Abstract
Non-syndromic mitral valve prolapse (MVP) is a common degenerative cardiac valvulopathy of unknown aetiology that predisposes to mitral regurgitation, heart failure and sudden death1. Previous family and pathophysiological studies suggest a complex pattern of inheritance2–5. We performed a meta-analysis of two genome-wide association studies in 1,442 cases and 2,439 controls. We identified and replicated in 1,422 cases and 6,779 controls six loci and provide functional evidence for candidate genes. We highlight LMCD1 encoding a transcription factor6, for which morpholino knockdown in zebrafish results in atrioventricular (AV) valve regurgitation. A similar zebrafish phenotype was obtained for tensin1 (TNS1), a focal adhesion protein involved in cytoskeleton organization. We also show the expression of tensin1 during valve morphogenesis and describe enlarged posterior mitral leaflets in Tns1−/− mice. This study identifies the first risk loci for MVP and suggests new mechanisms involved in mitral valve regurgitation, the most common indication for mitral valve repair7.
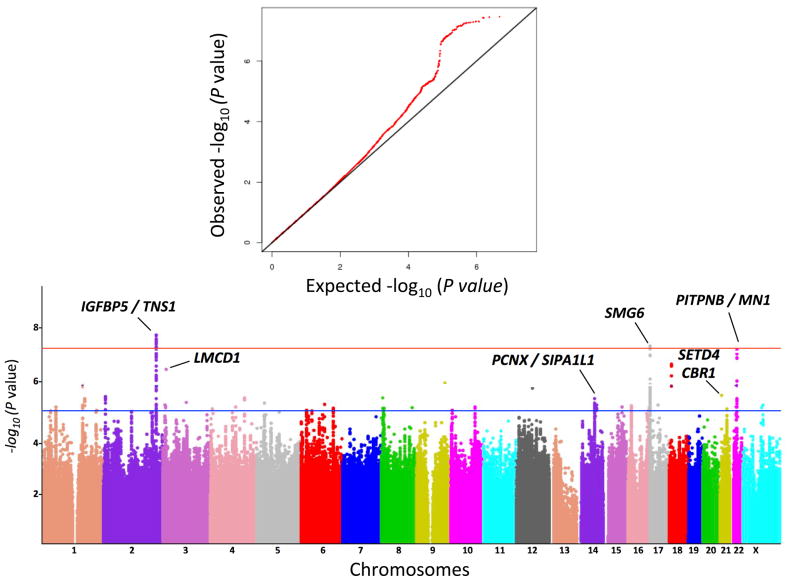
The prevalence of non-syndromic MVP has been estimated as 2.4% in the general population8. Family aggregation9,10, presence in rare connective tissue syndromes11 as well as the identification of four linked loci2–5 indicate genetic heterogeneity for MVP. Additional factors such as age- and sex-dependent penetrance, with possible association with myocardial structural and functional abnormalities, suggest additional genetic complexity12. We conducted an initial discovery meta-analysis on two independent French genome-wide association studies (GWAS) including 1,412 MVP cases and 2,439 controls (Supplementary Table 1), all of European ancestry, for ~4.8 million genotyped or imputed common (MAF > 0.1) single nucleotide polymorphisms (SNPs) (Supplementary Figure 1). Three loci showed genome-wide (GW) significant associations with MVP (P < 5×10−8) (Table 1). The strongest association (rs12465515; OR=1.33, P=1.08×10−8) was observed on Chr2q35 in a ~424 Kb gene-desert region where the nearest genes are TNP1, IGFBP5 and IGFBP2 (upstream) and DIRC3 and TNS1 (downstream) (Table 1). The two other GW-significant loci were at Chr17p13 (lead SNP rs216205, OR=1.35, P=3.02 × 10−8) in an intron of SMG6 and at Chr22q12 near MN1 and PITPNB (rs11705555 OR=1.34, P=4.47 × 10−8) (Table 1). We followed-up 23 loci with evidence of a suggestive association (P<1×10−5) in a first replication panel that included European Americans and European Spanish cases and controls (Set 1 and Set 2, Supplementary Figure 1).
Table 1. Genome-wide significant associations of single nucleotide polymorphisms with mitral valve prolapse.
OR: odds ratio; 95%CI: 95% confidence interval; CC: case control. Locus designed for nearest or the best candidate gene. RA: risk allele. Alleles are indexed to the forward strand of NCBI Build 37. P-values reported are two-sided and based on an inverse-variance weighted meta-analysis model (fixed effects). P-value for heterogeneity corresponds to Cochran Q statistics.
|
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery GWAS 1,442 cases vs. 2,439 controls |
Follow-up 1,422 cases vs. 6,779 controls |
Combined 2,864 Cases vs. 9,218 Controls |
||||||||||||
|
|
||||||||||||||
| MVP-Fr. vs SU.VI.MAX (953/1,566) |
MVP-N vs. D.E.S.I.R 1 (489/873) |
Discovery meta- analysis |
MGH+FHS cases vs. FHS (699/5,575) |
CNIC CC Study (171/282) |
Canada CC Study (102/102) |
Surgery vs.DESIR2 (450/820) |
Follow-up meta- analysis |
|||||||
|
|
||||||||||||||
| Chr | SNP | Locus | RA | Freq. | OR (95%CI) |
OR (95%CI) |
OR (95%CI) |
OR (95%CI) |
OR (95%CI) |
OR (95%CI) |
OR (95%CI) |
OR (95%CI) |
OR (95%CI) |
Hetero- geneity |
| P-value | P-value | P-value | P-value | P-value | P-value | P-value | P-value | P-value | P-value | |||||
| 2 | rs12465515 | IGFBP5/TNS1 | C | 0.34 | 1.39 (1.23–1.56) | 1.22 (1.03–1.45) | 1.33 (1.2–1.47) | 1.28 (1.14–1.47) | 1.11 (0.84–1.47) | 0.87 (0.57–1.33) | 1.12 (0.95–1.33) | 1.19 (1.09–1.32) | 1.25 (1.18–1.33) | |
| 8.0 × 10−8 | 0.019 | 1.2 × 10−8 | 1.2 × 10−4 | 0.451 | 0.521 | 0.158 | 1.6 × 10−4 | 3.1 × 10−11 | 0.145 | |||||
| 3 | rs171408 | LMCD1 | G | 0.22 | 1.41 (1.22–1.61) | 1.14 (0.93–1.39) | 1.3 (1.16–1.47) | 1.19 (1.02–1.37) | 1.64 (1.19–2.27) | 1.18 (0.76–1.82) | 1.52 (1.25–1.85) | 1.32 (1.19–1.47) | 1.32 (1.22–1.43) | |
| 3.0× 10−6 | 0.188 | 5.4 × 10−6 | 0.023 | 0.002 | 0.464 | 3.6 × 10−5 | 3.7 × 10−7 | 1.3 × 10−11 | 0.108 | |||||
| 14 | rs17767392 | PCNX/SIPA1L1 | T | 0.25 | 1.33 (1.16–1.52) | 1.12 (0.93–1.35) | 1.26 (1.13–1.4) | 1.19 (1.04–1.37) | 1.04 (0.76–1.41) | 1.2 (0.77–1.89) | 1.32 (1.09–1.56) | 1.20 (1.10–1.33) | 1.23 (1.15–1.32) | |
| 2.0 × 10−5 | 0.224 | 3.1 × 10−5 | 0.009 | 0.814 | 0.424 | 0.004 | 1.7 × 10−4 | 2.3 × 10−8 | 0.522 | |||||
