Abstract
Aim:
To determine associations between methylation of NR3C1, HSD11B2, FKBP5 and ADCYAP1R1 and newborn neurobehavioral outcomes.
Methods:
In 537 newborns, placental methylation was quantified using bisulfite pyrosequencing. Profiles of neurobehavior were derived via the Neonatal Intensive Care Unit Network Neurobehavioral Scales. Using exploratory factor analysis, the relationships between methylation factor scores and neurobehavioral profiles were examined.
Results:
Increased scores of the factor characterized by NR3C1 methylation were associated with membership in a reactive, poorly regulated profile (odds ratio: 1.47; 95% CI: 1.00–2.18), while increased scores of the factor characterized by HSD11B2 methylation reduced this risk.
Conclusion:
These results suggest that coordinated regulation of these genes influences neurobehavior and demonstrates the importance of examining these alterations in a harmonized fashion.
Keywords: : ADCYAP1R1, fetal basis of adult disease, FKBP5, glucocorticoids, HSD11B2, neurobehavior, NR3C1, placenta
Codified in the developmental origins of health and disease framework, the in utero environment transmits signals to the fetus during critical developmental windows that have long-term impacts on various aspects of health including neurodevelopment. Maternal cortisol levels increase over the course of gestation and appropriate control of glucocorticoid exposure is necessary for proper fetal development. Excessive exposure can contribute to dysregulation of the fetal cortisol response pathway, and may be detrimental to appropriate neurodevelopment [1,2]. The placenta is an endocrine organ that regulates the fetal environment and specifically fetal glucocorticoid exposure by expressing glucocorticoid uptake and inactivation genes, which shield the developing infant from excessive glucocorticoid exposure [3,4]. Appropriate functioning of the placenta is crucial to fetal development, and can be perturbed by epigenetic regulation. DNA methylation is a form of epigenetic variation that alters gene transcription potential [5]. This methylation may be susceptible to alteration by the environment, and emerging research has highlighted DNA methylation during the in utero period as a potential mechanism underlying prenatal programming [3].
NR3C1 encodes the glucocorticoid receptor, the primary nuclear receptor that elicits transcriptional activation of cortisol response pathways, and is highly expressed in the developing placenta through the end of gestation. In rodents, poor maternal care was associated with increased hippocampal NR3C1 exon 17 (equivalent to human exon 1F) methylation and increased anxiety-like behavior in the offspring [6]. Placental methylation of the same region (13 CpGs within exon 1F) has been associated with infant cortisol reactivity quantified through salivary cortisol measurements [7], suggesting that methylation of this region plays a role in programming the developing stress response. Placental NR3C1 exon 1 methylation has been associated with decreased expression, infant birth weight [8] and infant neurobehavioral outcomes [9,10]. As reviewed in Lester et al., [3], multiple studies have identified that aberrant methylation of this region disrupt infant neurobehavioral development and cortisol response throughout life.
Eleven beta hydroxysteroid dehydrogenase type 2 converts cortisol to its inactive form, cortisone, thereby limiting fetal exposure to maternal circulating active cortisol [11]. Placental methylation of four CpGs within the HSD11B2 promoter is negatively correlated with placental HSD11B2 expression [12], which may lead to increased fetal glucocorticoid exposure in utero. Implicating this alteration to fetal programming, placental HSD11B2 promoter methylation has been associated with prenatal adversity [13], reduced newborn quality of movement [12] and through an interaction with maternal anxiety to poor newborn muscle tone [10].
FKBP5 reduces cellular glucocorticoid response by impeding nuclear translocation of the glucocorticoid receptor [14]. FKBP5 is expressed in the placenta [15], and methylation of two CpGs within intron 7 has been associated with decreased placental gene expression [16]. In infants, placental FKBP5 methylation has been associated with increased arousal scores, which reflect inappropriate hyper-reactivity and sensitivity to the environment [16]. In peripheral blood from adults, methylation of this same region has been associated with post-traumatic stress disorder (PTSD), which is characterized by dysregulated stress response [17].
PACAP is neurotransmitter that stimulates cortisol secretion in adrenocortical cells when it binds to its receptor PAC1 [18]. PAC1 antagonism stimulates the hypothalamic–pituitary–adrenal (HPA) axis and attenuates corticosterone release in rodents [19]. There is increased placental expression of ADCYAP1 and ADCYAP1R1 over the course of gestation, which may promote placental growth [20]. Expression of ADCYAP1R1 is upregulated in rodents by both fear conditioning and estrogen [21], and methylation of one CpG in the ADCYAP1R1 promoter in white blood cells has been associated with PTSD in a sex-specific manner among adults [21], as well as asthma and exposure to violence in children [22]. This suggests that the environment can influence ADCYAP1R1 methylation at this region and potentially contribute to mental health outcomes.
FKBP5, HSD11B2, NR3C1 and PAC1 operate in a concerted fashion to regulate glucocorticoid secretion and cortisol response in the placenta and in other tissues. There is a growing interest in examining multiple factors as implied in the ‘genoset’ concept described in Bogdan et al. [23]. Previous work has examined the joint contribution of NR3C1 and HSD11B2 methylation in the placenta and components of neurodevelopment [24], and in this study we sought to provide a more integrated assessment of genes involved in cortisol response and their contribution to risk of neurobehavioral adversity. We examined the relationship between methylation of FKBP5, HSD11B2, NR3C1 and ADCYAP1R1 and newborn neurobehavioral outcomes using the Neonatal Intensive Care Unit Network Neurobehavioral Scales (NNNS), a validated, quantitative and prospectively predictive assessment of infant neurobehavior. The regions we have elected to sequence were selected because of prior biological evidence showing a relationship between methylation of these genes and cortisol response in other studies [6–10,12–13,16–17,21–22]. We summarized methylation data across these genes through exploratory factor analysis. This approach allowed us to distill this moderately high dimensional data into a smaller number of latent variables in order to appropriately represent variability of methylation in these regions preserving correlation structure and to overcome type I error related to a large number of comparisons being made. The factors generated are a data-driven summary statistic of the methylation status of glucocorticoid response genes in the placenta, and provide a more holistic assessment of methylation status of these candidate genes than using the mean of each gene individually. Factor analysis techniques are commonly used in psychology studies, and have been used to examine high dimensionality datasets including gene-expression data [25,26] and microarray data [27]. This is also among the first studies to conduct an integrated assessment of the relationship between placental glucocorticoid response genes and infant neurobehavior.
