Abstract
Mammals, including human beings, have evolved a unique viviparous reproductive system and a highly developed central nervous system. How did these unique characteristics emerge in mammalian evolution, and what kinds of changes did occur in the mammalian genomes as evolution proceeded? A key conceptual term in approaching these issues is “mammalian-specific genomic functions”, a concept covering both mammalian-specific epigenetics and genetics. Genomic imprinting and LTR retrotransposon-derived genes are reviewed as the representative, mammalian-specific genomic functions that are essential not only for the current mammalian developmental system, but also mammalian evolution itself. First, the essential roles of genomic imprinting in mammalian development, especially related to viviparous reproduction via placental function, as well as the emergence of genomic imprinting in mammalian evolution, are discussed. Second, we introduce the novel concept of “mammalian-specific traits generated by mammalian-specific genes from LTR retrotransposons”, based on the finding that LTR retrotransposons served as a critical driving force in the mammalian evolution via generating mammalian-specific genes.
Keywords: mammalian-specific genomic functions, evolution and development, genomic imprinting, newly acquired genes from LTR retrotransposon, placenta and viviparity, brain
Introduction
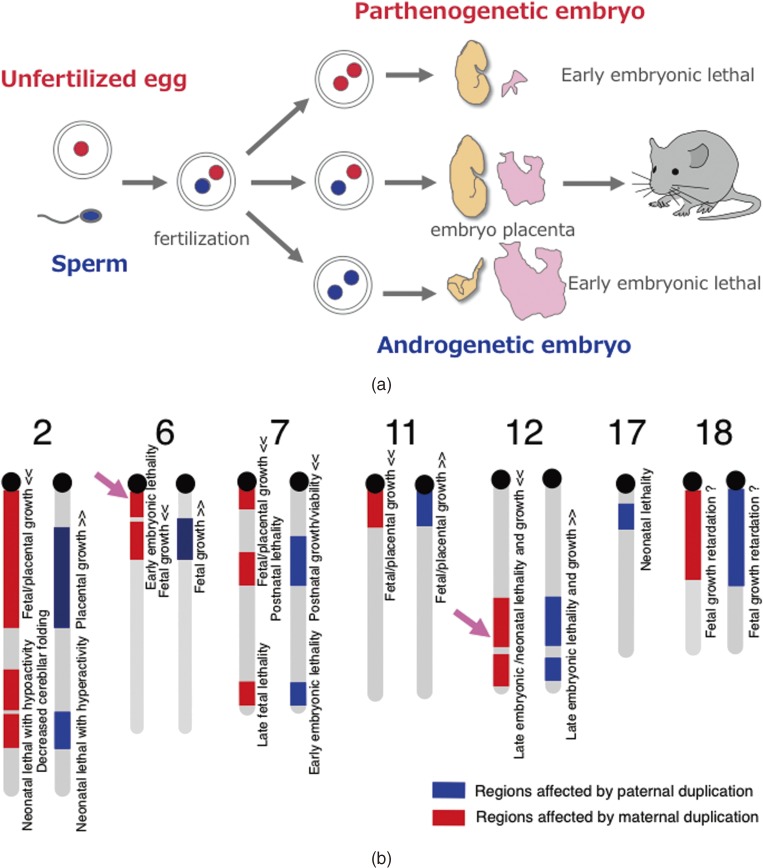
Mammals never develop to term without the cooperation of both of the parental genomes: embryos with either two maternally or paternally derived genomes, such as parthenogenetic or androgenetic embryos, exhibit early embryonic lethality (Fig. 1a).1–3) This is because of functional differences between the paternally and maternally derived genomes that are attributable to the presence of imprinted genes, paternally expressed genes (Peg) and maternally expressed genes (Meg), exhibiting parent-of-origin specific monoallelic expression.4–6) This mechanism is unique to mammals among the vertebrates, and is known as genomic imprinting (Fig. 1a).1–8) Therefore, it is of great interest to know how this mammalian-specific epigenetic mechanism emerged and why it has been widely conserved in mammals despites the apparent developmental disadvantage that arises because the limitation of the monoallelic expression of essential genes is quite likely to endanger the survival of individuals in both the pre- and postnatal periods. What is the advantage conferred by genomic imprinting to overcome this difficulty? Accidental or unexpected parthenogenesis may be life-threatening and undesirable in females, because the environmental conditions suitable for breeding pups, such as food, temperature and climate, are constrained by season. Therefore, a prohibition of parthenogenetic development might be advantageous for mammalian reproduction in terms of fitting with the seasonal conditions.4,9) Another biological advantage for mammals may be the absence of the placental trophoblast tissues in the parthenogenetic conceptus, because they have an infiltrative nature and a means of invading the maternal uterus. Mammalian females can avoid developing the malignant ovarian teratocarcinomas that arise during parthenogenetic development.10) These hypotheses suggest that there are certain advantages in genomic imprinting for mammalian development. However, this does not directly mean that genomic imprinting specifically evolved for the purpose of prohibiting parthenogenetic development and/or invasive trophoblast development. In general, it is difficult to prove such hypotheses by experiment. An important clue may be provided from the historical point of view. A relationship between placenta formation and genomic imprinting in mammals has also been proposed in the form of various “placenta hypotheses”.4,9,11) Thus, it is expected that the continuing investigation of genomic imprinting will provide clues in the molecular mechanism connecting these phenomena, as well as in the elucidation of mammalian evolution itself as one of the mammalian-specific genomic functions.
Figure 1.
Discovery of genomic imprinting. a. Pronuclear transplantation experiments. Both parthenogenetic and androgenetic embryos exhibit early embryonic lethality with totally different morphological defects on developmental day 10.5. b. Mouse genomic imprinting map deduced from a series of Robertosonian translocation experiments. The abnormal phenotypes observed in mice with partial uniparental duplications are shown. Modified from data in ref. 6. It should be noted that imprinted regions usually comprise both Pegs and Megs (see also Figs. 2 and 3a) and that most of them are conserved in eutherians, including humans, although some are lineage-specific (see also Fig. 6). Pink arrows indicate the imprinted regions associated with early embryonic lethality upon maternal duplication (left) and late embryonic lethality upon paternal duplication (right) where Peg10 and Peg11/Rtl1 were discovered afterwards, respectively.
Recent studies have clearly demonstrated the essential nature of genomic imprinting in the current form of the mammalian developmental system, where the reciprocal regulation of Pegs and Megs via DNA methylation on imprinting control regions (ICRs) plays a key role.12–14) Pegs and Megs cannot be expressed from the individual parental chromosomes simultaneously, therefore, two parental chromosomes with different expression profiles are necessary for normal development. Several lines of evidence also indicate that this mechanism emerged as a defense against exogenous DNA, because these ICRs themselves emerged in the mammalian genome as newly integrated DNA sequences during the course of evolution.15–19)
Moreover, genomic imprinting has brought into focus a new dimension to the model of mammalian evolution through the identification of two Pegs, Peg10 and Peg11/Rtl1, which play an essential role in mammalian development via the formation and maintenance of a mammalian-specific placenta, respectively, because these two genes are mammalian-specific genes derived from an LTR-retrotransposon.20–26) It is known that many housekeeping genes and genes related to various signaling pathways that are conserved in other organs are involved in placenta formation.27) In addition, several placenta-specific genes generated by gene duplication in a wide range of multigene families are known to play an essential role in this process. However, Peg10 and Peg11/Rtl1 were the first examples of mammalian-specific, essential placental genes acquired from a specific LTR-retrotransposon. Thus, a new concept established which says that the evolution of mammalian-specific traits occurred by the acquisition of mammalian-specific genes from LTR retrotransposons.24,25)
At least 30 LTR retrotransposon-derived genes exist in the human genome as therian- and eutherian-specific genes.25) A series of knockout mouse studies on these genes has gathered persuasive evidence that at least some of these genes are essential in the current form of the developmental and reproductive systems, which suggests that they made critical contributions to mammalian evolution in a variety of ways, such as the establishment of viviparity22–26) and presumably certain sophisticated brain functions. Thus, it is highly probable that the LTR retrotransposons served as one of the driving forces in mammalian evolution; the acquisition of novel genes from LTR retrotransposons is one of the important evolutionary mechanisms as well as being a gene/gemome duplicating mechanism.28,29) Importantly, from the viewpoint of the LTR retrotransposon-derived genes, eutherians and marsupials are entirely different animal groups, because most genes exist as eutherian-specific genes while a few are marsupial-specific except for a single exception, Peg10, which is common to both groups.25)
In the last part of this review, we also discuss a hypothetical two-step model from LTR retrotransposons to endogenous genes during mammalian evolution in which the nearly neutral theory of molecular evolution played an essential underlying role in the first part and then was succeeded by Darwinian evolution at the critical selection step.24,25,30–32)
1. The investigation of “mammalian-specific genomic functions”, including our own personal research history
Mammals are considered one of the most successful animals in the Cenozoic era because they have exhibited adaptive radiation since the extinction of the dinosaurs. They have evolved an efficient viviparous reproductive system using a placenta and a highly developed cerebral neocortex. In this review, we will endeavor to address how these unique characteristics have emerged in mammalian evolution, and what kinds of changes occurred in the mammalian genomes as evolution proceeded. A key conceptual term we will employ is “mammalian-specific genomic functions”, a concept covering both mammalian-specific epigenetics and genetics. This term is necessary, because mamsmals have unique epigenetic mechanisms, such as X chromosome inactivation (XCI) and genomic imprinting, as well as certain unique genes acquired from retroelements, such as retrotransposons and retroviruses.
Discoveries of XCI and genomic imprinting in mammals in the late 20th century provided an insight into how to approach these evolutionary issues from the perspective of mammalian-specific epigenetic mechanisms.1–11,33,34) Importantly, both XCI and genomic imprinting play an essential role in mammalian development. Female embryos with two active X chromosomes exhibit early embryonic lethality, so it is clearly evident that inactivation of one of the two X chromosomes in females is essential for mammalian development.33,34) As mentioned, parthenogenetic or androgenetic embryos also exhibit early embryonic lethality; therefore, both parental genomes are essential for mammalian development.1–3) Interestingly, in all of these cases, the developmental problems are associated with placental abnormalities. The genes involved in the mechanism underlying XCI as well as those causing a variety of imprinted phenotypes have been extensively examined using knockout mice and in human patients with diseases of genomic imprinting. It should be noted that genomic imprinting phenomena are also found in other organisms, such as certain insects (e.g. Sciara)35) and plants (e.g. Arabidopsis sp., Zea mays),36) but it is most widespread in mammals via its own, specific regulatory mechanism, and the same is true for XCI.
The completion in the early 21th century of the “human genome project” has dramatically changed the situation with regard to these evolutionary issues because this investigation has expanded to a wide range of subsequent genome projects. At present, sequencing of the whole genome of more than 100 mammalian species, including marsupial and monotreme species is available as well as that of many other vertebrate species including reptiles, birds and fish.37–42) Through comprehensive and comparative genomics, only 1.5% of the mammalian genome is comprised of protein coding genes, but it is occupied by abundant retrotransposons, such as short interspersed nuclear elements (SINEs), long interspersed nuclear elements (LINEs) and LTR retrotransposons/retroviruses. These retrotransposon-derived DNA sequences were thought to be so-called junk DNA and even potentially harmful for host organisms for a long time; however, we and others demonstrated by a series of knock-out mouse analyses that at least some of these sequence play essential roles in the current mammalian developmental system as endogenously functional genes specific to mammals.20–26,43–45) In other words, it turns out that certain LTR retrotransposon- and retrovirus-derived genes are actually responsible for a variety of mammalian-specific traits, such as the formation, maintenance and endocrinological regulation of the placenta and presumably for a variety of brain functions. The strategy of focusing on “mammalian-specific genomic functions” has thus been proven to be very successful thus far, and is expected to continue to be so in the future in terms of shedding light on the evolutionary issues related to mammals.
Two themes, genomic imprinting and the newly acquired genes from LTR retrotransposons, which serve as the representative examples of the mammalian-specific epigenetics and genetics, respectively, seem distinctly separated and independent of each other. However, the former investigation actually triggered the latter theme, because the first and second examples of LTR retrotransposon-derived genes were identified as paternally expressed imprinted genes, such as PEG10 and PEG11. Therefore, we would like to provide the behind-the-scenes story of the birth of the second theme of the acquired genes from LTR retrotransposons from the genomic imprinting work before getting into the main topic. When we started our genomic imprinting investigation in Surani’s laboratory in Cambridge in 1990, we hypothesized that functional differences between parental genomes were due to the presence of genes exhibiting parent-of-origin-specific monoallelic expression. Subsequently, we conducted a systematic screening for Pegs and Megs by developing a new method of subtracting the genes expressed in the parthenogenetic and androgenetic embryos from those in normally fertilized embryos.46–49)
One of us (T. K-I.) also had the idea that genomic imprinting was somehow induced by responses against exogenous DNA integrated into the mammalian genome. Therefore, her research target has long been the imprinted genes derived from exogenous DNA, such as retrotransposons. Eventually, a similar idea on the origin of genomic imprinting was proposed by Barlow in 1993.15) Meanwhile, the other one of us (F. I.) made a plan to identify the imprinted genes that are related to embryonic lethality, because such information would provide an important clue to help elucidate the biological significance of genomic imprinting. Therefore, his ultimate target became the genes responsible for the early embryonic lethality of the parthenogenetic embryos and the late fetal/neonatal lethality caused by paternal and maternal disomies of the mouse chromosome 12 (Chr12).8,50) Another reason to choose the latter was its medical importance, because paternal disomy of human orthologous chromosome 14 (upd(14)pat) causes a severe genomic imprinting disease associated with neonatal lethality by a respiratory problem, presumably related to a bell-shaped thorax and abdominal defects, placental overgrowth and polyhydramnios during gestation, with severe mental and postnatal growth retardation when the patients survived.51) Therefore, the development of a proper diagnosis and treatment of this syndrome are greatly anticipated.