| 17 | rs216205 | SMG6 | T | 0.74 | 1.41 (1.24–1.61) | 1.25 (1.04–1.5) | 1.35 (1.22–1.5) | 1.15 (1.00–1.33) | 1.06 (0.79–1.43) | 1.22 (0.80–1.86) | 1.15 (0.95–1.38) | 1.15 (1.03–1.27) | 1.24 (1.15–1.33) | |
| 3.0 × 10−7 | 0.018 | 3.0 × 10−8 | 0.044 | 0.697 | 0.364 | 0.141 | 0.009 | 1.5 × 10−8 | 0.273 | |||||
| 21 | rs62229266 | SETD4/CBR1 | T | 0.36 | 1.19 (1.05–1.35) | 1.37 (1.16–1.64) | 1.25 (1.13–1.38) | 1.35 (1.18–1.54) | 1.15 (0.88–1.52) | 0.86 (0.57–1.30) | 1.04 (0.88–1.23) | 1.19 (1.08–1.3) | 1.22 (1.14–1.30) | |
| 0.005 | 1.7 × 10−4 | 8.2 × 10−6 | 2.3 × 10−5 | 0.303 | 0.469 | 0.605 | 2.6 × 10−4 | 1.2 × 10−8 | 0.066 | |||||
| 22 | rs11705555 | PITPNB/MN1 | C | 0.26 | 1.32 (1.16–1.49) | 1.37 (1.15–1.64) | 1.34 (1.21–1.49) | 1.16 (1.01–1.35) | 1.09 (0.81–1.45) | 0.91 (0.58–1.41) | 1.19 (0.99–1.41) | 1.15 (1.04–1.27) | 1.23 (1.15–1.33) | |
| 2.8 × 10−5 | 4.0 × 10−4 | 4.5 × 10−8 | 0.032 | 0.594 | 0.657 | 0.059 | 0.007 | 1.4 × 10−8 | 0.313 | |||||
We genotyped or imputed a total of 47 SNPs (23 loci). An intermediate meta-analysis including the discovery and the follow-up Sets 1 and 2 (Ncases= 2,312 and Ncontrols=8,296) identified a subset of 24 SNPs (15 loci) with significant associations with MVP (P<0.01), which were genotyped or imputed in two additional case-control studies from Canada and France (Sets 3 and Set 4, Supplementary Figure 1). In the global meta-analysis that included 2,864 cases and 9,218 controls, three additional loci associate with MVP at the genomic level (Table 1, Supplementary Table 2). The overall strongest association was observed on Chr3p13 for rs171408 that maps in an intron of LMCD1 (OR=1.32, P =1.29 × 10−11, Table 1). Two additional signals were identified on Chr21q22 near CBR1 and SETD4 (rs62229266, OR=1.22, P=1.18×10−8) and on Chr14q24 near SIPA1L1 and PCNX (rs17767392, OR=1.23, P =2.27×10−8) (Table 1). We also confirmed the three GW-significant signals identified in the discovery samples with lead SNPs rs12465515 near IGFB55 and TNS1 (OR= 1.25, P =3.11×10−11), rs11705555 near PITPNB and MN1 (OR=1.23, P=1.39×10−8) and rs216205 in SMG6 (OR=1.24, P=1.46×10−8). Overall, we observed consistency in the direction of effects as well as nominal significant association in the follow-up meta-analysis and did not detect significant heterogeneity (P>0.05) among case control studies (Table 1).
Many patients in the general population with MVP show few clinical symptoms, if any1. Nonetheless, a substantial subset of patients are at risk of heart failure and cardiac death, and MVP is the most common cause of isolated mitral regurgitation requiring surgical repair7. To investigate if the confirmed MVP risks alleles could be more prevalent among the more severely affected patients who required valve repair or replacement, we analysed the 1,680 French patients who underwent surgical intervention and compared them to 3,259 French controls (Supplementary Table 3). We did not find a stronger effect of any MVP risk alleles, except a slight increase in the frequency of the risk allele of rs11705555 at the PITPNB/MN1 locus (OR=1.31, 95%CI (1.19–1.44), P=1.88 × 10−8). Overall, our findings support that MVP is under significant genetic control with susceptibility loci of relatively homogeneous effect sizes (OR from 1.22 to 1.33).
MVP-associated loci implicate four intergenic (IGFBP5/TNS1, SETD4/CBR1, PITBNB/MN1, and PCNX/SIPA1L1) and two intronic (LMCD1 and SMG6) regions. From an initial list of 53 genes (±500Kb to ±1Mb of the lead SNP), we identified candidate genes at each locus based on proximity to sentinel SNP, expression level in the heart, presence of eQTL signal in publically available databases (GTEX), proximity to previously identified GWAS signals for cardiovascular traits and a biological link with mitral valve or general cardiac development (Supplementary Table 5 and Supplementary Methods for full details of gene prioritization strategy per locus).
We then investigated the expression pattern of candidate genes during valve development in mouse embryos by immunohistochemistry (IHC) at three time points that represent: i) completion of endothelial-to-mesenchymal transformation (EMT; E13.5), ii) valve sculpting and elongation (E17.5) and iii) achievement of the mature adult form (9 months old). Functional antibodies were only available for Tns1 and Igfbp5 on Chr2 and Pitpnb on Chr22. We also prioritized candidate genes in the MVP risk loci using morpholino knockdown (KD) based on the presence of clear zebrafish orthologs which, after filtering (Supplementary note), limited our analysis to eight genes at three loci: igfbp2a, igfbp2b, igfbp5a, igfbp5b, and tns1 located at the chr2q35 locus; lmcd1 at the chr3p13 locus and smg6 and sgsm2 at the chr17p13 locus.
