Abstract
The Department of Veterans Affairs (VA) recognized the need to balance patient-centered care with responsible creation of generalizable knowledge on the effectiveness of molecular medicine tools. Embracing the principles of the rapid learning health-care system, a new clinical program called the Precision Oncology Program (POP) was created in New England. The POP integrates generalized knowledge about molecular medicine in cancer with a database of observations from previously treated veterans. The program assures access to modern genomic oncology practice in the veterans affairs (VA), removes disparities of access across the VA network of clinical centers, disseminates the products of learning that are generalizable to non-VA settings, and systematically presents opportunities for patients to participate in clinical trials of targeted therapeutics.
Keywords: veterans, precision oncology, learning health-care system, lung cancer, Bayesian
Introduction
Oncology clinical practice guidelines recommend more than 30 molecular tumor biomarkers across all cancers to aid treatment selection, a list of potential biomarkers that continues to grow.1–3 In addition to reimbursable, standard-of-care assays, physicians can order biomarker panel tests that sequence large regions of the tumor genome. The proximate goal of biomarker panel testing is to identify potential, even unproven, therapeutic agents that may offer longer survival and improved quality of life than existing regimens approved by the US Food and Drug Administration (FDA) and recommended by professional guidelines committees.4,5 Biomarker panel testing may be particularly useful in advanced cancers, where usual cytotoxic chemotherapy may at best lead to one-year survival. The best example is non-small cell lung cancer (NSCLC). A recent publication by Kris et al, reporting for the Lung Cancer Mutation Consortium, found that tumor genotype analysis aided physicians in selecting therapy and that patients with driver mutations who received matched targeted therapy lived longer.6
Some promising anecdotes and, in the past year, a few prospective studies suggest the potential of some biomarkers and treatment combinations,7 and there is emerging evidence that biomarker panel tests are safe and effective and may provide an efficient use of medical resources.8–10 Sponsors, such as manufacturers and government agencies, traditionally fund studies of the effects of particular treatments in patients with a specified molecular profile, with the notable exception of the new bucket and basket trials.11,12 The glacial pace of traditional clinical trial research has not kept up with rapid advancements in technology for biomarker discovery, targeted drug development, and the urgent need for patients faced with life-threatening illness.
Health-care systems see the need to understand how to efficiently use genomic technologies and verify their safety and effectiveness with timely evidence. Advances in informatics technology play a key role, enabling capture and storage of each patient’s clinical encounter and outcome data in a form that facilitates reuse in the care of other patients. The Institute of Medicine refers to this as the emergence of the rapid learning health-care system.13,14 The Department of Veterans Affairs Veterans Integrated Service Network 1 (VISN 1) embraces this paradigm and has developed a new clinically driven prototype, called the Point-of-Care Precision Oncology Program (POP), focused initially on lung cancer. It has been designed to seamlessly merge traditional clinical activities with a systematic approach to exploiting potential breakthroughs in genomic medicine, while generating credible, timely evidence in the clinical setting.
In this paper, we outline the program’s primary objectives and operating principles, the construction of the knowledge base, and the integration and analysis of data. We then describe how the information from the knowledge base can help guide individual decision-making.
Objectives and Core Principles
The POP has six primary objectives: (1) assure access to modern oncology genomic practice in the VA; (2) remove access disparities across the VISN 1 clinical centers; (3) increase the quality and speed of learning from new approaches to analyzing and interpreting the complex biological information used in real-world settings; (4) adapt clinical practice within the VA, based on lessons learnt from the program; (5) disseminate the products of learning and adaptation that are generalizable to non-VA settings and other diseases; and (6) systematically present opportunities for patients to participate in clinical trials of targeted therapeutics.
Putting the POP into action required establishing a set of core principles that would govern operations and address challenges that would arise in the course of creating a new program. The first and foremost principle is to place respect for patient autonomy and self-determination at the center of every decision (patient centeredness).15,16 The VA has a strong tradition of patient participation, from the bedside to program development, implementation, and monitoring. In addition to traditional committees (Institutional Review Board [IRB]), the POP will make use of an advisory human research committee comprised in part by veterans to oversee the ethical conduct of clinical studies and will participate directly in auditing the performance and modifying the direction of the program.
