Abstract
Cortical pulling forces on astral microtubules are essential to position the spindle. These forces are generated by cortical dynein, a minus‐end directed motor. Previously, another dynein regulator termed Spindly was proposed to regulate dynein‐dependent spindle positioning. However, the mechanism of how Spindly regulates spindle positioning has remained elusive. Here, we find that the misalignment of chromosomes caused by Spindly depletion is directly provoking spindle misorientation. Chromosome misalignments induced by CLIP‐170 or CENP‐E depletion or by noscapine treatment are similarly accompanied by severe spindle‐positioning defects. We find that cortical LGN is actively displaced from the cortex when misaligned chromosomes are in close proximity. Preventing the KT recruitment of Plk1 by the depletion of PBIP1 rescues cortical LGN enrichment near misaligned chromosomes and re‐establishes proper spindle orientation. Hence, KT‐enriched Plk1 is responsible for the negative regulation of cortical LGN localization. In summary, we uncovered a compelling molecular link between chromosome alignment and spindle orientation defects, both of which are implicated in tumorigenesis.
Keywords: spindle positioning, chromosome misalignment, LGN, PLK1, micropatterning
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Cell Cycle
Introduction
The bipolar spindle serves as the core structure mediating proper alignment and segregation of chromosomes 1. In addition, the spindle plays an important role in defining the position of the future cleavage plane, thus controlling daughter cell size and the positioning of the daughter cells in a polarized tissue context 2, 3. Proper positioning of the spindle is an important feature in many cell types and is essential for the maintenance of tissue organization and stem cell renewal.
Studies in various eukaryotic systems have revealed a highly conserved pathway for spindle positioning 4, 5, 6, 7, 8, 9, 10. This pathway involves a cortically localized protein complex consisting of Gαi, LGN, and NuMA, which ultimately recruits the microtubule (MT) minus‐end directed multi‐subunit motor, the dynein–dynactin complex 5, 10, 11. Cortically anchored dynein exerts forces on the astral microtubules emanating from the spindle poles 12, thereby moving the spindle toward the site with most dynein molecules.
Multiple lines of evidence suggest that cell extrinsic and intrinsic cues contribute to spatiotemporal regulation of cortical dynein. The presence of cell extrinsic cues is illustrated by the fact that the retraction fibers, actin filaments that connect the mitotic cell body to the substratum, dictate spindle orientation in cells grown on adhesive micropatterns 13. Laser cutting of retraction fibers leads to repositioning of the spindle along the cell axis where most force is exerted on the mitotic cell body, indicating a defined relation between external forces and internal regulation of spindle orientation cues 14. Concomitantly, a number of cell intrinsic signals control cortical dynein behavior. The guanine‐exchange factor (GEF) RCC1‐mediated generation of the Ran‐GTP gradient around mitotic chromatin was proposed to inhibit LGN‐NuMA localization to the cortex, thereby indirectly causing the polarized distribution of cortical dynein 11. In addition, the spindle pole‐enriched Plk1 negatively regulates dynein localization by disrupting its interaction with NuMA 11. Concurrently, astral MTs facilitate re‐depositing of dynein to the cortex facing the opposite spindle pole, resulting in a dynamic asymmetric dynein enrichment and spindle oscillations until the spindle is positioned in the center of the cell 11, 15.
Regulation of dynein throughout mitosis requires multiple adaptor proteins, some of which have been linked to the process of spindle orientation 16, 17, 18. Spindly is a coiled‐coil domain‐containing protein that recruits dynein to unattached kinetochores (KTs) in prometaphase 17, 19, 20, and was previously proposed to regulate dynein‐dependent spindle positioning 19. Spindly is required for efficient chromosome alignment 21, a function that is separate from its role in KT–dynein recruitment 22. Since cortical dynein recruitment is unaffected in the absence of Spindly 19, the mechanism of how Spindly regulates spindle positioning has remained elusive. Here, we showed that the spindle‐positioning defect seen after Spindly depletion is caused by chromosome misalignments. We showed that chromosome misalignments induced by several other means, including depletion of CLIP‐170, depletion and inhibition of CENP‐E, and treatment with the mild microtubule poison noscapine, all result in severe defects in spindle orientation. This establishes a direct causal link between chromosome misalignment and spindle misorientation, emphasizing how distinct mitotic defects can directly cause a spindle misorientation phenotype due to a spatial imbalance in the intracellular signals derived from the spindle poles and chromosomes.
Results and Discussion
Chromosome misalignments cause spindle misorientation
To study the role of Spindly in spindle positioning, we filmed cells grown on rectangular‐shaped micropatterns and measured the angle of the metaphase plate relative to the short cell axis (Fig 1A). Since Spindly depletion leads to chromosome alignment defects associated with a profound delay in mitosis (20 and Fig EV1A), we first determined the optimal time point for the analysis of spindle positioning. To this end, we filmed cells depleted of the APC/C co‐activator CDC20 to induce a mitotic delay without perturbing the processes involved in chromosome alignment (Fig EV2A). Cells depleted of CDC20 displayed correct spindle orientation at early time points comparable to GAPDH‐depleted cells, as determined by measuring the angle of the metaphase plate 32 min post‐nuclear envelope breakdown (NEB) (Figs 1B, EV2B and C and Video EV1). After a prolonged mitotic delay, CDC20‐depleted cells displayed spindle tilting and a more randomized spindle orientation as determined by measuring the spindle angle one frame before anaphase onset (Fig EV2C and Video EV2). Therefore, we decided to determine the spindle angle in all subsequent analyses at 32 min post‐NEB, unless indicated otherwise.
Figure 1. Chromosome misalignments cause spindle misorientation.
