Abstract
Damage caused by Phytophthora cinnamomi Rands remains an important concern on forest tree species. The pathogen causes root and collar rot, stem cankers, and dieback of various economically important Eucalyptus spp. In South Africa, susceptible cold tolerant Eucalyptus plantations have been affected by various Phytophthora spp. with P. cinnamomi considered one of the most virulent. The molecular basis of this compatible interaction is poorly understood. In this study, susceptible Eucalyptus nitens plants were stem inoculated with P. cinnamomi and tissue was harvested five days post inoculation. Dual RNA-sequencing, a technique which allows the concurrent detection of both pathogen and host transcripts during infection, was performed. Approximately 1% of the reads mapped to the draft genome of P. cinnamomi while 78% of the reads mapped to the Eucalyptus grandis genome. The highest expressed P. cinnamomi gene in planta was a putative crinkler effector (CRN1). Phylogenetic analysis indicated the high similarity of this P. cinnamomi CRN1 to that of Phytophthora infestans. Some CRN effectors are known to target host nuclei to suppress defense. In the host, over 1400 genes were significantly differentially expressed in comparison to mock inoculated trees, including suites of pathogenesis related (PR) genes. In particular, a PR-9 peroxidase gene with a high similarity to a Carica papaya PR-9 ortholog previously shown to be suppressed upon infection by Phytophthora palmivora was down-regulated two-fold. This PR-9 gene may represent a cross-species effector target during P. cinnamomi infection. This study identified pathogenicity factors, potential manipulation targets, and attempted host defense mechanisms activated by E. nitens that contributed to the susceptible outcome of the interaction.
Keywords: plant defense, RNA-seq, Eucalyptus nitens, pathogenesis related genes, crinkler
Introduction
Species of the Oomycete genus Phytophthora are the most economically important pathogens of plants worldwide (Zentmyer, 1983; Erwin and Ribeiro, 1996). Of critical concern to the forestry industry is Phytophthora cinnamomi Rands, one of the most pathogenic and damaging species that affects agriculture, forestry, and native forests worldwide (Linde et al., 1999; Brasier, 2008; Hansen, 2008; Oβwald et al., 2014). In Eucalyptus, P. cinnamomi has threatened the productivity of plantations (FAO, 2007; Wingfield et al., 2011) and caused extensive damage to native ecosystems including E. marginata forests in Western Australia (Podger et al., 1967; Burgess et al., 1999) and native vegetation of the Western Cape of South Africa (Vonbroembsen and Kruger, 1985). Eucalypt plantations contribute significantly to the economy of South Africa (Godsmark, 2009). Susceptible cold-tolerant Eucalyptus plantations have been affected by various Phytophthora species to such an extent that some valuable species such as E. fastigata and E. fraxinoides are no longer cultivated (Linde et al., 1994; Wingfield and Kemp, 1994). Eucalyptus nitens is considered resistant to P. cinnamomi in its native environment but succumbs to the pathogen in plantations (Cahill et al., 2008). P. cinnamomi, an introduced stramenopile pathogen (Adl et al., 2005, 2012) severely affected stands of E. nitens in South Africa (Maseko, 2010).
Plant-pathogen interactions in forest systems have not been as well studied at a genomic level as it has for herbaceous crops. The recent availability of the genome sequence of E. grandis (Myburg et al., 2014) has provided a valuable resource for transcriptomic studies to dissect defense responses in related Eucalyptus species. RNA sequencing (RNA-seq) has contributed knowledge towards several host responses during pathogen challenge (Xu et al., 2011, 2012; Dowen et al., 2012; Martinelli et al., 2012; Savory et al., 2012; Tremblay et al., 2012). Unlike previous probe based methods which require separation of host and pathogen cells, RNA-seq has allowed the study of both host and pathogen transcriptomics simultaneously. This technique, known as dual RNA-seq (reviewed in Westermann et al., 2012) allows the detection of minute amounts of pathogen RNA. It does not require predesigned species specific probes and is more sensitive than the previous methods of microarrays and northern blotting (Kunjeti et al., 2012; Tierney et al., 2012; Westermann et al., 2012; Camilios-Neto et al., 2014; Choi et al., 2014; Hayden et al., 2014). Pathogen RNA-seq data can be further mined for clues to pathogenicity (ability to cause disease) and virulence (degree of damage or pathology) factors based on functional genetics studies conducted in various host-pathogen interactions. The plant-host interactions database provides such data for comparative analysis (Winnenburg et al., 2006).
An enhanced understanding of the defense response of plants to P. cinnamomi will facilitate the production of resistant plants (Eshraghi et al., 2014a). Several RNA-seq host response studies to Phytophthora spp. have been undertaken to date (Kunjeti et al., 2012; Ali et al., 2014; Chen et al., 2014). The host responses of raspberry to P. rubi, and those of potato tubers to P. infestans, were successfully profiled using RNA-seq (Ward and Weber, 2012; Gao et al., 2013). This technique has also been applied to a native forest system, with the response of the oak Notholithocarpus densiflorus to P. ramorum elucidated (Kunjeti et al., 2012; Ward and Weber, 2012; Gao et al., 2013; Ali et al., 2014; Chen et al., 2014; Hayden et al., 2014).
The genome sequence of P. cinnamomi var. cinnamomi is currently available as a draft assembly of 77.97 Mbp and a sequence read coverage depth of 69.6x (Reeve, 2012, unpublished; JGI Project identity: 1003775). This is an invaluable resource that provides insight into pathogenicity determinants in this species. This has been demonstrated for other Phytophthora species sequenced genomes (e.g., P. infestans, P. sojae, P. ramorum, reviewed in Jiang and Tyler, 2012).
Phytophthora species are able to manipulate their hosts to their own advantage. For example, P. infestans manipulates its host to suit its life-style by suppressing the hypersensitive response (HR) in potato during its biotrophic phase, then manipulating the induction of HR during the necrotrophic phase (Bos et al., 2010; Gilroy et al., 2011). This type of manipulation could be a trend in other Phytophthora interactions (Belhaj et al., 2009; Porter et al., 2009). Effectors excreted during this interaction are primarily crinklers (CRN), a family of proteins expressed in all plant pathogenic oomycetes, and RxLRs which are confined to only Phytophthora species (Stam et al., 2013b; Chen et al., 2014).
Host defense responses are mediated through different mechanisms, and the timing and degree to which these are activated could determine the outcome of the interaction between a plant and pathogen (Tao et al., 2003). A factor driving compatible interactions has been revealed by several studies to involve a mass down-regulation of defense genes (Schlink et al., 2010). Pathways associated with these genes include the defense hormone salicylic acid (SA) which is associated with biotrophic defense responses, jasmonic acid (JA), and ethylene (ET) which are associated with necrotrophic defense responses, and abscisic acid (ABA) which is associated with abiotic stress as well as pathogen defense (Bari and Jones, 2009). The host responses elicited by P. cinnamomi have been studied in different woody species at various levels including anatomical, physiological, biochemical, and molecular levels (recently reviewed in Oβwald et al., 2014). Some of the major findings in these studies highlight the importance of correctly regulated HR and reactive oxygen species (ROS) and synthesis of phenylpropanoid pathway-related substances such as flavonoids, gibberellic acid (GA) and lignin. PR-1 and PR-5 feature in several interactions as possible resistance factors.
Multiple pathogenesis related (PR) gene classes are differentially regulated against Phytophthora and are thought to be important for successful defense (Moy et al., 2004; Schlink, 2009; Attard et al., 2010). The over-expression of specific PR genes have conferred tolerance against various Phytophthora spp. (Alexander et al., 1993; Fagoaga et al., 2001; Sarowar et al., 2009; Pushin et al., 2010; Acharya et al., 2013; He et al., 2013).
The E. nitens–P. cinnamomi interaction provided a novel system to study a compatible host-pathogen interaction using a dual RNA-sequencing approach. The aim of this study was to (i) discover pathogenicity factors produced by P. cinnamomi (ii) to determine the E. nitens defense response to the pathogen, and (iii) to identify host genes that could potentially be suppressed by the pathogen to promote susceptibility. We observed, among other responses, high expression of a putative P. cinnamomi crinkler effector in planta and the down-regulation of a PR-9 gene, which may represent a common host effector target in E. nitens; two factors possibly contributing to the susceptible outcome of the interaction.
Methods
Inoculated plant material
E. nitens seedlings were obtained from parents that were part of a third generation commercial breeding program (Sappi Forests Research, Shaw Research Centre, KwaZulu-Natal, South Africa) The criterion for selection was based on wood density gain. An MLRelate analysis, using the microsatellite markers developed by Faria et al. (2010), determined that the individuals from the Sappi breeding population showed higher relatedness to each other than those within the natural Australian E. nitens population (Melissa Reynolds, Forest Molecular Genetics, University of Pretoria). Previous sampling in E. nitens stands in South Africa have shown root rot due to P. cinnamomi (Maseko, 2010).
The E. nitens seedlings were grown in pine-bark seedling mix until stem thickness was >0.5 cm (~1 year), then stem inoculated with P. cinnamomi (CMW26310, Forestry and Agricultural Biotechnology Institute culture collection). A 4 mm cork borer and mycelial plug on cV8 agar [cV8A; modified from Erwin and Ribeiro (1996): 200 ml/L V8 juice (Campbell Soup Company, Camden, New Jersey), 20 g/L CaCO3 (Merck), 20 g/L agar)] was used for inoculations. Mock-inoculated plants were treated identically to infected trees, with a sterile cV8 agar plug. Inoculation sites were covered with damp sterile cheesecloth, tinfoil, and Parafilm (Parafilm, Chicago, IL).
Stems were inoculated at two sites, 10 cm apart. In the attempt to represent the typical responses to P. cinnamomi in commercially grown E. nitens, for both the mock-inoculated control and inoculated trees, stem tissue was harvested from 18 trees (which consisted of three pooled biological replicates of six trees each) at 5 days post inoculation (dpi). Three centimeters of stem tissue was harvested per inoculation site, with 1.5 cm of stem tissue below and above the center of the site. Harvested material was immediately frozen in liquid nitrogen.
Since sampling was destructive, nine extra trees were used to observe symptom development for 6 weeks following inoculation. Beneath-bark lesions were measured in other trials to statistically validate the effectivity of inoculation. For these lesions, a Shapiro-Wilk test was performed in GraphPad Prism 6 (Motulsky, 1999) and the non-parametric Mann-Whitney test was used to assess significance at a 95% confidence level.
