epithelium was scraped off and collected. To ensure isolation of pure epithelium, epithelial cells were incubated in 0.05% trypsin to obtain single cells, followed by incubation with an Alexa-488 mouse EpCAM (1:250; Biolegend, San Diego, CA) antibody, and sorted in a BD Influx instrument (BD Biosciences). The remaining mesenchymal tissue was washed to remove any epithelial contamination. From the epithelial and mesenchymal fractions, RNA was isolated and complementary DNA (cDNA) was synthesized. For mesenchymal and epithelial-specific markers as well as hDTR, reverse-transcription PCR was performed using GoTaq (Promega, Madison, WI). The primers used in this experiment are listed in Table 1.
Table 1.
Primer Sequences
| Cdh1 Fw | GGCTTCAGTTCCGAGGTCTA |
| Chd2 Rv | CTCTGGGTTGGATTCAGAGG |
| Vim Fw | GATCAGCTCACCAACGACAA |
| Vim Rv | TCCGGTACTCGTTTGACTCC |
| HBEGF Fw | ACTTTATCCTCCAAGCCACAAG |
| HBEGF Rv | CCCAGATACCATCGGACATACT |
Fw, forward; Rv, reverse.
Results
Foxl1 Is Expressed in a Subset of Mesenchymal Cells
We hypothesized that mesenchymal cells in close apposition to the crypt epithelium constitute the intestinal stem cell niche in vivo. We and others21, 22, 23 have shown that the winged-helix transcription factor Foxl1 (previously known as Fkh6) is expressed specifically in the gastrointestinal mesenchyme during fetal development and in adulthood. We developed new Foxl1-specific antibodies to map the location of this cell type in the mammalian gut more precisely. We established the specificity of these affinity-purified antibodies through immunoblot analysis of Foxl1-null gut tissue (Figure 1A). Foxl1 protein was confined to the nuclei of a subset of subepithelial cells that are tightly apposed to the epithelium of small intestinal crypts, with some cells extending into the villus tip (Figure 1B and C).
Figure 1.
Antibodies to the mouse Foxl1 protein are specific, and Foxl1 is expressed in a small subset of subepithelial fibroblasts in the intestine. (A) Immunoblot analysis for the Foxl1 protein in epithelial and mesenchymal fractions of mouse small intestine (jejunum) and colon, as indicated. Two different affinity-purified anti-Foxl1 antibodies were used. Note the absence of the Foxl1 protein band in mesenchymal fractions from Foxl1 null mice. Immunoblot analysis of α-smooth muscle actin (αSMA) confirms separation of epithelium and mesenchyme. mes, mesenchyme. (B) Immunostaining showing expression of Foxl1 (red) in the adult small intestine in a subset of mesenchymal cells located in close apposition to the epithelium (outlined with immunostaining for EpCAM, green). Nuclei are labeled with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Scale bar: 50 μm. (C) Cross-section of crypt region showing the localization of Foxl1+ cells (red) in pericryptal mesenchymal cells. Nuclei are labeled with DAPI (blue). Scale bar: 25 μm.
Foxl1+ Cells Are Distinct From Myofibroblasts and Express Potential Niche-Supporting Factors
To better define the relationship of Foxl1-expressing cells to established mesenchymal cell types, we used Foxl1–Cre;Rosa–mT/mG mice, in which Foxl1 promoter-dependent Cre activity results in the expression of a plasma membrane-bound green fluorescence protein (Figure 2A, scheme) (see Materials and Methods section for details), which allowed us to visualize the Foxl1-expressing cells in their full extension. As shown in Figure 2B, Foxl1+ cells elaborate long processes that span the width of multiple epithelial cells, suggesting that Foxl1+ cells are placed in an optimal position for short-range signaling to the epithelium. Dual-label immunofluorescence staining clearly showed that the Foxl1+ cell is distinct from mesenchymal cells expressing the smooth muscle markers α-smooth muscle actin (Figure 2B) and Myh11 (Figure 2C).
Figure 2.
