Abstract
Previously identified single nucleotide polymorphisms (SNPs) in genome wide association studies (GWAS) of cardiovascular disease (CVD) in participants of mostly European descent were tested for association with subclinical cardiovascular disease (sCVD), coronary artery calcium score (CAC) and carotid intima media thickness (CIMT) in the Multi-Ethnic Study of Atherosclerosis (MESA). The data in this data in brief article correspond to the article Common Genetic Variants and Subclinical Atherosclerosis: The Multi-Ethnic Study of Atherosclerosis [1]. This article includes the demographic information of the participants analyzed in the article as well as graphical displays and data tables of the association of the selected SNPs with CAC and of the meta-analysis across ethnicities of the association of CIMT-c (common carotid), CIMT-I (internal carotid), CAC-d (CAC as dichotomous variable with CAC>0) and CAC-c (CAC as continuous variable, the log of the raw CAC score plus one) and CVD. The data tables corresponding to the 9p21 fine mapping experiment as well as the power calculations referenced in the article are also included.
Keywords: Single nucleotide polymorphism (SNP), Common genetic variant, Subclincal atherosclerosis, Coronary artery calcium (CAC), Carotid intima-media thickness (CIMT)
Specifications Table
| Subject area | Genetics |
| More specific subject area | Cardiovascular genetics |
| Type of data | Tables and figures |
| How data was acquired | Cardiac CT, carotid ultrasound, genotyping |
| Data format | Analyzed |
| Experimental factors | Genetic association studies controlling for CVD risk factors |
| Experimental features | The program R was used to perform genetic association studies |
| Data source location | Multi-Ethnic Study of Atherosclerosis locations across the US |
| Data accessibility | Data is within this article |
Value of the data
-
•
Genetic variations play an important role in the atherosclerotic process.
-
•
The data shows novel associations between genetic variations and atherosclerosis.
-
•
The data also shows that previously described genetic associations with atherosclerosis vary considerably depending on ethnicity.
-
•
More research is needed to further elucidate the effect of ethnic-specific genetic variation in cardiovascular disease.
1. Data
Previously identified single nucleotide polymorphisms (SNPs) in genome wide association studies (GWAS) of cardiovascular disease (CVD) in participants of mostly European descent were tested for association with subclinical cardiovascular disease (sCVD), coronary artery calcium score (CAC) and carotid intima media thickness (CIMT) in the Multi-Ethnic Study of Atherosclerosis (MESA).
2. Experimental design, materials and methods
2.1. Study design
The MESA study has been previously described and it was designed to investigate the impact of sCVD and CVD risk factors on the development of clinically overt CVD [2]. Approximately 38% of the recruited participants are Caucasians (EUA), 12% Chinese (CHN), 28% African American (AFA) and 22% Hispanic (HIS). Table 1 describes the demographic characteristics of the participants.
Table 1.
Descriptive data for MESA participants whose data was used in this study. Data are presented as n (%) for binary measures or median [IQR] for continuous measure.
| Participant characteristicsa | EUA | CHN | AFA | HIS |
|---|---|---|---|---|
| No. subjects | 2329 | 691 | 2482 | 2012 |
| Women | 1212 (52.0) | 349 (50.5) | 1394 (56.2) | 1085 (53.9) |
| Age, years | 63 [54, 71] | 63 [54, 71] | 60 [53, 68] | 60 [53, 68] |
| BMI, kg/m2 | 27.0 [24.2, 30.3] | 23.7 [21.7, 26.0] | 29.4 [26.1, 33.8] | 28.6 [25.9, 32.0] |
| Fasting glucose, mg/dL | 87 [81, 95] | 92 [85, 101] | 92 [84, 102] | 93 [85, 105] |
| Hypertension | 899 (38.6) | 262 (37.9) | 1489 (60.0) | 830 (41.3) |
| Diabetes status | 128 (5.5) | 90 (13.0) | 423 (17.0) | 369 (18.3) |
| Lipid medication | 422 (18.1) | 94 (13.6) | 459 (18.5) | 333 (16.6) |
| Current smoking | 264 (11.3) | 37 (5.4) | 480 (19.3) | 272 (13.5) |
| Lipid levelsa | ||||
| HDL cholesterol, mg/dL | 50 [41, 61] | 48 [40, 56] | 50 [42, 61] | 46 [39, 55] |
| LDL cholesterol, mg/dL | 115 [95, 136] | 114 [96, 132] | 115 [95, 137] | 118 [96, 139] |
| Total cholesterol, mg/dL | 194 [172, 216] | 191 [171, 209] | 188 [165, 212] | 195 [171, 220] |
| Triglycerides, mg/dL | 113 [77, 162] | 122 [86, 170] | 88 [65, 121] | 135 [95, 189] |
| Subclinical atherosclerosis | ||||
| Common carotid IMT, mm | 0.84 [0.73, 0.97] | 0.81 [0.70, 0.92] | 0.86 [0.76, 0.99] | 0.81 [0.71, 0.93] |
| Internal carotid IMT, mm | 0.89 [0.72, 1.39] | 0.73 [0.60, 0.94] | 0.91 [0.70, 1.30] | 0.83 [0.68, 1.19] |
| CAC prevalence | 1325 (56.9) | 356 (51.5) | 1063 (43.2) | 926 (46.3) |
| CAC Agatston scoreb | 115.7 [23.9, 372.4] | 66.4 [21.2, 194.5] | 71.7 [17.9, 267.0] | 77.3 [20.1, 285.1] |
Sample sizes are reported for participants included in genetic analysis (e.g. participants with all covariates available).
Agatston score values are reported for participants with CAC>0.
2.2. Genotype data
The 66 single nucleotide polymorphisms (SNPs) included in this study (Table 2) were obtained from Affymetrix 6.0 GWAS dataset (MESA and MESA family data) on 8224 consenting MESA participants (2329 EUA, 691 CHN, 2482 AFA, and 2012 HIS) from the National Heart, Lung, and Blood Institute SNP Health Association Resource (SHARe) project. Absent SNPs were imputed using IMPUTE v2.2.2 [3] to the 1000 genomes cosmopolitan Phase 1 v3 as a reference. Genotypes were filtered for SNP level call rate <95% and individual level call rate <95%, and monomorphic SNPs as well as SNPs with heterozygosity >53% were removed. Allele frequencies were calculated separately within each racial/ethnic group, and only those SNPs with minor allele frequencies >0.01 were included in genetic association analyses. We further filtered imputed SNPs based on imputation quality >0.5, using the observed versus expected variance quality metric, and filtered genotyped SNPs for Hardy–Weinberg equilibrium P-value≥10−5.
Table 2.
SNPs previously associated with coronary artery disease (CAD), carotid intima media thickness (CIMT) and coronary artery calcium (CAC).