Despite the growing appeal of molecular versus tissue-of-origin stratification of tumor biology, we focused on a single indication (single cancer type). From the point of view of a system of screening, pathological diagnosis, care, and follow-up, tissue type works best as an organizing principle. However, as discussed below, such a system can use trials that enroll on the basis of molecular biomarker regardless of tissue type to offer molecularly guided treatments to patients. Criteria for selecting a cancer type included rapidly changing science on mutational tumor status, large interpatient variability in tumor mutation status, high likelihood that mutation status would affect treatment selection, and a relatively common cancer type. In the VA, adenocarcinoma (AC) and squamous cell carcinoma (SCC) of the lung satisfy these criteria.
Patients and physicians alike must come to see participation in learning as essential to their health-care system (participatory learning stance). Learning activities or research resides primarily in academic medical centers, funded by government agencies, philanthropy, or industry. We propose to transform the perception of learning and clinical practice as wholly separable activities and replace it with an active endorsement of participation in learning by patients and practitioners. The program intends to foster in all meetings and materials an increased awareness that each member of the group gains something from participating in the program.17 POP builds on existing VA programs, networks, and infrastructure to make the program economically viable and limit the disruption of existing high-quality care as the program continues to evolve (leverage existing structures). Prior efforts in the VA on clinical informatics and decision support include the Million Veteran Program,18 the Genomic Information System for Integrated Science, Veterans’ Informatics, Information, and Computing Infrastructure, and the Point-of-Care Clinical Trial Program.19 The POP engages participants and resources from VA’s network of New England clinical centers, clinicians, patients, and researchers, including methodologists and domain experts in laboratory medicine, genomics, and clinical research. Non-VA-affiliated researchers from academia, industry, and other health-care settings will provide invaluable insights. The program may on a case-by-case basis seek to provide access to drugs, which are not otherwise readily available (because of regulatory constraints or financial barriers), by seeking assistance from the FDA and industry on behalf of its patients.
The POP will provide regular and transparent dissemination of information about how the program is functioning and about the effects of genomics-driven cancer care (transparent dissemination of learnings). The learnings are to be disseminated so as to permit generalization to other parts of the VA and to interested parties outside the VA.
The final principle requires the POP to enable patients to find opportunities to participate (when it is appropriate) in clinical trials of targeted therapeutics, across the full spectrum of sponsoring agencies and companies. The pace of change forced upon the rapid Learning Healthcare System (rLFS) by the acceleration of basic science knowledge requires the reliability and inferential efficiency of the experimental approach, including participation in randomized trials. Therefore, the experiment must be integrated into the processes of care in the rLHS, rather than relegated to the periphery.
Into Action
Enrollment, consent, and tissue acquisition/analysis
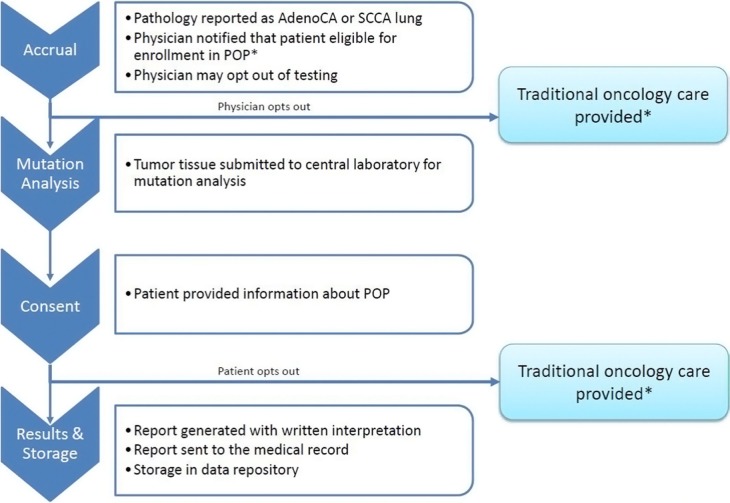
Patients with NSCLC are eligible to participate in the POP (Fig. 1) and are identified by a report in the existing electronic medical record system of a new diagnosis documented in a pathology report or by a physician notifying the POP center. Program coordinators approach the treating physician to present the option of molecular tumor profiling. The physician may exclude the patient from testing if testing is not clinically indicated or if the patient will not be actively treated at the VA facility. Reasons for opting out are recorded.