Methods
Study population
The infants involved in this analysis represent all infants enrolled in the Rhode Island Child Health Study (RICHS) from September 2010 until February 2013 that completed the neurobehavioral assessments (NNNS) at birth (n = 537). RICHS recruited mother-infant pairs from Women and Infants Hospital of Rhode Island. Newborns considered large for gestational age (LGA) and small for gestational age (SGA) were matched to adequate for gestational age (AGA) infants on sex, gestational age (±3 days) and maternal age (±2 years). Further Information about the cohort has been previously described [12]. All patients provided written informed consent for participation under protocols approved by the institutional review boards at Women and Infants Hospital and Dartmouth College.
Clinical measures of infant neurobehavior
Newborn neurobehavior was assessed via the NNNS, which was administered by certified psychometrists after the first 24 h of life, prior to hospital discharge. Individual components of the NNNS were compiled into a series of 13 summary scores [28]. A recursively partitioned mixed model algorithm (RPMM) [29] was previously used to hierarchically cluster these scores into seven unique neurobehavioral profiles, which are described in Lesseur et al. [30], and information about these profiles is provided in Supplementary Table 1.
Sample collection, DNA extraction & bisulfite modification
For each subject, placental parenchyma from standardized points 2 cm from the umbilical cord insertion site were excised from the fetal portion of the placenta, free of maternal decidua. Samples were placed immediately in RNAlater (Thermo Fisher, MA, USA) and stored at 4°C. At least 72 h later, samples were removed from RNAlater, blotted dry, snap frozen in liquid nitrogen, pulverized to a homogenous powder using a liquid nitrogen cooled stainless-steel mortar and pestle (Cellcrusher, OR, USA), and stored at -80°C. DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, CA, USA) and was quantified using the Nanodrop ND-2000 spectrophotometer (Thermo Fisher). DNA was bisulfite modified using the EZ-DNA methylation plate kit (Zymo Research, CA, USA) and stored at -20°C before analysis. All bisulfite converted DNA was used within 6 months of conversion. All procedures were performed following manufacturer’s instructions.
Bisulfite pyrosequencing
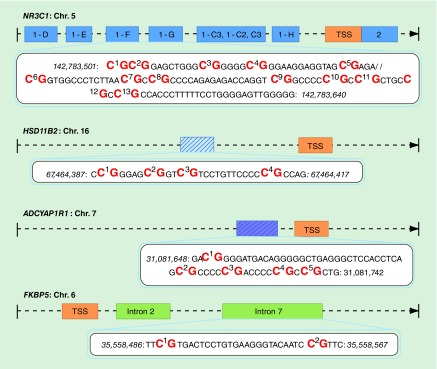
We quantified methylation of 24 CpGs from selected regions of NR3C1, HSD11B2, ADCYAP1R1 and FKBP5 (Figure 1 & Supplementary Table 2). This included 13 CpGs within the exon 1F of NR3C1 that reside in a region of epigenetic control region identified in rodents [6], and have been associated with infant and adult neurological outcomes [7–10,31–35]; four CpGs in the HSD11B2 promoter that were previously associated with infant neurobehavior [10,12–13]; two CpGs within intron 7 of FKBP5 that bind to the FKBP5 transcriptional start site to exhibit transcriptional control [16] and have been associated with phenotypes related to dysregulation of the HPA axis [16,17]; and five CpGs surrounding Chr 7: 31081720 (GRCh37/hg19), a location in the ADCYAP1R1 promoter that corresponds to Cg11218385 of the Infinium HumanMethylation 27K BeadChip array and has been associated with stress-related adversity [21,22]. Chromosomal locations and genomic annotation are provided in Supplementary Table 2.
Figure 1. . Sequencing strategy.
Regions sequenced for each of the four candidate genes in this study. Transcriptional start site (orange), exons are represented as dark blue boxes, and introns represented as green boxes.
TSS: Transcriptional start site.
All samples were amplified for each assay using a PyroMark PCR Kit (Qiagen). Primer design and cycling conditions described in Supplementary Table 2. PCR products were sequenced using a PyroMark MD system (Qiagen). Each sequencing run contained no template, genomic DNA negative controls to ensure there was no contamination, and methylated and unmethylated controls to assess the validity of the sequencing results (Qiagen). Conversion efficiency was assessed using a bisulfite conversion control within each assay. All PCR samples were pyrosequenced in triplicate, and methylation was defined as the mean of these technical triplicates. Outliers were defined as a deviation of greater than 5% from the mean of all samples, and samples with outliers were resequenced.
Statistical analysis
Methylation at individual CpGs were expressed as Z scores, and the relationship between CpGs examined using Pearson correlations. In this study we utilized factor analysis, which is a statistical technique that can describe variability among a number of measured, correlated variables (in this case specific CpG sites) as a lower number of latent factors, in order to reduce the number of comparisons made. Maximum likelihood factor analysis was performed with a varimax orthogonal rotation using the package Psych in R version 3.1.1. Factor significance was defined by an eigenvalue greater than 1 and explaining greater than 10% of the proportion of variation [36]. Factor loadings, which represent the correlation between methylation of individual CpGs and factor scores were obtained to identify contributions of individual CpGs to each factor. Factor scores, which represent an individual’s placing in a latent factor, from each of the three significant factors were used in subsequent analysis.
To explore the relationship between the latent methylation variables created through factor analysis and infant neurobehavioral profiles, polytomous regression was performed using NNET. Profile 2 was treated as the reference group in this analysis as this group displayed scores on each of the NNNS scales within a half a standard deviation of the overall population average, indicating average functioning across neurobehavioral domains (Supplementary Table 1). This model is adjusted for infant sex (male, female), as well as birth weight group (adequate for gestational age, large for gestational age and small for gestational age). We initially included maternal ethnicity (white, other), maternal education (high school or less, greater then high school), maternal smoking (yes, no) and maternal age group (less than 18–27, 27–32, greater than 32 years) as covariates in the model, and used backward selection to remove these covariates because they were not significantly associated with NNNS profile and did not alter the estimates of effect of the methylation factors on profile membership by greater than 10%.
