Abstract
Diacylglycerol lipase (DAGL)-α and -β are enzymes responsible for the biosynthesis of the endocannabinoid 2-arachidonoylglycerol (2-AG). Selective and reversible inhibitors are required to study the function of DAGLs in neuronal cells in an acute and temporal fashion, but they are currently lacking. Here, we describe the identification of a highly selective DAGL inhibitor using structure-guided and a chemoproteomics strategy to characterize the selectivity of the inhibitor in complex proteomes. Key to the success of this approach is the use of comparative and competitive activity-based proteome profiling (ABPP), in which broad-spectrum and tailor-made activity-based probes are combined to report on the inhibition of a protein family in its native environment. Competitive ABPP with broad-spectrum fluorophosphonate-based probes and specific β-lactone-based probes led to the discovery of α-ketoheterocycle LEI105 as a potent, highly selective and reversible dual DAGL-α/DAGL-β inhibitor. LEI105 did not affect other enzymes involved in endocannabinoid metabolism including abhydrolase domain-containing protein 6, abhydrolase domain-containing protein 12, monoacylglycerol lipase and fatty acid amide hydrolase and did not display affinity for the cannabinoid CB1 receptor. Targeted lipidomics revealed that LEI105 concentration-dependently reduced 2-AG levels, but not anandamide levels, in Neuro2A cells. We show that cannabinoid CB1-receptor-mediated short-term synaptic plasticity in a mouse hippocampal slice model can be reduced by LEI105. Thus, we have developed a highly selective DAGL inhibitor and provide new pharmacological evidence to support the hypothesis that ‘on demand biosynthesis’ of 2-AG is responsible for retrograde signaling.
Introduction
Endocannabinoids are endogenous signaling lipids that activate the cannabinoid CB1 and CB2 receptor. They play an essential role in human health and disease, regulating processes, such as immunomodulation, energy balance and neurotransmission.1 There are two main endocannabinoids: anandamide and 2-arachidonoylglycerol (2-AG).2-4 Both endocannabinoids are often found together, but their levels vary between species, tissue type, developmental stage and pathological condition.5 Although selective inhibitors of their metabolic pathways have provided information about the biological function of the endocannabinoids, it is still unclear to a large extent which endocannabinoid is responsible for specific cannabinoid CB1 receptor dependent (patho)physiological effects.6,7 Selective inhibition of the formation of anandamide and 2-AG would be instrumental to determine which endocannabinoid is responsible for specific CB1-mediated physiological effects. However, pathway-selective inhibitors for 2-AG and anandamide biosynthesis are currently lacking.
2-AG is mainly formed by the action of two diacylglycerol lipases (DAGL-α and DAGL-β).8 DAGLs are intracellular, multi-domain integral membrane proteins. The DAGLs share extensive homology, but differ in size: ~120 and ~70 kDa for DAGL-α and DAGL-β respectively.8,9 DAGLs belong to the class of serine hydrolases that employ the typical Ser-His-Asp catalytic triad to hydrolyze the ester bond of acyl chains from arachidonate-containing diacylglycerols in a sn-1 specific manner. Studies with DAGL knock-out mice have shown that DAGL-α controls to a large extent the formation of 2-AG in the central nervous system, whereas DAGL-β appears to partake in 2-AG production in the periphery during inflammation.5,10 Importantly, also basal anandamide levels were reduced in DAGL-α knock-out mice. Selective inhibitors for DAGLs, which can be used in an acute and temporal fashion and do not modulate anandamide levels, would, therefore, constitute an important counterpart of the DAGL knock-out mice, and allow the examination of acute versus congenital inhibition.
Although, several classes of DAGL inhibitors have been described in literature,8,11-15 these inhibitors are based on the natural substrates and/or have reactive chemical warheads, and are not selective over other serine hydrolases that modulate endocannabinoid signaling (e.g. abhydrolase domain-containing protein 6 and 12 (ABHD6 and ABHD12), monoacylglycerol lipase (MAGL) or fatty acid amide hydrolase (FAAH)), therefore highly selective DAGL inhibitors are warranted to study the cellular role of the biosynthetic enzymes.
