Summary
Non-human primate (NHP) models of tuberculosis (TB) immunity and pathogenesis, especially rhesus and cynomolgus macaques, are particularly attractive because of the high similarity of the human and macaque immune systems. However, little is known about the MHC class II epitopes recognized in macaques, thus hindering the establishment of immune correlates of immunopathology and protective vaccination. We characterized immune responses in rhesus macaques vaccinated against and/or infected with Mycobacterium tuberculosis (Mtb), to a panel of antigens currently in human vaccine trials. We defined 54 new immunodominant CD4+ T cell epitopes, and noted that antigens immunodominant in humans are also immunodominant in rhesus macaques, including Rv3875 (ESAT-6) and Rv3874 (CFP10). Pedigree and inferred restriction analysis demonstrated that this phenomenon was not due to common ancestry or inbreeding, but rather presentation by common alleles, as well as, promiscuous binding. Experiments using a second cohort of rhesus macaques demonstrated that a pool of epitopes defined in the previous experiments can be used to detect T cell responses in over 75% of individual monkeys. Additionally, 100% of cynomolgus macaques, irrespective of their latent or active TB status, responded to rhesus and human defined epitope pools. Thus, these findings reveal an unexpected general repertoire overlap between MHC class II epitopes recognized in both species of macaques and in humans, showing that epitope pools defined in humans can also be used to characterize macaque responses, despite differences in species and antigen exposure. The results have general implications for the evaluation of new vaccines and diagnostics in NHPs, and immediate applicability in the setting of macaque models of TB.
Keywords: Non-human primate, Epitopes, CD4+
1. Introduction
Biomedical research and vaccine testing depends in large part on the use of Non-human primate (NHP) models for testing and development of immunotherapeutic drugs, diagnostics and vaccines. In this context it is desirable to accurately measure immune responses by modern techniques, such as ELISPOT, intracellular cytokine staining (ICS) and tetramer staining assays. This quantification in turn requires definition of MHC class I and class II restricted epitopes recognized in the most widely used NHP species, such as the rhesus and cynomolgus macaques.
MHC class I and II genes from rhesus macaques of Indian and Chinese origin have been sequenced and several MHC molecules characterized for their binding specificity [1–39]. The specificity and epitope motifs associated with class I molecules expressed have been intensely studied, and revealed specific motifs, that in several cases overlapped in terms of epitope binding specificity with the motifs expressed by human HLA class I molecules [34,35,40]. However, in general the binding repertoires of rhesus and human class I appear to be for the most part distinct and non-overlapping [3,16,17,20,21,23,31,33,41,42]. By contrast, little is known about the specific epitopes presented by class II molecules expressed in NHP. In humans, a greater degree of promiscuity and repertoire overlap is associated with HLA class II molecules, as compared to class I molecules [43]. Until this study, it was not clear whether this repertoire overlap would also apply to NHP MHC class II, and whether any repertoire overlap could be exploited in practical terms for evaluation of NHP MHC class II restricted responses.
TB remains one of the greatest public health threats with over 2 billion people latently infected with Mycobacterium tuberculosis (Mtb) [44]. Approximately 10% of Mtb-infected individuals develop active TB, either primary disease or reactivation of latent infection [45–47]. Bacille Calmette–Guérin (BCG) is the only vaccine available against TB. BCG vaccination was developed about a century ago and its capacity to protect against TB is highly variable, with efficacy estimates ranging from 80% to no protection [48]. Development of alternative more efficacious vaccines is a complex task [49–54]. Significant hurdles include the complexity of efficacy trials that are of considerable duration [55], and since only a fraction of the individuals at risk do actually develop disease, require enrollment of large numbers of subjects [51].
In this context, reliable animal models to study TB pathogenesis, and to evaluate vaccine candidates and vaccination regimens are of significant importance. While humans are the only natural hosts of Mtb, several different animal models of TB vaccination have been extensively utilized, including murine, guinea pig, rabbits, cattle and NHPs [56–68]. Each animal species possesses advantages and caveats as each animal model reacts in its own particular fashion to Mtb infection [69] and differs in its ability to model human disease progression. Herein, we will point to some overarching characteristics of each. The murine model has advantages with its in-depth characterized immune system. However, mice are relatively resistant to infection with Mtb, and the disease process and pathology differs extensively [68]. Rabbits are also resistant to infection and only limited cytokine reagents are available [68,69]. Guinea pigs, on the other hand, are extremely susceptible to Mtb, as most infected animals succumb to disease, thus limiting the similarities to human disease process.
NHPs, e.g. rhesus and cynomolgus macaques, are appealing models of human disease and vaccination as the physiology, genetics, pathology and immunology are most similar to humans. In addition, there are many cross-reactive antibodies or macaque-specific antibodies, which allow for the characterization of specific cellular subsets, cytokines and chemokines [70,71]. The aerosol model of Mtb infection in rhesus macaques closely mimics human infections, allowing a more natural distribution of infecting bacilli onto the pulmonary surfaces [69,72]. Macaques develop latent infection characterized by the absence of any clinical signs of infection, similar to humans, but can be reactivated into an acute form [69]. However, despite their importance in Mtb infection [73–83], significant gaps in knowledge exist, particularly regarding CD4+ T cell responses. Only one Ag85B (Rv1886c)-specific macaque MHC class II tetramer has been described [84] and only two class II motifs has been characterized [9].
Thus, not surprisingly, the characterization of immune responses following Mtb infection and vaccination of macaques is incomplete. In this study, we characterized the MHC immunogenetic background and immunological response to antigens that have been used in human vaccine trials in NHPs that had been vaccinated against and/or infected with Mtb. We defined to an unprecedented level of detail epitopes recognized by macaque CD4+ T cells, including patterns of immunodominance and inferred restriction. The results suggest that while the CD4+ T cell response of NHPs to Mtb is broad and heterogeneous, epitope sets can be defined to broadly follow and characterize these responses in genetically heterogeneous cohorts of NHP. Furthermore, we reveal a striking repertoire overlap between epitopes recognized in NHPs and in humans, and show that epitopes previously defined in humans can be used to characterize NHP responses, despite these species differences, as well as, varying routes of antigen exposure. These results enable broad and effective characterization of immune responses in NHPs in general, and in particular in models of Mtb infection and vaccination.
2. Materials and methods
2.1. Infections and vaccinations of rhesus macaques
A total of 34 rhesus macaques were utilized in this study, 23 of which were infected and/or vaccinated as detailed in Table 1. All infections and vaccinations were performed and the animals housed in the Biosafety level 3 (BSL 3) Regional Biosafety Laboratory facility at Tulane National Primate Research Center in Covington, LA. The 23 animals described in Table 1 were vaccinated and challenged in support of two studies. All infected animals were positive for TST (tuberculin skin test) at weeks 3–4 post-experimental exposure. None of the naïve animals were positive for TST. Every animal prior to designation to these studies was checked and was TST negative. One of these studies was aimed at understanding the effectiveness of the Mtb:Δ-sigH mutant, delivered via aerosol, in the Mtb CDC1551 background [85,86] in response to lethal aerogenic challenge with homologous Mtb in rhesus macaques [140]. In this experiment, we used aerosolized BCG as a control. As shown by David Edwards and colleagues [87], BCG exhibits higher levels of protection when introduced via the inhalation route, relative to the intradermal route, in the context of guinea pigs. In a seminal study in 1973, aerosolized BCG was shown to be protective in monkeys [88]. Another study was aimed at assessing the effectiveness of a 3D-BCG recombinant strain, described by Douglas Kernodle and colleagues [89], where genes involved in both the secretion of antigens as well as anti-oxidant defense, including sigH, were deleted from the parental BCG strain; as a prime vaccine vehicle against aerogenic Mtb challenge in rhesus macaques. Animals that did not receive any immunization, were however challenged with comparable Mtb to serve as negative controls. Animals were euthanized and necropsied at least six weeks after the last infection. Spleen and whole blood samples obtained at necropsy were shipped to La Jolla Institute for Allergy and Immunology (LJI).
Table 1.
a. Immunization and infection regimens for Indian rhesus macaques that were initially tested with all pools of peptides from antigens.*
| Animal# | Vaccination | Booster | Infection | 2nd Infection | 3rd Infection |
|---|---|---|---|---|---|
| IB78 | 3D–BCG | MVA-Ag85B | Mtb CDC1551 | – | – |
| GP21, II69 | 3D–BCG | MVA control | Mtb CDC1551 | – | – |
| HN69, IJ42, HE06, II21 | BCG-Aero | – | Mtb | – | – |
| IJ23, IN85, IM73, CK53, II07 | SigH-Aero | – | Mtb | – | – |
| GI89, II43 | – | – | Mtb-Mutant Pools | Mtb-Mutant Pools | SIV-Mac239 |
| HB95 | – | – | Mtb-Pool 1,2,3 | – | – |
| b. Immunization and infection regimens for newly tested Indian rhesus macaques. | |||||
|---|---|---|---|---|---|
| Animal# | Vaccination | Booster | Infection | Infection | Infection |
| GK87, CL10 | – | – | Mtb CDC1551 | – | – |
| CR57, EA97 | BCG-Aero | – | Mtb CDC1551 | – | – |
| FM29, DF44 | SigH-Aero | – | Mtb CDC1551 | – | – |
| HK78 | 3D–BCG | MVA-AG858 | Mtb CDC1551 | – | – |
| IL33 | BCG-Aero | – | Mtb | – | – |
| c. Negative control Indian rhesus macaques. | ||||
|---|---|---|---|---|
| Animal# | Peptide pool tested | Vaccination | Booster | Infection |
| EA43, FD79, EP16 | Pool of 300 | None | None | None |
| IB62, DR27, HE68, FT88, IM14, HT41, CT52, BK01 |
Pool of 54 and 300 | None | None | None |
Each of the animals was infected with Mtb CDC1551. The animals were either vaccinated and/or immunized or considered a negative control. Immunizations consisted of the recombinant 3D–BCG strain provided by Dr. Doug Kernodle. The animals vaccinated with the 3D–BCG strain were boosted six weeks later with either a control MVA or MVA:Ag85B and then challenged six weeks after the boost with Mtb CDC1551 to deliver 1000 cfu into the lungs of macaques (86). Aerosol vaccination with a single, one time dose of BCG (Danish) (130, 131) as well as Mtb:Δ-sigH (CDC1551 background) [140] were performed in order to deliver ∼5000 cfu of bacilli into the lungs of macaques. The animals were challenged 8 weeks after the single pulmonary vaccination with the identical dose of Mtb CDC1551 (1000 cfu delivered into the lungs of macaques). SIV co-infection was performed 9 weeks after initial low-dose (25–50 cfu delivered to the lungs of macaques) Mtb CDC1551 infection.
2.2. Infections of cynomolgus macaques
Blood samples from 16 cynomolgus macaques from Chinese breeding facilities (Macaca fasciularis) (Valley Biosystems, Sacramento CA) were included in this study. These macaques were Mtb-infected and enrolled in unrelated ongoing studies in the Flynn laboratory. These animals were not vaccinated, undergoing anti-mycobacterial treatments, or experiencing immunomodulatory therapies. The macaques were infected with low dose (<25 CFU) or moderate dose (60–150 CFU) M. tuberculosis strain Erdman via bronchoscope, as described [90]. Samples were chosen from 8 animals that eventually developed active TB and 8 animals that developed latent infection, according to previously described criteria [90]. All macaques were at least 4 years of age and between 4 and 8 kg. The blood draws used for the samples were obtained at 12 weeks post-infection. Cynomolgus macaques were housed and maintained by the University of Pittsburgh’s Department of Laboratory Animals. All procedures were performed in accordance with protocols approved by the University of Pittsburgh’s Institutional Animal Care and Use committee.
2.3. Cell isolation
PBMCs were obtained by density gradient centrifugation (Ficoll for rhesus macaques, Percoll for cynomolgus macaques) from whole blood [5] and spleens were homogenized to a single cell suspension. Cells were isolated according to standard protocols in originating labs. All isolated cells, consisting of PBMCs and splenocytes, were cryopreserved in liquid nitrogen [91].
2.4. Peptides
Sets of 15mer peptides overlapping by 10 residues, were synthesized by A&A (San Diego, CA). The antigens studied correspond to antigens currently considered as vaccine candidates, or included in the QuantiFERON-TB Gold In-tube test [as shown in Table 2; including Rv0125 (Mtb32a), Rv1813c, Rv0288 (TB10.4), Rv1886c (Ag85B), Rv1196 (Mtb39A), Rv2608 (PPE42), Rv3619c (EsxV), Rv3620c (EsxW), Rv3874 (CFP-10) and Rv3875 (ESAT6)]. Pools of previously defined human epitopes were generated based on the information described in Lindestam Arlehamn et al. [92]. These pools therefore do not represent the entire TB proteome. Specific information regarding the patients included in this study includes the following [92]:
Table 2.
List of Antigens tested in the present study.*
| Antigen | Rv# | Vaccine trial | Reference |
|---|---|---|---|
| Mtb32A | Rv0125 | Mtb72F | [131] |
| TB10.4 | Rv0288 | HyVac4, AERAS-402 | [132] |
| Mtb39A | Rv1196 | Mtb72F | [131] |
| Rv1813c | ID93 | [133,134] | |
| Ag85B | Rv1886c | H1, HyVac4, H56, AERAS-402 | [135,136] |
| Rv2608 | ID93 | [133,134] | |
| EsxV | Rv3619c | ID93 | [133,134] |
| EsxW | Rv3620c | ID93 | [133,134] |
| CFP10 | Rv3874 | ||
| ESAT-6 | Rv3875 | H1, H56 | [137–139] |
List of Tuberculosis antigens tested in this study. The antigens studied correspond to antigens currently considered as vaccine candidates, or included in the QuantiFERON-TB Gold In-tube test.
Human LTBI subjects (age range 20–65 years) had a history of a positive tuberculin skin test (TST). LTBI was confirmed by a positive QuantiFERON-TB Gold In-Tube (Cellestis), as well as a physical exam and/or chest X-ray that was not consistent with active tuberculosis. None of the study subjects endorsed vaccination with BCG, or had laboratory evidence of HIV or Hepatitis B.