| SNP | Nearest Gene (s) | MAF (risk allele) | P-value | GWAS Phenotype | References |
|---|---|---|---|---|---|
| rs11781551 | ZHX2 | 0.48 (A) | 2.4×10−11 | CIMT | [7] |
| rs445925 | APOC1 | 0.11 (G) | 1.7×10−8 | CIMT | [7] |
| rs6601530 | PINX1, SOX7 | 0.45 (G) | 1.7×10−8 | CIMT | [7] |
| rs4712972 | SLC17A4, SCLC17A1, SLC17A3 | 0.12 (A) | 7.8×10−8 | CIMT | [7] |
| rs17398575 | PIK3CG | 0.25 (T) | 2.3×10−12 | Carotid Plaque | [7] |
| rs1878406 | EDNRA | 0.3 (T) | 6.9×10−12 | Carotid Plaque and CAD | [7] |
| rs1122608 | LDLR | 0.77(G) | 9.7×10−10 | CAD | [8] |
| rs6511720 | LDLR | 0.13 (T) | 1.0×10−7 | Carotid Plaque | [7] |
| rs2246833 | LIPA | 0.33 (T) | 4.4×10−8 | CAD | [9] |
| rs1412444 | LIPA | 0.33(T) | 3.7×10−8 | CAD | [9] |
| rs11206510 | PCSK9 | 0.82 (T) | 9.10×10−8 | CAD | [8] |
| rs6725887 | WRD12 | 0.15(C) | 1.2×10−9 | CAD | [8] |
| rs12526453 | PHACTR1 | 0.67(C) | 1.5×10−9 | CAD, CAC | [8] |
| rs9349379 | PHACTR1 | 0.59 (A) | 2.65×10−11 | CAC | [10] |
| rs2026458 | PHACTR1 | 0.46(T) | 1.78×10−7 | CAC | [10] |
| rs9982601 | MRPS6, SLC5A3, KCNE2 | 0.15(T) | 4.22×10−10 | CAD | [11] |
| rs9818870 | MRAS | 0.16(T) | 7.44×10−13 | CAD, CAC | [12] |
| rs3798220 | LPA | 0.02(C) | 3.0×10−11 | CAD, CAC | [13] |
| rs10455872 | LPA | 0.30(G) | 3.4×10−15 | CAC | [10] |
| rs3184504 | SH2B3 | 0.44 (T) | 8.6×10−8 | CAD | [14] |
| rs3739998 | KIAA1462 | 0.45 (C) | 1.27×10−11 | CAD | [15] |
| rs2505083 | KIAA1462 | 0.38 (C) | 3.87×10−8 | CAD | [16] |
| rs599839 | SORT1, PSRC1, CELSR2 | 0.78 (A) | 2.89×10−10 | CAD, LIPID | [8] |
| rs646776 | SORT1, PSRC1, CELSR2 | 0.81 (T) | 7.9×10−12 | CAD, LIPID, CAC | [17] |
| rs12740374 | SORT1, PSRC1, CELSR2 | 0.3 (T) | 1.8×10−42 | CAD, LIPID | [18] |
| rs1333049 | CDKN2A, CDKN2B | 0.46 (G) | 1.35×10−22 | CAD | [19] |
| rs4977574 | CDKN2A, CDKN2B | 0.46 (G) | 1.35×10−22 | CAD, CAC | [19] |
| rs16905644 | CDKN2A, CDKN2B | 0.036 (T) | 4.1×10−5 | CAC | [20] |
| rs17465637 | MIA3 | 0.74 (C) | 1.36×10−8 | CAD | [8] |
| rs1746048 | CXCL12 | 0.87 (C) | 2.93×10−10 | CAD, CAC | [8] |
| rs501120 | CXCL12 | 0.83 (A) | 7.13×10–5 | CAD | [21] |
| rs17114036 | PPAP2B | 0.91 (A) | 3.8×10−19 | CAD | [8] |
| rs17609940 | ANKS1A | 0.75 (G) | 1.36×10−8 | CAD | [8] |
| rs12190287 | TCF21 | 0.62 (C) | 1.07×10−12 | CAD | [8] |
| rs11556924 | ZC3HC1 | 0.62 (C) | 9.18×10−18 | CAD | [8] |
| rs579459 | ABO | 0.21 (C) | 4.08×10−14 | CAD | [8] |
| rs12413409 | CNNM2, NT5C2, CYP17A1 | 0.89 (G) | 1.03×10−9 | CAD | [8] |
| rs964184 | APOA5, APOA4, APOA1 | 0.13 (G) | 1.02×10−17 | CAD | [8] |
| rs9326246 | APOA5, APOA4, APOA1 | 0.10 © | 2.90×10–2 | CAD | [21] |
| rs4773144 | COL4A1, COL4A2 | 0.44 (G) | 3.84×10−9 | CAD | [8] |
| rs9515203 | COL4A1, COL4A2 | 0.74 (T) | 1.13×10–8 | CAD | [21] |
| rs2895811 | HHIPL1 | 0.43 (C) | 1.14×10−10 | CAD | [8] |
| rs1994016 | ADAMTS7 | 0.57 (A) | 1.07×10−12 | CAD | [8] |
| rs7173743 | ADAMTS7 | 0.58 (T) | 6.74×10–13 | CAD | [21] |
| rs216172 | SMG6, SSR | 0.37 (C) | 1.15×10−9 | CAD | [8] |
| rs12936587 | RASD1, SMCR5, PEMT | 0.56 (G) | 4.45×10−10 | CAD | [8] |
| rs46522 | UBE2Z, GIP, SNF8 | 0.53 (T) | 1.8×10−8 | CAD | [8] |
| rs974819 | PDGFD | 0.32 (T) | 2.41×10−9 | CAD | [16] |
| rs10953541 | PRKAR2B, HBP1 | 0.80 (C) | 3.12×10−8 | CAD | [16] |
| rs6922269 | MTHFD1L | 0.25 (A) | 2.90×10−8 | CAD | [19] |
| rs2123536 | LINC00954, TTC32, WDR35 | 0.39 (T) | 6.83×10−11 | CAD | [22] |
| rs9268402 | C6orf10 | 0.59 (G) | 2.77×10−15 | CAD | [22] |
| rs7136259 | LINC00936, ATP2B1 | 0.39 (T) | 5.68×10−10 | CAD | [22] |
| rs4845625 | IL6R | 0.47 (T) | 3.64×10–10 | CAD | [21] |
| rs515135 | APOB | 0.83 (G) | 2.56×10–10 | CAD | [22] |
| rs2252641 | ZEB2, TEX41, BC040861 | 0.46 (G) | 5.30×10–8 | CAD | [22] |
| rs1561198 | GGCX, RNF181, TMEM150A | 0.45 (A) | 1.22×10–10 | CAD | [22] |
| rs7692387 | GUCY1A3, GUCY1B3, TDO2 | 0.81 (G) | 2.65×10–11 | CAD | [22] |
| rs273909 | SLC22A4, SLC22A5, IRF1 | 0.14 (G) | 9.62×10–10 | CAD | [22] |
| rs10947789 | KCNK5 | 0.76 (T) | 9.81×10–9 | CAD | [22] |
| rs4252120 | PLG | 0.73 (T) | 4.88×10–10 | CAD | [22] |
| rs264 | LPL | 0.86 (G) | 2.88×10–9 | CAD | [22] |
| rs9319428 | FLT1 | 0.32 (A) | 7.32×10–11 | CAD | [22] |
| rs17514846 | FURIN | 0.44 (A) | 9.33×10–11 | CAD | [22] |
| rs2954029 | TRIB1, AK22787 | 0.55 (A) | 4.75×10–9 | CAD | [22] |
2.3. SCVD measurement
The imaging outcomes in the present study are coronary artery calcium [CAC, measured as a continuous variable as the raw Agatston CAC score plus one (CAC-c) or as a dichotomous variable (CAC-d) with CAC>0] and carotid artery intima-media thickness [CIMT; internal carotid intima media thickness (CIMT-i), common carotid intima media thickness (CIMT-c)].
CAC was measured by either electron-beam tomography or multi-detector computed tomography, as described previously [4]. All scans were read at the Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center. Measurements of CAC were adjusted between the different field centers and imaging machines by using a standard calcium phantom of known density, which was scanned with each participant and CAC calculated as described previously [5] and the mean value from two scans used for analysis.
CIMT measurements were performed by B-mode ultrasonography of the right and left, near and far walls, and images were recorded using a Logiq 700 ultrasound device (General Electric Medical Systems, Waukesha, WI). Maximal CIMT-i and CIMT-c was measured as the mean of the maximum values of the near and far wall of the right and left sides at a central ultrasound reading center (Department of Radiology, New England Medical Center, Boston, MA) as described previously [6].
