Abstract
Hallmarks of Yersinia pathogenesis include the ability to form biofilms on surfaces, the ability to establish close contact with eukaryotic target cells and the ability to hijack eukaryotic cell signaling and take over control of strategic cellular processes. Many of these virulence traits are already well-described. However, of equal importance is knowledge of both confined and global regulatory networks that collaborate together to dictate spatial and temporal control of virulence gene expression. This review has the purpose to incorporate historical observations with new discoveries to provide molecular insight into how some of these regulatory mechanisms respond rapidly to environmental flux to govern tight control of virulence gene expression by pathogenic Yersinia.
Keywords: acidity, temperature, metabolism, RovA, c-di-GMP, cAMP, extracytoplasmic stress, transition metals
Yersinia biology and classical virulence traits
Pathogenic Yersinia have been a long-standing model bacteria for furthering understanding of bacteria-host cell interplay. At center stage for well-over 100 years has been the highly virulent and obligate plague-causing pathogen Y. pestis. Having only recently evolved from ancestral Y. pseudotuberculosis, a mildly virulent enteric pathogen, has meant that the Yersiniae are a model genus to study active pathogen evolution (Wren, 2003; Drancourt, 2012; Rasmussen et al., 2015). Significant evolutionary events in the formation of Y. pestis as an obligate pathogen appear to be its genome reduction and the corresponding loss of functional coding potential (Sun et al., 2014; Bolotin and Hershberg, 2015), its re-wiring of regulatory circuitry that permits elevated virulence gene expression during host infections beyond the levels achieved by its close relative Y. pseudotuberculosis (Chauvaux et al., 2011; Ansong et al., 2013), as well as the gain of genetic information such as in the form of two additional plasmids pMT1 and pPCP1 encoding the murine toxin and the plasminogen activator, respectively (Chain et al., 2004).
In many cases, Y. pseudotuberculosis and Y. enterocolitica can serve as a convenient substitute for the studies of Y. pestis pathogenicity and this has meant that much has been learned about the Yersinia infectious cycle and how they react to contact with both non-immune and immune cells. Pathogenic Yersinia produce numerous surface located proteins that could possess auto-agglutinating properties, engage with host cell surface receptors or act as serum resistance factors that limit the action of complement-mediated opsonization and killing (Figure 1). The most prominent Yersinia adhesins studied to date are invasin, YadA, Ail, and pH 6 antigen (Kolodziejek et al., 2012; Zav'yalov, 2012; Mikula et al., 2013; Muhlenkamp et al., 2015). However, their relative importance to the biology of infection is pathogen-dependent, and in certain cases may not be required at all.
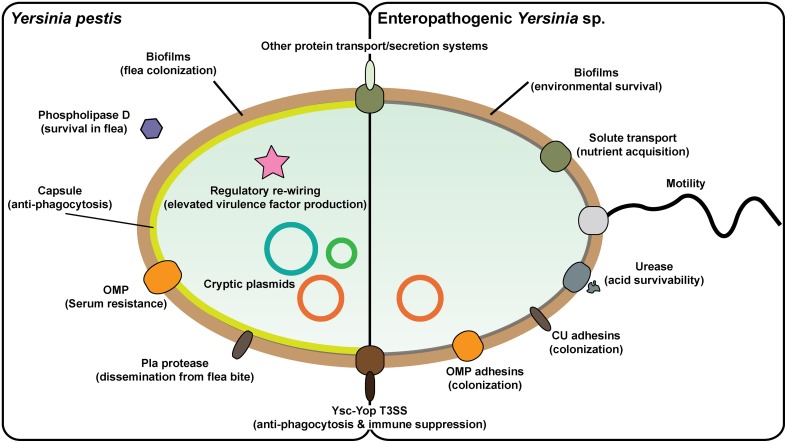
Figure 1.
Prominent Yersinia virulence factors. Yersinia pestis and enteropathogenic Y. pseudotuberculosis and Y. enterocolitica vary greatly in their pathogenicity and in aspects of their pathogenesis. This is reflected by the different repertoire of proven and potential virulence factors in their respective armories. In particular, Y. pestis has acquired additional plasmid DNA that encodes for factors that enable colonization and transmission via the flea vector and survival in blood. It is also apparent that the regulatory circuitry of Y. pestis has been rewired in ways that drive elevated in vivo expression of critical virulence associated factors. On the flip side, Y. pestis has lost flagella-mediated motility and cell-adhesive capacities that are otherwise critical for survival of the enterics both in the environment and in the GI tract, respectively. Yet commonalities between all three pathogens exist, such as the prominent virulence plasmid-encoded Ysc-Yop type III secretion system responsible for promoting an extracellular infection niche, along with other systems responsible for distributing de novo synthesized proteins into other extracytoplasmic compartments or even realized free from the bacteria.
Yersinia capitalizes on close contact with the host cell to employ a Ysc-Yop type III secretion system (T3SS) for the injection of anti-host effectors into the target cell (Keller et al., 2015; Figure 1). This system is encoded on a virulence plasmid common to all three human Yersinia pathogens, and contributes two major virulence traits to Yersinia—anti-phagocytic and immunosuppression activities (Plano and Schesser, 2013). Several additional protein secretion systems, especially including a chromosomal T3SS, a T2SS, multiple T5SSs, and T6SSs as well as chaperone-usher systems, are predicted in the genomes of pathogenic Yersinia (Yen et al., 2008), and the functionality of some of these have been verified experimentally (Haller et al., 2000; Venecia and Young, 2005; Yen et al., 2007; Felek et al., 2008, 2011; Lawrenz et al., 2009; Robinson et al., 2009; Hatkoff et al., 2012; Lenz et al., 2012; Pisano et al., 2012; Seo et al., 2012; Von Tils et al., 2012; Lane et al., 2013; Walker et al., 2013; Nair et al., 2015; Wang et al., 2015; Figure 1).
Biofilm formation by pathogenic Yersinia is another significant virulence trait (Figure 1). The ability of Y. pestis to form biofilms in fleas is considered a major evolutionary catalyst by providing a means of bacterial transmission from the flea to mammalian host (Darby, 2008; Hinnebusch and Erickson, 2008; Sun et al., 2014). The hms locus is encoded on the chromosome in a high pathogenicity island (HPI) and contributes to virulence of Y. pestis and environmental survival of the enteric Yersinia. The hms locus is responsible for the biosynthesis and secretion of an exopolysaccharide polymer (EPS) matrix material that helps to form highly aggregative biofilm (Darby et al., 2002; Jarrett et al., 2004; Kirillina et al., 2004).
With this knowledge of determinants contributing to bacterial survival in diverse environmental niches, the human pathogenic Yersinia represent ideal model systems for studying the environmental regulation of gene expression. Y. pestis has a strict lifecycle but still alternates between flea vector and mammalian host, while the food-borne enteropathogens establish environmental niches in soil and water along with intermittent mammalian host infections. As such, this bacterial family encounters many unique environments that all undergo continuous physical and chemical flux. With the capacity to sense this physicochemical flux, pathogenic Yersinia respond by utilizing impressive regulatory networks to coordinate the temporal and spatial control of collections of often unlinked genetic loci. This review aims to highlight some of these important sensory and regulatory networks that have capacity to facilitate rapid reprogramming of global gene expression profiles to enable Yersinia to adapt, survive, and prosper in selected environmental niches.
The mainstays of environmental sensing by Yersinia
Responsiveness to acidity
With pH values as low as 1.5–2.5, the acidic environment of the mammalian stomach is a natural barrier against infections of food-borne pathogens. Gastrointestinal bacterial pathogens have thus evolved elaborate mechanisms to cope with excursions into acidic environments. Acid survival mechanisms are remarkably different among different pathogens. To cope with different degrees of acidic environmental stress, several acid survival systems have evolved in enteric bacteria e.g., acid resistance (AR), acid tolerance response (ATR), and acid habituation (AH), and examples of these have been well-documented (Foster, 2004). In particular, at least four acid resistance (AR) systems have been documented. The glucose-repressed AR1 system is controlled by the regulators cAMP receptor protein (CRP) and RpoS (Foster, 2004). The other three AR systems (AR2, AR3, and AR4) are decarboxylase/antiporter-dependent systems that function in pH homeostasis by coupling extracellular glutamate (AR2), arginine (AR3), or lysine (AR4) and their corresponding amino acid decarboxylases GadA/B, AdiA, and CadA, with the cognate antiporters GadC, AdiC, and CadB, respectively (Foster, 2004; Song et al., 2015). The two gastrointestinal pathogens, Y. pseudotuberculosis and Y. enterocolitica transmit to humans after the ingestion of contaminated water or food. Like many food-borne pathogens, they have developed different survival systems that protect against acidic conditions for successful colonization and infection.
Carbohydrate metabolism and acid survival
The role of carbohydrate metabolism in acid survival of enteric bacteria remains largely unknown. Cyclic AMP receptor protein (CRP), which is a hallmark of glucose metabolism regulation, is a regulator of acid survival in Escherichia coli (Castanie-Cornet et al., 1999). Significantly, the global transcriptional regulator, Cra (cAMP-independent catabolite repressor/activator), also negatively regulates acid tolerance in Y. pseudotuberculosis (Hu et al., 2011). The Cra targets for acid survival regulation remain unknown as does its mechanism of action, but presumably Cra mediates this regulatory role via transcriptional regulation. Further experiments are needed to identify specific regulators to obtain more detailed information about carbohydrate metabolism and acid survival in Y. pseudotuberculosis.
Amino acid metabolism and acid survival
More is known about the important connections between amino acid metabolism and acid survival in bacteria. Several acid resistance systems, e.g., glutamate-, arginine-, or lysine-dependent, have been described in E. coli (Foster, 2004). Notably, several key genes in amino decarboxylase or antiporter-dependent acid resistance systems are absent in the genome of Y. pseudotuberculosis, which raises the question of whether other amino acids are involved in acid survival in this bacterium. Indeed, the enzyme aspartate ammonia lyase or aspartase (AspA), which is involved in aspartate metabolism by catalyzing the deamination of L-aspartate to form fumarate and ammonia, plays a role in acid survival in Y. pseudotuberculosis (Hu et al., 2010). AspA increases acid survivability of bacteria by producing ammonia from aspartate as demonstrated by mutational and in vitro enzyme activity studies. Interestingly, this aspartate-dependent acid survival pathway appears to exist in Y. enterocolitica as well as other food-borne pathogenic bacteria including E. coli O157:H7 and Salmonella enterica, given that the addition of aspartate into culture media also increases their survivability at low pH (Hu et al., 2010). This observation suggests that this enzyme could be a universal mechanism for acid survival of gastric bacteria and might therefore represent a notable target to develop new drugs for the control of bacterial infections. The reasons for why bacteria choose to couple different amino acid utilization pathways with acid responses is unknown, but is certainly worth further investigation.
Stress-related proteins and acid survival
The enzyme urease is a major player in the resistance to acidity and plays a central role in colonization and persistence in the host. Consistent with this, urease is constitutively active and comprises between 5 and 10% of the total cellular protein (Stingl and De Reuse, 2005). Urease catalyzes the hydrolysis of urea to yield ammonia, which neutralizes the presence of protons to mitigate acidity (Miller and Maier, 2014). Earlier studies have demonstrated that urease is responsible for an ATR in Y. enterocolitica (De Koning-Ward and Robins-Browne, 1995), and a urease mutant of Y. pseudotuberculosis has lost its ability to survive at pH 3.0 in the presence of urea (Riot et al., 1997; Figure 1). Using comparative proteomic analyses to identify global protein synthesis changes in Y. pseudotuberculosis that were induced by growth at pH 4.5 (a sub-lethal pH to this bacterium) compared to neutral pH, further highlighted the importance of urease in acid survival (Hu et al., 2009). Moreover, the OmpR response regulator of the EnvZ/OmpR two-component regulatory system (TCRS), was found to activate urease synthesis to enhance acid survival (Hu et al., 2009). This regulatory control appears to be direct, for the regulatory regions of the multiple urease polycistronic transcriptional units are all individually recognized by specific OmpR binding (Hu et al., 2009).
Other features of acid survival in Yersinia
Studies on acid survival in Yersinia mainly focus on mild acid conditions. Whether and how Yersinia survives within an extremely acidic environment (pH < 2.0) remains unclear. As a transcriptional regulator, OmpR may also regulate other acid survival pathways. In a recent study in Y. pseudotuberculosis, the production of a thermo-regulated type VI secretion system (T6SS4) was OmpR-regulated, and a direct relationship between this secretion system and an ATR was observed (Zhang et al., 2013a). The involvement of OmpR-regulated T6SS in pH homeostasis and acid tolerance is through proton efflux, and is dependent upon the ATPase activity of ClpV4—a core component of all T6SSs—that participates in proton extrusion (Zhang et al., 2013a). However, whether the T6SS4 is directly acting as a proton transporter remains obscure. Nevertheless, this is a novel acid survival strategy in which a protein secretion system associated with virulence has also an unexpected role in proton extrusion under acid conditions.
In Y. pseudotuberculosis, Song et al. recently demonstrated that RovM, a central regulator of the CsrABC-RovM-RovA cascade, inversely regulates two established acid survival systems (Song et al., 2015). In particular, RovM bound the promoters of T6SS4 genes to activate T6SS4 synthesis, but also bound to the −35 element in the arginine-dependent acid resistance system (AR3) promoter to repress AR3 synthesis. The authors proposed that RovM coordinately regulates the production of AR3 and T6SS4 in response to the availability of nutrients in the environment (Song et al., 2015).
In addition to OmpR, other TCRSs have also been reported to play a role in acid stress responsiveness by Yersinia. The response regulator PhoP of the PhoP-PhoQ TCRS is necessary for survival of Y. pseudotuberculosis in macrophages (Grabenstein et al., 2004; Bozue et al., 2011). These data were corroborated by a systematic analyses of two-component regulons in Y. pseudotuberculosis in which acid responsiveness depended upon intact phoP- and ompR-dependent regulatory pathways, as well as noting an involvement of the pmrA-dependent regulatory pathway (Flamez et al., 2008). The phoP gene of Y. pestis is also required for intracellular survival in macrophages and depending on the transmission route, also for virulence (Oyston et al., 2000; O'loughlin et al., 2010; Bozue et al., 2011). However, rather than being involved in acid responsiveness per se, it seems most likely that Y. pestis PhoP actually regulates essential survival genes (Grabenstein et al., 2006; O'loughlin et al., 2010) perhaps through its well-known ability to sense magnesium ions (Zhou et al., 2005; Li et al., 2008). Finally, the twin arginine translocation (Tat) pathway, which is essential for bacterial virulence, has also been demonstrated to contribute to acid survival in Y. pseudotuberculosis (Lavander et al., 2006). Though different acid survival systems have been documented in Yersinia, the molecular mechanisms integrating the coordination of these are not fully understood. No doubt an increase knowledge of acid survival mechanisms could benefit possibilities to develop broad-spectrum novel strategies for the prevention and treatment of infections by Yersinia and other food-borne pathogens.
Acid response systems in Yersinia pestis
Y. pestis does not utilize the intestinal route of infection, but is still likely to encounter acidic environments during the infectious process, especially upon internalization by immune cells (Lukaszewski et al., 2005; Spinner et al., 2014). Hence, it is anticipated that Y. pestis would require acid responsive systems for survival during infections. Indeed, in silico analyses suggests all known acid survival systems are complete and intact in Y. pestis, except for the established loss of urease activity (Table 1). Actually, all Y. pestis strains contain a mutated ureD gene that abolishes urease activity, and this is considered to be a key evolutionary step that facilitated the adaptation of Y. pestis to the flea-borne transmission route (Chouikha and Hinnebusch, 2014). Interestingly, the AR3 system is widespread in all three human pathogenic Yersinia, and these represent general acid survival/tolerance systems. However, no obvious acid survival related metabolic protein or AR system seems to be solely Y. pestis specific, which is consistent with the idea that these bacteria promote intracellular survival by dedicated mechanisms that avoid acidification of the Y. pestis-containing vacuoles (Pujol et al., 2009). By extension, the AR2 system is present only in Y. enterocolitica, and is most probably important for safe passage of these bacteria through the acidified environment of the mammalian stomach, thus offering an explanation as to why this bacterium seems to be more resistant to acid environments than is Y. pseudotuberculosis (Hu et al., 2010).
Table 1.
Comparison of acid survival and tolerance systems in all three human pathogenic Yersinia.
| Acid survival systems | Y. enterocolitica | Y. pseudotuberculosis | Y. pestis |
|---|---|---|---|
| METABOLIC FACTORS | |||
| Cra | ✓ | ✓ | ✓ |
| OmpR | ✓ | ✓ | ✓ |
| PhoP | ✓ | ✓ | ✓ |
| RovM | ✓ | ✓ | ✓ |
| Urease | ✓ | ✓ | Non-functional due to ureD mutation |
| AR2 SYSTEM | |||
| GadA | ✓ | ✘ | ✘ |
| GadB | ✓ | ✘ | ✘ |
| GadC | ✓ | ✘ | ✘ |
| AR3 system | |||
| AdiA | ✓ | ✓ | ✓ |
| AdiC | ✓ | ✓ | ✓ |
| AR4 SYSTEM | |||
| CadA | ✓ | ✘ | ✘ |
| CadB | ✘ | ✘ | ✘ |
| ClpV4 | ✘ | ✓ | ✓ |
✘: not detected (absent). ✓: present and genetically intact.
Alkalinity and Na+/H+ antiport
Precious little information describes the adaptation of Yersinia to alkaline environments. However, in silico evidence indicates that Y. pestis encodes the capacity to couple sodium ion cycling to energy metabolism, and this would constitute a bacterial adaptation strategy to maintain pH homeostasis particularly when exposed to alkaline environments (Hase et al., 2001; Mulkidjanian et al., 2008; Ganoth et al., 2011). To begin to understand the role of sodium ion cycling in Y. pestis physiology, Minato and colleagues established knockouts of the loci encoding the primary Na+ ion pump, NQR, and the secondary Na+ ion pumps known as the NhaA and NhaB Na+/H+ antiporters (Minato et al., 2013). They found that Y. pestis lacking both antiport systems were attenuated demonstrating clearly a role for Na+/H+ antiport in virulence. Interestingly, the NhaA Na+/H+ antiporter activity is pH dependent with maximal activity exhibited in alkaline conditions (Ganoth et al., 2011). Taken together, this suggests that the obligate lifecycle of Y. pestis demands that it has the potential to adapt to alkaline environments, and at least one of these adaptation mechanisms is via the NhaA antiport system.
Global effects of temperature
Temperature varies widely as microbes alternate between environmental, invertebrate, and/or vertebrate reservoirs. It is therefore commonly sensed by pathogens to recognize their environment and control virulence gene expression. Multiple molecular mechanisms of temperature-dependent control are well-described in the literature. In Yersinia, two prominent thermally controlled virulence properties are the plasmid encoded Ysc-Yop T3SS and the global regulator RovA.
Thermoregulation of the Ysc-Yop T3SS
It has been known for many years that temperature upshift from ambient temperature to 37°C and target cell contact are crucial to triggering Ysc secretion apparatus synthesis and the subsequent translocation of its specific cargo (Rosqvist et al., 1994; Persson et al., 1995; Pettersson et al., 1996). A cornerstone of this temperature response involves the AraC-like transcriptional activator, LcrF (Yother et al., 1986; Cornelis et al., 1989; Hoe et al., 1992; Lambert De Rouvroit et al., 1992; Hoe and Goguen, 1993; Figure 2A). Through a C-terminal helix-turn-helix motif, LcrF binds to DNA sequences overlapping the −35 region of σ70-dependent promoters to auto-activate lcrF expression as well as activate the expression of other ysc and yop genes (Wattiau and Cornelis, 1994; King et al., 2013). At ambient temperature, the DNA architecture of the lcrF promoter is conducive to binding by the small nucleoid associated protein YmoA and this impedes transcriptional output (Cornelis et al., 1991; Bohme et al., 2012). Furthermore, the presence of a complex stem loop structure in the 5-prime untranslated region of lcrF mRNA transcripts conceal the Shine-Dalgarno sequences from the ribosome to prevent its translation (Bohme et al., 2012). However, an elevation in the surrounding temperature sees a conformational change in the DNA curvature encompassing the lcrF promoter (Rohde et al., 1994, 1999) and also in the product of its transcription (Hoe and Goguen, 1993; Bohme et al., 2012; Figure 2B). Consequently, YmoA binding affinity is diminished and this free protein is degraded by the ClpP and Lon proteases (Jackson et al., 2004; Bohme et al., 2012). This allows RNA polymerase holoenzyme access to the lcrF promoter, and in collaboration with LcrF, will dramatically enhance transcription. In parallel, stem-loop structures in the lcrF mRNA denature, and this establishes ribosomal recognition and subsequent LcrF translation (Hoe and Goguen, 1993; Bohme et al., 2012). In this way, at least two levels of thermo-control regulate lcrF expression, which in turn impacts on the ability to synthesize Ysc proteins for assembly into a T3SS.
