Abstract
Population-based human in vitro models offer exceptional opportunities for evaluating the potential hazard and mode of action of chemicals, as well as variability in responses to toxic insults among individuals. This study was designed to test the hypothesis that comparative population genomics with efficient in vitro experimental design can be used for evaluation of the potential for hazard, mode of action, and the extent of population variability in responses to chemical mixtures. We selected 146 lymphoblast cell lines from 4 ancestrally and geographically diverse human populations based on the availability of genome sequence and basal RNA-seq data. Cells were exposed to two pesticide mixtures – an environmental surface water sample comprised primarily of organochlorine pesticides and a laboratory-prepared mixture of 36 currently used pesticides – in concentration response and evaluated for cytotoxicity. On average, the two mixtures exhibited a similar range of in vitro cytotoxicity and showed considerable inter-individual variability across screened cell lines. However, when in vitroto-in vivo extrapolation (IVIVE) coupled with reverse dosimetry was employed to convert the in vitro cytotoxic concentrations to oral equivalent doses and compared to the upper bound of predicted human exposure, we found that a nominally more cytotoxic chlorinated pesticide mixture is expected to have greater margin of safety (more than 5 orders of magnitude) as compared to the current use pesticide mixture (less than 2 orders of magnitude) due primarily to differences in exposure predictions. Multivariate genome-wide association mapping revealed an association between the toxicity of current use pesticide mixture and a polymorphism in rs1947825 in C17orf54. We conclude that a combination of in vitro human population-based cytotoxicity screening followed by dosimetric adjustment and comparative population genomics analyses enables quantitative evaluation of human health hazard from complex environmental mixtures. Additionally, such an approach yields testable hypotheses regarding potential toxicity mechanisms.
Keywords: Pesticide, Mixture, Population
1. Introduction
Pesticides are chemicals that are used to kill, repel, or control certain forms of plant or animal life that are considered to be pests (Krieger, 2010). Adverse health effects of pesticides can range from mild skin and mucous membrane irritation to more severe outcomes such as neurotoxicity and cancer (Bassil et al., 2007; Rother, 2014; Sanborn et al., 2007). Moreover, potential for adverse effects following exposure may be higher among vulnerable individuals, life stages or sub-populations (Jurewicz and Hanke, 2008; Perry et al., 2014). There are several challenges in the evaluation of the human health hazard of pesticides. First, pesticides have variable modes of action (MOA) dependent on use and activity, and are meant to be harmful and toxic to pests, but not humans. Second, because they are widely used in agricultural and household settings, people are frequently exposed to pesticide residues. Third, humans are typically exposed to mixtures of pesticides, creating challenges in hazard evaluation (Feron et al., 1998; Manikkam et al., 2012).
While safety testing of the individual pesticides is conducted according to established regulatory guidelines (Babut et al., 2013), evaluation of the toxicity of mixtures is less structured (U.S. EPA, 2002). The cumulative risk assessment is conducted for mixtures of chemicals with common mechanisms of toxicity, even though data are usually available only for individual chemicals. Indeed, current toxicity testing paradigms have been questioned for their failure to consider commonly occurring co-exposures and the magnitude of human population variability in response to chemicals (National Research Council, 2009).
Whole animal testing is difficult to employ for evaluating the hazards of chemical mixtures. In contrast, in vitro testing allows greater flexibility, as chemicals can be grouped according to their effects on key biologic pathways or tested over a broad range of concentrations to capture varied exposure scenarios in a rapid and inexpensive manner (Andersen and Krewski, 2009). The resulting data could enable an informed and focused approach to the problem of assessing hazard in risk-relevant manner in human populations that are exposed to mixtures. Furthermore, with an experimental in vitro design that represents a human population, we are able to explore not only the hazard, but also its intrinsic variability across different concentration ranges (Lock et al., 2012; O'Shea et al., 2011). Such information would be valuable to inform regulatory decisions that could more fully protect public health and sensitive subpopulations (Abdo et al., 2015).
In this study, we addressed the hypothesis that comparative population genomics with efficient in vitro experimental design can be used for evaluation of the potential for hazard, mode of action, and the extent of population variability in responses to chemical mixtures. Specifically, we aimed to address two important issues, mixtures toxicology and exploration of genetically-based inter-individual variation using in vitro cytotoxicity study design. While the emphasis was on genetic variability, the use of the two mixtures as testing agents allowed for a greater exploration of the genetic space because, for example, polymorphisms in different pathways may be the determinants of the variability for different chemicals. The overall study design is depicted in Supplemental Fig. 1. We screened 146 lymphoblast cell lines (LCLs) from four ancestrally and geographically diverse populations with publicly available genotypes and sequencing data from the 1000 Genomes Project (1000 Genomes Project Consortium, 2010). Cells were exposed to two pesticide mixtures (an environmental sample, comprised primarily of a mixture of organochlorines extracted from a passive surface water sampling device, and a mixture of 36 currently used pesticides) at 8 concentrations. Cytotoxic response was assessed using an effective concentration threshold of 10% (EC10), designed to be relevant to the dose– response evaluation commonly used in quantitative risk assessment practice and to meaningfully capture ranges of variation in response across individuals. Genome-wide association mapping was performed to evaluate the genetic determinants of susceptibility. Furthermore, in vitro-to-in-vivo extrapolation with reverse dosimetry was utilized to translate the in vitro concentrations to oral equivalents, which were then compared to predicted human cumulative exposures.
2. Materials and methods
2.1. Experimental design
2.1.1. Cell lines
A set of 146 immortalized LCLs was acquired from Coriell Cell Repositories (Camden, NJ). The 146 cell lines represent 4 ancestrally and geographically diverse populations (Table 1): Utah residents with Northern & European ancestry (CEU); Tuscan in Italy (TSI); Yoruban in Ibadan, Nigeria (YRI); and British from England & Scotland (GBR). Cell lines were chosen based on the availability of dense genotyping information (1000 Genomes Project Consortium et al., 2012). Screening was conducted in two batches, and cell lines were randomly divided into batches without regard to family structure, but with equal representation of population and gender. Cells were cultured in RPMI 1640 media (Gibco, Carlsbad, CA) supplemented with 15% fetal bovine serum (HyClone, South Logan, UT) and 1% penicillin-streptomycin (Gibco) and cultured at 37 °C with 5% CO2. Media was changed every 3 days. Cell count and viability were assessed once a day for five days for all cell lines using Cellometer Auto T4 Plus (Nexcelom Bioscience, Lawrence, MA). Cells were grown to a concentration of up to 106 cells/ml, volume of at least 100 ml, and viability of >85% before exposures. After centrifugation, the cells were re-suspended in fresh media. Cells (100 μl containing 104 cells) were aliquoted to each well in a 96-well treatment plate (following the addition of the chemicals) and mixed using the Biomek 3000 robot. Plates were incubated for 24 h after treatment at 37 °C and 0.5% CO2. To increase the robustness of the data and to evaluate reproducibility, each cell line was seeded in at least two plates so that each compound would be screened in each cell line on 2 or more plates.
Table 1.
