Abstract
Fungus-growing ants (Attini) are part of a complex symbiosis with Basidiomycetous fungi, which the ants cultivate for food, Ascomycetous fungal pathogens (Escovopsis), which parasitize cultivars, and Actinobacteria, which produce antibiotic compounds that suppress pathogen growth. Earlier studies that have characterized the association between attine ants and their bacterial symbionts have employed broad phylogenetic approaches, with conclusions ranging from a diffuse coevolved mutualism to no specificity being reported. However, the geographic mosaic theory of coevolution proposes that coevolved interactions likely occur at a level above local populations but within species. Moreover, the scale of population subdivision is likely to impact coevolutionary dynamics. Here, we describe the population structure of bacteria associated with the attine Apterostigma dentigerum across Central America using multilocus sequence typing (MLST) of six housekeeping genes. The majority (90%) of bacteria that were isolated grouped into a single clade within the genus Pseudonocardia. In contrast to studies that have suggested that Pseudonocardia dispersal is high and therefore unconstrained by ant associations, we found highly structured (FST = 0.39) and dispersal-limited (i.e., significant isolation by distance; r = 0.517, P = .05) populations over even a relatively small scale (e.g., within the Panama Canal Zone). Estimates of recombination versus mutation were uncharacteristically low compared with estimates for free-living Actinobacteria (e.g., ρ/θ = 0.028 in La Selva, Costa Rica), which suggests that recombination is constrained by association with ant hosts. Furthermore, Pseudonocardia population structure was correlated with that of Escovopsis species (r = 0.67, P = .02), supporting the bacteria's role in disease suppression. Overall, the population dynamics of symbiotic Pseudonocardia are more consistent with a specialized mutualistic association than with recently proposed models of low specificity and frequent horizontal acquisition.
Keywords: Apterostigma, Escovopsis, isolation by distance, population structure, Pseudonocardia, symbiosis
Introduction
Discerning how individuals within a species are structured over geographic space is essential to understanding how populations evolve. For example, allopatric speciation (divergence caused by geographic isolation) is typically regarded as the most common mechanism of speciation (Mayr 1963; Lande 1980; Coyne and Orr 2004), and migration can either facilitate or hinder local adaptation (Lenormand 2002; Kawecki and Ebert 2004). Furthermore, the scale of population subdivision plays an important role in shaping coevolutionary dynamics (Lively 1999; Thompson and Cunningham 2002; Thompson 2005, 2009). Within host-symbiont systems, symbiont populations can be structured by hosts (resulting in congruent population structure), although such congruence is more likely when specificity is maintained either through vertical transmission or adaptive mechanisms that determine symbiont selection (Wirth et al. 2005; Vollmer et al. 2010; Zanatta and Wilson 2011). In contrast, horizontal transmission and diffuse associations can decouple host-symbiont populations and constrain opportunities for pairwise coevolution.
The attine (Attini, Formicidae) ant-microbe system represents a complex community of well-characterized symbionts, which provides an opportunity to study the impact of multipartite symbiont associations on population structure. In addition to the ants, this symbiosis also includes mutualistic Basidiomycetous fungi, parasitic Ascomycetous fungi, and antibiotic-producing Actinobacteria (reviewed in Caldera et al. 2009). Attine ants are a monophyletic tribe of approximately 230 ant species, all of which cultivate fungi as their primary food source. The association between attine ants and their fungal cultivars is a textbook example of both obligate mutualism and agriculture. Like many agricultural crops, attine cultivars are susceptible to pathogens, the most common being Escovopsis (Ascomycota, Hypocreales) microfungi that persist as chronic infections within the fungal gardens (Currie et al. 1999a, 2003b; Currie 2001). If Escovopsis infection is not maintained at a low level, the pathogen can rapidly consume the cultivar, thus destroying the entire ant colony (Currie et al. 1999a; Currie 2001). To help combat Escovopsis infection, most attine species rely on antibiotic compounds produced by symbiotic Pseudonocardia (Actinobacteria, Actinomycetales) that grow on specialized regions of the ant cuticle and suppress Escovopsis growth (Currie et al. 1999b, 2003a, 2006; Oh et al. 2009; Cafaro et al. 2011).
Our current understanding of how symbiotic associations might shape population structure within the attine system begins with the ant-cultivar association. On nuptial flights, virgin queens carry in their mouth an inoculum of a fungal cultivar from the mother colony, which they use to start the fungal garden after mating (Hölldobler and Wilson 1990). This vertical transmission of cultivar clones from mother to daughter is believed to have contributed to the broad patterns of phylogenetic congruence between attine ants and their cultivars over evolutionary time (Chapela et al. 1994; Mikheyev et al. 2010). Despite the coupling of cultivar transmission with the reproductive lifecycle of attine queens, horizontal exchange among closely related ant species through cultivar sharing and/or stealing appears to be rampant (Adams et al. 2000; Green et al. 2002; Mikheyev et al. 2006, 2007; Poulsen et al. 2009), and some basal attine species may be capable of using free-living fungi (Mueller et al. 1998; Vo et al. 2009). Horizontal acquisition of cultivars occurs early during colony founding (Poulsen et al. 2009), and mature colonies appear to propagate only one cultivar clone, despite maintaining multiple garden chambers (Mueller et al. 2010b). To date, the geographic population genetic structure of an ant and its cultivar has been described in only one attine, Trachymyrmex septentrionalis (Mikheyev et al. 2008). Although both ants and cultivars exhibited significant isolation by distance (IBD) across the eastern United States (r = 0.23, P = .034, and r = 0.38, P = .003, respectively), there was no correlation between the ant's genetic and the fungal community similarity matrix, which suggests that cultivar population structure is not shaped by transmission with its ant host.
Although opportunities for both vertical and horizontal transmission exist for cultivars, Escovopsis pathogens are horizontally acquired, likely from invertebrates that associate with attine ant colonies, and are absent in the early stages of colony founding (Currie et al. 1999a). Although Escovopsis is acquired horizontally, broad phylogenetic congruence exists between fungal cultivars and Escovopsis, which suggests that additional mechanisms maintain pathogen specificity over evolutionary timescales (Currie et al. 2003b). The adaptive mechanisms governing cultivar-Escovopsis specificity are best understood in the basal attine genus Apterostigma. Among Apterostigma species, phylotype-specific patterns of Escovopsis attraction to cultivars through chemotaxis and cultivar resistance to Escovopsis reflect patterns of phylogenetic congruence (Gerardo et al. 2006a, b). A population-level study of Escovopsis-cultivar dynamics in the ant species Apterostigma dentigerum across Central America, however, found that cultivar resistance is ineffective against the most common pathogen phylotype (brown Escovopsis) of A. dentigerum, regardless of genotype or location (Gerardo and Caldera 2007). Moreover, although both cultivars and brown Escovopsis showed significant IBD (r = 0.34, P = .04, and r = 0.38, P = .03, respectively), their population genetic structure was not congruent, which indicates a lack of pairwise specificity at the population level. It remains possible that the seemingly labile association between A. dentigerum cultivars and brown Escovopsis persists because the role of specialization against Escovopsis is maintained by Pseudonocardia.
Less is known about the transmission and population dynamics of attine-associated Pseudonocardia than is known about cultivars or Escovopsis. As is the case for cultivars, a clear mechanism for vertical transmission of Pseudonocardia exists, but the frequency of horizontal acquisition or recombination among strains remains ambiguous. Virgin queens harbor a visible inoculum of Pseudonocardia on specialized cuticular structures before departing on mating flights (Currie et al. 2006); therefore, it is not necessary to acquire novel strains during colony founding. Doing so would also require the new bacteria to outcompete the original strain. Studies of Acromyrmex-associated Pseudonocardia have recovered only one genotype per colony (Poulsen et al. 2005), demonstrated sophisticated strain-level recognition by the ants (Zhang et al. 2007), and shown that acquisition from nonmaternal colonies results in less abundant bacterial cover (Armitage et al. 2011). This evidence together suggests that the ants transmit and maintain the same clone present during colony founding. Phylogenetic studies have shown a degree of congruence between attines and Pseudonocardia indicative of vertical transmission, but incongruences have also suggested that horizontal switches are perhaps more frequent than cultivar switches over evolutionary time (Cafaro and Currie 2005; Cafaro et al. 2011). Opportunity for horizontal acquisition could exist through contact with males during mating; however, Pseudonocardia are often absent or in low abundance on males, and males are not present during colony founding, so they are not likely to be the primary vector. Contact with the soil, foraging material, and other foraging fungus-growing ants may also provide opportunities for horizontal acquisition. Recently, a new model of attine-Pseudonocardia association (termed the acquisition model) has emerged, which argues that the ant-Pseudonocardia symbiosis is substantially less specialized than previously thought (Kost et al. 2007; Haeder et al. 2009; Sen et al. 2009) and may not be a mutualistic association but rather encompasses a range of associations that include parasitism (Mueller et al. 2008, 2010a). More specifically, the acquisition model emphasizes limited vertical transmission of Pseudonocardia across many ant generations and low potential (or absence) of Pseudonocardia-Escovopsis coevolution and specificity.
