Abstract
The contribution of rare and low-frequency variants to human traits is largely unexplored. Here we describe insights from sequencing whole genomes (low read depth, 7×) or exomes (high read depth, 80×) of nearly 10,000 individuals from population-based and disease collections. In extensively phenotyped cohorts we characterize over 24 million novel sequence variants, generate a highly accurate imputation reference panel and identify novel alleles associated with levels of triglycerides (APOB), adiponectin (ADIPOQ) and low-density lipoprotein cholesterol (LDLR and RGAG1) from single-marker and rare variant aggregation tests. We describe population structure and functional annotation of rare and low-frequency variants, use the data to estimate the benefits of sequencing for association studies, and summarize lessons from disease-specific collections. Finally, we make available an extensive resource, including individual-level genetic and phenotypic data and web-based tools to facilitate the exploration of association results.
Supplementary information
The online version of this article (doi:10.1038/nature14962) contains supplementary material, which is available to authorized users.
Subject terms: Next-generation sequencing, Genome-wide association studies
Low read depth sequencing of whole genomes and high read depth exomes of nearly 10,000 extensively phenotyped individuals are combined to help characterize novel sequence variants, generate a highly accurate imputation reference panel and identify novel alleles associated with lipid-related traits; in addition to describing population structure and providing functional annotation of rare and low-frequency variants the authors use the data to estimate the benefits of sequencing for association studies.
Supplementary information
The online version of this article (doi:10.1038/nature14962) contains supplementary material, which is available to authorized users.
Genome variation in health and disease
This paper, combining data and initial findings from the different arms of the UK10K project, describes insights from low-read-depth sequencing of whole genomes or high-read-depth exome sequencing of nearly 10,000 individuals sampled from a range of disease collections, as well as participants from healthy population based cohorts. The authors characterize novel sequence variants, generate a highly accurate imputation reference panel and identify novel alleles associated with lipid-related traits. In addition to describing population structure and providing functional annotation of rare and low frequency variants, they use the data to estimate the benefits of sequencing for association studies.
Supplementary information
The online version of this article (doi:10.1038/nature14962) contains supplementary material, which is available to authorized users.
Main
Assessment of the contribution of rare genetic variation to many human traits is still largely incomplete. In common and complex diseases, a lack of empirical data has to date hampered the systematic assessment of the contribution of rare and low-frequency genetic variants (defined throughout this paper as minor allele frequency (MAF) <1% and 1–5%, respectively). Rare variants are incompletely represented in genome-wide association (GWA) studies1 and custom genotyping arrays2,3, and impute poorly with current reference panels. Rare and low-frequency variants also tend to be population- or sample-specific, requiring direct ascertainment through resequencing4,5. Recent exome-wide resequencing studies have begun to explore the contribution of rare coding variants to complex traits6, but comparatively little is known of the non-coding part of the genome where most complex trait-associated loci lie7. At the other end of the human disease spectrum, the widespread application of exome-wide sequencing is accelerating the rate at which genes and variants causal for rare diseases are being identified. Despite this, many Mendelian diseases still lack a genetic diagnosis and the penetrance of apparently disease-causing loci remains inadequately assessed.
The UK10K project was designed to characterize rare and low-frequency variation in the UK population, and study its contribution to a broad spectrum of biomedically relevant quantitative traits and diseases with different predicted genetic architectures. Here we describe the data and initial findings generated by the different arms of the UK10K project. In addition to this paper, UK10K companion papers describe the utility of this resource for imputation8, association discovery for bone mineral density9, thyroid function10 and circulating lipid levels11 and provide access to the study results through novel web tools12.
Study designs in the UK10K project
The UK10K project includes two main project arms (Table 1). The UK10K-cohorts arm aimed to assess the contribution of genome-wide genetic variation to a range of quantitative traits in 3,781 healthy individuals from two intensively studied British cohorts of European ancestry, namely the Avon Longitudinal Study of Parents and Children (ALSPAC)13 and TwinsUK14. A low read depth (average 7×) whole-genome sequencing (WGS) strategy was employed in order to maximize total variation detected for a given total sequence quantity15 while allowing interrogation of noncoding variation. Sixty-four different phenotypes were analysed, including traits of primary clinical relevance in 11 major phenotypic groups (obesity, diabetes, cardiovascular and blood biochemistry, blood pressure, dynamic measurements of ageing, birth, heart, lung, liver and renal function; Supplementary Table 1). Of these, 31 phenotypes were available in both studies (referred to as ‘core’ and reported in association analyses), 18 were unique to TwinsUK and 15 were unique to ALSPAC.
Table 1.
Summary of sample collections and sequencing metrics for the four main studies of the UK10K project
| Study name and design | n | Sequencing strategy, mean read depth and Ts/Tv ratio | SNVs/INDELs | SNVs/INDELs by allele frequency |
|---|---|---|---|---|
| Cohorts. Unselected samples from two population-based cohorts | 3,781 |
WGS, 7× Ts/Tv = 2.15 |
42,001,210/3,490,825 | <1%: 34,247,969/2,296,962 1–5%: 2,298,220/412,168 >5%: 5,869,317/1,496,955 |
| Rare. Eight rare diseases with expected different allelic architectures (ciliopathy, coloboma, congenital heart disease, familial hypercholesterolaemia, intellectual disability, neuromuscular, severe insulin resistance and thyroid disease) | 961 (397) |
WES, 77× Ts/Tv = 3.02 |
252,809/ 1,621 |
<1%: 171,564/1,384 ≥1%: 81,245/237 |
| Obesity. Severely obese children (BMI > 3 s.d. from population mean) and adults with extreme obesity | 1,468 (1,359) |
WES, 82× Ts/Tv = 3.02 |
484,931/ 3,370 |
<1%: 403,684/3,133 ≥1%: 81,247/237 |
| Neurodevelopmental. Autism and schizophrenia (individual probands, families with one affected and other healthy individuals sampled, families with data from multiple affected individuals and individuals with comorbid intellectual disability and psychosis) | 2,753 (1,707) |
WES, 77× Ts/Tv = 3.02 |
538,526/ 3,826 |
<1%: 457,278/3,589 ≥1%: 81,248/237 |
For the cohorts arm, numbers are for the set of 3,781 samples passing quality control, while a subset of 3,621 was used for association testing. For the exome arm, numbers of sites are based on the joint call set, and are calculated for a subset of all individuals that represent the patient subset (in brackets). The total number of individuals sequenced in each study is also given (see Supplementary Methods). The transition to transversion ratio (Ts/Tv) was calculated for the final set of SNVs excluding multiallelic sites. WGS, whole-genome sequencing; WES, whole-exome sequencing.
The UK10K-exomes arm aimed to identify causal mutations through high read depth (mean ∼80× across studies) whole-exome sequencing of approximately 6,000 individuals from three different collections: rare disease, severe obesity and neurodevelopmental disorders. The disorders studied in the UK10K-exomes arm have been shown to have a substantial genetic component at least partially driven by very rare, highly penetrant coding mutations. The rare disease collection includes 125 patients and family members in each of eight rare disease areas (Table 1). Disease types were selected with different degrees of locus heterogeneity, prior evidence for monogenic causation and likely modes of inheritance (for example, dominant or recessive). The obesity collection comprises of samples with severe obesity phenotypes, including approximately 1,000 subjects from the Severe Childhood Onset Obesity Project (SCOOP)16, plus severely obese adults from several population cohorts. The neurodevelopmental collection comprises of ∼3,000 individuals selected to study two related neuropsychiatric disorders (autism spectrum disorder and schizophrenia).
Discovery of 24 million novel genetic variants
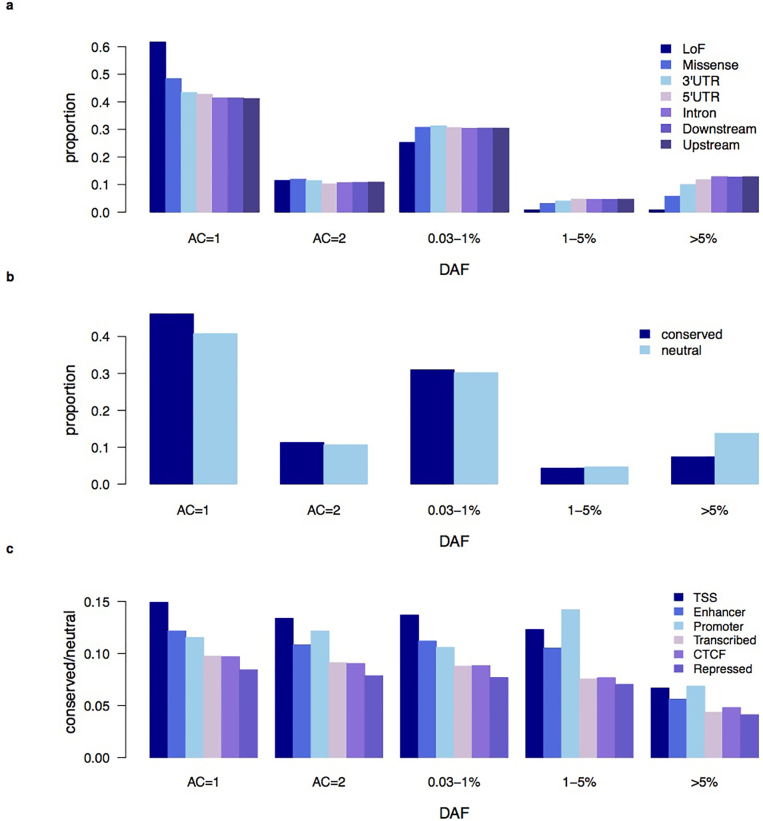
In total, 3,781 individuals were successfully whole-genome sequenced in the UK10K-cohorts arm. After conservative quality control filtering (Extended Data Figs 1 and 2 and Supplementary Table 2), the final call set contained over 42M single nucleotide variants (SNVs, 34.2M rare and 2.2M low-frequency), ∼3.5M insertion/deletion polymorphisms (INDELs; 2,291,553 rare and 415,735 low-frequency) and 18,739 large deletions (median size 3.7 kilobase). Each individual on average contained 3,222,597 SNVs (5,073 private), 705,684 INDELs (295 private) and 215 large deletions (less than 1 private). Of 18,903 analysed protein-coding genes, 576 genes contained at least one homozygous or compound heterozygous variant predicted to result in the loss of function of a protein (LoF, Supplementary Information, 14,516 variants in total). As previously shown5,17, variants predicted to have the greatest phenotypic impact (LoF and missense variants, and variants mapping to conserved regions), were depleted at the common end of the derived allele spectrum (Extended Data Fig. 3). There were 495 homozygous LoF variants, a subset of which associated with phenotypic outliers (Supplementary Table 3).