2.4. Statistical analyses
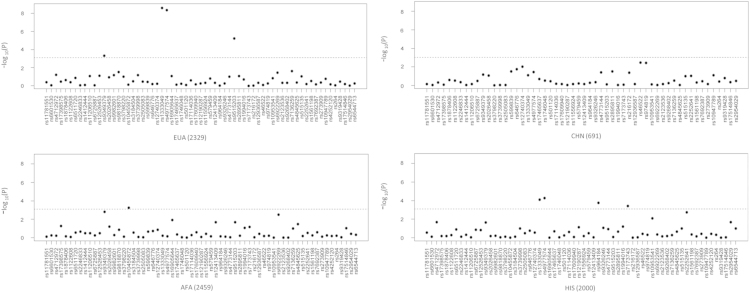
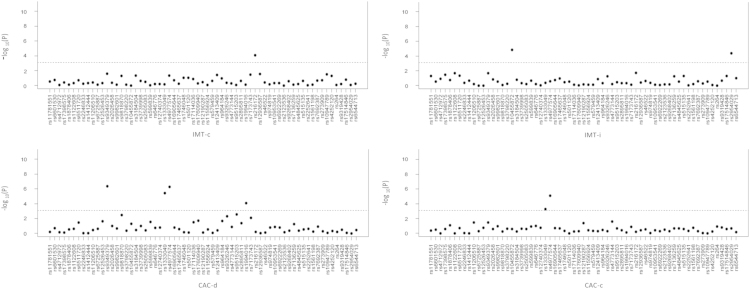
Given skewed distributions, the common (CIMT-c) and internal (CIMT-i) IMT values were log normalized. CAC was analyzed as a continuous variable by obtaining the log of the raw CAC score plus one (CAC-c) or as a dichotomous variable (CAC-d) with CAC>0. Analyses were first performed stratified within each racial/ethnic group. For analysis involving EUA and CHN, an unrelated subset of individuals was constructed by selecting at most one individual from each pedigree. For analysis of phenotypes with a substantial familial component, among AFA and HIS, the analysis was performed using a linear mixed-effects model (continuous variables) and by generalized estimating equations (dichotomous variables). Associations between each SNP and each individual phenotype was determined using separate multiple linear regressions (continuous variables) or logistic regressions (dichotomous variables) assuming an additive model. Two models were used to analyze the data. Model 1 accounted for age, sex, site of ascertainment, and principal components. Model 2 included Model 1 plus HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), triglycerides, body mass index (BMI), hypertension status (self-report of physician-diagnosed hypertension along with use of antihypertensive medication or systolic blood pressure of 140 mm Hg or greater and/or diastolic blood pressure of 90 mm Hg or greater), diabetes status (fasting blood glucose was 126 mg/dL or greater or use of diabetes medications), and current smoking use (self-reported current smoking use within the past 30 days). Fixed effect meta-analysis was used to combine results across all four race/ethnic groups, as implemented in METAL. [23] Fig. 1 shows associations of CAC-c by ethnicity. Fig. 2 shows SNP associations with sCVD in a meta-analysis across ethnicities.
Fig. 1.
Association of CAC-c (log of the raw CAC score plus one) with CVD and sCVD SNPs by ethnicity. Results from a linear regression assuming an additive model and controlling for age, gender, site of ascertainment, principal components, HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), triglycerides, BMI, hypertension status, diabetes status and tobacco. The dots represent previously identified CVD and sCVD SNPs in prior GWAS as detailed in Table 1. The y-axis represents the −log10 of the p-value and the dotted line the Bonferroni corrected significance threshold.
Fig. 2.
Meta-analysis across ethnicities of the association of CIMT-c (common carotid intima media thickness), CIMT-I (internal carotid intima media thickness), CAC-d (dichotomous variable, CAC>0) and CAC-c (log of the raw CAC score plus one) and CVD and sCVD SNPs. A linear regression assuming an additive model and controlling for age, gender, site of ascertainment, principal components, HDL cholesterol (HDL-C), LDL cholesterol (LDL-C), triglycerides, BMI, hypertension status, diabetes status and tobacco was performed in each ethnic group as described above. The program METAL was used to conduct a fixed effect meta-analysis to combine estimated effects and standard errors from stratified analyses. The dots represent previously identified CVD and sCVD SNPs in prior GWAS as detailed in Table 1. The y-axis represents the −log10 of the p-value and the dotted line the Bonferroni corrected significance threshold.
Fine mapping of the 9p21 region (100 kb upstream or downstream from SNPs rs1333049, rs4977574, and rs16905644) was performed for each ethnic group by selecting all SNPs on the chromosome 9 imputation set (NCBI Build 37) between positions 21997022–22225503. A total of 3282 SNPs were identified (598, 631, 1256 and 797 SNPs in EUA, CHN, AFA and HIS, respectively). This list of SNPs was supplemented by adding novel SNPs identified by deep sequencing efforts in this region [24], [25]. Given that each ethnicity has its own LD structure, to account for multiple comparisons in each of the race/ethnic-specific analyses, we use an eigen-decomposition to estimate the effective number of independent SNPs in each race/ethnic group [26]. Table 3 shows the association for SNPs in the 9p21 region and CAC-c in EUA and HIS. Table 4 shows the association for SNPs in the 9p21 region and sCVD across ethnicities.
Table 3.
Significant 9p21 SNP associations with CAC-c in EUA and HIS. There were no significant SNPs in AFA and CHN. SNPs were selected 100 kb upstream/downstream from SNPs rs1333049, rs4977574, and rs16905644. A total of 3282 SNPs were identified (598, 631, 1256 and 797 SNPs in EUA, CHN, AFA and HIS, respectively). The Bonferroni corrected p-value was determined by dividing 0.05 by the number of SNPs used in which ethnicity.
| SNP | Position | Beta | P-value | MAF |
|---|---|---|---|---|
| EUA | ||||
| rs3218020 | 21,997,872 | 0.342 | 2.09E−07 | 0.369 |
| rs3217992 | 22,003,223 | 0.310 | 1.58E−06 | 0.406 |
| rs1063192 | 22,003,367 | 0.283 | 8.43E−06 | 0.584 |
| rs2069418 | 22,009,698 | 0.271 | 1.80E−05 | 0.568 |
| rs2069416 | 22,010,004 | 0.282 | 2.50E−05 | 0.362 |
| rs10,811,641 | 22,014,137 | 0.324 | 5.54E−07 | 0.385 |
| rs523096 | 22,019,129 | −0.263 | 2.75E−05 | 0.423 |
| rs518394 | 22,019,673 | −0.258 | 3.90E−05 | 0.422 |
| rs568,447 | 22,021,615 | −0.273 | 2.06E−05 | 0.546 |
| rs10738604 | 22,025,493 | 0.324 | 5.75E−07 | 0.381 |
| rs613312 | 22,026,594 | −0.269 | 1.94E−05 | 0.398 |
| rs543830 | 22,026,639 | −0.269 | 1.94E−05 | 0.398 |
| rs1591136 | 22,026,834 | 0.247 | 7.82E−05 | 0.491 |
| rs599452 | 22,027,402 | −0.269 | 1.94E−05 | 0.398 |
| rs62560774 | 22,028,406 | −0.267 | 7.00E−05 | 0.301 |
| rs679038 | 22,029,080 | −0.269 | 1.92E−05 | 0.398 |
| rs10965215 | 22029445 | 0.245 | 8.36E−05 | 0.493 |
| rs564398 | 22029547 | −0.273 | 1.46E−05 | 0.399 |
| rs7865618 | 22,031,005 | 0.292 | 3.39E−06 | 0.594 |
| rs634537 | 22,032,152 | −0.275 | 1.29E−05 | 0.399 |
| rs2157719 | 22,033,366 | 0.292 | 3.45E−06 | 0.595 |