Results
Descriptive statistics & bivariate analyses
Demographic characteristics of the 537 infants in this study are shown in Table 1. Females and males are nearly evenly distributed in this study (48.8 vs 51.2%). The distribution of birth weight groups reflects the study sampling strategy, which oversampled for large for gestational age (LGA; >90th birth weight percentile, 25.2%) and small for gestational age (SGA, <10th birth weight percentile, 19.8%) infants. The majority of women were white (72%) and had achieved a high school degree or higher (72.67%).
Table 1. . Infant clinical and demographic information.
| Clinical and demographic information | n (%) |
|---|---|
| Birth weight group: | |
| – AGA | 290 (54.1) |
| – LGA | 137 (25.6) |
| – SGA |
109 (20.3) |
| Sex: | |
| – Female | 274 (51.1) |
| – Male |
262 (48.9) |
| Anxiety pregnancy: | |
| – No | 457 (85.3) |
| – Yes |
69 (12.9) |
| Depression pregnancy: | |
| – No | 452 (84.3) |
| – Yes |
74 (13.8) |
| Tobacco pregnancy: | |
| – No | 487 (92.1) |
| – Yes |
42 (7.9) |
| Maternal ethnicity: | |
| – White | 384(72) |
| – Other |
149 (28) |
| Maternal education: | |
| – High school graduate or less | 146 (27.4) |
| – Greater than high school education |
386 (72.6) |
| Maternal age (years): | |
| – 18–27 | 167 (31.2) |
| – 27–32 | 201 (37.5) |
| – >32 |
168 (31.3) |
| Infant gestational age (weeks): | |
| – 37–39 | 113 (21.1) |
| – 39 or higher | 423 (78.9) |
AGA: Adequate for gestational age; LGA: Large for gestational age; SGA: Small for gestational age.
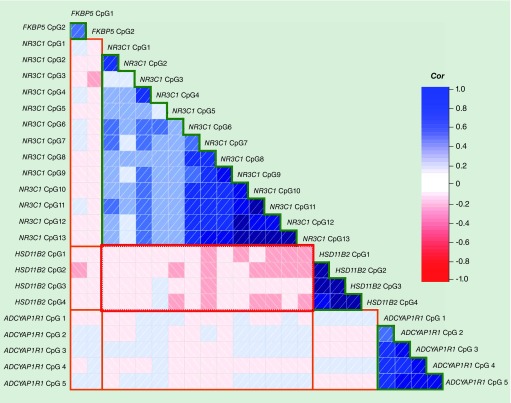
Average methylation of each of the 24 CpGs sequenced is shown in Supplementary Table 3. NR3C1, ADCYAP1R1 and HSD11B2 were relatively hypomethylated (less than 20%), whereas FKBP5 was hypermethylated (greater than 80%). As shown in Figure 2, adjacent CpGs within the same gene were mostly moderately to strongly correlated (r range 0.08–0.84; Supplementary Table 4). We observed mainly weak correlations between individual CpGs located on different genes that mostly did not reach statistical significance (r range -0.24–0.13). NR3C1 CpGs were weakly correlated with FKBP5 CpGs (correlation range -0.17–0.07), although this only reached statistical significance between FKBP5 CpG 2 and NR3C1 CpG 3 and 4. There were a series of statistically significant negative correlations between HSD11B2 CpGs and the majority of NR3C1 CpG sites (r range -0.24–0.02).
Figure 2. . Correlations between methylation of CpGs within and across candidate genes.
Correlogram of correlation coefficients derived from Pearson correlations between methylation of CpGs within the same gene (NR3C1, HSD11B2, FKBP5, ADCYAP1R1) (indicated by green lines) and across the six different combinations of genes (indicated by orange lines). Red squares with left justified slashes represent negative correlations, and blue squares with right justified slashes represent positive associations. The depth of the shading represents the strength of the association.
Factor analysis defines three factors that explain variability in methylation
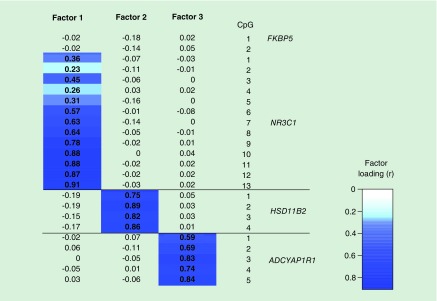
Using maximum likelihood factor analysis over the 24 normalized CpG sites, we identified three significant orthogonal factors with eigenvalues greater than 1. Figure 3 shows the loadings of each CpG onto individual factors, where the factor loading is the correlation coefficient between the factor scores and methylation at each of the CpGs. Correlations between factor scores and methylation at individual CpGs greater than 0.3 were considered significant loadings. Factor 1 explained 23% of the proportional variation in methylation, with an eigenvalue of 5.54. It was significantly loaded by methylation of NR3C1 exon 1F CpGs 3 and 5–13. Factor 2 was significantly loaded by HSD11B2 CpGs 1–4, and explained 12% of the proportional variation in methylation, with an eigenvalue of 2.91. Factor 3 (ADCYAP1R1 methylation factor) was significantly loaded by ADCYAP1R1 CpGs 1–5, and explained 12% of the proportional variation in methylation, with an eigenvalue of 2.80. Each factor was only loaded by CpGs from individual genes, and each CpG was significantly correlated and loaded onto only one factor. CpGs that did not load onto any factor, including FKBP5 CpGs 1 and 2, and NR3C1 CpGs 2 and 4, were not represented in subsequent analysis, which used these latent factors to represent methylation.
Figure 3. . Loading of individual CpGs onto latent methylation variables.
Heat map displaying the loading of each of the 24 CpGs across the three latent factors. Factor loadings (correlation) greater then 0.3 were considered significant, and are highlighted in dark blue.