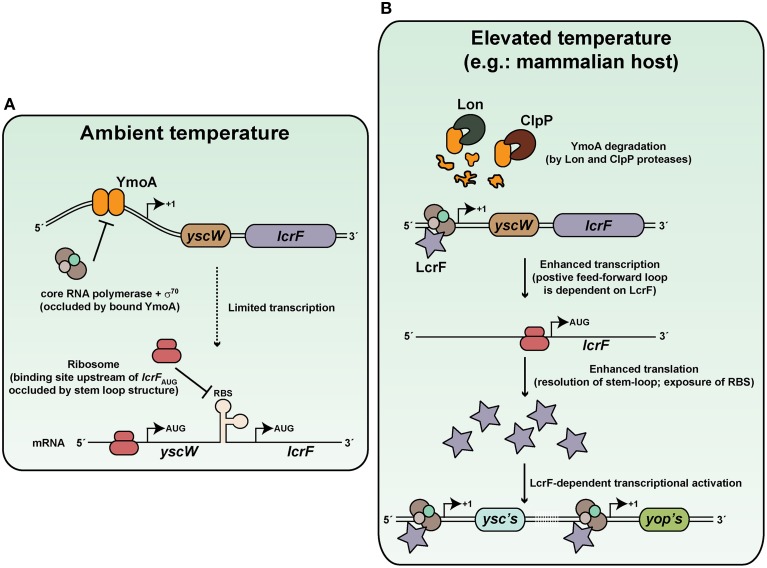
Figure 2.
Thermoregulation of Ysc-Yop type III secretion by Yersinia. Thermoregulation of Ysc-Yop type III secretion is mediated through control of the transcriptional activator, LcrF. (A) At ambient temperature, transcription of the yscW-lcrF operon is inhibited by the YmoA DNA binding protein. Furthermore, post-transcriptional inhibition occurs as a result of stem-loop formation of mRNA within the intergenic region between the two alleles. (B) De-regulation occurs at elevated temperature because YmoA affinity for the yscW-lcrF operon promoter is dramatically diminished, and this promotes operon transcription. Moreover, elevate temperature resolves the stem-loop structure in mRNA transcripts, so that translation into LcrF can proceed. This results in a positive auto-regulatory cascade that enhances lcrF transcription. Accumulated LcrF can then transcriptionally activate responsive ysc and yop promoters. This illustration is inspired in part from Bohme et al. (2012) and initial artistic work of Tiago Costa.
However, recent research has demonstrated that LcrF levels are directly controlled by additional regulatory factors that include the IscR iron-sulfur cluster regulator (Miller et al., 2014), the LysR-like transcriptional regulator YtxR (Axler-Diperte et al., 2009), CpxR of the CpxA-CpxR two-component system (Carlsson et al., 2007a; Liu et al., 2012) and RscB of the Rsc phosphorelay system (Li et al., 2015). Thus, despite Yersinia's responsiveness to temperature fluctuations having a major impact on Ysc-Yop T3SS control, other distinct regulators are clearly required to enable these bacteria to further fine-tune LcrF production in response to additional environmental cues. The precise interplay between these different regulators has not been investigated. However, it seems that YtxR functions to prevent engagement of a positive feed-forward loop by competing with LcrF for overlapping binding sites within target promoters in Y. enterocolitica (Axler-Diperte et al., 2009).
Thermoregulation of RovA
A second example of thermoregulation involves RovA, a MarR-type dimeric winged-helix DNA-binding protein (Ellison and Miller, 2006b). RovA is a master regulator of several physiological properties of pathogenic Yersinia, including metabolic and stress adaptation as well as virulence (Cathelyn et al., 2007; Yang et al., 2010). Transcription of rovA is positively auto-regulated in a temperature-dependent manner (Heroven et al., 2004; Zhang et al., 2011), while repression occurs largely through the DNA binding elements H-NS and/or YmoA in complex with RovM out-competing RovA for binding within the extended regulatory region upstream of rovA (Heroven et al., 2004; Tran et al., 2005; Ellison and Miller, 2006a; Figure 3). Intriguingly, thermoregulation of RovA occurs post-translationally, and is mechanistically defined by elevated temperature imparting intrinsic structural changes in the RovA homodimer that then specifically limits target DNA binding (Herbst et al., 2009; Quade et al., 2012). In turn, these temperature-induced conformational changes renders RovA more susceptible to proteolytic processing by the ClpP and Lon proteases (Herbst et al., 2009). Critically, thermosensing is inherent to the structural properties of RovA, for the close relative SlyA is thermotolerant (Quade et al., 2012). Hence, Yersinia has adapted a global regulator into a unique protein thermosensor that presumably affords these bacteria the capacity to rapidly adapt both environmental survival and virulence properties to the prevailing temperature conditions. However, consistent with its prominent role in coordinating multiple Yersinia physiological functions, the need to keep RovA levels closely checked has resulted in the integration of additional cues such as nutrient availability, bacterial growth phase and extracytoplasmic stress responsiveness (Heroven and Dersch, 2006; Heroven et al., 2008; Liu et al., 2011; Nuss et al., 2014). Some interesting aspects of these regulatory features are discussed later on in this review.
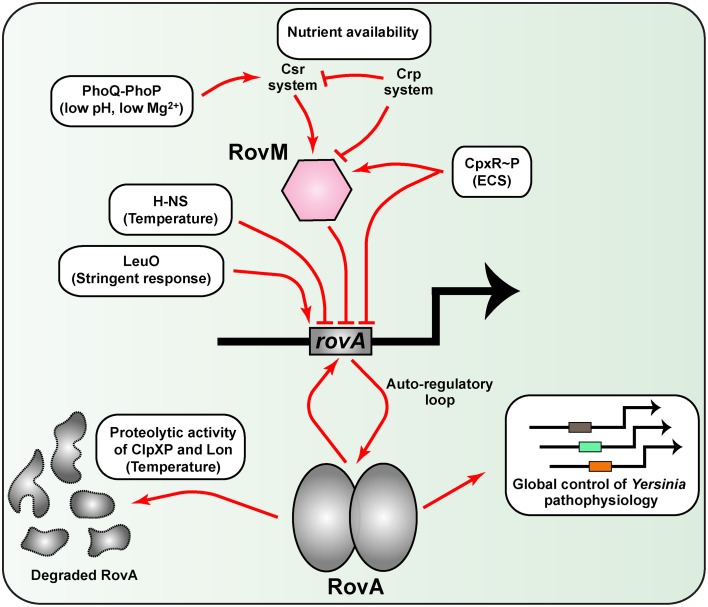
Figure 3.
A network of diverse regulatory inputs controls RovA transcriptional output in Yersinia. Available RovA is strictly controlled by cascade regulation at both the transcriptional and post-transcriptional levels in response to multiple environmental cues. The strongest influence on RovA production is through two opposing pathways. The first is an auto amplification loop, which in turn is responsive to thermo-regulated proteolysis of RovA by ClpXP and Lon proteases. The second is via RovM that is principally mediated by the prominent Csr and Crp pathways responsive to carbon and glucose availability. Other regulatory pathways are known, but the extent to which they alter RovM or RovA levels is less clear. In the diagram, induction of RovA expression is indicated by an arrow, while repression is indicated by a blunted line. For simplicity, information concerning whether the pathway is direct or indirect has been omitted on the basis that this is not always defined.
Survival in noxious environments
Protein content in the membrane of Gram negative bacteria contributes important patho-physiological functions necessary for viability. Exposure of membrane to damaging agents therefore poses a significant threat to bacterial survival, for when membrane integrity is compromised, so too is protein transport, and their folding and assembly in these compartments. To circumvent this, bacterial exposure to membrane damaging agents induce physiological responses that are termed extracytoplasmic stress (ECS) responses that ultimately serve to maintain the integrity of the bacterial envelope entity and ensure that the proteins residing in these compartments are functionally able to sustain life. The five known pathways responsive to ECS are the Bae, Cpx, Psp, Rcs, and σE pathways, most of which are well-characterized especially in E. coli (Figure 4). Crucially, accumulating evidence indicates that ECS responsiveness via some of these pathways are also a major regulator of bacterial virulence gene expression, and this is true of pathogenic Yersinia (Flores-Kim and Darwin, 2014).
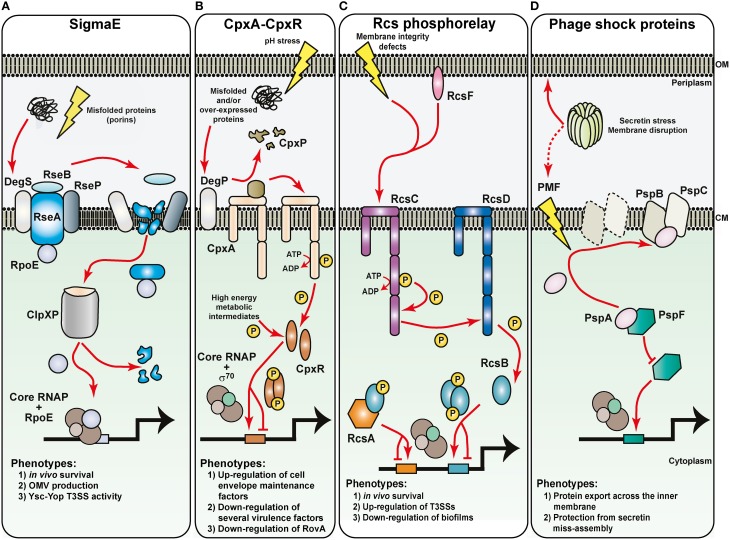
Figure 4.
Sensing of noxious extracytoplasmic stress by the bacteria envelope of Yersinia. The molecular basis for the activation of four prominent extracytoplasmic stress sensing sentinels are displayed, along with a summary of their respective phenotypic effects in pathogenic Yersinia. Maintaining the outer membrane (OM) are the RopE-, Cpx-, and Rcs-pathways. (A) Outer membrane protein misfolding initiates digestion of the anti-RpoE factor, RseA, through successive proteolytic actions of the DegS, RseP, and ClpXP proteases. Free RpoE is released into the cytoplasm, and when engaged with core RNA polymerase (RNAP), can establish controlled transcription of a large RpoE-regulon. The Cpx two-component system (B) and the Rcs phosphorelay system (C) both rely upon sensor kinase autophosphorylation (CpxA and RscC, respectively) to initiate the transduction of phosphate through to the cytoplasmic cognate response regulator (CpxR and RcsB, respectively). CpxA activation requires the DegP-dependent release of inhibitory CpxP, while RcsC activation might utilize an RcsF-dependent pathway. Inorganic phosphate can also be donated from unstable high-energy metabolic intermediates. Active phosphorylated response regulators dimerize and in concert with the house-keeping RNAP holoenzyme, target specific responsive promoters to influence transcriptional output. RcsB transcriptional control sometimes requires partners with RcsA. (D) Controlling the integrity of the cytoplasmic membrane (CM) are the phage shock proteins (psp). Secretin complex mislocalization to the cytoplasmic membrane risks dissipating the proton motive force (PMF). This is prevented by the PspB and PspC proteins actively sequestering the anti-PspF factor, PspA, to free up the PspF transcription factor to initiate promoter targeting and transcriptional output by the house-keeping RNAP holoenzyme.
σE-dependent cell envelope stress response
Arguably the most prominent sentinel of ECS is the σE (RpoE) pathway, which is also a model for regulated proteolysis (Barchinger and Ades, 2013; Guo and Gross, 2014; Paget, 2015). Reflecting this prominence, the σE-dependent cell envelope stress response in E. coli boasts the largest regulon of all the five pathways (Bury-Mone et al., 2009). The key players in this pathway are the transcription factor σE, the inner membrane-located anti-sigma factor RseA and RseB, the inner membrane proteases DegS and RseP and the cytoplasmic protease ClpXP (Figure 4A). In unstressed cells, a complex of RseA-RseB binds σE. Upon exposure to ECS, damaged intermediates that fall off the outer membrane protein assembly pathway accumulate in the periplasm, where they are sensed by, and activate the inner membrane protease DegS. Activated DegS forces RseB to detach from RseA, exposing the periplasmic domain of the latter to DegS-mediated proteolytic digestion. The remaining transmembrane portion of RseA then becomes a target for further digestion by the second inner membrane protease RseP. Now released into the cytoplasm, a soluble complex of σE bound to the cytoplasmic domain of RseA is specifically targeted by adapter proteins to the molecular chaperone ClpXP. ClpXP-mediated digestion of RseA enables release of σE. Free σE then competes with other sigma factors for core RNA polymerase (RNAP). The RNAP-σE holoenzyme then activates expression of the σE regulon that involves very many factors that contribute to the transport, assembly, and turn-over of outer membrane proteins.
In Yersinia, the rpoE gene is either essential (Heusipp et al., 2003) or essential for growth upon exposure to stress (Palonen et al., 2013). It follows that rpoE expression is inducible in Y. enterocolitica during a mouse model of infection (Young and Miller, 1997), as are known σE-regulon members when these bacteria are exposed to intracellular stress following phagocytosis by macrophages (Yamamoto et al., 1997). This indicates that RpoE might be responsible for controlling the synthesis of certain Yersinia determinants especially needed for in vivo survival (Figure 4A). This is not without precedent for the idea of a virulence specific regulatory role for rpoE in bacterial pathogens has been suggested already based upon analyses of in silico data (Rhodius et al., 2006). Consistent with this idea, genetically elevating σE levels via the removal of the anti-σE regulator, RseA, from Y. pestis resulted in the over-production of outer membrane vesicles (Eddy et al., 2014) that in other bacteria are a known contributor to pathogenicity (Ellis and Kuehn, 2010; Avila-Calderon et al., 2015). Using a similar genetic approach in Y. pseudotuberculosis, it was demonstrated that σE accumulation enhanced the synthesis of the virulence plasmid encoded Ysc-Yop T3SS (Carlsson et al., 2007a). Moreover, a gene deletion of rpoE resulted in lower production of Ysc-Yop components (Carlsson et al., 2007a; Figure 4A). Since ysc-yop gene expression is dependent on the housekeeping RNAP-σ70 holoenzyme, the effect of σE on Ysc-Yop production is probably indirect and points toward the possible involvement of one or more periplasmic protein quality control factors in the assembly of an Ysc-Yop T3SS in the Yersinia cell envelope. Thus, together these studies point toward vital regulatory roles of σE in pathogenic Yersinia that are central for ensuring survival under both extracellular environmental stress and intracellular stress.
Cpx two-component pathway
The CpxA-CpxR system is a classic TCRS, and responds to ECS (Hunke et al., 2012; Raivio, 2014). CpxA is located in the inner membrane and possesses auto-kinase activity. Additionally, it is both a kinase and phosphatase to the cognate CpxR response regulator located in the cytoplasm (Figure 4B). A third component, the periplasmic located CpxP, is responsible for interacting with CpxA to mediate ECS signal recognition (Tschauner et al., 2014). Upon sensing ECS, CpxA is freed from the clutches of CpxP, which is subsequently degraded by the DegP protease. CpxA then auto-phosphorylates and the phosphate group is readily passed on to CpxR. Active CpxR (CpxR~P) then acts as a transcription factor to activate or repress in the vicinity of 100 gene targets in non-pathogenic laboratory E. coli (Bury-Mone et al., 2009; Price and Raivio, 2009) or the pathogens Haemophilus ducreyi and Vibrio cholera (Labandeira-Rey et al., 2010; Gangaiah et al., 2013; Acosta et al., 2015). As ECS causes protein misfolding in the periplasm, this is counteracted by CpxR~P activating the production of protein folding and degradation factors that are destined to exert their function in the periplasm on these misfolded proteins. Additional regulon members also include LPS and phospholipid biosynthesis and transport operons. These are turned on by an active CpxR~P, which serves to maintain and enhance membrane integrity and barrier function. The Cpx pathway therefore functions to optimize bacterial fitness in harsh growth environments, and is a role conserved in many Gram negative bacteria (De Wulf et al., 2000).
Extensive work with this system in Y. pseudotuberculosis confirms the sentinel role of CpxR~P in maintaining cell envelope integrity. More significantly however, it was demonstrated that high CpxR~P levels accumulate in mutants devoid of CpxA phosphatase activity—most probably via the indiscriminate action of certain metabolic intermediates that can act as phosphodonors (Liu et al., 2011, 2012)—to repress gene transcription of essential Yersinia virulence determinants including the surface adhesin invasin and the plasmid encoded Ysc-Yop T3SS (Carlsson et al., 2007a,b; Liu et al., 2012; Figure 4B). This repression can be direct, for CpxR~P is able to bind to the promoter regions of the inv gene (encoding for invasin) and the rovA gene (encoding for the positive regulator of inv expression; Carlsson et al., 2007a,b; Liu et al., 2012) as well as to the promoter regions of numerous ysc-yop encoding operons including lcrF (encoding the AraC-like transcriptional activator of ysc-yop gene expression; Liu et al., 2012). As a consequence, CpxR~P accumulation leads to an acute reduction in Y. pseudotuberculosis toxicity toward infected human tissue culture cell lines (Carlsson et al., 2007b; Liu et al., 2011). Hence, it appears that the CpxA-CpxR signaling pathway functions in sensing noxious ECS to maintain envelope integrity while repressing virulence gene expression; processes crucial to Yersinia survivability in extreme environments.
With the ability to repress a large array of virulence determinants in several different bacterial pathogens, it is not surprising that genetically activated Cpx signaling results in virulence attenuation of bacteria in various in vivo infection models (Humphreys et al., 2004; Herbert Tran and Goodrich-Blair, 2009; Spinola et al., 2010; Leuko and Raivio, 2012; Debnath et al., 2013; Bontemps-Gallo et al., 2015; Thomassin et al., 2015). Yet, it remains to be seen if a fully intact Cpx system is actually activated in the in vivo environment of a eukaryotic host. After all, the primary role of the Cpx signaling pathway might be in free-living bacterial populations exposed to the naturally occurring membrane-damaging elements, and where the production of virulence determinants is seldom required.
Bacteria utilize cascade regulation to establish robust regulatory networks for the tight control of gene expression. Recent work indicates that Cpx signaling affects the levels of other regulatory factors, including non-coding regulatory RNAs, creating integrated regulatory networks (Vogt et al., 2014). In all three human pathogens of Yersinia, the RovA molecule is a key global regulator of gene expression (Cathelyn et al., 2006, 2007). The regulation of RovA is complex and involves autoregulation as well as global regulatory factors that sense products of central metabolism including the carbon storage regulatory system (Csr), cAMP receptor protein (Crp), and RovM (Heroven et al., 2008). Moreover, under genetically manipulated conditions where CpxR~P is known to accumulate to high levels, it was shown that CpxR~P also targets the rovA promoter and represses transcriptional output (Liu et al., 2011; Figure 4B). This suggests a scenario whereby Yersinia has evolved a network of integrated regulatory elements that work together to coordinate gene expression in response to both nutrient availability and ECS.
Rcs phosphorelay pathway
The complex Rcs signal transduction phosphorelay system is largely exclusive to the Enterobacteriaceae family (Majdalani and Gottesman, 2005; Huang et al., 2006). Membrane bound RcsC is the sensor kinase that upon auto-phosphorylation then transfers the phosphoryl group to the membrane-bound intermediate phosphotransfer protein RcsD, which then transfers it to RcsB, the response regulator. Phosphorylated RcsB monomers dimerize to establish binding to promoter targets, but this may instead require a monomer to first heterodimerize with the auxiliary protein RcsA. An outer membrane lipoprotein-like factor, RcsF, can also stimulate Rcs phosphorelay, but many RcsF-independent inputs also exist (Figure 4C). Most inputs are likely to have resulted from disruptions to cell envelope integrity or alterations in its composition (Majdalani and Gottesman, 2005; Huang et al., 2006). First identified as a regulator of capsule biosynthesis, it is now recognized as an important regulator of diverse processes such as cell division, flagella biosynthesis and motility, small regulatory RNA biosynthesis, biofilm formation, and pathogenicity (Majdalani and Gottesman, 2005; Huang et al., 2006).
The rcs loci are present in all Yersinia species, but in Y. pestis the rcsA allele is non-functional and the rcsD allele is frameshifted but a functional product is produced (Hinchliffe et al., 2008; Sun et al., 2008). For this reason, initial studies of the Rcs phosphorelay system focused mainly on the enteropathogenic Yersinia to reveal an important role in bacterial survival when grown under a variety of environment stresses (Hinchliffe et al., 2008) or during the initial stages of gastrointestinal colonization in a murine model of infection (Venecia and Young, 2005; Figure 4C). Consistent with this, transcriptional profiling indicated an extensive Rcs regulon in Y. pseudotuberculosis with many targets functionally linked to the bacterial envelope or in survival within the host or when free-living in the environmental (Hinchliffe et al., 2008). Thus, the finding that RcsB positively regulates the plasmid-encoded ysc-yop T3SS genes in Y. pseudotuberculosis (Li et al., 2015; Figure 4C) is significant for it establishes a direct involvement of the Rcs system as a player in systemic infections when Yersinia is in direct contact with phagocytic immune cells. Interestingly, the Rcs system also positively influences the chromosomal-encoded ysa-ysp T3SS genes that play an important role during the early gastrointestinal stage of murine infections by Y. enterocolitica (Venecia and Young, 2005). Control of ysa-ysp expression is further complicated by the involvement of a second regulatory element, the Ysr phosphorelay system, which ironically might be analogous to the Rcs system (Walker and Miller, 2004, 2009). This suggests that the Rcs and Ysr systems can respond to distinct environmental niches to impart spatial control on virulence gene expression as a strategy to promote both initial colonization and also systemic dissemination into deeper tissue. A deeper understanding of how these pathways coordinate the regulatory events in the different bacteria will therefore provide valuable insight into key aspects of enteric Yersinia pathogenicity.
