Abstract
Vitiligo is an idiopathic disorder characterized by depigmented patches on the skin due to a loss of melanocytes. The cause of melanocyte destruction is not fully understood. The aim of this study was to detect the potential pathways involved in the vitiligo pathogenesis to further understand the causes and entity of vitiligo. For that the transcriptome of peripheral blood mononuclear cells of 4 vitiligo patients and 4 control subjects was analyzed using the SOLiD System platform and whole transcriptome RNA sequencing application. Altogether 2,470 genes were expressed differently and GRID2IP showed the highest deviation in patients compared to controls. Using functional analysis, altogether 993 associations between the gene groups and diseases were found. The analysis revealed associations between vitiligo and diseases such as lichen planus, limb-girdle muscular dystrophy type 2B, and facioscapulohumeral muscular dystrophy. Additionally, the gene groups with an altered expression pattern are participating in processes such as cell death, survival and signaling, inflammation, and oxidative stress. In conclusion, vitiligo is rather a systemic than a local skin disease; the findings from an enormous amount of RNA sequencing data support the previous findings about vitiligo and should be further analyzed.
Key Words: Vitiligo, Gene expression, Transcriptome, RNA sequencing
Introduction
Vitiligo is characterized by progressive disappearance of skin pigment cells, with straightforward clinical translation – white macules or white hair appear, usually without any accompanying clinical symptoms. Vitiligo occurs worldwide with an estimated prevalence of 0.5-2% in most populations. In almost half of the patients, vitiligo starts before the age of 20 years, and males and females are affected with approximately equal frequency [1,2]. Vitiligo has been associated with the concomitant occurrence of a number of other autoimmune diseases, as well as a wide range of psychosocial difficulties significantly impacting the quality of life [3,4,5].
In vitiligo patients, skin melanocytes are partially or completely lost, and no melanin is synthesized in this area. The cause of the destruction of epidermal or follicular melanocytes is complex and not yet fully understood; however, there are several theories (autoimmune, neural, and biochemical hypotheses) [6,7]. Melanocyte death may occur due to factors from inside and/or outside the cell, and many potential systems could be involved. Histological data have demonstrated that on the perilesional skin of patients with nonsegmental vitiligo an inflammatory infiltrate of low intensity made of mononuclear cells occurs in the upper dermis and the dermal-epidermal interface [8]. The initiation mechanism of this microinflammatory reaction is still unknown, but it has been suggested that various local triggers alert the skin's innate immune system and may precede adaptive immune responses targeting melanocytes [9,10].
The genome-wide association studies account for a limited part of the heritability of the disease but may mediate a crucial part of the disease phenotype, such as progression or age at onset [11]. Most vitiligo susceptibility loci encode immunoregulatory proteins or melanocyte components that are likely to mediate immune targeting and the relationships among vitiligo, melanoma, and eye, skin, and hair coloration [12]. For example, associations have been established between vitiligo pathogenesis and polymorphisms in tyrosinase gene (TYR), catalase gene (CAT), melanocortin 1 receptor (MC1R), major histocompatibility complex genes (MHC), protein tyrosine phosphatase nonreceptor type 22 gene (PTPN22), NLR family pyrin domain containing 1 gene (NLRP1), and X-box binding protein 1 gene (XBP1). Both protective and susceptibility-increasing effects have been found in the case of different polymorphisms in these genes [13,14].
There is a wide range of evidence that vitiligo is not a local but rather a systemic disease. Abnormalities in both humoral and cell-mediated immunity have been documented in vitiligo patients [7,15]. Besides, increased local and systemic cytokine expression is observed [16,17,18,19,20,21]. Furthermore, in vitiligo skin, the elevated H2O2 level is the main cause of tetrahydrobiopterin (6BH4) recycling errors; the latter is essential for melanogenesis [22]. The activity of enzymes important to metabolize H2O2 has impaired on the patients' skin as well as systemically – the latter has been proven in the case of CAT and glutathione peroxidase (GPX1) [23,24,25].
The purpose of this study was to describe potential transcriptional changes in the peripheral blood mononuclear cells (PBMCs) of vitiligo patients. We aimed to find the changes in the patients' blood, which may help to detect the potential pathways participating in the vitiligo pathogenesis and to further understand the causes and entity of vitiligo.
Materials and Methods
Patients and Controls
The protocols and informed consent forms used in this study were approved by the Ethics Review Committee on Human Research of the University of Tartu, Estonia. All the participants signed a written informed consent and were Caucasians living in Estonia. Unrelated patients with vitiligo from the Department of Dermatology, University of Tartu were included in the study. The diagnosis of vitiligo was based on loss of pigmentation with typical localization and white color on the skin lesions under a Wood's lamp. All of the patients included in this study had vitiligo vulgaris and none of them had received specific therapy in the previous 6 months. The controls were volunteers without a positive family history of vitiligo, and they were recruited among medical students and health care personnel. The main characteristics of the participants are presented in table 1.
Table 1.
Characteristics of study participants
| Subjects | Age, years | Sex | Onset age, years (duration] | Vitiligo stage | Koebner's phenomenon | Hair Spontaneous involvement repigmentation | Presence of halo nevus | TPO AB positivity | |
|---|---|---|---|---|---|---|---|---|---|
| Control 1 | 27 | male | – | – | – | – | – | – | – |
| Control 2 | 29 | female | – | – | – | – | – | – | – |
| Control 3 | 40 | male | – | – | – | – | – | – | – |
| Control 4 | 27 | female | – | – | – | – | – | – | – |
| Patient 1 (V105) | 24 | male | 17 (7) | active | no | yes | yes | no | yes |
| Patient 2 (V142) | 43 | male | 21 (22) | active | yes | no | no | no | no |
| Patient 3 (V215) | 27 | female | 5 (22) | active | yes | yes | no | yes | yes |
| Patient 4 (V217) | 36 | female | 35 (1) | active | yes | no | no | no | no |
Sample Collection and RNA Extraction
Blood was collected from 4 volunteers and 4 vitiligo patients between 8:00 and 12:00 in the morning to limit the effect of circadian variation of cytokine production. BD Vacutainer CPT tubes (BD, New York, N.Y., USA) were used to separate PBMCs from other blood cells. The cells were centrifuged at 1,500 g for 30 min at 20°C. After that, blood sera were collected from the top of the PBMCs. Phosphate-buffered saline was used to wash the isolated PBMCs twice, after which they were centrifuged at 190 g for 10 min at 20°C. The supernatant was collected and the cells were stored at −80°C until RNA extraction.
Total RNA was extracted applying the mirVana miRNA Isolation Kit (Ambion, Life Technologies Corp., Carlsbad, Calif., USA) according to the manufacturer's protocol. RNA quality was assessed using Agilent 2100 Bioanalyzer and the RNA 6000 Nano Kit (Agilent Technologies Inc., Calif., USA). The RNA integrity number was over 9 for all samples.