Figure 1.
Workflow of accrual and consent into the VA’s Point-of-Care Precision Oncology Program.
Note: *Mutation analysis may be ordered by clinician outside of the POP structure.
Patients are then informed about the results of the biomarker panel test that is performed as part of routine POP clinical care and are provided written information about the additional research activities of the POP and the principles that govern the program. Through this process, they are given an opportunity to learn about how analyses are conducted, the algorithms used for data interpretation, and the implications for their care. The information makes it clear that participation in the research program is voluntary and that they may opt out at any point in the process. Informed consent and Health Insurance Privacy and Portability Act (HIPAA) authorization are requested from the patients to permit inclusion of their clinical data in the POP knowledge base, allow additional molecular profiling of residual biosample and blood (when scientifically useful), and recontact for scientific purposes, such as opportunities for enrollment into clinical trials.20
Tumor analysis and subsequent treatment decisions
Targeted sequencing of cancer genes (multiple biomarker panels) is performed on formalin-fixed, paraffin-embedded tissues from newly diagnosed lung cancers as part of routine POP cancer care. Samples are shipped within 48 hours of diagnosis to Personal Genome Diagnostics (PGD; CancerSelect88-targeted genome panel) or Personalis (ACE Extended Cancer Panel). Following sequencing the targeted gene regions for mutations, a formal report of identified genomic aberrations is collated, annotated, and transmitted for inclusion in patient medical records. Both PGD and Personalis use N-of-One to curate the medical literature and provide mutation annotations. The expected turnaround time for results is 14 days. The Ann Arbor VA Medical Center (VAMC) provides a molecular oncology consultation service to the POP to assist VA clinicians, as they make decisions to treat their patients based on the annotated results. This is accomplished through a virtual tumor board wherein the oncologist has access to patients’ clinical, anatomical pathological, and molecular diagnostic data. The POP will record how this information was used to decide the patient’s treatment, along with a detailed report of the therapy. If the patient is not treated according to the results from the molecular diagnostics, the reasons are recorded. Patients are followed up through the course of their therapy until death.
Until the POP knowledge base accrues a substantial body of information on outcomes in POP patients, treatment recommendations will be based on the results of the multiple biomarker panels and associated annotations and other relevant patient characteristics. In this section, we describe three broad categories of results and their possible consequences, with particular attention to those leading to the possibility of randomization. In a subsequent section, we describe how those possibilities can be realized.
In Table 1, panel A displays the mutations listed in authoritative sources (FDA labels, ASCO guidelines, NCCN guidelines and its compendium for lung cancer, and UptoDate® [Wolters Kluwers]), for which there is at least one therapy that has a proven efficacy for known end points (eg, progression-free survival and response rates) in patients with a particular biomarker. The following drugs are currently approved by the FDA: erlotinib and afatinib for epidermal growth factor receptor (EGFR)-mutant lung AC, crizotinib and ceritinib for anaplastic lymphoma kinase (ALK)-rearranged lung AC, and more recently nivolumab for SCC of lung. The FDA has approved eight other drugs for lung cancer, but without labeling for use as targeted therapy aligned with a mutation. Panel B lists the available cancer drugs that do not have a relationship with a known biomarker. Panel C lists the biomarkers that are thought to be important for cancer, but for which there is no current FDA-approved targeted therapy. Because of the rapidity of change and the difficulty in remaining current, Table 1 does not illustrate the large number of compounds being tested in the clinical trials of targeted therapies in solid tumors by an increasing number of sponsors. Table 1 is a snapshot of the current state of approved therapies and will require updating as new drugs and new associations with biomarkers are developed.
Table 1.