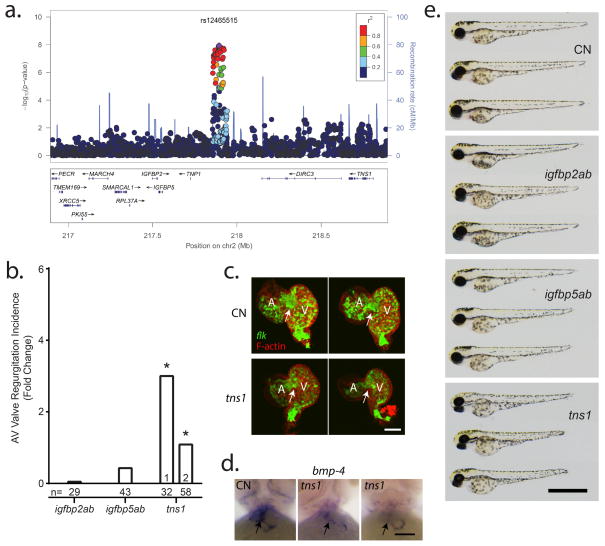
On Chr2q35, rs12465515 lies within a large intergenic region with TNS1 and IGFBP5 identified as the best two candidate genes (Supplementary Table 4). TNS1 maps 750 kb downstream from the association signal at the chr2q35 locus (Figure 2A). Tensin1, coded by TNS1, localizes to focal adhesions and interacts with actin, as does Filamin A, whose genetic variants can cause a rare X-linked form of MVP13. Tensin1 interacts with cytoplasmic tails of integrins to anchor stress fibers and plays an important role in metastatic capacities of cancer cells 14. On the other hand, IGFBP5, encoding insulin-like growth factor-binding protein 5 (IGFBP5), is known to modulate muscle differentiation and mediate high glucose-induced pro-fibrotic effects in cardiac fibroblasts15. IGFBP5 was also demonstrated to modulate migration and adhesion of cancer cells and could potentially be at play in valve development and valvular interstitial cell integrity.
Figure 2. Cardiac regurgitation in zebrafish morpholino knockdown for candidate genes on Chr2q35.
a) Genomic context of the association signal observed in the GWAS meta-analysis. The regional association plot was generated using locus zoom and displays surrounding genes, with TNS1, IGFBP2 and IGFBP5 identified as best potential candidates at this locus. b) Mitral regurgitation observed at 72 hours post fertilization (hpf) in zebrafish embryos after morpholino mediated knockdown. All results are presented as fold change compared to clutchmate controls. n=number of biological replicates per morpholino. (*) indicates p<0.05. c) 2-dimensional projections of z-series image stacks taken on excised control (CN) and tns1 knockdown zebrafish hearts. Green denotes EGFP expression, a marker of endothelium under the control of the flk promoter. Red staining indicates the distribution of F-actin, which is highly expressed in the functional myocardium. Scale bar represents 50μm. d) anti-bmp-4 probe labels the valve and surrounding myocardium in CN and tns1 knockdown embryos. Scale bar represents 50μm. e) Brightfield micrographs displaying gross morphology of 72hpf zebrafish morphants. Scale bar represents 1mm. Body axis length of morpholino-injected fish is slightly reduced compared to wild-types.
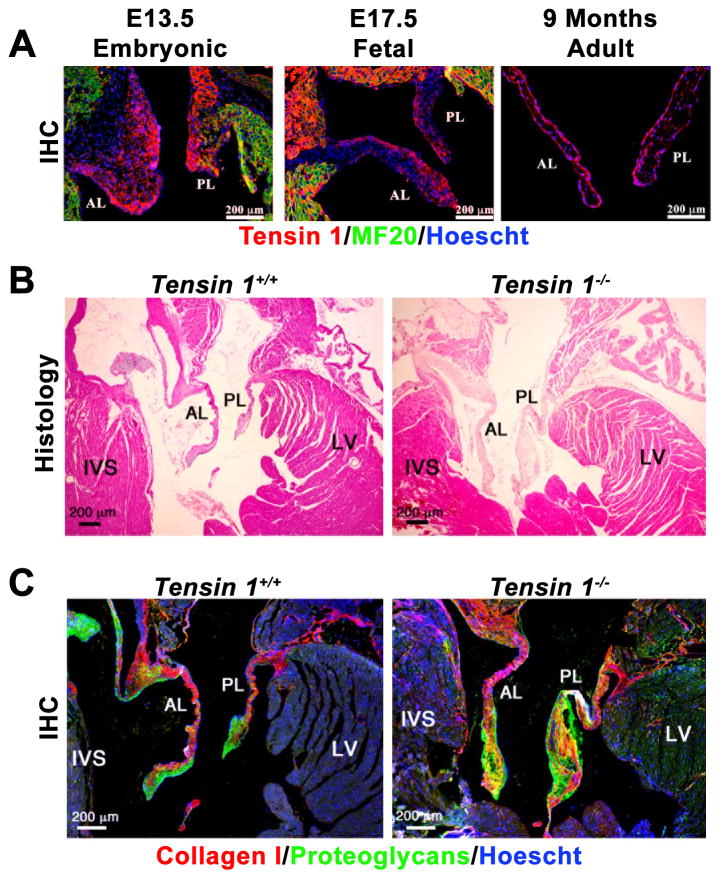
Only faint nonspecific staining was observed for Igfbp5 in valves of developing and adult mice (Supplementary Figure 2). In contrast, murine IHC data showed a sustained expression for Tensin1 during valve morphogenesis, being stronger along the atrialis aspect of the forming leaflet (Figure 3A). We also found that Tensin1 expression is maintained during adulthood and localized in the endothelial and valvular interstitial cells (Figure 3A). Hematoxylin and eosin (H&E) histological staining in 9-month old Tensin1−/− mice showed enlarged posterior mitral leaflets compared to wild-type littermates (Figure 3B). In addition, valves from tns1−/− mouse showed evidence of myxomatous degeneration, indicated by increased proteoglycan content and loss of normal matrix stratification as indicate the accumulation of proteoglycan in the valves (Figure 3C). Preliminary echographic exploration of Tns1−/− mice (n=2) showed slight leaflet displacement (0.4 mm) compared to wildtype (0.1 mm) consistent with larger leaflets but no mitral regurgitation (Supplementary Figure 3), indicating subtle anomalies of the mitral valve that deserve future confirmation. In zebrafish experiments, a significant increase in AV regurgitation incidence was observed in tns1 knockdown for both morpholinos (3- and 1.1-fold increase, respectively; P=0.02 and P=0.01) (Supplementary Video 2) but not in simultaneous knockdown of both igfbp2 and igfbp5 isoforms (Figure 2C). In situ hybridization identified high tns1 expression throughout the developing heart, and knockdown diminished the aggregation of endothelial cells at the developing valve (Supplementary Figure 4). Further, although notch1b expression remained normally localized to the developing valve, the distribution of the valve development marker bmp4 was highly disorganized (Supplementary Figure 5). Together, these results support Tensin 1 as the best candidate at the Chr2q35 locus for MVP pathogenesis.
Figure 3. Murine Tensin 1 expression during developing valves and knockout phenotype at 9 months.
