Abstract
Pyrosequencing is a technique that uses a sequencing-by-synthesis system which is designed to quantify single-nucleotide polymorphisms (SNPs). Artificial C/T SNP creation via bisulfite modification permits measurement of DNA methylation locally and globally in real time. Alteration in DNA methylation has been implicated in aging, as well as aging-related conditions such as cancer, as well as cardiovascular, neurodegenerative, and autoimmune diseases. Considering its ubiquitous presence in divergent clinical pathologies, quantitative analysis of DNA CpG methylation both globally and at individual genes helps to elucidate the regulation of genes involved in pathophysiological conditions. The ability to detect and quantify the methylation pattern of DNA has the potential to serve as an early detection marker and potential drug target for several diseases. Here, we provide a detailed technical protocol for pyrosequencing supplemented by critical information about assay design and nuances of the system that provides a strong foundation for beginners in the field.
Keywords: Pyrosequencing technique, DNA CpG methylation, Global methylation, Biomarker detection, SNPs, Bisulfide conversation, Assay design
1 Introduction
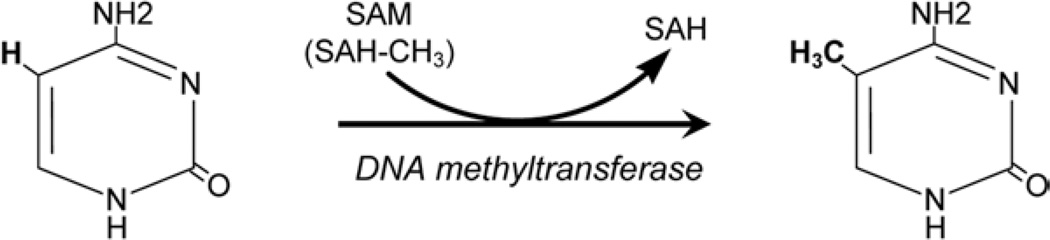
Epigenetic changes are heritable alterations in DNA that affect gene expression and function by mechanisms other than those from changes in DNA sequence. While each cell in an organism shares the same genetic material, epigenetic instructions define the expression of a gene that is conserved in mitosis. Epigenetic mechanisms regulate many cellular processes including development, differentiation, embryogenesis, X-chromosome inactivation, chromosomal stability, and genomic imprinting [1–4]. A number of epigenetic processes have been described including histone modification, chromatin remodeling, micro RNAs, and DNA methylation. Epigenetic “drift,” particularly T cell DNA demethylation, has been shown to contribute to immune dysfunction in aging. DNA methylation involves addition of a methyl group at the 5th carbon of cytosines preceding guanines (CpG dinucleotides), a modification catalyzed by DNA methyltransferases (DNMTs). S-adenosylmethionine (SAM), an intermediate product of methionine metabolism, acts as a methyl donor in the process (Fig. 1). DNA methylation has also been implicated in aging-associated diseases including cancer as well as neurodegenerative and cardiovascular disease and autoimmune syndromes such as lupus and rheumatoid arthritis (RA) [1, 5–7]. Therefore, it may be possible to use DNA methylation as a biomarker for disease risk [1, 8–12]. Among several established methods for measuring DNA methylation including HPLC, methylation-sensitive PCR, bisulfite sequencing, and next-generation sequencing, pyrosequencing offers a robust, versatile platform yielding rapid quantitative results without the onerous time commitment, high costs, and technical difficulty of alternative methods.
Fig. 1.
Methylation of cytosine to 5-methylcytosine. Cytosine preceding guanine (CpG sites) is methylated on carbon 5 (shown in bold) in the presence of DNA methyltransferase and SAM. SAM S-adenosyl methionine, SAH S-adenosyl homocysteine
1.1 Principle of Pyrosequencing
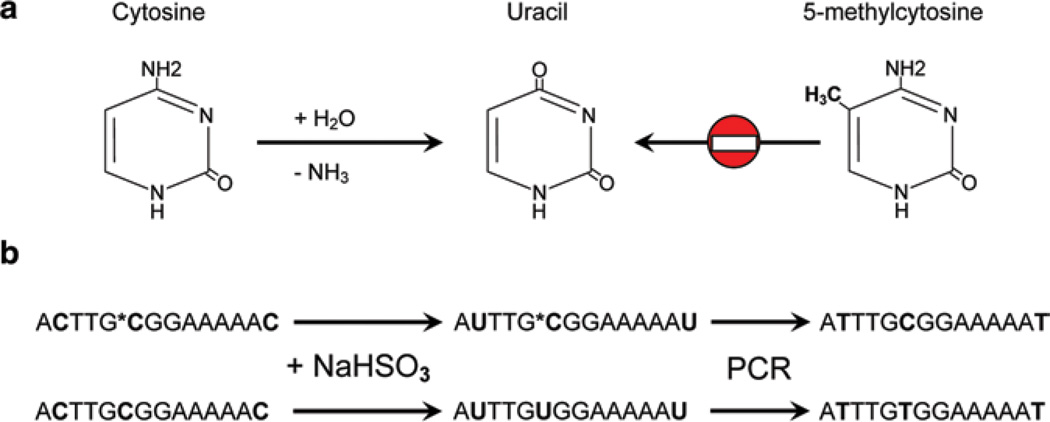
Pyrosequencing uses a high-throughput platform that can interrogate many CpG sites within an amplicon in real time. The pyrosequencing platform is designed to detect single-nucleotide polymorphisms, or SNPs, which can be artificially created at CpG sites through bisulfite modification. Treating genomic DNA with sodium bisulfite selectively converts cytosine to uracil; however, 5-methylcytosine is protected from deamination and the CG sequence is preserved in downstream reactions (Fig. 2). The technology is distinct from Sanger sequencing, in which labeled dideoxynucleotides are incorporated randomly in the reaction terminating extension of strands representative of each nucleotide position; rather, pyrosequencing uses a sequencing-by-synthesis system in which nucleotides are dispensed one at a time, incorporated into the extending strand and degraded prior to the next nucleotide dispensation (Fig. 3).