In highly virulent Y. pestis, the ability to form biofilms is a key in the initial colonization of the flea foregut and the subsequent flea-borne transmission of this pathogen. Initially, it had been observed that the Rcs pathway is a inhibitor of biofilm in both Y. pestis and Y. pseudotuberculosis (Sun et al., 2008). Subsequent studies have revealed that a complex of RcsAB is a direct repressor of Yersinia biofilm by targeting the hmsCDE, hmsT, and hmsHFRS loci that encode for enhancers of biofilm development (Sun et al., 2012; Fang et al., 2015; Guo et al., 2015; Figure 4C). Both HmsD and HmsT are diguanylate cyclases required for c-di-GMP production (Bobrov et al., 2011; Sun et al., 2011), an activity enhanced by the regulatory function of HmsC and HmsE (Ren et al., 2014; Bobrov et al., 2015). HmsR and HmsS might be responsible for sensing c-di-GMP and along with the remainder of the hmsHFRS operon are responsible for the biosynthesis and translocation of biofilm forming exopolysaccharide (Bobrov et al., 2008; Abu Khweek et al., 2010). Thus, it is the pseudogenization of rcsA during the evolution of Y. pseudotuberculosis into Y. pestis that has allowed the latter to actually form biofilms that enhances flea-borne transmission (Sun et al., 2008; Guo et al., 2015). This likely represents an example of positive Darwinian selection (Zhang, 2008).
Phage shock proteins and maintenance of proton motive force
Another ECS responsive element is the phage shock protein (Psp) pathway needed for bacterial survival when the inner membrane ion permeability barrier function has been breached (Joly et al., 2010; Yamaguchi and Darwin, 2012). Since proton motive force (PMF) dissolution would follow inner membrane disruption, the Psp pathway's main function is probably to reinstate a PMF to stress-damaged membranes (Jovanovic et al., 2006; Kobayashi et al., 2007; Figure 4D). Hence the efficiency of sec-dependent and tat-dependent protein secretion across the inner membrane is reliant on a stress-responsive Psp system (Jones et al., 2003; Delisa et al., 2004). A major component of the Psp system is the dynamic PspA protein encoded on the pspABCDE operon that is controlled by the PspF transcription factor. In unstressed bacteria, PspA acts as an anti-activator by sequestering PspF. Following stress exposure, PspA is targeted to the membrane to release free PspF to activate pspABCDE expression. At the inner membrane, stress alleviation is performed by PspA either alone or together with the integral inner membrane components PspB and PspC. For example, a chief inducer of the Psp response is the mislocalization of outer membrane secretin proteins, while the PspB and PspC proteins specifically work to prevent wrongful secretin insertion into the membrane (Lloyd et al., 2004; Guilvout et al., 2006; Seo et al., 2007; Figure 4D). Given that secretins form outer-membrane channels for the movement of macromolecules across the outer membranes of Gram-negative bacteria (Korotkov et al., 2011; Koo et al., 2012), the Psp response probably supports several aspects of bacterial virulence (Darwin, 2013).
Indeed, Y. enterocolitica has served as an excellent model pathogen to study the Psp system in bacterial virulence. An intact Psp system is required for Y. enterocolitica survival during active Ysc-Yop T3S when the YscC secretin is naturally over-produced (Darwin and Miller, 2001; Green and Darwin, 2004; Figure 4D). It is believed that this requirement stems from the need for the Psp system to prevent YscC-induced cytolethality upon any mislocalization to the inner membrane during T3S (Horstman and Darwin, 2012). Yet, the Psp system is not actually required for T3SS assembly and function per se (Darwin and Miller, 1999), which is a little perplexing considering that the Ysc-Yop T3SS is supposedly reliant on PMF for function (Wilharm et al., 2004). Hence, there is a clear need to better appreciate how the Psp system maintains the PMF in order to understand how energy supplies that drive T3S are preserved (Lee and Rietsch, 2015). It also seems prudent to explore what roles are played by the Psp system in other secretin-dependent protein export and secretion pathways of Yersinia.
Control of protein assembly at the bacterial surface
A prevailing theme during ECS responsiveness is the need to secure protein transport and assembly in and through the bacterial envelope. Quality control of protein folding in the bacterial envelope requires input from dedicated periplasmic protein folding and degradation factors. Hence, the transcriptional regulation of genes encoding these factors are usually wired to the σE and CpxA-CpxR regulatory networks responsive to ECS, and this ensures their elevated levels at acute times of ECS exposure. Such periplasmic-located quality control factors include: molecular chaperones such as Skp and Spy, folding catalysts such as the disulfide oxidoreductases and the peptidyl-prolyl isomerases (PPIases), and degradosomes such as the DegP/HtrA serine protease (Wick and Egli, 2004; Merdanovic et al., 2011; Mogk et al., 2011; Lyu and Zhao, 2015). While examples of these protein folding and degradation factors can be found in essentially all living organisms, in pathogenic bacteria they are particularly required for production of fully functional virulence determinants that ensure bacterial survival during transit in a host environment.
With this in mind, the contribution of PPIases in Yersinia pathogenicity was explored. PPIases are protein chaperones and folding factors that can catalyze proline isomerization in proteins (Gothel and Marahiel, 1999; Fanghanel and Fischer, 2004). Five periplasmic PPIases in Y. pseudotuberculosis: SurA, PpiD, PpiA, FkpA, and FklB, have recently been described (Obi et al., 2011), but they are essentially ubiquitous being widespread in other organisms. SurA is implicated in most phenotypic characteristics associated with this protein family (Behrens-Kneip, 2010). In Yersinia lacking the surA allele, outer membrane perturbations result that include aberrant cellular morphology, drastic alterations in OMP profile, susceptibility to detergents and antibiotics, altered fatty acid and phospholipid composition and leakiness of LPS into the extracellular environment (Obi et al., 2011; Southern et al., 2015). Not surprisingly therefore, the SurA PPIase and chaperone is essential for the virulence of Yersinia in a mouse infection models (Obi et al., 2011; Southern et al., 2015). Critically though, SurA+ Yersinia are also avirulent if they lack all four remaining periplasmic PPIases (i.e., PpiA, PpiD, FkpA, and FklB) (Obi et al., 2011). This suggests that SurA-dependent and SurA-independent pathways are responsible for trafficking essential virulence factors to the Yersinia surface. Consistent with this, recent studies have defined possible functions for the PPIases other than SurA in the bacterial cell envelope where under certain conditions they assume important SurA-independent functions in chaperoning, folding and assembly of outer surface proteins once they emerge from the Sec translocon (Antonoaea et al., 2008; Matern et al., 2010; Ge et al., 2014; Gotzke et al., 2014; Sachelaru et al., 2014). Significantly, in Y. pseudotuberculosis the PPIases are known to be necessary for proper outer membrane assembly of invasin and Ail, important Yersinia adhesins involved in attachment to eukaryotic cells (Obi and Francis, 2013). This is interesting for enteropathogenic Yersinia are famed for possessing an abundance of adhesins and secretion systems that are presumed to be important for host cell interactions, but the roles of several of these have not been experimentally proven (Francis, 2011; Mikula et al., 2013). Thus, proteomics approaches can be easily applied to Yersinia strains where SurA has been deleted and where PpiA, PpiD, FkpA, and FklB have all been deleted, with the goal to identify novel surface proteins that are essential for survival and/or virulence of Yersinia, and also describe the essential proteins in the trafficking and assembly pathways for these surface proteins. Both the surface protein and the trafficking pathway would be candidate targets for anti-infective drug development.
Metal homeostasis
Decisive for the survival of all organisms is an adequate supply of intracellular transition metals. Minor concentrations of iron, zinc, copper, and manganese are indispensable for countless cellular functions, but when in excess can lead to severe toxicity through disruption of cellular redox potential and production of deleterious reactive hydroxyl radicals that have in turn forced the cell to evolve resistance and detoxification strategies as a safeguard under certain environmental conditions (Hobman and Crossman, 2015; Imlay, 2015). Supply of these transition metals is influenced by oxygen levels and for bacterial pathogens in particular, metal availability is further restricted by the host's innate immune defense (Cassat and Skaar, 2013). In prokaryotes, examples from all major transporter families are known to contribute to metal homeostasis (Klein and Lewinson, 2011). In pathogenic Yersinia, most research has focused on mechanisms of iron acquisition, but this does not off-set the importance of other metal acquisition systems given how several of these are up-regulated during in vivo growth (Lathem et al., 2005; Sebbane et al., 2006; Table 2).
Table 2.
Prominent iron transport systems and associated regulatory factors in pathogenic Yersinia.
| System | Functional propertya |
|---|---|
| TonB-ExbB-ExbD | Energizer complex |
| Fur & RyhB | Iron homeostasis in response to iron |
| ArcA-ArcB & Fnr | Iron homeostasis in response to oxygen |
| FERRIC TRANSPORTERS | |
| Ybt | Yersiniabactin siderophore system |
| Ynp | Pseudochelin siderophore system |
| Ysu | Yersiniachelin siderophore system |
| Iuc | Aerobactin siderophore system |
| FERROUS TRANSPORTERS | |
| Yfe | ABC importer of iron and manganese |
| Feo | Non-ABC-importer |
| HEME TRANSPORTERS | |
| Hmu | ABC importer |
| Has | Hemophore system |
Whether these systems are contained in all three human pathogenic Yersinia has been addressed in part by the study by Forman et al. (2010). This study revealed significant variation in coding potential among the three species and even among strains of a species. Hence, mechanisms of iron transport can have redundant functions or operate only in a specific niche.
Iron transport systems in pathogenic Yersinia
In facultative anaerobic bacteria such as Yersinia, regulation of iron metabolism is linked closely to iron availability and to the levels of oxygen in the environment (Carpenter and Payne, 2014). In anoxic conditions, iron is present in a soluble ferrous (Fe2+) form, while in the presence of oxygen, is more commonly found in the insoluble ferric (Fe3+) form. Reflecting the importance of iron as a nutrient, bacteria including Yersinia have devised an impressive array of iron uptake mechanisms that enable utilization of the two redox forms.
In the presence of oxygen, highly virulent forms of pathogenic Yersinia all produce a high pathogenicity island (HPI) encoded yersiniabactin (Ybt) siderophore-based system for the acquisition of Fe3+ iron (Forman et al., 2010; Rakin et al., 2012; Table 2). Yersiniabactin production is considered to be an essential virulence determinant for both plague-causing Y. pestis and also the enteric Yersinia, and fitness profiling identifies it as a necessary requirement for optimal growth in vivo (Palace et al., 2014). Yet the fact that the ybt operon is truncated or even deleted in some strains of virulent Y. pestis and Y. pseudotuberculosis indicates other modes of Fe3+ uptake have evolved. Indeed, this may involve at least three possible alternative siderophore systems–the pseudochelin (Ynp) system, the yersiniachelin (Ysu) system, and the aerobactin (Iuc) system (Forman et al., 2010; Rakin et al., 2012) (Table 2). These particular siderophore-iron complexes are transported back into the cell via coupling to either cognate or generic ABC importers that all consist of a typical ATP binding protein, a periplasmic binding protein, and an outer membrane receptor (Forman et al., 2010) (Table 2). Transport is energized by the universal TonB/ExbBD energizer system and improved iron solubility requires ferric reductase activity. The general importance of the TonB/ExbBD energizer system is reflected in it being selected for optimal Yersinia fitness in vivo (Palace et al., 2014).
In oxygen limiting conditions, or when reducing agents are present, systems for the transport of Fe2+ iron are required. The Yfe and Feo systems appear to be the two predominant ferrous transporters utilized in Yersinia, and their functions share some partial redundancy (Perry et al., 2007; Table 2). The Yfe system is a typical ABC importer, which in some bacteria has demonstrated affinity for manganese transport. The Feo system is a non-ABC transporter, which is energized through GTP hydrolysis. Both systems are required for full Yersinia virulence in certain infection models (Fetherston et al., 2012). Additionally, when in a mammalian host environment, the potential for pathogenic Yersinia to utilize heme as an iron source is made possible via two transport systems—the Hmu ABC transporter and the Has hemophore system (Hornung et al., 1996; Perry and Fetherston, 2004; Forman et al., 2010; Table 2). However, in vivo studies suggest that the relative contributions of these two systems to pathogenicity is minimal (Rossi et al., 2001; Forman et al., 2010). Despite this, it seems likely that Yersinia have evolved multiple iron transport pathways to ensure their needs for iron are satisfied no matter the prevailing environment, although some of these may make relatively minor contributions to Yersinia pathophysiology, at least under the environmental conditions experimentally tested.
Regulating iron transport systems in pathogenic Yersinia
It is not surprising that the synthesis of iron transport systems is tightly controlled in response to available iron and oxygen levels. Responsiveness to iron is through the transcriptional repressor, ferric uptake regulator (Fur), a ubiquitous regulator in all prokaryotes (Troxell and Hassan, 2013; Fillat, 2014; Table 2). In fact, metal homeostasis is generally achieved by a large superfamily of Fur-like proteins that also specifically regulate the genes of other transition metal uptake systems (Fillat, 2014). In iron replete conditions, Fur binds Fe2+ to form homodimers that bind to DNA target sequences in iron-responsive gene promoters that represses their transcription through RNA polymerase occlusion. When concentrations of iron are low, Fe2+ is displaced from Fur, which now assumes a monomeric state that is unable to interact with DNA, allowing transcription from iron-responsive promoters to proceed. The iron-Fur regulon in Y. pestis has been studied by DNA microarray, biochemical and in silico analyses. Predictably, Fur and high iron concentrations repressed a number of operons that reinforced their involvement in iron homeostasis (Zhou et al., 2006; Gao et al., 2008). However, as in other bacteria the Fur protein was also observed to activate the transcription of a few genes (Zhou et al., 2006). It is assumed that this action occurs through the small non-coding RNA, RyhB (see below; Table 2). RyhB is a post-transcriptional repressor of gene expression and its expression is repressed by Fur (Salvail and Masse, 2012). Thus, when Fur is inactive at low iron concentrations, RyhB is generated to specifically promote the degradation of mRNA's that encode for a number of non-essential iron utilization genes that consequently liberates free iron for essential purposes (Salvail and Masse, 2012). In Y. pestis, the function of RyhB has been studied to some extent. Two Fur-regulated RyhB homologs are expressed, and while both are up-regulated during in vivo infections in a mouse model (Deng et al., 2012; Yan et al., 2013), neither of the two were essential for virulence under the inoculation routes tested (Deng et al., 2012).
Iron transport is also regulated in response to oxygen, which is necessary so that bacteria can adapt transport systems to the different forms of iron (Carpenter and Payne, 2014). Adaptation can occur by the direct action of global regulators of respiration at transport gene promoters or indirect through changes in Fur levels. In the latter, fur expression is repressed under iron-replete conditions through a feedback auto-regulatory loop, and in several systems is induced by the redox regulator OxyR in response to oxidative stress (Troxell and Hassan, 2013; Carpenter and Payne, 2014; Fillat, 2014). An increase in Fur levels in oxidative stress conditions will reduce iron uptake and subsequently the risk of hydroxyl radical formation by the Fenton reaction, but also lead to the upregulation of key strategies for the detoxification of these radicals (Troxell and Hassan, 2013; Carpenter and Payne, 2014; Fillat, 2014). Nevertheless, evidence for this Fur-OxyR regulatory pathway in Yersinia is still lacking (Fetherston et al., 2012).
Global regulators of respiration include the Fnr oxygen sensitive transcription factor and ArcA that is part of the ArcA-ArcB TCRS (Table 2). Fnr is a major global transcription factor, and uses self-contained iron-sulfur clusters to sense prevailing oxygen levels (Korner et al., 2003; Green et al., 2009). Homodimerization occurs in anaerobic conditions and these homodimers bind DNA to completely reprogram the cell from aerobic to anaerobic respiration. ArcA activation is linked to the ability of ArcB to sense redox potential and electron transfer among the soluble membrane electron carriers under anoxic and oxic conditions (Malpica et al., 2006). Among bacteria of the Enterobacteriaceae, both active Fnr and ArcA regulate the expression of genes belonging to iron transport systems using both Fur-dependent and—independent mechanisms. However, a whole genome promoter structure analysis to identify regulation during anaerobic respiration failed to predict these mechanisms in either the plague or enteric Yersinia (Ravcheev et al., 2007), and this is also supported by limited experimentation (Fetherston et al., 2012). Nevertheless, independent studies highlight the importance of both Fnr and Arc systems in the adaptation of Yersinia to in vivo growth environments (Palace et al., 2014; Avican et al., 2015), which makes them likely to be a cornerstone for metabolic adaptation during pathogenesis.
Other (non-iron) metal acquisition systems in pathogenic Yersinia
Due to their roles as cofactors in metal-containing proteins, many additional non-iron metals are also essential for cell function. Of these, zinc, manganese, copper, and nickel transporters are among the better characterized non-iron homeostasis systems in the Enterobacteriaceae (Porcheron et al., 2013). Higher organisms often limit the availability of these essential trace metals to lessen the threat of pathogen infection, in a process termed “nutritional immunity” (Becker and Skaar, 2014). As a counteractive measure, bacterial pathogens encode for host-inducible influx systems that are designed to sequester limiting trace metals from the host (Lathem et al., 2005; Sebbane et al., 2006). Although not studied to the same extent as iron scavenging systems, transporters for zinc and manganese are now established as an integral part of the Yersinia physiological makeup (Porcheron et al., 2013; Perry et al., 2015).
To date, there exists three known modes of zinc acquisition in Yersinia. The first is the ZnuABC importer common among many prokaryotes (Desrosiers et al., 2010). The second is the iron siderophore, yersiniabactin (Ybt; Bobrov et al., 2014), and the third is YezP, a zinc binding protein surprisingly secreted by one of the type 6 secretion systems of Y. pseudotuberculosis, designated T6SS4 (Wang et al., 2015). Despite this knowledge, the actually manner in which Zn2+ crosses the outer membrane is not known for any of these three systems. Moreover, it is not known how the Zn2+-laden Ybt and YezP then moves across the inner membrane for entry back into the Yersinia cytoplasm, although the utilization of Zn2+-Ybt is dependent on the inner membrane permease YbtX (Bobrov et al., 2014). As these two systems are unusual, this suggests that truly novel uptake mechanisms are waiting to be uncovered. As alluded to already in this section, zinc homeostasis operons in Yersinia are under the typical control of Zur, a zinc-responsive transcriptional repressor belonging to the Fur-like superfamily (Li et al., 2009).
Manganese in the form of Mn2+ is preferentially used by all biological systems. So far, two Mn2+ importers have been characterized in pathogenic Yersinia. The first is a rather common proton-dependent symporter termed MntH (Champion et al., 2011; Perry et al., 2012), an ortholog to the eukaryotic Nramp1 symporter that assists in the elimination of pathogens by restricting their access to divalent cations in the phagosome (Cellier et al., 2007). It follows that MntH enables intracellular Y. pseudotuberculosis to accumulate manganese and to resist antimicrobial killing by phagocytes producing Nramp1 (Champion et al., 2011). In Y. pestis, manganese transport could well be niche specific for bacteria lacking mntH were attenuated in a bubonic, but not pneumonic, mouse model of infection (Perry et al., 2012). The second influx system is the ABC transporter YfeABCD, which has dual specificity for both iron and manganese (Bearden and Perry, 1999). In some organisms, manganese homeostasis is controlled by a Fur-like transcriptional repressor termed Mur (Fillat, 2014), although the most common form of control in the Enterobacteriaceae involves the MntR regulator (Porcheron et al., 2013). Yet Y. pestis has evolved without the Mur or MntR regulators (Perry et al., 2015), so the manganese transporters Yfe and MntH are actually Fur-regulated (Perry et al., 2012).
Metabolism and virulence
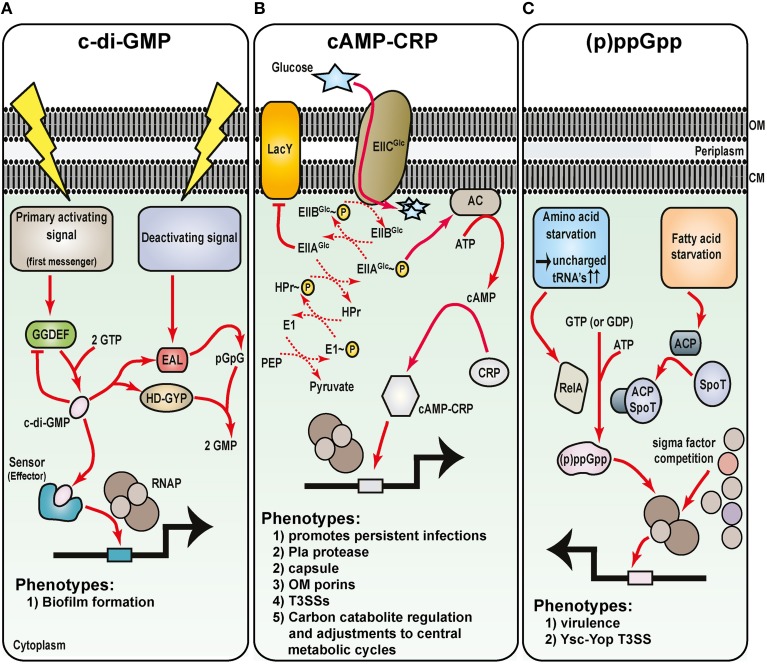
It has long been appreciated that bacterial pathogens produce an array of virulence factors to counteract the antibacterial immune responses from the host, and in turn this creates a favorable niche for bacterial colonization and survival. However, underappreciated until relatively recently is the need for bacteria to seek the necessary nutrients required to maintain energy levels to sustain bacterial replication in a host environment. Although, eukaryotic cells or tissues can provide an extensive source of nutrients, the variety and degree of available nutrients is often both spatially and temporally restricted. Hence, as bacteria transit through different host compartments and tissues, they must rapidly adjust their central metabolism to make efficient use of what nutrients are available. Interestingly, new evidence is emerging to indicate that pathogens have streamlined adaptive processes to couple metabolic activity to a defined program of virulence gene expression (Eisenreich et al., 2010; Rohmer et al., 2011; Heroven and Dersch, 2014; Wilharm and Heider, 2014). To some extent, this has been long forecast by the knowledge that common nucleotide metabolic derivatives—cyclic AMP, cyclic di-GMP, and the guanosine polyphosphate molecules ppGpp and pppGpp (Figure 5) play profound roles as signaling molecules in the control of bacterial gene expression (Kalia et al., 2013).
