Abstract
Epithelial-derived tumor cells acquire the capacity for epithelial-to-mesenchymal transition (EMT), which enables them to invade adjacent tissues and/or metastasize to distant organs. Cancer metastasis is the main cause of cancer-related death. Molecular mechanisms involved in the switch from an epithelial phenotype to mesenchymal status are complicated and are controlled by a variety of signaling pathways. Recently, a set of noncoding RNAs (ncRNAs), including miRNAs and long noncoding RNAs (lncRNAs), were found to modulate gene expressions at either transcriptional or posttranscriptional levels. These ncRNAs are involved in EMT through their interplay with EMT-related transcription factors (EMT-TFs) and EMT-associated signaling. Reciprocal regulatory interactions between lncRNAs and miRNAs further increase the complexity of the regulation of gene expression and protein translation. In this review, we discuss recent findings regarding EMT-regulating ncRNAs and their associated signaling pathways involved in cancer progression.
1. Introduction
Epithelial-to-mesenchymal transition (EMT) is a critical step in both embryonic development and tumor metastasis. EMT is composed of serial phenotypic changes through which epithelial cells lose their apical-basal polarity and tight cellular adhesions, while acquiring protease-producing properties that increase cell motility [1]. EMT is a well-recognized process in tumor metastasis through which tumor cells seed and colonize areas distant from their primary sites. The process of EMT is sophisticatedly regulated and requires the acquisition of variable genetic alterations among tumor cells and their microenvironment [2, 3]. Important cellular components of the tumor microenvironment (TME) include tumor-infiltrating immune cells, cancer-associated fibroblasts, and endothelial cells. In addition, hypoxic conditions, which alter the composition of extracellular matrix (ECM), cytokines, chemokines, and growth factors, are critical in the development of EMT [4, 5].
Among important TME-associated cytokines are members of the transforming growth factor-β (TGF-β) family, which paradoxically suppress tumor metastasis in early-stage cancers but drive the metastatic process in advanced disease. TGF-β signaling initiates EMT by activating EMT-inducing transcription factors (EMT-TFs), such as Snail/Slug, zinc-finger E-box-binding homeobox 1/2 (ZEB1/2), basic helix-loop-helix (bHLH) protein, E47, and Twist, or by transcriptionally repressing epithelial-specific genes via members of the histone deacetylase (HDAC) family [6–10]. Epithelial-specific genes, such as E-cadherin (CDH1), zona occludens 1 (ZO-1), and occludin (OCLN), are substantially downregulated at the transcriptional level during the EMT process [11–13]. Notably, promoter regions of CDH1 and OCLN genes contain EMT-TF binding sites, termed E-boxes. CDH1 and OCLN are frequently downregulated in high-grade malignancies with poor clinical outcomes [14–17], whereas mesenchymal markers, such as N-cadherin, vimentin, fibronectin, and α-smooth muscle actin (α-SMA), are upregulated [18]. Dynamic expression of these proteins results in alterations in cytoskeleton arrangements and cellular polarity, as well as changes in the ability of cells to degrade ECM.
Recent cancer genomic studies have identified numerous RNAs that do not encode proteins. These noncoding RNAs (ncRNAs), including snRNAs, snoRNAs, rRNAs, tRNAs, piRNAs, microRNAs (miRNAs), and long noncoding RNAs (lncRNAs), regulate biological functions through interactions between their specific structural domains and DNA, RNA, or proteins [19]. Of these ncRNAs, miRNA and lncRNAs have been found to serve as important gene expression regulators that fine-tune cell transcriptomes and adjust proteomes in response to extracellular stimulation [20]. In addition, these noncoding RNAs could be transported from primary site to another cell or distant organ through extracellular vesicles and alter the gene expression profile as well as their morphology and functions within the target sites [21, 22]. Furthermore, mutations and dysregulations of miRNAs and/or lncRNAs are associated with a diverse array of human diseases, including cancer [23–27].
In this review, we discuss recent findings on the roles of miRNAs and lncRNAs in regulating EMT-TFs (Tables 1 and 2). We also discuss the multilayered regulatory circuits among miRNAs, lncRNAs, and protein-coding genes that are associated with cancer EMT (Figure 1).
Table 1.
miRNAs and other molecules involved in EMT.
| miRNA | Expression levels in cancer | Upstream regulator | Known targets | References |
|---|---|---|---|---|
| miR-200 family | Breast cancer; prostate cancer | ZEB1/2, miR-22, Slug, GATA3, TGF-β, Foxf2 | ZEB1/2, Slug, GATA3, Maml2/3, Foxf2 | [69–72] |
|
| ||||
| miR-1 | Breast cancer; prostate cancer | ZEB1/2, miR-22, Slug, GATA3, TGF-β | ZEB1/2, Slug, GATA3, Maml2/3 | [69–72] |
|
| ||||
| miR-203 | Breast cancer, pancreatic cancer | Slug, Snail, TGF-β | Bmi-1, Snail, ZEB1/2 | [73, 74] |
|
| ||||
| miR-34 family | Pancreatic stem cell, neuroblastoma | p53, epigenetic regulation | Snail, ZEB1 | [75–77] |
|
| ||||
| miR-9 | Breast cancer | c-myc | E-cadherin, LIFR | [78, 79] |
|
| ||||
| miR-135b | Colon cancer, NSCLC, HNSCC | Epigenetic regulation, NF-κB, hypoxia | APC, LATS2, β-TrCP, NDR2, MOB1B | [80–82] |
|
| ||||
| miR-210 | Breast cancer | Hypoxia | E2F3, HOXA1, FGFLR1, EFNA3, PTP1B, VMP1 | [83–85] |
|
| ||||
| miR-103/107 | CRC, breast cancer | Hypoxia | DAPK, KLF4, Dicer | [78, 86] |
|
| ||||
| miR-10b | Breast cancer | Twist | HOXD-10 | [87] |
|
| ||||
| miR-21 | NSCLC, CRC, breast cancer | TGF-β/BMP, HER2/neu, hypoxia | Pdcd4, TGFBR2, PTEN, TAp63 | [88–92] |
|
| ||||
| miR-205 | Breast cancer | ΔNp63α | ZEB1/2, Jagged1 | [69, 93, 94] |
|
| ||||
| miR-23b | Colon cancer; bladder cancer | n/a | Src, ZEB1 | [95–97] |
|
| ||||
| miR-138 | Ovarian cancer; HNSCC | n/a | SOX4, HIF-1α, vimentin | [98–100] |
|
| ||||
| miR-7 | Gastric cancer; breast cancer | WISP | IGF1R, Snail, SETDB1 | [27, 101, 102] |
HNSCC: head and neck squamous cell carcinoma; NSCLC: non-small-cell lung carcinoma; CRC: colorectal cancer; ZEB1/2: zinc-finger E-box binding homeobox 1/2; LIFR: leukemia inhibitory factor receptor alpha; APC: adenomatous polyposis coli; LATS2: large tumor-suppressor kinase 2; β-TrCP: beta-transducin repeat-containing protein; NDR2: nuclear-Dbf2-related 2; MOB1B: Mps one binder 1b; E2F3: E2F transcription factor 3; HOXA1: homeobox A1; FGFLR1: fibroblast growth factor receptor like-1; EFNA3: ephrin-A3; PTP1B: protein-tyrosine phosphatase 1B; VMP1: vacuole membrane protein 1; DAPK: death-associated protein kinase; KLF4: Krüppel-like factor 4; HOXD10: homeobox D10; Pdcd4: programmed cell death protein 4; TGFBR2: TGF beta receptor 2; PTEN: phosphatase and tensin homolog; WISP: WNT1-inducible signaling pathway protein 2; IGF1R: insulin-like growth factor 1 receptor; SETDB1: SET domain, bifurcated 1; n/a: not available.