| rs1008878 | 22,036,112 | 0.293 | 3.31E−06 | 0.594 |
| rs1556515 | 22,036,367 | 0.293 | 3.30E−06 | 0.594 |
| rs1333037 | 22,040,765 | 0.290 | 4.56E−06 | 0.596 |
| rs1412830 | 22,043,612 | −0.268 | 4.38E−05 | 0.355 |
| rs1412829 | 22,043,926 | −0.278 | 1.10E−05 | 0.400 |
| rs1360589 | 22,045,317 | 0.298 | 2.61E−06 | 0.596 |
| rs7028268 | 22,048,414 | 0.333 | 2.21E−07 | 0.407 |
| rs10757265 | 22,048,859 | 0.249 | 7.69E−05 | 0.491 |
| rs944800 | 22,050,898 | 0.298 | 9.66E−06 | 0.690 |
| rs944801 | 22,051,670 | 0.299 | 2.34E−06 | 0.596 |
| rs6475604 | 22,052,734 | 0.300 | 2.26E−06 | 0.596 |
| rs7030641 | 22,054,040 | 0.299 | 2.41E−06 | 0.595 |
| rs7039467 | 22,056,213 | 0.315 | 3.49E−06 | 0.526 |
| rs7853090 | 22,056,295 | 0.324 | 1.39E−06 | 0.555 |
| rs7866783 | 22,056,359 | 0.293 | 3.63E−06 | 0.588 |
| rs10757268 | 22,059,905 | 0.275 | 6.16E−05 | 0.709 |
| rs2095144 | 22,060,136 | 0.275 | 6.20E−05 | 0.709 |
| rs2383205 | 22,060,935 | 0.269 | 2.81E−05 | 0.616 |
| rs2184061 | 22,061,562 | 0.269 | 2.83E−05 | 0.613 |
| rs1537378 | 22,061,614 | 0.268 | 2.96E−05 | 0.616 |
| rs8181050 | 22,064,391 | 0.265 | 3.42E−05 | 0.617 |
| rs10811647 | 22,065,002 | 0.317 | 7.24E−07 | 0.450 |
| rs1333039 | 22,065,657 | 0.266 | 3.09E−05 | 0.614 |
| rs4977755 | 22,066,363 | 0.270 | 2.58E−05 | 0.613 |
| rs10965223 | 22,067,004 | 0.267 | 3.25E−05 | 0.612 |
| rs10965224 | 22,067,276 | 0.264 | 3.52E−05 | 0.613 |
| rs10811648 | 22,067,542 | 0.265 | 3.39E−05 | 0.614 |
| rs10811649 | 22,067,554 | 0.265 | 3.26E−05 | 0.614 |
| rs10811650 | 22,067,593 | 0.329 | 2.26E−07 | 0.447 |
| rs10811651 | 22,067,830 | 0.264 | 3.55E−05 | 0.614 |
| rs4977756 | 22,068,652 | 0.265 | 3.30E−05 | 0.615 |
| rs4451405 | 22,071,750 | 0.280 | 1.90E−05 | 0.583 |
| rs4645630 | 22,071,751 | 0.278 | 2.21E−05 | 0.580 |
| rs10757269 | 22,072,264 | 0.381 | 1.33E−09 | 0.517 |
| rs9632884 | 22,072,301 | 0.366 | 1.05E−08 | 0.524 |
| rs9632885 | 22,072,638 | 0.376 | 2.00E−09 | 0.509 |
| rs10757270 | 22,072,719 | 0.320 | 3.68E−07 | 0.448 |
| rs1831733 | 22,076,071 | 0.388 | 1.98E−09 | 0.492 |
| rs10757271 | 22,076,795 | 0.410 | 8.83E−11 | 0.519 |
| rs10811652 | 22,077,085 | 0.412 | 6.79E−11 | 0.517 |
| rs1412832 | 22,077,543 | 0.292 | 2.11E−05 | 0.701 |
| rs10116277 | 22,081,397 | 0.406 | 9.13E−11 | 0.509 |
| rs6475606 | 22,081,850 | 0.406 | 7.71E−11 | 0.510 |
| rs1547705 | 22,082,375 | 0.576 | 1.31E−05 | 0.118 |
| rs1333040 | 22,083,404 | 0.388 | 1.12E−09 | 0.600 |
| rs1537370 | 22,084,310 | 0.408 | 8.66E−11 | 0.507 |
| rs1,970,112 | 22,085,598 | 0.395 | 3.33E−10 | 0.500 |
| rs7,857,345 | 22,087,473 | 0.311 | 1.39E−05 | 0.721 |
| rs10,738,606 | 22,088,090 | 0.363 | 4.56E−09 | 0.511 |
| rs10,738,607 | 22,088,094 | 0.363 | 4.56E−09 | 0.511 |
| rs10,757,272 | 22,088,260 | 0.360 | 6.01E−09 | 0.510 |
| rs10,757,273 | 22,090,301 | 0.402 | 1.89E−09 | 0.482 |
| rs9,644,859 | 22,090,521 | 0.453 | 3.66E−11 | 0.455 |
| rs9644860 | 22,090,603 | 0.426 | 1.06E−10 | 0.485 |
| rs9644862 | 22,090,936 | 0.436 | 1.83E−10 | 0.449 |
| rs10811653 | 22,091,069 | 0.407 | 9.51E−10 | 0.479 |
| rs7866503 | 22,091,924 | 0.397 | 4.90E−09 | 0.466 |
| rs2210538 | 22,092,257 | 0.400 | 7.12E−10 | 0.494 |
| rs141014318 | 22,092,924 | 0.361 | 8.43E−09 | 0.499 |
| rs4977757 | 22,094,330 | 0.386 | 1.17E−09 | 0.501 |
| rs10738608 | 22,094,796 | 0.403 | 1.54E−10 | 0.520 |
| rs10757274 | 22,096,055 | 0.368 | 3.18E−09 | 0.512 |
| rs4977574 | 22,098,574 | 0.366 | 3.84E−09 | 0.511 |
| rs2891168 | 22,098,619 | 0.358 | 7.58E−09 | 0.511 |
| rs1556516 | 22,100,176 | 0.392 | 3.34E−10 | 0.528 |
| rs7859727 | 22,102,165 | 0.368 | 3.15E−09 | 0.515 |
| rs1537372 | 22,103,183 | 0.329 | 2.93E−07 | 0.451 |
| rs1537373 | 22,103,341 | 0.391 | 3.68E−10 | 0.528 |
| rs1333042 | 22,103,813 | 0.387 | 5.38E−10 | 0.531 |
| rs7859362 | 22,105,927 | 0.386 | 8.58E−10 | 0.535 |
| rs10757275 | 22,106,225 | 0.367 | 5.35E−09 | 0.519 |
| rs6475609 | 22,106,271 | 0.386 | 8.57E−10 | 0.535 |
| rs1333043 | 22,106,731 | 0.386 | 8.56E−10 | 0.535 |
| rs1412834 | 22,110,131 | 0.380 | 1.60E−09 | 0.536 |
| rs7341786 | 22,112,241 | 0.380 | 1.75E−09 | 0.538 |
| rs7341791 | 22,112,427 | 0.381 | 1.63E−09 | 0.538 |
| rs10511701 | 22,112,599 | 0.355 | 1.68E−08 | 0.527 |
| rs10733376 | 22,114,469 | 0.381 | 1.75E−09 | 0.532 |
| rs10738609 | 22,114,495 | 0.358 | 1.13E−08 | 0.520 |
| rs2383206 | 22,115,026 | 0.387 | 8.47E−10 | 0.531 |
| rs944797 | 22,115,286 | 0.387 | 8.28E−10 | 0.531 |
| rs1004638 | 22,115,589 | 0.381 | 1.43E−09 | 0.536 |
| rs2383207 | 22,115,959 | 0.382 | 1.42E−09 | 0.536 |
| rs1537374 | 22,116,046 | 0.381 | 1.43E−09 | 0.536 |
| rs1537375 | 22,116,071 | 0.354 | 1.70E−08 | 0.525 |
| rs1537376 | 22,116,220 | 0.387 | 8.69E−10 | 0.531 |
| rs1333045 | 22,119,195 | 0.349 | 3.93E−08 | 0.530 |
| rs10217586 | 22,121,349 | 0.327 | 2.56E−07 | 0.549 |
| rs10738610 | 22,123,766 | 0.355 | 1.37E−08 | 0.517 |
| rs1333046 | 22,124,123 | 0.357 | 1.08E−08 | 0.517 |
| rs7857118 | 22,124,140 | 0.380 | 1.53E−09 | 0.533 |
| rs10757277 | 22,124,450 | 0.378 | 1.72E−09 | 0.498 |
| rs10811656 | 22,124,472 | 0.394 | 7.58E−10 | 0.491 |
| rs10757278 | 22,124,477 | 0.378 | 1.72E−09 | 0.498 |
| rs1333047 | 22,124,504 | 0.405 | 1.57E−10 | 0.515 |
| rs10757279 | 22,124,630 | 0.379 | 1.50E−09 | 0.498 |
| rs4977575 | 22,124,744 | 0.405 | 1.40E−10 | 0.514 |
| rs1333048 | 22,125,347 | 0.350 | 1.65E−08 | 0.519 |
| rs1333049 | 22,125,503 | 0.371 | 2.00E−09 | 0.493 |
| rs1333050 | 22,125,913 | 0.332 | 3.83E−05 | 0.670 |
| HIS | ||||
| rs10757270 | 22,072,719 | 0.278 | 5.28E−05 | 0.422 |
| rs1970112 | 22,085,598 | 0.288 | 3.24E−05 | 0.473 |
| rs9644860 | 22,090,603 | 0.297 | 5.52E−05 | 0.442 |
| rs9644862 | 22,090,936 | 0.331 | 7.94E−06 | 0.482 |
| rs10811653 | 22,091,069 | 0.303 | 4.04E−05 | 0.434 |
| rs141014318 | 22,092,924 | 0.289 | 5.09E−05 | 0.399 |
| rs10738608 | 22,094,796 | 0.289 | 4.98E−05 | 0.542 |
| rs4977574 | 22,098,574 | 0.283 | 5.24E−05 | 0.412 |
| rs2891168 | 22,098,619 | 0.280 | 5.81E−05 | 0.416 |
Table 4.