Whole Transcriptome RNA Sequencing Library Preparation
For whole transcriptome sequencing, 10 μg of total RNA was taken and treated with the RiboMinus Eukaryote Kit for RNA sequencing (Invitrogen Corp., Carlsbad, Calif., USA) to eliminate ribosomal RNA from the rest of the transcriptome. Five hundred nanograms of ribodepleted RNA and the SOLiD Total RNA-Seq Kit were used (according to the manufacturer's protocol; Life Technologies Corp.) for whole transcriptome RNA sequencing library preparation. The libraries were marked with different barcodes and pooled together for the following template preparation.
The automated SOLiD EZ Bead System and SOLiD EZ Bead E80 System Consumables (Life Technologies Corp.) were applied for the template preparation. For sequencing the controls' samples, the SOLiD 4 System and paired-end (50 bp forward and 35 bp reverse) chemistry for RNA sequencing were used (Life Technologies Corp.). For sequencing the patients' samples, the SOLiD 5500xl System and paired-end (75 bp forward and 35 bp reverse) chemistry for RNA sequencing were applied (Life Technologies Corp.).
Statistical and Functional Analysis
Raw reads were mapped using Lifescope 2.5.1 software (Life Technologies Corp.) and the whole transcriptome analysis workflow. This workflow generates a very complex output, from gene and exon counts to the alternative splicing and fusion transcripts. For further analysis, we only focused on the gene counts because our primary question was related to the abundance of gene-targeted transcripts. For differential expression analysis, we used the R Bioconductor package edgeR [26], which implemented exact statistical methods and generalized linear models for multigroup and multifactorial experiments. A particular feature of edgeR functionality is the empirical Bayes method that permits the estimation of gene-specific biological variation, even for experiments with minimal levels of biological replication. edgeR can be applied to examine differential expression at the gene, exon or tag level. In our study, we used model-based normalization and applied the negative binomial model. Testing for differential expression was done using the exact test.
A heat map was generated with the DESeq package in R. For the functional network analysis, the Ingenuity Pathway Analysis (IPA, www.ingenuity.com) software was used.
Results
To observe possible changes in the RNA expression pattern in vitiligo patients' blood cells (PBMCs) compared to the controls' cells, the whole transcriptome was sequenced.
RNA Expression Pattern in PBMCs
We found altogether 2,470 genes to be expressed differently (p < 0.05) in vitiligo patients compared to controls, after applying the false discovery rate correction (fig. 1). The gene with the most altered gene expression is glutamate receptor, ionotropic, delta 2 interacting protein (GRID2IP), and its expression has increased 7.2 times (p = 1.31 × 10−20) in vitiligo patients' PBMCs compared to controls' cells. GRID2IP is associated with synaptogenesis and synaptic plasticity; furthermore, the genes with an altered expression belong to different functional groups and participate in various processes. In table 2, the first 20 genes with the most altered RNA expression levels in patients versus controls are presented.
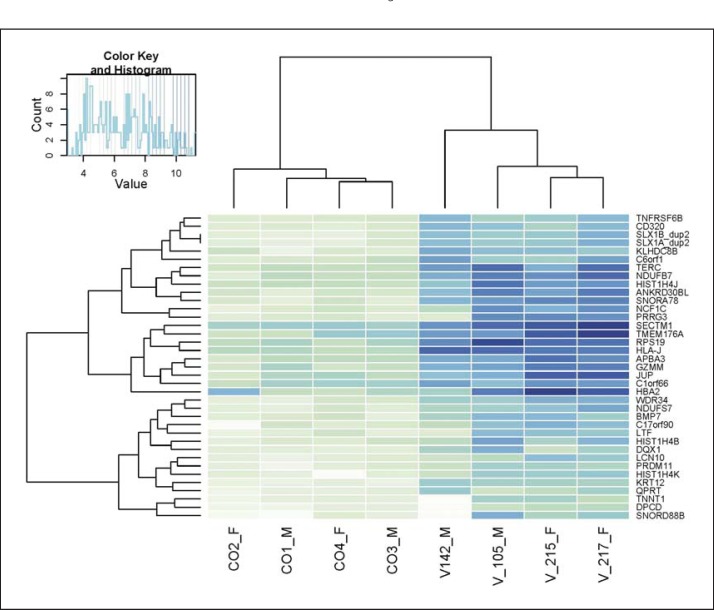
Fig. 1.
Heat map. Vitiligo patients have a distinct transcriptome profile in the blood. We clustered the 30 most significantly changed genes according to the expression values. For clustering, we transformed the count matrix to get sample-to-sample distances. The clustering correctly reflects the experimental design and the differential expression pattern between the study groups.
Table 2.
First 20 genes with the most altered RNA expression levels in vitiligo patients versus controls
| GeneID | Gene name | Fold change | FDR | Main function |
|---|---|---|---|---|
| GRID2IP | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein | 7.2 | 1.31E–20 | synaptogenesis and synaptic plasticity |
| MYO6 | myosin VI | 6.7 | 9.28E–20 | intracellular vesicle and organelle transport |
| LRRC48 | leucine rich repeat containing 48 | −7.5 | 1.32E–18 | cytoplasmatic protein |
| C3orf32 | ssu-2 homolog (C. elegans) | 4.6 | 5.62E–16 | cytoplasmatic protein |
| HMMR (HRAMM) | hyaluronan-mediated motility receptor (RHAMM) | 6.3 | 4.36E–15 | involved in cell motility |
| SNORD78 | small nucleolar RNA, C/D box 78 | −5.7 | 4.36E–15 | small nucleolar RNA |
| SPON1 | spondin 1, extracellular matrix protein | −5.9 | 4.56E–15 | cell adhesion |
| MIR103–A1 | microRNA 103a-1 | −5.4 | 6.46E–15 | microRNA |
| NAA11 | N(alpha)-acetyltransferase 11, NatA catalytic subunit | −6.4 | 1.01E–14 | peptide alpha-N-acetyltransferase |
| LOC285547 | uncharacterized LOC285547 | −5.5 | 9.87E–14 | miscellaneous small RNA |
| LOC283761 | uncharacterized LOC283761 | −5.8 | 2.65E–13 | miscellaneous small RNA |
| SNORD23 | small nucleolar RNA, C/D box 23 | 5.4 | 3.41E–13 | small nucleolar RNA |
| ARSJ | arylsulfatase family, member J | −5.6 | 4.05E–13 | sulfatase, hormone biosynthesis |
| FOLR3 | folate receptor 3 (gamma) | 5.4 | 3.77E–12 | folic acid transport |
| SNORD81 | small nucleolar RNA, C/D box 81 | −4.7 | 8.35E–12 | small nucleolar RNA |
| SNORA78 | small nucleolar RNA, H/ACA box 78 | 4.8 | 2.30E–11 | small nucleolar RNA |
| SPEM1 | spermatid maturation 1 | −4.6 | 2.30E–11 | spermatogenesis, cell differentiation |
| PRRG3 | proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) | 5.1 | 2.76E–11 | K-dependent transmembrane proteins |
| MIR181C | microRNA 181c | −6.7 | 3.99E–11 | microRNA |
| SLC2A14 | solute carrier family 2 (facilitated glucose transporter), member 14 | −6.7 | 7.69E–11 | glucose transporter |
FDR = False discovery rate.