Foxl1+cells have long processes and express potential niche-supporting factors. (A) Schema of Foxl1–Cre;Rosa–membrane-targeted dimer tomato protein membrane-targeted green fluorescent protein mouse used to drive expression of a membrane-bound green fluorescence protein (GFP) in Foxl1-Cre–positive cells. (B) Foxl1–Cre–driven expression of membrane-bound GFP is restricted to pericryptal mesenchymal cells, which elaborate long extensions into the villus tips and surrounding intestinal crypts. Immunofluorescence staining for GFP (green) and α-smooth muscle actin (αSMA) (red) shows very limited overlap between the 2 markers, indicating that Foxl1+ cells represent a unique cell population. αSMA expression is present in the core of the lamina propria, whereas GFP expression is located at the pericryptal sheath. (C) Immunofluorescence staining for Foxl1–Cre–driven GFP (green) and Myh11 (red) indicates very limited overlap between the 2 markers. (D) A histogram of fluorescence-activated cell sorting of intestinal mesenchymal cells isolated from C57BLl6 control and Foxl1–Cre;Rosa–YFP mice using YFP fluorescence. The x-axis shows YFP fluorescence intensity, and the gate indicated in the Foxl1–Cre;Rosa–YFP plot shows the sorted cells from which RNA was isolated for RNA sequence analysis. Note the absence of high-intensity YFP+ cells in the control histogram. (E) Relative mRNA levels of various markers and key signaling molecules in sorted Lgr5+-epithelial stem cells, differentiated villus epithelial cells, and sorted Foxl1+ cells.
Next, we used fluorescence-activated cell sorting to enrich for mesenchymal cells expressing Foxl1–Cre (Figure 2D). When we compared the mRNA levels of key genes between Foxl1+ mesenchymal cells and either Lgr5+ epithelial stem cells or differentiated intestinal villus cells (data from24), we found, as expected, that the Foxl1+ fraction did not express Lgr5 or the sodium/glucose cotransporter Sglt1, markers of epithelial stem and differentiated villus cells, respectively (Figure 2E). However, the mesenchymally expressed Wnt genes Wnt2b and Wnt5a were highly enriched in Foxl1+ pericryptal fibroblasts, suggesting that it is these cells that provide critical Wnt signals to the intestinal stem cell compartment. In addition, Foxl1+ cells express high levels of R-spondin 3, an activator of Went signaling,25 fibroblast growth factor 2, and gremlin 1 and 2, antagonists of the bone morphogenetic protein pathway that are expressed in intestinal reticular stem cells.26
Ablating Foxl1+ Cells Leads to Disrupted Epithelium
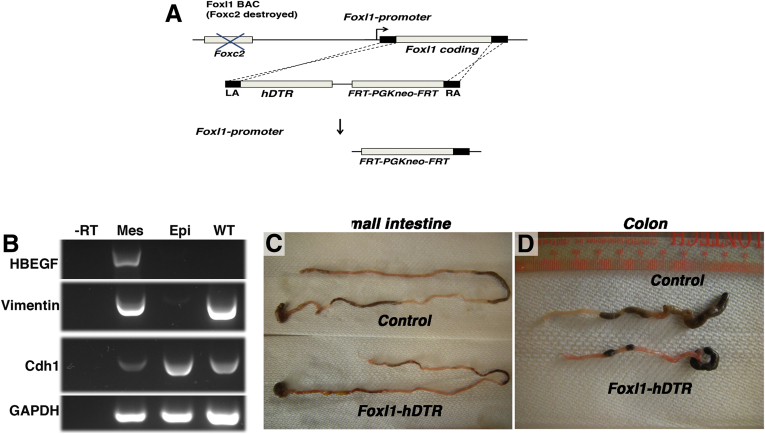
Because of this unique expression pattern, the elaboration of key signaling molecules, and because ablation of the Foxl1 gene itself causes altered Wnt signaling in stomach and intestine,27 we surmised that Foxl1-expressing mesenchymal cells might constitute the intestinal stem cell niche. To test this hypothesis, we derived mice transgenic for a bacterial artificial chromosome comprising 170 kb of the Foxl1 locus in which the Foxl1 coding region was replaced by the hDTR cDNA (Foxl1–hDTR mice) (Figure 3A). Although mice are normally resistant to diphtheria toxin, expression of the hDTR in Foxl1-expressing (Foxl1+) mesenchymal cells renders these cells diphtheria toxin–sensitive, leading to cell-specific apoptosis after toxin administration. As expected, expression of the human diphtheria toxin receptor was confined to the subepithelial mesenchyme (Figures 3B and 4A).
