Abstract
Both the prevalence of Helicobacter pylori infection and the incidence of gastric cancer are high in Bhutan. The high incidence of atrophic gastritis and gastric cancer suggest the phylogeographic origin of an infection with a more virulent strain of H. pylori. More than 90% of Bhutanese strains possessed the highly virulent East Asian-type CagA and all strains had the most virulent type of vacA (s1 type). More than half also had multiple repeats in East Asian-type CagA, which are rare in other countries and are reported characteristictly found in assciation with atrophic gastritis and gastric cancer consistent with Bhutanese strains having multiple H. pylori virulence factors associated with an increase in gastric cancer risk. Phylogeographic analyses showed that most Bhutanese strains belonged to the East Asian population type with some strains (17.5%) sharing East Asian and Amerindian components. Only 9.5% belonged to the European type consistant with H. pylori in Bhutan representing an intermediate evolutionary stage between H. pylori from European and East Asian countries.
Helicobacter pylori is a spiral-shaped, gram-negative bacterial pathogen infecting more than half of the world’s population. H. pylori plays a causative role in the pathogenesis of gastritis, peptic ulcer diseases, gastric cancer, and mucosa-associated lymphoid tissue lymphoma1,2. Although infection with H. pylori always results in histologic gastritis, the clinical outcome in different populations varies in relation H. pylori virulence, host genetic factors and environmental factors (especially diet)3,4. The cytotoxin-associated gene A (CagA) and vacuolating cytotoxin (VacA) are the most extensively studied H. pylori virulence factors5,6,7,8. cagA-positive strains are subdivided into an East Asian type and Western type depending on sequences in the 3′ region of the gene9,10,11 which contain a tyrosine phosphorylation site motif Glu-Pro-Ile-Tyr-Ala (EPIYA). The EPIYA motifs can be further subdivided into four distinct peptide segments, EPIYA-A, EPIYA-B, EPIYA-C, and EPIYA-D based on amino acid sequences flanking the EPIYA motif12. Most CagA-positive isolates possess EPIYA-A or EPIYA-B segments. The EPIYA-C genotype is characteristic of CagA from Western countries (Western-type CagA), and the EPIYA-D segment is characteristic of East Asian-type CagA9,10,11,13. CagA sequence type is based on all the EPIYA segments in the sequence (i.e., ABC, ABCC, ABCCC, etc. for Western-type CagA and ABD, etc. for East Asian-type CagA). The East Asian-type CagA is associated with a more robust mucosal inflammatory reaction than Western-type CagA and epidemiological studies in Thailand, South Korea, and Okinawa, Japan have shown East Asian-type CagA strains correlate with an increased risk of peptic ulcer or gastric cancer compared with those infected with Western-type CagA strains14,15,16.
Different genotypes of vacA have been described based on differences in the signal (s) region (s1 and s2) and the middle (m) region (m1 and m2)6. In epidemiological studies vacA has been classified into subtypes based on the combination of variants in the s and m regions. In vitro experiments have shown that s1m1 strains are the most cytotoxic, followed by s1m2 strains; s2m2 strains have no cytotoxic activity and do not produce VacA protein6; the s2m1 genotype is rare. Epidemiological studies have shown that infection with the vacA s1 or m1 strains correlates with an increased risk for peptic ulcers or gastric cancer compared with those with s2 or m2 strains, especially in Western countries6,17,18,19. Importantly, the presence of CagA and VacA are typically linked such that H. pylori either produce both or neither protein7.
H. pylori infection is one of those infectious diseasese that accompanied humans on their journey out of Africa. The diversity within H. pylori lineages examined by multi-locus sequence typing (MLST) using seven housekeeping genes20,21,22 identified seven population types based on geographical associations: hpEurope, hpEastAsia, hpAfrica1, hpAfrica2, hpAsia2, hpNEAfrica, and hpSahul20,21,22. Besides possessing East Asian-type cagA20,21,22, hpEastAsia type has been further divided into hspEAsia, hspAmerind, and hspMaori. Studies in Colombia suggested that European strain origin (hpEurope) was associated with premalignant histological lesions compared to infection with African strains (hpAfrica1)23,24. However no previous studies have focused on the relationship between the phylogeographic origin of H. pylori and gastric cancer risk in Asia despite the data regarding the more virulent type of CagA of hpEastAsia origin compared to strains of hpAsia2 and/or hpEurope origins.
Bhutan is a small, landlocked country in South Asia, located at the eastern end of the Himalayas, that shares borders in the south, east, and west with the Republic of India and to the north with the People’s Republic of China. The incidence of gastric cancer is reported to be greater than that of neighboring areas (17.2 cases per 100,000 individuals per year compared with 6.1 cases/100,000 individuals per year in India) (International Agency for Research on Cancer; GLOBOCAN2012, http://globocan.iarc.fr/). In India, Western-type CagA and the hpAsia2, and/or hpEurope types are predominant possibly reflecting the gene flow through Indo-Aryans that replaced endogenous strains in India20,21,22,25. We hypothesize that the difference in the incidence of gastric cancer between India and Bhutan could be in part related to differences in phylogeographic origin and virulence of H. pylori. In this study, we also examined the H. pylori virulence factors in Bhutan and their relationship with histological scores and clinical presentations.
Results
A total of 209 strains were isolated from H. pylori-positive Bhutanese volunteers (98 males, 16–92 years old, mean 35.6 years; 111 females, 16–75 years old, mean 37 years). Of these, 165 strains were isolated from subjects with histological gastritis without peptic ulcers or gastric cancer, 21 from gastric ulcer (GU) patients, 19 from duodenal ulcer (DU) subjects, 1 from a gastric cancer subject, and 3 from subjects with an unclear diagnosis (Table 1). The average age was significantly lower in DU subjects than in subjects with simple H. pylori gastritis (mean 29.4 years vs. mean 36.7 years; P = 0.03). The male/female ratio was significantly higher for the GU and DU subjects than for the gastritis subjects (male:female = 16:5 for GU, 13:6 for DU and 67:98 for gastritis; P < 0.002 and P = 0.02, respectively).