Studies both in support of and in conflict with the acquisition model have drawn heavily on phylogenetic and/or across-species approaches among many ant species (Cafaro and Currie 2005; Currie et al. 2006; Mueller et al. 2008, 2010a; Sen et al. 2009; Cafaro et al. 2011). The geographic mosaic concept of coevolution, however, posits that coevolved interactions likely occur at a scale above local populations, but below species (i.e. at the geographic population scale; Thompson 1994). Consequently, studies at this scale are likely to yield important insights into attine-microbe coevolution. To date, the geographic population structure of attine-associated Pseudonocardia has only been described in Trachymyrmex serptentrionalis (Mikheyev et al. 2008). Across the eastern United States, Pseudonocardia had little population structure at the EF-Tu locus and genetic distances were not correlated with the ant host, which did show substantial geographic structure. This led the authors to conclude that the bacteria's dispersal capacity is high and not constrained by association with the ants, although bacterial population structure might be shaped by environmental factors and/or interactions with Escovopsis. Currently, most genetic-based studies of symbiotic Pseudonocardia (Cafaro and Currie 2005; Poulsen et al. 2005; Zhang et al. 2007; Mikheyev et al. 2008; Mueller et al. 2008, 2010b; Sen et al. 2009), including the only description of Pseudonocardia population structure, have been limited by the use of a single, well-conserved housekeeping gene. Studies of microbial geographic distributions and niche evolution, however, strongly caution against interpretations drawn from a single locus, because evidence for finer geographic population structure tends to emerge as additional loci and molecular markers are added (Cho and Tiedje 2000; Whitaker et al. 2003; Hedlund and Staley 2004). Therefore, it remains possible that multilocus analysis of ant-associated Pseudonocardia may expose a finer degree of geographic structure yet unseen.
Here, we describe the population genetic structure of A. dentigerum–associated Pseudonocardia in Panama and Costa Rica using loci from six different housekeeping genes (determined with use of MLST). We use both phylogenetic and population genetic analyses to answer the following questions: (1) Are populations geographically structured within Central America? (2) Do populations conform to isolation by distance, and at what scale? (3) Are Pseudonocardia populations recombining, and to what extent? (4) Is Pseudonocardia population structure correlated with the population structure of the brown Escovopsis phylotype and/or cultivars? Finally, we discuss our findings in the context of differing models of attine-Pseudonocardia association.
Material and Methods
Collection of Apterostigma dentigerum
We collected colonies of A. dentigerum from La Selva Biological Station (LS), Costa Rica, during October 2006 and July 2007 and from seven locations in Panama during June 2008 (fig. 1A, 1B). Within Panama, sampling was greatest around the Panama Canal Zone, including the following locations: Gamboa Forest (GAM), Pipeline Road (PLR), and Barro Colorado Island (BCI). Collections on BCI were divided roughly into colonies collected in the northern and southern regions of the island, and collections along PLR were divided into colonies collected along the banks of rivers intersecting the northern part of the road (rivers Agua Salud, Guacharo, Pelon, and Macho) and the southern part of the road (rivers Limbo, Seda, Frijoles, and Frijolito). We collected additional ant colonies on the peninsulas surrounding BCI and in Bocas del Toro province (BT), near the Costa Rican border.
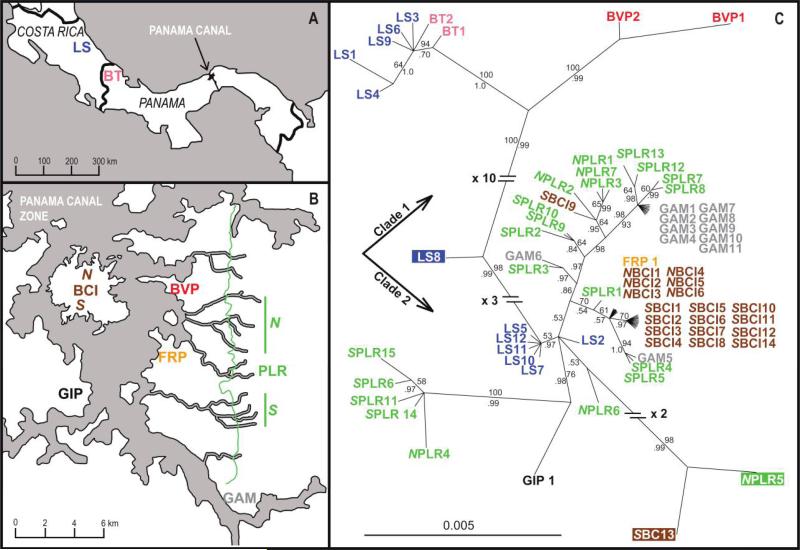
Figure 1.
Collection locations (A, B; see table 1 for definitions) and phylogenetic construction (C) of symbiotic Pseudonocardia. The phylogeny was based on a six-locus concatenated alignment and used Bayesian methods. The scale bar corresponds to 0.005 substitutions per site. Maximum likelihood methods yielded trees with similar topologies. Bayesian posterior probabilities are reported below branches, and bootstrap support values from maximum likelihood are reported above branches. Highlighted isolates are recombinant.
Symbiont Isolation
To obtain pure cultures of symbiotic Actinobacteria, we followed the methods of Poulsen et al. (2005) for Acromyrmex ants, but with the following modifications specific to A. dentigerum. Before scraping bacteria off of the ant cuticle, we first removed the head and forelegs from workers to expose the mesosternal lobe, where the bacteria are localized in A. dentigerum (Currie et al. 2006). Depending on the number of workers in a given nest, we scraped the mesosternal lobes of one to four ants. An additional 16 bacterial isolates collected during 2001–2003 by researchers from the University of Texas at Austin and the University of Kansas were included. These additional isolates were stored in 2-mL tubes at −80°C and contained either frozen cells or extracted DNA.
DNA Extraction and Sequencing
DNA was extracted from pure cultures following the methods described in Poulsen et al. (2005). To amplify the 16S rDNA gene, we used the polymerase chain reaction (PCR) primer sets 243F and 1378R (Heuer et al. 1997). MacVector 10.6 was used to design PCR primers for an additional five loci comprised of partial sequences from the following housekeeping genes: atpD, dnaA, EF-Tu, gyrB, and rpoB (table 2). DNA sequences for these five additional genes were extracted from a Pseudonocardia draft genome that was isolated from A. dentigerum (C. R. Currie, unpublished data). Amplifications were conducted using standard PCR methods, and positive amplifications were sequenced at the University of Wisconsin–Madison Biotechnology Center using Sanger sequencing technology. DNA sequence chromatograms were edited using Sequencher 4.6 (Gene Codes; Ann Arbor, MI). Because the 16S rDNA PCR primers are specific to Actinobacteria (Heuer et al. 1997), whereas primers for the additional five loci are potentially specific to a Pseudonocardia strain associated with A. dentigerum, isolates were first sequenced at the16S locus. Samples that were named Pseudonocardia species according to searches of the Basic Local Alignment Search Tool (BLAST) database (available at http://www.blast.ncbi.nlm.nih.gov) were subsequently genotyped for the remaining five housekeeping genes. Of the isolates identified as Pseudonocardia according to BLAST searches (i.e., those with >98% sequence identity), 71 were sequenced for all six loci (table 1). MLST sequences have been deposited in GenBank (accession numbers HQ540730–HQ541155).