A) Tensin1 expression in the mouse developing heart. IHC was performed for Tensin1 (red) at E13.5 (complete epithelial mesenchymal transformation), E17.5 (valve sculpting and elongation) and 9 months of age. MF20 (green) labels myocytes, Hoescht (Blue) labels nuclei. (B) Tensin1 knockout mice exhibit enlarged mitral leaflets. Hematoxylin and Eosin (H&E) histological staining was performed on Wild-type (Tensin+/+) and Tensin knockout (Tensin1−/−) mice. Scale bars are denoted. (C) Tensin1 knockout mice exhibit myxomatous mitral leaflets. Immunohistochemistry (IHC) for collagen (red), proteoglycans (green) show failure of normal matrix stratification and expansion of proteoglycan expression in the tensin1−/− mitral leaflets indicative of a myxomatous phenotype. AL= Anterior Leaflet, PL= Posterior Leaflet, LV=Left Ventricle, IVS=interventricular septum. Scale bars are denoted.
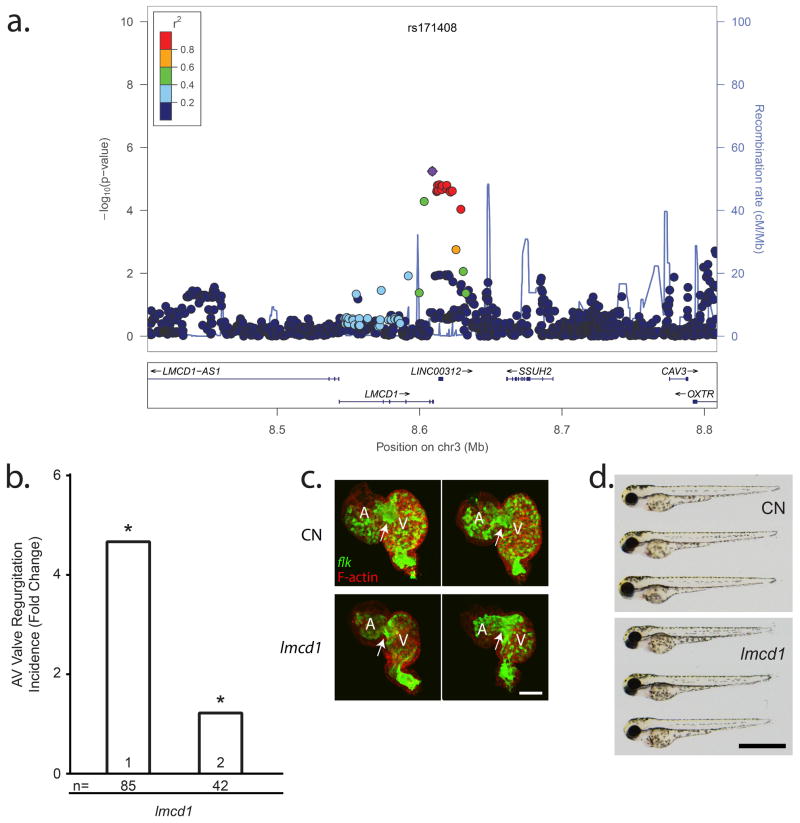
The association signal at the Chr3p13 is intronic to LMCD1. Also named Dyxin, LMCD1 is a member of the LIM domain family of zinc finger proteins that act as co-regulators of transcription. It is highly expressed in mouse cardiac tissue and was demonstrated to be a direct repressor of GATA6, an important regulator of cardiac development6. Somatic mutations in LMCD1 were described as potential oncogenic events in hepatocellular carcinoma metastasis by promoting cell migration16. The zebrafish knockdown of lmcd1 results in significantly increased AV regurgitation for both morpholinos (4.7 and 1.2 fold increase in AV regurgitation; P=0.001 and P=0.009) (Figure 4C, Supplementary Video 3). In addition, morphological analysis of the developing myocardium revealed lmcd1 morphants exhibited a moderate reduction in cardiac looping (Supplementary Video 3). However, although lmcd1 is expressed throughout the heart, after lmcd1 knockdown, expression patterns of valve development markers notch1b and bmp4 displayed no abnormalities (Supplementary Figure 5) and no mis-localization of endothelial cell aggregation was observed (Supplementary Figure 4). Further investigation will be required to determine if the AV regurgitation is due to a primary valve defect or a more general defect in cardiac development. Previous in vitro and in vivo studies showed that Lmcd1/Dyxin augment calcineurin17. Calcineurin signalling is required for AV endocardium EMT and subsequent valve morphogenesis in zebrafish18. rs355134, a highly correlated variant to the top SNP rs171408 (r2=0.84 according to 1000 genomes data), is located in a predicted myocyte enhancer factor 2A (MEF2A) binding site, a key transcription factor in cardiac development19. We have previously shown that Mef2C regulated matrix production in mouse valves20. Our data extend the role of LMCD1 to valve development, and the putative implication of LMCD1 in matrix production regulated by MEF2A deserves future investigation.
Figure 4. Cardiac regurgitation in zebrafish morpholino knockdown for Lmcd1 on Chr3p13.
a) Genomic context of the association signal observed in the GWAS meta-analysis. The regional association plot was generated using locus zoom and displays surrounding genes, with LMCD1 identified as best potential candidate as the signal is intronic to LMCD1. b) Mitral regurgitation observed at 72 hours post fertilization (hpf) in zebrafish embryos after morpholino mediated knockdown. All results are presented as fold change compared to clutchmate controls. n=number of biological replicates per morpholino. (*) indicates p<0.05.
c) 2-dimensional projections of z-series image stacks taken on excised 72hpf control (CN) and lmcd1 knockdown zebrafish hearts. Green denotes EGFP expression, a marker of endothelium under the control of the flk promoter. Red staining indicates the distribution of F-actin, which is highly expressed in the functional myocardium. Scale bar represents 50μm. d) Brightfield micrographs displaying gross morphology of 72hpf embryos following lmcd1 knockdown. Scale bar represents 1mm. CN=control morpholino injected embryos. No detectable morphological difference is observed between morpholino-injected fish and wild-types.
Amongst the remaining MVP loci, we detected expression for Pitpnb, candidate gene on Chr22q12, in valve endothelial and interstitial cells within the mouse mitral leaflets at each of the time points investigated (Supplementary Figure 6). At the Chr17p13 locus, knockdown of candidate genes included smg6 and sgsm2, neither of which led to a valvular phenotype in the zebrafish (Supplementary Figure 7). Additional candidate genes need to be explored at this locus, which has been associated with aortic root size21 and coronary heart disease19.