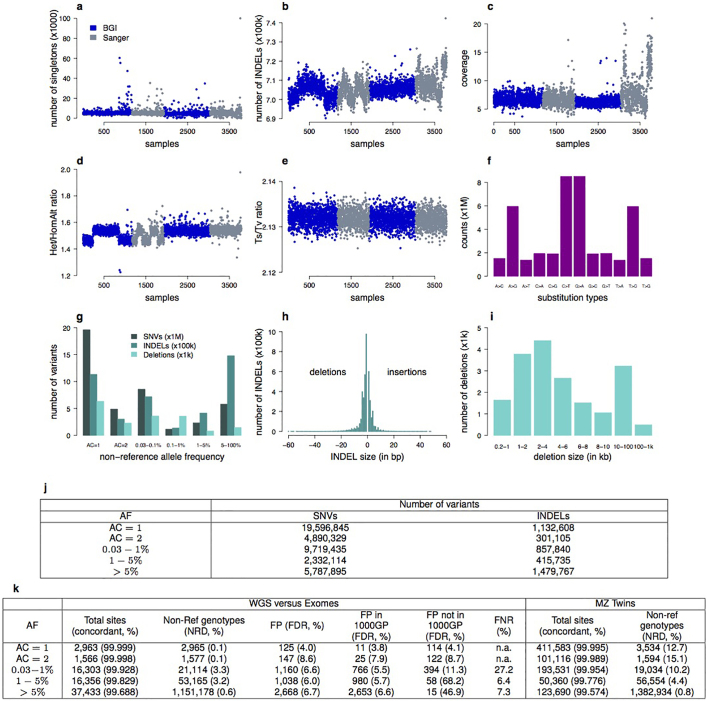
Extended Data Figure 1. UK10K-cohorts, sequence and sample quality and variation metrics.
a–e, Sample quality metrics for UK10K-cohorts (n = 3,781) where n = 1–1,927 corresponds to ALSPAC and 1,928 to 3,781 to TwinsUK. This sample includes all individuals passing sample quality control, including related pairs and non-European individuals that were later removed from association tests. A subset of 3,621 individuals was included in association analyses. Samples sequenced at BGI are coloured in blue and samples sequenced at Sanger are coloured in grey. a, Number of singletons (AC = 1) by sample (×103). b, Number of INDELs by sample (×105). c, Read depth (sequence coverage) by sample. d, Ratio of heterozygous and homozygous non-reference (=homozygous alternative) SNV genotypes (mean for females = 1.54, mean for males = 1.47). e, Transition to transversion ratio (Ts/Tv) by sample. f–i, Sequence variation metrics for UK10K-cohorts. f, Types of substitution (×106). g, Number of SNVs (×106), INDELs (×105) and large deletions (×103) by non-overlapping non-reference allele frequency (AF) bins. h, Size distribution of INDELs. Negative INDEL lengths represent deletions and positive INDEL lengths represent insertions. i, Large deletion size distribution in unequal bin sizes where the smallest deletions were 200 bp to 1 kb long and the largest deletions 100 kb to 1 Mb. In total 18,739 deletions were called with GenomeSTRiP14. The average deletion size was ˜13 kb and the median size was ˜3.7 kb. j, Total number of SNVs and INDELs by AF bin (based on 3,781 samples), multi-allelic variants are treated as separate variants. k, Sequence quality and variation metrics for UK10K-cohorts. For 61 overlapping TwinsUK individuals we compared the variant sites and genotypes of the low-coverage sequences with high-coverage exome data by non-overlapping AF bins (WGS versus Exomes). We considered 74,621 shared sites in non-overlapping AF bins. We calculated the fraction of concordant over total sites, the number of non-reference genotypes and non-reference genotype discordance (NRD, in %) between WGS and Exomes; false discovery rate (FDR = FP/(FP + TP); TP, true positive; FP, false positive), where we consider the exomes as the truth set; number of false positives (FP) and FDR for sites that are or not shared with the 1000 Genomes Project, phase I (1000GP); false negative rate (FNR = FN/(FN + TP); FN, false negative; TP, true positive), where AF bins were defined based on the 61 exomes. Furthermore, we compared 22 monozygotic twin pairs at 880,280 bi-allelic SNV sites on chromosome 20, reporting the percentage of concordant genotypes, non-reference genotypes and NRD. AFs are from the set of 3,621 samples, which contains at most one of the two monozygotic twins from each pair. We note that discrepancies can be caused by errors in either twin, so the expected NRD to the truth would be half the NRD value given.
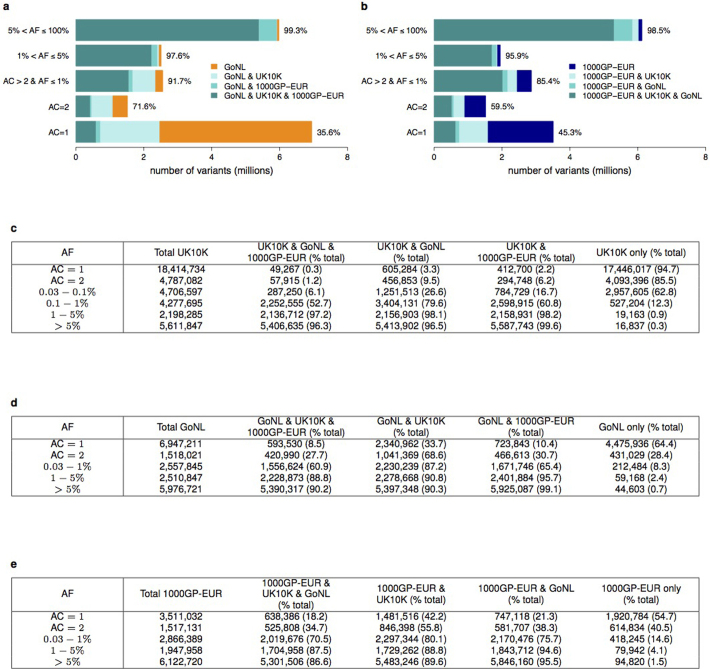
Extended Data Figure 2. UK10K-cohorts, comparison with GoNL and 1000GP-EUR.
Percentage of autosomal SNVs that are either shared between UK10K (n = 3,781), GoNL (n = 499) and 1000GP-EUR (n = 379), or unique to each set, for allele counts (AC) AC = 1, AC = 2, and non-overlapping allele frequency (AF) bins for higher AC. a, Shared and unique variants for GoNL with AF based on GoNL, and b, for 1000GP-EUR. AF bins are not directly comparable owing to the different sample sizes in each call set. The x-axis shows the number of variants in millions. The percentages next to the bars represent the percentage of variants from GoNL (a) and 1000GP-EUR (b) that are shared with at least one of the other data sets. All numerical values used in a can be found in d and for b in e. c, Numerical values for Fig. 1.
Extended Data Figure 3. UK10K-cohorts, derived allele frequency spectrum by functional annotation.
Derived allele frequency (DAF) spectrum for UK10K-cohorts chromosome 20 variants divided by functional class. a, Proportion of total variants (standardized across DAF bins) as a function of DAF for different genic elements. b, Standardized proportion of all variants by DAF bin, and divided into conserved (GERP > 2) versus neutral (GERP ≤ 2) sites. c, Ratio of conserved versus neutral variants by DAF bin, and classified by chromatin segmentation domains defined by ENCODE as detailed in the methods.
We assessed sequence data quality by comparison with an exome sequencing data set (WES, ∼50 × coverage)18 and in 22 pairs of monozygotic twins (Extended Data Fig. 1). The non-reference discordance (NRD, or the fraction of discordant genotypes for non-reference homozygous or heterozygous alleles) was 0.6% for common variants and 3.2% (range 0.1–3.3%; Extended Data Fig. 1) for low-frequency and rare variants. False discovery rates (FDR) were comparable between newly discovered sites and sites previously reported in the 1000 Genomes Project phase 1 (1000GP) data set5.
When compared to two large-scale European sequencing repositories, 1000GP and the Genome of the Netherlands (GoNL, 12 × read depth19), UK10K-cohorts discovered over 24M novel SNVs. Overall, 96.5% of variants with MAF > 1% were shared, reflecting a common reservoir within Europe (Fig. 1 and Extended Data Fig. 2). Conversely, 94.7% of singleton (allele count (AC) = 1) and 55.0% of rare (AC > 1 and MAF < 1%) SNVs were study-specific. In a similar comparison, 64.4% (AC = 1) and 15.8% of variants (AC > 1 and MAF < 1%) found in GoNL were found to be study-specific compared to 1.2% of variants above 1% MAF.
Figure 1. The UK10K-cohorts resource for variation discovery.
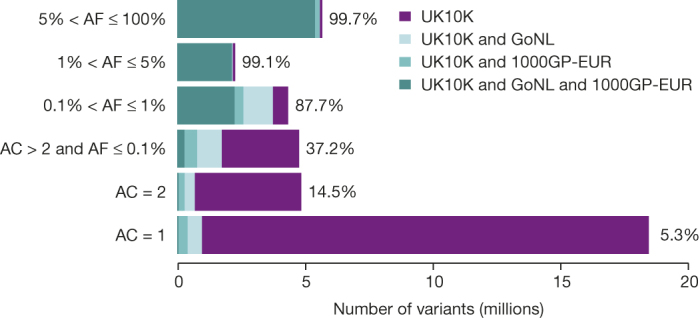
Number of SNVs identified in the UK10K-cohorts data set in all autosomal regions in different allele frequency (AF) bins, and percentages that were shared with samples of European ancestry from the 1000 Genomes Project (phase I, EUR n = 379) and/or the Genomes of the Netherlands (GoNL, n = 499) study, or unique to the UK10K-cohorts data set. AF bins were calculated using the UK10K data set, for allele count (AC) = 1, AC = 2, and non-overlapping AF bins for higher AC. All numerical values are in Extended Data Fig. 2.
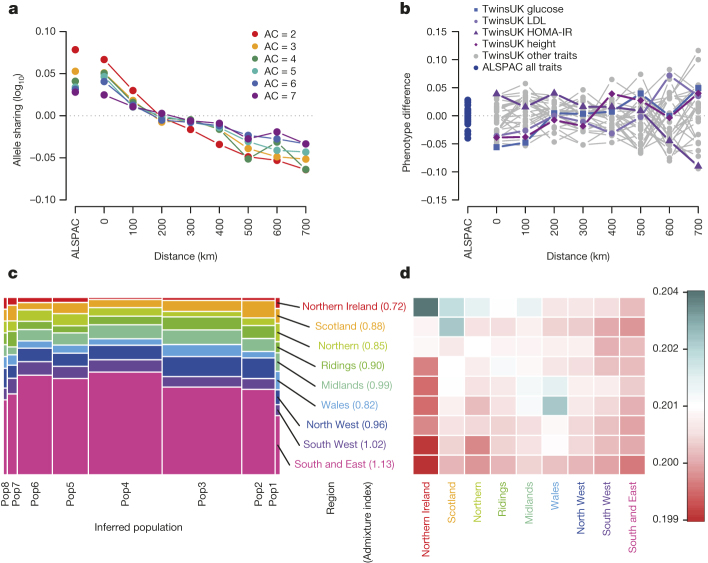
This deeper characterization of European genetic and haplotype diversity will benefit future studies by creating a novel genotype imputation panel with substantially increased coverage and accuracy compared to the 1000GP reference panel8 (see ref. 9 and the next section for its application). It further informs a detailed empirical assessment of the geographical structure of rare variation in the UK where we detected geographical structure for very rare alleles (AC = 2–7) in Northern and Western UK regions, although this did not show evidence of substantial correlation with variation in phenotype (Box 1).