Functional Analysis
Additionally, taken from RNA expression data, we demonstrated associations between the genes with an altered expression pattern and diseases (table 3). Using IPA software, altogether 993 associations between the gene groups and diseases were found. Among the first 20 diseases, the terms ‘dermatological’, ‘immunological’ and ‘inflammation’ are found, which are all related to vitiligo [27,28].
Table 3.
First 20 diseases which are associated with genes with an altered expression pattern in vitiligo patients’ PBMCs
| Functions annotation | Category | p value | Molecules |
|---|---|---|---|
| Lichen planus |
|
2,24E–09 | 24 |
| Limb-girdle muscular dystrophy type 2B |
|
3,26E–08 | 14 |
| Facioscapulohumeral muscular dystrophy |
|
5,61E–08 | 12 |
| Idiopathic pulmonary fibrosis |
|
5,67E–08 | 21 |
| Miyoshi myopathy |
|
6,61E–08 | 13 |
| Limb-girdle muscular dystrophy type 2A |
|
1,39E–07 | 13 |
| Cell death |
|
4,07E–07 | 287 |
| Congenital myopathy |
|
6,12E–07 | 18 |
| Nemaline myopathy |
|
9,53E–07 | 13 |
| Viral infection |
|
1,66E–06 | 169 |
| Limb-girdle muscular dystrophy |
|
1,89E–06 | 15 |
| Apoptosis | Cell death and survival | 2,27E–06 | 234 |
| Necrosis | Cell death and survival | 2,99E–06 | 220 |
| Function of blood cells | Cellular function and maintenance | 3,96E–06 | 68 |
| Pneumonia |
|
4,72E–06 | 23 |
| Proliferation of cells | Cellular growth and proliferation | 5,99E–06 | 301 |
| Function of leukocytes | Cellular function and maintenance | 6,03E–06 | 64 |
| Cancer | Cancer | 1,84E–05 | 330 |
| Production of reactive oxygen species | Free radical scavenging | 3,65E–05 | 38 |
| Breast cancer | Cancer | 3,92E–05 | 103 |
We also performed an analysis showing the top functions of the genes with an altered expression pattern in vitiligo patients' PBMCs (table 4). The genes are participating in processes such as cell death, survival and signaling, inflammation, and oxidative stress among others, which are all associated with vitiligo pathogenesis [27,28,29]. Furthermore, with this analysis, a group of 23 genes associated with dermatological diseases and conditions, immunological disease and inflammatory disease was found (fig. 2).
Table 4.
The top functions of genes with an altered expression pattern in vitiligo patients’ PBMCs
| Molecules in network | Score | Focus molecules | Top functions |
|---|---|---|---|
| ADRM1, AKT1S1, Ant, CORO1A, CYC1, DOK3, EDA, INO80E, KSR2, MAGEC1, MIB2, MICAL1, NCF1C, NFkB (complex), NFKBID, NMRAL1, PAM16 (includes EG:100042179), PIDD, PKN3, RAB1B, RUSC2, RUVBL2, SLC2A6, SPATA20, ST18, ST6GALNAC4, TELO2, TIMM13, TIMM17B, TNFRSF10C, TSPO, UQCRC1, VDAC2, WDR34, ZNHIT2 | 50 | 33 | Cell cycle, connective tissue disorders, dental disease |
| ABTB1, APBA3, ATP13A2, C1QA, C1QB, CA1, CCS, CD320, CFD, CLEC10A, creatine kinase, CTSD, DBF4 (includes EG:10926), DDAH2, DYSF, EGLN2, Ferritin, FTL, HBA1/HBA2, HBB, hemoglobin, LRWD1, MCM2, MYL4, MZF1, NFE2, PAPPA, PRDX2, SCAND1, SERPINF2, SIGLEC1, TBKBP1, TMSB10/TMSB4X, TNFRSF6B, Vegf | 43 | 31 | Protein synthesis, hematological disease, hereditary disorder |
| AES, AGTRAP, ASGR1, BCAT2, CSK, Eif4g, ERK, ETS, FAU, FERMT3, GP9, GP1BB, HK3, IL23R, ITGA2B, LIME1, Mac1, MATK, MBD3 (includes EG:17192), MVP, Ribosomal 40s subunit, RNF181, Rnr, RPS9, RPS10, RPS11, RPS16, RPS19, RPS26, RPS15A, SRCIN1, TRAF, USF2, ZBTB17, ZGPAT | 38 | 28 | Developmental disorder, hematological disease, hereditary disorder |
| ADAM15, ARHGAP4, c-Src, CAMK1, CLCF1, CYBA, Cyba-Ncf1c-Ncf2-Nox-Ncf4, EFNB2, ERK1/2, G6PC3, HMMR, IgG2b, IgG2c, KLRC1, MTUS1, NCF1, NCF4, NOXA1, Phox, RAB4B, Rap1, Rap1-gp91-p22 phox-p40 phox-p47 phox-p67 phox, RASGRP4, RGS14, RIN1, RRAS, SCRIB, SHARPIN, SIPA1, SORBS2, SYTL1, TMEM176B, TXN2, TYMP, VEGFB | 35 | 27 | Hereditary disorder, inflammatory disease, free radical scavenging |
| Akt, C12orf44, CD19, CD22, CD79A, CD79B, CDON, COL6A2, COLEC12, Collagen Type VI, FCGRT, HCST, IgD, IgG1, Igg3, IgG2a, INPP5E, KLKB1, LGALS3BP, MARCO, mir-26, mir-29, mir-221, MIR17HG, PFKL, Ppp2c, PPP2R5B, PVRL2, PVRL3, SCAVENGER RECEPTOR CLASS A, SPHK2, Sphk, TCL1A, TNFRSF13C, TYROBP | 32 | 26 | Cell death and survival, cellular development, hematological system development and function |
| ABCD1, adenosine-tetraphosphatase, APEX2, ATP5D, ATP5G1, ATP5I, ATP6V0D1, ATPase, BLVRB, CARNS1, CD40, DQX1, EVI5L, FBL, FCER2, FCRLA, H+-transporting two-sector ATPase, Iga, IgG, Igm, IL12 (complex), Immunoglobulin, JINK1/2, LILRB3, LST1, MHC Class II (complex), NAT14, NINL, NUDT18, RABAC1, SMOX, SYP, TCIRG1, TRAF2, UBXN11 | 31 | 25 | Energy production, nucleic acid metabolism, small molecule biochemistry |
| Actin, ADAM8, Alpha Actinin, Alpha catenin, Cadherin, Calcineurin protein(s), Calmodulin, CAMK2B, CBWD2, CDC42EP2, CORO6, CORO2B, CPNE5, DCTN6, DVL1, F Actin, FKBP8, Gamma tubulin, KCNN4, KLF5, LYPLA2, MARK4, MYL9, Myosin, ORAI1, OSCAR, PARVB, PDLIM7, PITPNM1, PLEC, PPP1R12C, RHOC, Rock, S100A9, TMOD1 | 31 | 25 | Cell morphology, cellular assembly and organization, cellular function and maintenance |