In 2006 and 2008, we finally identified Peg10 and Peg11/Rtl1 as the major imprinted genes responsible for the parthenogenetic death and the uniparental disomy of mouse Chr12/human upd(14)pat, and elucidated their essential roles in placenta formation and maintenance, respectively.22,23,52) Interestingly, both Peg10 and Peg11/Rtl1 are derived from an LTR retrotransposon and in fact are actually newly acquired genes specific to the mammals.16,20,21,53) At that time, we realized that our targets we had been pursuing were the same genes all along. To our great pleasure, this has personal meaning for us at the same time leading to a new concept of “mammalian-specific traits generated by mammalian-specific genes newly acquired from LTR retrotransposons”.24,25) Since then, we have devoted a tremendous amount of effort to elucidating the biological functions of the LTR retrotransposon-derived genes, as will be described in section 3.
2. Genomic imprinting as a mammalian-specific epigenetic mechanism
2-1. Discovery of genomic imprinting and the imprinted genes, Pegs and Megs.
Genomic imprinting was discovered by pronuclear exchange experiments in mice which showed that both parthenogenetic and androgenetic embryos with either two maternal or two paternal pronuclei exhibited early embryonic lethality, albeit with different morphological anomalies (Fig. 1a).1–3) In the former case, embryos were small but appeared normal yet died due to severe placental defects. The latter embryos exhibited severe growth retardation associated with abnormal placenta overgrowth. This clearly demonstrated the functional differences between the paternal and maternal genomes and critical requirement of both for normal development in mammals. As parthenogenesis is often observed naturally or experimentally among vertebrates, such as fish, amphibians, birds and reptiles, the complete absence of parthenogenesis is a unique feature of mammals.
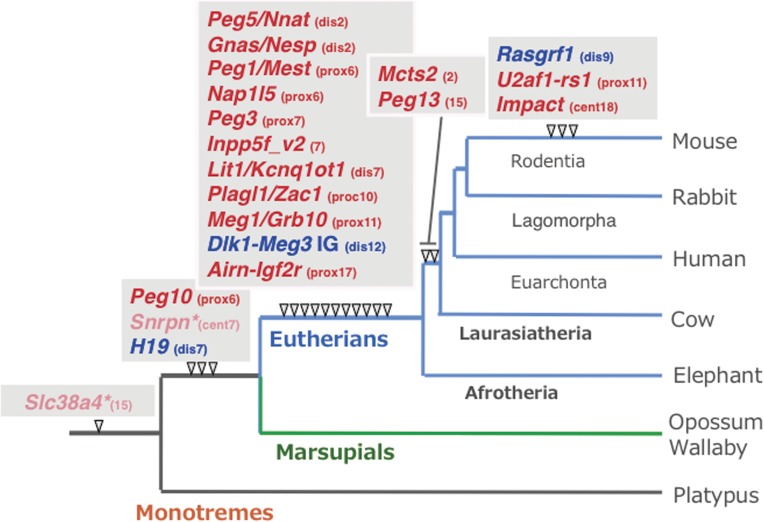
A series of experiments using mice with Robertsonian translocations also provided strong genetic evidence for the functional differences between the paternally and maternally derived chromosomes, because mice with uniparental duplication of certain specific chromosomal regions exhibited a variety of defects in development, growth and/or behavior (Fig. 1b).6–8) As a result, approximately 20 chromosomal imprinted regions in the mouse genome have been identified. Subsequently, genomic imprinting has attracted considerable attention as a mammalian-specific epigenetic mechanism. These phenomena are an apparent exception to Mendelian genetics in mammals, so humans suffer from a variety of non-Mendelian genetic disorders by the breakdown of this mechanism.
In 1991, the first three imprinted genes, Insulin-like growth factor 2 (Igf2), Insulin-like growth factor 2 receptor (Igf2r) and H19, imprinted maternal expressed transcript (H19) were identified.54–57) Igf2 and Igf2r are related functionally because the Igf2 protein promotes embryonic and placental growth as a growth factor, while the Igf2r protein inhibits embryonic and placental growth by degrading Igf2 through its ability to bind Igf2 and transport it to the lysosome.54–56) As Igf2 exhibits paternal expression (Peg) while Igf2r exhibits maternal expression (Meg), these two genes fit very well with the weak version of ‘the conflict hypothesis’ that states that paternally expressed genes promote embryonic growth, while maternally expressed genes inhibit embryonic growth as a consequence of the genetic conflict between the paternal and maternal alleles during mammalian evolution.58) It proposes their developmental advantages were conferred on the viviparous system that females are able to accept more than two males (polygamy). Not all, but nevertheless a substantial number of imprinted genes fit with this prediction, so the concept of “parental conflict” in the genome has come to be widely accepted. On the other hand, the strong version of “the conflict hypothesis” further predicted that the genomic imprinting mechanism itself originated under continuous pressure exerted by this genetic conflict during the course of evolution. We will discuss this issue, the origin of genomic imprinting, later in section 2-4.
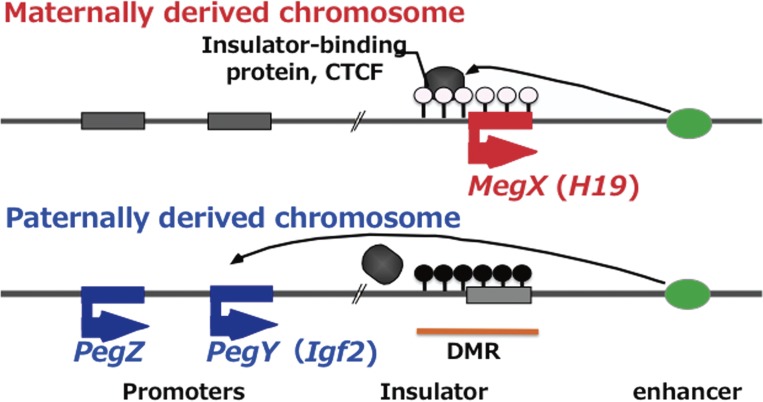
H19 is located next to Igf2 on the distal portion of mouse chromosome 7, therefore, these two genes are positionally related.57) Retrospectively, this finding can be seen to have been very important, because this was the first report demonstrating that imprinted genes exist in clusters. It subsequently led to the concept of a critical regulatory mechanism in which Pegs (such as Igf2) and Megs (such as H19) are reciprocally regulated by the DNA methylation status in the differentially methylated regions (DMRs) in each imprinted region.4,5,12,13) Thus, DMRs are regarded as the imprinting control region (ICR). In “the insulator model” independently proposed by Tilghman’s and Felsenfeld’s groups,12,13) H19-DMR extends from the H19 promoter region to an upstream insulator sequence where insulator-binding proteins, such as CTCF, bind and block several downstream enhancers when H19-DMR is not DNA methylated, after which the repression of the upstream Igf2 occurs. In contrast, when it is DNA methylated, the CTCF proteins cannot bind to the insulator sequence, so Igf2 becomes actively expressed instead of the repression of H19 due to promoter methylation (Fig. 2). An antisense RNA model and a bipartite regulation model have been proposed in other imprinted regions, but the reciprocal regulation of Pegs and Megs by DMR methylation is the common consequence of all of the imprinted regions.4,5,14) It is of critical importance to take this aspect of genomic imprinting into account, as will be discussed in section 2-3.
Figure 2.
Reciprocal regulation of Pegs and Megs in imprinted gene clusters. An insulator model is shown. In this example, paternal allele of DMR is fully methylated while maternal allele is none methylated (paternally imprinted region) like IGF2-H19 region. In the case of IGF2-H19 region, there are several cell-lineage-specific enhancers downstream of H19 but one of them is shown.
Several systematic methods of screening for imprinted genes, including ours, have contributed to the further identification of novel imprinted genes and a determination of the precise location of the imprinted regions in the estimated chromosomal regions.46–49,59–61) By our subtraction-hybridization method using parthenogenetic and androgenetic embryos combination with genome walking from the newly identified imprinted genes, we succeeded in isolating more than 20 imprinted genes (Peg1–12 and Meg1–5, 8–10 etc. including 4 of the known imprinted genes) in 8 imprinted regions, such as the mouse sub-distal chromosome 2 (Chr2), proximal Chr6, sub-proximal Chr6, proximal Chr7, central Chr7, distal Chr7, proximal Chr11 and distal Chr12. The mapping results were ultimately in good agreement with those of the previous genetic experiments using mice with uniparental chromosomal duplications, as shown in Fig. 1b. Thus, this series of our experiments united the two original observations of genomic imprinting, the pronuclear transplantation1–3) and the Robertsonian translocation experiments,6–8) thereby contributing to the establishment of the current concept that genomic imprinting phenomena are attributable to the existence of Pegs and Megs.4,5,14)
2-2. Roles of Peg10 and Peg11/Rtl1 in the placenta.
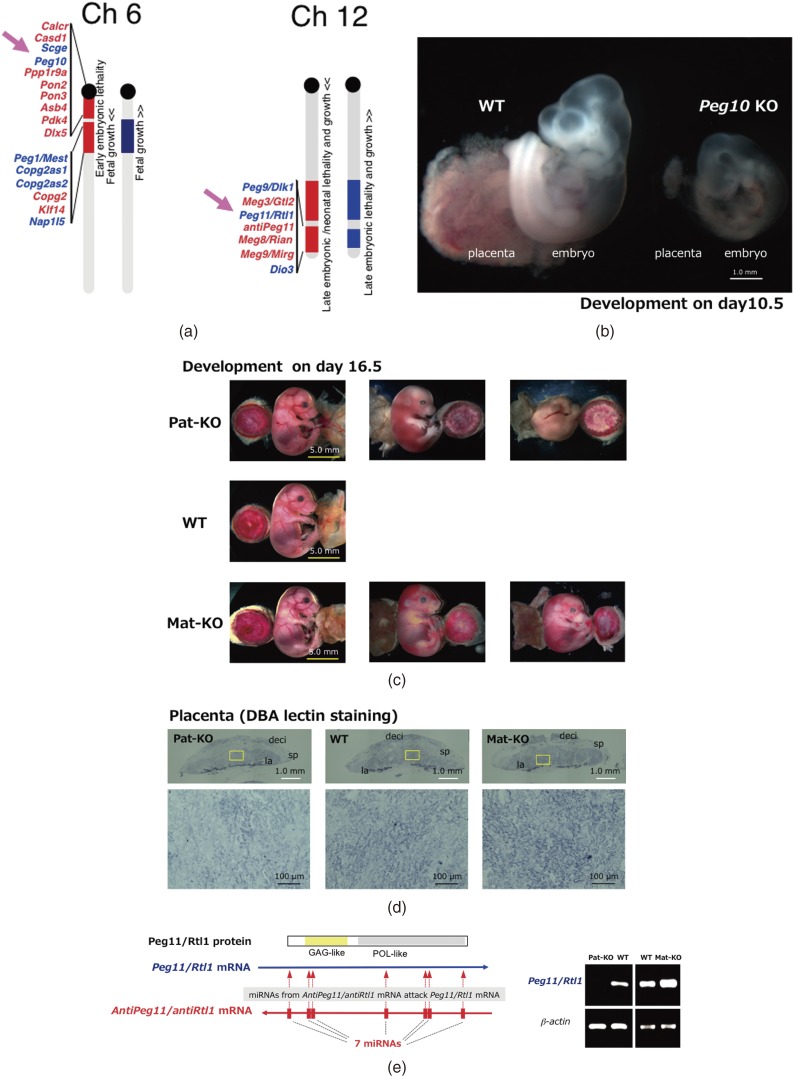
According to the genomic imprinting map, it appeared highly probable that the responsible gene for the parthenogenetic death should be located on mouse proximal Chr6, because this is the only imprinted region that causes early embryonic lethality upon maternal duplication, like the parthenogenetic embryos, with two maternally derived pronuclei (Fig. 1b).6,8) We then searched in the region near Sarcoglycan epsilon (Sgce), the first imprinted gene identified in this region.61) Using the human genome sequence information on chromosome 7q21 which is syntenic to the mouse proximal Chr6, we identified PEG10/Peg10 next to SGCE/Sgce in both humans and mice in 2001 (Fig. 3a, left).20) In the case of the mouse distal Chr12, we extensively sequenced around Meg3/Gtl2,49) the first imprinted gene in this cluster, by genome walking and determined the entire imprinted gene cluster comprising Peg9/Dlk1, Meg3/Gtl2, Peg11/Rtl1, Meg10/antiPeg11, Meg8/Rian and Meg9/Mirg in mice (Fig. 3a, right). Almost the same result was published by Micheal Georges’ group analyzing the same imprinted region in the ovine and human genomes including DLK1, GTL2, PEG11, antiPEG11 and MEG8 in 2001.21)
Figure 3.
Two essential imprinted genes from LTR retrotransposons, Peg10 and Peg11/Rtl1. a. Locations of mouse Peg10 and Peg11/Rtl1 (indicated by arrows) in the imprinted regions on proximal Chr6 and distal Chr12, respectively. b. Development of Peg10 KO on embryonic day 10.5. Severe developmental failure of Peg10 KO placenta and related embryonic retardation were clearly observed. The yolk sac was removed from both of the samples. A photograph was kindly provided by R Ono. c. Development of Peg11 Pat-KO (upper), wild type (middle) and Peg11 Mat-KO (lower) on embryonic day 16.5. Note that Pat-KO and Mat-KO exhibit late embryonic lethality. Placenta (left), embryo (middle) and yolk sack (right) were shown in each figure. d. Upper: DBA-lectin staining of whole placentas of Peg11 Pat-KO (left), wild type (center) and Peg11 Mat-KO (right). la: labyrinth layer, sp: spongiotrophoblast layer, de: maternal decidua. Lower: Magnified views of the labyrinth regions. Fetal capillary endothelial cells were stained in the labyrinth regions. Peg11 Pat-KO placenta was much weaker stained than wild type placenta while Peg11 Mat-KO placenta exhibited stronger signals. e. Regulation of Peg11/Rtl1 mRNA level by 7 miRNAs derived from maternally expressed AntiPeg11/AntiRtl1 non-coding RNA via an RNAi mechanism (left). Peg11/Rtl1 mRNA levels in Peg11 Pat-KO and Peg11 Mat-KO placentas on embryonic day 16.5 (right).