Fig. 2.
Deamination of cytosine via sodium bisulfide conversion. (a) Deamination of cytosine to uracil is prevented by methylation of the 5-carbon position of cytosine. (b) Methylated (above) and unmethylated (below) CpG-containing DNA undergoes bisulfite conversion. Methylated cytosines are unchanged while unmethylated cytosines are converted to uracil. Following PCR the cytosine is retained while uracil is converted to thymine. *C denotes methylated cytosine. Pyrimidines involved in bisulfite conversion are bolded. NaHSO3 sodium bisulfite, PCR polymerase chain reaction
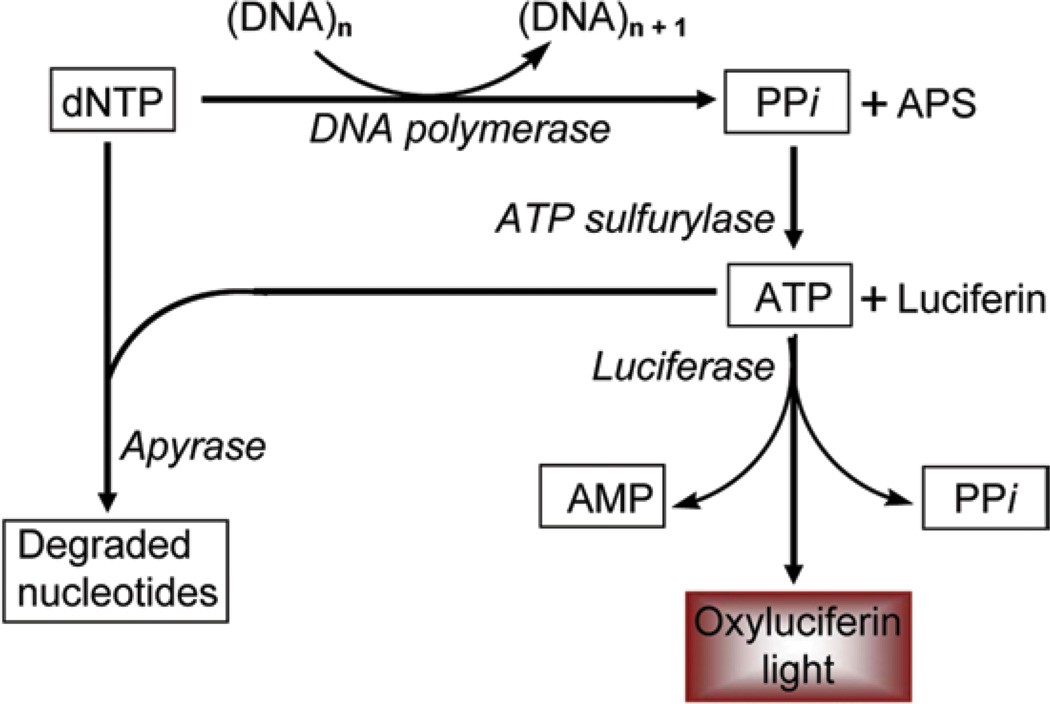
Fig. 3.