Human populations from which lymphoblast cell lines were selected for this study.
| Population | # of cell lines screened | % of total | N males | N females |
|---|---|---|---|---|
| CEU: Utah residents with Northern & Western European ancestry | 47 | 32.2 | 24 | 23 |
| YRI: Yoruban in Ibadan, Nigeria | 40 | 27.4 | 19 | 21 |
| TSI: Tuscan in Italy | 32 | 21.9 | 16 | 16 |
| GBR: British from England & Scotland | 27 | 18.5 | 14 | 13 |
| Total | 146 | 100 | 73 | 73 |
2.1.2. Chemical mixtures
Cells were exposed to two environmental chemical mixtures. First mixture, referred to as “chlorinated pesticide mixture” throughout the manuscript, is an environmental sample obtained from a universal passive sampling device deployed for 30 days in surface water next to a chlorinated pesticide storage facility. In this extract, 10 pesticides were present in detectable quantities in the post-collection laboratory analysis (see Table 2 for a complete list of pesticide chemicals identified by mass spectrometry). The second mixture, referred to as “current use pesticide mixture”, was a laboratory-generated mixture of 36 currently used pesticides with relative concentrations selected to mimic fractional composition of the pesticide exposures in Eastern North Carolina (Table 3). Stock solutions of each mixture were further diluted with dimethyl sulfoxide (DMSO) 8-fold in ½-log step-wise manner. Final cumulative concentrations ranged from 0.032 to 370.4 μM for the current use pesticide mixture and from 0.022 to 65.7 μM for the chlorinated pesticide mixture in 0.5% (vol/vol) DMSO. The mixtures were aliquoted to 96-well plate format using Biomek 3000 robot (Beckman Coulter, Inc., Brea CA). The negative control was DMSO at 0.5%; the positive control was tetra-octyl ammonium bromide at 46 μM.
Table 2.
Chemicals contained in the chlorinated pesticide mixture.
| Constituent name | MW | Constituent CAS# | μg in 1 ml | μM | % |
|---|---|---|---|---|---|
| α-Benzene hexachloride (BHC) | 290.8 | 319-84-6 | 107 | 0.368 | 5.60 |
| β-Benzene hexachloride (BHC) | 290.8 | 319-85-7 | 55 | 0.189 | 2.88 |
| γ-Benzene hexachloride (Lindane) | 290.8 | 58-899/55963-79-6 | 151 | 0.519 | 7.90 |
| δ-Benzene hexachloride (BHC) | 290.8 | 319-86-8 | 41 | 0.141 | 2.15 |
| cis-Chlordane | 409.8 | 5103-71-9 | 18 | 0.044 | 0.67 |
| trans-Chlordane | 409.8 | 5103-74-2 | 15 | 0.037 | 0.56 |
| 4,4′-DDD (Dichlorodiphenyldichloro ethane) | 320.1 | 72-54-8 | 293 | 0.915 | 13.94 |
| 4,4′-DDE (Dichlorodiphenyldichloro ethylene) | 318.0 | 72-55-9 | 1193 | 3.75 | 57.11 |
| 4,4′-DDT (dichlorodiphenyltrichloro ethane) | 354.5 | 50-29-3 | 176 | 0.496 | 7.56 |
| Dieldrin | 380.9 | 60-57-1 | 41 | 0.108 | 1.64 |
| Cumulative value | 2090 | 6.57 | 100 |
Table 3.
Chemicals contained in the current use pesticide mixture.
| Constituent name | MW | Constituent CAS# | μg in 1 ml | μM | % |
|---|---|---|---|---|---|
| Metolachlor | 283.8 | 94449-58-8/51218-45-2 | 115 | 0.405 | 22.77 |
| 2,6-Diethylaniline | 149.2 | 579-66-8 | 1259 | 8.44 | 19.76 |
| Molinate | 187.3 | 2212-67-1 | 139 | 0.742 | 19.45 |
| Tebuthiuron | 228.3 | 34014-18-1 | 65 | 0.285 | 14.74 |
| Trifluralin | 335.5 | 1582-09-8/75635-23-3 | 78 | 0.232 | 2.40 |
| Chlorothalonil | 265.9 | 1897-45-6 | 29 | 0.109 | 2.00 |
| Prometon | 225.3 | 1610-18-0 | 74 | 0.328 | 1.42 |
| Butylate | 217.4 | 2008-41-5 | 193 | 0.888 | 1.35 |
| Benfluralin | 335.3 | 1861-40-1 | 76 | 0.227 | 1.31 |
| Alachlor | 269.8 | 15972-60-8 | 37 | 0.137 | 1.23 |
| Ethoprop | 242.3 | 13194-48-4 | 45 | 0.186 | 1.09 |
| Desisopropyl atrazine | 173.6 | 1007-28-9 | 1,271 | 7.32 | 1.04 |
| Metribuzin | 214.3 | 21087-64-9 | 98 | 0.457 | 0.89 |
| Diazinon | 304.4 | 333-41-5 | 89 | 0.292 | 0.79 |
| Disulfoton | 274.4 | 298-04-4 | 26 | 0.095 | 0.77 |
| Aldicarb | 190.3 | 116-06-3 | 92 | 0.484 | 0.74 |
| Methyl parathion | 263.2 | 298-00-0 | 36 | 0.137 | 0.66 |
| Ethalfluralin | 333.3 | 55283-68-6 | 82 | 0.246 | 0.63 |
| Pebulate (Tilliam) | 203.4 | 1114-71-2 | 56 | 0.275 | 0.61 |
| Cyanazine | 240.7 | 21725-46-2/11096-88-1 | 31 | 0.129 | 0.55 |
| Permethrin | 391.3 | 52645-53-1 | 39 | 0.1 | 0.52 |
| Carbofuran | 221.3 | 1563-66-2 | 85 | 0.384 | 0.50 |
| Chlorpyrifos (Dursban) | 350.6 | 2921-88-2 | 71 | 0.202 | 0.48 |
| Prometryne | 241.4 | 7287-19-6/83653-07-0 | 42 | 0.174 | 0.47 |
| Carbaryl | 201.2 | 63-25-2 | 106 | 0.527 | 0.39 |
| Desethyl atrazine | 187.6 | 6190-65-4 | 1,352 | 7.21 | 0.37 |
| Flumetralin | 421.7 | 62924-70-3 | 81 | 0.192 | 0.37 |
| Dacthal | 332 | 65862-98-8/1861-32-1 | 15 | 0.045 | 0.37 |
| Atrazine | 215.7 | 1912-24-9 | 1178 | 5.46 | 0.35 |
| Simazine | 201.7 | 122-34-9 | 101 | 0.501 | 0.35 |
| Terbufos | 288.4 | 13071-79-9 | 42 | 0.146 | 0.35 |
| Fonofos (Dyfonate) | 246.3 | 944-22-9 | 32 | 0.13 | 0.32 |
| Pendimethalin | 281.3 | 40487-42-1 | 33 | 0.117 | 0.29 |
| Fenamiphos | 303.4 | 22224-92-6 | 54 | 0.178 | 0.27 |
| Tribufos (DEF 6) | 314.5 | 78-48-8 | 41 | 0.13 | 0.26 |
| Napropamide | 271.4 | 15299-99-7 | 37 | 0.136 | 0.12 |
| Cumulative value | 7200 | 37.0 | 100 |
2.1.3. Cytotoxicity profiling
The CellTiter-Glo Luminescent Cell Viability (Promega, Madison, WI) assay was used to assess intracellular ATP concentration, a marker for cytotoxicity, 40 h post treatment. Time points were selected based on previous experiments at the National Institutes of Health Chemical Genomics Center (Xia et al., 2008). A ViewLux plate reader (PerkinElmer, Shelton, CT) was used to detect luminescent intensity.