Figure 3.
Ablation of Foxl1+mesenchymal cells in diphtheria toxin–treated Foxl1–hDTR mice results in shortening of the gut tube. (A) Schema for the generation of Foxl1–hDTR mice using BAC recombineering. The coding sequence of exon 1 of Foxl1 was targeted by the sequence of the hDTR after destruction of the upstream FoxC2 coding sequence. FRT, flippase recognition target; LA, left homology arm; RA, right homology arm. (B) Expression of the human diphtheria toxin receptor (the HBEGF gene) is confined to the mesenchyme in Foxl1–hDTR mice. Mesenchyme and epithelium of the colon from Foxl1–hDTR mice were separated and cDNA was prepared. Reverse-transcription PCR analysis was performed to detect mesenchymal vimentin, epithelial E-cadherin, and ubiquitous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts. Note that the HBEGF/hDTR mRNA is present exclusively in the mesenchyme. -RT, without reverse transcriptase; Mes, mesenchyme; Epi, epithelium; WT, whole colon from control nontransgenic mice. (C and D) Representative images of small intestine and colon from DT-treated Foxl1–hDTR and control mice. Note the shortening of the gut tube in the DT-treated Foxl1–hDTR mice.
Figure 4.
Intestinal epithelial architecture and proliferation are dependent on Foxl1+mesenchymal cells. (A) Representative immunofluorescence images of human diphtheria toxin receptor (green) staining in the jejunum of control and Foxl1–hDTR mice, counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei (blue). (B and C) Intestinal morphology of the jejunum of (B) DT-treated control mice or (C) Foxl1–hDTR mice as assessed by H&E staining. Note the reduced number and length of intestinal villi in Foxl1–hDTR mice. (D) Quantification of small intestinal villus length (n = 4; in each animal > 20 villi were measured; ***P < .001). (E and F) Intestinal morphology of the proximal colon from (E) DT-treated control and (F) Foxl1–hDTR mice. Note the decreased depths of the colonic crypts and the irregular localization of epithelial nuclei in Foxl1–hDTR mice. (G) Quantification of colonic crypt depth (n = 4; in each animal > 20 crypts were measured; **P < .005). (H and I) Representative immunofluorescence images of the proliferation marker Ki67 (red) counterstained with DAPI to label nuclei (blue) in the jejunum of (H) control and (I) Foxl1–hDTR mice after toxin injection. (J) Quantification of the number of proliferating cells per crypt unit in jejunum of DT-treated control and Foxl1–hDTR mice (n = 20 crypts each). ***P < .001. (K and L) Ki67 labeling (red) overlaid with DAPI for nuclei (blue) in the colon of (K) DT-treated control and (L) Foxl1–hDTR mice. The number of Ki67-positive cells is reduced dramatically in Foxl1–hDTR mice. Scale bars: 50 μm. (M) Quantification of the number of proliferating cells per crypt unit in the proximal colon of DT-treated control and Foxl1–hDTR mice (n = 20 crypts each). ***P < .001.
Strikingly, after 3 days of exposure to diphtheria toxin, the length of the small and large intestines was reduced in Foxl1–hDTR mice (Figure 3C and D).
Foxl1–hDTR mice showed dramatic changes to the architecture of the small and large intestine (Figure 4B, C, E, and F). The length of the villi was reduced dramatically in the jejunum (Figure 4D), and epithelial architecture was abnormal in colonic crypts, which showed reduced depth (Figure 4G). To test our hypothesis that Foxl1+ mesenchymal cells constitute an essential part of the intestinal stem and progenitor cell niche, we next analyzed gut tissue for epithelial proliferation. Immunofluorescence labeling for the proliferation marker Ki67 showed that the deletion of Foxl1+ mesenchymal cells resulted in a dramatic and highly significant reduction of epithelial cell proliferation in the jejunum (Figure 4H–J) and colon (Figure 4K–M).