Methylation of cortisol regulation genes & infant neurobehavior
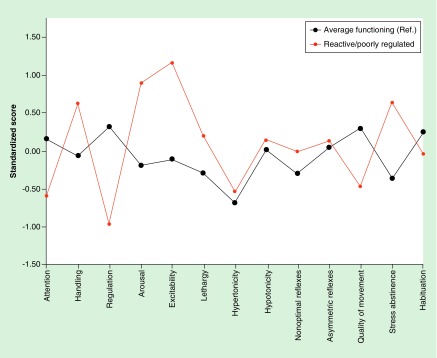
Next, we sought to identify how these latent methylation factors altered membership in profiles of infant behavior. The unadjusted model linking the methylation factors to the seven neurobehavioral profiles is shown in Supplementary Table 5, while the model adjusted sex and birth weight group is displayed as Table 2. Figure 4 shows the differences in standardized NNNS scores between only profiles 2 (average/ref) and 6 (reactive/poorly regulated), Compared to profile 2, infants in profile 6 had decreased quality of movement and self-regulation, increased arousal and excitability, and increased nonoptimal refluxes and stress abstinence scores, and thus are referred to as ‘poorly regulated/reactive.’ In the adjusted model, a 1 unit increase in factor 1 resulted in a 47% increased odds of membership in profile 6 compared with profile 2 (odds ratio [OR]: 1.47; 95% CI: 1.00–2.21). We observed that latent factor 2 was associated with reduced odds of membership in profile 6, although with wide confidence intervals (OR: 0.76; 95% CI: 0.55–1.05; Table 2). We found no significant relationships between Factor 3, characterized by methylation of the ADCYAP1R1, and any neurobehavioral profile.
Table 2. . Adjusted polytomous regression models for the association between latent methylation factors and profiles of infant neurodevelopment.
| Predictors | Profile 1 (n = 57), | Profile 2 (n = 83), | Profile 3 (n = 62), | Profile 4 (n = 78), | Profile 5 (n = 93), | Profile 6 (n = 96), | Profile 7 (n = 43), |
|---|---|---|---|---|---|---|---|
| OR (CI) | OR (CI) | OR (CI) | OR (CI) | OR (CI) | OR (CI) | OR (CI) | |
| Latent methylation variables: | |||||||
| – Factor 1 | 1.38 (0.89–2.12) | Ref. | 1.43 (0.93–2.21) | 1.30 (0.85–2.00) | 1.02 (0.66–1.59) | 1.47 (1.00–2.18) | 1.16 (0.7–1.95) |
| – Factor 2 | 0.75 (0.52–1.08) | Ref. | 0.73 (0.50–1.06) | 0.81 (0.58–1.15) | 0.74 (0.53–1.03) | 0.76 (0.55–1.05) | 0.80 (0.53–1.23) |
| – Factor 3 |
0.942 (0.64–1.33) |
Ref. |
0.90 (0.62–1.31) |
0.94 (0.67–1.32) |
0.84 (0.60–1.18) |
1.03 (0.76–1.41) |
0.66 (0.40–1.09) |
| Gender: | |||||||
| – Female | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. |
| – Male |
1.66 (0.80–3.48) |
Ref. |
1.29 (0.61–2.70) |
0.94 (0.67–1.32) |
1.23 (0.64–2.38) |
0.84 (0.44–1.61) |
1.29 (0.57–2.92) |
| Birth weight group: | |||||||
| – AGA | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. | Ref. |
| – LGA | 1.10 (0.46–2.63) | Ref. | 0.75 (0.30–1.87) | 0.71 (0.31–1.63) | 0.98 (0.45–2.12) | 0.90 (0.42–1.96) | 1.87 (0.76–4.60) |
| – SGA | 1.87 (0.73–4.77) | Ref. | 1.37 (0.53–3.55) | 0.60 (0.22–1.65) | 1.26 (0.52–3.04) | 1.26 (0.54–2.97) | 1.04 (0.31–3.48) |
AGA: Adequate for gestational age; LGA: Large for gestational age; SGA: Small for gestational age; OR: Odds ratio.
Figure 4. . Standardized neonatal intensive care unit network neurobehavioral scales scores across neurobehavioral profiles of interest.
Standardized NICU Network Neurobehavioral scales in relation to the mean across all groups for profile 2 (average functioning: n = 83) and profile 6 (reactive/poorly regulated: n = 96).
Discussion
The control of fetal exposure to glucocorticoids is critical to the normal programming of the developing infant’s neural circuitry. The placenta modulates fetal exposure to maternal cortisol, and this function is linked, in part, to the activities of the four glucocorticoid response genes examined in this study. We examined the impact of this methylation in a concerted fashion across these genes on risk of neurobehavioral adversity in a population of healthy, term newborns and identified an opposing relationship between methylation of the glucocorticoid receptor (NR3C1) and HSD11B2 and risk of membership in a neurobehavioral profile of infants who are highly excitable, highly aroused and reactive and as a consequence are poorly regulated, have poor quality of movement and poor attention requiring increased handling and demonstrate an elevated number of signs of stress.
In order to look at contributions of individual CpGs from multiple genes simultaneously, we conducted exploratory factor analysis to generate orthogonal latent methylation factors that loaded differently onto individual CpG sites. Factor 1 strongly loaded on NR3C1 exon IF CpG 3 and 6–13, and mean methylation of this region has previously been associated with decreased NR3C1 expression in the placenta [9]. Similarly, factor 2 strongly loaded on the four CpGs in the HSD11B2 promoter, and mean methylation of these four CpGs has previously been associated with decreased HSD11B2 expression in the placenta [12]. Factor 3 strongly loaded on the five CpGs in the ADCYAP1R1 promoter region. All CpGs loaded significantly onto only one factor, and FKBP5 CpGs did not significantly load onto any of the factors generated by our analysis, which may be due to the small number of CpGs examined, as FKBP5 CpGs were only correlated with each other. As a result, we were unable to observe any associations between neurobehavioral outcomes and FKBP5 methylation using this strategy.