Activity-based protein profiling (ABPP) has emerged as a powerful technique for discovering selective enzyme inhibitors acting in their native physiological context.16 ABPP hinges on the use of activity-based probes (ABPs) to report on enzyme activity in cells, tissue or animals.17 An ABP normally consists of a covalent, irreversible enzyme inhibitor featuring a reporter entity (fluorophore, biotin, bioorthogonal tag) to label the active site of the enzyme or enzyme family at hand. ABPP is unique in its ability to rapidly identify inhibitor activity and selectivity within large enzyme families in complex proteome samples. The prototypical ABP for serine hydrolases is based on a fluorophosphonate (FP)-warhead.18 However, this probe does not recognize DAGL-α and the signals for DAGL-β, MAGL and ABHD6 cannot be unequivocally established because not all gel bands can be clearly resolved in the brain proteome.19 Previously, we have reported on the design, synthesis and characterization of a specific β-lactone-containing probe (MB064), which was tailor-made for the detection of DAGL-α activity.20,21 Using this probe we identified 1-(oxazolo[4,5-b]pyridin-2-yl)-6-phenylhexan-1-one (LEI104, Figure 1), which belongs to the class of α-ketoheterocycles, as the first reversible inhibitor for DAGL-α. LEI104 was, however, weakly active in a cellular assay and was not selective over FAAH, the enzyme responsible for the metabolism of the other endocannabinoid anandamide (AEA).22,23 Moreover, activity of LEI104 on DAGL-β was not studied, but in view of the high homology between DAGL-α and DAGL-β, it is likely that there is cross-reactivity. In order to apply α-ketoheterocycles as chemical tools to study 2-AG signaling, it is important to increase their cellular activity, to have selectivity over FAAH and to assess their activity on endogenous DAGL-β.
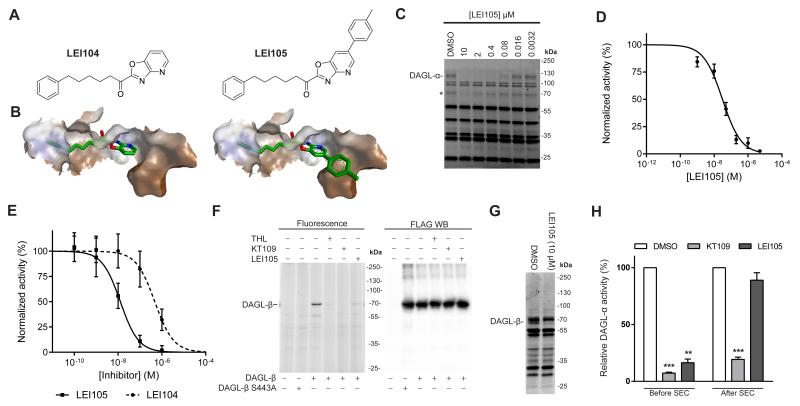
Figure 1.
Structure-guided modeling and biochemical characterization of LEI105. (a) Structures of α-ketoheterocycle based DAGL inhibitors LEI104 and LEI105. (b) LEI104 and LEI105 in a homology model of DAGL-α. (c) Representative fluorescent ABPP gel showing dose dependent inhibition of MB064 (250 nM) labeling of endogenous DAGL-α labeling by LEI105 in the mouse brain membrane proteome. (* = DAGL-α breakdown product) (d) Dose response curve of DAGL-α inhibition as determined with competitive ABPP (pIC50 7.5 ± 0.07 (IC50 = 32 nM); n = 3). (e) Dose response curve of DAGL-α inhibition by LEI104 (pIC50 6.3 ± 0.1 (IC50 = 501 nM); n = 4) and LEI105 (pIC50 7.9 ± 0.08 nM (IC50 = 13 nM); n = 4) as determined with a glycerol based natural substrate assay. (f) ABPP using MB064 (1 μM) with different hDAGL-β constructs and anti-FLAG western blot of the same gel. (g) Competitive ABPP in the mouse spleen membrane proteome using MB064 (1.0 μM) in competition with LEI105 (10 μM), LEI105 can block labeling of endogenously expressed DAGL-β in the mouse spleen membrane proteome. (H) Schematic representation of the size exclusion chromatography (SEC) experiment that shows reversibility of LEI105 in recombinant DAGL-α (n = 3, full fluorescent gel and western blot are given in the supporting information) Statistical analysis: 2-way ANOVA (*** = p < 0.001; ** = p < 0.01 vs vehicle)
Here, we report a structure-guided approach to optimize LEI104 employing a homology-model of DAGL-α. In addition, we discovered that our tailor-made β-lactone probe MB064 could also label DAGL-β in cells and tissues. Using these tools we characterized LEI105 as a cellular active, dual DAGL-α/β inhibitor. Comparative chemoproteomics revealed that LEI105 is selective over ABHD6, ABHD12, MAGL and FAAH. Furthermore, targeted lipidomics revealed that LEI105 is able to concentration-dependently reduce 2-AG levels in neuronal cells without affecting AEA levels. We showed that cannabinoid CB1-receptor-dependent short-term synaptic plasticity in a hippocampal slice model can be reduced by the selective DAGL-inhibitor LEI105. In summary, comparative and competitive chemoproteomics was applied to characterize the most selective DAGL inhibitor to date, that can be used to study DAGL function in an acute and temporal manner in a neuronal context.