2.5. Ex vivo IFN-γ ELISPOT assay
IFN-γ ELISPOT assays were performed as previously described [93,94]. Briefly, macaque PBMCs or splenocytes at a density of 2 × 106 cells/ml were stimulated with peptide pools (2–10 µg/ml) or individual peptides (20 µg/ml), ConA (20 µg/ml) or medium in flat-bottom 96-well nitrocellulose plate (Immobilon-P membrane; Millipore, Bedford, MA), coated with 15 µg/ml anti-human/monkey IFN-γ (GZ-4; Mabtech, Inc., Mairemont, OH). Each peptide or pool was tested in triplicate. After 20 h incubation at 37 °C, plates were washed with PBS/0.05% Tween 20 and incubated with 2 µg/ml biotinylated anti-human IFN-γ (7-B6-1; Mabtech) for 2 h at 37 °C. After additional washes with PBS-0.05% Tween 20, spots were developed using Vectastain ABC peroxidase (Vector Laboratories, Burlingame, CA) followed by 3-amino-9-ethylcarbazole solution (Sigma-Aldrich, St. Louis, MO) and counted by computer-assisted image analysis (Zeiss KS ELISPOT reader).
Responses are expressed as mean net spots per million cells for each peptide pool or individual peptide. Responses were considered positive if (i) the number of spot-forming cells (SFCs)/106 splenocytes or PBMCs exceeded the absolute value of the mean negative control wells (effectors without peptide) by twofold, (ii) the value exceeded 20 SFCs/106 splenocytes or PBMCs, and (iii) were significantly different from the negative (no peptide) control by either a Poisson distribution test or a Student’s t-test with p ≤ 0.05.
The response frequency was calculated by dividing the no. of donors responding with the no. of donors tested. The magnitude of response (total SFC) was calculated by summation of SFC from responding donors [92].
2.6. MHC sequencing
Genomic DNA (gDNA) was extracted using a QIAmp blood mini kit from splenocytes from the rhesus macaque samples and used as templates for PCR with a series of primer mixes that allow for parallel amplification of MHC class I, DRB, DQA, DQB, DPA & DPB sequences with the Fluidigm Access Array. These amplicons targeted the highly polymorphic peptide binding domain encoded by exon 2 of the MHC class I and II loci. Next, the PCR products were purified using Ampure beads and sequenced using Illumina MiSeq. Mamu-A, Mamu-B, Mamu-DR, Mamu-DQ and Mamu-DP haplotypes were determined by aligning the resulting sequence reads from each animal against a custom database of macaque MHC class I and class II alleles sequences.
2.7. Relative frequency (RF) calculations to infer MHC restriction
Standard Odd Ratios and Relative Frequency (RF) allele frequency calculations were used to infer MHC restriction. RF values were calculated as the ratio of the response in donors expressing a specific allele to the response in all donors and was calculated according to the formula:
Where: A+ = Subject expresses a specific allele; A− = Subject not expressing the specific allele; R+ = Subject has a positive immune response against the specific peptide; R− = Subject does not have a positive immune response against the specific peptide; A+ R+ indicates number of Subjects expressing the specific allele AND having a positive response against a specific peptide. RF values greater > 1.0 indicate a positive association, i.e. expressing the specific allele increases the “odds” of having positive immune response. Significance is calculated by standard Fisher’s exact test.
2.8. Intracellular cytokine analysis
Splenocytes were stimulated for 6 h in complete RPMI medium at 37 °C in 5% CO2 in the presence of 2 µg/ml peptide pool and 4 µg/ ml Brefeldin A (last 4 h). Unstimulated cells were used to assess nonspecific/background cytokine production. After 6 h, cells were harvested and stained for cell surface markers anti-CD4-PE (L200), anti-CD3-BV421 (SP34-2, both from BD Biosciences), anti-CD8-APC (RPA-T8), and Live/Dead Aqua (both from eBioscience). After washing, cells were fixed and permeabilized using 4% paraformaldehyde (PFA) and saponin buffer and then stained for cytokine using anti- IFN-γ-FITC (4S.B3, eBiosciences). Samples were acquired on a BD LSR II flow cytometer. The frequency of cells responding to the Mtb-specific peptides was quantified by determining the total number of cytokine+ cells and background values subtracted (as determined from the medium alone control) using FlowJo software (Tree Star). Lymphocytes, gated based on forward and side scatter, were gated for live CD3+ T cells. Live CD3+ T cells were then gated based on CD4 and CD8 expression. Then the IFN-γ+ cells were identified from these populations.
3. Results
3.1. NHP ex vivo T cell responses to TB antigens used in human vaccine trials vary in magnitude and frequency
A set of fifteen rhesus macaques was utilized in a first survey of immune responses to TB antigens. These macaques were associated with a rather diverse immunization and infection regimens, reflecting cell sample availability, detailed in Table 1. These samples enable the characterization of responses in a diverse set of conditions. Splenocytes from each of the fifteen animals were tested in ex vivo IFN-γ ELISPOT against pools of overlapping peptides spanning various TB antigens currently being tested for vaccines in clinical trials or used in diagnostic assays (Table 2).
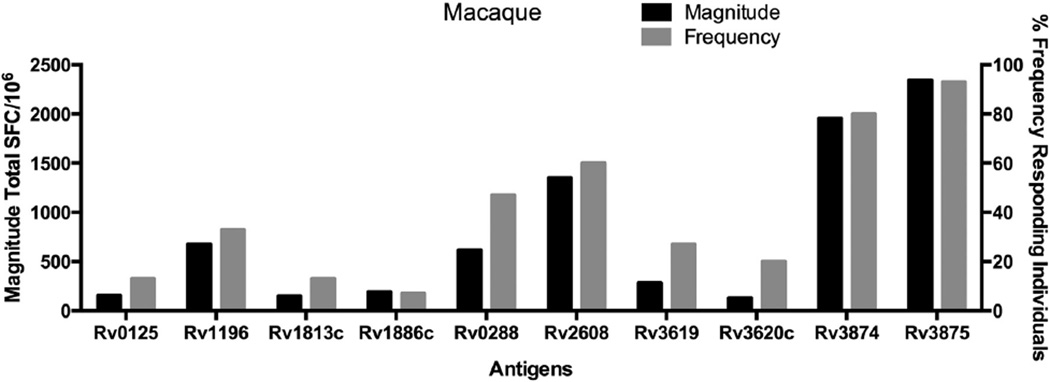
A wide spectrum of reactivity was apparent (Figure 1). Several antigens were immunodominant both in terms of total magnitude, measured by total SFC/106 across all animals, and frequency, measured by percentage of animals yielding positive responses, of responses. Rv3875 was the most dominant, with a total magnitude of 2343 SFC/106 and 93% of the animals responding to this antigen, followed by antigen Rv3874, with a total SFC/106 of 1955 and 80% of the animals responding. Rv2608, was associated with a magnitude of 1350 SFC/106 and a 60% response frequency. Two additional antigens, Rv0288 (613 SFC/106, 47%) and Rv1196 (673 SFC/106, 33%), were associated with intermediate immunogenicity. The remaining antigens had lower magnitudes and frequencies of less than 30%.
Figure 1.
T Cell Responses Detected for TB Antigens in Macaques. Splenocytes from 15 individual macaques (Table 1), were tested in ELISPOT assays to determine immunological responses to pools of overlapping peptides spanning ten common TB antigens. Splenocytes were incubated with 10 µg/ml per peptide, after which the number of IFN-γ+ producing cells were enumerated in an ELISPOT assay. The total magnitude of response, as measured by total spot forming cells (SFC) is shown on the left y-axis, depicted in black. Student’s T-Test, p < 0.05. The frequency of animals (right y-axis) that responded to each specific antigen is depicted in gray. Each individual peptide was tested in triplicate.
3.2. Identification of specific epitopes
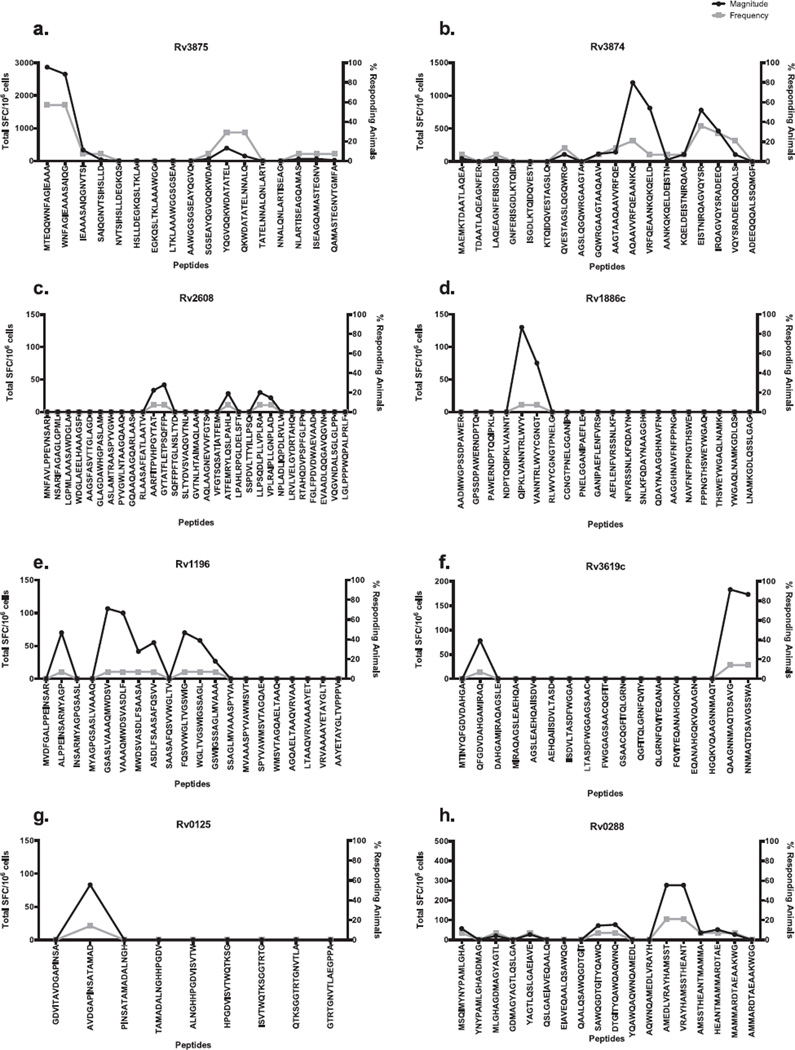
Next, positive pools were deconvoluted to identify individual epitopes. Figure 2a–h shows the magnitude and frequency of responses associated with each individual peptide contained within an antigen. Antigen Rv3875, which was the most immunogenic, contained two overlapping peptides, Rv38751–15MA15 and Rv38756–20WG15, with magnitudes of 2868 and 2655 SFC/106, respectively. These peptides were positive in 57% of the animals tested for this antigen. Overlapping peptides Rv387551–65YL15 and Rv387556–70QQ15 were positive in 29% of the animals tested with a magnitude of 392 and 150 SFC/106, respectively (Figure 2a).
Figure 2.
Epitope profiles for each of the antigens. Splenocytes from macaques (Table 1) were incubated with 20 mg/ml per peptide, after which the number of IFN-γ+ cells were enumerated in an ELISPOT assay. Each panel represents the peptides from the peptides from a unique antigen: a) Rv3875, b) Rv3874, c) Rv2608, d) Rv1886c, e) Rv1196, f) Rv3619c, g) Rv0125 and h) Rv0288. The total magnitude of response, as measured by total spot forming cells (SFC) is shown on the left y-axis, depicted in black. Student’s T-Test, p < 0.05. The x-axis represents the overlapping peptides tested (except for 2c, which depicts every other peptide). The frequency of animals (right y-axis) that responded to each specific antigen is depicted in gray. Each individual peptide was tested in triplicate.
A number of epitopes were also identified from antigen Rv3874 (Figure 2b), which also yielded a strong response with the corresponding peptide pools. The Rv387451–65AQ15 epitope was associated with positive responses in 21% of the subjects tested (total SFC/ 106 of 1198). The Rv387471–85ER15 epitope elicited a response in 36% of individuals with a magnitude of 780 SFC/106, and overlapped with Rv387476–90IQ15, which also elicited appreciable responses (465 SFC/106 in 29% of the animals tested).
In the case of antigen Rv2608, deconvolution experiments identified positive responses in two animals. Specifically, animal GP21 was positive for sequences Rv2608286–300PP15, Rv2608291–305AT15, Rv2608301–315GP15,, Rv2608326–340VM15, and Rv2608366–380RD15. Animal IN85 responded to peptides Rv2608486–500LA15, Rv2608496–510VD15, and Rv2608561–575DA15 (Figure 2c). The remainder of the antigens, including Rv1886 (Figure 2d), Rv1196 (Figure 2e), Rv3619c (Figure 2f), Rv0125 (Figure 2g) and Rv0288 (Figure 2h) yielded between 1 and 3 positive peptides, with magnitudes between 27 and 277 SFC/106, and response frequencies of 7%-21%. No specific peptide was identified from Rv1813c (not shown).
In summary, deconvolution of specific peptide pools identified 54 epitopes (Supplemental Table I). Elimination of redundant/ overlapping sequences by selecting the best response in the case of overlapping peptides, reduced this list to 37 epitopes. Of these, 13 were positive in two or more animals, and encompassed 81% of the overall sum of SFC per million, indicating their immunodominant nature (Table 3).
Table 3.
Epitopes detected in 2 or more animals.*
| Antigen | Position | Sequence | Magnitude | Frequency | #Animals |
|---|---|---|---|---|---|
| Rv0125 | 306–320 | AVDGAPINSATAMAD | 83.3 | 14% | 2 |
| Rv3619c | 76–90 | QAAGNNMAQTDSAVG | 183.3 | 14% | 2 |
| Rv3620c | 16–30 | AGRFEVHAQTVEDEA | 103.3 | 14% | 2 |
| Rv3620c | 61–75 | NQAFRNIVNMLHGVR | 80.0 | 14% | 2 |
| Rv3620c | 76–90 | DGLVRDANNYEQQEQ | 75.0 | 14% | 2 |
| Rv3874 | 31–45 | QVESTAGSLQGQWRG | 83.3 | 14% | 2 |
| Rv3874 | 51–65 | AQAAVVRFQEAANKQ | 1146.6 | 21% | 3 |
| Rv3874 | 71–85 | EISTNIRQAGVQYSR | 745.0 | 36% | 5 |
| Rv0288 | 61–75 | AMEDLVRAYHAMSST | 265.0 | 21% | 3 |
| Rv3620c | 6–20 | MTDPHAMRDMAGRFE | 81.7 | 21% | 3 |
| Rv3620c | 51–65 | ATSLDTMTQMNQAFR | 106.7 | 21% | 3 |
| Rv3875 | 1–15 | MTEQQWNFAGIEAAA | 2836.7 | 57% | 8 |
| Rv3875 | 51–65 | YQGVQQKWDATATEL | 375.0 | 29% | 4 |
Epitope responses detected in at least two animals are listed, with specific information including antigen source, position, as well as specific information about the response elicited, including total magnitude as measured by SFCs in ELISPOT, frequency of positive animals and specific numbers of animals responding to epitope.