Table 2.
lncRNAs and EMT.
| lncRNAs | Expression levels in cancer | Upstream regulator | Targets | References |
|---|---|---|---|---|
| ZEB1-AS1 | HCC | n/a | ZEB1↑ | [103] |
|
| ||||
| lncRNA-ATB | HCC | TGF-β | ZEB1/2↑, IL-11↑, miR-200↓ |
[104] |
|
| ||||
| lncRNA-HIT | Breast cancer | TGF-β | E-cadherin↓ | [105] |
|
| ||||
| MEG3 | HCC | TGFBR1↑, TGFB2↑, SMAD2↑ | [106] | |
|
| ||||
| lncRNA-Hh | Breast cancer | Twist | GAS1↑ | [107] |
|
| ||||
| lncTCF7 | Liver cancer | IL-6 | TCF↑ (Wnt signaling) | [108, 109] |
|
| ||||
| treRNA | Breast cancer | E-cadherin↓ | [110] | |
|
| ||||
| H19 | n/a | CTCF | IGF1R↓, NOMO1↓, Twist↓, TGF- β1/SMAD↓, miR-138↓, miR-200↓, Let-7↓, | [111–117] |
|
| ||||
| MALAT1 | Lung cancer, breast cancer, liver cancer, prostate cancer, renal cell carcinoma | TGF-β1, EZH2↑ | miR-205↓ | [118–120] |
|
| ||||
| Hotair | n/a | TGF-β1, miR-141 |
miR-34↓, miR-141↓, miR-7 |
[27, 121–124] |
HCC: hepatocellular carcinoma; TGFBR1: transforming growth factor beta receptor 1; TGFB2: transforming growth factor beta 2; GAS1: growth arrest-specific 1; TCF: transcription factor; CTCF: CCCTC-binding factor; IGF1R: insulin-like growth factor 1 receptor; NOMO1: NODAL modulator 1; n/a: not available.
Figure 1.
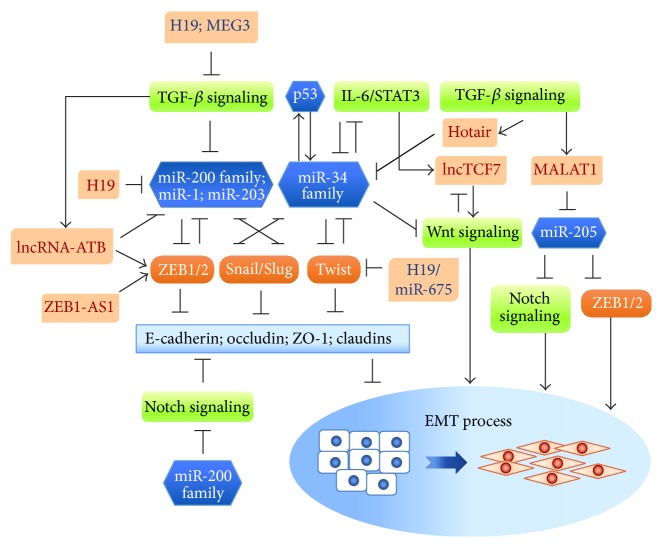
The reciprocally regulatory feedback loop between miRNAs and EMT-TFs that are involved in EMT. miRNAs form regulatory networks with EMT-TFs and EMT-associated signaling pathways that individually or cooperatively modulate EMT. EMT-suppressing miRNAs, such as the miR-200 family, miR-1, miR-203, and the miR-34 family (in blue), reciprocally suppress EMT-TFs (ZEB1/2, Snail/Slug, and Twist) and consequently downregulate the expression of epithelial markers (E-cadherin, occludin, ZO-1, and claudins). This negative feedback loop can be broken by TGF-β or IL-6/STAT3 signaling, and p53.
2. EMT-Related Signaling Pathways and the Tumor Microenvironment
The TME is a niche composed of various growth factors secreted by tumor cells or adjacent tissues, cytokines released by lymphoid cells, molecular components of the ECM, and intratumor hypoxia. The expression of EMT-TFs in cancer cells can be turned on in response to changes in the extracellular microenvironment. Signaling pathways, including those mediated by TGF-β, bone morphogenetic protein (BMP), Wnt, Notch, integrin, epidermal growth factor (EGF), fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), and sonic hedgehog (SHH), are overactivated during carcinogenesis [28, 29]. In addition, tumor hypoxia is responsible for the expression of a subset of EMT-TFs and activation of a category of EMT-related signaling pathways. Here, we discuss several signaling pathways that participate in the initiation of cancer EMT.
2.1. TGF-β Signaling Pathway
TGF-β signaling is a core pathway that tightly controls the process of cell proliferation and EMT during organ development, tissue fibrosis, and cancer progression [30]. This signaling pathway is typically initiated by ligands belonging to the TGF-β superfamily, which includes three isoforms of TGF-β (TGF-β1, TGF-β2, and TGF-β3) and six isoforms of BMP (BMP2–7). These ligands are expressed and secreted in different cellular contexts and in response to various stimuli [31]. TGF-β receptors are single-pass serine/threonine kinases that exist in different isoforms, including seven Type I (TGF-βRI) and five Type II (TGF-βRII) receptors that can form homo- or heterodimers. Combinatorial dimerization enables TGF-β receptors to differentially activate intracellular signaling pathways that are broadly distinguished by their SMAD dependence or independence. In response to phosphorylation of TGF-β receptors, a SMADs ternary complex, composed of SMAD2/3, R-SMAD and SMAD4, forms and translocates from the cytoplasm to the nucleus [32].
Several EMT-TFs, including members of the ZEB family, Snail/Slug and Twist, are transcriptionally upregulated in cancer cells by TGF-β signaling through conserved response elements on the promoters of the corresponding genes [10, 33–36]. TGF-β-SMADs was also shown to indirectly induce expression of EMT-TFs by enhancing the expression of its downstream effector, high mobility group A2 (HMGA2) [37, 38]. Activated TGF-β signaling is sustained by an autocrine loop, which in turn reinforces the EMT process [39–41]. Furthermore, Snail and SMAD3/4 form a transcriptional repressor complex, which synergistically suppresses the expression of coxsackie and adenovirus receptor (CAR), OCLN, and CDH1, and thus promotes EMT [42]. These results suggest the importance of TGF-β signaling in cancer EMT and its potential to serve as a therapeutic target.
2.2. Wnt, Notch, and MAPK Signaling Pathways
The Wnt signaling pathway is an important regulator of EMT-TF expression and the EMT process. WNT couples with the membrane protein Frizzled and low-density lipoprotein receptor (LRP), promoting translocation of β-catenin from the cytoplasm to the nucleus. In the nucleus, β-catenin acts as a coactivator of TCF/LEF1 (T cell factor/lymphoid-enhancing factor-1) and upregulates the transcription of SNAIL1/2 and TWIST, which in turn repress E-cadherin [43–45].
Notch signaling is activated by cell-cell contact. Interactions between JAG1/2 (Jagged-1/2), Notch ligand, and Notch receptors facilitate nuclear translocation of the Notch intracellular domain (NICD), which subsequently activates Notch effector genes [46]. Notch signaling not only enhances SNAIL transcription but also enhances SNAIL1/2 function through upregulation of hypoxia-inducible factor 1α (HIF-1α), thereby promoting tumor invasion and/or metastasis [47, 48].
Additional pathways are also involved in cancer EMT. For example, hepatocyte growth factors (HGFs) and insulin-like growth factor-1 (IGF-1) upregulate expression of SNAIL and ZEB1, respectively, through the mitogen-activated protein kinase (MAPK) pathway [49–51]. Collectively, these observations suggest that EMT-TF regulatory circuits are tightly controlled.
2.3. Tumor Microenvironment and Hypoxia
Hypoxic microenvironments, defined as those with a pO2 level less than 10 mmHg, trigger signaling cascades and immune responses that drive cancer progression [52, 53]. The hypoxic microenvironment contributes to the immune escape of tumors as well as tumor neovascularization and also promotes EMT [4]. Tumor hypoxia-dependent signaling is predominantly mediated by hypoxia-inducible factors (HIFs)—important protein complexes that regulate tumor progression and metastasis [52]. HIFs, which consist of an unstable α-subunit and a stable β-subunit [54], bind to promoters of target genes that contain the hypoxia response element (HRE) and promote the recruitment of transcriptional coactivators. In HIF-1α-mediated canonical hypoxia signaling, expression levels of Twist, Snail, ZEB1, and E12/E47 are upregulated [55, 56].
Studies have shown that a hypoxic TME contributes to the stabilization of HIF-1α, which functions to activate TGF-β signaling [57, 58]. TGF-β, in turn, assists in the maintenance of HIF-regulated vascular homeostasis and angiogenesis [59, 60]. The promoter region of VEGF (vascular endothelial growth factor), encoding a secretory factor involved in vasculogenesis and angiogenesis, harbors both HIF-1α and SMAD binding sites, suggesting the possibility that both hypoxia and TGF-β signaling pathways regulate VEGF expression [61]. The positive feedback loop between HIF-1α and TGF-β functions in the regulation of cancer EMT and angiogenesis [62].