Significant SNP associations in the 9p21 in meta-analysis across ethnicities. SNPs were selected 100 kb upstream/downstream from SNPs rs1333049, rs4977574, and rs16905644. A total of 3282 SNPs were identified (598, 631, 1256 and 797 SNPs in EUA, CHN, AFA and HIS, respectively). Chr=Chromosone, Gene=Closest gene, P. Beta=published beta (ethnicity), MAF=Minor allele frequency, H. P-value=Heterogeneity p-value. I2=Heterogeneity metric. P-values meeting Bonferroni correction are highlighted (the average number of SNPs per ethnicity was used for to derive the Bonferroni corrected p-value 0.05/820=6.1E−5).
| SNP | Position | Beta | P-value | EUA | CHN | AFA | HIS | I2 | H. P-value |
|---|---|---|---|---|---|---|---|---|---|
| CAC-D | |||||||||
| rs10757269 | 22,072,264 | 0.190 | 2.38E−05 | + | + | − | + | 87.4 | 2.68E−05 |
| rs9632884 | 22,072,301 | −0.194 | 2.25E−05 | − | − | + | − | 82.3 | 7.10E−04 |
| rs9632885 | 22,072,638 | −0.186 | 7.02E−06 | − | − | + | − | 77.9 | 3.50E−03 |
| rs10757270 | 22,072,719 | 0.168 | 2.93E−05 | + | + | − | + | 74.9 | 7.59E−03 |
| rs1831733 | 22,076,071 | 0.182 | 3.05E−05 | + | + | − | + | 83.1 | 4.95E−04 |
| rs10757271 | 22,076,795 | 0.225 | 1.46E−06 | + | + | − | + | 85.1 | 1.62E−04 |
| rs10811652 | 22,077,085 | 0.232 | 4.10E−07 | + | + | − | + | 83.1 | 4.85E−04 |
| rs10116277 | 22,081,397 | −0.247 | 4.71E−08 | − | − | + | − | 76.5 | 5.20E−03 |
| rs6475606 | 22,081,850 | −0.247 | 4.13E−08 | − | − | + | − | 77.6 | 3.90E−03 |
| rs1333040 | 22,083,404 | −0.176 | 1.70E−05 | − | − | + | − | 83.2 | 4.75E−04 |
| rs1537370 | 22,084,310 | −0.226 | 3.92E−08 | − | − | − | − | 78.3 | 3.11E−03 |
| rs1970112 | 22,085,598 | 0.220 | 4.09E−08 | + | + | + | + | 75.6 | 6.46E−03 |
| rs10738606 | 22,088,090 | 0.187 | 8.17E−06 | + | + | − | + | 78.6 | 2.85E−03 |
| rs10738607 | 22,088,094 | 0.187 | 8.17E−06 | + | + | − | + | 78.6 | 2.86E−03 |
| rs10757272 | 22,088,260 | −0.190 | 6.03E−06 | − | − | + | − | 78.5 | 3.00E−03 |
| rs9644860 | 22,090,603 | −0.176 | 3.11E−05 | − | − | + | − | 87.5 | 2.42E−05 |
| rs9644862 | 22,090,936 | 0.261 | 5.58E−09 | + | + | + | + | 70.3 | 1.77E−02 |
| rs141014318 | 22,092,924 | 0.215 | 6.22E−07 | + | + | − | + | 72.9 | 1.13E−02 |
| rs4977757 | 22,094,330 | 0.220 | 5.82E−07 | + | + | + | + | 69.6 | 1.97E−02 |
| rs10738608 | 22,094,796 | 0.255 | 7.84E−09 | + | + | + | + | 64.9 | 3.62E−02 |
| rs10757274 | 22,096,055 | 0.192 | 4.26E−06 | + | + | + | + | 77.5 | 3.94E−03 |
| rs4977574 | 22,098,574 | 0.212 | 5.33E−07 | + | + | − | + | 71.2 | 1.52E−02 |
| rs2891168 | 22,098,619 | 0.211 | 4.80E−07 | + | + | + | + | 69.3 | 2.06E−02 |
| rs1537371 | 22,099,568 | −0.253 | 2.27E−08 | − | − | + | − | 68.6 | 2.28E−02 |
| rs1556516 | 22,100,176 | −0.252 | 2.43E−08 | − | − | + | − | 68.8 | 2.22E−02 |
| rs7859727 | 22,102,165 | −0.221 | 1.28E−07 | − | − | − | − | 71.5 | 1.46E−02 |
| rs1537372 | 22,103,183 | −0.208 | 3.87E−06 | − | − | + | − | 65.7 | 3.29E−02 |
| rs1537373 | 22,103,341 | 0.252 | 2.39E−08 | + | + | − | + | 68.8 | 2.21E−02 |
| rs1333042 | 22,103,813 | 0.258 | 1.11E−08 | + | + | − | + | 68.1 | 2.42E−02 |
| rs7859362 | 22,105,927 | 0.250 | 4.43E−08 | + | + | − | + | 65.8 | 3.24E−02 |
| rs10757275 | 22,106,225 | −0.197 | 3.69E−06 | − | − | + | − | 76.9 | 4.70E−03 |
| rs6475609 | 22,106,271 | 0.250 | 4.01E−08 | + | + | − | + | 65.5 | 3.37E−02 |
| rs1333043 | 22,106,731 | −0.250 | 4.10E−08 | − | − | + | − | 65.5 | 3.37E−02 |
| rs1412834 | 22,110,131 | 0.249 | 5.00E−08 | + | + | − | + | 65.1 | 3.50E−02 |
| rs7341786 | 22,112,241 | 0.250 | 5.23E−08 | + | + | − | + | 64.8 | 3.65E−02 |
| rs7341791 | 22,112,427 | 0.250 | 5.25E−08 | + | + | − | + | 64.8 | 3.63E−02 |
| rs10511701 | 22,112,599 | 0.228 | 8.75E−08 | + | + | + | + | 37.0 | 1.90E−01 |
| rs10733376 | 22,114,469 | −0.251 | 5.36E−08 | − | − | + | − | 67.4 | 2.67E−02 |
| rs10738609 | 22,114,495 | 0.201 | 2.13E−06 | + | + | − | + | 69.8 | 1.92E−02 |
| rs1004638 | 22,115,589 | 0.251 | 4.21E−08 | + | + | − | + | 65.7 | 3.28E−02 |
| rs2383207 | 22,115,959 | 0.252 | 3.72E−08 | + | + | − | + | 65.9 | 3.19E−02 |
| rs1537374 | 22,116,046 | 0.252 | 3.77E−08 | + | + | − | + | 65.8 | 3.23E−02 |
| rs1537375 | 22,116,071 | 0.240 | 8.42E−09 | + | + | + | + | 26.9 | 2.51E−01 |
| rs10738610 | 22,123,766 | 0.207 | 9.77E−07 | + | + | − | + | 69.3 | 2.07E−02 |
| rs1333046 | 22,124,123 | −0.179 | 1.14E−05 | − | − | + | − | 79.4 | 2.25E−03 |
| rs7857118 | 22,124,140 | 0.240 | 1.53E−07 | + | + | − | + | 74.8 | 7.71E−03 |
| rs10757277 | 22,124,450 | 0.216 | 3.84E−07 | + | + | + | + | 71.1 | 1.55E−02 |
| rs10757278 | 22,124,477 | 0.216 | 3.84E−07 | + | + | + | + | 71.1 | 1.55E−02 |
| rs1333047 | 22,124,504 | 0.264 | 1.12E−08 | + | + | − | + | 67.1 | 2.77E−02 |