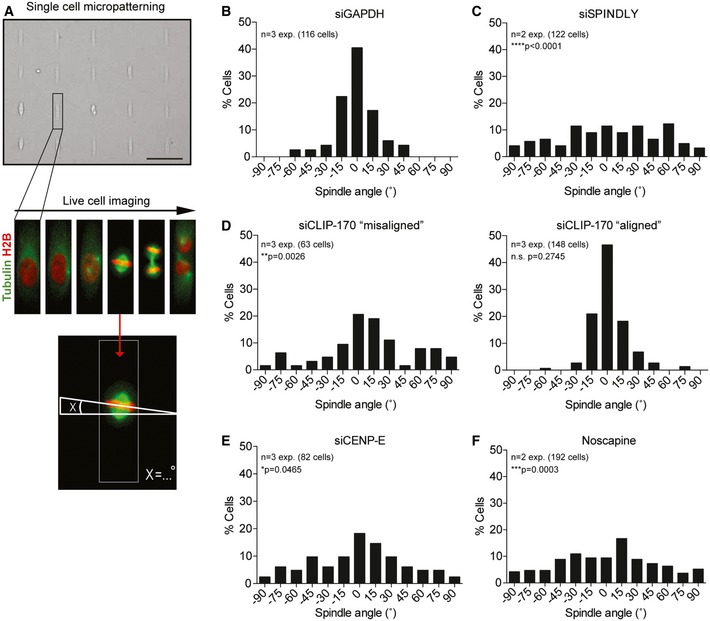
-
ASchematic showing the live single‐cell patterning setup. U2OS cells stably expressing H2B‐GFP and mCherry‐tubulin are synchronized in thymidine and released after 16 h into fresh medium. Cells are seeded on rectangular‐shaped micropatterns coated with fibronectin and incubated for additional 8 h before the start of the imaging. Images were taken every 8 min and the angle of the spindle was determined by measuring the angle of the metaphase relative to the short length of the rectangle at 32 min after NEB. Scale bar represents 100 μm.
-
B, CHistogram of spindle angles in cells transfected with GAPDH siRNA for 48 h (B) and Spindly siRNA for 48 h (C).
-
DHistograms showing spindle angles of CLIP‐170‐depleted cells. Due to low efficiency in generating a chromosome misalignment phenotype after 72 h of CLIP‐170 siRNA transfection, cells are split into two categories: “aligned” and “misaligned”, relating to the observed chromosome misalignment phenotype.
-
E, FSpindle angle histograms of CENP‐E‐depleted cells (48 h) (E) and noscapine‐treated cells (F).
Figure EV1. Spindly, CLIP‐170, and CENP‐E are required for chromosome alignment, but not for cortical dynein recruitment.
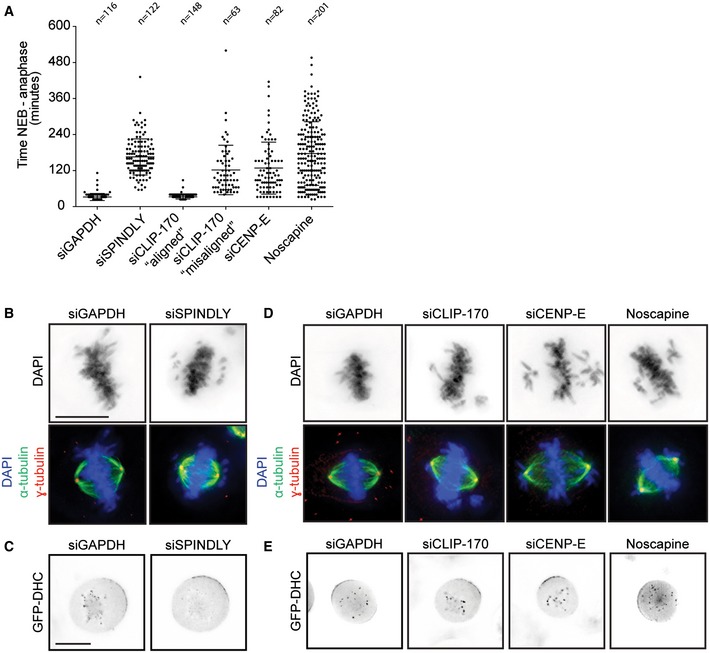
- Average time in mitosis (nuclear envelope breakdown till anaphase) of U2OS cells transfected with GAPDH, Spindly, CLIP‐170, and CENP‐E siRNA or treated with noscapine. Data adapted from experiments shown in Fig 1. Graph shows mean ± SD. (B, C) Scale bars represent 10 μm.
- Representative image of the mitotic phenotype observed after Spindly depletion. Cells were stained with α‐tubulin, γ‐tubulin, and DAPI.
- Representative live‐cell microscopic images of HeLa cells stably expressing GFP‐DHC from a bacterial artificial chromosome [49]. Cells were transfected with the indicated siRNAs for 48 h. One hour before images were taken, cells were treated with the Eg5 inhibitor STLC to arrest them in mitosis with a monopolar spindle and to allow the visualization of KT‐ and cortical dynein. Scale bar represents 10 μm.
- Cells were transfected with CLIP‐170 or CENP‐E siRNA for 48 h or treated for 30 min with noscapine and fixed and stained as in (B).
- Same experiment as described in (C) to examine GFP‐DHC localization after CLIP‐170 and CENP‐E depletion and noscapine treatment.
Figure EV2. Chromosome misalignments induce spindle misorientation independent of SAC activity.
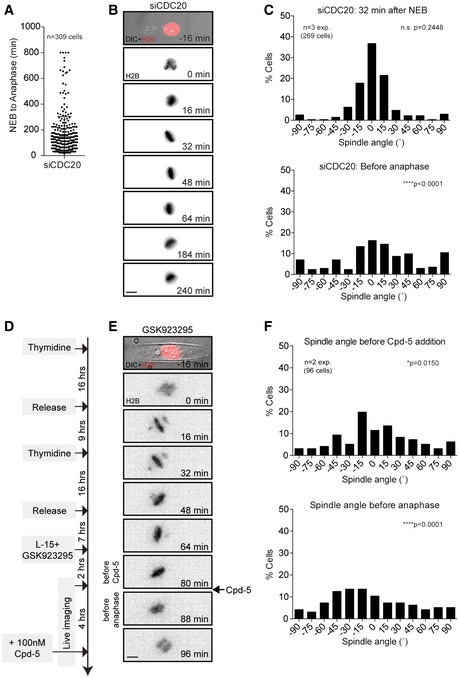
-
AAverage time in mitosis of U2OS cells transfected with CDC20 siRNA for 24 h. Error bar represents mean ± SD.
-
B, CLive‐cell images and histograms of GFP‐H2B U2OS cells filmed on rectangular micropatterns after CDC20 depletion. The spindle angles were scored at 32 min after NEB (top) and one frame before cells eventually exit mitosis (bottom).
-
DSchematic showing the experimental setup for the synchronization and live‐cell imaging of micropatterned U2OS cells that enter mitosis in the presence of a CENP‐E inhibitor to induce chromosome misalignments and subsequently are forced out of mitosis by the addition of the Mps1 inhibitor Cpd‐5.
-
E, FExample cell and spindle angle histograms of cells one frame before the addition of Cpd‐5 (top) and one frame before mitotic exit (bottom).
