Abstract
Objectives: Pathological biomarkers and mechanisms of dengue infection are poorly understood. We investigated a new serum biomarker using miRNAs and performed further correlation analysis in dengue-infected patients.
Methods: Expression levels of broad-spectrum miRNAs in serum samples from three patients with dengue virus type 1 (DENV-1) and three healthy volunteers were separately analyzed using miRNA PCR arrays. The expressions of the five selected miRNAs were verified by qRT-PCR in the sera of 40 DENV-1 patients and compared with those from 32 healthy controls. Receiver operating characteristic (ROC) curve and correlation analyses were performed to evaluate the potential of these miRNAs for the diagnosis of dengue infection.
Results: MiRNA PCR arrays revealed that 41 miRNAs were upregulated, whereas 12 miRNAs were down-regulated in the sera of DENV-1 patients compared with those in healthy controls. Among these miRNAs, qRT-PCR validation showed that serum hsa-miR-21-5p, hsa-miR-590-5p, hsa-miR-188-5p, and hsa-miR-152-3p were upregulated, whereas hsa-miR-146a-5p was down-regulated in dengue-infected patients compared with healthy controls. ROC curves showed serum hsa-miR-21-5p and hsa-miR-146a-5p could distinguish dengue-infected patients with preferable sensitivity and specificity. Correlation analysis indicated that expression levels of serum hsa-miR-21-5p and hsa-miR-146a-5p were negative and positively correlated with the number of white blood cells and neutrophils, respectively. Functional analysis of target proteins of these miRNAs in silico indicated their involvement in inflammation and cell proliferation.
Conclusion: Dengue-infected patients have a broad “fingerprint” profile with dysregulated serum miRNAs. Among these miRNAs, serum hsa-miR-21-5p, hsa-miR-146a-5p, hsa-miR-590-5p, hsa-miR-188-5p, and hsa-miR-152-3p were identified as promising serum indicators for dengue infection.
Keywords: microRNA, dengue, miRNA-21-5p, miRNA-146a-5p, miR-590-5p, miR-188-5p, miR-152-3p.
Introduction
Dengue is a mosquito-borne disease caused by dengue virus. Dengue virus, which is an RNA virus of the Flaviviridae family, has four serotypes, namely, DENV-1, DENV-2, DENV-3, and DENV-4. This virus is primarily transmitted by Aedes mosquitoes. Global incidence of dengue has grown dramatically in recent decades. Recent WHO report showed that more than 390 million people suffer from dengue virus infections annually worldwide 1. The majority of dengue-infected patients manifest flu-like symptoms, such as fever and headache 2. Meanwhile, some severe cases result in life-threatening dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS). The pathogenic mechanism of dengue is complex and remains ambiguous. Thus, no specific treatment or licensed vaccine against dengue is available. Early detection and access to proper medical care can help reduce death rate due to dengue.
Numerous assay methods or platforms have been developed for the early diagnosis of dengue infections, but these assays remain suboptimal. Viral isolation and identification is the gold standard for dengue infection but this process is time-consuming 3. Moreover, the sensitivity and specificity of the methods for detecting anti-DENV IgM /IgG antibody or NS1 antigen vary in many publications 4. Real time RT-PCR is sensitive, but this approach occasionally exhibits false negative or false positive results 5. Thus, new biomarkers for the diagnosis, treatment, and prognosis of dengue infection are still urgently needed.
MicroRNAs (miRNAs) are endogenous non-coding single-stranded RNA molecules made of approximately 22 nucleotides. They can regulate the post-transcriptional expression of target genes by targeting mRNAs for cleavage or translational repression and play an important role in the gene regulation of organism 6. Many studies showed that the abnormal expression of miRNA is closely related with the occurrence and development of diseases 7-11. Moreover, numerous miRNAs have been associated with infectious diseases. These miRNAs either play an important role in delayed disease progression or help virus escape the host defense 12-16.
Circulating miRNAs have recently attracted considerable attention because of their potential as noninvasive biomarkers for diagnosing various diseases, such as infectious diseases. MiRNAs are highly stable in both fresh serum and plasma. Circulating miRNAs have been associated with pathophysiological states 17. Several miRNAs have been reported to participate in the interaction between dengue virus and host 18. For example, miR-146a facilitates replication of DENV-2 in primary human monocytes and THP-1 cells upon DENV infection by targeting TRAF6 and inhibiting IFN-beta production 19. The expression of miR-24-1-5p, miR-512-5p, and miR-4640-3p in blood is recently reported to be beneficial in distinguishing mild dengue from those exhibiting liver complications 20. This characteristic indicates that miRNAs may serve as biomarkers for dengue infection.
In this study, 752 miRNAs in the sera of DENV-1 patients and healthy controls were screened for different expression profiles to explore the potential biomarkers for dengue infection. The selected miRNAs were further verified by qRT-PCR. Then, diagnostic potentials and biological functions were analyzed using statistical software or bioinformatics. Moreover, correlation analysis associated with clinical parameters was performed.
Materials and Methods
Sample collection
A total of 72 serum samples (40 patients with active DENV-1 replication and 32 healthy volunteers) were obtained from the Third People's Hospital of Nanhai District in Foshan City, Guangdong Province, China. Healthy controls were recruited randomly from individuals who had no clinical symptoms of infectious diseases after regular physical examination. Dengue patients enrolled in this study were confirmed to have no other infectious diseases, such as influenza A, influenza B, and HCV, and have no drug treatment. Serum samples were isolated within 1 h after receiving whole blood and then immediately stored at -80 °C until further use. This study was approved by the Ethics Committee of the Third People's Hospital of Nanhai District in Foshan City, Guangdong Province, China. Written informed consents were obtained from all participants. All assays were conducted according to the principles expressed in the Declaration of Helsinki.