The TME-associated HIF-1α-mediated hypoxia pathway also regulates cancer EMT through Notch signaling [48, 63, 64]. It has been shown that interaction between NICD and HIF-1α increases the expression of Snail and Slug, which enhance cancer invasion and migration [48, 65]. In addition, a hypoxic TME augments the nuclear translocation of β-catenin, which promotes activation of Wnt signaling [66]. Collectively, these findings demonstrate that a hypoxic TME acts as a driving force for cancer EMT, both directly, through stabilization of HIFs, and indirectly, through paracrine/autocrine stimulation.
2.4. Chemoresistance
Recent findings by in vivo mesenchymal lineage tracing showed that EMT might not be essential for tumor metastasis, and interestingly the phenotype of EMT in tumor cells was resistant to CTX (cyclophosphamide) and gemcitabine treatment [67, 68]. Despite the fact that there may be other EMT-inducing factors function to compensate for the genes that were manipulated in these studies, the discovery of EMT tumors displayed chemoresistance may provide a new insight for developing novel therapy targeting tumor metastasis.
3. miRNAs and EMT
miRNAs are a group of small (~22 nucleotides) ncRNAs that mediate destabilization and translational suppression of downstream RNAs at the posttranscriptional level. The expression of miRNAs can be ubiquitous or context-specific (e.g., during development or within certain tissues). miRNAs participate in a broad range of physiological functions. Therefore, miRNA dysregulation may break the harmony of normal genetic activity and result in a diverse array of diseases, including cancer. Recent studies have revealed that more than 50% of miRNAs are dysregulated in human cancer. Moreover, prognostic and predictive miRNA signatures have been reported in different types of cancer [125–127].
miRNAs serve both positive and negative roles in regulating cancer EMT (Figure 1 and Table 1) [128]. The regulation of miRNAs is complicated, as highlighted by the fact that some miRNA targets can, in turn, regulate the expression of miRNAs, forming regulatory loops. Two EMT-related regulatory feedback loops formed by miRNAs and EMT-TFs will be discussed here: the miR-200 family and ZEB1/2, and miR-203/miR-34 and Snail/Slug.
3.1. Reciprocal Regulation between miRNAs and EMT-TFs: The miR-200 Family and ZEB1/2
The miR-200 family consists of five members, including miR-200a/b/c, miR-429, and miR-141, all of which contain a similar seed sequence that targets a large common subset of genes [129]. The miR-200 family directly suppresses ZEB1/2 translation, consequently upregulating the expression of E-cadherin and maintaining an epithelial cellular morphology [69, 129, 130]. Conversely, ZEB1 has been shown to strongly promote tumorigenesis and cancer metastasis by inhibiting miR-200c and miR-203 transcription. These data suggest that the miR-200 family and ZEB1/2 form a negative regulatory feedback loop [73].
In pancreatic and breast cancer models, the miR-200 family is reported to suppress Notch-mediated ZEB1 activation by directly targeting the Notch coactivators MAML2 and MAML3 and the Notch ligand, JAG1 [70]. In lung cancer, miR-200 and GATA binding protein 3 (GATA3), a direct downstream target of Notch involved in lung cancer metastasis, have been shown to mutually inhibit each other. This inhibitory loop between miR-200 and GATA3 is perturbed by the Notch ligand, JAG2 [131]. These results suggest that Notch signaling broadens the spectrum of the miR-200/ZEB1 negative feedback loop in regulating cancer EMT and metastasis.
It has also been reported that miR-200 is associated with the reverse EMT process—mesenchymal-epithelial transition (MET)—in prostate cancer. miR-200 and miR-1 directly target the SLUG 3′-UTR (untranslated region), and Slug in turn inhibits miR-200/miR-1 expression [71]. In addition, prolonged TGF-β signaling increases miR-200 promoter methylation and leads to miR-200 suppression. This suggests that the induction and maintenance of a mesenchymal state require autocrine TGF-β signaling to sustain expression of EMT-TFs and inhibition of the miR-200 family [132, 133].
3.2. Reciprocal Regulation between miRNAs and EMT-TFs: miR-203/miR-34 and Snail/Slug
Similar to the ZEB/miR-200 negative feedback loop, Snail together with the miR-34 family (miR-34a, miR-34b, and miR-34c) and miR-203 constitutes another negative feedback loop. This negative feedback loop regulates epithelial plasticity [75]. Snail and miR-34a/b/c control ZNF281/ZBP-99, a Krüppel-type zinc-finger domain-containing transcription factor, acting as an integral component of an EMT-related feed-forward loop [134]. A negative feedback loop between miR-203 and Snail controls the dynamic transition between epithelial and mesenchymal phenotypes [135, 136]. miR-203 has been shown to suppress Slug expression in breast cancer cells, whereas TGF-β-mediated Slug activation reciprocally downregulates miR-203 expression [136, 137].
A feedback loop also exists between p53 and miR34. The tumor-suppressor p53 upregulates the expression of miR-34, which subsequently suppresses EMT. Mutated p53 proteins, in contrast, are unable to induce miR-34 expression, thus shifting the equilibrium toward a mesenchymal phenotype [76, 138–140]. In addition, the p53/miR-34 axis suppresses Wnt signaling, both in development and during cancer progression [138].
These results illustrate how miRNAs create networks that connect different EMT-associated signaling pathways. EMT-related signaling not only upregulates EMT-TFs but also suppresses miRNAs; this, in turn, breaks down miR-200/ZEB and/or miR-203/Snail/Slug feedback loops and facilitates cancer EMT (Figure 1).
3.3. Other EMT-Related miRNAs
Several other miRNAs are reported to be involved in EMT (Table 1). For example, miR-10b is transcriptionally upregulated by Twist and induces tumor invasion and metastasis in breast cancers by targeting homeobox D10 (HOXD10) [87]. miR-9 directly targets E-cadherin mRNA, resulting in activation of β-catenin signaling, which promotes EMT and metastasis [78]. miR-9 upregulation has also been observed in c-myc-induced mouse mammary tumors [141]. In addition, miR-9 has been shown to downregulate leukemia inhibitory factor receptor (LIFR). LIFR suppresses breast cancer metastasis by activating the Hippo kinase cascade, which in turn results in YAP (YES-associated protein) inactivation [79].
A recent study revealed that miR-9-3p negatively regulates the expression of TAZ, a YAP homolog [142]. A recent study by our laboratory also showed that miR-135b increases the levels of nuclear TAZ by directly suppressing multiple components of the Hippo pathway, including LATS2 (large tumor-suppressor kinase 2), MOB1B (Mps one binder 1b), NDR2 (nuclear-Dbf2-related 2), and β-TrCP (β-transducin repeat-containing protein) [80]. Notably, miR-135b expression level, LATS2 protein, and nuclear TAZ protein levels correlate with disease prognosis in non-small-cell lung cancer patients [80]. Furthermore, YAP was found to physically interact with p72, a RNA helicase that plays roles in miRNA processing [143]. These results may suggest how the Hippo pathway suppresses proliferation in response to contact inhibition.
3.4. miRNAs Involved in Hypoxia-Induced EMT
Some miRNAs are involved in hypoxia-induced EMT. Expression of miR-205 and miR-124, which regulate EMT by targeting ZEB1/2 and MMP2 (matrix metallopeptidase 2), respectively, is suppressed by hypoxia [69, 144, 145]. Hypoxia also downregulates the expression of miR-34a, which acts as a suppressor of Snail and ZEB1. miR-34a suppression results in upregulation of Notch1 and JAG1, and activated Notch signaling promotes EMT [146]. Forced expression of miR-34a under conditions of hypoxia not only reduces Notch1 and JAG1 expression but also abolishes Snail expression, suggesting that the interplay between hypoxia and Notch signaling is important in EMT modulation (Figure 1).
4. Long Noncoding RNAs and EMT
lncRNAs are RNA transcripts longer than 200 nucleotides that do not encode proteins [147]. The FANTOM project revealed that, in mammals, the number of lncRNAs is at least four times that of protein-coding RNAs [148, 149]. Although the functions of lncRNAs are largely unknown, accumulating evidence suggests that they are key regulators of a number of important biological processes, possibly exerting tissue-specific imprinting patterns and functioning in embryogenesis, development, lineage differentiation, tumorigenesis, and EMT regulation [150–152].
