Abstract
Males and females show differences in the prevalence of many major diseases that have important inflammatory components to their etiology. These gender-specific diseases, which include autoimmune diseases, hepatocellular carcinoma, diabetes, and osteoporosis, are largely considered to reflect the actions of sex hormones on the susceptibility to inflammatory stimuli. However, inflammation reflects a balance between pro- and anti-inflammatory signals, and investigation of gender-specific responses to the latter has been neglected. Glucocorticoids are the primary physiological anti-inflammatory hormones in mammals, and synthetic derivatives of these hormones are prescribed as anti-inflammatory agents, irrespective of patient gender. We explored the possibility that sexually dimorphic actions of glucocorticoid regulation of gene expression may contribute to the dimorphic basis of inflammatory disease by evaluating the rat liver, a classic glucocorticoid-responsive organ. Surprisingly, glucocorticoid administration expanded the set of hepatic sexually dimorphic genes. Eight distinct patterns of glucocorticoid-regulated gene expression were identified, which included sex-specific genes. Our experiments also defined specific genes with altered expression in response to glucocorticoid treatment in both sexes, but in opposite directions. Pathway analysis identified sex-specific glucocorticoid-regulated gene expression in several canonical pathways involved in susceptibility to and progression of diseases with gender differences in prevalence. Moreover, a comparison of the number of genes involved in inflammatory disorders between sexes revealed 84 additional glucocorticoid-responsive genes in the male, suggesting that the anti-inflammatory actions of glucocorticoids are more effective in males. These gender-specific actions of glucocorticoids in liver were substantiated in vivo with a sepsis model of systemic inflammation.
INTRODUCTION
Gender-based biology has identified physiological and pharmacological differences between women and men at the subcellular, cellular, tissue, organ, and system levels (1), as well as differences in the propensity for specific diseases. Indeed, gender emerges as an important epidemiological risk factor for the incidence, presentation, and progression of several diseases with known inflammatory components. For example, osteoporosis, rheumatoid arthritis, systemic lupus erythematosus, and Cushing’s disease are more prevalent in females (2–5). Conversely, males are about three to five times more likely than females to develop hepatocellular carcinoma, kidney diseases (6, 7), and Parkinson’s disease (8). Although sex hormones (including estrogens and androgens) have been implicated as key determinants of this observed dimorphism, the mechanisms responsible for differences in susceptibility, incidence, and progression of gender-prevalent disease are not fully understood.
Glucocorticoids are stress-induced steroids that regulate intermediary metabolism, vascular tone, central nervous system function, development, and programmed cell death (9, 10). These hormones also function as anti-inflammatory and immunosuppressive effectors, interfering with virtually every step of the immune response. Synthetic glucocorticoids are a mainstay in the treatment of many inflammatory and autoimmune diseases (10), for which they are prescribed largely without consideration for patient gender. Both endogenous and exogenous glucocorticoids inhibit inflammation by binding to the glucocorticoid receptor and regulating gene expression through various mechanisms (10). Because inflammation appears to be a common feature underlying many diseases that exhibit gender-based differences in prevalence, we considered the possibility that gender may influence the anti-inflammatory actions of glucocorticoids. With the rat liver as a model, genome-wide complementary DNA (cDNA) microarray studies revealed that glucocorticoid treatment expands the set of liver genes that exhibit sexually dimorphic expression. These findings, supported by in silico analyses and in vivo experiments, suggest that glucocorticoids selectively modulate the inflammatory response in male and female livers through distinct signaling pathways. Based on these findings, we speculate that these gender-specific actions of glucocorticoids might contribute to the susceptibility to, development, or progression of inflammatory diseases with differences in gender prevalence.
RESULTS
Liver gene expression in male and female rats
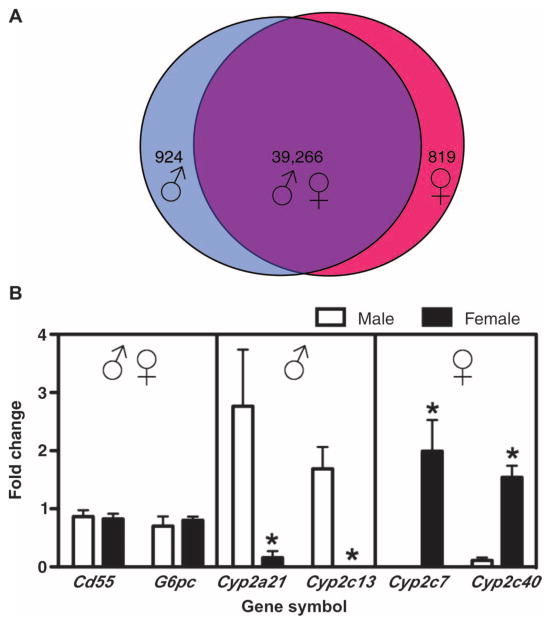
We used a whole-genome microarray approach in male and female adrenalectomized rats to evaluate the potential for sexually dimorphic gene expression in liver, a classical target for glucocorticoids. Comparison of significantly hybridized probe sets between the male and female vehicle groups (female/male, P < 0.001) revealed sex specificity for 1743 probe sets. Of these, 924 showed higher abundance in males (female/male, <1-fold change), and 819 showed higher abundance in females (female/male, >1-fold change) (Fig. 1A). We confirmed several of these sex-specific gene expression profiles by quantitative real-time polymerase chain reaction (qRT-PCR) analysis (Fig. 1B). In agreement with the microarray analysis, we detected greater messenger RNA (mRNA) abundance of cytochrome P450 2a1 (Cyp2a21) and cytochrome P450 2c13 (Cyp2c13) in males and greater mRNA abundance of cytochrome P450 2c7 (Cyp2c7) and cytochrome P450 2c40 (Cyp2c40) in females. In addition, genes that did not show statistical differences in expression between sexes (39,266 probe sets; female/male, P > 0.001) were confirmed by detection of CD55 molecule (CD55) and glucose-6-phosphatase, catalytic (G6pc) mRNA. Thus, our genome-wide analyses of control livers from adrenalectomized rats identified numerous genes that show sexual dimorphism in mRNA abundance, findings that are consistent with previous publications showing different liver gene expression patterns in male and female non-adrenalectomized mammals (11–14).
Fig. 1.
Comparison of liver gene expression profiles in male and female rats. (A) A total of 924 probe sets showed higher abundance in adult male (female vehicle compared to male vehicle, P < 0.001), 819 probe sets showed higher abundance in adult female (female vehicle compared to male vehicle, P < 0.001), and 39,266 probe sets showed similar abundance in males or females (female vehicle compared to male vehicle, P > 0.001). (B) Microarray results of representative sex-specific gene expression profiles in male (♂) and female (♀) rat liver were confirmed by qRT-PCR. Independent t tests were performed to compare values from male vehicle compared to female vehicle (*P < 0.05).