Ablating Foxl1+ Cells by a Separate Model Leads to Similar Epithelial Disruption
Next, we used an independent paradigm (Figure 5A) (Foxl1–Cre;Rosa–iDTR mice; see the Materials and Methods section for experimental details) in which expression of the simian DTR is activated in Foxl1-positive cells through Cre-mediated deletion of a stop cassette, again making cells susceptible to diphtheria toxin.28, 29 Remarkably similar results were obtained with this second cell-ablation model, in which again villus length and crypt depth were reduced significantly in the jejunum and colon, respectively (Figure 5B–G). Likewise, epithelial proliferation was reduced dramatically in diphtheria toxin–treated Foxl1–Cre, Rosa–iDTR mice (Figure 5H–M). Taken together, these results show that Foxl1-expressing mesenchymal cells are required for maintaining proliferation of the intestinal epithelium, both in stem cells and in the derived transit-amplifying cell population.
Figure 5.
Targeted ablation of Foxl1+mesenchymal cells using the iDTR model causes loss of epithelial proliferation. (A) Schema for the generation of Foxl1–iDTR mice. Foxl1–Cre mice were crossed to mice carrying a loxP-stop-loxP simian diphtheria toxin–receptor construct in the Rosa26 locus (RosaiDTR mice). (B and C) Small intestinal (jejunum) morphology was assessed by H&E staining of (B) DT-treated control and (C) Foxl1–iDTR mice. Note the reduced number and length of intestinal villi. (D) Quantification of small intestinal villus length (n = 4; in each animal > 20 villi were evaluated per mouse; ***P < .001). (E and F) Colonic morphology of (E) DT-treated control and (F) Foxl1–iDTR mice. Decreased depths of the colonic crypts is apparent in Foxl1–iDTR mice. (G) Quantification of colonic crypt depth (n = 4; in each animal >20 crypts were evaluated per mouse; *P < .05). (H and I) Representative immunofluorescence images of small intestinal (jejunum) sections stained for the proliferation marker Ki67 (red) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to label nuclei (blue) of (H) control and (I) Foxl1–iDTR mice. The number of Ki67-positive epithelial cells is reduced dramatically in Foxl1–iDTR mice. (J) Quantification of the number of proliferating cells per crypt unit in the jejunum of DT-treated control and Foxl1–iDTR mice (n = 20 crypts each). ***P < .001. (K and L) Ki67 labeling (red) overlaid with DAPI to mark nuclei (blue) in the colon of (K) DT-treated control and (L) Foxl1–iDTR mice. The number of Ki67-positive cells is reduced dramatically in Foxl1–iDTR mice. (M) Quantification of the number of proliferating cells per crypt unit in the colon of DT-treated control and Foxl1–iDTR mice (n = 20 crypts each). ***P < .001. Scale bars: 100 μm in the small intestine, 50 μm in the colon.
Stem Cells but Not Paneth Cells Are Affected by Ablating Foxl1+ Cells
In addition to loss of proliferation, expression of the stem cell marker Olfm4 was reduced dramatically in mice with ablation of Foxl1+ subepithelial mesenchymal cells (Figure 6A–C). However, the long-lived lysozyme-expressing Paneth cells were retained in the small intestinal epithelium of diphtheria toxin–treated Foxl1–hDTR mice, indicating that loss of epithelial proliferation was not secondary to loss of Paneth cells (Figure 6D–F).
Figure 6.
Foxl1+mesenchymal cells are required for intestinal stem cell marker expression independent of Paneth cells. (A and B) Expression of the intestinal stem cell marker Olfm4 is reduced in (B) DT-treated Foxl1–hDTR mice compared with (A) nontransgenic controls as determined by in situ hybridization. Insets show crypts at high magnification. (C) Quantification of the percentage of small intestinal crypts that are positive for Olfm4 mRNA (>200 crypts were counted for both DT-treated control and Foxl1–hDTR mice). *P < .01. (D and E) Expression of lysozyme (red), a Paneth cell marker, is not affected by ablating Foxl1+ cells. Scale bar: 50 μm. (F) Quantification of lysozyme-positive cells per crypt unit (n = 3 per genotype; >20 crypts were evaluated per animal). DAPI, 4′,6-diamidino-2-phenylindole.