| rs10757279 | 22,124,630 | 0.220 | 2.55E−07 | + | + | + | + | 71.8 | 1.38E−02 |
| rs4977575 | 22,124,744 | 0.265 | 9.46E−09 | + | + | − | + | 67.4 | 2.68E−02 |
| rs1333049 | 22,125,503 | −0.189 | 3.83E−06 | − | − | + | − | 81.2 | 1.17E−03 |
| CAC-C | |||||||||
| rs3218020 | 21,997,872 | −0.173 | 1.26E−05 | − | − | + | − | 82.4 | 6.77E−04 |
| rs1063192 | 22,003,367 | −0.185 | 2.47E−05 | − | − | + | − | 67.1 | 2.78E−02 |
| rs2069418 | 22,009,698 | −0.197 | 9.38E−06 | − | − | − | − | 66.6 | 2.94E−02 |
| rs10811641 | 22,014,137 | 0.160 | 4.08E−05 | + | + | − | + | 81.4 | 1.08E−03 |
| rs523096 | 22,019,129 | −0.185 | 2.87E−05 | − | − | + | − | 67.0 | 2.82E−02 |
| rs518394 | 22,019,673 | 0.182 | 3.76E−05 | + | + | − | + | 65.7 | 3.29E−02 |
| rs10738604 | 22,025,493 | −0.185 | 8.13E−06 | − | − | + | − | 81.6 | 9.87E−04 |
| rs615552 | 22,026,077 | −0.181 | 4.62E−05 | − | − | − | − | 61.2 | 5.17E−02 |
| rs613312 | 22,026,594 | 0.185 | 3.43E−05 | + | + | − | + | 72.9 | 1.14E−02 |
| rs543830 | 22,026,639 | −0.185 | 3.44E−05 | − | − | + | − | 72.9 | 1.14E−02 |
| rs599452 | 22,027,402 | 0.185 | 3.47E−05 | + | + | − | + | 72.8 | 1.15E−02 |
| rs62560774 | 22,028,406 | 0.196 | 5.60E−05 | + | + | − | + | 68.5 | 2.30E−02 |
| rs679038 | 22,029,080 | 0.183 | 4.12E−05 | + | + | − | + | 74.3 | 8.53E−03 |
| rs564398 | 22,029,547 | −0.182 | 4.53E−05 | − | − | + | − | 75.4 | 6.79E−03 |
| rs7865618 | 22,031,005 | −0.205 | 3.88E−06 | − | − | + | − | 74.2 | 8.81E−03 |
| rs634537 | 22,032,152 | −0.180 | 4.85E−05 | − | − | + | − | 76.9 | 4.61E−03 |
| rs2157719 | 22,033,366 | −0.205 | 4.30E−06 | − | − | + | − | 73.4 | 1.04E−02 |
| rs1008878 | 22,036,112 | −0.201 | 5.03E−06 | − | − | + | − | 75.8 | 6.10E−03 |
| rs1556515 | 22,036,367 | −0.201 | 5.24E−06 | − | − | + | − | 75.8 | 6.15E−03 |
| rs1333037 | 22,040,765 | −0.200 | 7.52E−06 | − | − | + | − | 73.8 | 9.52E−03 |
| rs1412829 | 22,043,926 | −0.188 | 2.69E−05 | − | − | + | − | 75.0 | 7.46E−03 |
| rs1360589 | 22,045,317 | −0.201 | 7.78E−06 | − | − | + | − | 75.2 | 7.04E−03 |
| rs7028268 | 22,048,414 | −0.169 | 2.40E−05 | − | + | + | − | 85.2 | 1.50E−04 |
| rs944800 | 22,050,898 | 0.218 | 7.26E−06 | + | + | − | + | 69.5 | 2.00E−02 |
| rs944801 | 22,051,670 | −0.200 | 8.51E−06 | − | − | + | − | 75.3 | 6.88E−03 |
| rs6475604 | 22,052,734 | 0.199 | 8.88E−06 | + | + | − | + | 75.9 | 6.04E−03 |
| rs7030641 | 22,054,040 | −0.199 | 9.32E−06 | − | − | + | − | 75.7 | 6.28E−03 |
| rs7039467 | 22,056,213 | 0.184 | 4.88E−05 | + | + | + | + | 64.8 | 3.62E−02 |
| rs7853090 | 22,056,295 | 0.214 | 2.92E−06 | + | + | − | + | 82.1 | 7.99E−04 |
| rs7866783 | 22,056,359 | 0.190 | 2.22E−05 | + | + | − | + | 78.1 | 3.33E−03 |
| rs10757268 | 22,059,905 | 0.206 | 3.17E−05 | + | + | − | + | 73.4 | 1.04E−02 |
| rs2095144 | 22,060,136 | 0.206 | 3.16E−05 | + | + | − | + | 73.3 | 1.06E−02 |
| rs2383205 | 22,060,935 | 0.184 | 5.07E−05 | + | + | − | + | 77.2 | 4.36E−03 |
| rs1537378 | 22,061,614 | 0.184 | 5.01E−05 | + | + | − | + | 76.8 | 4.78E−03 |
| rs8181050 | 22,064,391 | −0.183 | 5.23E−05 | − | − | + | − | 75.9 | 6.05E−03 |
| rs10811647 | 22,065,002 | 0.161 | 3.76E−05 | + | + | − | + | 87.9 | 1.74E−05 |
| rs10811650 | 22,067,593 | 0.169 | 1.03E−05 | + | + | − | + | 86.3 | 7.02E−05 |
| rs10757269 | 22,072,264 | 0.248 | 4.59E−10 | + | + | − | + | 86.9 | 4.22E−05 |
| rs9632884 | 22,072,301 | −0.244 | 1.81E−09 | − | − | + | − | 80.7 | 1.38E−03 |
| rs9632885 | 22,072,638 | −0.228 | 7.82E−10 | − | − | + | − | 81.8 | 9.04E−04 |
| rs10757270 | 22,072,719 | 0.206 | 1.15E−08 | + | + | − | + | 78.9 | 2.66E−03 |
| rs1831733 | 22,076,071 | 0.236 | 1.31E−09 | + | + | − | + | 84.5 | 2.32E−04 |
| rs10757271 | 22,076,795 | 0.280 | 9.53E−12 | + | + | − | + | 85.7 | 1.05E−04 |
| rs10811652 | 22,077,085 | 0.282 | 2.78E−12 | + | + | − | + | 83.0 | 5.10E−04 |
| rs1412832 | 22,077,543 | −0.204 | 1.99E−05 | − | − | + | − | 69.5 | 2.01E−02 |
| rs10116277 | 22,081,397 | −0.290 | 3.25E−13 | − | − | + | − | 76.5 | 5.16E−03 |
| rs6475606 | 22,081,850 | −0.291 | 2.29E−13 | − | − | + | − | 77.6 | 3.81E−03 |
| rs1333040 | 22,083,404 | −0.229 | 4.35E−10 | − | − | − | − | 83.6 | 3.87E−04 |
| rs1537370 | 22,084,310 | −0.259 | 1.86E−12 | − | − | − | − | 81.6 | 9.79E−04 |
| rs1970112 | 22,085,598 | 0.264 | 1.81E−13 | + | + | + | + | 75.0 | 7.40E−03 |
| rs66478960 | 22,086,826 | 0.225 | 6.00E−05 | + | ? | + | + | 0.0 | 4.33E−01 |
| rs7857345 | 22,087,473 | 0.229 | 3.16E−06 | + | + | + | + | 38.1 | 1.83E−01 |
| rs10738606 | 22,088,090 | 0.233 | 4.84E−10 | + | + | − | + | 80.4 | 1.60E−03 |
| rs10738607 | 22,088,094 | 0.233 | 4.84E−10 | + | + | − | + | 80.4 | 1.60E−03 |
| rs10757272 | 22,088,260 | −0.233 | 5.18E−10 | − | − | + | − | 80.0 | 1.82E−03 |
| rs10757273 | 22,090,301 | −0.201 | 9.63E−08 | − | − | + | − | 88.7 | 7.52E−06 |