Glucocorticoid-induced increase in sexually dimorphic gene expression in the rat liver
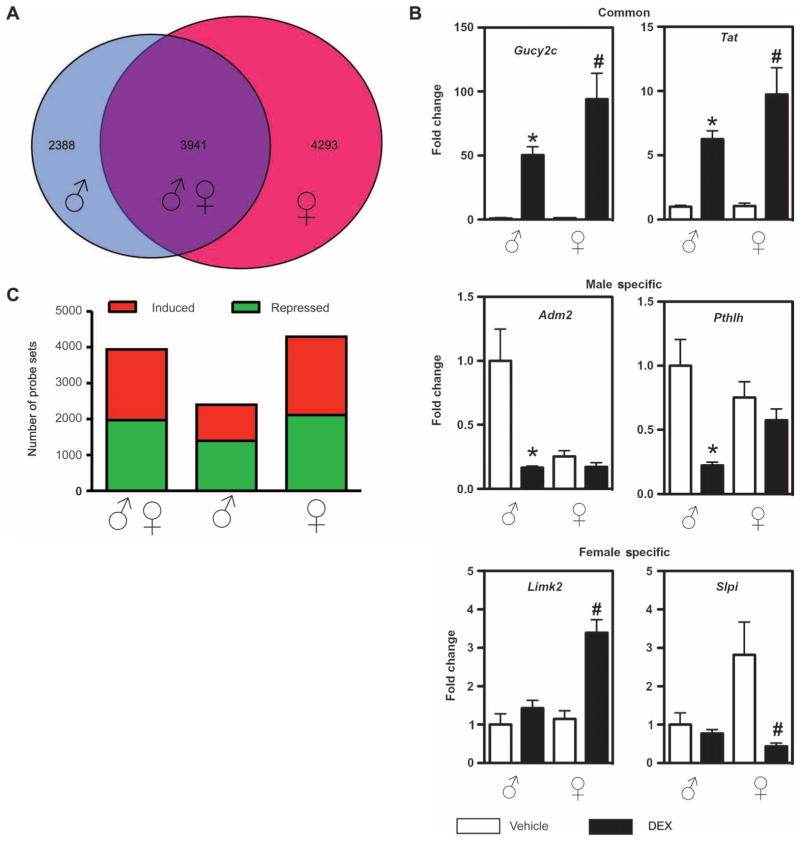
To investigate potential sexual dimorphism in glucocorticoid-regulated liver gene expression, we also performed microarray analysis of global gene expression on livers of male and female rats treated with the glucocorticoid dexamethasone (DEX). Surprisingly, we identified a higher number of significantly changed probe sets in females compared to males (8234 and 6329 probe sets, respectively). By comparing the two sets of data (male DEX/vehicle compared to female DEX/vehicle; P < 0.001), we detected sex-specific differences in glucocorticoid-regulated gene expression (Fig. 2A). We identified 4293 probe sets specifically regulated by glucocorticoids in females and 2388 distinct probe sets specifically regulated by glucocorticoids in males. The remaining probe sets were regulated in both sexes (3941 probe sets). Differential expression of sex-specific genes was confirmed by qRT-PCR of DEX-responsive genes (Fig. 2B). The genes adrenomedullin 2 (Adm2) and parathyroid hormone-like hormone (Pthlh) confirmed the patterns of expression for genes that are DEX responsive in males but not in females (male-specific genes). The genes LIM motif-containing protein kinase 2 (Limk2) and secretory leukocyte peptidase inhibitor (Slpi) confirmed the pattern of expression for genes that are DEX responsive in females but not in males (female-specific genes). The genes guanylate cyclase 2C (Gucy2c) and tyrosine aminotransferase (Tat) confirmed the pattern of expression for genes that are DEX responsive in males and females (common genes). About half of the common and half of the female-specific probe sets were altered by glucocorticoid treatment in the same direction (increased or decreased abundance). However, in males, we identified significantly more probe sets that were repressed by glucocorticoid treatment (Fig. 2C). Preliminary experiments with PCR primers that quantify nascent rather than mature mRNAs yielded similar results, suggesting that at least some of these alterations in mRNA abundance reflect a transcriptional response to glucocorticoids (fig. S1). This increase in sexually dimorphic gene expression in liver in response to glucocorticoid treatment has not been previously described and demonstrates that glucocorticoids regulate common and sex-specific genes. In male rats, glucocorticoids appear to act predominantly by repressing gene expression in the liver.
Fig. 2.
Selective expression of glucocorticoid-regulated genes in male and female rat liver. (A) Probe sets regulated by DEX in male (male DEX compared to male vehicle) (left-hand circle) are compared with probe sets regulated by DEX in female (female DEX compared to female vehicle) (right-hand circle). The overlapping circle represents probe sets that are common to males and females. (B) The number of genes with increased abundance (red) or decreased abundance (green) by each sex is represented. From left to right, the first bar represents probe sets that were DEX responsive in males extracted from the list of probe sets regulated by DEX treatment in males and females (common; ♂♀); the second bar represents probe sets that were DEX responsive only in males (male-specific; ♂); and the third bar represents probe sets that were DEX responsive only in females (female-specific; ♀). (C) Microarray results of representative glucocorticoid-regulated genes in male and female rat liver were confirmed by qRT-PCR. Independent t tests were performed to compare values between male vehicle and male DEX (*P < 0.05) and between female vehicle and female DEX (#P < 0.05).
Canonical pathway analysis of glucocorticoid-regulated genes in male and female
To investigate the possible biological implications of sex-specific regulation of genes by glucocorticoids, we used the Ingenuity Pathways Analysis (IPA) library to identify the most significantly affected signaling pathways in males and females (Table 1). The significantly regulated pathways were ranked by ratio (those with the highest percentage of genes regulated by DEX treatment) and the highest-ranked pathways are displayed. Only one [phosphatase and tensin homolog signaling (PTEN)] of the top five predicted glucocorticoid-regulated pathways was the same between males and females. In addition, several canonical pathways identified in our data set were significantly more affected by glucocorticoids in males or in females. For example, 25 genes in the apoptosis signaling pathway were regulated by glucocorticoids in both sexes. However, glucocorticoids regulated 12 additional genes in males (male-specific genes) and 4 additional genes in females (female-specific genes), suggesting that apoptosis signaling in livers in males may be more affected by glucocorticoid treatment than livers in females. Several canonical pathways associated with cell death, cellular growth and proliferation, immune response, tissue morphology, cancer, and inflammatory and immunological diseases were differentially regulated by glucocorticoid treatment in male and female rat livers. The canonical pathway analysis of our microarray data suggests that distinct differences exist in glucocorticoid signaling in males and females.
Table 1.
Identification of canonical pathways from the IPA library in male and female gene expression data sets.