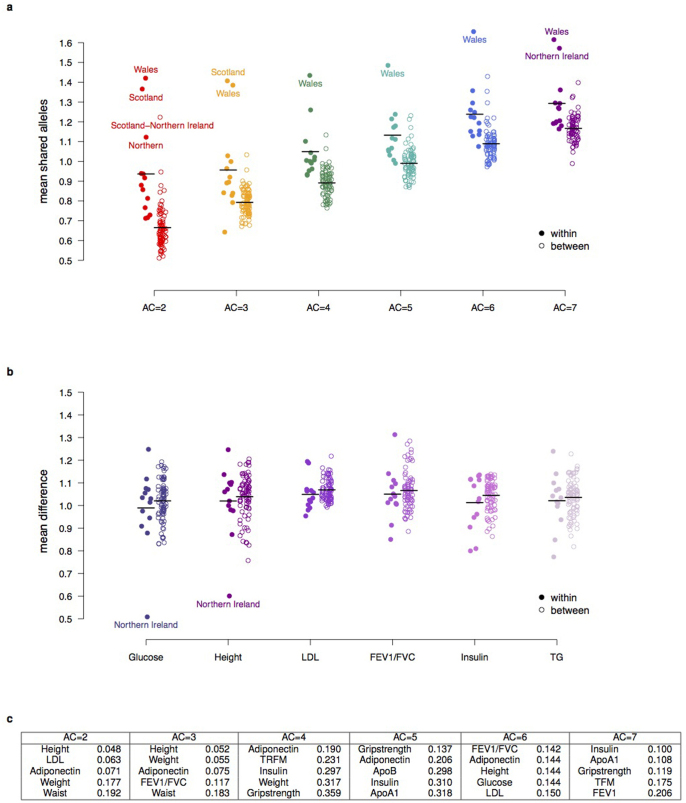
Box 1: Genetic structure of rare variation within the UK.
We used the ALSPAC cohort (from the Bristol region) and a subset of TwinsUK individuals (UK-wide origin) to investigate the spatial structure of rare genetic variants (Supplementary Table 16). We first sought to define the extent to which variants of different MAF were geographically structured. We estimated the excess of allele sharing between pairs of individuals as a function of their physical distance, as compared to expectations under a neutral model (Supplementary Information)46. Rare genetic variants showed excess allele sharing at distances smaller than about 200 km, and reduced sharing for more than about 300 km. There was a steeper geographical cline for doubletons (AC = 2), which decreased with increasing allele counts (3 up to 7, equivalent to a MAF of ∼0.1–0.3%; a). No corresponding geographical structure was observed for phenotypic variation (b).
We next assessed the extent to which the non-random distribution of rare SNVs could be accounted for by regional differences at the level of 13 main regions within the UK47. Overall, patterns of allele sharing were indicative of a larger degree of genetic homogeneity in Southern and Eastern England compared to individuals of Welsh, Northern, Scottish or Northern Irish origin. Doubletons were the most structured both within and between regions (Wilcoxon rank sum P value <0.05, Extended Data Fig. 8).
Finally, we used “chromosome painting”48 to gain insights into possible demographic events underlying the observed genetic structure. We first estimated the average length of DNA tracts shared between individuals, and used the number of such tracts to identify fine population structure in our data set. The tract length distribution showed weak geographic structure reflecting the rare variant analysis. A fine structure analysis suggested that the identified populations were not strongly geographically defined, indicative of a large degree of movement between regions compared to the samples in the Peoples of the British Isles study45, which were chosen to have all four grandparents born in the same location (Extended Data Fig. 9).
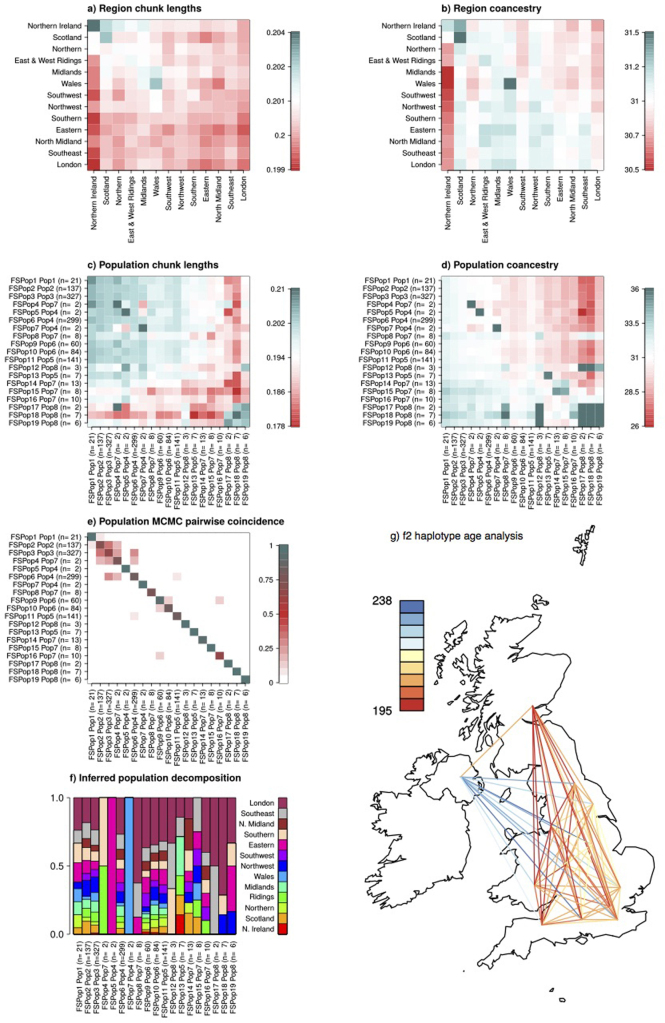
Population structure in UK10K-cohorts.
All ALSPAC (from Bristol), and 1,139 TwinsUK (UK-wide) participants with a complete set of genotype, phenotype and place of birth data.a, Excess of allele sharing as a function of geographical distance, expressed as the proportion of shared alleles between sample pairs for AC from 2 to 7 against their geographical distance. b, Phenotypic sharing, estimated for the 31 core phenotypes as the absolute difference between pairs of individuals, averaged within distance bins, rescaled and plotted against their geographical distance. The four traits with the most extreme structure are highlighted. HOMA-IR, homeostatic model assessment for insulin. c, Geographical decomposition of each population. Populations are shown proportional to size; historically ‘Celtic’ and ‘Briton’ regions are closer to the edges, whereas ‘Anglo-Saxon’ England is more homogeneous and at the centre (see ref. 45). Ridings refers to East and West Ridings, Yorkshire. d, Average length of DNA tracts shared between individuals when clustered by sampling location. The ‘admixture’ index is given in brackets, with one-third corresponding to regions containing completely unadmixed populations and infinity to completely admixed populations. See also Extended Data Fig. 9.
Findings from single-marker association tests
A main aim of the UK10K-cohorts project was to assess associations of low-frequency and rare variants under different analytical strategies (Fig. 2). We used a unified analysis strategy for the parallel evaluation of all quantitative traits (Supplementary Information, Supplementary Table 4). Here we describe results for the 31 core traits shared in ALSPAC and TwinsUK, with other results reported elsewhere12.
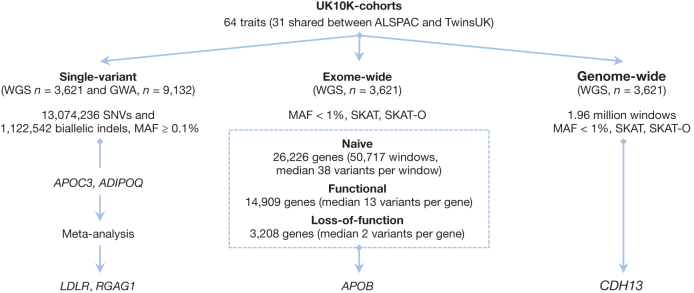
Figure 2. Study design for associations tested in the UK10K-cohorts study.
Summary of phenotype–genotype association testing strategies employed in the UK10K-cohorts study.
We first carried out single-marker association tests, as in standard genome-wide association studies of common variants20. Assuming an additive genetic model, we used standard approaches to model relationships between standardised traits, residualized for relevant covariates, and allele dosages of 13,074,236 SNVs, 1,122,542 biallelic INDELs (MAF ≥ 0.1%) and 18,739 large deletions in whole-genome sequenced samples (‘WGS sample’). We further assessed associations in an independent study sample of genome-wide genotyped individuals (‘GWA’ sample) including up to 6,557 ALSPAC and 2,575 TwinsUK participants who were not part of UK10K (actual numbers per trait are given in Supplementary Table 1). In the GWA sample, genotypes were imputed from genome-wide single nucleotide polymorphism (SNP) data using the UK10K haplotype reference panel, described in a companion manuscript8. The combined WGS+GWA sample had 80% power to detect associations of SNVs of low-frequency and rare down to ∼MAF 0.5%, for a per-alleles trait change (the regression beta coefficient or Beta) of ∼1.2 standard deviations or greater (Fig. 3). To combine WGS and GWA data we carried out a fixed effect meta-analysis using the inverse variance method, which showed no evidence of inflation of summary statistics at the traits investigated (GC lambda ≈ 1). We used a conservative stepwise procedure for reporting loci from single-variant analysis (Supplementary Table 5), and we discuss elsewhere replication and technical validation of associations of rare variants not supported in the combined WGS+GWA sample (Supplementary Information, Supplementary Table 6).
Figure 3. Summary of association results across the UK10K-cohorts study.
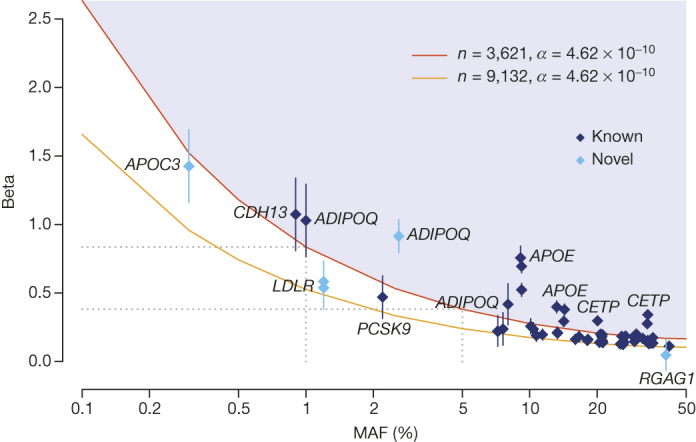
Allelic spectrum for single-marker association results for independent variants identified in the single-variant analysis (Supplementary Table 5). A variant’s effect (absolute value of Beta, expressed in standard deviation units) is given as a function of minor allele frequency (MAF, x axis). Error bars are proportional to the standard error of the beta, variants identifying known loci are dark blue and variants identifying novel signals replicated in independent studies are coloured in light blue. The red and orange lines indicate 80% power at experiment-wide significance level (t-test; P value ≤4.62 × 10−10) for the maximum theoretical sample size for the WGS sample and WGS+GWA, respectively.