Moreover, cell-lineage allocation among the differentiated lineages was not affected globally in the 3 days from toxin administration to analysis, as evidenced by the presence of mucin 2 and Alcian Blue–positive goblet cells (Figure 7A–D).
Figure 7.
The frequency of mucin-secreting goblet cells is unaffected by ablation of Foxl1+cells. (A and B) Detection of goblet cells by immunostaining for Muc2 (mucin 2; green) and (C and D) Alcian Blue histochemistry (blue) indicates that the frequency of goblet cells in (B and D) DT-treated Foxl1–hDTR mice is not different from that present in (A and C) control mice. DAPI, 4′,6-diamidino-2-phenylindole.
Ablating Foxl1+ Cells Leads to Reduced Wnt Signaling
Proliferation of the intestinal epithelium is driven by Wnt signaling,9, 10, 30 and we hypothesized that Foxl1-expressing subepithelial cells serve as an essential source of these signals. To determine if Wnt signaling is affected by loss of Foxl1+ cells, we evaluated nuclear β-catenin staining. As shown in Figure 8A and B, nuclear β-catenin expression was greatly reduced in the crypts of Foxl1–hDTR mice after diphtheria-toxin administration, which was indicative of loss of canonical Wnt signaling activity. Next, we investigated the expression of the Wnt target gene Sox9 in Foxl1–hDTR mice after diphtheria toxin administration. Sox9 protein normally is present in the entire crypt, including intestinal stem cells and Paneth cells.31 After diphtheria toxin treatment, we noted a striking decrease in the number of Sox9-expressing cells in Foxl1–hDTR crypts as compared with controls (Figure 8C and D), again supporting reduced Wnt signaling activity.
Figure 8.
Foxl1+mesenchymal cells provide essential Wnt ligands to the intestinal stem cell compartment. (A and B) Activation of the Wnt pathway in epithelial cells of the jejunum was analyzed by immunohistochemistry for ß-catenin. Reduced nuclear ß-catenin localization is apparent in (B) toxin-treated Foxl–hDTR mice compared with (A) toxin-treated controls. Yellow arrows point to nuclei positive for ß-catenin. Scale bars: 25 μm. (C and D) Immunofluorescence staining of the Wnt target Sox9 (red) shows decreased expression in the jejunum of (D) DT-treated Foxl1–hDTR mice compared with (C) nontransgenic controls. The epithelium is delineated with E-cadherin immunostaining (green). Scale bars: 50 μm. (E and F) Mesenchymal Wnt2b mRNA was detected by in situ hybridization (red) and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue) Wnt2b mRNA is reduced dramatically in the mesenchyme of (F) DT-treated Foxl1–hDTR mice compared with (E) controls. Scale bars: 50 μm. (G and H) Mesenchymal Wnt4 and (I and J) Wnt5a mRNAs were detected by in situ hybridization (red) and nuclei were counterstained with DAPI (blue). Wnt4 and Wnt5a mRNA levels were reduced dramatically in the mesenchyme of (H and J) DT-treated Foxl1–hDTR mice compared with (G and I) controls.
Both canonical and noncanonical Wnt factors are produced by the intestinal mesenchyme.15, 19 Farin et al15 established Wnt2b as a mesenchymal Wnt protein that is sufficient to support intestinal organoid growth in culture. Strikingly, Wnt2b, Wnt4, and Wnt5a signals that are normally detected in the subepithelial mesenchyme were decreased dramatically in Foxl1–hDTR mice after diphtheria toxin administration (Figure 8E–J). These data suggest that production of Wnt2b, Wnt4, and Wnt5a by Foxl1-expressing mesenchymal cells, which are located in close proximity to the basement membrane of the epithelial crypt, is part of the signal required to maintain the intestinal stem cell niche.