Figure 5.
Metabolic intermediates regulate Yersinia virulence. Understanding the role of nucleotide-based second messengers in the regulation of Yersinia survival and virulence is still in its infancy. As determined from many studies in E. coli systems, three major regulatory molecules are known: (A) c-di-GMP, (B) cAMP and its receptor Crp, and (C) ppGpp and pppGpp [collectively known as (p)ppGpp]. Primary activation signals control diguanylate cyclase (GGDEF domain containing proteins) to generate c-di-GMP (A). Various effector molecules then interact with c-di-GMP to regulate gene expression, including Hms-mediated biofilm formation. Deactivating signals stimulate phosphodiesterase activity of proteins containing either EAL or HD-GYP domains, and this degradation pathway ensures that c-di-GMP levels are stringently controlled. Glucose availability and the phosphoenolpyruvate (PEP)—carbohydrate phosphotransferase system (PTS) control cAMP production by adenylate cyclase (AC) (B). Upon cAMP production when glucose is limiting, cAMP-CRP complexes form and this enhances RNAP holoenzyme binding to vast numbers of sensitive promoters including several secreted Yersinia virulence factors. Finally, various forms of starvation stimulates RelA-dependent and SpoT/acyl carrier protein (ACP)-dependent synthesis of (p)ppGpp (C). By direct binding to the RNA polymerase, (p)ppGpp can influence sigma factor competition for core RNAP, and through this affects transcription of a plethora of genes that in Yersinia includes the prominent Ysc-Yop plasmid-encoded T3SS.
c-di-GMP signaling network in Yersinia biofilm formation
Cyclic dimeric guanosine monophosphate (c-di-GMP; cyclic diguanylate) is a ubiquitous nucleotide-second messenger in bacteria that regulates many physiological process. Most often this culminates in transitioning between a single-cell planktonic lifestyle to a multicellular sessile lifestyle in biofilm communities (Romling et al., 2013). Impacted by various environmental cues, such as oxygen, nitric oxide, light, nutrients, and temperature (Kalia et al., 2013), c-di-GMP is synthesized from two GTP molecules by diguanylate cyclases containing GGDEF domains (Figure 5A). Specific phosphodiesterases associated with EAL or HD-GYP domains are responsible for c-di-GMP degradation (Romling et al., 2013; Figure 5A). The biochemistry of c-di-GMP synthesis and degradation is quite obviously complex. This is brought about by the fact that a bacterial genome encodes for quite a few different proteins that harbor either a GGDEF or EAL domain, and there exits even examples of both domains being present in the same protein. Moreover, c-di-GMP is bound by diverse receptors that for example, includes PilZ-domain containing proteins and riboswitches (Romling et al., 2013). These findings have important functional consequences. Clearly, c-di-GMP levels must be in constant flux, and this creates critical concentration gradients in various foci throughout the cytoplasm. Hence, at a particular threshold concentration in a given location within the cytoplasm, c-di-GMP is capable of exerting its regulatory activity at multiple all levels ranging from transcription to post-translation.
Bioinformatics prediction suggests as many as 8 GGDEF and/or EAL domain-containing proteins encoded by the Y. pestis genome (Darby, 2008). In the Y. enterocolitica genome, this number swells to a possible 22 proteins (Heermann and Fuchs, 2008). These observations point to a complex c-di-GMP-mediated regulatory network in the control of biofilm formation at ambient temperature in pathogenic Yersinia (Zhou and Yang, 2011). In addition to the major hmsHFRS locus responsible for EPS synthesis and transport, Y. pestis harbors the hmsT, hmsCDE, and hmsP operons that encode for regulatory components. Among them are the two digualylate cyclases HmsT and HmsD responsible for c-di-GMP synthesis, and the HmsP phosphodiesterase responsible for c-di-GMP turnover (Jones et al., 1999; Kirillina et al., 2004; Bobrov et al., 2005, 2011; Simm et al., 2005; Sun et al., 2011; Ren et al., 2014). In addition, HmsB, a temperature dependent small non-coding regulatory RNA stimulates c-di-GMP levels by enhancing the expression of hmsB, hmsT, hmsCDE, and hmsHFRS, and repressing hmsP (Fang et al., 2014; Figure 5A). Consistent with HmsB involvement is the observation that Hfq, the premier RNA chaperone (see later), is also described to influence Yersinia biofilm formation, although some disparity exists between the two studies concerning the manner in which Hfq is thought to exert its affect (Bellows et al., 2012; Rempe et al., 2012). Nevertheless, it is still very evident that Y. pestis biofilms form following a build-up of c-di-GMP, but are repressed by the metabolism of c-di-GMP. In fact, the ability to accumulate c-di-GMP through incremental gene loss was a dominant force driving the evolution of Y. pestis, for it enabled biofilm formation in the flea foregut, which enhanced transmissibility from the flea vector (Sun et al., 2014).
The receptors of c-di-GMP are not yet experimentally defined. Hence, it is not really clear how direct c-di-GMP signaling is in regulating Y. pestis biofilm formation. Indeed, multiple factors controlling Yersinia biofilm formation are known, so the regulatory network is extremely complex. As mentioned earlier in this review, the Rcs phosphorelay system directly regulates Yersinia biofilm formation through repression of various hms loci (Hinchliffe et al., 2008; Sun et al., 2012; Fang et al., 2015; Guo et al., 2015). The classical PhoQ-PhoP two component regulatory system is also required for controlled biofilm formation (Sun et al., 2009b; Rebeil et al., 2013), as is the newly described LysR-type transcriptional regulator, yfbA (Tam et al., 2014). Additionally, deficiency of the polyamine putrescine restricts biofilm formation due to post-transcriptional defects in the synthesis of key Hms components that are required for biosynthesis of EPS (Patel et al., 2006; Wortham et al., 2010). Moreover, the NghA glycosyl hydrolase reduces biofilm formation by degrading extracellular matrix constituents (Erickson et al., 2008). Finally, a role for quorum sensing in enhancing biofilm formation by enteropathogenic Yersinia has been reported, but this appears indirect through quorum sensing-dependent repression of type III secretion (Atkinson et al., 2011). Yet, with all the varied positive and negative regulators of Yersinia biofilm formation described, the real challenge now is to integrate these into a cohesive and dynamic regulatory network.
Carbon catabolite repression—cAMP-CRP
The Enterobacteriaceae utilize glucose as the preferred carbon source. Glucose limiting environments promote cross-talk between the phosphoenolpyruvate-carbohydrate phosphotransferase system, adenylate cyclase (producer of the second messenger cyclic AMP—cAMP), and CRP (cAMP receptor protein; Deutscher, 2008; Gorke and Stulke, 2008; Figure 5B). This cross-talk specifically reprograms bacteria to activate secondary carbon scavenging and utilization pathways to maintain growth. Referred to as carbon catabolite repression, this regulatory process enables bacterial to preferentially use glucose prior to the metabolism of other non-preferred carbon nutrients (Deutscher, 2008; Gorke and Stulke, 2008). The regulatory mechanism relies upon enhanced production of cAMP by adenylate cyclase when glucose is limiting. When cAMP is bound by CRP, a global transcription activator is formed that is competent to bind DNA promoter sequences to reprogram gene expression so that bacteria can make use of new carbon nutrient sources (Lawson et al., 2004; Won et al., 2009).
It is quite common for cAMP-Crp dependent reprograming of gene expression to include alterations in virulence gene expression. In fact, the cAMP-Crp regulatory system of Yersinia is a vital player for pathogenicity (Zhan et al., 2008; Heroven et al., 2012b; Lathem et al., 2014; Avican et al., 2015), and this includes specific control of the Pla protease, components of T3SSs, the F1 capsule, outer membrane porins, and virulence associate carbon storage regulatory (Csr) system small non-coding RNAs (Petersen and Young, 2002; Kim et al., 2007; Zhan et al., 2008, 2009; Gao et al., 2011; Heroven et al., 2012b) as well as non-specific control over biofilm formation (Willias et al., 2015; Figure 5B). However, recent work has begun to shed light on the shear scope of cAMP-Crp involvement in Yersinia pathophysiology. Working in concert with the Csr system and the major transcriptional regulator RovA (Heroven et al., 2008, 2012a,b), this Crp-Csr-RovA regulatory cascade functions to control the sophisticated carbon metabolism network at the level of the pyruvate- tricarboxylic acid cycle, and these core metabolic adjustments are necessary for Yersinia to successfully transition from free-living to infectious state (Bucker et al., 2014). This process includes modulating virulence gene expression so that Yersinia can switch from acute infection to chronic persistent infection in an in vivo mouse model (Avican et al., 2015; Figure 5B). Reflecting this transition from free-living to infectious state, temperature up-shift from 26 to 37°C results in a dramatic reprogramming of the cAMP-Crp regulon (Nuss et al., 2015) and an adjustment of catabolic pathways in readiness to utilize nutrients derived from the host (Motin et al., 2004; Chromy et al., 2005). Several small non-coding regulatory RNAs are among the vast cAMP-Crp regulon, which suggests coupling between metabolism and virulence occurs at the post-transcriptional level (Nuss et al., 2015). This idea is supported by a multi-omics systems approach that revealed mechanisms of post-transcriptional control of metabolism that are conserved between Y. pestis and Y. pseudotuberculosis (Ansong et al., 2013).
It is obvious that the regulation of crp gene expression is then a significant process in Yersinia. In Y. pestis, the PhoQ-PhoP system directly regulates crp expression at the transcriptional level (Qu et al., 2013; Zhang et al., 2013b), whereas in Y. pseudotuberculosis it is the csr system that is transcriptionally regulated by PhoP (Nuss et al., 2014). Recently, a mechanism of positive post-transcriptional regulation of Crp production was found to involve the 5′ untranslated region of crp mRNA, temperature and the RNA binding protein Hfq (Lathem et al., 2014). This finding goes a long way to explaining the contributions of Hfq to the metabolic fitness and virulence of pathogenic Yersinia (Geng et al., 2009; Bai et al., 2010; Schiano et al., 2010; Bellows et al., 2012; Kakoschke et al., 2014). Further investigation of crp regulation will benefit understanding of how Yersinia adapts metabolic and virulence capacity during host infections.
Stringent response and (p)ppGpp
The bacterial stringent response is starvation induced and leads to accumulation of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp). Commonly referred to as (p)ppGpp, these second messengers coordinate multiple physiological processes within the bacterial cell in response to nutrient and environmental stress (Gaca et al., 2015; Hauryliuk et al., 2015). Regulation primarily occurs at the transcriptional level through alterations of promoter selection by RNA polymerase, but can also occur at the post-transcriptional level (Dalebroux and Swanson, 2012). The RelA-SpoT homolog (RSH) family enzymes are responsible for synthesis and hydrolysis of (p)ppGpp (Figure 5C). The RelA-SpoT pathway functions in β- and γ-proteobacteria, whereas a Rel pathway is much more widely distributed among prokaryotes (Gaca et al., 2015; Hauryliuk et al., 2015). At least in the Enterbacteriaceae, the RelA pathway is responsive to limiting amino acids and uncharged tRNA accumulation. The SpoT pathway collaborates with acyl carrier protein and is responsive to limiting phosphate, carbon, or fatty acids. Not surprisingly, (p)ppGpp is an active player in the regulation of virulence gene expression (Dalebroux et al., 2010). Although not well-studied in pathogenic Yersinia, a Y. pestis relA, spoT double mutant cannot accumulate (p)ppGpp and is attenuated in virulence (Sun et al., 2009a). One reason for this attenuation could well be an inactive Ysc-Yop T3SS (Sun et al., 2009a). Given the importance of this T3SS to Yersinia virulence, follow-up studies to understand how (p)ppGpp impacts on its activity would be beneficial.
New concepts in metabolism and virulence circuitry
With new technologies advancing knowledge of bacterial metabolism in vivo, it is now apparent that novel bacterial factors can be deployed to profoundly affect the metabolic status of the host which in turn augments bacterial virulence (Abu Kwaik and Bumann, 2013; Staib and Fuchs, 2014). A seminal paper in 2009, revealed a distinct physical cross-talk between metabolism and the production of type III secretion by Yersinia (Schmid et al., 2009). In particular, this study demonstrated that virulence plasmid-encoded Ysc-Yop T3SS synthesis and activity was inversely correlated to the levels of available oxaloacetate derived amino acids, via direct T3SS-dependent control of phosphoenol pyruvate carboxylase activity. This study served to garner the then fleeting concept that the activity of classical virulence determinants was purposefully linked to the control of nutrients essential for niche-specific in vivo growth.
Now with the routine use of systems level-based applications in infection biology research, the identification of metabolic factors with direct coupling to virulence is increasing steadily. For example, transcriptomics data from several studies designed to assess how Yersinia adapts to a mammalian host environment revealed an obvious and substantial commitment to reprogram carbon uptake and utilization strategies for the purpose of optimizing in vivo growth to suit prevailing nutritional availability (Lathem et al., 2005; Rosso et al., 2008; Fukuto et al., 2010; Ansong et al., 2013; Pradel et al., 2014), and these observations corroborated independent in vivo fitness profiling data of bacteria deficient in core metabolic pathways (Palace et al., 2014; Pradel et al., 2014; Avican et al., 2015; Deng et al., 2015). Within this context, Sasikaran and colleagues have recently demonstrated that Y. pestis and Pseudomonas aeruginosa possessed the capacity to metabolize itaconate, a mammalian metabolite with potent anti-bacterial properties (Sasikaran et al., 2014). This is significant because possession of functional itaconate degradation genes in Y. pestis is required for full pathogenicity (Pujol et al., 2005). The reality is that control of carbon utilization pathways to exploit prevailing environmental niches is believed to be a common phenomenon in diverse bacteria as they transition through a host. It follows that a new paradigm “nutritional virulence” has begun to take hold, and is designed to encapsulate the importance to pathogenicity of pathogen-directed exploitation of host nutrients (Abu Kwaik and Bumann, 2013). Hence, an increased understanding of bacterial metabolic mechanisms supporting optimal in vivo growth may eventually lead to novel strategies for the treatment and prevention of bacterial diseases. With biologically relevant acute and chronic in vivo infection models in place, studies designed to interrogate Yersinia pathophysiology are well-placed to shed further light on this nutritional virulence concept.
Post-transcriptional control by small non-coding regulatory RNA's
Small regulatory RNAs have a tremendous impact upon physiological and metabolic circuits in bacterial model organisms, such as E. coli K12 and Bacillus subtilis. The overall impact of small regulatory RNAs (sRNAs) on the pathogenesis and environmental adaptation of bacteria of medical importance cannot be understated. While investigations into Yersinia sRNAs are not completely novel, their magnitude and frequency are increasing. The initial foray into global sRNA biology in Yersinia species began with bioinformatics searches for homologs of small RNAs characterized in E. coli (Delihas, 2003). In general, small RNAs can be categorized by their dependence upon the RNA-chaperone Hfq. In fact, Hfq is a co-factor for the largest class of sRNAs in E. coli. These sRNAs require Hfq for their activity, as they bind to Hfq and facilitate interaction with mRNA targets. The Hfq-dependent small regulatory RNAs are trans-acting and act by complementary base pairing to a distal mRNA target. The Hfq-independent small RNAs act as cis-antisense RNAs or protein-binding RNAs.
Interestingly, Nakao and colleagues reported already in 1995 that the Y. enterocolitica heat-stable toxin (Y-ST) required the E. coli Hfq homolog for maximal production (Nakao et al., 1995); an observation suggesting that Y. enterocolitica heat-stable toxin (Y-ST) may be regulated by an as yet unknown sRNA. In fact, it is now known that Hfq is required for full virulence of all three pathogenic Yersinia species (Geng et al., 2009; Schiano et al., 2010; Kakoschke et al., 2014), and this reflects clearly on the importance of sRNA's and numerous post-transcriptional regulatory mechanisms in the control of Yersinia pathogenic mechanisms. Consequently, there are now numerous investigations into Yersinia small RNA biology. Many of these have identified unique Yersinia specific sRNAs that are not homologous to other sRNA sequences within other enteric bacteria. In this section, we will review the literature on both Hfq-dependent, Hfq-independent sRNA, and unique Yersinia specific sRNAs (Ysrs). We will also highlight functional characterization that was executed using surrogate genetics in E. coli vs. studies within the native Yersinia organisms.
Genome-wide searches for small regulatory RNAs in Yersinia sp.
Genome wide searches in E. coli led to the discovery of a significant amount of prokaryotic model small RNAs. Once these small RNAs were characterized, significant insights into physiology were uncovered in the areas of outer membrane metabolism, iron metabolism, envelope stress, and carbon metabolism. Inevitably, these genome wide searches were expanded into other bacteria including Yersinia species, yielding the identification of sRNAs homologous to many of these small RNAs. Deep sequencing of total RNA libraries from Y. pseudotuberculosis resulted in the discovery of approximately 150 previously unannotated sRNAs, defined as Yersinia small RNAs or Ysr (Koo et al., 2011). Thirty two of these Ysr's were orthologous to previously characterized sRNAs encoded within the E. coli and S. typhimurium genomes. The remaining 118 Ysr sequences were unique to specific Y. pseudotuberculosis and Y. pestis genomes (Koo et al., 2011). Ysr expression was confirmed by Northern Blot analysis and animal studies demonstrated that selected Ysr deletions were attenuated for virulence in a mouse model of infection (Koo et al., 2011).
Investigations and insights into Yersinia small RNA homologs using surrogate genetics in E. coli
GcvB
E. coli GcvB is a 205 nucleotide long sRNA regulated by the Glycine cleavage system regulators GcvA and GcvR. It post-transcriptionally regulates periplasmic transport proteins OppA and DppA. In contrast to the E. coli gcvB gene, Y. pestis KIM6 gcvB encodes for two small RNAs (Table 3). Using surrogate genetics in E. coli, McArthur and colleagues demonstrated that expression of Y. pestis KIM6 gcvB sRNAs leads to repression of E. coli dppA expression in vivo (McArthur et al., 2006). Furthermore, the expression of Y. pestis KIM6 gcvB sRNAs are regulated by Y. pestis KIM6 gcvA and gcvB gene products, with GcvA acting as an activator and GcvB acting as a repressor (McArthur et al., 2006). A Y. pestis KIM6 gcvB mutant has a different growth rate and colony morphology than wild type cells, with gcvB- cells appearing dry and compact as opposed to smooth and sticky (McArthur et al., 2006).
Table 3.
Investigations of non-Yersinia Specific sRNA in different species.
| Surrogate genetic investigations in E. coli | Investigations in Yersinia species | ||
|---|---|---|---|
| Y. pseudotuberculosis | Y. enterocolitica | Y. pestis | |
| GcvB (Y.pestis) | SraG | MicF | RyhB1 |
| SgrS (Y. pestis) | CsrB | RyhB2 | |
| GlmY (Y. pseudotuberculosis) | CsrC | CyaR | |
| GlmZ (Y. pseudotuberculosis) | SsrA | ||
SgrS
E. coli SgrS is a sRNA that modulates sugar-phosphate stress (Vanderpool and Gottesman, 2007). Y. pestis sgrS homologs were functionally analyzed using a surrogate genetics approach (Wadler and Vanderpool, 2009; Table 3). It was demonstrated that the over-expression of Y. pestis SgrS in E. coli ΔsgrS mutants resulted in sgrS complementation (Wadler and Vanderpool, 2009). Specifically, Y. pestis SgrS expression resulted in repression of ptsG expression and abrogation of sugar-phosphate toxicity, consistent with the function of E. coli SgrS (Wadler and Vanderpool, 2009).
GlmY and GlmZ
GlmY and GlmZ are two small regulatory RNAs in E. coli involved in the post-transcriptional regulation of glmS, encoding the L-glutamine:D-fructose-6-phosphate aminotransferase (GFAT), or Glucosamine-6-phosphate transferase (Urban et al., 2007; Urban and Vogel, 2008). The transcriptional regulation of Y. pseudotuberculosis glmY and glmZ was characterized using a surrogate genetic approach in E. coli (Gopel et al., 2011; Table 3). In this study it was demonstrated that Y. pseudotuberculosis glmY and glmZ expression are regulated by σ54 (σN), GlrR, and IHF suggesting that GlmY and GlmZ are regulated by nitrogen metabolism in vivo (Gopel et al., 2011).
Yersinia-specific investigations using genetics and molecular biology
SraG
SraG is a 146-174 nt sRNA originally found in a genome wide search for novel sRNAs (Argaman et al., 2001). The SraG homolog in Y. pseudotuberculosis YPIII was characterized in vivo using genetics and proteomic analyses (Lu et al., 2012; Table 3). A proteomic screen of YPIII identified 16 proteins that were modulated by SraG depletion (Lu et al., 2012). The putative protein targets with the strongest positive modulation include flagellar motor switch protein G (fliG–YPK_2392), maltose regulon periplasmic protein (malM–YPK_0382), 50S ribosomal protein L9 (rpll–YPK_3781), Glutamine ABC transporter (glnH–YPK_1600), and an uncharacterized protein likely involved in high-affinity Fe2+ transport (YPK_2251; Lu et al., 2012). The putative protein targets with the strongest negative modulation include polynucleotide phosphorylase / polyadenylase (pnp–YPK_3726) and protein of unknown function YPK_1205 (Lu et al., 2012). Subsequent analyses confirmed that YPIII SraG sRNA regulates post-transcriptional production of YPK_1205 (Lu et al., 2012). Specifically, mutation in sraG resulted in increased YPK_1205 transcript levels as measured by ß-galactosidase activity and over-expression of sraG led to decreased YPK_1205 cDNA levels (Lu et al., 2012).