Enzyme cascade system in pyrosequencing. An ssDNA template is first hybridized with the sequencing primer and mixed with enzymes (written in italics) and two substrates (APS and luciferin). After successful incorporation of a nucleotide by DNA polymerase into a growing DNA strands, the released PPi reacts with APS in the presence of ATP sulfurylase giving rise to ATP. ATP in the presence of substrate luciferin and enzyme luciferase produces oxyluciferin that generates visible light, which can be detected by inbuilt CCD camera. Any unincorporated nucleotides and ATP are degraded into its building blocks by enzyme apyrase prior to the next nucleotide dispensation. Cascade reactions repeat for every dispensation. ATP adenosine triphosphate, APS Adenosine 5′ phosphosulfate, PPi pyrophosphate
1.2 The Pyrosequencing Enzyme Cascade
Pyrosequencing requires a single-stranded PCR amplicon that serves as DNA template, four different enzymes including DNA polymerase, ATP sulfurylase, luciferase, and apyrase, and two different substrates including adenosine 5′ phosphosulfate (APS) and luciferin [13]. First, a sequencing primer is annealed to a single-stranded DNA (ssDNA) template. Upon addition of a single nucleotide, the DNA polymerase incorporates the dNTP into the growing strand, releasing pyrophosphate (PPi). ATP sulfurylase then generates ATP from the PPi and substrate APS, which activates luciferase-mediated conversion of luciferin to the light-emitting oxyluciferin. Light is given off proportionate to the amount of nucleotide added to the elongating strand and recorded by an inbuilt CCD camera. Excess nucleotide is degraded by apyrase, after which the next nucleotide is dispensed. Comparing the peak light emission of incorporation of C or T at a CpG site within the amplicon gives a precise measure of the amount of methylation at that position within the sample.
1.3 Technical Overview of Pyrosequencing
Genomic DNA is bisulfite converted, and then the region of interest is amplified via PCR. Incorporation of a single biotinylated PCR primer allows separation of the two strands of the amplicon to create a ssDNA template for annealing of a pyrosequencing primer and extension of the complementary strand by discrete dispensation of nucleotides. This platform is PCR-based, yields rapid results (i.e., within a single day if starting with PCR), and is highly quantitative. Many different assays can be performed simultaneously (i.e., in a 96-well format, 96 different assays could be performed on one sample, 96 samples could be analyzed with one assay, or anything in between) and the time required is entirely dependent on the number of dispensations needed to cover the region of interest (approximately 5 min + 1 min/dispensation). In addition, pyrosequencing technology can be used to interrogate regulatory elements of specific genes [12, 14–16] or as a means of estimating global methylation [17–19]. However, pyrosequencing assays can be more difficult to design and extensive optimization of these assays is required (see Subheading 4). Also, an emerging pitfall of the system is that Bisulfite modification cannot discriminate between 5-methylcytosine and the novel modification 5-hydroxymethylcytosine. Nevertheless, pyrosequencing is a validated means of estimating both global methylation and specific regulatory loci in mammalian samples.
2 Materials
2.1 Consumables
Bisulfite conversion kit (available from multiple suppliers).
PyroPCR kit (Qiagen) or any reliable PCR kit.
96-Well skirted PCR plate, PCR plate stickers.
Agarose.
Ethidium bromide.
Streptavidin Sepharose High Performance beads (GE Healthcare).
PyroMark Gold Q96 Reagent Kit (Qiagen) contains enzymes, substrates, and dNTPs for pyrosequencing reaction.
PyroMark Q96 HS Reagent Dispensing Tip (Qiagen).
PyroMark Q96 HS Nucleotide Tip (Qiagen)—for longer sequencing reads >50 dispensations.
PyroMark Q96 HS Capillary Tip (Qiagen)—for short reads <50 dispensations.
PyroMark Q96 HS Plate (Qiagen).
gDNA of interest.
Control low-methylated gDNA.
Control high-methylated gDNA.
Sss1 methylase (NEB).
5-Azacytidine (Sigma).
PCR primers, one biotinylated and HPLC purified: 100 µM stock in water or TE. Store at −20 °C.
Pyrosequencing primer(s): 0.5 µM in annealing buffer. Store at 4 °C.
2.2 Equipment
PCR machine.
Agarose gel electrophoresis cell, power supply, UV imaging system.
Vacuum Prep work station.
96-Well plate heating block (not a PCR machine).
PyroMark MD pyrosequencer or equivalent.
2.3 Buffers
1× TAE: 40 mM Tris–Acetate, 1 mM EDTA, pH 8.
70 % EtOH.
Binding buffer: 10 mM Tris–HCl, 2 M NaCl, 1 mM EDTA, 0.1 % Tween 20, pH 7.6. Store at 4 °C.
Annealing buffer: 20 mM Tris–Acetate, 2 mM MgAc2. Store at 4 °C.
Denaturation buffer: 0.2 N NaOH. Store at RT.
1× wash buffer: 10 mM Tris–acetate pH 7.6. Store at RT.
ddH2O.
3 Methods
3.1 Generating PCR Amplicon for Pyrosequencing
Isolate genomic DNA of interest.
Bisulfite modification of DNA of interest: Treating genomic DNA with sodium bisulfite selectively converts cytosine to uracil; however, 5-methylcytosine is protected from deamination and the CG sequence is preserved in downstream reactions (Fig. 2). Many commercial bisulfite modification kits are available for purchase. Follow the manufacturer’s instructions. Use 250–1000 ng per conversion reaction and elute with 10–40 µL as appropriate.
PCR region of interest (i.e., regulatory element/promoter/enhancer/etc.): Use primers designed specifically to bisulfite-modified DNA. Use 25–100 ng DNA per reaction, 0.1 µM biotinylated, and 0.2 µM non-biotinylated PCR primers. Designing primers for bisulfite-converted DNA may be more difficult than unmodified DNA because the loss of cytosine increases the degeneracy of the DNA and increases the likelihood of mispriming (see Note 1).