Through factor analysis, we generated a data-driven summary statistic that quantified methylation of these complicated regulatory regions across several genes, while avoiding multiple comparisons and generalizations that would have been made by using a simple summary statistic such as the mean across these entire regions, which may not take into account the more complex correlation structure of the methylation of these CpGs. Dimension reduction techniques such as exploratory factor analysis and principle components analysis are commonly used tools in psychology [37], and can be applied to high dimensionality biological data to identifying patterns of association, associating gene and expression patterns and uncovering underlying factors responsible for cellular phenotype [25]. There are a number of examples of these techniques applied to gene expression datasets [38] as well as microRNA data [39], but there are limited applications to the study of DNA methylation, and most prior studies have used this technique to summarize methylation across single genes [31,40], or across complex microarray data [39]. However, factor analysis may be more appropriate in hypothesis-driven gene studies, particularly involving multiple candidate genes [41], due to the dimensionality of the data as well as the likelihood of correlations among CpGs. In this example, factor analysis provided us with a more data-driven summary statistic than the mean alone, providing a clearer understanding of the contributions of related CpGs to neurobehavior.
We found that increasing scores of the methylation factor loaded by NR3C1 was associated with increased odds of membership in a reactive, poorly regulated profile. This profile bears resemblance to a profile in an independent population of infants exposed to a higher degree of prenatal adversity. This prior study showed that infants with these NNNS scores were more likely to experience medical and behavioral problems at 4.5 years of age [42]. These poorly regulated, reactive infants exhibit characteristics suggestive of altered glucocorticoid signaling. Specifically, infants in this profile have poor motor control, which has been linked to inadequate exposure to maternal cortisol [43]. In addition, the high arousal, and excitability of these infants, coupled with their poor self-regulation may be reflective of increased stress and a hypervigilant response to the environment. In adults, hypervigilant responses have been linked to reduced levels of cortisol and alterations to the glucocorticoid response pathway [44], and are traditionally associated with affective disorders such as post-traumatic stress disorder. Although it remains unclear how the NNNS scores observed in infancy directly correspond to adult health outcomes, these scores are suggestive of poor adaptability and increased stress in response to the postnatal environment.
We observed significant negative correlations between NR3C1 and HSD11B2 CpGs, which aligns with the opposing roles of cortisol inactivation and response played by these gene products. The negative correlation between methylation of NR3C1 and HSD11B2 CpGs indicates that individuals with increased NR3C1 methylation would likely have lower HSD11B2 methylation. In line with these correlations, we observed opposing relationships between HSD11B2 and NR3C1 methylation factors and membership in the high-risk profile that displays signs of glucocorticoid dysregulation and inappropriate stress response. Infants with increased factor 1 (loaded by methylation of NR3C1 CpGs) are more likely to be in this risk profile, and infants with increased loading of factor 2 (loaded by HSD11B2 CpGs) are less likely to be in this risk profile. We speculate that increased NR3C1 methylation may lead to reduced glucocorticoid receptor density of the placenta and inadequate placental cortisol response and signaling in utero. Decreased methylation of HSD11B2 may result in increased inactivation of cortisol by the placenta. We speculate that infants with decreased HSD11B2 methylation would likely have decreased cortisol levels in utero. Thus, infants in the reactive, poorly regulated profile display a pattern of methylation that may contribute to limited glucocorticoid signaling in utero.
Although prenatal stress and elevated cortisol levels are traditionally associated with adverse neurological outcomes [45], some level of cortisol exposure is necessary for development, as glucocorticoids help shape the development of neuronal systems, including the HPA axis [2,46]. These results are concurrent with prior studies involving NR3C1 methylation, where increased methylation was associated with increased anxiety in animals (methylation measured in hippocampus) [6], increased cortisol reactivity in infants (cord blood methylation) and attenuated cortisol response in adults (leukocyte methylation) [33]. Infants in profiles that present with deficiencies across different neurobehavioral domains may experience reduced glucocorticoid signaling in utero due to epigenetic patterning of these cortisol response genes, which appears to manifest as a reduced adaptability to the stresses of the postnatal environment. In contrast, infants in our reference profile with a pattern of methylation reflective of increased cortisol signaling in utero, have average functioning across multiple domains exhibit increased resiliency to these postnatal stressors. These observations are concurrent with the fetal programming theory, where a blunted cortisol response in infants has been hypothesized to lead to inadequate stress response over time, and increased risk of affective disorders [3,47].
Prior work in a subset of individuals from this cohort identified associations between mean methylation of NR3C1 and HSD11B2 and singular NNNS measurements [24]. Infants with high NR3C1 methylation and low HSD11B2 methylation had significantly higher asymmetrical refluxes, and infants with lower NR3C1 methylation and high HSD11B2 methylation exhibited lower excitability. We sought to expand this analysis including additional genes and using a more meaningful neurobehavioral outcome which may denote a subset of infants with a potential for later childhood mental and physical health risk. The NNNS profiles allow us to examine multiple NNNS scores that represent different cognitive motor controls simultaneously, while reducing the number of comparisons, and we have expanded to a larger panel of genes and utilize an approach that takes into account differential DNA methylation levels across genes in a more sophisticated technique then use of the mean alone, distilling the data to clearly identify the contribution of different CpGs. This work compliments our previous findings [24], as infants in our poorly regulated/reactive profile exhibit increased excitability compared with infants in the reference profile. These complimentary results suggest that exploratory factor analysis can be utilized to identify meaningful associations, between DNA methylation and neurobehavioral outcomes. It also highlights the role of NR3C1 and HSD11B2, as methylation of these genes emerge as the most important contributors to membership in neurobehavioral profiles in the context of a larger panel of genes.
We were unable to observe any associations between infant neurobehavioral outcomes and latent factor 3, which was loaded by ADCYAP1R1 methylation and represented 12% of the underlying variation in methylation across 24 CpGs. Our inability to observe associations suggests that this approach may not fully characterize the relationships between methylation of this gene and these neurobehavioral outcomes, and suggests that more traditional analyses specifically examining ADCYAP1R1, may be warranted. At the same time, it may not be surprising that we could not link methylation of this region to neurobehavioral outcomes in this relatively low-risk infant population, as prior studies have shown ADCYAP1R1 promoter methylation associated with adverse outcomes in high-risk infants experiencing extreme adversity [21,22]. Our null findings are concurrent with prior studies identifying no association between cord blood PACAP expression and other infant health outcomes [48].