Biomarkers and associate drugs by category.
| BIOMARKER | THERAPEUTIC IMPLICATIONS | BRAND NAME | NCCN GUIDELINES | NCCN COMPENDIUM | FDA-APPROVED LABELING | |
|---|---|---|---|---|---|---|
| LUNG | WITH BIOMARKER | |||||
| Category A | Known biomarker; FDA-approved biomarker-related drug for lung cancer | |||||
| EGFR mutation | Erlotinib | Tarceva | x | x | x | x |
| Gefitinib | Iressa | x | ||||
| Afatinib | Gilotrif | x | x | x | ||
| ALK translocation | Crizotinib | Xalkori | x | x | x | x |
| Category B | Biomarker unknown or not approved by FDA; FDA-approved drug for lung cancer | |||||
| Bevacizumab | Avastin | x | x | x | ||
| Carboplatin | Generic | x | x | x | ||
| EGFR expression | Cetuximab | Erbitux | x | x | x | |
| ERCC1 | Cisplatin | Generic | x | x | x | |
| ROS1 | Crizotinib | Xalkori | ||||
| Docetaxel | Taxotere | x | x | x | ||
| Erlotinib | [see above] | x | x | |||
| Etoposide | Generic | x | x | x | ||
| RRM-1 | Gemcitabine | [see above] | x | |||
| Ifosfamide | Ifex | x | x | |||
| Irinotecan | Camptosar | x | x | x | ||
| Mesna | Generic | x | ||||
| Mitomycin | Generic | x | x | |||
| PDL-1 | Nivolumab | Opdivo | ||||
| Paclitaxel | Generic | x | x | x | ||
| Pemetrexed | [see above] | x | ||||
| Vinblastine | Generic | x | x | x | ||
| Vinorelbine | Navelbine | x | x | |||
| Category C | Potential biomarker, no FDA-approved drug available | |||||
| MEK1 mutation | ||||||
| PIK3ca, AKT1, PTEN alterations | ||||||
| FGFR1 amplifications | ||||||
| Beta-catenin mutation | ||||||
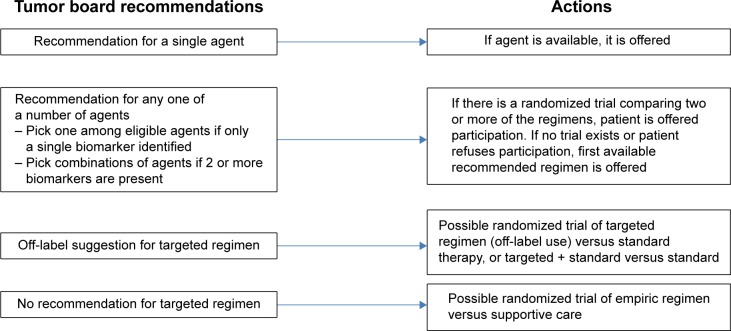
The annotated assays provided reflect the curation and updating of such information and are converted by the clinician into one of only a few possible recommendations, as depicted in Figure 2. Figure 2 illustrates the sequence of decision-making and possible actions that follow from the review of the patient’s data, including the results of the tumor sequencing. Despite the possible complexity of the molecular assay, it is expected that the tumor board’s recommendations will impose simplification, so that only the scenarios shown in Figure 2 will occur with any frequency. When the tumor board’s recommendation is unique and firm (first scenario), the treating physician will decide whether to agree to offer the targeted treatment (eg, crizotinib for ALK-rearranged lung AC). In the second scenario, mutations may be identified for which more than one targeted therapy could be recommended by the tumor board (eg, erlotinib or afatinib for initial therapy of EGFR-mutant lung AC). If the tumor board believes that available evidence for these approved drugs is insufficient to recommend one or the other (referred to as scientific equipoise), then either drug may be used, depending on availability in VA formulary.
Figure 2.
Tumor board recommendations and associated actions.
As shown in Table 1, there are genotypic subtypes (eg, RET mutation) for which there are available drugs (vandetanib) that may be effective in this subtype of lung AC (based on case reports) but not yet approved by the FDA for lung cancer (this drug has FDA approval for thyroid cancer). We anticipate creating an off-label use protocol, so that the treating physician may decide to use this drug for the particular patient, provided all permissions and IRB approval are obtained for this scenario or utilize a standard therapy. A more common scenario for patients with lung AC is the identification of an ROS-1 rearrangement. Recent data reveal dramatic responses to crizotinib, which is currently the only drug approved by the FDA for ALK-rearranged lung AC. In this latter instance, off-label use of crizotinib can be better justified, based on the level of evidence. Therefore, the decision to utilize targeted off-label therapy requires patient and physician education and IRB approval.
Access to trials
There are currently several entities (Southwest Oncology Group, pharmaceutical companies) offering bucket trials that offer a unique trial design with intermediate well-defined end points that match the patient’s tumor target to therapy (eg, access to phosphoinositide 3-kinase [PI3K] inhibitor for patients with a PI3K mutation).