RNA extraction
Total RNA was isolated from serum by Trizol method according to the previous method 21. Briefly, 300 µL of serum was thawed on ice and centrifuged at 12000 rev/min and at 4 °C for 10 min to remove pellet and cell debris. Then, 750 μL of Trizol LS reagent were added to 250 µL supernatant. After incubation for 5 min at room temperature, 25 fmol synthetic cel-miRNA-39-3p (Syn-cel-miR-39-3p miScript miRNA Mimic, Ribobio, China) was spiked-in. The total RNA, including small RNA, was extracted using Trizol LS reagent following the manufacturer's instructions (Invitrogen, USA) and dissolved in 20 μL of RNase-free water. RNA was stored at -80 °C until further use.
MiRNA expression profiles using miRNA PCR arrays
Sera from three dengue patients and 3 healthy control samples were used for miRNA PCR arrays (Human panel I+II, V4.M, KangChen Bio-tech, Shanghai, China). This assay was conducted according to the previous method with slight modification 21. Relative expressions of 752 miRNAs in sera from the DENV positive group and healthy control were screened using miRNA PCR arrays. The total RNA sample was diluted to 1.5-1.8 ng/µL in nuclease-free water. Reverse transcription (RT) was conducted in an RT reaction mix (Exiqon, Denmark). cDNA was diluted by 110-fold in nuclease-free water and amplified using SYBR™ Green master mix (Exiqon, Denmark) with an ABI PRISM 7900 Real-time PCR System (Applied Biosystems, USA) according to manufacturer's instructions. Relative miRNA expression levels were calculated using ΔCt method per manufacturer's recommendations. Three miRNAs (miR-191-5p, miR-423-5p, and miR-425-5p), which show low standard deviation, were selected to normalize the 752 miRNAs from PCR array. Fold changes of >2-fold at p < 0.05 were used to select significantly altered miRNAs.
Verifying miRNA array data by real-time quantitative reverse transcription PCR (qRT-PCR)
Total RNAs of sera in 40 dengue patients and 32 healthy controls were subjected to further miRNA validation assay via quantitative RT-PCR (qRT-PCR) 21. An miRNA assay kit (GenePharma, Shanghai, China) was used for miRNA detection and quantification. The RT reaction was performed using a PrimeScriptTM First Strand cDNA Synthesis Kit (Takara, Dalian, China) with an AlphaTM Unit Block Assembly for DNA EngineH systems (Bio-Rad, USA) under the following reaction conditions: 30 min at 25 °C, 30 min at 42 °C, 5 min at 85 °C, and maintained at 4 °C. Selected miRNAs were confirmed by SYBR Green I dye (Takara, Dalian, China) with an ABI PRISM 7300 Real-time PCR System (Applied Biosystems, USA) under 95 °C for 3 min, followed by 40 cycles of 95 °C for 12 s, and 62 °C for 40 s. This analysis was limited to miRNAs altered significantly.
Cel-miRNA-39-3p, a non-human miRNA, was spiked into RNA samples as a control for extraction and amplification steps. MiR-191-5p and miR-16 were used as internal reference for normalization. MiRNA-16 was used for normalization of serum samples based on previous publication 22, and miR-191-5P was used for normalization of serum samples following the miRNA PCR arrays.
Data analysis
Initial data analysis was performed using the software supplied with the real-time PCR instrument to obtain raw Ct values. Relative expression of miRNA was calculated by the 2-ΔΔCt formula, in which ΔCt = Ct(miRNA) - Ct (average of house-keeping miRNAs) and ΔΔCt = ΔCt(Dengue patients) - ΔCt(Healthy controls). Subsequently, the relative quantification value underwent log 2 transformation to compare the expression levels of candidate miRNAs between healthy controls and patients. Data were expressed as mean ± SD. Statistical significance of the data was evaluated using one-way ANOVA via SPSS software. P < 0.05 was considered statistically significant. Receiver operating characteristic (ROC) curve analysis was performed for selected miRNAs. In addition, area under the curve (AUC) values and 95% confidence intervals (CIs) were calculated to evaluate the specificity and sensitivity for detecting dengue infection. Correlation and significance analysis were conducted by SPSS software, and P < 0.05 was considered statistically significant.
Target prediction and functional analysis in silico
MiRecords (http://c1.accurascience.com/miRecords/prediction_query.php) was selected to collect the validated targets of the selected miRNAs or predicted targets if no targets were validated. MiRecords is an integration platform of miRNA target prediction produced by 11 established miRNA target prediction programs:DIANAmicroT,MicroInspector, miRanda, MirTarget2,miTarget, NBmiRTar, PicTar, PITA,RNA22, RNAhybrid, and TargetScan/TargertScanS) to guarantee good specificity for target prediction. The functional analysis in silico about the validated or predicted targets of selected miRNA was performed on STRING 10 (http://string-db.org/).
Results
Sample characteristics
Six serum samples (3 controls and 3 patients) were enrolled in this study for miRNA arrays. Meanwhile, 72 serum samples (32 controls and 40 patients) were enrolled for q-PCR verification. Age and sex distribution of the two groups showed no statistically significant differences (P > 0.05, Table 1). However, serum aspartate transarninase (AST) and alanine aminotransferase (ALT) activities, as well as C-response protein (CRP) levels, were significantly increased in dengue-infected patients, whereas the number of white blood cells (WBC), platelets (PLT), neutrophils (NEU), and lymphocytes were significantly reduced in dengue-infected patients compared with healthy controls. No other infectious diseases were involved. Moreover, these patients belonged to newly diagnosed cases and were not subjected to any drug treatment. The results indicated that dengue-infected patients showed acute inflammatory responses in organs or tissues.
Table 1.