| Acox, Alp, AMPK, CAPN10, CCDC99, CDC34, Cdk, CORO7/CORO7-PAM16, Cyclin A, Cyclin D, Cyclin E, EIF4E, FADS2, FGF9, FRMD6, GADD45GIP1, Insulin, mir-30, mir-130, mir-181, mir-368, MTORC1, PNKP, POLD1, POP7 (includes EG:100004610), RASSF8, Rb, RPPH1, SLC30A10, SLC6A8, STARD10, STK11, TFEB, TOB1, TUBE1 | 31 | 25 | Endocrine system disorders, reproductive system disease connective tissue disorders |
| AATK, Adaptor protein 1, alcohol group acceptor phosphotransferase, Casein, Caspase 3/7, CDC37, CDK5, DIXDC1, ECSIT, GCDH, GRK6, GZMH, Ikk (family), IL-1R, IL1R2, IRAK1, IRAK, IRF, IRS, Jnk, LLGL2, LONP1, MAP2K2, MAP3K10, MAP3K11, MAP3K, mir-199, NEFH, NEFL, PAWR, PKN1, PPP5C, SIGIRR, SPSB2, TRAF7 | 28 | 24 | Cell-to-cell signaling and interaction, cellular assembly and organization, cellular function and maintenance |
| ABCC9, ACD, ARL4A, BATF, Cbp/p300, CDA, COBRA1, EDF1, FKBP14, GAPDH, HDAC10, HIST1H3A (includes others), HIST1H4A (includes others), HISTONE, Histone h4, Holo RNA polymerase II, IFI35, IFI27L2, IFITM2, IFN ALPHA RECEPTOR, IFN alpha/beta, Ifnar, Importin alpha, Interferon-alpha induced, MED16, mediator, MX1, POLR2E, PPARG, PRMT1, RNA polymerase II, SSBP4, TERC, THAP7, ZBTB7B | 28 | 24 | Cellular assembly and organization, cardiovascular system development and function, cellular function and maintenance |
| 60S ribosomal subunit, ADAMTS1, Ap1, C1q, Cg, CK1, Creb, DSCC1, FGF2, Fibrinogen, GAS1 (includes EG:14451), glutathione peroxidase, glutathione transferase, GPX1 (includes EG:14775), GST, GSTM3, GSTP1, GSTZ1, KLHL4, KPTN, Laminin, Mek, mir-23, NKG7, PTX3, RPL3, RPL8, RPL18, RPL23, RPL24, RPL18A, RPL37A, RPL7A, SPI1 (includes EG:20375), TNFRSF8 | 28 | 23 | Drug metabolism, protein synthesis, cardiovascular system development and function |
| ARHGEF10L, CD3-TCR, CTDSP1, DHX58, DLEU1, DNASE2, DRAP1, E2f, GAS2L1, Hat, HIST1H2BJ/HIST1H2BK, HLA Class I, HLA-A, Hla-abc, HLA-G, HLA-J, IFN TYPE 1, IMPA2, IRF7, LILRA5, MHC, MHC CLASS I (family), Nc2, NCLN, NFAT (complex), NFkB (family), NLRP12, NOMO1 (includes others), OAS1, PSMB10, Ras, RFXANK, SLC25A22, SPCS2, TNFRSF14 | 27 | 23 | Dermatological diseases and conditions, immunological disease, inflammatory disease |
The first 12 functional groups are presented, altogether 21 groups were found.
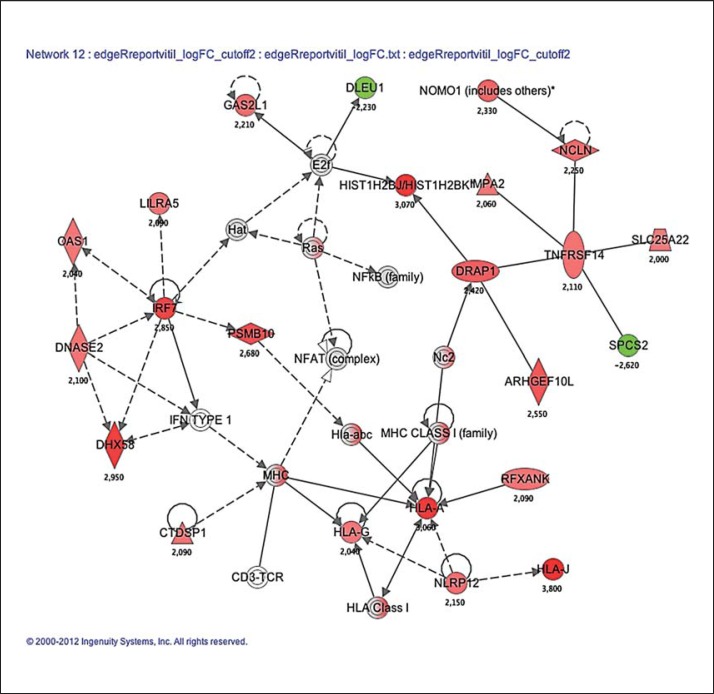
Fig. 2.
The illustration of genes in a pathway associated with dermatological diseases and conditions, immunological disease and inflammatory disease (table 4) (filter set to p < 0.05 after false discovery rate correction) in vitiligo patients compared to controls. Gene expression profiles were uploaded to the Ingenuity Pathway Analysis software and based on the differential expression of these genes, the most relevant biochemical network with functional links is found. Green: decreased expression level in patients vs. controls; red: increased expression level; gray: similar expression level; white: no associations between vitiligo and the gene were found. The numbers reflect the t value of the statistical comparison with the Bayes moderated t test. The figure was generated through the use of the Ingenuity Pathway Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com).
Discussion
The theory that vitiligo is a systemic disease affecting the whole organism rather than a local disorder of certain skin areas is best supported by the autoimmune nature of vitiligo. Additionally, other autoimmune diseases may appear in vitiligo patients, such as autoimmune thyroid disease, psoriasis, rheumatoid arthritis, pernicious anemia, and alopecia areata [30,31,32,33].
In the present study, whole transcriptome RNA sequencing was performed using 4 vitiligo patients' and 4 control individuals' PBMC samples to observe the possible systemic changes at the transcriptional level. The primary results demonstrated that among 2,470 differentially expressed genes (p < 0.05), the first 20 genes, whose mRNA expression has statistically altered most significantly, have distinctive functions. When looking more thoroughly at some of these genes, GRID2IP is the one with the most altered expression level if comparing vitiligo patients and controls, i.e. the level is higher in patients. GRID2IP is predominantly expressed at parallel fiber-Purkinje cell postsynapses, where it may control GRIP2 signaling in Purkinje cells [34,35]. Vitiligo has been demonstrated to be present in addition to several neuropathologies (spastic paraplegia, spastic paraparesis, and peripheral neuropathy), which are associated for example with myelin sheet defects, neuron damages, and scarring [36,37]. Thus, we see additional signs that the mechanisms participating in these diseases may be associated.
The expression of MYO6 (myosin VI) has significantly increased in vitiligo patients' blood cells. It has been demonstrated to be associated with lysosome-related organelles such as melanosomes – it regulates the production of melanin and the size of maturing pigmented melanosomes. Depletion of MYO6 accumulates TYRP1 and increases the melanin content in enlarged premature melanosomes [38]. Thus, overexpression of MYO6 in patients may have a controversial effect and inhibit melanogenesis; however, this also needs to be observed on the skin.