Importantly, both Peg10 (the former region) and Peg11 (the latter region) are genes acquired from a sushi-ichi related retrotransposon because they encode proteins exhibiting a high homology with the Gag and Pol proteins of a sushi-ichi retrotransposon, respectively.20,21) PEG11 was later officially renamed Retrotransposon-like 1 (RTL1).62) They were actually TK-I’s original target of imprinted genes, obtained ten years after the project had been started. The fact that their origin was an LTR retrotransposon is very interesting, at that time nobody knew whether they were functional, and it would require a period of another five to seven years to elucidate their biological functions in mammalian development and evolution.
It was big surprise when our knockout mouse experiments on Peg10 and Peg11/Rtl1 clearly demonstrated that both are essential genes in development: Peg10 is responsible for the early embryonic lethality caused by maternal duplication of the mouse proximal Chr6 (Fig. 3b)6,8,22) and Peg11/Rtl1 is responsible for the late fetal/neonatal lethality caused by the maternal disomy of mouse Chr12 (Fig. 3c).6,8,23) At that point, the two concepts, the retrotransposon-derived imprinted genes and the responsible genes for embryonic lethality became united and thus brought a new dimension to mammalian evolution: that LTR retrotransposon-derived genes exerted a critical impact on mammalian evolution (see section 3).24,25) Our first presentation on the roles of retrotrasposon-derived Peg10 and Peg11/Rtl1 in placental formation and functions had an impact at the genomic imprinting meeting in 2004 in Montpellier and stimulated considerable work on the retrotransposon-derived genes in mammals thereafter.63,64)
Peg10 KO mice exhibited early embryonic lethality due to poor placental growth associated with complete lack of labyrinth and spongiotrophoblast layers (Fig. 3b).22) The labyrinth layer is an essential part of the placenta, where nutrient and gas exchange occurs between fetal and maternal blood cells, therefore, Peg10 KO embryos stopped growth on day 9.5 and could not grow any further. Importantly, the phenotypes of the Peg10 KO embryos and placentas resemble those of parthenogenetic embryos and placenta closely, strongly suggesting that Peg10 is a major gene responsible for parthenogenetic death.1–3,22)
One half of the Peg11/Rtl1 KO mice exhibited late fetal lethality while the other half exhibited neonatal lethality associated with late fetal growth retardation (Figs. 3c and d).23) This corresponds very well with what is observed in mice with maternal duplication of the mouse distal Chr12.8,50) In the labirynth layer of the Peg11/Rtl1 KO placenta, severe abnormality of the fetal capillaries was observed: they were clogged at many sites because endothelial cells had been phagocytosed by surrounding trophoblast cells, indicating that Peg11/Rtl1 plays an essential role in the maintenance of the feto-maternal interface of the placenta during gestation.
Peg11/Rtl1 is overlapped with the maternally expressed non-coding RNA antiPeg11/antiRtl1 that comprises seven micro RNAs (miRNAs) that target Peg11/Rtl1 mRNA by an RNAi mechanism23,62,65) (Fig. 3e). Our Peg11/Rtl1 KO mice lacked most portions of both Peg11/Rtl1 and antiPeg11/antiRtl1, therefore, upon maternal transmission, antiPeg11/antiRtl1 KO mice were generated exhibiting neonatal lethality associated with placental overgrowth due to a 2–3 fold accumulation of Peg11/Rtl1 mRNA, resulting in an overproduction of the Peg11/Rtl1 protein. In the labyrinth layer of antiPeg11/antiRtl1 KO mice, fetal capillary size expansion associated with severely damaged surrounding trophoblast cell layers was observed. Thus, both the lack and overexpression of Peg11/Rtl11 cause lethality around birth, just by different mechanisms (Figs. 3c and d).
The AntiPeg11/antiRtl1 KO mouse is a good model for paternal duplication (or paternal disomy) of mouse Chr12 as well as human Chr 14.23,50–52) It should be noted that one important difference is that a double dose of Peg11/Rtl1 without maternally expressed antiPeg11/antiRtl1 leads to a 4–6 fold increment of Peg11/Rtl1 mRNA, causing more severe abnormal phenotypes in the case of paternal duplication (or paternal disomy) than antiPeg11/antiRtl1 KO with only one normal Peg11/Rtl1 allele.23,52) In patients with upd(14)pat-like phenotypes due to deletion of this locus that contains an imprinting control region (IG-DMR), symptom severity correlated well with the degree of PEG11/RTL1 overexpression, indicating that the overexpression of PEG11/RTL1 is attributable to the major phenotypes of upd(14)pat.52) In the case of upd(14)mat, both PEG11/RTL1 and PEG9/DLK1 are responsible for growth deficiency in an additive manner.52)
2-3. The Essential nature of genomic imprinting.
Why is genomic imprinting widely conserved in mammals? An important key to answering this question lies in the reciprocal regulation mechanism of Pegs and Megs, as mentioned earlier, because the two parental chromosomes have different epigenotypes (different DNA methylation status in the DMR) that are necessary for the expression of both Pegs and Megs. As shown in the insulator model, it is apparent that Pegs and Megs cannot be expressed from the individual parental chromosomes simultaneously (Fig. 2).12–14) In this model, the essential components comprising the reciprocal regulation mechanism are four kinds of functional genomic units, such as promoters, an insulator, enhancer(s) and a DMR, as well as insulator binding proteins, such as CTCF, which interact with the insulator sequence. All of the components except the DMR are ubiquitous in eukaryotes, so the DMR may be a key factor in this mechanism, with the important constraint that the DMR overlaps with the insulator sequence as well as the neighboring promoter in this model. However, it is apparent that the reciprocal expression of Pegs and Megs is intrinsically controlled by their own genomic DNA; in other words, the mechanism is already integrated into the mammalian genome.
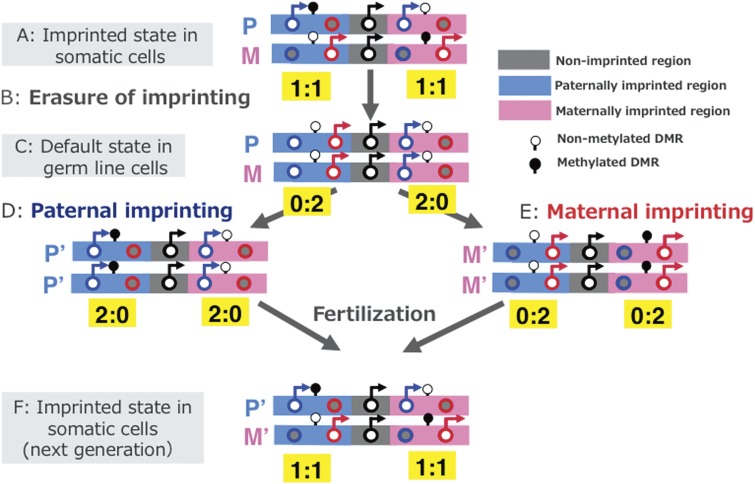
This provides a good explanation for why genomic imprinting, a mechanism which promotes the monoallelic expression of Pegs and Megs, is highly conserved in mammals so as to ensure the current form of the mammalian development system, even though it seems disadvantageous upon first consideration (complementation hypothesis) (Fig. 4).14) In other words, monoallelic expression of Pegs and Megs in the imprinted gene cluster would appear to be a good means for avoiding a situation in which one half (Pegs or Megs) is completely repressed throughout the course of life, even though the other half (Megs or Pegs, respectively) exhibits normal biallelic expression. There are two kinds of imprinted regions, paternally imprinted regions (defined as the regions where paternal allele is methylated in sperm: D in Fig. 4) and maternally imprinted regions (where maternal allele is methylated in oocyte: E in Fig. 4).14) In either case, the situation is the same and the Pegs and Megs are reciprocally regulated by the DMR DNA mehylation status.
Figure 4.
Paternal and maternal imprinted regions and their default states of expression. Expression profiles of imprinted genes in somatic cells (A and F), day 12.5 PGC cloned embryos (C), androgenetic (D) and parthenogenetic embryos (E) are illustrated. The black, blue and red circles represent normal biallelic, paternally and maternally expressed genes, respectively. There are two types of imprinted regions, paternally (right blue) and maternally (pink) imprinted regions, where the paternally and maternally-derived DMRs are methylated, respectively. Erasure of imprinted memories occurred in PGCs and the paternal and maternal imprints established during spermatogonia development around the time of birth and oocyte maturation after birth in the male and female germ lines, respectively. Modified from Fig. 2 in ref. 14.
This conclusion is deduced from studies which generated embryos without genomic imprinting memories, such as cloned mice produced from primordial germ cells (PGCs) that give rise to eggs and sperm in females and males, respectively. Imprinted memories (maternal and paternal imprints) are already established in both eggs and sperm as DNA methylation marks on the DMRs, respectively, and are stably maintained after fertilization in the somatic cell lineages. However, these parental memories are erased in the PGCs from embryonic day 9.5 to 12.5 (B in Fig. 4) and reestablished again in oocytes after birth (during oocyte maturation) in females (E in Fig. 4) and in spermatogonia cells just around the time of birth in males (D in Fig. 4).66–70) When using d10.5 PGCs with almost complete parental imprinted memories remained, like somatic cells, as donors, the clones (d10.5 PGC clones) developed to term.71) In contrast, d12.5 PGC clones exhibited early embryonic lethality because they had completely erased imprinted memories (B and C in Fig. 4b).67) In this case, only half of the Pegs and half of the Megs are active, while the other halves are repressed. In explicit detail, only Pegs are active in the maternally imprinted regions where Megs are repressed, and only Megs are active in the paternally imprinted regions where Pegs are repressed in the default state of genomic imprinting (C in Fig. 4).14) It should be noted that the repressed Pegs in the latter regions as well as the repressed Megs in the former regions need paternal and maternal imprints (DNA methylation) for their induction, respectively (D and E in Fig. 4).14,67,72–75) In addition, it is also reported that embryos without any maternally imprinted memory (combination of normal sperm (C in Fig. 4) and oocyte in the default status (B in Fig. 4)) cannot develop to term and exhibit early embryonic lethality in both humans and mice, indicating the genomic imprinting mechanism is essential in the current form of the mammalian developmental system.74,76) It also should be noted that the imprinted gene expression pattern in Fig. 4C is illustrated based on the result of d12.5 PGC cloned embryos,67) and not that of PGCs themselves. However, it seems consistent with the previous results showing that migrating PGCs (d9.5–10.5) basically exhibit monoallelic expression,77) while d11.5 PGCs exhibit biallelic expression.78) Similarly, Figs. 4D and E represent the imprinted gene expression pattern of androgenetic and parthenogenetic embryos,46,48) respectively, and not those of the sperm or oocyte themselves.
2-4. Establishment of genomic imprinting regions during mammalian evolution.
How did genomic imprinting emerge in the course of mammalian evolution? As mentioned previously, the differential DNA methylation of the DMRs is the most critical factor in this regulatory mechanism. Therefore, the emergence of DMRs in the genome must be a pivotal event in the emergence of genomic imprinting in the mammals. How did these regions come to be differentially methylated in the male and female germ lines? Are there any conserved recognition sequences among them? Unfortunately, such sequence homology has never been identified in the DMRs in either the paternally nor the maternally imprinted regions.
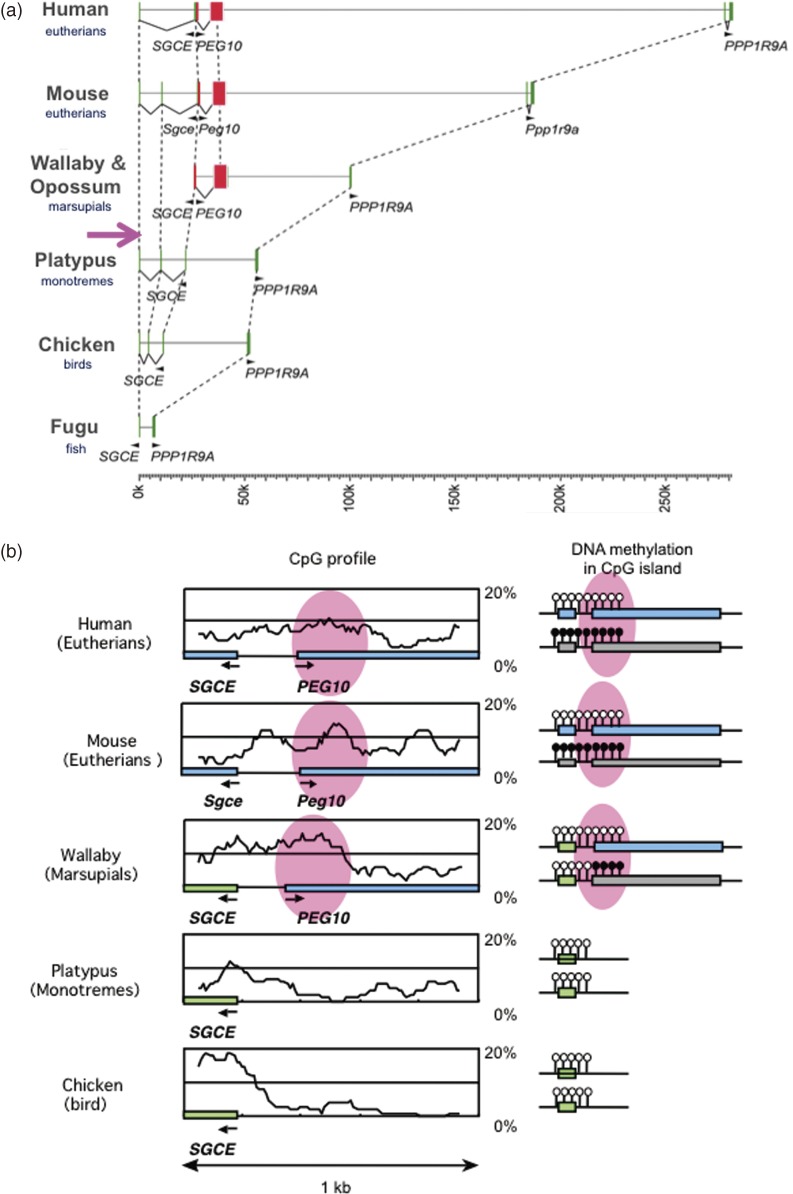
An important clue to a way to overcome this problem came from the analysis performed on PEG10. We found that PEG10 is also present in both Australian and South American marsupial species, it is absent from the platypus, an Australian monotreme species, work done in collaboration with Renfree at the University of Melbourne and Graves at the Australian National University. This demonstrated that the putative PEG10 retrotransposon was inserted into the genome of a common therian ancestor and became an endogenous gene before the divergence of the eutherians and marsupials 160 million years ago (Ma) (Fig. 5a).16) An important fact is that PEG10 is also imprinted and paternally expressed in the tammer wallaby, an Australian marsupial species, and there is a PEG10-DMR overlapped with the PEG10 promoter within the 5′ non-coding region of PEG10 mRNA, suggesting that the PEG10 promoter was derived from an LTR of the ancient retrotransposon from which PEG10 originated. What is clear is that the PEG10-DMR also emerged in the therian ancestor together with PEG10, implying that the DMRs originated from inserted DNA sequences exogenous origin (Figs. 5b and 6). The PEG10 imprinted region is the first imprinted region with a common DMR in the eutherians and marsupials, demonstrating that it is at least one of the oldest imprinted regions in mammalian history.16)
Figure 5.