Despite the widespread prevalence, the molecular basis of MVP has largely been elusive. The first molecular pathways implicated in MVP arose from the observation of the disease in patients with Marfan or Ehlers-Danlos syndromes, findings that highlight the importance of extracellular matrix composition22, the TGF-beta growth factor pathway12,23 and valve cell proliferation and differentiation24. Several structural mechanisms have also been proposed, such as enlargement and flattening of the mitral annulus25 that can impose additional stresses on genetically susceptible valves and chordae26. This first GWAS of non-syndromic MVP reveals several susceptibility loci supporting the concept that genetic variants affecting the expression of proteins during valve development can progressively affect mitral valve function into adult life, as was recently shown for Filamin-A27. In particular, we provide genetic and functional evidence that TNS1 and LMCD1 both implicated in cell proliferation and migration are contributing to mitral valve degeneration possibly during valve development, thus revealing new pathways as possible innovative therapeutic targets.
Online methods
Leducq Transatlantic MITRAL Network
The majority of patients were recruited as a major project of the Leducq Mitral Network, a transatlantic consortium investigating the physiopathology of mitral valve disease with basic and clinical investigators from 10 clinical and research centres. Six centres recruited MVP patients, MVP-Nantes and MVP-France for the initial GWAS effort, MVP-USA, Framingham Heart Study (FHS) and PROlapso Mitral en cEntros eSpAñoles (PROMESA) at the Centro Nacional de Investigaciones Cardiovasculares (CNIC) for initial replication and HEGP-Surgical Cases with QCCMRC data-sets in the last replication stage. Cases were compared with controls (Framingham Heart Study, PROMESA-CNIC and QCCMRC) or general population (D.E.S.I.R for initial GWAS and one replication stage), SU.VI.MAX (GWAS) (Supplementary Figure 1).
Cases recruitment criteria
We used consensus inclusion criteria of adult (≥ 18 years) patients with idiopathic MVP if they presented displacement into the left atrium of any part of the mitral valve leaflet(s) ≥ 2 mm beyond a line connecting the annular hinge points on the parasternal long-axis view of the left ventricle by two-dimensional (2D) echocardiography1,2. We also included patients with previous surgery for pure severe MR due to MVP supported by an operative report and written confirmation of the diagnosis by the surgeon (MVP-France, MVP-Nantes and Surgery Cases). All cases were validated by a local, experienced team of cardiologists on the basis of clinical and echocardiography records. Recruitments excluded patients with MVP associated with other heart disease (coronary artery disease with papillary muscle disruption, hypertrophic cardiomyopathy or rheumatic disease) or known syndromes (e.g. Marfan and Ehlers-Danlos). Local ethics committees approved all studies and all patients and controls provided written informed consent. Recruitment procedures of DNA collection are detailed per cohort in Supplementary Note.
Analytical methods
GWAS genotyping and quality control
Genotyping of the discovery cohorts was independently performed by different genetic platforms that included standard quality control measures of genotyping and data acquisition from diverse high-density genotyping arrays (Supplementary Table 1). We excluded participants with genotype call rate < 97% and individual heterozygosity (IHe) level < 10,000 (determined as outlier limit after visual inspection). We excluded SNPs with a minor allele frequency (MAF) <0.1, call rate <95%, monomorphic, and with an exact Hardy Weinberg Equilibrium (HWE) p < 0.0001 in controls and p < 10−7 in demographically homogenous cases to exclude SNPs that show very large deviations.
Imputation
To complement directly genotyped SNPs we performed large-scale imputation in the four discovery cohorts. First, genotyped SNPs in cases and controls were phased using the SHAPE-IT (v1) program3. Then, the imputation of 4.8 million common SNPs (MAF>0.1 in 1000G Europeans, proper-info > 0.4) was carried out using IMPUTE v24 in ~7 Mb chunks. The reference panel used was Phase I integrated variant set release (v3), in NCBI build 37 (hg19). We used similar procedures to impute non-genotyped SNPs in the replication cohort FHS using MACH software (0.3 r2_hat)5.
Direct genotyping in the replication sets
MGH cases from the follow-up Set1 and all cases and controls of Set2 were genotyped at the Massachusetts General Hospital PNGU Core Lab using the Sequenom iPLEX Gold® application and MassARRAY® system. Follow-up Sets 3 and 4 were genotyped at the LGC genomics company using the KASP® genotyping chemistry. We excluded 9 individuals that failed genotyping for all SNPs, and SNPs with call rate <0.90. No SNP deviated from HWE (P>0.05).
Demographic analyses
The ancestry of participants was assessed using a multi-dimensional scaling technique implemented in PLINK6. SNPs were selected for short-range linkage disequilibrium (LD) independence (r2 > 0.2). Multi-dimensional scaling method was applied on the Identity-By-State matrix and we excluded outliers on the first two components (Supplementary Figure 8) using an expectation-maximization (EM)-fitted Gaussian mixture clustering method implemented in the R package M-CLUST, assuming one cluster and noise (Supplementary Note).
Statistical Analyses
Genome-wide and replication association with MVP status
We applied a logistic regression (additive model) as implemented in SNPTEST4 to test the association with MVP in the GWAS discovery adjusted for the five first principal components as covariates. We also used SNPTEST and/or logistic regression on allele dosage in replication sets when cases and/or controls were imputed for genotypes (FHS in Set 1 and D.E.S.I.R. 2 in Set 4, Supplementary Figure 1) and took into account for relatedness among (FHS). For directly genotyped cases control studies (Set 2 and Set 3) we used logistic regression as implemented in PLINK.
For the GWAS meta-analysis, we applied the inverse normal strategy7. Because the number of controls greatly exceeds the number of cases in all studies, we used the effective sample size as advised in the METAL software 8:W= 4/(1/Ncases+1/Ncontrols).
Regional association plots for Chr2q35, Chr3p13 and Chr17p13 were created using Locus Zoom9.
Protein detection in mouse embryos and adult hearts
Standard histological and immunochemical procedures were used as previously described10. For all immunohistochemistry (IHC) experiments, 5-min antigen retrieval was performed with VectaStain and Pressure Cooker (Cuisinart). Antibodies used for immunological experiments were: Tensin1 (Novus), MF20 (Developmental Hybridoma Banks). Primary antibodies were used for IHC at a 1:100 dilution, Hoescht 33342 (nuclear stain) was used at a 1:10,000 dilution. Appropriate secondary antibodies were used for detection.
Histology and expression studies were performed on adult (9-month) wild-type (Tensin1+/+) and knockout (Tensin1−/−) hearts. For Histology: Adult (9-month) hearts were processed for hematoxylin and eosin stainings and immunohistochemistry (IHC) as previously described 11. For all analyses male mice were used and N=3 for each genotype. Antibodies used for IHC were: Hyaluronan Binding Protein (HABP) to stain proteoglycans (1:100) (Callbiochem), collagen I (1:100) (MDbio), and Hoescht to stain nuclei (1:10,000) (Invitrogen).
