Abstract
In this chapter the basic premises, the recent findings and the future challenges in the use of amelogenin for enamel tissue engineering are being discoursed on. Results emerging from the experiments performed to assess the fundamental physicochemical mechanisms of the interaction of amelogenin, the main protein of the enamel matrix, and the growing crystals of apatite, are mentioned, alongside a moderately comprehensive literature review of the subject at hand. The clinical importance of understanding this protein/mineral interaction at the nanoscale are highlighted as well as the potential for tooth enamel to act as an excellent model system for studying some of the essential aspects of biomineralization processes in general. The dominant paradigm stating that amelogenin directs the uniaxial growth of apatite crystals in enamel by slowing down the growth of (hk0) faces on which it adheres is being questioned based on the results demonstrating the ability of amelogenin to promote the nucleation and crystal growth of apatite under constant titration conditions designed to mimic those present in the developing enamel matrix. The role of numerous minor components of the enamel matrix is being highlighted as essential and impossible to compensate for by utilizing its more abundant ingredients only. It is concluded that the three major aspects of amelogenesis outlined hereby – (1) the assembly of amelogenin and other enamel matrix proteins, (2) the proteolytic activity, and (3) crystallization – need to be in precise synergy with each other in order for the grounds for the proper imitation of amelogenesis in the lab to be created.
Keywords: Apatite, Bone, Calcium phosphate, Enamel, Tooth, Nanoparticle, Hard tissue engineering
13.1 Introduction
Progress in the war against disease results from discoveries in remote and unexpected fields of medicine and the underlying sciences. (Bush 1945) A Report to the U.S. President by Vannevar Bush, Director of the Office of Scientific Research and Development, July 1945.
Understanding the interaction of organic and inorganic phases during biomineralization events at the atomic scale would present a milestone with repercussions for an array of biomedical fields revolving around teeth, bone and other organs composed of mineralized tissues. Thanks to its relative structural simplicity in the realm of mammalian hard tissues, tooth enamel presents an excellent model system for studying this interaction, even though reasonable concerns exist that the genesis of it may be governed by fundamentally different mechanisms compared to those present in bone. Namely, while nanoparticulate mineral particles comprising bone form by nucleation on the active surface of highly phosphorylated bone-specific proteins, exceptionally long crystals of the mineral phase form in enamel by the action of proteins with significantly lesser nucleation and growth potential.
As it is usually the case, fundamental insights of any nature create either an immediate or delayed effect on the way certain issues are practically solved and hopes exist that understanding the interaction between amelogenin, the main protein of the enamel matrix, and the crystals forming with a specific and highly defined structure from within its gelatinous volume, would revolutionize the clinical approach to dental restoration and, possibly, change the mainstream approach to orthopedic therapies too.
What follows is a short discourse on the current state of our knowledge on the interaction between these two species, amelogenin and apatite, in the course of which the strongest and, as it usually is, the most brittle of all mammalian tissues forms: dental enamel (Uskoković et al. 2010). Results of the recent studies set up to simultaneously yield an insight into the fundamental nature of this interaction and utilize it for the purpose of growing enamel in vitro will be mentioned too.
13.2 The Structure and Composition of Mature Enamel
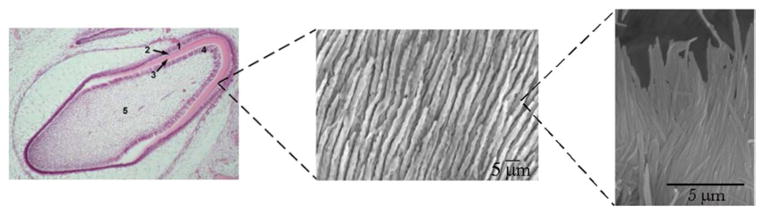
Enamel is composed of 4–8 3μ wide rod-shaped bundles of apatite fibers whose diameter is in the range of 40–60 nm and whose aspect ratio reaches up to 3 · 104 (Fig. 13.1). Apatite is the least soluble phase of calcium phosphates, with the crystal structure adopting pseudo-hexagonal P63/m space group. Owing to its structural flexibility, it allows for a moderate amount of substitution of its dominant, Ca2+ and PO43− ions with a variety of biological microelements, so that its composition is most accurately given as (Ca,Z)10 (PO4,Y)6(OH,X)2, where Z = Na+, Mg2+, K+, Sr2+, etc., Y = CO32−, HPO42−, and X = Cl−, F−. Some of these ions, such as Na+ or Mg2+, increase the solubility of the compound, while others, such as F−, decrease it.
Fig. 13.1.
Histological section of the developing human tooth in the maturation stage (left) and micrographs showing the parallel arrangement of enamel rods (middle) and the parallel arrangement of apatite nanofibers within each enamel rod (right). 1 ameloblasts, 2 enamel, 3 dentin, 4 odontoblasts, 5 pulp
The great majority of enamel, 96–98 wt.%, is of mineral composition, which is more than in any other mammalian hard tissue. Water, fatty acids and various peptides account for the rest 2–4 wt.%. Discussions have been sparked recently about the nature of this miniscule amount of impurities. Namely, after it was found out that only 0.02 wt.% of glycoprotein in the spine of sea urchin (i.e., ~10 proteins per 106 unit cells) is enough to efficiently absorb the energy from propagating cracks and markedly increase the strength of the material (Stupp and Braun 1997), the long-lasting paradigm stating that these impurities present accidental remnants of incomplete proteolytic digestion of the enamel matrix has been questioned and challenged with a hypothesis that these peptides are purposefully left in the tissue so as to provide it with greater resistance to fracture under compression or shear.
Approximately one thousand apatite fibers are assembled in bundles within each enamel rod, 5–12 million of which are found lined up in rows per single tooth crown. The size and the packing density of the crystals of apatite comprising enamel are highly different from those comprising bone. Whereas bone consists of plate-shaped nanoscopic crystals with 20°×°10°×°2 nm in size on average (Eppell et al. 2001), the crystals of enamel, albeit of the same composition, are approximately 1,000 times longer along their [001], c-axis. In part, this has been made possible by the fact that enamel is a tissue that does not depend on intrinsic cellular proliferation in the course of its lifetime, the reason for which bone regeneration materials are nowadays designed to be porous so as to allow for the proliferation of bone cells across its volume (Cai et al. 2007). These structural dissimilarities between enamel and bone suggest that the mechanisms of their respective formation may be vastly different.
13.3 The Basic Model of Amelogenesis and a Question Mark Over It
The process of enamel growth, a.k.a. amelogenesis, is one of the slowest morphogenetic processes, taking more time to complete than it is needed for the embryo to form in utero, which speaks well in favor of its extraordinary complexity. Growing at the appositional rate of ~2–4 μm per day, enamel forms over a period of approximately 4 years in a process that involves a controlled crystal growth through gelatinous enamel matrix composed of a number of proteins at the overall concentration of 200–300mg/ml, 90% of which has been identified as a single protein: amelogenin. The remaining 10% is comprised of other proteins: ameloblastin, enamelin, serum albumin, amelotin, and proteolytic enzymes. Together, they assemble into a scaffold that serves as a template for the uniaxial growth of apatite crystals.
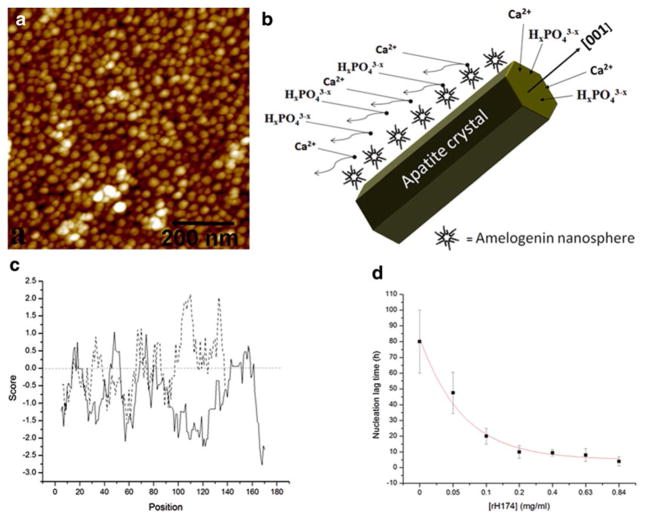
The reigning model of enamel growth is built on the assumption that amelogenin self-assembles into narrowly disperse nanospheres with ~20 nm in diameter (Fig. 13.2a), which then align onto (hk0) faces of apatite crystals, blocking the adherence of the ionic growth units, Ca2+, HxPO4x−3 and OH−, onto those faces and allowing for the crystal growth to occur only in the direction of [001] axis (Fig. 13.2b).
Fig. 13.2.