Emergence of PEG10 in a common ancestor of marsupials and eutherians. a. Comparison of genome sequences among the higher vertebrates. The insertion of the PEG10 retrotransposon occurred after the spilt of the therians and monotremes, and endogenization of the PEG10 gene was completed before the eutherian-marsupial split. b. Emergence of PEG10-DMR from a newly inserted DNA. Modified from Figs. 1 and 5 in ref. 16.
Figure 6.
Emergence of DMRs in imprinted regions in mammals. Paternally and maternally methylated DMRs are shown in blue and red, respectively. The locations of the mouse chromosomes are shown. Only PEG10 and H19-IGF2 DMRs are common to the eutherians and marsupials. *The imprinted regulation of these two regions started in the eutherians associated with chromosome rearrangements near the DMRs. Modified from Fig. 6 in ref. 18.
Another imprinted region that has a common DMR in the therians is the IGF2-H19 region. Reik’s group collaborated with Renfree to show that H19 and H19-DMR exist, and that IGF2 and H19 are reciprocally expressed in the tamer wallaby in the same manner as in eutherian species.17) Although, no information is available on the presence or absence of H19 in the monotremes, it is highly likely that there is no H19 (and H19-DMR) in the platypus, because it was reported that IGF2 is biallelically expressed.79) Therefore, this case further suggests that the H19-DMRs originated from inserted DNA sequences of exogenous origin together with H19 (Fig. 6). In addition, there are also several reports that the small imprinted genes that reside in the introns of other genes, such as Mcts2, Nap1l5, Inpp5f_v2, U2af1-rs1 and Nnat, are thought to have been inserted into their present positions by cDNA retrotransposition.80,81) In every case, the DMR likely emerged as novel CpG ilands (CGI) at the same time as the retrotranspositioning of each gene occurred.
We examined the generality of this hypothesis by reviewing the time of novel CGI emergence for all the maternal DMR loci obtained in collaboration with Renfree. The comprehensive and comparative analyses demonstrated that the emergence of the novel CGIs occurred universally in the maternal DMR loci at different time points during mammalian evolution, presumably in correspondence with the time when the imprinting regulation started in each locus,18,19) and the same seems true for all the three paternal DMRs (Fig. 6).17,53,82) The evidence suggests that the acquisition of the DMRs, such as insertion events from the CpG rich exogenous DNA sequences and also by cDNA retrotransposition, is a common evolutionary pathway for generating novel imprinted regions,16,18,19,80,81) and that genomic imprinting originated as the defense mechanism against the insertion of exogenous DNA into the mammalian genome.4,15)
As mentioned above, imprinting arose at many different time points during mammalian evolution, presumably due to different selective pressures at different loci, and it is still continuing to evolve.19,83) It should be noted that the PEG10 and IGF2-H19 regions are the only two imprinted regions with common DMRs in the therians identified so far,16,17) implying that genomic imprinting was originally related to the prevention of parthenogenetic growth as well as the regulation of the growth of both the placenta and embryo from the beginning, that is, they may be the fundamental biological significance of this mechanism, as initially proposed by the placental hypotheses,4,9,11) complementary hypothesis4,14) and the weak version of the conflict hypothesis,58) despite the great diversity of roles that imprinted genes play at present.
3. LTR retrotransposon-derived genes in mammals
3-1. Evolution of mammalian-specific traits by acquisition of SIRH genes.
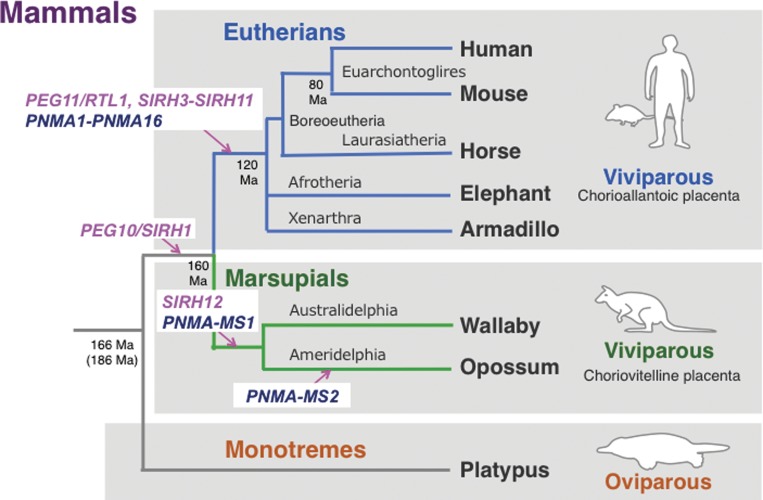
As stated in section 2-2, the acquisition of novel genes took place via a conversion from the LTR retrotransposons to endogenous genes during mammalian evolution. According to the definition originally proposed by Gould and his colleageus,84,85) PEG10 and PEG11/RTL1 are very good examples of “exapted” (domesticated) genes that exhibit novel functions that are different from the original functions in the original organisms. We demonstrated that PEG10 is specific to the therian mammals,16) while Ferguson-Smith’s group demonstrated that PEG11/RTL1 is a eutherian-specific gene that is absent from marsupials.53) These facts clearly demonstrate that PEG10 was acquired after the split of the therians from the monotremes 166 or 186 Ma and before the eutherian/marsurpial split 160 Ma.16,25) Furthermore, PEG11/RTL1 was acquired after the eutherian/marsurpial split and before the split of the three major eutherian lineages, boreoeutheria (including euarchontoglires and laurasiatheria), afrotheria and xenarthra 120 Ma (see Fig. 9).25,53,86) According to the Darwinian theory of evolution, it is highly probable that PEG10 and PEG11/RTL1 were once positively selected and thus presumably contributed to the establishment of the subclasses and infraclasses of the mammals, the therians and the eutherians, respectively.14,24,25) While this supports the previous hypothesis that retrotransposons in the genome provide genetic materials to generate novel genes,85) it has also led to the novel concept that at least in some cases the LTR retrotransposon-derived genes contributed to mammalian evolution, such as the emergence of viviparity.
Figure 9.
Emergence of the SIRH and PNMA genes in two mammalian lineages. Most of the SIRH and PNMA genes are eutherian-specific, with a few being marsupial-specific, while the single exception of PEG10 is conserved in both the eutherians and marsupials. Interestingly, no SIRH or PNMA genes are present in egg-lying montremes. Modified from Fig. 3 in ref. 25.
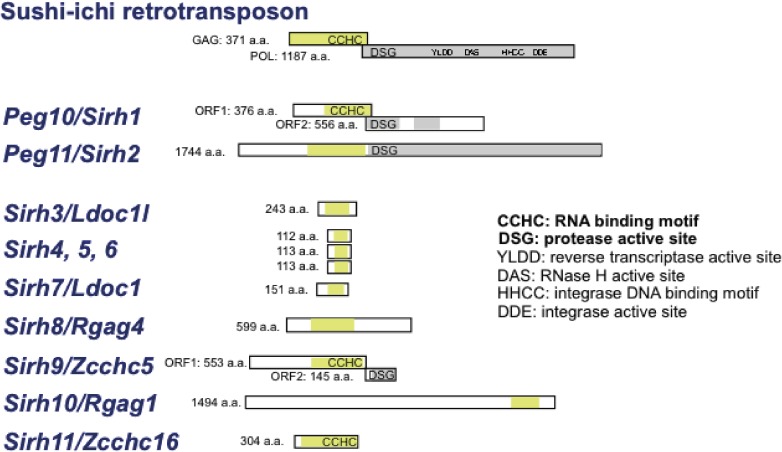
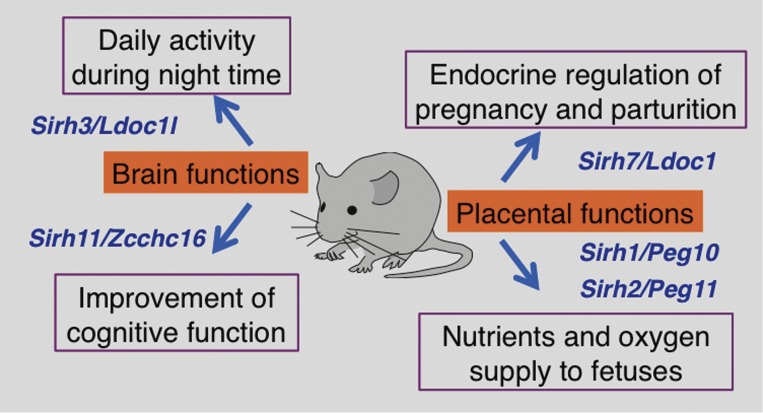
PEG10 and PEG11/RTL1 exhibit a high homology with the same LTR retrotransposon, a sushi-ichi retrotransposon isolated from the pufferfish (Takifugu rubripes).20,21,87,88) Using a sushi-ichi Gag amino acid sequence as a query, genes with high homology were screened from the human and mouse genomes. Eleven genes, including PEG10 and PEG11/RTL1, were identified and named sushi-ichi-related retrotransposon homologue (SIRH)1-11 (also called mammalian retrotransposon-derived (MART) or SUSHI genes.22,63,64) Most of the SIRH genes exhibit homology with Gag, but not with Pol, except for PEG10/SIRH1, PEG11/RTL1/SIRH2 and SIRH9/ZCCHC5 (Zinc finger CCHC domain-containing 5) (Fig. 7). Interestingly, 8 out of the 11 genes, i.e. SIRH4 to SIRH11, locate on the X chromosome. Therefore, they exhibit monoallelic expression like the imprinted genes PEG10/SIRH1 and PEG11/RTL1/SIRH2. A series of knockout mouse experiments on Sirh genes has been carried out in an effort to elucidate their biological functions in development, reproduction, growth and behavior, as well as their presumed contribution to eutherian evolution.
Figure 7.
SIRH family genes in eutherians. Mouse gene examples are shown. These genes are basically conserved in all of the eutherian species, but some may be mutated in a lineage- and species-specific manner.
Female Sirh7/Ldoc1 (leucine zipper, downregulated in cancer 1, also called Mart7) KO mice had structural and related endocrinological problems in the placenta and exhibited delayed parturition along with a low pup weaning rate.26) The placenta is a major endocrine organ during gestation, and in the case of mice, a variety of placental cells, such as giant trophoblast, spongiotrophoblast and cytotrophoblast cells, produce progesterone (P4) and placental lactogen I/II (PL1 and 2) as well as several prolactin-like proteins in order to maintain pregnancy and determine the timing of parturition. Sirh7/Ldoc1 encodes a small Gag-like protein comprising only 151 amino acids, but plays an essential role in controlling the differentiation and maturation of a wide variety of placental cells, thereby regulating both the amount and timing of placental hormones (Fig. 8).26) Thus, Sirh7/Ldoc1 is another essential gene in development as well as reproduction, suggesting that it was also positively selected during the eutherian evolution because of the reproductive advantage it conferred. Together with the results on Peg10 and Peg11/Rtl1 KO mice, this additional evidence provides further support for the critical role of the LTR retrotransposon-derived genes in the evolution of the mammalian viviparous reproduction system (Fig. 8).
Figure 8.
Biological functions of the Sirh genes in mice. The biological functions deduced from five KO mice experiments on Peg10, Peg11/Rtl1, Sirh7/Ldoc1, Sirh3/Ldoc1l and Sirh11/Zcchc16 are shown. Only the major phenotypes of each KO mouse are shown.
In addition to viviparity, another unique characteristic in mammals is higher brain function. Among the SIRH family members, Sirh11/Zcchc16 (also called Mart4) is unique in that it does not exhibit any placental expression during mouse development, but is expressed in the brain, testis, ovary and kidney. Sirh11/Zcchc16 KO mice exhibited abnormal behaviors related to cognition, including attention, impulsivity and working memory, conferring a critically important advantage in both the competition of daily life and eutherian evolution (Fig. 8).89) It also suggests that human SIRH11/ZCCHC16 may be a good candidate for X-linked intellectual disability (XLID). Another example of brain-related phenotypes was observed in Sirh3/Ldoc1l (Ldoc 1 like, also called Mart6) KO mice, such as lower activity during the night period (Irie, in preparation). As mice are nocturnal animals and the same seems likely true for the eutherian ancestors 160 Ma because they lived under the rule of dinosaurs, it is highly probable that acquisition of Sirh3/Ldoc1l in the eutherian ancestors was quite advantageous for survival in the field, thus it was positively selected in eutherian evolution (Fig. 8).
3-2. LTR retrotransposon-derived genes in eutherians and marsupials.