The POP will identify and activate a number of clinical trials relevant for patients with NSCLC and will match participant characteristics contained in the knowledge base to inclusion and exclusion criteria to determine eligibility. Patients and care providers are informed of available trials to which they have matched and invited to enroll. A number of unique VA features including a central IRB, Federal Wide Accreditation to perform research at most facilities, credentialed research oncologists at key facilities, and pharmacies experienced in handling investigational drugs make it possible to treat most patients at their home facility. Facilities participating in the POP will be eligible to join SWOG as a member of a VA consortium approved through an SWOG Storefront mechanism. It is anticipated that clinical trials ranging from phase II to IV will be sponsored by a variety of entities (including the NCI study groups as well as pharma companies) and made available to participating VA facilities based on the study complexity and infrastructure requirements.
Knowledge base
In this example of an rLHS, learning is accomplished by fitting statistical models to the data from previous patients, as discussed below. It is then put into action by introducing the predictions from those models into the decision-making process. An informatics platform tracks specimen status and includes findings from the targeted sequencing of the tumor tissue, as described earlier. The information is part of a comprehensive knowledge base that can help guide clinical management decisions. The knowledge base includes (1) general background understanding; (2) observed patient-level data, including baseline characteristics, treatment, disease outcomes, and side effects; and (3) model-based actuarial summaries of observed outcomes of patients enrolled in the POP. Background understanding includes biological knowledge culled, collated, and curated from many public data sources. Examples include findings of published studies of the effect of a mutational variant upregulating a known pathway and how certain treatments interfere with a downstream product of that variant. Updates to this background (externally derived) knowledge are provided by N-of-One (see above) each time a mutation is identified in a patient sample, thus assuring that the knowledge base is current for the patient under consideration. In addition, the background knowledge may include results of patient-level data analysis from other programs and databases. Observed patient-level data from POP patients include demographics, mutations, management choices, outcomes, and resource use, which are drawn from various VA informatics platforms. The actuarial summaries of observed outcomes are the product of statistical analyses (models), which predict the outcomes that are likely to occur in the patient treated one way or the other.
Examples of statistical analyses are survival curves, Cox models, and logistic regressions. Such analyses of patient-level data are invaluable tools for identifying and testing clinically meaningful associations between outcomes and potential predictive factors, such as patient characteristics and treatments. Reports on point-of-care informatics support for clinical decision-making, where Cancer Commons21 and The Green Button22 propose to provide customized predictions for individual patients by fitting statistical models of outcomes to data on treatments and outcomes in patients similar to the current patients. One can define the notion of similarity between patients by considering only the predictive features, without using the outcomes, by means of methods, such as cluster analysis and principal components analysis, which are referred to as unsupervised. Otherwise, one can use a statistical model that relates the predictors to outcomes to define similarity, giving differences in influential features more weight in the distance metric.23 In either case, once a similarity metric is defined, the cohort of similar patients can be constructed and a customized model fit to outcomes in that cluster. The choice of general approach and specific techniques is the focus of intensive research in the emerging field of data science.
There are two challenges to statistical modeling in our context: (1) there will be few patient subgroups with similar molecular profiles, while (2) the space of possible available therapy to target combinations may be large and sparsely filled by observed instances of use of those combinations and their outcomes. Thus, the number of patients similar to the current patients may not be adequate to support a conventional statistical model comparing response to various treatments, unless the notion of similarity is relaxed enough to lose the advantage of customization. Because of the challenges posed by modern high-dimensional data, this is a well-studied statistical problem and there are ways to make the most of such data, by means of Empirical Bayes and related techniques.24,25 These methods are based on the recognition that when there are not enough direct data, one must borrow strength from related information.
