Abstract
Mitochondria carry their own genetic material and gene-expression machinery, including ribosomes, which are responsible for synthesizing polypeptides that form essential components of the complexes involved in oxidative phosphorylation (or ATP generation) for the eukaryotic cell. Mitochondrial ribosomes (mitoribosomes) are quite divergent from cytoplasmic ribosomes in both composition and structure even as their main functional cores, such as the mRNA decoding and peptidyl transferase sites, are highly conserved. Remarkable progress has been made recently towards understanding the structure of mitoribosomes, by obtaining high-resolution cryo-electron microscopic (cryo-EM) maps. These studies confirm previous structural findings that had revealed that a significant reduction in size of ribosomal RNAs has caused topological changes in some of the functionally relevant regions, including the transfer RNA (tRNA)-binding sites and the nascent polypeptide-exit tunnel, within the structure of the mammalian mitoribosome. In addition, these studies provide unprecedented detailed views of the molecular architecture of those regions. In this review, we summarize the current state of knowledge of the structure of the mammalian mitoribosome and describe the molecular environment of its tRNA-exit region.
1. Introduction
The mammalian mitochondrial genome consists of multiple copies of a 16.8 kb circular DNA, which encodes 37 genes, including 2 ribosomal RNAs (rRNAs), 22 mitochondrial tRNAs (tRNAmt) and 13 polypeptide chains, which form essential components of the complexes involved in oxidative phosphorylation (OXPHOS). The OXPHOS complexes reside in the mitochondrial inner membrane (mtIM) and are responsible for generating about 90 % of the energy (ATP) required by the cell. All proteins required for mammalian mitochondrial translation (or protein synthesis), including the mitochondrial ribosomal proteins (MRPs), are encoded by the nuclear genome, translated in the cytoplasm and then imported into the mitochondria. Mitochondria are thought to have originated through an early endosymbiotic event between an α-protobacterium and a primitive host cell [1], and therefore, mitoribosomes were expected to be structurally similar to their widely studied eubacterial counterparts (see ref. [2]).
However, the mitoribosomes are quite divergent from bacterial ribosomes in their overall composition and physical properties [3, 4]. Mammalian mitoribosomes have lower sedimentation coefficients (~55S) and, like all ribosomes, consist of two subunits (the 28S small subunit, or SSU; and the 39S large subunit, or LSU) [5]. The two mitochondrially-encoded rRNA species, 12S (~955 nts) in the SSU and 16S (~1571 nts) in the LSU, account for ~30% of mass of the mammalian mitoribosome. In contrast, eubacterial ribosomes consist of 60-70% RNA [6, 7] and eukaryotic cytoplasmic ribosomes are 50-60% RNA [8-10]. Since the majority of the mass of the mammalian mitoribosome consists of MRPs [11], the RNA-to-protein ratio is inverted in the mitoribosome relative to other ribosomes (for recent reviews, see [3, 4, 12, 13].
The mitochondrial rRNAs show only a small amount of sequence identity with cytoplasmic rRNAs [14]. Unlike cytoplasmic ribosomes, whose X-ray crystallographic structures are known for several bacterial (e.g., [6, 7], archaeal [15] and eukaryotic [8, 9] species, the three-dimensional (3D) structures of mitoribosomes have been studied primarily using the single-particle cryo-EM. These studies have revealed that the structures of critical core regions of mito-rRNAs have been preserved [16-20]. The very existence of a 5S rRNA species in the 39S LSU has been debated for some time [21, 22], and a previous cryo-EM study at ~5 Å resolution [19] had detected discrete densities that could be tentatively assigned only to a small segment of a potential 5S rRNA molecule. However, more recently, high-resolution (~ 3Å) cryo-EM structures [23, 24] have revealed that the RNA-like density within the central protuberance of the mitoribosomal 39S LSU corresponds to a tRNAmt that partially occupies the position of the 5S rRNA in the bacterial 50S LSU.
There are eighty-two MRPs in the mammalian mitoribosome, and about half of these are homologues of bacterial ribosomal proteins. However, most of the homologous MRPs carry mito-specific extensions; and therefore are generally larger in size as compared to their bacterial counterparts [25-28]. Thirty-nine MRPs that are specific to mammalian mitoribosomes [29] have no apparent homologues in bacterial, chloroplast, archaeal, or cytosolic ribosomes. Our first cryo-EM study had revealed the overall locations of the mito-specific MRPs within the map of the 55S mammalian mitoribosome [16]. Recent cryo-EM studies at higher (3-7 Å) resolutions in our [20] and other [19, 23, 24] laboratories not only confirmed those locations but also assigned positions of most mito-specific MRPs and mito-specific extensions in homologous MRPs within the cryo-EM maps of two subunits of the mitoribosome.