Further comparative genomics demonstrated that all of the SIRH genes (PEG11/RTL1/SIRH2 and SIRH3-11) are eutherian-specific except for PEG10/SIRH1, strongly supporting our hypothesis that the SIRH genes exerted a major impact on therian and eutherian evolution in a variety of ways, but also in a therian- and eutherian-specific manner.24,25) We subsequently found SIRH12 only in marsupial species, such as the tammer wallaby and opossum, an Australian and a South American marsupial species, respectively, although the opossum SIRH12 is degenerated and has surely lost its function (Fig. 9).90)
A similar result was obtained by the analysis of another LTR retrotransposon-derived gene family in mammals, the paraneoplastic Ma antigen (PNMA) family, which exhibits a high homology with gypsey_12 DR retrotransposon isolated from zebra fish (Danio Renio).91–94) At least 19 and 15 PNMA/Pnma genes have been confirmed in humans and mice, respectively, with the difference in number due to rodent-specific deletion of PNMA6A-D.93,95) All of the PNMA genes encode only a Gag-related and not a Pol-related region, and 11 out of 19 are X-linked genes. Moreover, it is also highly likely that they are all eutherian-specific. In contrast, PNMA-MS1 (PNMA marsupial-specific 1) is only present in the marsupial species (both in the tammer wallaby and opossum), while PNMA-MS2 is specific to the opossum (Fig. 9).95) Thus, the number of LTR retrotransposon-derived genes exhibits a difference between the eutharians and marsupials, for example, comprising 11 and 2 genes in the case of the SIRH family, respectively, including one common gene (SIRH1/PEG10), and 19 and 2 genes in the case of the PNMA family, with no common genes.25) The reason for this difference remains unclear, but it should be noted that many pseudogenes are observed in the marsupial genomes and almost none in the eutherian genomes, suggesting that the eutherians have a better ability to use the LTR retrotransposons for novel purposes.90,94) An important conclusion deduced from these results is that the eutherians and marsupials are totally different animal groups in terms of the LTR retrotransposon-derived genes, suggesting that these genes contributed to the diversification of the two viviparous mammalian groups via a generation of eutherian- and marsupial-specific traits like PEG11/RTL1, SIRH7/LDOC1, SIRH3/LDOC1L and SIRH11/ZCCHC16.25) Therefore, analyses on the remaining Sirh KO as well as Pnma KO mice are expected to provide further insight into the impact of the LTR retrotransposons on the acquisition of a series of novel traits during the course of eutherian evolution.
3-3. How did retrotransposons become endogenous genes?
According to the Darwinian theory of evolution, two essential factors in biological evolution are “mutation” and “the environment”. Organisms change their traits by the mutations and the environment select the fittest by means of natural selection. In the light of alteration of the genomic information, the creation of novel genes by a gene/genome duplication mechanism28,29,96,97) is regarded as a “mutation” in addition to the known types of a simple point mutation, insertion or deletion of DNA, and chromosomal rearrangement via homologous and non-homologous recombination as well as crossbreeding between closely related species (homoploid hybrid speciation)98,99) and symbiosis between and/or among different organisms.100–103) It is now evident that the acquisition of novel genes from exogenous DNA, such as LTR retrotransposons/retroviruses, is another important mechanism in the creation of novel genes.20–26,43–45,63,64,88–95) It is an important task to understand how this process has proceeded or is still proceeding in the course of mammalian evolution.
As retrotranposons are usually intrinsically harmful to host organisms, host organisms possess several defense mechanisms for regulating their transcription, thereby preventing their excessive propagation. Mammals use epigenetic mechanisms for this purpose, such as DNA methylation and histone modifications:104) the integrated retrotransposons are usually heavily DNA methylated and transcriptionally silenced in somatic cells72,105) while some are repressed in ES cells, mainly by histone H3K9 methylation.106) As a result, they behave like neutral genes and can be transmitted to the next generation (Fig. 10a).
Figure 10.
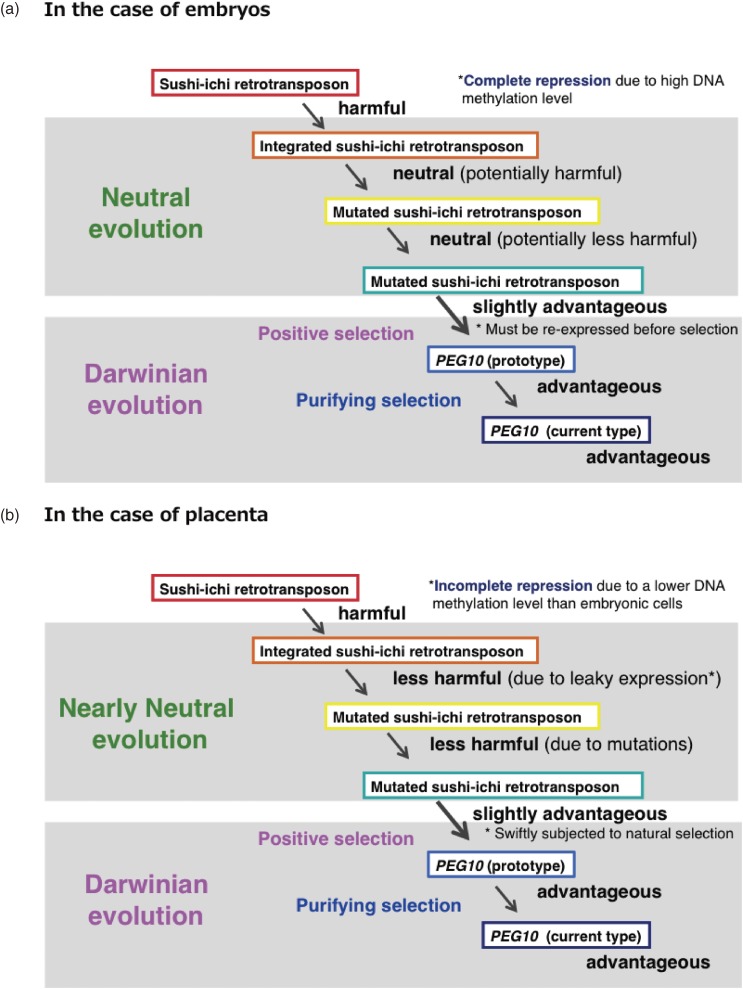
Acquisition of LTR retrotransposon-derived genes (a two-step model). Hypothetical scenarios for the embryonic (a) and extraembryonic tissues (b) are shown. Inserted retroelements are usually fully DNA methylated in somatic cells, while leaky expression of these elements are observed in extraembryonic tissues, such as the placenta, due to the lower DNA methylation level. As a result, they behave as neutral (a) and nearly neutral (less harmful) (b) genes and become fixed in a population according to the mechanisms described in the neutral and nearly neutral theory of molecular evolution, respectively. A key point is the rapid shift to the pressures of Darwinian evolution occurs when they become slightly advantageous.
According to the “neutral theory of molecular evolution” proposed by Kimura, neutral mutations can become fixed in a population by the mechanism of random drift.30,31) Ohta extended this theory and proposed the “nearly neutral theory of molecular evolution” that states that, even nearly neutral (less harmful) mutations can become fixed in a population when the population size is small enough.32) We think it highly probable that the neutral and nearly neutral modes of evolution played an essential background role by inactivating and neutralizing the integrated retrotransposons so as to generate the potential genetic material in the first place. A gradual conversion from being potentially harmful to slightly advantageous must have taken place as the result of multiple mutations of the genes during this period. At this point, Darwinian forces came into play, and a fraction of these genes were selected and became more advantageous by natural selection so as to generate the prototype and present type of novel endogenous genes for the host organisms, and subsequently have been maintained by a purifying selection process (Two step model, Fig. 10a).14,24) However, there are two apparent problems with this hypothetical scenario. First, how is the gene that confers this slightly advantage recognized in somatic cells, since it was completely repressed by DNA methylation? Second, is it possible that random mutation could have generated such a slightly advantageous gene from a potentially harmful gene without selection on multiple occasions?
In extraembryonic tissues, such as the yolk sac and placenta, a leaky expression of retrotransposons and retroviruses occurs because of a lower level of DNA methylation (Fig. 10b). It should be pointed out that this two-step process may be accelerated in the placenta, because the placenta is vulnerable to whether the integrated retrotransposons and its mutated forms turn out to be harmful or advantageous. In such a situation, a swift transition from the first (the state of nearly neutral evolution) to the second step (that of Darwinian evolution) would take place more readily. In this light, the placenta may be regarded as a natural laboratory for mammalian evolution.24,25) It should also be noted that the nearly neutral evolution proposed by Ohta makes a contribution to phenotypical evolution as well as Darwinian evolution, while the neutral evolution originally proposed by Kimua actively contributes to phenotypical evolution, as in the case of gene duplication.30)
This idea is consistent with the fact that the acquired PEG10, PEG11/RTL1 and SIRH7/LDOC1 actually play essential roles in the placenta. However, what about the cases of SIRH3/LDOC1L and SIRH11/ZCCHC16? Sirh3/Ldoc1l expression is observed in the mouse placenta although its major functional site seems to be in the brain, while Sirh11/Zcchc16 does not exhibit any placental expression, as mentioned. It seems that the original SIRH genes in the eutherian ancestors might have some placental function, but subsequently shifted their central expression site to the brain after they became endogenous genes. Alternatively, such SIRH genes might be generated by the gene duplication from other SIRH genes playing a major role in the placenta, with the newly differentiated copy taking on a new role in the brain during evolution.
4. Discussion and future prospects
In this review, we have discussed two themes, genomic imprinting and the LTR retrotransposon-derived genes, as mammalian-specific genomic functions. We would like to also emphasize that genomic imprinting is a very important issue, not only for genomic imprinting diseases, but also for future regenerative medicine and reproductive technology, because genomic imprinting (parental DNA methylation) memories are labile in ES, iPS and even in somatic cells in vitro, although they are stably maintained throughout life in vivo.107–116) Recently, PGC-like cells that give rise to germ cells, such as sperm and oocytes, were reportedly induced from human and mouse iPS cells in vitro.117–121) Morphologically, such oocytes look normal, but it remains unknown whether the imprinted memories are in fact normally erased and reestablished in these artificial processes in vitro. It is well known that both a lack of and abnormally imprinted memories lead to severe developmental failure and genomic imprinting diseases, respectively.74,76) Thus, genomic imprinting issue may be a big hurdle for the application of ES and iPS cells to human regenerative medicine and reproductive technology.
The mechanism of imprint erasure in PGCs is a very important part of mammalian-specific epigenetics. The recent discovery of hydroxylmethyl cytosine (hmC) as a key intermediate of DNA demethylation has completely changed the previous view.122,123) It is now clear that mC is first converted to hmC by the Tet enzyme. However, it is now under intense debate which plays the major role in this process, the passive (dilution) DNA demethylation mechanism via continuous cell divisions under a condition of no hemimethylation activity of DNMT1,124–127) or the active DNA demethylation mechanism proceeded without cell division.128) Alternatively, both may play different but essential roles in this process, respectively. It is possible that the erasing mechanism is much more complex than generally thought.
Another major remaining question with regard to genomic imprinting is how the maternal and paternal DNA methylation imprints are established in the female and male germ cells in a locus-specific manner. The de novo DNA methytransferases DNMT3A and DNMT3B, as well as the co-factor DNMT3L, are major players in the general mechanisms underlying DNA methylation,72–75) but the locus specificity is not explained by this alone. What is the specific targeting mechanism of DNA methylation? Histone H3K4 methylation may be a key involved in this process, because histone H3K4 methylation blocks the association of DNMT3L with the N-terminal of hisone H3, thus preventing the DNMT3A-DNMT3L complexes from accessing DNA.129) Importantly, the deletion of KDMB1, one of histone H3K4 demethylation enzymes, causes the loss of maternal imprints at certain maternal DMRs, presumably due to a substantial increment of H3K4 methylation, but without any abnormality in any of the three paternal and the remaining maternal DMRs.130) These data demonstrate that both DNA methylation and histone modification play crucial roles in establishing the parental imprints and suggest that certain histone modification enzymes function as locus- and parental-specific targeting of the de novo DNA methylation machinery.131)
However, recent genome-wide DNA methylation analyses on oocyte and sperm have presented a completely different perspective on this issue. It is now clear that the oocyte and the sperm are at the outset poorly and highly DNA methylated, respectively, so the difference in the DMR number between the two is far more than that of the known imprinted DMRs.132,133) Although most of the DMRs disappear during the preimplantation period by DNA demethylation reaction, the known imprinted DMRs remaining as a result, indicating that the mechanism of escaping DNA demethylation is critical in the DNA methylation differences between the parental DNAs in somatic cells in postimplantation embryos. Interestingly, DPPA3 (also called STELLA or PGC7) plays a crucial role in protecting certain maternal and paternal DMRs against DNA demethylation,134) although DPPA3 appears to have a general role in protecting the maternal genome against the conversion of 5mC to 5hmC.135)
It should be noted that the interaction between DNA methylation and histone modification also play crucial roles in maintaining the parental imprints in the preimplantation stage.124) The loss of ZFP57, a KRAB domain zinc finger protein, induces somatic loss of DNA methylation at multiple DMRs.136) It functions together with its cofactor, KRAB-associated protein 1 (KAP1, also called TRIM28). ZFP57 brings KAP1 to chromatin, thereby promoting the recruitment of repressive chromatin modifiers, including ESET/SETDB1, which controls H3K9 trimethylation.137,138) ZFP57/KAP1 complexes also interact with DNMTs and the hemimethylated DNA binding protein UHRF1 (also called NP95), which are essential for the maintenance of DNA methylation.139,140) It is also known that maternal KAP1 in the oocyte is essential for the maintenance of DMR methylation at several DMRs in the first round of cell division.141) Thus, many epigenetic factors are known to be involved in the maintenance of the DNA methylation of DMRs in the preimplantation stage, but the elucidation of the mechanisms of the locus- and parental origin-specific DMRs of genomic imprinting awaits further investigation.