Table 1. Association between Helicobacter pylori virulence factors and clinical presentations.
| Total |
Gastritis |
Gastric ulcer |
Duodenal ulcer |
Gastric cancer |
Unclear diagnosis |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 209 | % | n = 165 | % | n = 21 | % | n = 19 | % | n = 1 | % | n = 3 | % | |
| cagA positive | 206 | (98.6) | 162 | (98.2) | 21 | (100) | 19 | (100) | 1 | (100) | 3 | (100) |
| cagA undetermined | 3 | (1.4) | 3 | (1.8) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
| cagA genotype undetermined | 2 | (1.0) | 2 | (1.2) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
| East Asian-type cagA | 189 | (91.7) | 149 | (90.1) | 18 | (85.7) | 18 | (94.7) | 1 | (100) | 3 | (100) |
| Western-type cagA | 15 | (7.3) | 11 | (6.8) | 3 | (14.3) | 1 | (5.3) | 0 | (0.0) | 0 | (0.0) |
| vacA s1 | 209 | (100) | 165 | (100) | 21 | (100) | 19 | (100) | 1 | (100) | 3 | (100) |
| s2 | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
| vacA m1 | 80 | (38.3) | 63 | (38.2) | 5 | (23.8) | 9 | (47.4) | 1 | (100) | 2 | (66.7) |
| m2 | 42 | (20.1) | 32 | (19.4) | 4 | (19.0) | 5 | (26.3) | 0 | (0.0) | 1 | (33.3) |
| m1m2 chimera (m12) | 83 | (39.7) | 66 | (40.0) | 12 | (57.1) | 5 | (26.3) | 0 | (100) | 0 | (14.3) |
| m genotype undetermined | 4 | (1.9) | 4 | (2.4) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
| East Asian cagA/vacA s1m1 | 69 | (33.0) | 54 | (32.7) | 3 | (14.3) | 9 | (47.4) | 1 | (100) | 2 | (66.7) |
| East Asian cagA/vacA s1m2 | 37 | (17.7) | 28 | (17.0) | 4 | (19.0) | 4 | (21.1) | 0 | (0.0) | 1 | (33.3) |
| East Asian cagA/vacA s1m12 | 80 | (38.3) | 64 | (38.8) | 11 | (52.4) | 5 | (26.3) | 0 | (0.0) | 0 | (0.0) |
| Western cagA/vacA s1m1 | 8 | (3.3) | 6 | (3.7) | 2 | (9.5) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
| Western cagA/vacA s1m2 | 5 | (2.4) | 4 | (2.5) | 0 | (0.0) | 1 | (5.3) | 0 | (0.0) | 0 | (0.0) |
| Western cagA/vacA s1m12 | 1 | (0.5) | 0 | (0.0) | 1 | (0.48) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
| Others | 9 | (4.3) | 9 | (4.3) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) | 0 | (0.0) |
Others: cagA genotype undetermined and/or vacA m genotype undetermined.
cagA genotypes
cagA was present in 98.6% (206/209) of H. pylori strains cultured (Table 1). The 3 strains negative for cagA by PCR showed negative band using the cag pathogenicity island (PAI) empty site PCR indicating the presence of at least a partial cag PAI; therefore, we considered them cagA-undetermined. Two strains were cagA–positive based on PCR; however, the cagA genotypes were undetermined by sequencing because the primer pair did not amplify the gene. Sequencing showed that among cagA-positive strains, the East Asian-type CagA was predominant (189/206, 91.7%); Western-type CagA was found in 7.3% of strains (15/206) Sequencing analysis revealed that only 82 (43.4%) of the 189 East Asian-type CagA strains possessed the typical East Asian genotype (ABD) (Table 2). For Western-type CagA, 13 of 15 had the ABC genotype (86.7%). The two remaining strains were AB- and AC-type.
Table 2. Association between EPIYA segment type of CagA and clinical presentations.
| Total | Gastritis | Gastric ulcer | Duodenal ulcer | Gastric cancer | others | |
|---|---|---|---|---|---|---|
| Western-type CagA | ||||||
| AB* | 1 | 1 | 0 | 0 | 0 | 0 |
| AC | 1 | 1 | 0 | 0 | 0 | 0 |
| ABC | 13 | 9 | 3 | 1 | 0 | 0 |
| Total | 15 | 11 | 3 | 1 | 0 | 0 |
| East Asian-type CagA | ||||||
| ABD | 82 | 68 | 5 | 7 | 1 | 1 |
| AB’D | 1 | 1 | 0 | 0 | 0 | 0 |
| ABBD | 5 | 5 | 0 | 0 | 0 | 0 |
| AB’BD | 97 | 72 | 13 | 10 | 0 | 2 |
| AB’B’BD | 2 | 2 | 0 | 0 | 0 | 0 |
| BD | 2 | 1 | 0 | 1 | 0 | 0 |
| Total | 189 | 149 | 18 | 18 | 1 | 3 |
*AB type was defined as Western-type CagA since the sequence of the B segment was mostly identical with the Western-type B segment (TGQVASPEEPIYAQVAKKVKAKIDRLDQIASGLGGVGQAG).
In addition to the four major segments originally designated as EPIYA-A, -B, -C, and -D (i.e., A, B, C, and D), we previously designated several minor segments, including EPIYA-B’ and –B”26. EPIYA segments were classified according to these groups. The AB’BD type was most prominent among the East Asian strains (97/189; 51.3%). This type represents only 0.7% [2/280] of East Asian-type CagA strains deposited in GenBank26.
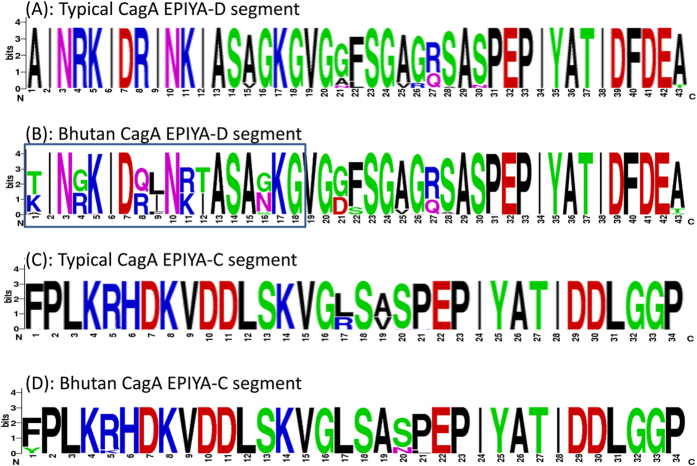
Interestingly, the sequences of the EPIYA-D segments in Bhutanese strains showed large variation (Fig. 1) which differed from strains deposited in GenBank where more than 90% of EPIYA-D segments had the same amino acid sequences26. Reflecting this variation, pair-wise genetic distances (Kimura-2-parameter model) among Bhutanese East Asian-type cagA were large (median 0.078, mean 0.074, and variance 0.025) with average sequence identity 91.6%. In contrast, the sequences of the EPIYA-C segments in Bhutanese strains were very similar (Fig. 1) and were identical to typical sequences of EPIYA-C segments deposited in GenBank (median, mean, variance of pair-wise genetic distances and average sequence identity among Bhutanese Western-type cagA were 0.01, 0.015, 0.0002, and 98.4%, respectively).
Figure 1. Variation in the CagA amino acid sequence of East Asian-type and Western-type CagA.