Basic characteristics of healthy controls and patients enrolled in the study.
| Sample Characteristics | miRNA PCR Array | PCR Validation | ||
|---|---|---|---|---|
| Controls | Patients | Controls | Patients | |
| Number | 3 | 3 | 32 | 40 |
| Genotype(DENV-1/other) | No | 3/0 | No | 37/2 |
| DENV Ct | No Ct | 20.65±4.84 | No Ct | 28.62±4.83 |
| DENV-1 Ct | No Ct | 20.88±1.49 | No Ct | 29.86±5.65 |
| Sex (male/female) | 3/0 | 3/0 | 10/22 | 20/20 |
| Age (Mean ± SD) | 28.3±6.4 | 29.3±8.7 | 36.6±8.5 | 40.7±12.7 |
| ALT(U/L) | 20.3±8.6 | 31.3±6.7 | 19.4±9.8 | 61.9±54.1 |
| AST(U/L) | 21.3±0.6 | 50.3±16.2 | 19.3±4.1 | 89.9±103.2 |
| WBC ( 10E9/L) | 6.92±0.42 | 3.97±1.43 | 7.27±2.13 | 2.88±1.59 |
| LYM (10E/L) | 2.45±0.0.21 | 0.56±0.49 | 2.11±0.49 | 0.62±0.25 |
| NEU (10E9/L) | 3.74±0.27 | 2.65±1.18 | 4.59±2.05 | 1.78±1.48 |
| PLT (10E9/L) | 237±81.2 | 105±21.9 | 262±72 | 119±78.1 |
| HCT (%) | 47.5±3.0 | 41.3±4.8 | 41.2±4.8 | 41.7±5.0 |
| CRP(mg/L) | 0.51±0.45 | 17.47±10.18 | 2.23±4.8 | 5.5±7.43 |
| Infectious Diseases | No | DENV Only | No | DENV Only |
| Anti-virus treatment | No | No | No | No |
Global analysis of serum miRNA expression profiles by miRNA PCR array
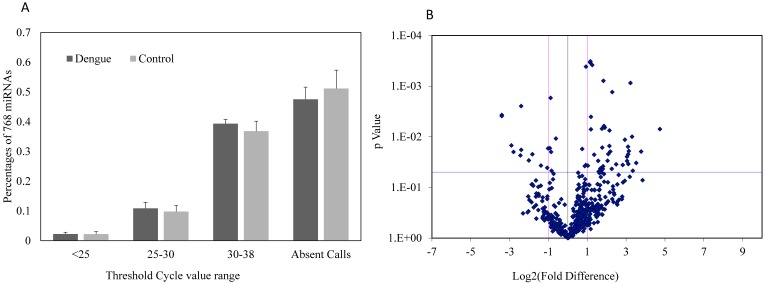
Expressions of 752 miRNAs were conducted using miRNA PCR panels to obtain a broad profile of relative miRNA expression levels in sera between dengue patients and healthy controls. Approximately 48.5% of the 752 miRNAs were undetectable in dengue patients, whereas about 52.3% of miRNAs were undetectable in healthy controls (Fig. 1A). No significant differences were found in the detectable or undetectable serum miRNA number observed between dengue patients and healthy control (n=3). However, volcano plots of this miRNA PCR panel showed significant fold change differences in the serum miRNA levels between dengue patients and healthy controls (Fig. 1B). The majority of miRNAs with significant fold changes (P<0.05) showed an increase in dengue patients compared with healthy controls.
Figure 1.
(A) Percentage composition of 752 miRNAs with different threshold cycle ranges (Ct values); (B) Fold difference and significance levels with statistics in sera of dengue patients and healthy controls using miRNA PCR array (n=3).
Aberrantly expressed miRNAs associated with dengue infection were defined to meet the following requirements: 1) Ct values <35 either in patients or controls to ensure stable detection; 2) relative fold change ≥2 (upregulation) or ≤-2 (downregulation) between the patient and control groups; and 3) acceptable statistical significance levels for fold change, p values <0.05. After screening, 41 upregulated and 12 downregulated miRNAs met the aforementioned conditions and might be associated with dengue infection (Tables 2 and 3).
Table 2.
Upregulated miRNA in sera of dengue patients compared with healthy control (n=3).
| miRNA ID | Fold Difference | P value | miRNA ID | Fold Difference | P value |
|---|---|---|---|---|---|
| hsa-miR-1972 | 26.7 | 0.0071 | hsa-miR-424-5p | 4.1 | 0.0298 |
| hsa-miR-518a-3p | 13.7 | 0.0196 | hsa-miR-335-5p | 3.7 | 0.0066 |
| hsa-miR-1538 | 11.5 | 0.0329 | hsa-miR-548a-3p | 3.6 | 0.0061 |
| hsa-miR-300 | 10.2 | 0.0471 | hsa-miR-320d | 3.6 | 0.0008 |
| hsa-miR-21-5p | 9.9 | 0.0100 | hsa-miR-1203 | 3.5 | 0.0383 |
| hsa-miR-589-5p | 9.4 | 0.0009 | hsa-miR-148b-3p | 3.5 | 0.0433 |
| hsa-miR-1537-3p | 8.8 | 0.0192 | hsa-miR-22-5p | 3.4 | 0.0164 |
| hsa-miR-340-5p | 8.7 | 0.0346 | hsa-miR-338-5p | 3.4 | 0.0069 |
| hsa-miR-1207-5p | 8.6 | 0.0222 | hsa-miR-1271-5p | 3.2 | 0.0455 |
| hsa-miR-632 | 8.6 | 0.0157 | hsa-miR-497-5p | 3.2 | 0.0425 |
| hsa-miR-934 | 8.3 | 0.0300 | hsa-miR-551a | 3.1 | 0.0496 |
| hsa-miR-1227-3p | 8.3 | 0.0248 | hsa-miR-362-3p | 3.0 | 0.0487 |
| hsa-miR-152-3p | 7.7 | 0.0114 | hsa-miR-223-5p | 3.0 | 0.0300 |
| hsa-miR-596 | 7.1 | 0.0361 | hsa-miR-708-3p | 2.6 | 0.0226 |
| hsa-miR-149-3p | 5.6 | 0.0351 | hsa-miR-339-3p | 2.4 | 0.0004 |
| hsa-miR-629-5p | 4.9 | 0.0013 | hsa-miR-425-3p | 2.3 | 0.0040 |
| hsa-miR-188-5p | 4.8 | 0.0431 | hsa-miR-877-5p | 2.3 | 0.0071 |
| hsa-miR-23a-5p | 4.6 | 0.0196 | hsa-miR-28-3p | 2.2 | 0.0003 |
| hsa-miR-590-5p | 4.4 | 0.0075 | hsa-let-7d-3p | 2.2 | 0.0003 |
| hsa-let-7g-3p | 4.4 | 0.0152 | hsa-miR-185-3p | 2.0 | 0.0373 |
| hsa-miR-421 | 4.2 | 0.0220 |
Table 3.