In the second part of this review, we discussed the emergence of the LTR retrotransposon-derived genes in mammalian-lineages. From the viewpoint of mammalian evolution, endogenous retroviruses (ERV) may be another important factor. Retroviruses are structurally related to the LTR retrotransposons: both have LTR sequences at both ends and encode GAG and POL proteins, while the former also encodes an additional ENV protein for infection into cells. It is known that ENV genes have also become essential endogenous genes in a lineage-specific manner in the eutherians and marsupials, e.g. several syncytins and FEMATRIN-1, that is, they were acquired independently from different retroviruses.43–45,142–146) The capacity of human syncytin to fuse cells to produce multinucleated cells was first demonstrated in vitro.43) As it exhibits a high level of expression in syncytiotrophoblast cells in the human placenta, it was proposed that syncytin is essential for formation of these cells.43) Out of 18 ENV related DNA sequences in the human genome, only syncytin-1 and -2 that are derived from human-specific ERVs, HERV-W and HERV-FRD, and were integrated into a primate lineage 25 and >40 Ma, respectively, exhibited the fusogenic activity in cell fusion assays.142)
Mice have two syncytin genes, syncytin-A and syncytin-B, derived from Muridae family-specific integrations of HERV-F/H related ERV(s) approximately 20 Ma.143) Importantly, syncytin-A knockout mice exhibit mid-fetal lethality, presumably due to a functional abnormality of one of the two syncytiotrophoblast layers, and double knockout of both syncytin-A and -B was shown to lead to a more severe phenotype and early embryonic lethality, demonstrating that they are clearly essential for syncytiotrophoblast cell formation in vivo, as expected.44,45) Among the eutherians, placental morphology have substantially diverged. Therefore, it is highly probable that other syncytins also have important roles in the placenta in an order- or family-specific manner during the time of mammalian radiation (Fig. 11).
Figure 11.
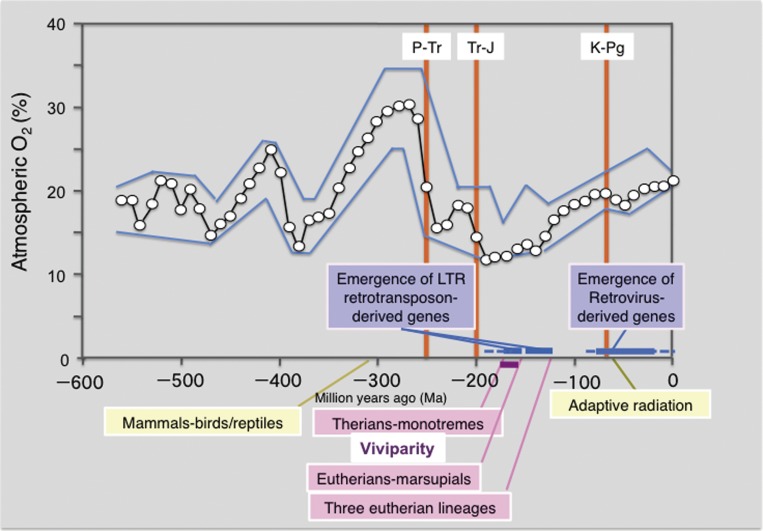
Atmospheric O2 and mammalian evolution. The emergence of the LTR retrotransposon-derived genes and retrovirus-derived genes exhibits close relation with the establishment of viviparous eutherian and marsupial mammals and the radial adaptation of mammals, respectively. The oxygen concentration data are cited from ref. 112: Open circles indicate calculated atmospheric O2 concentration and the upper and lower boundaries are estimates of error in modeling atmospheric O2 concentration. The timing of the divergence between mammals and birds/reptiles (310 Ma), therians and monotremes (186–166 Ma), eutherians and marsupials (160 Ma) and three eutherian lineages (120 Ma), such as boreoeutheria (including euarchontoglires and laurasiatheria), afrotheria and xenarthra, is shown (see Fig. 9). The three major mass extinctions deeply related to mammalian evolution, such as Permian-Triassic (P-Tr: 250 Ma), Triassic-Jurassic (Tr-J: 200 Ma) and Kreide (Cretaceous)-Paleogene (K-Pg: 65 Ma) extinctions are shown.
According to the mammalian genome record, PEG10 is a subclass- and other SIRH and PNMA genes are infraclass-specific genes: PEG10 emerged in the periods when the therian ancestors appeared, and other SIRH and PNMA genes in the eutherian and marsupial ancestors appeared. This is the time when mammalian viviparity had begun and two different types of viviparous systems using chorioallantoic and choriovitelline (yolk sac) placentas were established in the eutherians and marsupials, respectively (Figs. 9 and 11).
What were the environmental condition(s) promoted these radical changes in the mammalian reproductive system? It is reported that in the Phanerozoic eon (from 550 Ma to the present), the highest atmospheric oxygen (30%) was observed in the Carboniferous period 300 Ma, however, it dramatically decreased to 15% around one of five major extinctions that occurred at the Permian-Triassic (P-Tr) boundary 250 Ma (Fig. 11).147) The period with the lowest atmospheric oxygen (12–13%) was estimated to have taken place in a period from 190 to 150 Ma after the next mass extinction of Triassic-Jurassic (Tr-J) boundary approximately 200 Ma. This scenario correlates well with the timing of the divergence of the therians from the monotremes (186 or 166 Ma), and also with the eutherian and marsupial spilt (160 Ma). Therefore, a low oxygen concentration might be a major driving force in the establishment of a prototype for the current viviparous reproductive system because of the advantage of an efficient oxygen supply to embryos via the placental blood circulating system along with an effective nutrient supply. This would promote embryo maturation in the uterus and thus contribute to a higher viable ratio at the perinatal stage. Frequent recruitment of LTR retrotransposon-derived genes in this period may have allowed our therian and eutherian ancestors to undergo an acceleration of this evolutionary process.
It is well known that an adaptive radiation of mammals took place after the extinction of the dinosaurs at the Kreide (Cretaceous)-Paleogene (K-Pg) boundary 65 Ma. At that time, atmospheric oxygen concentration was gradually increased to a point near the present concentration (∼20%) despite a slight lag just after K-Pg mass extinction, therefore, the low oxygen concentration was no longer a major selective pressure. It is highly probable that large vacant ecological niches on both land and sea allowed the ancestors of eutherian and marsupial lineages to become diversified by continuing the acquisition of a variety of retrovirus-derived genes, suggesting that release from powerful selective pressure also serves as a major driving force in macroevolution.30,148)
Interestingly, Koala retrovirus (KoRV) is currently undergoing endogenization in the koala genome and this process possibly started within the last 200 years.149–151) It is reported that KoRV infection has expanded in the Northern Australia Koala population to reportedly cause leukemia, lymphoma and immunosuppression, and the integrated KoRV DNAs are actually transmitted to the next generation in the manner of endogenous genes. Therefore, retrotransposon and retroviral endogenization may be a fairly ordinary process in the long course of evolution. Novel genes may continuously appear by this mechanism, both at present and in the future, although this may depend upon a mechanism of random gene drift such that it remains to be fixed in the koala population. Therefore the strategy we have introduced in this review can also be applied to elucidate human evolution by identifying human-lineage-specific acquired genes derived from the different kinds of retroviruses that have infected us and become endogenized at different stages of the ongoing process toward the primate and finally the human in evolution.
Acknowledgements
We thank all the collaborators and all the laboratory members in both Ishino’s and Kaneko-Ishino’s laboratories, especially, Azim Surani, Marilyn Renfree, Atsuo Ogura, Takashi Kohda, Yoshimi Kuroiwa, Naoki Miyoshi, Shin Kobayashi, Tomohisa Okutsu, Hirotaka Wagatsuma, Takafusa Hikichi, Jiyoung Lee, Ryu-ichi Ono, Masayuki Kai, Yoichi Sekita, Shunsuke Suzuki, Hirosuke Shiura, Daisuke Endo, Noriko Wakisaka, Mie Naruse, Masahito Irie, Hirotaka Iwafune, Sawa Iwasaki, Yuki Kawasaki, Saori Takahashi, Mami Oikawa, Miki Soma and Moe Kitazawa. The work has long been supported by a number of grants, Grants-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan, Creative Science Research from the Japan Society for the Promotion of Science (JSPS), Core Research for Evolutional Science and Technology (CREST) from Japan Science and Technology Agency (JST), Bilateral Program on Joint Research Project from JSPS, Joint Usage/Research Program of Medical Research Institute Tokyo Medical and Dental University (FI and TK-I), Funding Program for Next Generation World-Leading Researchers (NEXT Program) and The Asahi Glass Foundation (TK-I), Precursory Research for Embryonic Science and Technology (PRESTO) from JST and The Uehara Memorial Foundation and The Mitsubishi Foundation (FI).
Abbreviations
- LTR
long terminal repeat
- Peg
paternally expressed gene
- Meg
maternally expressed gene
- ICR
imprinting control center
- DMR
differentially methylated region
- upd14pat
uniparental disomy of paternal chromosome 14
- XCI
X chromosome inactivation
- CTCF
CCCTC-binding factor
- PGC
primordial germ cell
- miRNA
micro RNA
- SINE
short interspersed nuclear element
- LINE
long interspersed nuclear element
- Ma
million years ago
- SIRH
sushi-ichi related retrotransposon homologue
- PNMA
paraneoplastic Ma antigen
- XLID
X-linked intellectual disability
- mC
methyl cytosine
- hmC
hydroxylmethyl cytosine
- HERV
human endogenous retrovirus
- P-Tr
Permian-Triassic
- Tr-J
Triassic-Jurassic
- K-Pg
Kreide-Paleogene
Biographies
Profile
Fumitoshi Ishino was born in Yokohama in 1955 and graduated from the University of Tokyo in 1978. He completed his Ph.D. under the supervision of Prof. Michio Matsuhashi and worked as an Assistant Professor at Institute for Applied Microbiology, the University of Tokyo from 1983 to 1991. His research theme at that time was “genetic and biochemical approaches for elucidation of bacterial cell elongation, morphogenesis and division by analyzing penicillin-binding proteins”. As an Overseas Fellow from the Ministry of Education, he visited Dr. Azim Surani at AFRC Animal Physiology and Genetics Institute in Babraham, Cambridge in UK with his wife, Tomoko Kaneko-Ishino (a British Council Fellow), and started their research on genomic imprinting in mammals. In 1991, he moved to the Gene Research Center, Tokyo Institute for Technology as an Assistant Professor. He became a Professor of Medical Research Institute, Tokyo Medical and Dental University in 2003 and has been the Director of this Institute from 2014. He and Tomoko Kaneko-Ishino have proposed the concept of mammalian-specific genes from LTR retrotransposons playing essential roles in mammalian development and growth from a series of their knockout mouse studies.
Tomoko Kaneko-Ishino was born in Tokyo in 1956 and graduated from the University of Tokyo in 1980. She completed her Ph.D. under the supervision of Prof. Michio Oishi at Institute for Applied Microbiology, the University of Tokyo. Her research theme was “Molecular mechanism on terminal differentiation of Friend cells”. After she worked as a researcher in the Industrial Research Institute, Japan, she studied in the Dr. Azim Surani’s laboratory at AFRC Animal Physiology and Genetics Institute in Babraham, Cambridge in UK as a British Council Fellow. Together with her husband, Fumitoshi Ishino, she started her research on genomic imprinting in mammals. After 4 years in the Gene Research Center, Tokyo Institute for Technology, she moved to Tokai University, School of Health Sciences, as an Associate Professor in 1995 and became a Professor in 1999. She and Fumitoshi Ishino have proposed the concept of mammalian-specific genes from LTR retrotransposons playing essential roles in mammalian development and growth from a series of their knockout mouse studies.