3.3. Cohort inbreeding does not explain epitope dominance in the animal cohort
As presented above, a total of 13 epitopes were recognized in multiple animals. Frequently recognized peptides may be uniquely presented by certain MHC class II allele(s), abnormally frequent in the specific cohort tested, because of common pedigrees and inbreeding. To address this possibility we assessed the pedigree of these animals to determine the level of inbreeding in this particular cohort.
Review of pedigree information from the colony at Tulane Primate Center revealed only minor inbreeding. Specifically, only 3 pairs of animals shared a parent or grandparent: GI89 shares one grandparent and one parent with II21 (shown in red in Supplemental Table II); animals IM73 and HE06 both share a grandparent (shown in green in Supplemental Table II); IB78 and II69 both share a grandparent (shown in blue in Supplemental Table II). Of a total of 54 peptide/macaque combinations associated with the peptides,13 were recognized by multiple animals and only 8 were represented by instances in which the responding macaque was a relative of another macaque responding to the same peptide (Supplemental Table II). Hence these epitopes are not dominant because of inbreeding and common ancestry.
3.4. Inferred MHC alleles potentially restricting detected immune responses
Frequently recognized peptides may be uniquely presented by certain MHC class II allele(s) commonly expressed in the general macaque population. Alternatively, the more dominant epitopes might be restricted by more than one MHC class II molecule (promiscuous restriction). Indeed, in humans it has been reported that promiscuous epitopes account for a large fraction of responses [95], and promiscuous binding predictions have been used to perform genome-wide analysis of Mtb class II responses in humans [92]. To address these possibilities, we first determined which specific MHC class II allele(s) were most frequently expressed by next generation sequencing (Supplemental Table II). The frequency of the most common alleles is summarized in Table 4.
Table 4.
MHC class II alleles present in more than two animals.*
| Allele or cluster of alleles | Frequency |
|---|---|
| DRB1*03:03 | 33% |
| DRB1*03:09 | 20% |
| DRB1*04:06 | 20% |
| DRB*W21:04 | 20% |
| DQA1*26:g11 | 80% |
| DQA1*01:g2 | 40% |
| DQA1*01:g3 | 20% |
| DQA1*01:04 | 33% |
| DQB1*18:g1 | 60% |
| DQB1*06:01 | 40% |
| DQB1*06:g1 | 20% |
| DPA1*02:g2 | 40% |
| DPA1*02:g1 | 67% |
| DPA1*06 | 40% |
| DPB1*07:01 | 40% |
Allele nomenclature including a:g designation reflects that a unique allele was not identified, that this includes a cluster of alleles.
To gain more insight into potential allelic restriction for the epitopes recognized by multiple animals, we examined the relative frequency in which different alleles are expressed in animals that responded or did not respond to each epitope. Classical genetic calculations calculating Odds Ratios, Relative Frequencies (RF) and associated p values were utilized to infer MHC allelic variants potential restricting the epitope-specific responses. Four epitope/ MHC allele combinations with a significant association were revealed by this analysis (Table 5).
Table 5.
Predicted restrictions of TB epitopes in Macaques.*
| Antigen | Sequence | Allele | A+R+ | A−R+ | A+R− | A−R− | RF | P-value |
|---|---|---|---|---|---|---|---|---|
| Rv0288 | AMEDLVRAYHAMSST | DPA1*02:g2 | 3 | 0 | 3 | 9 | 2.5 | 0.044 |
| DPB1*07:01 | 3 | 0 | 3 | 9 | 2.5 | 0.044 | ||
| Rv3620c | ATSLDTMTQMNQAFR | DQB1*18:g2 | 2 | 1 | 0 | 12 | 5.0 | 0.029 |
| DRB*W1:g1 | 2 | 1 | 0 | 12 | 5.0 | 0.029 | ||
| DRB*W6:02 | 2 | 1 | 0 | 12 | 5.0 | 0.029 | ||
| DRB*W6:g2 | 2 | 1 | 0 | 12 | 5.0 | 0.029 | ||
| DRB1*03:g2 | 2 | 1 | 0 | 12 | 5.0 | 0.029 | ||
| Rv3874 | EISTNIRQAGVQYSR | DQB1*18:g1 | 5 | 0 | 4 | 6 | 1.7 | 0.044 |
| Rv3620c | MTDPHAMRDMAGRFE | DQB1*18:g2 | 2 | 1 | 0 | 12 | 5.0 | 0.029 |
| DRB*W1:g1 | 2 | 1 | 0 | 12 | 5.0 | 0.029 | ||
| DRB*W6:02 | 2 | 1 | 0 | 12 | 5.0 | 0.029 | ||
| DRB*W6:g2 | 2 | 1 | 0 | 12 | 5.0 | 0.029 | ||
| DRB1*03:g2 | 2 | 1 | 0 | 12 | 5.0 | 0.029 |
A+: genotyped HLA allele positive; A−: genotyped HLA allele negative; R+: epitope response positive; R−: epitope response negative. P value is calculated from Fisher exact test from A+R+, A−R+. A+R−, and A−R−.
The Rv387471–85ER epitope was exclusively recognized by animals expressing DQB1*18:g1. In total, five of nine animals expressing this allele responded. In contrast, none of the six animals that did not express DQB1*18:g1 responded to this peptide. Thus, this epitope is likely restricted by DQB1*18:g1 (RF = 1.7; p = 0.044) (Table 5). Similarly, the Rv028861–75AT epitope was predicted to be restricted to DPA1*02g2/B1*07:01. Three of the six animals expressing these alpha and beta DP chain alleles responded to the peptide, whereas none of remaining nine animals, which did not express this DP chain gene combination, responded to the epitope (RF = 2.5; p = 0.044) (Table 5).
By contrast, in the case of the Rv3620c31–65 AR and Rv3620c6–20ME epitopes, both animals expressing the haplotype DQB1*18:g2, DRB*W1:g1, DRB*W6:02, DRB*W6:g2, DRB1*03:g2 responded to these epitopes, and thus one of these alleles is likely to restrict the response (RF = 5.0; p = 0.029) (Table 5). However, these peptides are also recognized by an additional animal not carrying this haplotype or sharing any other MHC expressed in the other two animals, and therefore are associated, by definition, with promiscuous restriction.
We selected representative macaque alleles and performed a BLAST analysis (using the HLA/MHC specific tool http://www.ebi.ac.uk/ipd/imgt/hla/) to test whether the macaque alleles of interest would have a human match. The following macaque/human MHC allele pairs were found with the listed percentage identity in parenthesis: Mamu-DPA1*02:01/HLA-DPA1*01:03 (93%), Mamu-DPB1*07:01/HLA-DPB1*106:01 (93%), Mamu-DQB1*18:01/HLA-DQB1*03:03 (93%), Mamu-DQB1*18:04/HLA-DQB1*03:96 (92%) and Mamu-DRB*W1:01/HLA-DRB1*14:07 (91%), with an overall mean of 92% of all the pair of Mamu/HLA alleles.
3.5. The epitopes identified allow general coverage and analysis of T cell responses in rhesus macaques
The results presented above identify a total of 54 different peptide epitopes, recognized in a set of 15 genetically diverse animals. We hypothesized that these epitope set might allow uniform coverage of Indian rhesus macaques in general. Accordingly, we generated a pool of these epitopes and tested it with splenocytes derived from a set of 8 new macaques (Table 1), in which only one animal was related with one of the 15 animals used for epitope discovery and an additional 8 naïve animals (Table 1; Supplemental Table II) that had not been vaccinated or infected with Mtb.
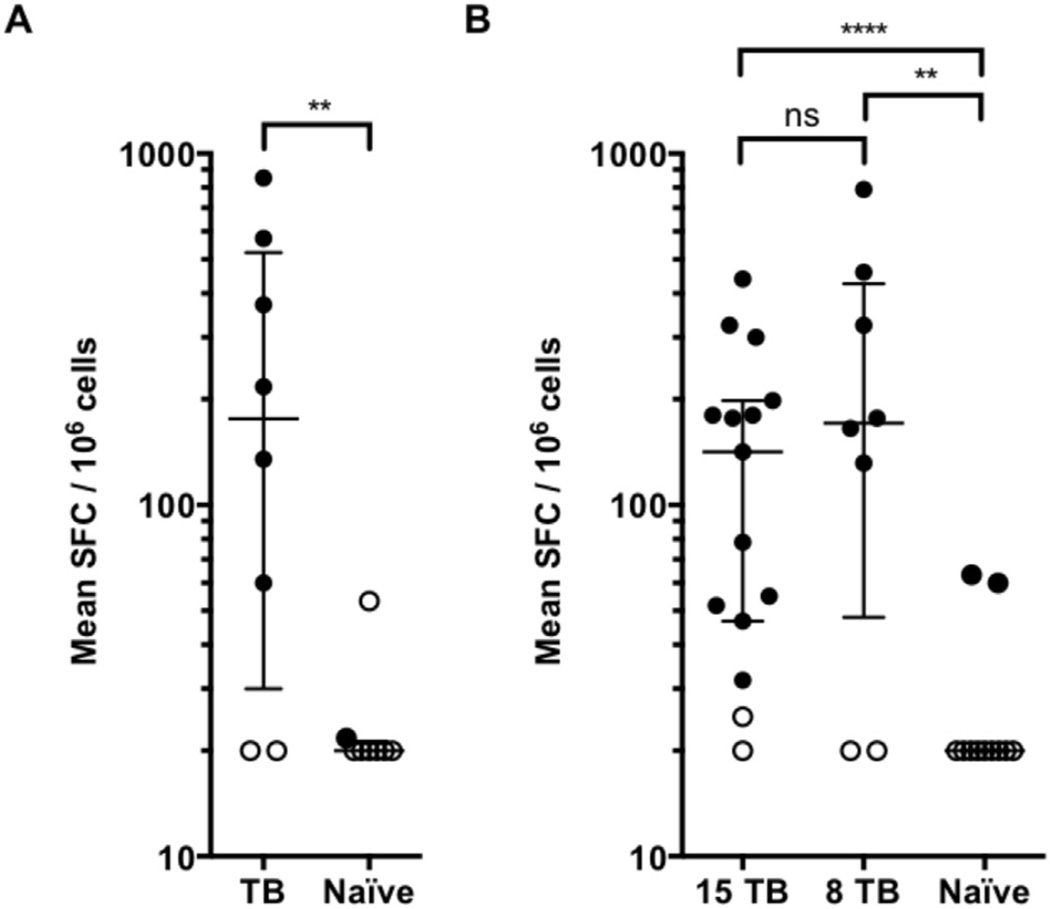
Consistent with the notion that this epitope set would allow general coverage in Indian rhesus macaques, we found that 6 of the 8 new animals were associated with strong responses, with median of 176 SFC/106 cells (interquartile range 30–523). By contrast, no or weak, responses were observed in splenocytes from the 8 naïve animals tested (Figure 3A). These data suggest that the 54 epitopes may allow general coverage and analysis of T cell responses in Indian rhesus macaques.
Figure 3.
Human epitopes allow general coverage and analysis of T cell responses in Indian rhesus macaques. Splenocytes from Indian rhesus macaques (black dots, positive responses; white dots, negative responses as per criteria described in the materials and methods section, Table 1) were incubated with 2 µg/ml per peptide in pool, after which the number of IFN-γ+ cells were enumerated in an ELISPOT assay. Each dot represents mean SFC/106 cells in one animal. Each individual pool was tested in triplicate. One-tailed Mann Whitney test, ****, p < 0.0001, **, p < 0.001, two-tailed Mann Whitney test, ns, no significant difference. A) Pool of 54 epitopes, defined in macaques, tested in TB animals (n = 8) and naïve animals (n = 8). B) Pool of 300 epitopes, defined in humans, tested in TB animals (n = 15 + 8) and naïve animals (n = 11).
3.6. Extensive similarity between human and NHP epitope repertoires
The MHC class II molecules of NHPs bear very significant degrees of sequence homology to human HLA class II molecules. A functional overlap between HLA DR and macaque DR molecules has indeed been reported [8].
Based on these observations we hypothesized that at least some of the dominant epitopes detected in NHPs might have been previously detected in humans. Strikingly, we found that in the antigens Rv3875 (ESAT-6), Rv0288 (TB10.4), Rv1886c (85B) and Rv3874 (CFP10), every single epitope we identified has also been identified in human studies (Supplemental Table III).
For the epitopes in antigen Rv3619c (EsxV), only epitope Rv3619c6–20QQ15 was positive in humans [96](Supplemental Table III). The only epitope in Rv0125 (MTb32a), Rv0125306–320AD15, as well as the epitopes in Rv1196 (mtb39a) and Rv2608 (PPE42) have not been reported in the setting of human infection. Hence, while some epitope macaque-specific epitope responses are noted, the majority of epitopes in our study have also been detected in human Mtb infection.
3.7. Human Mtb epitopes also allow general coverage and analysis of T cell responses in Indian rhesus macaques
The results presented above demonstrate that the majority of epitopes in our study have also been detected in human Mtb infection. We, therefore, hypothesized that conversely epitopes detected in humans might also allow uniform coverage of Indian macaques in general. Accordingly, we generated a pool of 300 epitopes previously recognized in human LTBIs [92]. This pool contained 25 of the 54 epitopes seen in macaque and 9 epitopes with partial overlap (10aa) to the 54 epitopes. This human epitope pool was first tested in splenocytes from the original 15 animals that were immunized and/or infected with tuberculosis (Table 1). In addition, we subsequently tested this pool in splenocytes derived from the set of 8 new animals that were also immunized and infected with tuberculosis (Table 1), and an additional 11 naïve animals (Table 1).