Downregulated miRNA in sera of dengue patients compared with healthy control (n=3).
| miRNA ID | Fold Difference | P value | miRNA ID | Fold Difference | P value |
|---|---|---|---|---|---|
| hsa-miR-200a-3p | -10.5 | 0.0037 | hsa-miR-885-5p | -4.0 | 0.0293 |
| hsa-miR-375 | -7.5 | 0.0148 | hsa-miR-942-5p | -3.5 | 0.0222 |
| hsa-miR-370-3p | -6.9 | 0.0200 | hsa-miR-146a-5p | -3.4 | 0.0039 |
| hsa-miR-20b-5p | -5.4 | 0.0234 | hsa-miR-379-5p | -2.6 | 0.0371 |
| hsa-let-7b-5p | -5.3 | 0.0025 | hsa-miR-769-5p | -2.1 | 0.0412 |
| hsa-miR-365a-3p | -5.3 | 0.0181 | hsa-miR-409-3p | -2.0 | 0.0170 |
Confirmation of miRNA array data by qRT-PCR
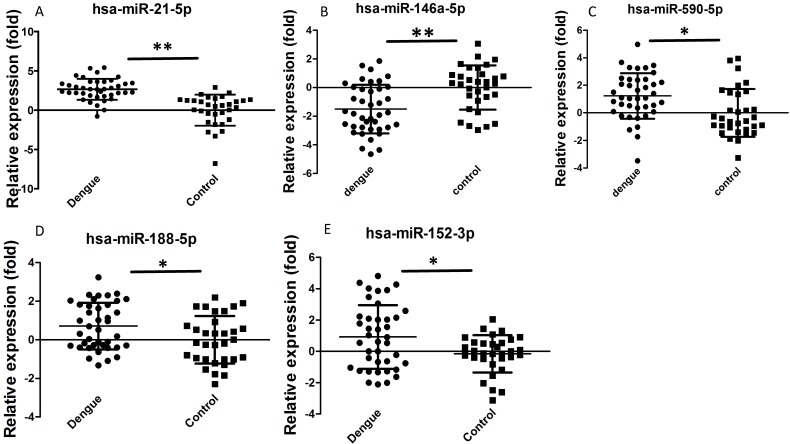
Dengue-infected patients showed acute inflammatory responses, such as reduced WBC, NEU, or lymphocyte number. We selected some miRNAs for further validation based on previous studies and bioinformatics analysis to determine whether these miRNAs are involved in the dengue pathological process. Hsa-miR-21-5p can regulate inflammation and cell proliferation 23. Hsa-miR-146a-5p is mainly associated with anti-inflammation function 24. Hsa-miR-590-5p is also related with inflammation and cell proliferation 25. Hsa-miR-188-5p 26 and hsa-miR-152-3p 27 inhibit cell proliferation. These functions might be useful to elucidate why dengue patients manifest increased inflammation and reduced WBC, NEU, and lymphocyte number. In addition, these miRNAs showed good Ct values (average Ct value<35) and were easily detected in sera. In this study, validation results by qRT-PCR showed that hsa-miR-21-5p, hsa-miR-590-5p, hsa-miR-188-5p, and hsa-miR-152-3p were significantly increased in dengue patients, whereas hsa-miR-146a-5p was significantly decreased (Fig. 2). Statistical analysis showed that these differences were significant (P<0.05). Hsa-miR-21-5p in sera of dengue-infected patients showed the best change compared with healthy controls.
Figure 2.
Relative expression of selected miRNA in sera of dengue patients (n=40) and healthy controls (n=32) validated by qRT-PCR method. **P<0.01, *P<0.05 following Student t-test between dengue patients and healthy controls.
Diagnostic potential of miRNAs
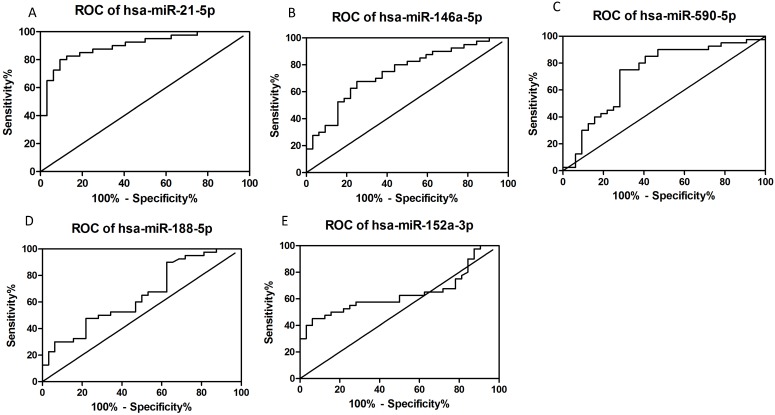
ROC curves were plotted according to the software of GraphPad Prism 5 to explore the diagnostic potential of verified miRNAs for dengue infection (Figs. 3A-E). The AUCs for hsa-miR-21-5p, hsa-miR-146a-5p, hsa-miR-590-5p, hsa-miR-188-5p, and miR-152-3p were as follows: 0.9008 (95% CI: 0.8303-0.9712, P<0.05), 0.7422 (95% CI: 0.6282-0.8561, P<0.05), 0.7219 (95% CI: 0.5974-0.8464, P<0.05), 0.6488 (95% CI: 0.5213-0.7764, P<0.05), and 0.6371 (95% CI: 0.5064-0.77678, P<0.05), respectively. Thus, hsa-miR-21-5p and hsa-miR-146a-5p seemed to have better diagnostic potential than the others.