(a) Monodisperse recombinant full-length human amelogenin nanospheres forming in water. Forty to sixty amelogenin molecules form a single nanospherical aggregate with 20–40 nm in size. (b) Schematic depiction of the crystal growth during amelogenesis according to the nowadays questionable dominant paradigm. (c) Hydrophobicity plots obtained using ExPASy ProtScale Kyte & Doolittle model (window size = 9; linear weight variation model) for human amelogenin (straight line) and human hemoglobin alpha chain (dashed line). The positive score on the diagram denotes hydrophobic sequences. (d) Nucleation lag time for the precipitation of apatite from aqueous suspensions of human recombinant full-length amelogenin in the concentration range of 0 840 μg/ml ([KH2PO4] = 1.0 M; [CaCl2] = 1.67 M; pH = 7.4, T = 37 °C)
There are multiple grounds on which this paradigmatic explanation can be questioned. Firstly, recombinant amelogenin forms such nanospherical entities when suspended in water, but their existence in vivo has not been accurately pinpointed to this date. DNA molecules assemble into a variety of morphologies, from cubes to triangles to pentagons to hexagons to octahedrons (Aldaye et al. 2008) and could be used for the assembly of nanoparticles into superlattices (Young et al. 2014) and other sophisticated geometries that are otherwise difficult to obtain (Liu et al. 2013) wherefrom their use in organic electronics has begun to be intensely researched as well (Hamedi et al. 2012). None of these potentially practical potentials of DNA need be necessarily tied to its biological function as a storage place for the genetic content of the cell. Similarly, detection of morphologies adopted by amelogenin assemblies in vitro, be they nanospheres, nanobeads or nanofilaments (Martinez-Avila et al. 2012), may be irrelevant for explaining the biologically relevant forms and functions thereof.
Secondly, the abovementioned model of amelogenesis at the level of organic/inorganic interface assumes hydrophobicity of amelogenin, as the direct result of which it is supposed to act as an inhibitor rather than a promoter of crystallization of apatite. This common assumption is, however, incorrect, as amelogenin, like every other protein, contains alternately changing hydrophilic and hydrophobic sequences along its primary structure (Uskoković et al. 2011a). As shown in Fig. 13.2c, although amelogenin as a whole is still more hydrophobic than most proteins, it is, for example, more hydrophilic than human hemoglobin alpha chain. Moreover, the intensely hydrophilic 13-amino-acid-long segment towards the C-terminus of the protein suggests its amphiphilicity, which may be crucial in endowing it with the ability to form nano-spherical assemblies in water, similar in form to reverse micelles (Uskoković et al. 2005), with the hydrophilic ends exposed to the polar environment and the rest of the protein folded internally. In view of this, it is logical to expect that amelogenin is capable of promoting the nucleation of apatite in vitro. As shown in Fig. 13.2d, the nucleation lag time for precipitation of apatite from metastable solutions of KH2PO4 and CaCl2 at the physiological pH decreases in direct proportion with the concentration of human recombinant full-length amelogenin (Uskoković et al. 2011b). Compared to recombinant human amelogenin used in these studies (rH174) (Uskoković et al. 2008), which lacks phosphorylation on 16Ser residue, the biological variant of it is expected to have an even more pronounced propensity to stimulate the nucleation of apatite, considering the apatite-nucleation potential of abundantly phosphorylated extracellular bone and dentin matrix proteins. Other studies, having elucidated the conditions under which amelogenin can promote the nucleation of apatite (Wang et al. 2008; Tarasevich et al. 2007), came to a similar conclusion, thus implicitly questioning the correctness of the dominant paradigm in this field.
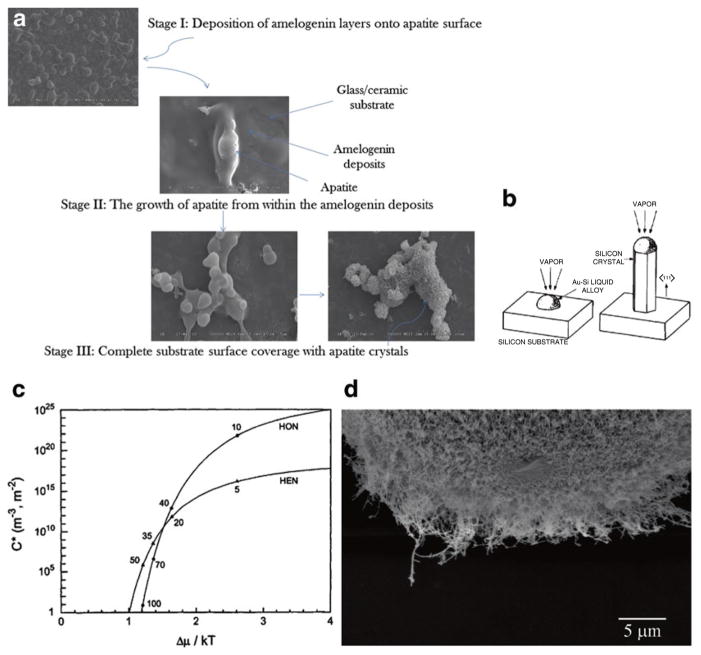
Another cue in terms of inverting the paradigm comes from the fact that adsorption of amelogenin onto a growing crystal surface appears to be the first step prior to the induction of surface- specific, controlled crystal growth (Uskoković et al. 2011c; Habelitz et al. 2004, 2005a). Figure 13.3a demonstrates a typical surface growth of apatite crystals immersed in amelogenin sols under low supersaturation ratios and a constant titration regimen. The overall process could be divided to three stages. In the first stage, amelogenin nanospheres from the solution adsorb onto the crystal growth substrates, forming miniature islands on them. In the second stage, the growth is observed to occur exclusively from inside of the amelogenin deposits, an effect that would be virtually impossible had it not been for the ability of amelogenin to promote apatite nucleation and growth. Finally, in the third stage, amelogenin deposits are fully replaced by the elongated crystals of apatite (Fig. 13.3a). The idea that adsorption of the protein implies the hindrance of the crystal growth on the binding sites is thus directly refuted. Osteocalcin, a protein involved in mineralization of bone, for example, does not constrain the growth of crystal planes, even though it binds to them (Robinson 2006).
Fig. 13.3.
(a) Evolution of the surface layers on crystal growth substrates in the course of a typical, 7-day long continuous titration experiment, during which amelogenin sols at the concentration of 400 μg/ml and a relatively high initial concentration of KH2PO4 are titrated with CaCl2 and KH2PO4. (b) This process bears resemblance to the mechanism for the formation of silicon nanowires via the action of gold nanodroplets sputtered over the substrate surface and used as a means for ensuring sufficiently slow increase in the supersaturation ratio of silicon atoms introduced to them through vapor (Reproduced with permission from Wagner and Ellis (1964)). (c) As the supersaturation ratio for active species in the system is gradually increased and exceeds 1, the conditions for heterogeneous nucleation (HEN) are hit before those for homogeneous one (HON) (Reprinted with permission from Kashchiev (2000)). (d) Silicon nanowires outgrown from silica beads using the gold nanodroplet-assisted chemical vapor deposition process whose mechanism is similar to the growth of apatite from the surface of amelogenin-covered apatite
Amelogenin may be thus said to act not as an inhibitor of crystal growth, but as a bridge between the ionic solutes or semisolid complexes and the crystalline surface that they are anchored to. One model based on hypothesized β-spirals formed by a series of β-turns in the secondary structure of folded amelogenin and their channeling of Ca2+ ions to the mineralization front was previously proposed (Renugopalakrishnan et al. 1989; Zheng et al. 1987). This view of amelogenin as an ion-channeling molecular entity also bears a resemblance to the model describing the formation of silicon nanowires in the so-called vapor-liquid-solid process, during which nano-droplets of gold deposited on top of silicon wafers attract silicon atoms from the vapor. Under sufficiently slowly increased supersaturation (Fig. 13.3c), the conditions for heterogeneous nucleation are approached without crossing the boundary for homogeneous nucleation too, leading to a highly specific growth from the underlying surface and, in this case, resulting in well-aligned nanowires oriented perpendicular to the substrate and perfectly parallel to each other. Nanowires obtained in one such process are shown in Fig. 13.3d.