Recent studies have shown that lncRNA dysregulation is associated with cancer progression [153]. lncRNAs may interact with their nearby protein-coding genes, for example, TAL1, SNAIL, SLUG, and the master regulator of hematopoiesis, SCL/TAL1 (T Cell Acute Lymphocytic Leukemia 1), as evidenced by the fact that depletion of certain lncRNAs results in upregulation of these genes [154]. In addition, some lncRNAs encompass miRNA binding sites. These lncRNAs function as ceRNAs (competing endogenous RNA) that antagonize miRNAs. lncRNAs may positively or negatively regulate EMT-associated proteins and miRNAs, as discussed below (Figure 2 and Table 2).
Figure 2.
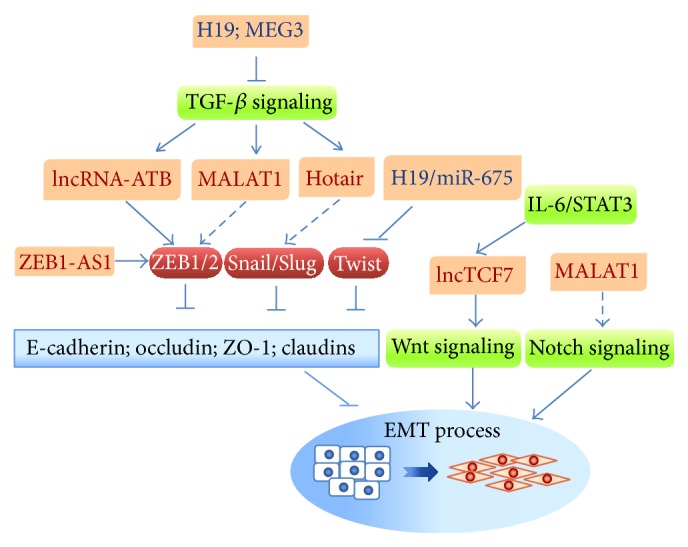
The reciprocally regulatory feedback loop between lncRNAs and EMT-TFs that are involved in EMT. lncRNAs form regulatory networks with EMT-TFs and EMT-associated signaling pathways that individually or cooperatively modulate EMT. The EMT-suppressive lncRNAs, H19 and MEG3, can downregulate TGF-β signaling. H19, lncRNA-ATB, and ZEB-AS1 promote EMT-TFs through direct or indirect regulation. In addition, H19 is reported to possess a controversial ability to downregulate Twist though its intergenic miRNA, miR-675. TGF-β and IL-6/STAT3 signaling pathways also promote activity of the lncRNAs, Hotair, lncTCF7, and MALAT1, and thus crosstalk with Notch signaling and Wnt signaling, to modulate EMT process. Dysregulation of these miRNAs and lncRNAs may lead to tumor progression.
4.1. lncRNAs Regulate EMT through EMT-TFs
Most lncRNAs act as transcription inducers or form RNA-protein complexes that promote expression of EMT-associated genes at transcriptional or posttranslational levels [105]. The lncRNA, ZEB1-AS1, was found to be frequently upregulated in hepatocellular carcinoma (HCC) [103]. ZEB1-AS1, whose transcription locus is close to ZEB1, increases ZEB1 promoter activity through an unknown mechanism. ZEB1, in turn, suppresses proteins that maintain the epithelial phenotype, such as E-cadherin, ZO-1, and occludin [103]. Thus, targeting ZEB1-AS1 may inhibit ZEB1-related EMT.
Expression of lncRNA-ATB, another lncRNA that upregulates ZEB1/2 expression and functions as a ceRNA, is enhanced by TGF-β. lncRNA-ATB promotes a metastatic cascade by competitively binding to members of the miR-200 family, thereby attenuating the inhibitory function of miR-200 on ZEB1/2. At the same time, lncRNA-ATB upregulates interleukin-11 (IL-11) and activates IL-11/STAT3 (signal transducer and activator of transcription 3) signaling, which enables cancer cell colonization [104].
lncRNA-HIT, a HOXA transcript induced by TGF-β, is a newly identified lncRNA upregulated by TGF-β that also mediates TGF-β-induced EMT [105]. It has been found that lncRNA-MEG3 associates with a PRC2 (polycomb repressive complex 2) complex through interactions with EZH2 (enhancer of zeste 2 polycomb repressive complex 2 subunit). lncRNA-MEG3 is recruited to a GA-rich sequence in target genes, forming a RNA-DNA triplex structure that regulates expression of the TGF-β receptor genes, TGFBR1 and TGFB2, as well as SMAD2 [106]. lncRNA-Hh, activated by Twist at the transcriptional level, directly targets GAS1 (growth arrest-specific 1) and activates hedgehog signaling [103]. The Twist/lncRNA-Hh signaling cascade enhances the stemness property of cells, suggesting a connection between EMT and stemness [107].
4.2. lncRNAs Regulate EMT through Their Interplay with miRNAs
EMT is a tightly controlled physiological and pathological process. Not only proteins but also lncRNAs and miRNAs are involved in fine-tuning EMT regulation. lncRNAs may serve as ceRNAs, which act as molecular “sponges” to regulate the harmony of miRNA pools and the biological signaling regulated by them. Interestingly, ceRNAs absorb target miRNAs without altering their total amount. Therefore, it is important to note that biological functions of miRNAs are simply determined not only by their measured abundance but also by their interactions with lncRNAs. lncRNAs are thus attractive therapeutic targets in miRNA-mediated diseases. Three lncRNAs—H19, MALAT1, and Hotair—will be discussed in this section.
4.2.1. H19
H19, which is highly expressed at the embryonic stage in mesodermal and endodermal tissues [155], and insulin-like growth factor II (IGF2) are reciprocally imprinted. After the early gestation period, H19 is solely expressed from the maternal-inherited allele whereas IGF2 is exclusively expressed from the paternal-inherited allele [156, 157]. Loss of IGF2 or H19 imprinting leads to IGF2 upregulation and subsequent H19 promoter hypermethylation, a phenomenon commonly found in cancers [158–160].
Studies have suggested that H19 possesses tumor-suppressor functions and have implicated chromatin insulator protein CCCTC-binding factor (CTCF) in methylating the promoter region of the H19 gene [161, 162]. How H19 functions in cells has grown clearer with the introduction miRNAs [111, 163]. miR-675 is an intergenic miRNA embedded in the first exon of H19 and coexpressed with H19. H19/miR-675 negatively regulates insulin-like growth factor 1 receptor (IGF1R), nodal modulator 1 (NOMO1), and Twist proteins and suppresses TGF-β1/SMADs signaling [111–114, 164, 165]. Since TGF-β/SMAD signaling is a well-known EMT pathway, it is possible that H19/miR-675 plays a role in EMT regulation.
On the other hand, a cancer-promoting role of H19 has been suggested in colorectal and gastric cancers [115, 166]. In colorectal cancer, overexpressed H19 serves as a ceRNA that antagonizes miR-138 and miR-200a, leading to derepression of their endogenous targets, vimentin, ZEB1, and ZEB2 [115]. In addition, H19 was found to harbor several binding sites for Let-7 family miRNAs and act as a sponge that negatively regulates their activity [167]. Notably, expression of Let-7 miRNAs, which inhibit EMT by suppressing HMGA2, is frequently downregulated in cancers with a mesenchymal phenotype. Thus, H19 may exert an EMT-promoting function through its role as a miR-200 and Let-7 family sponge [116, 117].
These results suggest that the dual roles of H19 in cancer EMT regulation are deciphered by cellular context, in which different sets of miRNAs are involved in disease pathogenesis.
4.2.2. MALAT1
MALAT1 (metastasis associated in lung adenocarcinoma transcript 1) has been reported to be a prognostic marker in several cancers, including lung, breast, pancreas, liver, colon, uterus, cervix, and prostate cancers [118]. In bladder cancer, TGF-β1 induces MALAT1 expression, whereas silencing of endogenous MALAT1 and its binding partner, SUZ12, suppresses TGF-β1-induced EMT [119]. In renal cancer, reciprocal crosstalk among MALAT1, miR-205, and EZH2 suppresses the expression of E-cadherin and enhances Wnt signaling activity, thereby promoting cancer metastasis [120]. EZH2 and SUZ12 are subunits of PRC2, which is responsible for the repressive histone 3 lysine 27 trimethylation (H3K27me3) chromatin modification. Previous studies have suggested that MALAT1 might be associated with the PRC2 complex and promote cancer EMT.