| Gender | Order | Canonical pathway | Common genes | Male-specific genes | Female-specific genes |
|---|---|---|---|---|---|
| Male | 1 | Endoplasmic reticulum stress pathway | 5 | 3 | 2 |
| 2 | Death receptor signaling | 18 | 9 | 3 | |
| 3 | Apoptosis signaling | 25 | 12 | 4 | |
| 4 | PTEN signaling | 23 | 15 | 12 | |
| 5 | Interferon signaling | 10 | 2 | 0 | |
| Female | 1 | Circadian rhythm signaling | 4 | 2 | 9 |
| 2 | IL-6 signaling | 23 | 12 | 14 | |
| 3 | Hepatic fibrosis or hepatic stellate cell activation | 33 | 11 | 18 | |
| 4 | PTEN signaling | 23 | 15 | 12 | |
| 5 | Hypoxia signaling in the cardiovascular system | 13 | 5 | 13 |
Sex-distinctive signature patterns of gene expression and gene networks
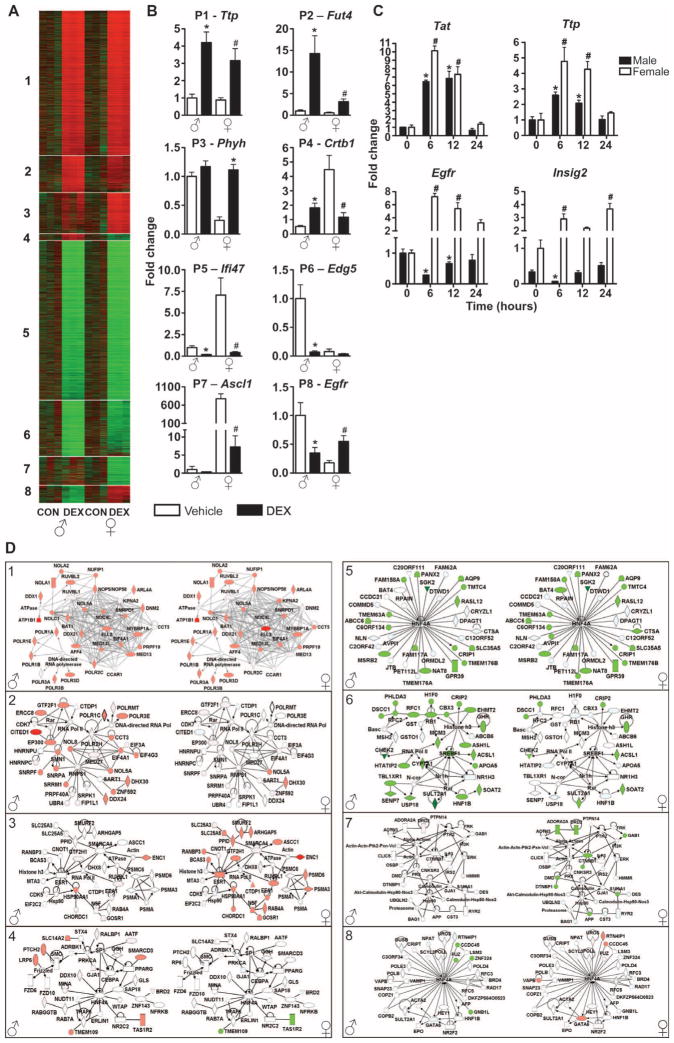
An alternative approach to evaluate changes in liver gene expression in response to glucocorticoids is to identify patterns of expression that are distinct between sexes. We applied a profile-based method called EPIG (extracting microarray gene expression patterns and identifying coexpressed genes) to our data sets (15). EPIG identified eight distinct patterns of gene expression (Fig. 3A and table S1). As expected, many liver genes are regulated by glucocorticoids in both sexes in a similar manner. In pattern 1, 1667 probe sets showed increased abundance, and in pattern 5, 1827 probe sets showed decreased abundance in both males and females in response to glucocorticoid treatment. These first two described patterns were validated by measurement of mRNA abundance of tritestrapolin (Ttp) and interferon γ inducible protein 47 (Ifi47) by qRT-PCR (Fig. 3B). Thus, patterns 1 and 5 represent the classic actions of glucocorticoids on gene expression, in which genes are regulated by glucocorticoid treatment in both sexes in the same direction. However, we also identified gene sets that showed gender-specific profiles of regulation. For example, patterns 2 and 6 are representative of probe sets with more robust regulation by glucocorticoids in males than females. In pattern 2, 417 probe sets showed glucocorticoid-induced increases in abundance in males [validated by myeloid α-3-fucosyltransferase (Fut4) gene], and in pattern 6, 640 probe sets showed glucocorticoid-induced decreases in abundance in males [validated by sphingolipid G protein–coupled receptor, 5 (Edg5) gene]. Another distinctive pattern of gene expression is shown in patterns 3 and 7. These probe sets were preferentially regulated by glucocorticoids in females. In pattern 3, EPIG identified 463 female-specific probe sets with glucocorticoid-induced increases in abundance, and in pattern 7, EPIG identified 323 female-specific probe sets with glucocorticoid-induced decreases in abundance. Pattern 3 was validated by qRT-PCR for phytanoyl-CoA hydroxylase (Phyh), whereas pattern 7 was validated by qRT-PCR for achaetescute complex homolog-like 1 (Ascl1). Finally, patterns 4 and 8 have probe sets that exhibited unexpected profiles. In both patterns, each probe set selected was altered in response to glucocorticoids in males and females, but in opposite directions (anticorrelated). Pattern 4 represents a cluster of 73 probe sets with increased abundance in males, but decreased abundance in females, and pattern 8 represents a cluster of 199 glucocorticoid-responsive rat liver probe sets with decreased abundance in male, but increased abundance in females. Detection of chymotrypsinogen B1 (Crtb1) mRNA abundance by qRT-PCR confirmed pattern 4. Crtb1 mRNA abundance was increased in males and decreased in females in response to glucocorticoids. Pattern 8 was validated by qRT-PCR for the Egfr gene (Fig. 3B). In this pattern of anticorrelated genes, Egfr mRNA abundance was decreased in males and increased in females in response to glucocorticoid treatment. Because the phenomenon described in patterns 4 and 8 has not been previously described, the mRNA abundance of Egfr and insulin-induced gene 2 (Insig-2) was also evaluated at later time points (12 and 24 hours after DEX treatment). The mRNA abundance of correlated (Tat and Ttp in pattern 1) and anticorrelated genes (Egfr and Insig-2 in pattern 8) confirmed that the phenomena persisted up to 12 hours after DEX treatment (Fig. 3C).
Fig. 3.
Patterns of DEX-responsive genes in male and female rat liver. (A) The patterns were extracted by EPIG and are displayed in the heat map. From top to bottom are the 5609 probe sets selected by EPIG listed in order from patterns 1 to 8. The experimental groups are indicated in the bottom part of the figure. Red and green colors correspond to increased and decreased abundance, respectively, with a darker color denoting smaller differences. (B) Microarray results of representative glucocorticoid-regulated genes for each pattern of gene expression were confirmed by qRT-PCR. Independent t tests were performed to compare values between male vehicle and male DEX (*P < 0.05) and between female vehicle and female DEX (#P < 0.05). (C) Quantitative RT-PCR confirmation of glucocorticoid-regulated correlated and anticor-related genes in male and female rat livers 6, 12, and 24 hours after DEX treatment (1 mg/kg ip). Statistical analysis was performed by analysis of variance (ANOVA) followed by t test subjected to the Dunnett’s correction. *P < 0.05 compared to time 0 in males; #P < 0.05 compared to time 0 in females. (D) Network representations of the focus genes presented in each pattern of gene expression. Networks 1 to 8 correspond to patterns 1 to 8, respectively. Genes are represented as nodes and the biological relationship between two nodes is represented as an edge. The color of the node indicates increased (red) or decreased (green) abundance. The name and expression values of each gene presented in the network are shown in table S2.
To predict the effect on cellular function, we generated a gene correlation network for each of the eight patterns of gene expression identified by EPIG analyses. Networks were generated on the basis of direct pairwise interactions between glucocorticoid-responsive genes. Figure 3D displays the networks with the highest number of direct pairwise interactions generated by IPA with a maximum of 35 molecules per network. As expected, the interaction between genes that are regulated by glucocorticoids in both sexes generated the same network in males and females (represented by networks 1, 4, 5, and 8). As a reflection of EPIG analysis, glucocorticoids regulated networks 1 and 5 in the same direction in both sexes (compare networks 1 and 5 with patterns 1 and 5, respectively) and networks 4 and 8 in opposite directions (compare networks 4 and 8 with patterns 4 and 8, respectively). Networks 2, 3, 6, and 7 represent interactions between sex-specific glucocorticoid-responsive genes and therefore are generated in only one gender. For example, networks can only be generated in males for patterns 2 and 6 because genes in these patterns are regulated by glucocorticoids preferentially in males. The same gene network generated for males is shown for females to illustrate that genes in networks 2 and 6 are regulated weakly or not detectably by glucocorticoids in females. Finally, networks 3 and 7 represent interactions between female-specific glucocorticoid-responsive genes. The expression values of the genes that constitute network 3 were increased, whereas those in network 7 were decreased by glucocorticoid treatment in females. Thus, the gene networks generated for each pattern of gene expression represent groups of glucocorticoid-responsive genes that are either driving the expression of each other or responding similarly to regulatory factors. In three of the eight gene networks (networks 4, 5, and 8), the transcription factor hepatocyte nuclear factor 4 α (Hnf4α) is the gene with the highest number of connections (Fig. 3D), suggesting that direct or indirect glucocorticoid effects on Hnf4α signaling might be important for establishing differences in cellular functions between sexes.
Thus, by using this stringent approach for analysis and comparison of DEX-regulated genes between sexes, we were able to identify several probe sets with similar profiles of expression in each of the signature patterns extracted by EPIG. The eight patterns identified are also organized in three major groups of probe sets (compare to Fig. 2A): (i) one that is commonly regulated by glucocorticoids in male and female (correlated and anticorrelated genes), (ii) another that is preferentially regulated by glucocorticoids in females, and (iii) a group that is preferentially regulated by glucocorticoids in males. These data demonstrate that glucocorticoids have sex-specific effects in the livers of male and female rats, suggesting that glucocorticoids orchestrate the activity of many components that interact with each other through gene networks in a sex-specific manner.