We found that the pool of human epitopes elicited significant responses in 13 of the 15 original animals (Median 142 SFC/106, IQR 47–198) (Figure 3B). Consistent with the notion that this epitope set would allow general coverage in Rhesus macaques, we found that 6 out of 8 new animals were associated with vigorous responses, with median of 171 SFC/106 cells (IQR 48–426; Figure 3B). No responses were observed in splenocytes from 9 of the 11 naïve animals tested, and weak responses (<70 SFC/106 cells) were observed for the remaining two (Figure 3B). These data suggest that the human epitope pool represents an alternative strategy to allow general coverage and analysis of T cell responses in Indian rhesus macaques.
3.8. T cell responses associated with the human and macaque Mtb epitope pools
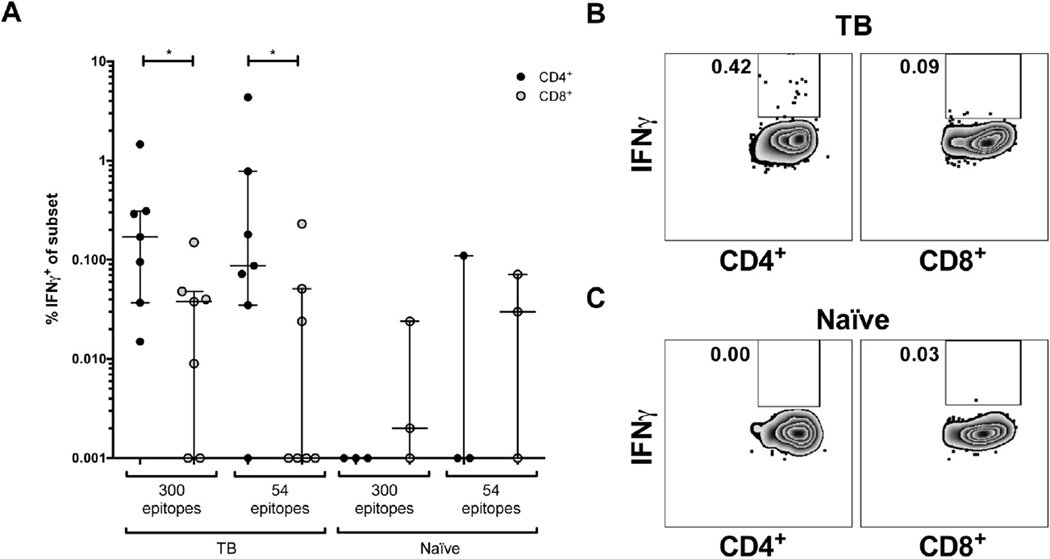
As shown above, Mtb-specific responses in Indian macaques can be investigated using either macaque or human-derived epitopes. As a way to further illustrate this point, we used intracellular cytokine staining (ICS) assays. Specifically, we wanted to determine the CD4/CD8 phenotype of responding T cells and to compare the magnitude of responses between the two epitope pools. Accordingly, we tested the two pools in 7 animals that had been exposed to TB, and 3 naïve controls.
As expected, the responses were almost entirely associated with CD4+ T cells (Figure 4A–C). This is an important point, as until these experiments were performed the CD4/Class II phenotype was inferred on the basis of the epitope size and the fact that in humans ex vivo responses are vastly dominated by this phenotype.
Figure 4.
Mtb-specific CD4+ T cell responses can be analyzed using epitopes derived from human studies. Epitope-specific IFN-γ production by splenocytes from Indian rhesus macaques (Table 1) measured after 6 h peptide pool stimulation. A) % IFN-γ-producing CD4+ (black dots) or CD8+ (grey dots) T cells in response to the pool of 54 and 300 epitopes. TB animals (IB78, II69, GP21, IJ42, IM73, II43, GI89, n = 7, left) and naïve animals (IB62, DR27, HE68, n = 3, right). B and C) Representative dot plots from one TB animal (B) and one naïve animal (C). Plots are gated on total CD4+ (left panel) or CD8+ (right panel) T cells. Numbers indicate percentage of IFN-γ-producing cells.
When the average magnitude of ex vivo responses to the two pools was compared, 0.1% (IQR 0.04–0.8%) of the total CD4+ T cells secreted IFN-γ in response to the pool of macaque epitopes, while 0.2% (IQR 0.04–0.3%) of the total CD4 cells responded in response to the pool of human epitopes. Weak or no IFN-γ responses were detected from CD8+ T cells and naïve animals (Figure 4A). The difference in response between the 54 epitope and the 300 epitope pools was not significant, but nevertheless the 300 epitope pool tended to have a stronger response and lower background in naïve animals. These data strengthens the feasibility of utilizing the human 300 epitope pool as an alternative to analyze T cell responses against Mtb in Indian rhesus macaques.
3.9. Rhesus and human Mtb epitopes also allow general coverage and analysis of T cell responses in cynomolgus macaques
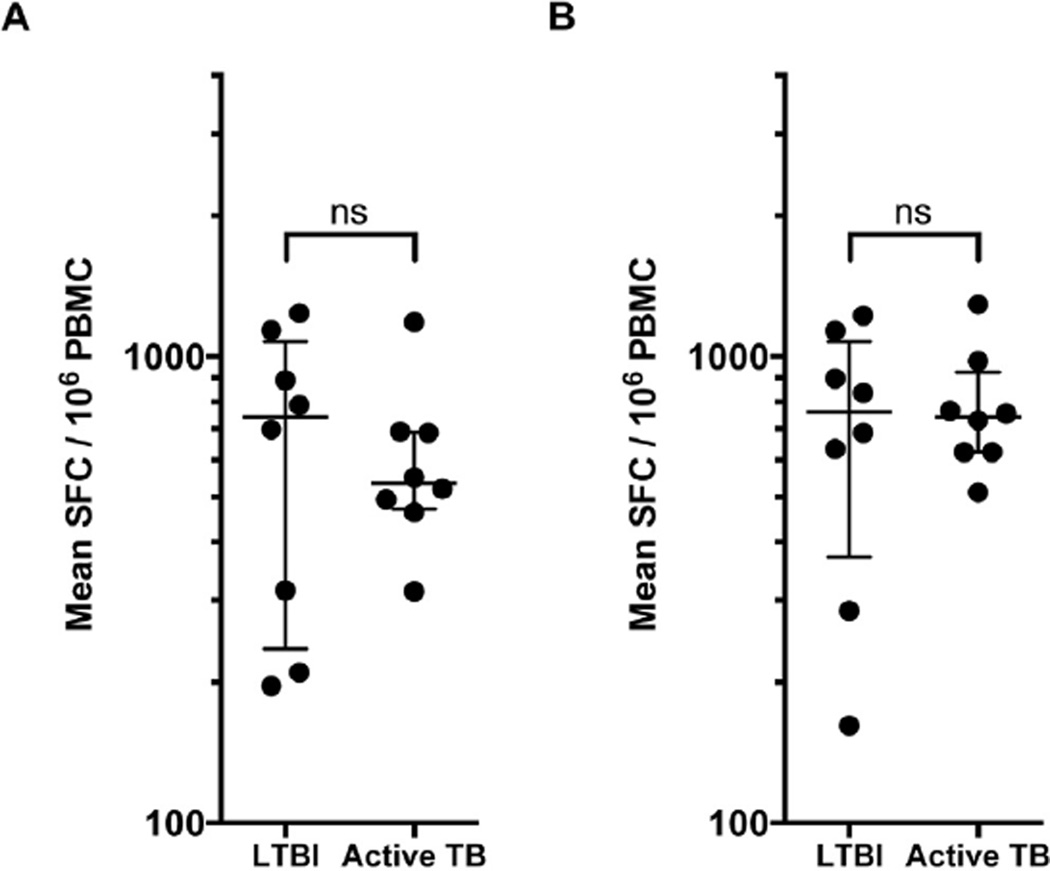
The previous data demonstrated a high cross-reactivity between human and rhesus class II molecules. Based on this observation, we reasoned that both the rhesus and human epitopes might also be cross-reactively recognized by cynomolgus macaques, the other widely used NHP species. To test this hypothesis, we tested a set of cynomolgus macaques with latent TB infection (LTBI; n = 8) and active TB infection (n = 8) with the pool of 54 rhesus epitopes and the 300 human epitope pool.
All of the cynomolgus macaques, irrespective of latent TB infection or active TB, tested responded strongly to the pool of rhesus epitopes (Median 743 SFC/106, IQR 236–1076 for LTBI and Median 535 SFC/106, IQR 471–689 for active TB) (Figure 5A). As shown in Figure 5B, responses of LTBI and active TB individuals against the 300 human epitopes pool were also high, with median 761 SFC/106 (372e1075) and 743 SFC/106 (623e925), respectively. For both peptide pools tested, there was no significant difference between the two groups (Mann Whitney test). In conclusion, these data suggest that these epitope sets allow for broad coverage across multiple NHP species.
Figure 5.
Human epitopes allow general coverage and analysis of T cell responses in cynomolgus macaques. PBMC from cynomolgus macaques (Materials and Methods) were incubated with 2 µg/ml per peptide in pool, after which the number of IFN-γ-producing cells were enumerated in an ELISPOT assay. Each dot represents mean SFC/ 106 cells in one animal. Each individual pool was tested in triplicate. Two-tailed Mann Whitney test, ns, no significant difference. Peptide pools tested in LTBI animals (n = 8) and animals with active TB (n = 8). A) Pool of 54 epitopes B) Pool of 300 epitopes.
4. Discussion
While NHP class I responses have been rather extensively characterized, much less is known at level of MHC class II restricted responses. To our knowledge, this is the first comprehensive epitope identification effort for NHPs in general, and in the context of Mtb in particular. We identified 54 new epitopes, significantly expanding the number of specific epitopes that can be used to monitor immune responses after vaccination and experimental infection. In fact, before this study, only one CD4+ T cell epitope had been extensively characterized in NHP [84]. Of general importance, in the present study we report that epitope repertoire of human and NHP class II overlaps significantly, to the point of allowing use of human epitopes to detect responses in both rhesus and cynomolgus macaques, and the use of rhesus epitopes to detect responses in cynomolgus macaques.
It is important to consider the molecular basis of the general epitope overlap described herein. The most likely explanation resides in the extreme degree of sequence similarity between different class II alleles in various primates. In many cases, different human and macaque alleles are more similar to each other than the same human or macaque allele is to other alleles of the same species. For example, the macaque class II alleles Mamu DRB*W1:01 and DRB*21:04 share 89.5% identity. Yet, DRB*W1:01 shares 91.4% identity to HLA DRB1*14:07, while DRB*21:04 is 91.7% identical to HLA DRB1*04:01. In fact, more similarity exist between different HLA DR and macaque DR alleles, than between HLA DR and HLA DQ molecules [9,97–99]. Furthermore, previous studies highlighted a significant degree of overlap in the repertoire of peptides bound by different alleles of the same human HLA class II locus, and even across HLA class II loci [43]. Accordingly promiscuous epitope recognition across different alleles of a given locus or even across different HLA class II loci is not uncommon [43,83,95]. In addition, a functional overlap between HLA DR and macaque DR molecules has also been reported [8].
It is also possible that peptides that are cross-presented and recognized by CD4+ T cells are associated with special unique features. A recent study from our laboratory specifically investigated prediction of epitopes promiscuously cross-recognized in humans (Paul et al., J. Imm. Methods, in press). Future studies will address whether these features also extend to human-primate cross-reactive epitopes.
Here, we found that the majority of TB epitopes we identified in NHPs has also been identified in human studies [77,83,84,100–106]. Indeed, we further show that a pool of epitopes previously identified in humans could be used to measure immune responses after vaccination and experimental infection in NHP. These results demonstrate that these reagents can be used to broadly characterize TB-specific NHP responses. They further suggest that this approach might be generally used to characterize NHP responses in other pathogen systems.
The observation that the repertoire of peptides recognized by CD4+ T cells restricted by specific alleles is similar in macaques and humans could have suggest potential approaches to rapidly defining epitope specificity and restriction of macaque responses. Though it is beyond the scope of this paper, we predict that it should be feasible to find macaque class II molecules with the same binding motif as a human class II molecule and show that the two consistently elicit the same T cell responses. Indeed, we previously pointed out extensive similarities between Mamu-DRB*w201 and HLA-DRB1*0101 peptide binding repertoires [9].
In our study, we evaluated the mechanism underlying the observation that certain epitopes were frequently recognized in the cohort of rhesus macaques analyzed. We concluded that the shared responses were not due specific MHC class II allele(s), unexpectedly common due to common pedigrees and inbreeding. Rather, the analysis demonstrates that in some cases these shared responses are likely restricted by alleles commonly expressed in the general macaque population. In other instances, based on the fact that responding macaques shared no MHC class II molecules, it is most likely that promiscuous binding and restriction is associated with epitope recognition. While promiscuous epitopes account for a large fraction of responses in humans [95], this is the first account of this phenomenon in the NHP in general, and in the rhesus macaque Mtb infection model in particular.
The large overlap in repertoire observed between macaques and humans is particularly striking. While some precedence for this has been noted for MHC class I molecules [14], this has not been generally noted for MHC class II because macaques epitopes have been most studied in the context of SIV/HIV and HIV is used in humans and SIV in monkeys. This can also be interpreted in the context of research addressing the coevolution of humans and Mtb. The results suggest that human T cell epitopes within MTB are highly conserved, suggesting purifying selection acting on T cell epitopes in MTB [107]. This has led to the hypothesis that MTB is benefiting from being recognized by T cells, possibly because this response may drive lung inflammation and transmission of MTB. A possible explanation for this resides in the fact that HLA and macaque MHC class II molecules dictate a similar repertoire of epitope presentation. Indeed, the motifs and repertoires of human and macaque DR are very similar [9].