Table 2.
Housekeeping genes used in multilocus genotyping, including polymerase chain reaction primer pairs, dN/dS ratios, and Tajima's D
| Target | Forward primer | Reverse primer | Size (bp) | dN/dS | Tajima's D |
|---|---|---|---|---|---|
| 16S rRNA | GGATGAGCCCGCGGCCTA | CGGTGTGTACAAGGCCCGGGAACG | 1,175 | ... | ... |
| atpD | AGGAGATGATCCAGCGTGTC | GTTCTTCTCCAGGTCGTCCA | 890 | .0313 | –.483 |
| dnaA | GTCGACGACATCCAGTTCCT | CTCGTTGCGGATCTTCTTGT | 645 | 0 | 1.038 |
| EF-Tu | CACGACAAGTACCCGAACCT | AGTTGTTGAAGAACGGGGTG | 880 | .1959 | .090 |
| gyrB | AGCTACACGCTGGAGACGA | GGTGATGATCGACTGGACCT | 999 | .1916 | .479 |
| rpoB | AACAAGAAGCTCGGTCTGGA | CATGACGGTGATGTAGTCGG | 1,038 | .127 | .274 |
Table 1.
Location and number of Pseudonocardia symbionts isolated from the mesosternal lobes of Apterostigma dentigerum and successfully amplified with polymerase chain reaction at six loci
| Location | Abbreviation | Distancea | No. isolates (n = 71) |
|---|---|---|---|
| La Selva Biological Station, Costa Rica | LS | 482 | 12 |
| Bocas del Toro province, Panama | BT | 273 | 2 |
| Gigante Peninsula, Panama | GIP | 6.3 | 1 |
| Frijoles Peninsula, Panama | FRP | 6.8 | 1 |
| Buena Vista Peninsula, Panama | BVP | 4.7 | 2 |
| North Barro Colorado Island, Panama | NBCI | 0 | 6 |
| South Barro Colorado Island, Panama | SBCI | 3.5 | 14 |
| Gamboa Forest, Panama | GAM | 17.9 | 11 |
| North Pipeline Road, Panama | NPLR | 7 | 7 |
| South Pipeline Road, Panama | SPLR | 21.9 | 15 |
Distance in kilometers from North Barro Colorado Island.
Phylogenetic Analysis
Nucleotide sequences were aligned using CLUSTALX 2.0.11 (Larkin et al. 2007), and models of nucleotide substitution were determined using jMODELTEST (Guindon and Gascuel 2003; Posada 2008). We constructed phylogenetic trees independently for each of six genes, and we also used a concatenation of all genes. Single-locus phylogenetic relationships were determined using the maximum-likelihood (ML) criterion, and the concatenated alignment used both ML and Bayesian methods implemented in the programs GARLI 0.951 (Zwickl 2006; available at http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html) and MrBayes 3 (Ronquist and Huelsenbeck 2003). To ensure that parameter space was thoroughly explored, MrBayes was run four separate times. Each run used four heated chains, and each chain featured 4,000,000 generations with sampling every 100 generations, following a burn-in period of 100,000 generations. GARLI was run 10 times using default parameters. One hundred bootstrap replicates were generated with the generation threshold for topology termination (genthreshfortopoterm) parameter set to 5,000, as recommended by the GARLI manual. Finally, to confirm that phylogenetic inferences were not impacted by recombination among isolates, we constructed additional phylograms in ClonalFrame (Didelot and Falush 2007), which removes recombination, and we visualized multiple phylograms using SplitsTree (Huson and Bryant 2006).
Population Genetic Analyses
To test whether the housekeeping genes are under selection, we compared synonymous and nonsynonymous substitutions by calculating overall dN/dS ratios and Tajima's D for each translated locus using MEGA (Kumar et al. 2008). Significance was assessed with a Z-test that used 500 bootstraps and the Nei-Gojobori method. To test for significant population structure among geographically defined regions, we calculated the overall fixation index, FST (Weir and Cockerham 1984) using ARLEQUIN 3.11 (Excoffier et al. 2005). Genetic distances among populations were calculated using FST and Nei's net DA (Nei et al. 1983). Genetic diversity within populations was assessed using π (mean number of nucleotide differences). The P values represent the proportion of permutations leading to a value larger than or equal to the observed value.
Our tests for IBD involved exploring correlation between matrices of genetic distance (FST/(1 – FST)) and natural log (geographic distance) and were conducted using a Mantel test with a Pearson's product moment correlation coefficient (Slatkin 1993). Geographic distances were determined using Mapsource 6. Because of low sampling, the following locations were excluded from IBD and FST analyses: BT, BVP, and FRP (see table 1).
Finally, to examine whether Pseudonocardia population genetic structure is geographically congruent with A. dentigerum fungal symbionts, we obtained pairwise ΦST (an analog of FST; Excoffier 2001) values for cultivars and brown Escovopsis among the following populations (with corresponding sample sizes): BCI = 10, GAM = 7, PLR = 12, and LS = 9. Fungal isolates were genotyped by N. Gerardo using amplified fragment-length polymorphism genotyping in an earlier study of cultivar-Escovopsis interactions in A. dentigerum (Gerardo and Caldera 2007). Isolation of fungal symbionts was conducted before this study, so fungal and bacterial isolates derive from different individual ant colonies, although they are from the same geographic locations and host ant species. Correlation between the pairwise matrices of Pseudonocardia FST values and ΦST in fungal symbionts was also assessed with a Mantel test.
Tests for Recombination
To assess whether Pseudonocardia populations are clonal or recombining, we began with the a priori assumption that populations are clonal and subsequently tested for evidence of recombination events. Recombination was initially examined by looking for phylogenetic incongruence among six loci, in addition to performing tests for recombination using two methods implemented in the Recombination Detection Program (RDP) 3.44 software package (Heath et al. 2006): RDP (Martin and Rybicki 2000) and GENECONV (Padidam et al. 1999). Estimates of the population recombination parameter ρ = 2Ner and the population mutation rate Waterson's θ = 2Neμ were determined using the coalescent method LDhat (McVean et al. 2002). LDhat simulations used 1,000,000 Markov chain Monte Carlo (MCMC) iterations after a burn-in of 100,000. As recommended by the RDP documentation, we conducted an initial estimate of ρ and used the output as the starting ρ for subsequent simulations. Employing a range of block penalties (5–50) had little impact on ρ and θ estimates. We calculated ρ and θ both within geographic regions and incorporating all isolates.
Results
Phylogenetic Inferences
A concatenated phylogeny across six loci revealed multiple, well-supported clades of A. dentigerum–associated Pseudonocardia that corresponded to geographic locations (fig. 1). ClonalFrame analyses confirmed that topologies resulting from phylogenetic reconstructions were not impacted by recombination (figs. A1 and A2, available online). Bayesian and ML approaches yielded virtually identical topologies. Isolates grouped into two major clades (clades 1 and 2). Clade 1 consisted of two subclades, the first of which contained five isolates from LS, Costa Rica, and the only two isolates from BT, Panama, which is located near the border of Costa Rica. Clade 1 also contained a subclade comprised of the only two isolates from BVP in the Panama Canal Zone. With the exception of the two isolates from BVP, most isolates collected in the Panama Canal Zone were contained in clade 2. Although approximately half of the Costa Rican isolates (from LS) were also found in clade 2, the majority of these isolates formed a single group. One isolate (LS 8) fell between clade 1 and clade 2 because of a recombination event among isolates from the two clades (see “Recombination”). Within the Panama Canal Zone, the majority of isolates from SBCI, NBCI, and GAM (see table 1) formed distinct clades. In contrast, isolates collected along PLR formed multiple subclades throughout clade 1 and thus appeared to be less geographically distinct. Many of the subclades were, however, limited to either the northern or southern regions of PLR. All six locus-specific phylogenies included isolates from clade 1 and clade 2 (fig. 2), but each phylogeny individually lacked variation to capture many of the geographically distinct subclades of the concatenated tree. The BVP clade was recovered in four loci, and a PLR specific clade was recovered in five loci.
