Abstract
Chikungunya virus (CHIKV) represents a pandemic threat with no approved vaccine available. Recently, we described a novel vaccination strategy based on iDNA® infectious clone designed to launch a live-attenuated CHIKV vaccine from plasmid DNA in vitro or in vivo. As a proof of concept, we prepared iDNA plasmid pCHIKV-7 encoding the full-length cDNA of the 181/25 vaccine. The DNA-launched CHIKV-7 virus was prepared and compared to the 181/25 virus. Illumina HiSeq2000 sequencing revealed that with the exception of the 3’ untranslated region, CHIKV-7 viral RNA consistently showed a lower frequency of single-nucleotide polymorphisms than the 181/25 RNA including at the E2-12 and E2-82 residues previously identified as attenuating mutations. In the CHIKV-7, frequencies of reversions at E2-12 and E2-82 were 0.064% and 0.086%, while in the 181/25, frequencies were 0.179% and 0.133%, respectively. We conclude that the DNA-launched virus has a reduced probability of reversion mutations, thereby enhancing vaccine safety.
Keywords: Chikungunya virus, Chikungunya fever, CHIKV, DNA vaccine, live attenuated vaccine, alphavirus
INTRODUCTION
Chikungunya virus (CHIKV) virus belongs to the Alphavirus genus of the Togaviridae family (Schwartz and Albert, 2010; Strauss and Strauss, 1994). CHIKV is transmitted to humans primarily by Aedes aegypti and A.albopictus mosquitoes (Arankalle et al., 2007; Couderc et al., 2009; Weaver and Reisen, 2010). The virus causes chikungunya fever, an infectious disease with a major health impact. Symptoms include arthralgia, respiratory failure, cardiovascular disease, hepatitis and central nervous system complications, especially in the elderly and children (Burt et al., 2012; Long and Heise, 2015; Queyriaux et al., 2008b). CHIKV is found nearly worldwide, with approximately 40 countries affected, mostly in warm climates in Asia, Africa and recently, in the Americas (Halstead, 2015; Rolph et al., 2015). Cases of human infections in Europe were also detected (Queyriaux et al., 2008a). Recent epidemics included outbreaks in India with an estimated 1.3 million affected people; the 2005–2006 outbreak on La Reunion islands in the Indian Ocean that caused 284 deaths; and an ongoing epidemic in the Caribbean and Latin America (Enserink, 2008; Halstead, 2015; Weaver et al., 2012). Climate change, urbanization, and global travel favor the geographical expansion of CHIKV (Dhiman et al., 2010; Pistone et al., 2009; Randolph and Rogers, 2010; Thiboutot et al., 2010; Weaver, 2013). Given the current epidemics and nearly worldwide presence of A.aegypti and A.albopictus, there is a risk of CHIKV pandemic (Petersen et al., 2010). Currently there is no approved CHIKV vaccine, in part due to the challenge of balancing vaccine safety and immunogenicity (Weaver et al., 2012). Experimental vaccines have been developed including live-attenuated vaccine strain 181/25 (TSI-GSD-218), which was tested in Phase I – II clinical trials (Edelman et al., 2000; Hoke et al., 2012) . The phase II trial enrolling 59 healthy volunteers resulted in the successful seroconversion in 98% of the volunteers, with mild transient adverse reactions in 8% of patients (Edelman et al., 2000). Recent studies revealed that attenuation of the 181/25 vaccine relies on two independently attenuating mutations in residues E2-12 and E2-82 of the envelope glycoprotein, with adverse events linked to genetic reversions (Glass, 2007; Gorchakov et al., 2012). Thus, although the 181/25 vaccine can be useful for an emergency response (Hoke et al., 2012), an improved CHIKV vaccine is needed. Previously, we described a novel iDNA® infectious clone technology, which allows launching vaccine virus in vitro or in vivo from plasmid DNA (Tretyakova et al., 2014; Tretyakova et al., 2013). For vaccination purposes, this technology combines the advantages of DNA immunization with the efficacy of a live attenuated vaccine. As a proof of concept, we prepared iDNA plasmid pCHIKV-7 encoding the full-length cDNA of the 181/25 vaccine strain (Tretyakova et al., 2014). Here we hypothesized that despite the fact that pCHIKV-7 iDNA encodes the full-length cDNA of the 181/25 vaccine strain, the genetic composition of iDNA-launched CHIKV-7 virus differs from the standard 181/25 virus due to the genetic uniformity of the infectious clone and a lower number of replication cycles needed to amplify the virus from the plasmid. To test this hypothesis, we characterized iDNA-launched CHIKV-7 virus and standard 181/25 virus in vitro. Furthermore, we performed next generation sequencing (NGS) of viral RNAs isolated from the iDNA-derived virus CHIKV-7 and from the standard 181/25 virus. Implications of these data on vaccine safety are discussed.
MATERIALS AND METHODS
Cell Lines, Plasmid and Viruses
African green monkey Vero cell lines (American Type Culture Collection, ATCC, Manassas, VA) were maintained in a humidified incubator at 37°C and 5% CO2 in aMEM supplemented with 10% fetal bovine serum (FBS) and gentamicin sulfate (10 μg/ml) (ThermoFisher Scientific (Thermo), Carlsbad, CA). Preparation of iDNA® infectious clone pCHIKV-7 (p181/25-7) was described previously (Tretyakova et al., 2014). Briefly, in pCHIKV-7, the full-length cDNA of 181/25 viral RNA was placed downstream from a CMV promoter within the modified, pcDNA3.1-based high-copy plasmid containing pBR322 origin of replication. The distance between the CMV promoter and the 5’ of cDNA was optimized to ensure correct 5’ terminus of transcribed CHIKV genomic RNA. To generate CHIKV-7 virus, Vero cells were transfected with pCHIKV-7 plasmid. Transcription of this plasmid in the transfected cells resulted in synthesis of the full-length infectious RNA and launching replication of vaccine virus CHIKV-7.
The CHIKV 181/25 live-attenuated vaccine strain TSI-GSD-218 (GenBank accession L37661) was obtained from the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) and used as a control. The TSI-GSD-218 vaccine virus was passed once in Vero cells in 75 cm2 flask to generate P1 virus. At 48 hr post infection, the virus was harvested, clarified, and frozen at −80°C.
Transfection with DNA, Infection with CHIKV and Assays In Vitro
To prepare DNA-launched virus, the pCHIKV-7 iDNA plasmid was isolated from E. coli resulting in a sterile DNA with a 95% supercoiled fraction and an A260/A280 ratio of ~1.9. In order to prepare live CHIKV-7 virus in vitro, 1 μg of iDNA plasmid pCHIKV-7 (~103 infectious centers, ICs) was transfected into Vero cells using electroporation. Previously, we used the same preparation of DNA to launch the virus in vivo for experimental vaccination of BALB/c mice (Tretyakova et al., 2014). Transfection in Vero cells was carried out essentially as described previously (Messer et al., 2012; Tretyakova et al., 2014; Tretyakova et al., 2013). Briefly, Vero cells were grown to 70–80% confluency, then harvested using TrypLE Express enzyme (Thermo) and electroporated using a square wave electroporator (ECM 830, BTX Genetronics, San Diego, CA) in 0.8 ml volume of cell suspension. Transfected cells were seeded into 75 cm2 flasks. To remove free plasmid DNA, at 5 hours post-electroporation, medium was removed and each flask containing adherent electroporated cells was rinsed 3 times with 1x PBS. This procedure was done to eliminate untransfected free plasmid DNA from cells. The removal of plasmid was monitored by PCR analysis. To prepare virus for NGS, transfection supernatants were harvested at indicated times post-transfection and the virus titer was determined by standard plaque assay in Vero cells. Aliquots of transfected cell suspension were taken for CHIKV expression tests by immunofluorescence assay (IFA), infectious center assay (ICA) and western blot using CHIKV hyperimmune mouse ascitic fluid (HMAF) VR-64 (ATCC VR-1241AF) prepared against Chikungunya strain S-27. For ICA, electroporated cells were seeded into 6-well plates, allowed to adhere for 4 h, covered with 1% low-melting agarose in complete medium (aMEM containing 10% FBS and 10 μg/ml gentamicin), and incubated for 72 h. To visualize plaques, monolayers were stained using neutral red and counted the following day. For IFA, aliquots of transfected Vero cells were seeded in 8-well chamber slides and incubated for 24 h in aMEM containing 10% FBS. IFA was carried out using mouse primary antibody VR-64 for 30 min, slides were rinsed with PBS, and incubated for 30 min with secondary FITC-conjugated antibody to mouse IgG (H+L). After a PBS rinse, slides were air-dried and covered with mounting medium containing the propidium iodide (PI) nuclear counterstain to visualize cell nuclei.