In line with previous results, we could confirm a critical role for Spindly in spindle positioning (Fig 1B and C, and 19). Functional inactivation of Spindly after siRNA‐mediated knockdown was confirmed by an increase in mitotic timing (Fig EV1A), a defect in chromosome alignment (Fig EV1B), and the loss of dynein from KTs (Fig EV1C). Consistent with previous data, we observed normal recruitment of GFP‐tagged dynein heavy chain (DHC) to the cortex after Spindly depletion (Fig EV1C and 19).
So how can loss of Spindly, a KT protein, affect spindle orientation? The most striking phenotype of a Spindly‐depleted cell is a defect in chromosome alignment (Fig EV1B). Since chromosome‐derived signals have been shown to perturb the localization of cortical factors involved in spindle positioning 11, we asked whether misaligned chromosomes can promote misorientation of the mitotic spindle. Therefore, we depleted CLIP‐170 and CENP‐E, which both have important functions in chromosome alignment 23, 24, 25, 26. Depletion of either factor leads to relatively mild chromosome misalignment phenotypes without disrupting normal bipolar spindle assembly (Fig EV1D). Furthermore, depletion of CLIP‐170 or CENP‐E did not lead to a defect in dynein localization to the KTs and, more importantly, both factors were found to be dispensable for cortical dynein recruitment (Fig EV1E). The phenotype in CLIP‐170‐depleted cells was not very penetrant with only 33% of cells presenting a clear phenotype in chromosome alignment, marked by an increase in mitotic timing (Fig EV1A). We therefore separated the CLIP‐170‐depleted cells into two categories, “misaligned” and “aligned”. The “aligned” cells displayed no defect in spindle orientation, whereas the “misaligned” cells did not position their spindles correctly (Fig 1D). Loss of CENP‐E also produced chromosome alignment defects accompanied by an increase in mitotic timing (Fig EV1A and D and 24, 27). Consistently, CENP‐E‐depleted cells displayed an almost completely random distribution of spindle angle (Fig 1E). Finally, we generated chromosome misalignments with a low dose of noscapine, an opium alkaloid that interferes with microtubule dynamics, resulting in a relatively mild chromosome misalignment phenotype (Fig EV1A and D and 28, 29). This drug treatment also resulted in random spindle orientation, while dynein recruitment to the cell cortex was unaffected (Figs 1F and EV1E).
Next, we wondered whether chromosome misalignments only have a transient effect on spindle orientation in mitosis or whether this defect affects the position of the cell division plane upon mitotic exit. To this end, we treated cells with the CENP‐E inhibitor GSK923295 and subsequently forced them out of mitosis by the addition of an inhibitor of the mitotic checkpoint kinase Mps1 (Cpd‐5) (Fig EV2D and 30). Consistent with our previous results with RNAi‐mediated depletion of CENP‐E (Fig 1E), the addition of GSK923295 resulted in chromosome misalignments associated with random spindle orientation (Fig EV2D–F). Addition of a high dose of Cpd‐5 caused the cells to exit within the following 8–16 min. Importantly, cells with misaligned chromosomes entered anaphase with misoriented spindles leading to altered division planes (Fig EV2F and Videos EV3 and EV4).
Taken together, we found that chromosome misalignments induced by four different methods all result in spindle‐positioning defects in mitosis.
Misaligned chromosomes negatively regulate cortical localization of LGN
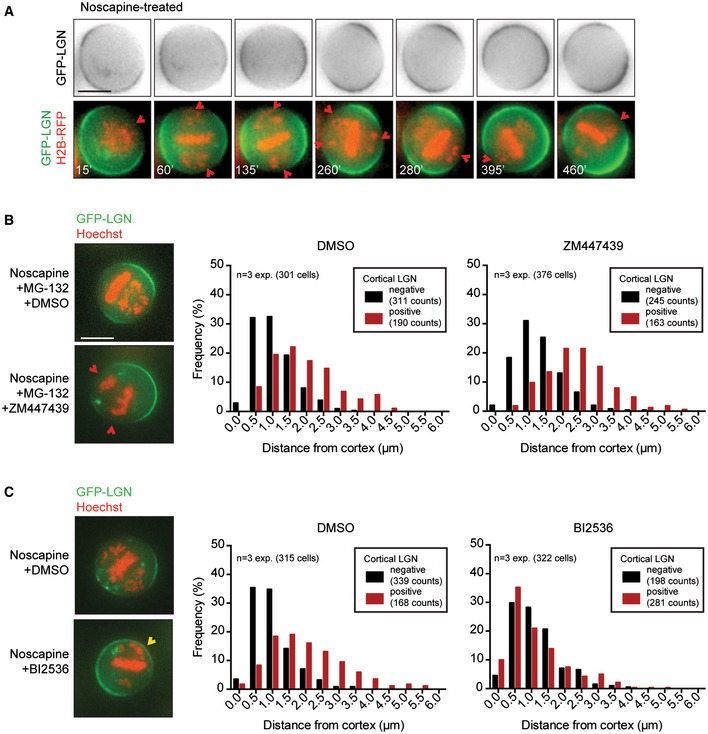
We next tested the behavior of cortically associated LGN, the cortical docking factor for dynein, by live‐cell imaging of cells expressing GFP‐LGN and H2B‐RFP. Cells treated with noscapine showed a clear negative correlation between the presence of chromosomes near the cell cortex and the enrichment of LGN at these sites (Fig 2A). The distribution of LGN was extremely dynamic in these cells and appeared to circulate around the cortex. This movement of LGN was accompanied by continuous rotation of the metaphase plate (formed by alignment of the majority of chromosomes), indicating that the spindle continuously re‐orients toward the site with highest cortical LGN concentration. Most likely, this phenomenon is caused by the pulling forces on astral MTs mediated by dynein and concomitant displacement of its upstream cortical docking factor LGN by a negative signal emitted from misaligned chromosomes in its proximity. In line with the result we obtained with Spindly, CENP‐E, and CLIP‐170 depletion, this indicates that misaligned chromosomes create a negative feedback to cortically localized LGN, thereby preventing steady spindle positioning.
Figure 2. Plk1 restricts cortical LGN localization near misaligned chromosomes.
-
AHeLa cell stably expressing GFP‐LGN and transiently expressing RFP‐H2B was treated with noscapine to induce chromosome misalignments. Cells were imaged every 5 min. The time is relative to NEB. The red arrows indicate cortical regions where the chromosomes are in close proximity to the cell boundaries, accompanied by the displacement of LGN from the cortex. Scale bar represents 10 μm.
-
B, CSimilar assay as described in Fig EV3E was carried out in the presence of a small molecule inhibitor against Aurora B (ZM447439) in combination with the proteasome inhibitor MG‐132 to keep cells arrested in mitosis (B) and with an inhibitor against Plk1 (BI2536) (C). Drugs were added at the same timing as Hoechst, 30 min prior to imaging. Scale bar represents 10 μm.