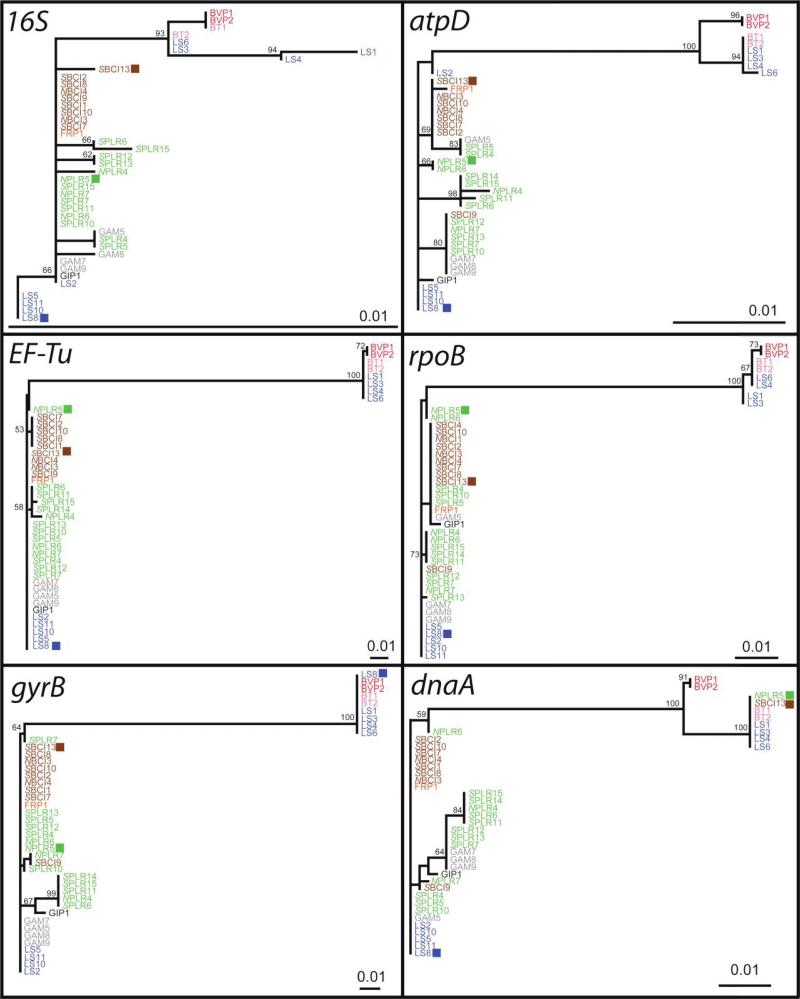
Figure 2.
Phylogenetic analysis (maximum likelihood) of partial sequences from each of six housekeeping genes. The scale bar corresponds to 0.01 substitutions per site, and values at nodes report percentage bootstrap support. Topologies were largely congruent with the exception of three isolates that showed incongruence across clade 1 and clade 2, which suggests that they are recombinant. Note that 29 isolates, none of which showed incongruence, were removed for visual purposes. LS8 shows incongruence within gyrB, and isolates NPLR5 and SBCI13 show incongruence in dnaA. These isolates were also identified as recombinant using the Recombination Detection Program 3.44 software package and GENECONV (see “Material and Methods”); however, the overall recombination to mutation rate was marginal (ρ/θ = 0.09).
Population Genetic Structure
All translated housekeeping genes showed dN/dS ratios below 1 (the mean dN/dS ratio was 0.109; table 2) and thus are not under positive selection. Consistent with conserved housekeeping genes, the results of Z-tests were highly significant for purifying selection (dN < dS). Tajima's D values were relatively low, ranging from −0.48 to 1.04, further showing that positive selection is not occurring on these loci. Because of the high divergence between clade 1 and clade 2, it is possible that these groups may reflect species level differences, so population genetic analyses were restricted to clade 2. The overall FST = 0.390 (P < .0001) revealed significant structure among the populations LS, BCI, GAM, and PLR. A second FST further dividing BCI and PLR into northern and southern sampling sites returned a similar level of structure (FST = 0.391, P < .0001) across all populations, with significant pairwise FST between northern and southern BCI (NBCI-SBCI FST = 0.367, P < .05; table 3). In contrast, PLR was not structured between north-south sampling sites (NPLR-SPLR FST = 0.0297, P > .05; table 3). Population specific genetic diversity (π) and pairwise Nei's genetic distances DA are also presented in table 3.
Table 3.
Pairwise population genetic distances
| Location |
||||||
|---|---|---|---|---|---|---|
| Location | NBCI | SBCI | GAM | NPLR | SPLR | LS |
| NBCI | <.001 | .846** | 7.036** | 6.881** | 3.417** | 6.667** |
| SBCI | .367** | 1.692 | 6.931** | 6.617** | 3.562** | 6.897** |
| GAM | .698** | .713** | 4.109 | 2.502** | 1.281* | 6.612** |
| NPLR | .329** | .472** | .221** | 21.095 | .456 | 3.833** |
| SPLR | .204* | .344** | .132** | .030 | 15.611 | 4.083** |
Note: Pairwise FST is presented below the diagonal, DA is presented above the diagonal, and mean pairwise genetic differences (π) are presented in the diagonal. For abbreviations of locations, see table 1.
P < .1.
P < .05.
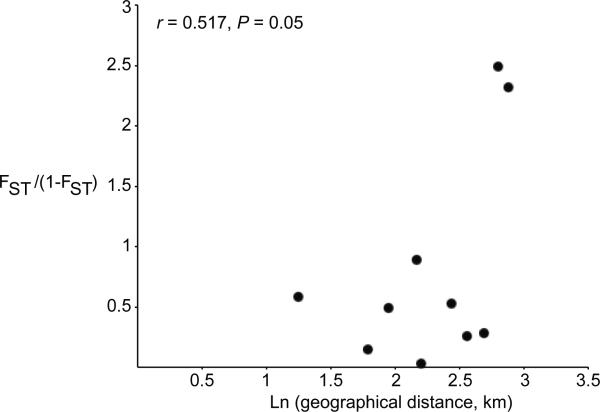
Mantel tests showed significant IBD across Costa Rica and Panama (r = 0.4711, P = .03); however, because the LS population is the only well-sampled population outside of the Panama Canal Zone, the sampling scheme does not thoroughly address IBD across the two countries. Thus, we conducted a subsequent test for IBD at a smaller scale (i.e., within the Panama Canal Zone) that also showed a significant correlation between genetic and geographic distances among populations (r = 0.517, P = .05). Figure 3 presents the relationship between genetic divergence (FST) and geographic distance (km) in the Panama Canal Zone.
Figure 3.
Isolation by distance in symbiotic Pseudonocardia in the Panama Canal Zone. Correlation between genetic and geographic distance was assessed using a Mantel test.
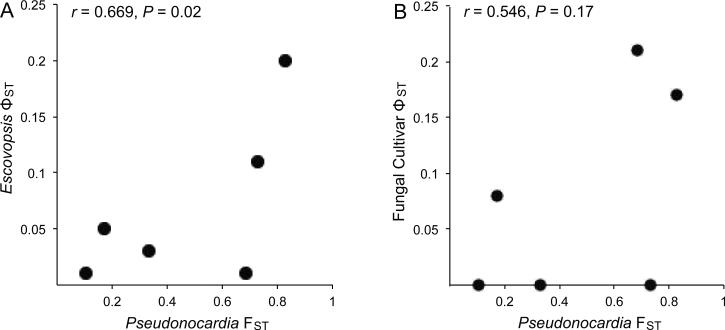
Pseudonocardia population genetic structure was congruent with the genetic structure of brown Escovopsis (r = 0.669, P = .02; fig. 4A). In contrast, there was no significant association between fungal cultivars and Pseudonocardia (r = 0.546, P = .17; fig. 4B).
Figure 4.