Overall, across the 31 traits 27 independent loci reached our experiment-wide significance threshold21 P value ≤ 4.62 × 10−10 in the combined WGS+GWA sample (Fig. 3 and Supplementary Table 5). Two associations have been newly discovered by this project, and were conditionally independent of other variants previously reported at the same loci. The first was a low-frequency intronic variant in ADIPOQ associated with decreased adiponectin levels (rs74577862-A, effect allele frequency (EAF) = 2.6%, P value = 3.04 × 10−64). The second was a rare splice variant (rs138326449) in APOC3 described in advance of this manuscript11,22,23. The remaining 25 loci reaching experiment-wide significance in the combined WGS+GWA sample included common, low-frequency and rare variants tagging known associations with adiponectin levels (CDH13 and ADIPOQ), lipid traits (APOB, APOC3-APOA1, APOE, CETP, LIPC, LPL, PCSK9, SORT1-PSRC1-CELSR2), C-reactive protein (LEPR), haemoglobin levels (HFE) and fasting glycaemic traits (G6PC2-ABCB11, Supplementary Table 5). In contrast to previous projections24, from this analysis of a wide range of biomedical traits there was no evidence of low-frequency alleles with large effects upon traits (Fig. 3)25, with classical lipid alleles identifying extremes of single-variant genetic contributions for these traits. This suggests that few, if any, low-frequency variants with stronger effects than those we see are likely to be detected in the general European population for the wide range of traits that we considered.
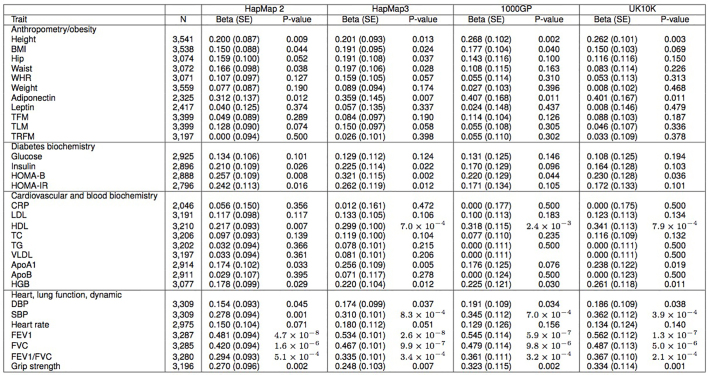
Increasing sample size may identify additional moderate effect variants, or variants with rarer frequency. We therefore sought to assess the extent to which the more accurate imputation offered by the UK10K reference panel, applied to larger study samples, could discover additional associations. A restricted maximum likelihood (REML)26 analysis suggested that using the UK10K data could increase the estimated variance explained, compared to the sparser HapMap2, HapMap3 and 1000GP data sets (Extended Data Table 1). We tested four lipid traits (high-density and low-density lipoprotein cholesterol, total cholesterol and triglycerides) in up to 22,082 additional samples from 14 cohorts imputed to the combined UK10K+1000GP phase I panel (Supplementary Table 7).
Extended Data Table 1.
UK10K-cohorts, estimated variance explained by SNVs across the 31 UK10K traits shared by both cohorts
We used the restricted maximum likelihood (REML) method implemented in GCTA to estimate phenotypic variance explained by SNV sets (MAF ≤ 1%) in our discovery sequence data (n = 3,621 individuals). SNVs were selected from the WGS data to correspond to the content of four different reference panels: HapMap2 (n = 2,331,713 SNVs), Hapmap3 (n = 1,168,695), 1000 Genomes (n = 7,475,230) and the entire UK10K reference panel (n = 8,317,582). Each GRM was individually tested against the 31 traits with phenotypic values present in both cohort studies, producing a beta, s.e. and P value for total trait variance explained by the given SNV set. Full trait names are given in Supplementary Table 1.
This effort identified two novel associations with low-density lipoprotein cholesterol (Fig. 3, Supplementary Table 8), which we further replicated in an independent imputation data set of 15,586 samples from 8 cohorts and through genotyping in 95,067 samples from the Copenhagen General Population Study (CGPS27). The first was a rare intronic variant in LDLR (rs72658867-A, c.2140 + 5G > A; EAF = 0.01, combined sample P value = 1.27 × 10−46); per allele effect Beta (s.e.m.) = −0.23 mmol l−1 (0.02), P value = 7.63 × 10−30 (CGPS, n = 95,079). The second was a common, X-linked variant near RGAG1 (rs5985471-T, EAF = 0.403, P value = 1.53 × 10−12); per allele effect Beta (s.e.m.) = −0.02 mmol l−1 (0.004), P value = 1.8 × 10−5 (CGPS, n = 93,639). The LDLR variant was previously classified to be of uncertain impact in ClinVar, and reported to have no effect on plasma cholesterol levels in a small sample of familial hypercholesterolaemia patients28. The LDLR-A allele is almost perfectly imputed in our sample (info = 0.96), but absent in previous imputation panels29; the RGAG1-T allele is common but was missed in previous studies, which focused predominantly on autosomal variation29. Within CGPS, these variants were weakly associated with ischaemic heart disease (odds ratio (OR) = 0.77(0.66, 0.92), P = 0.003 for rs72658867; 0.96(0.94, 0.99), P = 0.005 for rs5985471) and rs72658867 with myocardial infarction (OR = 0.65(0.49, 0.87), P = 0.003; Supplementary Table 8). These results demonstrate the value of our expanded haplotype reference panel for discovery of trait associations driven by low-frequency and rare variants, as also shown in refs 9, 10.
Findings from rare variant association tests
Single-marker association tests are typically underpowered for rare variants30. Many questions remain regarding the optimal choice of test, owing to the unknown allelic architecture of rare variant contribution to traits, in particular outside protein-coding regions. We first evaluated associations by considering genes (GENCODE v15) as functional units of analysis using three separate variant selection strategies. Naive tests considered all variants in exons, untranslated regions (UTRs) and essential splice sites, weighted equally. Functional tests considered missense and LoF variants, the latter defined as being predicted to cause essential splice site changes, stop codon gains or frameshifts. For each scenario we applied two separate statistical models with different properties, sequence kernel association tests (SKAT) and burden tests implemented in SKAT and SKAT-O31,32, to rare variants (MAF < 1%).
Overall, there was an excess of test statistics with P values ≤10−4 for functional and loss-of-function tests (Extended Data Figs 4 and 5), with a total of 9, 70 and 196 genes associated with the 31 core traits with the LoF, functional and naive tests, respectively (Supplementary Table 9). A signal driven by loss-of-function variants in the APOB gene (encoding apolipoprotein B) achieved our threshold for experiment-wide significance (P value ≤1.97 × 10−7), in a burden-type test (min P value for TG = 7.02 × 10−9). Overall, 3 singleton LoF variants were responsible for this signal, of which two were not previously reported (rs141422999 and Chr2:21260958). Examples of novel rare variants in complex trait-associated loci (for example, G6PC2 associated with fasting glucose) were also seen for genes reaching suggestive levels of association (P value ≤10−4). Lastly, we tested the value of a genome-wide naive approach to explore associations outside protein-coding genes by combining variants across ∼1.8 million genome-wide tiled windows of 3 kb in size (median 37 SNVs per window, MAF < 1%, assigning an equal weight to all variants in the window). Overall association statistics appeared underpowered to detect true signals, apart from an association signal for adiponectin driven by a known rare intronic variant at the CDH13 locus (rs12051272, EAF = 0.09%, P value = 6.52 × 10−12; Supplementary Table 10)33,34. As previously shown for single-variant tests, in this study adiponectin and lipid traits yielded the greatest evidence for associations for region-based tests.
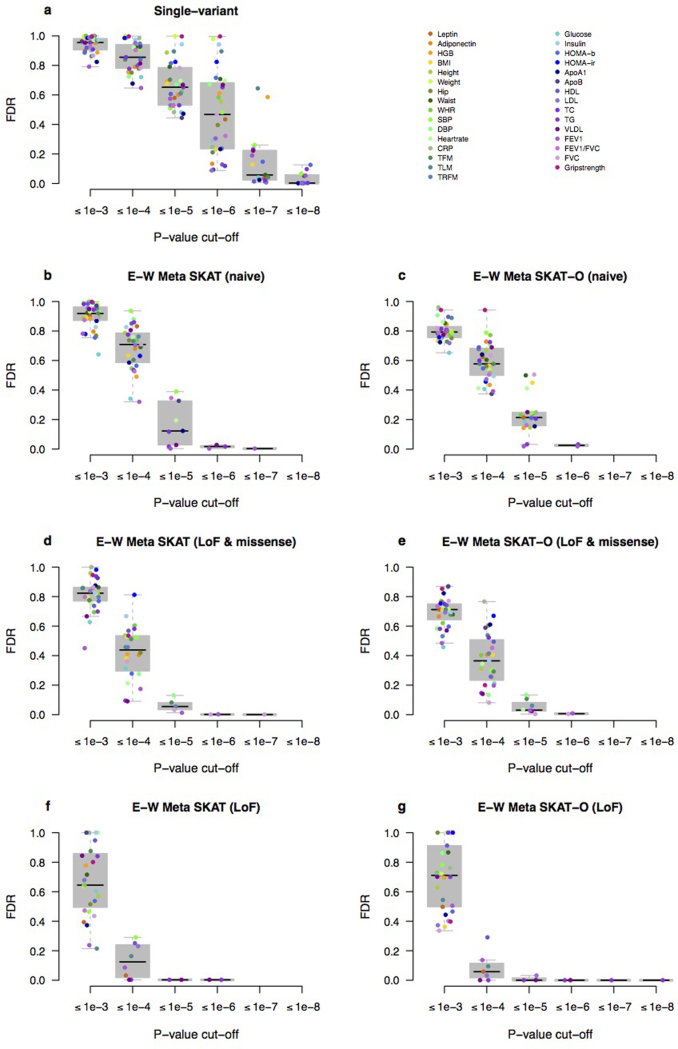
Extended Data Figure 4. UK10K-cohorts, false discovery rate (FDR).
a–g, FDR values for reporting associations at different P value cut-offs for all analyses reported in this study and the 31 core traits for single-variant analysis (a); naive exome-wide Meta SKAT (b); naive exome-wide Meta SKAT-O (c); functional exome-wide Meta SKAT (LoF and missense) (d); functional exome-wide Meta SKAT-O (LoF and missense) (e); functional exome-wide Meta SKAT (LoF) (f); functional exome-wide Meta SKAT-O (LoF) (g).