The second challenge is more profound, relating to the usual disconnect between background knowledge and statistical modeling. In this context, the background knowledge includes basic and preclinical science on the pathways affected by the mutations, the targeting of therapies to those pathways, and similar information. It also includes early clinical information on pharmacokinetics and pharmacodynamics parameters of treatments, class effects, and observations of responses and surrogates. It includes information on clinical predictors of outcome, such as histopathology, size and location of tumors, and others. Traditional statistical models seldom bring the background knowledge into the analysis in an explicit way. Instead, the background knowledge is used to guide the statistical approach to patient-level data. Background knowledge is essential to patient and feature selection, choice of outcome (overall survival or progression-free survival), and choice of predictors (in particular, what driver mutations should be considered, which histopathological and clinical predictors, and which candidate treatments should be included in the model). The background knowledge helps cut the problem down to size by proposing only those analyses for which there is biological plausibility, avoiding a blind search.
As the relevance and complexity of the background knowledge grow, it is difficult to use it informally as described earlier. The complexity of cancer biology is so great, and the body of published background knowledge is growing at such a fast pace that experts have begun to question whether even the most experienced panel, such as would be convened in a tumor board, could capture the information, retain it in their memories, and render a timely interpretation for routine clinical care. This line of reasoning motivates a proposal to develop the necessary theory and methods for a more automated approach, likely based on the Bayesian paradigm, but with data-based priors. The construction of data-based priors is straightforward when the data are of the same type as the observations in the POP database, for example, response or survival rates in non-POP lung cancer patients along with their individual driver mutations, other clinical features, and treatment information. Currently, much of this information is dispersed across the individual health-care systems that are beginning to collect and use tumor genotype information and record outcomes. Access to such information may be facilitated by methods for distributed model building without aggregation of data.26 If participation in the program grows faster than evidence from external sources, which is continually updated in the knowledge base, model-based predictions will be dominated by data from patients in the program.
As the POP begins, there will not be a great deal of patient-level data available for use in constructing data-based priors. We do not know if there is enough information in other kinds of data (eg, cell line and animal work) to be of use in defining data-based priors for patient-based modeling. Therefore, we expect that such data will be used implicitly, as described earlier, to shape the modeling approaches.
From learning to adaptation
Patient information will grow eventually to an appropriate level for analysis and interpretation. At that point, it must be introduced into the decision-making process of the tumor board, and this step implements the learning, which is reified in the statistical models discussed earlier. The models built on the knowledge base will generate a list of possible treatment assignments and the model-based estimates of expected outcomes, as described earlier, with a range of statistical uncertainties for the individual patient. All potential assignments are subjected to clinical review by the tumor board and the prescribing physician. (Interpretation of the predictions generated from the knowledge base is not on autopilot.) As described earlier, there are instances where the output from the knowledge base proposes several treatments and where the statistical uncertainty is so large that the tumor board may choose to recommend a set of therapies with scientific equipoise among them. Methods to introduce randomization in the clinical setting are being tested by the Office of Research and Development of the Department of Veterans Affairs in the Point-of-Care Clinical Trials Program.27 As the data accumulate, the updated knowledge base recommendations will become more definite, enabling the transition from learning to adaptation from what has been learned.
That transition need not be abrupt. An example of a gradual transition is the randomized play the winner strategy, where the probability of randomization to a treatment is dependent on the strength of the evidence favoring that treatment. Ultimately, the goal is to find optimal strategies that efficiently improve the health and well-being of the entire group of patients in the learning system.
Ethical issues
As an instance of an rLHS, POP operates across the ill-defined border between clinical quality improvement and clinical research. As long as regulatory conflicts remain unresolved, it is necessary for each rLHS to find an ethical way to operate efficiently. Examples of complicating issues concern the permissions required to share patient data with researchers who in turn not only provide treatment recommendations for clinical consideration but also produce generalizable knowledge. Another example is patient permission to perform additional research-level tissue assays (such as whole exome sequencing), where the results of such assays could impact clinical care (by suggesting a preferred treatment option) or improve our understanding of cancer biology. The approach taken by the POP and approved by the VA Boston Healthcare System IRB is to separate what can be considered purely clinical activities (tumor analysis and treatment recommendations) from research activities (data sharing for discovery and validation, expanded tumor sequencing) and obtaining informed consent from the patients and HIPAA authorization for the latter. Therefore, patients can benefit from the clinical program even if they choose not to participate in the research offerings.