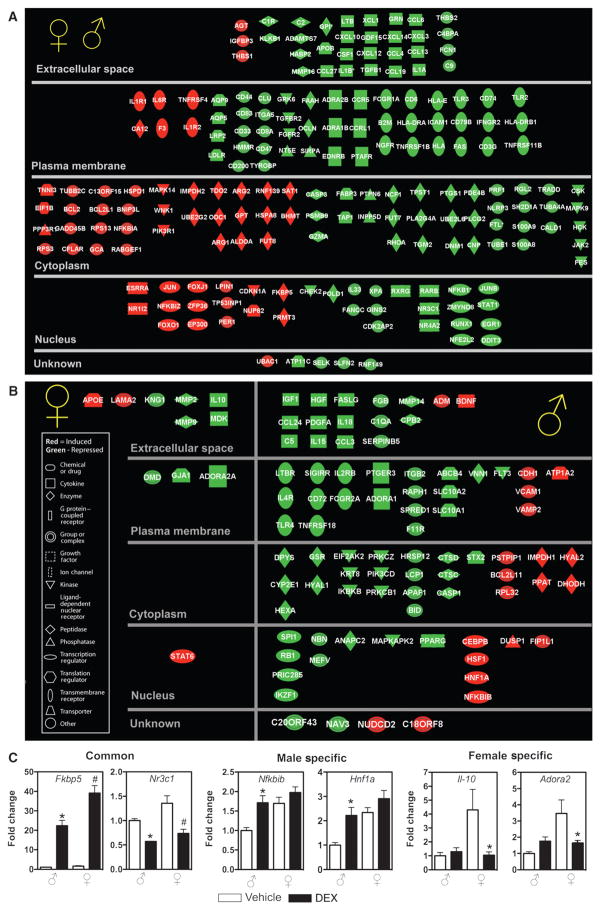
Comparison of common and sex-specific glucocorticoid-responsive genes implicated in inflammatory disorders
Because glucocorticoids are key physiological anti-inflammatory hormones, and many diseases with differences in gender prevalence have an inflammatory component, we compared the anti-inflammatory effect of glucocorticoids in our model system. With IPA, we identified genes implicated in inflammatory processes that are commonly and sex-specifically regulated by glucocorticoids. Genes from each group (common, male-specific, and female-specific DEX-responsive genes) that were significantly associated with biological functions or diseases in the Ingenuity Pathways Knowledge Base (or both) were considered for the analysis. Many genes were regulated by glucocorticoids in a similar manner in male and female rats (189 genes). These included several cytokines [chemokine (C-X-C motif ) ligand 10 (Cclx10) and chemokine (C-C motif ) ligand 4], pattern recognition receptors (Toll-like receptor 2 and Toll-like receptor 3), and transcription factors (nuclear factor of κ light polypeptide gene enhancer in B cells 1 and signal transducer and activator of transcription 1) that showed decreased abundance in response to DEX treatment, and are considered classical mediators of glucocorticoid anti-inflammatory effects (Fig. 4A). We also analyzed the genes that are regulated by glucocorticoids in a gender-specific manner. We identified 11 additional genes that were specifically regulated in livers in female rats by glucocorticoid treatment (Fig. 4B). In contrast, analysis of the male-specific glucocorticoid-regulated liver genes identified 84 additional genes implicated in inflammatory disorders, most of which showed decreased abundance in response to glucocorticoids (Fig. 4B). We confirmed several of these sex-specific glucocorticoid-regulated genes implicated in inflammatory disorders by qRT-PCR analysis (Fig. 4C). The genes Fk506 binding protein 5 (Fkbp5) and nuclear receptor subfamily 3, group C, member 1 (Nr3c1) confirmed the pattern of expression for genes that were DEX responsive in both males and females, the genes nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor, β (Nfkbib) and HNF1 homeobox A (Hnf1a) confirmed the pattern of expression of genes that were DEX responsive only in males, and the genes interleukin-10 (Il-10) and adenosine A2a receptor (Adora2) confirmed the pattern of expression for genes that were DEX responsive only in females. Thus, glucocorticoids regulate significantly more genes implicated in inflammatory disorders in livers from male rats than in those from female animals. This finding raises the possibility that the anti-inflammatory effect of glucocorticoids may be greater in males than females and perhaps explain why females have a higher risk of developing some autoimmune diseases than do males.
Fig. 4.
Comparison of common and sex-specific glucocorticoid-regulated genes implicated in inflammatory disorders. Genes from the data sets (common, male-specific, and female-specific) that met the P < 0.001 cutoff and are associated with inflammatory disorders in the Ingenuity Pathways Knowledge Base are displayed. (A and B) Common DEX-responsive genes implicated in inflammatory disorders regulated in male and female (A), female-specific DEX-responsive genes implicated in inflammatory disorders, and male-specific DEX-responsive genes implicated in inflammatory disorders (B). Each gene is represented by a node and is arranged on the basis of cellular localization. The color of the node indicates increased (red) or decreased (green) abundance. The name and expression value of each gene are shown in table S3. (C) Microarray results of representative common, male-specific, and female-specific glucocorticoid-regulated genes implicated in inflammatory disorders were confirmed by qRT-PCR. Independent t tests were performed to compare values from male vehicle to male DEX (*P < 0.05), and values from female vehicle to female DEX (#P < 0.05).
Anti-inflammatory effect of DEX in male and female rats during systemic inflammatory responses
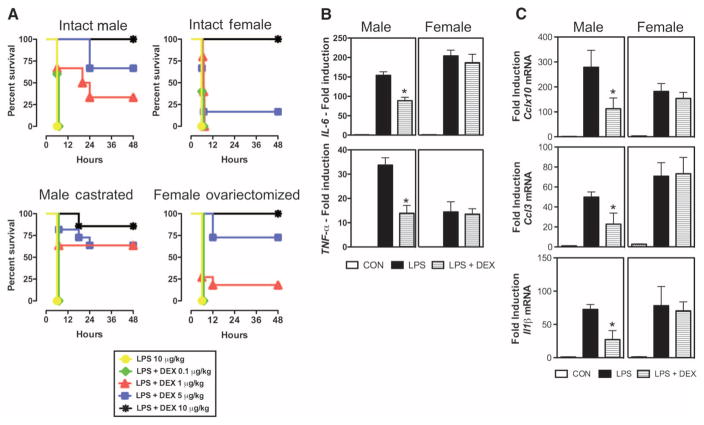
To evaluate the hypothesis that the anti-inflammatory actions of glucocorticoids are more effective in males than in females, we injected rats with lipopolysaccharide (LPS) intraperitoneally to trigger a systemic inflammatory response. Different doses of DEX were injected 2 hours after LPS treatment to counteract the proinflammatory response induced by LPS. More males (Fig. 5A, upper left panel) survived LPS injection after DEX treatment than females (Fig. 5A, upper right panel). For example, 67% of males survived the LPS injection when animals were treated with DEX (5 μg/kg). In contrast, only 17% of females survived LPS injection when treated with the same dose of glucocorticoid. In this model, many events other than inflammation also contribute to the development of a systemic inflammatory response and, consequently, to mortality; thus, we also evaluated the ability of DEX to prevent the induction of classical proinflammatory parameters in the plasma and in the liver of endotoxemic animals. In agreement with the survival studies, glucocorticoid treatment abrogated increases in the plasma concentrations of interleukin-6 (IL-6) and tumor necrosis factor–α (TNF-α) and in the mRNA abundance of Cclx10, chemokine (C-C motif ) ligand 3 (Ccl3), and interleukin-1β (Il-1β) only in livers of males (Fig. 5, B and C). Thus, by comparing both the survival as well as the cytokine concentrations of male and female endotoxemic rats treated with DEX, we confirmed that the anti-inflammatory actions of glucocorticoids are more effective in males.