Association between the population genetic structure of symbiotic, antibiotic-producing Pseudonocardia associated with Apterostigma dentigerum in Central America (La Selva Biological Station, Costa Rica [LS-CR]; Barro Colorado Island, Panama [BCI-PA]; Pipeline Road, Panama [PLR-PA]; and Gamboa Forest, Panama [GAM-PA]) and two fungal symbionts (Escovopsis-pathogen and cultivar-mutualist). Population structure was congruent between Pseudonocardia and the Escovopsis pathogens they inhibit (A). No significant association was detected between Pseudonocardia and the cultivars that serve as the ant food source (B). Significance was assessed using a Mantel test. Escovopsis and cultivar ΦST values were taken from Gerardo and Caldera (2007).
Recombination
The topologies of the phylogenies constructed for each of the six loci were largely congruent, which suggests that recombination is relatively infrequent among Pseudonocardia isolates (fig. 2). Three of 71 isolates (LS8, NPLR5, and SBCI13), however, showed incongruence between clade 1 and clade 2. LS8 grouped with clade 2 across five loci but fell within clade 1 for the gyrB locus. Similarly, NPLR5 and SBCI13 clustered with clade 2 across five loci but associated with clade 1 within the dnaA phylogeny, which indicated that some recombination events have taken place. The same three recombination events were identified using the RDP and GENECONV methods (P < 1 × 10−22).
The population recombination rate (ρ = 2Ner) across all isolates was estimated as 5.069 (95% confidence interval, 3.953–6.273) with a per-site estimate of 9.886 × 10−4 (7.712 × 10−4 – 1.224 × 10−3). The mutation rate, Watterson's θ = 2Ne μ, was 56.45, with a per-site rate of 1.1 × 10−2. Region-specific estimates of ρ and θ are presented in table 4. We do not report estimates for BCI because the 95% confidence intervals spanned five orders of magnitude.
Table 4.
Population-scaled recombination rates, mutation rates, and r/u ratios
| Region | ρ | 95% confidence interval | θ | ρ/θ |
|---|---|---|---|---|
| All | 5.069 | 3.953–6.273 | 56.45 | .0898 |
| LS | 2.096 | 2.018–2.295 | 74.435 | .0280 |
| GAM | 2.079 | .213–4.921 | 9.273 | .224 |
| PLR | 8.369 | 3.971–16.918 | 22.48 | .372 |
Note: For abbreviations of locations, see table 1.
Discussion
In contrast to previous studies that have suggested that Pseudonocardia dispersal is far greater than that of its ant host (Mikheyev et al. 2008; Mueller et al. 2008, 2010a) we found highly structured and dispersal-limited populations across Central America and within the Panama Canal Zone. Estimates of recombination versus mutation were uncharacteristically low compared with those for free-living Actinobacteria (Pérez-Losada et al. 2006; Deletoile et al. 2010; Doroghazi and Buckley 2010), which suggests that recombination is constrained by association with its ant host. Further, Pseudonocardia population structure was correlated with Escovopsis but not with cultivar across Central America, which supports the bacteria's role in disease suppression. These findings may reflect the geographic mosaic coevolutionary concept that coevolved dynamics occur at a geographic scale above local populations but below the species level (Thompson 1994).
Geographic Structure
Despite the extraordinary dispersal capacity of many bacterial species, A. dentigerum–associated Pseudonocardia populations are both highly structured and dispersal-limited over even a relatively small geographic space. Multilocus phylogenetic constructions (fig. 1) revealed several clades that correspond to specific locations (e.g., GAM and BCI in the Panama Canal Zone), and the overall FST = 0.39 showed substantial structure. In addition, we found a significant correlation between genetic distance and geographic distance over even the relatively small geographic scale of the Panama Canal Zone (r = 0.517, P = .05).
Given the substantial amount of structure in Pseudonocardia, it seems likely that host associations play a role in shaping the bacteria's populations. A previous description of Pseudonocardia geographic structure in the attine Trachymyrmex septentrionalis ruled out vertical transmission with ant hosts as a likely force structuring populations but hypothesized that interactions with Escovopsis might structure populations (Mikheyev et al. 2008). A single locus (EF-Tu) analysis revealed little geographic structure and slight IBD in Pseudonocardia across the eastern United States (whereas the ants showed a clear east-west divide), which led the authors to conclude that the bacteria are likely capable of existence outside of the symbiosis or are better dispersers than the ants. That finding was later cited in support of the recruitment hypothesis (Mueller et al. 2008) as evidence that bacterial symbionts are not closely tied to their ant hosts.
Although it does appear that Escovopsis interactions do shape Pseudonocardia genetic structure (fig. 4), our findings suggest that the lack of geographic structure in T. septentrionalis–associated Pseudonocardia was attributable to low variation in the EF-Tu locus and not to greater dispersal by the bacteria. Our EF-Tu locus alone also recovered little phylogeographic structure (as was the case with each individual locus; fig. 2); however, the concatenation of all six loci revealed substantial geographic structure and significant IBD (r = 0.471, P = .03). Thus, it is likely that additional loci will reveal additional structure in T. septentrionalis–associated Pseudonocardia. Consequently, the assumption that Pseudonocardia dispersal is not tied to ant hosts should be revisited. Future work should incorporate the population structure of A. dentigerum to address this further. We now know that the scale of Pseudonocardia population subdivision is finer than that of Escovopsis and cultivars and thus perhaps more closely mirrors that of A. dentigerum, whose dispersal capacity is likely under 500 m per year, based on flight distances of the much larger and more powerful leaf-cutter species Acromyrmex octospinosus (Mikheyev 2008).
Pseudonocardia and Fungal Symbionts
It appears that interactions with Escovopsis play a role in shaping Pseudonocardia population structure; there was a significant correlation between the population structures of Pseudonocardia and brown Escovopsis (r = 0.669, P = .02; fig. 4). Although congruence between Pseudonocardia and cultivars was similarly correlated (r = 0.546, P = .17), the correlation lacked significance, so it remains possible that the lack of difference is the result of a Type II error. An earlier study of cultivar-Escovopsis (brown) population dynamics in A. dentigerum found no congruence between the population structures of the two fungal symbionts, and bioassay experiments demonstrated that the secondary metabolites of fungal cultivars are not effective in combating brown Escovopsis, regardless of genotype or geographic location (Gerardo and Caldera 2007). This finding was interesting given that cultivar secondary metabolites are known to inhibit other Escovopsis phylotypes that associate with different, sympatric Apterostigma species (Gerardo et al. 2006a). Together, these findings illustrate two important points about attine-microbe interactions: (1) The “indirect” association between Pseudonocardia and Escovopsis is more specialized than the “direct” cultivar-Escovopsis interaction at the population level, and (2) this pairwise specialization arose within the seemingly diffuse associations attines have with multiple cultivar and Escovopsis phylotypes (Gerardo et al. 2006b; Mikheyev et al. 2007; Taerum et al. 2007). Although this seems counterintuitive, the emergence of specialized interactions within seemingly diffuse associations is consistent with coevolutionary theory (Thompson 1994), and we can envision how these dynamics occur in nature. Attine colonies encounter several Escovopsis phylotypes. The cultivar is capable of suppressing many of these phylotypes; however, in the case of A. dentigerum cultivars, they do not suppress brown Escovopsis (Gerardo et al. 2006a). In this scenario, there would be strong selection for colonies whose Actinobacteria inhibit the brown phylotype. This selection for Escovopsis resistance could then be a force structuring bacterial populations (e.g., Buckling and Rainey 2002). Indeed, the correlated population structure between the fungal pathogens and bacterial symbionts implies that such dynamics are occurring between Pseudonocardia and Escovopsis. To better understand these interactions, future work should include bioassay experiments to detect coadaptation by Pseudonocardia to Escovopsis, perhaps through local adaptation experiments across the geographically structured populations recovered here.