4.2.3. Hotair
Hotair (Hox transcript antisense intergenic RNA) epigenetically regulates its target sequences by recruiting PRC2, which in turn results in gene silencing [168, 169]. Hotair upregulation was found to be a prognostic indicator of poor outcome in various types of cancers [121, 122]. In addition, Hotair was shown to be required for TGF-β-mediated EMT in colon cancer [170]. Hotair epigenetically silences miR-34 transcription, resulting in augmentation of C-Met and Snail expression [123]. It was also found that miR-141, an EMT suppressor, decreases the expression of Hotair through complementary binding and thereby inhibits its oncogenic functions [124]. miR-141 negatively regulates Hotair target genes, including SNAIL, the nonreceptor tyrosine kinase ABL2, and PCDH10 (protocadherin 10). In addition, the expression levels of miR-141 and Hotair were found to be inversely correlated in renal cancer cells [124]. These results suggest that crosstalk between Hotair and miRNAs play important roles in cancer EMT regulation.
5. Conclusion
EMT is recognized as the first step of cancer metastasis—the main cause of cancer mortality. Cancer EMT is a tightly controlled pathological process. Multilayered regulatory elements, including proteins, miRNAs, and lncRNAs, are involved in the complex EMT regulatory networks through RNA-protein, RNA-miRNAs, and RNA-DNA interactions at pretranscriptional, posttranscriptional, and posttranslational levels. ncRNAs modulate epithelial plasticity by targeting different signaling pathways, EMT-TFs, and/or EMT-associated proteins. Several important reciprocal feedback loops, composed of ncRNAs and EMT-TFs, are involved in establishing flexible control over EMT and MET. Therefore, peeling back the mysteries surrounding ncRNAs in EMT regulation will be important in furthering advances in cancer therapy strategies (Figure 3).
Figure 3.
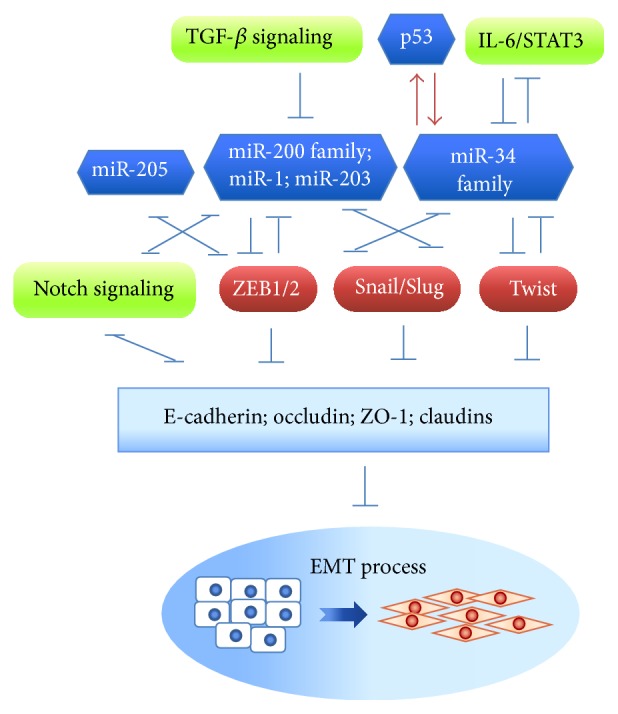
The molecular network composed of miRNAs/lncRNAs and EMT-TFs. miRNAs and lncRNAs form regulatory networks with EMT-TFs and EMT-associated signaling pathways that individually or cooperatively modulate EMT. EMT-suppressing miRNAs reciprocally suppress EMT-TFs and consequently downregulate the expression of epithelial markers. This negative feedback loop can be broken by TGF-β or IL-6/STAT3 signaling, p53, and lncRNAs (e.g., H19, lncRNA-ATB, and ZEB-AS1). Dysregulation of these miRNAs and lncRNAs may lead to tumor progression.
Acknowledgments
The authors apologize to their colleagues whose work could not be cited owing to space limitations. The work in the author's laboratory was supported by Ministry of Science and Technology, Taiwan (MOST 103-2321-B-002-022; MOST 104-2314-B-002-228-MY3), and National Taiwan University (NTU103R7601-2; NTU104R7601-2).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Thiery J. P., Acloque H., Huang R. Y. J., Nieto M. A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Nieto M. A. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342(6159) doi: 10.1126/science.1234850.1234850 [DOI] [PubMed] [Google Scholar]
- 3.De Craene B., Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nature Reviews Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 4.Philip B., Ito K., Moreno-Sánchez R., Ralph S. J. HIF expression and the role of hypoxic microenvironments within primary tumours as protective sites driving cancer stem cell renewal and metastatic progression. Carcinogenesis. 2013;34(8):1699–1707. doi: 10.1093/carcin/bgt209. [DOI] [PubMed] [Google Scholar]
- 5.Taddei M. L., Giannoni E., Comito G., Chiarugi P. Microenvironment and tumor cell plasticity: an easy way out. Cancer Letters. 2013;341(1):80–96. doi: 10.1016/j.canlet.2013.01.042. [DOI] [PubMed] [Google Scholar]
- 6.Yang J., Mani S. A., Donaher J. L., et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z.-F., Behringer R. R. Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes & Development. 1995;9(6):686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 8.Cano A., Pérez-Moreno M. A., Rodrigo I., et al. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nature Cell Biology. 2000;2(2):76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 9.Nieto M. A. The snail superfamily of zinc-finger transcription factors. Nature Reviews Molecular Cell Biology. 2002;3(3):155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 10.Peinado H., Olmeda D., Cano A. Snail, ZEB and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nature Reviews Cancer. 2007;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 11.Ikenouchi J., Matsuda M., Furuse M., Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. Journal of Cell Science. 2003;116(10):1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 12.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends in Genetics. 1993;9(9):317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 13.Puisieux A., Brabletz T., Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nature Cell Biology. 2014;16(6):488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 14.Huber G. F., Züllig L., Soltermann A., et al. Down regulation of E-Cadherin (ECAD)—a predictor for occult metastatic disease in sentinel node biopsy of early squamous cell carcinomas of the oral cavity and oropharynx. BMC Cancer. 2011;11(217):211–218. doi: 10.1186/1471-2407-11-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birchmeier W., Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochimica et Biophysica Acta (BBA)—Reviews on Cancer. 1994;1198(1):11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 16.Tsukita S., Yamazaki Y., Katsuno T., Tamura A., Tsukita S. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene. 2008;27(55):6930–6938. doi: 10.1038/onc.2008.344. [DOI] [PubMed] [Google Scholar]
- 17.Chao Y.-C., Pan S.-H., Yang S.-C., et al. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. American Journal of Respiratory and Critical Care Medicine. 2009;179(2):123–133. doi: 10.1164/rccm.200803-456OC. [DOI] [PubMed] [Google Scholar]
- 18.Kalluri R., Weinberg R. A. The basics of epithelial-mesenchymal transition. The Journal of Clinical Investigation. 2009;119(6):1420–1428. doi: 10.1172/jci39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cech T. R., Steitz J. A. The noncoding RNA revolution—trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Kapranov P., Cheng J., Dike S., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 21.Collino F., Bruno S., Incarnato D., et al. AKI recovery induced by mesenchymal stromal cell-derived extracellular vesicles carrying microRNAs. Journal of the American Society of Nephrology. 2015;26(10):2349–2360. doi: 10.1681/asn.2014070710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camussi G., Quesenberry P. J. Perspectives on the potential therapeutic uses of vesicles. Exosomes and Microvesicles. 2013;1(6) doi: 10.5772/57393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diederichs S. The four dimensions of noncoding RNA conservation. Trends in Genetics. 2014;30(4):121–123. doi: 10.1016/j.tig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Braconi C., Kogure T., Valeri N., et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ergun S., Oztuzcu S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumor Biology. 2015;36(5):3129–3136. doi: 10.1007/s13277-015-3346-x. [DOI] [PubMed] [Google Scholar]
- 26.Cui M., Xiao Z., Wang Y., et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Research. 2015;75(5):846–857. doi: 10.1158/0008-5472.can-14-1192. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H., Cai K., Wang J., et al. MiR-7, inhibited indirectly by LincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. STEM CELLS. 2014;32(11):2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez D. M., Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Science Signaling. 2014;7(344, article re8) doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Timmerman L. A., Grego-Bessa J., Raya A., et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes & Development. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tirado-Rodriguez B., Ortega E., Segura-Medina P., Huerta-Yepez S. TGF-β: an important mediator of allergic disease and a molecule with dual activity in cancer development. Journal of Immunology Research. 2014;2014:15. doi: 10.1155/2014/318481.318481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akhurst R. J., Derynck R. TGF-β signaling in cancer—a double-edged sword. Trends in Cell Biology. 2001;11(11):S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 32.Derynck R., Zhang Y., Feng X.-H. Smads: transcriptional activators of TGF-β responses. Cell. 1998;95(6):737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 33.Peinado H., Quintanilla M., Cano A. Transforming growth factor β-1 induces Snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. The Journal of Biological Chemistry. 2003;278(23):21113–21123. doi: 10.1074/jbc.m211304200. [DOI] [PubMed] [Google Scholar]
- 34.Slabáková E., Pernicová Z., Slavíčková E., Staršíchová A., Kozubík A., Souček K. TGF-β1-induced EMT of non-transformed prostate hyperplasia cells is characterized by early induction of SNAI2/Slug. Prostate. 2011;71(12):1332–1343. doi: 10.1002/pros.21350. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y.-Y., Peck K., Chang Y.-L., et al. SCUBE3 is an endogenous TGF-β receptor ligand and regulates the epithelial-mesenchymal transition in lung cancer. Oncogene. 2011;30(34):3682–3693. doi: 10.1038/onc.2011.85. [DOI] [PubMed] [Google Scholar]