Moreover, a model based on an analogy between (a) the role of amelogenin assemblies in channeling the controlled transfer of ions from the solution onto the growing faces, and (b) the ion-tunneling effect through the hydrophobic center of ion channel proteins located at the cell membrane (Murakami 1995), could thus be proposed. Namely, ionophores need to be hydrophobic in order to be soluble in the lipid membrane layers, whereas this internal hydrophobicity is also crucial in terms of enabling the ion- channeling effect through their core (Nelson and Cox 2004). The presence of hydrophobic domains within amelogenin structure may be similarly important in ensuring the proper “gating” of the units of growth, as it occurs in ion channels on cell membranes (Zhaohua et al. 2008). In that sense, the 100–150 kDa lipoprotein ATPase complex that simultaneously releases the bound Ca2+ ions on one side and protons on the other may serve as a model for the possible role that amelogenin may play in the transport of ions onto the growing apatite faces that it is physisorbed to. This effect is particularly relevant since, as it could be seen from Eq. (13.1), an increase in the acidity of the medium is entailed by the formation of apatite. This implies that the conditions for simultaneous controlled delivery of ions to the mineralization front and dissipation of the released H+/H3O+ ions to the surrounding amelogenin gel need to be ensured for the conditions for the interaction between amelogenin and apatite to be set properly.
| (13.1) |
The idea that amelogenin hinders the crystal growth has found its support in the observations of disorganized apatite fibers in the amelogenin knockout mouse (Gibson et al. 2001). The fact that enamel formed in the absence of amelogenin is pathologically thin, however, could counteract this idea by indicating that amelogenin might be involved in the process of extension of the primary crystals by means of its ability to promote uniaxial crystal growth. Finally, the ability to hinder or foster crystallization oftentimes depends on the protein concentration (Gower 2008) and other structural modifications it may undergo, so that around a single protein wrapped around a single crystal could be expected to play the role of inhibitor of the growth of one and of promoter of the growth of other faces. This brings us over to the proteolytic aspect of amelogenesis, without the mention of which no truly consistent model thereof could be proposed.
13.4 The Role of Proteases or How Amelogenin Needs to Disappear in Order for Apatite to Appear
Those with a developed sense for aesthetics may agree that the interaction between amelogenin and apatite has a poetic beauty intrinsic to it. Namely, enamel is the only tissue in the human body whose formation is conditioned by the gradual disappearance of the agents that direct this process. One of the most intriguing features of amelogenesis comes from the fact that not only does its final product, the tooth enamel, present the hardest tissue in the vertebrate body, but its high mineral content coupled with an ultrafine architecture implies that in this process the extracellular matrix directs not only the crystal growth, but its own constructive degradation too. In that sense, the enamel protein matrix is unique in the realm of biomineralization as it fulfills the old truism of biology: “Intercellular matrix exists to be destroyed”. Its role could also be described by the ancient Biblical verses: “Verily, verily, I say unto you, except a corn of wheat fall into the ground and die, it abideth alone: but if it die, it bringeth forth much fruit” (The Holy Bible 1609). One could even argue that this also makes amelogenesis somewhat a more intricate mineralization process compared to dentinogenesis during which the collagenous protein matrix essentially remains intact and kept in the same place. In view of this, understanding amelogenesis becomes directly conditioned by understanding the effects of the enzymatic hydrolysis of amelogenin on the crystal formation.
The major proteases of the enamel matrix include matrix metalloproteinase-20 (MMP-20, a.k.a. enamelysin), enamel matrix serine protease 1 (EMSP1, a.k.a. kallikrein-4), and cathepsin B. They are secreted into the extracellular space by ameloblasts with the role of catalyzing hydrolysis of specific peptide bonds in amelogenin molecules. An increasing amount of evidence suggests that the initial cleavage products carry out an array of assembly-related functions in the developing enamel matrix (Bartlett and Simmer 1999). The main support for this idea comes from the fact that enamel matrix proteases are expressed early during development. In fact, the initially secreted nascent proteins are present in the enamel matrix in a transient form and are relatively quickly processed to generate a wide spectrum of smaller peptides. The nascent amelogenin is thus broken down to several fragments that serve specific roles in the protein assembly of protein and the mineral growth.
For example, the expression of MMP-20, the enamel matrix protease hydrolyzing amelogenin in a highly controlled manner, peaks during the secretory stage and then gradually drops during the maturation, just as it is the case with amelogenin (Bartlett et al. 1998). The constancy of the ratio between enamel matrix components throughout relatively long periods of time (Simmer and Hu 2002) implies that the rate of generation and secretion of amelogenin corresponds to the rate of its cleavage. In view of this, enamel proteases might carry out not only the function of degrading amelogenin so as to provide free space for the sideway growth of enamel crystals, but also act as essential regulators of the activity of amelogenin and other enamel matrix proteins. The structure of amelogenin may thus be such that it contains several functional domains that become activated for different purposes and at different stages of amelogenesis (Snead 2003).
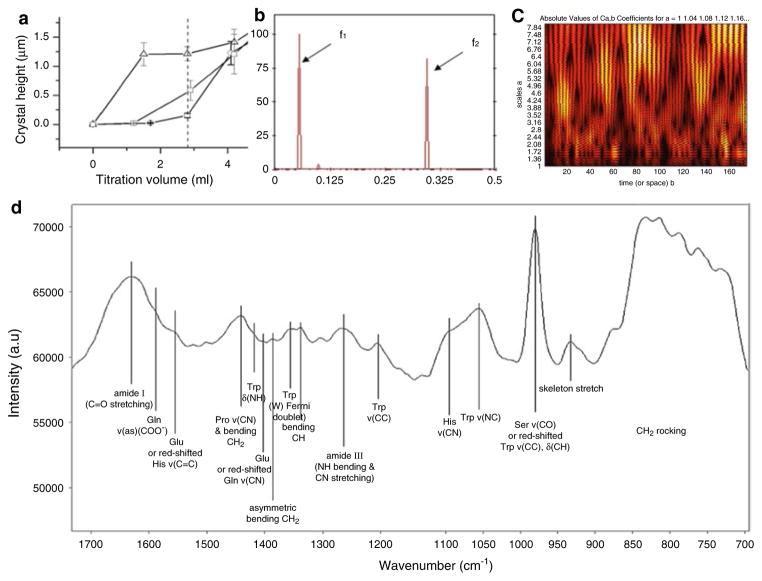
It has been shown that coupling the proteoly-sis using MMP-20 to apatite growth in the presence of amelogenin has the same effect on increasing the rate of crystal formation as quadrupling the concentration of amelogenin (Fig. 13.4a). These and similar findings have spoken in favor of the immense importance of MMP- 20 for the process of amelogenesis (Uskoković et al. 2011d). The essentiality of the role of enamel matrix proteases is supported by studies that have shown that the mutations not only in amelogenin genes, but in those that encode MMP-20 cause amelogenesis imperfecta, i.e., a pathological state typified by abnormal and significantly weakened enamel (Bartlett et al. 2006; Caterina et al. 2002). Inhibition of the activity of MMP-2, MMP-9 and MMP-20 by marimastat similarly led to an impairment of the mineralization of dental tissues in mice (Bourd-Boittin et al. 2005). KLK4 is another major protease in amelogenesis, known for its ability to rather aggressively degrade amelogenin towards the end of the maturation stage, similar to cathepsin B. That its role is equally crucial is known since mutation g.2142G>A on the gene coding for this protease causes an abnormal enzymatic activity, resulting in the enamel crystals of normal length but of insufficient thickness (Hart et al. 2004).
Fig. 13.4.
(a) The average height of apatite crystals grown after different titration volumes compared between 0.4 mg/ml rH174 sample without MMP-20 (-○-), with MMP-20 in 105:1 weight ratio (-□-), and with MMP-20 in 103:1 weight ratio (-△-) with respect to rH174. (b) Raman spectrum of recombinant full-length human ame-logenin dispersed in water at room temperature. (c) Multiple cross-spectral function and (d) wavelet scalo-gram of human amelogenin, showing the two characteristic radio frequencies for the protein to lie at f1 = 0.0547 and f2 =0.3438 and the two “hottest spots" to be 70Val–90 His and the C-terminal sequence from 145Phe to 165Thr, respectively, along with the primary sequence of the protein. For the relationship between the numerical values given here and the electron-ion interaction potential that describes the average energy states of all valence electrons in an amino acid, the variable used as a basis for the given calculations (See Ćosić and Pirogova 2007). The amino acid sequence of human amelogenin (h174) is the following: PLPPHPGHPGYINFSYEVLTPLKWYQ SIRPPYPSYGYEPMGGWLHHQIIPVLSQQHPP THTLQPHHHIPVVPAQQPVIPQQPMMPVPG QHSMTPIQHHQPNLPPPAQQPYQPQPVQPQPHQP MQPQP
Hence, whereas the full-length amelogenin is only present at the surface, in the outer enamel layer, its cleavage products are exclusively found in the deeper, inner enamel layers where they also tend to organize into specific compartments. The C-terminal-containing cleavage products also tend to position at the enamel surface and are hardly found in the deeper layers, suggesting that the full-length molecules might be involved in the crystal growth only in the first stage during which the formation of elongated particles is initiated and is followed by the reorganization of the fibrous crystals into rods through finer peptide- mineral interaction mediated by the C-terminal- lacking peptides, small enough to protrude and line up in the inner enamel regions.