The target α-EAS sequences “AINRKIDRINKIASAGKG” in EPIYA segment D, based on the sequences of Japanese strains, are shown in frame. A sequence analysis revealed that the typical target sequences in Bhutanese strains was “(T/K)IN(G/R)KID(Q/R)(L/I)N(R/K)(T/I)ASA(G/N)KG,” where (X/Y) means X and Y as the two major amino acids. The sequences of the EPIYA-D segments in Bhutanese strains were highly variable compared to strains deposited in GenBank. In contrast, the East Asian-type and Western-type sequences of EPIYA-C segments in Bhutanese strains were largely identical, similar to the typical sequences of EPIYA-C segments deposited in GenBank. Reference strains used in Fig. 1A (strain name [accession number]) were 103a (AB110966.1), 105a (AB110967.1), 106a (AB110968.1), 108a (AB110969.1), 113b (AB110970.1), 120a (AB110971.1), 122b (AB110972.1), 125b (AB110973.1), 128a (AB110974.1), FJT77 (KF028580.1), 04-518 (AB267252.1), 03-166 (AB267253.1), 04-264 (AB267254.1), THP1477 (AB116744.1), 04-334 (AB267249.1), 03-292 (AB267250.1), 04-366 (AB267251.1), THP1260 (AB116742.1), M3 (AB116740.1), THP463 (AB116735.1), Korea23 (AB057044.1), Korea 12 (AB057043.1), K69 (FJ458129.1), Korea2-3 (AB057040.1), k266 (FJ458163.1), K265 (FJ458162.1), K264 (FJ458161.1), K261 (FJ458158.1), K260 (FJ458157.1), K259 (FJ458156.1). Reference strains used in Fig. 1C (strain name [accession number]) were India41 (AF222807.1), India99 (AF222809.1), OSC40A (EU089774.1), OSC42B (EU089775.1), PCR-156i (EU368669.1), PCR218vi (EU089766.1). RIGLD-OC149 (JX428784.1), SAN53 (EU089771.1), PD682 (EF450167.1), PD636 (EF450165.1), PD308 (EF450162.1), PD488 (EF450161.1), PD537 (EF450160.1), PD501 (EF450159.1), PD351 (EF450158.1), PD348 (EF450157.1), PD6481K (EF450153.1), 216G (GQ899171.1), 1407 (GU143415.1), HPI-14 (FJ849792.1), HPI-13 (FJ849791.1), HPI-11 (FJ849789.1), USA2791 (AB057099.1), Kazak3 (AB057098.1), USA35 (AB057095.1), Italy329 (AB057094.1), Arizona2 (AB057075.1), Arizona1 (AB057074.1).
The EPIYA motif in these strains is shown in Table 3. We obtained 11 types of EPIYA or EPIYA-like sequences. In total, 710 EPIYA motifs were obtained from 206 CagA sequences. On average, each CagA sequence contained approximately 3 EPIYA motifs. The 3 most frequent EPIYA motifs were EPIYA (552/710 = 77.8%), EPIYT (14.8%), and ESIYT (6.2%). This result differs from our previous study examining 560 CagA loci deposited in GenBank (92.3% had EPIYA, 5.1% EPIYT, and 0.4% ESIYT)26. The EPIYA-B motif had a high degree of variation in the five amino acids (e.g., EPIYA, EPIYT and ESIYT) (Table 3). Among 190 EPIYA-B motifs in East Asian-type CagA, EPIYT was more frequent than EPIYA (53.7% vs. 20.0%). However, there was no association between EPIYA-like sequence type and disease presentation.
Table 3. Frequencies of the 11 EPIYA motif types.
| All motifs | A motif | B’ motif | B motif | C or D motif | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All CagA types | EPIYA | 552 | EPIYA | 201 | EPIYA | 99 | EPIYA | 50 | EPIYA | 202 |
| EPIYT | 105 | EPIYT | 2 | EPIYT | 103 | |||||
| ESIYT | 44 | ESIYT | 44 | |||||||
| ESIYA | 2 | ESIYA | 2 | |||||||
| EYIYA | 1 | EYIYA | 1 | |||||||
| ETIYT | 1 | ETIYT | 1 | |||||||
| KSIYT | 1 | KSIYT | 1 | |||||||
| EPIYV | 1 | EPIYV | 1 | |||||||
| EPIYS | 1 | EPIYS | 1 | |||||||
| GPIYA | 1 | GPIYA | 1 | |||||||
| EPLYA | 1 | EPLYA | 1 | |||||||
| Total | 710 | 202 | 101 | 204 | 203 | |||||
| Western-type CagA | EPIYA | 41 | EPIYA | 15 | EPIYA | 12 | EPIYA | 14 | ||
| EPIYT | 1 | EPIYT | 1 | |||||||
| EPIYS | 1 | EPIYS | 1 | |||||||
| Total | 43 | 15 | 14 | 14 | ||||||
| East-Asian-type CagA | EPIYA | 511 | EPIYA | 186 | EPIYA | 99 | EPIYA | 38 | EPIYA | 188 |
| EPIYT | 104 | EPIYT | 2 | EPIYT | 102 | |||||
| ESIYT | 44 | ESIYT | 44 | |||||||
| ESIYA | 2 | ESIYA | 2 | |||||||
| EYIYA | 1 | EYIYA | 1 | |||||||
| ETIYT | 1 | ETIYT | 1 | |||||||
| KSIYT | 1 | KSIYT | 1 | |||||||
| EPIYV | 1 | EPIYV | 1 | |||||||
| GPIYA | 1 | GPIYA | 1 | |||||||
| EPLYA | 1 | EPLYA | 1 | |||||||
| Total | 667 | 187 | 101 | 190 | 189 | |||||
cagA genotypes and histological findings
The degree of inflammation (monocyte infiltration), neutrophil activity, atrophy, and intestinal metaplasia were classified into four grades according to the updated Sydney system (0 to 3)27. Chronic inflammation and neutrophil activity are characteristic of H. pylori infection. Antral predominant gastritis is associated with DU whereas pangastritis with atrophy and intestinal metaplasia is predominant in determining gastric cancer risk27.
Individuals with East Asian-type CagA, either single-repeat (ABD) or multiple-repeat (AB’BD and ABBBD) had a slightly higher atrophy score in the antrum than individuals with Western-type CagA (mean score [median score]; 1.45 [1], and 1.43 [1] vs. 1.07 [1]; both P = 0.03), although the statistical differences were disappeared after using multivariate analysis adjusting for age and sex (odds ratios [OR] = 3.30, 95% confidence interval [CI] = 0.61 to 26.39 and OR = 3.79, 95% CI = 0.71 to 28.34). Individuals with the multiple-repeat East Asian-type CagA tended to have higher scores for intestinal metaplasia in the antrum than those with single-repeat East Asian-type CagA (mean [median]; 0.22 [0] vs. 0.11 [0], P = 0.10). Subjects infected with H. pylori with the EPIYT motif tended to have higher activity score in the antrum than those infected with EPIYA motif strains (mean [median]; 1.58 [2] vs. 1.37 [1], P = 0.06).