References
- 1).Surani M.A., Barton S.C., Norris M.L. (1984) Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550. [DOI] [PubMed] [Google Scholar]
- 2).McGrath J., Solter D. (1984) Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183. [DOI] [PubMed] [Google Scholar]
- 3).Mann J.R., Lovell-Badge R.H. (1984) Inviability of parthenogenones is determined by pronuclei, not egg cytoplasm. Nature 310, 66–67. [DOI] [PubMed] [Google Scholar]
- 4).Kaneko-Ishino T., Kohda T., Ishino F. (2003) The regulation and biological significance of genomic imprinting in mammals. J. Biochem. 133, 699–711. [DOI] [PubMed] [Google Scholar]
- 5).Barlow D.P., Bartolomei M.S. (2004) Genomic imprinting in Mammals. Cold Spring Harb. Symp. Quant. Biol. 69, 357–375. [DOI] [PubMed] [Google Scholar]
- 6).Blake A., Pickford K., Greenaway S., Thomas S., Pickard A., Williamson C.M., Adams N.C., Walling A., Beck T., Fray M., Peters J., Weaver T., Brown S.D.M., Hancock J.M., Mallon A.-M. (2010) MouseBook: an integrated portal of mouse resources. Nucleic Acids Res. 38 (suppl 1), D593–D599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Cattanach B.M., Kirk M. (1985) Differential activity of maternally and paternally derived chromosome regions in mice. Nature 315, 496–498. [DOI] [PubMed] [Google Scholar]
- 8).Cattanach B.M., Beechey C.V. (1990) Autosomal and X-chromosome imprinting. Development 108 Suppl., 63–72. [PubMed] [Google Scholar]
- 9).Solter D. (1988) Differential imprinting and expression of maternal and paternal genomes. Annu. Rev. Genet. 22, 127–146. [DOI] [PubMed] [Google Scholar]
- 10).Varmuza S., Mann M. (1994) Genomic imprinting–defusing the ovarian time bomb. Trends Genet. 10, 118–123. [DOI] [PubMed] [Google Scholar]
- 11).Hall J.G. (1990) Genomic imprinting: review and relevance to human diseases. Am. J. Hum. Genet. 46, 857–873. [PMC free article] [PubMed] [Google Scholar]
- 12).Bell A.C., Felsenfeld G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485. [DOI] [PubMed] [Google Scholar]
- 13).Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489. [DOI] [PubMed] [Google Scholar]
- 14).Kaneko-Ishino T., Kohda T., Ono R., Ishino F. (2006) Complementation hypothesis: the necessity of a monoallelic gene expression mechanism in mammalian development. Cytogenet. Genome Res. 113, 24–30. [DOI] [PubMed] [Google Scholar]
- 15).Barlow D.P. (1993) Methylation and imprinting: from host defense to gene regulation? Science 260, 309–310. [DOI] [PubMed] [Google Scholar]
- 16).Suzuki S., Ono R., Narita T., Pask A.J., Shaw G., Wang C., Kohda T., Alsop A.E., Graves M.J.A., Kohara Y., Ishino F., Renfree M.B., Kaneko-Ishino T. (2007) Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Smits G., Mungall A.J., Griffiths-Jones S., Smith P., Beury D., Matthews L., Rogers J., Pask A.J., Shaw G., VandeBerg J.L., McCarrey J.R., SAVOIR Consortium. Renfree M.B., Reik W., Dunham I. (2008) Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat. Genet. 40, 971–976. [DOI] [PubMed] [Google Scholar]
- 18).Suzuki S., Shaw G., Kaneko-Ishino T., Ishino F., Renfree M.B. (2011) The evolution of mammalian genomic imprinting was accompanied by the acquisition of novel CpG islands. Genome Biol. Evol. 3, 1276–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Renfree M.B., Suzuki S., Kaneko-Ishino T. (2013) The origin and evolution of genomic imprinting and viviparity in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Ono R., Kobayashi S., Wagatsuma H., Aisaka K., Kohda T., Kaneko-Ishino T., Ishino F. (2001) A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237. [DOI] [PubMed] [Google Scholar]
- 21).Charlier C., Segers K., Wagenaar D., Karim L., Berghmans S., Jaillon O., Shay T., Weissenbach J., Cockett N., Gyapay G., Georges M. (2001) Human–Ovine Comparative Sequencing of a 250-kb Imprinted Domain Encompassing the Callipyge (clpg) Locus and Identification of Six Imprinted Transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 11, 850–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Ono R., Nakamura K., Inoue K., Naruse M., Usami T., Wakisaka-Saito N., Hino T., Suzuki-Migishima R., Ogonuki N., Miki H., Kohda T., Ogura A., Yokoyama M., Kaneko-Ishino T., Ishino F. (2006) Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106. [DOI] [PubMed] [Google Scholar]
- 23).Sekita Y., Wagatsuma H., Nakamura K., Ono R., Kagami M., Wakisaka N., Hino T., Suzuki-Migishima R., Kohda T., Ogura A., Ogata T., Yokoyama M., Kaneko-Ishino T., Ishino F. (2008) Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40, 243–248. [DOI] [PubMed] [Google Scholar]
- 24).Kaneko-Ishino T., Ishino F. (2010) Retrotransposon silencing by DNA methylation contributed to the evolution of placentation and genomic imprinting in mammals. Dev. Growth Differ. 52, 533–543. [DOI] [PubMed] [Google Scholar]
- 25).Kaneko-Ishino T., Ishino F. (2012) The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front. Microbiol. 3, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Naruse M., Ono R., Irie M., Nakamura K., Furuse T., Hino T., Oda K., Kashimura M., Yamada I., Wakana S., Yokoyama M., Ishino F., Kaneko-Ishino T. (2014) Sirh7/Ldoc1 knockout mice exhibit placental P4 overproduction and delayed parturition. Development 141, 4763–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Rawn S.M., Cross J.C. (2008) The evolution, regulation, and function of placenta-specific genes. Annu. Rev. Cell Dev. Biol. 24, 159–181. [DOI] [PubMed] [Google Scholar]
- 28).Ohno, S. (1970) Evolution by Gene Duplication. Springer-Verlag, Berlin-Heidelberg-New York, pp. 1–2, 59–88. [Google Scholar]
- 29).Ohno S. (1999) Gene duplication and the uniqueness of vertebrate genomes circa 1970–1999. Semin. Cell Dev. Biol. 10, 517–522. [DOI] [PubMed] [Google Scholar]
- 30).Kimura, M. (1983) The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, pp. 98–116, 305–327. [Google Scholar]
- 31).Kimura M. (1968) Evolutionary rate at the molecular level. Nature 217, 624–626. [DOI] [PubMed] [Google Scholar]
- 32).Ohta T. (2002) Near-neutrality in evolution of genes and gene regulation. Proc. Natl. Acad. Sci. U.S.A. 99, 16134–16137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Lyon M.F. (1963) Lyonization of the X chromosome. Lancet 2, 1120–1121. [DOI] [PubMed] [Google Scholar]
- 34).Lyon M.F. (1986) X chromosomes and dosage compensation. Nature 320, 313. [DOI] [PubMed] [Google Scholar]
- 35).Crouse H.V., Brown A., Mumford B.C. (1971) –chromosome inheritance and the problem of chromosome “imprinting” in Sciara (Sciaridae, Diptera). Chromosoma 34, 324–398. [DOI] [PubMed] [Google Scholar]
- 36).Alleman M., Doctor J. (2000) Genomic imprinting in plants: observations and evolutionary implications. Plant Mol. Biol. 43, 147–161. [DOI] [PubMed] [Google Scholar]
- 37).International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431, 931–945. [DOI] [PubMed] [Google Scholar]
- 38).Mouse Genome Sequencing Consortium (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 39).Mikkelsen T.S., et al. (2007) Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447, 167–177. [DOI] [PubMed] [Google Scholar]
- 40).Warren W.C., et al. (2008) Genome analysis of the platypus reveals unique signatures of evolution. Nature 453, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41).Renfree M.B., et al. (2011) Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 12, R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42).International Chicken Genome Sequencing Consortium (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716. [DOI] [PubMed] [Google Scholar]
- 43).Mi S., Lee X., Li X.-P., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.-Y., Edouard P., Howes S., Keith J.C., Jr., McCoy J.M. (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785–789. [DOI] [PubMed] [Google Scholar]
- 44).Dupressoir A., Vernochet C., Bawa O., Harper F., Pierron G., Opolon P., Heidmann T. (2009) Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. U.S.A. 106, 12127–12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Dupressoir A., Vernochet C., Harper F., Guégan J., Dessen P., Pierron G., Heidmann T. (2011) A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. U.S.A. 108, 1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Kaneko-Ishino T., Kuroiwa Y., Miyoshi N., Kohda T., Suzuki R., Yokoyama M., Viville S., Barton S.C., Ishino F., Surani A. (1995) Peg1/Mest imprinted gene on Chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 11, 52–59. [DOI] [PubMed] [Google Scholar]
- 47).Kuroiwa Y., Kaneko-Ishino T., Kagitani F., Kohda T., Li L.-L., Tada M., Suzuki R., Yokoyama M., Shiroishi T., Wakana S., Barton S.C., Ishino F., Surani A. (1996) Peg3 imprinted gene on proximal chromosome 7, encodes for a zinc finger protein. Nat. Genet. 12, 186–190. [DOI] [PubMed] [Google Scholar]
- 48).Miyoshi N., Kuroiwa Y., Kohda T., Shitara H., Yonekawa H., Kawabe T., Hasegawa H., Barton S.C., Surani M.A., Kaneko-Ishino T., Ishino F. (1998) Identification of the Meg1/Grb10 imprinted gene on mouse proximal chromosome 11, a candidate for the Silver-Russell syndrome gene. Proc. Natl. Acad. Sci. U.S.A. 95, 1102–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49).Miyoshi N., Wagatsuma H., Wakana S., Shiroishi T., Nomura M., Aisaka K., Kohda T., Surani M.A., Kaneko-Ishino T., Ishino F. (2000) Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5, 211–220. [DOI] [PubMed] [Google Scholar]
- 50).Georgiades P., Watkins M., Surani M.A., Ferguson-Smith A.C. (2000) Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 127, 4719–4728. [DOI] [PubMed] [Google Scholar]
- 51).Kagami M., Kurosawa K., Miyazaki O., Ishino F., Matsuoka K., Ogata T. (2015) Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami-Ogata syndrome). Eur. J. Hum. Genet. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52).Kagami M., Sekita Y., Nishimura G., Irie M., Kato F., Okada M., Yamamori S., Kishimoto H., Nakayama M., Tanaka Y., Matsuoka K., Takahashi T., Noguchi M., Tanaka Y., Masumoto K., Utsunomiya T., Kouzan H., Komatsu Y., Ohashi H., Kurosawa K., Kosaka K., Ferguson-Smith A.C., Ishino F., Ogata T. (2008) Deletions and epimutations affecting the human chromosome 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat. Genet. 40, 237–242. [DOI] [PubMed] [Google Scholar]
- 53).Edwards C.A., Mungall A.J., Matthews L., Ryder E., Gray D.J., Pask A.J., Shaw G., Graves J.A., Rogers J., SAVOIR consortium. Dunham I., Renfree M.B., Ferguson-Smith A.C. (2008) The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 6, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).DeChiara T.M., Efstratiadis A., Robertson E.J. (1990) A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345, 78–80. [DOI] [PubMed] [Google Scholar]
- 55).DeChiara T.M., Robertson E.J., Efstratiadis A. (1991) Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859. [DOI] [PubMed] [Google Scholar]
- 56).Barlow D.P., Stoger R., Herrmann B.G., Saito K., Schweifer N. (1991) The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349, 84–87. [DOI] [PubMed] [Google Scholar]
- 57).Bartolomei M.S., Zemel S., Tilghman S.M. (1991) Parental imprinting of the mouse H19 gene. Nature 351, 153–155. [DOI] [PubMed] [Google Scholar]
- 58).Moore T., Haig D. (1991) Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7, 45–49. [DOI] [PubMed] [Google Scholar]
- 59).Hayashizaki Y., Shibata H., Hirotsune S., Sugino H., Okazaki Y., Sasaki N., Hirose K., Imoto H., Okuizumi H., Muramatsu M., Komatsubara H., Shiroishi T., Moriwaki K., Katsuki M., Chapman V.M. (1994) Identification of an imprinted U2af binding-protein-related sequence on mouse chromosome 11 using the RLGS method. Nat. Genet. 6, 33–40. [DOI] [PubMed] [Google Scholar]
- 60).Hagiwara Y., Hirai M., Nishiyama K., Kanazawa I., Ueda T., Sakaki Y., Ito T. (1997) Screening for imprinted genes by allelic message display: identification of a paternally expressed gene impact on mouse chromosome 18. Proc. Natl. Acad. Sci. U.S.A. 94, 9249–9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).Piras G., El Kharroubi A., Kozlov S., Escalante-Alcalde D., Hernandez L., Copeland N.G., Gilbert D.J., Jenkins N.A., Stewart C.L. (2000) Zac1 (Lot1), a potential tumor suppressor gene, and the gene for epsilon-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol. Cell. Biol. 20, 3308–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62).Seitz H., Youngson N., Lin S.P., Dalbert S., Paulsen M., Bachellerie J.P., Ferguson-Smith A.C., Cavaillé J. (2003) Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34, 261–262. [DOI] [PubMed] [Google Scholar]
- 63).Brandt J., Schrauth S., Veith A.M., Froschauer A., Haneke T., Schultheis C., Gessler M., Leimeister C., Volff J.N. (2005) Transposable elements as a source of genetic innovation: expression and evolution of a family of retrotransposon-derived neogenes in mammals. Gene 345, 101–111. [DOI] [PubMed] [Google Scholar]
- 64).Youngson N.A., Kocialkowski S., Peel N., Ferguson-Smith A.C. (2005) A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 61, 481–490. [DOI] [PubMed] [Google Scholar]
- 65).Davis E., Caiment F., Tordoir X., Cavaillé J., Ferguson-Smith A., Cockett N., Georges M., Charlier C. (2005) RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 15, 743–749. [DOI] [PubMed] [Google Scholar]
- 66).Ueda T., Abe K., Miura A., Yuzuriha M., Zubair M., Noguchi M., Niwa K., Kawase Y., Kono T., Matsuda Y., Fujimoto H., Shibata H., Hayashizaki Y., Sasaki H. (2000) The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells 5, 649–659. [DOI] [PubMed] [Google Scholar]
- 67).Lee J., Inoue K., Ono R., Ogonuki N., Kohda T., Kaneko-Ishino T., Ogura A., Ishino F. (2002) Erasing genomic imprinting memory in mouse clone embryos produced from day-11.5 primordial germ cells. Development 129, 1807–1817. [DOI] [PubMed] [Google Scholar]
- 68).Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M.A. (2002) Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117, 15–23. [DOI] [PubMed] [Google Scholar]
- 69).Obata Y., Kono T. (2002) Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J. Biol. Chem. 277, 5285–5289. [DOI] [PubMed] [Google Scholar]
- 70).Kamimura S., Hatanaka Y., Hirasawa R., Matsumoto K., Oikawa M., Lee J., Matoba S., Mizutani E., Ogonuki N., Inoue K., Kohda T., Ishino F., Ogura A. (2014) Establishment of paternal genomic imprinting in mouse prospermatogonia analyzed by nuclear transfer. Biol. Reprod. 91, 120. [DOI] [PubMed] [Google Scholar]
- 71).Miki H., Inoue K., Kohda T., Honda A., Ogonuki N., Yuzuriha M., Mise N., Matsui Y., Baba T., Abe K., Ishino F., Ogura A. (2005) Birth of mice produced by germ cell nuclear transfer. Genesis 41, 81–86. [DOI] [PubMed] [Google Scholar]
- 72).Okano M., Bell D.W., Haber D.A., Li E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 73).Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. (2004) Essential role for de novo DNA methyltransperase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903. [DOI] [PubMed] [Google Scholar]
- 74).Bourc’his D., Xu G.L., Lin C.S., Bollman B., Bestor T.H. (2001) Dnmt3l and the establishment of maternal genomic imprints. Science 294, 2536–2539. [DOI] [PubMed] [Google Scholar]
- 75).Hata K., Okano M., Lei H., Li E. (2002) Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129, 1983–1993. [DOI] [PubMed] [Google Scholar]