Recombination
Recombination rates in bacterial populations can range from highly clonal to reaching linkage equilibrium (Spratt et al. 1999, 2001). If Pseudonocardia dispersal is restricted by vertical transmission with ant hosts, one predicts restricted opportunities for recombination with other strains. This restriction would be consistent with mutualism evolutionary theory, which predicts that hosts are selected to prevent mixing of genetically different symbionts when competition among symbionts negatively impacts the mutualism (Hoekstra 1987; Frank 1996; Herre et al. 1999; Poulsen and Boomsma 2005). Indeed, such antagonistic interactions between Pseudonocardia lineages have been demonstrated in leaf-cutter ants (Poulsen et al. 2007) and would likely have similar impacts in Apterostigma colonies. Vertical transmission also aligns host-symbiont interests, because both partners benefit from successful host transmission, in addition to ensuring that offspring initially acquire a single symbiont genotype (Ewald 1987; Bull and Rice 1991; Frank 1996, 2003; Herre et al. 1999). Consistent with these predictions, A. dentigerum–associated Pseudonocardia showed restricted recombination. Only three recombination events were detected among 71 isolates and across six loci (fig. 2 and RDP/GENCOVE results). The low ratio of estimated recombination ρ = 5.065 to mutation θ = 56.43 (ρ/θ = 0.0898) across all isolates also indicates mutation occurs more frequently than recombination. Within regions, LS provided the most robust estimates of recombination (because of higher sequence variation at that site) and returned an even lower ρ/θ ratio (0.028). These ρ/θ estimates are also very low in comparison to those for other bacterial species. Among 19 non-Actinobacteria species for which we could obtain estimates of ρ/θ, only one species had lower recombination ρ/θ (Pérez-Losada et al. 2006; de Bakker et al. 2008; Yan et al. 2008; Deletoile et al. 2010; Doroghazi and Buckley 2010). Unfortunately, recent surveys of recombination rates in bacteria have included little or no Actinobacteria, so direct comparisons within this phylum are limited (Pérez-Losada et al. 2006; Vos and Didelot 2009). Nonetheless, among Actinobacteria species in the genera Bifidobacterium and Streptomyces, ρ/θ values were either one or (in most cases) two orders of magnitude higher than they were for Pseudonocardia (Deletoile et al. 2010; Doroghazi and Buckley 2010). Overall, Pseudonocardia recombination is substantially restricted compared with that of other bacterial species.
At this time, we can only conjecture that this restriction is attributable to association with ant host, because we do not have estimates of recombination in free-living Pseudonocardia. We can, however, draw useful comparisons with free-living Streptomyces Actinobacteria, because ρ/θ calculations used the same six loci as were used in our study (Doroghazi and Buckley 2010). Moreover, this genus has been suggested as an additional associate of attines under the acquisition model (Kost et al. 2007; Haeder et al. 2009). The acquisition model posits that Streptomyces bacteria could be acquired regularly from the environment (as could be the case for other groups, such as Pseudonocardia), and thus recombination rates among ant-associated isolates would mirror those among free-living populations. Interestingly, along the spectrum of recombination, Streptomyces and A. dentigerum–associated Pseudonocardia are on opposite ends; free-living Streptomyces reaches linkage equilibrium. To explain this difference under the acquisition model, free-living Pseudonocardia populations are predicted to have a similarly low ρ/θ ratio. Alternatively, it is also possible that Streptomyces-attine associations are transient, whereas Pseudonocardia associations are specialized. We could not describe the population genetic structure of A. dentigerum–associated Streptomyces here, because we only obtained one isolate, further suggesting that attine-Streptomyces associations are only transient.
Finally, the rarity of recombination could be driven by a lack of genotype diversity within individual ant nests, thus masking the detection of recombination. The dominance of particular Pseudonocardia genotypes recovered here in Apterostigma, and a previous study in Acromyrmex leaf-cutter ants that found only one genotype per ant nest lends credence to this idea (Poulsen et al. 2005). However, if the low rates of recombination were indeed caused by low detection due to low sequence diversity, that bottleneck in diversity would arise due to restricted transmission imposed by association with hosts and would remain inconsistent with the acquisition model.
Reexamining Horizontal Acquisition: Methodological Considerations
Support for the acquisition model stems, in large part, from the observation that ant-associated 16S rDNA Pseudonocardia genotypes are often identical to cosmopolitan isolates, implying they are horizontally acquired from the environment and not limited to dispersal by their ant hosts. For example, Mueller et al. (2008) and Mueller et al. (2010a) showed that ant-associated genotypes have close affinities with free-living Pseudonocardia, including isolates from the soil, plants, geographically distant locations, and disparate environments, such as marine sediment near China, industrial sludge from France, Panamanian attine ants, and Argentinean attine ants. Given the lack of sequence variation in 16S recovered here (fig. 2) and what is known about the low level of variation in 16S (i.e., that it should not be used for identifications below the genus level; Cohan 2006; Staley 2006), it is not surprising that ant-associated genotypes cluster with environmental isolates. These affinities, however, may not reflect exchange with the environment but, rather, may indicate lower phylogenetic resolution than is necessary to detect specificity. Additional genomic analyses could reveal higher specificity, just as our multilocus analysis revealed geographic structure not detectable with a single locus. A recent two-locus (16S and EF-Tu) phylogenetic study of attine-associated Pseudonocardia (Cafaro et al. 2011) has revealed higher specificity to attine lineages than was found in the single-locus studies that support the recruitment hypothesis (Mueller et al. 2008, 2010a). Overall, these differences illustrate the importance of genome-wide analyses for both phylogenetic and population genetic analyses.
Just as single versus multilocus analyses have lead to different conclusions about attine-Pseudoncoardia specificity, different microbial isolation techniques have also led to different conclusions. Studies that have detected diverse Actinobacteria genera (e.g., Streptomyces) in addition to Pseudonocardia have argued that the presence of these additional bacteria negates specificity in the ant-Pseudonocardia association (Kost et al. 2007; Mueller et al. 2008). In this study, 90% of Actinobacteria isolated from the mesosternal lobes of A. dentigerum were Pseudonocardia, and we take this strong bias as evidence that attines maintain a specialized association with this lineage. Attines house their bacterial symbionts on specialized, genus-specific, locations on the cuticle, and these sites often contain crypts and glands that likely facilitate bacterial growth (Currie et al. 2006). Actinobacteria are highly concentrated at these cuticular sites, such that they are visible to the naked eye, and these high concentrations likely facilitate the accumulation of antibiotic compounds. Here, we obtained isolates from the mesosternal lobe because it is the site of visibly high concentrations of Actinobacteria in Apterostigma. Studies that did not find a similar bias toward Pseudonocardia, however, used less targeted approaches. For example, the original study that proposed the acquisition model isolated Actinobacteria bacteria by smearing the entire ventral side of Acromyrmex ants across soy agar plates (Kost et al. 2007), although the specialized crypt and gland cells are concentrated on the propleural plate in Acromyrmex species (Currie et al. 2006). A subsequent phylogenetic study in support of the acquisition model did not isolate Actinobacteria from the ant's cuticle but instead used the infrabuccal pellets and fungus gardens of Atta texana (Mueller et al. 2008).
An additional argument against the specificity of the attine-Pseudonocardia association includes the potential for laboratory-specific biases in the isolation of particular Pseudonocardia lineages, which have created false phylo-genetic clusters that imply specificity (Mueller et al. 2008, 2010a). In consideration of these potential biases, we incorporated Pseudonocardia isolates that were (1) cultured by different individuals, (2) cultured at different locations, and (3) PCR-amplified and sequenced at different facilities. None of these factors determined the phylogenetic or population genetic placement of isolates; thus, laboratory-specific biases have not impacted this study.
Conclusions
Since the discovery of attine-associated bacteria over a decade ago, several studies have examined the coevolutionary dynamics between symbiotic Pseudonocardia and their ant host, and over the past 5 years, support for a model of association lacking pairwise specificity has grown. Thus far, these conclusions have been drawn with little to no information about the population-level dynamics of Pseudonocardia. Our multilocus population genetic analysis of Apterostigma dentigerum–associated Pseudonocardia shows that the geographic scale of population subdivision is smaller than that of fungal symbionts and overturns previous ideas that Pseudonocardia dispersal is greater than that of its ant host. Uncharacteristically low recombination rates suggest that horizontal interactions are restricted by a symbiotic lifestyle, which is consistent with coevolutionary theory for mutualistic associations. Finally, correlated population structure with the Escovopsis phylotype found most commonly in A. dentigerum gardens suggests that Pseudonocardia population structure is shaped, in part, by its role in disease suppression. Our findings support the geographic mosaic of coevolution concept that coevolved dynamics operate within species but across local populations. Our findings have helped identify local populations and define the geographic scale at which attinemicobe dynamics occur. Future studies of attine-microbe coevolution should seek to operate at this scale.