The specific system we have chosen to perform our studies is TB infection and vaccination. The current, commercially available, BCG vaccine is associated with variable effectiveness against Mtb infection and disease, and is not indicated for immunocompromised individuals [108–110] . Testing new TB vaccines in NHP models is of considerable interest, to optimize schedule and administration regimens, as well as comparison of different adjuvant and delivery strategies. Furthermore, accurate quantification of immune responses in the context of vaccinations associated with varied degrees of protection would allow identification of immunological correlates of protection. NHP models with the specific use of aerosolized Mtb have yielded the best parallel infection by modeling human outcomes [69,72]. However, most studies in have focused on the molecular biology of the pathogen after infection and whether vaccination had a direct impact on subsequent infection, with lesser emphasis on immunological characterizations [72,111–115].
Several Mtb antigens immunodominant in humans [81–83,103,110,116–120] are included in the design of new TB vaccines. Herein, we tested the immunogenicity in rhesus macaques of a set of antigens that have been used in human clinical trials and diagnostics. Of these antigens, a subset has been shown to be protective in animal models (Rv3619c, Rv3620c, Rv2608), highly immunogenic in humans (Rv3875, Rv3874, Rv0288, Rv1196) and strongly recognized in the initial stages of infection in humans (Rv0288 and Rv3875) [121,122]. We show that several of these antigens, including Rv3875 (ESAT-6) and Rv3874 (CFP10), which are immunodominant in humans [123–130], are also immunodominant in rhesus macaques. Our results therefore support the feasibility of evaluation in NHPs of vaccines based on these antigens. By the same token, we detected weak and inconsistent responses in the case of other vaccine antigen candidates, such as Rv0125, Rv1886c, Rv1813c, Rv3619c and Rv3620c. Even though the present study has expanded our understanding of the cellular immune response to selected antigens, the analysis is limited to Mtb antigens that are either immunodominant or that were considered potentially protective [92]. This is a limitation of the study especially given that protection is not defined in the context of these studies. Therefore, caution should be exercised in interpreting results of vaccines based on these antigens in NHP, as the low immunogenicity in macaques might not allow extrapolating the results to humans.
In conclusion, this study highlights a previously unappreciated extensive overlap in epitope recognition at the level of class II responses in humans, rhesus macaques and cynomolgus macaques. This is particularly unique given the species differences and the varying routes of antigen exposure. The observation provides the basis for the development of epitope-based tools to study the immunodominance patterns in the context of novel vaccines as well as characterizing different outcomes of infection in NHP in any pathogen system, and potentially also in the context of immune intervention in cancer, autoimmunity and allergies. In particular, our findings have immediate applicability for the evaluation of new vaccines and diagnostics in the setting of non-human primate model of TB.
Supplementary Material
Acknowledgments
We thank Charles Scanga for coordinating the cynomolgus macaque shipments and Carolyn Bigbee for laboratory assistance with the cynomolgus macaque sample processing.
Funding: This study was supported by the National Institutes of Health (NIH) 2 R15 AI064175 03A1 (BRM), R01AI089323 (DK), R01HL106790 (DK) and contract HHSN272200900044C (AS) , Ciencia sem Fronteiras 400938/2012-0 (BRM) and Bill and Melinda Gates Foundation Global Health Grant OPP1066265 (TJS).
Appendix A. Supplementary data.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.tube.2015.07.005
Footnotes
Competing interests: None declared.
Ethical approval: Not required.
References
- 1.de Groot NG, Otting N, Robinson J, Blancher A, Lafont BA, Marsh SG, O’Connor DH, Shiina T, Walter L, Watkins DI, Bontrop RE. Nomenclature report on the major histocompatibility complex genes and alleles of great ape, old and new world monkey species. Immunogenetics. 2012;64:615–631. doi: 10.1007/s00251-012-0617-1. http://dx.doi.org/10.1007/s00251-012-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. http://dx.doi.org/10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen TM, Mothe BR, Sidney J, Jing P, Dzuris JL, Liebl ME, Vogel TU, O’Connor DH, Wang X, Wussow MC, Thomson JA, Altman JD, Watkins DI, Sette A. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule Mamu-A*01: implications for vaccine design and testing. J Virol. 2001;75:738–749. doi: 10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen TM, Sidney J, del Guercio MF, Glickman RL, Lensmeyer GL, Wiebe DA, DeMars R, Pauza CD, Johnson RP, Sette A, Watkins DI. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 5.Allen TM, Vogel TU, Fuller DH, Mothe BR, Steffen S, Boyson JE, Shipley T, Fuller J, Hanke T, Sette A, Altman JD, Moss B, McMichael AJ, Watkins DI. Induction of AIDS virus-specific CTL activity in fresh, unstimulated peripheral blood lymphocytes from rhesus macaques vaccinated with a DNA prime/ modified vaccinia virus Ankara boost regimen. J Immunol. 2000;164:4968–4978. doi: 10.4049/jimmunol.164.9.4968. [DOI] [PubMed] [Google Scholar]
- 6.Blasky AJ, Karl JA, Wiseman RW, Read DS, O’Connor DH. Rapid high-resolution MHC class I genotyping of Chinese rhesus macaques by capillary reference strand-mediated conformational analysis. Immunogenetics. 2008;60:575–584. doi: 10.1007/s00251-008-0315-1. http://dx.doi.org/10.1007/s00251-008-0315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrington M, Bontrop RE. Effects of MHC class I on HIV/SIV disease in primates. AIDS. 2002;16(Suppl. 4):S105–S114. doi: 10.1097/00002030-200216004-00015. [DOI] [PubMed] [Google Scholar]
- 8.Dzuris JL, Sidney J, Appella E, Chesnut RW, Watkins DI, Sette A. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J Immunol. 2000;164:283–291. doi: 10.4049/jimmunol.164.1.283. [DOI] [PubMed] [Google Scholar]
- 9.Dzuris JL, Sidney J, Horton H, Correa R, Carter D, Chesnut RW, Watkins DI, Sette A. Molecular determinants of peptide binding to two common rhesus macaque major histocompatibility complex class II molecules. J Virol. 2001;75:10958–10968. doi: 10.1128/JVI.75.22.10958-10968.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joag SV. Primate models of AIDS. Microb Infect. 2000;2:223–229. doi: 10.1016/s1286-4579(00)00266-5. [DOI] [PubMed] [Google Scholar]
- 11.Karl JA, Wiseman RW, Campbell KJ, Blasky AJ, Hughes AL, Ferguson B, Read DS, O’Connor DH. Identification of MHC class I sequences in Chineseorigin rhesus macaques. Immunogenetics. 2008;60:37–46. doi: 10.1007/s00251-007-0267-x. http://dx.doi.org/10.1007/s00251-007-0267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li A, Wang X, Liu Y, Zhao Y, Liu B, Sui L, Zeng L, Sun Z. Preliminary observations of MHC class I A region polymorphism in three populations of Chinese-origin rhesus macaques. Immunogenetics. 2012;64:887–894. doi: 10.1007/s00251-012-0645-x. http://dx.doi.org/10.1007/s00251-012-0645-x. [DOI] [PubMed] [Google Scholar]
- 13.Loffredo JT, Maxwell J, Qi Y, Glidden CE, Borchardt GJ, Soma T, Bean AT, Beal DR, Wilson NA, Rehrauer WM, Lifson JD, Carrington M, Watkins DI. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, Hawkins OE, Piaskowski S, Wilson NA, Hildebrand WH, Watkins DI, Sette A. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loffredo JT, Sidney J, Piaskowski S, Szymanski A, Furlott J, Rudersdorf R, Reed J, Peters B, Hickman-Miller HD, Bardet W, Rehrauer WM, O’Connor DH, Wilson NA, Hildebrand WH, Sette A, Watkins DI. The high frequency Indian rhesus macaque MHC class I molecule, Mamu-B*01, does not appear to be involved in CD8+ T lymphocyte responses to SIVmac239. J Immunol. 2005;175:5986–5997. doi: 10.4049/jimmunol.175.9.5986. [DOI] [PubMed] [Google Scholar]
- 16.Loffredo JT, Sidney J, Wojewoda C, Dodds E, Reynolds MR, Napoe G, Mothe BR, O’Connor DH, Wilson NA, Watkins DI, Sette A. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J Immunol. 2004;173:5064–5076. doi: 10.4049/jimmunol.173.8.5064. [DOI] [PubMed] [Google Scholar]
- 17.Loffredo JT, Sidney J, Piaskowski S, Szymanski A, Furlott J, Rudersdorf R, Reed J, Peters B, Hickman-Miller HD, Bardet W, Rehrauer WM, O’Connor DH, Wilson NA, Hildebrand WH, Sette A, Watkins DI. The high frequency Indian rhesus macaque MHC class I molecule, Mamu-B*01, does not appear to be involved in CD8+ T lymphocyte responses to SIVmac239. J Immunol. 2005;175:5986–5997. doi: 10.4049/jimmunol.175.9.5986. [DOI] [PubMed] [Google Scholar]
- 18.Maness NJ, Walsh AD, Rudersdorf RA, Erickson PA, Piaskowski SM, Wilson NA, Watkins DI. Chinese origin rhesus macaque major histocom-patibility complex class I molecules promiscuously present epitopes from SIV associated with molecules of Indian origin; implications for immunodomi-nance and viral escape. Immunogenetics. 2011;63:587–597. doi: 10.1007/s00251-011-0538-4. http://dx.doi.org/10.1007/s00251-011-0538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mothe BR, Weinfurter J, Wang C, Rehrauer W, Wilson N, Allen TM, Allison DB, Watkins DI. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2003;77:2736–2740. doi: 10.1128/JVI.77.4.2736-2740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mothe BR, Horton H, Carter DK, Allen TM, Liebl ME, Skinner P, Vogel TU, Fuenger S, Vielhuber K, Rehrauer W, Wilson N, Franchini G, Altman JD, Haase A, Picker LJ, Allison DB, Watkins DI. Dominance of CD8 responses specific for epitopes bound by a single major histocompatibility complex class I molecule during the acute phase of viral infection. J Virol. 2002;76:875–884. doi: 10.1128/JVI.76.2.875-884.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mothe BR, Sidney J, Dzuris JL, Liebl ME, Fuenger S, Watkins DI, Sette A. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J Immunol. 2002;169:210–219. doi: 10.4049/jimmunol.169.1.210. [DOI] [PubMed] [Google Scholar]
- 22.Muhl T, Krawczak M, Ten Haaft P, Hunsmann G, Sauermann U. MHC class I alleles influence set-point viral load and survival time in simian immunodeficiency virus-infected rhesus monkeys. J Immunol. 2002;169:3438–3446. doi: 10.4049/jimmunol.169.6.3438. [DOI] [PubMed] [Google Scholar]
- 23.O’Connor DH, Mothe BR, Weinfurter JT, Fuenger S, Rehrauer WM, Jing P, Rudersdorf RR, Liebl ME, Krebs K, Vasquez J, Dodds E, Loffredo J, Martin S, McDermott AB, Allen TM, Wang C, Doxiadis GG, Montefiori DC, Hughes A, Burton DR, Allison DB, Wolinsky SM, Bontrop R, Picker LJ, Watkins DI. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J Virol. 2003;77:9029–9040. doi: 10.1128/JVI.77.16.9029-9040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otting N, de Vos-Rouweler AJ, Heijmans CM, de Groot NG, Doxiadis GG, Bontrop RE. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics. 2007;59:367–375. doi: 10.1007/s00251-007-0201-2. http://dx.doi.org/10.1007/s00251-007-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otting N, Heijmans CM, van der Wiel M, de Groot NG, Doxiadis GG, Bontrop RE. A snapshot of the Mamu-B genes and their allelic repertoire in rhesus macaques of Chinese origin. Immunogenetics. 2008;60:507–514. doi: 10.1007/s00251-008-0311-5. http://dx.doi.org/10.1007/s00251-008-0311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouyang D, Wang X, He X, Xu L, Shi H, Gao Q, Guo H. Construction of soluble Mamu-b*1703, a class I major histocompatibility complex of Chinese rhesus macaques, monomer and tetramer loaded with a simian immunodeficiency virus peptide. Cell Mol Immunol. 2009;6:117–122. doi: 10.1038/cmi.2009.16. http://dx.doi.org/10.1038/cmi.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang D, Xu L, Dai Z, Shi H, Zhang G, Zheng Y, He X. Identification of major histocompatibility complex class I alleles in Chinese rhesus macaques. Acta Biochim Biophys Sin (Shanghai) 2008;40:919–927. doi: 10.1111/j.1745-7270.2008.00474.x. [DOI] [PubMed] [Google Scholar]
- 28.Ouyang DY, Xu LH, Shi HJ, Zheng YT, He XH. Eight novel MHC class I alleles identified in Chinese-origin rhesus macaques. Tissue Antigens. 2009;73:285–287. doi: 10.1111/j.1399-0039.2008.01198.x. http://dx.doi.org/10.1111/j.1399-0039.2008.01198.x. [DOI] [PubMed] [Google Scholar]