Microscopy
Preparation of plant material
Approximately 1.5–2 cm of stem tissue surrounding the inoculation site of P. cinnamomi inoculated and mock-inoculated E. nitens tissue was harvested in triplicate at 24 h post inoculation (hpi), 48 hpi, 96 hpi, and 1 week post inoculation (wpi). The harvested tissue was fixed in formalin-acetic acid-alcohol [FAA; 100 ml/L formalin, 50 ml/L glacial acetic acid, 500 ml/L 95% ethanol]. For confocal microscopy, thin longitudinal- and cross-sections were made of stem tissue away from the immediate vicinity of the inoculation site. These sections were stained in 0.01% Calcofluor white fluorescent brightener 28 (Sigma-Aldrich®).
Visualization
Representative stem tissue samples from each time-point were visualized under a Stemi SV6 stereo microscope (Zeiss, Munchen, Germany). Images were captured using an AxioCamMRc digital camera (Zeiss) and Axiovision 4.7 software (Zeiss). The presence of hyphae inside the sampled tissue at various time-points was verified through use of a confocal laser scanning microscope (CLSM 510 Meta, Zeiss). To view Calcofluor-stained tissue, a wavelength of 405 nm was used, and a 543 nm wavelength was used to visualize autofluorescence. LSM Image Browser v4.2.0.121 (Zeiss) was used to view images generated from the confocal microscopy.
RNA extraction and quality analysis
RNA was isolated using a modified cetyl-trimethyl-ammonium-bromide (CTAB) method (Naidoo et al., 2013). On-column DNAse treatment with 10 units DNaseI (Fermentas, Ontario, Canada) and RNA purification was performed with the RNeasy® Mini kit (Qiagen, Valencia, California). Quality analysis was done on a 2100 Bioanalyzer (Agilent, Santa Clara, California).
RNA sequencing
Approximately 20 μg total RNA for each sample was submitted for sequencing at Beijing Genomics Institute (BGI, Beijing, China). Sequencing of mRNA was performed with random fragmentation of the mRNA and adapter ligation. Fifty bp paired end reads were obtained using an Illumina HiSeq 2000 (Illumina, San Diego, CA).
Bioinformatic analysis
Quality analysis and filtering
Adaptors, low quality reads, and reads with more than 10% unknown nucleotides were removed from the dataset by BGI. The Galaxy platform (Giardine et al., 2005; Blankenberg et al., 2010b; Goecks et al., 2010) was used to analyze and process the RNA-seq reads. FASTQ Groomer (Blankenberg et al., 2010a) and FASTQC (Babraham Bioinformatics, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) were used to format reads and to assess read quality.
Mapping and transcript expression analysis of the host
Mapping to the E. grandis v1.1 genome was done with Bowtie (Langmead et al., 2009) and TopHat v1.3.1 (Trapnell et al., 2009), allowing for 2 bp mismatches per 50 bp read and a maximum intron length of 10,000 bp. Mapping statistics were verified using SAMtools flagstat (Li et al., 2009). Assembly of mapped reads and calculation of expression values of predicted E. grandis transcripts as fragments per kilobase of transcript per million fragments (FPKM) was performed by Cufflinks (Trapnell et al., 2010). Significant differential expression analysis was done with Cuffdiff (Trapnell et al., 2010), where parameter settings were changed to allow for a minimum alignment count of 1000, a false discovery rate of 0.05, as well as quartile normalization and bias correction. Additional parameter settings were set to an average fragment length of 49 bp and a standard deviation of 10 bp for the fragment lengths.
Mapping and transcript expression analysis of the pathogen
Reads from the three uninoculated samples and the three inoculated samples were mapped to the P. cinnamomi var cinnamomi draft genome downloaded from the Joint Genome Institute (Reeve, 2012, unpublished; JGI Project identity: 1003775). The genome assembly, based on Illumina, 454 and Sanger sequencing, is 77.97 Mbp and the sequence read coverage depth is 69.6x (Grigoriev et al., 2012). Mapping was performed as described above and assembly of reads and FPKM value were generated using Cufflinks. Any genes or transcripts showing an FPKM value above 100 in the control samples were considered conserved eukaryotic genes and removed from further analysis. Remaining genes or transcripts which showed FPKM values >100 in the three inoculated samples with a coefficient of variation < 0.2 were considered further. Approximately 280 genes satisfied this criteria and were subsequently analyzed using the pathogen host interactions (PHI) database (http://www.phi-base.org/ Winnenburg et al., 2006) to determine pathogenicity or virulence factors expressed in planta using a local BLASTP search in CLCBio main workbench (version 6.1; Qiagen). The database was downloaded in July 2014 and hits with the lowest e-value were considered. Selected genes were also subjected to Blast2GO® analysis to annotate the gene models.
Gene ontology over-representation analysis of host transcripts
Microsoft Excel 2007 (Microsoft, Redmond, WA) was used to match significantly differentially expressed E. grandis v1.1 gene and transcript models to TAIR10 and TAIR9 identifiers based on a reciprocal BLAST analysis. The significantly differentially expressed genes (and transcripts for which the representative gene model was not differentially expressed) with TAIR10 putative orthologs were divided into up- and down-regulated datasets, which were analyzed for gene ontology (GO) over-representation in BiNGO v2.44 (Maere et al., 2005) using the Cytoscape v2.8.3a platform (Shannon et al., 2003). Over-representation was evaluated against the Arabidopsis thaliana genome in the categories for “biological processes,” “molecular function,” and “cellular component.” A hypergeometric test with a Benjamini & Hochberg FDR correction of 0.05 was used. Understanding of biological pathways was aided by MapMan v3.5.1R2 (Thimm et al., 2004).
RT-qPCR
Genomic contamination in total RNA samples was removed by treating extracted total RNA samples with RNase-free DNaseI enzyme (Qiagen Inc., Valencia, CA). Total RNA samples were then purified using the RNeasy® MinElute Kit (Qiagen Inc.) and subsequently analyzed using a Bio-Rad Experion automated electrophoresis system (Bio-Rad Laboratories, Hercules, CA, USA), to determine RNA integrity. The Improm-IITM Reverse Transcription System (Promega, Wisconsin, USA) was used to synthesize first strand cDNA from purified RNA samples. Primers were designed using Primer Designer 4 v4.20 (Sci Ed Central, Cary, North Carolina, USA). Primer pairs are indicated in Table 1.
Table 1.
Primer sequences for Eucalyptus nitens RT-qPCR target and reference genes.
| Eucalyptus ID* | Gene name | Forward primer | Reverse primer |
|---|---|---|---|
| Eucgr.B03520 | EgrWRKY75 | AAGCGCCAGCAGCGGTGGATGAGAA | TGCAGCCGTGGAACGTGTCAACGGTA |
| Eucgr.F02181 | EgrLRR-RLK7 | TTGGTGAATCTCTGGCGACTTGAGC | GACAGATTGACGAGAGCCTCTGGAACT |
| Eucgr.H02533 | EgrNRT2.5 | TGTCCGAATGGAGCGACAAGGAGAA | ACACGGTGCACGAGTACATGAACAG |
| Eucgr.I01495 | EgrPR-3 | GTATTGCTCTCCTAATCC | CATTGCCCGTAGTTATAG |
| Eucgr.J01100 | EgrMLO | GTCAAGAGGTCATTAGAAG | TAGAAGCAAGAAGATAACG |
| Eucgr.I01779 | EgrARF | TGCGTACCGAGTTGTTGAGG | GTTGCACAGGTGCTCTGGAT |
| Eucgr.B02864 | EgrFBA | TGAAGACATGGCAAGGAAGG | GTACCGAAGTTGCTCCGAAT |
| Eucgr.G01186 | EgrTUB | TGAGGTCTTCTCGCGCATTG | AGAGATCTGGCGCAGACAC |
Eucalyptus grandis identities according to www.phytozome.net.
Real-time quantitative reverse transcriptase PCR (RT-qPCR) was conducted according to the Minimum Information for Publication of RT-qPCR Experiments (MIQE) guidelines (Bustin et al., 2009) using a LightCycler® 480 Real-Time PCR system (Roche Diagnostics, GmBh, Basa, Switzerland) following parameters described in Naidoo et al. (2013). The qBASEplus v1.0 (Biogazelle NV, Belgium) software package was used to perform normalization and relative quantification. Significance was determined using a two-tailed Student's t-test in Microsoft® Office Excel 2010.
Phylogenetic analysis
Protein sequences were retrieved from GenBank, Phytozome and JGI, and aligned using MUSCLE (Edgar, 2004). RAxML (Stamatakis, 2006) was used to search for the best scoring maximum likelihood tree with rapid bootstrapping, with GAMMA BLOSUM62 as an evolutionary model. A Bayesian inference analysis was conducted in MrBayes 3.2.2 (Ronquist and Huelsenbeck, 2003). Two chains were sampled once every thousand generations out of a total of one million generations. Trees were summarized with a 10% burn-in. All programs used were housed in Geneious software package version 7.1.5.
Results
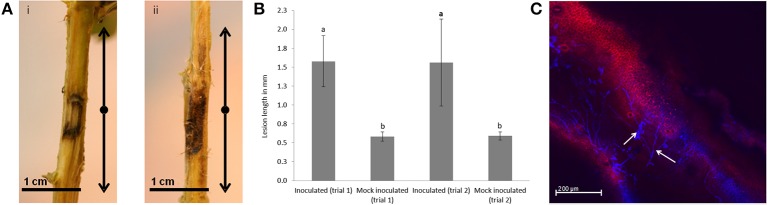
P. cinnamomi infected tissue for RNA-seq profiling was obtained by inoculating the stems of E. nitens seedlings and harvesting at 5 dpi. At this time-point, slight browning was visible around inoculation sites of control plants, whereas lesions of inoculated plants extended to the boundaries of the 3 cm sampled area (Figure 1A). At 6 wpi lesion length was pronounced compared to the mock-inoculated controls (Figure 1B). At 4 wpi, ~50% of the inoculated plants showed mortality. Lesion length was measured for live plants only. The efficacy of inoculation was verified by the presence of hyphae in stem tissue and lesion development over a 4 day time-course (Figure 1C).
Figure 1.
Symptom development in Eucalyptus nitens following challenge with Phytophthora cinnamomi. (A) A section of 1.5 cm stem tissue was harvested below and above the site of inoculation at 5 dpi. (i) Mock-inoculated and (ii) inoculated. (B) Lesions on E. nitens seedlings 6 wpi with P. cinnamomi. The small letters indicate that lesions on inoculated seedlings were significantly larger than the mock-inoculated negative control at p < 0.05 using the Mann–Whitney test for non-parametric data. Error bars show standard error based on n = 12 replicates. (C) Confocal microscopy of a longitudinal stem section showing P. cinnamomi hyphae (white arrows) at 4 dpi.