- 36.Zavadil J., Böttinger E. P. TGF-β and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 37.Thuault S., Valcourt U., Petersen M., Manfioletti G., Heldin C.-H., Moustakas A. Transforming growth factor-β employs HMGA2 to elicit epithelial-mesenchymal transition. Journal of Cell Biology. 2006;174(2):175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Zhang H., Liu L., et al. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. Journal of Cancer Research and Clinical Oncology. 2013;139(6):1033–1042. doi: 10.1007/s00432-012-1363-3. [DOI] [PubMed] [Google Scholar]
- 39.Wu J., Ru N.-Y., Zhang Y., et al. HAb18G/CD147 promotes epithelial-mesenchymal transition through TGF-β signaling and is transcriptionally regulated by Slug. Oncogene. 2011;30(43):4410–4427. doi: 10.1038/onc.2011.149. [DOI] [PubMed] [Google Scholar]
- 40.Dhasarathy A., Phadke D., Mav D., Shah R. R., Wade P. A. The transcription factors snail and slug activate the transforming growth factor-beta signaling pathway in breast cancer. PLoS ONE. 2011;6(10) doi: 10.1371/journal.pone.0026514.e26514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., Tian X.-J., Zhang H., et al. TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Science Signaling. 2014;7(345, article ra91) doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- 42.Vincent T., Neve E. P. A., Johnson J. R., et al. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-β mediated epithelial-mesenchymal transition. Nature Cell Biology. 2009;11(8):943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conacci-Sorrell M., Simcha I., Ben-Yedidia T., Blechman J., Savagner P., Ben-Ze'Ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of β-catenin signaling, Slug, and MAPK. The Journal of Cell Biology. 2003;163(4):847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yook J. I., Li X.-Y., Ota I., et al. A Wnt-Axin2-GSK3β cascade regulates Snail1 activity in breast cancer cells. Nature Cell Biology. 2006;8(12):1398–1406. doi: 10.1038/ncb1508. [DOI] [PubMed] [Google Scholar]
- 45.Howe L. R., Watanabe O., Leonard J., Brown A. M. C. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Research. 2003;63(8):1906–1913. [PubMed] [Google Scholar]
- 46.Kopan R. Notch: a membrane-bound transcription factor. Journal of Cell Science. 2002;115(6):1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- 47.Leong K. G., Niessen K., Kulic I., et al. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. The Journal of Experimental Medicine. 2007;204(12):2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahlgren C., Gustafsson M. V., Jin S., Poellinger L., Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagai T., Arao T., Furuta K., et al. Sorafenib inhibits the hepatocyte growth factor-mediated epithelial mesenchymal transition in hepatocellular carcinoma. Molecular Cancer Therapeutics. 2011;10(1):169–177. doi: 10.1158/1535-7163.mct-10-0544. [DOI] [PubMed] [Google Scholar]
- 50.Graham T. R., Zhau H. E., Odero-Marah V. A., et al. Insulin-like growth factor-I-dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Research. 2008;68(7):2479–2488. doi: 10.1158/0008-5472.can-07-2559. [DOI] [PubMed] [Google Scholar]
- 51.Grotegut S., von Schweinitz D., Christofori G., Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. The EMBO Journal. 2006;25(15):3534–3545. doi: 10.1038/sj.emboj.7601213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai Y.-P., Wu K.-J. Hypoxia-regulated target genes implicated in tumor metastasis. Journal of Biomedical Science. 2012;19, article 102 doi: 10.1186/1423-0127-19-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilkes D. M., Semenza G. L., Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nature Reviews Cancer. 2014;14(6):430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaelin W. G., Jr., Ratcliffe P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Molecular Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Yang M.-H., Wu M.-Z., Chiou S.-H., et al. Direct regulation of TWIST by HIF-1α promotes metastasis. Nature Cell Biology. 2008;10(3):295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 56.Lundgren K., Nordenskjöld B., Landberg G. Hypoxia, Snail and incomplete epithelial-mesenchymal transition in breast cancer. British Journal of Cancer. 2009;101(10):1769–1781. doi: 10.1038/sj.bjc.6605369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orphanides C., Fine L. G., Norman J. T. Hypoxia stimulates proximal tubular cell matrix production via a TGF-β1-independent mechanism. Kidney International. 1997;52(3):637–647. doi: 10.1038/ki.1997.377. [DOI] [PubMed] [Google Scholar]
- 58.Toomey D., Condron C., Di Wu Q., et al. TGF-β1 is elevated in breast cancer tissue and regulates nitric oxide production from a number of cellular sources during hypoxia re-oxygenation injury. British Journal of Biomedical Science. 2001;58(3):177–183. [PubMed] [Google Scholar]
- 59.Harada H., Itasaka S., Zhu Y., et al. Treatment regimen determines whether an HIF-1 inhibitor enhances or inhibits the effect of radiation therapy. British Journal of Cancer. 2009;100(5):747–757. doi: 10.1038/sj.bjc.6604939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furuta C., Miyamoto T., Takagi T., et al. Transforming growth factor-beta signaling enhancement by long-term exposure to hypoxia in a tumor microenvironment composed of Lewis lung carcinoma cells. Cancer Science. 2015;106(11):1524–1533. doi: 10.1111/cas.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sánchez-Elsner T., Botella L. M., Velasco B., Corbí A., Attisano L., Bernabéu C. Synergistic cooperation between hypoxia and transforming growth factor-β pathways on human vascular endothelial growth factor gene expression. The Journal of Biological Chemistry. 2001;276(42):38527–38535. doi: 10.1074/jbc.m104536200. [DOI] [PubMed] [Google Scholar]
- 62.Lu X., Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clinical Cancer Research. 2010;16(24):5928–5935. doi: 10.1158/1078-0432.ccr-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y., De Marco M. A., Graziani I., et al. Oxygen concentration determines the biological effects of NOTCH-1 signaling in adenocarcinoma of the lung. Cancer Research. 2007;67(17):7954–7959. doi: 10.1158/0008-5472.CAN-07-1229. [DOI] [PubMed] [Google Scholar]
- 64.Zheng X., Linke S., Dias J. M., et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(9):3368–3373. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J., Imanaka N., Chen J., Griffin J. D. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. British Journal of Cancer. 2010;102(2):351–360. doi: 10.1038/sj.bjc.6605486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cannito S., Novo E., Compagnone A., et al. Redox mechanisms switch on hypoxia-dependent epithelial-mesenchymal transition in cancer cells. Carcinogenesis. 2008;29(12):2267–2278. doi: 10.1093/carcin/bgn216. [DOI] [PubMed] [Google Scholar]
- 67.Zheng X., Carstens J. L., Kim J., et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fischer K. R., Durrans A., Lee S., et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gregory P. A., Bert A. G., Paterson E. L., et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 70.Brabletz S., Bajdak K., Meidhof S., et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. The EMBO Journal. 2011;30(4):770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y.-N., Yin J. J., Abou-Kheir W., et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32(3):296–306. doi: 10.1038/onc.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song S. J., Poliseno L., Song M. S., et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154(2):311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wellner U., Schubert J., Burk U. C., et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nature Cell Biology. 2009;11(12):1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 74.Qu Y., Li W.-C., Hellem M. R., et al. MiR-182 and miR-203 induce mesenchymal to epithelial transition and self-sufficiency of growth signals via repressing SNAI2 in prostate cells. International Journal of Cancer. 2013;133(3):544–555. doi: 10.1002/ijc.28056. [DOI] [PubMed] [Google Scholar]
- 75.Kim N. H., Kim H. S., Li X.-Y., et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. The Journal of Cell Biology. 2011;195(3):417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He L., He X., Lim L. P., et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nalls D., Tang S.-N., Rodova M., Srivastava R. K., Shankar S. Targeting epigenetic regulation of mir-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE. 2011;6(8) doi: 10.1371/journal.pone.0024099.e24099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martello G., Rosato A., Ferrari F., et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 79.Chen D., Sun Y., Wei Y., et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nature Medicine. 2012;18(10):1511–1517. doi: 10.1038/nm.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin C.-W., Chang Y.-L., Chang Y.-C., et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nature Communications. 2013;4, article 1877 doi: 10.1038/ncomms2876. [DOI] [PubMed] [Google Scholar]
