Abstract
Brain-derived neurotrophic factor (BDNF) is a neurotrophin family member that is highly expressed and widely distributed in the brain. BDNF is critical for neural survival and plasticity both during development and in adulthood, and dysfunction in its signaling may contribute to a number of neurodegenerative disorders. Deep understanding of the BDNF-activated molecular cascade may thus help to find new biomarkers and therapeutic targets. One interesting direction is related to the early phase of BDNF-dependent gene expression regulation, which is responsible for the activation of selective gene programs that lead to stable functional and structural remodeling of neurons. Immediate-early coding genes activated by BDNF are under investigation, but the involvement of the non-coding RNAs is largely unexplored, especially the long non-coding RNAs (lncRNAs). lncRNAs are emerging as key regulators that can orchestrate different aspects of nervous system development, homeostasis, and plasticity, making them attractive candidate markers and therapeutic targets for brain diseases. We used microarray technology to identify differentially expressed lncRNAs in the immediate response phase of BDNF stimulation in a neuronal cell model. Our observations on the putative functional role of lncRNAs provide clues to their involvement as master regulators of gene expression cascade triggered by BDNF.
Keywords: long non-coding RNA, BDNF, immediate-early genes, neuronal gene expression, brain diseases
The Essential Role Of Bdnf In Neuron Life
Brain-Derived Neurotrophic Factor (BDNF) has been shown to have a central function in both neuronal development and in the adult nervous system. Knockout mice for BDNF usually die soon after birth and suffer developmental defects in the brain and sensory nervous system (Ernfors et al., 1995). Suppression of BDNF expression results in defective long-term potentiation (LTP) and memory formation (Korte et al., 1995; Linnarsson et al., 1997; Ma et al., 1998; Mu et al., 1999). In contrast, treatment of hippocampal slices from BDNF knockout mice with recombinant BDNF completely reversed deficits in LTP and significantly improved deficits in basal synaptic transmission (Patterson et al., 1996). BDNF interaction with NTRK2 receptor activates three signaling pathways: PI3K-Akt (PI3K, phosphatidylinositol-3 kinase), Ras-MAPK (MAPK, mitogen-activated protein kinase), and PLCγ-Ca+ (PLC, phospholipase C; Duman and Voleti, 2012). It is also known to regulate a large spectrum of processes of the nervous system, including cell survival, growth, and differentiation (Casaccia-Bonnefil et al., 1999; Bibel and Barde, 2000; Huang and Reichardt, 2003; Park and Poo, 2013; Suliman et al., 2013; Zagrebelsky and Korte, 2014), synaptic plasticity of neurons, and LTP (Xu et al., 2000; Bramham and Messaoudi, 2005; Gottmann et al., 2009; Minichiello, 2009; Mizui et al., 2014; Zagrebelsky and Korte, 2014; Leal et al., 2015). BDNF’s roles in neuronal processes during development and adulthood, support its potential role in the pathogenesis and treatment of both neurological and psychiatric disorders (Pruunsild et al., 2007; Nagahara and Tuszynski, 2011; Weissmiller and Wu, 2012). In fact, BDNF exerts potent pro-survival and functional effects in models of neurological diseases such as Parkinson’s (Howells et al., 2000; van der Kolk et al., 2015), Huntington’s (Zuccato et al., 2008; Jiang et al., 2013), and Alzheimer’s diseases (Holsinger et al., 2000; Michalski and Fahnestock, 2003; Peng et al., 2005; Faria et al., 2014), as well as depression and other psychiatric disorders (Karege et al., 2002; Aydemir et al., 2005; Gonul et al., 2005; Cunha et al., 2006).
Bdnf: The Molecular Mechanism Of Action
Understanding the molecular mechanisms of BDNF’s functions may help to develop efficient therapeutic strategies for brain diseases. The molecular cascade triggered by BDNF has largely been investigated in the adult hippocampus, which retains a high degree of synaptic plasticity into adulthood. The effects of BDNF on LTP have been largely assigned to effects on modulation of receptor trafficking and local protein synthesis at the synapse by local activation of the translation machinery (Leal et al., 2014). BDNF acts at different levels to increase translation activity by altering the phosphorylation of proteins involved in the initiation step of protein synthesis, such as the guanine nucleotide exchange factor eIF2B (Takei et al., 2001) and eIF4E and 4EBP1 (Takei et al., 2004). In addition, BDNF can affect the elongation step, as shown in cultured cortical neurons, where BDNF stimulation was shown to increase the phosphorylation (activation) of eukaryotic elongation factor 1A (eEF1A; Inamura et al., 2005; Leal et al., 2014).
Bdnf And De Novo Gene Expression
Long-term effects of BDNF on synaptogenesis, synaptic plasticity, and cell survival often depend on de novo gene expression. BDNF first affects the expression of genes (immediate-early genes, IEGs) that are regulated directly downstream of its signal transduction pathways and couple early signals to late expression of the downstream target responsible for persistent changes in neuronal phenotype. Some BDNF-induced IEGs involved in synaptic plasticity have been identified. Among them, activity-regulated cytoskeleton-associated protein (ARC), salt-inducible kinase 1 (SIK1), and transcription factor NR4A1 (Nur77), have been shown to be up-regulated early by BDNF stimulation of neurons (Ying et al., 2002; Rao et al., 2006; Zheng et al., 2009; Finsterwald et al., 2013). Beyond the classical protein-coding IEGs, micro-RNAs (miRNAs) have also been singled out in the early response to BDNF stimulation. For example, miR-132 has been shown to be markedly up-regulated early after BDNF treatment of cortical neurons and involved in neuron morphogenesis likely triggering a rapid and persistent downregulation of protein levels (Vo et al., 2005). Both the proteins and miRNA classes of regulators therefore seem to participate in the early response to BDNF stimulation to induce the expression of downstream effectors involved in long-term synaptic changes and cell survival. Mechanisms regulating the initial molecular cascade are likely even more intricate when considering that a new class of non-coding RNA has recently been emerging as a key regulator of gene expression: the long non-coding RNA (lncRNA).