2.2. Data processing
2.2.1. Cytotoxicity EC10 estimation and outlier detection
Cytotoxicity data were normalized relative to positive/negative controls as described elsewhere (Abdo et al., 2015). We derived an effective concentration 10th percentile (EC10) to provide a single cytotoxicity dose summary per chemical and cell line. The derivation of EC10 was based on the logit model:
with y=η+ε,ε~N(0,σ2), where y is the observed normalized signal representing proportion of surviving cells (which we term the “cytotoxicity value”), d is the log(concentration) for each chemical, and θmax is the limiting mean cytotoxicity value for the zero concentration. θmin was set to zero, to avoid difficulties in estimating the minimum cytotoxicity value for chemicals with low cytotoxicity. An exception was made for chemicals in which the cytotoxicity value at the highest concentration was higher than 0.4, as a very few number of plates/chemicals did not reliably reach maximum cytotoxicity. In those instances θmin was set at the observed cytotoxicity at the maximum concentration. Inspection of these data revealed good fits in such instances. Although in principle θmax should have been 1.0, a number of plates exhibited a drift from this value, and thus the parameter was estimated from the data.
Fitting for the parameters [β0,β1,σ2,θmax] proceeded by maximum likelihood using numerical optimization in R v2.15. An automatic outlier detection algorithm was devised by considering the impact of dropping each concentration value in succession, and removing those values for which the maximum likelihood improved by a factor of 10 or more and refitting the model using the non-outlying observations.
2.2.2. Normalizing for batch effects
Batch effects were evaluated by running principal component analysis. EC10 values were adjusted for batch effect using the ComBat method (Johnson et al., 2007).
2.2.3. Concentration response for populations and individuals
For each pesticide mixture, the three-parameter logistic regression described above in EC10 estimation was fit to concentration–response data for each cell line. The variation in the EC10 estimates was used as illustrative of population variation in true EC10 values, although additional sampling variation underlies each EC10 estimate. An overall logistic concentration–response curve was fit to the aggregated data across all individuals (Fig. 1).
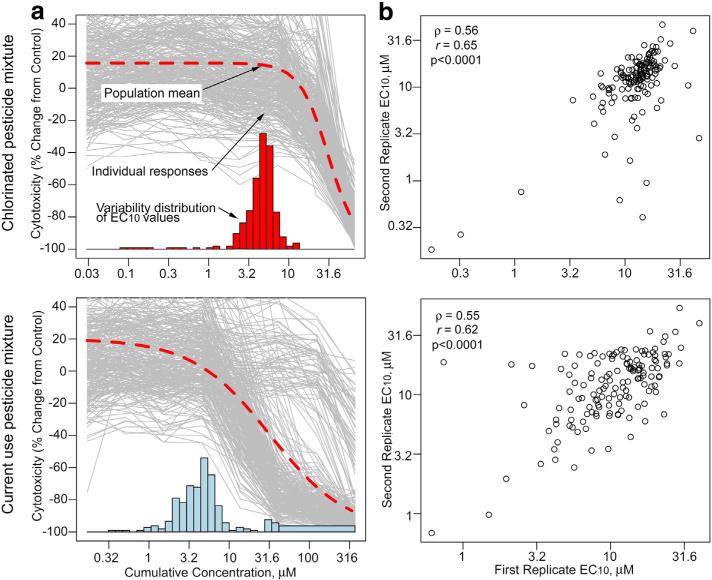
Fig. 1.
Inter-individual and population variability and reproducibility of the cytotoxicity of pesticide-containing mixtures in human lymphoblast cell lines. (a) A population concentration response was modeled using in vitro cytotoxicity of the chlorinated pesticide mixture (top) and the current use pesticide mixture (bottom). Logistic dose–response modeling was applied to each individual cell line, with individual data shown by thin gray lines. Bars represent a histogram of the individual EC10 values, and the dashed curve represents the fit of the logistic model to the pooled data. A histogram in each graph depicts a frequency distribution (y-axis) for the cell lines with a corresponding EC10 (x-axis is identical to that already displayed). (b) Intra-experimental reproducibility of EC10 values for within-batch replicate plates for cell lines for the chlorinated pesticide mixture (top) and the current use pesticide mixture (bottom). Spearman and Pearson's correlation coefficients are shown.
2.2.4. Reproducibility and correlation between mixtures
Pearson and Spearman correlation coefficients (r) between pairs of replicate plates were used to assess experimental reproducibility and the correlation between the two mixtures. For this analysis, the two replicate plates were selected for each mixture and cell line pair.
2.2.5. Chemical/mixture specific toxicodynamic variability factor (VFd)
Variability in response for each mixture across the 146 cell lines was derived as the longest tail of the variability distribution (in our case the ratio of the 50th percentile to the 5th percentile was greater than the ratio of the 95th to the 50th percentile) using the World Health Organization guidance (World Health Organization, 2005).
2.2.6. Chemical descriptors
Chemical descriptors were calculated using Dragon version 5.5 (Mauri et al., 2006). Constant and near constant descriptors as well as highly correlated descriptors were excluded and descriptor values were normalized on a scale from 0 to 1.
2.2.7. Differences in cytotoxicity across different populations
Analysis of Variance (ANOVA) was performed to assess population differences in cytotoxicity between the four screened populations for each mixture.
2.2.8. Genotypes
The primary source of genotypes was obtained as described in Abdo et al. (2015). SNPs with a call rate below 99%, minor allele frequency (MAF) < 0.05, or Hardy–Weinberg equilibrium p-value < 1 × 10−3 were excluded.
2.2.9. Multivariate association analysis (MAGWAS)
The MAGWAS analysis of covariance model (Brown et al., 2012) was used for association mapping. The approach allows for use of the full concentration–response profile, as opposed to a univariate summary (such as EC10) as a single response, with the advantage of robustness and power under a wide variety of association patterns. The model used for association for the jth individual and genotype i for the chemical/SNP was:
where Yij is the vector of responses (across the eight concentrations) for the jth individual having genotype i, Xij is the design matrix of covariates, including sex, indicator variables for laboratory batch, and the first ten genotype principal components, and μi is the eight-vector of parameters modeling the effects of genotype i on the response. The model assumes that the error terms are multivariate normally distributed, with mean vector 0 and variance-covariance matrix Σ, allowing for dependencies in the observations. p-Values were obtained using Pillai's trace (Pillai, 1955). Because this method makes use of asymptotic theory, markers with fewer than 20 individuals representing any genotype were removed, leaving 692,013 SNPs for analysis.
2.2.10. Estimation of Css using in vitro in vivo extrapolation (IVIVE) and Monte Carlo simulation
Key determinants of steady-state pharmacokinetics were experimentally measured for chemicals and published previously (Wetmore et al., 2012, Wetmore et al., in press). Briefly, plasma protein binding was measured using rapid equilibrium dialysis (Wetmore et al., 2012) and the rate of hepatic metabolism of the parent compound was determined using the substrate depletion approach (Rotroff et al., 2010; Wetmore et al., 2012). See flow chart for these analyses in Supplemental Fig. 2.
These data were then used to calculate chemical steady-state blood concentrations (Css) as previously described, with modification (Wetmore et al., 2012, Wetmore et al., in press). The base equation used to calculate static Css is based on constant uptake of a daily oral dose and factors in blood binding, hepatic clearance and non-metabolic renal clearance. The daily oral dose was set to 1 μg/kg/day to reflect ambient environmental exposures. A correlated Monte Carlo approach was employed (Jamei et al., 2009) using Simcyp (Simcyp v.1.3; Certara, Sheffield, UK) to simulate variability across a population of 10,000 individuals equally comprised of males and females, 20– 50 years of age. A coefficient of variation of 30% was used for intrinsic and renal clearance. The median, upper and lower fifth percentiles for the Css were obtained as output.