Besides its mitoribosome, several other components of the mammalian mitochondrial translation system have also acquired some unusual features. The mammalian mitochondrial mRNAs have an almost complete lack of 5′ and 3′ untranslated nucleotides. The translational start codon is generally located within 3 nucleotides of the 5′ end of the mRNA [30, 31]. There is no Shine-Dalgarno sequence and no cap structure. The mechanism by which the mitoribosome recognizes the start site on the mRNA is unknown. Mammalian mitochondrial tRNAs (tRNAmt) are generally shorter (59-75 nucleotides in length) than bacterial or eukaryotic cytoplasmic tRNAs (76-90 nucleotides in length). Many tRNAmts lack the D-loop/T-loop interactions in the elbow region that stabilize the L-shape in the “more conventional” tRNAs (reviewed in [32]). The first average structure of the tRNAmt, revealed in our cryo-EM study of the mammalian mitoribosome [16], showed an L-shape, but with a “caved-in” elbow region. In addition, most mammalian mitochondrial translation factors appear to carry mito-specific sequences (reviewed in [13]). Finally, several components of the mammalian mitochondrial translational machinery, including mito-rRNAs, MRPs, tRNAmts, tRNAmt synthetases and translational factors, have been associated with a number of human genetic diseases (see [32-37]). Therefore, detailed knowledge of each of the above components of the mitochondrial translational machinery is essential for understanding not only the mechanism of mitochondrial protein synthesis but also of molecular mechanism underlying these diseases.
Owing to significantly reduced size of rRNAs in the mammalian mitoribosomes, as compared to their prokaryotic and cytoplasmic counterparts, topologies of some of the functional-ligand binding sites are significantly altered in the mitoribosome. One such site involves one of the tRNA-binding sites, called the tRNA-exit site (E site). The E site plays a vital role in maintaining the mRNA reading frame in bacterial and cytoplasmic ribosomes [38], as it receives the deacylated tRNA along with the mRNA codon from the peptidyl site (P site) during the EF-G (or EF-G1mt in mammalian mitochondrion) – dependent tRNA translocation reaction. In this article, we briefly describe the overall structure of the mammalian mitoribosome, by outlining what new structural information has been gathered from recently published structures, and discuss the role of the E site in context of the altered microenvironment of the mitoribosome and tRNAmts and its possible implication on mammalian mitochondrial protein synthesis.
2. Structure of the mammalian mitoribosome
Previous low-resolution cryo-EM studies had already revealed some of the unique features of the mammalian mitoribosome [16, 17]. These features include (i) a significantly modified and porous overall structure of the mitoribosome that is predominantly shielded by MRPs with fewer exposed rRNA regions as compared to its bacterial counterpart. This is primarily due to the occupation of new spatial positions by mito-specific MRPs and by extensions and insertions within several of MRPs that are homologous to their bacterial counterparts; (ii) a gate-like structural feature at the mRNA entrance within the 28S SSU that was implicated in the recruitment of leaderless mitochondrial mRNAs to initiate the process of protein synthesis on the mitoribosome; (iii) a significantly modified lower two-third portion of the nascent polypeptide-exit tunnel within the 39S LSU, apparently to streamline the co-translational insertion of all mitochondrially-encoded proteins into the mtIM ; and (iv) a finger-like MRP structure emerging from the central protuberance region of the 39S LSU to the mitoribosomal P-site, referred to as the P-site finger.
Recent technological developments in the cryo-EM field, specifically single-particle data collection using the direct-electron detectors and image reconstruction using movie processing techniques [39, 40], have provided far more refined structural models of the mammalian 39S LSU [19, 23, 24] and yeast 54S LSU [18] mitoribosomes at close to atomic resolution (~3 Å); whereas the structure of the mito-SSU has been reported at ~7 Å resolution, using conventional cryo-EM techniques [20] (Figure 1). Besides providing important insights into structural evolution of protein-rich mitoriboosmes, these studies significantly aid to our current understanding of the mitochondrial translation. In the remainder of this section, we summarize some of the main findings of the recent cryo-EM studies of the mammalian mitoribosomes.
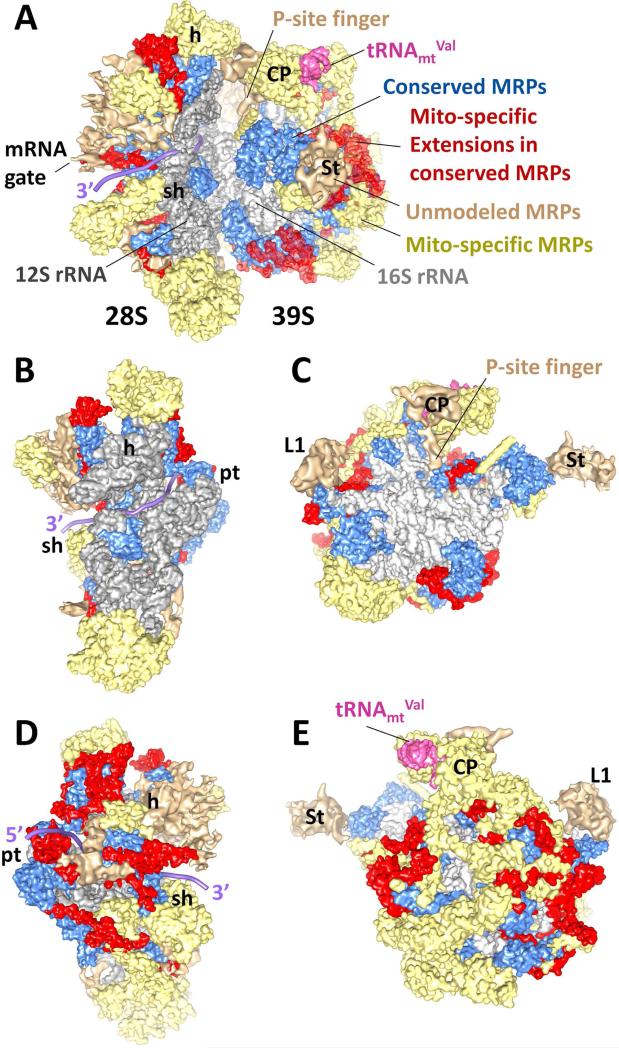
Figure 1. Structure of the 55S mammalian mitochondrial ribosome.