For 181/25 TSI-GSD-218 control vaccine virus production, Vero cells were infected in triplicate with 103 PFU of the TSI-GSD-218 181/25 P1 seed virus, multiplicity of infection (m.o.i) <0.1. The virus was harvested from the medium of infected cells at the indicated times post infection. To characterize growth and plaque morphology of the CHIKV-7 and 181/25 viruses in the transfected or infected cells, respectively, medium samples were taken every 12 h and quantitated in 12 well plates in duplicates by a plaque assay in Vero cell monolayers. Images of plates were digitally magnified and plaques were counted, with their diameters measured. Average and standard deviation were determined. Each experiment was done at least three times to ensure reproducibility of the results.
RNA preparation and Sequencing Using Illumina HiSeq2000
For RNA preparation, CHIKV-7 virus from the growth supernatant was harvested 50 h post transfection, then concentrated ~200-fold by ultracentrifugation using an SW28 rotor for 5 h at 100000 x g. Viral RNA was isolated by TRIzol LS extraction (Thermo). After extraction, viral RNA was treated with DpnI to remove the residual plasmid DNA, resuspended in 50 μl of ultrapure, nuclease-free water, then split into 2×20 μl aliquots and frozen at −80°C before cDNA preparation and NGS by using Illumina HiSeq2000. RNA concentration was monitored by Qubit 2.0 fluorometer using RNA quantitation protocol (Thermo). RNA isolation from the 181/25 P1 virus was performed similarly. The virus was grown by infecting Vero cells at m.o.i <0.1 with the original CHIKV 181/25 vaccine virus TSI-GSD-218. Virus was propagated and the medium supernatant was harvested at 30 h, concentrated by ultracentrifugation and viral RNA was isolated using Trizol LS.
Sequencing of viral RNA was done by using Illumina HiSeq2000 protocol (Axeq Technologies, Rockville, MD). For each RNA, a random primer cDNA library was initially prepared. The cDNA preparation was monitored for quality using the standard Illumina protocol. The resulting library was sequenced in order to obtain 1 Gb of sequencing data per sample. The cDNA library preparations were sequenced using Illumina’s HiSeq2000 method with 100 bp paired-end reads. Raw data, as well as contig assembly, alignment to reference sequence, and comparison analysis between the two samples were carried out. Single-nucleotide polymorphism (SNP) analysis was also carried out. As a reference sequence, we used sequence of TSI-GSD-218 virus, GenBank accession number L37661.
Sequencing data processing and analysis
NGS data were saved in FASTQ format, and processed further using the web-based cloud computing Galaxy software suite (https://usegalaxy.org). Briefly, the sequencing data sets of CHIKV-7 and 181/25 were uploaded to the server. The reference sequences, TSI-GSD-218 GenBank L37661 virus and the plasmid backbone, were also uploaded in FASTA format, which were then used to align the NGS sequences. Prior to alignment, the FASTQ files were converted to Sanger format using FASTQ Groomer, and subsequently aligned to the reference sequence, either TSI-GSD-218 or the plasmid vector backbone sequence. The output was in Sequence Alignment/Map (SAM) format, and subsequently converted to the compressed binary (BAM) format using SAM tools. The resulting BAM files were downloaded to a local computer for analysis using Integrative Genomics Viewer (IGV) from the Broad Institute. SNPs were analyzed using IGV and BAM file and the frequencies of each nucleotide at each specific site of interest were determined including at nsP3 opal codon and attenuating mutations E2-12 and E2-82.
RESULTS
Characterization of iDNA-derived CHIKV-7 virus
To prepare iDNA-derived CHIKV-7 vaccine virus in vitro, plasmid pCHIKV-7 was transfected in Vero cells using pCHIKV-7 plasmid by using electroporation. In the pCHIKV-7, the full-length cDNA of 181/25 RNA has been cloned downstream from the CMV major immediate-early promoter (Fig. 1a). Transcription from the CMV promoter in eukaryotic cell resulted in the synthesis of the full-length “infectious” RNA capable of launching live CHIKV-7 vaccine virus (Fig. 1a). Transfected Vero cells were observed for expression of CHIKV-7 virus by ICA, IFA and western blot, while the medium supernatant was examined for the presence of progeny CHIKV-7 virus by plaque assay. The ICA revealed formation of infectious centers (ICs), each presumably reflecting viral progeny originating from a single transfected Vero cell (Fig. 1b). Calculations of ICs from ICA showed that one μg of transfected plasmid resulted in approximately 500–1000 ICs suggesting specific infectivity <1.0 IC/ng of pCHIKV-7. This result is consistent with the previous observation that CHIKV vaccine virus could be successfully launched using transfection with 10 ng but not 1 ng of iDNA plasmid (Tretyakova et al., 2014). Expression of CHIKV-7 antigens in transfected Vero cells was confirmed by IFA using mouse HMAF (Fig. 1c). At 18 h (day 1 post-transfection), mostly single fluorescent cells were detected. At 30 h (day 2), the majority of cells were positive for CHIKV antigens, while all cells became positive at 42 h post-transfection indicating replication of live CHIKV-7 virus. As expected, expression of CHIKV antigen was observed in the cytoplasm of transfected cells by IFA. SDS-PAGE and western blot confirmed similarities in the expression of structural proteins in pCHIKV-7 -transfected cells and in 181/25-infected cells (Fig. 1d). The major protein band corresponded to the expected size of capsid protein, consistent with the previous study (Tretyakova et al., 2014). Minor bands likely represented proteolytic processing of the structural polyprotein and glycosylation variants (Metz et al., 2011). In the cell culture media from transfected Vero cells, the titer of the iDNA-derived CHIKV-7 virus reached 107–108 PFU/ml on day 3 post-transfection (Fig. 2a). Typical plaque morphology of CHIKV-7 iDNA-derived virus is shown in Fig. 2b.
Figure 1.