MicF
The MicF sRNA was identified and annotated in Yersinia enterocolitica by bioinformatics (Delihas, 2003; Table 3). In this study was also identified predicted interactions between Y. enterocolitica MicF and OmpF. Experimental analyses of Yersinia small RNAs was executed in Y. pestis, where the predicted interaction was likely conserved (Liu et al., 2015). Unlike E. coli, Y. pestis MicF over-expression resulted in stimulation rather than repression of OmpF. In a strain of Y. pestis cured of plasmid pPCP1, MicF repression of OmpF was restored (1.8- vs. 5-fold in E. coli), suggesting that stimulation was due to the presence of endogenous plasmid pPCP1 (Liu et al., 2015).
RyhB
RyhB is an 80 nt sRNA originally discovered in E. coli (Masse and Gottesman, 2002). It is regulated by the Ferric uptake regulator, Fur, and its expression is induced under iron-limited conditions (Masse and Gottesman, 2002). RyhB post-transcriptionally regulates the production of several iron-sulfur cluster proteins in E. coli (Masse and Gottesman, 2002; Masse et al., 2003). E. coli RyhB has a relatively large regulon and significant impact on the physiology of the cell (Masse and Gottesman, 2002; Masse et al., 2005). RyhB homologs have been discovered in several phylogenetically divergent species, starting with those most closely related to E. coli, as RyhB is highly conserved amongst enteric bacteria like many of the Hfq-dependent sRNAs in E. coli (Masse and Gottesman, 2002). Comparative genomic analyses led to the identification of two RyhB homologs in Y. pestis, RyhB1 and RyhB2 (Deng et al., 2012; Table 3). Both RyhB1 and RyhB2 levels are responsive to iron depletion, the presence of hfq, and the presence of fur (Deng et al., 2012). However, RyhB2 is less stable in the absence of hfq whereas the stability of RyhB1 is not affected by the absence of hfq (Deng et al., 2012). Analyses of Y. pestis RyhB expression in a mouse model of infection demonstrated a significant induction of RyhB1 and RyhB2 expression in lung tissue compared to spleen and growth in vitro in BHI (Deng et al., 2012; Yan et al., 2013). However, neither the ryhB1 or ryhB2 mutants demonstrated attenuation for virulence (Deng et al., 2012). The steady-state levels of Y. pestis RyhB1 and RyhB2 are modulated by the activity of ribonucleases (Deng et al., 2014). Specifically, RNaseIII is necessary for the accumulation of both RyhB1 and RyhB2 via repression of PNPase levels in the absence of Hfq (Deng et al., 2014). The half-life of RyhB1 and RyhB2 levels were increased in the absence of pnp, confirming that PNPase affects stability of Y. pestis RyhB1 and RyhB2 (Deng et al., 2014).
The cAMP receptor protein (Crp) regulatory circuit and small RNAs
The global regulator involved in carbon metabolism, Crp, regulates and is regulated by several small RNAs in Yersinia species. In E. coli and Salmonella, CyaR is a Crp regulated sRNA. CyaR is widely conserved amongst Gram-negative enteric bacteria. The genome wide search excuted by Yan and colleagues, and discussed above, yielded the identification of Y. pestis CyaR as well as three small RNAs as new members of the Y. pestis Crp regulon (Yan et al., 2013; Table 3). Those three small RNAs, sR065, sR066, and sR084, are encoded within intergenic regions of the pPCP1 plasmid (Yan et al., 2013). Primer extension, northern blot analyses, and computational analyses suggested that sR084 is positively regulated by Crp and sR066 is negatively regulated by Crp (Yan et al., 2013).
Hfq was picked up in a screen to identify factors that regulate Y. pestis Pla activity (Plasminogen activator; Lathem et al., 2014). In this study it was demonstrated that Hfq regulated Pla synthesis post-transcriptionallly in a manner independent of the pla 5′ UTR. This led to the hypothesis that Y. pestis Hfq post-transcriptionally regulated crp transcript levels. Post-transcriptional expression of crp was decreased in the absence of hfq (Lathem et al., 2014). The authors further demonstrated that multi-copy crp expression leads to induction of pla and partial suppression of the Δhfq growth defect (Lathem et al., 2014).
RNA sequencing analyses of Y. pestis grown in vitro and in an animal model of infection led to the identification of over 10 Crp-regulated sRNAs (Nuss et al., 2015). Three of these were novel sRNAs, not identified in previous screens (Nuss et al., 2015). The Crp-dependent regulation of these sRNAs were temperature variable, with the vast majority of the regulation observed at 25 vs. 37°C (Nuss et al., 2015). The authors identified 1 novel trans-encoded RNA (Ysr206) and two cis-antisense RNAs (Ysr232 and Ysr114) (Nuss et al., 2015). Crp also regulates the Hfq-independent sRNAs CsrB and CsrC, and these RNAs are discussed further down.
Yop-Ysc Type III secretion and small regulatory RNAs
In E. coli and other bacteria, the SmpB (small protein B)–SsrA (small stable RNA A) system is used for protein quality control. SsrA (or tmRNA) is a very unique RNA molecule that relieves ribosome stalling through its combined action as both an mRNA and a tRNA. SsrA enters stalled ribosomes as an alanine-charged tRNA mimic, the stalled polypeptide chain is then transferred to the alanine charged SsrA molecule via transpeptidation (Roche and Sauer, 1999). SsrA then enters the ribosome as an mRNA molecule and is translated resulting in the addition of an AANDENYALAA peptide tag to the stalled polypeptide. C-terminally AANDENYALAA tagged proteins are marked for degradation by the ATP-dependent protease ClpXP (Keiler et al., 1996; Gottesman et al., 1998). SmpB binds to SsrA and acts as a cofactor for SsrA, assisting its SsrA binding to ribosomes and ClpXP (Karzai et al., 1999; Wah et al., 2002). In contrast to the post-transcriptional regulators of RNA stability or translational initiation exhibited by the vast majority of small regulatory RNAs, SsrA is unique amongst small regulatory RNAs and was identified prior to the contemporary genome wide screens were executed. The SmpB-SsrA system is active and contributes to the physiology of Y. pseudotuberculosis (Okan et al., 2006; Table 3). Specifically, Y. pseudotuberculosis deleted for SmpB and SsrA (designated ΔBA) is attenuated for virulence in a mouse model of infection and is defective for proliferation in macrophages (Okan et al., 2006). The SmpB-SsrA locus is also necessary for the secretion of Yops (Okan et al., 2006). Specifically, the ΔBA mutant exhibited decreased mRNA and protein levels of the secreted substrates YopB, YopD, YopE, and LcrV (Okan et al., 2006).
The identification of Hfq in the regulation of T3SS in Y. pseudotuberculosis (Schiano et al., 2010) led to the hypothesis that Hfq and small RNAs may be involved in the regulation of T3SS in Y. pestis (Schiano et al., 2014). RNA sequencing led to the identification of 63 previously unidentified sRNAs. One of those sRNA, the Yersinia-specific sRNA, Ysr141, was encoded on the T3SS plasmid pCD1. In this study the authors demonstrated post-transcriptional activation of Y. pestis yopJ, via direct interaction of Ysr141 with the yopJ 5′UTR (Schiano et al., 2014).
Quorum sensing
Quorum sensing is the regulation of gene expression in a population or cell density-dependent manner. Small RNA regulation of quorum sensing has been well-characterized in Vibrio harveyi and Vibrio cholerae. Global regulators of quorum sensing in V. fischeri LuxR and LuxI are homologous to YenR and YenI proteins of Y. enterocolitica (Tsai and Winans, 2011). These authors could demonstrate a role for YenR in quorum sensing and swarming motility. YenS, a 169 nt YenR-regulated sRNA, post-transcriptionally regulates expression of the yenI gene that encodes for a protein responsible for the synthesis of the 3-oxohexanoylhomoserine lactone (OHHL) pheromone (Tsai and Winans, 2011). Epistatic analyses of yenI, yenR, and yenS mutations, on swarming motility demonstrated that yenS mutants suppressed the hypermotility seen in yenI mutants (Tsai and Winans, 2011). This suggests that, in addition to post-transcriptionally inhibiting yenI expression, yenS directly stimulates swarming motility of Y. enterocolitica downstream of yenI (Tsai and Winans, 2011).
Csr sRNAs
Csr sRNAs, along with 6S RNA, are amongst the most distinct non-Hfq dependent small RNAs in E. coli. These particular non-Hfq dependent small RNAs exert their regulatory function via direct binding and sequestration of the target proteins. There are two major Csr RNAs in E. coli, CsrB, and CsrC (Liu et al., 1997; Weilbacher et al., 2003). In E. coli CsrB and CsrC exert their regulatory effect by binding to, and thereby preventing the regulatory action of CsrA (Liu et al., 1997; Weilbacher et al., 2003). CsrA is a post-transcriptional regulator of genes necessary for carbon metabolism, motility, and biofilm formation in E. coli and other bacteria.
Y. pseudotuberculosis has a functional Csr regulatory system, including CsrB and CsrC sRNAs, involved in virulence (Heroven et al., 2008, 2012a; Table 3). CsrB and CsrC levels are differentially regulated based on genetic and environmental conditions, with CsrC levels elevated in complex media and absent in minimal media (Heroven et al., 2008). CsrB levels, which or normally low, are activated by the response regulator UvrY (Heroven et al., 2008). As already mentioned, Crp regulates the expression of the Csr RNAs (Heroven et al., 2012b). The Crp regulation of RovM-RovA-InvA regulatory cascade is through the Csr RNAs (Heroven et al., 2012b; Figure 3). Crp regulation of flagellar synthesis genes, flhDC, is through CsrA (Heroven et al., 2012b). CsrB and CsrC levels are repressed in Y. pestis isolated from the lungs of a mouse model of infection, suggesting that CsrB and CsrC expression is detrimental for virulence (Yan et al., 2013). The PhoP response regulator that senses environmental Mg2+, low pH, and antimicrobial peptides, through direct interaction between PhoP and the CsrC upstream region, also stimulates csrC transcription (Nuss et al., 2014; Figure 3). The stability of CsrC is variable between strains of Y. pseudotuberculosis YPIII and IP32953 due to the presence of a 20 nucleotide insertion mutation in the IP32953 csrC gene (Nuss et al., 2014). The Y. pseudotuberculosis IP32953 CsrC is significantly less stable than the YPIII CsrC, with t1∕2 of 90.5 and 41.7 min, respectively. When Y. pseudotuberculosis YPIII csrC gene was engineered to include the 20 nucleotide insertion from the Y. pseudotuberculosis IP32953 csrC gene, the YPIII CsrC transcript was destabilized to levels seen in IP32953 (Nuss et al., 2014).
As described in earlier sections, RovA is a premier global regulator of virulence gene expression in Y. pseudotuberculosis (Nagel et al., 2001; Figure 3). Y. pseudotuberculosis CsrA overexpression leads to the repression of RovA. CsrA plays a role in motility in both Y. pseudotuberculosis and Y. enterocolitica (Heroven et al., 2008; Legrand et al., 2015). RNA sequencing analyses of Y. pseudotuberculosis obtained using a mouse model of persistent infection revealed an overlap between genes modulated during persistent infection and the Crp/CsrA/RovA regulons, suggesting a role for CsrA in Y. pseudotuberculosis persistent infections (Avican et al., 2015). CsrA plays a role in resistance to several osmolytes (NaCl, KCl, CaCl2, and Rhamnose), temperature extremes (4 and 42°C), and antibiotic susceptibility (ampicillin and spectinomycin; Legrand et al., 2015).
Potential targets for anti-bacterial development
Infectious disease is a major cause of mortality world-wide, and is now exacerbated by the rapid spread of antibiotic resistance. New approaches to treat infectious diseases are urgently needed, but persisting with traditional bacteriocidal antibiotics is pointless since bacteria are too adept at rapidly evolving resistance mechanisms toward them. One new approach being investigated to potentially limit mechanisms selecting for drug resistant bacteria is to identify bacteriostatic molecules that neutralize classical pathogen virulence factors or the mechanisms controlling their synthesis—the so-called “anti-infective” molecules (National Research Council, 2006). Many examples exist of high throughput screens designed to identify inhibitory molecules derived from natural or chemical libraries. In fact, studies using Yersinia as a model pathogen have demonstrated a potentially useful broad spectrum therapeutic target to be regulatory, assembly or functional mechanisms associated with the Ysc-Yop T3SS or the activity of its cargo (Kauppi et al., 2003; Tautz et al., 2005; Kim et al., 2009; Pan et al., 2009; Harmon et al., 2010; Wang et al., 2011; Jessen et al., 2014). This proved that identifying chemical inhibitors of classical virulence determinants (i.e.: anti-infectives) is a feasible approach to novel antibacterial innovation.
Moreover, ongoing methodological developments should benefit our future knowledge of the complex regulation of metabolism and virulence that underscores bacterial fitness during host infections. Greater understanding of these regulatory mechanisms will heighten selection of the most appropriate targets for the design and development of alternative novel anti-bacterials. For example, reduced in vivo fitness of several Yersinia mutants defective in metabolic pathways and transition metal transport pathways suggests that similar effects could be generated with the use of inhibitory anti-infectives preventing a pathogens quest for available carbon and trace element supplies (Klein and Lewinson, 2011; Palace et al., 2014; Pradel et al., 2014; Avican et al., 2015; Deng et al., 2015). A recent study has also successfully demonstrated the potential for targeting the CpxA-CpxR two-component regulatory system as a means to de-weaponize bacterial pathogens (Van Rensburg et al., 2015), which is consistent with our own observations that CpxA mutants deficient in phosphatase activity, exhibit over-active CpxA-CpxR signaling, and this leads to a dramatic reduction in virulence gene expression by Y. pseudotuberculosis (Carlsson et al., 2007a,b; Liu et al., 2011, 2012). Signaling molecules in the form of c-di-GMP and (p)ppGpp have also been the target for novel therapies against infectious disease. As these molecules have profound effects on bacterial pathophysiology, the synthesis of membrane permeable analogs might serve as useful antibacterial agents (Kalia et al., 2013). Much investment is still needed to uncover effective novel anti-infectives for use in the clinic, but studies on model bacterial systems such as pathogenic Yersinia give clear indication that new methods of infectious disease control will be discovered once again.
Author contributions
SC, KT, and MF designed, drafted and revised the manuscript, and approved of its final content. SC, KT, and MF agree to be accountable for all aspects of the content.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research in the author's own laboratories has been possible through the generous past or present funding support of the National Science Foundation of China (#31170133 and #31570132; SC), National Institute of General Medical Sciences of the National Institutes of Health (#SC2 GM105419; KT), Howard University Medical Alumni Association (KT), Medical Research Foundation of Umeå University (MF), Swedish Research Council (#2014-2105; MF), and Swedish Research Council framework grant—antibiotics and infection (#2014-6652; SC, KT, and MF). Work in the laboratory of MF is performed within the framework of the Umeå Centre for Microbial Research—Linnaeus Program. The author's apologize to all those researchers whose valuable contributions were not cited in this review due to length limitations.
References
- Abu Khweek A., Fetherston J. D., Perry R. D. (2010). Analysis of HmsH and its role in plague biofilm formation. Microbiology 156, 1424–1438. 10.1099/mic.0.036640-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Kwaik Y., Bumann D. (2013). Microbial quest for food in vivo: ‘nutritional virulence’ as an emerging paradigm. Cell. Microbiol. 15, 882–890. 10.1111/cmi.12138 [DOI] [PubMed] [Google Scholar]
- Acosta N., Pukatzki S., Raivio T. L. (2015). The Vibrio cholerae Cpx envelope stress response senses and mediates adaptation to low iron. J. Bacteriol. 197, 262–276. 10.1128/JB.01957-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansong C., Schrimpe-Rutledge A. C., Mitchell H. D., Chauhan S., Jones M. B., Kim Y. M., et al. (2013). A multi-omic systems approach to elucidating Yersinia virulence mechanisms. Mol. Biosyst. 9, 44–54. 10.1039/C2MB25287B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonoaea R., Furst M., Nishiyama K., Muller M. (2008). The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry 47, 5649–5656. 10.1021/bi800233w [DOI] [PubMed] [Google Scholar]
- Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E. G., Margalit H., et al. (2001). Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11, 941–950. 10.1016/S0960-9822(01)00270-6 [DOI] [PubMed] [Google Scholar]
- Atkinson S., Goldstone R. J., Joshua G. W., Chang C. Y., Patrick H. L., Camara M., et al. (2011). Biofilm development on Caenorhabditis elegans by Yersinia is facilitated by quorum sensing-dependent repression of type III secretion. PLoS Pathog. 7:e1001250. 10.1371/journal.ppat.1001250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avican K., Fahlgren A., Huss M., Heroven A. K., Beckstette M., Dersch P., et al. (2015). Reprogramming of Yersinia from virulent to persistent mode revealed by complex in vivo RNA-seq analysis. PLoS Pathog. 11:e1004600. 10.1371/journal.ppat.1004600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Calderon E. D., Araiza-Villanueva M. G., Cancino-Diaz J. C., Lopez-Villegas E. O., Sriranganathan N., Boyle S. M., et al. (2015). Roles of bacterial membrane vesicles. Arch. Microbiol. 197, 1–10. 10.1007/s00203-014-1042-7 [DOI] [PubMed] [Google Scholar]
- Axler-Diperte G. L., Hinchliffe S. J., Wren B. W., Darwin A. J. (2009). YtxR acts as an overriding transcriptional off switch for the Yersinia enterocolitica Ysc-Yop type 3 secretion system. J. Bacteriol. 191, 514–524. 10.1128/JB.01305-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G., Golubov A., Smith E. A., Mcdonough K. A. (2010). The importance of the small RNA chaperone Hfq for growth of epidemic Yersinia pestis, but not Yersinia pseudotuberculosis, with implications for plague biology. J. Bacteriol. 192, 4239–4245. 10.1128/JB.00504-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchinger S. E., Ades S. E. (2013). Regulated proteolysis: control of the Escherichia coli sigma(E)-dependent cell envelope stress response. Subcell. Biochem. 66, 129–160. 10.1007/978-94-007-5940-4_6 [DOI] [PubMed] [Google Scholar]
- Bearden S. W., Perry R. D. (1999). The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32, 403–414. 10.1046/j.1365-2958.1999.01360.x [DOI] [PubMed] [Google Scholar]
- Becker K. W., Skaar E. P. (2014). Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 38, 1235–1249. 10.1111/1574-6976.12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens-Kneip S. (2010). The role of SurA factor in outer membrane protein transport and virulence. Int. J. Med. Microbiol. 300, 421–428. 10.1016/j.ijmm.2010.04.012 [DOI] [PubMed] [Google Scholar]
- Bellows L. E., Koestler B. J., Karaba S. M., Waters C. M., Lathem W. W. (2012). Hfq-dependent, co-ordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol. 86, 661–674. 10.1111/mmi.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Fetherston J. D., Miller M. C., Burlison J. A., Perry R. D. (2014). The Yersinia pestis siderophore, yersiniabactin, and the ZnuABC system both contribute to zinc acquisition and the development of lethal septicaemic plague in mice. Mol. Microbiol. 93, 759–775. 10.1111/mmi.12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Forman S., Mack D., Perry R. D. (2008). Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ. Microbiol. 10, 1419–1432. 10.1111/j.1462-2920.2007.01554.x [DOI] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Perry R. D. (2005). The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247, 123–130. 10.1016/j.femsle.2005.04.036 [DOI] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Ryjenkov D. A., Waters C. M., Price P. A., Fetherston J. D., et al. (2011). Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79, 533–551. 10.1111/j.1365-2958.2010.07470.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrov A. G., Kirillina O., Vadyvaloo V., Koestler B. J., Hinz A. K., Mack D., et al. (2015). The Yersinia pestis HmsCDE regulatory system is essential for blockage of the oriental rat flea (Xenopsylla cheopis), a classic plague vector. Environ. Microbiol. 17, 947–959. 10.1111/1462-2920.12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohme K., Steinmann R., Kortmann J., Seekircher S., Heroven A. K., Berger E., et al. (2012). Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518. 10.1371/journal.ppat.1002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin E., Hershberg R. (2015). Gene loss dominates as a source of genetic variation within clonal pathogenic bacterial species. Genome Biol. Evol. 7, 2173–2187. 10.1093/gbe/evv135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontemps-Gallo S., Madec E., Lacroix J. M. (2015). The two-component system CpxAR is essential for virulence in the phytopathogen bacteria Dickeya dadantii EC3937. Environ. Microbiol. 17, 4415–4428. 10.1111/1462-2920.12874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozue J., Mou S., Moody K. L., Cote C. K., Trevino S., Fritz D., et al. (2011). The role of the phoPQ operon in the pathogenesis of the fully virulent CO92 strain of Yersinia pestis and the IP32953 strain of Yersinia pseudotuberculosis. Microb. Pathog. 50, 314–321. 10.1016/j.micpath.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Bucker R., Heroven A. K., Becker J., Dersch P., Wittmann C. (2014). The pyruvate-tricarboxylic acid cycle node: a focal point of virulence control in the enteric pathogen Yersinia pseudotuberculosis. J. Biol. Chem. 289, 30114–30132. 10.1074/jbc.M114.581348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury-Mone S., Nomane Y., Reymond N., Barbet R., Jacquet E., Imbeaud S., et al. (2009). Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 5:e1000651. 10.1371/journal.pgen.1000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson K. E., Liu J., Edqvist P. J., Francis M. S. (2007a). Extracytoplasmic-stress-responsive pathways modulate type III secretion in Yersinia pseudotuberculosis. Infect. Immun. 75, 3913–3924. 10.1128/IAI.01346-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson K. E., Liu J., Edqvist P. J., Francis M. S. (2007b). Influence of the Cpx Extracytoplasmic-stress-responsive pathway on Yersinia sp.-eukaryotic cell contact. Infect. Immun. 75, 4386–4399. 10.1128/IAI.01450-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C., Payne S. M. (2014). Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J. Inorg. Biochem. 133, 110–117. 10.1016/j.jinorgbio.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat J. E., Skaar E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13, 509–519. 10.1016/j.chom.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanie-Cornet M. P., Penfound T. A., Smith D., Elliott J. F., Foster J. W. (1999). Control of acid resistance in Escherichia coli. J. Bacteriol. 181, 3525–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathelyn J. S., Crosby S. D., Lathem W. W., Goldman W. E., Miller V. L. (2006). RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. U.S.A. 103, 13514–13519. 10.1073/pnas.0603456103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathelyn J. S., Ellison D. W., Hinchliffe S. J., Wren B. W., Miller V. L. (2007). The RovA regulons of Yersinia enterocolitica and Yersinia pestis are distinct: evidence that many RovA-regulated genes were acquired more recently than the core genome. Mol. Microbiol. 66, 189–205. 10.1111/j.1365-2958.2007.05907.x [DOI] [PubMed] [Google Scholar]