The relatively high content of small peptides resulting from the enzymatic hydrolysis of amelogenin unequivocally suggests their important role in conducting the crystal growth. A detailed analysis of the crystal growth effects of smaller polypeptides as the cleavage products of the full- length amelogenin could correspondingly present the logical next step in the investigation of the mechanism of amelogenesis. One of such molecules is tyrosine-rich amelogenin peptide (TRAP) obtained by cleaving a short sequence of amino acids (44) at the N-terminal of the nascent molecule (Ravindranath et al. 2007). Engineering of de novo peptides with compacted functionalities corresponding to their bigger biological counterparts presents another approach that is yet to be meticulously explored in the context of amelogenesis in particular and biomineralization in general. Phage peptide library screening may be an experimental method of choice to assess this, while, as far as theoretical methods are concerned, the evaluation of protein “hot spots” by means of the Continuous Wavelet Transform Resonant Recognition Model (CWT-RRM) presents one possibility too (De Trad et al. 2000). “Hot spot” sequences are usually found clustered in and around the active site of the folded protein and CWT-RRM analysis of human amelogenin resulted in the detection of two such sequences, one centrally located (70Val–90His) and one in the vicinity of the C-terminal (145Phe–165Thr). The main mammalian lineages, in fact, display highly conserved residues in the hydrophilic C-terminal region, while the central region of amelogenin molecules is more variable (Delgado et al. 2005), suggesting that C-terminal plays a major role in the protein-guided crystal growth. Usually proteins exhibit a single frequency peak during multiple cross-spectral RRM analysis, but in the case of amelogenin, two such peaks were detected. A single biological function of a protein is expected to correspond to a single frequency on this diagram and the doublet in this case suggests two different protein functions. The ambiguous and intrinsically antagonistic role of amelogenin, already hypothesized to be present in its presumed ability to act as a hinderer and a fosterer of crystal nucleation and growth, is thus being reaffirmed by means of one such analysis.
13.5 Attempts to Probe the Higher Orders of the Structure of Amelogenin
What we know today about the structure of amelogenin assemblies is far more versatile than what we know about its molecular structure. Namely, the typically observed morphology of amelogenin aggregates in vitro is the one of nanospheres with the size at the order of tens of nanometers. Combined small-angle X-ray scattering (SAXS) and dynamic light scattering (DLS) experiments indicated that a certain ellipticity (with the aspect ratio in the range of 0.45–0.5) may be attributed to amelogenin assemblies (Aichmayer et al. 2010). Furthermore, limited proteolysis studies and experiments performed on polyelectrolyte multilayers have indicated that regions at both C- and N- termini are exposed on the surface of the nano-spheres (Moradian-Oldak et al. 2002a; Gergely et al. 2007). Experiments in which C-terminal was cleaved prior to the interaction with apatite have demonstrated a reduced ability of amelogenin cleavage products to interact with apatite (Aoba et al. 1987; Moradian-Oldak et al. 2002b), suggesting that the hydrophilic C-terminal, naturally, should be the region of the protein in direct contact with apatite (Shaw et al. 2004). With both C- and N- terminals exposed on the nanosphere surface, it is expected that C- terminal would be involved in the attachment onto the mineral surface, while N- terminal and the hydrophobic core of the protein would be involved in protein-protein interactions.
The knowledge on secondary and tertiary structures of amelogenin molecules is, on the other hand, still very poor. Diffraction studies have been impeded by the pronounced hydrophobicity of the protein, which tends to clump the molecules together and prevent the monomers from adopting a crystalline arrangement in space. Tens of thousands of serendipitous crystallization attempts by numerous research groups are informally said to have failed. Only the amino acid sequence of amelogenin is currently known, although there is a prospect that both evolutionary structural alignment simulations (Sire et al. 2005) and ab initio modeling will provide an insight into other structural levels of this protein. Despite the fact that the sequence of amelogenin is 90 % evolutionarily conserved, its primary structure is rather unique in the animal kingdom, with only 24% similarity to the closest structurally neighboring protein in the human body. Of course, although there are examples of exceptionally high structural similarity between proteins that share only 20% of sequence similarity (e.g., hemoglobins), substitution of one or a few out of hundreds of residues in a protein sequence often results in drastic changes in its secondary and tertiary structures (Horst and Samudrala 2009). The main challenge for computational studies aimed to assess the higher orders of the structure of amelogenin, however, comes from a relatively high proportion of Pro residues: 49 out of 175 in the complete X chromosome sequence of human amelogenin (including the exon 4 otherwise missing in the full-length amelogenin secreted in the enamel matrix) and 42 out of 175 in the complete Y chromosome sequence. The large number of Pro residues along the primary structure of amelogenin presents a considerable limitation due to their structure-breaking role and deviations from the regular secondary structure elements that they induce. The Raman Amide I band of recombinant full-length human amelogenin detected at 1,620 cm−1 indicated intermolecular extended chains (Fig. 13.6), and is in agreement with the results of circular dichroism (CD) studies, which have suggested the existence of polyproline type II structure in porcine amelogenin (Lakshminarayanan et al. 2009; Delak et al. 2009).
Fig. 13.6.
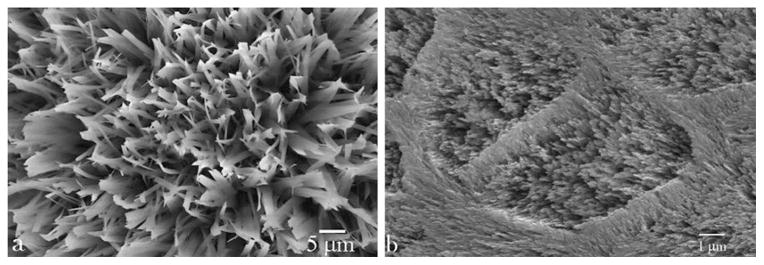
(a) Microstructure of apatite fibers outgrown from fluoroapatite/glass substrates immersed in amelogenin sols at high initial phosphate concentration and pH 6.5, the conditions under which the interaction between amelogenin and apatite is expected to more intense owing to opposite surface charges, negative for apatite and positive for amelogenin. (b) Natural enamel displaying structural similarity to that synthesized in the lab (a), though composed of apatite fibers finer in diameter
A single 41Pro→Thr mutation in recombinant full-length human amelogenin has been shown to result in significantly lower rates of apatite growth compared with the wild-type (Zhu et al. 2011). In view of the fact that the nearest proteo-lytic cleavage site lies between the residues 45Trp and 46Leu and that this mutation significantly reduces the enzymatic hydrolysis of amelogenin in the reaction with MMP-20, it has been suggested that proline residues might play a major role in aligning the cleavage-site residues along the active site of the enzyme (Tanimoto et al. 2008a) . In fact, the concentration of proline residues along the amelogenin sequence typically increases in the vicinity of the sites that are subject to proteolytic cleavage, suggesting that the hindered enzymatic interaction between amelogenin and MMP-20 may be the major cause of amelogenesis imperfecta.
High content of proline residues, however, does not necessarily predispose a protein for adopting poly-L-proline helix of type II in aqueous solution, similar to the one adopted by native collagen or many globular proteins (10% of individual amino acid residues in proteins exist in form of the polyproline conformation, and each protein on average contains one polyproline helix, although most of them are short, ranging from 4 to 6 residues in length) (Stapley and Creamer 1999). Whereas the sequence of collagen is composed of the repeating sequence of Gly-Pro-Tyr (with Pro residues preventing collagen from adopting α-helix and instead imposing a left-handed helix with ~3 residues per turn), proline residues in amelogenin are not positioned in such a periodic manner. Despite that, there are certain structural insights that can be derived from the high content of Pro residues. First of all, the side chains of residues in the polyproline helix protrude outward from the axis of the helix and are considerably separated by the extended nature of the helix, thus precluding hydrogen bonding interactions between adjacent side chains. As a result, both hydrophilic and hydrophobic side chains become exposed on the surface, providing favorable conditions for protein-protein interactions. The majority of side chains and backbone carbonyl and amide groups are thus also solvent-exposed, which is readily visible as kinks or bulges produced by a Pro residue in the middle of an α-helix or β–sheet, respectively (Eswar etal. 2003). Unlike secondary structures with intensive intramolecular hydrogen bonding, such as α-helix, the backbone carbonyl oxygen atoms are free to participate in hydrogen bonds across protein surfaces. Polyproline secondary structures also exhibit a significant conformational stability, which additionally contributes to their exploitation as binding sites. Proline-rich sequences are, in fact, common recognition sites for protein-protein interaction modules (Rath et al. 2005). An intrinsic predisposition of amelogenin for intermolecular interactions and for the formation of functional assemblies naturally follows.
Amelogenin sequence also has a relatively high content of glutamine: 26 out of 174 residues. The only exception among side chains that preclude the formation of intramolecular interactions between side chains of a polyproline protein is exactly glutamine, as it can participate in hydrogen bonding with the backbone carbonyl oxygen of the preceding residue. On the other hand, just as proline residues tend to participate in the formation of isolated extended strands that are conformationally distinct from polyproline helices, glutamines have also been implicated in the formation of aggregates through the extended strand formation. Polyglutamines are also some of the peptides that readily adopt the polyproline helical structure. Most proteins in human parotid and submandibular saliva, in fact, belong to the family of proline-rich proteins. On average, proline, glycine and glutamine account for 70–80% of all the amino acids within these proteins that are, however, not unique to salivary glands in the oral cavity, but are found in the respiratory tract and pancreas (Bennick 1987). These proline-rich proteins are known for their ability to bind calcium and thus presumably assist in buffering the concentration of ionic Ca2+ in saliva. They have also been shown to adhere strongly to apatite, exhibiting a lubricating effect and contributing to the formation of dental pellicle. However, owing to a high content of the three amino acids, their sequence is, unlike the one of amelogenin, highly repetitive.