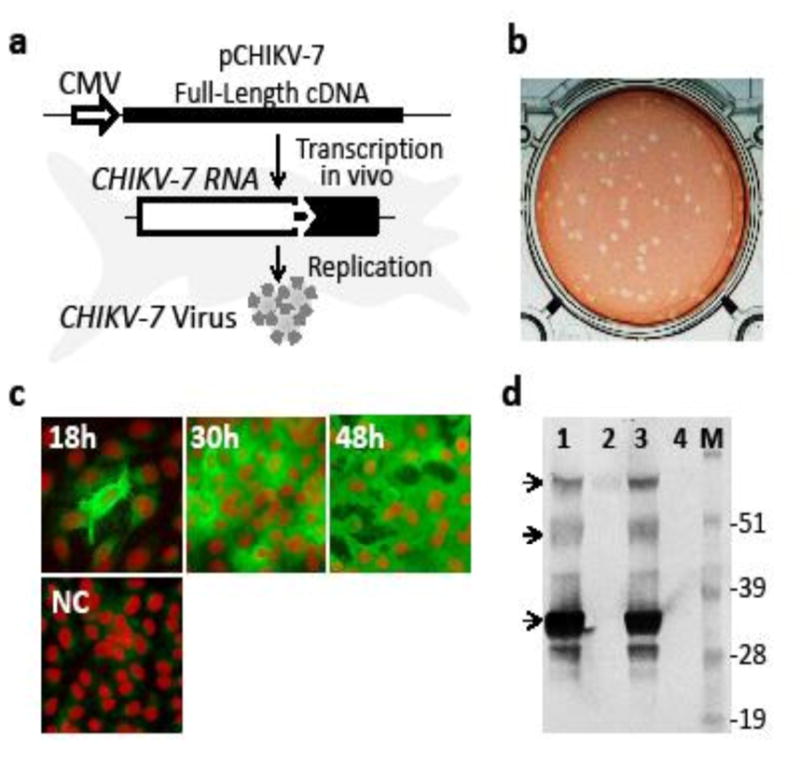
Preparation and characterization of iDNA-launched CHIKV-7 vaccine virus. (a) Schematic depiction of the CHIKV-7 virus production in Vero cells transfected with iDNA plasmid. The full-length cDNA copy of CHIKV genome (solid line) was placed downstream from the CMV promoter (open arrow) to transcribe the full-length viral RNA. Within the RNA, indicated are the non-structural genes (open box), 26S promoter (solid arrow), and structural genes (filled box). (b) Infectious center assay (ICA) of transfected Vero cells. Vero cells were transfected by electroporation using 1μg of iDNA plasmid pCHIKV-7. Aliquot of transfected cells was seeded into 6-well plates for ICA as described in Materials and Methods. (c) Expression of CHIKV antigens in iDNA-transfected Vero cells by immunofluorescence assay (IFA) at 18, 30 and 48 h post-transfection. Sham-transfected cells are shown as negative control (NC). Aliquots of transfected cells were seeded in 8-well chamber slides, fixed at indicated times in cold acetone and processed by IFA using mouse CHIKV-specific antibody VR-64, followed by FITC-conjugated secondary antibody to mouse IgG (H+L). Propidium iodide (PI) nuclear counterstain was used to visualize cell nuclei (Jones and Kniss, 1987). Red fluorescence indicates nuclear staining with the PI. (d) Expression of CHIKV antigens in Vero cells transfected with pCHIKV-7 DNA or infected with 181/25 P1 virus, by western blot. Cells were harvested at 60 h. Proteins in cells lysates were separated by SDS-PAGE. Western blot was done using mouse antibody VR-64 followed by AP-conjugated secondary antibody to mouse IgG (H+L). Lane 1, pCHIKV-7 transfected Vero cells; lane 2, medium from untreated Vero cells; lane 3, 181/25-infected cells; lane 4, untreated Vero cells; M, See BluePlus2 protein molecular weight marker (Thermo). Arrows indicate bands resulting from the CHIKV structural protein expression.
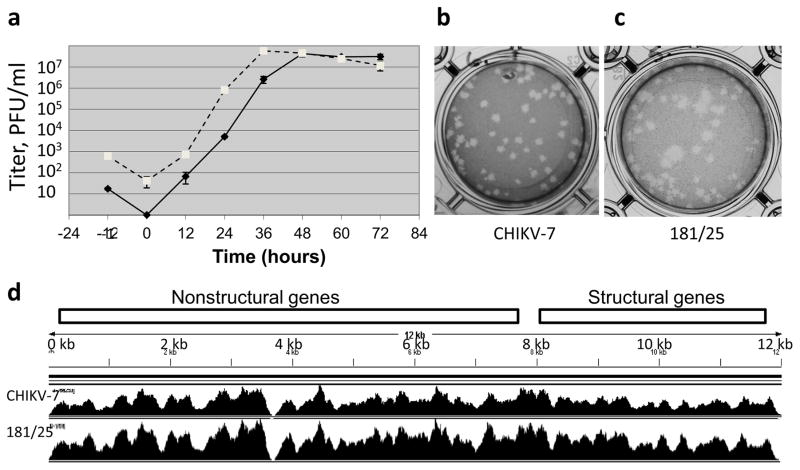
Figure 2.
Characterization of the CHIKV-7 vaccine virus and 181/25 virus by growth curve and next generation sequencing. (a) Growth curves of CHIKV-7 virus in iDNA-transfected Vero cells (solid line) and of 181/25 virus in infected Vero cells (dashed line). Vero cells were transfected with 1 μg of pCHIKV-7 iDNA plasmid or infected with 103 PFU of 181/25 virus. Aliquots of growth medium supernatants were collected and the virus was quantitated by direct plaque assay. Virus presence in the growth medium was determined by plaque assay in duplicates. Each data point represents average of three measurements. Standard deviations are indicated. Data at -1 h indicate PFU of 181/25 virus at the start of infection and IC number corresponding to 1 μg iDNA at the start of transfection, respectively. Data from 0 h show PFU/ml. (b) Plaque morphology of CHIKV-7 virus from transfected Vero cells. Average size of total 51 plaques is 4.30±0.93 mm, as measured using magnified image; with five largest plaques 1.37 times larger than the average size. (c) Plaque morphology of 181/25 P1 virus from transfected Vero cells. Average size of total 48 plaques is 5.01±1.78 mm, as measured using magnified image, with five largest plaques 1.78 times larger than the average size. (d) Comparison of sequence coverage and depth of the virus genomes. RNA was isolated from each virus by Trizol LS and sequenced using Illumina HiSeq2000. Sequencing products were assembled to a reference CHIKV 181/25 sequence. The locations of the nonstructural and structural polyproteins are indicated.
For comparison, Vero cells were infected with the CHIKV live-attenuated vaccine TSI-GSD-218 (clone 181/25) P1 virus. The titer and plaque morphology of the 181/25 virus in infected cell culture supernatants was determined by direct plaque assay. The titer reached 107–108 PFU/ml on day 2 post-transfection (Fig. 2a). By plaque assay of medium supernatants, both small and larger plaques were detected (Fig. 2c). The average diameters of plaques as measured using magnified images shown on Fig. 2b,c were 4.30±0.93 mm and 5.01±1.78 mm for CHIKV-7 and 181/25 viruses, respectively. The ratio of the average diameter of five largest plaques (1%) to the average plaque size for the same virus was 1.37 and 1.78 for CHIKV-7 and 181/25, respectively. Some of the largest plaques for the 181/25 virus were 2 times larger in diameter than average plaques indicating apparent phenotypic variation within the 181/25 virus population. The peak titer of 181/25 virus was reached one day earlier as compared to the iDNA-derived CHIKV-7 virus, in agreement with the previous report (Tretyakova et al., 2014).
NGS Analysis
Analysis of plaque assays showed variations in plaque size, especially in the 181/25 virus (Fig. 2c) suggesting the potential presence of genetic variants within the virus population. The presence of genetic variants within the virus population may contribute to a phenotypic heterogeneity of the 181/25 vaccine virus and observed rare adverse reactions (Glass, 2007).