Long Non-Coding Rna: A New Class Of Regulatory Rna
Long non-coding RNA are a heterogeneous class of numerous transcripts mainly produced by RNA polymerase II and defined as RNA molecules of more than 200 bases in length with no protein-coding capacity. Although the functional role of the vast majority of lncRNAs transcribed in a cell has been subject to much debate (Palazzo and Lee, 2015), the literature shows many examples of their effects on gene expression regulation. The mechanism of action is based on their ability to interact with other molecules, such as DNA, RNA, and proteins. In this way, they can act at different stages of gene expression and in processes ranging from chromatin remodeling to transcriptional, post-transcriptional, and translational regulation (Vance and Ponting, 2014). In addition, a recent analysis of Ruiz-Orera et al. (2014) on ribosome profiling experiments provided important evidence that lncRNAs associated with ribosomes may play an important role in de novo protein evolution by encoding short peptides. A large fraction of tissue-specific lncRNAs is expressed in the brain (Derrien et al., 2012), and many reviews have emphasized their role in neurodevelopment, brain function, and a wide range of neurodevelopmental, neurodegenerative and psychiatric diseases (for instance, see St Laurent and Wahlestedt, 2007; van de Vondervoort et al., 2013; Wu et al., 2013; Barry, 2014; Roberts et al., 2014; Tushir and Akbarian, 2014). An emblematic example is the antisense transcript, Bdnf-AS (Pruunsild et al., 2007). Recently, Modarresi et al. (2012) demonstrated that Bdnf-AS downregulates Bdnf expression through a role in the guidance, introduction, and maintenance of H3K27me3 involving PRC2-mediated repressive chromatin remodeling. Single-stranded oligonucleotides and siRNA-mediated depletion of Bdnf-AS in adult mouse brain resulted in a several-fold increase in Bdnf transcript and protein in the hippocampus and frontal cortex (Modarresi et al., 2012). These results paved the way for considering lncRNA as potential drug targets and to search for new strategies to inhibit BDNF-AS function to treat a number of neurological diseases in which BDNF is downregulated. Due to their growing relevance in the regulation of neuronal gene expression, we pondered whether lncRNAs may have a role in the regulatory cascade triggered by BDNF stimulation. In particular, we focused on immediate early response to the neurotrophin stimulation to gain insight into the potential involvement of lncRNAs as a master regulator in BDNF-induced neuronal transcriptional changes.
Rationale, Materials And Methods
Neuronal Cell Model and Culture Conditions
We used the popular SHSY-5Y cell line as a neuronal cell model for BDNF stimulation. SHSY-5Y is a neuroblastoma cell line that can be propagated by easy and low-cost methods when in its native undifferentiated status (Kovalevich and Langford, 2013). SHSY-5Y can be differentiated into cells with a more mature and neuron-like phenotype (Kovalevich and Langford, 2013). In particular, for our study, SH-SY5Y cells (ATCC®) were grown and propagated in Dulbecco’s modified Eagle’s medium (DMEM, Microtech®), supplemented with 2 mM L-glutamine (Lonza BioWhittakerTM), and a solution of 1% penicillin/streptomycin (Lonza BioWhittakerTM) and 15% Fetal Bovine Serum (FBS, Gibco®). The SHSY-5Y line comprises at least two morphologically and biochemically distinct phenotypes: neuroblastic (N-type) and a low proportion of epithelial-like (S-type; Encinas et al., 2000) phenotypes. For BDNF treatment on a more homogeneous neuronal cell population, we performed an enrichment procedure based on the different substrate adherence between the two cell phenotypes. The obtained N-enriched population of SHSY-5Y was differentiated by decreasing FBS concentration from 15 to 1.5% and adding 10 μM of RA (retinoic acid-RA, Sigma–Aldrich®) for 6 days (the medium was refreshed every 2 days). This produced differentiated cells responsive to BDNF, since RA treatment induces the expression of NTRK2 receptor in SH-SY5Y cells (Kaplan et al., 1993; Encinas et al., 2000). After 6 days of differentiation, the medium containing 1.5% FBS and RA was removed and substituted with a medium without FBS for two groups of cells. One of these groups was treated with 10 ng/mL of BDNF (PeproTech®) for a specific time, whereas the second group was not treated and used as a control for the gene expression analysis. We carried out a preliminary investigation to choose the most appropriate time for analyzing the immediate early response to BDNF treatment. We based this analysis on the expression pattern of three immediate early genes downstream of BDNF signaling activation: ARC, NR4A1, and SIK1. The mRNA level of all these genes peaked at 1 h of BDNF treatment (data not shown). Based on these results, microarray analysis was carried out on RNA extracted after 1 h BDNF-treatment.