Figure 3.
Constructed ROC curves to evaluate the diagnostic potential of serum miRNAs for dengue infection (n=72).
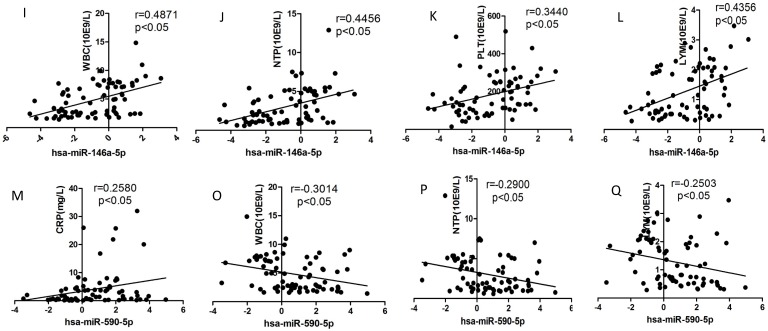
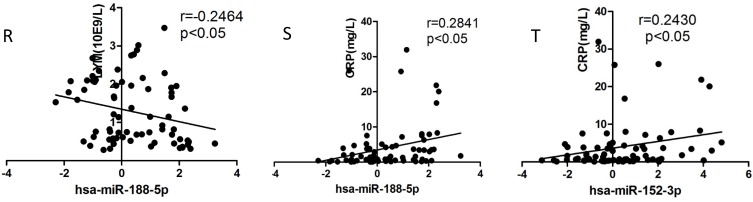
Correlation analysis
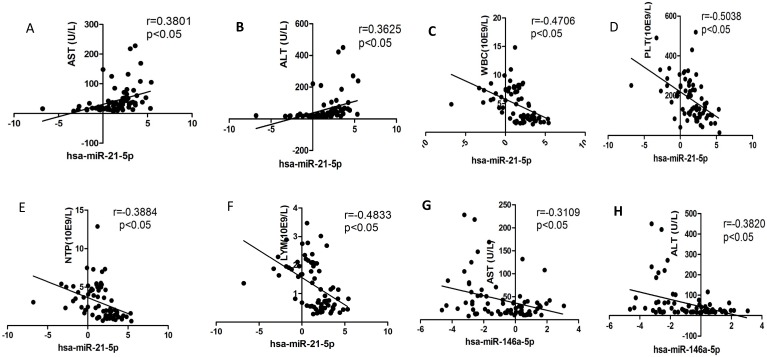
We selected the aforementioned miRNAs for further correlation analysis among clinical parameters because of their preferable diagnostic potential. Serum AST and ALT activities were significantly increased, whereas WBC, PLT, and NEU numbers significantly decreased in dengue patients compared with healthy controls. Interestingly, hsa-miR-21-5p showed positive correlation with serum AST and ALT activities but negative correlation with WBC, PLT, NEU, and lymphocyte numbers (Figs. 4A-F). However, hsa-miR-146a-5p showed negative correlation with serum AST and ALT activities, but positive correlation with WBC, PLT, NEU, and lymphocyte numbers (Figs. 4G-L). Hsa-miR-590-5p showed negative correlation with serum CRP level but positive correlation with WBC, NEU, and lymphocyte numbers (Figs. 4M-Q). Hsa-miR-188-5p and hsa-miR-152-3p showed positive correlation with serum lymphocyte number and CRP level (Figs. 4R-S). Hsa-miR-152-3p showed a positive correlation only with serum CRP levels (Fig. 4T). These results indicated that these miRNA might play key roles in regulating inflammatory response and dysfunction of blood cells.
Figure 4.
Correlation analysis between serum miRNAs and aspartate transarninase (AST) and alanine aminotransferase (ALT) activities, C-response protein (CRP) levels and the number of white blood cell (WBC), platelet (PLT), lymphocytes, and neutrophils (NEUs) in dengue patients and healthy controls (n=72).
Functional analysis in silico
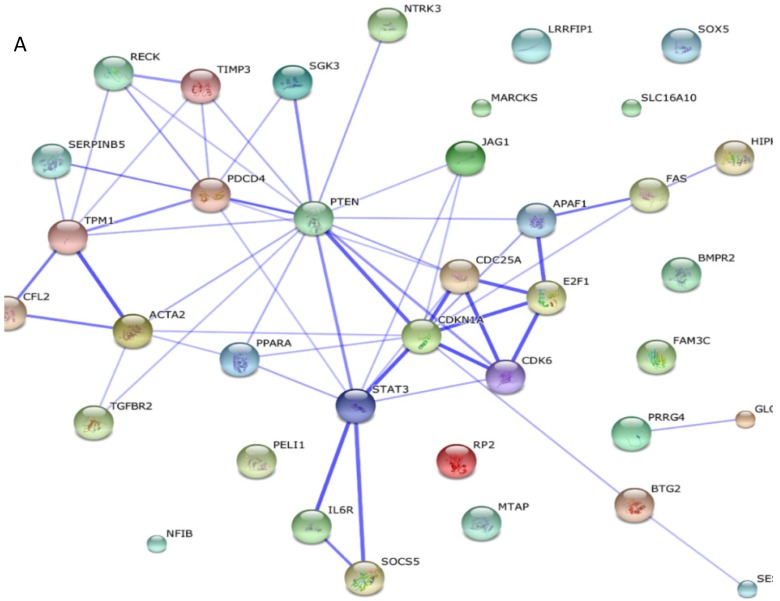
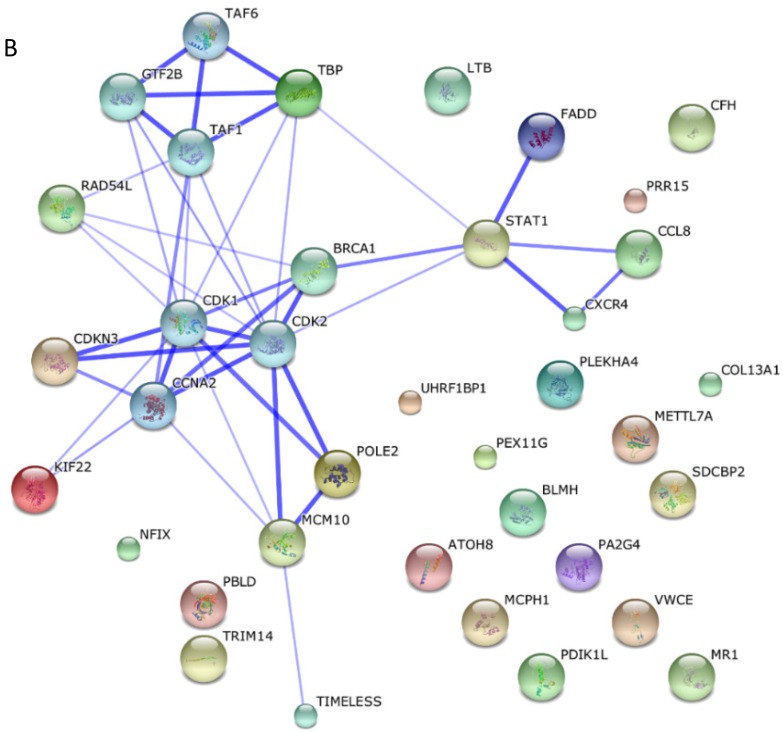
Among these miRNAs, miR-21 and miR-146a have well-defined functions. Hsa-miR-21-5p has 40 validated target proteins (Fig. 5A). These proteins form a network of interaction and may involve cell proliferation, motility, migration, locomotion, and inflammation by microRNA in cancer, p53 signaling pathway, pathway in cancer, and hepatitis B pathways (KEGG pathways). Hsa-miR-146a-5p has 35 validated target proteins (Fig. 5B). These proteins also form a network of interaction and may involve cell chemotaxis and inflammation by hepatitis B, herpes simplex infection, and chemokine signaling pathways (KEGG pathways). We predicted the targets of hsa-miR-590-5p, hsa-miR-188-5p, and hsa-miR-152-3p using the website of http://c1.accurascience.com/miRecords/. In the website, the selected targets should be predicted by at least 5 different miRNA software. We conducted functional analysis of the predicted targets of hsa-miR-590-5p, hsa-miR-188-5p, and hsa-miR-152-3p through STRING 10 (http://string-db.org). Hsa-miR-590-5p might involve Jak-STAT signaling pathway and inflammatory bowel disease. Hsa-miR-188-5p might invoke MAPK signaling pathway and microRNA in cancer. Hsa-miR-152-3p might involve TGF-beta signaling pathway and proteoglycans in cancer pathway. However, the majority of the predicted targets of hsa-miR-590-5p, hsa-miR-188-5p, and hsa-miR-152-3p require further validation. Nevertheless, these validated or predicted targets might indicate that the tested miRNAs mediate the inflammatory stress or cell survival and account for the complications of dengue infection.