Results & discussion
Structure-guided modeling to identify LEI105 as DAGL-α inhibitor
Previously, we have identified the α-ketoheterocycle, 1-(oxazolo[4,5-b]pyridin-2-yl)-6-phenylhexan-1-one (LEI104, Figure 1A), as an inhibitor for DAGL-α. To improve the potency of LEI104 (Figure 1A) we used a structure-guided modeling approach. We investigated in detail the binding pose of LEI104 in DAGL-α using a molecular dynamics simulation in our previously generated homology model.21 From this analysis we identified an additional hydrophobic pocket close to the catalytic site that did not appear to be occupied by LEI104 (Figure 1B). Introduction of a phenyl substituent at the 6-position of the oxazolopyridine would allow us to probe this pocket with the aim of increasing potency and/or selectivity. We synthesized compound (LEI105) (See supporting information for synthesis) and tested its activity on human DAGL-α in a colorimetric assay using para-nitrophenylbutyrate as a surrogate substrate.24 LEI105 proved to be a potent inhibitor with a pIC50 of 8.5 ± 0.06 (n=4), thus some 10-fold more potent than LEI104 (7.4 ± 0.05; n=4).21 Reduction of the α–keto group to the corresponding alcohol (compound 8) led to a ~150 fold drop of activity against DAGL-α, thereby indicating that the α–carbonyl in LEI105 reacts with the active site serine hydroxyl to form a covalent, though reversible, enzyme-inhibitor hemiketal adduct. To confirm that LEI105 was also able to block conversion of the natural substrate 1-stearoyl-2-arachidonoyl-sn-glycerol of DAGL-α to the endocannabinoid 2-AG, we employed our recently developed real-time, fluorescence-based assay.25 In this assay LEI105 inhibited recombinant human DAGL-α with a pIC50 of 7.9 ± 0.08 (n=4) (Figure 1E), which is a 40-fold increase compared to LEI104 (pIC50 6.3 ± 0.1 (n = 4)). The inhibitory activity of LEI105 was confirmed in a radiometric assay using 1-[14C]oleoyl-2-arachidonoyl-sn-glycerol (1.0 mCi mmol−1, 20 μm) as substrate (pIC50 of 6.6; n = 2).8
Determining endogenous DAGL activity using MB064 as ABP
To test the activity of LEI105 on endogenously expressed DAGL-α in mouse membrane proteome, we used our previously reported ABPP method with MB064.21 Eleven tissues from wild-type and DAGL-α knock-out mice were screened to obtain a tissue-wide profile of endogenous DAGL-α activity (supporting information). DAGL-α activity was found to be highest in the brain (which is in line with our previous reported results on a smaller set of tissues).21 LEI105 prevented DAGL-α labeling in the mouse brain membrane proteome by MB064 with a pIC50 of 7.5 ± 0.07 (n=3) (Figure 1C, D).