Plk1, not Ran, is required to delocalize LGN
A prominent candidate to mediate the displacement of cortical LGN from the proximity of chromosomes is Ran‐GTP 11. Therefore, we depleted Ran by siRNA‐mediated knockdown and confirmed functional inactivation of Ran by staining for HURP on k‐fibers (Fig EV3A and 31, 32). To our surprise, rather than promoting cortical LGN localization near chromosomes, Ran depletion led to a global reduction in cortical LGN recruitment (Fig EV3B). Consistently, the addition of importazole 33 to inhibit Ran‐GTP activity also resulted in a rapid reduction in cortical LGN localization (Fig EV3C). In addition, transient expression of a dominant‐negative mCherry‐RanT24N, but not of a GTP hydrolysis‐defective RanQ69L, mutant resulted in a globally reduced cortical LGN recruitment (Fig EV3D).
Figure EV3. Ran depletion causes a global reduction in cortical LGN recruitment.
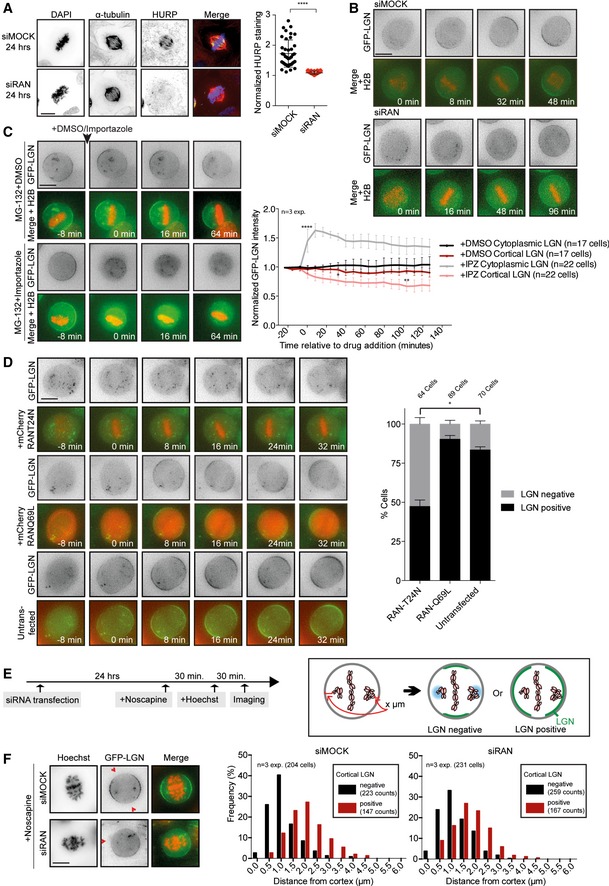
- Representative immunofluorescence image of a U2OS cell after Ran depletion. Cells were either mock‐transfected or with Ran siRNA for 24 h. After fixation, the cells were stained with α‐tubulin, HURP, and DAPI. For the quantification of the HURP staining, the spindle area was sectioned into two parts, one half in the vicinity of the chromosomes and the other half in the vicinity of the spindle pole. The mean HURP signal intensity within the section of the spindle close to the chromosomes was normalized to its mean intensity in the region close to the poles. n = 40 cells per condition from two experiments. ****P < 0.0001 and unpaired t‐test. Graph shows mean ± SD.
- Representative live‐time microscopy images of mitotic HeLa cells stably expressing GFP‐LGN and transiently expressing RFP‐H2B after mock and Ran depletion. Time is relative to NEB.
- HeLa cells stably expressing GFP‐LGN and transiently expressing RFP‐H2B were treated for 30 min with 5 μM MG‐132. Subsequently, DMSO or 40 μM importazole was added to the medium while filming. Images were taken every 8 min. The graph on the right shows the quantification of cortical and cytoplasmic GFP‐LGN levels over time. Intensity values are normalized over the background intensities measured around the cell. ANOVA Dunnett's multiple comparisons test. Graph shows mean ± SEM. (****P < 0.0001, **P < 0.003, *P < 0.05).
- HeLa cells stably expressing GFP‐LGN were transfected for 48 h with mCherry‐RanT24N or mCherry‐RanQ69L constructs and subsequently filmed at 8‐min intervals. The presence or absence of cortical LGN was scored at 32 min after NEB. n = 3 experiments, *P = 0.0014, multiple t‐test. Graph shows mean ± SEM.
- HeLa cells stably expressing GFP‐LGN and transiently expressing RFP‐H2B were mock‐ or siRNA‐transfected for 24 h. Cells were treated with 12.5 μg/ml noscapine to induce chromosome misalignments. After 30 min, Hoechst 33342 was added to the medium to visualize the chromosomes. Another 30 min later, live images of mitotic cells with misaligned chromosomes were taken within 1 h. Images were later used to measure the distance of misaligned chromosomes to the closest cortical region and to score for cortical LGN presence or absence in the same region.
- Representative images of mock‐ and Ran‐depleted cells from the experiment described in (D). The histograms show the effect of misaligned chromosome position on cortical LGN enrichment. The frequency of incidents was plotted where cortical LGN was excluded (black bars) or enriched (red bars) from the nearest misaligned chromosome.
Next, we treated Ran‐depleted cells with noscapine to induce chromosome misalignments in GFP‐LGN‐expressing cells. Although general GFP‐LGN levels at the cell cortex were reduced in the Ran‐depleted cells, low levels of LGN could still be observed. The presence or absence of cortical LGN was scored and plotted against the distance of the most cortex‐proximal misaligned chromosome (Fig EV3E, assay described in 11). In mock‐depleted cells, misaligned chromosomes restricted LGN from localizing to cortical regions when positioned less than ~1.0 μm away from the cortex (Fig EV3F). This negative effect of misaligned chromosome proximity on cortical LGN localization persisted when cells were depleted of Ran (Fig EV3F). Based on these results, we conclude that Ran is unlikely the factor that mediates the displacement of LGN from the cortex near misaligned chromosomes.
Although Ran‐GTP was previously proposed to be a negative regulator of LGN at the cell cortex 11, we find that Ran acts as a positive regulator using three independent methods of Ran inhibition (Fig EV3B–D). A potential explanation for this discrepancy might lie in the timing at which the analyses were conducted. We routinely analyze LGN recruitment in cells that have been in mitosis for < 1 h, but can observe appearance of LGN at cortical regions in Ran‐inhibited cells that were delayed in mitosis for longer periods (for example, see Fig EV3B lower panel). This appearance of LGN at later time points might reflect the presence of distinct pathways of cortical LGN recruitment and could be a consequence of lowered Cdk1 activity, similar to what was shown for the recruitment of NuMA in metaphase versus anaphase 34, 35, and might involve differential regulation by Ran‐GTP.