- Cellier M. F., Courville P., Campion C. (2007). Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 9, 1662–1670. 10.1016/j.micinf.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Chain P. S., Carniel E., Larimer F. W., Lamerdin J., Stoutland P. O., Regala W. M., et al. (2004). Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U.S.A. 101, 13826–13831. 10.1073/pnas.0404012101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion O. L., Karlyshev A., Cooper I. A., Ford D. C., Wren B. W., Duffield M., et al. (2011). Yersinia pseudotuberculosis mntH functions in intracellular manganese accumulation, which is essential for virulence and survival in cells expressing functional Nramp1. Microbiology 157, 1115–1122. 10.1099/mic.0.045807-0 [DOI] [PubMed] [Google Scholar]
- Chauvaux S., Dillies M. A., Marceau M., Rosso M. L., Rousseau S., Moszer I., et al. (2011). In silico comparison of Yersinia pestis and Yersinia pseudotuberculosis transcriptomes reveals a higher expression level of crucial virulence determinants in the plague bacillus. Int. J. Med. Microbiol. 301, 105–116. 10.1016/j.ijmm.2010.08.013 [DOI] [PubMed] [Google Scholar]
- Chouikha I., Hinnebusch B. J. (2014). Silencing urease: a key evolutionary step that facilitated the adaptation of Yersinia pestis to the flea-borne transmission route. Proc. Natl. Acad. Sci. U.S.A. 111, 18709–18714. 10.1073/pnas.1413209111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromy B. A., Choi M. W., Murphy G. A., Gonzales A. D., Corzett C. H., Chang B. C., et al. (2005). Proteomic characterization of Yersinia pestis virulence. J. Bacteriol. 187, 8172–8180. 10.1128/JB.187.23.8172-8180.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G., Sluiters C., De Rouvroit C. L., Michiels T. (1989). Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171, 254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G. R., Sluiters C., Delor I., Geib D., Kaniga K., Lambert De Rouvroit C., et al. (1991). ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5, 1023–1034. 10.1111/j.1365-2958.1991.tb01875.x [DOI] [PubMed] [Google Scholar]
- Dalebroux Z. D., Svensson S. L., Gaynor E. C., Swanson M. S. (2010). ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199. 10.1128/MMBR.00046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z. D., Swanson M. S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. 10.1038/nrmicro2720 [DOI] [PubMed] [Google Scholar]
- Darby C., Hsu J. W., Ghori N., Falkow S. (2002). Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417, 243–244. 10.1038/417243a [DOI] [PubMed] [Google Scholar]
- Darby C. (2008). Uniquely insidious: Yersinia pestis biofilms. Trends Microbiol. 16, 158–164. 10.1016/j.tim.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Darwin A. J., Miller V. L. (1999). Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32, 51–62. 10.1046/j.1365-2958.1999.01324.x [DOI] [PubMed] [Google Scholar]
- Darwin A. J., Miller V. L. (2001). The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced. Mol. Microbiol. 39, 429–444. 10.1046/j.1365-2958.2001.02235.x [DOI] [PubMed] [Google Scholar]
- Darwin A. J. (2013). Stress relief during host infection: the phage shock protein response supports bacterial virulence in various ways. PLoS Pathog. 9:e1003388. 10.1371/journal.ppat.1003388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koning-Ward T. F., Robins-Browne R. M. (1995). Contribution of urease to acid tolerance in Yersinia enterocolitica. Infect. Immun. 63, 3790–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P., Akerley B. J., Lin E. C. (2000). Presence of the Cpx system in bacteria. Microbiology 146(Pt 2), 247–248. 10.1099/00221287-146-2-247 [DOI] [PubMed] [Google Scholar]
- Debnath I., Norton J. P., Barber A. E., Ott E. M., Dhakal B. K., Kulesus R. R., et al. (2013). The Cpx stress response system potentiates the fitness and virulence of uropathogenic Escherichia coli. Infect. Immun. 81, 1450–1459. 10.1128/IAI.01213-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delihas N. (2003). Annotation and evolutionary relationships of a small regulatory RNA gene micF and its target ompF in Yersinia species. BMC Microbiol. 3:13. 10.1186/1471-2180-3-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisa M. P., Lee P., Palmer T., Georgiou G. (2004). Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J. Bacteriol. 186, 366–373. 10.1128/JB.186.2.366-373.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Liu Z., Bi Y., Wang X., Zhou D., Yang R., et al. (2014). Rapid degradation of Hfq-free RyhB in Yersinia pestis by PNPase independent of putative ribonucleolytic complexes. Biomed. Res. Int. 2014:798918. 10.1155/2014/798918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Liu Z., He J., Wang J., Yan Y., Wang X., et al. (2015). TyrR, the regulator of aromatic amino acid metabolism, is required for mice infection of Yersinia pestis. Front. Microbiol. 6:110. 10.3389/fmicb.2015.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z., Meng X., Su S., Liu Z., Ji X., Zhang Y., et al. (2012). Two sRNA RyhB homologs from Yersinia pestis biovar microtus expressed in vivo have differential Hfq-dependent stability. Res. Microbiol. 163, 413–418. 10.1016/j.resmic.2012.05.006 [DOI] [PubMed] [Google Scholar]
- Desrosiers D. C., Bearden S. W., Mier I., Jr., Abney J., Paulley J. T., Fetherston J. D., et al. (2010). Znu is the predominant zinc importer in Yersinia pestis during in vitro growth but is not essential for virulence. Infect. Immun. 78, 5163–5177. 10.1128/IAI.00732-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J. (2008). The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11, 87–93. 10.1016/j.mib.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Drancourt M. (2012). Plague in the genomic area. Clin. Microbiol. Infect. 18, 224–230. 10.1111/j.1469-0691.2012.03774.x [DOI] [PubMed] [Google Scholar]
- Eddy J. L., Gielda L. M., Caulfield A. J., Rangel S. M., Lathem W. W. (2014). Production of outer membrane vesicles by the plague pathogen Yersinia pestis. PLoS ONE 9:e107002. 10.1371/journal.pone.0107002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenreich W., Dandekar T., Heesemann J., Goebel W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412. 10.1038/nrmicro2351 [DOI] [PubMed] [Google Scholar]
- Ellis T. N., Kuehn M. J. (2010). Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74, 81–94. 10.1128/MMBR.00031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison D. W., Miller V. L. (2006a). H-NS represses inv transcription in Yersinia enterocolitica through competition with RovA and interaction with YmoA. J. Bacteriol. 188, 5101–5112. 10.1128/JB.00862-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison D. W., Miller V. L. (2006b). Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9, 153–159. 10.1016/j.mib.2006.02.003 [DOI] [PubMed] [Google Scholar]
- Erickson D. L., Jarrett C. O., Callison J. A., Fischer E. R., Hinnebusch B. J. (2008). Loss of a biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J. Bacteriol. 190, 8163–8170. 10.1128/JB.01181-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N., Qu S., Yang H., Fang H., Liu L., Zhang Y., et al. (2014). HmsB enhances biofilm formation in Yersinia pestis. Front. Microbiol. 5:685. 10.3389/fmicb.2014.00685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N., Yang H., Fang H., Liu L., Zhang Y., Wang L., et al. (2015). RcsAB is a major repressor of Yersinia biofilm development through directly acting on hmsCDE, hmsT, and hmsHFRS. Sci. Rep. 5:9566. 10.1038/srep09566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanghanel J., Fischer G. (2004). Insights into the catalytic mechanism of peptidyl prolyl cis/trans isomerases. Front. Biosci. 9:1494. 10.2741/1494 [DOI] [PubMed] [Google Scholar]
- Felek S., Jeong J. J., Runco L. M., Murray S., Thanassi D. G., Krukonis E. S. (2011). Contributions of chaperone/usher systems to cell binding, biofilm formation and Yersinia pestis virulence. Microbiology 157, 805–818. 10.1099/mic.0.044826-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felek S., Lawrenz M. B., Krukonis E. S. (2008). The Yersinia pestis autotransporter YapC mediates host cell binding, autoaggregation and biofilm formation. Microbiology 154, 1802–1812. 10.1099/mic.0.2007/010918-0 [DOI] [PubMed] [Google Scholar]
- Fetherston J. D., Mier I., Jr., Truszczynska H., Perry R. D. (2012). The Yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect. Immun. 80, 3880–3891. 10.1128/IAI.00086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillat M. F. (2014). The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546, 41–52. 10.1016/j.abb.2014.01.029 [DOI] [PubMed] [Google Scholar]
- Flamez C., Ricard I., Arafah S., Simonet M., Marceau M. (2008). Phenotypic analysis of Yersinia pseudotuberculosis 32777 response regulator mutants: new insights into two-component system regulon plasticity in bacteria. Int. J. Med. Microbiol. 298, 193–207. 10.1016/j.ijmm.2007.05.005 [DOI] [PubMed] [Google Scholar]
- Flores-Kim J., Darwin A. J. (2014). Regulation of bacterial virulence gene expression by cell envelope stress responses. Virulence 5, 835–851. 10.4161/21505594.2014.965580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman S., Paulley J. T., Fetherston J. D., Cheng Y. Q., Perry R. D. (2010). Yersinia ironomics: comparison of iron transporters among Yersinia pestis biotypes and its nearest neighbor, Yersinia pseudotuberculosis. Biometals 23, 275–294. 10.1007/s10534-009-9286-4 [DOI] [PubMed] [Google Scholar]
- Foster J. W. (2004). Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907. 10.1038/nrmicro1021 [DOI] [PubMed] [Google Scholar]
- Francis M. S. (2011). Secretion systems and metabolism in the pathogenic Yersiniae, in Stress Response in Pathogenic Bacteria, ed Kidd S. P. (Wallingford, CT: CABI Publishing; ), 185–220. [Google Scholar]
- Fukuto H. S., Svetlanov A., Palmer L. E., Karzai A. W., Bliska J. B. (2010). Global gene expression profiling of Yersinia pestis replicating inside macrophages reveals the roles of a putative stress-induced operon in regulating type III secretion and intracellular cell division. Infect. Immun. 78, 3700–3715. 10.1128/IAI.00062-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaca A. O., Colomer-Winter C., Lemos J. A. (2015). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156. 10.1128/JB.02577-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaiah D., Zhang X., Fortney K. R., Baker B., Liu Y., Munson R. S., Jr., et al. (2013). Activation of CpxRA in Haemophilus ducreyi primarily inhibits the expression of its targets, including major virulence determinants. J. Bacteriol. 195, 3486–3502. 10.1128/JB.00372-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoth A., Alhadeff R., Kohen D., Arkin I. T. (2011). Characterization of the Na(+)/H(+) antiporter from Yersinia pestis. PLoS ONE 6:e26115. 10.1371/journal.pone.0026115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Zhang Y., Yang L., Liu X., Guo Z., Tan Y., et al. (2011). Regulatory effects of cAMP receptor protein (CRP) on porin genes and its own gene in Yersinia pestis. BMC Microbiol. 11:40. 10.1186/1471-2180-11-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Zhou D., Li Y., Guo Z., Han Y., Song Y., et al. (2008). The iron-responsive Fur regulon in Yersinia pestis. J. Bacteriol. 190, 3063–3075. 10.1128/JB.01910-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Lyu Z. X., Liu Y., Wang R., Zhao X. S., Fu X., et al. (2014). Identification of FkpA as a key quality control factor for the biogenesis of outer membrane proteins under heat shock conditions. J. Bacteriol. 196, 672–680. 10.1128/JB.01069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Song Y., Yang L., Feng Y., Qiu Y., Li G., et al. (2009). Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS ONE 4:e6213. 10.1371/journal.pone.0006213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopel Y., Luttmann D., Heroven A. K., Reichenbach B., Dersch P., Gorke B. (2011). Common and divergent features in transcriptional control of the homologous small RNAs GlmY and GlmZ in Enterobacteriaceae. Nucleic Acids Res. 39, 1294–1309. 10.1093/nar/gkq986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorke B., Stulke J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- Gothel S. F., Marahiel M. A. (1999). Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell. Mol. Life Sci. 55, 423–436. 10.1007/s000180050299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Roche E., Zhou Y., Sauer R. T. (1998). The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12, 1338–1347. 10.1101/gad.12.9.1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotzke H., Palombo I., Muheim C., Perrody E., Genevaux P., Kudva R., et al. (2014). YfgM is an ancillary subunit of the SecYEG translocon in Escherichia coli. J. Biol. Chem. 289, 19089–19097. 10.1074/jbc.M113.541672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabenstein J. P., Fukuto H. S., Palmer L. E., Bliska J. B. (2006). Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect. Immun. 74, 3727–3741. 10.1128/IAI.00255-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabenstein J. P., Marceau M., Pujol C., Simonet M., Bliska J. B. (2004). The response regulator PhoP of Yersinia pseudotuberculosis is important for replication in macrophages and for virulence. Infect. Immun. 72, 4973–4984. 10.1128/IAI.72.9.4973-4984.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Crack J. C., Thomson A. J., Lebrun N. E. (2009). Bacterial sensors of oxygen. Curr. Opin. Microbiol. 12, 145–151. 10.1016/j.mib.2009.01.008 [DOI] [PubMed] [Google Scholar]
- Green R. C., Darwin A. J. (2004). PspG, a new member of the Yersinia enterocolitica phage shock protein regulon. J. Bacteriol. 186, 4910–4920. 10.1128/JB.186.15.4910-4920.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilvout I., Chami M., Engel A., Pugsley A. P., Bayan N. (2006). Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 25, 5241–5249. 10.1038/sj.emboj.7601402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. S., Gross C. A. (2014). Stress-induced remodeling of the bacterial proteome. Curr. Biol. 24, R424–R434. 10.1016/j.cub.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. P., Ren G. X., Zhu H., Mao X. J., Sun Y. C. (2015). Differential regulation of the hmsCDE operon in Yersinia pestis and Yersinia pseudotuberculosis by the Rcs phosphorelay system. Sci. Rep. 5, 8412. 10.1038/srep08412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller J. C., Carlson S., Pederson K. J., Pierson D. E. (2000). A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36, 1436–1446. 10.1046/j.1365-2958.2000.01964.x [DOI] [PubMed] [Google Scholar]
- Harmon D. E., Davis A. J., Castillo C., Mecsas J. (2010). Identification and characterization of small-molecule inhibitors of Yop translocation in Yersinia pseudotuberculosis. Antimicrob. Agents Chemother. 54, 3241–3254. 10.1128/AAC.00364-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase C. C., Fedorova N. D., Galperin M. Y., Dibrov P. A. (2001). Sodium ion cycle in bacterial pathogens: evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 65, 353–370. 10.1128/MMBR.65.3.353-370.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkoff M., Runco L. M., Pujol C., Jayatilaka I., Furie M. B., Bliska J. B., et al. (2012). Roles of chaperone/usher pathways of Yersinia pestis in a murine model of plague and adhesion to host cells. Infect. Immun. 80, 3490–3500. 10.1128/IAI.00434-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V., Atkinson G. C., Murakami K. S., Tenson T., Gerdes K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. 10.1038/nrmicro3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heermann R., Fuchs T. M. (2008). Comparative analysis of the Photorhabdus luminescens and the Yersinia enterocolitica genomes: uncovering candidate genes involved in insect pathogenicity. BMC Genomics 9:40. 10.1186/1471-2164-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert Tran E. E., Goodrich-Blair H. (2009). CpxRA contributes to Xenorhabdus nematophila virulence through regulation of lrhA and modulation of insect immunity. Appl. Environ. Microbiol. 75, 3998–4006. 10.1128/AEM.02657-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst K., Bujara M., Heroven A. K., Opitz W., Weichert M., Zimmermann A., et al. (2009). Intrinsic thermal sensing controls proteolysis of Yersinia virulence regulator RovA. PLoS Pathog. 5:e1000435. 10.1371/journal.ppat.1000435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroven A. K., Bohme K., Dersch P. (2012a). The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol. 9, 379–391. 10.4161/rna.19333 [DOI] [PubMed] [Google Scholar]
- Heroven A. K., Bohme K., Rohde M., Dersch P. (2008). A Csr-type regulatory system, including small non-coding RNAs, regulates the global virulence regulator RovA of Yersinia pseudotuberculosis through RovM. Mol. Microbiol. 68, 1179–1195. 10.1111/j.1365-2958.2008.06218.x [DOI] [PubMed] [Google Scholar]
- Heroven A. K., Dersch P. (2006). RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol. Microbiol. 62, 1469–1483. 10.1111/j.1365-2958.2006.05458.x [DOI] [PubMed] [Google Scholar]
- Heroven A. K., Dersch P. (2014). Coregulation of host-adapted metabolism and virulence by pathogenic yersiniae. Front. Cell. Infect. Microbiol. 4:146. 10.3389/fcimb.2014.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroven A. K., Nagel G., Tran H. J., Parr S., Dersch P. (2004). RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 53, 871–888. 10.1111/j.1365-2958.2004.04162.x [DOI] [PubMed] [Google Scholar]
- Heroven A. K., Sest M., Pisano F., Scheb-Wetzel M., Steinmann R., Bohme K., et al. (2012b). Crp induces switching of the CsrB and CsrC RNAs in Yersinia pseudotuberculosis and links nutritional status to virulence. Front. Cell. Infect. Microbiol. 2:158. 10.3389/fcimb.2012.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusipp G., Schmidt M. A., Miller V. L. (2003). Identification of rpoE and nadB as host responsive elements of Yersinia enterocolitica. FEMS Microbiol. Lett. 226, 291–298. 10.1016/S0378-1097(03)00613-X [DOI] [PubMed] [Google Scholar]
- Hinchliffe S. J., Howard S. L., Huang Y. H., Clarke D. J., Wren B. W. (2008). The importance of the Rcs phosphorelay in the survival and pathogenesis of the enteropathogenic yersiniae. Microbiology 154, 1117–1131. 10.1099/mic.0.2007/012534-0 [DOI] [PubMed] [Google Scholar]
- Hinnebusch B. J., Erickson D. L. (2008). Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr. Top. Microbiol. Immunol. 322, 229–248. 10.1007/978-3-540-75418-3_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobman J. L., Crossman L. C. (2015). Bacterial antimicrobial metal ion resistance. J. Med. Microbiol. 64, 471–497. 10.1099/jmm.0.023036-0 [DOI] [PubMed] [Google Scholar]
- Hoe N. P., Goguen J. D. (1993). Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175, 7901–7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe N. P., Minion F. C., Goguen J. D. (1992). Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J. Bacteriol. 174, 4275–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung J. M., Jones H. A., Perry R. D. (1996). The hmu locus of Yersinia pestis is essential for utilization of free haemin and haem–protein complexes as iron sources. Mol. Microbiol. 20, 725–739. 10.1111/j.1365-2958.1996.tb02512.x [DOI] [PubMed] [Google Scholar]
- Horstman N. K., Darwin A. J. (2012). Phage shock proteins B and C prevent lethal cytoplasmic membrane permeability in Yersinia enterocolitica. Mol. Microbiol. 85, 445–460. 10.1111/j.1365-2958.2012.08120.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Lu P., Wang Y., Ding L., Atkinson S., Chen S. (2009). OmpR positively regulates urease expression to enhance acid survival of Yersinia pseudotuberculosis. Microbiology 155, 2522–2531. 10.1099/mic.0.028381-0 [DOI] [PubMed] [Google Scholar]
- Hu Y., Lu P., Zhang Y., Li L., Chen S. (2010). Characterization of an aspartate-dependent acid survival system in Yersinia pseudotuberculosis. FEBS Lett. 584, 2311–2314. 10.1016/j.febslet.2010.03.045 [DOI] [PubMed] [Google Scholar]
- Hu Y., Lu P., Zhang Y., Li Y., Li L., Huang L., et al. (2011). Cra negatively regulates acid survival in Yersinia pseudotuberculosis. FEMS Microbiol. Lett. 317, 190–195. 10.1111/j.1574-6968.2011.02227.x [DOI] [PubMed] [Google Scholar]