Agarose gel verification of amplicon. Verify that you have a single, strong band and no unincorporated primers. A robust amplicon with no primer dimer is critical for success (see Note 2).
3.2 Isolation of Biotinylated ssDNA from PCR Amplicon for Pyrosequencing Template
Add 14 µL of 0.5 µM pyrosequencing primer in annealing buffer into each appropriate well of a pyrosequencing plate (white, with clear bottoms).
- Isolate single-stranded pyrosequencing template.
- In a skirted 96-well PCR plate mix together DNA + H2O to a volume of 40 µL (amplicon DNA usually 5–10 µL per well).
- In a tube combine 40 µL binding buffer and 2 µL streptavidin beads per well (e.g., 10 wells = 400 µL binding buffer + 20 µL beads). For 96-well plate (4 mL + 200 µL beads). Vortex.
- Add 40 µL of the WELL-MIXED binding buffer/bead suspension to each well containing 40 µL of DNA/H2O in PCR plate.
- Cover plate with plastic PCR sticker to prevent spilling. Shake plate at RT at 1400 rpm for a minimum of 10 min to allow streptavidin beads to bind biotin-labeled strand.
- While plate is shaking, prepare vacuum prep station (Fig. 4).
- Add ~200 mL of water, 70 % ethanol, denaturation buffer, and 1× wash buffer to the appropriate plastic trays.
- To activate the vacuum, turn on the vacuum pump, and then turn on the switch on the vacuum station.
- Pass water through the filter probe vacuum system to prepare the probes for separation.
- In a 96-well plate, add water in each of the wells that correspond to the same position as your test samples to make sure that the appropriate probes have good suction. A well-functioning probe will clear a full well in about 10 s. If probe fails to suck out water, it is likely blocked by salt buildup and should be replaced. Note the color of the filter tip can be indicative of function. Brighter white tips are usually newer and functional, while duller tips tend to be older and less functional.
After vacuum prep station is ready and at least 10 min of shaking has elapsed, stop the shaker. Immediately upon removing the plate from the shaker, place the probes into the plate and watch to make sure all fluid is sucked out of the wells. Delay will give beads time to settle out of solution, which will lower your recovery of amplicon.
Transfer probes to the tray containing 70 % EtOH. Once fluid is observed passing through the vacuum hose, count to 10 s to wash away residual salts and unlabeled DNA.
Transfer probes to the tray containing denaturation buffer and count to 10 as above. Amplicons are denatured and the unlabeled DNA strands are removed from the sample, leaving a ssDNA pyrosequencing template strand.
Transfer probes to the tray containing 1× wash buffer and count to 10 to let the fluid drain through. The base in the previous step is neutralized, allowing the pyrosequencing reaction to proceed at proper conditions.
Position probes directly above the pyrosequencing plate that has been seated in the appropriate orientation on the vacuum prep station. Do not drop probes into the pyrosequencing plate until the vacuum is disengaged lest the annealing primer/buffer be suctioned out of the well. Turn off the vacuum and as soon as pressure gauge has dropped to zero, lower the probes into the pyrosequencing plate. Vigorously agitate the probes to facilitate beads dropping from the filters into the annealing buffer and pyrosequencing primer.
Remove the filter probes and place them in the waste water. Shake as above to remove any excess beads/salts, and then pass another 200 mL water through probes to rinse away salts. Store the filter probes dry in an empty 96 tip rack.
Fig. 4.
A representative picture of vacuum prep station showing four different trays, probe connected to vacuum system, and appropriate places for the 96-well plates
3.3 Denaturation and Annealing of Sequencing Primer
Place pyrosequencing plate on a heating block pre-heated to 90 °C for 2 min. This eliminates any secondary structure in the single-stranded template that may interfere with primer annealing or enzymatic addition of nucleotides.
Remove plate from heat and let cool to RT. (Optional) Turn off heater, but leave pyrosequencing plate on the block for 10 min. This step may facilitate sequencing primer binding by allowing a slower cooldown.
3.4 Preparing the Pyrosequencer
Note that these steps can and should be performed when time permits in the above protocol to eliminate unwanted lag between preparation of the sample and running the pyrosequencing reaction. It is advisable to input the assay into the software prior to strand separation.
Create a new assay within a folder in the CpG assay folder by right-clicking the folder and selecting New Assay. Enter the sequence to analyze in the appropriate field and click “Generate Dispensation Order.” A theoretical program is displayed (see Note 3).
Turn on machine. It will take about 2 min to warm up and establish communication with the computer. Wait for the info light to begin blinking as an indicator of successful data exchange.
Open PyroCpG software.
Under the CpG run folder, select the appropriate subfolder (if applicable), right-click, and select “New Run.” Name the run.
Enter the relevant information (sample ID, assay, notes about experiment, and type of dispensing tips used) into the PyroCpG software for each well you are using for this run. Save often to prevent data loss if software crashes before run commences.