Fig. 5.
Anti-inflammatory effect of DEX in male and female rats during systemic inflammatory response. (A) Survival curves of male and female rats treated with LPS and LPS + DEX. Adrenalectomized animals are displayed in the upper panels, and adrenalectomized and gonadectomized animals are displayed in the lower panels. (B and C) Cytokine plasma concentrations and mRNA abundance in livers of male and female rats treated with LPS and LPS + DEX. Two hours after the treatment with LPS, animals were injected with DEX (5 μg/kg ip). Blood and liver were harvested 3 hours after DEX treatment for ELISA of plasma IL-6 and TNF-α (B) and for qRT-PCR measurement of Cclx10, Ccl3, and Il-1β mRNA abundance in the liver (C). Statistical analysis was performed by Wilcoxon signed-rank test for survival studies (P < 0.05) and by ANOVA followed by t test subjected to the Tukey’s multiple comparison test. *P < 0.05 compared to male LPS; #P < 0.05 compared to female LPS. CAST, castrated; OVA, ovariectomized; CON, control.
The influence of sex hormones in the immune response has been extensively studied (16, 17). Estrogens, progestins, and androgens can promote or interfere with the progression of the inflammatory response, depending on the experimental model and tissue studied (17). To evaluate the influence of sex hormones on the anti-inflammatory actions of glucocorticoids in our model of systemic inflammatory response, we determined the survival rate of male and female gonadectomized rats (Fig. 5A, lower panels) using the same experimental protocol as in the previous survival studies. The absence of endogenous testicular hormones did not affect the survival rate of endotoxemic male rats treated with glucocorticoids, and similarly to uncastrated male rats (Fig. 5A, left upper panels), ~60% of castrated rats treated with DEX (5 μg/kg) survived LPS injection (Fig. 5A, left lower panel). In contrast, ovariectomy increased the survival rate of female endotoxemic rats treated with DEX (Fig. 5A, compare upper right and lower right panels). Seventy-three percent of females treated with DEX (5 μg/kg) survived LPS injection in the absence of endogenous ovarian hormones, a four times higher survival rate than nongonadectomized females. Thus, in this model of systemic inflammatory response, ovariectomy potentiated the anti-inflammatory actions of glucocorticoids in females, suggesting antagonism between ovarian hormones and glucocorticoids. However, the sexually dimorphic, anti-inflammatory effect of glucocorticoids persisted in gonadectomized animals that were treated with a lower dose of glucocorticoid (Fig. 5A, compare male castrated and female ovariectomized panels). About 60% of males treated with DEX (1 μg/kg) survived LPS injection, whereas <20% of females survived in the same experimental protocol. Together, these findings support the hypothesis that the anti-inflammatory actions of glucocorticoids are more effective in males. Furthermore, they suggest that ovarian hormones can influence the anti-inflammatory actions of glucocorticoids in the context of a rat adrenalectomized model of sepsis.
DISCUSSION
Sexual dimorphism is ubiquitous among higher eukaryotes. In mammals, sexual differentiation is initiated by the presence or absence of the testis-determining factor encoded on the Y chromosome (SRY in humans and Sry in mice) and mediated by the effects of sex hormones (18–21) and growth hormone (20, 21). These hormones produce unique cellular environments in males and females that lead to gender-specific differences in gene expression in many rodent tissues, including liver, adipose tissue, muscle, kidney, brain, blastocysts, and lacrimal gland (22). These genes exhibit tissue-specific patterns of expression and are enriched for distinct pathways represented in the Gene Ontology database. In the rat liver, ~1000 genes (representing almost 5% of the entire rat genome) have been reported to show sexually dimorphic expression patterns (23). Sex-specific temporal patterns of circulating growth hormones secreted by the pituitary contribute to the gender-specific patterns of liver gene expression through the actions of signal transducer and activator of transcription 5B (Stat5b) (23).
Because the liver is a classic target for glucocorticoids, we explored the possibility that glucocorticoids may regulate distinct patterns of gene expression in the livers of male and female rats. We discovered that glucocorticoids expanded the normal set of genes that showed sexually dimorphic expression in the liver. These genes clustered into eight patterns of glucocorticoid-regulated genes that were specifically regulated by glucocorticoids in livers of either male or female rats. There was also a significant number of genes that showed glucocorticoid-induced alterations in abundance in opposite directions in both sexes (anticorrelated genes). One gene with anticorrelated changes in abundance was Egfr, which encodes a receptor that is abundant in the adult liver and has been proposed to play an important role during liver development, function, and regeneration after partial hepatectomy (24). Glucocorticoid-regulated Egfr expression in opposite directions may contribute to the gender-specific differences in liver regeneration after partial hepatectomy (25).
To reduce the complexity of our gene expression data and to compare the behavior of glucocorticoid-responsive genes between sexes, we determined the dynamic interactions among genes for each pattern of gene expression. Hnf4α was identified as the node with the highest number of interactions in three different gene networks; a large fraction of the liver transcriptome is thought to be regulated by Hnf4α (26). Upon binding to DNA, Hnf4α recruits transcriptional coactivators and other accessory proteins, leading to alterations in the expression of target genes involved in critical metabolic pathways (such as gluconeogenesis, glycolysis, and fatty acid metabolism) (27) that are also regulated by glucocorticoids. Thus, based on theories of cellular network generation in which a few general transcription factors serve as master regulators (28), Hnf4α appears to play a critical role in glucocorticoid regulation of liver gene expression in terms of both transcriptional activation and repression. Glucocorticoids, through direct or indirect interactions with Hnf4α, may contribute to establishing differences in cellular stress responses between sexes.
Sexual dimorphism in the rat liver has been previously attributed to gender-specific differences in the hypothalamic-pituitary-adrenal (HPA) axis. In addition, depending on the estrous cycle stage, female rats can have higher plasma concentrations of corticosterone than males (29). To avoid interference of fluctuating concentrations of endogenous glucocorticoids in our experiments, we previously adrenalectomized all animals and cycled females to the diestrous 1 phase of the estrous cycle (when sex steroids are low in concentration). No differences in liver morphology between intact and adrenalectomized rats were observed (in either sex) (fig. S2), and we found no gender- or cycle-related differences in the mRNA or protein abundance for glucocorticoid receptor (figs. S3 and S4). Additionally, glucocorticoid receptor binding sites and affinity were nearly identical in male and female livers (fig. S3). These findings suggest that the unique profiles of glucocorticoid-regulated gene expression observed between sexes were most likely due to gender-specific mechanisms controlling glucocorticoid receptor–mediated gene expression in the liver, rather than interference of estrous cycle or variation in the concentrations of endogenous glucocorticoids.
Because growth hormone is a major determinant of sexual dimorphism observed in rodent liver, we also explored the possibility that growth hormone contributed to the sexually dimorphic response to glucocorticoids by studying the effects of glucocorticoids on isolated hepatocytes from male and female rats (fig. S5). As expected, the profile of gene expression in isolated hepatocytes under serum-free conditions differed from the profile in the whole liver; however, glucocorticoid administration still elicited sex-specific differences in gene expression. This finding suggests that the rapid effect we observed in our studies (within 6 hours) reflects direct actions of glucocorticoids on the liver rather than indirect actions mediated by alterations in growth hormone signaling. Additionally, our preliminary studies also show that sexually dimorphic regulation of gene expression by glucocorticoids is not exclusive to the rat liver but is also observed in the mouse liver (fig. S6). Thus, under various experimental paradigms, glucocorticoids regulate both common and sex-specific genes.