For a detailed analysis of virus population, viruses were harvested from Vero cells on day 2 when they reached peak titers (Fig. 2a). This recapitulates viremia in vivo, which was observed on days 2–4 after virus administration (McClain et al., 1998). Viral RNAs were isolated from both the iDNA-derived CHIKV-7 and the 181/25 P1 viruses and NGS was performed using Illumina HiSeq2000 NGS. Viral RNAs were prepared using standard Trizol LS method. A total of 4,539,717 sequences were generated for CHIKV-7 RNA preparation, while for the 181/25 virus, a total of 5,897,193 sequences were generated. As a reference sequence, we used sequence of TSI-GSD-218 virus, GenBank L37661. Coverage was achieved for the entire genome (Fig. 2d) except for very 5’ and 3’ ends. From nucleotides 1-12036 of the published L37661 TSI-GSD-218 sequence, the NGS sequencing reads started at approximately nucleotide position 10 (89 and 135 sequencing reads for CHIKV-7 and CHIKV-P1, respectively) and ended around nucleotide position 12013 (140 and 94 reads for CHIKV-7 and CHIKV-P1, respectively). Overall, the coverage at the 5’ and 3’ ends were comparable for both viral samples. Relatively low coverage depth was obtained in either virus RNA within genomic positions 3694-3716. These regions were covered by approximately 1000 sequences per nucleotide in each virus. As NGS quality control, alignments were also carried out in the CHIKV-7 NGS library for any sequences with similarities to the plasmid vector backbone that might have been present and co-purified together with the viral RNA from the medium of transfected Vero cells. For this purpose, the NGS library was compared to the pcDNA3.1 backbone vector part of the pCHIKV-7 plasmid. Only 0.004% (<200 reads) were partially aligned with the vector (data not shown) suggesting that most of the NGS library was generated from the viral cDNA.
Next, we analyzed SNPs in the NGS libraries for both iDNA-derived CHIKV-7 virus and 181/25 virus. For each virus, Table 1 shows a summary of SNPs that were present at a greater than 3% frequency among all sequence reads. The stringent 3% cutoff was chosen to minimize interference resulting from the polymerase and other potential sources of errors (Laehnemann et al., 2015; Schirmer et al., 2015). Interestingly enough, many SNPs were detected in the 3’untranslated region (UTR) of CHIKV-7 virus. Ten SNPs with frequencies ranging from 3% to 17.36% were detected in the 3’-UTR of CHIKV-7, while only two SNPs were detected in the 3’-UTR of 181/25 virus, with frequencies 7.19% and 15.48% (Table 1). Lower SNP cutoff of 1% showed even more SNPs in the 3’-UTR of CHIKV-7 virus (data not shown).
Table 1.
Summary of SNPs (3% cutoff) in the CHKV-7 and 181/25 viruses.
| Subject | Position | Nucleotide | SNP | %Frequency | Coverage | Length | Region |
|---|---|---|---|---|---|---|---|
| CHIKV-7 | |||||||
|
| |||||||
| 646 | T | G | 4.83 | 18259 | 12076 | nsP1 | |
|
| |||||||
| 11407 | C | T | 4.54 | 39436 | 12076 | 3'UTR | |
| 11514 | T | C | 3.11 | 30988 | 12076 | ||
| 11559 | T | C | 3.74 | 31925 | 12076 | ||
| 11593 | T | C | 5.94 | 30838 | 12076 | ||
| 11617 | T | G | 3.47 | 27459 | 12076 | ||
| 12034 | T | C | 17.36 | 2627 | 12076 | ||
| 12035 | T | C | 3.2 | 2592 | 12076 | ||
| 12066 | A | G | 3.11 | 804 | 12076 | ||
| 12070 | A | G | 3.6 | 694 | 12076 | ||
| 12075 | A | G | 3.0 | 434 | 12076 | ||
|
| |||||||
| 181/25 | |||||||
|
| |||||||
| 646 | T | G | 5.55 | 48630 | 12076 | nsP1 | |
|
| |||||||
| 11593 | T | C | 7.19 | 71031 | 12076 | 3'UTR | |
| 12034 | T | C | 15.48 | 5736 | 12076 | ||
Furthermore, one SNP was detected at nsP1 gene position 646T G of each virus, resulting in the synonymous Gly codon change from GGT to GGG. This SNP was detected at ~4.8% frequency in the CHIKV-7 virus, while the same SNP was detected at 5.6% frequency in the 181/25 virus (Table 1).
In summary, the highest polymorphism was detected at the same positions 646 T G (nsP1), 11593 T C (3’UTR) and 12034T C (3’UTR) in both iDNA-derived CHIKV-7 and 181/25 virus suggesting similarities of both iDNA-derived CHIKV-7 and 181/25 viruses. The reasons for higher rate of SNPs in the 3’ UTR of the CHIKV-7 virus are not clear.
Low frequency SNPs at nsP3 opal codon
SNPs play a critical role in the alphavirus life cycle. The nsP3 UGA opal stop codon at nt 5645-5647 requires readthrough in order to synthesize nsP1234 polyprotein. Readthrough occurs with 5 to 20% efficiency as found by using previous in vitro translation studies (de Groot et al., 1990; Shirako and Strauss, 1994). In our NGS library for CHIKV-7, SNPs were found at nt 5645-5648 UGAC positions with frequencies from 0.02 to 0.09% (Table 2). Generally, CHIKV-7 had lower frequency of SNPs as compared to the 181/25 virus. For example, the frequency of UGA to UGG (opal Trp) was 0.062%. At the same time, in the 181/25 virus, UGA to UGG was found at a frequency 0.146% (Table 3). Other SNPs that can lead to readthrough were also found at lower frequencies in CHIKV-7 at these nucleotide positions. These data suggest that SNPs responsible for readthrough of an opal stop codon and expression of nsP1234 polyprotein occur at a lower frequency in iDNA-derived CHIKV-7 virus than in standard 181/25 virus.
Table 2.
SNPs at the nsP3 opal stop codon in the CHIKV-7 and 181/25 viruses*.
| 5645 | 5646 | 5647 | 5648 | |||||
|---|---|---|---|---|---|---|---|---|
| CHIKV-7 |
|
C | ||||||
| Count | % | Count | % | Count | % | Count | % | |
|
|
||||||||
| Total | 35465 | 100.000 | 35554 | 100.000 | 35601 | 100.000 | 36206 | 100.000 |
| A | 27 | 0.076 | 22 | 0.062 | 35521 | 99.775 | 36 | 0.099 |
| C | 7 | 0.020 | 14 | 0.039 | 25 | 0.070 | 36135 | 99.804 |
| G | 10 | 0.028 | 35492 | 99.826 | 22 | 0.062 | 11 | 0.030 |
| U | 35421 | 99.876 | 25 | 0.070 | 33 | 0.093 | 21 | 0.058 |
| N | 0 | 0.000 | 1 | 0.003 | 0 | 0.000 | 3 | 0.008 |
| 181/25
| ||||||||
| Total | 51894 | 100.000 | 51974 | 100.000 | 51972 | 100.000 | 52376 | 100.000 |
| A | 102 | 0.197 | 41 | 0.079 | 51748 | 99.569 | 62 | 0.118 |
| C | 37 | 0.071 | 30 | 0.058 | 91 | 0.175 | 52201 | 99.666 |
| G | 80 | 0.154 | 51850 | 99.761 | 76 | 0.146 | 61 | 0.116 |
| U | 51674 | 99.576 | 52 | 0.100 | 56 | 0.108 | 48 | 0.092 |
| N | 1 | 0.002 | 1 | 0.002 | 1 | 0.002 | 4 | 0.008 |
Columns show nucleotides at indicated genomic positions. Rows indicate frequencies of SNPs for each possible nucleotide. Opal codon UGAis boxed.