RNA Isolation and Quality
Total cellular RNA was isolated using an RNeasy®Mini Kit (Qiagen) according to the RNeasy®Mini Handbook (Qiagen). DNA contamination was efficiently removed by on-column DNAse digestion (Qiagen). The concentration and the purity of the RNA sample were assessed using NanoDrop®1000 (Thermo Scientific). Total RNA quality was assessed by an Agilent 2100 Bioanalyzer (Agilent Technologies).
Microarray Experiment Analysis
Microarray experiments were performed on biological triplicate samples. Microarray hybridizations were performed by the Transcriptomics and Genomics core facility of the Department of Emergency and Organ Transplants (DETO) – Nephrology Unit – of the University of Bari ‘Aldo Moro’ Italy1. The labeled cRNA was produced using a Low Input Quick Amp Labeling (LIQA) kit (Agilent Technologies) and hybridized for 17 h at 65°C on an Agilent SurePrint G3 8 × 60K custom lncRNA expression array (Agilent Technologies). This array contains two probes for 22,001 lncRNAs targeting the Gencode v15 human lncRNA annotation, together with one probe for 17,535 randomly chosen protein-coding transcripts. After hybridization, the slide was washed according to Agilent protocols and scanned using a High-Resolution Microarray C Scanner (Agilent Technologies). The image file was processed using Agilent Feature Extraction software (v10.7.3). The microarray grid was correctly placed, and outlier pixels (which were rejected) and inlier pixels were identified. Normalization was performed according to the Quantile method. The differentially expressed probes were selected using a moderated t-test with a p-value cut-off of 0.05.
Quantitative RT-PCR
qPCR validation was performed on independent biological replicates in triplicate. cDNA was synthesized from 1 μg of total RNA using an Invitrogen SuperScript III®kit. Real-time PCR was performed using the SYBR green method and an Applied Biosystems 7500 System. The PCR conditions included a denaturation step (95°C for 10 min) followed by 40 cycles of amplification and quantification (95°C for 35 s, 60°C for 1 min). Relative gene expression levels were normalized to the reference gene hypoxanthine phosphoribosyl transferase 1 (HPRT1) and calculated by the 2-ΔΔCt method. The sequences of the primers used are reported in Supplementary Table S1.
lncRNA Classification and Functional Analysis
The LNCipedia database2 (Volders et al., 2015) was used for retrieving the transcript ID, gene ID, and alternative gene name of the differentially expressed lncRNAs. Differentially expressed lncRNAs were classified by considering their position relative to adjacent protein-coding genes as reported by Mattick and Rinn (2015). Protein-coding potential of the differentially expressed lncRNAs was assessed by CPAT (Coding-Potential Assessment Tool) software3 (Wang et al., 2013). For the functional analysis of the differentially expressed lncRNAs, a list of nearby potentially regulated genes was retrieved using the computational tool GREAT (Genomic Regions Enrichment of Annotations Tool4; McLean et al., 2010). Functional enrichment analysis for the predicted target genes and differentially expressed coding genes was performed using the DAVID system (Database for Annotation, Visualization, and Integrated Discovery5), which uses Gene Ontology (GO) to identify the molecular function represented in the gene profile (Dennis et al., 2003). We obtained a list of potentially regulated miRNAs from lnCeDB6, a database that provides human lncRNAs (version Gencode 19) that can potentially act as competitive endogenous RNAs (ceRNAs) and interfere with the pathway of miRNAs (Das et al., 2014). These miRNAs were analyzed by miR2Disease7 to find miRNAs deregulated in human diseases (Jiang et al., 2009). The potentially regulated miRNAs were also analyzed by miRTarBase8 to find their experimentally validated mRNA targets (Chou et al., 2016). The list of the retrieved mRNA was analyzed by the DAVID tool. For all the DAVID analyses, the significance of enrichment of each GO term was assessed by a p-value of <0.05 and ranked by the number of differentially expressed genes (count).
Results and Discussion
Summary of Microarray Experiment Results
Long non-coding RNA expression profiles are summarized in Table 1. We found that 155 lncRNAs and 238 mRNAs were significantly differentially expressed (p-value <0.05). A fold change of >1.5 was found in 41 lncRNAs (24 up and 17 down regulated) and 40 mRNAs (31 up and nine down regulated; Table 1, Supplementary Table S2). A panel of both differentially expressed lncRNAs and mRNAs with fold change above 1.5 was validated by qPCR (Supplementary Figure S1). We analyzed the 41 differentially expressed lncRNAs by CPAT software to assess their protein-coding potential. We found that 37 lncRNAs have a coding probability value below the cutoff (0.364), while the remaining 3 lncRNAs have a coding probability value above the cutoff (Supplementary Table S3). Among these three transcripts we found C6orf176, the RNA with the highest fold change in our analysis (Supplementary Tables S2 and S3). The vast majority of the top regulated coding transcripts with fold change above 2 are classical immediate early genes involved in different biological processes in response to various neural stimuli, such as NR4A3, ARC, EGR1, and DUSP5 (Table 2). A number of lncRNAs showed a fold change above 2, including the previously reported C6orf176 and MIAT (also known as Gomafu; Table 2). C6orf176 has been shown to be readily upregulated with a peak at the 2-h time point of treating human ocular ciliary smooth muscle cells with an EP2- and EP4-specific agonist (Reitmair et al., 2012). Notably, there was rapid and pronounced transcriptional upregulation followed by a brisk decline which resembles the kinetics of immediate early response genes (Reitmair et al., 2012). MIAT was recently shown to be acutely regulated in response to neuronal activation in mouse primary cortical neurons (Barry et al., 2014). In particular, this lncRNA is strongly downregulated after 1 h and 3 h of KCl depolarization (Barry et al., 2014). Interestingly, our data are in accordance with that of Barry et al. (2014), since we detected downregulation of MIAT transcript after 1 h of stimulation with BDNF, which is a well known activity-dependent factor.