Our interpretation of the data generated through this analysis is limited by common methodological and study design issues commonly encountered in epidemiological studies of DNA methylation. We attempted to limit the number of multiple comparisons made in this study by utilizing infant profiles instead of individual NNNS scores through our polytomous regression models. We recognize that the placental parenchyma examined is composed of a variety of different cell types, and since DNA methylation is cell type-specific, cell type proportions may confound results of both candidate gene and genome-wide methylation analyses. In genome-wide studies, there are statistical techniques available to correct for these problems [49], but these techniques cannot be applied to candidate gene studies, and so future candidate gene analyses may consider isolation of highly purified cell types in order to isolate these effects, which is beyond the scope of this study. We also did not demonstrate, specifically, the relationship between the factor scores identified and gene expression of these candidate genes, although the relationship between mean methylation and expression of these genes has been reported previously [9,12,16]. Finally, this is an observational study and we cannot directly measure how DNA methylation influences the complex neurobehavioral traits described in this analysis, and can only make assumptions on the underlying biological mechanism based on previous studies and the functional significance of these genes. We would encourage further analysis of the relationship between methylation of these genes in and infant cortisol response in other populations, including an analysis of maternal or fetal cortisol reactivity or cortisol levels in the placenta if possible, to further elucidate the biological relevance of these observations.
Strengths of this study are the unique application of the NNNS assessment, which has shown efficacy in predicting motor outcomes in older infants as quantified through the Bayley psychomotor developmental index [50], and identifying changes in brain physiology quantified through MRI [51,52]. This study utilizes established profiles generated through NNNS scores, which provides an integrated assessment of multiple NNNS scores across different cognitive domains, and reduces the number of comparisons. Multiple studies from different centers analyzing both high- and low-risk infants have utilized clustering based approaches to derive profiles, and these profiles are relatively similar [42,53], indicating that the NNNS profiles may be generalizable to multiple populations and allows easier identification of at-risk individuals. .The cohort examined represents a relatively healthy population and the in utero adversity experienced by these infants is likely similar to the population at large, enhancing the generalizability of our findings. Another strength of this study lies within the use of placenta as a relevant biomarker, as this tissue plays a key functional role in controlling fetal glucocorticoid exposures, something that studies of other tissues like blood may not represent. The placenta has emerged as a key biomarker to examine the in utero environment because of its functional relevance, environmentally altered epigenetic signatures and accessibility [54].
Conclusion
This study is among the first to apply exploratory factor analysis as a data reduction technique to methylation data across multiple candidate genes, and we encourage broader application to explore interactions between methylation of multiple candidate genes. Our findings suggest that methylation patterning of glucocorticoid response genes results in a degree of inability to adapt to the stresses in the postnatal environment among healthy populations of infants exposed to low-to-moderate prenatal stress. Further analysis is needed to identify predictors of methylation of glucocorticoid response genes, and the mechanism by which methylation of cortisol response genes influences lifelong cortisol response and disorders associated with dysregulation of the HPA axis.
Future perspective
This study compliments the growing body of evidence implicating aberrant methylation of glucocorticoid response genes in infant neurobehavioral outcomes, which have implications for long-term mental health and cognitive outcomes. Longitudinal studies are necessary to determine the influence of placental epigenetics on these outcomes directly. This study provides an important impetus for mechanistic research to further define the role of factors that respond to glucocorticoids in the placenta and infant brain development. Altogether, identification of sites with epigenetic dysregulation may provide us with valuable biomarkers that may predict risk of mental health adversity, which would be useful for clinical intervention.
Executive summary.
Background
Glucocorticoids play an important role in fetal brain development, and cortisol exposure is associated with infant neurobehavioral outcomes.
DNA methylation of four genes involved in glucocorticoid response, NR3C1, HSD11B2, FKBP5, ADCYAP1R1 has been associated with newborn neurobehavior, or psychological outcomes in adults.
We sought to perform an integrated assessment of methylation patterning of these four genes and its association with infant neurobehavioral outcomes.
We hypothesized that underlying factors related to methylation of these genes would be associated with different infant neurobehavioral profiles.
Descriptive statistics & bivariate analyses
Methylation of adjacent CpGs from the same gene were strongly positively correlated, and methylation of CpGs from different genes were mostly not correlated.
Methylation of HSD11B2 CpGs were negatively correlated with methylation of NR3C1 CpGs.
Factor analysis defines three factors that explain variability in methylation
Using maximum likelihood factor analysis over the 24 normalized CpG sites, we identified three significant orthogonal factors which were individually loaded from CpGs from NR3C1, HSD11B2 and ADCYAP1R1.
Methylation of cortisol regulation genes & infant neurobehavior
Through multinomial regression, we observed differences in latent methylation factors that influenced membership in predefined neurobehavioral profile.
In this fully adjusted model, increased factor 1 (loaded by NR3C1 methylation) resulted in a 47% increased odds of membership in a reactive, poorly regulated profile of infants.
Factor 2 (loaded by HSD11B2 methylation) was associated with reduced odds of membership in the same reactive, poorly regulated infant profile.
Conclusion
Epigenetic placental glucocorticoid regulation influences reactivity and self-regulation.
Developmental epigenetics may impact post-natal stress response.
Supplementary Material
Acknowledgements
The authors would like to thank the staff at Women and Infants Hospital, particularly J Lee and E Oliveira for their help with the data collection, as well as A Roberts, R John and L Kwan for their assistance with biological processing and data entry. We would also like to acknowledge M Karagas, B Christensen and L Stroud for their guidance with this paper.
Footnotes
Disclaimer
The articles contents are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA. Further, the US EPA does not endorse the purchase of any commercial products or services mentioned in the publication. The sponsors of this research had no role in study design, analysis and interpretation of data, in writing of the report or decision to submit the report for publication.