Zebrafish experiments
Zebrafish experiments were performed in accordance with approved Institutional Animal Care and Use Committee (IACUC) protocols. TuAB zebrafish strains were reared according to standard techniques. Minimal effective doses of antisense morpholino oligonucleotides were injected at the single cell stage and compared to non-targeting morpholino injected controls. Nucleotides sequences are indicated in Supplementary Table 6. Embryos were scored for presence of AV regurgitation at 72 hours post-fertilization (hpf) using high speed videography. Semiquantitative PCR was used to demonstrate morpholino knockdown efficacy (Supplementary Figure 9). In situ hybridizations for tissue specific expression of lmcd1, tns1, bmp4, and notch1b were performed as described12. In order to visualize the localization of the developing cardiac cushions, flk-EGFP reporter fish were microinjected with anti-tns1 or anti-lmcd1 morpholinos. After manual excision of the heart at 72hpf, hearts were counterstained with rhodamine labelled phalloidin and mounted using Vectashield. Confocal micrographs were acquired on a Zeiss 510 LSM with a 20X air lens. Final Images represent 2D projections of a z-series (ImageJ).
Supplementary Material
Supplementary Table 1. Clinical and genotyping features of the study populations.
Supplementary Table 2. Association with MVP of loci carried for follow-up and genotyped in all the study populations (N=13 loci). One best-associated SNP is indicated per locus. Alleles are indexed to the forward strand of NCBI Build 37; P-values reported are two-sided and based on an inverse-variance weighted meta-analysis model (fixed-effects). P-values are for heterogeneity test between all individual studies (Cochran Q statistics); *: OR and p values in the global met-analysis are indicated for the proxy genotyped in the follow-up.
Supplementary Table 3. Associations with MVP among surgery patients.
Supplementary Table 4. Short literature descriptive of candidate genes near MVP confirmed loci. References are listed in Supplementary note.
Supplementary Table 5. Initial list of genes for candidates prioritization for zebrafish knockdown.
Supplementary Table 6. Nucleotide sequences used for morpholinos and morpholino efficacy 72 hours post fertilization (hpf). * E(X)I(X) indicates the exon and intron border which the morpholino targets.
Supplementary Figure 1. Study design with cohorts in discovery and follow-up and filtration strategy of SNPs.
Supplementary Figure 2. Igfbp5 expression in mouse developing heart. Cardiac expression of Igfbp5 (red) was analysed at embryonic (E13.5), foetal (E17.5) and adult time points. At all time points investigated very little Igfbp5 protein was detected in the valves. Expression was observed in the myocardium of the left and right atria as well and the primary atrial septum (PAS) at E13.5 and E17.5 (arrows) as well as the coronary endothelium in the adult (arrow heads). AL, PL, IVS, LV= anterior and posterior mitral leaflets, interventricular septum, and left ventricle, respectively. Green=MF20 (sarcomeric myosin-myocytes), Blue= hoescht (nuclei)
Supplementary Figure 3. In vivo assessment of mitral valve phenotypes in Tns1 total knockout mice using echocardiographs. Tensin1+/+: wildtype; Tensin1−/−: Tns1 knockout mice. Images were obtained in a four-chamber view of the heart.
Supplementary Figure 4. In situ hybridizations of tns1 and lmcd1 mRNA distribution in zebrafish embryos. Top panel: tns1 expression pattern at 96 hours post fertilization. Expression found throughout the myocardium with highest levels in the outflow tract (right). Lower panel: lmcd1 expression at 72 hours post fertilization. Expression is found in the entire myocardium with enhanced expression in the ventricular chamber.
Supplementary Figure 5. In situ hybridizations for developmental markers of valvulogenesis lmcd1 and tns knockdown embryos. Top panel: anti-notch1b probe labels the developing valve in 72 hours post fertilization embryos. Lower panel: anti-bmp-4 probe labels the valve and surrounding myocardium in control (CN) and lmcd1 knockdown embryos. Arrows in both panels point to the approximate location of the AV valve.
Supplementary Figure 6. Pitpnb expression in mouse developing heart. Cardiac expression of Pitpnb (red) was analysed at embryonic (E13.5), foetal (E17.5) and adult time points. Pitpnb was detected in valve endothelial and interstitial cells within the mitral leaflets at each of the time points investigated. Expression was also observed in the epicardium (arrow head) and coronary endothelium (arrow) at E17.5. AL, PL, IVS, LV= anterior and posterior mitral leaflets, interventricular septum, and left ventricle, respectively. Green=MF20 (sarcomeric myosin-myocytes), Blue= hoescht (nuclei)
Supplementary Figure 7. Assessment of cardiac regurgitation in zebrafish morpholino knockdown for genes on Chr17p13. a) Genomic context of the association signal observed in the GWAS meta-analysis. b) Morpholino-mediated knockdown efficacy. Efficacy for smg6 and sgsm2 in embryonic zebrafish was measured by RT-PCR. c) Fold change in observed mitral regurgitation in 72 hpf zebrafish embryos after morpholino mediated knockdown. All results are relative to clutchmate controls. n=number of biological replicates per morpholino. d) Brightfield micrographs displaying gross morphology of 72hpf embryos following smg6 or sgsm2 knockdown. Scale bar represents 1mm. CN=control morpholino injected embryos.
Supplementary Figure 8. Multidimensional scale-based principal component analysis. Cases and controls positions on first and second components axis are presented for the two discovery samples (GWAS 1 and GWAS 2). C1: first principal component. C2: second principal component. All individuals plotted are those included in the discovery GWAS who are all French with European ancestry origin. Cases and controls with non-European ancestry based on the principal component analyses were excluded before association tests.
Supplementary Figure 9: Representative gel images from analysis of morpholino efficacy. rsID indicates the sentinel SNP at the analyzed locus. CN indicates samples amplified from control injected embryos whereas MO indicates amplicons from samples obtained following microinjection of gene-specific morpholino. All samples were obtained from 72 hours post fertilization embryos.
Figure 1. Quantile-quantile (A) and manhattan (B) plots representing the association of 4.8 million SNPs at the GWAS meta-analysis.
Red line indicates genome-wide significant threshold (P < 5 × 10−8) and blue line the p-value thershold used for follow-up (P < 10−5).