| rs9644859 | 22,090,521 | −0.217 | 2.68E−08 | − | − | + | − | 90.5 | 6.80E−07 |
| rs9644860 | 22,090,603 | −0.221 | 5.39E−09 | − | − | + | − | 89.6 | 2.41E−06 |
| rs9644862 | 22,090,936 | 0.304 | 3.19E−14 | + | + | + | + | 70.8 | 1.65E−02 |
| rs10811653 | 22,091,069 | −0.205 | 7.62E−08 | − | − | + | − | 90.6 | 5.20E−07 |
| rs7866503 | 22,091,924 | −0.206 | 1.08E−07 | − | − | + | − | 87.9 | 1.68E−05 |
| rs2210538 | 22,092,257 | −0.211 | 1.87E−08 | − | − | + | − | 89.6 | 2.40E−06 |
| rs141014318 | 22,092,924 | 0.249 | 8.90E−11 | + | + | + | + | 73.4 | 1.04E−02 |
| rs4977757 | 22,094,330 | 0.251 | 1.44E−10 | + | + | − | + | 81.7 | 9.47E−04 |
| rs10738608 | 22,094,796 | 0.289 | 1.76E−13 | + | + | + | + | 70.8 | 1.65E−02 |
| rs4977574 | 22,098,574 | 0.252 | 2.55E−11 | + | + | + | + | 73.3 | 1.05E−02 |
| rs2891168 | 22,098,619 | 0.244 | 7.19E−11 | + | + | + | + | 74.8 | 7.69E−03 |
| rs1537371 | 22,099,568 | −0.284 | 1.01E−12 | − | − | − | − | 73.1 | 1.09E−02 |
| rs1556516 | 22,100,176 | −0.283 | 1.17E−12 | − | − | − | − | 73.5 | 1.01E−02 |
| rs7859727 | 22,102,165 | −0.245 | 4.40E−11 | − | − | − | − | 77.9 | 3.55E−03 |
| rs1537372 | 22,103,183 | −0.248 | 6.93E−10 | − | − | + | − | 65.5 | 3.37E−02 |
| rs1537373 | 22,103,341 | 0.283 | 1.22E−12 | + | + | + | + | 72.9 | 1.14E−02 |
| rs1333042 | 22,103,813 | 0.283 | 1.10E−12 | + | + | + | + | 72.7 | 1.18E−02 |
| rs7859362 | 22,105,927 | 0.276 | 6.92E−12 | + | + | + | + | 70.5 | 1.72E−02 |
| rs10757275 | 22,106,225 | −0.235 | 5.68E−10 | − | − | + | − | 78.4 | 3.08E−03 |
| rs6475609 | 22,106,271 | 0.277 | 5.91E−12 | + | + | + | + | 69.9 | 1.88E−02 |
| rs1333043 | 22,106,731 | −0.277 | 6.27E−12 | − | − | − | − | 69.9 | 1.89E−02 |
| rs1412834 | 22,110,131 | 0.274 | 1.09E−11 | + | + | + | + | 69.4 | 2.04E−02 |
| rs7341786 | 22,112,241 | 0.276 | 1.03E−11 | + | + | + | + | 68.2 | 2.39E−02 |
| rs7341791 | 22,112,427 | 0.276 | 9.76E−12 | + | + | + | + | 68.4 | 2.33E−02 |
| rs10511701 | 22,112,599 | 0.255 | 2.01E−11 | + | + | + | + | 56.9 | 7.34E−02 |
| rs10733376 | 22,114,469 | −0.279 | 7.26E−12 | − | − | − | − | 69.7 | 1.93E−02 |
| rs10738609 | 22,114,495 | 0.235 | 5.10E−10 | + | + | + | + | 72.2 | 1.30E−02 |
| rs2383206 | 22,115,026 | 0.185 | 3.13E−07 | + | + | − | + | 89.4 | 2.97E−06 |
| rs944797 | 22,115,286 | 0.186 | 2.69E−07 | + | + | − | + | 89.5 | 2.92E−06 |
| rs1004638 | 22,115,589 | 0.275 | 9.00E−12 | + | + | + | + | 69.8 | 1.91E−02 |
| rs2383207 | 22,115,959 | 0.275 | 8.92E−12 | + | + | + | + | 69.8 | 1.92E−02 |
| rs1537374 | 22,116,046 | 0.276 | 8.19E−12 | + | + | + | + | 69.8 | 1.91E−02 |
| rs1537375 | 22,116,071 | 0.266 | 8.73E−13 | + | + | + | + | 43.6 | 1.50E−01 |
| rs1537376 | 22,116,220 | 0.185 | 3.29E−07 | + | + | − | + | 89.4 | 2.96E−06 |
| rs1333045 | 22,119,195 | 0.182 | 2.89E−07 | + | + | − | + | 86.4 | 6.24E−05 |
| rs10217586 | 22,121,349 | 0.172 | 1.21E−06 | + | + | − | + | 84.9 | 1.77E−04 |
| rs10738610 | 22,123,766 | 0.235 | 4.94E−10 | + | + | + | + | 72.4 | 1.25E−02 |
| rs1333046 | 22,124,123 | −0.210 | 9.85E−09 | − | − | + | − | 82.0 | 8.27E−04 |
| rs7857118 | 22,124,140 | 0.270 | 2.30E−11 | + | + | − | + | 77.5 | 4.02E−03 |
| rs10757277 | 22,124,450 | 0.248 | 6.77E−11 | + | + | + | + | 75.4 | 6.68E−03 |
| rs10811656 | 22,124,472 | −0.198 | 6.29E−08 | − | − | + | − | 88.6 | 8.51E−06 |
| rs10757278 | 22,124,477 | 0.248 | 6.78E−11 | + | + | + | + | 75.4 | 6.67E−03 |
| rs10757277 | 22,124,450 | 0.248 | 6.77E−11 | + | + | + | + | 75.4 | 6.68E−03 |
| rs10811656 | 22,124,472 | −0.198 | 6.29E−08 | − | − | + | − | 88.6 | 8.51E−06 |
| rs10757278 | 22,124,477 | 0.248 | 6.78E−11 | + | + | + | + | 75.4 | 6.67E−03 |
| rs1333047 | 22,124,504 | 0.296 | 3.23E−13 | + | + | + | + | 70.4 | 1.76E−02 |
| rs10757279 | 22,124,630 | 0.250 | 4.65E−11 | + | + | + | + | 75.6 | 6.46E−03 |
| rs4977575 | 22,124,744 | 0.297 | 2.65E−13 | + | + | + | + | 70.5 | 1.72E−02 |
| rs1333048 | 22,125,347 | 0.186 | 2.32E−07 | + | + | − | + | 85.0 | 1.66E−04 |
| rs1333049 | 22,125,503 | −0.218 | 2.42E−09 | − | − | + | − | 84.6 | 2.18E−04 |
| rs1333050 | 22,125,913 | −0.199 | 8.65E−06 | − | − | + | − | 69.3 | 2.05E−02 |
Significance was defined by Bonferroni correction by dividing an alpha of 0.05 by the number of SNPs tested (p<7.6×10−4 given 66 SNPs tested (0.05/66) for the initial analysis, with greater number of SNPs used for the correction for the fine mapping effort). To assess genetic heterogeneity seen in stratified analyses of the four MESA race/ethnic groups, we used the I2 heterogeneity metric to quantify the proportion of total variation across studies attributable to heterogeneity rather than chance [27]. Table 5, Table 6 shows power calculations for dichotomous and quantitative traits.