- Huang Y. H., Ferrieres L., Clarke D. J. (2006). The role of the Rcs phosphorelay in Enterobacteriaceae. Res. Microbiol. 157, 206–212. 10.1016/j.resmic.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Humphreys S., Rowley G., Stevenson A., Anjum M. F., Woodward M. J., Gilbert S., et al. (2004). Role of the two-component regulator CpxAR in the virulence of Salmonella enterica serotype Typhimurium. Infect. Immun. 72, 4654–4661. 10.1128/IAI.72.8.4654-4661.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunke S., Keller R., Muller V. S. (2012). Signal integration by the Cpx-envelope stress system. FEMS Microbiol. Lett. 326, 12–22. 10.1111/j.1574-6968.2011.02436.x [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (2015). Transcription factors that defend bacteria against reactive oxygen species. Annu. Rev. Microbiol. 69, 93–108. 10.1146/annurev-micro-091014-104322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. W., Silva-Herzog E., Plano G. V. (2004). The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54, 1364–1378. 10.1111/j.1365-2958.2004.04353.x [DOI] [PubMed] [Google Scholar]
- Jarrett C. O., Deak E., Isherwood K. E., Oyston P. C., Fischer E. R., Whitney A. R., et al. (2004). Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190, 783–792. 10.1086/422695 [DOI] [PubMed] [Google Scholar]
- Jessen D. L., Bradley D. S., Nilles M. L. (2014). A type III secretion system inhibitor targets YopD while revealing differential regulation of secretion in calcium-blind mutants of Yersinia pestis. Antimicrob. Agents Chemother. 58, 839–850. 10.1128/AAC.01170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly N., Engl C., Jovanovic G., Huvet M., Toni T., Sheng X., et al. (2010). Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34, 797–827. 10.1111/j.1574-6976.2010.00240.x [DOI] [PubMed] [Google Scholar]
- Jones H. A., Lillard J. W., Jr., Perry R. D. (1999). HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology 145 (Pt. 8), 2117–2128. 10.1099/13500872-145-8-2117 [DOI] [PubMed] [Google Scholar]
- Jones S. E., Lloyd L. J., Tan K. K., Buck M. (2003). Secretion defects that activate the phage shock response of Escherichia coli. J. Bacteriol. 185, 6707–6711. 10.1128/JB.185.22.6707-6711.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic G., Lloyd L. J., Stumpf M. P., Mayhew A. J., Buck M. (2006). Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J. Biol. Chem. 281, 21147–21161. 10.1074/jbc.M602323200 [DOI] [PubMed] [Google Scholar]
- Kakoschke T., Kakoschke S., Magistro G., Schubert S., Borath M., Heesemann J., et al. (2014). The RNA chaperone Hfq impacts growth, metabolism and production of virulence factors in Yersinia enterocolitica. PLoS ONE 9:e86113. 10.1371/journal.pone.0086113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia D., Merey G., Nakayama S., Zheng Y., Zhou J., Luo Y., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341. 10.1039/C2CS35206K [DOI] [PubMed] [Google Scholar]
- Karzai A. W., Susskind M. M., Sauer R. T. (1999). SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18, 3793–3799. 10.1093/emboj/18.13.3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi A. M., Nordfelth R., Uvell H., Wolf-Watz H., Elofsson M. (2003). Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10, 241–249. 10.1016/S1074-5521(03)00046-2 [DOI] [PubMed] [Google Scholar]
- Keiler K. C., Waller P. R., Sauer R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. 10.1126/science.271.5251.990 [DOI] [PubMed] [Google Scholar]
- Keller B., Muhlenkamp M., Deuschle E., Siegfried A., Mossner S., Schade J., et al. (2015). Yersinia enterocolitica exploits different pathways to accomplish adhesion and toxin injection into host cells. Cell. Microbiol. 17, 1179–1204. 10.1111/cmi.12429 [DOI] [PubMed] [Google Scholar]
- Kim O. K., Garrity-Ryan L. K., Bartlett V. J., Grier M. C., Verma A. K., Medjanis G., et al. (2009). N-hydroxybenzimidazole inhibitors of the transcription factor LcrF in Yersinia: novel antivirulence agents. J. Med. Chem. 52, 5626–5634. 10.1021/jm9006577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. J., Chauhan S., Motin V. L., Goh E. B., Igo M. M., Young G. M. (2007). Direct transcriptional control of the plasminogen activator gene of Yersinia pestis by the cyclic AMP receptor protein. J. Bacteriol. 189, 8890–8900. 10.1128/JB.00972-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. M., Schesser Bartra S., Plano G., Yahr T. L. (2013). ExsA and LcrF recognize similar consensus binding sites, but differences in their oligomeric state influence interactions with promoter DNA. J. Bacteriol. 195, 5639–5650. 10.1128/JB.00990-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirillina O., Fetherston J. D., Bobrov A. G., Abney J., Perry R. D. (2004). HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54, 75–88. 10.1111/j.1365-2958.2004.04253.x [DOI] [PubMed] [Google Scholar]
- Klein J. S., Lewinson O. (2011). Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metallomics 3, 1098–1108. 10.1039/c1mt00073j [DOI] [PubMed] [Google Scholar]
- Kobayashi R., Suzuki T., Yoshida M. (2007). Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66, 100–109. 10.1111/j.1365-2958.2007.05893.x [DOI] [PubMed] [Google Scholar]
- Kolodziejek A. M., Hovde C. J., Minnich S. A. (2012). Yersinia pestis Ail: multiple roles of a single protein. Front. Cell. Infect. Microbiol. 2:103. 10.3389/fcimb.2012.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo J., Burrows L. L., Howell P. L. (2012). Decoding the roles of pilotins and accessory proteins in secretin escort services. FEMS Microbiol. Lett. 328, 1–12. 10.1111/j.1574-6968.2011.02464.x [DOI] [PubMed] [Google Scholar]
- Koo J. T., Alleyne T. M., Schiano C. A., Jafari N., Lathem W. W. (2011). Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence. Proc. Natl. Acad. Sci. U.S.A. 108, E709–E717. 10.1073/pnas.1101655108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner H., Sofia H. J., Zumft W. G. (2003). Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27, 559–592. 10.1016/S0168-6445(03)00066-4 [DOI] [PubMed] [Google Scholar]
- Korotkov K. V., Gonen T., Hol W. G. (2011). Secretins: dynamic channels for protein transport across membranes. Trends Biochem. Sci. 36, 433–443. 10.1016/j.tibs.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labandeira-Rey M., Brautigam C. A., Hansen E. J. (2010). Characterization of the CpxRA regulon in Haemophilus ducreyi. Infect. Immun. 78, 4779–4791. 10.1128/IAI.00678-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert De Rouvroit C., Sluiters C., Cornelis G. R. (1992). Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6, 395–409. 10.1111/j.1365-2958.1992.tb01483.x [DOI] [PubMed] [Google Scholar]
- Lane M. C., Lenz J. D., Miller V. L. (2013). Proteolytic processing of the Yersinia pestis YapG autotransporter by the omptin protease Pla and the contribution of YapG to murine plague pathogenesis. J. Med. Microbiol. 62, 1124–1134. 10.1099/jmm.0.056275-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathem W. W., Crosby S. D., Miller V. L., Goldman W. E. (2005). Progression of primary pneumonic plague: a mouse model of infection, pathology, and bacterial transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 102, 17786–17791. 10.1073/pnas.0506840102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathem W. W., Schroeder J. A., Bellows L. E., Ritzert J. T., Koo J. T., Price P. A., et al. (2014). Posttranscriptional regulation of the Yersinia pestis cyclic AMP receptor protein Crp and impact on virulence. MBio 5:e01038–13. 10.1128/mBio.01038-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavander M., Ericsson S. K., Broms J. E., Forsberg A. (2006). The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infect. Immun. 74, 1768–1776. 10.1128/IAI.74.3.1768-1776.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrenz M. B., Lenz J. D., Miller V. L. (2009). A novel autotransporter adhesin is required for efficient colonization during bubonic plague. Infect. Immun. 77, 317–326. 10.1128/IAI.01206-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C. L., Swigon D., Murakami K. S., Darst S. A., Berman H. M., Ebright R. H. (2004). Catabolite activator protein: DNA binding and transcription activation. Curr. Opin. Struct. Biol. 14, 10–20. 10.1016/j.sbi.2004.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. C., Rietsch A. (2015). Fueling type III secretion. Trends Microbiol. 23, 296–300. 10.1016/j.tim.2015.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand K., Petersen S., Zheng Y., Liu K. K., Ozturk G., Chen J., et al. (2015). CsrA impacts survival of Yersinia enterolitica by affecting a myriad of physiological activities. BMC Microbiol. 15:31. 10.1186/s12866-015-0343-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J. D., Temple B. R., Miller V. L. (2012). Evolution and virulence contributions of the autotransporter proteins YapJ and YapK of Yersinia pestis CO92 and their homologs in Y. pseudotuberculosis IP32953. Infect. Immun. 80, 3693–3705. 10.1128/IAI.00529-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuko S., Raivio T. L. (2012). Mutations that impact the enteropathogenic Escherichia coli Cpx envelope stress response attenuate virulence in Galleria mellonella. Infect. Immun. 80, 3077–3085. 10.1128/IAI.00081-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gao H., Qin L., Li B., Han Y., Guo Z., et al. (2008). Identification and characterization of PhoP regulon members in Yersinia pestis biovar Microtus. BMC Genomics 9:143. 10.1186/1471-2164-9-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hu Y., Francis M. S., Chen S. (2015). RcsB positively regulates the Yersinia Ysc-Yop type III secretion system by activating expression of the master transcriptional regulator LcrF. Environ. Microbiol. 17, 1219–1233. 10.1111/1462-2920.12556 [DOI] [PubMed] [Google Scholar]
- Li Y., Qiu Y., Gao H., Guo Z., Han Y., Song Y., et al. (2009). Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. 9:128. 10.1186/1471-2180-9-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Obi I. R., Thanikkal E. J., Kieselbach T., Francis M. S. (2011). Phosphorylated CpxR restricts production of the RovA global regulator in Yersinia pseudotuberculosis. PLoS ONE 6:e23314. 10.1371/journal.pone.0023314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Thanikkal E. J., Obi I. R., Francis M. S. (2012). Elevated CpxR~P levels repress the Ysc-Yop type III secretion system of Yersinia pseudotuberculosis. Res. Microbiol. 163, 518–530. 10.1016/j.resmic.2012.07.010 [DOI] [PubMed] [Google Scholar]
- Liu M. Y., Gui G., Wei B., Preston J. F., III, Oakford L., Yuksel U., et al. (1997). The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizaes its activity in Escherichia coli. J. Biol. Chem. 272, 17502–17510. 10.1074/jbc.272.28.17502 [DOI] [PubMed] [Google Scholar]
- Liu Z., Wang H., Wang H., Wang J., Bi Y., Wang X., et al. (2015). Intrinsic plasmids influence MicF-mediated translational repression of ompF in Yersinia pestis. Front. Microbiol. 6:862. 10.3389/fmicb.2015.00862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd L. J., Jones S. E., Jovanovic G., Gyaneshwar P., Rolfe M. D., Thompson A., et al. (2004). Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein G (PspG). J. Biol. Chem. 279, 55707–55714. 10.1074/jbc.M408994200 [DOI] [PubMed] [Google Scholar]
- Lu P., Zhang Y., Li L., Hu Y., Huang L., Li Y., et al. (2012). Small non-coding RNA SraG regulates the operon YPK_1206-1205 in Yersinia pseudotuberculosis. FEMS Microbiol. Lett. 331, 37–43. 10.1111/j.1574-6968.2012.02548.x [DOI] [PubMed] [Google Scholar]
- Lukaszewski R. A., Kenny D. J., Taylor R., Rees D. G., Hartley M. G., Oyston P. C. (2005). Pathogenesis of Yersinia pestis infection in BALB/c mice: effects on host macrophages and neutrophils. Infect. Immun. 73, 7142–7150. 10.1128/IAI.73.11.7142-7150.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Z. X., Zhao X. S. (2015). Periplasmic quality control in biogenesis of outer membrane proteins. Biochem. Soc. Trans. 43, 133–138. 10.1042/BST20140217 [DOI] [PubMed] [Google Scholar]
- Majdalani N., Gottesman S. (2005). The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59, 379–405. 10.1146/annurev.micro.59.050405.101230 [DOI] [PubMed] [Google Scholar]
- Malpica R., Sandoval G. R., Rodriguez C., Franco B., Georgellis D. (2006). Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8, 781–795. 10.1089/ars.2006.8.781 [DOI] [PubMed] [Google Scholar]
- Masse E., Escorcia F. E., Gottesman S. (2003). Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383. 10.1101/gad.1127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E., Gottesman S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E., Vanderpool C. K., Gottesman S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. 10.1128/JB.187.20.6962-6971.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern Y., Barion B., Behrens-Kneip S. (2010). PpiD is a player in the network of periplasmic chaperones in Escherichia coli. BMC Microbiol. 10:251. 10.1186/1471-2180-10-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur S. D., Pulvermacher S. C., Stauffer G. V. (2006). The Yersinia pestis gcvB gene encodes two small regulatory RNA molecules. BMC Microbiol. 6:52. 10.1186/1471-2180-6-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdanovic M., Clausen T., Kaiser M., Huber R., Ehrmann M. (2011). Protein quality control in the bacterial periplasm. Annu. Rev. Microbiol. 65, 149–168. 10.1146/annurev-micro-090110-102925 [DOI] [PubMed] [Google Scholar]
- Mikula K. M., Kolodziejczyk R., Goldman A. (2013). Yersinia infection tools-characterization of structure and function of adhesins. Front. Cell. Infect. Microbiol. 2:169. 10.3389/fcimb.2012.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. F., Maier R. J. (2014). Ammonium metabolism enzymes aid Helicobacter pylori acid resistance. J. Bacteriol. 196, 3074–3081. 10.1128/JB.01423-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. K., Kwuan L., Schwiesow L., Bernick D. L., Mettert E., Ramirez H. A., et al. (2014). IscR is essential for Yersinia pseudotuberculosis type III secretion and virulence. PLoS Pathog. 10:e1004194. 10.1371/journal.ppat.1004194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minato Y., Ghosh A., Faulkner W. J., Lind E. J., Schesser Bartra S., Plano G. V., et al. (2013). Na+/H+ antiport is essential for Yersinia pestis virulence. Infect. Immun. 81, 3163–3172. 10.1128/IAI.00071-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A., Huber D., Bukau B. (2011). Integrating protein homeostasis strategies in prokaryotes. Cold Spring Harb. Perspect. Biol. 3:a004366. 10.1101/cshperspect.a004366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motin V. L., Georgescu A. M., Fitch J. P., Gu P. P., Nelson D. O., Mabery S. L., et al. (2004). Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186, 6298–6305. 10.1128/JB.186.18.6298-6305.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenkamp M., Oberhettinger P., Leo J. C., Linke D., Schutz M. S. (2015). Yersinia adhesin A (YadA)–beauty & beast. Int. J. Med. Microbiol. 305, 252–258. 10.1016/j.ijmm.2014.12.008 [DOI] [PubMed] [Google Scholar]
- Mulkidjanian A. Y., Dibrov P., Galperin M. Y. (2008). The past and present of sodium energetics: may the sodium-motive force be with you. Biochim. Biophys. Acta 1777, 985–992. 10.1016/j.bbabio.2008.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G., Lahrz A., Dersch P. (2001). Environmental control of invasin expression in Yersinia pseudotuberculosis is mediated by regulation of RovA, a transcriptional activator of the SlyA/Hor family. Mol. Microbiol. 41, 1249–1269. 10.1046/j.1365-2958.2001.02522.x [DOI] [PubMed] [Google Scholar]
- Nair M. K., De Masi L., Yue M., Galvan E. M., Chen H., Wang F., et al. (2015). Adhesive properties of YapV and paralogous autotransporter proteins of Yersinia pestis. Infect. Immun. 83, 1809–1819. 10.1128/IAI.00094-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao H., Watanabe H., Nakayama S., Takeda T. (1995). yst gene expression in Yersinia enterocolitica is positively regulated by a chromosomal region that is highly homologous to Escherichia coli host factor 1 gene (hfq). Mol. Microbiol. 18, 859–865. 10.1111/j.1365-2958.1995.18050859.x [DOI] [PubMed] [Google Scholar]
- National Research Council (2006). Treating Infectious Diseases in a Microbial World: Report of Two Workshops on Novel Antimicrobial Therapeutics. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- Nuss A. M., Heroven A. K., Waldmann B., Reinkensmeier J., Jarek M., Beckstette M., et al. (2015). Transcriptomic profiling of Yersinia pseudotuberculosis reveals reprogramming of the Crp regulon by temperature and uncovers Crp as a master regulator of small RNAs. PLoS Genet. 11:e1005087. 10.1371/journal.pgen.1005087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuss A. M., Schuster F., Kathrin Heroven A., Heine W., Pisano F., Dersch P. (2014). A direct link between the global regulator PhoP and the Csr regulon in Y. pseudotuberculosis through the small regulatory RNA CsrC. RNA Biol. 11, 580–593. 10.4161/rna.28676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obi I. R., Francis M. S. (2013). Demarcating SurA activities required for outer membrane targeting of Yersinia pseudotuberculosis adhesins. Infect. Immun. 81, 2296–2308. 10.1128/IAI.01208-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obi I. R., Nordfelth R., Francis M. S. (2011). Varying dependency of periplasmic peptidylprolyl cis-trans isomerases in promoting Yersinia pseudotuberculosis stress tolerance and pathogenicity. Biochem. J. 439, 321–332. 10.1042/BJ20110767 [DOI] [PubMed] [Google Scholar]
- Okan N. A., Bliska J. B., Karzai A. W. (2006). A Role for the SmpB-SsrA System in Yersinia pseudotuberculosis Pathogenesis. PLoS Pathog. 2:e6. 10.1371/journal.ppat.0020006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'loughlin J. L., Spinner J. L., Minnich S. A., Kobayashi S. D. (2010). Yersinia pestis two-component gene regulatory systems promote survival in human neutrophils. Infect. Immun. 78, 773–782. 10.1128/IAI.00718-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyston P. C., Dorrell N., Williams K., Li S. R., Green M., Titball R. W., et al. (2000). The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68, 3419–3425. 10.1128/IAI.68.6.3419-3425.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget M. S. (2015). Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5, 1245–1265. 10.3390/biom5031245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palace S. G., Proulx M. K., Lu S., Baker R. E., Goguen J. D. (2014). Genome-wide mutant fitness profiling identifies nutritional requirements for optimal growth of Yersinia pestis in deep tissue. MBio 5:e01385–14. 10.1128/mBio.01385-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palonen E., Lindstrom M., Somervuo P., Korkeala H. (2013). Alternative sigma factor sigmaE has an important role in stress tolerance of Yersinia pseudotuberculosis IP32953. Appl. Environ. Microbiol. 79, 5970–5977. 10.1128/AEM.01891-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N. J., Brady M. J., Leong J. M., Goguen J. D. (2009). Targeting type III secretion in Yersinia pestis. Antimicrob. Agents Chemother. 53, 385–392. 10.1128/AAC.00670-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel C. N., Wortham B. W., Lines J. L., Fetherston J. D., Perry R. D., Oliveira M. A. (2006). Polyamines are essential for the formation of plague biofilm. J. Bacteriol. 188, 2355–2363. 10.1128/JB.188.7.2355-2363.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. D., Bobrov A. G., Fetherston J. D. (2015). The role of transition metal transporters for iron, zinc, manganese, and copper in the pathogenesis of Yersinia pestis. Metallomics 7, 965–978. 10.1039/C4MT00332B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. D., Craig S. K., Abney J., Bobrov A. G., Kirillina O., Mier I., Jr., et al. (2012). Manganese transporters Yfe and MntH are Fur-regulated and important for the virulence of Yersinia pestis. Microbiology 158, 804–815. 10.1099/mic.0.053710-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. D., Fetherston J. D. (2004). Iron and heme uptake systems, in Molecular and Cellular Biology of Pathogenic Yersinia, eds Carniel E., Hinnebusch B. J. (Wymondham: Horizon Scientific Press/Caister Academic Press; ), 257–283. [Google Scholar]
- Perry R. D., Mier I., Jr., Fetherston J. D. (2007). Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. Biometals 20, 699–703. 10.1007/s10534-006-9051-x [DOI] [PubMed] [Google Scholar]
- Persson C., Nordfelth R., Holmström A., Håkansson S., Rosqvist R., Wolf-Watz H. (1995). Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol. Microbiol. 18, 135–150. 10.1111/j.1365-2958.1995.mmi_18010135.x [DOI] [PubMed] [Google Scholar]
- Petersen S., Young G. M. (2002). Essential role for cyclic AMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 70, 3665–3672. 10.1128/IAI.70.7.3665-3672.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson J., Nordfelth R., Dubinina E., Bergman T., Gustafsson M., Magnusson K. E., et al. (1996). Modulation of virulence factor expression by pathogen target cell contact. Science 273, 1231–1233. 10.1126/science.273.5279.1231 [DOI] [PubMed] [Google Scholar]
- Pisano F., Kochut A., Uliczka F., Geyer R., Stolz T., Thiermann T., et al. (2012). In vivo-induced InvA-like autotransporters Ifp and InvC of Yersinia pseudotuberculosis promote interactions with intestinal epithelial cells and contribute to virulence. Infect. Immun. 80, 1050–1064. 10.1128/IAI.05715-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plano G. V., Schesser K. (2013). The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol. Res. 57, 237–245. 10.1007/s12026-013-8454-3 [DOI] [PubMed] [Google Scholar]