RNA-sequencing and mapping to Phytophthora cinnamomi and Eucalyptus grandis genomes
Approximately 36 million reads were obtained per sample, and reads were mapped to the E. grandis genome version 1.1 (Table 2). In addition to host transcripts, we mapped P. cinnamomi transcripts expressed in planta, based on the draft P. cinnamomi assembly. A low percentage of mapping (0.08%) was observed in the control (mock inoculated) samples, which were considered conserved eukaryotic gene sequences. On average, 1% of the reads mapped to the P. cinnamomi genome in the inoculated samples. Approximately 78% of the transcripts derived from both inoculated and mock-inoculated E. nitens samples mapped to the E. grandis genome (Table 2). The number of expressed genes and the average FPKM values were similar across the three biological replicates of each treatment which provided confidence that the samples were treated in a consistent manner and the results were comparable across data sets (Table 2).
Table 2.
Flagstat and FastQC RNA-seq mapping statistics of Eucalyptus nitens reads to the v1.1 Eucalyptus grandis and Phytophthora cinnamomi var cinnamomi genomes.
| Sample name | Total reads mapped to E. grandis | Properly paired to E. grandis (%)a | Singletons mapped to E. grandis (%)b | % GC content | Expressed genes in host | Average FPKM in host | Total reads mapped to pathogen | % reads mapped to pathogen |
|---|---|---|---|---|---|---|---|---|
| Control 1 | 37444809 | 66.44 | 10.24 | 50 | 29024 | 493598 | 30756 | 0.08 |
| Control 2 | 36111678 | 68.02 | 10.16 | 50 | 29250 | 492972 | 29274 | 0.08 |
| Control 3 | 37060251 | 68.76 | 8.90 | 49 | 29135 | 467006 | 29116 | 0.08 |
| Inoculated 1 | 37234371 | 66.13 | 11.65 | 49 | 29429 | 471923 | 444935 | 1.19 |
| Inoculated 2 | 36622434 | 67.30 | 12.48 | 49 | 29407 | 497171 | 552202 | 1.51 |
| Inoculated 3 | 36022978 | 68.19 | 10.18 | 49 | 29576 | 473466 | 196534 | 0.55 |
Number of proper pairs in proportion to the total reads mapped.
Number reads where one from a pair in proportion to the total mapped.
Phytophthora cinnamomi genes expressed in planta
The list of transcripts expressed in the three inoculated E. nitens samples with CV < 0.2 are provided as a Supplementary File (Table S1). Of these 283 genes, the genes with hits (E-value ~0) to the plant host interactions database described as loss in pathogenicity, avirulence determinants, or reduced virulence are indicated in Table 3. Several of these have homology to transcripts with known roles in pathogenicity and virulence based on functional genetics experiments in other pathogen species. The highest expressed gene was a member of the CRN family protein. The P. cinnamomi putative CRN (JGI: Phyci261170) was aligned with other putative and described CRN family members and a maximum likelihood phylogenetic tree describing the relationship was produced. A congruent topology was recovered from Bayesian inference. The closest relationship was to a putative CRN protein from P. ramorum. Both the P. ramorum and P. cinammomi putative CRN proteins are closely related to the better characterized P. infestans CRN proteins (Figure 2).
Table 3.
Phytophthora cinnamomi genes expressed in planta implicated in pathogenicity or virulence (E-value < 0.01) based on comparison to the plant-host interactions database.
| Gene identity | Average FPKM | Gene ontology | E-value | Gene name | Knock-out phenotype | PHI base accession |
|---|---|---|---|---|---|---|
| e_gw1.822.3.1 | 1443.83 | gi|301096130|ref|XP_002897163.1|Crinkler (CRN) family protein | 3.85E-91 | crn1 | Effector plant avirulence determinant | 656 |
| estExt_fgenesh1_pg.C_680034 | 1323.48 | hydrogen-transporting ATPase activity, rotational mechanism | 9.04E-26 | invC | Reduced virulence | 645 |
| estExt_Genewise1.C_2370044 | 1096.91 | DNA binding | 5.16E-11 | GzLam002 | Reduced virulence | 1533 |
| e_gw1.40.157.1 | 950 | zinc ion binding, glutathione peroxidase activity, response to oxidative stress | 8.14E-35 | MoHYR1 | Reduced virulence | 2356 |
| fgenesh1_kg.2_#_22_#_Locus405v1rpkm794.38 | 915.58 | protein binding, transcription factor binding, GTP binding, ATP binding | 1.40E-67 | CLPT1 | Reduced virulence | 339 |
| e_gw1.76.61.1 | 853.95 | catalytic activity, hydrolase activity, ATP binding, ATPase activity, ATPase activity | 7.85E-55 | PMR1 | Reduced virulence | 440 |
| estExt_Genewise1.C_610064 | 741.59 | protein kinase activity, GTP binding, protein-tyrosine kinase activity | 1.27E-22 | MoSNF1 | Reduced virulence | 1058 |
| fgenesh1_kg.16_#_102_#_Locus840v1rpkm311.24 | 731.62 | catalytic activity, FAD binding, oxidoreductase activity | 3.59E-04 | ALO1 | Reduced virulence | 197 |
| fgenesh1_kg.9_#_117_#_Locus4954v1rpkm35.55 | 647.81 | catalytic activity, cofactor binding, oxidoreductase activity | 4.47E-32 | MGG | Reduced virulence | 881 |
| fgenesh1_pg.124_#_4 | 620.76 | FAD binding, oxidoreductase activity, cell redox homeostasis, electron transport | 7.82E-06 | SID1 | Reduced virulence | 1010 |
| gm1.2704_g | 421.24 | catalytic activity, ATP binding, metabolism | 0.01 | ACL2 | Loss of pathogenicity | 2387 |
| gm1.12073_g | 324.79 | nucleoside triphosphatase activity, nucleotide binding, hydrolase activity | 2.38E-66 | PEX6 | Loss of pathogenicity | 226 |
| gm1.7056_g | 315.86 | catalytic activity, metabolism | 6.30E-17 | SidI | Reduced virulence | 2321 |
| gm1.272_g | 294.34 | catalytic activity, acetate-CoA ligase activity, AMP binding, etabolism | 3.33E-22 | AKT1 | Loss of pathogenicity | 133 |
| e_gw1.31.72.1 | 268.44 | microtubule motor activity, ATP binding, microtubule-based movement | 2.50E-32 | KIN2 | Reduced virulence | 465 |
| e_gw1.1.234.1 | 254.89 | catalytic activity, metabolism | 9.49E-12 | SidI | Reduced virulence | 2321 |
| e_gw1.93.22.1 | 254.29 | protein binding, protein kinase activity, protein-tyrosine kinase activity | 5.50E-22 | Ste11 | Loss of pathogenicity | 2484 |
| e_gw1.1.500.1 | 204.35 | antioxidant activity, oxidoreductase activity | 5.61E-04 | TSA1 | Reduced virulence | 386 |
| estExt_Genemark1.C_2810025 | 203.63 | electron-transferring-flavoprotein dehydrogenase activity, electron transport | 0 | SIDA | Loss of pathogenicity | 486 |
| e_gw1.67.108.1 | 199.31 | ATP binding | 1.08E-57 | LHS1 | Reduced virulence | 2058 |
| e_gw1.82.257.1 | 198.02 | ATP binding | 3.65E-13 | LHS1 | Reduced virulence | 2058 |
| e_gw1.108.166.1 | 195.13 | protein kinase activity, protein-tyrosine kinase activity | 2.83E-66 | MoCMK1 | Reduced virulence | 2158 |
| e_gw1.2.24.1 | 187.23 | helicase activity, nucleic acid binding, ATP dependent helicase activity, ATP binding | 9.73E-70 | VAD1 | Reduced virulence | 423 |
| e_gw1.184.44.1 | 185.29 | protein kinase activity, protein-tyrosine kinase activity, protein serine/threonine kinase activity | 1.67E-51 | SNF1 | Reduced virulence | 188 |
| fgenesh1_pg.86_#_13 | 184.79 | catalytic activity, aspartic-type endopeptidase activity, metabolism | 1.15E-31 | SidI | Reduced virulence | 2321 |
| MIX7251_264_83 | 180.35 | polygalacturonase activity, carbohydrate metabolism | 0 | Pcipg2 | Reduced virulence | 2343 |
| gm1.8946_g | 164.67 | ATP binding, nucleotide binding, nucleoside triphosphatase activity, tRNA ligase activity | 2.70E-47 | ABC4 | Loss of pathogenicity | 2067 |
| e_gw1.2.738.1 | 153.76 | hydrolase activity, cellulose binding, serine-type endopeptidase activity, blood coagulation | 1.21E-46 | CBEL | Effector plant avirulence determinant | 660 |
| fgenesh1_pg.112_#_14 | 142.08 | gi|325187184|emb|CCA21725.1|bromodomain containing 2 putative [Albugo laibachii Nc14] | 1.63E-09 | GzBrom002 | Reduced virulence | 1317 |
| e_gw1.11.45.1 | 137.55 | phosphotransferase activity, alcohol group as acceptor | 1.02E-31 | VPS34 | Loss of pathogenicity | 195 |
| e_gw1.74.48.1 | 116.37 | protein binding, transcription factor binding, GTP binding, ATP binding, GTPase activity | 5.62E-40 | CLPT1 | Reduced virulence | 339 |
| estExt_fgenesh1_pg.C_1470005 | 110.97 | transporter activity, binding, ATPase activity, ATP binding, transport | 1.15E-26 | MgAtr4 | Reduced virulence | 310 |
| gm1.14921_g | 103.14 | motor activity, ATP binding, myosin | 3.16E-114 | GzWing020 | Reduced virulence | 1648 |
Figure 2.