(A) the 55S mitoribosome is shown from the L7/L12 stalk (St) side, with components of both 28S SSU (PDB ID # 3J6V) and 39S LSU (PDB ID # 3J7Y) docked into a cryo-EM map. (B) and (D), 28S SSU shown from the interface and solvent sides, respectively. (C) and (E), 39S LSU shown from the interface and solvent sides, respectively. Color codes: light grey, 16S rRNA of the LSU; dark grey, 12S rRNA of the SSU; pink, tRNAmtVal as a structural component of the LSU; blue, conserved portion of the homologous MRPs; red, mito-specific extensions in the homologous MRPs; yellow, mito-specific MRPs; tan, unmodeled mito-specific MRP mass. Based on previous x-ray crystallographic study of the bacterial ribosome ([54], PDB ID # 2HGR), the mRNA path (purple) is modeled on the 28S SSU and its further extended 5’ and 3’ ends are depicted. Labels on the SSU: h, head; pt, platform; and sh, shoulder. Labels on the LSU: CP, central protuberance; L1, L1 stalk; and St, L7/L12 stalk. Chimera software [55] was used for visualization.
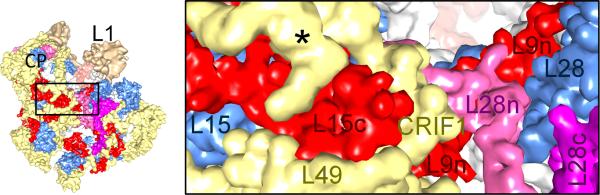
The new structures reveal that a majority of the mito-specific extensions in homologous MRPs are involved either in inter-MRP contacts or in contacts with mito-specific MRPs [20]. This observation suggests a stepwise evolution of the current architecture of the mitoribosome, in which the mito-specific extensions first help stabilizing the homologous MRPs, some of which have lost binding partners in the absence of bacterial proteins and rRNA segments, and then create a new binding surface for the binding of mito-specific MRPs [20]. For example, in the mito-SSU the N- and C-terminus extensions of MRP S15 and C-terminus extension of MRP S17 make extensive interactions with each other and with mito-specific MRPs tentatively assigned as S26 and S28 (Figure 2A). For a similar example in the mito-LSU, the mito-specific extensions of MRPs L9, L15, L28 interact with each other and with mito-specific MRPs L49, CRIF1 and the α-helix of an unidentified MRP (Figure 2B).
Figure 2.
An example of interactions among mito-specific extensions in homologous MRPs L9, L15, and L28 and several mito-specific MRPs in the 39S LSU ([24] PDB ID: 3J7Y). N- and CTEs of MRPs are indicated by a suffix “n” or “c”, respectively, in matching colors. As yet unknown mito-specific MRP is marked by an asterisk (*). The rRNA regions (semitransparent gray) are not labeled. A thumbnail to the left is shown for overall orientation of the mito-LSU.
Several of mito-specific MRPs and mito-specific extensions in some of the homologous MRPs form of the lining of the mRNA-entrance and -exit paths, suggesting a significant structural reorganization in these regions of the mito-SSU, perhaps to facilitate the recruitment of mito-specific mRNAs most of which do not possess a 5’ leader sequence [20]. For example, some of crucial components of the mRNA entrance in bacterial SSU, such as protein S4 and C-terminus domain of protein S3, are absent in the mito-SSU. A portion of S4 is replaced by a mito-specific MRP, tentatively assigned to MRP S37 [20], while the N-terminal domain of bacterial S3 is replaced by a structurally homologous MRP S24. MRP S5 is significantly enlarged in the mito-SSU, and its mito-specific N-terminal extension contributes to the formation of a portion of the gate-like feature at the mRNA entrance. In addition, three other mito-specific MRPs, tentatively identified as S22, S33, and S39 [20], encircle the mRNA entrance in the mito-SSU (Figure 3A). Similarly, the mito-specific extensions in MRPs S7, S11, S18, and S21 significantly change the landscape of the mRNA-exit region in the mito-SSU (Figure 3B). Whether these structural changes directly contribute to recruitment of leaderless mitochondrial mRNAs remains unclear.
Figure 3. Molecular architectures of the mRNA entrance and exit regions.
(A) The mRNA-entrance, and (B) the mRNA-exit regions, with identified MRPs in the immediate vicinity. N- and CTEs of MRPs are indicated by a suffix “n” or “c”, respectively, in matching colors. Tentatively assigned mito-specific MRPs [20] are identified with asterisks (*) and brackets. The 12S rRNA regions (semitransparent gray) are not labeled. The 5’ and 3’ ends of the modeled mRNA are shown as thick purple Ribbon. Thumbnails to the left are shown for overall orientation of the mito-SSU.