Once all wells and assays are entered, select Volume Information from the drop down Tools menu. Note the amount of enzyme, substrate, and nucleotides to add to the appropriate dispensing tips. Enzyme and substrate always go in black RDTs, while nucleotides in CDTs or NDTs. Tips may be reused several times, so label each tip with its contents (see Note 4).
Add enzyme, substrate, and nucleotides to their respective dispensing tips, and place those tips in the correct slot of the appropriate tip holder. See diagram for proper placement.
Place tip holder in upper chamber of pyrosequencer manually, with enzyme and substrate tips toward the back of the machine.
Using the software, click “Open Process Chamber Lid” either from the shortcut icon or from the Instrument drop-down menu.
Cover a new pyrosequencing plate with clear plastic PCR sticker to use to test the dispensation tips. This plate can be reused ad infinitum if consistently maintained.
Place plate inside the lower chamber with the notch in the upper left corner. Click “Close Process Chamber Lid” command either from the shortcut icon or from the Instrument drop-down menu.
Test the dispensing tips by clicking “Test dispensing tips” icon or from the Instrument drop-down menu. Make sure that your stickered test plate is in position or the dispensations will damage the camera. Each tip will dispense a small volume of fluid, which will be visible on top of the sticker, validating that the tips are functional. Do not click “Done” after the dispensation test finishes until the hissing stops or the software may crash!
If all six drops are visible on the film, proceed with pyrosequencing. If one or more drops are absent, check to see if there are bubbles in the tips by flicking them gently. Residual salts from repeated use eventually will clog the tips. If tips are blocked, discard and use a new tip. It is recommended to test the dispensation tips twice, once the tips are ready and immediately prior to starting the run, ensuring the tips have not clogged in the interim.
3.5 Run the Pyrosequencing Reaction
Once the plate has cooled to RT, open the process chamber door using the software, insert the plate in the correct orientation, and close the process chamber lid using the software.
Click “Run.” The length of the run is determined only by the number of dispensations in the longest assay selected and is not dependent on the number of wells used.
3.6 Cleanup
Immediately after all runs are finished, clean out dispensing tips to prevent salt buildup and tip blockage. RDTs and CDTs can be “milked”; that is, rinse and fill them with water, and then apply pressure to squeeze water in a stream through the tip. NEVER milk NDTs as the bore size of the tip is too small; rather, gently rinse the tip inside and out with water. Store tips upright in a 5 mL tube rack and avoid contact with the delicate tips, which are easily bent/damaged.
Rinse out plastic trays from the vacuum prep station and allow to dry.
3.7 Data Analysis
While the pyrosequencer is running, the light trace for each well detected by the camera is presented in real time, generating a pyrogram of peaks, the height of which indicate the stoichiometric incorporation of nucleotides. Each non CpG peak becomes a reference peak that the software uses to calculate the percent methylation of the sample (Fig. 5). However, quantitative analysis cannot be performed until the run is finished.
Once the run is completed, the 96-well plate map is displayed. Click wells of interest to see the pyrogram.
-
Click “Analyze all” found in the lower right corner of the display. The software will measure the percent methylation at each CpG site and perform quality control analysis of each run. Quality control consists of using the non-CpG dispensations as reference peaks and measuring how well they conform to the theoretical pyrogram generated from the original sequence to analyze input into the assay file. Tolerances are typically set by the manufacturer. CpG sites that pass quality control are indicated in blue, sites that are questionable due to deviation from expected peak heights are yellow, and sites that fail are marked red. It is common to observe different indicated results within the same run and even within the same well at different CpG sites.
To facilitate data analysis, right click on the pyrogram and select “Show Histogram,” which underlays the theoretical pyrogram beneath the real result. Also, “Show Reference Peaks” allows elimination of problematic peaks from contributing to quality control analysis; for example, a long run of nucleotide (>5) may not reach the theoretical peak height due to limitations of detection.
Once the run is analyzed, export the raw data to a .txt file by selecting Reports → Analysis Results → Save. The exported file may be opened in spreadsheet format (semicolon delimited) for further analysis, and includes percent methylation of each CpG site, whether that site received a Pass/Check/Fail and any applicable warnings, the mean methylation across the entire sequence to analyze and various statistics.
To ensure that the assay is not biased toward methylated or unmethylated DNA, validate the assay using a standard curve of DNA with known methylation (see Note 5).