- 76).Judson H., Hayward B.E., Sheridan E., Bonthron D.B. (2002) A global disorder of imprinting in the human female germ line. Nature 416, 539–542. [DOI] [PubMed] [Google Scholar]
- 77).Szabó P.E., Mann J.R. (1995) Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 9, 1857–1868. [DOI] [PubMed] [Google Scholar]
- 78).Szabó P.E., Hübner K., Schöler H., Mann J.R. (2002) Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech. Dev. 115, 157–160. [DOI] [PubMed] [Google Scholar]
- 79).Killian J.K., Nolan C.M., Stewart N., Munday B.L., Andersen N.A., Nicol S., Jirtle R.L. (2001) Monotreme IGF2 expression and ancestral origin of genomic imprinting. J. Exp. Zool. 291, 205–212. [DOI] [PubMed] [Google Scholar]
- 80).Evans H.K., Weidman J.R., Cowley D.O., Jirtle R.L. (2005) Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene. Mol. Biol. Evol. 22, 1740–1748. [DOI] [PubMed] [Google Scholar]
- 81).Wood A.J., Roberts R.G., Monk D., Moore G.E., Schulz R., Oakey R.J. (2007) A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 3, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82).Watanabe T., Tomizawa S., Mitsuya K., Totoki Y., Yamamoto Y., Kuramochi-Miyagawa S., Iida N., Hoki Y., Murphy P.J., Toyoda A., Gotoh K., Hiura H., Arima T., Fujiyama A., Sado T., Shibata T., Nakano T., Lin H., Ichiyanagi K., Soloway P.D., Sasaki H. (2011) Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332, 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83).Das R., Anderson N., Koran M.I., Weidman J.R., Mikkelsen T.S., Kamal M., Murphy S.K., Linblad-Toh K., Greally J.M., Jirtle R.L. (2012) Convergent and divergent evolution of genomic imprinting in the marsupial Monodelphis domestica. BMC Genomics 13, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84).Gould S.J., Vrba E.S. (1982) Exaptation; a missing term in the science of form. Paleobiology 8, 4–15. [Google Scholar]
- 85).Brosius J., Gould S.J. (1992) On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc. Natl. Acad. Sci. U.S.A. 89, 10706–10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86).Nishihara H., Maruyama S., Okada N. (2009) Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc. Natl. Acad. Sci. U.S.A. 106, 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87).Poulter R., Butler M. (1998) A retrotransposon family from the pufferfish (fugu) Fugu rubripes. Gene 215, 241–249. [DOI] [PubMed] [Google Scholar]
- 88).Volff J., Körting C., Schartl M. (2001) Ty3/Gypsy retrotransposon fossils in mammalian genomes: did they evolve into new cellular functions? Mol. Biol. Evol. 18, 266–270. [DOI] [PubMed] [Google Scholar]
- 89).Irie M., Yoshikawa M., Ono R., Iwafune H., Furuse T., Yamada I., Wakana S., Yamashita Y., Abe T., Ishino F., Kaneko-Ishino T. (2015) Cognitive function related to the Sirh11/Zcchc16 gene acquired from an LTR retrotransposon in eutherians. PLoS Genet. 11, e1005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90).Ono R., Kuroki Y., Naruse M., Ishii M., Iwasaki S., Toyoda A., Fujiyama A., Shaw G., Renfree M.B., Kaneko-Ishino T., Ishino F. (2011) Identification of SIRH12, a retrotransposon-derived gene specific to marsupial mammals. DNA Res. 18, 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91).Voltz R., Gultekin S.H., Rosenfeld M.R., Gerstner E., Eichen J., Posner J.B., Dalmau J. (1999) A serologic marker of paraneoplastic limbic and brain-stem encephalitis in patients with testicular cancer. N. Engl. J. Med. 340, 1788–1795. [DOI] [PubMed] [Google Scholar]
- 92).Rosenfeld M.R., Eichen J.G., Wade D.F., Posner J.B., Dalmau J. (2001) Molecular and clinical diversity in paraneoplastic immunity to Ma proteins. Ann. Neurol. 50, 339–348. [PubMed] [Google Scholar]
- 93).Schüller M., Jenne D., Voltz R. (2005) The human PNMA family: Novel neuronal proteins implicated in paraneoplastic neurological disease. J. Neuroimmunol. 169, 172–176. [DOI] [PubMed] [Google Scholar]
- 94).Campillos M., Doerks T., Shah P.K., Bork P. (2006) Computational characterization of multiple Gag-like human proteins. Trends Genet. 22, 585–589. [DOI] [PubMed] [Google Scholar]
- 95).Iwasaki S., Suzuki S., Clark H., Ono R., Shaw G., Renfree M.B., Kaneko-Ishino T., Ishino F. (2013) Identification of novel PNMA-MS1 in marsupials suggests LTR retrotransposon-derived PNMA genes differently expanded in marsupials and eutherians. DNA Res. 20, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96).Putnam N.H., et al. (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071. [DOI] [PubMed] [Google Scholar]
- 97).Smith J.J., et al. (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 45, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98).Rieseberg L.H., Raymond O., Rosenthal D.M., Lai Z., Livingstone K., Nakazato T., Durphy J.L., Schwarzbach A.E., Donovan L.A., Lexer C. (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301, 1211–1216. [DOI] [PubMed] [Google Scholar]
- 99).Mavárez J., Salazar C.A., Bermingham E., Salcedo C., Jiggins C.D., Linares M. (2006) Speciation by hybridization in Heliconius butterflies. Nature 441, 868–871. [DOI] [PubMed] [Google Scholar]
- 100).Margulis, L. (1981) Symbiosis in Cell Evolution. W. H. Freeman and Company, New York. [Google Scholar]
- 101).Margulis, L. and Fester, R. (1991) Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. The MIT Press, Cambridge. [PubMed] [Google Scholar]
- 102).Pierce S.K., Curtis N.E. (2012) Cell biology of the chloroplast symbiosis in sacoglossan sea slugs. Int. Rev. Cell. Mol. Biol. 293, 123–148. [DOI] [PubMed] [Google Scholar]
- 103).Márquez L.M., Redman R.S., Rodriguez R.J., Roossinck M.J. (2007) A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315, 513–515. [DOI] [PubMed] [Google Scholar]
- 104).Rowe H.M., Trono D. (2011) Dynamic control of endogenous retroviruses during development. Virology 411, 273–287. [DOI] [PubMed] [Google Scholar]
- 105).Walsh C.P., Chaillet J.R., Bestor T.H. (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117. [DOI] [PubMed] [Google Scholar]
- 106).Matsui T., Leung D., Miyashita H., Maksakova I.A., Miyachi H., Kimura H., Tachibana M., Lorincz M.C., Shinkai Y. (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931. [DOI] [PubMed] [Google Scholar]
- 107).Eversole-Cire P., Ferguson-Smith A.C., Sasaki H., Brown K.D., Cattanach B.M., Gonzales F.A., Surani M.A., Jones P.A. (1993) Activation of an imprinted Igf 2 gene in mouse somatic cell cultures. Mol. Cell. Biol. 13, 4928–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108).Dean W., Bowden L., Aitchison A., Klose J., Moore T., Meneses J.J., Reik W., Feil R. (1998) Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 125, 2273–2282. [DOI] [PubMed] [Google Scholar]
- 109).Humpherys D., Eggan K., Akutsu H., Hochedlinger K., Rideout W.M., Biniszkiewicz D., Yanagimachi R., Jaenisch R. (2001) Epigenetic instability in ES cells and cloned mice. Science 293, 95–97. [DOI] [PubMed] [Google Scholar]
- 110).Humpherys D., Eggan K., Akutsu H., Friedman A., Hochedlinger K., Yanagimachi R., Lander E.S., Golub T.R., Jaenisch R. (2002) Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc. Natl. Acad. Sci. U.S.A. 99, 12889–12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111).Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. (2010) Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112).Rugg-Gunn P.J., Ferguson-Smith A.C., Pedersen R.A. (2007) Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum. Mol. Genet. 16, R243–R251. [DOI] [PubMed] [Google Scholar]
- 113).Kim K.P., Thurston A., Mummery C., Ward-van Oostwaard D., Priddle H., Allegrucci C., Denning C., Young L. (2007) Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 17, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114).Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., Popp C., Thienpont B., Dean W., Reik W. (2012) The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115).Dawlaty M.M., Breiling A., Le T., Raddatz G., Barrasa M.I., Cheng A.W., Gao Q., Powell B.E., Li Z., Xu M., Faull K.F., Lyko F., Jaenisch R. (2013) Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 24, 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116).Yamaguchi S., Shen L., Liu Y., Sendler D., Zhang Y. (2013) Role of Tet1 in erasure of genomic imprinting. Nature 504, 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117).Nakaki F., Hayashi K., Ohta H., Kurimoto K., Yabuta Y., Saitou M. (2013) Induction of mouse germ-cell fate by transcription factors in vitro. Nature 501, 222–226. [DOI] [PubMed] [Google Scholar]
- 118).Hayashi K., Saitou M. (2013) Generation of eggs from mouse embryonic stem cells and induced pluripotent stem cells. Nat. Protoc. 8, 1513–1524. [DOI] [PubMed] [Google Scholar]
- 119).Irie N., Weinberger L., Tang W.W., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J.H., Surani M.A. (2015) SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120).Kurimoto K., Yabuta Y., Hayashi K., Ohta H., Kiyonari H., Mitani T., Moritoki Y., Kohri K., Kimura H., Yamamoto T., Katou Y., Shirahige K., Saitou M. (2015) Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell Stem Cell 16, 517–532. [DOI] [PubMed] [Google Scholar]
- 121).Sasaki K., Yokobayashi S., Nakamura T., Okamoto I., Yabuta Y., Kurimoto K., Ohta H., Moritoki Y., Iwatani C., Tsuchiya H., Nakamura S., Sekiguchi K., Sakuma T., Yamamoto T., Mori T., Woltjen K., Nakagawa M., Yamamoto T., Takahashi K., Yamanaka S., Saitou M. (2015) Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 17, 178–194. [DOI] [PubMed] [Google Scholar]
- 122).Kriaucionis S., Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123).Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124).Hackett J.A., Sengupta R., Zylicz J.J., Murakami K., Lee C., Down T.A., Surani M.A. (2013) Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339, 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125).Yamaguchi S., Hong K., Liu R., Inoue A., Shen L., Zhang K., Zhang Y. (2013) Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 23, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126).Kagiwada S., Kurimoto K., Hirota T., Yamaji M., Saitou M. (2013) Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. EMBO J. 32, 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127).Ohno R., Nakayama M., Naruse C., Okashita N., Takano O., Tachibana M., Asano M., Saitou M., Seki Y. (2013) A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development 140, 2892–2903. [DOI] [PubMed] [Google Scholar]
- 128).Kawasaki Y., Lee J., Matsuzawa A., Kohda T., Kaneko-Ishino T., Ishino F. (2014) Active DNA demethylation is required for complete imprint erasure in primordial germ cells. Sci. Rep. 4, 3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129).Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.P., Allis C.D., Cheng X., Bestor T.H. (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130).Ciccone D.N., Su H., Hevi S., Gay F., Lei H., Bajko J., Xu G., Li E., Chen T. (2009) KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418. [DOI] [PubMed] [Google Scholar]
- 131).Kelsey G., Feil R. (2013) New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132).Smallwood S.A., Tomizawa S., Krueger F., Ruf N., Carli N., Segonds-Pichon A., Sato S., Hata K., Andrews S.R., Kelsey G. (2011) Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133).Kobayashi H., Sakurai T., Imai M., Takahashi N., Fukuda A., Yayoi O., Sato S., Nakabayashi K., Hata K., Sotomaru Y., Suzuki Y., Kono T. (2012) Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 8, e1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134).Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Tanaka S., Shiota K., Nakano T. (2007) PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9, 64–71. [DOI] [PubMed] [Google Scholar]
- 135).Wossidlo M., Nakamura T., Lepikhov K., Marques C.J., Zakhartchenko V., Boiani M., Arand J., Nakano T., Reik W., Walter J. (2011) 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241. [DOI] [PubMed] [Google Scholar]
- 136).Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P., Ferguson-Smith A.C. (2008) A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137).Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S., Baglivo I., Pedone P.V., Grimaldi G., Riccio A., Trono D. (2011) In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 44, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138).Yuan P., Han J., Guo G., Orlov Y.L., Huss M., Loh Y.H., Yaw L.P., Robson P., Lim B., Ng H.H. (2009) Eset partners with Oct4 to restrict extraembryonic trophoblast lineage potential in embryonic stem cells. Genes Dev. 23, 2507–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139).Sharif J., Muto M., Takebayashi S., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., Tajima S., Mitsuya K., Okano M., Koseki H. (2007) The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. [DOI] [PubMed] [Google Scholar]
- 140).Bostick M., Kim J.K., Esteve P.O., Clark A., Pradhan S., Jacobsen S.E. (2007) UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764. [DOI] [PubMed] [Google Scholar]
- 141).Messerschmidt D.M., de Vries W., Ito M., Solter D., Ferguson-Smith A., Knowles B.B. (2012) Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science 335, 1499–1502. [DOI] [PubMed] [Google Scholar]
- 142).Blaise S., de Parseval N., Bénit L., Heidmann T. (2003) Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 13013–13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143).Dupressoir A., Vernochet C., Harper F., Guégan J., Dessen P., Pierron G., Heidmann T. (2011) A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. U.S.A. 108, 1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144).Lavialle C., Cornelis G., Dupressoir A., Esnault C., Heidmann O., Vernochet C., Heidmann T. (2013) Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145).Cornelis G., Vernochet C., Carradec Q., Souquere S., Mulot B., Catzeflis F., Nilsson M.A., Menzies B.R., Renfree M.B., Pierron G., Zeller U., Heidmann O., Dupressoir A., Heidmann T. (2015) Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc. Natl. Acad. Sci. U.S.A. 112, E487–E496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146).Nakaya Y., Koshi K., Nakagawa S., Hashizume K., Miyazawa T. (2013) Fematrin-1 is involved in fetomaternal cell-to-cell fusion in Bovinae placenta and has contributed to diversity of ruminant placentation. J. Virol. 87, 10563–10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147).Berner R.A., Vandenbrooks J.M., Ward P.D. (2007) Evolution. Oxygen and evolution. Science 316, 557–558. [DOI] [PubMed] [Google Scholar]
- 148).Ho M.W., Saunders P.T. (1979) Beyond neo-Darwinism — an epigenetic approach to evolution. J. Theor. Biol. 78, 573–591. [DOI] [PubMed] [Google Scholar]
- 149).Tarlinton R.E., Meers J., Young P.R. (2006) Retroviral invasion of the koala genome. Nature 442, 79–81. [DOI] [PubMed] [Google Scholar]
- 150).Tarlinton R., Meers J., Young P. (2008) Biology and evolution of the endogenous koala retrovirus. Cell. Mol. Life Sci. 65, 3413–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151).Stoye J.P. (2006) Koala retrovirus: a genome invasion in real time. Genome Biol. 7, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]