Table 3.
SNPs at attenuating mutations E2-12 and E2-82 in CHIKV-7 and 181/25 viruses*.
| E2-12 | 8575 | 8576 | 8577 | |||
|---|---|---|---|---|---|---|
| CHIKV-7 | A | U | A | |||
|
| ||||||
| Count | % | Count | % | Count | % | |
| Total | 32737 | 100.000 | 32820 | 100.000 | 32474 | 100.000 |
| A | 32689 | 99.853 | 45 | 0.137 | 32426 | 99.852 |
| C | 17 | 0.052 |
|
10 | 0.031 | |
| G | 16 | 0.049 | 22 | 0.067 | 31 | 0.095 |
| U | 14 | 0.043 | 32729 | 99.723 | 7 | 0.022 |
| N | 1 | 0.003 | 3 | 0.009 | 0 | 0.000 |
|
| ||||||
| 181/25 | ||||||
|
| ||||||
| Total | 46481 | 100.000 | 46351 | 100.000 | 45665 | 100.000 |
| A | 46332 | 99.679 | 242 | 0.522 | 45540 | 99.726 |
| C | 38 | 0.082 |
|
29 | 0.064 | |
| G | 77 | 0.166 | 136 | 0.293 | 75 | 0.164 |
| U | 30 | 0.065 | 45885 | 98.995 | 17 | 0.037 |
| N | 4 | 0.009 | 5 | 0.011 | 4 | 0.009 |
| E2-82 | 8785 | 8786 | 8787 | |||
|---|---|---|---|---|---|---|
| CHIKV-7 | A | G | G | |||
|
| ||||||
| Count | % | Count | % | Count | % | |
| Total | 33856 | 33703 | 33551 | |||
| A | 33789 | 99.802 | 8 | 0.024 | 5 | 0.015 |
| C | 16 | 0.047 | 7 | 0.021 | 6 | 0.018 |
| G |
|
33654 | 99.855 | 33478 | 99.782 | |
| U | 22 | 0.065 | 34 | 0.101 | 62 | 0.185 |
| N | 0 | 0.000 | 0 | 0.000 | 0 | 0.000 |
|
| ||||||
| 181/25 | ||||||
|
| ||||||
| Total | 55660 | 55573 | 56070 | |||
| A | 55494 | 99.702 | 40 | 0.072 | 29 | 0.052 |
| C | 40 | 0.072 | 23 | 0.041 | 45 | 0.080 |
| G |
|
55372 | 99.638 | 55818 | 99.551 | |
| U | 42 | 0.075 | 130 | 0.234 | 174 | 0.310 |
| N | 10 | 0.018 | 8 | 0.014 | 4 | 0.007 |
Amino acid position E2-12: the AUA codon encodes attenuated Ile, while ACA codon encodes reversion mutation to the wild-type Thr. Amino acid position E2-82: the AGG codon encodes attenuated Arg, while GGG codon encodes reversion mutation to the wild-type Gly. Columns show nucleotides at indicated genomic positions. Rows indicate frequencies of SNPs for each possible nucleotide. Reversion SNPs are boxed.
Low frequency SNPs at E2-12 and E2-82 attenuating mutations
Next, we studied low frequency SNPs at amino acid residues E2-12 and E2-82 corresponding to Ile and Arg, respectively. These amino acid residues have been identified as attenuating loci (Gorchakov et al., 2012). These residues are encoded by the translational codons ATA (8575-8677) and AGG (8785-8787), respectively. Reversion mutation Ile Thr from ATA to ACA at E2-12 depends on a single nucleotide 8576 change. Likewise, reversion mutation Arg Gly from AGG to GGG at E2-82 locus requires a single mutation (nt 8785). We have analyzed the SNP at these codon positions in CHIKV-7 and 181/25 viruses (Table 3). Frequencies of all SNPs were below 1%, with CHIKV-7virus consistently showing lower frequencies of SNPs as compared to the 181/25 virus. SNP frequencies at E2-12 (nt 8576) and E2-82 in CHIKV-7 were 0.064% and 0.086%, respectively, while corresponding SNP frequencies at E2-12 and E2-82 in 181/25 were 0.179% and 0.133%, respectively (boxed entries in Table 3). Thus, the frequencies of reversion at E2-12 and E2-82 in CHIKV-7 are 2.8 and 1.6 times lower, respectively, than in 181/25 virus. 2). In summary, it appears that iDNA-launched CHIKV-7 virus represents a more homogenous virus population, which may have an advantage for vaccine applications. In particular, lower frequencies of SNPs at E2-12 and E2-82 in CHIKV-7 virus, which represent revertants to the wild-type, virulent parent strain sequence, may play an important role in maintaining attenuated phenotype as compared to the standard 181/25 strain vaccine.
DISCUSSION
There is currently no approved vaccine or specific antiviral therapy for CHIKV. Current treatments are mostly supportive, with anti-inflammatory drugs, steroids and fluids. Experimental treatments include ribavirin, interferon-a and furin inhibitor dec-RVKR-cmk (de Lamballerie et al., 2009; Ozden et al., 2008; Rudd et al., 2012). Candidate vaccines include standard DNA-based vaccines expressing CHIKV genes (Mallilankaraman et al., 2011; Muthumani et al., 2008), formalin-inactivated virus in combination with aluminum hydroxide (alhydrogel) adjuvant (Tiwari et al., 2009), as well as live-attenuated viruses (Plante et al., 2011; Plante et al., 2015; Wang et al., 2008), virus-like particles (Akahata et al., 2010), viral vectors (Ramsauer et al., 2015; Wang et al., 2011a) and subunit vaccines (Khan et al., 2012). Elicitation of protective immunity with a single-dose vaccination is expected for an emergency vaccine, especially in resource deficient environments (Hoke et al., 2012; Levine, 2011). Live-attenuated CHIKV strain 181/25 has been confirmed to be highly immunogenic in the clinic as a single-dose experimental human vaccine (Edelman et al., 2000; Hoke et al., 2012). In phase II clinical trial, the 181/25 vaccine showed favorable safety and immunogenicity with only minor transient adverse effects detected (Edelman et al., 2000). In the absence of licensed vaccines, this vaccine can be an option for emergency vaccinations, as well as a good starting point for CHIKV vaccine development (Hoke et al., 2012). Recently, we adapted iDNA® technology (Tretyakova et al., 2013) to the 181/25 vaccine to prepare a proof-of-concept novel CHIKV vaccine. The iDNA approach resembles traditional “infectious clone” technology except it does not involve in vitro RNA transcription using a bacteriophage polymerase and in vitro transfection to produce the virus. Instead, CHIKV iDNA clone employs a CMV promoter that transcribes the full-length genomic RNA from a plasmid in the eukaryotic cells either in vitro or in vivo. The 181/25 iDNA plasmid allowed for the production of live vaccine virus in vitro (Tretyakova et al., 2014). Furthermore, a single-dose direct immunization with iDNA plasmid resulted in the successful protection of mice from CHIKV, thereby combining the advantages of DNA immunization and efficacy of live attenuated vaccine (Tretyakova et al., 2014). Thus, the CHIKV iDNA vaccine may offer certain advantages over both the traditional DNA vaccines and the live attenuated vaccines and could represent a feasible option for vaccination. DNA is easy to prepare and formulate to be stable at ambient temperatures. Therefore, an iDNA vaccine would not be a subject of cold chain limitations of traditional live vaccines such as 181/25 that may account for up to 80% of costs in warm climates (Levine, 2011). However, because iDNA is a novel approach, more research is still needed.