13.6 Combining Protein Assembly, Crystal Growth and Proteolysis in Experiments Attempting to Engineer the Artificial Enamel
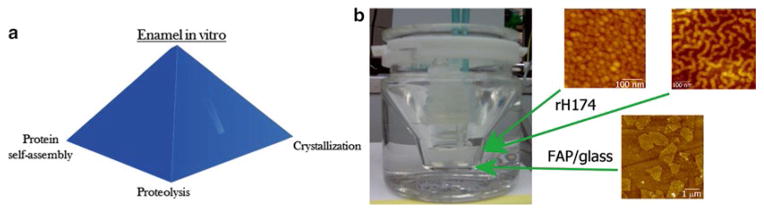
From the previous sections of the discourse, it could be concluded that assembly of amelogenin and its proteolytic products into dynamically evolving geometries able to guide the crystal growth along the right directions presents the central challenge for the attempts to engineer enamel in the lab using amelogenin as the crystal growth agent. In view of interrelated (a) amelogenin assembly, (b) proteolytic hydrolysis and (c) the crystal growth, a triadic nature of amelogenesis as the basis for its biomimicry could be proposed (Fig. 13.5a) and an experimental setting aimed at accomplishing this is shown in Fig. 13.5b. According to this model, the biologically relevant assembly of amelogenin is presumed to depend on its proteolytic hydrolysis, whereas the assembly of amelogenin nano-spheres, naturally, affects the rate and selectivity of proteolysis by exposing specific active groups to the surface. Similarly, no uniaxial and accurately orchestrated growth of apatite fibers could be possible without the assembly of amelogenin into biologically relevant forms, while this assembly may occur only when coupled to the crystal growth through the amelogenin matrix, as proposed by Cölfen and Mann (2003). According to this model, the aggregation of primary particles of apatite in the form of filamentous crystals modifies the thixotropic gelatinous environment around them and produces conditions for the transformation of the protein nanospheres into soft filaments anchored on the surface of the growing crystals. For example, crystallites precipitated in the presence of monomeric rM179 and rM166 comprised acicular morphologies, whereas the pre-assembled full-length rM179 had no influence on the crystal morphology (Beniash et al. 2005), indicating that conditions for a co-assembly in which both phases would structurally change need to be established for a successful protein-crystal interaction to be promoted instead of attempting to use one phase as a static structural template for the transition of another. Finally, while proteolytic digestion is necessary to clear the space for the filling of the protein-occupied space by the newly grown crystals, the ongoing crystal growth may be involved in shifting the balance of active species in the system in favor of selective proteolysis.
Fig. 13.5.
(a) Biomimicry of amelogenesis as based on well understood and utilized three essential aspects of the process: protein self-assembly, proteolysis and crystallization. (b) The image of a borosilicate glass vessel for the continuous and computerized (Dosimat 755 and Tiamo 1.2, Brinkmann–Methrohm) titration of amelogenin (rH174) sols with different protein assembly geometries for the purpose of controlled growth of finely polished substrates containing apatite (FAP) crystals with (001) faces exposed on the surface and interspersed with a glass matrix
The low levels of supersaturation, bordering the metastable state, appear to be crucial for providing the right conditions for amelogenin- guided crystal growth and the fabrication of enamel-like crystals (Fig. 13.6). Low rates of nucleation and crystal growth naturally favor the formation of elongated crystals. For example, when controlled degradation of urea is used to slowly increase alkalinity of the solution and provide conditions for precipitation, apatite crystals formed are either plate-shaped or needle-shaped (Jevtić and Uskoković 2007) . Single-crystal apatite fibers with 20–60 μm in length and 100–300 nm in diameter were thus obtained by precipitation using decomposition of urea (Aizawa et al. 2005). Although elongated apatite crystals have been obtained by methods involving rapid crystallization (Ashok et al. 2007), attempts to initiate nucleation and crystal growth at a higher rate than optimal by increasing the supersaturation ratio is expected to disrupt the continuity of amelogenin-guided crystal growth (Habelitz et al. 2005b).
Another essential requirement for the properly conducted amelogenesis is to increase the supersaturation ratio sufficiently slowly as well as with setting the precise ratio between Ca2+ and HxPO4x−3 species. The concentrations of Ca2+ and HxPO4x−3 ions in the fluid of developing enamel are 0.5 mM on average, and 2–5 mM, respectively, and the high initial concentrations of HxPO4x−3 in amelogenin suspensions, together with the absence of Ca2+ prior to the onset of titration, which gradually raises its concentration in the system, proved best for the controlled surface growth of apatite fibers. Curiously enough, the same ratio between the concentration of calcium and phosphate ions (markedly different from the one within hydroxyapatite crystals, i.e., Ca/P = 1.667) is present in saliva, suggesting its favorableness for both the natural regeneration of enamel in the presence of proteins that would mimic the role play by amelogenin in the course of amelogenesis.
13.7 The Role of Other Protein Species, Fluoride, pH, Water and Dentin
By now we must have been convinced that amelogenin is an absolutely essential element for the proper replication of amelogenesis in vitro and engineering of artificial enamel. Not only have studies on transgenic mice shown that the missing C- or N- terminals in amalogenin induce severe defects in the resulting enamel (Paine et al. 2000, 2002; Fong et al. 2003), but a single point mutation (41Pro→Thr) in the amelogenin gene causes severe dental enamel malformation known as amelogenesis imperfecta (Collier et al. 1997). However, amelogenin still constitutes 90, not 100 % of the composition of the enamel matrix. The prospect of attempts to engineer artificial enamel by means of harnessing only the right interaction between amelogenin and apatite, while discarding all other protein species of the enamel matrix, is dubious, to say the least. In that sense, despite the fact that reports on the role of enamel matrix proteins other than amelogenin in the enamel formation are comparatively scarce (Wang et al. 2005), evidence exists of the essentiality of macromolecular species present in minor amounts in the developing and maturating enamel matrix for the proper formation of the tissue. For example, mutations on the enamelin gene resulted in severe phenotypic amelogenesis imperfecta (Sawada et al. 2011; Lindemeyer and Gibson 2010; Masuya et al. 2005), demonstrating its essential role in the process of amelogenesis. Although its low concentration in the enamel matrix could easily trick us into thinking that we could do without it as well, this need not be necessarily so. For, there are many examples of macromolecular or amphiphilic additives that exhibit a cooperative effect on the assembly of the precipitated phase at low concentrations only (Mann et al. 2001). Polymeric or aliphatic additives introduced to repel colloidal entities, for one, oftentimes undergo aggregation at higher concentrations, leading to the loss of individuality or particles in the colloid and its irreversible destabilization (Uskoković 2013).
Ameloblastin is another protein of the enamel matrix expected to have a significant function, not only because of its localization at the secretory end of ameloblasts where the crystal growth is initiated, but because of both an augmented and inhibited expression of ameloblastin has been shown to result in amelogenesis imperfecta (Paine et al. 2003; Margolis et al. 2006). The roles of even less abundant components of the enamel matrix, such as KLK4, keratin K14, DLX3 or biglycan proteins, the mutant expressions of which are also known to produce the conditions of amelogenesis imperfecta (Stephanopoulos et al. 2005), have all but been investigated thoroughly and it is doubtful whether there would be any room for functionless ingredients in biosynthetic pathways.
Although adding fluoride to biomimetic experiments aimed toward replicating amelogenesis would also be a natural approach in view of its presence in natural enamel apatite, exceeding amounts thereof are known to result in increased porosity and weakening of the enamel structure (Lyaruu et al. 2014). It was also shown that increased levels of fluoride in developing enamel decrease the activity of MMP-20 (Zhang et al. 2006), resulting in the condition known as fluorosis. The role of fluoride ions in promoting elongation of apatite crystals has, however, been well documented. In a set of experiments, only the combination of amelogenin and fluoride led to formation of rod-like apatite crystals, while merely octacalcium phosphate precipitated in the absence of fluoride (Iijima and Moradian-Oldak 2005; Iijima et al. 2006). On the other hand, it was demonstrated that fluoride ions do not directly interact with amelogenins, but limit their effect on the process of amelogenesis to their incorporation into the apatite crystal lattice (Tanimoto et al. 2008b).
pH during amelogenesis varies within the range of more than a single unit, that is, from 6.0 to 7.2 (Sasaki et al. 1991), exhibiting variations from one end of ameloblasts to another. For this reason, pH is often considered to be one of the most important parameters to control during biomineralization events (Weaver et al. 2009) and phosphate, carbonate and protein species altogether work to buffer the system and prevent catastrophic drops or soars in acidity or alkalinity. Although the unavailability of techniques for measuring pH variations at the nanometer scale is to be blamed for the enigmatic status of this variable in the process of amelogenesis, its importance is beyond question. The need for precise orchestration of pH, even if not at the local scale, as it is most probably the case, certainly place an additional burden on biomimeticians of amelogenesis.