Table 1.
Summary of microarray analysis.
| Transcript | Number | Differentially expressed | FC ≥ 1.5 |
|---|---|---|---|
| lncRNA | 22,001 | 155 | 41 |
| mRNA | 17,535 | 238 | 40 |
Table 2.
Top regulated coding and long non-coding transcripts with fold change above 2.
| Coding | Regulation | FC | lncRNA | Regulation | FC |
|---|---|---|---|---|---|
| NR4A3 | Up | 9.2 | C6orf176 | Up | 3.8 |
| ARC | Up | 8.3 | lnc-NPAS4-1 | Up | 3.1 |
| RHOB | Up | 4.3 | lnc-WDR1-1 | Up | 2.8 |
| fam46a | Up | 3.6 | IGFBP7-AS1 | Up | 2.1 |
| EGR1 | Up | 3.0 | lnc-ZSCAN10-4 | Up | 2.1 |
| DUSP5 | Up | 3.0 | MIAT-003 | Down | 2.1 |
| KLF10 | Up | 2.3 | MIAT-001 | Down | 2.0 |
| MAP3K14 | Up | 2.0 | lnc-RHOF-1 | Down | 2.0 |
| F3 | Up | 2.0 | |||
Classification and Functional Analysis of lncRNA Regulated by BDNF
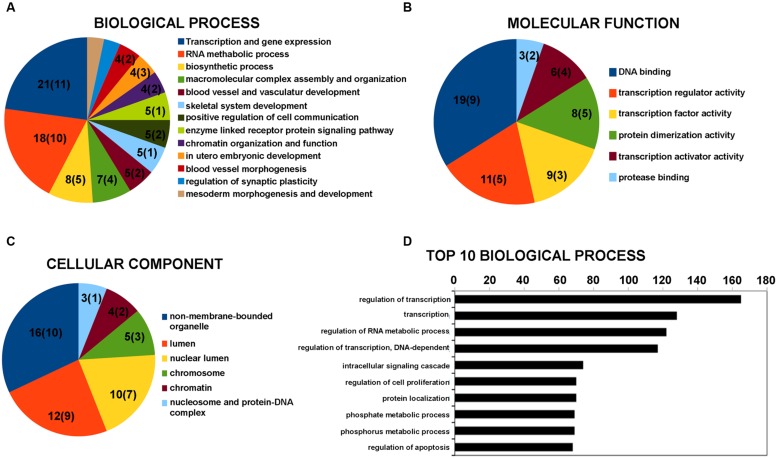
The lncRNAs were classified in accordance with the definition discussed in “Materials and Methods” Section. The vast majority of differentially expressed lncRNAs were included in the intergenic and antisense classes (Supplementary Table S2). Five lncRNAs with a fold change above 1.5 were hypothesized to have a role in physiological and pathological processes in neuronal cells by regulating gene expression (Table 3). This supports the hypothesis of an involvement of lncRNA in BDNF-mediated molecular effects. To assess the potential function of the vast majority of the differentially expressed lncRNAs, we examined the function of genes located near the lncRNAs in the genome. The list of these coding genes and the list of differentially expressed coding RNAs found in our microarray survey were used for GO and pathway enrichment analysis with the DAVID tool. GO terms that shared the same genes were grouped together as shown in Figure 1 with the ID of single GO terms. We report the number of genes associated with the GO group in a pie chart with the number of genes located near the lncRNAs in brackets. The most enriched biological processes are related to transcription regulation and RNA metabolic process (Figure 1A). In addition, there are also processes related to chromatin organization and function (Figure 1A). The involvement of genomic loci of lncRNA in transcription control was also revealed by GO terms of molecular function (Figure 1B). Regarding cellular component analysis, enriched GO are related to non-membrane-bound organelle, lumen and chromatin structure (Figure 1C).
Table 3.
Differentially expressed lncRNA that have been identified in literature.
| Gene symbol | Regulation | Function |
|---|---|---|
| MALAT1 (Neat2) | Down | Control of the expression of genes involved in synapse function (Bernard et al., 2010). Down-regulation led to cell arrest in the G1/S or G2/M phase (Yang et al., 2013). |
| MIAT (Gomafu) | Down | Down-regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing (Barry et al., 2014). Decreased in the medial prefrontal complex following fear conditioning and knockdown of promoted stress reactivity and anxiety-like behavior (Spadaro et al., 2015). Neurogenic commitment and neuronal survival, sustained overexpression of Miat promoted neuronal death (Aprea et al., 2013). |
| HAND2-AS1 (Dein) | Down | Highly expressed in stage IVS neuroblastoma (Voth et al., 2007). Expression is neuroblastoma is co-regulated together with HAND2 (Voth et al., 2009). |
| C6orf176 | Up | A possible regulatory function in response to cAMP signaling (Reitmair et al., 2012). |
| HOXD-AS1 | Up | It is induced by RA, could be regulated via PI3K/Akt pathway and controls genes involved in RA signaling, angiogenesis and inflammation (Yarmishyn et al., 2014). |
FIGURE 1.