We next set out to investigate the involvement of two crucial regulators of mitotic progression, Aurora B and Plk1 36, 37, in the cortical regulation of LGN. Aurora B localizes along entire chromosomes at the onset of mitosis and becomes concentrated at the inner centromeric chromatin in prometaphase 38, whereas Plk1 localizes to spindle poles and KTs throughout mitosis 39. We induced chromosome misalignments by noscapine treatment, and subsequently, cells were treated with ZM447439 or BI2536, specific inhibitors of Aurora B and Plk1, respectively 40, 41. We observed no difference between DMSO treatment and Aurora B inhibition with regard to LGN restriction from cortical regions in the proximity of misaligned chromosomes (Fig 2B). In contrast, LGN localization persisted near chromosomes that were positioned in close proximity of the cortex (< 0.5 μm) in Plk1‐inhibited cells (Fig 2C). Thus, Plk1 activity on misaligned chromosomes is required to displace LGN from the cortex. Another centromere‐ and KT‐localized kinase NDR1 was recently reported to regulate spindle orientation through Plk1 42. To test the involvement of NDR1, we treated cells with BI2536 after siRNA‐mediated knockdown of NDR1 (Fig EV4A and B). However, cortical LGN enrichment near misaligned chromosomes persisted in the absence of NDR1, suggesting that Plk1‐mediated displacement of cortical LGN occurs through as yet unknown factors.
Figure EV4. Plk1 does not function through NDR1 to displace cortical LGN.
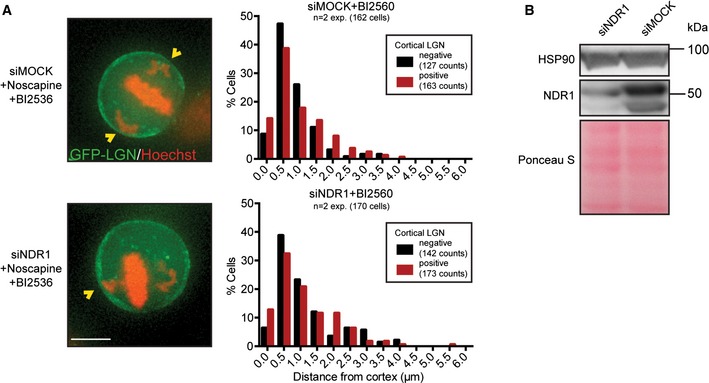
- Representative images and histograms showing the effect of chromosome position on cortical LGN enrichment after 48 h of mock and NDR1 depletion and co‐inhibition of Plk1 with BI2560. Scale bar represents 10 μm.
- Western blot showing the depletion level of NDR1 after transfection of cells with indicated siRNAs for 48 h.
Spindle pole‐localized Plk1 restricts cortical LGN localization
Pole‐localized Plk1 was previously shown to negatively regulate dynein localization at the cell cortex 11. However, this negative regulation was proposed to act by disrupting the interaction of dynein/dynactin with its upstream recruitment factors LGN and NuMA without disrupting the binding of LGN to the cell cortex. To investigate the effect of spindle pole‐localized Plk1 on LGN recruitment to the cortex, we filmed HeLa cells expressing GFP‐tagged LGN and mCherry‐tagged Arp1 (a dynactin subunit) (Fig EV5A and B). Spindle pole association of Arp1 allowed the tracking of spindle movements and revealed clear spindle rocking throughout mitosis. In line with previous data, we observed switching of cortical Arp1 enrichment when the spindle pole came in close proximity of the cell cortex (Fig EV5B and 11. However, in contrast to previous observations, live‐cell imaging at high time resolution (5 min/frame) revealed that LGN also switches from one side of the cell cortex to the other. These switching events coincided with those of Arp1, and cortical LGN was displaced whenever the spindle pole approached the cell periphery (Fig EV5A). To test whether Plk1 activity at the pole contributes to this dynamic re‐localization, we made use of the Plk1 inhibitor BI2536. Plk1 inhibition causes the cells to form monopolar spindles 41, 43. Thus, in order to establish comparable spindle morphology in control DMSO‐ and BI2536‐treated cells, we first induced monopolar spindles in all cells with the Eg5 inhibitor STLC. In control cells treated with STLC, we observed continuous oscillating movements of the monopolar spindle, accompanied by dynamic re‐localization of cortical LGN when spindle poles came in proximity (Fig EV5C). In contrast, when cells were treated with a combination of STLC and BI2536, cortical LGN switching and spindle movement were strongly reduced (Fig EV5D). These observations indicate that Plk1 at the spindle poles acts as a negative regulator of cortical LGN localization. Furthermore, switching of dynein at the cell cortex during mitosis is regulated by Plk1 upstream or at the level of LGN rather than downstream of LGN.
Figure EV5. Spindle pole‐localized Plk1 restricts cortical LGN localization.
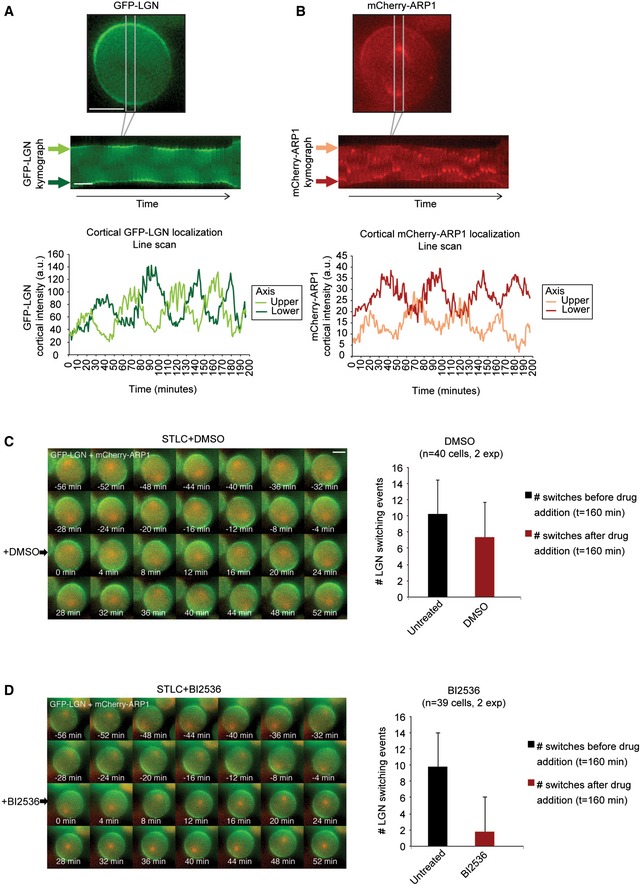
-
A, BKymographs showing the cortical enrichment sites of GFP‐LGN and mCherry‐Arp1 switching in a single HeLa cell. Images were taken every 5 min. As an example, a cell was selected that took almost 200 min to complete mitosis to be able to show multiple switching events. Line scans were made from the kymographs of both the “upper” and the “lower” cortical regions. Fluorescence intensity values are corrected for background fluorescence.