We have considered several potential mechanisms to explain how glucocorticoids exhibit their sexually dimorphic actions in regulating liver gene expression. One possible explanation is the presence of gender-specific profiles of co-regulatory molecules that contribute to the function of nuclear hormone receptors and other transcription factors. Analysis of our microarray data for mRNA abundance of the known co-regulators (30) reveals dimorphic patterns of gene expression for 12 co-regulators (fig. S7 and table S4). Of these, the abundance of histone deacetylase 2 (HDAC2), TATA box binding protein (Tbp), and protein arginine methyltransferase 2 (Prmt2) is higher in male livers, whereas the abundance of prohibitin 2 (Phb2), tripartite motif-containing 24 (Trim 24), and cyclin-dependent kinase inhibitor 1C (Cdknic) is higher in female livers (31–36). These co-regulators have been implicated in glucocorticoid receptor signaling. Thus, dimorphic co-regulator expression remains a viable candidate for a potential mechanism underlying the sexual dimorphic regulation of liver gene expression by glucocorticoids. Furthermore, the abundance of numerous co-regulators appears to be altered by gluco-corticoid treatment, and glucocorticoids also expand the dimorphic nature of co-regulator gene expression at the mRNA level (fig. S7 and table S4). Given the 6-hour time frame of our experiments, it is unlikely, however, that any of these glucocorticoid-regulated co-regulators contribute to the dimorphic response in rat liver.
We have also performed dose-response studies that evaluate the ability of glucocorticoids to modulate the expression of some genes discovered in our microarray analysis, which were all performed with a glucocorticoid dose that saturates glucocorticoid receptors. Although males and females had equivalent numbers of glucocorticoid receptors with similar affinity for DEX (fig. S8), we observed a significant increase in the EC50 (median effective concentration) in female animals for nondimorphic genes (fig. S8). This finding correlates well with the requirement for higher doses of glucocorticoids to rescue female adrenalectomized animals from an LPS challenge. Given that the males and females have similar pharmacokinetic properties for clearance of DEX (37), it is unlikely that metabolism is responsible for this finding. One possible explanation for the difference may reside in the dimorphic expression of co-regulators, which can alter the potency of glucocorticoids (38). Alternatively, estrogens have been suggested to antagonize glucocorticoid action through alterations in phosphorylation of the glucocorticoid receptor (39). Other potential mechanisms that could account for this gender-specific action of glucocorticoids include RNA splicing, RNA stability, microRNAs, and posttranslational modification of the glucocorticoid receptor.
Several canonical pathways identified in male and female rat liver are associated with cell death, cellular growth and proliferation, immune response, tissue morphology, cancer, and inflammatory and immunological diseases. We detected sexual dimorphism in all the canonical pathways significantly present in our analysis, and several pathways are implicated in diseases in which susceptibility is sex-biased. For example, ablation of IL-6 signaling in Kupffer cells abolishes sex differences in hepatocarcinogenesis in mice (40), and our data indicate that glucocorticoids regulate sex-specific genes that encode components of the IL-6 signaling pathway (Table 1). We also identified several pathways implicated in innate immune response that are altered by glucocorticoids in a gender-specific manner, such as interferon signaling and endoplasmic reticulum pathway signaling, suggesting an important role for glucocorticoids in the liver in inflammatory stress responses (Table 1).
Emerging evidence suggests that the liver is an important part of the body’s immune response and should therefore be considered an immune organ (41, 42). Hepatocytes are responsible for the biosynthesis of 80 to 90% of complement components, secreted pattern recognition receptors, and chemokines. Our analysis of inflammatory pathways identified significantly more liver genes regulated by glucocorticoids in males compared to females, raising the possibility that the anti-inflammatory effect of glucocorticoids may be greater in males. To evaluate this hypothesis, we compared the ability of glucocorticoids to elicit an anti-inflammatory response in a sepsis model of liver inflammation. These in vivo experiments not only confirmed the in silico predictions made by genome-wide analysis but also suggest that the sex-specific anti-inflammatory actions of glucocorticoids occur in the liver. Based on our in vivo sepsis rescue experiment, the anti-inflammatory effects of glucocorticoids in females seem to also involve some interplay with ovarian hormones, because their presence, even at the low concentrations, alters the ability of glucocorticoids to abrogate the inflammatory responses to LPS. Thus, we speculate that sex-based differences in the predisposition to autoimmune diseases in females, such as rheumatoid arthritis and systemic lupus erythematosus, might reflect differences in glucocorticoid anti-inflammatory actions between sexes. Failure to mount an adequate glucocorticoid response to autoimmune challenge may result in altered incidence of autoimmune diseases observed in females.
MATERIALS AND METHODS
Animals
Adult male and female adrenalectomized Sprague-Dawley rats (8 to 10 weeks of age) were purchased from Charles River Laboratories. After operation, the animals were fed ad libitum and were given 0.154 M sodium chloride to drink for 7 days. The estrous cycle of female adrenalectomized rats was determined by vaginal wash (43). Females in estrous were selected for treatment that was performed the following morning.
Glucocorticoid treatment
Male and female (diestrous 1) adrenalectomized rats were treated with vehicle (phosphate-buffered saline) or DEX [1 mg/kg intraperitoneally (ip)] (Steraloids) on the same day. The dose of steroid was given to the animals in a single injection to mimic situations when plasma concentrations of endogenous glucocorticoids rapidly increase 300- to 400-fold in response to stress (such as trauma and systemic inflammation) (44). Based on previous gene expression studies in the rat liver evaluated from 3 to 72 hours after glucocorticoid treatment, most genes showed altered abundance 6 hours after treatment (45). Thus, for our microarray analyses, livers were harvested 6 hours after DEX treatment and kept in RNAlater solution at 4°C overnight. For time course experiments, livers were harvested 6, 12, and 24 hours after treatment and kept in RNAlater solution at 4°C overnight. All samples were frozen and stored at −80°C.
Anti-inflammatory effect of DEX in vivo
Male and female (diestrous 1) rats were injected with endotoxin (Escherichia coli LPS 0111:B4; Sigma) as a model of systemic inflammatory response. Two hours after LPS (10 μg/kg ip) injection, animals were treated with different doses of DEX (0.1, 1.0, 5.0, and 10 μg/kg ip). The mortality was recorded four times per day for up to 48 hours after LPS challenge (n = 6 to 9 animals per group). A second group of animals was treated with LPS and LPS + DEX following the protocol described above. Animals were euthanized (while under anesthesia) 5 hours after LPS treatment in the LPS group and 3 hours after DEX treatment in the LPS + DEX group. Blood was obtained for analysis of cytokine plasma concentrations [enzyme-linked immunosorbent assay (ELISA); Thermo Fisher Scientific], and tissue from liver was harvested for analysis of cytokine mRNA abundance by qRT-PCR (n = 4 to 6 animals per group). To evaluate the influence of sex hormones in the anti-inflammatory actions of DEX in vivo, we treated adrenalectomized and gonadectomized male and female adult (8 to 10 weeks) Sprague-Dawley rats (Charles River Laboratories) with LPS and LPS + DEX following the protocol described above. The mortality was recorded four times per day for up to 48 hours after LPS challenge (n = 6 to 9 animals per group). Statistical differences between survival curves from different animal groups are described in the text (P < 0.05). Analysis was performed by Wilcoxon signed-rank test in SAS 9.1 (SAS Institute Inc.).
RNA isolation
Livers were homogenized and total RNA from the liver was extracted with the RNeasy Midi Kit (Qiagen) according to the manufacturer’s instructions. All samples were treated with the RNase-Free DNase Set (Qiagen). Samples were aliquoted and frozen at −80°C.
Microarray analysis
Three replicate microarrays were completed for each group (male control, male treated, female control, and female treated) for a total of 12 microarrays. Each microarray was hybridized with a pooled RNA sample, and each pool was created from equal amounts of total RNA from three independent RNA samples, representing individual animals (n = 9 animals per group). This approach provides appropriate biological replication within the constraints of a limiting number of microarrays (46). Gene expression analysis was conducted with Agilent Whole Genome Rat 4 × 44 multiplex format oligo arrays (014879; Agilent Technologies).