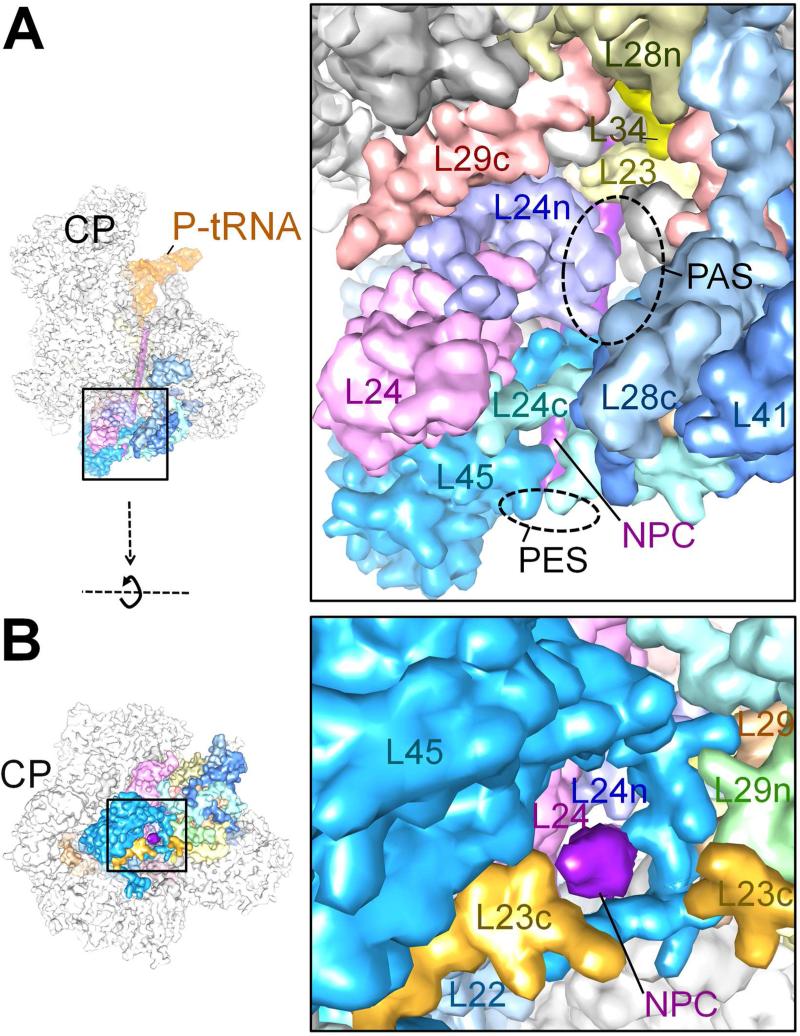
In contrast to the situation with cytoplasmic ribosomes, where a variety of possible destinations for the synthesized nascent polypeptide chains are possible, all of the nascent chains synthesized by the mammalian mitoribosome are inserted into the mtIM, and it is most likely that these polypeptides are inserted co-translationally into mtIM [41, 42]. Therefore, detailed knowledge of the architecture and composition the polypeptide-exit tunnel in the mito-LSU has been of particular interest. High-resolution cryo-EM studies of the mito-LSU provide a clear and detailed view of the nascent polypeptide-exit tunnel (PET), with identification of some of the hydrophobic MRP components of the 39S LSU that are involved in direct interaction with the predominantly hydrophobic mitochondrial nascent-polypeptide chains [19, 23, 24]. The overall topology of PET in the mammalian mito-LSU is known to be significantly diverged from its bacterial counterpart as it carries two openings exposed to the solvent: a conventional polypeptide-exit site (PES) and a polypeptide-accessible site (PAS) [16]. Recent high-resolution structures of the mito-LSU have now identified specific MRPs that line and encircle the PET. However, in the two high-resolution 39S LSU structures the description of the PAS is not the same. It is not clear, however, if the difference in description partly relates to the source of the 39S LSU, porcine tissues [23] and a human cell line [24], used in these two studies. While Greber and coworkers confirm the existence of PAS [19, 23] (Figure 4A), Brown and coworkers [24] do not mention about this structural feature. However, the deposited structure by Brown and coworkers (EMD ID: 2762) suggests that the PAS-like feature does exist in human 39S LSU as well, but MRP region that separates PAS and PES is not modeled into the coordinates (PDB ID # 3J7Y) submitted by Brown and coworkers, and therefore, a larger continuous opening appears from PAS to PES. A closer inspection of the PAS/PES regions between the two structures reveals that a loop region (corresponding to amino-acid residues 159 -168) of MRP L24 and an N-terminus region (corresponding to amino-acid residues 56-156) are disordered and absent in the Brown and coworker's PDB (# 3J7Y). However, the finding of a continuous density corresponding to a nascent-polypeptide chain from the peptidyl-transferase center up to the PES [43] by Brown and coworkers [24] suggests that PAS is unlikely to provide an alternative path for the nascent polypeptide exit, but that it provides a regulatory conduit cannot be ruled out. Interestingly, while the PAS/PES region is spanned by a set of common but larger sized bacterial homologues, including MRPs L22, L23, L24 and L29, both openings also carry mito-specific MRPs (Figure 4A,B). At the PES, a mito-specific MRP L45 extends from L24, while a homologous MRP L34 and a mito-specific MRP L41 are present in the immediate neighborhood of PAS. MRP L45 protrudes from the rest of the mito-LSU and is strategically located to be involved in direct interaction with, or anchoring of mitoribosome to, mtIM [19, 23, 24]. Furthermore, being a homologue of two membrane associated proteins in Saccharomyces cerevisiae, Mba1 [44] and carboxy-terminal domain of TIM44 [45] that are known to interact with the mtIM, involvement of MRP L45 in such interaction is further strengthened in the recent cryo-tomographic studies of the yeast mitoribosome attached to the mtIM [46].