-
C, DGFP‐LGN‐ and mCherry‐Arp1‐expressing HeLa cells were treated with STLC to induce monopolar spindles and filmed for 160 min. Subsequently, DMSO or BI2536 was added (at t = 0 min) and filmed for an additional 160 min. The number of switching events observed for cortical GFP‐LGN before and after the addition of the secondary drug is plotted on the right. Graphs show mean + SD.
Loss of KT‐localized Plk1 rescues cortical LGN enrichment in the proximity of misaligned chromosomes
Next, we investigated whether the displacement of LGN/dynein by misaligned chromosomes was mediated by KT‐bound Plk1. KT recruitment of Plk1 can be reduced upon siRNA‐mediated depletion of PBIP1 (Fig 3A and 44). Consistent with a role of KT‐bound Plk1 in the displacement of LGN from the cortex, we could observe a large fraction of PBIP1‐depleted cells where the proximity of misaligned chromosomes (< 1.0 μm) no longer restricted LGN from the cortex (Fig 3B).
Figure 3. Loss of KT‐localized Plk1 rescues cortical LGN enrichment in the proximity of misaligned chromosomes and spindle misorientation.
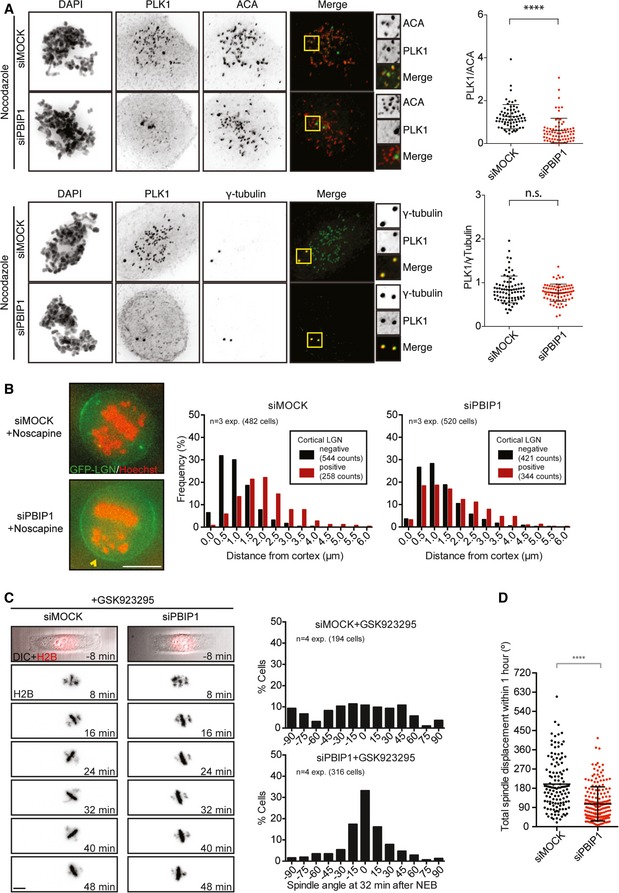
- Immunofluorescence images and quantifications of KT‐ and centrosome‐localized Plk1 in U2OS cells after PBIP1 depletion. Cells were treated with nocodazole for 30 min and fixed. Cells were stained with γ‐tubulin or the centromere marker ACA, Plk1, and DAPI. Top: siMock n = 67, siPBIP1 n = 72 and bottom: siMock n = 40, siPBIP1 n = 40 cells from three experiments. ****P < 0.0001, and Mann–Whitney test, mean ± SD.
- Representative images and histograms showing the effect of chromosome position on cortical LGN enrichment after 72 h of mock and PBIP1 depletion. Bar represents 10 μm.
- Live‐cell images and histograms of GFP‐H2B U2OS cells mock‐transfected or with siPBIP1 filmed on rectangular micropatterns in the presence of a CENP‐E inhibitor. The spindle angles for both conditions were scored at 32 min after NEB.
- The total spindle angle displacement within the first hour of mitosis was determined by measuring the spindle angle at each time point and calculating the sum of the Δ spindle angle between each time point. Dataset from (C) and cells exiting mitosis within 1 h were excluded. siMock n = 127 and siPBIP1 n = 187 cells from four experiments. ****P < 0.0001, and Mann–Whitney test, mean ± SD.
We next wanted to investigate whether KT‐localized Plk1 is indeed responsible for the spindle misorientation provoked by misaligned chromosomes. We induced chromosome misalignments using the CENP‐E inhibitor in cells expressing GFP‐H2B (Fig 3C). As expected, CENP‐E inhibition resulted in random spindle orientation in early mitosis (32 min after NEB). The spindles of CENP‐E‐inhibited cells displayed highly dynamic behavior with a mean spindle displacement of 180° in the first hour after NEB (Fig 3D, Video EV5). In contrast, the combination of CENP‐E inhibition and PBIP1 depletion resulted in a rescue of the spindle misorientation phenotype (Fig 3C and Video EV6) and the dampening of the spindle movements as compared to CENP‐E inhibition alone (Fig 3D). Combined, our data show that Plk1 is a negative regulator of cortical LGN localization and that the KT‐bound pool of Plk1 on misaligned chromosomes is responsible for the displacement of LGN from cortical sites, when placed in its proximity. Collectively, our results indicate that chromosome misalignment is causally linked to spindle misorientation due to a spatial imbalance in local Plk1 concentration.
Plk1 was previously implicated as a regulator of spindle orientation through the displacement of cortical dynein near spindle poles 11. We find that Plk1‐mediated regulation of cortical dynein occurs at the level of its docking factor LGN, rather than at the level of dynein/NuMA. Thus, it is likely that the main function of Plk1 is the asymmetric distribution of LGN and the centering of the mitotic spindle during unperturbed mitosis. Since Plk1 levels at KTs reduce drastically upon the alignment of chromosomes 43, 45, its contribution in the restricting of cortical LGN near the metaphase plate is expected to be minor. Possibly other factors are required for the regulation of cortical LGN dissociation at these cortical regions. Nonetheless, our results underscore the importance of the proper spatial distribution of intracellular signaling factors in accurate spindle positioning.