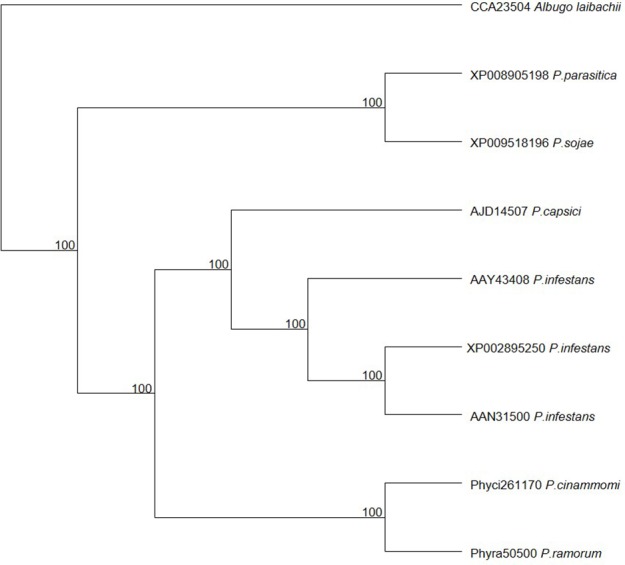
Maximum likelihood phylogenetic tree of the putative Phytophthora cinnamomi crinkler (CRN) protein in relation to CRN proteins in other Phytophthora species.
Differentially expressed genes in Eucalyptus nitens
After mapping to the genome, Cuffdiff was used to compare pathogen-inoculated E. nitens samples with the mock-inoculated controls. A total of 890 up-regulated and 585 down-regulated gene models were observed at a false discovery rate (FDR) of 0.05. The expression of a sub-set of differentially expressed genes EgrWRKY75, EgrPR-3, EgrNRT2.5, EgrMLO, and EgrLRR-RLK7 were validated using RT-qPCR (Figure 3). The tissue for the RT-qPCR validation was sourced from a separate trial that was set up identically to the first trial, with three biological repeats. The expression patterns were comparable to RNA-seq results, with the correlation coefficient between the RNA-seq and the RT-qPCR expression being 0.73.
Figure 3.
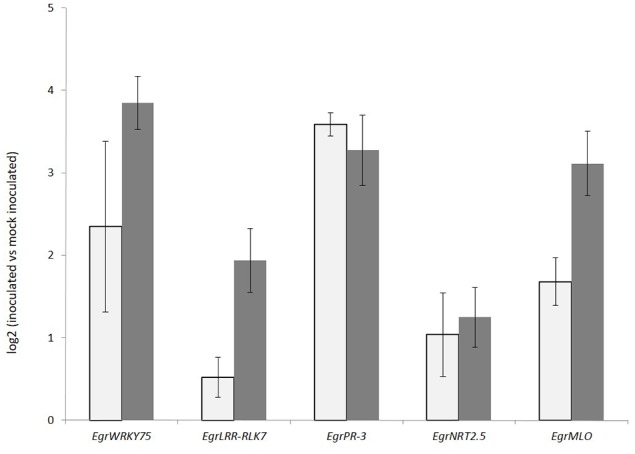
Expression validation of selected genes in Eucalyptus nitens under Phytophthora cinnamomi inoculated compared to mock inoculated conditions 5 days post inoculation. Gray bars represent RT-qPCR expression patterns and dark gray bars, RNA-seq expression patterns.
Over-represented gene ontologies
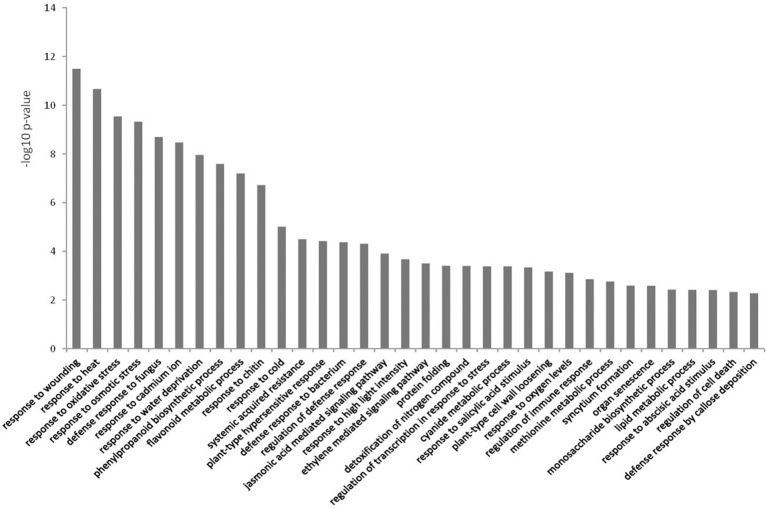
Differentially expressed genes with matching TAIR10 IDs were used in BiNGO to test for over-representation against the A. thaliana genome as background. In the up-regulated dataset for biological processes, the majority of over-represented GO terms were related to defense (Figure 4). Several of the categories involved JA and ET signaling, and there were a few terms related to SA (Figure 4 and Table S1). Phenylpropanoid pathway terms and aromatic compound synthesis related to flavonoid biosynthesis were also found in this dataset (Figure 4 and Table S2). Another prominent term in the up-regulated dataset was response to water deprivation/water stress terms.
Figure 4.
Over-represented Gene Ontologies in the category biological process for the genes up-regulated in response to Phytophthora cinnamomi challenge in Eucalyptus nitens at 5 dpi.
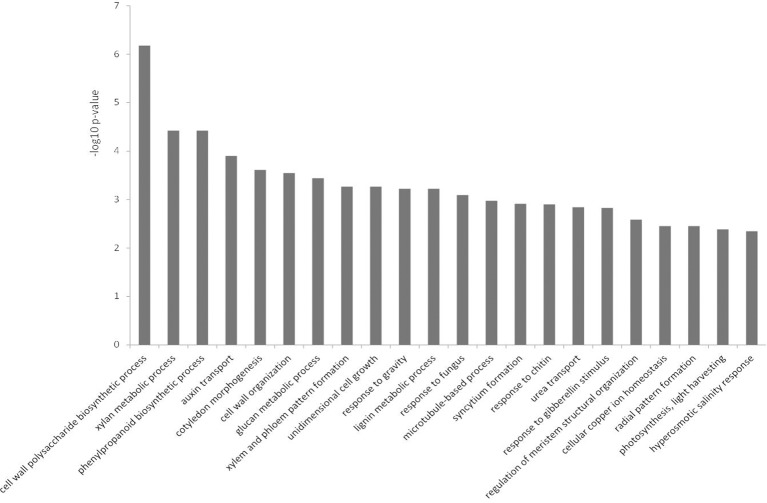
Over-represented GO terms in the down-regulated dataset for biological processes (Figure 5) were predominantly related to growth, cell wall modifications, and cell wall chemistry. The phenylpropanoid pathway terms were related to lignin biosynthesis, as opposed to flavonoid synthesis in the up-regulated dataset (Figure 5 and Table S2). There were minimal biotic stress-related terms, and hormone-related terms in this dataset such as auxin and gibberellin were apparent (Figure 5 and Table S1). Photosynthesis-related terms were also over-represented.
Figure 5.
Over-represented Gene Ontologies in the category biological process for the genes down-regulated in response to Phytophthora cinnamomi challenge in Eucalyptus nitens at 5 dpi.
One of the noteworthy aspects of the RNA-seq data obtained was that several putative PR genes displayed particularly high fold-change values, mostly in the up-regulated dataset. This is summarized in Table 4. Within the E. nitens- P. cinnamomi interaction, the most prominent putative PR genes were PR-1, PR-3 (chitinase), and PR-5 (thaumatin-like and osmotin). These were not only consistently highly up-regulated in inoculated tissue, but there were multiple E. nitens putative orthologs per A. thaliana gene that are all regulated at similar fold-change levels. Putative E. nitens orthologs of PR-4 (chitin-binding), PR-8 (chitinase class III), and PR-12 (defensins) were up-regulated. The PR-9 (peroxidase) and PR-10 (ribonuclease-like) classes contained a mix of up- and down-regulated putative orthologs. PR-14 (lipid transfer proteins) and PR-15 (oxalate oxidase/germin) putative orthologs were all down-regulated.
Table 4.
Expression of pathogenesis related genes in response to Phytophthora cinnamomi inoculation in Eucalyptus nitens.