The glycosaminoglycan hyaluronic acid is a ubiquitous component of the extracellular matrix and the level of hyaluronic acid synthesis is increased in highly metastatic melanoma cell lines [39]. The capacity of hyaluronic acid to induce cell growth, differentiation and motility has been shown in a number of experimental systems [40,41]. Hyaluronan-mediated motility receptor is a cell surface receptor, and its expression has increased during melanoma progression [42]. We also found it to be overexpressed in vitiligo patients' blood cells, which correlates with an increased level of cytotoxic T cells in patients [43,44]. However, associations between vitiligo and hyaluronan-mediated motility receptor have not been demonstrated before, and it needs to be further analyzed to understand the possible effect.
Additionally, there are several small nucleolar, miscellaneous and microRNA genes, whose expression has systemically changed in vitiligo patients. Previously, using microRNA expression profiling possible serum biomarkers for vitiligo has been found [45]. microRNAs possibly derived from melanocytes, immune cells and other related cells were observed in nonsegmental vitiligo patients and controls. As Shi et al. [45] presented in their 2013 study, there were microRNAs with a higher and lower expression level in blood sera, and miR-16, miR-19b and miR-720 were suggested as potential biomarkers for vitiligo. Also, associations between intergenic SNP sequences, where small transregulatory RNAs bind, and vitiligo pathogenesis have been demonstrated [46]. The present study provides an enormous amount of data on the expression of small RNA genes in patients, which should be further analyzed.
The present study showed that genes with an altered mRNA expression level in vitiligo patients are also associated with other diseases. The functional analysis revealed the gene groups to be linked to dermatological conditions, immunological and inflammatory diseases as well as cell survival, which all fit the vitiligo profile [27,28]. However, the genes which gave the highest statistical significance in the analysis are also associated with lichen planus. The latter is a mucocutaneous disease, which affects the skin, but also oral mucosa. The etiology is unknown; however, similarly to vitiligo, it is possibly associated with oxidative stress and autoimmunity [47,48] as well as inflammatory processes [49]. Furthermore, these two diseases have demonstrated to be present at the same time together with other autoimmune diseases [50,51,52]. Thus, lichen planus and vitiligo both seem to be rather systemic than local diseases, and they may have similar onset and progress causes.
Additionally, the genes expressed differently in vitiligo patients are also associated with limb-girdle muscular dystrophy type 2B, which is an autosomal recessive disease caused by the diminution or absence of dysferlin. The patients may have perivascular and endomysial inflammatory infiltrate on muscle biopsy [53]. In addition, limb-girdle muscular dystrophy type 2B patients may have other autoimmune diseases [54]. The facioscapulohumeral muscular dystrophy is another example; it is an autosomal dominant disease caused by integral deletion of D4Z4 repeats on the 4q35, and it has also been associated with autoimmune diseases and inflammatory processes [55,56,57]. There are no direct associations demonstrated between these two muscular dystrophies and vitiligo, and in contrast to vitiligo, these dystrophies are inherited diseases. Still, the processes which are activated during the pathogeneses are similar to vitiligo and possibly use the same pathways to some extent.
The functional analysis revealed that the genes with an altered mRNA expression pattern in vitiligo patients participate in processes associated with cell cycle, signaling, death and survival. For example, it has been proven that during the vitiligo pathogenesis, in addition to melanocytes, keratinocytes are also demonstrated to be affected – apoptotic keratinocytes are not able to produce enough supportive factors essential for melanocyte survival [58]. The inflammatory processes are common to vitiligo – the level of inflammatory cytokines has increased both locally and systemically [16,17,18,19,20,21]. The level of cytotoxic T cells has also increased [44]. The pathways for free radical scavenging have been altered in patients, and the activity of enzymes important to metabolize H2O2 has been impaired [23,24,25]. Additionally, genes associated with nervous system functioning appeared on the list. This may be related to the neural hypothesis regarding vitiligo – the amount of neuromediators such as dopamine and norepinephrine has altered in the blood and skin of vitiligo patients [59,60,61,62,63,64,65].
Additionally, functional analysis revealed that in vitiligo patients' blood, most of the 23 molecules associated with dermatological diseases and conditions, immunological and inflammatory diseases were upregulated. These data again refer to the activated processes in patients, which may both cause and try to compensate the deviation from local and systemic homeostasis. Some genes in this pathway, like major histocompatibility complex (HLA), class I genes, interferon (IFN) type I genes, nuclear factor of activated T-cells (NFAT), nuclear factor of kappa B (NF-κB), and NLR family, pyrin domain containing 12 (NLRP12), have already been demonstrated to be associated with vitiligo pathogenesis in previous studies [66,67,68,69,70,71,72,73,74,75,76]. Taken altogether, the results from the functional analysis rather support the previous findings about vitiligo pathogenesis.
In conclusion, the present work provided supportive evidence that vitiligo is rather a systemic than a local disease. The RNA expression pattern in vitiligo patients' blood cells relates to dermatological, immune, inflammatory, and cell survival-associated processes, which all support the previous findings about vitiligo. Additionally, the present work points out possible links between different diseases, such as lichen planus, limb-girdle muscular dystrophy type 2B, facioscapulohumeral muscular dystrophy and vitiligo. Furthermore, the study gives an enormous amount of data, which should be used for more thorough analyses, e.g. the behavior of different types of small RNA genes in vitiligo pathogenesis.
Disclosure Statement
The authors declare that there is no conflict of interest in this paper.
Acknowledgements
This study was financially supported by the target-based funding from the Estonian Ministry of Education grants SF0180043s07 and SF0180148s08, by the University of Tartu research grant PARFS 07915, by the Estonian Science Foundation grants No. 6576, 7549 and 7479, by the Estonian Research Council grant PUT177, Centre of Translational Genomics of University of Tartu, by the European Union through the European Regional Development Fund (Centre of Translational Medicine, University of Tartu) and by the Archimedes Foundation.