Taken together, we identify a causal link between two mitotic defects, chromosome misalignment and spindle misorientation. This finding may have important relevance to studies on cancer progression, in particular that of chromosomally instable (CIN) tumors. For example, in CENP‐E +/− 46 or Hec1‐overexpression 47 mouse models, where CIN is induced by interfering with chromosome alignment, the coinciding spindle orientation defects might potentially contribute to enhanced tumorigenesis.
Materials and Methods
Detailed descriptions of immunofluorescence and live imaging methods used in this study can be found in the Appendix.
Cell lines and culture conditions
HeLa and U2OS cells were cultured in DMEM (Gibco) with 6% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Noscapine was dissolved in DMSO and used with a final concentration of 12.5 μM. Nocodazole was used at 250 ng/ml, importazole at 40 μM, BI2536 at 200 nM, MG132 at 5 μM, ZM447439 at 2 μM, GSK923295 at 200 nM, STLC at 20 μM, and Cpd‐5 at 100 nM. Hoechst 33342 was used at a concentration of 50 ng/ml. Transient expression of H2B‐RFP was achieved by transducing HeLa cells with a modified baculovirus expressing histone 2B‐RFP (CellLight® BacMam 2.0, Molecular Probes) following the manufacturer's guidelines.
Plasmids and siRNAs
siRNA transfections were performed using RNAiMax (Invitrogen) in a reverse transfection protocol following the manufacturer's guidelines. The following siRNAs were used: GAPDH OTP SMARTpool, Spindly OTP SMARTpool, CLIP‐170 custom based on 48, CENP‐E custom: AACACGGAUGCUGGUGACCUC, RAN OTP: CUAGGAAGCUCAUUGGAGA, PBIP1 OTP SMARTpool. STK38 OTP SMARTpool. siRNAs were purchased at Thermo Scientific and were used at a final concentration of 20 nM. Constructs with mCherry‐RanT24N (#37396) and mCherry‐RanQ69L (#30309) were purchased from Addgene.
Antibodies
The following antibodies were used for immunofluorescence microscopy experiments: anti‐α‐tubulin (1:10,000; Sigma‐Aldrich), anti‐HURP (1:500 custom made 32), anti‐pH3 (1:1,000; EMD Millipore), anti‐centromere antibody (1:1,000; Antibodies Incorporated, Davis, CA), anti‐γ‐tubulin (1:300; Sigma, T5192), and anti‐Plk1 (1:1,000; Santa Cruz, sc‐17783). Secondary antibodies used were Alexa Fluor 488 and Alexa Fluor 568 (Molecular Probes). Anti‐NDR1/STK38 (M04) was purchased from Abnova.
Author contributions
MAT and JAR performed experiments and analyzed data. PA carried out data analysis. MAT, JAR, and RHM designed experiments and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Video EV1
Video EV2
Video EV3
Video EV4
Video EV5
Video EV6
Review Process File
Acknowledgements
We thank R. Klompmaker and L. Kleij for maintenance of the microscopes and A. Lindqvist for setting up and providing help with the micropatterning. We also thank B.v.d. Broek for providing the ImageJ macro for the immunofluorescence quantification of Plk1. Furthermore, we thank I. Poser and A. Hyman for the GFP‐DHC HeLa cell line, E. Nigg for the Hurp antibody, and T. Kiyomitsu and I. Cheeseman for the GFP‐LGN‐ and mCherry‐ARP1‐expressing HeLa cell line and for helpful discussion. We thank our colleagues for support and valuable discussions. This research was supported by the Zwaartekracht programma (Cancer Genomics Center CGC.nl, project # 58588).
EMBO Reports (2016) 17: 317–325
References
- 1. Walczak CE, Cai S, Khodjakov A (2010) Mechanisms of chromosome behaviour during mitosis. Nat Rev Mol Cell Biol 11: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knoblich JA (2010) Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol 11: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morin X, Bellaïche Y (2011) Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell 21: 102–119 [DOI] [PubMed] [Google Scholar]
- 4. Couwenbergs C, Labbe JC, Goulding M, Marty T, Bowerman B, Gotta M (2007) Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol 179: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen‐Ngoc T, Afshar K, Gönczy P (2007) Coupling of cortical dynein and Gα proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol 9: 1294–1302 [DOI] [PubMed] [Google Scholar]
- 6. Lechler T, Fuchs E (2005) Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437: 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams SE, Ratliff LA, Postiglione MP, Knoblich JA, Fuchs E (2014) Par3–mInsc and Gαi3 cooperate to promote oriented epidermal cell divisions through LGN. Nat Cell Biol 16: 758–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Du Q, Stukenberg PT, Macara IG (2001) A mammalian Partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat Cell Biol 3: 1069–1075 [DOI] [PubMed] [Google Scholar]
- 9. Du Q, Macara IG (2004) Mammalian pins is a conformational switch that links NuMA to Heterotrimeric G proteins. Cell 119: 503–516 [DOI] [PubMed] [Google Scholar]
- 10. Kotak S, Busso C, Gonczy P (2012) Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol 199: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kiyomitsu T, Cheeseman IM (2012) Chromosome‐ and spindle‐pole‐derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol 14: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laan L, Pavin N, Husson J, Romet‐Lemonne G, van Duijn M, López MP, Vale RD, Jülicher F, Reck‐Peterson SL, Dogterom M (2012) Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell 148: 502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Théry M, Racine V, Pépin A, Piel M, Chen Y, Sibarita J‐B, Bornens M (2005) The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol 7: 947–953 [DOI] [PubMed] [Google Scholar]
- 14. Fink J, Carpi N, Betz T, Bétard A, Chebah M, Azioune A, Bornens M, Sykes C, Fetler L, Cuvelier D et al (2011) External forces control mitotic spindle positioning. Nat Cell Biol 13: 771–778 [DOI] [PubMed] [Google Scholar]