| Gene family | TAIR ID | E. grandis ID | Description | log2 expression |
|---|---|---|---|---|
| PR-1 | AT2G14580.1 | Eucgr.D01552 | Basic pathogenesis-related protein 1 | 4.56 |
| AT2G14580.1 | Eucgr.G01140 | Basic pathogenesis-related protein 1 | 4.40 | |
| AT2G14580.1 | Eucgr.G01148 | Basic pathogenesis-related protein 1 | 4.37 | |
| AT2G14580.1 | Eucgr.D01560 | Basic pathogenesis-related protein 1 | 4.36 | |
| AT2G14580.1 | Eucgr.G01171 | Pathogenesis-related gene 1 | 4.31 | |
| AT2G14610.1 | Eucgr.G01134 | Basic pathogenesis-related protein 1 | 3.34 | |
| AT2G14610.1 | Eucgr.G01137 | Pathogenesis-related gene 1 | 3.24 | |
| AT2G14610.1 | Eucgr.L02505 | Pathogenesis-related gene 1 | 3.14 | |
| AT2G14610.1 | Eucgr.L01707 | Pathogenesis-related gene 1 | 3.13 | |
| PR-3—Chitinase class I, II, IV, VI, VII | AT3G12500.1 | Eucgr.L00941 | Basic chitinase | 4.02 |
| AT3G12500.1 | Eucgr.J02519 | Basic chitinase | 3.94 | |
| AT3G12500.1 | Eucgr.L00938 | Basic chitinase | 3.93 | |
| AT3G54420.1 | Eucgr.H00326 | Homolog of carrot EP3-3 chitinase | 3.75 | |
| AT3G54420.1 | Eucgr.H00321 | Homolog of carrot EP3-3 chitinase | 3.75 | |
| AT3G12500.1 | Eucgr.L00939 | Basic chitinase | 3.73 | |
| AT3G54420.1 | Eucgr.H00328 | Homolog of carrot EP3-3 chitinase | 3.57 | |
| AT3G12500.1 | Eucgr.L00937 | Basic chitinase | 3.47 | |
| AT3G12500.1 | Eucgr.I01495 | Basic chitinase | 3.11 | |
| AT3G54420.1 | Eucgr.K02166 | Homolog of carrot EP3-3 chitinase | 2.53 | |
| AT3G54420.1 | Eucgr.K02166 | Homolog of carrot EP3-3 chitinase | 2.21 | |
| AT3G54420.1 | Eucgr.A00020 | Homolog of carrot EP3-3 chitinase | 1.43 | |
| AT1G05850.1 | Eucgr.H00455 | Chitinase family protein (TAIR 9) | −0.75 | |
| AT3G16920.1 | Eucgr.H04034 | Chitinase-like protein 2 | −1.33 | |
| PR-4—Chitin-binding | AT3G04720.1 | Eucgr.B02124 | Pathogenesis-related 4 | 3.42 |
| AT3G04720.1 | Eucgr.L03258 | Pathogenesis-related 4 | 3.32 | |
| AT3G04720.1 | Eucgr.B02122 | Pathogenesis-related 4 | 3.06 | |
| PR-5—Thaumatin-like and osmotin | AT1G20030.2 | Eucgr.E01382 | Pathogenesis-related thaumatin superfam | 5.42 |
| AT1G20030.2 | Eucgr.E01384 | Pathogenesis-related thaumatin superfam | 5.31 | |
| AT1G20030.2 | Eucgr.E01389 | Pathogenesis-related thaumatin superfam | 5.23 | |
| AT1G20030.2 | Eucgr.E01385 | Pathogenesis-related thaumatin superfam | 5.13 | |
| AT4G11650.1 | Eucgr.H03863 | Osmotin 34 | 5.09 | |
| AT1G20030.2 | Eucgr.E01381 | Pathogenesis-related thaumatin superfam | 4.96 | |
| AT4G11650.1 | Eucgr.H03865 | Osmotin 34 | 4.93 | |
| AT4G11650.1 | Eucgr.H03864 | Osmotin 34 | 4.77 | |
| AT4G11650.1 | Eucgr.L01962 | Osmotin 34 | 4.69 | |
| AT4G11650.1 | Eucgr.E00557 | Osmotin 34 | 4.43 | |
| AT4G11650.1 | Eucgr.D01888 | Osmotin 34 | 3.57 | |
| AT4G11650.1 | Eucgr.D01892 | Osmotin 34 | 3.34 | |
| AT4G11650.8 | Eucgr.D01887 | Osmotin 34 | 3.31 | |
| AT4G11650.9 | Eucgr.E00560 | Osmotin 34 | 2.97 | |
| AT5G38280.1 | Eucgr.A01474 | PR5-like receptor kinase | 1.03 | |
| AT5G38280.1 | Eucgr.A01470 | PR5-like receptor kinase | 0.96 | |
| AT5G38280.1 | Eucgr.A01478 | PR5-like receptor kinase | 0.78 | |
| AT2G28790.1 | Eucgr.J02061 | Pathogenesis-related thaumatin superfam | −1.18 | |
| AT4G38660.1 | Eucgr.G01772 | Pathogenesis-related thaumatin superfam | −1.33 | |
| AT1G73620.1 | Eucgr.B00944 | Pathogenesis-related thaumatin superfam | −1.36 | |
| PR-8—Chitinase class III | AT5G24090.1 | Eucgr.E00091 | Chitinase A | 2.40 |
| AT5G24090.1 | Eucgr.L03478 | Chitinase A | 1.52 | |
| PR-9—Peroxidase | AT4G37530.1 | Eucgr.J02352 | Peroxidase superfamily protein | 3.65 |
| AT4G11600.1 | Eucgr.D01857 | Glutathione peroxidase 6 (TAIR 9) | 3.08 | |
| AT1G71695.1 | Eucgr.F04198 | Peroxidase superfamily protein | 2.34 | |
| AT1G05260.1 | Eucgr.A01385 | Peroxidase superfamily protein | 1.83 | |
| AT1G71695.1 | Eucgr.F04195 | Peroxidase superfamily protein | 1.46 | |
| AT1G71695.1 | Eucgr.L02740 | Peroxidase superfamily protein | 1.39 | |
| AT5G40150.1 | Eucgr.J02173 | Peroxidase superfamily protein | −1.23 | |
| AT5G42180.1 | Eucgr.F03724 | Peroxidase superfamily protein | −1.76 | |
| AT4G21960.1 | Eucgr.E04056 | Peroxidase superfamily protein | −1.85 | |
| PR-10—Ribonuclease-like | AT1G80780.3 | Eucgr.F03953 | Polynucleotidyl transferase, ribonuclease | 0.83 |
| AT5G22250.1 | Eucgr.J00535 | Polynucleotidyl transferase, ribonuclease | −1.46 | |
| PR-12—Defensins | AT4G11393.1 | Eucgr.K03440 | Defensin-like (DEFL) family protein | 2.72 |
| PR-14—Lipid transfer proteins | AT5G48485.1 | Eucgr.H00727 | Bifunctional inhibitor/lipid-transfer protein | −0.85 |
| AT5G64080.1 | Eucgr.I02679 | Bifunctional inhibitor/lipid-transfer protein | −0.95 | |
| AT3G18280.1 | Eucgr.B00824 | Bifunctional inhibitor/lipid-transfer protein | −1.00 | |
| AT5G59320.1 | Eucgr.K01283 | Lipid transfer protein 3 | −1.20 | |
| AT5G55460.1 | Eucgr.F03514 | Bifunctional inhibitor/lipid-transfer protein | −1.31 | |
| AT5G59320.1 | Eucgr.A00746 | Lipid transfer protein 4 | −1.60 | |
| AT5G05960.1 | Eucgr.K03041 | Bifunctional inhibitor/lipid-transfer protein | −1.66 | |
| AT5G59320.1 | Eucgr.K01282 | Lipid transfer protein 5 | −1.76 | |
| PR-15—Oxalate oxidases (Germin) | AT3G62020.1 | Eucgr.A00990 | Germin-like protein 10 | −1.18 |
Up and down regulated genes are indicated as a gradient from bright red to bright green.
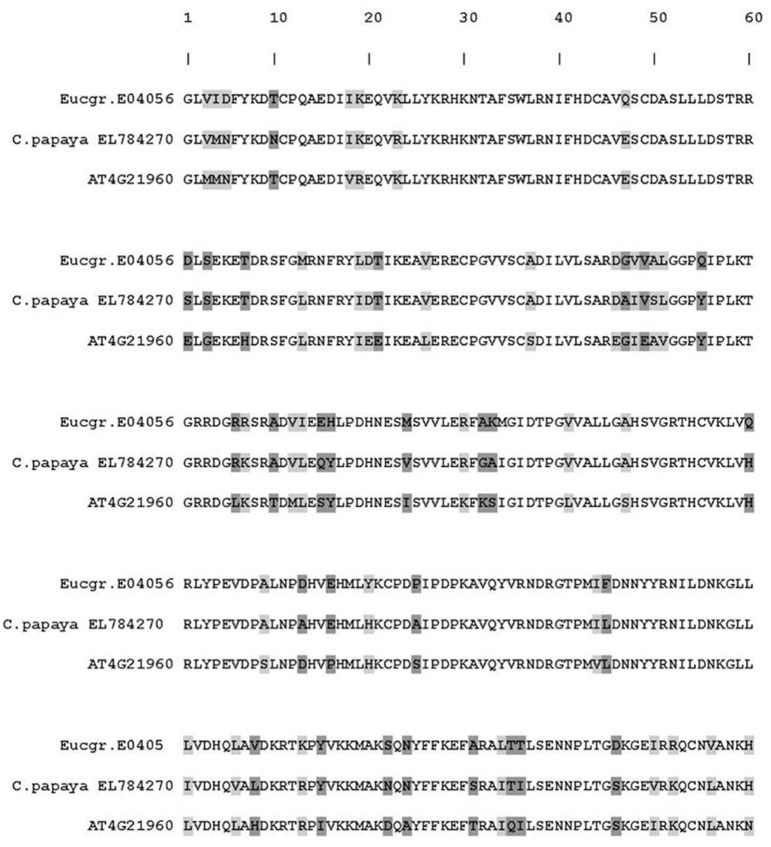
The E. nitens putative PR-9 ortholog (Eucgr.E04056) was down regulated by a 1.85 fold-change on a log2 scale in the P. cinnamomi-inoculated tissue (indicated in bold in Table 4). The sequence of this ortholog was compared to peroxidase from Carica papaya (EL784270) and A. thaliana. The amino acid alignment is indicated in Figure 6. The E. nitens PR-9 sequence shared 80% amino acid identity with the C. papaya PR-9 sequence.
Figure 6.
Amino acid alignment of the putative PR-9 peroxidase ortholog of Eucalyptus nitens (Phytozome: Eucgr.E04056) in comparison to the Arabidopsis thaliana (TAIR number AT4G21960) and Carica papaya (Genbank: EL784270) orthologs. Light gray highlights conservative amino acid substitutions and dark gray indicates non-conservative substitutions.
Discussion
The compatible interaction between E. nitens and P. cinnamomi provided a model system to study the compatible interaction between Eucalyptus spp. and Phytophthora spp. One year old E. nitens plants were stem inoculated using P. cinnamomi and pronounced lesions were obtained (Figure 1) suggesting that successful infection had occurred. This was corroborated with the presence of hyphae in the stems as early as 48 hpi in some instances and subsequently consistently observed at 5 dpi. In order to gain a better understanding of this compatible pathosystem, a dual RNA sequencing approach, as described previously for other plant-pathogen interactions (Hayden et al., 2014), was undertaken to concurrently detect pathogenicity factors and host responses.
Analysis of transcripts mapping to the P. cinnamomi genome confirmed the presence of ~1% pathogen in the tissue profiled. We obtained 78% mapping of the E. nitens transcripts to the E. grandis genome which was similar to that obtained in a study by Ward and Weber (2012) where raspberry transcripts were mapped to the strawberry genome. This substantiates the use of cross-species resources in the event that no such genomic resources are available for the species of interest.
Table 3 indicates possible determinants expressed in planta which may contribute to the susceptible outcome of the interaction. A pathogen transcript encoding a CRN family protein was highly expressed in planta. In Phytophthora species, the CRNs are a complex family of large proteins and various experiments suggest that some CRNs are able to target host factors to suppress plant defenses (Adhikari et al., 2013). The P. cinnamomi CRN is closely related to the CRN1 protein from P. infestans, suggesting that it may have the same role as described in P. infestans. Torto et al. (2003) showed that the P. infestans CRN1 and CRN2 effectors were expressed during infection of tomato and that CRN1 and CRN2 were able to cause necrosis in tobacco and CRN2 induced PR1a expression in tomato. While some CRN effectors are known to target host nuclei (Stam et al., 2013a) the role in virulence may be diverse in various life stages of P. infestans (Resjö et al., 2014). Liu et al. (2011) described two CRN effectors from P. sojae where one induced cell death and the other suppressed cell death in soybean. More recently, it was shown that the two effectors together, or one alone, can suppress host defenses by interacting with catalases and in so doing, modulate the HR and H2O2 levels in planta (Zhang et al., 2015). Another well characterized example of an apoplastic effector is that of the Cellulose Binding, Elicitor, and Lectin-like (CBEL) transcript, which elicits necrosis and defense gene expression in hosts and is necessary for attachment onto plant surfaces (Mateos et al., 1997; Séjalon-Delmas et al., 1997). Further examination of the virulence factors and the host targets they affect, will provide insight into the cause of the host responses observed in E. nitens.