CagA immunohistochemistry
We performed immunohistochemistry for 205 H. pylori culture-positive cases to detect immunoreactivity with CagA and anti-East Asian-type CagA-specific antibody (α-EAS Ab) (Table 4). α-EAS Ab is immunoreactive with only the East Asian-type CagA28 and proved useful for typing CagA immunohistochemically in Japan29, Vietnam and Thailand30. One biopsy specimen for an H. pylori culture-positive case (infected with the East Asian-type CagA strain) was unavailable and was excluded from the analyses. Three samples infected with cagA-undetermined strains showed negative immunoreactivity with both anti-CagA and α-EAS Abs. The remaining 205 samples showed positive immunoreactivity with the anti-CagA Ab. As expected, all 15 samples infected with Western-type cagA strains showed negative immunoreactivity with α-EAS Ab. Surprisingly, only 68 out of 188 samples (36.2%) infected with East Asian-type CagA strains tested positive for α-EAS Ab. The target α-EAS sequences in EPIYA segment D (Fig. 1) was designed based on the sequences of Japanese strains28. Only 6 Bhutanese strains had identical sequences for the α-EAS Ab designed amino acid sequence and showed positive immunoreactivity. When one or two amino acid sequences were different from the designed amino acid sequences (70 cases), positive α-EAS Ab results were detected in 72.9% of cases (51/70). When more than three amino acid sequences were different from the designed amino acid sequence (112 cases), positive α-EAS Ab results decreased to 11.6% of cases (13/112). Using PCR-based sequencing as the gold standard, the sensitivity, specificity, negative predictive value, and positive predictive value of the α-EAS Ab were 36.2%, 94.1%, 11.8%, and 98.6%, respectively in Bhutan. Overall accuracy rate was only 41%. Therefore, the α-EAS Ab was not useful to distinguish East Asian-type CagA strains from other CagA strains in the Bhutanese population. There were no definite relationship between the rate of α-EAS Ab positivity and clinical presentation (42.6% [69/162] for gastritis, 42.9% [9/21] for GU, 47.4% [9/19] for DU and 100% [1/1] for gastric cancer).
Table 4. cagA genotypes and CagA immunoreactivity.
| anti-CagA Ab-positive | α-EAS Ab-positive | |
|---|---|---|
| cagA positive (n = 205) | 205 (100%) | |
| cagA undetermined (n = 3) | 0 (0%) | 0 (0%) |
| cagA genotype undetermined (n = 2) | 2 (100%) | 1 (50%) |
| East Asian-type cagA (n = 188) | 188 (100%) | 68 (36.2%) |
| Western-type cagA (n = 15) | 15 (100%) | 0 (0%) |
vacA genotypes
All 209 strains had the vacA s1 genotype. PCR results showed that 80 strains (38.3%) possessed the vacA m1 genotype and 42 strains (20.1%) possessed the m2 genotype. The genotype of the remaining 87 strains could not be distinguished based on the length of the PCR product. Therefore, we examined the vacA genotypes by DNA sequencing. A total of 205 strains were successfully sequenced. The m1 type can be further subdivided into m1a, m1b, and m1c6. Among 80 Bhutanese m1 strains, 46 were classified as m1b, 34 as m1c, and none as m1a. Forty-two strains were classified as m2. Interestingly, 83 strains could not be typed based on PCR and were located on a branch between m1 and m2. These strains contained special sequences with m1-m2 chimeric patterns (Fig. 2). Around 85% of the gene by length was consistent with the vacA m1 sequence, and the remaining portion was similar to vacA m2. The predominant East Asian-type cagA had the vacA s1 m1-m2 chimeric genotype (80/209, 38.3%) or s1m1 (69/209, 33.0%). Only one Western-type cagA strain had the s1 m1-m2 chimeric genotype. There was no relationship between vacA and cagA types in Bhutan (P = 0.65).
Figure 2. The vacA m1-m2 chimeric genotypes.
Approximately 85% of the gene sequences for m1-m2 chimeric genotypes were similar to vacA m1, and the remaining nucleotides were similar to vacA m2.
vacA genotypes and histological findings
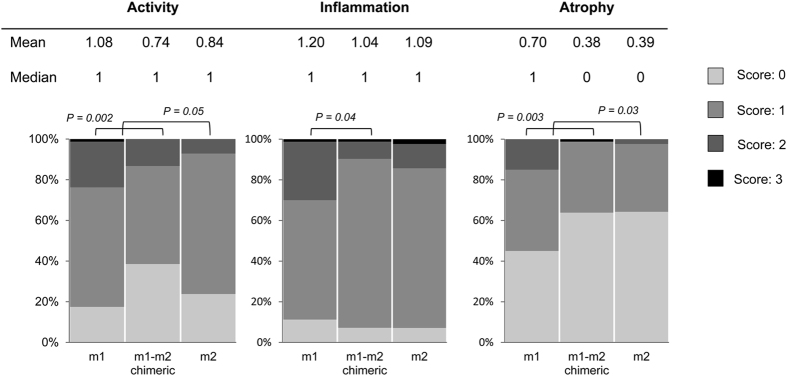
Histological findings of vacA m genotypes showed that subjects infected with m1 strains had a higher score of corporal mucosal atrophy than those with m2 genotypes (mean score [median score]; 0.70 [1] vs. 0.39 [0], P = 0.03) (Fig. 3). Subjects infected with vacA m1 genotypes also had a higher scores of corporal activity, inflammation, and atrophy than those with m1-m2 chimeric genotypes (mean [median]; 1.08 [1] vs. 0.74 [1], 1.20 [1] vs. 1.04 [1], and 0.70 [1] vs. 0.38 [0], P = 0.002, P = 0.04, and P = 0.003, respectively) (Fig. 3). Subjects infected with vacA m1 also had a significantly higher risk of activity and atrophy in the corpus after adjusting for age and sex (OR = 4.14, 95% CI = 1.87 to 9.17 and OR = 2.74, 95% CI = 1.39 to 5.42, respectively) than those with the m1-m2 chimeric type. However, there were no significant differences in histological scores between vacA m1-m2 chimeric type and m2.
Figure 3. The vacA m region genotypes and histological findings in the corpus.
Subjects infected with the vacA m1 genotype showed higher mucosal atrophy score than those with m2 genotype. Subjects infected with vacA m1 genotype also had higher histological severity scores than those with m1-m2 chimeric genotypes.