Acknowledgments
We are grateful to J. J. Scott for valuable assistance with field collection, G. Suen for identifying housekeeping loci, G. Emmerich for help with DNA sequencing, M. Poulsen for insights into Pseudonocardia isolation, members of the Currie Lab for support and discussion, Carol Lee for comments on the manuscript, the Smithsonian Tropical Research Institute for providing logistical support in Panama and facilities to work in Gamboa, the Autoridad Nacional del Ambiente y el Mar for collection and export permits in Panama, the Organization for Tropical Studies (OTS) for logistical support and facilities at La Selva Biological Station, and Ministerio del Ambiente y Energia for collection and export permits in Costa Rica. This research was supported by a National Science Foundation (NSF) Career Award (DEB-74702) and an NSF Microbial Genome Sequencing grant (MCB-0731822) to C. R. Currie. Support to E. J. Caldera included an NSF Graduate Research Fellowship, a National Institutes of Health (NIH) Microbes in Health and Disease Fellowship (NIH National Research Service Award AI55397), an OTS Christiane and Christopher Tyson Award, and a University of Wisconsin–Madison Department of Zoology Graduate Research Grant.
Literature Cited
- Adams RMM, Mueller UG, Holloway AK, Green AM, Narozniak J. Garden sharing and garden stealing in fungus-growing ants. Naturwissenschaften. 2000;87:491–493. doi: 10.1007/s001140050765. [DOI] [PubMed] [Google Scholar]
- Armitage SAO, Broch JF, Marin HF, Nash DR, Boomsma JJ. Immune defense in leaf-cutting ants: a cross-fostering approach. Evolution. 2011;65:1791–1799. doi: 10.1111/j.1558-5646.2011.01241.x. [DOI] [PubMed] [Google Scholar]
- Buckling A, Rainey PB. The role of parasites in sympatric and allopatric host diversification. Nature. 2002;420:496–499. doi: 10.1038/nature01164. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Rice WR. Distinguishing mechanisms for the evolution of cooperation. Journal of Theoretical Biology. 1991;149:63–74. doi: 10.1016/s0022-5193(05)80072-4. [DOI] [PubMed] [Google Scholar]
- Cafaro MJ, Currie CR. Phylogenetic analysis of mu tualistic filamentous bacteria associated with fungus-growing ants. Canadian Journal of Microbiology. 2005;51:441–446. doi: 10.1139/w05-023. [DOI] [PubMed] [Google Scholar]
- Cafaro MJ, Poulsen M, Little AEF, Price SL, Gerardo NM, Wong B, Stuart AE, et al. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proceedings of the Royal Society B: Biological Sciences. 2011;278:1814–1822. doi: 10.1098/rspb.2010.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldera EJ, Poulsen M, Suen G, Currie CR. Insect symbioses: a case study of past, present, and future fungus-growing ant research. Environmental Entomology. 2009;38:78–92. doi: 10.1603/022.038.0110. [DOI] [PubMed] [Google Scholar]
- Chapela IH, Rehner SA, Schultz TR, Mueller UG. Evolutionary history of the symbiosis between fungus-growing ants and their fungi. Science. 1994;266:1691–1694. doi: 10.1126/science.266.5191.1691. [DOI] [PubMed] [Google Scholar]
- Cho JC, Tiedje JM. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Applied and Environmental Microbiology. 2000;66:5448–5456. doi: 10.1128/aem.66.12.5448-5456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan FM. Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Philosophical Transactions of the Royal Society B: Biological Sciences. 2006;361:1985–1996. doi: 10.1098/rstb.2006.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr AH. Speciation. Sinauer; Sunderland, MA.: 2004. [Google Scholar]
- Currie CR. Prevalence and impact of a virulent parasite on a tripartite mutualism. Oecologia (Berlin) 2001;128:99–106. doi: 10.1007/s004420100630. [DOI] [PubMed] [Google Scholar]
- Currie CR, Bot ANM, Boomsma JJ. Experimental evidence of a tripartite mutualism: bacteria protect ant fungus gardens from specialized parasites. Oikos. 2003a;101:91–102. [Google Scholar]
- Currie CR, Mueller UG, Malloch D. The agricultural pathology of ant fungus gardens. Proceedings of the National Academy of Sciences of the USA. 1999a;96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311:81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999b;398:701–704. [Google Scholar]
- Currie CR, Wong B, Stuart AE, Schultz TR, Rehner SA, Mueller UG, Sung GH, et al. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science. 2003b;299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deletoile A, Passet V, Aires J, Chambaud I, Butel MJ, Smokvina T, Brisse S. Species delineation and clonal diversity in four Bifidobacterium species as revealed by multilocus sequencing. Research in Microbiology. 2010;161:82–90. doi: 10.1016/j.resmic.2009.12.006. [DOI] [PubMed] [Google Scholar]
- den Bakker HC, Didelot X, Fortes ED, Nightingale KK, Wiedmann M. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC Evolutionary Biology. 2008;8:13. doi: 10.1186/1471-2148-8-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi JR, Buckley DH. Widespread homologous recombination within and between Streptomyces species. ISME Journal. 2010;4:1136–1143. doi: 10.1038/ismej.2010.45. [DOI] [PubMed] [Google Scholar]
- Ewald PW. Transmission modes and evolution of the parasitism-mutualism continuum. Annals of the New York Academy of Sciences. 1987;503:295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L. Analysis of population subdivision. In: Balding AR, Bishiop M, Cannings C, editors. Handbook of statistical genetics. Wiley; New York: 2001. pp. 271–307. [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Frank SA. Host-symbiont conflict over the mixing of symbiotic lineages. Proceedings of the Royal Society B: Biological Sciences. 1996;263:339–344. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- Frank SA. Perspective: repression of competition and the evolution of cooperation. Evolution. 2003;57:693–705. doi: 10.1111/j.0014-3820.2003.tb00283.x. [DOI] [PubMed] [Google Scholar]
- Gerardo NM, Caldera EJ. Labile associations between fungus-growing ant cultivars and their garden pathogens. ISME Journal. 2007;1:373–384. doi: 10.1038/ismej.2007.57. [DOI] [PubMed] [Google Scholar]
- Gerardo NM, Jacobs SR, Currie CR, Mueller UG. Ancient host-pathogen associations maintained by specificity of chemotaxis and antibiosis. PLoS Biology. 2006a;4:1358–1363. doi: 10.1371/journal.pbio.0040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerardo NM, Mueller UG, Currie CR. Complex host-pathogen coevolution in the Apterostigma fungus-growing ant-microbe symbiosis. BMB Evolutionary Biology. 2006b;6:88–97. doi: 10.1186/1471-2148-6-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AM, Mueller UG, Adams RMM. Extensive exchange of fungal cultivars between sympatric species of fungus-growing ants. Molecular Ecology. 2002;11:191–195. doi: 10.1046/j.1365-294x.2002.01433.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Haeder S, Wirth R, Herz H, Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proceedings of the National Academy of Sciences of the USA. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath L, van der Walt E, Varsani A, Martin DP. Recombination patterns in aphthoviruses mirror those found in other picornaviruses. Journal of Virology. 2006;80:11827–11832. doi: 10.1128/JVI.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedlund BP, Staley JT. Microbial endemism and bio-geography. In: Bull AT, editor. Microbial diversity and bioprospecting. American Society for Microbiology; Washington DC.: 2004. pp. 225–231. [Google Scholar]
- Herre EA, Knowlton N, Mueller UG, Rehner SA. The evolution of mutualisms: exploring the paths between conflict and cooperation. Trends in Ecology & Evolution. 1999;14:49–53. doi: 10.1016/s0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Applied and Environmental Microbiology. 1997;63:3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra RF. The evolution of sexes. In: Stearns SC, editor. Evolution of sex and its consequences. Birkhäuser; Basel: 1987. pp. 59–91. [Google Scholar]
- Hölldobler B, Wilson EO. The ants. Harvard University Press; Cambridge, MA.: 1990. [Google Scholar]