Labeling and hybridization
Starting with a 500-ng aliquot of total RNA from each pool, Cy3-labeled complementary RNA (cRNA) was produced with the Agilent-1 color microarray-based gene expression protocol. For each sample, 1.65 μg of Cy3-labeled cRNAs was fragmented and hybridized for 17 hours in a rotating hybridization oven. Slides were washed and then scanned with an Agilent Scanner. Data were obtained with the Agilent Feature Extraction software (v9.1), using the one-color defaults for all parameters.
Identification of significantly expressed genes
The Agilent Feature Extraction Software performed error modeling and adjusting for additive and multiplicative noise. The resulting data were processed with the Rosetta Resolver system (version 7.1) (Rosetta Biosoftware). Replicate hybridizations were combined into intensity experiments with an error-weighted average as described by Weng et al. (47). Next, ratios (male DEX/vehicle, female DEX/vehicle, and vehicle female/male) were built, and P values were generated representing the probability that a given gene was significantly, differentially expressed. A filter (P < 0.001) was applied to the P values obtained from the ratios described above. Thus, from the 41,012 features (probes) included on the array, 10,622 met the P value (P < 0.001) for at least one of the comparisons. All microarray data are available from the Gene Expression Omnibus database with series accession number GSE13461.
Biological pathway analysis
The lists of probe sets generated in Rosetta Resolver that were DEX responsive in males and females were analyzed in the IPA tool (version 6.5) (Ingenuity Systems). The average expression value of duplicate identifiers for the same molecule was used in the analyses to eliminate redundancy. Canonical pathway analysis identified pathways from the IPA library of canonical pathways and ranked them by ratio (number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway). The five highest ranked pathways that met P < 0.05 (Fisher’s exact test) are displayed.
Signature pattern extraction
The extraction of gene expression patterns in male and female glucocorticoid-regulated genes (through EPIG) was performed in the data set generated by microarray analysis. Briefly, ratio intensity values from all of the probe sets on the arrays were log2-transformed and adjusted by systematic variation normalization (15). Distinct patterns of gene expression were extracted on the basis of the expression profile correlation values, the minimum cluster size for the patterns, and the cluster-partitioning resolution (48). From the patterns and with a signal-to-noise ratio of 3 (P < 0.006), magnitude of 0.5 (1.4-fold change), and a correlation r value of 0.64 (P < 0.02) of the gene profiles, probe sets were selected. In this data set, the vehicle-treated group was used as a reference state for each of the sexes. The average of the replicates of vehicle-treated group arrays was aligned to log 0 as a baseline, with the DEX-treated samples adjusted by the same amount.
Network generation
EPIG-selected genes that directly interact with at least one other molecule in Ingenuity Pathways Knowledge Base were identified and overlaid onto a global molecular network developed from information in the Ingenuity Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated on the basis of their connectivity. Duplicate identifiers were eliminated during the process of network generation. By default, when networks are generated in the absence of expressionvalues, the first instance of the molecule is used in the analysis. The mRNA abundance for various genes in males and females was then overlaid for each network generated.
qRT-PCR analysis
Microarray results were confirmed by comparison with mRNA abundance obtained by qRT-PCR with selected predeveloped primer sets (Applied Biosystems) and following the manufacturer’s instructions. The genes that met the signal intensity value greater than or equal to 50 or had a significant glucocorticoid-induced fold change in abundance (or both) were selected for validation. Each primer set was analyzed in duplicate at least twice and normalized to the unregulated housekeeping gene TATA box binding protein primer set. The RNA was isolated as described above. Reverse transcription was carried out with 2 μg of total RNA following the protocol for the Taqman Reverse Transcription Master Mix (Applied Biosystems), and 100 ng of RNA converted into cDNA of each sample was used. Quantification was achieved with the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Statistical analysis was performed in Prism 5.0 (GraphPad).
Supplementary Material
Acknowledgments
We thank J. Revollo for discussion and assistance with data analysis, R. Oakley for discussion and assistance with manuscript preparation, and D. Andrews and H. Harris for technical assistance.
Funding: This research was supported by the Intramural Research Program of the NIH National Institute of Environmental Health Sciences.
Footnotes
Author contributions: D.D. designed and performed the experiments, analyzed the data, and wrote the manuscript. J.B.C. and J.W.C. performed microarray analysis. J.A.C. designed the experiments, analyzed the data, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Accession numbers: The microarray data have been deposited in the Gene Expression Omnibus Database (http://www.ncbi.nlm.nih.gov/geo) under accession
www.sciencesignaling.org/cgi/content/full/3/143/ra74/DC1
Methods
Fig. S1. Nascent RNA analysis of glucocorticoid-regulated genes.
Fig. S2. Comparison of male and female rat liver cross sections stained with hematoxylin and eosin.
Fig. S3. Comparison of glucocorticoid receptor mRNA and protein abundance and binding to [3H]dexamethasone in male and female rat liver.
Fig. S4. Comparison of glucocorticoid receptor mRNA and protein abundance in the liver of female rats in different phases of estrous cycle.
Fig. S5. Sexually dimorphic regulation of gene expression by glucocorticoids in isolated hepatocytes.
Fig. S6. Sexually dimorphic regulation of gene expression by glucocorticoids in mouse liver.
Fig. S7. Analyses of gender-specific expression patterns of nuclear receptor co-regulators.
Fig. S8. Analysis of dose-response curves for dexamethasone in male and female rat livers.
References
Table S1. List of glucocorticoid-responsive genes in male and female rat liver for each pattern of gene expression identified by EPIG analyses.
Table S2. List of genes contained in gene networks generated by IPA for each pattern of gene expression.
Table S3. List of common, male-specific, and female-specific glucocorticoid-regulated genes involved in inflammatory disorders.
Table S4. List of gender-specific nuclear receptor co-regulators.