Figure 5.
Interactions of targeting proteins of (A) hsa-miR-21-5p and (B) hsa-miR-146a-5p.
Discussion
Dengue is caused by mosquito-derived virus. This disease can lead to severe hemorrhagic fever or even lethal shock. Effective biomarkers that mirror the pathological process of dengue infectious should be established. A mature method for identifying dengue patients has been established. However, numerous problems in dengue identification, such as time-consuming or false negative/positive results, remain. Thus, more unique assay methods as alternatives for the identification of dengue patients should be formulated. Nevertheless, dengue-infected patients usually show significant reduction in the number of WBC, PLT, and NEUs 1, 28. The mechanisms that mediate these changes remain unclear 29. Circulating miRNAs can be released from various organs or tissues 30. When these organs or tissues are in a pathological state, circulating miRNA can vary with this change. These released miRNAs can form vesicles and prevent degradation by nuclease 30. Thus, these miRNAs can remain stable in serum or other body fluids. Emerging evidence showed that circulating miRNAs can serve as good biomarkers for various diseases 12, 31, 32. In this study, we first conducted serum miRNA PCR array and found many interesting miRNAs with significant differences between dengue patients and healthy controls. Approximately half of the 752 miRNAs can be effectively detected in sera, which is consistent with the results of a previous study in which half one miRNAs can be detected 21. Furthermore, a total of 53 aberrantly expressed miRNA were observed in sera of dengue-infected patients, which indicated that dengue patients showed a broad profile of serum miRNA dysfunction. These findings of dysregulated miRNAs in sera might supply important hints not only for effective biomarkers but also for elucidating the pathological process of dengue infection.
Thus, we selected hsa-miR-21-5p, hsa-miR-146a-5p, hsa-miR-590-5p, hsa-miR-188-5p, and hsa-miR-152-3p for further validation using qRT-PCR. These miRNAs were selected because of the following factors: 1) remarkable differences (more than 2-fold or less than 0.5) between the two groups; 2) good Ct values (<35), which indicates that they might be highly abundant and can be easily assayed in sera; 3) most importantly, their association with inflammatory responses or WBC death or survival frequently observed in dengue patients based on bioinformatics analysis or published data. This characteristic might supply a key reference for the pathological process of dengue infection. ROC curve analysis showed that hsa-miR-21-5p and hsa-miR-146a-5p could well distinguish between dengue patients and healthy controls. Furthermore, hsa-miR-21-5p and hsa-miR-146a-5p had highly negative or positive correlation with the number of WBC, PLT, NEUs and lymphocytes, as well as serum ALT and AST activities, in newly diagnosed patients. Thus, these results indicate that miRNAs may serve as early biomarkers in dengue-infected patients and also likely function in the development of the disease and its complication, e.g., liver inflammation. However, exact mechanism of these miRNA-mediated dengue infection and complications need to be further elucidated.