In this study, we initiated analysis of genetic composition of iDNA-based CHIKV-7 vaccine virus by NGS. Generally, we observed that CHIKV-7 has high genetic stability as compared to the 181/25 virus. The exception was the 3’ UTR of CHIKV-7, in which we found higher number of SNPs. Hypothetically, the higher SNP rate in the 3’UTR of CHIKV-7 could result from the fact that viral RNA derived from the iDNA clone may have different 3’ sequence than that in the 181/25 virus due to the presence of additional sequences in the plasmid such as the ribozyme; therefore, plasmid-derived RNA may undergo intracellular optimization during the first rounds of replication, which can lead to the heterogeneity of 3’ terminal sequences. The effects of the SNPs in 3‘ UTR on the CHIKV-7 virus characteristics remain to be studied.
Consistent with the previous report (Tretyakova et al., 2014), we observed a delay in virus production from iDNA as compared to the control 181/25 virus (Fig. 2a). The reason for the delay is not clear. One explanation could be the need for RNA transcription and transport from the nucleus into the cytoplasm for the production of CHIKV-7 vaccine virus. Transfected cells that receive plasmid in the cytoplasm may need to undergo a division stage of the cell cycle to allow plasmid into the nucleus for transcription. In contrast, the standard 181/25 virus introduces genomic RNA directly in the cytoplasm, where it can readily be translated and replicate. Another hypothesis is that higher genetic stability of the vaccine virus derived from the iDNA clone may contribute to the delay in virus replication. In our NGS analysis, we detected lower frequencies of SNPs in CHIKV-7 virus (except the 3’-UTR), including a lower frequency of SNPs at the site of the nsP3 opal codon. The polyprotein nsP123 is made when translation terminates at this codon, while nsP1234 is generated when readthrough of the stop codon occurs with 5 to 20% efficiency as determined by using in vitro translation reactions (de Groot et al., 1990; Shirako and Strauss, 1994; Strauss et al., 1983). Sequence information indicated that at least some alphavirus populations possess both sense and stop codons in equilibrium at this position (Lanciotti et al., 1998; Myles et al., 2006) and selection for replication in certain cell lines can result in mutations in wild-type codons at this position (Weaver et al., 1999). Mosquito cell adaptation resulted in replacement of the stop codon with arginine or cysteine (Weaver et al., 1999). Our NGS analysis indicates that the majority of CHIKV-7 and 181/25 RNAs have an opal codon and that the frequency of sense mutations are low at this locus, with CHIKV-7 showing lower level SNPs as compared to the 181/25 virus. The lower frequency of the nsP3 opal codon read-through potentially can inhibit expression of viral nsP1234 protein and contribute to the observed delay of replication of CHIKV-7 virus and improved attenuation as compared to the 181/25 virus. However, this hypothesis requires additional research out of scope of the current study. Interestingly enough, it has been previously reported that homogenous virus prepared using another technology, high fidelity RNA polymerase, has reduced viral fitness in invertebrate and vertebrate hosts (Coffey et al., 2011). In newborn mice, high fidelity CHIKV produced truncated viremias and lower organ titers. Low fidelity polymerase CHIKV mutants were also found to be attenuated (Rozen-Gagnon et al., 2014). Similarly, high fidelity variants of human enterovirus 71 exhibited an attenuated phenotype in AG129 mice (Meng and Kwang, 2014).
Higher genetic stability can also play a role in preventing reversion mutations and maintaining attenuated phenotype. Adverse reactions to the 181/25 vaccine have been linked to reversion mutations (Glass, 2007), and experimental studies in mice have recapitulated these reversions (Gorchakov et al., 2012). In general, for live attenuated vaccines such as 181/25, poliovirus, and other live vaccines it is expected that reversion mutations may be present as a small fraction of bulk vaccine preparations. It is important to maintain reversion mutations at a minimal level to ensure vaccine safety. For example, it has been well-established in international collaborative studies with serotype 3 Sabin oral poliomyelitis vaccine that vaccine lots containing revertant genomes below a critical threshold successfully pass the in vivo monkey neurovirulence test (MNVT), while vaccine lots containing more revertants fail the MNVT (Dorsam et al., 2000). The WHO established Standard Operating Procedure for poliovirus (Sabin) vaccine types 1, 2 or 3 that allows mutant analysis by PCR and restriction enzyme cleavage (MAPREC), as well as calculation of the percentage of revertants (WHO, 2012). Therefore, it is not always clear if a reversion mutation occurred during live vaccine immunization in the vaccine recipient, or reversion mutation was already present as an SNP in the formulated vaccine. The 181/25 vaccine was derived by 18 plaque-to-plaque passages in MRC-5 cell culture (Levitt et al., 1986). Plaque purification is expected to result in a high homogeneity of the virus. Subsequent passages increase probability of mutations. The NGS analysis showed relatively high homogeneity of the 181/25 virus. However, our data indicated that except the 3’ UTR, the overall level of SNPs was lower in the iDNA-derived CHIKV-7 virus than in the 181/25 virus. Both E2-12 and E2-82 attenuating mutations contained lower number of SNP reversions in the CHIKV-7 virus as compared to the 181/25 virus. Although simultaneous reversions at both E2-18 and E2-82 showed highest reactogenicity, reversion of either site resulted in a detectable reactogenicity (Gorchakov et al., 2012). Given the proximity of the E2-12 and E2-82 codons (~210 nucleotides), we attempted to determine if mutations at these positions are occurring in the same genomic reads. However, as the average read of HiSeq2000 is ~100nt (Koboldt et al., 2012; Liu et al., 2012), we were unable to identify any genomic reads that co-localized both these codons. This can be achieved in the future studies by using NGS methods that generate longer sequencing reads. In summary, data suggest that CHIKV-7 virus may have safety advantage due to lower total number of the reversions at E2-18 and E2-82 sites (Table 3). 3). This study shows that advantages of iDNA vaccine may include higher genetic homogeneity resulting in lower frequency of the nsP3 opal codon and reduced rate of virulence reversions. Potentially, these features can result in enhanced vaccine safety. However, this deserves additional studies in vivo. The DNA launched virus will presumably undergo several rounds of replication and potentially, selective pressure in vivo may favor reversion mutations, which remains to be investigated.
Thus, CHIKV iDNA represents a genetically defined molecular clone with higher viral sequence homogeneity, which may have a safety advantage of iDNA-derived CHIKV-7 virus for vaccine applications. The iDNA clone can serve as a reference source for the production of live or killed CHIKV vaccines with improved safety characteristics. Furthermore, vaccination can be done directly with iDNA in vivo (Tretyakova et al., 2014). Such direct vaccination with iDNA is expected to minimize the number of replication cycles of vaccine virus, thus further reducing the probability of reversion mutations. Finally, as a platform technology and a reverse genetics system, iDNA can also be adapted to prepare other CHIKV vaccines including live chimeric alphaviruses and other attenuated forms (Plante et al., 2011; Plante et al., 2015; Wang et al., 2011b; Wang et al., 2008) or to develop novel vaccines, as well as for studying CHIKV biology.
Research Highlights.