- 29.Qiu CL, Yang GB, Yu K, Li Y, Li XL, Liu Q, Zhao H, Xing H, Shao Y. Characterization of the major histocompatibility complex class II DQB (MhcMamu-DQB1) alleles in a cohort of Chinese rhesus macaques (Macaca mulatta) Hum Immunol. 2008;69:513–521. doi: 10.1016/j.humimm.2008.05.014. http://dx.doi.org/10.1016/j.humimm.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 30.Sauermann U. Making the animal model for AIDS research more precise: the impact of major histocompatibility complex (MHC) genes on pathogenesis and disease progression in SIV-infected monkeys. Curr Mol Med. 2001;1:515–522. doi: 10.2174/1566524013363555. [DOI] [PubMed] [Google Scholar]
- 31.Sette A, Sidney J, Bui HH, Del Guercio MF, Alexander J, Loffredo J, Watkins DI, Mothe BR. Characterization of the peptide-binding specificity of Mamu-A*11 results in the identification of SIV-derived epitopes and interspecies cross-reactivity. Immunogenetics. 2005;57:53–68. doi: 10.1007/s00251-004-0749-z. [DOI] [PubMed] [Google Scholar]
- 32.Sette A, Sidney J, Livingston B, Dzuris J, Crimi C, Walker CM, Southwood S, Collins EJ, Hughes A. Class I molecules with similar peptide binding specificities are the result of both common ancestry and convergent evolution. Immunogenetics. 2003;54:830–841. doi: 10.1007/s00251-002-0530-0. [DOI] [PubMed] [Google Scholar]
- 33.Sidney J, Dzuris JL, Newman MJ, Johnson RP, Kaur A, Amitinder K, Walker CM, Appella E, Mothe B, Watkins DI, Sette A. Definition of the Mamu A*01 peptide binding specificity: application to the identification of wild-type and optimized ligands from simian immunodeficiency virus regulatory proteins. J Immunol. 2000;165:6387–6399. doi: 10.4049/jimmunol.165.11.6387. [DOI] [PubMed] [Google Scholar]
- 34.Solomon C, Southwood S, Hoof I, Rudersdorf R, Peters B, Sidney J, Pinilla C, Marcondes MC, Ling B, Marx P, Sette A, Mothe BR. The most common Chinese rhesus macaque MHC class I molecule shares peptide binding repertoire with the HLA-B7 supertype. Immunogenetics. 2010;62:451–464. doi: 10.1007/s00251-010-0450-3. http://dx.doi.org/10.1007/s00251-010-0450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Southwood S, Solomon C, Hoof I, Rudersdorf R, Sidney J, Peters B, Wahl A, Hawkins O, Hildebrand W, Mothe BR, Sette A. Functional analysis of frequently expressed Chinese rhesus macaque MHC class I molecules Mamu-A1*02601 and Mamu-B*08301 reveals HLA-A2 and HLA-A3 supertypic specificities. Immunogenetics. 2011;63:275–290. doi: 10.1007/s00251-010-0502-8. http://dx.doi.org/10.1007/s00251-010-0502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel T, Norley S, Beer B, Kurth R. Rapid screening for Mamu-A1-positive rhesus macaques using a SIVmac Gag peptide-specific cytotoxic T-lymphocyte assay. Immunology. 1995;84:482–487. [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel TU, Evans DT, Urvater JA, O’Connor DH, Hughes AL, Watkins DI. Major histocompatibility complex class I genes in primates: co-evolution with pathogens. Immunol Rev. 1999;167:327–337. doi: 10.1111/j.1600-065x.1999.tb01402.x. [DOI] [PubMed] [Google Scholar]
- 38.Vogel TU, Friedrich TC, O’Connor DH, Rehrauer W, Dodds EJ, Hickman H, Hildebrand W, Sidney J, Sette A, Hughes A, Horton H, Vielhuber K, Rudersdorf R, De Souza IP, Reynolds MR, Allen TM, Wilson N, Watkins DI. Escape in one of two cytotoxic T-lymphocyte epitopes bound by a high-frequency major histocompatibility complex class I molecule, Mamu-A*02: a paradigm for virus evolution and persistence? J Virol. 2002;76:11623–11636. doi: 10.1128/JVI.76.22.11623-11636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E, Jr, Wright C, Harkins T, O’Connor DH. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15:1322–1326. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mothe BR, Southwood S, Sidney J, English AM, Wriston A, Hoof I, Shabanowitz J, Hunt DF, Sette A. Peptide-binding motifs associated with MHC molecules common in Chinese rhesus macaques are analogous to those of human HLA supertypes and include HLA-B27-like alleles. Immunogenetics. 2013;65:371–386. doi: 10.1007/s00251-013-0686-9. http://dx.doi.org/10.1007/s00251-013-0686-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sette A, Sidney J, Southwood S, Moore C, Berry J, Dow C, Bradley K, Hoof I, Lewis MG, Hildebrand WH, McMurtrey CP, Wilson NA, Watkins DI, Mothe BR. A shared MHC supertype motif emerges by convergent evolution in macaques and mice, but is totally absent in human MHC molecules. Immunogenetics. 2012 doi: 10.1007/s00251-011-0598-5. http://dx.doi.org/10.1007/s00251-011-0598-5. [DOI] [PMC free article] [PubMed]
- 42.Allen TM, Jing P, Calore B, Horton H, O’Connor DH, Hanke T, Piekarczyk M, Ruddersdorf R, Mothe BR, Emerson C, Wilson N, Lifson JD, Belyakov IM, Berzofsky JA, Wang C, Allison DB, Montefiori DC, Desrosiers RC, Wolinsky S, Kunstman KJ, Altman JD, Sette A, McMichael AJ, Watkins DI. Effects of cytotoxic T lymphocytes (CTL) directed against a single simian immunodeficiency virus (SIV) Gag CTL epitope on the course of SIVmac239 infection. J Virol. 2002;76:10507–10511. doi: 10.1128/JVI.76.20.10507-10511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenbaum J, Sidney J, Chung J, Brander C, Peters B, Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. http://dx.doi.org/10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zumla A, George A, Sharma V, Herbert N. Baroness Masham of I. WHO’s 2013 global report on tuberculosis: successes, threats, and opportunities. Lancet. 2013;382:1765–1767. doi: 10.1016/S0140-6736(13)62078-4. http://dx.doi.org/10.1016/S0140-6736(13)62078-4. [DOI] [PubMed] [Google Scholar]
- 45.Comstock GW. Epidemiology of tuberculosis. Am Rev Respir Dis. 1982;125:8–15. doi: 10.1164/arrd.1982.125.3P2.8. [DOI] [PubMed] [Google Scholar]
- 46.Kaufmann SH. Is the development of a new tuberculosis vaccine possible? Nat Med. 2000;6:955–960. doi: 10.1038/79631. http://dx.doi.org/10.1038/79631. [DOI] [PubMed] [Google Scholar]
- 47.Walter ND, Painter J, Parker M, Lowenthal P, Flood J, Fu Y, Asis R, Reves R. Tuberculosis epidemiologic studies C. Persistent latent tuberculosis reactivation risk in United States immigrants. Am J Respir Crit Care Med. 2014;189:88–95. doi: 10.1164/rccm.201308-1480OC. http://dx.doi.org/10.1164/rccm.201308-1480OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersen P, Doherty TM. The success and failure of BCG e implications for a novel tuberculosis vaccine. Nat Rev Microbiol. 2005;3:656–662. doi: 10.1038/nrmicro1211. http://dx.doi.org/10.1038/nrmicro1211. [DOI] [PubMed] [Google Scholar]
- 49.Doherty TM. Real world TB vaccines: clinical trials in TB-endemic regions. Vaccine. 2005;23:2109–2114. doi: 10.1016/j.vaccine.2005.01.060. http://dx.doi.org/10.1016/j.vaccine.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 50.Bonah C. Packaging BCG: standardizing an anti-tuberculosis vaccine in interwar Europe. Sci Context. 2008;21:279–310. doi: 10.1017/s0269889708001725. [DOI] [PubMed] [Google Scholar]
- 51.Tameris M, Geldenhuys H, Luabeya AK, Smit E, Hughes JE, Vermaak S, Hanekom WA, Hatherill M, Mahomed H, McShane H, Scriba TJ. The candidate TB vaccine, MVA85A, induces highly durable Th1 responses. PLoS One. 2014;9:e87340. doi: 10.1371/journal.pone.0087340. http://dx.doi.org/10.1371/journal.pone.0087340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colditz GA, Berkey CS, Mosteller F, Brewer TF, Wilson ME, Burdick E, Fineberg HV. The efficacy of bacillus Calmette-Guerin vaccination of newborns and infants in the prevention of tuberculosis: meta-analyses of the published literature. Pediatrics. 1995;96:29–35. [PubMed] [Google Scholar]
- 53.Calmette A. Preventive vaccination against tuberculosis with BCG. Proc R Soc Med. 1931;24:1481–1490. doi: 10.1177/003591573102401109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, Shea JE, McClain JB, Hussey GD, Hanekom WA, Mahomed H, McShane H. Team MATS. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet. 2013;381:1021–1028. doi: 10.1016/S0140-6736(13)60177-4. http://dx.doi.org/10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rustomjee R, Lockhart S, Shea J, Fourie PB, Hindle Z, Steel G, Hussey G, Ginsberg A, Brennan MJ. Novel licensure pathways for expeditious introduction of new tuberculosis vaccines: a discussion of the adaptive licensure concept. Tuberc Edinb Scotl. 2014;94:178–182. doi: 10.1016/j.tube.2013.11.002. http://dx.doi.org/10.1016/j.tube.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 56.Orme I, Gonzalez-Juarrero M. Animal models of M. tuberculosis infection. Curr Protoc Microbiol. 2007 doi: 10.1002/9780471729259.mc10a05s7. http://dx.doi.org/10.1002/9780471729259.mc10a05s7 [Chapter 10]:Unit 10A 15. [DOI] [PubMed]
- 57.Orme I. Cellular and genetic mechanisms underlying susceptibility of animal models to tuberculosis infection. Novartis Found Symp. 1998;217:112–117. doi: 10.1002/0470846526.ch8. discussion 117–119. [DOI] [PubMed] [Google Scholar]
- 58.McShane H, Williams A. A review of preclinical animal models utilised for TB vaccine evaluation in the context of recent human efficacy data. Tuberc Edinb Scotl. 2014;94:105–110. doi: 10.1016/j.tube.2013.11.003. http://dx.doi.org/10.1016/j.tube.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ordway D, Palanisamy G, Henao-Tamayo M, Smith EE, Shanley C, Orme IM, Basaraba RJ. The cellular immune response to Mycobacterium tuberculosis infection in the guinea pig. J Immunol. 2007;179:2532–2541. doi: 10.4049/jimmunol.179.4.2532. [DOI] [PubMed] [Google Scholar]
- 60.Shang S, Harton M, Tamayo MH, Shanley C, Palanisamy GS, Caraway M, Chan ED, Basaraba RJ, Orme IM, Ordway DJ. Increased Foxp3 expression in guinea pigs infected with W-Beijing strains of M. tuberculosis. Tuberc Edinb Scotl. 2011;91:378–385. doi: 10.1016/j.tube.2011.06.001. http://dx.doi.org/10.1016/j.tube.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tree JA, Smith S, Baker N, Clark S, Aldwell FE, Chambers M, Williams A, Marsh PD. Method for assessing IFN-gamma responses in guinea pigs during TB vaccine trials. Lett Appl Microbiol. 2012;55:295–300. doi: 10.1111/j.1472-765X.2012.03292.x. http://dx.doi.org/10.1111/j.1472-765X.2012.03292.x. [DOI] [PubMed] [Google Scholar]
- 62.Goonetilleke NP, McShane H, Hannan CM, Anderson RJ, Brookes RH, Hill AV. Enhanced immunogenicity and protective efficacy against Mycobacterium tuberculosis of bacille Calmette-Guerin vaccine using mucosal administration and boosting with a recombinant modified vaccinia virus Ankara. J Immunol. 2003;171:1602–1609. doi: 10.4049/jimmunol.171.3.1602. [DOI] [PubMed] [Google Scholar]
- 63.Williams A, Goonetilleke NP, McShane H, Clark SO, Hatch G, Gilbert SC, Hill AV. Boosting with poxviruses enhances Mycobacterium bovis BCG efficacy against tuberculosis in guinea pigs. Infect Immun. 2005;73:3814–3816. doi: 10.1128/IAI.73.6.3814-3816.2005. http://dx.doi.org/10.1128/IAI.73.6.3814-3816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams A, Hatch GJ, Clark SO, Gooch KE, Hatch KA, Hall GA, Huygen K, Ottenhoff TH, Franken KL, Andersen P, Doherty TM, Kaufmann SH, Grode L, Seiler P, Martin C, Gicquel B, Cole ST, Brodin P, Pym AS, Dalemans W, Cohen J, Lobet Y, Goonetilleke N, McShane H, Hill A, Parish T, Smith D, Stoker NG, Lowrie DB, Kallenius G, Svenson S, Pawlowski A, Blake K, Marsh PD. Evaluation of vaccines in the EU TB vaccine cluster using a guinea pig aerosol infection model of tuberculosis. Tuberc Edinb Scotl. 2005;85:29–38. doi: 10.1016/j.tube.2004.09.009. http://dx.doi.org/10.1016/j.tube.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 65.Verreck FA, Vervenne RA, Kondova I, van Kralingen KW, Remarque EJ, Braskamp G, van der Werff NM, Kersbergen A, Ottenhoff TH, Heidt PJ, Gilbert SC, Gicquel B, Hill AV, Martin C, McShane H, Thomas AW. MVA.85A boosting of BCG and an attenuated, phoP deficient M. tuberculosis vaccine both show protective efficacy against tuberculosis in rhesus macaques. PLoS One. 2009;4:e5264. doi: 10.1371/journal.pone.0005264. http://dx.doi.org/10.1371/journal.pone.0005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waters WR, Palmer MV, Buddle BM, Vordermeier HM. Bovine tuberculosis vaccine research: historical perspectives and recent advances. Vaccine. 2012;30:2611–2622. doi: 10.1016/j.vaccine.2012.02.018. http://dx.doi.org/10.1016/j.vaccine.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 67.Vordermeier HM, Villarreal-Ramos B, Cockle PJ, McAulay M, Rhodes SG, Thacker T, Gilbert SC, McShane H, Hill AV, Xing Z, Hewinson RG. Viral booster vaccines improve Mycobacterium bovis BCG-induced protection against bovine tuberculosis. Infect Immun. 2009;77:3364–3373. doi: 10.1128/IAI.00287-09. http://dx.doi.org/10.1128/IAI.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McMurray DN. Disease model: pulmonary tuberculosis. Trends Mol Med. 2001;7:135–137. doi: 10.1016/s1471-4914(00)01901-8. [DOI] [PubMed] [Google Scholar]