References
- 1.Taieb A, Picardo M. Clinical practice. Vitiligo. N Engl J Med. 2009;360:160–169. doi: 10.1056/NEJMcp0804388. [DOI] [PubMed] [Google Scholar]
- 2.Huggins RH, Schwartz RA, Janniger CK. Vitiligo. Acta Dermatovenerol Alp Panonica Adriat. 2005;14:137–142, 144-135. [PubMed] [Google Scholar]
- 3.Kostopoulou P, Jouary T, Quintard B, Ezzedine K, Marques S, Boutchnei S, Taieb A. Objective versus subjective factors in the psychological impact of vitiligo: the experience from a French referral centre. Br J Dermatol. 2009;161:128–133. doi: 10.1111/j.1365-2133.2009.09077.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang KY, Wang KH, Zhang ZP. Health-related quality of life and marital quality of vitiligo patients in China. J Eur Acad Dermatol Venereol. 2011;25:429–435. doi: 10.1111/j.1468-3083.2010.03808.x. [DOI] [PubMed] [Google Scholar]
- 5.Mashayekhi V, Javidi Z, Kiafar B, Manteghi AA, Saadatian V, Esmaeili HA, Hosseinalizadeh S. Quality of life in patients with vitiligo: a descriptive study on 83 patients attending a PUVA therapy unit in Imam Reza Hospital, Mashad. Indian J Dermatol Venereol Leprol. 2010;76:592. doi: 10.4103/0378-6323.69097. [DOI] [PubMed] [Google Scholar]
- 6.Kemp EH, Waterman EA, Weetman AP. Autoimmune aspects of vitiligo. Autoimmunity. 2001;34:65–77. doi: 10.3109/08916930108994127. [DOI] [PubMed] [Google Scholar]
- 7.Ongenae K, Van Geel N, Naeyaert JM. Evidence for an autoimmune pathogenesis of vitiligo. Pigment Cell Res. 2003;16:90–100. doi: 10.1034/j.1600-0749.2003.00023.x. [DOI] [PubMed] [Google Scholar]
- 8.Picardo M, Taieb A. ed 1. Heidelberg: Springer; 2010. Vitiligo. [Google Scholar]
- 9.van den Boorn JG, Picavet DI, van Swieten PF, van Veen HA, Konijnenberg D, van Veelen PA, van Capel T, Jong EC, Reits EA, Drijfhout JW, et al. Skin-depigmenting agent monobenzone induces potent T-cell autoimmunity toward pigmented cells by tyrosinase haptenation and melanosome autophagy. J Invest Dermatol. 2011;131:1240–1251. doi: 10.1038/jid.2011.16. [DOI] [PubMed] [Google Scholar]
- 10.Kroll TM, Bommiasamy H, Boissy RE, Hernandez C, Nickoloff BJ, Mestril R, Le Poole CI. 4-Tertiary butyl phenol exposure sensitizes human melanocytes to dendritic cell-mediated killing: relevance to vitiligo. J Invest Dermatol. 2005;124:798–806. doi: 10.1111/j.0022-202X.2005.23653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, Bennett DC, Herbstman DM, Wallace MR, McCormack WT, et al. Genome-wide analysis identifies a quantitative trait locus in the MHC class II region associated with generalized vitiligo age of onset. J Invest Dermatol. 2011;131:1308–1312. doi: 10.1038/jid.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, Cole JB, Gowan K, Holland PJ, Bennett DC, et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44:676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spritz RA. Six decades of vitiligo genetics: genome-wide studies provide insights into autoimmune pathogenesis. J Invest Dermatol. 2012;132:268–273. doi: 10.1038/jid.2011.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulut H, Pehlivan M, Alper S, Tomatir AG, Onay H, Yuksel SE, Ozkinay F, Pehlivan S. Lack of association between catalase gene polymorphism (T/C exon 9) and susceptibility to vitiligo in a Turkish population. Genet Mol Res. 2011;10:4126–4132. doi: 10.4238/2011.October.31.12. [DOI] [PubMed] [Google Scholar]
- 15.Castanet J, Ortonne JP. Pathophysiology of vitiligo. Clin Dermatol. 1997;15:845–851. doi: 10.1016/s0738-081x(97)00125-9. [DOI] [PubMed] [Google Scholar]
- 16.Honda Y, Okubo Y, Koga M. Relationship between levels of soluble interleukin-2 receptors and the types and activity of vitiligo. J Dermatol. 1997;24:561–563. doi: 10.1111/j.1346-8138.1997.tb02292.x. [DOI] [PubMed] [Google Scholar]
- 17.Yu HS, Chang KL, Yu CL, Li HF, Wu MT, Wu CS, Wu CS. Alterations in IL-6, IL-8, GM-CSF, TNF-alpha, and IFN-gamma release by peripheral mononuclear cells in patients with active vitiligo. J Invest Dermatol. 1997;108:527–529. doi: 10.1111/1523-1747.ep12289743. [DOI] [PubMed] [Google Scholar]
- 18.Caixia T, Hongwen F, Xiran L. Levels of soluble interleukin-2 receptor in the sera and skin tissue fluids of patients with vitiligo. J Dermatol Sci. 1999;21:59–62. doi: 10.1016/s0923-1811(99)00029-8. [DOI] [PubMed] [Google Scholar]
- 19.Moretti S, Spallanzani A, Amato L, Hautmann G, Gallerani I, Fabiani M, Fabbri P. New insights into the pathogenesis of vitiligo: imbalance of epidermal cytokines at sites of lesions. Pigment Cell Res. 2002;15:87–92. doi: 10.1034/j.1600-0749.2002.1o049.x. [DOI] [PubMed] [Google Scholar]
- 20.Zailaie MZ. Decreased proinflammatory cytokine production by peripheral blood mononuclear cells from vitiligo patients following aspirin treatment. Saudi Med J. 2005;26:799–805. [PubMed] [Google Scholar]
- 21.Grimes PE. White patches and bruised souls: advances in the pathogenesis and treatment of vitiligo. J Am Acad Dermatol. 2004;51((1 suppl)):S5–S7. doi: 10.1016/j.jaad.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Schallreuter KU, Moore J, Wood JM, Beazley WD, Peters EM, Marles LK, Behrens-Williams SC, Dummer R, Blau N, Thony B. Epidermal H(2)O(2) accumulation alters tetrahydrobiopterin (6BH4) recycling in vitiligo: identification of a general mechanism in regulation of all 6BH4-dependent processes? J Invest Dermatol. 2001;116:167–174. doi: 10.1046/j.1523-1747.2001.00220.x. [DOI] [PubMed] [Google Scholar]
- 23.Maresca V, Roccella M, Roccella F, Camera E, Del Porto G, Passi S, Grammatico P, Picardo M. Increased sensitivity to peroxidative agents as a possible pathogenic factor of melanocyte damage in vitiligo. J Invest Dermatol. 1997;109:310–313. doi: 10.1111/1523-1747.ep12335801. [DOI] [PubMed] [Google Scholar]
- 24.Schallreuter KU, Wood JM, Berger J. Low catalase levels in the epidermis of patients with vitiligo. J Invest Dermatol. 1991;97:1081–1085. doi: 10.1111/1523-1747.ep12492612. [DOI] [PubMed] [Google Scholar]
- 25.Beazley WD, Gaze D, Panske A, Panzig E, Schallreuter KU. Serum selenium levels and blood glutathione peroxidase activities in vitiligo. Br J Dermatol. 1999;141:301–303. doi: 10.1046/j.1365-2133.1999.02980.x. [DOI] [PubMed] [Google Scholar]