Chikungunya virus (CHIKV) is an emerging pandemic threat
In vivo DNA-launched attenuated CHIKV is a novel vaccine technology
DNA-launched virus was sequenced using HiSeq2000 and compared to the 181/25 virus
DNA-launched virus has lower frequency of SNPs at E2-12 and E2-82 attenuation loci
Acknowledgments
This study was supported in part by the NIH NIAD grant 1R03AI094159. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, Nabel GJ. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nature medicine. 2010;16:334–338. doi: 10.1038/nm.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, Sudeep AB, Mishra AC. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. The Journal of general virology. 2007;88:1967–1976. doi: 10.1099/vir.0.82714-0. [DOI] [PubMed] [Google Scholar]
- Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. Chikungunya: a re-emerging virus. Lancet. 2012;379:662–671. doi: 10.1016/S0140-6736(11)60281-X. [DOI] [PubMed] [Google Scholar]
- Coffey LL, Beeharry Y, Borderia AV, Blanc H, Vignuzzi M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci U S A. 2011;108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couderc T, Khandoudi N, Grandadam M, Visse C, Gangneux N, Bagot S, Prost JF, Lecuit M. Prophylaxis and therapy for Chikungunya virus infection. The Journal of infectious diseases. 2009;200:516–523. doi: 10.1086/600381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot RJ, Hardy WR, Shirako Y, Strauss JH. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J. 1990;9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lamballerie X, Ninove L, Charrel RN. Antiviral treatment of chikungunya virus infection. Infectious disorders drug targets. 2009;9:101–104. doi: 10.2174/187152609787847712. [DOI] [PubMed] [Google Scholar]
- Dhiman RC, Pahwa S, Dhillon GP, Dash AP. Climate change and threat of vector-borne diseases in India: are we prepared? Parasitology research. 2010;106:763–773. doi: 10.1007/s00436-010-1767-4. [DOI] [PubMed] [Google Scholar]
- Dorsam V, Weimer T, Schmeel A, Hein B, Enssle K, Chumakov KM, Fibi MR. Increased safety level of serotype 3 Sabin oral poliomyelitis vaccine lots by improved seed virus, and tissue culture and virus infection conditions. Vaccine. 2000;18:2435–2443. doi: 10.1016/s0264-410x(99)00531-9. [DOI] [PubMed] [Google Scholar]
- Edelman R, Tacket CO, Wasserman SS, Bodison SA, Perry JG, Mangiafico JA. Phase II safety and immunogenicity study of live chikungunya virus vaccine TSI-GSD-218. The American journal of tropical medicine and hygiene. 2000;62:681–685. doi: 10.4269/ajtmh.2000.62.681. [DOI] [PubMed] [Google Scholar]
- Enserink M. Entomology. A mosquito goes global. Science (New York, NY. 2008;320:864–866. doi: 10.1126/science.320.5878.864. [DOI] [PubMed] [Google Scholar]
- Glass PJ, Lybarger EA, Teehee ML, Parker MD. Chikungunya virus strain TSI-GSD-218-VR1, complete genome. 2007 http://www.ncbi.nlm.nih.gov/nuccore/EF452494.1.
- Gorchakov R, Wang E, Leal G, Forrester NL, Plante K, Rossi SL, Partidos CD, Adams AP, Seymour RL, Weger J, Borland EM, Sherman MB, Powers AM, Osorio JE, Weaver SC. Attenuation of Chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. Journal of virology. 2012;86:6084–6096. doi: 10.1128/JVI.06449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead SB. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerg Infect Dis. 2015;21:557–561. doi: 10.3201/eid2104.141723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke CH, Jr, Pace-Templeton J, Pittman P, Malinoski FJ, Gibbs P, Ulderich T, Mathers M, Fogtman B, Glass P, Vaughn DW. US Military contributions to the global response to pandemic chikungunya. Vaccine. 2012;30:6713–6720. doi: 10.1016/j.vaccine.2012.08.025. [DOI] [PubMed] [Google Scholar]
- Jones KH, Kniss DA. Propidium iodide as a nuclear counterstain for immunofluorescence studies on cells in culture. J Histochem Cytochem. 1987;35:123–125. doi: 10.1177/35.1.2432112. [DOI] [PubMed] [Google Scholar]
- Khan M, Dhanwani R, Rao PV, Parida M. Subunit vaccine formulations based on recombinant envelope proteins of Chikungunya virus elicit balanced Th1/Th2 response and virus-neutralizing antibodies in mice. Virus research. 2012;167:236–246. doi: 10.1016/j.virusres.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Larson DE, Chen K, Ding L, Wilson RK. Massively parallel sequencing approaches for characterization of structural variation. Methods Mol Biol. 2012;838:369–384. doi: 10.1007/978-1-61779-507-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laehnemann D, Borkhardt A, McHardy AC. Denoising DNA deep sequencing data-high-throughput sequencing errors and their correction. Brief Bioinform. 2015 doi: 10.1093/bib/bbv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti RS, Ludwig ML, Rwaguma EB, Lutwama JJ, Kram TM, Karabatsos N, Cropp BC, Miller BR. Emergence of epidemic O'nyong-nyong fever in Uganda after a 35-year absence: genetic characterization of the virus. Virology. 1998;252:258–268. doi: 10.1006/viro.1998.9437. [DOI] [PubMed] [Google Scholar]
- Levine MM. "IDEAL" vaccines for resource poor settings. Vaccine. 2011;29(Suppl 4):D116–125. doi: 10.1016/j.vaccine.2011.11.090. [DOI] [PubMed] [Google Scholar]
- Levitt NH, Ramsburg HH, Hasty SE, Repik PM, Cole FE, Jr, Lupton HW. Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine. 1986;4:157–162. doi: 10.1016/0264-410x(86)90003-4. [DOI] [PubMed] [Google Scholar]
- Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. doi: 10.1155/2012/251364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KM, Heise MT. Protective and Pathogenic Responses to Chikungunya Virus Infection. Curr Trop Med Rep. 2015;2:13–21. doi: 10.1007/s40475-015-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K, Shedlock DJ, Bao H, Kawalekar OU, Fagone P, Ramanathan AA, Ferraro B, Stabenow J, Vijayachari P, Sundaram SG, Muruganandam N, Sarangan G, Srikanth P, Khan AS, Lewis MG, Kim JJ, Sardesai NY, Muthumani K, Weiner DB. A DNA vaccine against chikungunya virus is protective in mice and induces neutralizing antibodies in mice and nonhuman primates. PLoS neglected tropical diseases. 2011;5:e928. doi: 10.1371/journal.pntd.0000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain DJ, Pittman PR, Ramsburg HH, Nelson GO, Rossi CA, Mangiafico JA, Schmaljohn AL, Malinoski FJ. Immunologic interference from sequential administration of live attenuated alphavirus vaccines. J Infect Dis. 1998;177:634–641. doi: 10.1086/514240. [DOI] [PubMed] [Google Scholar]