Population structure and phylogenetic position
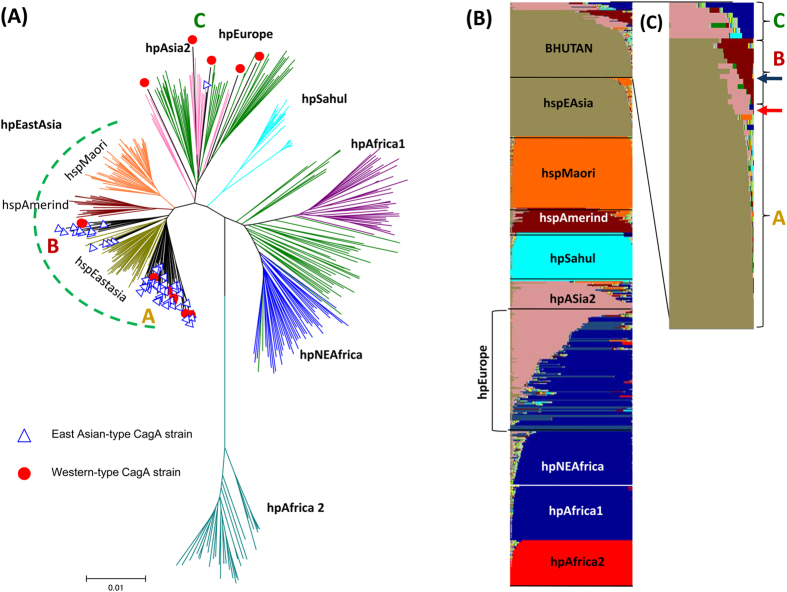
Concordant with the evidence that the Bhutanese strains contained virulent H. pylori genotypes (East Asian-type CagA and vacA s1), population analyses showed that the main branch belonged to the hspEAsia strains (blue triangle) (Fig. 4A: group A). Interestingly some East Asian-type CagA strains were located between the hspEAsia and hspAmerind (group B). The AB’BD and ABBD-type CagA was evenly distributed among groups A and B (Table 5).
Figure 4. MLST phylogeny and population structure of Bhutanese strains.
The population types of 13 Western-type CagA and 50 East Asian-type CagA strains were analyzed by MLST. The strains belonged to three groups: the main branch included hspEAsia strains (group A) and was between hspEAsia and hspAmerind (group B) and hpEurope/hpAsia2 strains (group C) (Fig. 4A). An MLST analysis revealed that most Western-type CagA strains belonged to sub-branch group A or C (red dots). In contrast, only one East Asian-type CagA strain belonged to group C (blue triangle). Figure 4B,C shows the results of a STRUCTURE analysis assuming K = 15, which had the highest posterior probability of the five runs. Each vertical line of the bar chart represents a single strain, and the line colors indicate populations to which the strain may belong. The lengths of the colors in a line are proportional to the probabilities that the strain belongs to the particular population. When the bars were magnified and aligned from top to botom in descending order with respect to the dark yellow color, only two strains were not concordant between the phylogeny and population structure analyses (blue arrow [group A in the phylogeny] in opposite with red arrow) (Fig. 4C).
Table 5. Association between MLST phylogeny type and cagA and vacA type.
| Group | n | AB’BD | ABD | Western-type CagA | others | m1 | m2 | hybrid |
|---|---|---|---|---|---|---|---|---|
| A | 46 | 17 (73.9%) | 18 (85.7%) | 6 (46.2%) | 5 (83.3%) | 20 (74.1%) | 13 (61.9%) | 13 (86.7%) |
| B | 11 | 5 (21.7%) | 3 (14.3%) | 2 (15.4%) | 1 (16.7%) | 5 (18.5%) | 4 (19.0%) | 2 (13.3%) |
| C | 6 | 1 (4.3%) | 0 (0.0%) | 5 (38.5%) | 0 (0.0%) | 2 (7.4%) | 4 (19.0%) | 0 (0.0%) |
| Total | 63 | 23 (100.0%) | 21 (100.0%) | 13 (100.0%) | 6 (100.0%) | 27 (100%) | 21 (100.0%) | 15 (100.0%) |
To confirm these results, we investigated the population structure of the Bhutanese strains using the highest posterior probability of the five runs (K = 15) by STRUCTURE. Consistent with MLST phylogeny, the Bhutanese strains showed the most color commonality with hspEAsia strains (dark yellow) and some of which shared components with hspAmerind (dark red color) which might represent an intermediate evolutionary stage between H. pylori from European and East Asian countries. The hspAmerind subgroup of hpEastAsia was isolated from Inuits and Amerinds in North and South America21. Only the results for two strains were not concordant between phylogenetic and population structure analyses; however both shared an hspEAsia component (blue arrow [group A in the phylogeny] in opposite with red arrow). Group C had a high proportion of m2 and no m1-m2 chimeric types, which suggests a little recombination in the hpEurope Bhutanese strains (Table 5). However, all m1-m2 chimeric type strains were categorized as group A (80%, 12/15) or B (20%, 3/15).
Phylogenetic origin and histological findings
Subjects infected with hspEAsia strains (group A) had a significantly higher inflammation score in the antrum than those with hpEurope strains (group C) (mean score [median score]; 1.91 [2] vs. 1.33 [1], P = 0.03). Subjects infected with hspEAsia had a significantly higher risk of inflammation in the antrum after adjusting for age and sex (OR = 6.10, 95% CI = 1.00 to 38.13) than those with the hpEurope strains. Moreover, those infected by group A strains tended to have more inflammation than those infected by group B strains (mean [median]; 1.91 [2] vs. 1.55 [2], P = 0.08).
Nucleotide and amino acid sequences
All nucleotide sequence data are available under the DDBJ accession numbers LC067575–LC068428.
Discussion
We confirmed that almost all Bhutanese H. pylori strains possessed cagA of the East Asian-type CagA type, which is associated with a more robust mucosal inflammatory reaction than Western-type CagA14,15,16. Histological analsysis of gastric mucosa from Bhutanese showed that infection with East Asian-type CagA (AB’BD and AB’BBD) tended to increase the risk of atrophy compared to those infected with Western-type CagA strains. Therefore, in addition to host and environmental factors, the presence of a CagA type associated with an increased inflammatory response might in part be responsible for the higher risk of gastric cancer in Bhutan than in India.