Figure 4. Molecular architecture of the lower portion of the polypeptide-exit tunnel.
(A) Surface view of the nascent polypeptide chain (NPC)-exit tunnel as seen from the L1 protein side, showing both the polypeptide-accessible site (PAS) and polypeptide-exit site (PES); and (B) the PES, as seen from the bottom of the mito-LSU. MRPs in the immediate vicinity are identified. N- and CTEs of MRPs are indicated by a suffix “n” or “c”, respectively, in matching colors. The 16S rRNA regions (semitransparent grey) are not labeled. PDB IDs : 4V1A and 4V19 were used to generate the figure. NPC (purple) has been modeled based on ref. 24. Thumbnails to the left are shown for overall orientation of the mito-LSU.
The central protuberance (CP) of the mammalian mito-LSU is also significantly remodeled and is larger in size as compared to that in its bacterial counterpart [16]. The fact that homologous bacterial proteins L5 and L25 are absent in the CP of the mito-LSU is known. However, the existence of the 5S rRNA in mito-LSU has been debated for some time [21, 22]. The two high-resolution cryo-EM structures [23, 24] reveal almost complete composition of the CP. Surprisingly, both structures show that the mito-LSU has acquired a tRNA as one of its structural components in addition to several mito-specific MRPs including L38, L40, L46, L48, L52 and ICT1 [23, 24]. This finding has effectively ended a long-standing debate about the presence of 5S rRNA in the mitoribosome [21, 22]. However, while Greber and coworkers [23] tentatively identified the RNA component of the central protuberance as tRNAmtPhe, Brown and coworker's study [24] positively identified it as tRNAmtVal. It is not clear, however, if the difference in tRNAmt species relates to the source of the 39S LSU, porcine tissues [23] and a human cell line [24], used in these two studies. Furthermore, it remains to be seen if structural tRNAmtVal identified by the Brown and coworkers carries the same set of base modifications as an elongator tRNAmtVal [47, 48].
By combining the results of recent cryo-EM studies of the SSU and LSU, we are now able to better understand the interaction between the two mitoribosomal subunits by identifying the participating ribonucleoprotein components of the inter-subunit bridges and topology of the inter-subunit space [20], where mRNAs, tRNAs and translational factors interact with the ribosome. As previously suggested (16), in contrast to the situation in bacterial ribosomes, many bridges between the two mitoribosomal subunits are formed by MRPs than by rRNAs [20]. However, despite unprecedented progress made in structural studies of mammalian mitoribosomes, some of the functionally relevant regions --such as the L1 stalk, the L7/L12 stalk, and the composition of the interface-side of the central protuberance involving the P-site finger in the 39S LSU, and a high-resolution identification of mito-specific MRPs including the composition of the gate-like feature at the mRNA entrance on the 28S SSU--remain to be resolved.
tRNAmt binding in the E-site region
Previous cryo-EM studies combined with molecular modeling [16, 17] had indicated that the deacylated tRNA may not reside in the putative tRNA-exit site (E-site) within the mammalian mitoribosome, and that instead deacylated tRNAmt may exit the mitoribosome directly from the P site after the peptide-bond formation and subsequent EF-G-dependent translocation. It was further suggested that existence of such a tRNA-binding site would offer only few interactions to the deacylated tRNAmt [17]. These suggestions were based on following facts: (i) absence of a strong density corresponding to the E-site tRNA in those cryo-EM reconstructions, and (ii) a significantly altered molecular landscape of the putative E site; that is, deletion of 11 of the 12 interaction sites of tRNA involving 23S rRNA segments of the bacterial ribosomal E site in the mito-LSU [17]. It should be noted that the E site is found almost 100 % filled with deacylated tRNAs in purified and undissociated cytoplasmic ribosomes. Furthermore, the fact that the overall size of mitochondrial tRNAmts is smaller [32, 47-49] (Figure 5) than their cytoplasmic counterparts would require greater, not less, structural complementarity from the mitoribosome structure.
Figure 5. Secondary and tertiary structures of the mitochondrial tRNAPhe and tRNASer/AGY.
(A) and (B) Secondary structures of tRNAPhe and tRNASer/AGY, respectively, shown without posttranscriptional modification. (C) and (D) Three dimensionally folded structural models of tRNAPhe and tRNASer/AGY, respectively. tRNAmtPhe represents a typical tRNAmt with cloverleaf structure but with shorter versions of each loop structure, whereas tRNAmtSer/AGY is highly unusual and lacks the entire D stem. Color coded D and T refer to D- and T-loop regions, respectively, on the 3D model. Sequences and secondary structure information of tRNAmts were obtained from MamittRNA database [56] and their 3D structures were generated using RNAComposer [57].