Table 5.
Power to detect a genetic additive effect assuming a type I error rate of <7.6×10−4 given 66 SNPs tested (0.05/66) for a dichotomous trait with a prevalence of 50% as a function of minor allele frequency (MAF) and genetic relative risk (GRR). The prevalence of CAC in MESA varies according to age, gender and ethnicity and could be either slightly above or below 50% depending on these factors. The samples sizes used in the power calculation encompass those of the different ethnic groups in MESA (European Americans 2329, African Americans 2482, Hispanic Americans 2012 and Chinese Americans 691).
| MAF | GRR | Power (n=800) | Power (n=1700) | Power (n=2600) |
|---|---|---|---|---|
| 0.06 | 1.1 | 0.0031 | 0.0058 | 0.0093 |
| 1.2 | 0.0104 | 0.0296 | 0.0586 | |
| 1.3 | 0.0275 | 0.0943 | 0.1950 | |
| 0.11 | 1.1 | 0.0048 | 0.0109 | 0.0191 |
| 1.2 | 0.0220 | 0.0731 | 0.1514 | |
| 1.3 | 0.00675 | 0.2430 | 0.4622 | |
| 0.16 | 1.1 | 0.0066 | 0.0165 | 0.0307 |
| 1.2 | 0.0359 | 0.1267 | 0.2592 | |
| 1.3 | 0.1172 | 0.4009 | 0.6759 | |
| 0.21 | 1.1 | 0.0083 | 0.02223 | 0.0431 |
| 1.2 | 0.0508 | 0.1830 | 0.3627 | |
| 1.3 | 0.1700 | 0.5365 | 0.8105 |
Table 6.
Power to detect a genetic additive effect assuming a type I error rate of 7.6×10−4 given 66 SNPs tested (0.05/66) for a quantitative trait with a population standard deviation of 0.11 as a function of SNP effect size (beta) and minor allele frequency (MAF). The estimation of standard deviation as well as SNP effect size are based on published IMT and genetic association data. The samples sizes used in the power calculation encompass those of the different ethnic groups in MESA (European Americans 2329, African Americans 2482, Hispanic Americans 2012 and Chinese Americans 691).
| MAF | Beta | Power (n=800) | Power (n=1700) | Power (n=2600) |
|---|---|---|---|---|
| 0.06 | 0.0100 | 0.0062 | 0.0175 | 0.0351 |
| 0.0160 | 0.0236 | 0.0881 | 0.1907 | |
| 0.0220 | 0.0714 | 0.2759 | 0.5244 | |
| 0.11 | 0.0100 | 0.0129 | 0.0438 | 0.0941 |
| 0.0160 | 0.0611 | 0.2385 | 0.4672 | |
| 0.0220 | 0.1950 | 0.6135 | 0.8757 | |
| 0.16 | 0.0100 | 0.0210 | 0.0773 | 0.1678 |
| 0.0160 | 0.1090 | 0.3998 | 0.6855 | |
| 0.0220 | 0.3347 | 0.8210 | 0.9734 | |
| 0.21 | 0.0100 | 0.0297 | 0.1137 | 0.2433 |
| 0.0160 | 0.1602 | 0.5370 | 0.8191 | |
| 0.0220 | 0.4608 | 0.9190 | 0.9942 |
Sources of funding
MESA and the MESA SHARe project are conducted and supported by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079 and UL1-TR-000040 from the National Heart, Lung, and Blood Institute (NHLBI, http://www.nhlbi.nih.gov). MESA Family is conducted and supported in collaboration with MESA investigators; support is provided by grants and contracts R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258, R01HL071259, M01-RR00425, UL1RR033176, and UL1TR000124. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02‐HL‐6‐4278. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. This manuscript was approved for submission by the Presentations and Publications Committee.
Acknowledgments
The authors thank the MESA investigators, the staff, and the participants of the MESA study for their valuable contributions. A full-list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2016.01.048.
Appendix A. Supplementary material
Supplementary material
References
- 1.Vargas J.D. Common genetic variants and subclinical atherosclerosis: the multi-ethnic study of atherosclerosis (MESA) Atherosclerosis. 2015 doi: 10.1016/j.atherosclerosis.2015.11.034. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bild D.E. Multi-ethnic study of atherosclerosis: objectives and design. Am. J. Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 3.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Detrano R.C. Coronary calcium measurements: effect of CT scanner type and calcium measure on rescan reproducibility – MESA study. Radiology. 2005;236:477–484. doi: 10.1148/radiol.2362040513. [DOI] [PubMed] [Google Scholar]
- 5.Agatston A.S. Quantification of coronary artery calcium using ultrafast computed tomography. J. Am. Coll. Cardiol. 1990;15:827–832. doi: 10.1016/0735-1097(90)90282-t. [DOI] [PubMed] [Google Scholar]
- 6.O׳Leary D.H. Use of sonography to evaluate carotid atherosclerosis in the elderly. The cardiovascular health study. CHS Collaborative Research Group. Stroke. 1991;22:1155–1163. doi: 10.1161/01.str.22.9.1155. [DOI] [PubMed] [Google Scholar]
- 7.Bis J.C., Kavousi M., Franceschini N., Isaacs A., Abecasis G.R., Schminke U. Meta-analysis of genome-wide association studies from the charge consortium identifies common variants associated with carotid intima media thickness and plaque. Nat. Genet. 2011;43:940–947. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T.L., Thompson J.R. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdmann J., Grosshennig A., Braund P.S., Konig I.R., Hengstenberg C., Hall A.S. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat. Genet. 2009;41:280–282. doi: 10.1038/ng.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdmann J., Willenborg C., Nahrstaedt J., Preuss M., Konig I.R., Baumert J. Genome-wide association study identifies a new locus for coronary artery disease on chromosome 10p11.23. Eur. Heart J. 2011;32:158–168. doi: 10.1093/eurheartj/ehq405. [DOI] [PubMed] [Google Scholar]
- 11.Gudbjartsson D.F., Bjornsdottir U.S., Halapi E., Helgadottir A., Sulem P., Jonsdottir G.M. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 12.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X., Wang L., Chen S., He L., Yang X., Shi Y. Genome-wide association study in han chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V. From noncoding variant to phenotype via sort1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Donnell C.J., Kavousi M., Smith A.V., Kardia S.L., Feitosa M.F., Hwang S.J. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124:2855–2864. doi: 10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schunkert H., Konig I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tregouet D.A., Konig I.R., Erdmann J., Munteanu A., Braund P.S., Hall A.S. Genome-wide haplotype association study identifies the slc22a3-lpal2-lpa gene cluster as a risk locus for coronary artery disease. Nat Genet. 2009;41:283–285. doi: 10.1038/ng.314. [DOI] [PubMed] [Google Scholar]
- 20.Wild P.S., Zeller T., Schillert A., Szymczak S., Sinning C.R., Deiseroth A. A genome-wide association study identifies lipa as a susceptibility gene for coronary artery disease. Circ. Cardiovasc. Genet. 2011;4:403–412. doi: 10.1161/CIRCGENETICS.110.958728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wojczynski M.K., Li M., Bielak L.F., Kerr K.F., Reiner A.P., Wong N.D. Genetics of coronary artery calcification among african americans, a meta-analysis. BMC Med. Genet. 2013;14:75. doi: 10.1186/1471-2350-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peden J.F., Hopewell J.C., Saleheen D., Chambers J.C., Hager J., Soranzo N. A genome-wide association study in europeans and south asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 23.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson A.D. Resequencing and clinical associations of the 9p21.3 region: a comprehensive investigation in the Framingham heart study. Circulation. 2013;127:799–810. doi: 10.1161/CIRCULATIONAHA.112.111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shea J. Comparing strategies to fine-map the association of common SNPs at chromosome 9p21 with type 2 diabetes and myocardial infarction. Nat. Genet. 2011;43:801–805. doi: 10.1038/ng.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao X. Multiple testing corrections for imputed SNPs. Genet. Epidemiol. 2011;35:154–158. doi: 10.1002/gepi.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material