Intriguingly, the very rare AB’BD CagA type was dominant among East Asian-type CagA (51.3%) in Bhutan. There are data that the biological activity of CagA is related in part to the number of CagA phosphorylation sites. In vitro studies have suggested that phosphorylation of EPIYA-C is necessary, but not sufficient, to induce the epithelial cell elongation in vitro, a morphology was originally referred to as the “hummingbird phenotype” which reflects changes in host cell signaling pathways31. It contributes to the epithelial proliferation and pro-inflammatory processes as well as the disruption of cell-to-cell junctions, or loss of cell polarity, all of which are seen in gastric cancer32. A previous study31 showed that EPIYA-B is also important because at least two phosphorylated EPIYAs are necessary for the elongation phenotype. It was found that East Asian H. pylori expressing CagA EPIYA-A/EPIYA-D or EPIYA-B/EPIYA-D, but not EPIYA-A/EPIYA-B, induced moderate epithelial cell elongation31. In the case of Western-type CagA, the incidence of gastric cancer was higher in patients infected with strains carrying multiple EPIYA-C repeats than in those infected with strains with a single repeat9,10,26,33,34. However, there are considerable data to suggest that multiple EPIYA-C repeats are a response to atrophic gastritis with hypochlorhydria rather than its cause10. Because gastric cancer is associated with atrophic gastritis and hypochlorhydria, it is not surprising that even in Japanese individuals infected with East Asian-type CagA strains, most strains with multiple EPIYA-B repeats are isolated from patients with gastric cancer9. Therefore, the high prevalence of Bhutanese strains with CagA containing multiple EPIYA-B segments is consistent with a high prevalence of atrophic gastritis in Bhutan. Based on pepsinogen levels, the Bhutanese population has a higher incidence of advanced mucosal atrophy, even in the younger population, than other populations (e.g., Japan, Singapore, and the USA)35.
Interestingly, the structure of East Asian-type CagA in Bhutan differed from the typical East Asian-type CagA. In particular, the first 18 amino acids of EPIYA-D had what appears to be a population-specific variation in Bhutan which was responsible for the low accuracy of α-EAS Ab. Furthermore, the variance in EPIYA motifs also differed. In Bhutan, EPIYA (77.8%) was the predominant type, followed by EPIYT (14.8%) and ESIYT (6.2%). Previous studies reported that EPIYT was the second most common EPIYA-B sequence of Western-type cagA, but was very rare in East Asian-type cagA14,26,36. In our previous study of 1,796 EPIYA motifs, including 274 Western and 286 East Asian strains, found 92.2% were EPIYA. EPIYT and ESIYT were only found in 5.1% and 0.3% of strains, respectively26. Zhang et al. analyzed 364 Western-type cagA strains and reported that gastric cancer was significantly associated with EPIYA sequences compared with gastritis alone, whereas EPIYT sequences was significantly associated with DU36. The role of the EPIYT-B motif in East Asian-type CagA remains unclear and future studies are necessary to determine whether specific CagA sequences are involved in the pathogenesis of Hp-associated disease.
In Bhutan, all strains had the vacA s1 genotype, but the vacA m region could not be distinguished for 83 samples using PCR because they contained m1-m2 chimeric sequences. Although the prevalence of the vacA m1-m2 chimeric genotype tended to be higher in strains obtained from GU patients than that in those obtained from DU patients, the sample sizes of those with ulcers were small and further studies are needed to evaluate the significance of this observation. m1-m2 chimeric genotype have been rarely found37,38,39. Bhutanese strains possessed specific genotypes for both cagA and vacA that are very rare in other countries. In vitro level of vacuolating activity in strains with the m1-m2 chimeric genotype have been reported to be comparable to those of m1 strains, and were higher than those of m2 strains39. In contrast, histological findings showed that vacA m1 genotypes was associated with higher activity, inflammation, and atrophy than m1-m2 chimeric genotypes in Bhutan. Further studies are necessary to clarify the whether there is biological importance of the m1-m2 chimeric type.
The Bhutanese strains with East Asian-type CagA were primarily categorized as the hpEAsia population type which had severer histological scores than other populations in this study. This association is consistent with the ASR of gastric cancer in the East Asia region (24.2/100.000) being greater than in the European continent and the South-Central Asia region (9.4 and 6.7/100,000, respectively). Interestingly some also shared an hspAmerind component based on population genetic analyses suggesting that Bhutanese share part of lineage of hspAmerind ancestry. A previous human DNA study concluded that the Africans being the first group of people from which the rest of the human populations split are most divergent from other human populations40,41. The second major split separated the North Eurasian supercluster (Caucasians, Northeast Asians, and Amerindians) from the Southeast Asian supercluster (Southeast Asians, Australians, Papua New Guineans, and Pacific Islanders)42. Amerindians and the current Northeast Asians (Tibetans, Koreans, Japanese, and Mongolians) separated before Amerindians migrated to North America43. Although far less likely, an alternative hypothesis is that hspAmerind arose after crossing the Bering Strait, and shared ancestry with the Bhutan population. Further analyses are necessary to confirm the origin of the Bhutan strains. Interestingly, Furuta et al. have suggested that Amerind CagA could be intermediate between Western- and East Asian-type CagA; segment C of Amerind CagA contained part of segment D44. This remains highly speculative.
The data suggest that Bhutanese strains were influenced by contact with or gene flow from India where some strains are hpEurope45,46. There are many ethnic groups in Bhutan; however most people included in this study are major ethnic groups “Ngalops” and “Sharchops”; the “Ngalops” are people of Tibetan origin, and the “Sharchops” are the population of mixed Tibetan and Southeast Asian descent. It remains possible that some Bhutanese and Indian strains share a common ancestor.
In conclusion, the higher prevalence of H. pylori-associated atrophic gastritis in Bhutanese is consistent with a higher risk of gastric cancer and the high prevalence of the more virulent East Asian-type CagA. The presence of a very rare and population-specific sequence in Bhutanese strains might represent a turning point in the evolution of H. pylori and help explain differences between strains from European countries and those from East Asian countries.
Materials and Methods
Patients and H. pylori
H. pylori strains were obtained from the gastric mucosa of Bhutanese volunteers who underwent endoscopy in December 2010 in three cities in Bhutan (Thimphu, Punakha, and Wangdue), as described in our previous study47. Presentations for H. pylori-infected subjects included gastritis, DU, GU, and gastric cancer. DU, GU, and gastric cancer were identified by endoscopy, and gastric cancer was further confirmed by histopathology. Experienced endoscopists (RV, TR, LT, VM and YY) performed all endoscopy procedures and determined the clinical data. Written informed consent was obtained from all participants, and the protocol was approved by the Ethics Committees of Jigme Dorji Wangchuck National Referral Hospital, Bhutan, Chulalongkorn University, Thailand, and Oita University Faculty of Medicine, Japan. We declare that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
Isolation and genotyping of H. pylori
H. pylori colonies were cultured from antral biopsy specimens using standard methods37. H. pylori DNA was extracted from these colonies for genotyping using the QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. The cagA status was determined by PCR amplification and direct sequencing of the 3′ repeat region of cagA, as described previously11. The absence of cagA was confirmed by the presence of a cag PAI empty site, as described previously48. The EPIYA segment types of CagA were compared with previous data obtained from GenBank using the program WebLogo (version 3) (http://weblogo.threeplusone.com/).