Gene Ontology (GO) enrichment analysis for lncRNAs by DAVID bioinformatics tool. (A) GO analysis of lncRNA-target genes + differentially expressed coding genes according to biological process. (B) GO analysis of lncRNA-target genes + differentially expressed coding genes according to molecular function. (C) GO analysis of lncRNA-target genes + differentially expressed coding genes according to cell component. (D) Top 10 biological processes for coding genes that are target of miRNA potentially regulated by the differentially expressed lncRNAs. Grouped GO terms are reported as following with the ID of the single GO term. Transcription and gene expression: GO:0045449 + GO:0006350 + GO:0006355 + GO:0006357 + GO:0045893 + GO:0045941 + GO:0010628 + GO:0045944; RNA metabolic process: GO:0051252 + GO:0051254 + GO:0045935 + GO:0051173 + GO:0010604; biosynthetic process: GO:0010557 + GO:0031328 + GO:0009891; macromolecular complex assembly and organization: GO:0065003 + GO:0043933 + GO:0034622 + GO:0034621; blood vessel and vasculature development: GO:0001568 + GO:0001944; chromatin organization and function: GO:0006334 + GO:0031497 + GO:0065004 + GO:0034728 + GO:0006323 + GO:0006333; regulation of synaptic plasticity: GO:0048168 + GO:0048167; mesoderm morphogenesis and development: GO:0048332 + GO:0007498. The number on the pie chart indicates the number of differentially expressed coding genes associated to the GO term (or GO group), while the number in brackets indicates the number of genes located near the differentially expressed lncRNAs.
miRNA Targets on lncRNA
Long non-coding RNAs can affect gene expression by interfering with the microRNA pathways and acting as competing endogenous RNA. We therefore used the lnCeDB database to unravel microRNA (miRNA)–lncRNA putative functional interactions by identifying miRNAs with binding sites in differently expressed lncRNAs. Seven of the miRNAs found are associated with neuropathologies reported in the mir2disease database (Table 4). To acquire information on biological processes affected by the putative lncRNA-miRNA interaction, we retrieved a list of experimentally validated mRNA targets of all the previously identified miRNAs and carried out a functional analysis with the DAVID tool. In the top 10 biological processes, the most represented categories are related to the regulation of transcription and RNA metabolic processes (Figure 1D).
Table 4.
Putative miRNA targets on lncRNA and their involvement in neuropsychiatric diseases.
| miRNA symbol | Neuropathology |
|---|---|
| hsa-miR-339-5p | Neurodegeneration |
| hsa-miR-433 | Parkinson’s disease |
| hsa-miR-133b | Parkinson’s disease |
| hsa-miR-346 | Schizophrenia |
| hsa-miR-328 | Alzheimer’s disease |
| hsa-miR-299-3p | Alzheimer’s disease |
| hsa-miR-422a | Multiple sclerosis |
Conclusion
The gene expression occurring immediately after neural stimuli plays critical roles in long-lasting neuronal changes. Further efforts are needed to isolate novel IEGs, with the hope of finding “master genes” for neuronal processes and subsequent therapeutic targets for brain diseases. The molecular mechanisms underlying the gene expression regulation in the initial phase of the stimulus-induced molecular cascade comprise the product of coding genes, miRNAs, and very recently, lncRNAs (Aitken et al., 2015). We sought to evaluate the potential role of lncRNAs in early gene expression regulation triggered by the neurotrophin BDNF. In particular, using microarray technology, we found that hundreds of lncRNAs and coding IEGs changed their transcript level after 1 h of BDNF treatment on neuronal cells. Some of the differentially expressed lncRNAs are already known. These are strictly associated with neuronal cellular processes and are involved in gene expression regulation. However, the biological functions of a vast majority of lncRNAs identified in this study are not currently understood. We therefore used computational approaches to provide preliminary insights into their potential functionality. The most enriched ontology terms were related to the transcription regulation processes, which highlights the putative role of lncRNAs in orchestrating the immediate response to BDNF. Overall, this study presents an interesting area for further investigations into lncRNAs with essential roles in molecular and cellular processes triggered by neurotrophins.
Author Contributions
Conceived and designed the experiments: AD. Performed the experiments: VA and AD. Analyzed the data: VA and AD. Wrote the paper: AD.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Matteo Accetturo (matteo.accetturo@uniba.it) of Transcriptomics and Genomics core facility of the Department of Emergency and Organ Transplants (DETO) – Nephrology Unit – University of Bari ‘Aldo Moro’ for execution of microarray hybridizations and analysis.