Acknowledgments
We acknowledge the major contribution of the Leducq Foundation, Paris, for supporting a transatlantic consortium investigating the physiopathology of mitral valve disease, for which this genome-wide association study was a major project (Coordinators: RA. Levine and AA. Hagège). We thank Jordan Leyton-Mange, Xinh-Xinh Nguyen and Michael McLellan, for help with the zebrafish experiments and Patrick Mathieu as one of the main investigators of the PROGRAM (Determinants of the Progression and Outcomes of Organic Mitral Regurgitation) study, from which were ascertained the Canadian MVP case control study. C. Dina, T. Le Tourneau and JJ. Schott acknowledge a translational research grant on genetics of mitral valve funded by the Nantes Hospital (CHU Nantes). RA Levine acknowledges grant support from the National Institutes of Health (K24 HL67434, R01 HL72265 and HL109506). N. Bouatia-Naji is recipient of a French young investigator fund (ANR-13-ISV1-0006-0). DJ. Milan acknowledges the support of the Hassenfeld Scholar Program. PT. Ellinor is supported by grants from the National Institutes of Health (HL092577, HL104156, K24HL105780, HL065962), by an Established Investigator Award from the American Heart Association (13EIA14220013) and by support from the Fondation Leducq (14CVD01). P. Pibarot holds the Canada Research Chair in Valvular Heart Diseases and is supported by grants from the Canadian Institutes of Health Research (MOP-102737, MOP-114997, and MOP-126072). Y. Bossé is the recipient of a Junior 2 Research Scholar award from the Fonds de recherche Québec – Santé (FRQS) and is supported by the Canadian Institutes of Health Research (MOP-102481 and MOP-137058). L. Fernandez-Friera and J. Solis received financial support from the Spanish Society of Cardiology. R. Durst was supported by fellowships from the fund for medical discovery, Massachusetts General Hospital, executive committee of research, and Marie Curie reintegration award from the European Commission.
Footnotes
Contributions
CD, NBN, and XJ organized and designed the genome-wide association study and the manuscript preparation. AAH, RAL, SAS, HLM, PP, JS and LFF organized and coordinated the network efforts and recruitment of patients. RD, VP, TLT, FK, PM and YB participated to the recruitment of patients and the interpretation of the echocardiographs. JJS and XJ coordinated the collection of patient samples. MP and SL managed the collection of patient samples and performed DNA extraction and genotyping. CD and NBN conceptualized the statistical analyses. CD supervised the statistical analyses. CD, MHC and FS performed the statistical analyses. DJM conceptualized and supervised the zebrafish experiments. NT performed zebrafish experiments interpreted the data and wrote parts of the manuscript. PTE and ED participated in zebrafish experiments and interpretation of data. RAN conceptualized and supervised the mouse studies and interpreted the data. KT performed immunohistochemistry staining of mouse organs. SHL generated the mice. FND, EB, DZ, ML, SH, RR, FB, R Redon, PF and RSV conducted the epidemiological studies in control cohorts and/or contributed samples to the GWAS and/or follow-up genotyping. PB supervised valve tissues DNA extraction. SAS contributed patient samples and organized follow-up genotyping and interpreted the GWAS and follow-up data. CD, NBN, and XJ wrote the manuscript. FND, RAN, DJL, SAS, RAL, JJS and AAH edited the manuscript and all authors approved its content.
Competing financial interests.
The authors declare no competing financial interests.
References
- 1.Freed LA, et al. Mitral valve prolapse in the general population: the benign nature of echocardiographic features in the Framingham Heart Study. J Am Coll Cardiol. 2002;40:1298–304. doi: 10.1016/s0735-1097(02)02161-7. [DOI] [PubMed] [Google Scholar]
- 2.Disse S, et al. Mapping of a first locus for autosomal dominant myxomatous mitral-valve prolapse to chromosome 16p11.2-p12.1. Am J Hum Genet. 1999;65:1242–51. doi: 10.1086/302624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freed LA, et al. A locus for autosomal dominant mitral valve prolapse on chromosome 11p15.4. Am J Hum Genet. 2003;72:1551–9. doi: 10.1086/375452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyndt F, et al. Mapping of X-linked myxomatous valvular dystrophy to chromosome Xq28. Am J Hum Genet. 1998;62:627–32. doi: 10.1086/301747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nesta F, et al. New locus for autosomal dominant mitral valve prolapse on chromosome 13: clinical insights from genetic studies. Circulation. 2005;112:2022–30. doi: 10.1161/CIRCULATIONAHA.104.516930. [DOI] [PubMed] [Google Scholar]
- 6.Rath N, Wang Z, Lu MM, Morrisey EE. LMCD1/Dyxin is a novel transcriptional cofactor that restricts GATA6 function by inhibiting DNA binding. Mol Cell Biol. 2005;25:8864–73. doi: 10.1128/MCB.25.20.8864-8873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avierinos JF, et al. Natural history of asymptomatic mitral valve prolapse in the community. Circulation. 2002;106:1355–61. doi: 10.1161/01.cir.0000028933.34260.09. [DOI] [PubMed] [Google Scholar]
- 8.Freed LA, et al. Prevalence and clinical outcome of mitral-valve prolapse. N Engl J Med. 1999;341:1–7. doi: 10.1056/NEJM199907013410101. [DOI] [PubMed] [Google Scholar]
- 9.Devereux RB, Brown WT, Kramer-Fox R, Sachs I. Inheritance of mitral valve prolapse: effect of age and sex on gene expression. Ann Intern Med. 1982;97:826–32. doi: 10.7326/0003-4819-97-6-826. [DOI] [PubMed] [Google Scholar]
- 10.Delling FN, et al. Mild expression of mitral valve prolapse in the Framingham offspring: expanding the phenotypic spectrum. J Am Soc Echocardiogr. 2014;27:17–23. doi: 10.1016/j.echo.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glesby MJ, Pyeritz RE. Association of mitral valve prolapse and systemic abnormalities of connective tissue. A phenotypic continuum. JAMA. 1989;262:523–8. [PubMed] [Google Scholar]
- 12.Hagege AA, et al. The mitral valve in hypertrophic cardiomyopathy: old versus new concepts. J Cardiovasc Transl Res. 2011;4:757–66. doi: 10.1007/s12265-011-9319-6. [DOI] [PubMed] [Google Scholar]
- 13.Kyndt F, et al. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 2007;115:40–9. doi: 10.1161/CIRCULATIONAHA.106.622621. [DOI] [PubMed] [Google Scholar]
- 14.Hall EH, Daugherty AE, Choi CK, Horwitz AF, Brautigan DL. Tensin1 requires protein phosphatase-1alpha in addition to RhoGAP DLC-1 to control cell polarization, migration, and invasion. J Biol Chem. 2009;284:34713–22. doi: 10.1074/jbc.M109.059592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sureshbabu A, et al. IGFBP5 induces cell adhesion, increases cell survival and inhibits cell migration in MCF-7 human breast cancer cells. J Cell Sci. 2012;125:1693–705. doi: 10.1242/jcs.092882. [DOI] [PubMed] [Google Scholar]
- 16.Chang CY, Lin SC, Su WH, Ho CM, Jou YS. Somatic LMCD1 mutations promoted cell migration and tumor metastasis in hepatocellular carcinoma. Oncogene. 2012;31:2640–52. doi: 10.1038/onc.2011.440. [DOI] [PubMed] [Google Scholar]