The over-represented GO terms in E. nitens challenged with P. cinnamomi (Figures 4, 5) were indicative of a host actively attempting to combat infection, albeit at a late stage of a compatible interaction. We observed differential expression of various defense responses, but highlight two possible factors contributing to susceptibility.
Some physiological responses to P. cinnamomi include the down-regulation of photosynthesis-related terms and up-regulation of water stress terms. Various tree species inoculated with P. cinnamomi show declines in stomatal conductance and photosynthesis. In Eucalyptus sieberi, this decline is associated with susceptibility, since these factors decreased less severely in resistant Eucalyptus sideroxylon (Dempsey et al., 2012). In Quercus suber, photosynthesis and stomatal conductance also decreased after inoculation (Medeira et al., 2012). Manter et al. (2007) noted the prevalence of photosynthetic and stomatal conductance decreases in Phytophthora-host interactions. They showed that photosynthetic decline could be caused by elicitins in the absence of water stress. Maintaining adequate photosynthetic levels may assist tolerance or resistance, which is a possible explanation of why lower photosynthetic rates and stomatal conductance is associated with an increase in pathogen (Portz et al., 2007) and lower tolerance to Phytophthora spp. (Reeksting et al., 2014).
Distinct components of the phenylpropanoid pathway are present in both the up- and down-regulated datasets. There are several up-regulated genes with GO annotations associated with flavonoid biosynthesis. Susceptible Lupinus angustifolius up-regulated the flavonoid genistein in response to P. cinnamomi (Gunning et al., 2013) however, certain Citrus flavonoids have an antimicrobial action against Phytophthora citrophthora (del Río et al., 2004). In the down-regulated dataset, GO terms associated with lignin synthesis via the phenylpropanoid pathway are over-represented. For lignin synthesis, most genes encoding enzymes involved in biosynthesis of the coniferyl alcohol (G subunit) and sinapyl alcohol (S subunit) are down-regulated, although genes encoding enzymes catalyzing the synthesis of coumaryl alcohol (H subunit) are up-regulated (Table S2). Since synthesis of the S and G subunits is possibly suppressed, lignin biosynthesis could be down-regulated in E. nitens. Lignin is associated with strengthening of cell-walls and helps prevent penetration by a pathogen (Bechinger et al., 1999), and down-regulation of monolignols can compromise host resistance (Naoumkina et al., 2010). Lignin synthesis plays a role in raspberry responses to challenge with P. rubi (Ward and Weber, 2012) and transgenic potato plants with limited phenylpropanoid substrates had increased susceptibility to P. infestans (Yao et al., 1995).
Gene ontology terms pertaining to JA, SA, and ET pathways were over-represented in the up-regulated dataset in E. nitens, suggesting that they could play a role in defense signaling for this interaction. Other studies involving hosts inoculated with Phytophthora spp. have also shown mixed hormone responses (Attard et al., 2010; Shibata et al., 2010). JA may be needed for successful defense against P. cinnamomi in maize, a resistant monocot (Allardyce et al., 2013). Terms related to GA and auxin were over-represented in the down-regulated dataset. Treatment of soybean with GA increased susceptibility to P. sojae (Sugano et al., 2013) and it has been proposed that GA influences defense against necrotrophic fungi by repressing resistance (Mengiste, 2012). The role of auxin in defense against P. cinnamomi has recently been described (Eshraghi et al., 2014b), where Arabidopsis auxin sensitivity and transport mutants were shown to be highly susceptible to the pathogen. Treatment of L. angustifolius with an inhibitor of auxin transport increased susceptibility to P. cinnamomi.
Since PR genes are markers of defense hormone signaling, the different putative PR genes expressed reflect the mix of JA and SA signaling noted in the over-representation analysis.
Gene models identified as PR genes in this dataset are putative orthologs of A. thaliana PR genes. For many of these genes, there are multiple differentially expressed E. grandis gene models matching to one A. thaliana putative ortholog and an expansion of several PR genes in E. grandis has been noted (Naidoo et al., 2014). Expression of these multiple PR gene transcripts in E. nitens could indicate that some of the orthologs have slightly different functions and are all used during a defense response.
Transcription of PR-1, chitinase (PR-3), chitin-binding protein (PR-4), and thaumatin-like protein/osmotin (PR-5) putative orthologs appears to be highly up-regulated in E. nitens (Table 4). A Phytophthora-resistant potato expresses PR-1 constitutively (Ali et al., 2012) and constitutive expression of PR-1 in transgenic tobacco confers resistance to P. parasitica (Alexander et al., 1993). Transgenic plants over-expressing PR-5 genes have been shown to increase resistance to P. citrophthora and P. infestans (Fagoaga et al., 2001; Pushin et al., 2010; Acharya et al., 2013).
Peroxidases (PR-9) are potential cross-species Phytophthora effector targets, since a certain C. papaya peroxidase (EL784270) and its putative orthologs have been suppressed in different hosts upon inoculation with P. sojae, P. palmivora, and P. infestans (Moy et al., 2004; Restrepo et al., 2005; Porter et al., 2009). An E. grandis gene, Eucgr.E04056, is highly similar to the C. papaya ortholog, and is also strongly suppressed in the current interaction. Putative PR-14 and PR-15 orthologs were down-regulated in E. nitens and while these orthologs would have to be characterized further, it is tempting to speculate that their suppression may be driving susceptibility. For example, enhanced resistance to P. nicotianae was conferred by a pepper lipid transfer protein over-expressed in tobacco (Sarowar et al., 2009) and PR-15 may encode for a germin-like oxalate oxidase known to produce hydrogen peroxide that is toxic to pathogens (van Loon et al., 2006; Ferreira et al., 2007).
Conclusion
The outcomes of this dual RNA-seq study provided valuable insights into P. cinnamomi pathogenicity and virulence factors and E. nitens defense mechanisms utilized against P. cinnamomi. Several factors may contribute to the compatibility however, we have used further sequence and functional genetics support to motivate that the P. cinnamomi CRN and the E. nitens PR-9 genes are important contributors to the susceptible outcome. Future work involving comparison with a resistant interaction over a time-course is required to provide an indication of host targets manipulated by P. cinnamomi and to enhance understanding of the defense pathways required for resistance.
Author contributions
FM, LS, SiN, and TM performed the experimental work, conducted and interpreted data analyses. SaN conceived the study, obtained funding to support the research. AM provided input into the experimental design and technical aspects of RNA-sequencing and assisted with critical evaluation of the manuscript. FM, LS, and SaN wrote the manuscript with input from DB and NV, who supervised aspects of this research.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Charles Hefer for creating the TopHat and FastQGroomer pipeline on Galaxy, Melissa Reynolds, and Christy Marais for conducting the relatedness analysis of the plant material, and Eshchar Mizrachi, Burger van Jaarsveld, and Karen van der Merwe for bioinformatics support. This work is based on the research supported in part by the National Research Foundation of South Africa for the grant no. 76225 and 86936; the Forest Molecular Genetics Program by Mondi and Sappi, the Technology and Human Resources for Industry Program (80118), and Department of Science and Technology, The Eucalyptus Genomics Platform: Tree genomics and biotechnology for wood fibre, bioenergy and biomaterials. Any opinion, finding and conclusion or recommendation expressed in this material is that of the author(s) and the NRF does not accept any liability in this regard.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00191
References
- Acharya K., Pal A. K., Gulati A., Kumar S., Singh A. K., Ahuja P. S. (2013). Overexpression of Camellia sinensis thaumatin-like protein, CsTLP in potato confers enhanced resistance to Macrophomina phaseolina and Phytophthora infestans infection. Mol. Biotechnol. 54, 609–622. 10.1007/s12033-012-9603-y [DOI] [PubMed] [Google Scholar]
- Adhikari B. N., Hamilton J. P., Zerillo M. M., Tisserat N., Lévesque C. A., Buell C. R. (2013). Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes. PLoS ONE 8:e75072. 10.1371/journal.pone.0075072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adl S. M., Simpson A. G. B., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., et al. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451. 10.1111/j.1550-7408.2005.00053.x [DOI] [PubMed] [Google Scholar]
- Adl S. M., Simpson A. G. B., Lane C. E., Lukeš J., Bass D., Bowser S. S., et al. (2012). The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–493. 10.1111/j.1550-7408.2012.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D., Goodman R. M., Gut-Rella M., Glascock C., Weymann K., Friedrich L., et al. (1993). Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. U.S.A. 90, 7327–7331. 10.1073/pnas.90.15.7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A., Alexandersson E., Sandin M., Resjö S., Lenman M., Hedley P., et al. (2014). Quantitative proteomics and transcriptomics of potato in response to Phytophthora infestans in compatible and incompatible interactions. BMC Genomics 15:497. 10.1186/1471-2164-15-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A., Moushib L. I., Lenman M., Levander F., Olsson K., Carlson-Nilson U., et al. (2012). Paranoid potato: phytophthora-resistant genotype shows constitutively activated defense. Plant Signal. Behav. 7, 400–408. 10.4161/psb.19149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allardyce J. A., Rookes J. E., Hussain H. I., Cahill D. M. (2013). Transcriptional profiling of Zea mays roots reveals roles for jasmonic acid and terpenoids in resistance against Phytophthora cinnamomi. Funct. Integr. Genomics 13, 217–228. 10.1007/s10142-013-0314-7 [DOI] [PubMed] [Google Scholar]