In view of the high homology between DAGL-α and DAGL-β, we assessed the activity of LEI105 also on native DAGL-β. To this end, we tested whether our ABP MB064 was also able to label DAGL-β. We incubated MB064 with membranes from mock and hDAGL-β transfected HEK293T cells. MB064 labeled a protein at the expected molecular weight of DAGL-β, which was not present in the control membranes, or in S443A-hDAGL-β transfected cells, in which the catalytic serine is replaced by alanine using site-directed mutagenesis (Figure 1F). Thus, our tailor-made probe can also detect DAGL-β. Labeling of hDAGL-β was inhibited by LEI105 with a pIC50 of 7.4 ± 0.07 (n=3) (supporting information). We confirmed the activity of LEI105 on human DAGL-β using a biochemical assay with para-nitrophenylbutyrate as a surrogate substrate (pIC50 of 8.1 ± 0.07, n=4; supporting information). Next, we used the MB064 ABP to profile endogenous DAGL-β activity in twelve tissues from wild-type and DAGL-β knock-out mice. Spleen tissue was found to display the highest DAGL-β activity. This activity could be inhibited by LEI105 (Figure 1G). Of note, we could not detect DAGL-β activity with MB064 in the brain.
To determine the mode of action (reversible versus irreversible) of LEI105, we performed an experiment in which human DAGL-α membranes were pre-incubated at the IC80 concentration of LEI105 or the irreversible DAGL inhibitor KT-109.15 The protein was separated from small molecule inhibitors via size exclusion column chromatography and remaining enzyme activity was visualized with MB064. No recovery of DAGL-α activity was found for KT-109, whereas MB064 labeled DAGL-α exposed to LEI105 with similar intensity to DMSO treated DAGL-α (Figure 1H). This indicates that LEI105 is a reversible inhibitor of DAGL-α.
Determining proteome-wide selectivity of LEI105 using comparative and competitive chemoproteomics
To investigate the selectivity of LEI105 in the mouse brain proteome we have used comparative and competitive ABPP with a broad-spectrum FP-based probe (TAMRA-FP) and the tailored DAGL-α ABP MB064. As a reference we first tested the widely used β-lactone-based DAGL-inhibitors THL and OMDM-188, two highly potent non-selective covalent and irreversible serine hydrolase inhibitors.8,14 In our experimental setup, both compounds blocked labeling of at least 4 serine hydrolases at concentrations as low as 1 μM, including ABHD6 and ABHD12, enzymes involved in 2-AG metabolism in the brain (Figure 2 A,B).
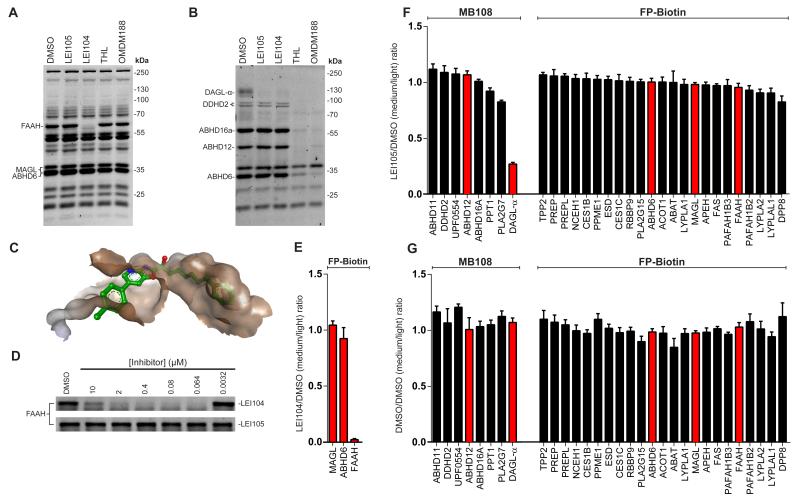
Figure 2.