Funding. This work was supported by the MIUR FIRB Grant RBFR12QW4I.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fnmol.2016.00015
References
- Aitken S., Magi S., Alhendi A. M., Itoh M., Kawaji H., Lassmann T., et al. (2015). Transcriptional dynamics reveal critical roles for non-coding RNAs in the immediate-early response. PLoS Comput. Biol. 11:e1004217 10.1371/journal.pcbi.1004217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea J., Prenninger S., Dori M., Ghosh T., Monasor L. S., Wessendorf E., et al. (2013). Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 32 3145–3160. 10.1038/emboj.2013.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydemir O., Deveci A., Taneli F. (2005). The effect of chronic antidepressant treatment on serum brain-derived neurotrophic factor levels in depressed patients: a preliminary study. Prog. Neuropsychopharmacol. Biol. Psychiatry. 29 261–265. 10.1016/j.pnpbp.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Barry G. (2014). Integrating the roles of long and small non-coding RNA in brain function and disease. Mol. Psychiatry 19 410–416. 10.1038/mp.2013.196 [DOI] [PubMed] [Google Scholar]
- Barry G., Briggs J. A., Vanichkina D. P., Poth E. M., Beveridge N. J., Ratnu V. S., et al. (2014). The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19 486–494. 10.1038/mp.2013.45 [DOI] [PubMed] [Google Scholar]
- Bernard D., Prasanth K. V., Tripathi V., Colasse S., Nakamura T., Xuan Z., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29 3082–3093. 10.1038/emboj.2010.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibel M., Barde Y. A. (2000). Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev. 14 2919–2937. 10.1101/gad.841400 [DOI] [PubMed] [Google Scholar]
- Bramham C. R., Messaoudi E. (2005). BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog. Neurobiol. 76 99–125. 10.1016/j.pneurobio.2005.06.003 [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P., Gu C., Chao M. V. (1999). Neurotrophins in cell survival/death decisions. Adv. Exp. Med. Biol. 468 275–282. 10.1007/978-1-4615-4685-6_22 [DOI] [PubMed] [Google Scholar]
- Chou C. H., Chang N. W., Shrestha S., Hsu S. D., Lin Y. L., Lee W. H., et al. (2016). miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 44 D239–D247. 10.1093/nar/gkv1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha A. B., Frey B. N., Andreazza A. C., Goi J. D., Rosa A. R., Gonçalves C. A., et al. (2006). Serum brain-derived neurotrophic factor is decreased in bipolar disorder during depressive and manic episodes. Neurosci. Lett. 398 215–219. 10.1016/j.neulet.2005.12.085 [DOI] [PubMed] [Google Scholar]
- Das S., Ghosal S., Sen R., Chakrabarti J. (2014). lnCeDB: database of human long noncoding RNA acting as competing endogenous RNA. PLoS ONE 9:e98965 10.1371/journal.pone.0098965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G. J., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., et al. (2003). DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4:P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22 1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman R. S., Voleti B. (2012). Signaling pathways underlying the pathophysiology and treatment of depression: novel mechanisms for rapid-acting agents. Trends Neurosci. 35 47–56. 10.1016/j.tins.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas M., Iglesias M., Liu Y., Wang H., Muhaisen A., Ceña V., et al. (2000). Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 75 991–1003. 10.1046/j.1471-4159.2000.0750991.x [DOI] [PubMed] [Google Scholar]
- Ernfors P., Kucera J., Lee K. F., Loring J., Jaenisch R. (1995). Studies on the physiological role of brain-derived neurotrophic factor and neurotrophin-3 in knockout mice. Int. J. Dev. Biol. 39 799–807. [PubMed] [Google Scholar]
- Faria M. C., Gonçalves G. S., Rocha N. P., Moraes E. N., Bicalho M. A., Gualberto Cintra M. T., et al. (2014). Increased plasma levels of BDNF and inflammatory markers in Alzheimer’s disease. J. Psychiatr. Res. 53 166–172. 10.1016/j.jpsychires.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Finsterwald C., Carrard A., Martin J. L. (2013). Role of salt-inducible kinase 1 in the activation of MEF2-dependent transcription by BDNF. PLoS ONE 8:e54545 10.1371/journal.pone.0054545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonul A. S., Akdeniz F., Taneli F., Donat O., Eker C., Vahip S. (2005). Effect of treatment on serum brain-derived neurotrophic factor levels in depressed patients. Eur. Arch. Psychiatry Clin. Neurosci. 255 381–386. 10.1007/s00406-005-0578-6 [DOI] [PubMed] [Google Scholar]
- Gottmann K., Mittmann T., Lessmann V. (2009). BDNF signaling in the formation, maturation and plasticity of glutamatergic and GABAergic synapses. Exp. Brain Res. 199 203–234. 10.1007/s00221-009-1994-z [DOI] [PubMed] [Google Scholar]
- Holsinger R. M., Schnarr J., Henry P., Castelo V. T., Fahnestock M. (2000). Quantitation of BDNF mRNA in human parietal cortex by competitive reverse transcription-polymerase chain reaction: decreased levels in Alzheimer’s disease. Brain Res. Mol. Brain Res. 76 347–354. 10.1016/S0169-328X(00)00023-1 [DOI] [PubMed] [Google Scholar]
- Howells D. W., Porritt M. J., Wong J. Y., Batchelor P. E., Kalnins R., Hughes A. J., et al. (2000). Reduced BDNF mRNA expression in the Parkinson’s disease substantia nigra. Exp. Neurol. 166 127–135. 10.1006/exnr.2000.7483 [DOI] [PubMed] [Google Scholar]
- Huang E. J., Reichardt L. F. (2003). Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 72 609–642. 10.1146/annurev.biochem.72.121801.161629 [DOI] [PubMed] [Google Scholar]