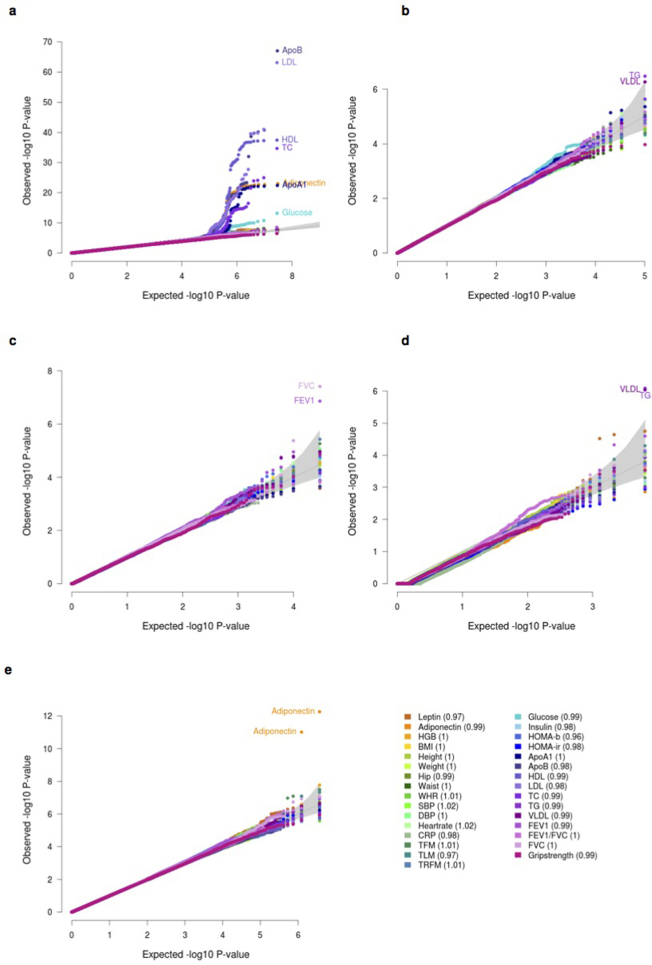
Extended Data Figure 5. UK10K-cohorts, QQ plots.
QQ plots for the association tests of the 31 core traits in the WGS data set (n = 3,621 individuals). a, Single-variant analysis (˜14 million variants with MAF ≥ 0.1%); b, naive exome-wide Meta SKAT (1,783,548 variants with MAF < 1% in 50,717 windows); c, functional exome-wide Meta SKAT (LoF and missense; 256,733 variants with MAF < 1% in 14,909 windows); d, loss-of-function functional exome-wide Meta SKAT (LoF; 9,113 variants with MAF < 1% in 3,208 windows); e, genome-wide Meta SKAT (35,858,684 variants with MAF < 1% in 1,845,982 windows).
Informing studies of low-frequency and rare variants
The UK10K-cohorts data allow an empirical evaluation of the relative importance of increasing sample size, genotyping accuracy or variant coverage for increasing power of genetic discoveries across the allele frequency spectrum. In a companion paper8 we show that common variants are exhaustively and accurately imputed using current haplotype reference panels, so increasing sample size is likely to be the single most beneficial approach for discovering novel loci driven by common variants. We further show that the UK10K haplotype reference panel, with tenfold more European samples compared to 1000GP, yields substantial improvements in imputation accuracy and coverage for low-frequency and rare variants. To obtain realistic estimates of the power benefit due to imputation with 1000GP+UK10K compared to 1000GP alone, we averaged the smallest value of Beta (the magnitude of a per-allele effect measured in standard deviations) detectable at 80% power, across variants imputable from both reference panels on chromosome 20. Fig. 4a shows sizable reductions in the magnitude of the effect sizes that can be identified at any sample size through use of the UK10K reference panel, compared to the 1000GP panel alone. For instance, for a variant of MAF = 0.3% we have equivalent power when imputing from UK10K+1000GP into a 3,621 sample as we have when using the 1000GP imputation panel alone with 10,000 samples.
Figure 4. Power for single-variant and region-based tests.
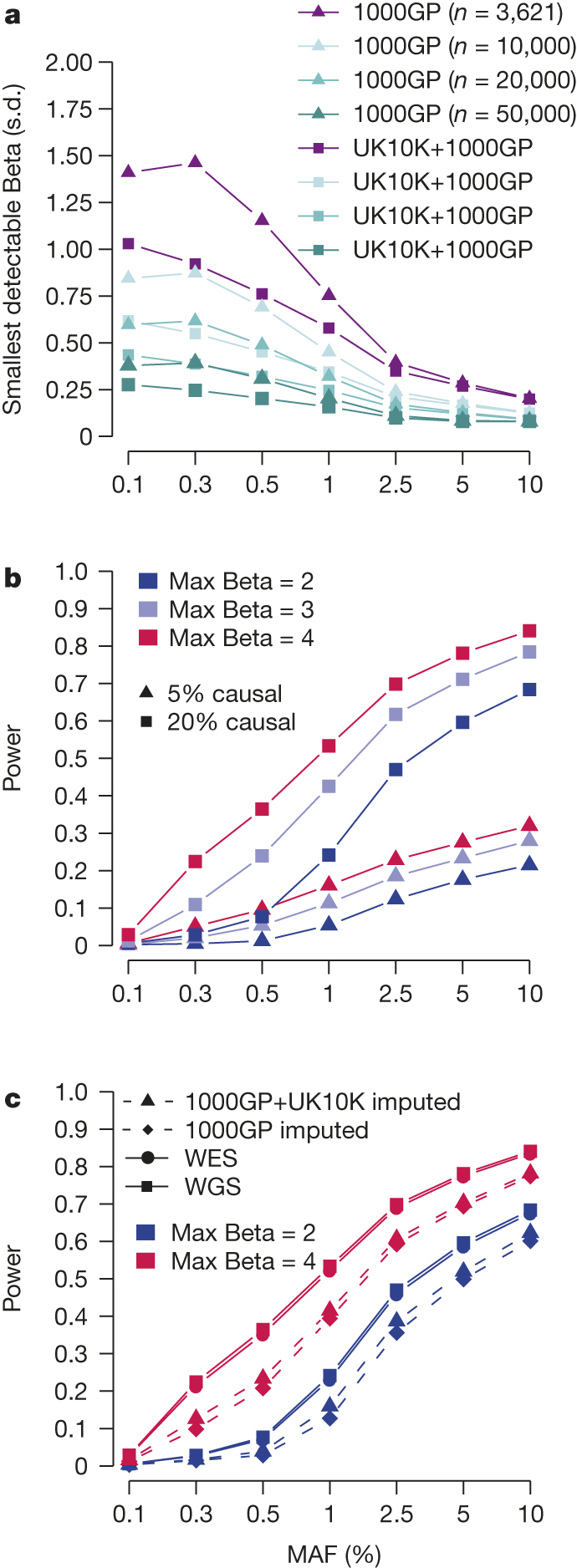
a, Strength of single-variant associations detectable at 80% power as a function of MAF and sample size. Using data from chromosome 208, we calculated the smallest value of the strength of association Beta (measured in standard deviations), that would be detectable under a linear dosage model, given the MAF and r2 of each variant imputable from both the 1000GP and the UK10K+1000GP reference panels, for various sample sizes, n. The averages of these minimum detectable beta values by MAF and sample size are shown. b, Power of region-based tests in the UK10K-cohorts sample. Evaluations assume n = 3,621, α = 6.7 × 10−8 and that the proportion of causal variants in the regions is either 5% or 20%, for maximum association (Max Beta) in a region = 2, 3, 4 s.d. c, Power of region-based tests and the impact of genotype imputation. Ten regions of 30 variants were randomly sampled from each autosome, and then genotype errors were randomly added to the data following observed r2 values between genotypes from data imputed from different sources (WGS, high depth WES, GWAS imputed against 1000GP, GWAS imputed against the combined reference panel of 1000GP and UK10K; Supplementary Table 11), and matching the MAF of each variant using the same parameters as in b, with the proportion of causal variants in the regions set to 20%.
Similar, although weaker, increases in power were seen for region-based tests of rare variants. Using the WGS autosome data from UK10K, we used simulation to introduce genotype errors into 220 randomly selected regions of 30 variants each. For each variant, errors were simulated to match the MAF and the observed r2 values between imputation and sequencing, and between whole-exome and whole-genome sequencing (Supplementary Table 11). We modified the SKAT power calculator35 to estimate power both for the true genotypes in a region and the data containing error, and averaged results across the 220 regions (see Supplementary Information). Although absolute power in Fig. 4b is generally poor, we can also see demonstrable power improvements when data are better imputed or are directly sequenced (Fig. 4c).
Tests involving non-coding rare variants may further benefit from aggregation strategies driven by biological annotation that takes into consideration the context- and trait-specific impact of non-coding variation36,37,38. Exploiting the denser sequence ascertainment of the UK10K-cohorts, we developed a robust approach to quantify fold-enrichment statistics for different categories of non-coding variants compared to null sets matched for minor allele frequency, local linkage disequilibrium and gene density (Supplementary Information). We used this approach to assess the relative contribution of low-frequency and common variants to associations with five exemplar lipid measures (the study did not have sufficient signal for rarer variants). We considered twelve different functional annotation domains, five in or near protein-coding regions and seven main chromatin segmentation states, defined using data from a cell line informative for lipid traits (HepG2; Supplementary Table 12). Low-frequency variants in exonic regions displayed the strongest degree of enrichment (25-fold, compared to fivefold for common variants, Fig. 5), compatible with the effect of purifying selection39. Importantly, however, we showed nearly as strong levels of functional enrichment at both sets of variants for several non-coding domains (∼10- to 20-fold for transcription start sites, DNase I hotspots and 3′ UTRs of genes), confirming the important contribution of non-coding low-frequency alleles to phenotypic trait variance.
Figure 5. Enrichment of single-marker associations by functional annotation in the UK10K-cohorts study.
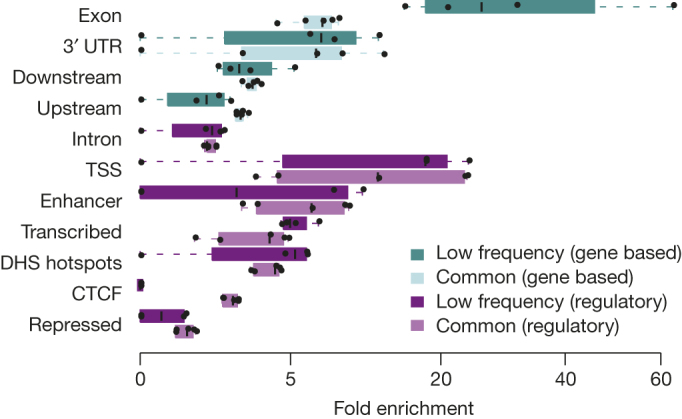
Distribution of fold enrichment statistics for single-variant associations of low-frequency (MAF 1–5%) and common (MAF ≥ 5%) SNVs in near-genic elements or selected chromatin states and DNase I hotspots (DHS). Boxplots represent distributions of fold enrichment statistics estimated across the five (out of 31 core) traits where at least 10 independent SNVs were associated with the trait at 10−7 P value (permutation test) threshold (HDL, LDL, TC, APOA1 and APOB). Chromatin state and DHS regions were inferred from ENCODE data in a liver cell line, HepG2, which is informative for lipids. Promoter and 5′ UTR are not shown, but corresponding statistics are given in Supplementary Table 12.