- Meng T, Kwang J. Attenuation of human enterovirus 71 high-replication-fidelity variants in AG129 mice. J Virol. 2014;88:5803–5815. doi: 10.1128/JVI.00289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer WB, Yount B, Hacker KE, Donaldson EF, Huynh JP, de Silva AM, Baric RS. Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS neglected tropical diseases. 2012;6:e1486. doi: 10.1371/journal.pntd.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz SW, Geertsema C, Martina BE, Andrade P, Heldens JG, van Oers MM, Goldbach RW, Vlak JM, Pijlman GP. Functional processing and secretion of Chikungunya virus E1 and E2 glycoproteins in insect cells. Virol J. 2011;8:353. doi: 10.1186/1743-422X-8-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthumani K, Lankaraman KM, Laddy DJ, Sundaram SG, Chung CW, Sako E, Wu L, Khan A, Sardesai N, Kim JJ, Vijayachari P, Weiner DB. Immunogenicity of novel consensus-based DNA vaccines against Chikungunya virus. Vaccine. 2008;26:5128–5134. doi: 10.1016/j.vaccine.2008.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles KM, Kelly CL, Ledermann JP, Powers AM. Effects of an opal termination codon preceding the nsP4 gene sequence in the O'Nyong-Nyong virus genome on Anopheles gambiae infectivity. J Virol. 2006;80:4992–4997. doi: 10.1128/JVI.80.10.4992-4997.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden S, Lucas-Hourani M, Ceccaldi PE, Basak A, Valentine M, Benjannet S, Hamelin J, Jacob Y, Mamchaoui K, Mouly V, Despres P, Gessain A, Butler-Browne G, Chretien M, Tangy F, Vidalain PO, Seidah NG. Inhibition of Chikungunya virus infection in cultured human muscle cells by furin inhibitors: impairment of the maturation of the E2 surface glycoprotein. The Journal of biological chemistry. 2008;283:21899–21908. doi: 10.1074/jbc.M802444200. [DOI] [PubMed] [Google Scholar]
- Petersen LR, Stramer SL, Powers AM. Chikungunya virus: possible impact on transfusion medicine. Transfus Med Rev. 2010;24:15–21. doi: 10.1016/j.tmrv.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Pistone T, Ezzedine K, Schuffenecker I, Receveur MC, Malvy D. An imported case of Chikungunya fever from Madagascar: use of the sentinel traveller for detecting emerging arboviral infections in tropical and European countries. Travel medicine and infectious disease. 2009;7:52–54. doi: 10.1016/j.tmaid.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Plante K, Wang E, Partidos CD, Weger J, Gorchakov R, Tsetsarkin K, Borland EM, Powers AM, Seymour R, Stinchcomb DT, Osorio JE, Frolov I, Weaver SC. Novel chikungunya vaccine candidate with an IRES-based attenuation and host range alteration mechanism. PLoS Pathog. 2011;7:e1002142. doi: 10.1371/journal.ppat.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante KS, Rossi SL, Bergren NA, Seymour RL, Weaver SC. Extended Preclinical Safety, Efficacy and Stability Testing of a Live-attenuated Chikungunya Vaccine Candidate. PLoS Negl Trop Dis. 2015;9:e0004007. doi: 10.1371/journal.pntd.0004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queyriaux B, Armengaud A, Jeannin C, Couturier E, Peloux-Petiot F. Chikungunya in Europe. Lancet. 2008a;371:723–724. doi: 10.1016/S0140-6736(08)60337-2. [DOI] [PubMed] [Google Scholar]
- Queyriaux B, Simon F, Grandadam M, Michel R, Tolou H, Boutin JP. Clinical burden of chikungunya virus infection. The Lancet infectious diseases. 2008b;8:2–3. doi: 10.1016/S1473-3099(07)70294-3. [DOI] [PubMed] [Google Scholar]
- Ramsauer K, Schwameis M, Firbas C, Mullner M, Putnak RJ, Thomas SJ, Despres P, Tauber E, Jilma B, Tangy F. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: a randomised, double-blind, placebo-controlled, active–comparator, first-in-man trial. Lancet Infect Dis. 2015;15:519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- Randolph SE, Rogers DJ. The arrival, establishment and spread of exotic diseases: patterns and predictions. Nature reviews Microbiology. 2010;8:361–371. doi: 10.1038/nrmicro2336. [DOI] [PubMed] [Google Scholar]
- Rolph MS, Foo SS, Mahalingam S. Emergent chikungunya virus and arthritis in the Americas. Lancet Infect Dis. 2015;15:1007–1008. doi: 10.1016/S1473-3099(15)00231-5. [DOI] [PubMed] [Google Scholar]
- Rozen-Gagnon K, Stapleford KA, Mongelli V, Blanc H, Failloux AB, Saleh MC, Vignuzzi M. Alphavirus mutator variants present host-specific defects and attenuation in mammalian and insect models. PLoS Pathog. 2014;10:e1003877. doi: 10.1371/journal.ppat.1003877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd PA, Wilson J, Gardner J, Larcher T, Babarit C, Le TT, Anraku I, Kumagai Y, Loo YM, Gale M, Jr, Akira S, Khromykh AA, Suhrbier A. Interferon response factors 3 and 7 protect against Chikungunya virus hemorrhagic fever and shock. Journal of virology. 2012;86:9888–9898. doi: 10.1128/JVI.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Ijaz UZ, D'Amore R, Hall N, Sloan WT, Quince C. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 2015;43:e37. doi: 10.1093/nar/gku1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nature reviews. 2010;8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- Shirako Y, Strauss JH. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss EG, Rice CM, Strauss JH. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A. 1983;80:5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiological reviews. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiboutot MM, Kannan S, Kawalekar OU, Shedlock DJ, Khan AS, Sarangan G, Srikanth P, Weiner DB, Muthumani K. Chikungunya: a potentially emerging epidemic? PLoS neglected tropical diseases. 2010;4:e623. doi: 10.1371/journal.pntd.0000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari M, Parida M, Santhosh SR, Khan M, Dash PK, Rao PV. Assessment of immunogenic potential of Vero adapted formalin inactivated vaccine derived from novel ECSA genotype of Chikungunya virus. Vaccine. 2009;27:2513–2522. doi: 10.1016/j.vaccine.2009.02.062. [DOI] [PubMed] [Google Scholar]
- Tretyakova I, Hearn J, Wang E, Weaver S, Pushko P. DNA vaccine initiates replication of live attenuated chikungunya virus in vitro and elicits protective immune response in mice. J Infect Dis. 2014;209:1882–1890. doi: 10.1093/infdis/jiu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova I, Lukashevich IS, Glass P, Wang E, Weaver S, Pushko P. Novel vaccine against Venezuelan equine encephalitis combines advantages of DNA immunization and a live attenuated vaccine. Vaccine. 2013;31:1019–1025. doi: 10.1016/j.vaccine.2012.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Suhrbier A, Penn-Nicholson A, Woraratanadharm J, Gardner J, Luo M, Le TT, Anraku I, Sakalian M, Einfeld D, Dong JY. A complex adenovirus vaccine against chikungunya virus provides complete protection against viraemia and arthritis. Vaccine. 2011a;29:2803–2809. doi: 10.1016/j.vaccine.2011.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Kim DY, Weaver SC, Frolov I. Chimeric Chikungunya viruses are nonpathogenic in highly sensitive mouse models but efficiently induce a protective immune response. Journal of virology. 2011b;85:9249–9252. doi: 10.1128/JVI.00844-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Volkova E, Adams AP, Forrester N, Xiao SY, Frolov I, Weaver SC. Chimeric alphavirus vaccine candidates for chikungunya. Vaccine. 2008;26:5030–5039. doi: 10.1016/j.vaccine.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC. Urbanization and geographic expansion of zoonotic arboviral diseases: mechanisms and potential strategies for prevention. Trends Microbiol. 2013;21:360–363. doi: 10.1016/j.tim.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Brault AC, Kang W, Holland JJ. Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J Virol. 1999;73:4316–4326. doi: 10.1128/jvi.73.5.4316-4326.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Osorio JE, Livengood JA, Chen R, Stinchcomb DT. Chikungunya virus and prospects for a vaccine. Expert review of vaccines. 2012;11:1087–1101. doi: 10.1586/erv.12.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral research. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Mutant analysis by PCR and restriction enzyme cleavage (MAPREC) for oral poliovirus (Sabin) vaccine types 1, 2 or 3 Standard Operating Procedure. 2012 [Google Scholar]