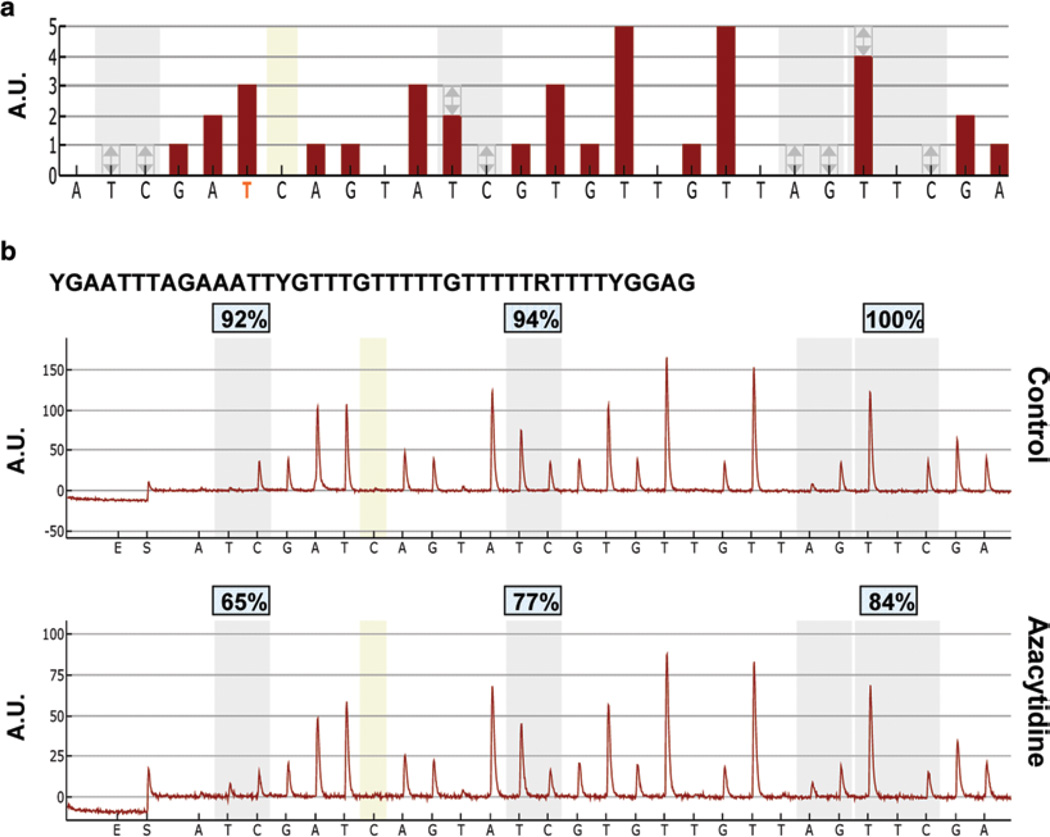
Fig. 5.
Representative pyrograms showing hypomethylation of the B1 element following 5-azacytidine treatment. (a) Theoretical pyrogram generated by analytical software based on the input sequence to analyze for B1 element pyrosequencing primer 2 (see table). (b) Pyrograms of DNA isolated from T cells cultured in the absence (control) or presence of cytosine analog 5-azacytidine, a known hypomethylating agent. The B1 “sequence to analyze 2” is shown bolded. Grey shaded areas indicate CpG sites, tan shaded bars indicate bisulfite control dispensations. Percent methylation is indicated above each CpG site
5 Conclusion
Pyrosequencing provides a rapid, high-throughput means of detecting methylation levels at individual loci or estimating global methylation changes. Commercially available bisulfite conversion kits and straightforward PCR amplification step make this technology accessible at reasonable cost while avoiding onerous technical challenges of next-generation sequencing or the delay and labor inherent in cloning fragments for Sanger sequencing. The pyrosequencing platform has been demonstrated to be a versatile means of quantifying DNA methylation globally and at regulatory elements of methylation sensitive genes addition to its broader uses in SNP analysis, association studies, and mutation screening. Thus, pyrosequencing can help elucidate pathogenic dysregulation of gene expression in methylation sensitive genes and advance the pace of biomedical science. Recent advances have focused on high-throughput whole-genome methylation analyses. However, the sensitivity of these assays has not been compared with pyrosequencing.
Table 1.