The vacA genotyping (s1a, s1b, s1c, or s2; and m1a, m1b, m1c, or m2) was performed following previously described methods6,10,49. Genetic distances for the vacA m region were estimated by the six-parameter method, and phylogenetic trees were constructed using the neighbor-joining method including 112 previously published vacA m region sequences as a reference50. DNA sequencing was performed using the Big Dye Terminator v3.1 Cycle Sequencing Kit on an AB 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Multiple sequence alignments of the vacA sequences were generated using MAFFT version 7 (available in http://mafft.cbrc.jp/alignment/server/) and confirmed by visual inspection.
Population and phylogenetic analysis based on MLST data
Thirteen Bhutan strains possessing the Western-type CagA and 50 Bhutan strains possessing the East Asian-type CagA were randomly selected and seven MLST genes were sequenced in each of these strains by PCR-based sequencing as previously described51. Additionally, MLST sequence data were downloaded from the PubMLST database (http://pubmlst.org/) and 430 strains that are representative of each H. pylori population were chosen. Then, the Bhutan and selected PubMLST data were combined and the population type of the Bhutanese strains was analyzed using the population analysis software STRUCTURE (v.2.3.2)52. Markov chain Monte Carlo (MCMC) simulations were run in STRUCTURE using the admixture model with a burn-in of 20,000, followed by 30,000 iterations for each run. The analysis was repeated for a population number (K) ranging from 7 to 15, and 5 runs were performed for each K. For a given K, STRUCTURE determines K population components, which are represented using K colors. A neighbor-joining tree (Kimura’s two-parameter model) was constructed using the same dataset.
Histology and immunohistochemistry
Biopsy specimens were collected from the antrum and corpus and fixed in 10% buffered formalin for 24 h and then embedded in paraffin. Immunohistochemistry to determine CagA status and the status of East Asian-type CagA was performed as described previously28. Briefly, after antigen retrieval and inactivation of endogenous peroxidase activity, tissue sections were incubated with α-H. pylori antibody (DAKO, Glostrup, Denmark), anti-CagA antibody (b-300; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or α-EAS Ab diluted 1:2,000 with diluting solution (DAKO) overnight at 4 °C. After washing, the sections were incubated with biotinylated goat anti-rabbit or anti-rat IgG (Nichirei Co., Tokyo, Japan), followed by incubation with a solution of avidin-conjugated horseradish peroxidase (Vectastain Elite ABC kit; Vector Laboratories Inc., Burlingame, CA, USA). Peroxidase activity was detected using H2O2/diaminobenzidine substrate solution. H. pylori were identified by Giemsa staining and positively immunostained with anti-H. pylori antibody. The degree of inflammation, neutrophil activity, atrophy, intestinal metaplasia, and bacterial density were classified into four grades according to the updated Sydney system: 0, ‘normal’; 1, ‘mild’; 2, ‘moderate’; and 3, ‘marked’27. Bacterial loads greater than or equal to grade 1 were considered positive for H. pylori. Samples classified as grade 1 or higher were considered atrophy-positive53. The bacterial load was classified into four grades: 0, ‘none’; 1, ‘mild’; 2, ‘moderate’; and 3, ‘marked’ based on specimens stained with May-Giemsa27.
Statistical analysis
Discrete variables were tested using the chi-square test and Fisher’s exact probability test (average age and sex ratio vs. diagnosis, relationship between genotypes); The difference of histologic score between genotypes were tested using the Mann-Whitney U test. A multivariate logistic regression model was used to calculate the OR and 95% CI of the clinical presentations including age, sex, and H. pylori genotype. All determinants with P-values of less than 0.10 were combined in the full model for the logistic regression, and the model was reduced by excluding variables with P-values of greater than 0.10. A P-value of < 0.05 was accepted as statistically significant. SPSS statistical software package version 19.0 (SPSS, Inc., Chicago, IL, USA) was used for all statistical analyses.
Additional Information
How to cite this article: Matsunari, O. et al. Rare Helicobacter pylori Virulence Genotypes in Bhutan. Sci. Rep. 6, 22584; doi: 10.1038/srep22584 (2016).
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (DK62813) and the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (24406015, 24659200, 25293104, 26640114 and 15H02657) (YY). It was also supported by the Japan Society for the Promotion of Science (JSPS) Institutional Program for Young Researcher Overseas Visits (YY), the Strategic Funds for the Promotion of Science and Technology from Japan Science and Technology Agency (JST) (YY).
Footnotes
Author Contributions Conceived and designed the experiments: O.M. and Y.Y. Performed the experiments: O.M. and T.U. Analyzed the data: O.M., S.S., M.M., R.S. and Y.Y. Contributed reagents/materials/analysis tools: R.V., T.U., T.R., L.T. and V.M. Wrote the paper: O.M., S.S., M.M. and Y.Y.