Findings from the exome arm of UK10K
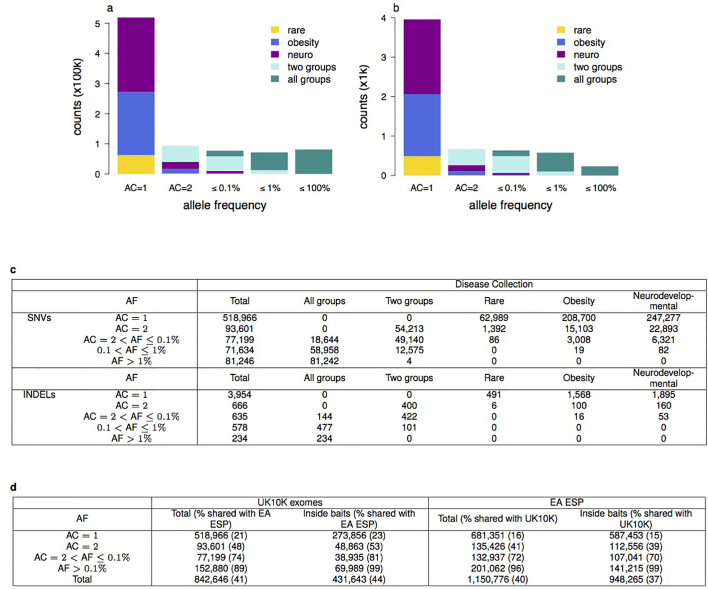
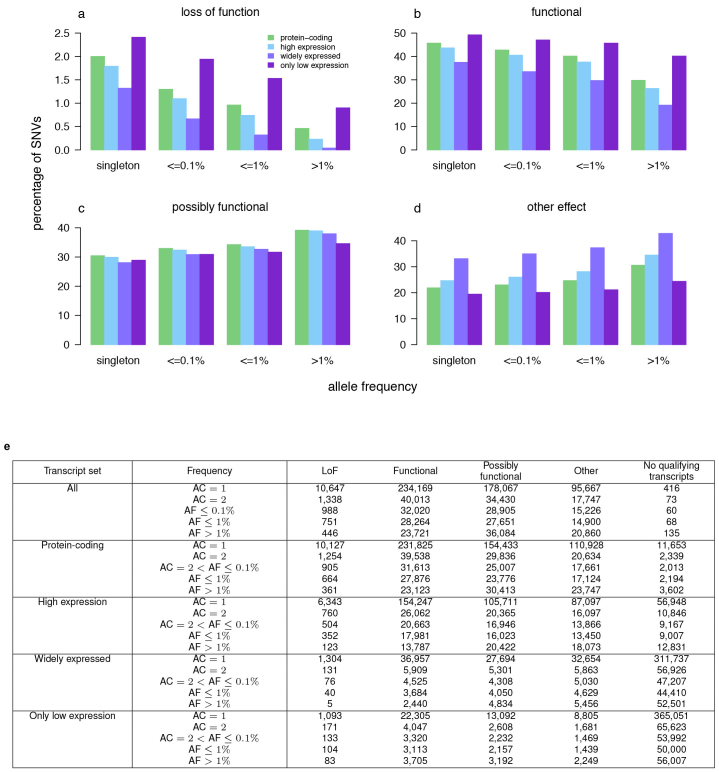
In the UK10K-exomes arm studies (see Supplementary Table 13), 5,182 individuals passed sequencing quality control with an average read depth of 80× in the bait regions. We analysed variation discovered in 3,463 disease-affected, unrelated, European-ancestry samples (Supplementary Information). We discovered 842,646 SNVs (of which 1.6% were multiallelic) and 6,067 INDELs. Both variant types were dominated by very rare variants, with more than 60% observed in only one individual. (Extended Data Fig. 6). When compared to European-American samples from the NHLBI Exome Sequencing Project (ESP)39, we found near-complete overlap at sites with MAF ≥ 1%: 99% of SNVs that are well covered by both projects and pass quality control are present in both data sets. By contrast, 72% of well-covered SNVs seen only once or twice in UK10K are present in ESP. To inform the functional annotation of these variants, we used the Illumina Body Map to determine if the frequency of LoF and functional variants changed when transcripts are selected based on their expression level (Extended Data Fig. 7). When only consequences from highly expressed transcripts and especially those highly expressed in all the Body Map tissues were considered, LoF and functional changes declined. This demonstrates that the choice of transcript can affect the consequence and this should be taken into account when annotating patient exomes.
Extended Data Figure 6. UK10K-exomes, sequence variant statistics.
Number of variants (×103) that are found in one or more of the three UK10K-exomes disease data sets, as a function of allele frequency (AF) of the non-reference allele. Variants are split into allele counts (AC) AC = 1, AC = 2 and non-overlapping AF bins for AC > 2. Allele frequency is the frequency of the alternative allele. The distributions of SNVs and INDELs across frequencies and disease collections are similar, except that there is a lower proportion of INDELs with AF > 1% compared to SNVs. a, SNVs. Multiallelic sites are included (1.6%), and non-reference alleles at the same site are treated as separate variants. b, INDELs. Counts are given in c. c, Variants are classed by whether they were found in more than one disease collection or unique to a specific group. d, Comparison of UK10K patient set with European-Americans individuals from the NHLBI Exome Sequencing project (EA ESP). The left panel shows the variants identified in UK10K and the percentage shared with EA ESP. Both the total number of variants and the number within the EA ESP bait regions (intersection of bait sets) are given. The right panel shows the variants identified in EA ESP and the percentage shared with UK10K. Both the total number of variants, and the number within the UK10K baits after removing any that failed UK10K quality control, are given. There is some overlap in the ranges of AC and AF for EA ESP variants because different numbers of individuals were included.
Extended Data Figure 7. UK10K-exomes, functional consequences.
a–d, Percentage of SNVs in each allele frequency bin that are loss of function (a), functional (b), possibly functional (c) and other (d), when consequences are restricted to given subsets of transcripts, and where the most severe consequence in qualifying transcripts is used. Values are percentages of SNVs that have transcripts of a given type. Protein-coding is transcripts with a biotype of protein coding. High expression is transcripts with FPKM (fragments per kilobase of transcript per million mapped reads) ≥1 in any tissue. Widely expressed is transcripts with FPKM ≥ 1 in 16 tissues. Only low expression is transcripts expressed at FPKM < 1 in all 16 tissues where there were no transcripts with high expression in that variant. Expression was determined from the Illumina Body Map data set. Variants mapping to protein-coding transcripts <300-bp long or with missing or low quality expression data were excluded. Frequency bins are singletons and non-overlapping allele frequency ranges for allele counts above 1. Allele frequency is the frequency of the alternative allele. Multi-allelic sites were included with alternative alleles at the same site treated as separate variants. e, Counts of single nucleotide polymorphisms in each consequence class by allele frequency and transcript subset.
The rare disease collection studied 1,000 exomes, or ∼125 from each of eight rare diseases. Thus far, 25 novel genetic causes have been identified for five of the eight diseases: ciliopathies (n = 14), neuromuscular disorders (n = 7), eye malformations (n = 2), congenital heart defects (n = 1) and intellectual disability (n = 1; Supplementary Table 14). Notably, there was marked variation in our ability to identify causal variants based on familial recurrence risk, with the primary factors appearing to be: (1) the proportion of patients with a monogenic cause, (2) the strength of prior information about the mode of inheritance (for example, dominant, recessive), and (3) the extent of prior knowledge of the relevant functional pathways. In contrast with our success identifying single-diagnostic variants in these rare diseases, our analysis of three complex diseases (obesity, autism spectrum disorder and schizophrenia) on their own did not yield replicating disease-associated loci. This is perhaps unsurprising given expected locus and allelic heterogeneity, and modest sample size40. We therefore engaged in a collaborative meta-analysis as part of the Autism Sequencing Consortium41 which identified 13 associated genes (FDR < 0.01), many of which have been previously shown to cause intellectual disability or developmental disorders. This suggests that rare variation in single genes can have a large role causing a subset of autism spectrum disorder, but these effects only become apparent when large numbers of individuals are studied.
We also used the UK10K-exomes sequence data to explore the occurrence of incidental findings. We focused on disease-specific genes identified in current guidelines for the analysis of exome/whole-genome data by the American College of Medical Genetics and Genomics (ACMG)42, and used objective criteria described in the Supplementary Information. We identified a total of 29 distinct reportable variants affecting a total of 2.3% of the UK10K cases considered in this analysis (42 out of 1,805 individuals), a number similar to previous estimates (2% estimate in adults of European ancestry43). The incidental findings were predominantly associated with cardiovascular disorders (Supplementary Table 15).
Two main challenges of reporting incidental findings from whole-exome surveys emerge. The need for clinical expertise, the difficulty of interpreting a fraction of variants, and the lack of completeness of the ClinVar database44 all highlighted the need to further consolidate knowledge from the community into freely accessible and more exhaustive databases. Furthermore, for some disorders, the frequency of carriers is likely to be too high compared to the disease frequency, despite our strict assessment criteria. This suggests that reported estimates of the penetrance of recognized variants for specific disorders are too high. Given these challenges, we suggest that, in the absence of additional evidence, scientific publications describing proposed penetrant associations for rare variants need to be complemented by accurate estimates of population frequencies.
Conclusions
In summary we have generated a high-quality whole-genome sequence data repository including 24 million novel variants from nearly 4,000 European-ancestry individuals. We showed that the UK10K haplotype reference panel greatly increases accuracy and coverage of low-frequency and rare variants compared to existing panels such as the 1000GP phase 1 panel. We carried out a large-scale empirical exploration of association testing of common, low-frequency and rare genetic variants with a large variety of biomedically important quantitative traits. For each of the different association scenarios tested, we report first examples of novel alleles associated with lipid and adiponectin traits. This provides proof-of-principle evidence on the value of the large-scale sequencing data for complex traits, while also indicating that there are few low-frequency large effect ‘quick wins’ that make substantial contributions to population trait variation and that can be discovered from sequencing studies of few thousands individuals. Our power calculations, informed by the sequence data, provide realistic estimates of the benefit of sequencing versus imputation in future association studies. Finally, rare variation tests showed limited evidence for confounding owing to population stratification at the traits investigated, likely to be due to a weakening of historical patterns of population structure in the current general UK population45.
Overall, this effort has given us both new genomic tools12 and insights into the role of low-frequency and rare variation on human complex traits, and will inform strategies for future association studies. Our exploration of non-coding variants supports the need for incorporating functional genome information in association tests of rare variants outside protein-coding regions. Improved study power through larger numbers, and a better understanding of the observed heterogeneity in allelic architecture between different loci, are likely to provide the best route forward to describe the contribution of rare variants to phenotypic variance in health and disease, and for assessing their utility in healthcare.
Supplementary information
This file contains Supplementary Text and Data, Supplementary References and Acknowledgements – see contents pages for details. (PDF 3453 kb)
This file contains Supplementary Tables 1-19. (XLSX 432 kb)
This file contains a full list of authors and affiliations for The UCLEB Consortium. (PDF 37 kb)
Acknowledgements
This study makes use of data generated by the UK10K Consortium. The Wellcome Trust provided funding for UK10K (WT091310). Additional grant support and acknowledgements can be found in the Supplementary Information.