- Attard A., Gourgues M., Callemeyn-Torre N., Keller H. (2010). The immediate activation of defense responses in Arabidopsis roots is not sufficient to prevent Phytophthora parasitica infection. New Phytol. 187, 449–460. 10.1111/j.1469-8137.2010.03272.x [DOI] [PubMed] [Google Scholar]
- Bari R., Jones J. D. G. (2009). Role of plant hormones in plant defense responses. Plant Mol. Biol. 69, 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Bechinger C., Giebel K.-F., Schnell M., Leiderer P., Deising H. B., Bastmeyer M. (1999). Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 285, 1896–1899. 10.1126/science.285.5435.1896 [DOI] [PubMed] [Google Scholar]
- Belhaj K., Lin B., Mauch F. (2009). The chloroplast protein RPH1 plays a role in the immune response of Arabidopsis to Phytophthora brassicae. Plant J. 58, 287–298. 10.1111/j.1365-313X.2008.03779.x [DOI] [PubMed] [Google Scholar]
- Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., Nekrutenko A. (2010a). Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783–1785. 10.1093/bioinformatics/btq281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D., Kuster G. V., Coraor N., Ananda G., Lazarus R., Mangan M., et al. (2010b). Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 89, 19.10.11–19.10.21. 10.1002/0471142727.mb1910s89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. I. B., Armstrong M. R., Gilroy E. M., Boevink P. C., Hein I., Taylor R. M., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. U.S.A. 107, 9909–9914. 10.1073/pnas.0914408107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier C. M. (2008). The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 57, 792–808. 10.1111/j.1365-3059.2008.01886.x [DOI] [Google Scholar]
- Burgess T., McComb J. A., Colquhoun I., Hardy G. E. S. (1999). Increased susceptibility of Eucalyptus marginata to stem infection by Phytophthora cinnamomi resulting from root hypoxia. Plant Pathol. 48, 797–806. 10.1046/j.1365-3059.1999.00396.x [DOI] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Cahill D. M., Rookes J. E., Wilson B. A., Gibson L., McDougall K. L. (2008). Turner Review No. 17. Phytophthora cinnamomi and Australia's biodiversity: impacts, predictions and progress towards control. Aust. J. Bot. 56, 279–310. 10.1071/BT07159 [DOI] [Google Scholar]
- Camilios-Neto D., Bonato P., Wassem R., Tadra-Sfeir M. Z., Brusamarello-Santos L. C. C., Valdameri G., et al. (2014). Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Genomics 15:378. 10.1186/1471-2164-15-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-R., Zhang B.-Y., Xing Y.-P., Li Q.-Y., Li Y.-P., Tong Y.-H., et al. (2014). Transcriptomic analysis of the phytopathogenic oomycete Phytophthora cactorum provides insights into infection-related effectors. BMC Genomics 15:980. 10.1186/1471-2164-15-980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. J., Aliota M. T., Mayhew G. F., Erickson S. M., Christensen B. M. (2014). Dual RNA-seq of parasite and host reveals gene expression dynamics during filarial worm-mosquito interactions. PLoS Negl. Trop. Dis. 8:e2905. 10.1371/journal.pntd.0002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río J., Gómez P., Baidez A., Arcas M., Botia J., Ortuno A. (2004). Changes in the levels of polymethoxyflavones and flavanones as part of the defense mechanism of Citrus sinensis (cv. Valencia Late) fruits against Phytophthora citrophthora. J. Agric. Food Chem. 52, 1913–1917. 10.1021/jf035038k [DOI] [PubMed] [Google Scholar]
- Dempsey R. W., Merchant A., Tausz M. (2012). Differences in ascorbate and glutathione levels as indicators of resistance and susceptibility in Eucalyptus trees infected with Phytophthora cinnamomi. Tree Physiol. 32, 1148–1160. 10.1093/treephys/tps076 [DOI] [PubMed] [Google Scholar]
- Dowen R. H., Pelizzola M., Schmitz R. J., Lister R., Dowen J. M., Nery J. R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U.S.A. 109, E2183–E2191 10.1073/pnas.1209329109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin D. C., Ribeiro O. K. (1996). Phytophthora Diseases Worldwide. St. Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Eshraghi L., Anderson J. P., Aryamanesh N., McComb J. A., Shearer B., Hardy G. E. S. J. (2014a). Defence signalling pathways involved in plant resistance and phosphite-mediated control of Phytophthora cinnamomi. Plant Mol. Biol. Rep. 32, 342–356. 10.1007/s11105-013-0645-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshraghi L., Anderson J. P., Aryamanesh N., McComb J. A., Shearer B., Hardy G. E. S. J. (2014b). Suppression of the auxin response pathway enhances susceptibility to Phytophthora cinnamomi while phosphite-mediated resistance stimulates the auxin signalling pathway. BMC Plant Biol. 14:68. 10.1186/1471-2229-14-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagoaga C., Rodrigo I., Conejero V., Hinarejos C., Tuset J. J., Arnau J., et al. (2001). Increased tolerance to Phytophthora citrophthora in transgenic orange plants constitutively expressing a tomato pathogenesis related protein PR-5. Mol. Breed. 7, 175–185. 10.1023/A:1011358005054 [DOI] [Google Scholar]
- FAO (2007). Overview of forests pests South Africa, in Forest Health & Biosecurity Working Papers, ed Allard G. (Rome: Food and Agriculture Organization of the United Nations; ), 4–20. [Google Scholar]
- Faria D. A., Mamani E. M. C., Pappas G. J., Grattapaglia D. (2010). Genotyping systems for Eucalyptus based on tetra-, penta-, and hexanucleotide repeat EST microsatellites and their use for individual fingerprinting and assignment tests. Tree Genet. Genomes 7, 63–77. 10.1007/s11295-010-0315-9 [DOI] [Google Scholar]
- Ferreira R. B., Monteiro S., Freitas R., Santos C. N., Chen Z., Batista L. M., et al. (2007). The role of plant defense proteins in fungal pathogenesis. Mol. Plant Pathol. 8, 677–700. 10.1111/j.1364-3703.2007.00419.x [DOI] [PubMed] [Google Scholar]
- Gao L., Tu Z. J., Millett B. P., Bradeen J. M. (2013). Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions. BMC Genomics 14:340. 10.1186/1471-2164-14-340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., Shah P., et al. (2005). Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455. 10.1101/gr.4086505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy E. M., Taylor R. M., Hein I., Boevink P., Sadanandom A., Birch P. R. J. (2011). CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 190, 653–666. 10.1111/j.1469-8137.2011.03643.x [DOI] [PubMed] [Google Scholar]
- Godsmark R. (2009). The South African forestry and forest products industry 2009, in Forestry South Africa. Available Online at: http://www.forestry.co.za/statistical-data/ (Accessed May 31 2012).
- Goecks J., Nekrutenko A., Taylor J. (2010). Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86. 10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I. V., Nordberg H., Shabalov I., Aerts A., Cantor M., Goodstein D., et al. (2012). The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 40, D26–D32. 10.1093/nar/gkr947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning T. K., Conlan X. A., Parker R. M., Dyson G. A., Adams M. J., Barnett N. W., et al. (2013). Profiling of secondary metabolites in blue lupin inoculated with Phytophthora cinnamomi following phosphite treatment. Funct. Plant Biol. 40, 1089–1097. 10.1071/FP13023 [DOI] [PubMed] [Google Scholar]
- Hansen E. M. (2008). Alien forest pathogens: Phytophthora species are changing world forests. Boreal Environ. Res. 13, 33–41. [Google Scholar]
- Hayden K. J., Garbelotto M., Knaus B. J., Cronn R. C., Rai H., Wright J. W. (2014). Dual RNA-seq of the plant pathogen Phytophthora ramorum and its tanoak host. Tree Genet. Genomes 10, 489–502. 10.1007/s11295-014-0698-0 [DOI] [Google Scholar]
- He X., Miyasaka S. C., Fitch M. M., Khuri S., Zhu Y. J. (2013). Taro (Colocasia esculenta) transformed with a wheat oxalate oxidase gene for improved resistance to taro pathogen Phytophthora colocasiae. HortSci. 48, 22–27. [Google Scholar]
- Jiang R. H., Tyler B. M. (2012). Mechanisms and evolution of virulence in oomycetes. Annu. Rev. Phytopathol. 50, 295–318. 10.1146/annurev-phyto-081211-172912 [DOI] [PubMed] [Google Scholar]
- Kunjeti S. G., Evans T. A., Marsh A. G., Gregory N. F., Kunjeti S., Meyers B. C., et al. (2012). RNA-Seq reveals infection-related global gene changes in Phytophthora phaseoli, the causal agent of lima bean downy mildew. Mol. Plant Pathol. 13, 454–466. 10.1111/j.1364-3703.2011.00761.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde C., Drenth A., Wingfield M. J. (1999). Gene and genotypic diversity of Phytophthora cinnamomi in South Africa and Australia revealed by DNA polymorphisms. Eur. J. Plant Pathol. 105, 667–680. 10.1023/A:1008755532135 [DOI] [Google Scholar]
- Linde C., Kemp G. H. J., Wingfield M. J. (1994). Diseases of pines and eucalypts in South Africa associated with Pythium and Phytophthora species. South Afr. For. J. 169, 25–32. 10.1080/00382167.1994.9629663 [DOI] [Google Scholar]
- Liu T., Ye W., Ru Y., Yang X., Gu B., Tao K., et al. (2011). Two host cytoplasmic effectors are required for pathogenesis of Phytophthora sojae by suppression of host defenses. Plant Physiol. 155, 490–501. 10.1104/pp.110.166470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- Manter D. K., Kelsey R. G., Karchesy J. J. (2007). Photosynthetic declines in Phytophthora ramorum-infected plants develop prior to water stress and in response to exogenous application of elicitins. Phytopathology 97, 850–856. 10.1094/PHYTO-97-7-0850 [DOI] [PubMed] [Google Scholar]
- Martinelli F., Uratsu S. L., Albrecht U., Reagan R. L., Phu M. L., Britton M., et al. (2012). Transcriptome profiling of citrus fruit response to huanglongbing disease. PLoS ONE 7:e38039. 10.1371/journal.pone.0038039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maseko B. (2010). Dieback of Cold Tolerant Eucalypts Associated with Phytophthora spp. in South Africa. Ph.D. thesis, University of Pretoria. [Google Scholar]
- Mateos F. V., Rickauer M., Esquerretugaye M. T. (1997). Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol. Plant-Microbe Interact. 10, 1045–1053. 10.1094/MPMI.1997.10.9.1045 [DOI] [PubMed] [Google Scholar]
- Medeira C., Quartin V., Maia I., Diniz I., Matos M. C., Semedo J. N., et al. (2012). Cryptogein and capsicein promote defense responses in Quercus suber against Phytophthora cinnamomi infection. Eur. J. Plant Pathol. 134, 145–159. 10.1007/s10658-012-9972-x [DOI] [Google Scholar]