Financial & competing interests disclosure
This work is supported by NIH-NIMH R01MH094609, NIH-NIEHS R01ES022223 and NIH-NIEHS P01 ES022832/EPA RD83544201. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Sandman CA, Davis EP, Buss C, Glynn LM. Exposure to prenatal psychobiological stress exerts programming influences on the mother and her fetus. Neuroendocrinology. 2012;95(1):7–21. doi: 10.1159/000327017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cottrell EC, Seckl JR. Prenatal stress, glucocorticoids and the programming of adult disease. Front. Behav. Neurosci. 2009;3:19. doi: 10.3389/neuro.08.019.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lester BM, Marsit CJ, Conradt E, Bromer C, Padbury JF. Behavioral epigenetics and the developmental origins of child mental health disorders. J. Dev. Orig. Health Dis. 2012;3(6):395–408. doi: 10.1017/S2040174412000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bzoskie L, Blount L, Kashiwai K, Tseng YT, Hay WW, Jr, Padbury JF. Placental norepinephrine clearance: in vivo measurement and physiological role. Am. J. Physiol. 1995;269(1 Pt 1):E145–E149. doi: 10.1152/ajpendo.1995.269.1.E145. [DOI] [PubMed] [Google Scholar]
- 5.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 6.Weaver ICG, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 7.Stroud LR, Papandonatos GD, Rodriguez D, et al. Maternal smoking during pregnancy and infant stress response: test of a prenatal programming hypothesis. Psychoneuroendocrinology. 2014;48:29–40. doi: 10.1016/j.psyneuen.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filiberto AC, Maccani MA, Koestler D, et al. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6(5):566–572. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bromer C, Marsit CJ, Armstrong DA, Padbury JF, Lester B. Genetic and epigenetic variation of the glucocorticoid receptor (NR3C1) in placenta and infant neurobehavior. Dev. Psychobiol. 2013;55(7):673–683. doi: 10.1002/dev.21061. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study identified a significant negative relationship between NR3C1 mean methylation and expression. It also found assocations between methylation of individual CpGs within the NR3C1 exon 1F promoter and infant neurobehavior as quantified thorugh the NNNS (NICU [neonatal intensive care unit] Network Neurobehavioral Scales) assesment.
- 10.Conradt E, Lester BM, Appleton AA, Armstrong DA, Marsit CJ. The roles of DNA methylation of NR3C1 and 11beta-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics. 2013;8(12):1321–1329. doi: 10.4161/epi.26634. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study examined interactions between methylation of individual CpGs from NR3C1 exon 1–F and the HSD11B2 promoter and maternal anxiety and depression independently.
- 11.López Bernal A, Craft IL. Corticosteroid metabolism in vitro by humanplacenta, fetal membranes and decidua in early and late gestation. Placenta. 1981;2(4):279–285. doi: 10.1016/s0143-4004(81)80025-2. [DOI] [PubMed] [Google Scholar]
- 12.Marsit CJ, Maccani MA, Padbury JF, Lester BM. Placental 11-beta hydroxysteroid dehydrogenase methylation is associated with newborn growth and a measure of neurobehavioral outcome. PLoS ONE. 2012;7(3):e33794. doi: 10.1371/journal.pone.0033794. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study identified a significant negative relationship between mean HSD11B2 methylation and expression. It also identified associations between mean methylation and infant neurobehavioral outcomes.
- 13.Appleton AA, Armstrong DA, Lesseur C, et al. Patterning in placental 11-B hydroxysteroid dehydrogenase methylation according to prenatal socioeconomic adversity. PLoS ONE. 2013;8(9):e74691. doi: 10.1371/journal.pone.0074691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology. 2009;34(Suppl. 1):S186–S195. doi: 10.1016/j.psyneuen.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Baughman G, Wiederrecht GJ, Chang F, Martin MM, Bourgeois S. Tissue distribution and abundance of human FKBP51, and FK506-binding protein that can mediate calcineurin inhibition. Biochem. Biophys. Res. Commun. 1997;232(2):437–443. doi: 10.1006/bbrc.1997.6307. [DOI] [PubMed] [Google Scholar]
- 16.Paquette AG, Lester BM, Koestler DC, Lesseur C, Armstrong DA, Marsit CJ. Placental FKBP5 genetic and epigenetic variation is associated with infant neurobehavioral outcomes in the RICHS cohort. PLoS ONE. 2014;9(8):e104913. doi: 10.1371/journal.pone.0104913. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study identified a negative assocation between FKBP5 intron 7 methylation and FKBP5 expression in the placenta, and found associations between this methylation as well as genetic variation that contributes to infant behavioral outcomes.
- 17.Klengel T, Mehta D, Anacker C, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicol MR, Cobb V, Williams B, Morley S, Walker S, Mason J. Vasoactive intestinal peptide (VIP) stimulates cortisol secretion from the H295 human adrenocortical tumour cell line via VPAC1 receptors. J. Mol. Endocrinol. 2004;32(3):869–877. doi: 10.1677/jme.0.0320869. [DOI] [PubMed] [Google Scholar]
- 19.Roman CW, Lezak KR, Hartsock MJ, et al. PAC1 receptor antagonism in the bed nucleus of the stria terminalis (BNST) attenuates the endocrine and behavioral consequences of chronic stress. Psychoneuroendocrinology. 2014;47(0):151–165. doi: 10.1016/j.psyneuen.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh PO, Kwak SD, Kim HJ, et al. Expression patterns of pituitary adenylate cyclase activating polypeptide and its type I receptor mRNAs in the rat placenta. Mol. Reprod. Dev. 2003;64(1):27–31. doi: 10.1002/mrd.10221. [DOI] [PubMed] [Google Scholar]
- 21.Ressler KJ, Mercer KB, Bradley B, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470(7335):492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study identified assocation between white blood cell ADCYAP1R1 methylation and post-traumatic stress disorder in adults.
- 22.Chen W, Boutaoui N, Brehm JM, et al. ADCYAP1R1 and asthma in Puerto Rican children. Am. J. Respir. Crit. Care Med. 2013;187(6):584–588. doi: 10.1164/rccm.201210-1789OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogdan R, Hyde LW, Hariri AR. A neurogenetics approach to understanding individual differences in brain, behavior, and risk for psychopathology. Mol. Psychiatry. 2013;18(3):288–299. doi: 10.1038/mp.2012.35. [DOI] [PubMed] [Google Scholar]
- 24.Appleton AA, Lester BM, Armstrong DA, Lesseur C, Marsit CJ. Examining the joint contribution of placental NR3C1 and HSD11B2 methylation for infant neurobehavior. Psychoneuroendocrinology. 2014;52c:32–42. doi: 10.1016/j.psyneuen.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study examined methylation patterning of NR3C1 and HSD11B2 dichotomized as low and high and its assocation with indivdual NNNS scores in a subset of infants from the RICHS cohort.