More recently, we have obtained several cryo-EM reconstructions by classifying the 55S mitoribosome dataset using Relion [50]. A 7.8 Å resolution 3D reconstructions obatined for one of the classes, represented by a fraction (~24 %) of the 55S population, shows a clear but fragmented tRNA-like density in the E-site region (Figure 6A), indicating that the tRNAmt binding is weaker in this region of the mammalian mitoribosome, as compared to that in its aminoacyl (A) and peptidyl (P) sites that are much more prominantly occupied (unpublished results). It should be noted that Brown and coworkers [24] also reported a tRNA density in the E-site region in one of their low-resolution maps, apparently also presented only in a small fraction of the 55S population. The fact that ~76% of the mitoribosomal population in our study does not show the presence of a tRNA-like density in the E-site region further suggests that the E site is indeed one of the weaker tRNA-binding sites in the mammalian mitoribosome, unlike the E site in the cytoplasmic ribosomes.
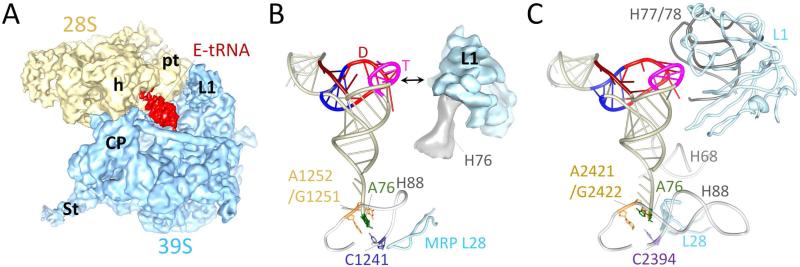
Figure 6. Averaged tRNA density and micro-environment of a tRNA in the E-site region of the mammalian mitoribosome.
(A) A 7.8 Å resolution cryo-EM reconstruction obtained from only 24% of the total population of the 55S mitoribosome that contained density for an E-site tRNAmt. (B) and (C) Micro-environments of E-site regions in mammalian mitochondrial and bacterial ribosomes, respectively. The model of tRNAPhe is shown with ladders whereas rRNA segments are shown as backbone structures. Since H76 of the mito-16S rRNA has not been modeled in high-resolution structures, the cryo-EM density corresponding to that helix is shown in panel B. H followed by a number identifies helix of the LSU rRNA (according to bacterial numbering of helices). A double headed arrow in panel B indicates the gap between the L1 stalk and the elbow region (D and T loop regions) of bacterial tRNAPhe. This gap would be even larger when bacterial tRNAPhe is replaced with a smaller mitochondrial tRNA species such as tRNASer/AGY (see Figure 2). Structure of the T. thermophiles 70S ribosome ([7], PDB ID # 2J00 and 2J01) was used to illustrate the E-site microenvironment in panel C.
The tRNA in the bacterial ribosomal E site makes extensive interactions with the 23S rRNA helices 68 and 88, and helices 77 and 78 (according to bacterial 23S rRNA numbering) of the L1-stalk region [7]. While helices 77 and 78 of the L1 stalk are absent, helices 68 and 88 are present but partially truncated in the mitoribosome [17]. However, the mito-16S rRNA has retained the binding cleft within the helix 88 that is known to hold the 3’-terminal adenine residue of a tRNA at the ribosomal E site. This segment of helix 88 includes the universally conserved residue C1241 (C2394 in bacterial 23S rRNA; [51] (Figure 6B, C). The terminal adenine residue of the E-site tRNA also makes the base stacking interactions with G2421 and A2422 of the 23S rRNA helix 88. The corresponding residues G1251 and A1252 and the overall base-stacking architecture are also conserved in the mito-16S rRNA. However, the β-hairpin of the ribosomal protein L28 that is also known to interact with the accepter end of the E-site tRNA in bacterial ribosome adapts a slightly different conformation in the mitoribosome, leading to loss of its potential interaction with the accepter end of tRNAmt.
From the SSU side, ribosomal protein S7 and S11 interact with the E-site tRNA in the bacterial ribosome. Both corresponding MRPs are present in the mitoribosome and carry mito-specific extensions that are situated mostly on the solvent side of the 28S SSU [20]. Despite the fact that the overall E-site architecture on the mito-SSU is conserved, we find that the density corresponding to the anticodon stem-loop (ASL) region is even weaker than the rest of the E-site tRNAmt. Since our map represents an average density corresponding to a mixed population of indigenously-bound tRNAmts, there may be two different reasons for a weak E-site ASL density: (i) not all tRNAmts bind stably at the mitoribosomal E site. For example, smaller sized tRNAmts like tRNAmtSer/AGY that have lost most of its D-loop structure (Figure 5) would have even less chance of interaction with the L1-stalk region of the mitoribosome and therefore are less likely to spend the same amount of the time at the E site as other canonical tRNAs; and (ii) some of the smaller tRNAmt species do not conform to a canonical tRNA structure, and therefore would have to stretch a bit to maintain interactions at both of its anticodon and CCA ends (Figure 5), and this could lead to severance of their interaction with the SSU side of E site including the mRNA.
In clear contrast to the situation in cytoplasmic ribosome preparations, where tRNA is invariably found at the E site, the binding of tRNAmt in the mitoribosmal E site is certainly non-stoichiometric (only ~24% carrying a tRNA in that region). However, the conservation of the CCA-end interaction site on the mito-LSU strongly suggests that, following the peptide-bond formation, a hybrid tRNA-binding state [52] would be also formed on the mitoribosome, where the CCA ends of A- and P-site tRNAs move to P and E sites, respectively, while their anticodon ends remain in A and P sites. The question however remains whether the E site (or an E/E state) is a canonical tRNA-binding site, or is a tRNAmt binding in this region of the mitoribosome a conditional event dependent upon the tRNAmt species (size) that translocates from the P site following the EF-G1mt-dependent tRNAmt translocation reaction during the translation elongation cycle.