- Inamura N., Nawa H., Takei N. (2005). Enhancement of translation elongation in neurons by brain-derived neurotrophic factor: implications for mammalian target of rapamycin signaling. J. Neurochem. 95 1438–1445. 10.1111/j.1471-4159.2005.03466.x [DOI] [PubMed] [Google Scholar]
- Jiang M., Peng Q., Liu X., Jin J., Hou Z., Zhang J., et al. (2013). Small-molecule TrkB receptor agonists improve motor function and extend survival in a mouse model of Huntington’s disease. Hum. Mol. Genet. 22 2462–2470. 10.1093/hmg/ddt098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., et al. (2009). miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 37 D98–D104. 10.1093/nar/gkn714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. R., Matsumoto K., Lucarelli E., Thiele C. J. (1993). Induction of TrkB by retinoic acid mediates biologic responsiveness to BDNF and differentiation of human neuroblastoma cells. Neuron 11 321–331. 10.1016/0896-6273(93)90187-V [DOI] [PubMed] [Google Scholar]
- Karege F., Perret G., Bondolfi G., Schwald M., Bertschy G., Aubry J. M. (2002). Decreased serum brain-derived neurotrophic factor levels in major depressed patients. Psychiatry Res. 109 143–148. 10.1016/S0165-1781(02)00005-7 [DOI] [PubMed] [Google Scholar]
- Korte M., Carroll P., Wolf E., Brem G., Thoenen H., Bonhoeffer T. (1995). Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc. Natl. Acad. Sci. U.S.A. 92 8856–8860. 10.1073/pnas.92.19.8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalevich J., Langford D. (2013). Considerations for the use of SH-SY5Y neuroblastoma cells in neurobiology. Methods Mol. Biol. 1078 9–21. 10.1007/978-1-62703-640-5_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal G., Afonso P. M., Salazar I. L., Duarte C. B. (2015). Regulation of hippocampal synaptic plasticity by BDNF. Brain Res. 1621 82–101. 10.1016/j.brainres.2014.10.019 [DOI] [PubMed] [Google Scholar]
- Leal G., Comprido D., Duarte C. B. (2014). BDNF-induced local protein synthesis and synaptic plasticity. Neuropharmacology 76 639–656. 10.1016/j.neuropharm.2013.04.005 [DOI] [PubMed] [Google Scholar]
- Linnarsson S., Bjorklund A., Ernfors P. (1997). Learning deficit in BDNF mutant mice. Eur. J. Neurosci. 9 2581–2587. 10.1111/j.1460-9568.1997.tb01687.x [DOI] [PubMed] [Google Scholar]
- Ma Y. L., Wang H. L., Wu H. C., Wei C. L., Lee E. H. (1998). Brain-derived neurotrophic factor antisense oligonucleotide impairs memory retention and inhibits long-term potentiation in rats. Neuroscience 82 957–967. 10.1016/S0306-4522(97)00325-4 [DOI] [PubMed] [Google Scholar]
- Mattick J. S., Rinn J. L. (2015). Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 22 5–7. 10.1038/nsmb.2942 [DOI] [PubMed] [Google Scholar]
- McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., et al. (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski B., Fahnestock M. (2003). Pro-brain-derived neurotrophic factor is decreased in parietal cortex in Alzheimer’s disease. Brain Res. Mol. Brain Res. 111 148–154. 10.1016/S0169-328X(03)00003-2 [DOI] [PubMed] [Google Scholar]
- Minichiello L. (2009). TrkB signalling pathways in LTP and learning. Nat. Rev. Neurosci. 10 850–860. 10.1038/nrn2738 [DOI] [PubMed] [Google Scholar]
- Mizui T., Tanima Y., Komatsu H., Kumanogoh H., Kojima M. (2014). The biological actions and mechanisms of brain-derived neurotrophic factor in healthy and disordered brains. Neurosci. Med. 5 183–195. 10.4236/nm.2014.54021 [DOI] [Google Scholar]
- Modarresi F., Faghihi M. A., Lopez-Toledano M. A., Fatemi R. P., Magistri M., Brothers S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30 453–459. 10.1038/nbt.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J. S., Li W. P., Yao Z. B., Zhou X. F. (1999). Deprivation of endogenous brain-derived neurotrophic factor results in impairment of spatial learning and memory in adult rats. Brain Res. 835 259–265. 10.1016/S0006-8993(99)01592-9 [DOI] [PubMed] [Google Scholar]
- Nagahara A. H., Tuszynski M. H. (2011). Potential therapeutic uses of BDNF in neurological and psychiatric disorders. Nat. Rev. Drug Discov. 10 209–219. 10.1038/nrd3366 [DOI] [PubMed] [Google Scholar]
- Palazzo A. F., Lee E. S. (2015). Non-coding RNA: what is functional and what is junk? Front. Genet. 6:2 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Poo M. M. (2013). Neurotrophin regulation of neural circuit development and function. Nat. Rev. Neurosci. 14 7–23. 10.1038/nrn3379 [DOI] [PubMed] [Google Scholar]
- Patterson S. L., Abel T., Deuel T. A. S., Martin K. C., Rose J. C., Kandel E. R. (1996). Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron 16 1137–1145. 10.1016/S0896-6273(00)80140-3 [DOI] [PubMed] [Google Scholar]
- Peng S., Wuu J., Mufson E. J., Fahnestock M. (2005). Precursor form of brain-derived neurotrophic factor and mature brain-derived neurotrophic factor are decreased in the pre-clinical stages of Alzheimer’s disease. J. Neurochem. 93 1412–1421. 10.1111/j.1471-4159.2005.03135.x [DOI] [PubMed] [Google Scholar]
- Pruunsild P., Kazantseva A., Aid T., Palm K., Timmusk T. (2007). Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 90 397–406. 10.1016/j.ygeno.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V. R., Pintchovski S. A., Chin J., Peebles C. L., Mitra S., Finkbeiner S. (2006). AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nat. Neurosci. 9 887–895. 10.1038/nn1708 [DOI] [PubMed] [Google Scholar]
- Reitmair A., Sachs G., Im W. B., Wheeler L. (2012). C6orf176: a novel possible regulator of cAMP-mediated gene expression. Physiol. Genomics 44 152–161. 10.1152/physiolgenomics.00089.2011 [DOI] [PubMed] [Google Scholar]