2.2.11. Calculation of oral equivalent dose values
In conventional use, pharmacokinetic models are used to relate exposure concentrations to a blood or tissue concentration. This is typically referred to as “forward dosimetry”. In contrast, the models can also be reversed to relate blood or tissue concentrations to an exposure concentration, which is referred to as “reverse dosimetry” (Tan et al., 2007). Based on the principal of reverse dosimetry, the median, upper and lower 5th percentiles for the Css were used as conversion factors to generate oral equivalent doses according to the following formula:
In the equation above, the oral equivalent dose value is lin μ early related to the in vitro EC10 and inversely related to Css. This equation is valid only for first-order metabolism that is expected at ambient exposure levels. An oral equivalent value was generated for each chemical-cell line combination and summed to provide a cumulative oral equivalent value for each cell line.
2.2.12. Predicted exposure limits
Pesticide specific predicted exposures were obtained as previously detailed in (Wambaugh et al., 2013). The pesticide specific exposure limit was available for 35 out of the 36 pesticides in the current-use pesticide mixture and for 6 out of 10 pesticides in the chlorinated pesticide mixture. Missing values were replaced by the highest exposure within each mixture. Then, a cumulative exposure was computed for each mixture from the upper 95th percentile (see flow chart in Supplemental Fig. 3).
3. Results
3.1. Cytotoxicity of pesticide mixtures in vitro
Screening was conducted in a 96-well plate format using a robotic system to facilitate reproducibility and throughput. The 146 cell lines were randomly assigned to two batches with blocking to achieve balancing by sex and population. Each cell line was plated on two plates to evaluate technical reproducibility and pesticide mixtures were added at 8 different concentrations ranging from 0.032 to 370.4 μM for current use pesticide mixture, and from 0.022 to 65.7 μM for chlorinated pesticide mixture. Positive and negative controls for cytotoxicity, as assessed by intracellular ATP concentrations, were included on each plate. Normalization to the control for each plate was performed as described in the Materials and Methods section separately for each cell line. EC10s were derived, batch-corrected and averaged across replicate plates for each cell line.
To visualize “individual” vs. “population” response to each pesticide mixture, we fitted a 3-parametric logistic regression to each cell line's concentration–response, as well a single concentration–response curve for the entire population, as illustrated in Fig. 1. Population variability in cytotoxicity of each mixture is shown as a histogram of EC10 values. Both mixtures demonstrated considerable inter-individual variability in cytotoxicity. To evaluate the reproducibility of the EC10 values, pair-wise correlations among duplicate plate pairs were calculated for each mixture. Highly significant correlations were observed for both mixtures (p < 0.0001). For current pesticides mixtures r[Pearson's] = 0.62 and ρ[Spearman] = 0.55. For chlorinated pesticides mixture, r [Pearson's] = 0.65 and ρ[Spearman] = 0.56. Overall reproducibility for both mixtures was also significant (p < 0.0001) with r[Pearson's] = 0.62 and ρ[Spearman] = 0.54.
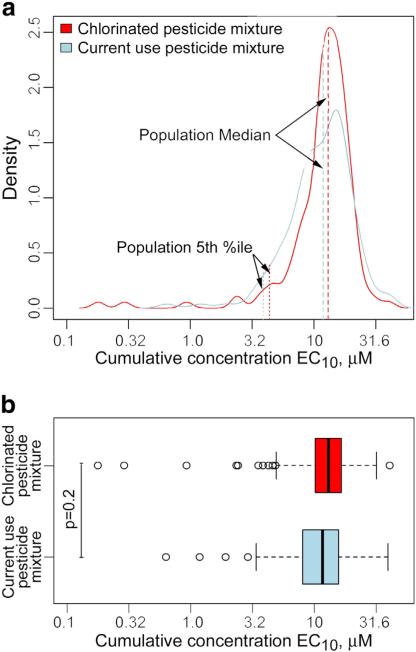
We found that both mean and median EC10 values for in vitro cytotoxicity, as well as the range among cell lines tested, were not significantly different between the two mixtures (Fig. 2). Using these data, the extent of population variation in in vitro cytotoxicity may be derived to serve as a surrogate for cellular variation in the toxicodynamic relationship between systemically available concentrations and toxic responses (Zeise et al., 2013). We calculated a toxicodynamic variability factor (VFd) for these human cell lines as 10, analogous to a chemical-specific toxicodynamic uncertainty factor for inter-individual variability (World Health Organization, 2005), and found it to be around 3-fold for either mixture (Table 4).
Fig. 2.
Distribution of EC10s across 146 cell lines for each mixture. (a) A density plot for the distribution and mean of EC10 of each pesticide mixture (red: chlorinated pesticide mixture, blue: current use pesticide mixture) across 146 cell lines. (b) Box plots (box represents first and third quartiles; vertical line inside the box, the median; whiskers are the 1.5 inter-quantile range; circles are outliers with >1.5 IQR above minimum or maximum). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 4.
Summary statistics for the range in EC10 values for each mixture.
| Pesticide mixture | Meana | STDb | Range | Median | Q05c | Q95d | VFde |
|---|---|---|---|---|---|---|---|
| Chlorinated pesticides | 11.6 | 1.96 | (0.180-40.6) | 13.1 | 4.36 | 21.7 | 3.00 |
| Current use pesticides | 11.1 | 1.85 | (0.649-39.9) | 11.9 | 3.89 | 24.7 | 3.05 |
All values (except for VFd column) are in μM.
The standard deviation of the mean EC10.
The value corresponding to the 5th percentile of EC10 across 146 averaged values for each mixture.
The value corresponding the 95th percentile of EC10 across 146 averaged values for each mixture.
The range of inter-individual variability [10(q95 – q50)] for each mixture.
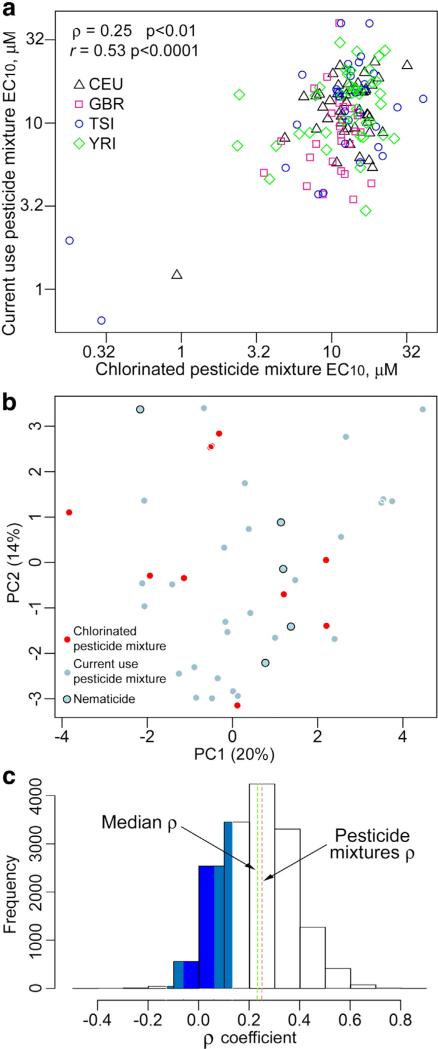
Next, we evaluated the extent of the similarity of cytotoxic responses to the mixtures across cell lines. Strong (significant even after removal of the three outlier cell lines) correlation (r[Pearson] = 0.53, p < 0.0001; ρ[Spearman] = 0.25, p < 0.01) was observed between the mixtures, illustrating appreciable degree of concordance in individual cell line responses (Fig. 3a). There were no suggestive patterns of population clustering in the correlation between the mixtures and neither mixture exhibited significant differences among the populations tested (Supplemental Fig. 4). It is of note, however, that GBR cell lines were the most sensitive, while YRI cell lines the least sensitive to in vitro cytotoxicity of these mixtures. Moreover, within-population variability was greater for the current use pesticide mixture as compared to the chlorinated pesticide mixture, especially when considering the range of the upper quartile to the lower quartiles.