- 25.Ringner M. What is principal component analysis? Nat. Biotechnol. 2008;26(3):303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 26.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415(6870):436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 27.Elovitz MA, Anton L, Bastek J, Brown AG. Can microRNA profiling in maternal blood identify women at risk for preterm birth? Am. J. Obstet. Gynecol. 2015 doi: 10.1016/j.ajog.2015.01.023. doi:10.1016/j.ajog.2015.01.023. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 28.Lester BM, Tronick EZ. History and description of the Neonatal Intensive Care Unit Network Neurobehavioral Scale. Pediatrics. 2004;113(3 Pt. 2):634–640. [PubMed] [Google Scholar]
- 29.Koestler DC, Marsit CJ, Christensen BC, et al. Semi-supervised recursively partitioned mixture models for identifying cancer subtypes. Bioinformatics. 2010;26(20):2578–2585. doi: 10.1093/bioinformatics/btq470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lesseur C, Armstrong DA, Murphy MA, et al. Sex-specific associations between placental leptin promoter DNA methylation and infant neurobehavior. Psychoneuroendocrinology. 2014;40(0):1–9. doi: 10.1016/j.psyneuen.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study describes the rationale and derivation of the NNNS profiles from summary scores in the RICHS cohort.
- 31.Mulligan CJ, D’errico NC, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics. 2012;7(8):853–857. doi: 10.4161/epi.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 33.Tyrka AR, Price LH, Marsit C, Walters OC, Carpenter LL. Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PLoS ONE. 2012;7(1):e30148. doi: 10.1371/journal.pone.0030148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perroud N, Paoloni-Giacobino A, Prada P, et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl. Psychiatry. 2011;1:e59. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melas PA, Wei Y, Wong CCY, et al. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int. J. Neuropsychopharmacol. 2013;16(07):1513–1528. doi: 10.1017/S1461145713000102. [DOI] [PubMed] [Google Scholar]
- 36.Ruscio J, Roche B. Determining the number of factors to retain in an exploratory factor analysis using comparison data of known factorial structure. Psychol. Assessment. 2012;24(2):282–292. doi: 10.1037/a0025697. [DOI] [PubMed] [Google Scholar]
- 37.Fabrigar LR, Wegener DT, Maccallum RC, Strahan EJ. Evaluating the use of exploratory factor analysis in psychological research. Psychol. Methods. 1999;4(3):272. [Google Scholar]
- 38.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartenhagen C, Klein H-U, Ruckert C, Jiang X, Dugas M. Comparative study of unsupervised dimension reduction techniques for the visualization of microarray gene expression data. BMC Bioinformatics. 2010;11(1):567. doi: 10.1186/1471-2105-11-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum. Mol. Gen. 2007;16(5):547–554. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 41.Houseman EA. Biostatistical methods in epigenetic epidemiology. In: Michels KB, editor. Epigenetic Epidemiology. Springer; The Netherlands: 2012. pp. 57–76. [Google Scholar]
- 42.Liu J, Bann C, Lester B, et al. Neonatal neurobehavior predicts medical and behavioral outcome. Pediatrics. 2010;125(1):e90–e98. doi: 10.1542/peds.2009-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dipietro JA, Kivlighan KT, Costigan KA, Laudenslager ML. Fetal motor activity and maternal cortisol. Dev. Psychobiol. 2009;51(6):505–512. doi: 10.1002/dev.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yehuda R. Status of glucocorticoid alterations in post-traumatic stress disorder. Ann. NY Acad. Sci. 2009;1179(1):56–69. doi: 10.1111/j.1749-6632.2009.04979.x. [DOI] [PubMed] [Google Scholar]
- 45.Kapoor A, Dunn E, Kostaki A, Andrews MH, Matthews SG. Fetal programming of hypothalamo-pituitary-adrenal function: prenatal stress and glucocorticoids. J. Physiol. 2006;572(Pt. 1):31–44. doi: 10.1113/jphysiol.2006.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis EP, Sandman CA. The timing of prenatal exposure to maternal cortisol and psychosocial stress is associated with human infant cognitive development. Child Dev. 2010;81(1):131–148. doi: 10.1111/j.1467-8624.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lester BM, Conradt E, Marsit CJ. Epigenetic basis for the development of depression in children. Clin. Obstet. Gynecol. 2013;56(3):556–565. doi: 10.1097/GRF.0b013e318299d2a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winters SJ, King JC, Brees CK, Moore JP., Jr Pituitary adenylate cyclase-activating polypeptide (PACAP) in fetal cord blood. Early Hum. Dev. 2014;90(9):451–453. doi: 10.1016/j.earlhumdev.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 49.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics (Oxford, England) 2014;30(10):1431–1439. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens BE, Liu J, Lester B, et al. Neurobehavioral assessment predicts motor outcome in preterm infants. J. Pediatr. 2010;156(3):366–371. doi: 10.1016/j.jpeds.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown NC, Inder TE, Bear MJ, Hunt RW, Anderson PJ, Doyle LW. Neurobehavior at term and white and gray matter abnormalities in very preterm infants. J. Pediatr. 2009;155(1):32–38. doi: 10.1016/j.jpeds.2009.01.038. [DOI] [PubMed] [Google Scholar]
- 52.Coleman MB, Glass P, Brown J, et al. Neonatal neurobehavioral abnormalities and MRI brain injury in encephalopathic newborns treated with hypothermia. Early Hum. Dev. 2013;89(9):733–737. doi: 10.1016/j.earlhumdev.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sucharew H, Khoury JC, Xu Y, Succop P, Yolton K. NICU Network Neurobehavioral Scale profiles predict developmental outcomes in a low-risk sample. Paediatr. Perinat. Epidemiol. 2012;26(4):344–352. doi: 10.1111/j.1365-3016.2012.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schroeder DI, Blair JD, Lott P, et al. The human placenta methylome. Proc. Natl Acad. Sci. USA. 2013;110(15):6037–6042. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.