Further dissemination to reduce disparities of care
Once the feasibility testing of the process is completed at the VISN 1 oncology consortium sites, the program will be rolled out to a wider circle of VAMCs, based on their interest and ability to participate. As the program continues to be refined and improved, it will be offered to all VAMCs. In this way, the program will provide uniform access to modern precision oncology to all VA patients who wish to participate and who are seeking care at all VAMCs that join. This uniform access is the first critical step in the process of reducing disparities. Subsequent steps include detecting anomalies in the process of care that might suggest remediable disparities and alerting responsible parties to take action.
Conclusion
By creating the POP, VISN 1 seeks an efficient approach to provide access to potential breakthroughs in genomic medicine, while generating credible evidence in real-world settings and in real time. In addition, it already has fostered valuable partnerships between providers, pharmaceutical companies, and cooperative study groups working toward a common goal of enhanced cancer treatment. The POP directly influences patient care and is forming a knowledge base to improve the standard of care, create parity in the treatment of cases across VA hospitals, and improve cancer outcomes for veterans. In 2015, the VA is well placed to attempt a test of the participatory learning stance advanced by POP because its members’ military service and training have instilled the principle that loyalty to the group is essential for the success of individual members and the group as a whole. However, the learnings from the VA may be relevant in the future to non-VA sites creating programs built along similar principles but adapted to their clinical cultures and specific needs of their patients.
Future Directions
Success of the POP will be determined largely by the extent to which targeted therapies are adopted by the clinical community. Entry into the program is determined by the clinical decision to test patients, which is driven by an understanding of the costs and benefits of both testing and treatments. While the cost of tumor profiling is dropping, there is concern that the price tag of novel targeted therapies will remain prohibitively high. Partnerships between health-care systems and drug researchers have the promise of reducing the cost of drug discovery and accelerating the pace of discoveries for the mutual benefit of research and health care.
Executive Summary
– Molecular profiling of tumors identifies mutations against which targeted therapies are effective.
– The great diversity of driver mutations within and across tumor types results in enrollment difficulties for clinical trials studying novel treatments and makes it difficult to understand the impact of specific mutations.
– The Department of Veterans Affairs has launched a pilot program of lung cancer sequencing that provides for optimal clinical care and presents research and learning opportunities as by-products.
– The POP enables a learning health-care system by the reuse of clinical data to determine significant mutational status and to match patients to ongoing trials.
Acknowledgments
Lavori acknowledges support from the grant UL1 TR001085 from The National Institutes of Health to Stanford University.
Footnotes
ACADEMIC EDITOR: Barbara Guinn, Editor in Chief
PEER REVIEW: Two peer reviewers contributed to the peer review report. Reviewers’ reports totaled 598 words, excluding any confidential comments to the academic editor.
FUNDING: This study was funded by the Department of Veterans Affairs Veterans Integrated Service Network 1. PL received support from the grant UL1 TR001085 from The National Institutes of Health to Stanford University. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: LDF, MTB, PL. Wrote the first draft of the manuscript: LDF, JH, PL. Contributed to the writing of the manuscript: LDF, JH, PL. Agree with manuscript results and conclusions: LDF, MTB, ST, VK, NR, CS, RF, SP, MAF, JH, PL. Jointly developed the structure and arguments for the paper: CS, ST, VK, RF, MAF, NR. Made critical revisions and approved final version: LDF. All authors reviewed and approved of the final manuscript
REFERENCES
- 1.Fan YS. Companion diagnostic testing for targeted cancer therapies: an overview. Genet Test Mol Biomarkers. 2013;17(7):515–523. doi: 10.1089/gtmb.2012.0510. [DOI] [PubMed] [Google Scholar]