Fig. 3.
Comparative analysis of the mixtures. (a) Scatter plot comparison of EC10 values of each cell line between pesticide mixtures. Symbols represent populations as shown in the inset. Pearson and Spearman correlations are also shown. (b) Scatter plot of 1st and 2nd principal components of the molecular descriptors of the individual chemicals in each pesticide mixture. (c) Frequency histogram of 15,931 pair-wise correlation values (Spearman) among 179 chemicals screened in (Abdo et al., 2015). The green dashed line represents a median ρ value for all correlations, and the red dashed line represents pairwise correlation of pesticide mixtures. Blue shading represents non-significant correlations after correction for false discoveries. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The finding of the significant concordance in responses to both mixtures is of interest because there is no individual chemical overlap (Tables 2 and 3). These results may suggest potential shared mechanisms for cytotoxicity. To further explore chemical similarity among compounds in each mixture, we performed principal components analysis using chemical descriptors. We found that two mixtures overlap in their chemical descriptor space (Fig. 3b), which may partially explain the correlation between two mixtures. While some of the individual components in both mixtures are closely related isomers, no clustering of compounds based on the known pesticidal mode of action (http://www.irac-online.org/documents/moa-classification/?ext=pdf) was observed.
We also compared the strength of the correlation between two mixtures to that of a pair-wise comparison between any pair of compounds in another study that evaluated cytotoxicity of 179 diverse environmental compounds and drugs in a population of lymphoblast cell lines (Abdo et al., 2015). The correlation between two mixtures tested in this study (ρ[Spearman] = 0.25) was comparable to the median correlation of a randomly chosen pair from 15,931 possible combinations in the previous cytotoxicity experiment (Fig. 3c).
3.2. In vitro-to-in vivo extrapolation of cytotoxicity of pesticide mixtures in a population-based model to oral human equivalents and predicted human exposure levels
To conduct a comparative analysis of in vitro cytotoxicity measures of pesticide mixtures with potential human exposures, we computed oral equivalent doses for both mixtures using the reverse dosimetry approach (Wetmore, 2015). In vitro pharmacokinetic data (Wetmore et al., 2012; Wetmore et al., in press) were available for 31 of the 36 chemicals present in the current use pesticide mixture, and for 4 of the 10 chemicals in the chlorinated pesticide mixture. In comparison to 180 ToxCast Phase II chemicals similarly assessed for in vitro pharmacokinetics, the Css values for the 31 current use pesticides had a similar distribution (a median Css < 1 μM and 95th percentile ≈ 200 μM). Only two of the chemicals in the current use pesticide mixture had very high Css values, ethalfluralin (350 μM) and flumetralin (277 μM), the rest were below 8 μM. The distribution of the values for 4 compounds in the chlorinated pesticide mixture was different, the maximum Css value was 58.5 μM.
Because there is no standard approach for evaluation of pharmacokinetics of mixtures, for the purposes of pharmacokinetic modeling in this study we assumed that pharmacokinetics of each chemical will not be significantly affected by the presence of other chemicals in the mixture. Given that cytotoxicity was measured across 146 individual cell lines, separate oral equivalents were calculated for each individual based on the percentage of a given chemical in the mixture (Tables 2 and 3). Furthermore, because some chemicals in each mixture were without in vitro pharmacokinetic parameters, oral equivalent doses were computed based on four different scenarios (see Supplemental Fig. 2 for the workflow). We substituted missing Css values with either median or largest (based on the most conservative simulation assuming no hepatic clearance, high blood binding and only renal clearance, referred to as a “worst-case-scenario”) Css value of other chemicals in the mixture. In addition, oral equivalents were calculated with and without weighting of the EC10 by the percentage of chemical in the mixture.
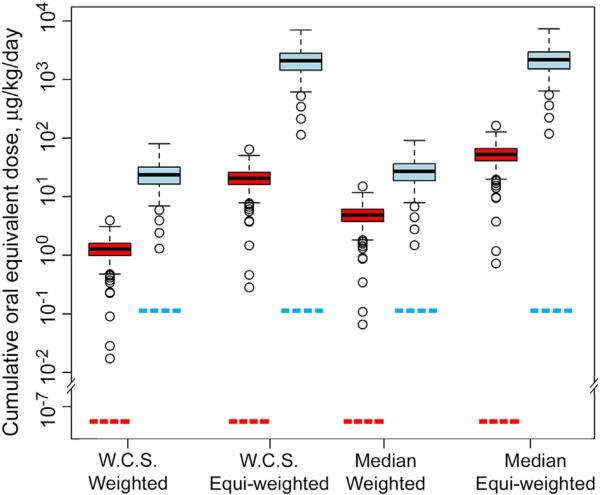
The Css values were derived using the Simcyp software with Monte Carlo simulations to account for the population variability in pharmacokinetics in healthy individuals (Northern European, 20–50 years of age, equally mixed sex). To be reasonably conservative, the upper 95th percentile values from a series 10 simulation (1000 individuals each for estimating pharmacokinetics variability) per trial were used to determine the oral equivalents. This analysis showed that highly significant differences (p < 0.01 or greater) arise when oral equivalents are computed from in vitro EC10 values (Fig. 4). The chlorinated pesticide mixture was predicted as about an order of magnitude more toxic than the current use pesticide mixture, regardless of the methodology that was utilized to account for missing values in calculating Css (see Supplemental Fig. 2).
Fig. 4.
In vitro-to-in vivo extrapolation of cytotoxicity EC10 values. Box plots (box represents first and third quartiles; horizontal line inside the box is the median; whiskers are the 1.5 inter-quantile range; circles are outliers with >1.5 IQR above minimum or maximum) of the cumulative oral doses for each pesticide mixture (red: chlorinated pesticide mixture, blue: current use pesticide mixture) across 146 cell lines in four different scenarios for handling missing data, weighted by chemical percentage in the mixture or not (“equi-weighted”), and assuming the “worst case scenario” (WCS) vs median for missing values. Red and blue dotted horizontal lines indicate the estimated cumulative human oral exposure levels to each pesticide mixture. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To further interpret the outcome of these experiments in the context of human health risk, we examined the relationship of the calculated oral dose equivalent with estimated human exposures to these mixtures. First, we computed a cumulative exposure value for each mixture based on the exposure estimates for each individual chemical obtained from ExpoCast (Wambaugh et al., 2013), a framework that estimated human exposure potential for 1936 chemicals. Predicted estimates of exposure were available for 35 of the 36 chemicals present in the current use pesticides mixture, and 6 of the 10 chemicals in the chlorinated pesticide mixture. To remain conservative, missing values were substituted with the highest predicted exposure from ExpoCast data for a chemical in the respective mixture (see Supplemental Fig. 3). Next, cumulative exposure for each mixture was computed as the upper 95th percentile and compared to oral equivalent doses for in vitro cytotoxicity (Fig. 4). While human exposure estimates were lower than oral dose equivalent in vitro cytotoxic doses for both mixtures, a much greater margin of safety is evident for the chlorinated pesticide mixture than for the current use pesticide (5-fold or greater vs less than 2-fold, respectively). This indicates a wider margin of safety for the chlorinated pesticide mixture than the current-use pesticide mixture.