- Mengiste T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. 10.1146/annurev-phyto-081211-172955 [DOI] [PubMed] [Google Scholar]
- Motulsky H. (1999). Prism 5 Statistics Guide. San Diego, CA: GraphPad Software Incorporated; Available online at: www.graphpad.com [Google Scholar]
- Moy P., Qutob D., Chapman B. P., Atkinson I., Gijzen M. (2004). Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol. Plant-Microbe Interact. 17, 1051–1062. 10.1094/MPMI.2004.17.10.1051 [DOI] [PubMed] [Google Scholar]
- Myburg A. A., Grattapaglia D., Tuskan G. A., Hellsten U., Hayes R. D., Grimwood J., et al. (2014). The genome of Eucalyptus grandis. Nature 510, 356–362. 10.1038/nature13308 [DOI] [PubMed] [Google Scholar]
- Naidoo R., Ferreira L., Berger D. K., Myburg A. A., Naidoo S. (2013). The identification and differential expression of Eucalyptus grandis pathogenesis-related genes in response to salicylic acid and methyl jasmonate. Front. Plant Science 4:43. 10.3389/fpls.2013.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo S., Kulheim C., Zwart L., Mangwanda R., Oates C. N., Visser E. A., et al. (2014). Uncovering the defence responses of Eucalyptus to pests and pathogens in the genomics age. Tree Physiol. 34, 931–943. 10.1093/treephys/tpu075 [DOI] [PubMed] [Google Scholar]
- Naoumkina M. A., Zhao Q., Gallego-Giraldo L., Dai X., Zhao P. X., Dixon R. A. (2010). Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 11, 829–846. 10.1111/j.1364-3703.2010.00648.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oβwald W., Fleischmann F., Rigling D., Coelho A. C., Cravador A., Diez J., et al. (2014). Strategies of attack and defence in woody plant-Phytophthora interactions. For. Pathol. 44, 169–190. 10.1111/efp.12096 [DOI] [Google Scholar]
- Podger F. D., Palzer C. R., Batini F. E. (1967). Phytophthora cinnamomi in the Jarrah forests of Western Australia. Commonwealth Phytopathol. News 4, 1–2. [Google Scholar]
- Porter B. W., Zhu Y. J., Christopher D. A. (2009). Carica papaya genes regulated by Phytophthora palmivora: a model system for genomic studies of compatible Phytophthora-plant interactions. Trop. Plant Biol. 2, 84–97. 10.1007/s12042-009-9030-9 [DOI] [Google Scholar]
- Portz R. L., Koehl J., Fleischmann F., Oßwald W. (2007). Physiological changes and gene expression on European beech (Fagus sylvatica) infected with Phytophthora citricola, in Fourth Meeting of IUFRO Working Party S07.02.09, Phytophthoras in Forests and Natural Ecosystems (Monterey, CA: ), 310. [Google Scholar]
- Pushin A. S., Firsov A. P., Dolgov S. V., Monakhos G. F., Motamedi Shalamzari A., Dzhalilov F. S., et al. (2010). Transgenic tomato plants expressing pr-5 protein genes demonstrated disease resistance against Phytophthora infestans and xanthomonas vesicatoria, in III International Symposium on Tomato Diseases (Naples: ), 415–418. [Google Scholar]
- Reeksting B. J., Taylor N. J., Van Den Berg N. (2014). Flooding and phytophthora cinnamomi: effects on photosynthesis and chlorophyll fluorescence in shoots of non-grafted persea americana (Mill.) rootstocks differing in tolerance to Phytophthora root rot. South Afr. J. Bot. 95, 40–53. 10.1016/j.sajb.2014.08.004 [DOI] [Google Scholar]
- Resjö S., Ali A., Meijer H. J. G., Seidl M. F., Snel B., Sandin M., et al. (2014). Quantitative label-free phosphoproteomics of six different life stages of the late blight pathogen Phytophthora infestans reveals abundant phosphorylation of members of the CRN effector family. J. Proteome Res. 13, 1848–1859. 10.1021/pr4009095 [DOI] [PubMed] [Google Scholar]
- Restrepo S., Myers K. L., del Pozo O., Martin G. B., Hart A. L., Buell C. R., et al. (2005). Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Mol. Plant-Microbe Interact. 18, 913–922. 10.1094/MPMI-18-0913 [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sarowar S., Kim Y. J., Kim K. D., Hwang B. K., Ok S. H., Shin J. S. (2009). Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep. 28, 419–427. 10.1007/s00299-008-0653-3 [DOI] [PubMed] [Google Scholar]
- Savory E. A., Adhikari B. N., Hamilton J. P., Vaillancourt B., Buell C. R., Day B. (2012). mRNA-Seq analysis of the Pseudoperonospora cubensis transcriptome during cucumber (Cucumis sativus L.) infection. PLoS ONE 7:e35796. 10.1371/journal.pone.0035796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlink K. (2009). Identification and characterization of differentially expressed genes from Fagus sylvatica roots after infection with Phytophthora citricola. Plant Cell Rep. 28, 873–882. 10.1007/s00299-009-0694-2 [DOI] [PubMed] [Google Scholar]
- Schlink K. (2010). Down-regulation of defense genes and resource allocation into infected roots as factors for compatibility between Fagus sylvatica and Phytophthora citricola. Funct. Integr. Genom. 10, 253–264. 10.1007/s10142-009-0143-x [DOI] [PubMed] [Google Scholar]
- Séjalon-Delmas N., Mateos F. V., Bottin A., Rickauer M., Dargent R., Esquerretugaye M. T. (1997). Purification, elicitor activity, and cell wall localization of a glycoprotein from Phytophthora parasitica var. nicotianae, a fungal pathogen of tobacco. Phytopathology 87, 899–909. 10.1094/PHYTO.1997.87.9.899 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y., Kawakita K., Takemoto D. (2010). Age-related resistance of Nicotiana benthamiana against hemibiotrophic pathogen Phytophthora infestans requires both ethylene-and salicylic acid-mediated signaling pathways. Mol. Plant-Microbe Interact. 23, 1130–1142. 10.1094/MPMI-23-9-1130 [DOI] [PubMed] [Google Scholar]
- Stam R., Howden A. J. M., Delgado-Cerezo M., Amaro T. M. M. M., Motion G. B., Pham J., et al. (2013a). Characterization of cell death inducing Phytophthora capsici CRN effectors suggests diverse activities in the host nucleus. Front. Plant Sci. 4:387. 10.3389/fpls.2013.00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam R., Jupe J., Howden A. J., Morris J. A., Boevink P. C., Hedley P. E., et al. (2013b). Identification and characterisation CRN effectors in Phytophthora capsici shows modularity and functional diversity. PLoS ONE 8:e59517. 10.1371/journal.pone.0059517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Sugano S., Sugimoto T., Takatsuji H., Jiang C. J. (2013). Induction of resistance to Phytophthora sojae in soyabean (Glycine max) by salicylic acid and ethylene. Plant Pathol. 62, 1048–1056. 10.1111/ppa.12011 [DOI] [Google Scholar]
- Tao Y., Xie Z., Chen W., Glazebrook J., Chang H. S., Han B., et al. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell Online 15, 317–330. 10.1105/tpc.007591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., et al. (2004). MapMan: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. 10.1111/j.1365-313X.2004.02016.x [DOI] [PubMed] [Google Scholar]
- Tierney L., Linde J., Mueller S., Brunke S., Molina J. C., Hube B., et al. (2012). A dual-systems RNA-seq approach to predict and decipher mechanisms of host-fungus interactions. Mycoses 55, 8–9. 10.1111/j.1439-0507.2012.02204.x [DOI] [Google Scholar]
- Torto T. A., Li S. A., Styer A., Huitema E., Testa A., Gow N., et al. (2003). EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 13, 1675–1685. 10.1101/gr.910003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., Van Baren M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay A., Hosseini P., Alkharouf N. W., Li S., Matthews B. F. (2012). Gene expression in leaves of susceptible Glycine max during infection with Phakopsora pachyrhizi using next generation sequencing. Sequencing 2011:827250 10.1155/2011/827250 [DOI] [Google Scholar]
- van Loon L. C., Rep M., Pieterse C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. 10.1146/annurev.phyto.44.070505.143425 [DOI] [PubMed] [Google Scholar]
- Vonbroembsen S. L., Kruger F. J. (1985). Phytophthora cinnamomi associated with mortality of native vegetation in South Africa. Plant Dis. 69, 715–717. 10.1094/PD-69-715 [DOI] [Google Scholar]
- Ward J., Weber C. (2012). Comparative RNA-seq for the investigation of resistance to Phytophthora root rot in the red raspberry ‘Latham’. Acta Hortic. 946, 67–72. 10.17660/ActaHortic.2012.946.7 [DOI] [Google Scholar]
- Westermann A. J., Gorski S. A., Vogel J. (2012). Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 10, 618–630. 10.1038/nrmicro2852 [DOI] [PubMed] [Google Scholar]
- Wingfield M. J., Kemp G. H. J. (1994). Diseases of pines, eucalypts and wattle, in South African Forestry Hand-Book, ed Van Der Sijde H. A. (Pretoria: South African Institute of Forestry; ), 231–249. [Google Scholar]
- Wingfield M. J., Roux J., Heath R., Dyer C., Feely J. (2011). Forest diseases, in Sappi Tree Farming Guidelines for Private Growers. Available Online at: http://www.sappi.com/regions/sa/SappiSouthernAfrica/Sappi%20Forests/Pages/Tree-Farming-Guidelines.aspx (Accessed May 31 2012).
- Winnenburg R., Baldwin T. K., Urban M., Rawlings C., Kohler J., Hammond-Kosack K. E. (2006). PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 34, D459–D464. 10.1093/nar/gkj047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D. L., Long H., Liang J. J., Zhang J., Chen X., Li J. L., et al. (2012). De novo assembly and characterization of the root transcriptome of Aegilops variabilis during an interaction with the cereal cyst nematode. BMC Genomics 13:133. 10.1186/1471-2164-13-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Zhu L., Tu L., Liu L., Yuan D., Jin L., et al. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. 10.1093/jxb/err245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K., De Luca V., Brisson N. (1995). Creation of a metabolic sink for tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to Phytophthora infestans. Plant Cell Online 7, 1787–1799. 10.1105/tpc.7.11.1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentmyer G. A. (1983). The world of Phytophthora, in Phytophthora. Its Biology, Taxonomy, Ecological and Physiology, eds Erwin D. C., Bartnicki-Garcia S., Tsao P. H. (St. Paul, MN: The American Phytopathological Society; ), 1–8. [Google Scholar]
- Zhang M., Li Q., Liu T., Liu L., Shen D., Zhu Y., et al. (2015). Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases. Plant Physiol. 167, 164–175. 10.1104/pp.114.252437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.