- 2.Keedy VL, Temin S, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29(15):2121–2127. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- 3.Topol EJ. Individualized medicine from prewomb to tomb. Cell. 2014;157(1):241–253. doi: 10.1016/j.cell.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiore LD, D’Avolio LW. Detours on the road to personalized medicine: barriers to biomarker validation and implementation. JAMA. 2011;306(17):1914–1915. doi: 10.1001/jama.2011.1605. [DOI] [PubMed] [Google Scholar]
- 5.Garraway LA, Verweij J, Ballman KV. Precision oncology: an overview. J Clin Oncol. 2013;31(15):1803–1805. doi: 10.1200/JCO.2013.49.4799. [DOI] [PubMed] [Google Scholar]
- 6.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted therapies. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez-Chavez A, Thomas A, Rajan A, et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol. 2015;33(9):1000–1007. doi: 10.1200/JCO.2014.58.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadauld L. Implementation of a precision cancer program in an integrated health care system. J Clin Oncol. 2015;33(suppl):abstre17647. [Google Scholar]
- 9.Nadauld L. Precision medicine to improve survival without increasing costs in advanced cancer patients. J Clin Oncol. 2015;33(suppl):abstre17641. [Google Scholar]
- 10.Sparano JA, Gray RJ, Makower DF, et al. Validation of a 21-gene expression assay in breast cancer. N Engl J Med. 2015;373(21):2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasche B, Grant SC. Non-small cell lung cancer and precision medicine. A model for the incorporation of genomic features into clinical trial design. JAMA. 2014;311(19):1975–1976. doi: 10.1001/jama.2014.3742. [DOI] [PubMed] [Google Scholar]
- 12.Redig AJ, Janne PA. Basket trials and the evolution of clinical trial design in an era of genomic medicine. J Clin Oncol. 2015;33(9):975–977. doi: 10.1200/JCO.2014.59.8433. [DOI] [PubMed] [Google Scholar]
- 13.Institute of Medicine . The Learning Healthcare System: Workshop Summary (IOM Roundtable on Evidence-Based Medicine) Washington, DC: The National Academies Press; 2007. [PubMed] [Google Scholar]
- 14.Mason AR, Barton AJ. The emergence of a learning healthcare system. Clin Nurse Spec. 2013;27(1):7–9. doi: 10.1097/NUR.0b013e3182776dcb. [DOI] [PubMed] [Google Scholar]
- 15.Beauchamp TL, Childress JF. Principles of Biomedical Ethics. 2nd ed. New York, NY: Oxford University Press; 1983. [Google Scholar]
- 16.Thompson J, Bissell P, Cooper CL, Armitage CJ, Barber R. Exploring the impact of patient and public involvement in a cancer research setting. Qual Health Res. 2014;24(1):46–54. doi: 10.1177/1049732313514482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu CG, Mitchell TL, Fitch MI. From patient to participant: enhancing the validity and ethics of cancer research through participatory research. J Cancer Educ. 2013;28(2):237–246. doi: 10.1007/s13187-013-0464-2. [DOI] [PubMed] [Google Scholar]
- 18.Million Veteran Program (MVP) VA Office of Research and Development. 2014. [Accessed April 22, 2014]. Available at: http://www.research.va.gov/MVP/researchers.cfm.
- 19.D’Avolio L, Ferguson R, Goryachev S, et al. Implementation of the Department of Veterans Affairs’ first point-of-care clinical trial. J Am Med Inform Assoc. 2012;19(e1):e170–e176. doi: 10.1136/amiajnl-2011-000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shrager J, Tenenbaum JM. Rapid learning for precision oncology. Nat Rev Clin Oncol. 2014;11(2):109–118. doi: 10.1038/nrclinonc.2013.244. [DOI] [PubMed] [Google Scholar]
- 22.Longhurst CA, Harrington RA, Shah NH. A ‘Green Button’ for using aggregate patient data at the point of care. Health Aff. 2014;33(7):1229–1235. doi: 10.1377/hlthaff.2014.0099. [DOI] [PubMed] [Google Scholar]
- 23.Wang F, Sun J, Li T, Anerousis N. Two heads better than one: Metric + active learning and its applications for IT service classification; ICDM. Ninth IEEE International Conference on Data Mining; Miami, FL, USA: 2009. pp. 1022–1027. [Google Scholar]
- 24.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference and Prediction. 2nd ed. New York, NY: Springer; 2009. [Google Scholar]
- 25.Efron B. Large Scale Inference. IMS Monographs. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- 26.Narasimhan B, Rubin DL, Gross SM, Bendersky M, Lavori PW. Software for distributed computation on medical databases: a demonstration project. 2015. arXiv:1412.6890. http://arxiv.org/abs/1412.6890 (last accessed 5 February 2016)
- 27.Fiore LD, Brophy M, Ferguson RE, et al. A point-of-care clinical trial comparing insulin administered using a sliding scale versus a weight-based regimen. Clin Trials. 2011;8(2):183–195. doi: 10.1177/1740774511398368. [DOI] [PMC free article] [PubMed] [Google Scholar]