Selectivity of LEI105. (a) Competitive ABPP in the mouse brain membrane proteome with LEI104 (10 μM), LEI105 (10 μM), OMDM188 (1 μM) and THL (1 μM) using TAMRA-FP (500 nM). (b) Same experiment as in (a) with THL based ABP MB064 (250 nM) showing excellent selectivity of LEI105 at 10 μM compared to THL and OMDM188. (c) One possible LEI105-like conformer in a previously reported co-crystal structure of FAAH, showing a steric clash between the toluyl group and FAAH. (d) Activity of LEI104 and LEI105 against FAAH in a concentration response experiment in the mouse brain membrane proteome using TAMRA-FP (500 nM, full gels are given in the supporting information) (e) Chemoproteomic competition between LEI104 (10 μM) and FP-biotin (5 μm) for FAAH, MAGL and ABHD6, showing that the chemoproteomic settings using FP-biotin are compatible with reversible α-ketoheterocycles. (f) MB108 and FP-biotin based chemoproteomic analysis of serine hydrolase activities in the mouse brain membrane proteome treated with LEI105 (10 μM). DAGL-α labeling is reduced to over 70%. (g) Chemoproteomic control experiment shows equal isotopic labeling and detection of peptides from MB108 and FP-biotin labeled serine hydrolases. Enzymes known to be directly involved in endocannabinoid biosynthesis and metabolism are highlighted in red. (n = 3-4 independent chemoproteomics experiments; Error bars represent ± s.e.m. of medium over light ratios of quantified peptides (minimum of two unique peptides per enzyme). Only unique proteins are given for MB108, all detected proteins including proteins overlapping between the probes are given in the supporting information. See supplementary data set 1 for complete proteomic data.
In our previous studies, we identified fatty acid amide hydrolase (FAAH) as a major off-target for LEI104. In stark contrast to LEI104, which inhibited FAAH labeling with a pIC50 of 7.8 ± 0.04 (n=3) (Figure 2D, supporting information), LEI105 did not block FAAH labeling up to a concentration of 10 μM (n=3) (Figure 2A, D). Of note, LEI105 did not inhibit labeling of any other band either, and we conclude that LEI105 is highly selective at least within the panel of serine hydrolases labeled by TAMRA-FP and MB064. To explain this remarkable selectivity of LEI105 over FAAH, we looked in detail into a previously published co-crystal structure of FAAH with OL-135 (pdb: 2WJ1 and 2WJ2).26 In silico modification of the heterocyclic part of OL-135, in the presence of the protein receptor, to a LEI105 like conformer revealed a steric clash of the toluoyl moiety of LEI105 with the substrate channel of FAAH, thereby possibly explaining the high selectivity of LEI105 over FAAH compared to its analog LEI104 (Figure 2C).
Since, the human serine hydrolase family contains approximately 200 family members,27 we wanted to examine the selectivity of LEI105 in the brain proteome in a broader and more detailed manner. To this end we adapted a semi-quantitative chemoproteomics protocol, which was previously applied to determine the selectivity profile of the irreversible inhibitor KT-109, using MB108, a biotinylated version of MB064 and the FP-probe (see supporting information for synthesis and characterization). This methodology allows for a more accurate quantification avoiding band overlap (as observed with gel-based assay) and enables screening over a broader range of specified serine hydrolases. We first validated equal isotopic labeling and detection of light and medium peptides from proteins targeted by both ABPs (Figure 2G). Next, we set out to investigate the selectivity of LEI105 in this chemoproteomic assay. LEI105 (10 μM, 30 min) did not reduce labeling of the detected serine hydrolases by more than 20%, except for DAGL-α (Figure 2F). In addition, we tested LEI105 in biochemical assays using recombinant human ABHD6 and MAGL and in a cannabinoid CB1 receptor radioligand displacement assay. We did not find any significant interaction (pKi < 5) with these proteins of the endocannabinoid system (supporting information). Together these results indicate that LEI105 is a highly selective DAGL-inhibitor.