4. Concluding Remarks
Recent progress made in high-resolution cryo-EM structure determination of mitoribosomes has not only confirmed the unique features of the mammalian mitoribosome studies reported previously, but has provided an unprecedented detailed view of its molecular architecture. However, these studies are only the first steps in understanding the basic functioning of this highly complex macromolecular machine that is made of about 85 component molecules but is responsible for synthesizing only 13 but crucial polypeptide chains. The overall complexity of the mitoribosome structure, where a multitude of new mito-specific MRPs are added to it at locations away from its central core structure, suggests that some of these MRPs may have functions that are not directly related to protein translation. Moreover, these high-resolution structures will now allow us to map the previously identified disease causing mutations in both mito-rRNAs and MRPs in order to better understand molecular mechanism of those diseases and function of some of the MRPs that may not be directly involved in the translational process. Furthermore, the finding of a tRNAmtVal as one of the structural components of the mammalian mitoribosomal large subunit is quite intriguing, although its functional implication if any is unclear. It should be noted that a mutation in tRNAmtVal is involved human genetic disease (see for review [53]). However, it remains to be seen if the disease is caused by the mutation in structural tRNAmtVal or in the functional (translation elongator) tRNAmtVal.
With recent technological advancement in the cryo-EM field, it is now feasible to obtain structures of functional complexes of the mitoribosome at ever increasing resolution to understand the functions of various insertion and extension sequences in mitochondrial translation factors, and to unravel mechanistic and molecular details of mitochondrial protein synthesis. Further structural studies with defined ribosome-binding ligands, such as leaderless mito-mRNAs, tRNAmts and mito-translational factors, will be necessary to develop molecular models of various steps of mitochondrial translation. Among some of the immediate questions for the mitochondrial translation community are: (i) How are the leaderless mRNAs recruited to the mitoribosome to initiate the process of protein synthesis on the mitoribosome? (ii) Now that we have a detailed view of the molecular architecture of the nascent polypeptide-exit tunnel [23, 24], the next frontier would be understand how the mammalian mitoribosome interacts with the intact mtIM to facilitate the co-translational insertion into the mtIM. (iii) Functionally defined complexes with differently-sized tRNAmts will be required to understand if and how the smaller sized tRNAmts are complemented by mitochondrial translational factors and by the components of the mitoribosome architecture itself. Structures of these complexes should be able to address the question of whether the E site does play an integral functional role in mammalian mitochondrial translation, and whether all tRNAmts have similar affinity at the mitoribosomal E site. Unfortunately, because of limited mitoribosomal quantities and inherent compositional heterogeneity in mitoribosomes isolated from animal tissues, a robust in vitro biochemical assay system for mammalian mitochondrial protein synthesis has been lacking, but would be essential to complement the rapidly progressing structural studies of the mammalian mitoribosomes and to better address the aforementioned basic mechanistic questions.
Highlights.
Recent progress in structure determination of the mammalian mitoribosomes
Overall molecular architecture of both subunits of the mammalian mitoribosome
A mitochondrial tRNA as a structural component of the large mitoribosomal subunit
A protein-rich mRNA path could be relevant to recruitment of leaderless mito-mRNAs
A significantly diverged architecture and composition of the polypeptide-exit tunnel
Ribosomal RNA-depleted tRNA-exit region of the mammalian mitoribosome
Acknowledgements
This work was supported by the National Institutes of Health grant R01 GM61576 (to R.K.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
After this article went into press, two papers describing high-resolution cryo-EM structures (at 3.5 e 3.8 Å) by Venki Ramakrishnan (Amunts et al., Science, vol. 348, 2015, 95-98) and Nenad Ban (Greber et al., Science, 2015, in press, DOI: 10.1126/science.aaa3872) of the complete 55S mitoribosome have appeared. Based on these two papers, some of the annotations in Figure 3 of this article may change.