- 69.Kaushal D, Mehra S, Didier PJ, Lackner AA. The non-human primate model of tuberculosis. J Med Primatol. 2012;41:191–201. doi: 10.1111/j.1600-0684.2012.00536.x. http://dx.doi.org/10.1111/j.1600-0684.2012.00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pahar B, Li J, Rourke T, Miller CJ, McChesney MB. Detection of antigen-specific T cell interferon gamma expression by ELISPOT and cytokine flow cytometry assays in rhesus macaques. J Immunol Methods. 2003;282:103–115. doi: 10.1016/j.jim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 71.Amara RR. Methods for quantitating antigen-specific T cell responses using functional assays in rhesus macaques. Methods Mol Biol. 2009;485:417–424. doi: 10.1007/978-1-59745-170-3_28. http://dx.doi.org/10.1007/978-1-59745-170-3_28. [DOI] [PubMed] [Google Scholar]
- 72.Sharpe SA, McShane H, Dennis MJ, Basaraba RJ, Gleeson F, Hall G, McIntyre A, Gooch K, Clark S, Beveridge NE, Nuth E, White A, Marriott A, Dowall S, Hill AV, Williams A, Marsh PD. Establishment of an aerosol challenge model of tuberculosis in rhesus macaques and an evaluation of endpoints for vaccine testing. Clin Vaccin Immunol. 2010;17:1170–1182. doi: 10.1128/CVI.00079-10. http://dx.doi.org/10.1128/CVI.00079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harriff MJ, Cansler ME, Toren KG, Canfield ET, Kwak S, Gold MC, Lewinsohn DM. Human lung epithelial cells contain Mycobacterium tuberculosis in a late endosomal vacuole and are efficiently recognized by CD8(+) T cells. PloS One. 2014;9:e97515. doi: 10.1371/journal.pone.0097515. http://dx.doi.org/10.1371/journal.pone.0097515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prezzemolo T, Guggino G, La Manna MP, Di Liberto D, Dieli F, Caccamo N. Functional signatures of human CD4 and CD8 T Cell responses to Mycobacterium tuberculosis. Front Immunol. 2014;5:180. doi: 10.3389/fimmu.2014.00180. http://dx.doi.org/10.3389/fimmu.2014.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ussher JE, Klenerman P, Willberg CB. Mucosal-associated invariant T-cells: new players in anti-bacterial immunity. Front Immunol. 2014;5:450. doi: 10.3389/fimmu.2014.00450. http://dx.doi.org/10.3389/fimmu.2014.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boer MC, van Meijgaarden KE, Joosten SA, Ottenhoff TH. CD8+ regulatory T cells, and not CD4+ T cells, dominate suppressive phenotype and function after in vitro live Mycobacterium bovis-BCG activation of human cells. PloS One. 2014;9:e94192. doi: 10.1371/journal.pone.0094192. http://dx.doi.org/10.1371/journal.pone.0094192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Commandeur S, Lin MY, van Meijgaarden KE, Friggen AH, Franken KL, Drijfhout JW, Korsvold GE, Oftung F, Geluk A, Ottenhoff TH. Double- and monofunctional CD4(+) and CD8(+) T-cell responses to Mycobacterium tuberculosis DosR antigens and peptides in long-term latently infected individuals. Eur J Immunol. 2011;41:2925–2936. doi: 10.1002/eji.201141602. http://dx.doi.org/10.1002/eji.201141602. [DOI] [PubMed] [Google Scholar]
- 78.Young JM, Adetifa IM, Ota MO, Sutherland JS. Expanded polyfunctional T cell response to mycobacterial antigens in TB disease and contraction post-treatment. PloS One. 2010;5:e11237. doi: 10.1371/journal.pone.0011237. http://dx.doi.org/10.1371/journal.pone.0011237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nunes-Alves C, Booty MG, Carpenter SM, Jayaraman P, Rothchild AC, Behar SM. In search of a new paradigm for protective immunity to TB. Nat Rev Microbiol. 2014;12:289–299. doi: 10.1038/nrmicro3230. http://dx.doi.org/10.1038/nrmicro3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caccamo N, Guggino G, Joosten SA, Gelsomino G, Di Carlo P, Titone L, Galati D, Bocchino M, Matarese A, Salerno A, Sanduzzi A, Franken WP, Ottenhoff TH, Dieli F. Multifunctional CD4(+) T cells correlate with active Mycobacterium tuberculosis infection. Eur J Immunol. 2010;40:2211–2220. doi: 10.1002/eji.201040455. http://dx.doi.org/10.1002/eji.201040455. [DOI] [PubMed] [Google Scholar]
- 81.Arlehamn CL, Seumois G, Gerasimova A, Huang C, Fu Z, Yue X, Sette A, Vijayanand P, Peters B. Transcriptional profile of tuberculosis antigen-specific T cells reveals novel multifunctional features. J Immunol. 2014;193:2931–2940. doi: 10.4049/jimmunol.1401151. http://dx.doi.org/10.4049/jimmunol.1401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewinsohn DA, Winata E, Swarbrick GM, Tanner KE, Cook MS, Null MD, Cansler ME, Sette A, Sidney J, Lewinsohn DM. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog. 2007;3:1240–1249. doi: 10.1371/journal.ppat.0030127. http://dx.doi.org/10.1371/journal.ppat.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arlehamn CS, Sidney J, Henderson R, Greenbaum JA, James EA, Moutaftsi M, Coler R, McKinney DM, Park D, Taplitz R, Kwok WW, Grey H, Peters B, Sette A. Dissecting mechanisms of immunodominance to the common tuberculosis antigens ESAT-6, CFP10, Rv2031c (hspX), Rv2654c (TB7.7), and Rv1038c (EsxJ) J Immunol. 2012;188:5020–5031. doi: 10.4049/jimmunol.1103556. http://dx.doi.org/10.4049/jimmunol.1103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei H, Wang R, Yuan Z, Chen CY, Huang D, Halliday L, Zhong W, Zeng G, Shen Y, Shen L, Wang Y, Chen ZW. DR*W201/P65 tetramer visualization of epitope-specific CD4 T-cell during M. tuberculosis infection and its resting memory pool after BCG vaccination. PLoS One. 2009;4:e6905. doi: 10.1371/journal.pone.0006905. http://dx.doi.org/10.1371/journal.pone.0006905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaushal D, Schroeder BG, Tyagi S, Yoshimatsu T, Scott C, Ko C, Carpenter L, Mehrotra J, Manabe YC, Fleischmann RD, Bishai WR. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc Natl Acad Sci U S A. 2002;99:8330–8335. doi: 10.1073/pnas.102055799. http://dx.doi.org/10.1073/pnas.102055799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mehra S, Kaushal D. Functional genomics reveals extended roles of the Mycobacterium tuberculosis stress response factor sigmaH. J Bacteriol. 2009;191:3965–3980. doi: 10.1128/JB.00064-09. http://dx.doi.org/10.1128/JB.00064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garcia-Contreras L, Wong YL, Muttil P, Padilla D, Sadoff J, Derousse J, Germishuizen WA, Goonesekera S, Elbert K, Bloom BR, Miller R, Fourie PB, Hickey A, Edwards D. Immunization by a bacterial aerosol. Proc Natl Acad Sci U S A. 2008;105:4656–4660. doi: 10.1073/pnas.0800043105. http://dx.doi.org/10.1073/pnas.0800043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barclay WR, Busey WM, Dalgard DW, Good RC, Janicki BW, Kasik JE, Ribi E, Ulrich CE, Wolinsky E. Protection of monkeys against airborne tuberculosis by aerosol vaccination with bacillus Calmette-Guerin. Am Rev Respir Dis. 1973;107:351–358. doi: 10.1164/arrd.1973.107.3.351. [DOI] [PubMed] [Google Scholar]
- 89.Sadagopal S, Braunstein M, Hager CC, Wei J, Daniel AK, Bochan MR, Crozier I, Smith NE, Gates HO, Barnett L, Van Kaer L, Price JO, Blackwell TS, Kalams SA, Kernodle DS. Reducing the activity and secretion of microbial antioxidants enhances the immunogenicity of BCG. PLoS One. 2009;4:e5531. doi: 10.1371/journal.pone.0005531. http://dx.doi.org/10.1371/journal.pone.0005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin PL, Rodgers M, Smith L, Bigbee M, Myers A, Bigbee C, Chiosea I, Capuano SV, Fuhrman C, Klein E, Flynn JL. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect Immun. 2009;77:4631–4642. doi: 10.1128/IAI.00592-09. http://dx.doi.org/10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hel Z, Nacsa J, Kelsall B, Tsai WP, Letvin N, Parks RW, Tryniszewska E, Picker L, Lewis MG, Edghill-Smith Y, Moniuszko M, Pal R, Stevceva L, Altman JD, Allen TM, Watkins D, Torres JV, Berzofsky JA, Belyakov IM, Strober W, Franchini G. Impairment of Gag-specific CD8(+) T-cell function in mucosal and systemic compartments of simian immunodeficiency virus mac251- and simian-human immunodeficiency virus KU2-infected macaques. J Virol. 2001;75:11483–11495. doi: 10.1128/JVI.75.23.11483-11495.2001. http://dx.doi.org/10.1128/JVI.75.23.11483-11495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, Kim Y, Sidney J, James EA, Taplitz R, McKinney DM, Kwok WW, Grey H, Sallusto F, Peters B, Sette A. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog. 2013;9:e1003130. doi: 10.1371/journal.ppat.1003130. http://dx.doi.org/10.1371/journal.ppat.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mothe BR, Stewart BS, Oseroff C, Bui HH, Stogiera S, Garcia Z, Dow C, Rodriguez-Carreno MP, Kotturi M, Pasquetto V, Botten J, Crotty S, Janssen E, Buchmeier MJ, Sette A. Chronic lymphocytic choriomeningitis virus infection actively down-regulates CD4+ T cell responses directed against a broad range of epitopes. J Immunol. 2007;179:1058–1067. doi: 10.4049/jimmunol.179.2.1058. [DOI] [PubMed] [Google Scholar]
- 94.Tangri S, Mothe BR, Eisenbraun J, Sidney J, Southwood S, Briggs K, Zinckgraf J, Bilsel P, Newman M, Chesnut R, Licalsi C, Sette A. Rationally engineered therapeutic proteins with reduced immunogenicity. J Immunol. 2005;174:3187–3196. doi: 10.4049/jimmunol.174.6.3187. [DOI] [PubMed] [Google Scholar]
- 95.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A. Molecular determinants of T cell epitope recognition to the common Timothy grass allergen. J Immunol. 2010;185:943–955. doi: 10.4049/jimmunol.1000405. http://dx.doi.org/10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Alderson MR, Bement T, Day CH, Zhu L, Molesh D, Skeiky YA, Coler R, Lewinsohn DM, Reed SG, Dillon DC. Expression cloning of an immunodo-minant family of Mycobacterium tuberculosis antigens using human CD4(+) T cells. J Exp Med. 2000;191:551–560. doi: 10.1084/jem.191.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Doxiadis GG, Otting N, de Groot NG, Bontrop RE. Differential evolutionary MHC class II strategies in humans and rhesus macaques: relevance for biomedical studies. Immunol Rev. 2001;183:76–85. doi: 10.1034/j.1600-065x.2001.1830106.x. [DOI] [PubMed] [Google Scholar]
- 98.Otting N, de Groot NG, Noort MC, Doxiadis GG, Bontrop RE. Allelic diversity of Mhc-DRB alleles in rhesus macaques. Tissue Antigens. 2000;56:58–68. doi: 10.1034/j.1399-0039.2000.560108.x. [DOI] [PubMed] [Google Scholar]
- 99.de Groot N, Doxiadis GG, De Groot NG, Otting N, Heijmans C, Rouweler AJ, Bontrop RE. Genetic makeup of the DR region in rhesus macaques: gene content, transcripts, and pseudogenes. J Immunol. 2004;172:6152–6157. doi: 10.4049/jimmunol.172.10.6152. [DOI] [PubMed] [Google Scholar]
- 100.Li Pira G, Ivaldi F, Dentone C, Righi E, Del Bono V, Viscoli C, Koopman G, Manca F. Evaluation of antigen-specific T-cell responses with a miniaturized and automated method. Clin Vaccin Immunol. 2008;15:1811–1818. doi: 10.1128/CVI.00322-08. http://dx.doi.org/10.1128/CVI.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ulrichs T, Munk ME, Mollenkopf H, Behr-Perst S, Colangeli R, Gennaro ML, Kaufmann SH. Differential T cell responses to Mycobacterium tuberculosis ESAT6 in tuberculosis patients and healthy donors. Eur J Immunol. 1998;28:3949–3958. doi: 10.1002/(SICI)1521-4141(199812)28:12<3949::AID-IMMU3949>3.0.CO;2-4. http://dx.doi.org/10.1002/(SICI)1521-4141(199812)28 12<3949:: AID-IMMU3949>3.0.CO;2–4. [DOI] [PubMed] [Google Scholar]
- 102.Day CL, Mkhwanazi N, Reddy S, Mncube Z, van der Stok M, Klenerman P, Walker BD. Detection of polyfunctional Mycobacterium tuberculosis-specific T cells and association with viral load in HIV-1-infected persons. J Infect Dis. 2008;197:990–999. doi: 10.1086/529048. http://dx.doi.org/10.1086/529048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Axelsson-Robertson R, Loxton AG, Walzl G, Ehlers MM, Kock MM, Zumla A, Maeurer M. A broad profile of co-dominant epitopes shapes the peripheral Mycobacterium tuberculosis specific CD8+ T-cell immune response in South African patients with active tuberculosis. PloS One. 2013;8:e58309. doi: 10.1371/journal.pone.0058309. http://dx.doi.org/10.1371/journal.pone.0058309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tully G, Kortsik C, Hohn H, Zehbe I, Hitzler WE, Neukirch C, Freitag K, Kayser K, Maeurer MJ. Highly focused T cell responses in latent human pulmonary Mycobacterium tuberculosis infection. J Immunol. 2005;174:2174–2184. doi: 10.4049/jimmunol.174.4.2174. [DOI] [PubMed] [Google Scholar]
- 105.Shams H, Klucar P, Weis SE, Lalvani A, Moonan PK, Safi H, Wizel B, Ewer K, Nepom GT, Lewinsohn DM, Andersen P, Barnes PF. Characterization of a Mycobacterium tuberculosis peptide that is recognized by human CD4+ and CD8+ T cells in the context of multiple HLA alleles. J Immunol. 2004;173:1966–1977. doi: 10.4049/jimmunol.173.3.1966. [DOI] [PubMed] [Google Scholar]