Set of primers both for PCR and pyrosequencing and reaction conditions to analyze Line 1 (human) and B1 element (mouse)
| Line 1 (Human) | |
| PCR primer (F): TTTTGAGTTAGGTGTGTGGGATATA PCR primer (R): biotin-AAAATCAAAAAATTCCCTTTC |
Pyrosequencing primer (F): AGTTAGGTGTGGGATATAGT |
| Reaction conditions: 95 °C for 5 min; (95 °C 30 s, 50 °C 30 s, 72 °C 30 s) × 45 cycles; 72 °C for 5 min | |
| Sequence to analyze: TTYGTGGTGYGTYGTTTTTTAAGTYGGTTTGAAAAGYGTA | |
| B1 element (Mouse) | |
| PCR primer (F): TGGTGGTGGTGGTTGAGAT | Pyrosequencing primer 1 (F): TGGTGGTGGTTGAGAT |
| PCR primer (R): biotin-AATAACACACACCTTTAATCCCAA | Pyrosequencing primer 1 (R): TTTGTAGATTAGGTTGGTTT |
| Reaction conditions: 95 °C for 15 min; (95 °C 30 s, 63 °C 30 s, 72 °C 30 s) × 45 cycles; 72 °C for 10 min | |
| Sequence to analyze 1: AGYGTTTTTTTGTGTAGTTTTGGTTATTTTGGAATTTATTTTGTAGATTAGGTTGGTTTYGAATTT | |
| Sequence to analyze 2: YGAATTTAGAAATTYGTTTGTTTTTGTTTTTRTTTTYGGAG | |
Acknowledgments
This work was supported by National Institutes of Health National Institute on Aging (AG020628, AG028268), National Institute of Environmental Health Science (P30 ES017885), University of Michigan (Claude D. Pepper Older American Independence Center, Nathan Shock Center for the Basic Biology of Aging, Rheumatic Disease Clinical Center, Caner Center Microarray Core, Michigan Diabetes and Research Training Center Animal Phenotyping Core), Geriatrics Research, Education and Clinical Care Center (GRECC), and the VA Ann Arbor Healthcare System. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Assay design: Pyrosequencing assays require three steps—bisulfite conversion of unmethylated cytosine to uracil, PCR to generate an amplicon biotin-labeled on one of the two strands, and a pyrosequencing reaction to analyze the nucleotide content of the amplified fragment. Following amplification, the PCR product is purified and denatured, at which time the unlabeled strand is removed to allow the pyrosequencing primer to anneal efficiently to the single-strand template. Typically, assays require at least three primers, a forward and reverse PCR primer as well as a pyrosequencing primer. One and only one of the PCR primers must be biotinylated so that the single-stranded template can be isolated prior to the pyrosequencing reaction, and the pyrosequencing primer must be complementary to the biotin-labeled strand (i.e., if reverse PCR primer is labeled, the pyrosequencing primer would be oriented in the “forward” direction, and vice versa). Alternatively, it is more economical and feasible to perform a three-primer PCR incorporating a universal biotinylated primer [20]. Also, to interrogate distant CpG sites on the amplicon (greater than ~60 dispensations), multiple pyrosequencing primers may be raised against a single amplicon. There are several software options available to assist in designing pyrosequencing assays as well as companies willing to design and validate custom assays.
Successful PCR conditions: In order to efficiently capture the biotinylated strand from the amplicon, all biotinylated primer must be incorporated into the PCR product. Unincorporated biotinylated primer will compete with the amplicon during the ssDNA isolation step prior to the pyrosequencing reaction and interfere with the quantification of C/T ratios during the reaction. Therefore, it is common to increase cycles in the PCR step to 45 or greater and use lower concentration of labeled primer to unlabeled primer (e.g., 0.1 µM biotin-primer vs. 0.2 µM unlabeled primer). Increasing the cycles also increases the risk of amplification of unwanted DNA from the environment and care should be taken to minimize exposure to sources of contamination.
Understanding and creating proper pyrosequencing assay files: In many ways “pyrosequencing” is an unfortunate term because it leads to confusion in scientists familiar with Sanger sequencing. In contrast to Sanger method, pyrosequencing uses sequencing by synthesis of an already known DNA sequence that contains a SNP at a known position. A proper pyrosequencing run requires entering this “sequence to analyze” into the operating software and indicating in that sequence where the CpG sites are. For CpG methylation detection, inputting the bisulfite converted sequence is necessary. Typically, indicating the cytosines of CpG sites with either a “Y” (for pyrimidine) or “C/T” is understood by the software. For example, a possible bisulfite-converted sequence to analyze could be ATTTGYGGAAAAAT, derived from the gDNA sequence ACTTGCGGAAAAAC (Fig. 2). Note that if a biotinylated forward primer is used, the pyrosequencing primer is in the reverse orientation and the reverse complement sequence must be input (e.g., in the case of the above sequence, the appropriate sequence to analyze would be ATTTTTCCRCAAAT, where the “R” stands for purine or “A/G”). From the sequence to analyze, the software generates a “dispensation order” that takes into account the order of nucleotides in the sequence to analyze, the length of runs of a single nucleotide, and controls to verify successful bisulfite conversion and integrity of nucleotides. For example, the sequence to analyze ATTTGYGGAAAAAT may generate a dispensation order GATCGCTGAAT. The first G is a negative control, followed by A producing a single-base peak, the run of 3 T’s can be measured with a single dispensation of T nucleotide, the first C is a negative control that verifies complete bisulfite conversion, the G is a single-nucleotide peak, the C followed by T is quantifying the CpG site of interest, the next G produces a double-nucleotide peak, and the two A dispensations ensure that the long run of A nucleotide is completely filled in. Incomplete synthesis interferes with downstream nucleotide incorporation, making data analysis difficult or impossible.
Because nucleotide droplets are being added sequentially, as the number of dispensations increases the volume of the reaction increases accordingly. This volume effect dilutes the enzyme and substrate reagents, lowering the signal to noise ratio over time. It follows that there is a finite amount of dispensations per run that will produce usable data. Often data generated after 90–100 dispensations is not usable. It is advisable to use multiple sequencing primers in separate wells to cover a large amplicon rather than attempt to analyze a large region with one reaction.