- 15. Tame MA, Raaijmakers JA, van den Broek B, Lindqvist A, Jalink K, Medema RH (2014) Astral microtubules control redistribution of dynein at the cell cortex to facilitate spindle positioning. Cell Cycle 13: 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kardon JR, Vale RD (2009) Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raaijmakers JA, Tanenbaum ME, Medema RH (2013) Systematic dissection of dynein regulators in mitosis. J Cell Biol 201: 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dunsch AK, Hammond D, Lloyd J, Schermelleh L, Gruneberg U, Barr FA (2012) Dynein light chain 1 and a spindle‐associated adaptor promote dynein asymmetry and spindle orientation. J Cell Biol 198: 1039–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chan YW, Fava LL, Uldschmid A, Schmitz MHA, Gerlich DW, Nigg EA, Santamaria A (2009) Mitotic control of kinetochore‐associated dynein and spindle orientation by human Spindly. J Cell Biol 185: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Griffis ER, Stuurman N, Vale RD (2007) Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177: 1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barisic M, Sohm B, Mikolcevic P, Wandke C, Rauch V, Ringer T, Hess M, Bonn G, Geley S (2010) Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol Biol Cell 21: 1968–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A (2010) Removal of Spindly from microtubule‐attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev 24: 957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW (2002) Unstable kinetochore‐microtubule capture and chromosomal instability following deletion of CENP‐E. Dev Cell 3: 351–365 [DOI] [PubMed] [Google Scholar]
- 24. Wood KW, Sakowicz R, Goldstein LS, Cleveland DW (1997) CENP‐E is a plus end‐directed kinetochore motor required for metaphase chromosome alignment. Cell 91: 357–366 [DOI] [PubMed] [Google Scholar]
- 25. Tanenbaum ME, Galjart N, van Vugt MATM, Medema RH (2006) CLIP‐170 facilitates the formation of kinetochore‐microtubule attachments. EMBO J 25: 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dujardin D, Wacker UI, Moreau A, Schroer TA, Rickard JE, De Mey JR (1998) Evidence for a role of CLIP‐170 in the establishment of metaphase chromosome alignment. J Cell Biol 141: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW (2000) CENP‐E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol 2: 484–491 [DOI] [PubMed] [Google Scholar]
- 28. Ye K, Ke Y, Keshava N, Shanks J, Kapp JA, Tekmal RR, Petros J, Joshi HC (1998) Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells. Proc Natl Acad Sci 95: 1601–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Landen JW, Lang R, McMahon SJ, Rusan NM, Yvon A‐M, Adams AW, Sorcinelli MD, Campbell R, Bonaccorsi P, Ansel JC et al (2002) Noscapine alters microtubule dynamics in living cells and inhibits the progression of melanoma. Cancer Res 62: 4109–4114 [PubMed] [Google Scholar]
- 30. Koch A, Maia A, Janssen A, Medema RH (2015) Molecular basis underlying resistance to Mps1/TTK inhibitors. Oncogene doi: 10.1038/onc.2015.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koffa MD, Casanova CM, Santarella R, Köcher T, Wilm M, Mattaj IW (2006) HURP is part of a Ran‐dependent complex involved in spindle formation. Curr Biol 16: 743–754 [DOI] [PubMed] [Google Scholar]
- 32. Silljé HHW, Nagel S, Körner R, Nigg EA (2006) HURP is a Ran‐importin beta‐regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol 16: 731–742 [DOI] [PubMed] [Google Scholar]
- 33. Soderholm JF, Bird SL, Kaláb P, Sampathkumar Y, Hasegawa K, Uehara‐Bingen M, Weis K, Heald R (2011) Importazole, a small molecule inhibitor of the transport receptor importin‐β. ACS Chem Biol 6: 700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kotak S, Busso C, Gönczy P (2014) NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J 33: 1815–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng Z, Wan Q, Meixiong G, Du Q (2014) Cell cycle‐regulated membrane binding of NuMA contributes to efficient anaphase chromosome separation. Mol Biol Cell 25: 606–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carmena M, Earnshaw WC (2003) The cellular geography of aurora kinases. Nat Rev Mol Cell Biol 4: 842–854 [DOI] [PubMed] [Google Scholar]
- 37. van Vugt MATM, Medema RH (2005) Getting in and out of mitosis with Polo‐like kinase‐1. Oncogene 24: 2844–2859 [DOI] [PubMed] [Google Scholar]
- 38. Vader G, Medema RH, Lens SMA (2006) The chromosomal passenger complex: guiding Aurora‐B through mitosis. J Cell Biol 173: 833–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bruinsma W, Raaijmakers JA, Medema RH (2012) Switching Polo‐like kinase‐1 on and off in time and space. Trends Biochem Sci 37: 534–541 [DOI] [PubMed] [Google Scholar]
- 40. Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS (2003) Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp‐E to kinetochores. J Cell Biol 161: 267–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Steegmaier M, Hoffmann M, Baum A, Lénárt P, Petronczki M, Krššák M, Gürtler U, Garin‐Chesa P, Lieb S, Quant J et al (2007) BI 2536, a potent and selective inhibitor of polo‐like kinase 1, inhibits tumor growth in vivo. Curr Biol 17: 316–322 [DOI] [PubMed] [Google Scholar]
- 42. Yan M, Chu L, Qin B, Wang Z, Liu X, Jin C, Zhang G, Gomez M, Hergovich A, Chen Z et al (2015) Regulation of NDR1 activity by PLK1 ensures proper spindle orientation in mitosis. Sci Rep 5: 10449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lénárt P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters J‐M (2007) The small‐molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo‐like kinase 1. Curr Biol 17: 304–315 [DOI] [PubMed] [Google Scholar]
- 44. Kang YH, Park J‐E, Yu L‐R, Soung N‐K, Yun S‐M, Bang JK, Seong Y‐S, Yu H, Garfield S, Veenstra TD et al (2006) Self‐regulated Plk1 recruitment to kinetochores by the Plk1‐PBIP1 interaction is critical for proper chromosome segregation. Mol Cell 24: 409–422 [DOI] [PubMed] [Google Scholar]
- 45. Liu D, Davydenko O, Lampson MA (2012) Polo‐like kinase‐1 regulates kinetochore‐microtubule dynamics and spindle checkpoint silencing. J Cell Biol 198: 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weaver BAA, Silk AD, Montagna C, Verdier‐Pinard P, Cleveland DW (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36 [DOI] [PubMed] [Google Scholar]
- 47. Diaz‐Rodríguez E, Sotillo R, Schvartzman J‐M, Benezra R (2008) Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA 105: 16719–16724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P (2004) Functional complementation of human centromere protein A (CENP‐A) by Cse4p from Saccharomyces cerevisiae . Mol Cell Biol 24: 6620–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poser I, Sarov M, Hutchins JR, Hériché J‐K, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW (2008) BAC TransgeneOmics: a high‐throughput method for exploration of protein function in mammals. Nat Methods 5: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Video EV1
Video EV2
Video EV3
Video EV4
Video EV5
Video EV6
Review Process File