- 106.Nagai H, Suzukawa M, Sakakibara Y, Ohta K, Reche PA, Suzuki K, Hoshino Y. Immunological responses and epitope mapping by tuberculosis-associated antigens within the RD1 region in Japanese patients. J Immunol Res. 2014;2014:764028. doi: 10.1155/2014/764028. http://dx.doi.org/10.1155/2014/764028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. http://dx.doi.org/10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Andersen P, Woodworth JS. Tuberculosis vaccineserethinking the current paradigm. Trends Immunol. 2014;35:387–395. doi: 10.1016/j.it.2014.04.006. http://dx.doi.org/10.1016/j.it.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 109.Roy A, Eisenhut M, Harris RJ, Rodrigues LC, Sridhar S, Habermann S, Snell L, Mangtani P, Adetifa I, Lalvani A, Abubakar I. Effect of BCG vaccination against Mycobacterium tuberculosis infection in children: systematic review and meta-analysis. BMJ Clin Res Ed. 2014:349. doi: 10.1136/bmj.g4643. http://dx.doi.org/10.1136/bmj.g4643.g4643. [DOI] [PMC free article] [PubMed]
- 110.Yuk JM, Jo EK. Host immune responses to mycobacterial antigens and their implications for the development of a vaccine to control tuberculosis. Clin Exp Vaccin Res. 2014;3:155–167. doi: 10.7774/cevr.2014.3.2.155. http://dx.doi.org/10.7774/cevr.2014.3.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang J, Xian Q, Guo M, Huang Z, Rao Y, Wang Y, Wang X, Bao R, Evans TG, Hokey D, Sizemore D, Ho WZ. Mycobacterium tuberculosis Erdman infection of rhesus macaques of Chinese origin. Tuberc Edinb Scotl. 2014;94:634–643. doi: 10.1016/j.tube.2014.08.005. http://dx.doi.org/10.1016/j.tube.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 112.Ravindran R, Krishnan VV, Dhawan R, Wunderlich ML, Lerche NW, Flynn JL, Luciw PA, Khan IH. Plasma antibody profiles in non-human primate tuberculosis. J Med Primatol. 2014;43:59–71. doi: 10.1111/jmp.12097. http://dx.doi.org/10.1111/jmp.12097. [DOI] [PubMed] [Google Scholar]
- 113.Min F, Zhang Y, Pan J, Wang J, Yuan W. Mycobacterium tuberculosis infection in rhesus monkeys (Macaca mulatta) and evaluation of ESAT-6 and CFP10 as immunodiagnostic antigens. Exp Anim Jpn Assoc Lab Anim Sci. 2013;62:281–293. doi: 10.1538/expanim.62.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.White AD, Sibley L, Dennis MJ, Gooch K, Betts G, Edwards N, Reyes-Sandoval A, Carroll MW, Williams A, Marsh PD, McShane H, Sharpe SA. Evaluation of the safety and immunogenicity of a candidate tuberculosis vaccine, MVA85A, delivered by aerosol to the lungs of macaques. Clin Vaccin Immunol. 2013;20:663–672. doi: 10.1128/CVI.00690-12. http://dx.doi.org/10.1128/CVI.00690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lewinsohn DM, Tydeman IS, Frieder M, Grotzke JE, Lines RA, Ahmed S, Prongay KD, Primack SL, Colgin LM, Lewis AD, Lewinsohn DA. High resolution radiographic and fine immunologic definition of TB disease progression in the rhesus macaque. Microb Infect. 2006;8:2587–2598. doi: 10.1016/j.micinf.2006.07.007. http://dx.doi.org/10.1016/j.micinf.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 116.Clark SA, Martin SL, Pozniak A, Steel A, Ward B, Dunning J, Henderson DC, Nelson M, Gazzard B, Kelleher P. Tuberculosis antigen-specific immune responses can be detected using enzyme-linked immunospot technology in human immunodeficiency virus (HIV)-1 patients with advanced disease. Clin Exp Immunol. 2007;150:238–244. doi: 10.1111/j.1365-2249.2007.03477.x. http://dx.doi.org/10.1111/j.1365-2249.2007.03477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Won DI, Park JR. Flow cytometric measurements of TB-specific T cells comparing with QuantiFERON-TB gold. Cytom Part B Clin Cytom. 2010;78:71–80. doi: 10.1002/cyto.b.20503. http://dx.doi.org/10.1002/cyto.b.20503. [DOI] [PubMed] [Google Scholar]
- 118.Lindestam Arlehamn CS, Lewinsohn D, Sette A, Lewinsohn D. Antigens for CD4 and CD8 T cells in tuberculosis. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.a018465. http://dx.doi.org/10.1101/cshperspect.a018465.a018465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lindestam Arlehamn CS, Sette A. Definition of CD4 immunosignatures associated with MTB. Front Immunol. 2014;5:124. doi: 10.3389/fimmu.2014.00124. http://dx.doi.org/10.3389/fimmu.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cho S, Mehra V, Thoma-Uszynski S, Stenger S, Serbina N, Mazzaccaro RJ, Flynn JL, Barnes PF, Southwood S, Celis E, Bloom BR, Modlin RL, Sette A. Antimicrobial activity of MHC class I-restricted CD8+ T cells in human tuberculosis. Proc Natl Acad Sci U S A. 2000;97:12210–12225. doi: 10.1073/pnas.210391497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Girard MP, Fruth U, Kieny MP. A review of vaccine research and development: tuberculosis. Vaccine. 2005;23:5725–5731. doi: 10.1016/j.vaccine.2005.07.045. http://dx.doi.org/10.1016/j.vaccine.2005.07.045. [DOI] [PubMed] [Google Scholar]
- 122.Reed S, Lobet Y. Tuberculosis vaccine development; from mouse to man. Microb Infect. 2005;7:922–931. doi: 10.1016/j.micinf.2005.03.011. http://dx.doi.org/10.1016/j.micinf.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 123.Mamo G, Mihret A, Taffesse M, Gebru G, Afework M, Yamuah LK, Wassie L, Abebe M, Aseffa A, Parida SK. T cell response to alpha crystalline and Mycobacterium tuberculosis specific antigens using ex-vivo elispot assay for detecting latent tuberculosis infection in Addis Ababa, Ethiopia. Ethiop Med J. 2014;(Suppl. 1):15–22. [PubMed] [Google Scholar]
- 124.Nyendak MR, Park B, Null MD, Baseke J, Swarbrick G, Mayanja-Kizza H, Nsereko M, Johnson DF, Gitta P, Okwera A, Goldberg S, Bozeman L, Johnson JL, Boom WH, Lewinsohn DA, Lewinsohn DM. Tuberculosis Research U. the Tuberculosis Trials C. Mycobacterium tuberculosis specific CD8(+) T cells rapidly decline with antituberculosis treatment. PLoS One. 2013;8:e81564. doi: 10.1371/journal.pone.0081564. http://dx.doi.org/10.1371/journal.pone.0081564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sutherland JS, Lalor MK, Black GF, Ambrose LR, Loxton AG, Chegou NN, Kassa D, Mihret A, Howe R, Mayanja-Kizza H, Gomez MP, Donkor S, Franken K, Hanekom W, Klein MR, Parida SK, Boom WH, Thiel BA, Crampin AC, Ota M, Walzl G, Ottenhoff TH, Dockrell HM, Kaufmann SH. Consortium GBfT. Analysis of host responses to Mycobacterium tuberculosis antigens in a multi-site study of subjects with different TB and HIV infection states in sub-Saharan Africa. PLoS One. 2013;8:e74080. doi: 10.1371/journal.pone.0074080. http://dx.doi.org/10.1371/journal.pone.0074080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yamashita Y, Hoshino Y, Oka M, Matsumoto S, Ariga H, Nagai H, Makino M, Ariyoshi K, Tsunetsugu-Yokota Y. Multicolor flow cytometric analyses of CD4+ T cell responses to Mycobacterium tuberculosis-related latent antigens. Jpn J Infect Dis. 2013;66:207–215. doi: 10.7883/yoken.66.207. [DOI] [PubMed] [Google Scholar]
- 127.Yuan K, Wu X, Zhang Q, Zhong Z, Chen J. Enzyme-linked immunospot assay response to recombinant CFP-10/ESAT-6 fusion protein among patients with spinal tuberculosis: implications for diagnosis and monitoring of surgical therapy. Int J Infect Dis IJID Off Publ Int Soc Infect Dis. 2013;17:e733–e738. doi: 10.1016/j.ijid.2013.02.027. http://dx.doi.org/10.1016/j.ijid.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 128.Borgstrom E, Andersen P, Atterfelt F, Julander I, Kallenius G, Maeurer M, Rosenkrands I, Widfeldt M, Bruchfeld J, Gaines H. Immune responses to ESAT-6 and CFP-10 by FASCIA and multiplex technology for diagnosis of M. tuberculosis infection; IP-10 is a promising marker. PloS One. 2012:7. doi: 10.1371/journal.pone.0043438. http://dx.doi.org/10.1371/journal.pone.0043438.e43438. [DOI] [PMC free article] [PubMed]
- 129.van Altena R, Duggirala S, Groschel MI, van der Werf TS. Immunology in tuberculosis: challenges in monitoring of disease activity and identifying correlates of protection. Curr Pharm Des. 2011;17:2853–2862. doi: 10.2174/138161211797470228. [DOI] [PubMed] [Google Scholar]
- 130.Van-Lume DS, De Souza JR, Cabral MM, Rego JC, Balbino V, Saad MH, Schindler HC, Abath FG, Montenegro SM. Immunological diagnosis of tuberculosis based on recombinant antigens ESAT-6 and CFP-10 in children from an endemic area in northeast Brazil. Scand J Immunol. 2010;72:460–468. doi: 10.1111/j.1365-3083.2010.02459.x. http://dx.doi.org/10.1111/j.1365-3083.2010.02459.x. [DOI] [PubMed] [Google Scholar]
- 131.Leroux-Roels I, Forgus S, De Boever F, Clement F, Demoitie MA, Mettens P, Moris P, Ledent E, Leroux-Roels G, Ofori-Anyinam O, Group MS. Improved CD4(+) T cell responses to Mycobacterium tuberculosis in PPD-negative adults by M72/AS01 as compared to the M72/AS02 and Mtb72F/AS02 tuberculosis candidate vaccine formulations: a randomized trial. Vaccine. 2013;31:2196–2206. doi: 10.1016/j.vaccine.2012.05.035. http://dx.doi.org/10.1016/j.vaccine.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 132.Dietrich J, Aagaard C, Leah R, Olsen AW, Stryhn A, Doherty TM, Andersen P. Exchanging ESAT6 with TB10.4 in an Ag85B fusion molecule-based tuberculosis subunit vaccine: efficient protection and ESAT6-based sensitive monitoring of vaccine efficacy. J Immunol. 2005;174:6332–6339. doi: 10.4049/jimmunol.174.10.6332. [DOI] [PubMed] [Google Scholar]
- 133.Bertholet S, Ireton GC, Ordway DJ, Windish HP, Pine SO, Kahn M, Phan T, Orme IM, Vedvick TS, Baldwin SL, Coler RN, Reed SG. A defined tuberculosis vaccine candidate boosts BCG and protects against multidrug-resistant Mycobacterium tuberculosis. Sci Transl Med. 2010;2:53ra74. doi: 10.1126/scitranslmed.3001094. http://dx.doi.org/10.1126/scitranslmed.3001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Coler RN, Bertholet S, Pine SO, Orr MT, Reese V, Windish HP, Davis C, Kahn M, Baldwin SL, Reed SG. Therapeutic immunization against Mycobacterium tuberculosis is an effective adjunct to antibiotic treatment. J Infect Dis. 2013;207:1242–1252. doi: 10.1093/infdis/jis425. http://dx.doi.org/10.1093/infdis/jis425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Abel B, Tameris M, Mansoor N, Gelderbloem S, Hughes J, Abrahams D, Makhethe L, Erasmus M, de Kock M, van der Merwe L, Hawkridge A, Veldsman A, Hatherill M, Schirru G, Pau MG, Hendriks J, Weverling GJ, Goudsmit J, Sizemore D, McClain JB, Goetz M, Gearhart J, Mahomed H, Hussey GD, Sadoff JC, Hanekom WA. The novel tuberculosis vaccine, AERAS-402, induces robust and polyfunctional CD4+ and CD8+ T cells in adults. Am J Respir Crit Care Med. 2010;181:1407–1417. doi: 10.1164/rccm.200910-1484OC. http://dx.doi.org/10.1164/rccm.200910-1484OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Havenga M, Vogels R, Zuijdgeest D, Radosevic K, Mueller S, Sieuwerts M, Weichold F, Damen I, Kaspers J, Lemckert A, van Meerendonk M, van der Vlugt R, Holterman L, Hone D, Skeiky Y, Mintardjo R, Gillissen G, Barouch D, Sadoff J, Goudsmit J. Novel replication-incompetent adenoviral B-group vectors: high vector stability and yield in PER.C6 cells. J Gen Virol. 2006;87:2135–2143. doi: 10.1099/vir.0.81956-0. http://dx.doi.org/10.1099/vir.0.81956-0. [DOI] [PubMed] [Google Scholar]
- 137.Agger EM, Rosenkrands I, Olsen AW, Hatch G, Williams A, Kritsch C, Lingnau K, von Gabain A, Andersen CS, Korsholm KS, Andersen P. Protective immunity to tuberculosis with Ag85B–ESAT-6 in a synthetic cationic adjuvant system IC31. Vaccine. 2006;24:5452–5460. doi: 10.1016/j.vaccine.2006.03.072. http://dx.doi.org/10.1016/j.vaccine.2006.03.072. [DOI] [PubMed] [Google Scholar]
- 138.Dietrich J, Andersen C, Rappuoli R, Doherty TM, Jensen CG, Andersen P. Mucosal administration of Ag85B–ESAT-6 protects against infection with Mycobacterium tuberculosis and boosts prior bacillus Calmette-Guerin immunity. J Immunol. 2006;177:6353–6360. doi: 10.4049/jimmunol.177.9.6353. [DOI] [PubMed] [Google Scholar]
- 139.Weinrich Olsen A, van Pinxteren LA, Meng Okkels L, Birk Rasmussen P, Andersen P. Protection of mice with a tuberculosis subunit vaccine based on a fusion protein of antigen 85b and esat-6. Infect Immun. 2001;69:2773–2778. doi: 10.1128/IAI.69.5.2773-2778.2001. http://dx.doi.org/10.1128/iai.69.5.2773-2778.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kaushal D, Foreman TW, Gautam US, Adekambi T, Rangel-Moreno J, Golden NA, Johnson-May AF, Roy CJ, Didier PJ, Doyle LA, Russell-Lodrigue KE, Blanchard JL, Rengarajan J, Lackner AA, Khader SA, Mehra S. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat Commun. 2015;6:8533. doi: 10.1038/ncomms9533. http://dx.doi.org/10.1038/ncomms9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.