- 17.Bian ZY, et al. LIM and cysteine-rich domains 1 regulates cardiac hypertrophy by targeting calcineurin/nuclear factor of activated T cells signaling. Hypertension. 2010;55:257–63. doi: 10.1161/HYPERTENSIONAHA.109.135665. [DOI] [PubMed] [Google Scholar]
- 18.Beis D, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 19.Schunkert H, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lockhart MM, et al. Mef2c regulates transcription of the extracellular matrix protein cartilage link protein 1 in the developing murine heart. PLoS One. 2013;8:e57073. doi: 10.1371/journal.pone.0057073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasan RS, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302:168–78. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pyeritz RE, Wappel MA. Mitral valve dysfunction in the Marfan syndrome. Clinical and echocardiographic study of prevalence and natural history. Am J Med. 1983;74:797–807. doi: 10.1016/0002-9343(83)91070-7. [DOI] [PubMed] [Google Scholar]
- 23.Ng CM, et al. TGF-beta-dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J Clin Invest. 2004;114:1586–92. doi: 10.1172/JCI22715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabkin E, et al. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation. 2001;104:2525–32. doi: 10.1161/hc4601.099489. [DOI] [PubMed] [Google Scholar]
- 25.Lee AP, et al. Quantitative analysis of mitral valve morphology in mitral valve prolapse with real-time 3-dimensional echocardiography: importance of annular saddle shape in the pathogenesis of mitral regurgitation. Circulation. 2013;127:832–41. doi: 10.1161/CIRCULATIONAHA.112.118083. [DOI] [PubMed] [Google Scholar]
- 26.Jensen MO, Hagege AA, Otsuji Y, Levine RA, Leducq Transatlantic MN. The unsaddled annulus: biomechanical culprit in mitral valve prolapse? Circulation. 2013;127:766–8. doi: 10.1161/CIRCULATIONAHA.112.000628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauls K, et al. Developmental basis for filamin-A-associated myxomatous mitral valve disease. Cardiovasc Res. 2012;96:109–19. doi: 10.1093/cvr/cvs238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Clinical and genotyping features of the study populations.
Supplementary Table 2. Association with MVP of loci carried for follow-up and genotyped in all the study populations (N=13 loci). One best-associated SNP is indicated per locus. Alleles are indexed to the forward strand of NCBI Build 37; P-values reported are two-sided and based on an inverse-variance weighted meta-analysis model (fixed-effects). P-values are for heterogeneity test between all individual studies (Cochran Q statistics); *: OR and p values in the global met-analysis are indicated for the proxy genotyped in the follow-up.
Supplementary Table 3. Associations with MVP among surgery patients.
Supplementary Table 4. Short literature descriptive of candidate genes near MVP confirmed loci. References are listed in Supplementary note.
Supplementary Table 5. Initial list of genes for candidates prioritization for zebrafish knockdown.
Supplementary Table 6. Nucleotide sequences used for morpholinos and morpholino efficacy 72 hours post fertilization (hpf). * E(X)I(X) indicates the exon and intron border which the morpholino targets.
Supplementary Figure 1. Study design with cohorts in discovery and follow-up and filtration strategy of SNPs.
Supplementary Figure 2. Igfbp5 expression in mouse developing heart. Cardiac expression of Igfbp5 (red) was analysed at embryonic (E13.5), foetal (E17.5) and adult time points. At all time points investigated very little Igfbp5 protein was detected in the valves. Expression was observed in the myocardium of the left and right atria as well and the primary atrial septum (PAS) at E13.5 and E17.5 (arrows) as well as the coronary endothelium in the adult (arrow heads). AL, PL, IVS, LV= anterior and posterior mitral leaflets, interventricular septum, and left ventricle, respectively. Green=MF20 (sarcomeric myosin-myocytes), Blue= hoescht (nuclei)
Supplementary Figure 3. In vivo assessment of mitral valve phenotypes in Tns1 total knockout mice using echocardiographs. Tensin1+/+: wildtype; Tensin1−/−: Tns1 knockout mice. Images were obtained in a four-chamber view of the heart.
Supplementary Figure 4. In situ hybridizations of tns1 and lmcd1 mRNA distribution in zebrafish embryos. Top panel: tns1 expression pattern at 96 hours post fertilization. Expression found throughout the myocardium with highest levels in the outflow tract (right). Lower panel: lmcd1 expression at 72 hours post fertilization. Expression is found in the entire myocardium with enhanced expression in the ventricular chamber.
Supplementary Figure 5. In situ hybridizations for developmental markers of valvulogenesis lmcd1 and tns knockdown embryos. Top panel: anti-notch1b probe labels the developing valve in 72 hours post fertilization embryos. Lower panel: anti-bmp-4 probe labels the valve and surrounding myocardium in control (CN) and lmcd1 knockdown embryos. Arrows in both panels point to the approximate location of the AV valve.
Supplementary Figure 6. Pitpnb expression in mouse developing heart. Cardiac expression of Pitpnb (red) was analysed at embryonic (E13.5), foetal (E17.5) and adult time points. Pitpnb was detected in valve endothelial and interstitial cells within the mitral leaflets at each of the time points investigated. Expression was also observed in the epicardium (arrow head) and coronary endothelium (arrow) at E17.5. AL, PL, IVS, LV= anterior and posterior mitral leaflets, interventricular septum, and left ventricle, respectively. Green=MF20 (sarcomeric myosin-myocytes), Blue= hoescht (nuclei)
Supplementary Figure 7. Assessment of cardiac regurgitation in zebrafish morpholino knockdown for genes on Chr17p13. a) Genomic context of the association signal observed in the GWAS meta-analysis. b) Morpholino-mediated knockdown efficacy. Efficacy for smg6 and sgsm2 in embryonic zebrafish was measured by RT-PCR. c) Fold change in observed mitral regurgitation in 72 hpf zebrafish embryos after morpholino mediated knockdown. All results are relative to clutchmate controls. n=number of biological replicates per morpholino. d) Brightfield micrographs displaying gross morphology of 72hpf embryos following smg6 or sgsm2 knockdown. Scale bar represents 1mm. CN=control morpholino injected embryos.
Supplementary Figure 8. Multidimensional scale-based principal component analysis. Cases and controls positions on first and second components axis are presented for the two discovery samples (GWAS 1 and GWAS 2). C1: first principal component. C2: second principal component. All individuals plotted are those included in the discovery GWAS who are all French with European ancestry origin. Cases and controls with non-European ancestry based on the principal component analyses were excluded before association tests.
Supplementary Figure 9: Representative gel images from analysis of morpholino efficacy. rsID indicates the sentinel SNP at the analyzed locus. CN indicates samples amplified from control injected embryos whereas MO indicates amplicons from samples obtained following microinjection of gene-specific morpholino. All samples were obtained from 72 hours post fertilization embryos.