Early secretory enamel consists of 50–60 vol.% of water, 20–30 vol.% of protein, and about 15–20 vol.% of mineral. High concentrations of amelogenin (~200–300 mg/ml) in the developing enamel matrix imply that the latter resembles a gel more than an aqueous solution. Growing apatite in gelatinous media rather than in ordinary aqueous solutions thus presents a natural biomimetic choice (Wen et al. 2000; Petta et al. 2006). Crystallization of apatite from such dense media may favor the slow and controlled growth. Precipitation of fluoroapatite in gelatin per se, without the presence of amelogenin, thus resulted in spherical composites consisting of needle-shaped crystals and around 2% of organic matter (Busch et al. 2001; Busch 2004). Density of the aqueous medium is larger compared to ordinary aqueous solutions not only in the enamel matrix, but in biological environments per se. Under such circumstances, water exhibits modified structure and properties. Cytoplasm typically contains about 400 g/dm3 of macromolecules, which as such occupy 5–40% of the total cell volume with an average separation between them of 1–2 nm. Within such nanoscopically confined conditions, water possesses an altered hydrogen bonding structure in comparison with the bulk water. Also, by playing various structural roles, water presents an essential component of a fully functional protein. Although it has been shown that structure and functionality of some enzymes can be preserved in non-polar media or even in vacuum (albeit the preservation of bound water even under such circumstances), it is suggested that water “lubricates” the peptide chains and provides conditions for favorable molecular recognition effects. Consequently, the concepts of diffusion and solubility limits should be redefined with the transition to complex and dense media such as those from which enamel crystal grow.
The initial enamel crystals are nucleated along the dentin-enamel junction and a proper substrate is therefore of vital importance in ensuring the right final structure of the material. Epitaxial effects were many times proven as essential in self-assembly procedures (Uskoković 2008), and many biomineralization mechanisms (e.g., crystallization of thin flakes of nacre in the mollusk shells) depend on the interfacial structural matching between an organic substrate and an inorganic phase. Despite the fact that the hardness of enamel is a result of its nanoscale superstructural organization, the strength of enamel is also highly dependent on the supporting dentin. This interaction between the dentin substrate and superstructurally organized enamel crystals may be another factor of critical importance for replication of the assembly of fibrous apatite crystals in vitro. It is also known that signals originating from the dental papilla are required to activate the expression of amelogenin (Garant 2003), which points to an even wider scope of amelogenesis, in view of which the prospects of replicating the process by focusing only on a selected number of species and control parameters can be subjected to reasonable scrutiny.
13.8 Conclusion and Future Prospects
It may have become clear by now that the replication of amelogenesis in vitro stands for a daunting task that requires knowledge on the ability to orchestrate interactions between a multitude of polypeptides in precise correlation with setting the right conditions for diffusion of the ionic growth units and their precipitation in form of uniaxial crystals. In the end, it is logical to expect that the three major aforementioned aspects of amelogenesis – (1) the assembly of amelogenin and other enamel matrix proteins, (2) the proteolytic activity, and (3) crystallization – need to be in precise synergy with each other in order to produce the desired outcome. These endeavors are additionally made difficult because they are being designed to yield fundamental insights regarding amelogenesis, while at the same time to be harnessed for practical purposes. As much as it is natural, this entwinement of the practical and the fundamental aspects of the biomimetic settings aimed at replicating biomineralization in a beaker is also inherently illogical. For, how could we be expected to create a desired product without knowing the chemical mechanisms intrinsic to its formation and how could we be expected to understand the fundamental features of a process if we do not know how to replicate it? As of today, however, it is difficult to estimate which aspect of the process is more difficult to penetrate into: fundamental or practical. In any case, conceiving original experimental approaches to mimic amelogenesis presents the key, although two eyes need to be used to analyze the outcomes. The proteomic, life science eye would follow the protein-related aspects of the process, whereas the materials science eye would follow the crystal formation facets of it. Needless to add, these two eyes need to look in the same direction and in synergy from the top of the aforementioned pyramid (Fig. 13.5a) in order for the path of biomimetics of tooth enamel to be walked on successfully. In such a way, there is a chance that the future development of this field will transcend the broad speculations that dominate the contemporary literature reports on amelogenesis-related studies, though remain receptive to the effects of some of the most minor components of this fascinating biological process. For, if the science of the enamel growth teaches us something profound, it is that “small is beautiful” and that a tiny detail of this Universe, such as the enamel, hides many mysterious patterns, diligent plunging in the research of which may open the doors to understanding of much greater secrets of the physical reality in which we abide.
Acknowledgments
Writing of this chapter was supported by the National Institute of Health grant R00-DE021416. The author thanks Irena Ćosić and Elena Pigorova of the Royal Melbourne Institute of Technology for performing the CWT-RRM analysis of human amelogenin.
References
- Aichmayer B, Wiedemann-Bidlack FB, Gilow C, Simmer JP, Yamakoshi Y, Emmerling F, Margolis HC, Fratzl P. Amelogenin nanoparticles in suspension: deviations from spherical shape and pH-dependent aggregation. Biomacromolecules. 2010;11(2):369–376. doi: 10.1021/bm900983b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa M, Porter AE, Best SM, Bonifide W. Ultrastructural observation of single-crystal apatite fibres. Biomaterials. 2005;26:3427–3433. doi: 10.1016/j.biomaterials.2004.09.044. [DOI] [PubMed] [Google Scholar]
- Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- Aoba T, Fukae M, Tanabe T, Shimizu M, Moreno EC. Selective adsorption of porcine amelogenins onto hydroxyapatite and their inhibitory activity on seeded crystal growth of hydroxyapatite. Calcif Tissue Int. 1987;41:281–289. doi: 10.1007/BF02555230. [DOI] [PubMed] [Google Scholar]
- Ashok M, Kalkura SN, Sundaram NM, Arivuoli D. Growth and characterization of hydroxyapatite crystals by hydrothermal method. J Mater Sci Mater Med. 2007;18:895–898. doi: 10.1007/s10856-006-0070-5. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Simmer JP. Proteinases in developing enamel. Crit Rev Oral Biol Med. 1999;10(4):425–441. doi: 10.1177/10454411990100040101. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Ryu OH, Xue J, Simmer JP, Margolis HC. Enamelysin mRNA displays a developmentally defined pattern of expression and encodes a protein which degrades amelogenin. Connect Tissue Res. 1998;39:405–413. doi: 10.3109/03008209809023916. [DOI] [PubMed] [Google Scholar]
- Bartlett JD, Skobe Z, Lee DH, Wright JT, Li Y, Kulkarni AB, Gibson CW. A developmental comparison of matrix metalloproteinase-20 and amelogenin null mouse enamel. Eur J Oral Sci. 2006;114(Suppl 1):18–23. doi: 10.1111/j.1600-0722.2006.00292.x. [DOI] [PubMed] [Google Scholar]
- Beniash E, Simmer JP, Margolis HC. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol. 2005;149(2):182–190. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Bennick A. Structural and genetic aspects of proline-rich proteins. J Dent Res. 1987;66(2):457–461. doi: 10.1177/00220345870660021201. [DOI] [PubMed] [Google Scholar]
- Bourd-Boittin K, Fridman R, Fanchon S, Septier D, Goldberg M, Menashi S. Matrix metalloproteinase inhibition impairs the processing, formation and mineralization of dental tissues during mouse molar development. Exp Cell Res. 2005;304(2):493–505. doi: 10.1016/j.yexcr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Busch S. Regeneration of human tooth enamel. Angew Chem Int Ed Engl. 2004;43:1428–1431. doi: 10.1002/anie.200352183. [DOI] [PubMed] [Google Scholar]
- Busch S, Schwarz U, Kniep R. Morphogenesis and structure of human teeth in relation to biomimetically grown fluorapatite-gelatine composites. Chem Mater. 2001;13:3260–3271. [Google Scholar]