- Huson DH, Bryant D. Application of phylogentic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004;7:1225–1241. [Google Scholar]
- Kost C, Lakatos T, Bottcher I, Arendholz WR, Redenbach M, Wirth R. Non-specific association between filamentous bacteria and fungus-growing ants. Naturwissenschaften. 2007;94:821–828. doi: 10.1007/s00114-007-0262-y. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Genetic variation and phenotypic evolution during allopatric speciation. American Naturalist. 1980;116:463–479. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lenormand T. Gene flow and the limits to natural selection. Trends in Ecology & Evolution. 2002;17:183–189. [Google Scholar]
- Lively CM. Migration, virulence, and the geographic mosaic of adaptation by parasites. American Naturalist. 1999;153(suppl.):S34–S47. doi: 10.1086/303210. [DOI] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Harvard University Press; Cambridge, MA.: 1963. [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheyev AS. History, genetics and pathology of a leaf-cutting ant introduction: a case study of the Guadeloupe invasion. Biological Invasions. 2008;10:467–473. [Google Scholar]
- Mikheyev AS, Mueller UG, Abbot P. Cryptic sex and many-to-one colevolution in the fungus-growing ant symbiosis. Proceedings of the National Academy of Sciences of the USA. 2006;103:10702–10706. doi: 10.1073/pnas.0601441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheyev AS, Mueller UG, Abbot P. Comparative dating of attine ant and lepiotaceous cultivar phylogenies reveals coevolutionary synchrony and discord. American Naturalist. 2010;175:E126–E133. doi: 10.1086/652472. [DOI] [PubMed] [Google Scholar]
- Mikheyev AS, Mueller UG, Boomsma JJ. Population genetic signatures of diffuse co-evolution between leaf-cutting ants and their cultivar fungi. Molecular Ecology. 2007;16:209–216. doi: 10.1111/j.1365-294X.2006.03134.x. [DOI] [PubMed] [Google Scholar]
- Mikheyev AS, Vo T, Mueller UG. Phylogeography of post-Pleistocene population expansion in a fungus-gardening ant and its microbial mutualists. Molecular Ecology. 2008;17:4480–4488. doi: 10.1111/j.1365-294X.2008.03940.x. [DOI] [PubMed] [Google Scholar]
- Mueller UG, Dash D, Rabeling C, Rodrigues A. Co-evolution between attine ants and actinomycete bacteria: a reevaluation. Evolution. 2008;62:2894–2912. doi: 10.1111/j.1558-5646.2008.00501.x. [DOI] [PubMed] [Google Scholar]
- Mueller UG, Ishak H, Lee JC, Sen R, Gutell RR. Placement of attine ant-associated Pseudonocardia in a global Pseudonocardia phylogeny (Pseudonocardiaceae, Actinomycetales): a test of two symbiont-association models. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology. 2010a;98:195–212. doi: 10.1007/s10482-010-9427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller UG, Rehner SA, Schultz TR. The evolution of agriculture in ants. Science. 1998;281:2034–2038. doi: 10.1126/science.281.5385.2034. [DOI] [PubMed] [Google Scholar]
- Mueller UG, Scott JJ, Ishak HD, Cooper M, Rodrigues A. Monoculture of leafcutter ant gardens. PLoS ONE. 2010b;5:7. doi: 10.1371/journal.pone.0012668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. 2. Gene-frequency data. Journal of Molecular Evolution. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Oh DC, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nature Chemical Biology. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- Perez-Losada M, Browne EB, Madsen A, Wirth T, Viscidi RP, Crandall KA. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infection Genetics and Evolution. 2006;6:97–112. doi: 10.1016/j.meegid.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Poulsen M, Boomsma JJ. Mutualistic fungi control crop diversity in fungus-growing ants. Science. 2005;307:741–744. doi: 10.1126/science.1106688. [DOI] [PubMed] [Google Scholar]
- Poulsen M, Cafaro M, Boomsma JJ, Currie CR. Specificity of the mutualistic association between actinomycete bacteria and two sympatric species of Acromyrmex leaf-cutting ants. Molecular Ecology. 2005;14:3597–3604. doi: 10.1111/j.1365-294X.2005.02695.x. [DOI] [PubMed] [Google Scholar]
- Poulsen M, Erhardt DP, Molinaro DJ, Lin TL, Currie CR. Antagonistic bacterial interactions help shape host-symbiont dynamics within the fungus-growing ant-microbe mutualism. PLoS ONE. 2007;2:e960. doi: 10.1371/journal.pone.0000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen M, Fernandez-Marin H, Currie CR, Boomsma JJ. Ephemeral windows of opportunity for horizontal transmission of fungal symbionts in leaf-cutting ants. Evolution. 2009;63:2235–2247. doi: 10.1111/j.1558-5646.2009.00704.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sen R, Ishak HD, Estrada D, Dowd SE, Hong EK, Mueller UG. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proceedings of the National Academy of Sciences of the USA. 2009;106:17805–17810. doi: 10.1073/pnas.0904827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Spratt BG, Hanage WP, Feil EJ. The relative contributions of recombination and point mutation to the diversification of bacterial clones. Current Opinion in Microbiology. 2001;4:602–606. doi: 10.1016/s1369-5274(00)00257-5. [DOI] [PubMed] [Google Scholar]
- Spratt BG, Maiden MCJ. Bacterial population genetics, evolution and epidemiology. Philosophical Transactions of the Royal Society B: Biological Sciences. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JT. The bacterial species dilemma and the genomicphylogenetic species concept. Philosophical Transactions of the Royal Society B: Biological Sciences. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taerum SJ, Cafaro MJ, Little AEF, Schultz TR, Currie CR. Low host-pathogen specificity in the leaf-cutting antmicrobe symbiosis. Proceedings of the Royal Society B: Biological Sciences. 2007;274:1971–1978. doi: 10.1098/rspb.2007.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. The coevolutionary process. University of Chicago Press; Chicago: 1994. [Google Scholar]
- Thompson JN. The geographic mosaic of coevolution. University of Chicago Press; Chicago: 2005. [Google Scholar]
- Thompson JN. The coevolving web of life. American Naturalist. 2009;173:125–140. doi: 10.1086/595752. [DOI] [PubMed] [Google Scholar]
- Thompson JN, Cunningham BM. Geographic structure and dynamics of coevolutionary selection. Nature. 2002;417:735–738. doi: 10.1038/nature00810. [DOI] [PubMed] [Google Scholar]
- Vo TL, Mueller UG, Mikheyev AS. Free-living fungal symbionts (Lepiotaceae) of fungus-growing ants (Attini: Formicidae). Mycologia. 2009;101:206–210. doi: 10.3852/07-055. [DOI] [PubMed] [Google Scholar]
- Vollmer SA, Bormane A, Dinnis RE, Seeling F, Dobson ADM, Aanensen DM, James MC, et al. Host migration impacts on the phylogeography of Lyme Borreliosis spirochaete species in Europe. Environmental Microbiology. 2010;13:184–192. doi: 10.1111/j.1462-2920.2010.02319.x. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME Journal. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- Wirth T, Meyer A, Achtman M. Deciphering host migrations and origins by means of their microbes. Molecular Ecology. 2005;14:3289–3306. doi: 10.1111/j.1365-294X.2005.02687.x. [DOI] [PubMed] [Google Scholar]
- Yan SC, Liu HJ, Mohr TJ, Jenrette J, Chiodini R, Zaccardelli M, Setubal JC, et al. Role of recombination in the evolution of the model plant pathogen Pseudomonas syringae pv. tomato DC3000, a very atypical tomato strain. Applied and Environmental Microbiology. 2008;74:3171–3181. doi: 10.1128/AEM.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanatta DT, Wilson CC. Testing congruency of geographic and genetic population structure for a freshwater mussel (Bivalvia: Unionoida) and its host fish. Biological Journal of the Linnean Society. 2011;102:669–685. [Google Scholar]
- Zhang MM, Poulsen M, Currie CR. Symbiont recognition of mutualistic bacteria by Acromyrmex leaf-cutting ants. ISME Journal. 2007;1:313–320. doi: 10.1038/ismej.2007.41. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequences datasets under the maximum likelihood criterion. University of Texas at Austin; 2006. [Google Scholar]