- 81.Zhang L., Sun Z.-J., Bian Y., Kulkarni A. B. MicroRNA-135b acts as a tumor promoter by targeting the hypoxia-inducible factor pathway in genetically defined mouse model of head and neck squamous cell carcinoma. Cancer Letters. 2013;331(2):230–238. doi: 10.1016/j.canlet.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nagel R., Le Sage C., Diosdado B., et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Research. 2008;68(14):5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 83.Huang X., Ding L., Bennewith K. L., et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Molecular Cell. 2009;35(6):856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan S. Y., Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9(6):1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Volinia S., Galasso M., Sana M. E., et al. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(8):3024–3029. doi: 10.1073/pnas.1200010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen H.-Y., Lin Y.-M., Chung H.-C., et al. MiR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Research. 2012;72(14):3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 87.Ma L., Teruya-Feldstein J., Weinberg R. A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 88.Toiyama Y., Takahashi M., Hur K., et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. Journal of the National Cancer Institute. 2013;105(12):849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Y., Li J., Tong L., et al. The prognostic value of miR-21 and miR-155 in non-small-cell lung cancer: a meta-analysis. Japanese Journal of Clinical Oncology. 2013;43(8):813–820. doi: 10.1093/jjco/hyt084. [DOI] [PubMed] [Google Scholar]
- 90.Davis B. N., Hilyard A. C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bornachea O., Santos M., Martínez-Cruz A. B., et al. EMT and induction of miR-21 mediate metastasis development in Trp53-deficient tumours. Scientific Reports. 2012;2, article 434 doi: 10.1038/srep00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang T.-H., Wu F., Loeb G. B., et al. Up-regulation of miR-21 by HER2/neu signaling promotes cell invasion. The Journal of Biological Chemistry. 2009;284(27):18515–18524. doi: 10.1074/jbc.m109.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dang T. T., Esparza M. A., Maine E. A., Westcott J. M., Pearson G. W. DeltaNp63alpha promotes breast cancer cell motility through the selective activation of components of the epithelial-to-mesenchymal transition program. Cancer Research. 2015;75:3925–3935. doi: 10.1158/0008-5472.CAN-14-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chao C.-H., Chang C.-C., Wu M.-J., et al. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. The Journal of Clinical Investigation. 2014;124(7):3093–3106. doi: 10.1172/jci73351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang H., Hao Y., Yang J., et al. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nature Communications. 2011;2(1, article 554) doi: 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
- 96.Majid S., Dar A. A., Saini S., et al. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Research. 2012;72(24):6435–6446. doi: 10.1158/0008-5472.CAN-12-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Majid S., Dar A. A., Saini S., et al. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0067686.e67686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yeh Y.-M., Chuang C.-M., Chao K.-C., Wang L.-H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1α . International Journal of Cancer. 2013;133(4):867–878. doi: 10.1002/ijc.28086. [DOI] [PubMed] [Google Scholar]
- 99.Liu X., Jiang L., Wang A., Yu J., Shi F., Zhou X. microRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Letters. 2009;286(2):217–222. doi: 10.1016/j.canlet.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu X., Wang C., Chen Z., et al. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochemical Journal. 2011;440(1):23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao X., Dou W., He L., et al. microRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2013;32(11):1363–1372. doi: 10.1038/onc.2012.156. [DOI] [PubMed] [Google Scholar]
- 102.Akalay I., Tan T. Z., Kumar P., et al. Targeting WNT1-inducible signaling pathway protein 2 alters human breast cancer cell susceptibility to specific lysis through regulation of KLF-4 and miR-7 expression. Oncogene. 2015;34(17):2261–2271. doi: 10.1038/onc.2014.151. [DOI] [PubMed] [Google Scholar]
- 103.Li T., Xie J., Shen C., et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2015 doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 104.Yuan J.-H., Yang F., Wang F., et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 105.Richards E. J., Zhang G., Li Z. P., et al. Long non-coding RNAs (LncRNA) regulated by transforming growth factor (TGF) β: LncRNA-hit-mediated TGFβ-induced epithelial to mesenchymal transition in mammary epithelia. The Journal of Biological Chemistry. 2015;290(11):6857–6867. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mondal T., Subhash S., Vaid R., et al. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nature Communications. 2015;6, article 7743 doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou M., Hou Y., Yang G., et al. LncRNA-Hh strengthen cancer stem cells generation in twist-positive breast cancer via activation of hedgehog signaling pathway. STEM CELLS. 2016;34(1):55–66. doi: 10.1002/stem.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Y., He L., Du Y., et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16(4):413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 109.Wu J., Zhang J., Shen B., et al. Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. Journal of Experimental & Clinical Cancer Research. 2015;34(1, article 116) doi: 10.1186/s13046-015-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gumireddy K., Li A., Yan J., et al. Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. The EMBO Journal. 2013;32(20):2672–2684. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smits G., Mungall A. J., Griffiths-Jones S., et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nature Genetics. 2008;40(8):971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- 112.Keniry A., Oxley D., Monnier P., et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nature Cell Biology. 2012;14(7):659–665. doi: 10.1038/ncb2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao W.-L., Liu M., Yang Y., et al. The imprinted H19 gene regulates human placental trophoblast cell proliferation via encoding miR-675 that targets Nodal Modulator 1 (NOMO1) RNA Biology. 2012;9(7):1002–1010. doi: 10.4161/rna.20807. [DOI] [PubMed] [Google Scholar]
- 114.Hernandez J. M., Elahi A., Clark C. W., et al. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Annals of Surgical Oncology. 2013;20(supplement 3):S625–S635. doi: 10.1245/s10434-013-3106-3. [DOI] [PubMed] [Google Scholar]
- 115.Liang W., Fu W., Wong C., et al. The LncRNA H19 promotes epithelial to mesenchymal transition by functioning as MiRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y., VandenBoom T. G., II, Kong D., et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Research. 2009;69(16):6704–6712. doi: 10.1158/0008-5472.can-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guo L., Chen C., Shi M., et al. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene. 2013;32(45):5272–5282. doi: 10.1038/onc.2012.573. [DOI] [PubMed] [Google Scholar]
- 118.Shi X., Sun M., Liu H., Yao Y., Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Letters. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 119.Fan Y., Shen B., Tan M., et al. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clinical Cancer Research. 2014;20(6):1531–1541. doi: 10.1158/1078-0432.ccr-13-1455. [DOI] [PubMed] [Google Scholar]
- 120.Hirata H., Hinoda Y., Shahryari V., et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Research. 2015;75(7):1322–1331. doi: 10.1158/0008-5472.can-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ono H., Motoi N., Nagano H., et al. Long noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. Cancer Medicine. 2014;3(3):632–642. doi: 10.1002/cam4.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu Y., Zhang L., Wang Y., et al. Long noncoding RNA HOTAIR involvement in cancer. Tumor Biology. 2014;35(10):9531–9538. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 123.Liu Y. W., Sun M., Xia R., et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death & Disease. 2015;6(7) doi: 10.1038/cddis.2015.150.e1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chiyomaru T., Fukuhara S., Saini S., et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. The Journal of Biological Chemistry. 2014;289(18):12550–12565. doi: 10.1074/jbc.m113.488593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Calin G. A., Croce C. M. MicroRNA signatures in human cancers. Nature Reviews Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 126.Lu J., Getz G., Miska E. A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 127.Roldo C., Missiaglia E., Hagan J. P., et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. Journal of Clinical Oncology. 2006;24(29):4677–4684. doi: 10.1200/jco.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 128.Lin C. W., Kao S. H., Yang P. C. The miRNAs and epithelial-mesenchymal transition in cancers. Current Pharmaceutical Design. 2014;20(33):5309–5318. doi: 10.2174/1381612820666140128204508. [DOI] [PubMed] [Google Scholar]