- 26.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 27.Lotti TM, Berti SF, Hercogova J, Huggins RH, Lee BW, Janniger CK, Schwartz RA. Vitiligo: recent insights and new therapeutic approaches. G Ital Dermatol Venereol. 2012;147:637–647. [PubMed] [Google Scholar]
- 28.Richetta A, D'Epiro S, Salvi M, Campoli M, Giancristoforo S, Mattozzi C, Macaluso L, Luci C, Cantisani C, Carboni V, et al. Serum levels of functional T-regs in vitiligo: our experience and mini-review of the literature. Eur J Dermatol. 2013;23:154–159. doi: 10.1684/ejd.2013.1986. [DOI] [PubMed] [Google Scholar]
- 29.Shah AA, Sinha AA. Oxidative stress and autoimmune skin disease. Eur J Dermatol. 2013;23:5–13. doi: 10.1684/ejd.2012.1884. [DOI] [PubMed] [Google Scholar]
- 30.Reimann E, Kingo K, Karelson M, Salum T, Aunin E, Reemann P, Abram K, Vasar E, Silm H, Koks S. Analysis of the expression profile of CRH-POMC system genes in vitiligo skin biopsies. J Dermatol Sci. 2010;60:125–128. doi: 10.1016/j.jdermsci.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Cui J, Arita Y, Bystryn JC. Cytolytic antibodies to melanocytes in vitiligo. J Invest Dermatol. 1993;100:812–815. doi: 10.1111/1523-1747.ep12476636. [DOI] [PubMed] [Google Scholar]
- 32.Harning R, Cui J, Bystryn JC. Relation between the incidence and level of pigment cell antibodies and disease activity in vitiligo. J Invest Dermatol. 1991;97:1078–1080. doi: 10.1111/1523-1747.ep12492607. [DOI] [PubMed] [Google Scholar]
- 33.Alkhateeb A, Fain PR, Thody T, Bennett DC, Spritz RA. Vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16:208–214. doi: 10.1034/j.1600-0749.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda K, Matsuda S, Gladding CM, Yuzaki M. Characterization of the delta2 glutamate receptor-binding protein delphilin: splicing variants with differential palmitoylation and an additional PDZ domain. J Biol Chem. 2006;281:25577–25587. doi: 10.1074/jbc.M602044200. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi T, Ohtsuki G, Yoshida T, Fukaya M, Wainai T, Yamashita M, Yamazaki Y, Mori H, Sakimura K, Kawamoto S, et al. Enhancement of both long-term depression induction and optokinetic response adaptation in mice lacking delphilin. PLoS One. 2008;3:e2297. doi: 10.1371/journal.pone.0002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdallat A, Davis SM, Farrage J, McDonald WI. Disordered pigmentation, spastic paraparesis and peripheral neuropathy in three siblings: a new neurocutaneous syndrome. J Neurol Neurosurg Psychiatry. 1980;43:962–966. doi: 10.1136/jnnp.43.11.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bamforth JS. Vitiligo-spasticity syndrome: new case. Clin Dysmorphology. 2003;12:137–139. doi: 10.1097/00019605-200304000-00013. [DOI] [PubMed] [Google Scholar]
- 38.Loubery S, Delevoye C, Louvard D, Raposo G, Coudrier E. Myosin VI regulates actin dynamics and melanosome biogenesis. Traffic. 2012;13:665–680. doi: 10.1111/j.1600-0854.2012.01342.x. [DOI] [PubMed] [Google Scholar]
- 39.van Muijen GN, Danen EH, Veerkamp JH, Ruiter DJ, Lesley J, van den Heuvel LP. Glycoconjugate profile and CD44 expression in human melanoma cell lines with different metastatic capacity. Int J Cancer. 1995;61:241–248. doi: 10.1002/ijc.2910610217. [DOI] [PubMed] [Google Scholar]
- 40.Kaya G, Rodriguez I, Jorcano JL, Vassalli P, Stamenkovic I. Selective suppression of CD44 in keratinocytes of mice bearing an antisense CD44 transgene driven by a tissue-specific promoter disrupts hyaluronate metabolism in the skin and impairs keratinocyte proliferation. Genes Dev. 1997;11:996–1007. doi: 10.1101/gad.11.8.996. [DOI] [PubMed] [Google Scholar]
- 41.Hamann KJ, Dowling TL, Neeley SP, Grant JA, Leff AR. Hyaluronic acid enhances cell proliferation during eosinopoiesis through the CD44 surface antigen. J Immunol. 1995;154:4073–4080. [PubMed] [Google Scholar]
- 42.Ahrens T, Assmann V, Fieber C, Termeer C, Herrlich P, Hofmann M, Simon JC. CD44 is the principal mediator of hyaluronic-acid-induced melanoma cell proliferation. J Invest Dermatol. 2001;116:93–101. doi: 10.1046/j.1523-1747.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- 43.Lili Y, Yi W, Ji Y, Yue S, Weimin S, Ming L. Global activation of CD8+ cytotoxic T lymphocytes correlates with an impairment in regulatory T cells in patients with generalized vitiligo. PLoS One. 2012;7:e37513. doi: 10.1371/journal.pone.0037513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogg GS, Rod Dunbar P, Romero P, Chen JL, Cerundolo V. High frequency of skin-homing melanocyte-specific cytotoxic T lymphocytes in autoimmune vitiligo. J Exp Med. 1998;188:1203–1208. doi: 10.1084/jem.188.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi YL, Weiland M, Li J, Hamzavi I, Henderson M, Huggins RH, Mahmoud BH, Agbai O, Mi X, Dong Z, et al. MicroRNA expression profiling identifies potential serum biomarkers for non-segmental vitiligo. Pigment Cell Melanoma Res. 2013;26:418–421. doi: 10.1111/pcmr.12086. [DOI] [PubMed] [Google Scholar]
- 46.Glinskii AB, Ma J, Ma S, Grant D, Lim CU, Sell S, Glinsky GV. Identification of intergenic trans-regulatory RNAs containing a disease-linked SNP sequence and targeting cell cycle progression/differentiation pathways in multiple common human disorders. Cell Cycle. 2009;8:3925–3942. doi: 10.4161/cc.8.23.10113. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi A, Shiono H, Okumura M. Thymoma accompanied by lichen planus. Interact Cardiovasc Thorac Surg. 2008;7:347–348. doi: 10.1510/icvts.2007.164178. [DOI] [PubMed] [Google Scholar]
- 48.Aly DG, Shahin RS. Oxidative stress in lichen planus. Acta Dermatovenerol Alp Panonica Adriat. 2010;19:3–11. [PubMed] [Google Scholar]
- 49.Roopashree MR, Gondhalekar RV, Shashikanth MC, George J, Thippeswamy SH, Shukla A. Pathogenesis of oral lichen planus – a review. J Oral Pathol Med. 2010;39:729–734. doi: 10.1111/j.1600-0714.2010.00946.x. [DOI] [PubMed] [Google Scholar]
- 50.Qiao J, Zhou G, Ding Y, Zhu D, Fang H. Multiple paraneoplastic syndromes: myasthenia gravis, vitiligo, alopecia areata, and oral lichen planus associated with thymoma. J Neurol Sci. 2011;308:177–179. doi: 10.1016/j.jns.2011.05.038. [DOI] [PubMed] [Google Scholar]
- 51.Tan RS. Ulcerative colitis, myasthenia gravis, atypical lichen planus, alopecia areata, vitiligo. Proc R Soc Med. 1974;67:195–196. [PMC free article] [PubMed] [Google Scholar]