Extended data figures and tables
Extended Data Figure 8. UK10K-cohorts, genotype and phenotype similarities within and between regions.
a, b, Dot plots show the genetic (a) and phenotypic distribution (b) of the relationships of 1,139 unrelated TwinsUK individuals by their regional place of birth. To determine the genetic relationships we used the mean number of shared alleles between two individuals within and between regions for allele counts (AC) 2 to 7, where AC is calculated from the whole data set of 3,781 samples. To determine phenotypic similarities we calculated the mean difference between the residualized phenotypes. Genetically-related individuals are more closely related within a region than between regions, while the phenotypic distance measure has similar distributions within and between regions. The mean shared alleles increase with increasing allele count, and simultaneously the within and between distributions converge. c, The five lowest P values for AC 2 to 7 obtained from Mantel tests to determine similarities between genotypes and phenotypes by region. P values were not significant after correcting for multiple testing using the FDR method49. Full trait names are given in Supplementary Table 1.
Extended Data Figure 9. UK10K-cohorts, population fine structure in the TwinsUK sample.
a, Chunk length matrix for all UK10K defined geographic regions, calculated as described in the methods. The bottom 5 regions are merged in Box 1 Figure. b, Coancestry matrix for all UK10K defined geographic regions, calculated as described in the methods. c, Chunk length matrix for all UK10K FineSTRUCTURE inferred populations, calculated as described in the methods. d, Coancestry matrix for all UK10K FineSTRUCTURE inferred populations. Details on calculation of these parameters are described in Methods. e, Pairwise coincidence matrix for the UK10K FineSTRUCTURE MCMC run, showing the fraction of the 1,000 retained iterations from the posterior in which each pair of individuals is in the same population, averaged for each pair of populations. The full posterior is extremely complex, which is indicative of a continuous admixture cline rather than discrete populations. f, Sources distribution for the FineSTRUCTURE inferred populations with the full set of inferred populations and geographic labels. Geographic labels of London, Southeast, North Midland, Southern and Eastern are merged into South and East for Box 1 Figure. FSPop labels are given to populations inferred by FineSTRUCTURE, which are merged into the Pop labels as shown in the main Box 1 Figure. g, The f2 haplotype age analysis estimates the time to the most recent common ancestor (tMRCA) between the two haplotypes underlying a given observed variant of allele count 2 in all of the TwinsUK samples. The observed IBD segment length around each f2 variant estimates the tMRCA, using an explicit model parameterized by the recombination and the mutation rates. Shown is the map of the UK with all regions used in this analysis depicted by their location, and lines colour-coding the observed median tMRCA of f2 haplotypes.
PowerPoint slides
Author Contributions
Project management: D.M., K.R.; designed individual studies and contributed data: A.A., A.Do., A.G.M., A.I., A.Ma., A.McI., A.McQ., A.Mor., A.O., A.R.F., A.T.H., A.Val., A.Var., B.H.S., B.N., C.B., C.C., C.M.v., C.W., Cl.L., D.A., D.B., D.B.S., D.Co., D.Cu., D.Ge., D.Gr., D.H., D.J.P., D.R.F., D.S.-C., D.S., D.T., E.M.v., E.St., E.Z., F.M., F.Z., G.B., G.Cl., G.D., G.G., G.L., G.Mal., G.S., G.Z., H.Gu., H.M.M., H.W., I.L., I.N.M.D., I.S.F., J.B., J.C., J.C.C., J.H., J.J., J.Keo., J.L.M., J.Lö., J.Lu., J.Mo., J.R.P., J.S.K., J.Suv., J.Wal., K.A.W., K.Ch., K.J.W., K.N., L.G., F.L.R., L.S., M.A., M.Be., M.C.O., M.Ca., M.Co., M.D.T., M.E.K., M.J.O., M.M., M.S., M.T., N.C., N.J.T., N.R., N.Sc., N.So., O.S., P.Be., P.Bo., P.G., P.Ho., P.M., P.Sc., P.W., R.A., R.B., R.K.S., R.M., Ri.S., S.Bh., S.Ci., S.Cu., S.E.H., S.G.W., S.I.S., S.O., S.R., T.D.S., T.G., T.P., T.W., V.I., V.Pa., W.M., UCLEB Consortium†; generated and/or quality controlled sequence data: A.Da., A.K.-K., C.J., Co.L., D.K.J., D.M., F.Z., G.Co., G.W., H.L., J.H., J.Li., J.Mas., J.St., J.Sun., J.T., K.Wo., M.A.Q., P.C., P.D., P.E., P.F., R.D., Ru.L., Ry.L., S.Ba., S.E., S.McC., T.C., T.K., Xi.G., Y.D.; designed new statistical or bioinformatics tools: A.C., A.H., B.H., C.M.T.G., C.X., E.Bi., E.Z., G.R.S.R., H.S., I.D., I.T., J.Mar., K.O., N.So., Ru.L., S.Me., T.D., T.H., V.I.; analysed the data and provided critical interpretation of results: A.D.-W., A.H., A.M.V., A.Moa., A.P., A.S., J.B.R., B.S., C.A., C.M.T.G., C.K.R., C.S.F., C.W., D.E., D.G.M., D.L., E.Bo., E.Se., E.W., E.Z., F.K., F.P., F.Z., G.Mar., G.R.S.R., H.Z., I.B., I.M., I.T., J.C.B., J.F., J.H., J.Kem., J.L.M., J.Mo., J.R.B.P., J.Y., K.Ca., K.P., K.S., K.Wa., K.Wo., L.Ch., L.Cr., L.P., L.Q., L.R.L., L.S., L.V.W., M.Co., M.E.H., M.F., M.G., M.Le., M.S.-A., M.v., N.J.T., N.M., N.So., O.P., P.D., P.Hy., P.M.V., P.Sy., P.V., R.C., R.C.P., R.D., R.E., R.L.R., R.T., Ri.S., S.-Y.S., S.A., S.E.H., S.G.W., S.McC., S.Me., S.P., S.S., T.G., V.I., V.Pl., Y.J., Y.M.; ethics: A.K., C.S., D.M., D.R.F., F.M., H.Gr., J.Ka., K.K., F.L.R., M.Bo., M.E.H., N.J.T., P.Bo., R.D., R.K.S., T.D.S.; designed and/or managed the study: A.P., J.B.R., Co.L., D.M., D.R.F., E.Z., G.D.-S., I.S.F., J.C.B., J.Ka., J.St., K.K., M.E.H., M.J.O., N.J.T., N.So., R.D., S.McC., T.D.S.; wrote the manuscript: A.H., J.B.R., C.M.T.G., C.X., D.L., E.Z., I.B., J.C.B., J.F., J.H., J.L.M., J.R.B.P., K.Wa., L.Cr., M.E.H., M.F., N.J.T., N.So., P.D., R.D., Ru.L., S.E.H., S.McC., S.S., V.I., V.Pl., Y.M.
Accession codes
Data deposits
Data access form is available at http://www.uk10k.org/data_access.html, raw and processed data files at https://www.ebi.ac.uk/ega/, imputation panel at https://www.ebi.ac.uk/ega/, UK10K Genome Browser at http://www.uk10k.org/dalliance.html, single-marker loci navigator at http://fathmm.biocompute.org.uk/UK10K_Browser/ and dynamic power calculator at http://fathmm.biocompute.org.uk/UK10K_Browser/Power.htm. All sequence and phenotype data were deposited to the European Genome-Phenome archive (EGA, https://www.ebi.ac.uk/ega/), with accession numbers EGAD00001000740, EGAD00001000789, EGAD00001000741, EGAD00001000790, EGAD00001000776, EGAD00001000433, EGAD00001000434, EGAD00001000435, EGAD00001000436, EGAD00001000613, EGAD00001000614, EGAD00001000437, EGAD00001000438, EGAD00001000615, EGAD00001000439, EGAD00001000440, EGAD00001000441, EGAD00001000442, EGAD00001000443, EGAD00001000430, EGAD00001000431, EGAD00001000432, EGAD00001000429, EGAD00001000413, EGAD00001000414, EGAD00001000415, EGAD00001000416, EGAD00001000417, EGAD00001000418, EGAD00001000419 and EGAD00001000420. A breakdown of studies is given in Supplementary Table 13. All study participants provided informed consent. Details of REC approvals are given in Supplementary Table 17.
Competing interests
P.F. is a member of the Scientific Advisory Board of Omicia, Inc.
Footnotes
A list of authors and affiliations appears in the Supplementary Information.
Hugh Gurling: Deceased
A list of authors and affiliations appears in the Supplementary Information.
Klaudia Walter, Josine L. Min, Jie Huang and Lucy Crooks: These authors contributed equally to this work.
Change history
9/30/2015
Four authors (A.Do., A.Mor., D.J.P. and B.H.S.) were added to the ‘Obesity group’ of the consortium, and the Author Contributions section was accordingly updated.