References
- 1.Gray MW, Burger G, Lang BF. The origin and early evolution of mitochondria. Genome Biol. 2001;2:REVIEWS1018. doi: 10.1186/gb-2001-2-6-reviews1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–42. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal RK, Sharma MR, Yassin AS, Lahiri I, Spremulli L. Structure and function of organellar ribosomes as revealed by cryo-EM. SpringerWien, New York. 2011:83–96. [Google Scholar]
- 4.Agrawal RK, Sharma MR. Structural aspects of mitochondrial translational apparatus. Curr Opin Struct Biol. 2012;22:797–803. doi: 10.1016/j.sbi.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Brien TW, Denslow ND, Faunce W, Anders J, Lui J, O'Brien B. In: The translational apparatus: Structure, function regulation and evolution, Nierhaus K, Franceschi F, Subramanian A, Erdmann V, Wittmann-Liebold B, editors. Vol. 354. Plenum Press; New York: 1993. pp. 575–86. [Google Scholar]
- 6.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 7.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–9. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 9.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–8. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 10.Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell. 2014;157:1632–43. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pietromonaco SF, Denslow ND, O'Brien TW. Proteins of mammalian mitochondrial ribosomes. Biochimie. 1991;73:827–35. doi: 10.1016/0300-9084(91)90062-6. [DOI] [PubMed] [Google Scholar]
- 12.Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta. 2012;1819:1035–54. doi: 10.1016/j.bbagrm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma MR, Kaushal PS, Gupta M, Banavali NK, Agrawal RK. Insights into Structural Basis of Mammalian Mitochondrial Translation. Translation in Mitochondria and Other Organelles. 2013:1–28. [Google Scholar]
- 14.Mears JA, Cannone JJ, Stagg SM, Gutell RR, Agrawal RK, Harvey SC. Modeling a minimal ribosome based on comparative sequence analysis. J Mol Biol. 2002;321:215–34. doi: 10.1016/s0022-2836(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 15.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 16.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 17.Mears JA, Sharma MR, Gutell RR, McCook AS, Richardson PE, Caulfield TR, et al. A structural model for the large subunit of the mammalian mitochondrial ribosome. J Mol Biol. 2006;358:193–212. doi: 10.1016/j.jmb.2006.01.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–9. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, et al. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;505:515–9. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- 20.Kaushal PS, Sharma MR, Booth TM, Haque EM, Tung CS, Sanbonmatsu KY, et al. Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc Natl Acad Sci U S A. 2014;111:7284–9. doi: 10.1073/pnas.1401657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smirnov A, Entelis N, Martin RP, Tarassov I. Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev. 2011;25:1289–305. doi: 10.1101/gad.624711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lightowlers RN, Rozanska A, Chrzanowska-Lightowlers ZM. Mitochondrial protein synthesis: figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation. FEBS Lett. 2014;588:2496–503. doi: 10.1016/j.febslet.2014.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, et al. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;515:283–6. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 24.Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, et al. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346:718–22. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, et al. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem. 2001;276:43958–69. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 26.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001;276:19363–74. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, et al. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J Biol Chem. 2001;276:21724–36. doi: 10.1074/jbc.M100432200. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, et al. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J Biol Chem. 2001;276:33181–95. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 29.Koc EC, Cimen H, Kumcuoglu B, Abu N, Akpinar G, Haque ME, et al. Identification and characterization of CHCHD1, AURKAIP1, and CRIF1 as new members of the mammalian mitochondrial ribosome. Front Physiol. 2013;4:183. doi: 10.3389/fphys.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs--like members of all families, similar but different. Biochim Biophys Acta. 2010;1797:1081–5. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallberg BM, Larsson NG. Making proteins in the powerhouse. Cell metabolism. 2014;20:226–40. doi: 10.1016/j.cmet.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe K. Unique features of animal mitochondrial translation systems. The non- universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:11–39. doi: 10.2183/pjab.86.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien TW. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene. 2002;286:73–9. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien TW, O'Brien BJ, Norman RA. Nuclear MRP genes and mitochondrial disease. Gene. 2005;354:147–51. doi: 10.1016/j.gene.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 35.Rotig A. Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta. 2011;1807:1198–205. doi: 10.1016/j.bbabio.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Pearce S, Nezich CL, Spinazzola A. Mitochondrial diseases: translation matters. Mol Cell Neurosci. 2013;55:1–12. doi: 10.1016/j.mcn.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annual review of genetics. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 38.Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia L, Kaur J, Stuart RA. Mapping of the Saccharomyces cerevisiae Oxa1-mitochondrial ribosome interface and identification of MrpL40, a ribosomal protein in close proximity to Oxa1 and critical for oxidative phosphorylation complex assembly. Eukaryot Cell. 2009;8:1792–802. doi: 10.1128/EC.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gruschke S, Ott M. The polypeptide tunnel exit of the mitochondrial ribosome is tailored to meet the specific requirements of the organelle. Bioessays. 2010;32:1050–1057. doi: 10.1002/bies.201000081. [DOI] [PubMed] [Google Scholar]
- 43.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–30. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 44.Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider HC, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, et al. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature. 1994;371:768–74. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- 46.Pfeffer S, Woellhaf MW, Herrmann JM, Forster F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nature communications. 2015;6:6019. doi: 10.1038/ncomms7019. [DOI] [PubMed] [Google Scholar]
- 47.Roe BA, Wong JFH, Chen EY, Armstrong PW. Sequence analysis of mammalian mitochondrial tRNAs in recombinant DNA. In: Walton AG, editor. Recombinant DNA: Proceedings of the Third Cleveland Symposium on Macromolecules. Cleveland, Ohio: Jun 22–26, 1981. 1981. pp. 167–176. [Google Scholar]
- 48.Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42:7346–57. doi: 10.1093/nar/gku390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wende S, Platzer EG, Juhling F, Putz J, Florentz C, Stadler PF, et al. Biological evidence for the world's smallest tRNAs. Biochimie. 2014;100:151–8. doi: 10.1016/j.biochi.2013.07.034. [DOI] [PubMed] [Google Scholar]
- 50.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. Rna. 2003;9:1345–52. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–8. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 53.Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nature reviews Genetics. 2007;8:711–23. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 54.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–4. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 55.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Putz J, Dupuis B, Sissler M, Florentz C. Mamit-tRNA, a database of mammalian mitochondrial tRNA primary and secondary structures. Rna. 2007;13:1184–90. doi: 10.1261/rna.588407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]