- Bush V. Science: the endless frontier: a report to the President by Vannevar Bush, Director of the Office of Scientific Research and Development. United States Government Printing Office; Washington, DC: 1945. [Google Scholar]
- Cai Y, Liu Y, Yan W, Hu Q, Tao J, Zhang M, Shi Z, Tang R. Role of hydroxyapatite nanoparticle size in bone cell proliferation. J Mater Chem. 2007;17:3780. [Google Scholar]
- Caterina JJ, Skobe Z, Shi J, Dang Y, Simmer JP, Birkedal-Hansen H, Bartlett JD. Enamelysin (MMP-20) deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277(51):49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- Cölfen H, Mann S. Higher-order organization by mesoscale self-assembly and transformation of hybrid nanostructures. Angew Chem Int Ed. 2003;42:2350–2365. doi: 10.1002/anie.200200562. [DOI] [PubMed] [Google Scholar]
- Collier PM, Sauk JJ, Rosenbloom SJ, Yuan ZA, Gibson CW. An amelogenin gene defect associated with human x-linked amelogenesis imperfecta. Arch Oral Biol. 1997;42:235–242. doi: 10.1016/s0003-9969(96)00099-4. [DOI] [PubMed] [Google Scholar]
- Ćosić I, Pirogova E. Bioactive peptide design using the resonant recognition model. Nonlinear Biomed Phys. 2007;1:1–17. doi: 10.1186/1753-4631-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Trad CH, Fang Q, Ćosić I. The resonant recognition model (RRM) predicts amino acid residues in highly conservative regions of the hormone prolactin (PRL) Biophys Chem. 2000;84(2):149–157. doi: 10.1016/s0301-4622(00)00109-5. [DOI] [PubMed] [Google Scholar]
- Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, Evans JS. The tooth enael protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48(10):2272–2281. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S, Girondot M, Sire JY. Molecular evolution of amelogenin in mammals. J Mol Evol. 2005;60:12–30. doi: 10.1007/s00239-003-0070-8. [DOI] [PubMed] [Google Scholar]
- Eppell SJ, Tong W, Katz JL, Kuhn L, Glimcher MJ. Shape and size of isolated bone mineralites measured using atomic force microscopy. J Orthop Res. 2001;19:1027–1034. doi: 10.1016/S0736-0266(01)00034-1. [DOI] [PubMed] [Google Scholar]
- Eswar N, Ramakrishnan C, Srinivasan N. Stranded in isolation: structural role of isolated extended strands in proteins. Protein Eng. 2003;16(5):331–339. doi: 10.1093/protein/gzg046. [DOI] [PubMed] [Google Scholar]
- Fong H, White SN, Paine ML, Luo W, Snead ML, Sarikaya M. Enamel structure properties controlled by engineered proteins in transgenic mice. J Bone Miner Res. 2003;18(11):2052–2059. doi: 10.1359/jbmr.2003.18.11.2052. [DOI] [PubMed] [Google Scholar]
- Garant PR. Quintessence, Carol Stream. 2003. Oral cells and tissues. [Google Scholar]
- Gergely C, Szalontai B, Moradian-Oldak J, Cuisinier F. Polyelectrolyte-mediated adsorption of amelogenin monomers and nanospheres forming mono- or multilayers. Biomacromolecules. 2007;8:2228–2236. doi: 10.1021/bm070088+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Yuan Z-A, Hall B, Longenecker G, Cheng E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkami AB. Molecular basis of cell and developmental biology. J Biol Chem. 2001;276(34):31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Gower LB. Biomimetic model systems for investigating the amorphous precursor pathway and its role in biomineralization. Chem Rev. 2008;108(11):4551–4627. doi: 10.1021/cr800443h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habelitz S, Kullar A, Marshall SJ, DenBesten PK, Balooch M, Marshall GW, Li W. Amelogenin- guided crystal growth on fluoroapatite glass-ceramics. J Dent Res. 2004;83(9):698–702. doi: 10.1177/154405910408300908. [DOI] [PubMed] [Google Scholar]
- Habelitz S, DenBesten PK, Marshall SJ, Marshall GW, Li W. Amelogenin control over apatite crystal growth is affected by the pH and degree of ionic saturation. Orthod Craniofac Res. 2005a;8:232–238. doi: 10.1111/j.1601-6343.2005.00343.x. [DOI] [PubMed] [Google Scholar]
- Habelitz S, DenBesten PK, Marshall SJ, Marshall GW, Li W. Amelogenin control over apatite crystal growth is affected by the pH and degree of ionic saturation. Orthod Craniofacial Res. 2005b;8:232–238. doi: 10.1111/j.1601-6343.2005.00343.x. [DOI] [PubMed] [Google Scholar]
- Hamedi M, Elfwing A, Gabrielsson R, Inganas O. Electronic polymers and DNA self-assembled in nanowire transistors. Small. 2012;9:363–368. doi: 10.1002/smll.201201771. [DOI] [PubMed] [Google Scholar]
- Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst J, Samudrala R. Diversity of protein structures and difficulties in fold recognition: the curious case of protein G. F111 Biol Reports. 2009;14(1):69. doi: 10.3410/B1-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M, Moradian-Oldak J. Control of apatite crystal growth in a fluoride containing amelogenin- rich matrix. Biomaterials. 2005;26(13):1595–1603. doi: 10.1016/j.biomaterials.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Iijima M, Du C, Abbott C, Doi Y, Moradian-Oldak J. Control of apatite crystal growth by the co- operative effect of a recombinant porcine amelogenin and fluoride. Eur J Oral Sci. 2006;114(Suppl 1):304–307. doi: 10.1111/j.1600-0722.2006.00324.x. [DOI] [PubMed] [Google Scholar]
- Jevtić M, Uskoković D. Influence of urea as a homogenous precipitation agent on sonochemical hydroxyapatite synthesis. Mater Sci Forum. 2007;555:285–290. [Google Scholar]
- Kashchiev D. Nucleation: basic theory with applications. Butterworth-Heinemann; Oxford: 2000. [Google Scholar]
- Lakshminarayanan R, Yoon I, Hegde BG, Fan D, Du C, Moradian-Oldak J. Analysis of secondary structure and self-assembly of amelogenin by variable temperature circular dichroism and isothermal titration calorimetry. Proteins. 2009;76(3):560–569. doi: 10.1002/prot.22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemeyer RG, Gibson CW, Wright TJ. Amelogenesis imperfecta due to a mutation of the enamelin gene: clinical case with genotype-phenotype correlations. Pediatr Dent. 2010;32(1):56–60. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Song C, Wang ZG, Li N, Ding B. Precise organization of metal nanoparticles on DNA origami template. Methods. 2013 doi: 10.1016/j.ymeth.2013.10.006. pii: S1046–2023(13)00402-7. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Lyaruu DM, Medina JF, Sarvide S, Bervoets TJ, Everts V, Denbesten P, Smith CE, Bronckers AL. Barrier formation: potential molecular mechanism of enamel fluorosis. J Dent Res. 2014 doi: 10.1177/0022034513510944. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S. Biomineralization: principles and concepts in bioinorganic materials chemistry. Oxford University Press; Oxford: 2001. [Google Scholar]
- Margolis HC, Beniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85(9):775–793. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- Martinez-Avila O, Wu S, Kim SJ, Cheng Y, Khan F, Samudrala R, Sali A, Horst JA, Habelitz S. Self- assembly of filamentous amelogenin requires calcium and phosphate: from dimers via nanoribbons to fibrils. Biomacromolecules. 2012;13(11):3494–3502. doi: 10.1021/bm300942c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuya H, Shimizu K, Sezutsu H, Sakuraba Y, Nagano J, Shimizu A, Fujimoto N, Kawai A, Miura I, Kaneda H, Kobayashi K, Ishijima J, Maeda T, Gondo Y, Noda T, Wakana S, Shiroishi T. Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI) Hum Mol Genet. 2005;14(5):575–583. doi: 10.1093/hmg/ddi054. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Gharakhanian N, Jimenez I. Limited proteolysis of amelogenin: toward understanding the proteolytic processes in enamel extracellular matrix. Connect Tissue Res. 2002a;43(2):450–455. doi: 10.1080/03008200290000835. [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Bouropoulos N, Wang L, Gharakhanain N. Analysis of self-assembly and apatite binding properties of amelogenin proteins lacking the hydrophilic C-terminal. Matrix Biol. 2002b;21(2):197–205. doi: 10.1016/s0945-053x(01)00190-1. [DOI] [PubMed] [Google Scholar]
- Murakami M. Critical amino acids responsible for conferring calcium channel characteristics are located on the surface and around beta-turn potentials of channel proteins. J Protein Chem. 1995;14(3):111–114. doi: 10.1007/BF01980322. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Cox MM. Lehninger principles of biochemistry. 4. W. H. Freeman; New York: 2004. [Google Scholar]
- Paine ML, Zhu DH, Luo W, Bringas P, Jr, Goldberg M, White SN, Lei YP, Sarikaya M, Fong HK, Snead ML. Enamel biomineralization defects result from alterations to amelogenin self-assembly. J Struct Biol. 2000;132(3):191–200. doi: 10.1006/jsbi.2000.4324. [DOI] [PubMed] [Google Scholar]