- 129.Park S.-M., Gaur A. B., Lengyel E., Peter M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & Development. 2008;22(7):894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Korpal M., Lee E. S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. The Journal of Biological Chemistry. 2008;283(22):14910–14914. doi: 10.1074/jbc.c800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang Y., Ahn Y.-H., Gibbons D. L., et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. The Journal of Clinical Investigation. 2011;121(4):1373–1385. doi: 10.1172/jci42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gill B. J., Gibbons D. L., Roudsari L. C., et al. A synthetic matrix with independently tunable biochemistry and mechanical properties to study epithelial morphogenesis and EMT in a lung adenocarcinoma model. Cancer Research. 2012;72(22):6013–6023. doi: 10.1158/0008-5472.CAN-12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gregory P. A., Bracken C. P., Smith E., et al. An autocrine TGF-β/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Molecular Biology of the Cell. 2011;22(10):1686–1698. doi: 10.1091/mbc.e11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hahn S., Jackstadt R., Siemens H., Hünten S., Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. The EMBO Journal. 2013;32(23):3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moes M., Le Béchec A., Crespo I., et al. A novel network integrating a miRNA-203/SNAI1 feedback loop which regulates epithelial to mesenchymal transition. PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0035440.e35440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ding X., Park S. I., McCauley L. K., Wang C.-Y. Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. The Journal of Biological Chemistry. 2013;288(15):10241–10253. doi: 10.1074/jbc.m112.443655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang Z., Zhang B., Li W., et al. Epigenetic silencing of miR-203 upregulates SNAI2 and contributes to the invasiveness of malignant breast cancer cells. Genes & Cancer. 2011;2(8):782–791. doi: 10.1177/1947601911429743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim N. H., Kim H. S., Kim N.-G., et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Science Signaling. 2011;4(197):p. ra71. doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Choi Y. J., Lin C.-P., Ho J. J., et al. MiR-34 miRNAs provide a barrier for somatic cell reprogramming. Nature Cell Biology. 2011;13(11):1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hwang C.-I., Matoso A., Corney D. C., et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(34):14240–14245. doi: 10.1073/pnas.1017536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sun Y., Wu J., Wu S.-H., et al. Expression profile of microRNAs in c-Myc induced mouse mammary tumors. Breast Cancer Research and Treatment. 2009;118(1):185–196. doi: 10.1007/s10549-008-0171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Higashi T., Hayashi H., Ishimoto T., et al. miR-9-3p plays a tumour-suppressor role by targeting TAZ (WWTR1) in hepatocellular carcinoma cells. British Journal of Cancer. 2015;113(2):252–258. doi: 10.1038/bjc.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mori M., Triboulet R., Mohseni M., et al. Hippo signaling regulates microprocessor and links cell-density-dependent mirna biogenesis to cancer. Cell. 2014;156(5):893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zell S., Schmitt R., Witting S., Kreipe H. H., Hussein K., Becker J. U. Hypoxia induces mesenchymal gene expression in renal tubular epithelial cells: an in vitro model of kidney transplant fibrosis. Nephron Extra. 2013;3(1):50–58. doi: 10.1159/000351046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.White K., Loscalzo J., Chan S. Y. Holding our breath: the emerging and anticipated roles of microRNA in pulmonary hypertension. Pulmonary Circulation. 2012;2(3):278–290. doi: 10.4103/2045-8932.101395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Du R., Sun W., Xia L., et al. Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030771.e30771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mercer T. R., Dinger M. E., Mattick J. S. Long non-coding RNAs: insights into functions. Nature Reviews Genetics. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 148.Carninci P., Kasukawa T., Katayama S., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 149.Kapranov P., Willingham A. T., Gingeras T. R. Genome-wide transcription and the implications for genomic organization. Nature Reviews Genetics. 2007;8(6):413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 150.Grote P., Herrmann B. G. Long noncoding RNAs in organogenesis: making the difference. Trends in Genetics. 2015;31(6):329–335. doi: 10.1016/j.tig.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 151.Dey B. K., Mueller A. C., Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4) doi: 10.4161/21541272.2014.944014.e944014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li C. H., Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. The International Journal of Biochemistry & Cell Biology. 2013;45(8):1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 153.Wapinski O., Chang H. Y. Long noncoding RNAs and human disease. Trends in Cell Biology. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 154.Derrien T., Guigó R. Long non-coding RNAs with enhancerlike function in human cells. Médecine/Sciences. 2011;27(4):359–361. doi: 10.1051/medsci/2011274009. [DOI] [PubMed] [Google Scholar]
- 155.Rugg-Gunn P. J., Ferguson-Smith A. C., Pedersen R. A. Epigenetic status of human embryonic stem cells. Nature Genetics. 2005;37(6):585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- 156.Sasaki H., Ishihara K., Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. The Journal of Biochemistry. 2000;127(5):711–715. doi: 10.1093/oxfordjournals.jbchem.a022661. [DOI] [PubMed] [Google Scholar]
- 157.Jinno Y., Ikeda Y., Yun K., et al. Establishment of functional imprinting of the H19 gene in human developing placentae. Nature Genetics. 1995;10(3):318–324. doi: 10.1038/ng0795-318. [DOI] [PubMed] [Google Scholar]
- 158.Casola S., Pedone P. V., Cavazzana A. O., et al. Expression and parental imprinting of the H19 gene in human rhabdomyosarcoma. Oncogene. 1997;14:1503–1510. doi: 10.1038/sj.onc.1200956. [DOI] [PubMed] [Google Scholar]
- 159.Li X., Kogner P., Sandstedt B., Haas O. A., Ekström T. J. Promoter-specific methylation and expression alterations of igf2 and h19 are involved in human hepatoblastoma. International Journal of Cancer. 1998;75(2):176–180. doi: 10.1002/(SICI)1097-0215(19980119)75:2<176::AID-IJC2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 160.Nakagawa H., Chadwick R. B., Peltomäki P., Plass C., Nakamura Y., De La Chapelle A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(2):591–596. doi: 10.1073/pnas.011528698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zambrano P., Segura-Pacheco B., Perez-Cardenas E., et al. A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes. BMC Cancer. 2005;5, article 44 doi: 10.1186/1471-2407-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Prawitt D., Enklaar T., Gärtner-Rupprecht B., et al. Microdeletion of target sites for insulator protein CTCF in a chromosome 11p15 imprinting center in Beckwith-Wiedemann syndrome and Wilms' tumor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(11):4085–4090. doi: 10.1073/pnas.0500037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dey B. K., Pfeifer K., Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes and Development. 2014;28(5):491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ochiai H., Okada S., Saito A., et al. Inhibition of insulin-like growth factor-1 (IGF-1) expression by prolonged transforming growth factor-β1 (TGF-β1) administration suppresses osteoblast differentiation. The Journal of Biological Chemistry. 2012;287(27):22654–22661. doi: 10.1074/jbc.m111.279091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang Y., Zheng Y., Jia L., Li W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. STEM CELLS. 2015;33(12):3481–3492. doi: 10.1002/stem.2225. [DOI] [PubMed] [Google Scholar]
- 166.Li H., Yu B., Li J., et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kallen A. N., Zhou X.-B., Xu J., et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Molecular Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gupta R. A., Shah N., Wang K. C., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kogo R., Shimamura T., Mimori K., et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Research. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.can-11-1021. [DOI] [PubMed] [Google Scholar]
- 170.Alves C. P., Fonseca A. S., Muys B. R., et al. Brief report: the lincRNA hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. STEM CELLS. 2013;31(12):2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]