- Roberts T. C., Morris K. V., Wood M. J. (2014). The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130507 10.1098/rstb.2013.0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J., Messeguer X., Subirana J. A., Alba M. M. (2014). Long non-coding RNAs as a source of new peptides. Elife 3:e03523 10.7554/eLife.03523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro P. A., Flavell C. R., Widagdo J., Ratnu V. S., Troup M., Ragan C., et al. (2015). Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol. Psychiatry 78 848–859. 10.1016/j.biopsych.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G., III, Wahlestedt C. (2007). Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 30 612–621. 10.1016/j.tins.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Suliman S., Hemmings S. M., Seedat S. (2013). Brain-derived neurotrophic factor (BDNF) protein levels in anxiety disorders: systematic review and meta-regression analysis. Front. Integr. Neurosci. 7:55 10.3389/fnint.2013.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N., Inamura N., Kawamura M., Namba H., Hara K., Yonezawa K., et al. (2004). Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J. Neurosci. 24 9760–9769. 10.1523/JNEUROSCI.1427-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N., Kawamura M., Hara K., Yonezawa K., Nawa H. (2001). Brain-derived neurotrophic factor enhances neuronal translation by activating multiple initiation processes: comparison with the effects of insulin. J. Biol. Chem. 276 42818–42825. 10.1074/jbc.M103237200 [DOI] [PubMed] [Google Scholar]
- Tushir J. S., Akbarian S. (2014). Chromatin-bound RNA and the neurobiology of psychiatric disease. Neuroscience 264 131–141. 10.1016/j.neuroscience.2013.06.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vondervoort I. I., Gordebeke P. M., Khoshab N., Tiesinga P. H., Buitelaar J. K., Kozicz T., et al. (2013). Long non-coding RNAs in neurodevelopmental disorders. Front. Mol. Neurosci. 6:53 10.3389/fnmol.2013.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kolk N. M., Speelman A. D., van Nimwegen M., Kessels R. P., IntHout J., Hakobjan M., et al. (2015). BDNF polymorphism associates with decline in set shifting in Parkinson’s disease. Neurobiol. Aging 36 1605.e1–1605.e6 10.1016/j.neurobiolaging.2014.08.023 [DOI] [PubMed] [Google Scholar]
- Vance K. W., Ponting C. P. (2014). Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 30 348–355. 10.1016/j.tig.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N., Klein M. E., Varlamova O., Keller D. M., Yamamoto T., Goodman R. H., et al. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102 16426–16431. 10.1073/pnas.0508448102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders P. J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., et al. (2015). An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 43 4363–4364. 10.1093/nar/gkv295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth H., Oberthuer A., Simon T., Kahlert Y., Berthold F., Fischer M. (2007). Identification of DEIN, a novel gene with high expression levels in stage IVS neuroblastoma. Mol. Cancer Res. 5 1276–1284. 10.1158/1541-7786.MCR-06-0258 [DOI] [PubMed] [Google Scholar]
- Voth H., Oberthuer A., Simon T., Kahlert Y., Berthold F., Fischer M. (2009). Co-regulated expression of HAND2 and DEIN by a bidirectional promoter with asymmetrical activity in neuroblastoma. BMC Mol. Biol. 10:28 10.1186/1471-2199-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Park H. J., Dasari S., Wang S., Kocher J. P., Li W. (2013). CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 41 e74 10.1093/nar/gkt006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmiller A. M., Wu C. (2012). Current advances in using neurotrophic factors to treat neurodegenerative disorders. Transl. Neurodegener. 1:14 10.1186/2047-9158-1-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Zuo X., Deng H., Liu X., Liu L., Ji A. (2013). Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 97 69–80. 10.1016/j.brainresbull.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Xu B., Gottschalk W., Chow A., Wilson R. I., Schnell E., Zang K., et al. (2000). The role of brain-derived neurotrophic factor receptors in the mature hippocampus: modulation of long-term potentiation through a presynaptic mechanism involving trkB. J. Neurosci. 20 6888–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Yi F., Han X., Du Q., Liang Z. (2013). MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 587 3175–3181. 10.1016/j.febslet.2013.07.048 [DOI] [PubMed] [Google Scholar]
- Yarmishyn A. A., Batagov A. O., Tan J. Z., Sundaram G. M., Sampath P., Kuznetsov V. A., et al. (2014). HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics 15:S7 10.1186/1471-2164-15-S9-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S. W., Futter M., Rosenblum K., Webber M. J., Hunt S. P., Bliss T. V., et al. (2002). Brain-derived neurotrophic factor induces longterm potentiation in intact adult hippocampus: requirement for ERK activation coupled to CREB and up-regulation of Arc synthesis. J. Neurosci. 22 1532–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrebelsky M., Korte M. (2014). Form follows function: BDNF and its involvement in sculpting the function and structure of synapses. Neuropharmacology 76 628–638. 10.1016/j.neuropharm.2013.05.029 [DOI] [PubMed] [Google Scholar]
- Zheng F., Luo Y., Wang H. (2009). Regulation of brain-derived neurotrophic factor-mediated transcription of the immediate early gene Arc by intracellular calcium and calmodulin. J. Neurosci. Res. 87 380–392. 10.1002/jnr.21863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C., Marullo M., Conforti P., MacDonald M. E., Tartari M., Cattaneo E. (2008). Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington’s disease. Brain Pathol. 18 225–238. 10.1111/j.1750-3639.2007.00111.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.