3.3. Relationships between cytotoxicity of pesticide mixtures and genotype
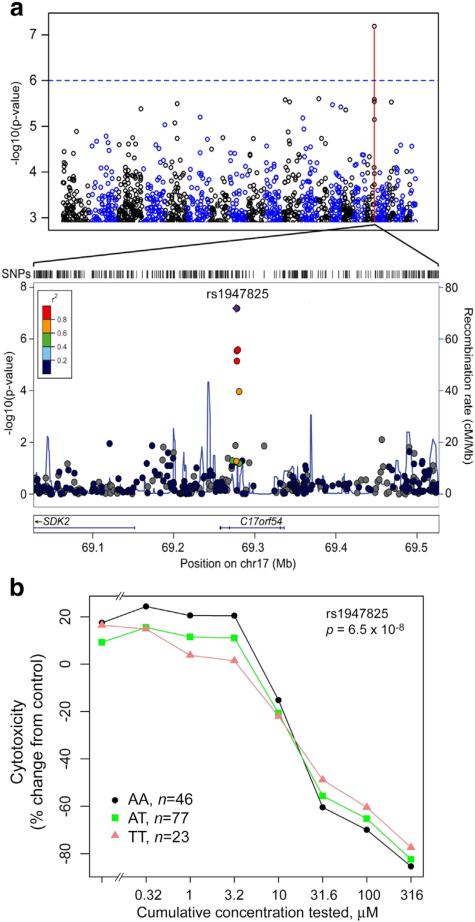
Because the cell lines used in this study are densely genotyped (1000 Genomes Project Consortium et al., 2012), association analysis was performed between the quantitative estimates of cytotoxicity and genetic variability among 146 individuals included in screening. Genotyping data was processed as detailed in Methods. Sex, experimental batch and date, population, and the first ten genotype principal components were included as covariates in multivariate ANCOVA genome-wide association analysis (Brown et al., 2012), a sensitive method designed for evaluating a pattern of variation of cytotoxicity measurements due to genotype. Despite a relatively small population of 146 cell lines, a highly suggestive (p < 6.5 × 10−08) association was observed between cytotoxicity of the current use pesticide mixture and a locus on Chr17 (Fig. 5a) [near the Bonferroni threshold, and genome-wide significant by the criterion of (Dudbridge and Gusnanto, 2008)]. The most highly associated SNP (rs1947825) is located in an open reading frame C17orf54 (Fig. 5b). When the cytotoxicity concentration–response patterns for cells with each of three genotypes for rs1947825 were examined (Fig. 5c), we found that the major allele (AA) confers greater sensitivity, with the heterozygous genotype (AT) falling consistently in the middle across all concentrations.
Fig. 5.
Genome-wide association analysis of population variability in cytotoxicity of the current use pesticide mixture. (a) Manhattan plot of MAGWAS −log10(p) vs. genomic position for association of genotype and cytotoxicity to current use pesticide mixture. The dashed blue line indicates suggestive association (expected once per genome scan). A LocusZoom plot of the most significant (p = 6.5 × 10−8) region at SNP rs1947825. (b) Average concentration–response profiles of cytotoxicity of current use pesticide mixture plotted separately for each genotype at rs1947825. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Following the advice from the U.S. National Academies on developing a long-range strategic plan to update and advance the way environmental agents are tested for toxicity (Krewski et al., 2011), substantial advancements in high-throughput approaches to characterize the biological activity that may be indicative of potential human health hazard of environmental chemicals in vitro have been implemented (Collins et al., 2008). Nonetheless, difficulties are many in conducting human health risk assessments from in vitro endpoints (Crump et al., 2010; Judson et al., 2011). A major challenge in human health assessments is developing a comprehensive understanding of population variability in susceptibility to chemical toxicity (Zeise et al., 2013). Regulatory risk assessment incorporates multiple uncertainty factors that are based on default assumptions and only recently experimental approaches have become available to provide scientific data to replace defaults in inter-individual variability in toxicokinetics (Wetmore et al., 2014) and toxicodynamics (Abdo et al., 2015).
Furthermore, no clear framework has been set to evaluate potential toxicity of chemical mixtures in non-animal alternative models. Few environmental chemical mixtures have been evaluated, especially at environmentally relevant concentrations (Carvalho et al., 2014), with regulatory decisions primarily based on a single compound evaluation. However, potentiation and synergistic interactions of chemicals in mixtures is of great concern (Cedergreen, 2014). It has been shown that exposure to chemical mixtures, including pesticides, often occurs with each chemical in the mixture present at respective safety limit concentrations (Carvalho et al., 2014). Moreover, evaluation of chemical mixtures with similar modes of action, without consideration of realistic exposure in the environment, might underestimate the toxicological risk associated with their exposure (Hadrup, 2014).
To address the challenges of assessment of potential hazard of complex mixtures while accounting for potential inter-individual variability, we aimed to provide quantitative measures for population-based in vitro toxicity of pesticide mixtures. We also used a reverse dosimetry approach to translate in vitro cytotoxicity estimates to oral equivalent doses and compared those to estimates of human exposure (Wetmore et al., 2012). Although investigation of population variability in toxicity of hundreds of individual chemicals is ongoing (Abdo et al., 2015; Lock et al., 2012), to our knowledge this study is first to examine inter-individual variability in response to mixtures.
This screening approach showed that both pesticide mixtures that were tested exhibited appreciable inter-individual variation in cytotoxicity. Interestingly, the toxicodynamic uncertainty factor for both pesticide mixtures (3.0 and 3.05) derived from the population variability in our present study was similar to the median inter-individual variability for the 179 individual chemicals previously tested. This finding is consistent with the default uncertainty factor for toxicodynamic difference among humans (100.5) that is used in risk assessments when no chemical-specific data are available (World Health Organization, 2005). On average, there was no significant difference between the in vitro cytotoxicity concentrations (i.e., EC10) of the current use pesticide mixture and the chlorinated pesticide mixture. However, incorporation of dosimetry with the in vitro data and conversion to an oral equivalent dose for each mixture revealed that a significantly lower dose of a chlorinated pesticide mixture would lead to an internal concentration equal to the cytotoxicity-eliciting EC10. Conversion of the in vitro data in this manner allows a risk-relevant ranking of the mixtures that considers chemical pharmacokinetic behavior along with additional exposure data to adjust the potencies. Incorporation of human dosimetry and predicted human exposure is necessary for greater confidence in the “presumed hazard” from in vitro high throughput screening alone (Gangwal et al., 2012).
It is not surprising that the cumulative human predicted exposure limit is much higher for the current-use pesticide mixture compared to the chlorinated pesticide mixture, which mostly consisted of pesticides withdrawn from the market. The current-use pesticide mixture included 36 currently used pesticides and mimicked real exposure levels in Eastern North Carolina, with atrazine pesticides being the most abundant. Atrazine is among the highest used (64–80 million pounds annually in the United States) agricultural pesticides (Barr et al., 2007). Therefore, the predicted exposure limit for the current-use pesticide mixture was expected to be high, and in our case it was very close to the calculated cytotoxic oral equivalent dose.
In addition to demonstrating how an in vitro human population-based model system may be used to evaluate potential hazard of complex mixtures, we also took advantage of the availability of genetic information on the cells to evaluate genotype-phenotype associations. Recognizing the genetic underpinning of cytotoxicity may offer valuable insights into the underlying casual physiological variation and biologically-associated pathways. The significant locus (C17orf54) identified in this study is in a presumably non-coding genomic region, consistent with 90% of the significant findings from human GWAS studies to date (Fraser, 2013). The long intergenic non-protein coding RNA 469 resides in the region. A critical role of non-coding RNAs in response to carcinogen and toxicant exposure is an emerging area of investigation in toxicology (Marrone et al., 2014), and a potential relationship between cytotoxicity of the current use pesticide mixture and the long non-coding RNA remains to be explored.