Discussion
Wnt signaling is required for the maintenance of the adult intestine; however, the cellular source of this ligand has not been defined satisfactorily. Although Sato et al11 deduced that Paneth cells are this source because they express and secrete Wnt ligands, and because co-culture of Paneth cells and Lgr5+ cells allows the formation of intestinal organoids, several lines of evidence challenged the view of the Paneth cell as an absolutely required in vivo source of signals required to maintain the intestinal stem cell. For example, loss of Paneth cells12, 13 or loss of the essential Wnt-processing protein Porcupine (Porcn) in Paneth cells32, 33 has no effect on intestinal stem cell maintenance.
An alternative source for the signals required for stem cell maintenance is the intestinal mesenchyme. Recently, however, San Roman et al33 showed that Porcn, and therefore the Wnt signal, do not originate solely from Myh11-expressing intestinal smooth muscle cells and myofibroblasts or, independently, from villin-expressing epithelial cells. Kabiri et al33 ablated Porcn expression from the entire intestinal epithelium using villin-Cre and showed normal intestinal morphogenesis. Together, these studies suggested that either redundant Wnt signaling from both epithelial and mesenchymal compartments or a rare Wnt-producing cell type in one of these compartments that had not been targeted with the existing Cre lines is responsible for crypt maintenance.
We show here that Foxl1+ mesenchymal cells exist independently of the Myh11-expressing subepithelial myofibroblasts targeted by San Roman et al33 (Figure 1). Thus, the rare Foxl1+ cells that reside in the intestinal mesenchyme, in close apposition to the intestinal crypt, are outstanding candidates to be the minor Wnt-producing cell type in the intestine postulated by San Roman et al.33
These Foxl1+ cells are located directly adjacent to the epithelium and extend long processes that span the length of multiple epithelial cells. They express Wnt2b, Wnt4, and Wnt5a, and the loss of these Foxl1+ cells results in decreased β-catenin nuclear localization and proliferation within the intestinal epithelium, reduced villi length, diminished crypts depths, and an overall shortening of the intestine. Therefore, Foxl1+ cells are required for intestinal stem cell maintenance and are not functionally redundant with Wnt-expressing Paneth cells.
We considered whether the diphtheria-toxin–mediated killing of Foxl1+ cells might have indirect effects on the intestinal stem cell niche not mediated by controlling physiological signaling components such as the Wnt proteins described earlier. For instance, one might envisage a bystander effect in which debris from dying Foxl1+ cells is taken up by intestinal stem cells, leading to their death. We consider this scenario exceedingly unlikely, considering that diphtheria toxin–mediated ablation of Paneth cells, which after all are abutting Lgr5+ stem cells directly, is without an effect on crypt health.14
Of note, mice deficient for the winged helix transcription factor Foxl1, although showing abnormal gastrointestinal development,23 are viable and maintain functioning crypts. In fact, the role of Foxl1 and its direct targets is in limiting Wnt signaling to the epithelial compartment.23 Of course, it is not surprising that the genetic ablation of one gene within a cell does not have the same consequences as ablating that cell type entirely because each transcription factor controls only part of the transcriptional program within the cell.
Although the cellular identity of the intestinal stem cell niche has been controversial until this point, by using 2 novel, independent, cell-ablation models, we show that Foxl1+ subepithelial mesenchymal cells are the source of essential Wnt signals and thus constitute the intestinal stem and progenitor cell niche (model in Figure 9). Positive identification of the cells that provide the intestinal stem cell niche might aid in the future development of regenerative strategies for intestinal disorders.
Figure 9.
Proposed model of Foxl1-positive subepithelial mesenchymal cells as the intestinal stem cellniche, providing essential Wnt signals, and possibly other signaling molecules, to the epithelium.
Acknowledgments
The authors thank Tia Bernard-Banks for technical assistance with the mouse colony.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by grants from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases R37-DK053839 (K.H.K.), and 5U01-DK089570 and 5U01-DK072473 (C.V.E.W.). Also supported by the Transgenic Mouse and Molecular Pathology and Imaging Cores of the Center for Molecular Studies in Digestive and Liver Diseases (National Institutes of Health P30 DK050306).