LEI105 reduces 2-AG levels in Neuro2A cells
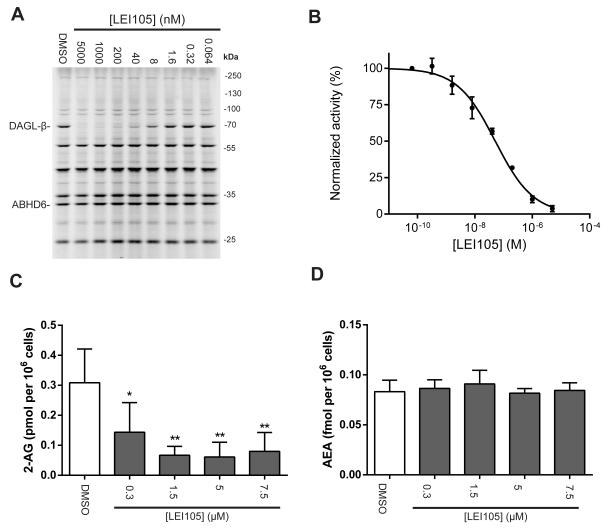
To test the cellular activity of LEI105, we used Neuro2A cells, which is a mouse neuroblastoma cell line known to express both DAGL-α and DAGL-β.28 First, we confirmed the presence of the mRNA transcripts of both DAGLs. Interestingly, we found that DAGL-β mRNA levels are ~128-fold higher, compared to DAGL-α, as determined by qPCR (supporting information). In line, DAGL-α protein activity was below the detection limit of our ABPP assay, whereas we detected a fluorescent band at the expected molecular weight of DAGL-β using MB064 (Figure 3A; supporting information for validation of DAGL-β labeling). LEI105 selectively reduced DAGL-β labeling in a concentration dependent manner with a pIC50 of 7.3 ± 0.07 (Figure 3A, B). Of note, no inhibition of ABHD6 or FAAH was observed (supporting information). Next, in a targeted lipidomics experiment we determined the effect of LEI105 on the cellular levels of 2-AG and anandamide. A concentration-dependent reduction of 2-AG was found in Neuro2A cells, whereas anandamide levels were unaffected (Figure 3C, D). To test the activity of LEI105 in a human cell line, we used human PC3 cells. LEI105 reduced 2-AG levels, but no change in substrate levels of DAGL was detected, which may suggest that DAG species are rapidly converted into other membrane constituents, such as phosphatidic acid by DAG-kinases (supporting information). Interestingly, arachidonic acid levels were reduced by LEI105 treatment of PC3 cells (supporting information). This may indicate that downstream metabolic pathways of 2-AG signaling are also affected and that the biosynthesis of 2-AG is the rate-limiting step. This is in line with a previously reported reduction of arachidonic acid levels in DAGL-α knock-out mice and by KT-109 in mouse liver.15, 5
Figure 3.
Cellular activity of LEI105. (a) Representative gel of concentration dependent inhibition of endogenous DAGL-β in vitro in the Neuro2A proteome as determined with ABP MB064. (b) Dose response curve of DAGL-β inhibition as determined with competitive ABP MB064 (± SEM, n = 3). (c) In situ treatment of Neuro2A cells (1h, 37 °C) dose dependently decreased basal 2-AG levels while keeping anandamide (AEA) levels (d) constant (mean ± SEM; n =4). Statistical analysis: 2-way ANOVA (*** = p < 0.001; ** = p < 0.01; * = p < 0.05 vs vehicle)
LEI105 reduces synaptic transmission in hippocampus
To demonstrate a physiological effect of the inhibition of 2-AG biosynthesis by LEI105, we focused our attention on the hippocampal slice preparation in which depolarization-induced suppression of inhibition (DSI) is thought to be mediated via 2-AG-induced activation of presynaptic cannabinoid CB1 receptors.29,30 Experimental evidence, which supports a role of DAGL-α in DSI, has been obtained through the use of genetically engineered mice that constitutively lack DAGL-α. DSI was absent in these DAGL-α knock-out mice, but not in their DAGL-β knock-out counter parts.5,10 Of note, anandamide levels were also reduced in these DAGL-α knock-out mice. Acute DAGL inhibition studies with chemical tools would, therefore, provide an additional line of evidence to verify the role of DAGL-α in DSI. Pharmacological intervention studies with the two highly potent non-selective DAGL inhibitors, THL and OMDM-188, however, have challenged the unequivocal role of the DAGLs in DSI. Multiple groups reported a suppression of DSI by THL and/or OMDM-188,31-36 whereas other groups did not find any inhibitory effect of THL and/or OMDM-188.37,38 The discrepancy between the genetic model and the pharmacological studies, led to the alternative hypothesis that 2-AG is released from pre-formed lipid stores instead of the ‘on demand’ production of 2-AG.39,40