- Paine ML, Lei YP, Dickerson K, Snead ML. Altered amelogenin self-assembly based on mutations observed in human X-linked Amelogenesis Imperfecta (AIH1) J Biol Chem. 2002;277(19):17112–17116. doi: 10.1074/jbc.M110473200. [DOI] [PubMed] [Google Scholar]
- Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML. A transgenic animal model resembling amelo-genesis imperfecta related to ameloblastin overexpression. J Biol Chem. 2003;278(21):19447–19452. doi: 10.1074/jbc.M300445200. [DOI] [PubMed] [Google Scholar]
- Petta V, Moradian-Oldak J, Yannopoulos SN, Bouropoulos N. Dynamic light scattering study of an amelogenin gel-like matrix in vitro. Eur J Oral Sci. 2006;114(Suppl 1):308–314. doi: 10.1111/j.1600-0722.2006.00325.x. [DOI] [PubMed] [Google Scholar]
- Rath A, Davidson AR, Deber CM. The structure of ‘Unstructured’ regions in peptides and proteins: role of the polyproline II helix in protein folding and recognition. Biopolymers. 2005;80:179–185. doi: 10.1002/bip.20227. [DOI] [PubMed] [Google Scholar]
- Ravindranath RM, Devarajan A, Bringas P., Jr Enamel formation in vitro in mouse molar explants exposed to amelogenin polypeptides ATMP and LRAP on enamel development. Arch Oral Biol. 2007;52(12):1161–1171. doi: 10.1016/j.archoralbio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Renugopalakrishnan V, Prabhakaran M, Huang SG, Balasubramaniam A, Strawich E, Glimcher MJ. Secondary structure and limited three-dimensional structure of bovine amelogenin. Connect Tissue Res. 1989;22(1–4):131–138. [PubMed] [Google Scholar]
- Robinson C., moderator Discussion of session 8: amelogenin assembly and function. Eur J Oral Sci. 2006;114(Suppl 1):327–329. doi: 10.1111/j.1600-0722.2006.00311.x. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Tagaki T, Suzuki M. Cyclical changes in pH in bovine developing enamel as sequential bands. Arch Oral Biol. 1991;36:227–231. doi: 10.1016/0003-9969(91)90090-h. [DOI] [PubMed] [Google Scholar]
- Sawada T, Sekiguchi H, Uchida T, Yamashita H, Shintani S, Yanagisawa T. Histological and immunohistochemical analyses of molar tooth germ in enamelin- deficient mouse. Acta Histochem. 2011;113(5):542–546. doi: 10.1016/j.acthis.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Shaw WJ, Campbell AA, Paine ML, Snead ML. The COOH terminus of the amelogenin, LRAP, is oriented next to the hydroxyapatite surface. J Biol Chem. 2004;279(39):40263–40266. doi: 10.1074/jbc.C400322200. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Hu JC-C. Expression, structure, and function of enamel proteinases. Connect Tissue Res. 2002;43(2):441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- Sire JY, Delgado S, Forementin D, Girondot M. Amelogenin: lessons from evolution. Arch Oral Biol. 2005;50(2):205–212. doi: 10.1016/j.archoralbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Snead ML. Amelogenin protein exhibits a modular design: implications for form and function. Connect Tissue Res. 2003;44(1):47–51. [PubMed] [Google Scholar]
- Stapley BJ, Creamer TP. A survey of left-handed polyproline II helices. Protein Sci. 1999;8(3):587–595. doi: 10.1110/ps.8.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos G, Garefalaki M-E, Lyroudia K. Genes and related proteins involved in amelogenesis imperfecta. J Dent Res. 2005;84(12):1117–1126. doi: 10.1177/154405910508401206. [DOI] [PubMed] [Google Scholar]
- Stupp SI, Braun PV. Molecular manipulation of microstructures: biomaterials, ceramics, and semiconductors. Science. 1997;277:1242–1248. doi: 10.1126/science.277.5330.1242. [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Le T, Zhu L, Witkowska HE, Robinson S, Hall S, Hwang P, DenBesten P, Li W. Reduced amelogenin-MMP20 interactions in amelogenesis imperfecta. J Dent Res. 2008a;87(5):451–455. doi: 10.1177/154405910808700516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K, Le T, Zhu L, Chen J, Featherstone JDB, Li W, DenBesten P. Effects of fluoride on the interactions between amelogenin and apatite crystals. J Dent Res. 2008b;87:39–44. doi: 10.1177/154405910808700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine M, Shaw WJ. The nucleation and growth of calcium phosphate by amelogenin. J Crystal Growth. 2007;304(2):407–415. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Holy Bible (1609) King James Edition, John 12:24
- Uskoković V. Insights into morphological nature of precipitation of cholesterol. Steriods. 2008;73:356–369. doi: 10.1016/j.steroids.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Uskoković V. Prospects and pits on the path of bio-mimetics: the case of tooth enamel. J Biomimetics Biomater Tissue Eng. 2010;8:45–78. doi: 10.4028/www.scientific.net/JBBTE.8.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskoković V. Entering the era of nanoscience: time to be so small. J Biomed Nanotechnol. 2013;9:1441–1470. doi: 10.1166/jbn.2013.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskoković V, Drofenik M. Synthesis of materials within reverse micelles. Surf Rev Lett. 2005;12(2):239–277. [Google Scholar]
- Uskoković V, Kim M-K, Li W, Habelitz S. Enzymatic processing of amelogenin during continuous crystallization of apatite. J Mater Res. 2008;32:3184–3195. doi: 10.1557/JMR.2008.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskoković V, Odsinada R, Djordjevic S, Habelitz S. Dynamic light scattering and zeta potential of colloidal mixtures of amelogenin and hydroxyapatite in calcium and phosphate rich ionic milieus. Arch Oral Biol. 2011a;56:521–532. doi: 10.1016/j.archoralbio.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskoković V, Li W, Habelitz S. Amelogenin as a promoter of nucleation and crystal growth of apatite. J Crystal Growth. 2011b;316:106–117. doi: 10.1016/j.jcrysgro.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskoković V, Li W, Habelitz S. Biomimetic precipitation of uniaxially grown calcium phosphate crystals from full-length human amelogenin sols. J Bionic Eng. 2011c;8(2):114–121. doi: 10.1016/S1672-6529(11)60017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uskoković V, Khan F, Liu H, Witkowska HE, Zhu L, Li W, Habelitz S. Proteolytic hydrolysis of amelogenin by means of matrix metalloprotease-20 accelerates mineralization in vitro. Arch Oral Biol. 2011d;56(12):1548–1559. doi: 10.1016/j.archoralbio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner RS, Ellis WC. Vapor-liquid-solid mechanism of single crystal growth. Appl Phys Lett. 1964;4(5):89–90. [Google Scholar]
- Wang HJ, Tannukit S, Zhu DH, Snead ML, Paine ML. Enamel matrix protein interactions. J Bone Miner Res. 2005;20(6):1032–1040. doi: 10.1359/JBMR.050111. [DOI] [PubMed] [Google Scholar]
- Wang L, Guan X, Yin H, Moradian-Oldak J, Nancollas GH. Mimicking the self-organized microstructure of tooth enamel. J Phys Chem C. 2008;112(15):5892–5899. doi: 10.1021/jp077105+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ML, Qiu SR, Hoyer JR, Casey WH, Nancollas GH, De Yoreo JJ. Surface aggregation of urinary proteins and aspartic Acid-rich peptides on the faces of calcium oxalate monohydrate investigated by in situ force microscopy. Calcif Tissue Int. 2009;84(6):462–473. doi: 10.1007/s00223-009-9223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen HB, Moradian-Oldak J, Fincham AG. Dose- dependent modulation of octacalcium phosphate crystal habit by amelogenins. J Dent Res. 2000;79(11):1902–1906. doi: 10.1177/00220345000790111501. [DOI] [PubMed] [Google Scholar]
- Young KL, Ross MB, Blaber MG, Rycenga M, Jones MR, Senesi AJ, Lee B, Schatz GC, Mirkin CA fluoride on Zhang C. Using DNA to design plasmonic metamaterials with tunable optical properties. Adv Mater. 2014;26:653–659. doi: 10.1002/adma.201302938. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan Q, Li W, DenBesten PK. Fluoride down-regulates the expression of matrix metalloproteinase-20 in human fetal tooth ameloblast-lineage cell in vitro. Eur J Oral Sci. 2006;114(Suppl 1):105–110. doi: 10.1111/j.1600-0722.2006.00303.x. [DOI] [PubMed] [Google Scholar]
- Zhaohua G, Caixia L, Hong Y, Yu X, Yingliang W, Wenxin L, Tao X, Jiuping D. A residue at the cytoplasmic entrance of BK-type channels regulating single-channel opening by its hydrophobicity. Biophys J. 2008;94(9):3714–3725. doi: 10.1529/biophysj.107.120022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Tu AT, Renugopalakrishnan V, Strawich E, Glimcher MJ. A mixed beta-turn and beta-sheet structure for bovine tooth enamel amelogenin: Raman spectroscopic evidence. Biopolymers. 1987;26(10):1809–1813. doi: 10.1002/bip.360261012. [DOI] [PubMed] [Google Scholar]
- Zhu L, Uskoković V, Le T, DenBesten P, Huang YL, Habelitz S, Li W. Altered self-assembly and apatite binding of amelogenin induced by N-terminal proline mutation. Arch Oral Biol. 2011;56(4):331–336. doi: 10.1016/j.archoralbio.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]