- 52.Gupta S, Gupta S, Aggarwal K, Jain VK. Becker nevus with vitiligo and lichen planus: cocktail of dermatoses. N Am J Med Sci. 2010;2:333–335. [PMC free article] [PubMed] [Google Scholar]
- 53.Gallardo E, Rojas-Garcia R, de Luna N, Pou A, Brown RH, Jr, Illa I. Inflammation in dysferlin myopathy: immunohistochemical characterization of 13 patients. Neurology. 2001;57:2136–2138. doi: 10.1212/wnl.57.11.2136. [DOI] [PubMed] [Google Scholar]
- 54.Selva-O'Callaghan A, Labrador-Horrillo M, Gallardo E, Herruzo A, Grau-Junyent JM, Vilardell-Tarres M. Muscle inflammation, autoimmune Addison's disease and sarcoidosis in a patient with dysferlin deficiency. Neuromuscul Disord. 2006;16:208–209. doi: 10.1016/j.nmd.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Coelho PC, Morgado F, Reis P, de Queiroz MV. Facioscapulohumeral dystrophy with myositis associated with rheumatoid arthritis. Clin Rheumatol. 1996;15:185–188. doi: 10.1007/BF02230338. [DOI] [PubMed] [Google Scholar]
- 56.Hauerslev S, Orngreen MC, Hertz JM, Vissing J, Krag TO. Muscle regeneration and inflammation in patients with facioscapulohumeral muscular dystrophy. Acta Neurol Scand. 2013;128:194–201. doi: 10.1111/ane.12109. [DOI] [PubMed] [Google Scholar]
- 57.Frisullo G, Frusciante R, Nociti V, Tasca G, Renna R, Iorio R, Patanella AK, Iannaccone E, Marti A, Rossi M, et al. CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J Clin Immunol. 2011;31:155–166. doi: 10.1007/s10875-010-9474-6. [DOI] [PubMed] [Google Scholar]
- 58.Lee AY. Role of keratinocytes in the development of vitiligo. Ann Dermatol. 2012;24:115–125. doi: 10.5021/ad.2012.24.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turnbull AV, Rivier CL. Regulation of the hypothalamic-pituitary-adrenal axis by cytokines: actions and mechanisms of action. Physiol Rev. 1999;79:1–71. doi: 10.1152/physrev.1999.79.1.1. [DOI] [PubMed] [Google Scholar]
- 60.Birol A, Kisa U, Kurtipek GS, Kara F, Kocak M, Erkek E, Caglayan O. Increased tumor necrosis factor alpha (TNF-alpha) and interleukin 1 alpha (IL1-alpha) levels in the lesional skin of patients with nonsegmental vitiligo. Int J Dermatol. 2006;45:992–993. doi: 10.1111/j.1365-4632.2006.02744.x. [DOI] [PubMed] [Google Scholar]
- 61.Toosi S, Orlow SJ, Manga P. Vitiligo-inducing phenols activate the unfolded protein response in melanocytes resulting in upregulation of IL6 and IL8. J Invest Dermatol. 2012;132:2601–2609. doi: 10.1038/jid.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sullivan RM, Dufresne MM. Mesocortical dopamine and HPA axis regulation: role of laterality and early environment. Brain Res. 2006;1076:49–59. doi: 10.1016/j.brainres.2005.12.100. [DOI] [PubMed] [Google Scholar]
- 63.Morrone A, Picardo M, de Luca C, Terminali O, Passi S, Ippolito F. Catecholamines and vitiligo. Pigment Cell Res. 1992;5:65–69. doi: 10.1111/j.1600-0749.1992.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 64.Salzer BA, Schallreuter KU. Investigation of the personality structure in patients with vitiligo and a possible association with impaired catecholamine metabolism. Dermatology. 1995;190:109–115. doi: 10.1159/000246657. [DOI] [PubMed] [Google Scholar]
- 65.Cucchi ML, Frattini P, Santagostino G, Orecchia G. Higher plasma catecholamine and metabolite levels in the early phase of nonsegmental vitiligo. Pigment Cell Res. 2000;13:28–32. doi: 10.1034/j.1600-0749.2000.130106.x. [DOI] [PubMed] [Google Scholar]
- 66.Jin Y, Ferrara T, Gowan K, Holcomb C, Rastrou M, Erlich HA, Fain PR, Spritz RA. Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of generalized vitiligo via antigen presentation. J Invest Dermatol. 2012;132:1730–1733. doi: 10.1038/jid.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh A, Sharma P, Kar HK, Sharma VK, Tembhre MK, Gupta S, Laddha NC, Dwivedi M, Begum R, Gokhale RS, et al. HLA alleles and amino-acid signatures of the peptide-binding pockets of HLA molecules in vitiligo. J Invest Dermatol. 2012;132:124–134. doi: 10.1038/jid.2011.240. [DOI] [PubMed] [Google Scholar]
- 68.Bowcock AM, Fernandez-Vina M. Targeting skin: vitiligo and autoimmunity. J Invest Dermatol. 2012;132:13–15. doi: 10.1038/jid.2011.353. [DOI] [PubMed] [Google Scholar]
- 69.Reimann E, Kingo K, Karelson M, Reemann P, Loite U, Sulakatko H, Keermann M, Raud K, Abram K, Vasar E, et al. The mRNA expression profile of cytokines connected to the regulation of melanocyte functioning in vitiligo skin biopsy samples and peripheral blood mononuclear cells. Hum Immunol. 2012;73:393–398. doi: 10.1016/j.humimm.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 70.Seckin D, Durusoy C, Sahin S. Concomitant vitiligo and psoriasis in a patient treated with interferon alfa-2a for chronic hepatitis B infection. Pediatr Dermatol. 2004;21:577–579. doi: 10.1111/j.0736-8046.2004.21512.x. [DOI] [PubMed] [Google Scholar]
- 71.Passeron T, Ortonne JP. Physiopathology and genetics of vitiligo. J Autoimmun. 2005;25((suppl)):63–68. doi: 10.1016/j.jaut.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 72.Kocer B, Nazliel B, Oztas M, Batur HZ. Vitiligo and multiple sclerosis in a patient treated with interferon beta-1a: a case report. Eur J Neurol. 2009;16:e78–e79. doi: 10.1111/j.1468-1331.2009.02563.x. [DOI] [PubMed] [Google Scholar]
- 73.Rodriguez-Cerdeira C, Sanchez-Blanco E, Molares-Vila A. Clinical application of development of nonantibiotic macrolides that correct inflammation-driven immune dysfunction in inflammatory skin diseases. Mediators Inflamm. 2012;2012:563709. doi: 10.1155/2012/563709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Becatti M, Prignano F, Fiorillo C, Pescitelli L, Nassi P, Lotti T, Taddei N. The involvement of Smac/DIABLO, p53, NF-kB, and MAPK pathways in apoptosis of keratinocytes from perilesional vitiligo skin: protective effects of curcumin and capsaicin. Antioxid Redox Signal. 2010;13:1309–1321. doi: 10.1089/ars.2009.2779. [DOI] [PubMed] [Google Scholar]
- 75.Kim NH, Jeon S, Lee HJ, Lee AY. Impaired PI3K/Akt activation-mediated NF-kappaB inactivation under elevated TNF-alpha is more vulnerable to apoptosis in vitiliginous keratinocytes. J Invest Dermatol. 2007;127:2612–2617. doi: 10.1038/sj.jid.5700900. [DOI] [PubMed] [Google Scholar]
- 76.Wilson SP, Cassel SL. Inflammasome-mediated autoinflammatory disorders. Postgrad Med. 2010;122:125–133. doi: 10.3810/pgm.2010.09.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]