In vitro cytotoxicity assays, while nonspecific measures of adverse effect of test chemicals, are widely considered to be informative of the potential in vivo health hazards (National Research Council, 2015). Since it was suggested that evaluation of the cytotoxic potential of chemicals with cell-based assays could provide an indication of their potential in vivo toxicity, several studies demonstrated correlation between in vivo acute toxicity and in vitro cytotoxicity (Ekwall et al., 1998; Kinsner-Ovaskainen et al., 2013). Thus, cytotoxicity assay-derived data have demonstrated success in predictive toxicology, both alone and in combination with the information on chemical structure (Lessigiarska et al., 2006; Zhu et al., 2008; Sedykh et al., 2011). In vitro cytotoxicity tests are also amenable to high-throughput screening (Xia et al., 2008) and have been recommended as an adjunct to animal tests to improve initial dose selection and reduce the number of animals used.
A key limitation of cytotoxicity assays is that they do not provide data on some of the most important toxic mechanisms, specifically ones that involve organ-specific or cell type-specific physiology. However, the choice of the cell type is largely irrelevant as cell lines derived from various tissues have relatively equal value in assessing general cytotoxicity and specific organ toxicity cannot be accurately predicted using in vitro cytotoxicity assays (Lin & Will, 2012). There are a number of additional limitations to extrapolating from in vitro cytotoxicity pro-filing using lymphoblasts to humans, including severe limitations in metabolic capacity of these cells, acute nature of exposure, and no opportunity to consider other important variables such as age, lifestyle factors and diet. It is also yet to be established how chemicals may interact with one another in mixtures, both in terms of pharmacokinetics and in terms of toxicity; the assumptions made in our work with regards to reverse dosimetry and treatment of missing values may constrain the interpretation of the data presented in this work. There remains a pressing need to screen individual pesticides, in addition to their mixtures, in order to test these assumptions. These limitations notwithstanding, our work highlights the value of a population-based in vitro cytotoxicity survey combined with assessment of oral equivalents and human exposures for pesticides and other chemicals. These experiments also advance our understanding of the genetic underpinnings of susceptibility-related regulatory networks in response to toxicants.
Supplementary Material
Acknowledgments
The authors wish to thank Philippe Marlot and Munir Pirmohamed (Institute for Translational Medicine, University of Liverpool, UK) for technical assistance and useful discussions on study design. This work was supported, in part, by grants from U.S. EPA (STAR RD83516601) and NIH (P42 ES005948). The views expressed are those of the authors and do not necessarily reflect the statements, opinions, views, conclusions, or policies of the U.S. EPA, NIH, or the United States government.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx. doi.org/10.1016/j.envint.2015.09.012.
The authors declare they have no competing financial interests.
References
- 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdo N, Xia M, Brown CC, Kosyk O, Huang R, Sakamuru S, Zhou YH, Jack JR, Gallins P, Xia K, Li Y, Chiu WA, Motsinger-Reif AA, Austin CP, Tice RR, Rusyn I, Wright FA. Population-based in vitro hazard and concentration–response assessment of chemicals: the 1000 genomes high-throughput screening study. Environ. Health Perspect. 2015;123(5):458–466. doi: 10.1289/ehp.1408775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen ME, Krewski D. Toxicity testing in the 21st century: bringing the vision to life. Toxicol. Sci. 2009;107(2):324–330. doi: 10.1093/toxsci/kfn255. [DOI] [PubMed] [Google Scholar]
- Babut M, Arts GH, Barra Caracciolo A, Carluer N, Domange N, Friberg N, Gouy V, Grung M, Lagadic L, Martin-Laurent F, Mazzella N, Pesce S, Real B, Reichenberger S, Roex EW, Romijn K, Rottele M, Stenrod M, Tournebize J, Vernier F, Vindimian E. Pesticide risk assessment and management in a globally changing world—report from a European interdisciplinary workshop. Environ. Sci. Pollut. Res. Int. 2013;20(11):8298–8312. doi: 10.1007/s11356-013-2004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr DB, Panuwet P, Nguyen JV, Udunka S, Needham LL. Assessing exposure to atrazine and its metabolites using biomonitoring. Environ. Health Perspect. 2007;115(10):1474–1478. doi: 10.1289/ehp.10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil KL, Vakil C, Sanborn M, Cole DC, Kaur JS, Kerr KJ. Cancer health effects of pesticides: systematic review. Can Fam Physician. 2007;53(10):1704–1711. [PMC free article] [PubMed] [Google Scholar]
- Brown CC, Havener TM, Medina MW, Krauss RM, McLeod HL, Motsinger-Reif AA. Multivariate methods and software for association mapping in dose–response genome-wide association studies. BioData Min. 2012;5(1):21. doi: 10.1186/1756-0381-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho RN, Arukwe A, Ait-Aissa S, Bado-Nilles A, Balzamo S, Baun A, Belkin S, Blaha L, Brion F, Conti D, Creusot N, Essig Y, Ferrero VEV, Flander-Putrle V, Furhacker M, Grillari-Voglauer R, Hogstrand C, Jona A, Kharlyngdoh JB, Loos R, Lundebye AK, Modig C, Olsson PE, Pillai S, Polak N, Potalivo M, Sanchez W, Schifferli A, Schirmer K, Sforzini S, Sturzenbaum SR, Softeland L, Turk V, Viarengo A, Werner I, Yagur-Kroll S, Zounkova R, Lettieri T. Mixtures of chemical pollutants at european legislation safety concentrations: how safe are they? Toxicol. Sci. 2014;141(1):218–233. doi: 10.1093/toxsci/kfu118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergreen N. Quantifying synergy: a systematic review of mixture toxicity studies within environmental toxicology. PLoS One. 2014;9(5):e96580. doi: 10.1371/journal.pone.0096580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology. Transforming environmental health protection. Science. 2008;319(5865):906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump KS, Chen C, Louis TA. The future use of in vitro data in risk assessment to set human exposure standards: challenging problems and familiar solutions. Environ. Health Perspect. 2010;118(10):1350–1354. doi: 10.1289/ehp.1001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 2008;32(3):227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feron VJ, Cassee FR, Groten JP. Toxicology of chemical mixtures: international perspective. Environ. Health Perspect. 1998;106(Suppl 6):1281–1289. doi: 10.1289/ehp.98106s61281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB. Gene expression drives local adaptation in humans. Genome Res. 2013;23(7):1089–1096. doi: 10.1101/gr.152710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwal S, Reif DM, Mosher S, Egeghy PP, Wambaugh JF, Judson RS, Hubal EA. Incorporating exposure information into the toxicological prioritization index decision support framework. Sci. Total Environ. 2012;435–436:316–325. doi: 10.1016/j.scitotenv.2012.06.086. [DOI] [PubMed] [Google Scholar]