References
- 1.Barker N., van Es J.H., Kuipers J. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Sangiorgi E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeda N., Jain R., LeBoeuf M.R. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Furstenberg R.J., Gulati A.S., Baxi A. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol. 2011;300:G409–G417. doi: 10.1152/ajpgi.00453.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell A.E., Wang Y., Li Y. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright N.A., Irwin M. The kinetics of villus cell populations in the mouse small intestine. I. Normal villi: the steady state requirement. Cell Tissue Kinet. 1982;15:595–609. doi: 10.1111/j.1365-2184.1982.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 7.van der Flier L.G., Haegebarth A., Stange D.E. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 8.Montgomery R.K., Carlone D.L., Richmond C.A. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto D., Gregorieff A., Begthel H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhnert F., Davis C.R., Wang H.T. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T., van Es J.H., Snippert H.J. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durand A., Donahue B., Peignon G. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc Natl Acad Sci U S A. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim T.H., Escudero S., Shivdasani R.A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc Natl Acad Sci U S A. 2012;109:3932–3937. doi: 10.1073/pnas.1113890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garabedian E.M., Roberts L.J., McNevin M.S. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J Biol Chem. 1997;272:23729–23740. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- 15.Farin H.F., Van Es J.H., Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143:1518–1529 e1517. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 16.Sackett S.D., Fulmer J.T., Friedman J.R. Foxl1-Cre BAC transgenic mice: a new tool for gene ablation in the gastrointestinal mesenchyme. Genesis. 2007;45:518–522. doi: 10.1002/dvg.20315. [DOI] [PubMed] [Google Scholar]
- 17.Guan K.L., Dixon J.E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 18.Candia A.F., Wright C.V. Differential localization of Mox-1 and Mox-2 proteins indicates distinct roles during development. Int J Dev Biol. 1996;40:1179–1184. [PubMed] [Google Scholar]
- 19.Gregorieff A., Pinto D., Begthel H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Silahtaroglu A.N. LNA-FISH for detection of microRNAs in frozen sections. Methods Mol Biol. 2010;659:165–171. doi: 10.1007/978-1-60761-789-1_11. [DOI] [PubMed] [Google Scholar]
- 21.Fukamachi H., Fukuda K., Suzuki M. Mesenchymal transcription factor Fkh6 is essential for the development and differentiation of parietal cells. Biochem Biophys Res Commun. 2001;280:1069–1076. doi: 10.1006/bbrc.2001.4247. [DOI] [PubMed] [Google Scholar]
- 22.Kaestner K.H., Bleckmann S.C., Monaghan A.P. Clustered arrangement of winged helix genes fkh-6 and MFH-1: possible implications for mesoderm development. Development. 1996;122:1751–1758. doi: 10.1242/dev.122.6.1751. [DOI] [PubMed] [Google Scholar]
- 23.Kaestner K.H., Silberg D.G., Traber P.G. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 24.Sheaffer K.L., Kim R., Aoki R. DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014;28:652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Lau W., Peng W.C., Gros P. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28:305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worthley D.L., Churchill M., Compton J.T. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perreault N., Sackett S.D., Katz J.P. Foxl1 is a mesenchymal Modifier of Min in carcinogenesis of stomach and colon. Genes Dev. 2005;19:311–315. doi: 10.1101/gad.1260605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buch T., Heppner F.L., Tertilt C. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 29.Saito M., Iwawaki T., Taya C. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol. 2001;19:746–750. doi: 10.1038/90795. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman J., Kuhnert F., Davis C.R. Wnts as essential growth factors for the adult small intestine and colon. Cell Cycle. 2004;3:554–557. [PubMed] [Google Scholar]
- 31.Blache P., van de Wetering M., Duluc I. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabiri Z., Greicius G., Madan B. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206–2215. doi: 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- 33.San Roman A.K., Jayewickreme C.D., Murtaugh L.C. Wnt secretion from epithelial cells and subepithelial myofibroblasts is not required in the mouse intestinal stem cell niche in vivo. Stem Cell Reports. 2014;2:127–134. doi: 10.1016/j.stemcr.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]