References
- Peek R. M. Jr. & Blaser M. J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2, 28–37, doi: 10.1038/nrc703 (2002). [DOI] [PubMed] [Google Scholar]
- Suerbaum S. & Michetti P. Helicobacter pylori infection. N Engl J Med 347, 1175–1186, doi: 10.1056/NEJMra020542 (2002). [DOI] [PubMed] [Google Scholar]
- McLean M. H. & El-Omar E. M. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol 11, 664–674, doi: 10.1038/nrgastro.2014.143 (2014). [DOI] [PubMed] [Google Scholar]
- Yamaoka Y. & Graham D. Y. Helicobacter pylori virulence and cancer pathogenesis. Future oncol 10, 1487–1500, doi: 10.2217/fon.14.29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A. et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA 90, 5791–5795 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton J. et al. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem 270, 17771–17777 (1995). [DOI] [PubMed] [Google Scholar]
- Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol 7, 629–641, doi: 10.1038/nrgastro.2010.154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota S., Suzuki R. & Yamaoka Y. The significance of virulence factors in Helicobacter pylori. J Dig Dis 14, 341–349, doi: 10.1111/1751-2980.12054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y., Kodama T., Kashima K., Graham D. & Sepulveda A. Variants of the 3′ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol 36, 2258–2263 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y. et al. Relationship between the cagA 3′ repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology 117, 342–349, doi: 10.1053/gast.1999.0029900342 (1999). [DOI] [PubMed] [Google Scholar]
- Yamaoka Y. et al. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol Infect 124, 91–96 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer 4, 688–694, doi: 10.1038/nrc1433 (2004). [DOI] [PubMed] [Google Scholar]
- Suzuki R. et al. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infect Genet Evol. 12, 203–13, doi: 10.1016/j.meegid.2011.12.002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunari O. et al. Association between Helicobacter pylori virulence factors and gastroduodenal diseases in Okinawa, Japan. J Clin Microbiol 50, 876–883, doi: 10.1128/JCM.05562-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaichone R. K. et al. Molecular epidemiology and outcome of Helicobacter pylori infection in Thailand: a cultural cross roads. Helicobacter 9, 453–459, doi: 10.1111/j.1083-4389.2004.00260.x (2004). [DOI] [PubMed] [Google Scholar]
- Jones K. R. et al. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J Clin Microbiol 47, 959–968, doi: 10.1128/JCM.02330-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover T. L. & Blaser M. J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem 267, 10570–10575 (1992). [PubMed] [Google Scholar]
- Sugimoto M., Zali M. R. & Yamaoka Y. The association of vacA genotypes and Helicobacter pylori-related gastroduodenal diseases in the Middle East. Eur J Clin Microbiol Infect Dis 28, 1227–1236, doi: 10.1007/s10096-009-0772-y (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M. & Yamaoka Y. The association of vacA genotype and Helicobacter pylori-related disease in Latin American and African populations. Clin Microbiol Infect 15, 835–842, doi: 10.1111/j.1469-0691.2009.02769.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley Y. et al. The peopling of the Pacific from a bacterial perspective. Science 323, 527–530, doi: 10.1126/science.1166083 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D. et al. Traces of human migrations in Helicobacter pylori populations. Science 299, 1582–1585, doi: 10.1126/science.1080857 (2003). [DOI] [PubMed] [Google Scholar]
- Linz B. et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918, doi: 10.1038/nature05562 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sablet T. et al. Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut 60, 1189–1195, doi: 10.1136/gut.2010.234468 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodaman N. et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci USA 111, 1455–1460, doi: 10.1073/pnas.1318093111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol 7, 629–641, doi: 10.1038/nrgastro.2010.154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Yamaoka Y., Zhu Q., Matha I. & Gao X. A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori. PloS one 4, e7736, doi: 10.1371/journal.pone.0007736 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M., Genta R., Yardley J. & Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol 20, 1161–1181 (1996). [DOI] [PubMed] [Google Scholar]
- Uchida T. et al. Immunohistochemical diagnosis of the cagA-gene genotype of Helicobacter pylori with anti-East Asian CagA-specific antibody. Cancer Sci 98, 521–528, doi: 10.1111/j.1349-7006.2007.00415.x (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanada R. et al. Genotyping of the cagA gene of Helicobacter pylori on immunohistochemistry with East Asian CagA-specific antibody. Pathol Int 58, 218–225, doi: 10.1111/j.1440-1827.2008.02214.x (2008). [DOI] [PubMed] [Google Scholar]
- Nguyen L. T. et al. Evaluation of the anti-East Asian CagA-specific antibody for CagA phenotyping. Clin Vaccine Immunol 16, 1687–1692, doi: 10.1128/CVI.00200-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D. et al. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J Clin Invest 122, 1553–1566, doi: 10.1172/JCI61143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong X. et al. Helicobacter pylori virulence factor CagA promotes tumorigenesis of gastric cancer via multiple signaling pathways. J Cell Commun Signal 13, 30, doi: 10.1186/s12964-015-0111-0 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argent R. et al. Determinants and consequences of different levels of CagA phosphorylation for clinical isolates of Helicobacter pylori. Gastroenterology 127, 514–523, doi: 10.1053/j.gastro.2004.06.006 (2004). [DOI] [PubMed] [Google Scholar]
- Azuma T. et al. Correlation between variation of the 3′ region of the cagA gene in Helicobacter pylori and disease outcome in Japan. J Infect Dis 186, 1621–1630, doi: 10.1086/345374 (2002). [DOI] [PubMed] [Google Scholar]
- Shiota S. et al. Seroprevalence of Helicobacter pylori infection and gastric mucosal atrophy in Bhutan, a country with a high prevalence of gastric cancer. J Med Microbiol 62, 1571–1578, doi: 10.1099/jmm.0.060905-0 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. S. et al. A specific A/T polymorphism in Western tyrosine phosphorylation B-motifs regulates Helicobacter pylori CagA epithelial cell interactions. PLoS pathogens 11, e1004621, doi: 10.1371/journal.ppat.1004621 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y. et al. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter 3, 241–253 (1998). [DOI] [PubMed] [Google Scholar]
- Wang H. J., Kuo C. H., Yeh A. A., Chang P. C. & Wang W. C. Vacuolating toxin production in clinical isolates of Helicobacter pylori with different vacA genotypes. J Infect Dis 178, 207–212 (1998). [DOI] [PubMed] [Google Scholar]
- Pan Z. J. et al. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J Infect Dis 178, 220–226 (1998). [DOI] [PubMed] [Google Scholar]
- Nei M. & Takezaki N. The root of the phylogenetic tree of human populations. Mol Biol Evol 13, 170–177 (1996). [DOI] [PubMed] [Google Scholar]
- Barbujani G., Magagni A., Minch E. & Cavalli-Sforza L. L. An apportionment of human DNA diversity. Proc Natl Acad Sci USA 94, 4516–4519 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. & Roychoudhury A. K. Evolutionary relationships of human populations on a global scale. Mol Biol Evol 10, 927–943 (1993). [DOI] [PubMed] [Google Scholar]
- Gayden T. et al. The Himalayas as a directional barrier to gene flow. Am J Hum Genet 80, 884–894, doi: 10.1086/516757 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y., Yahara K., Hatakeyama M. & Kobayashi I. Evolution of cagA oncogene of Helicobacter pylori through recombination. PloS one 6, e23499, doi: 10.1371/journal.pone.0023499 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi S. M. et al. Ancestral European roots of Helicobacter pylori in India. BMC genomics 8, 184, doi: 10.1186/1471-2164-8-184 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B. et al. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. J Hum Genet 107, 582–590 (2000). [DOI] [PubMed] [Google Scholar]
- Vilaichone R. K. et al. Extremely high prevalence of Helicobacter pylori infection in Bhutan. World J Gastroenterol 19, 2806–2810, doi: 10.3748/wjg.v19.i18.2806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyants N. S. et al. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol 28, 37–53 (1998). [DOI] [PubMed] [Google Scholar]
- Yamazaki S. et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J Clin Microbiol 43, 3906–3916, doi: 10.1128/JCM.43.8.3906-3916.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaoka Y. et al. Helicobacter pylori in North and South America before Columbus. FEBS Lett 517, 180–184, doi: 10.1016/S0014-5793(02)02617-0 (2002). [DOI] [PubMed] [Google Scholar]
- Achtman M. et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol 32, 459–470 (1999). [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M. & Pritchard J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornschein J., Selgrad M., Wex T., Kuester D. & Malfertheiner P. Serological assessment of gastric mucosal atrophy in gastric cancer. BMC Gastroenterol 12, 10, doi: 10.1186/1471-230X-12-10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]