Dispensing tip selection: Enzymes and substrates are always dispensed from Reagent Dispensing Tips (RDTs). However, there are different options for dispensing nucleotides, capillary dispensing tips (CDTs) and nucleotide dispensing tips (NDTs). CDTs are easier to maintain and as such may be better for beginners; however, they dispense a larger volume droplet and as such are not suited for longer dispensations >50. NDTs are ideal for longer runs and may produce cleaner results because they dispense smaller droplets, yet they are easier to clog and are harder to clean and maintain.
Bias testing of pyrosequencing assays: Once an assay is initially optimized to produce a robust amplicon and a clean pyrogram, it is important to validate that the efficiency of the PCR reaction is not altered by differing cytosine content at the CpG sites contained within the region amplified. Differences in efficiencies may skew results, potentially masking or overestimating differences between treatment groups. To check for PCR bias, perform the pyrosequencing assay with amplicons generated from DNA with known methylation content. Start with DNA from low methylated sources—whole genome amplified or cloned plasmid DNA. Treat that DNA with Sss1 methylase enzymes. Alternatively, low-methyl DNA and high-methyl DNA are available commercially. Mix these two DNA samples in known ratios to create a standard curve. If the results diverge from linearity, the assay may be biased and must be re-optimized.
Application: global methylation measurement: Global methylation analysis using pyrosequencing technology utilizes the ubiquity of specific repetitive elements randomly inserted throughout mammalian genomes. Often these elements number in the thousands. In humans, LINE1 and Alu elements have been shown to be useful in measuring changes in global methylation due to cancer, aging, and environmental stressors [17, 21–24]. In mice, the B1 element as well as the intracisternal alpha particle (IAP) can detect changes in methylation in cancer and/or cells treated with hypomethylating agents like the cytosine analog 5-azacytidine [18, 19]. See Table 1 for the primer sets and conditions reported to amplify these elements as an estimate of global methylation.
Contributor Information
Colin Delaney, Department of Internal Medicine, University of Michigan Medical School, 109 Zina Pitcher Place, Ann Arbor, MI, 48109, USA.
Sanjay K. Garg, Department of Internal Medicine, University of Michigan Medical School, 109 Zina Pitcher Place, Ann Arbor, MI, 48109, USA
Raymond Yung, Department of Internal Medicine, University of Michigan Medical School, 109 Zina Pitcher Place, Ann Arbor, MI, 48109, USA, ryung@umich.edu.
References
- 1.Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 2.Eden A, Gaudet F, Waghmare A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 3.Vera E, Canela A, Fraga MF, et al. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27:6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- 4.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 5.Ballestar E, Esteller M, Richardson BC. The epigenetic face of systemic lupus erythematosus. J Immunol. 2006;176:7143–7147. doi: 10.4049/jimmunol.176.12.7143. [DOI] [PubMed] [Google Scholar]
- 6.Brooks WH, Le Dantec C, Pers JO, et al. Epigenetics and autoimmunity. J Autoimmun. 2010;34:J207–J219. doi: 10.1016/j.jaut.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Kim M, Long TI, Arakawa K, et al. DNA methylation as a biomarker for cardiovascular disease risk. PLoS One. 2010;5:e9692. doi: 10.1371/journal.pone.0009692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lund G, Andersson L, Lauria M, et al. DNA methylation polymorphisms precede any histological sign of atherosclerosis in mice lacking apolipoprotein E. J Biol Chem. 2004;279:29147–29154. doi: 10.1074/jbc.M403618200. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warnecke PM, Bestor TH. Cytosine methylation and human cancer. Curr Opin Oncol. 2000;12:68–73. doi: 10.1097/00001622-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 11.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 12.Strickland FM, Hewagama A, Lu Q, et al. Environmental exposure, estrogen and two X chromosomes are required for disease development in an epigenetic model of lupus. J Autoimmun. 2012;38:J135–J143. doi: 10.1016/j.jaut.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 14.Polansky JK, Kretschmer K, Freyer J, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 15.Floess S, Freyer J, Siewert C, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoenborn JR, Dorschner MO, Sekimata M, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang AS, Estecio MR, Doshi K, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong KS, Lee S. Estimating the total mouse DNA methylation according to the B1 repetitive elements. Biochem Biophys Res Commun. 2005;335:1211–1216. doi: 10.1016/j.bbrc.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Delaney C, Hoeltzel M, Garg SK, et al. Maternal micronutrient supplementation suppresses T cell chemokine receptor expression and function in f1 mice. J Nutr. 2012;142:1329–1335. doi: 10.3945/jn.111.155903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Royo JL, Pascual MH, Salinas A, et al. Pyrosequencing protocol requiring a unique biotinylated primer. Clin Chem Lab Med. 2006;44:435–441. doi: 10.1515/CCLM.2006.072. [DOI] [PubMed] [Google Scholar]
- 21.Yang AS, Doshi KD, Choi SW, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 22.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 23.Bollati V, Schwartz J, Wright R, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–578. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]