Contributor Information
The UK10K Consortium:
Klaudia Walter, Josine L. Min, Jie Huang, Lucy Crooks, Yasin Memari, Shane McCarthy, John R. B. Perry, ChangJiang Xu, Marta Futema, Daniel Lawson, Valentina Iotchkova, Stephan Schiffels, Audrey E. Hendricks, Petr Danecek, Rui Li, James Floyd, Louise V. Wain, Inês Barroso, Steve E. Humphries, Matthew E. Hurles, Eleftheria Zeggini, Jeffrey C. Barrett, Vincent Plagnol, J. Brent Richards, Celia M. T. Greenwood, Nicholas J. Timpson, Richard Durbin, Nicole Soranzo, Senduran Bala, Peter Clapham, Guy Coates, Tony Cox, Allan Daly, Petr Danecek, Yuanping Du, Richard Durbin, Sarah Edkins, Peter Ellis, Paul Flicek, Xiaosen Guo, Xueqin Guo, Liren Huang, David K. Jackson, Chris Joyce, Thomas Keane, Anja Kolb-Kokocinski, Cordelia Langford, Yingrui Li, Jieqin Liang, Hong Lin, Ryan Liu, John Maslen, Shane McCarthy, (co-chair), Dawn Muddyman, Michael A. Quail, Jim Stalker, (co-chair), Jianping Sun, Jing Tian, Guangbiao Wang, Jun Wang, Yu Wang, Kim Wong, Pingbo Zhang, Inês Barroso, Ewan Birney, Chris Boustred, Lu Chen, Gail Clement, Massimiliano Cocca, Petr Danecek, George Davey Smith, Ian N. M. Day, Aaron Day-Williams, Thomas Down, Ian Dunham, Richard Durbin, David M. Evans, Tom R. Gaunt, Matthias Geihs, Celia M. T. Greenwood, Deborah Hart, Audrey E. Hendricks, Bryan Howie, Jie Huang, Tim Hubbard, Pirro Hysi, Valentina Iotchkova, Yalda Jamshidi, Konrad J. Karczewski, John P. Kemp, Genevieve Lachance, Daniel Lawson, Monkol Lek, Margarida Lopes, Daniel G. MacArthur, Jonathan Marchini, Massimo Mangino, Iain Mathieson, Shane McCarthy, Yasin Memari, Sarah Metrustry, Josine L. Min, Alireza Moayyeri, Dawn Muddyman, Kate Northstone, Kalliope Panoutsopoulou, Lavinia Paternoster, John R. B. Perry, Lydia Quaye, J. Brent Richards, (co-chair), Susan Ring, Graham R. S. Ritchie, Stephan Schiffels, Hashem A. Shihab, So-Youn Shin, Kerrin S. Small, María Soler Artigas, Nicole Soranzo, (co-chair), Lorraine Southam, Timothy D. Spector, Beate St Pourcain, Gabriela Surdulescu, Ioanna Tachmazidou, Nicholas J. Timpson, (co-chair), Martin D. Tobin, Ana M. Valdes, Peter M. Visscher, Louise V. Wain, Klaudia Walter, Kirsten Ward, Scott G. Wilson, Kim Wong, Jian Yang, Eleftheria Zeggini, Feng Zhang, Hou-Feng Zheng, Richard Anney, Muhammad Ayub, Jeffrey C. Barrett, Douglas Blackwood, Patrick F. Bolton, Gerome Breen, David A. Collier, Nick Craddock, Lucy Crooks, Sarah Curran, David Curtis, Richard Durbin, Louise Gallagher, Daniel Geschwind, Hugh Gurling, Peter Holmans, Irene Lee, Jouko Lönnqvist, Shane McCarthy, Peter McGuffin, Andrew M. McIntosh, Andrew G. McKechanie, Andrew McQuillin, James Morris, Dawn Muddyman, Michael C. O'Donovan, Michael J. Owen, (co-chair), Aarno Palotie, (co-chair), Jeremy R. Parr, Tiina Paunio, Olli Pietilainen, Karola Rehnström, Sally I. Sharp, David Skuse, David St Clair, Jaana Suvisaari, James T. R. Walters, Hywel J. Williams, Inês Barroso, (co-chair), Elena Bochukova, Rebecca Bounds, Anna Dominiczak, Richard Durbin, I. Sadaf Farooqi, (co-chair), Audrey E. Hendricks, Julia Keogh, Gaëlle Marenne, Shane McCarthy, Andrew Morris, Dawn Muddyman, Stephen O'Rahilly, David J. Porteous, Blair H. Smith, Ioanna Tachmazidou, Eleanor Wheeler, Eleftheria Zeggini, Saeed Al Turki, Carl A. Anderson, Dinu Antony, Inês Barroso, Phil Beales, Jamie Bentham, Shoumo Bhattacharya, Mattia Calissano, Keren Carss, Krishna Chatterjee, Sebahattin Cirak, Catherine Cosgrove, Richard Durbin, David R. Fitzpatrick, (co-chair), James Floyd, A. Reghan Foley, Christopher S. Franklin, Marta Futema, Detelina Grozeva, Steve E. Humphries, Matthew E. Hurles, (co-chair), Shane McCarthy, Hannah M. Mitchison, Dawn Muddyman, Francesco Muntoni, Stephen O'Rahilly, Alexandros Onoufriadis, Victoria Parker, Felicity Payne, Vincent Plagnol, F. Lucy Raymond, Nicola Roberts, David B. Savage, Peter Scambler, Miriam Schmidts, Nadia Schoenmakers, Robert K. Semple, Eva Serra, Olivera Spasic-Boskovic, Elizabeth Stevens, Margriet van Kogelenberg, Parthiban Vijayarangakannan, Klaudia Walter, Kathleen A. Williamson, Crispian Wilson, Tamieka Whyte, Antonio Ciampi, Celia M. T. Greenwood, (co-chair), Audrey E. Hendricks, Rui Li, Sarah Metrustry, Karim Oualkacha, Ioanna Tachmazidou, ChangJiang Xu, Eleftheria Zeggini, (co-chair), Martin Bobrow, Patrick F. Bolton, Richard Durbin, David R. Fitzpatrick, Heather Griffin, Matthew E. Hurles, (co-chair), Jane Kaye, (co-chair), Karen Kennedy, Alastair Kent, Dawn Muddyman, Francesco Muntoni, F. Lucy Raymond, Robert K. Semple, Carol Smee, Timothy D. Spector, Nicholas J. Timpson, Ruth Charlton, Rosemary Ekong, Marta Futema, Steve E. Humphries, Farrah Khawaja, Luis R. Lopes, Nicola Migone, Stewart J. Payne, Vincent Plagnol, (chair), Rebecca C. Pollitt, Sue Povey, Cheryl K. Ridout, Rachel L. Robinson, Richard H. Scott, Adam Shaw, Petros Syrris, Rohan Taylor, Anthony M. Vandersteen, Jeffrey C. Barrett, Inês Barroso, George Davey Smith, Richard Durbin, (chair), I. Sadaf Farooqi, David R. Fitzpatrick, Matthew E. Hurles, Jane Kaye, Karen Kennedy, Cordelia Langford, Shane McCarthy, Dawn Muddyman, Michael J. Owen, Aarno Palotie, J. Brent Richards, Nicole Soranzo, Timothy D. Spector, Jim Stalker, Nicholas J. Timpson, Eleftheria Zeggini, Antoinette Amuzu, Juan Pablo Casas, John C. Chambers, Massimiliano Cocca, George Dedoussis, Giovanni Gambaro, Paolo Gasparini, Tom R. Gaunt, Jie Huang, Valentina Iotchkova, Aaron Isaacs, Jon Johnson, Marcus E. Kleber, Jaspal S. Kooner, Claudia Langenberg, Jian'an Luan, Giovanni Malerba, Winfried März, Angela Matchan, Josine L. Min, Richard Morris, Børge G. Nordestgaard, Marianne Benn, Susan Ring, Robert A. Scott, Nicole Soranzo, Lorraine Southam, Nicholas J. Timpson, Daniela Toniolo, Michela Traglia, Anne Tybjaerg-Hansen, Cornelia M. van Duijn, Elisabeth M. van Leeuwen, Anette Varbo, Peter Whincup, Gianluigi Zaza, Eleftheria Zeggini, and Weihua Zhang
References
- 1.Manolio TA. Bringing genome-wide association findings into clinical use. Nature Rev. Genet. 2013;14:549–558. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 2.Voight BF, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nature Genet. 2014;46:220–224. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature467, 1061–1073 (2010) [DOI] [PMC free article] [PubMed]
- 6.Lange LA, et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am. J. Hum. Genet. 2014;94:233–245. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nature Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng, H. et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature10.1038/nature14878 (2015) [DOI] [PMC free article] [PubMed]
- 10.Taylor PN, et al. Whole-genome sequence-based analysis of thyroid function. Nature Commun. 2015;6:5681. doi: 10.1038/ncomms6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timpson NJ, et al. A rare variant in APOC3 is associated with plasma triglyceride and VLDL levels in Europeans. Nature Commun. 2014;5:4871. doi: 10.1038/ncomms5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geihs, M. et al. An interactive genome browser of association results from the UK10K cohorts project. Bioinformatics10.1093/bioinformatics/btv491 (2015) [DOI] [PMC free article] [PubMed]
- 13.Boyd A, et al. Cohort Profile: the ‘children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moayyeri A, Hammond CJ, Hart DJ, Spector TD. The UK Adult Twin Registry (TwinsUK Resource) Twin Res. Hum. Genet. 2013;16:144–149. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler E, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nature Genet. 2013;45:513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudbjartsson DF, et al. Large-scale whole-genome sequencing of the Icelandic population. Nature Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 18.Williams FM, et al. Genes contributing to pain sensitivity in the normal population: an exome sequencing study. PLoS Genet. 2012;8:e1003095. doi: 10.1371/journal.pgen.1003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nature Genet.46, 818–825 (2014) [DOI] [PubMed]
- 20.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu C, et al. Estimating genome-wide significance for whole-genome sequencing studies. Genet. Epidemiol. 2014;38:281–290. doi: 10.1002/gepi.21797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jørgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med. 2014;371:32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 23.The TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med.371, 22–31 (2014) [DOI] [PMC free article] [PubMed]
- 24.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 25.Park JH, et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc. Natl Acad. Sci. USA. 2011;108:18026–18031. doi: 10.1073/pnas.1114759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nature Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordestgaard BG, Benn M, Schnohr P, Tybjaerg-Hansen A. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. J. Am. Med. Assoc. 2007;298:299–308. doi: 10.1001/jama.298.3.299. [DOI] [PubMed] [Google Scholar]
- 28.Whittall RA, Matheus S, Cranston T, Miller GJ, Humphries SE. The intron 14 2140+5G>A variant in the low density lipoprotein receptor gene has no effect on plasma cholesterol levels. J. Med. Genet. 2002;39:e57. doi: 10.1136/jmg.39.9.e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annu. Rev. Genet. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- 31.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu DJ, Leal SM. Estimating genetic effects and quantifying missing heritability explained by identified rare-variant associations. Am. J. Hum. Genet. 2012;91:585–596. doi: 10.1016/j.ajhg.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morisaki H, et al. CDH13 gene coding T-cadherin influences variations in plasma adiponectin levels in the Japanese population. Hum. Mutat. 2012;33:402–410. doi: 10.1002/humu.21652. [DOI] [PubMed] [Google Scholar]
- 34.Dastani Z, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature488, 57–74 (2012) [DOI] [PMC free article] [PubMed]
- 37.Adams D, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nature Biotechnol. 2012;30:224–226. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 38.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature518, 317–330 (2015) [DOI] [PMC free article] [PubMed]
- 39.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuk O, et al. Searching for missing heritability: designing rare variant association studies. Proc. Natl Acad. Sci. USA. 2014;111:E455–E464. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green RC, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaye J, et al. Managing clinically significant findings in research: the UK10K example. Eur. J. Hum. Genet. 2014;22:1100–1104. doi: 10.1038/ejhg.2013.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amendola LM, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25:305–315. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leslie S, et al. The fine-scale genetic structure of the British population. Nature. 2015;519:309–314. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nature Genet. 2012;44:243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature447, 661–678 (2007) [DOI] [PMC free article] [PubMed]
- 48.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Text and Data, Supplementary References and Acknowledgements – see contents pages for details. (PDF 3453 kb)
This file contains Supplementary Tables 1-19. (XLSX 432 kb)
This file contains a full list of authors and affiliations for The UCLEB Consortium. (PDF 37 kb)
Data Availability Statement
Data deposits
Data access form is available at http://www.uk10k.org/data_access.html, raw and processed data files at https://www.ebi.ac.uk/ega/, imputation panel at https://www.ebi.ac.uk/ega/, UK10K Genome Browser at http://www.uk10k.org/dalliance.html, single-marker loci navigator at http://fathmm.biocompute.org.uk/UK10K_Browser/ and dynamic power calculator at http://fathmm.biocompute.org.uk/UK10K_Browser/Power.htm. All sequence and phenotype data were deposited to the European Genome-Phenome archive (EGA, https://www.ebi.ac.uk/ega/), with accession numbers EGAD00001000740, EGAD00001000789, EGAD00001000741, EGAD00001000790, EGAD00001000776, EGAD00001000433, EGAD00001000434, EGAD00001000435, EGAD00001000436, EGAD00001000613, EGAD00001000614, EGAD00001000437, EGAD00001000438, EGAD00001000615, EGAD00001000439, EGAD00001000440, EGAD00001000441, EGAD00001000442, EGAD00001000443, EGAD00001000430, EGAD00001000431, EGAD00001000432, EGAD00001000429, EGAD00001000413, EGAD00001000414, EGAD00001000415, EGAD00001000416, EGAD00001000417, EGAD00001000418, EGAD00001000419 and EGAD00001000420. A breakdown of studies is given in Supplementary Table 13. All study participants provided informed consent. Details of REC approvals are given in Supplementary Table 17.