MiR-146a basically plays an important role in regulating innate immune, inflammatory response, and virus infection 33. MiR-146a exerts inflammation-regulating effects by targeting inflammatory signaling pathway 34. However, the promoter of miR-146a gene has several transcription factor NF-kappaB (NF-κB) binding sites, and LPS, IL-1β, and TNF-α can induce expression of miR-146a 35. Thus, miR-146a participates in the development of endotoxin tolerance 36. Virus can also induce the miR-146a expression through NF-kB binding site in the miR-146a promoter 37. Dengue virus infection significantly induced the expression of miR-146a in primary monocytes and THP-1 cells in vitro 19. In this study, miR-146a apparently exhibited decreased extracellular secretion, which remains to be further investigated. Nevertheless, good negative correlation between miR-146a and serum AST/ALT were observed in dengue-infected patients, suggesting that miR-146a might mediate the development of liver inflammation. However, the physiological or pathological significance of reduced miR-146a in the serum of dengue-infected patients remain ambiguous. MiR-21 plays a key role in the resolution of inflammation and negative regulation of pro-inflammatory response 38. Many inflammatory stimuli can trigger miR-21 induction itself. Hence, miR-21 can serve as anti-inflammatory response to balance the inflammatory stimuli. In addition to the inflammatory regulation, miR-21 promotes cell invasiveness and acts as oncomiRs 39 by targeting RECK. However, WBCs, lymphocytes, and NEUs were significantly reduced, indicating that WBCs, NEUs, or lymphocytes may rapidly migrate into extra-vascular tissues or organs after dengue virus invasion. MiR-590-5p promotes cell proliferation and invasion 40 and is also linked to inflammatory and immune responses. MiR-188 and miR-152 are apparently associated with the inhibition of cell proliferation and invasion 40, 41. Increased miR-188 and miR-152 might inhibit the proliferation or production of WBCs, NEUs, or lymphocytes. Therefore, these results indicated that miR-146a and miR-21 may regulate inflammation response. MiR-21 and miR-590 may promote cell proliferation, whereas miR-188 and miR-152 inhibit cell proliferation. These miRNAs might form a complex network involving the delicate regulation of inflammatory stress and immune response induced by dengue viruses. These virus-stressed miRNAs indicated that they balance inflammatory or other pathological stress. Many viruses can hijack a host cell's machinery and evade antiviral immune responses by miRNA mechanisms 42. However, these functional analyses are conducted in silico. Further experimental studies should be conducted to confirm the functions and reasons for these dysregulated serum miRNAs.
MiRNAs derived from cell cytoplasm and cell-free miRNAs can be released from cell cytoplasm to extracellular fluid by forming vesicles 43. These cell-free miRNAs were relatively stable, which can well serve as new biomarkers. These miRNAs also play a key role in the regulation of physiological and pathological states in vivo. These secreted cell-free miRNAs can be taken up by other cells, so they can mediate cell-cell communications 44. However, the roles of these dysfunctions of serum miRNA in the pathological process of dengue infection or other diseases are still undefined, although we have conducted functional analysis in silico. Nevertheless, these serum miRNAs have been identified as promising biomarkers for early evaluation in dengue-infected patients.
Conclusion
In conclusion, miRNA PCR array assays revealed differential expression of 41 upregulated and 12 downregulated miRNAs in sera of dengue patients compared with healthy controls. Among these miRNAs, serum hsa-miR-21-5p, hsa-miR-146a-5p, hsa-miR-590-5p, hsa-miR-188-5p, and hsa-miR-152-3p were identified as potential non-invasive molecular markers for detecting dengue infection. Upregulation of serum hsa-miR-21-5p and downregulation of hsa-miR-146a-5p may serve as interesting biomarkers for early inflammation responses or other pathological process in DENV-1 patients. However, the biological functions of these miRNAs require further investigation.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81373460 and 81072680), Shenzhen Science and Technology R&D Foundation (Grant Nos. SGLH20121008144756945 and ZYC201105170341A), Natural Science Foundation of Guangdong Province (2014A030313744), China Scholarship Council (201308440130), ITF Grant (GHX/002/12SZ), State Quality Inspection Administration (2014IK042), and Program for Tackling Key Problems of the People's livelihood Science and Technology of Guangzhou Haizhu District (2013CG31).
Abbreviations
- DENV
dengue virus
- DENV-1
dengue virus type 1
- WBC
white blood cell
- ALT
alanine aminotransferase
- AST
aspartate transarninase
- NEU
neutrophil
- PLT
platelet
- HCT
hematocrit
- CRP
C-reactive protein
- LYM
lymphocyte count
- HCV
hepatitis c virus.