- Porcheron G., Garenaux A., Proulx J., Sabri M., Dozois C. M. (2013). Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front. Cell. Infect. Microbiol. 3:90. 10.3389/fcimb.2013.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E., Lemaitre N., Merchez M., Ricard I., Reboul A., Dewitte A., et al. (2014). New insights into how Yersinia pestis adapts to its mammalian host during bubonic plague. PLoS Pathog. 10:e1004029. 10.1371/journal.ppat.1004029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. L., Raivio T. L. (2009). Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191, 1798–1815. 10.1128/JB.00798-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C., Grabenstein J. P., Perry R. D., Bliska J. B. (2005). Replication of Yersinia pestis in interferon gamma-activated macrophages requires ripA, a gene encoded in the pigmentation locus. Proc. Natl. Acad. Sci. U.S.A. 102, 12909–12914. 10.1073/pnas.0502849102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C., Klein K. A., Romanov G. A., Palmer L. E., Cirota C., Zhao Z., et al. (2009). Yersinia pestis can reside in autophagosomes and avoid xenophagy in murine macrophages by preventing vacuole acidification. Infect. Immun. 77, 2251–2261. 10.1128/IAI.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Zhang Y., Liu L., Wang L., Han Y., Yang R., et al. (2013). Cyclic AMP receptor protein is a repressor of adenylyl cyclase gene cyaA in Yersinia pestis. Can. J. Microbiol. 59, 304–310. 10.1139/cjm-2012-0705 [DOI] [PubMed] [Google Scholar]
- Quade N., Mendonca C., Herbst K., Heroven A. K., Ritter C., Heinz D. W., et al. (2012). Structural basis for intrinsic thermosensing by the master virulence regulator RovA of Yersinia. J. Biol. Chem. 287, 35796–35803. 10.1074/jbc.M112.379156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio T. L. (2014). Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim. Biophys. Acta 1843, 1529–1541. 10.1016/j.bbamcr.2013.10.018 [DOI] [PubMed] [Google Scholar]
- Rakin A., Schneider L., Podladchikova O. (2012). Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia. Front. Cell. Infect. Microbiol. 2:151. 10.3389/fcimb.2012.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S., Allentoft M. E., Nielsen K., Orlando L., Sikora M., Sjogren K. G., et al. (2015). Early Divergent Strains of Yersinia pestis in Eurasia 5,000 Years Ago. Cell 163, 571–582. 10.1016/j.cell.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravcheev D. A., Gerasimova A. V., Mironov A. A., Gelfand M. S. (2007). Comparative genomic analysis of regulation of anaerobic respiration in ten genomes from three families of gamma-proteobacteria (Enterobacteriaceae, Pasteurellaceae, Vibrionaceae). BMC Genomics 8:54. 10.1186/1471-2164-8-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeil R., Jarrett C. O., Driver J. D., Ernst R. K., Oyston P. C., Hinnebusch B. J. (2013). Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J. Bacteriol. 195, 1920–1930. 10.1128/JB.02000-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempe K. A., Hinz A. K., Vadyvaloo V. (2012). Hfq regulates biofilm gut blockage that facilitates flea-borne transmission of Yersinia pestis. J. Bacteriol. 194, 2036–2040. 10.1128/JB.06568-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. X., Yan H. Q., Zhu H., Guo X. P., Sun Y. C. (2014). HmsC, a periplasmic protein, controls biofilm formation via repression of HmsD, a diguanylate cyclase in Yersinia pestis. Environ. Microbiol. 16, 1202–1216. 10.1111/1462-2920.12323 [DOI] [PubMed] [Google Scholar]
- Rhodius V. A., Suh W. C., Nonaka G., West J., Gross C. A. (2006). Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 4:e2. 10.1371/journal.pbio.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riot B., Berche P., Simonet M. (1997). Urease is not involved in the virulence of Yersinia pseudotuberculosis in mice. Infect. Immun. 65, 1985–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. B., Telepnev M. V., Zudina I. V., Bouyer D., Montenieri J. A., Bearden S. W., et al. (2009). Evaluation of a Yersinia pestis mutant impaired in a thermoregulated type VI-like secretion system in flea, macrophage and murine models. Microb. Pathog. 47, 243–251. 10.1016/j.micpath.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche E. D., Sauer R. T. (1999). SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 18, 4579–4589. 10.1093/emboj/18.16.4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J. R., Fox J. M., Minnich S. A. (1994). Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol. Microbiol. 12, 187–199. 10.1111/j.1365-2958.1994.tb01008.x [DOI] [PubMed] [Google Scholar]
- Rohde J. R., Luan X. S., Rohde H., Fox J. M., Minnich S. A. (1999). The Yersinia enterocolitica pYV virulence plasmid contains multiple intrinsic DNA bends which melt at 37 degrees C. J. Bacteriol. 181, 4198–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Hocquet D., Miller S. I. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348. 10.1016/j.tim.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U., Galperin M. Y., Gomelsky M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. 10.1128/MMBR.00043-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosqvist R., Magnusson K. E., Wolf-Watz H. (1994). Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13, 964–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M. S., Fetherston J. D., Letoffe S., Carniel E., Perry R. D., Ghigo J. M. (2001). Identification and characterization of the hemophore-dependent heme acquisition system of Yersinia pestis. Infect. Immun. 69, 6707–6717. 10.1128/IAI.69.11.6707-6717.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M. L., Chauvaux S., Dessein R., Laurans C., Frangeul L., Lacroix C., et al. (2008). Growth of Yersinia pseudotuberculosis in human plasma: impacts on virulence and metabolic gene expression. BMC Microbiol. 8:211. 10.1186/1471-2180-8-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachelaru I., Petriman N. A., Kudva R., Koch H. G. (2014). Dynamic interaction of the sec translocon with the chaperone PpiD. J. Biol. Chem. 289, 21706–21715. 10.1074/jbc.M114.577916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvail H., Masse E. (2012). Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip. Rev. RNA 3, 26–36. 10.1002/wrna.102 [DOI] [PubMed] [Google Scholar]
- Sasikaran J., Ziemski M., Zadora P. K., Fleig A., Berg I. A. (2014). Bacterial itaconate degradation promotes pathogenicity. Nat. Chem. Biol. 10, 371–377. 10.1038/nchembio.1482 [DOI] [PubMed] [Google Scholar]
- Schiano C. A., Bellows L. E., Lathem W. W. (2010). The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect. Immun. 78, 2034–2044. 10.1128/IAI.01046-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiano C. A., Koo J. T., Schipma M. J., Caulfield A. J., Jafari N., Lathem W. W. (2014). Genome-wide analysis of small RNAs expressed by Yersinia pestis identifies a regulator of the Yop-Ysc type III secretion system. J. Bacteriol. 196, 1659–1670. 10.1128/JB.01456-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A., Neumayer W., Trulzsch K., Israel L., Imhof A., Roessle M., et al. (2009). Cross-talk between type three secretion system and metabolism in Yersinia. J. Biol. Chem. 284, 12165–12177. 10.1074/jbc.M900773200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbane F., Lemaitre N., Sturdevant D. E., Rebeil R., Virtaneva K., Porcella S. F., et al. (2006). Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. U.S.A. 103, 11766–11771. 10.1073/pnas.0601182103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J., Savitzky D. C., Ford E., Darwin A. J. (2007). Global analysis of tolerance to secretin-induced stress in Yersinia enterocolitica suggests that the phage-shock-protein system may be a remarkably self-contained stress response. Mol. Microbiol. 65, 714–727. 10.1111/j.1365-2958.2007.05821.x [DOI] [PubMed] [Google Scholar]
- Seo K. S., Kim J. W., Park J. Y., Viall A. K., Minnich S. S., Rohde H. N., et al. (2012). Role of a new intimin/invasin-like protein in Yersinia pestis virulence. Infect. Immun. 80, 3559–3569. 10.1128/IAI.00294-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R., Fetherston J. D., Kader A., Romling U., Perry R. D. (2005). Phenotypic convergence mediated by GGDEF-domain-containing proteins. J. Bacteriol. 187, 6816–6823. 10.1128/JB.187.19.6816-6823.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Xiao X., Li C., Wang T., Zhao R., Zhang W., et al. (2015). The dual transcriptional regulator RovM regulates the expression of AR3- and T6SS4-dependent acid survival systems in response to nutritional status in Yersinia pseudotuberculosis. Environ. Microbiol. 17, 4631–4645. 10.1111/1462-2920.12996 [DOI] [PubMed] [Google Scholar]
- Southern S. J., Scott A. E., Jenner D. C., Ireland P. M., Norville I. H., Sarkar-Tyson M. (2015). Survival protein A is essential for virulence in Yersinia pestis. Microb. Pathog. 92, 50–53. 10.1016/j.micpath.2015.12.013 [DOI] [PubMed] [Google Scholar]
- Spinner J. L., Winfree S., Starr T., Shannon J. G., Nair V., Steele-Mortimer O., et al. (2014). Yersinia pestis survival and replication within human neutrophil phagosomes and uptake of infected neutrophils by macrophages. J. Leukoc. Biol. 95, 389–398. 10.1189/jlb.1112551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinola S. M., Fortney K. R., Baker B., Janowicz D. M., Zwickl B., Katz B. P., et al. (2010). Activation of the CpxRA system by deletion of cpxA impairs the ability of Haemophilus ducreyi to infect humans. Infect. Immun. 78, 3898–3904. 10.1128/IAI.00432-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib L., Fuchs T. M. (2014). From food to cell: nutrient exploitation strategies of enteropathogens. Microbiology 160, 1020–1039. 10.1099/mic.0.078105-0 [DOI] [PubMed] [Google Scholar]
- Stingl K., De Reuse H. (2005). Staying alive overdosed: how does Helicobacter pylori control urease activity? Int. J. Med. Microbiol. 295, 307–315. 10.1016/j.ijmm.2005.06.006 [DOI] [PubMed] [Google Scholar]
- Sun W., Roland K. L., Branger C. G., Kuang X., Curtiss R., III. (2009a). The role of relA and spoT in Yersinia pestis KIM5 pathogenicity. PLoS ONE 4:e6720. 10.1371/journal.pone.0006720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Guo X. P., Hinnebusch B. J., Darby C. (2012). The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J. Bacteriol. 194, 2020–2026. 10.1128/JB.06243-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Hinnebusch B. J., Darby C. (2008). Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. U.S.A. 105, 8097–8101. 10.1073/pnas.0803525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Jarrett C. O., Bosio C. F., Hinnebusch B. J. (2014). Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe 15, 578–586. 10.1016/j.chom.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Koumoutsi A., Darby C. (2009b). The response regulator PhoP negatively regulates Yersinia pseudotuberculosis and Yersinia pestis biofilms. FEMS Microbiol. Lett. 290, 85–90. 10.1111/j.1574-6968.2008.01409.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Koumoutsi A., Jarrett C., Lawrence K., Gherardini F. C., Darby C., et al. (2011). Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS ONE 6:e19267. 10.1371/journal.pone.0019267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam C., Demke O., Hermanas T., Mitchell A., Hendrickx A. P., Schneewind O. (2014). YfbA, a Yersinia pestis regulator required for colonization and biofilm formation in the gut of cat fleas. J. Bacteriol. 196, 1165–1173. 10.1128/JB.01187-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz L., Bruckner S., Sareth S., Alonso A., Bogetz J., Bottini N., et al. (2005). Inhibition of Yersinia tyrosine phosphatase by furanyl salicylate compounds. J. Biol. Chem. 280, 9400–9408. 10.1074/jbc.M413122200 [DOI] [PubMed] [Google Scholar]
- Thomassin J. L., Giannakopoulou N., Zhu L., Gross J., Salmon K., Leclerc J. M., et al. (2015). The CpxRA two-component system is essential for Citrobacter rodentium virulence. Infect. Immun. 83, 1919–1928. 10.1128/IAI.00194-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H. J., Heroven A. K., Winkler L., Spreter T., Beatrix B., Dersch P. (2005). Analysis of RovA, a transcriptional regulator of Yersinia pseudotuberculosis virulence that acts through antirepression and direct transcriptional activation. J. Biol. Chem. 280, 42423–42432. 10.1074/jbc.M504464200 [DOI] [PubMed] [Google Scholar]
- Troxell B., Hassan H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. 10.3389/fcimb.2013.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. S., Winans S. C. (2011). The quorum-hindered transcription factor YenR of Yersinia enterocolitica inhibits pheromone production and promotes motility via a small non-coding RNA. Mol. Microbiol. 80, 556–571. 10.1111/j.1365-2958.2011.07595.x [DOI] [PubMed] [Google Scholar]
- Tschauner K., Hornschemeyer P., Muller V. S., Hunke S. (2014). Dynamic interaction between the CpxA sensor kinase and the periplasmic accessory protein CpxP mediates signal recognition in E. coli. PLoS ONE 9:e107383. 10.1371/journal.pone.0107383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J. H., Papenfort K., Thomsen J., Schmitz R. A., Vogel J. (2007). A conserved small RNA promotes discoordinate expession of the glmUS operon mRNA to activate GlmS synthesis. J. Mol. Biol. 373, 521–528. 10.1016/j.jmb.2007.07.035 [DOI] [PubMed] [Google Scholar]
- Urban J. H., Vogel J. (2008). Two seemingly homologous noncoding RNAs act hierarchically to activate glmS translation. PLoS Biol. 18:e64. 10.1371/journal.pbio.0060064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rensburg J. J., Fortney K. R., Chen L., Krieger A. J., Lima B. P., Wolfe A. J., et al. (2015). Development and validation of a high-throughput cell-based screen to identify activators of a bacterial two-component signal transduction system. Antimicrob. Agents Chemother. 59, 3789–3799. 10.1128/AAC.00236-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool C. K., Gottesman S. (2007). The novel transcription factor SgrR coordinates the response to glucose-phosphate stress. J. Bacteriol. 189, 2238–2248. 10.1128/JB.01689-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venecia K., Young G. M. (2005). Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect. Immun. 73, 5961–5977. 10.1128/IAI.73.9.5961-5977.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt S. L., Evans A. D., Guest R. L., Raivio T. L. (2014). The Cpx envelope stress response regulates and is regulated by small noncoding RNAs. J. Bacteriol. 196, 4229–4238. 10.1128/JB.02138-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Tils D., Bladel I., Schmidt M. A., Heusipp G. (2012). Type II secretion in Yersinia-a secretion system for pathogenicity and environmental fitness. Front. Cell. Infect. Microbiol. 2:160. 10.3389/fcimb.2012.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadler C. S., Vanderpool C. K. (2009). Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 37, 5477–5485. 10.1093/nar/gkp591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wah D. A., Levchenko I., Baker T. A., Sauer R. T. (2002). Characterization of a specificity factor for an AAA+ ATPase: assembly of SspB dimers with ssrA-tagged proteins and the ClpX hexamer. Chem. Biol. 9, 1237–1245. 10.1016/S1074-5521(02)00268-5 [DOI] [PubMed] [Google Scholar]
- Walker K. A., Maltez V. I., Hall J. D., Vitko N. P., Miller V. L. (2013). A phenotype at last: essential role for the Yersinia enterocolitica Ysa type III secretion system in a Drosophila melanogaster S2 cell model. Infect. Immun. 81, 2478–2487. 10.1128/IAI.01454-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K. A., Miller V. L. (2004). Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186, 4056–4066. 10.1128/JB.186.13.4056-4066.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K. A., Miller V. L. (2009). Synchronous gene expression of the Yersinia enterocolitica Ysa type III secretion system and its effectors. J. Bacteriol. 191, 1816–1826. 10.1128/JB.01402-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zetterstrom C. E., Gabrielsen M., Beckham K. S., Tree J. J., Macdonald S. E., et al. (2011). Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence-blocking compounds. J. Biol. Chem. 286, 29922–29931. 10.1074/jbc.M111.233858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Si M., Song Y., Zhu W., Gao F., Wang Y., et al. (2015). Type VI secretion System transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. 10.1371/journal.ppat.1005020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattiau P., Cornelis G. R. (1994). Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J. Bacteriol. 176, 3878–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilbacher T., Suzuki K., Dubey A. K., Wang X., Gudapaty S., Morozov I., et al. (2003). A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48, 657–670. 10.1046/j.1365-2958.2003.03459.x [DOI] [PubMed] [Google Scholar]
- Wick L. M., Egli T. (2004). Molecular components of physiological stress responses in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 89, 1–45. 10.1007/b93957 [DOI] [PubMed] [Google Scholar]
- Wilharm G., Heider C. (2014). Interrelationship between type three secretion system and metabolism in pathogenic bacteria. Front. Cell. Infect. Microbiol. 4:150. 10.3389/fcimb.2014.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilharm G., Lehmann V., Krauss K., Lehnert B., Richter S., Ruckdeschel K., et al. (2004). Yersinia enterocolitica type III secretion depends on the proton motive force but not on the flagellar motor components MotA and MotB. Infect. Immun. 72, 4004–4009. 10.1128/IAI.72.7.4004-4009.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willias S. P., Chauhan S., Lo C. C., Chain P. S., Motin V. L. (2015). CRP-mediated carbon catabolite regulation of Yersinia pestis biofilm formation is enhanced by the carbon storage regulator protein, CsrA. PLoS ONE 10:e0135481. 10.1371/journal.pone.0135481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H. S., Lee Y. S., Lee S. H., Lee B. J. (2009). Structural overview on the allosteric activation of cyclic AMP receptor protein. Biochim. Biophys. Acta 1794, 1299–1308. 10.1016/j.bbapap.2009.04.015 [DOI] [PubMed] [Google Scholar]
- Wortham B. W., Oliveira M. A., Fetherston J. D., Perry R. D. (2010). Polyamines are required for the expression of key Hms proteins important for Yersinia pestis biofilm formation. Environ. Microbiol. 12, 2034–2047. 10.1111/j.1462-2920.2010.02219.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren B. W. (2003). The yersiniae–a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1, 55–64. 10.1038/nrmicro730 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Darwin A. J. (2012). Recent findings about the Yersinia enterocolitica phage shock protein response. J. Microbiol. 50, 1–7. 10.1007/s12275-012-1578-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Hanawa T., Ogata S., Kamiya S. (1997). The Yersinia enterocolitica GsrA stress protein, involved in intracellular survival, is induced by macrophage phagocytosis. Infect. Immun. 65, 2190–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Su S., Meng X., Ji X., Qu Y., Liu Z., et al. (2013). Determination of sRNA expressions by RNA-seq in Yersinia pestis grown in vitro and during infection. PLoS ONE 8:e74495. 10.1371/journal.pone.0074495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Ke Y., Tan Y., Bi Y., Shi Q., Yang H., et al. (2010). Cell membrane is impaired, accompanied by enhanced type III secretion system expression in Yersinia pestis deficient in RovA regulator. PLoS ONE 5:e12840. 10.1371/journal.pone.0012840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y. T., Bhattacharya M., Stathopoulos C. (2008). Genome-wide in silico mapping of the secretome in pathogenic Yersinia pestis KIM. FEMS Microbiol. Lett. 279, 56–63. 10.1111/j.1574-6968.2007.01008.x [DOI] [PubMed] [Google Scholar]
- Yen Y. T., Karkal A., Bhattacharya M., Fernandez R. C., Stathopoulos C. (2007). Identification and characterization of autotransporter proteins of Yersinia pestis KIM. Mol. Membr. Biol. 24, 28–40. 10.1080/09687860600927626 [DOI] [PubMed] [Google Scholar]
- Yother J., Chamness T. W., Goguen J. D. (1986). Temperature-controlled plasmid regulon associated with low calcium response in Yersinia pestis. J. Bacteriol. 165, 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G. M., Miller V. L. (1997). Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol. Microbiol. 25, 319–328. 10.1046/j.1365-2958.1997.4661829.x [DOI] [PubMed] [Google Scholar]
- Zav'yalov V. P. (2012). Fimbrial polyadhesins: anti-immune armament of Yersinia. Adv. Exp. Med. Biol. 954, 183–201. 10.1007/978-1-4614-3561-7_24 [DOI] [PubMed] [Google Scholar]
- Zhan L., Han Y., Yang L., Geng J., Li Y., Gao H., et al. (2008). The cyclic AMP receptor protein, CRP, is required for both virulence and expression of the minimal CRP regulon in Yersinia pestis biovar microtus. Infect. Immun. 76, 5028–5037. 10.1128/IAI.00370-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L., Yang L., Zhou L., Li Y., Gao H., Guo Z., et al. (2009). Direct and negative regulation of the sycO-ypkA-ypoJ operon by cyclic AMP receptor protein (CRP) in Yersinia pestis. BMC Microbiol. 9:178. 10.1186/1471-2180-9-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. (2008). Positive selection, not negative selection, in the pseudogenization of rcsA in Yersinia pestis. Proc. Natl. Acad. Sci. U.S.A. 105:E69; author reply E70. 10.1073/pnas.0806419105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wang Y., Song Y., Wang T., Xu S., Peng Z., et al. (2013a). A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ. Microbiol. 15, 557–569. 10.1111/1462-2920.12005 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gao H., Wang L., Xiao X., Tan Y., Guo Z., et al. (2011). Molecular characterization of transcriptional regulation of rovA by PhoP and RovA in Yersinia pestis. PLoS ONE 6:e25484. 10.1371/journal.pone.0025484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang L., Han Y., Yan Y., Tan Y., Zhou L., et al. (2013b). Autoregulation of PhoP/PhoQ and positive regulation of the cyclic AMP receptor protein-cyclic AMP complex by PhoP in Yersinia pestis. J. Bacteriol. 195, 1022–1030. 10.1128/JB.01530-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Han Y., Qin L., Chen Z., Qiu J., Song Y., et al. (2005). Transcriptome analysis of the Mg2+-responsive PhoP regulator in Yersinia pestis. FEMS Microbiol. Lett. 250, 85–95. 10.1016/j.femsle.2005.06.053 [DOI] [PubMed] [Google Scholar]
- Zhou D., Qin L., Han Y., Qiu J., Chen Z., Li B., et al. (2006). Global analysis of iron assimilation and fur regulation in Yersinia pestis. FEMS Microbiol. Lett. 258, 9–17. 10.1111/j.1574-6968.2006.00208.x [DOI] [PubMed] [Google Scholar]
- Zhou D., Yang R. (2011). Formation and regulation of Yersinia biofilms. Protein Cell 2, 173–179. 10.1007/s13238-011-1024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]