- Hadrup N. Evidence from pharmacology and pathophysiology suggests that chemicals with dissimilar mechanisms of action could be of bigger concern in the toxicological risk assessment of chemical mixtures than chemicals with a similar mechanism of action. Regul. Toxicol. Pharmacol. 2014;69(3):281–283. doi: 10.1016/j.yrtph.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Jamei M, Marciniak S, Feng K, Barnett A, Tucker G, Rostami-Hodjegan A. The Simcyp population-based ADME simulator. Expert Opin. Drug Metab. Toxicol. 2009;5(2):211–223. doi: 10.1517/17425250802691074. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Judson RS, Kavlock RJ, Setzer RW, Cohen Hubal EA, Martin MT, Knudsen TB, Houck KA, Thomas RS, Wetmore BA, Dix DJ. Estimating Toxicity-related biological pathway altering doses for high-throughput chemical risk assessment. Chem. Res. Toxicol. 2011;24(4):451–462. doi: 10.1021/tx100428e. [DOI] [PubMed] [Google Scholar]
- Jurewicz J, Hanke W. Prenatal and childhood exposure to pesticides and neurobehavioral development: review of epidemiological studies. Int. J. Occup. Med. Environ. Health. 2008;21(2):121–132. doi: 10.2478/v10001-008-0014-z. [DOI] [PubMed] [Google Scholar]
- Krewski D, Westphal M, Al Zoughool M, Croteau MC, Andersen ME. New directions in toxicity testing. Annu RevPublic Health. 2011;32:161–178. doi: 10.1146/annurev-publhealth-031210-101153. [DOI] [PubMed] [Google Scholar]
- Krieger R. Haye's Handbook of Pesticide Toxicology. 3rd ed. Academic Press; Waltham, MA.: 2010. [Google Scholar]
- Lin Z, Will Y. Evaluation of drugs with specific organ toxicities in organ-specific cell lines. Toxicological sciences : an official journal of the Society of Toxicology. 2012;126:114–127. doi: 10.1093/toxsci/kfr339. [DOI] [PubMed] [Google Scholar]
- Lock EF, Abdo N, Huang R, Xia M, Kosyk O, O'Shea SH, Zhou YH, Sedykh A, Tropsha A, Austin CP, Tice RR, Wright FA, Rusyn I. Quantitative high-throughput screening for chemical toxicity in a population-based in vitro model. Toxicol. Sci. 2012;126(2):578–588. doi: 10.1093/toxsci/kfs023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. Pesticide and insect repellent mixture (permethrin and DEET) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod. Toxicol. 2012;34(4):708–719. doi: 10.1016/j.reprotox.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone AK, Beland FA, Pogribny IP. Noncoding RNA response to xeno-biotic exposure: an indicator of toxicity and carcinogenicity. Expert Opin. Drug Metab. Toxicol. 2014;10(10):1409–1422. doi: 10.1517/17425255.2014.954312. [DOI] [PubMed] [Google Scholar]
- Mauri A, Consonni V, Pavan M, Todeschini R. DRAGON Software: an easy approach to molecular descriptor calculations. MATCH Commun Math Comput Chem. 2006;56:237–248. [Google Scholar]
- National Research Council . Science and Decisions: Advancing Risk Assessment. National Academies Press; Washington, DC.: 2009. [PubMed] [Google Scholar]
- National Research Council . Application of modern toxicology approaches for predicting acute toxicity for chemical defense. The National Academies Press; Washington, DC.: 2015. [PubMed] [Google Scholar]
- O'Shea SH, Schwarz J, Kosyk O, Ross PK, Ha MJ, Wright FA, Rusyn I. In vitro screening for population variability in chemical toxicity. Toxicol. Sci. 2011;119(2):398–407. doi: 10.1093/toxsci/kfq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry L, Adams RD, Bennett AR, Lupton DJ, Jackson G, Good AM, Thomas SH, Vale JA, Thompson JP, Bateman DN, Eddleston M. National toxicovigilance for pesticide exposures resulting in health care contact — an example from the UK's National Poisons Information Service. Clin. Toxicol. (Phila.) 2014;52(5):549–555. doi: 10.3109/15563650.2014.908203. [DOI] [PubMed] [Google Scholar]
- Pillai K. Some new test criteria in multivariate analysis. Ann Math Stat. 1955;26:117–121. [Google Scholar]
- Rother HA. Communicating pesticide neurotoxicity research findings and risks to decision-makers and the public. Neurotoxicology. 2014;45:327–337. doi: 10.1016/j.neuro.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Rotroff DM, Wetmore BA, Dix DJ, Ferguson SS, Clewell HJ, Houck KA, LeCluyse EL, Andersen ME, Judson RS, Smith CM, Sochaski MA, Kavlock RJ, Boellmann F, Martin MT, Reif DM, Wambaugh JF, Thomas RS. Incorporating human dosimetry and exposure into high-throughput in vitro toxicity screening. Toxicol. Sci. 2010;117(2):348–358. doi: 10.1093/toxsci/kfq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn M, Kerr KJ, Sanin LH, Cole DC, Bassil KL, Vakil C. Non-cancer health effects of pesticides: systematic review and implications for family doctors. Can Fam Physician. 2007;53(10):1712–1720. [PMC free article] [PubMed] [Google Scholar]
- Sedykh A, Zhu H, Tang H, Zhang L, Richard A, Rusyn I, Tropsha A. Use of in vitro HTS-derived concentration-response data as biological descriptors improves the accuracy of QSAR models of in vivo toxicity. Environ Health Perspect. 2011;119:364–370. doi: 10.1289/ehp.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YM, Liao KH, Clewell HJ., 3rd Reverse dosimetry: interpreting trihalomethanes biomonitoring data using physiologically based pharmacokinetic modeling. J. Expo. Sci. Environ. Epidemiol. 2007;17(7):591–603. doi: 10.1038/sj.jes.7500540. [DOI] [PubMed] [Google Scholar]
- EPA US. Guidance on Cumulative Risk Assessment of Pesticide Chemicals That Have a Common Mechanism of Toxicity. Office of Pesticide Programs; Washington, DC.: 2002. [Google Scholar]
- Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, Joliet O, Frame A, Rabinowitz J, Knudsen TB, Judson RS, Egeghy P, Vallero D, Cohen Hubal EA. High-throughput models for exposure-based chemical prioritization in the ExpoCast project. Environ. Sci. Technol. 2013;47(15):8479–8488. doi: 10.1021/es400482g. [DOI] [PubMed] [Google Scholar]
- Wetmore BA. Quantitative in vitro-to-in vivo extrapolation in a high-throughput environment. Toxicology. 2015;332:94–101. doi: 10.1016/j.tox.2014.05.012. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Allen B, Clewell HJ, 3rd, Parker T, Wambaugh JF, Almond LM, Sochaski MA, Thomas RS. Incorporating population variability and susceptible subpopulations into dosimetry for high-throughput toxicity testing. Toxicol. Sci. 2014;142(1):210–224. doi: 10.1093/toxsci/kfu169. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Ferguson SS, Sochaski MA, Rotroff DM, Freeman K, Clewell HJ, 3rd, Dix DJ, Andersen ME, Houck KA, Allen B, Judson RS, Singh R, Kavlock RJ, Richard AM, Thomas RS. Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment. Toxicol. Sci. 2012;125(1):157–174. doi: 10.1093/toxsci/kfr254. [DOI] [PubMed] [Google Scholar]
- Wetmore BA, Wambaugh JF, Allen B, Ferguson SS, Sochaski MA, Setzer RW, Houck KA, Strope CL, Cantwell K, Judson RS, LeCluyse E, Clewell HJ, 3rd, Thomas RS, Andersen ME. Incorporating High-Throughput Exposure Predictions with Dosimetry-Adjusted In Vitro Bioactivity to Inform Chemical Toxicity Testing. Toxicol Sci. 2015 doi: 10.1093/toxsci/kfv171. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Chemical-specific adjustment factors for interspecies differences in human variability: Guidance document for use of data in dose/concentration–response assessment. 2005 http://www.inchem.org/documents/harmproj/harmproj/harmproj2.pdf.
- Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, Tice RR, Austin CP. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ. Health Perspect. 2008;116(3):284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeise L, Bois FY, Chiu WA, Hattis D, Rusyn I, Guyton KZ. Addressing human variability in next-generation human health risk assessments of environmental chemicals. Environ. Health Perspect. 2013;121(1):23–31. doi: 10.1289/ehp.1205687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.