References
- 1.Guzman MG, Harris E. Dengue. Lancet. 2015;385:453–65. doi: 10.1016/S0140-6736(14)60572-9. [DOI] [PubMed] [Google Scholar]
- 2.Messina JP, Brady OJ, Pigott DM. et al. The many projected futures of dengue. Nat Rev Microbiol. 2015;13:230–9. doi: 10.1038/nrmicro3430. [DOI] [PubMed] [Google Scholar]
- 3.Fry SR, Meyer M, Semple MG. et al. The diagnostic sensitivity of dengue rapid test assays is significantly enhanced by using a combined antigen and antibody testing approach. PLoS Negl Trop Dis. 2011;5:e1199. doi: 10.1371/journal.pntd.0001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waggoner JJ, Abeynayake J, Sahoo MK. et al. Single-reaction, multiplex, real-time rt-PCR for the detection, quantitation, and serotyping of dengue viruses. PLoS Negl Trop Dis. 2013;7:e2116. doi: 10.1371/journal.pntd.0002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teoh BT, Sam SS, Tan KK. et al. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification. J Clin Microbiol. 2015;53:830–7. doi: 10.1128/JCM.02648-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–62. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 8.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 9.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell cycle. 2005;4:1179–84. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 11.Pekarsky Y, Calin GA, Aqeilan R. Chronic lymphocytic leukemia: molecular genetics and animal models. Curr Top Microbiol Immunol. 2005;294:51–70. doi: 10.1007/3-540-29933-5_4. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Li L, Zhou Z. et al. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med Microbiol Immun. 2012;201:389–95. doi: 10.1007/s00430-011-0223-0. [DOI] [PubMed] [Google Scholar]
- 13.Song L, Liu H, Gao S. et al. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J Virol. 2010;84:8849–60. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z, Qi Y, Ge A. et al. Comprehensive characterization of serum microRNA profile in response to the emerging avian influenza A (H7N9) virus infection in humans. Viruses. 2014;6:1525–39. doi: 10.3390/v6041525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Li WX, Xie D. et al. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microrna in host gene expression. Plant cell. 2004;16:1302–13. doi: 10.1105/tpc.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hariharan M, Scaria V, Pillai B. et al. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–8. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 17.Reid G, Kirschner MB, van Zandwijk N. Circulating microRNAs: Association with disease and potential use as biomarkers. Crit Rev Oncol Hematol. 2011;80:193–208. doi: 10.1016/j.critrevonc.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Qi Y, Li Y, Zhang L. et al. microRNA expression profiling and bioinformatic analysis of dengue virusinfected peripheral blood mononuclear cells. Mol Med Report. 2013;7:791–8. doi: 10.3892/mmr.2013.1288. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, He L, Li Y. et al. miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J Infect. 2013;67:329–41. doi: 10.1016/j.jinf.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Tambyah PA, Chai S, Sepramaniam S. et al. ANNALS EXPRESS: microRNA expression in blood of dengue patients. Ann Clin Biochem; 2015. Aug 19. pii: 0004563215604001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Ouyang X, Jiang X. et al. Dysregulated Serum MicroRNA Expression Profile and Potential Biomarkers in Hepatitis C Virus-infected Patients. Int J Med Sci. 2015;12:590–8. doi: 10.7150/ijms.11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDermott AM, Kerin MJ, Miller N. Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PloS one. 2013;8:e83718. doi: 10.1371/journal.pone.0083718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Yang H, Ren L. MiR-21 promoted proliferation and migration in hepatocellular carcinoma through negative regulation of Navigator-3. Biochem Biophys Res Commun. 2015;464:1228–34. doi: 10.1016/j.bbrc.2015.07.110. [DOI] [PubMed] [Google Scholar]
- 24.Dalbeth N, Pool B, Shaw OM. et al. Role of miR-146a in regulation of the acute inflammatory response to monosodium urate crystals. Ann Rheum Dis. 2015;74:786–90. doi: 10.1136/annrheumdis-2014-205409. [DOI] [PubMed] [Google Scholar]
- 25.Shan X, Miao Y, Fan R. et al. MiR-590-5P inhibits growth of HepG2 cells via decrease of S100A10 expression and Iihibition of the Wnt pathway. Int J Mol Sci. 2013;14:8556–69. doi: 10.3390/ijms14048556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Lv Q, He J. et al. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun Signal. 2014;12:66. doi: 10.1186/s12964-014-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Song YX, Wang ZN. The microRNA-148/152 family: multi-faceted players. Mol Cancer. 2013;12:43. doi: 10.1186/1476-4598-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilman JM, De Wolff J, Beards GM. et al. Dengue fever: a Wikipedia clinical review. Open Med. 2014;8:e105–15. [PMC free article] [PubMed] [Google Scholar]
- 29.Diamond MS, Pierson TC. Molecular Insight into Dengue Virus Pathogenesis and Its Implications for Disease Control. Cell. 2015;162:488–92. doi: 10.1016/j.cell.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber JA, Baxter DH, Zhang S. et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta A, Nagilla P, Le HS. et al. Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1) PloS one. 2011;6:e22730. doi: 10.1371/journal.pone.0022730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Zhang ZN, Xu JJ, Fu YJ. et al. Transcriptomic analysis of peripheral blood mononuclear cells in rapid progressors in early HIV infection identifies a signature closely correlated with disease progression. Clin Chem. 2013;59:1175–86. doi: 10.1373/clinchem.2012.197335. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Chen XP, Li YJ. MicroRNA-146a and human disease. Scand J Immunol. 2010;71:227–31. doi: 10.1111/j.1365-3083.2010.02383.x. [DOI] [PubMed] [Google Scholar]
- 34.Nahid MA, Satoh M, Chan EK. Mechanistic role of microRNA-146a in endotoxin-induced differential cross-regulation of TLR signaling. J Immunol. 2011;186:1723–34. doi: 10.4049/jimmunol.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taganov KD, Boldin MP, Chang KJ. et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nahid MA, Pauley KM, Satoh M. et al. miR-146a is critical for endotoxin-induced tolerance: IMPLICATION IN INNATE IMMUNITY. J Biol Chem. 2009;284:34590–9. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motsch N, Pfuhl T, Mrazek J. et al. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4:131–7. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- 38.Sheedy FJ. Turning 21: Induction of miR-21 as a Key Switch in the Inflammatory Response. Front Immun. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reis ST, Pontes-Junior J, Antunes AA. et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012;12:14. doi: 10.1186/1471-2490-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang X, Xiang G, Wang Y. et al. MicroRNA-590-5p regulates proliferation and invasion in human hepatocellular carcinoma cells by targeting TGF-beta RII. Mol Cells. 2012;33:545–51. doi: 10.1007/s10059-012-2267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Z, Ma R, Tan W. et al. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp Mol Med. 2014;46:e112. doi: 10.1038/emm.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swaminathan G, Martin-Garcia J, Navas-Martin S. RNA viruses and microRNAs: challenging discoveries for the 21st century. Physiol Genomics. 2013;45:1035–48. doi: 10.1152/physiolgenomics.00112.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koberle V, Pleli T, Schmithals C. et al. Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PloS one. 2013;8:e75184. doi: 10.1371/journal.pone.0075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rayner KJ, Hennessy EJ. Extracellular communication via microRNA: lipid particles have a new message. J Lipid Res. 2013;54:1174–81. doi: 10.1194/jlr.R034991. [DOI] [PMC free article] [PubMed] [Google Scholar]