References
- 1.Stotland N. Gender-based biology. Am J Psychiatry. 2001;158:161–162. doi: 10.1176/appi.ajp.158.2.161. [DOI] [PubMed] [Google Scholar]
- 2.Pecori Giraldi F, Moro M, Cavagnini F. Study Group on the Hypothalamo-Pituitary-Adrenal Axis of the Italian Society of Endocrinology, Gender-related differences in the presentation and course of Cushing’s disease. J Clin Endocrinol Metab. 2003;88:1554–1558. doi: 10.1210/jc.2002-021518. [DOI] [PubMed] [Google Scholar]
- 3.Ten S, New M, Maclaren N. Clinical review 130: Addison’s disease 2001. J Clin Endocrinol Metab. 2001;86:2909–2922. doi: 10.1210/jcem.86.7.7636. [DOI] [PubMed] [Google Scholar]
- 4.Schuurs AH, Verheul HA. Effects of gender and sex steroids on the immune response. J Steroid Biochem. 1990;35:157–172. doi: 10.1016/0022-4731(90)90270-3. [DOI] [PubMed] [Google Scholar]
- 5.Pietschmann P, Rauner M, Sipos W, Kerschan-Schindl K. Osteoporosis: An age-related and gender-specific disease—a mini-review. Gerontology. 2008;55:3–12. doi: 10.1159/000166209. [DOI] [PubMed] [Google Scholar]
- 6.Ishikawa I, Maeda K, Nakai S, Kawaguchi Y. Gender difference in the mean age at the induction of hemodialysis in patients with autosomal dominant polycystic kidney disease. Am J Kidney Dis. 2000;35:1072–1075. doi: 10.1016/s0272-6386(00)70042-4. [DOI] [PubMed] [Google Scholar]
- 7.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 8.Van Den Eeden SK, Tanner CM, Bernstein AL, Fross RD, Leimpeter A, Bloch DA, Nelson LM. Incidence of Parkinson’s disease: Variation by age, gender, and race/ethnicity. Am J Epidemiol. 2003;157:1015–1022. doi: 10.1093/aje/kwg068. [DOI] [PubMed] [Google Scholar]
- 9.Duma D, Jewell CM, Cidlowski JA. Multiple glucocorticoid receptor isoforms and mechanisms of post-translational modification. J Steroid Biochem Mol Biol. 2006;102:11–21. doi: 10.1016/j.jsbmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids—new mechanisms for old drugs. N Engl J Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 11.Eguchi H, Westin S, Strom A, Gustafsson JA, Zaphiropoulos PG. Gene structure and expression of the rat cytochrome P450IIC13, a polymorphic, male-specific cytochrome in the P450IIC subfamily. Biochemistry. 1991;30:10844–10849. doi: 10.1021/bi00109a006. [DOI] [PubMed] [Google Scholar]
- 12.Ahluwalia A, Clodfelter KH, Waxman DJ. Sexual dimorphism of rat liver gene expression: Regulatory role of growth hormone revealed by deoxyribonucleic acid microarray analysis. Mol Endocrinol. 2004;18:747–760. doi: 10.1210/me.2003-0138. [DOI] [PubMed] [Google Scholar]
- 13.Endo M, Takahashi Y, Sasaki Y, Saito T, Kamataki T. Novel gender-related regulation of CYP2C12 gene expression in rats. Mol Endocrinol. 2005;19:1181–1190. doi: 10.1210/me.2004-0063. [DOI] [PubMed] [Google Scholar]
- 14.Wiwi CA, Waxman DJ. Role of hepatocyte nuclear factors in transcriptional regulation of male-specific CYP2A2. J Biol Chem. 2005;280:3259–3268. doi: 10.1074/jbc.M409294200. [DOI] [PubMed] [Google Scholar]
- 15.Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, Malarkey DE, Houle CD, Ward SM, Wilson RE, Fannin RD, Russo MW, Watkins PB, Tennant RW, Paules RS. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci USA. 2007;104:18211–18216. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tait AS, Butts CL, Sternberg EM. The role of glucocorticoids and progestins in inflammatory, autoimmune, and infectious disease. J Leukoc Biol. 2008;84:924–931. doi: 10.1189/jlb.0208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stice JP, Knowlton AA. Estrogen NFκB and the heat shock response. Mol Med. 2008;14:517–527. doi: 10.2119/2008-00026.Stice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McElreavey K, Barbaux S, Ion A, Fellous M. The genetic basis of murine and human sex determination: A review. Heredity. 1995;75:599–611. doi: 10.1038/hdy.1995.179. [DOI] [PubMed] [Google Scholar]
- 19.Smith MJ. Sex determination. Turning on sex. Curr Biol. 1994;4:1003–1005. doi: 10.1016/s0960-9822(00)00226-8. [DOI] [PubMed] [Google Scholar]
- 20.Agrawal AK, Shapiro BH. Differential expression of gender-dependent hepatic isoforms of cytochrome P-450 by pulse signals in the circulating masculine episodic growth hormone profile of the rat. J Pharmacol Exp Ther. 2000;292:228–237. [PubMed] [Google Scholar]
- 21.Pampori NA, Shapiro BH. Feminization of hepatic cytochrome P450s by nominal levels of growth hormone in the feminine plasma profile. Mol Pharmacol. 1996;50:1148–1156. [PubMed] [Google Scholar]
- 22.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wauthier V, Waxman DJ. Sex-specific early growth hormone response genes in rat liver. Mol Endocrinol. 2008;22:1962–1974. doi: 10.1210/me.2007-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci USA. 2007;104:17081–17086. doi: 10.1073/pnas.0704126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tessitore L, Sesca E, Pani P, Dianzani MU. Sexual dimorphism in the regulation of cell turnover during liver hyperplasia. Chem Biol Interact. 1995;97:1–10. doi: 10.1016/0009-2797(94)03602-1. [DOI] [PubMed] [Google Scholar]
- 26.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez FJ. Regulation of hepatocyte nuclear factor 4 α-mediated transcription. Drug Metab Pharmacokinet. 2008;23:2–7. doi: 10.2133/dmpk.23.2. [DOI] [PubMed] [Google Scholar]
- 28.Barabási AL, Oltvai ZN. Network biology: Understanding the cells functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 29.Atkinson HC, Waddell BJ. Circadian variation in basal plasma corticosterone and adrenocorticotropin in the rat: Sexual dimorphism and changes across the estrous cycle. Endocrinology. 1997;138:3842–3848. doi: 10.1210/endo.138.9.5395. [DOI] [PubMed] [Google Scholar]
- 30.Database of Nuclear Receptor Signaling Atlas. 2010 Jul 15; http://www.nursa.org/datasets.cfm.
- 31.Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem. 2006;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer T, Carlstedt-Duke J, Starr DB. A weak TATA box is a prerequisite for glucocorticoid-dependent repression of the osteocalcin gene. J Biol Chem. 1997;272:30709–30714. doi: 10.1074/jbc.272.49.30709. [DOI] [PubMed] [Google Scholar]
- 33.Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, Adcock IM. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-κB suppression. J Exp Med. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambrou GI, Vlahopoulos S, Papathanasiou C, Papanikolaou M, Karpusas M, Zoumakis E, Tzortzatou-Stathopoulou F. Prednisolone exerts late mitogenic and bi-phasic effects on resistant acute lymphoblastic leukemia cells: Relation to early gene expression. Leuk Res. 2009;33:1684–1695. doi: 10.1016/j.leukres.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 35.Laz EV, Holloway MG, Chen CS, Waxman DJ. Characterization of three growth hormone-responsive transcription factors preferentially expressed in adult female liver. Endocrinology. 2007;148:3327–3337. doi: 10.1210/en.2006-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Y, Chen L, Wright GM, Pillai SR, Chellappan SP, Cress WD. CDKN1C negatively regulates RNA polymerase II C-terminal domain phosphorylation in an E2F1-dependent manner. J Biol Chem. 2010;285:9813–9822. doi: 10.1074/jbc.M109.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samtani MN, Jusko WJ. Comparison of dexamethasone pharmacokinetics in female rats after intravenous and intramuscular administration. Biopharm Drug Dispos. 2005;26:85–91. doi: 10.1002/bdd.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Blackford JA, Jr, Song LN, Huang Y, Cho S, Simons SS., Jr Equilibrium interactions of corepressors and coactivators with agonist and antagonist complexes of glucocorticoid receptors. Mol Endocrinol. 2004;18:1376–1395. doi: 10.1210/me.2003-0421. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Leung DY, Nordeen SK, Goleva E. Estrogen inhibits glucocorticoid action via protein phosphatase 5 (PP5)-mediated glucocorticoid receptor dephosphorylation. J Biol Chem. 2009;284:24542–24552. doi: 10.1074/jbc.M109.021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 41.Gao B, Jeong WI, Tian Z. Liver: An organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 42.Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology. 2006;43:S54–S62. doi: 10.1002/hep.21060. [DOI] [PubMed] [Google Scholar]
- 43.Montes GS, Luque EH. Effects of ovarian steroids on vaginal smears in the rat. Acta Anat. 1988;133:192–199. doi: 10.1159/000146639. [DOI] [PubMed] [Google Scholar]
- 44.Szabó C, Thiemermann C, Wu CC, Perretti M, Vane JR. Attenuation of the induction of nitric oxide synthase by endogenous glucocorticoids accounts for endotoxin tolerance in vivo. Proc Natl Acad Sci USA. 1994;91:271–275. doi: 10.1073/pnas.91.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almon RR, Dubois DC, Jin JY, Jusko WJ. Pharmacogenomic responses of rat liver to methylprednisolone: An approach to mining a rich microarray time series. AAPS J. 2005;7:E156–E194. doi: 10.1208/aapsj070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci USA. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, Bassett DE. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
- 48.Zhou T, Chou JW, Simpson DA, Zhou Y, Mullen TE, Medeiros M, Bushel PR, Paules RS, Yang X, Hurban P, Lobenhofer EK, Kaufmann WK. Profiles of global gene expression in ionizing-radiation-damaged human diploid fibroblasts reveal synchronization behind the G1 checkpoint in a G0-like state of quiescence. Environ Health Perspect. 2006;114:553–559. doi: 10.1289/ehp.8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.