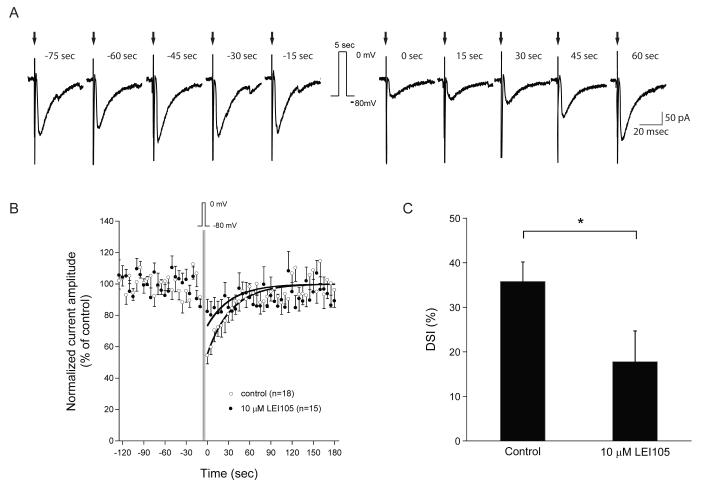
Here, we set out to investigate cannabinoid CB1 receptor-dependent DSI in CA1 pyramidal neurons in hippocampal slices with our selective DAGL inhibitor. Pre-incubation and continuous application of 10 μM LEI105 had no effect on the amplitude of the stimulus-evoked inhibitory post-synaptic currents (IPSC) (271 ± 22 pA) compared to control (276 ± 32 pA). In control conditions, a DSI response could be evoked which decayed back with an exponential time course (τ = 34 sec) to baseline levels. DSI was quantified as the reduction in IPSC amplitude averaged over the first 15 sec after its induction. DSI evoked in slices treated with LEI105 was smaller than DSI induced in control slices (18 ± 7%, n=15 and 36 ± 4%, n=18, respectively; p<0.05, Student’s t-test) (Figure 4). However, it is clear that DSI is not completely blocked in the presence of LEI105, which suggests that in addition to on demand 2-AG synthesis, the recruitment of 2-AG pools may contribute to DSI.40 Alternatively, other endocannabinoids (e.g. anandamide) may be involved. Of note, at 10 μM LEI105 was still selective in our experimental set up and did not displace 3H-CP55940 from the CB1 receptor (supporting information), a result that excludes direct antagonism of the CB1 receptor by LEI105. Thus, our data support the hypothesis of a major, but not exclusive, role of ‘on demand’ production of 2-AG, which is responsible for the cannabinoid CB1 receptor-mediated synaptic plasticity.33
Figure 4.
DSI is reduced in hippocampal slices treated with 10 μM LEI015. (a) Example of a typical whole-cell voltage-clamp recording of evoked inhibitory post-synaptic currents (IPSCs) in a CA1 pyramidal neuron under control conditions. IPSCs are evoked every 5 seconds, but for clarity, IPSCs shown here are representative traces recorded every 15 seconds. DSI is induced with a 5-sec duration depolarization from −80 mV to 0 mV and is observed as a marked and brief reduction of IPSC amplitude following the depolarization. (b) Averaged IPSC amplitude recorded every 5 seconds in CA1 neurons from vehicle-treated slices (DMSO) and from slices pre-incubated at least 30 minutes and in the continuous presence of LEI 105 (10 μM). The dark lines represent the single exponential fit of the recovery phase following the depolarizing step. (c) Initial DSI amplitude under control conditions (DMSO) and in the presence of LEI105 (10 μM). (* p < 0.05 vs vehicle)
In conclusion, we have applied comparative and competitive chemproteomics using a tailor made ABP (MB064) in combination with a broad-spectrum probe (TAMRA-FP) to identify and characterize the reversible DAGL inhibitor LEI105. This inhibitor was found to be an excellent tool to study the enzymatic role of DAGL-α in a neuronal setting and provides a critical counterpart to currently available DAGL knock-out mice. This was exemplified in Neuro2A cells (where LEI105 reduced 2-AG levels, but not AEA levels) and in intact hippocampal brain slices (where a clear suppression of DSI was observed in the presence of LEI105). Using this combination of activity-based probes in a chemoproteomic setting many lipases involved in the regulation of endocannabinoid levels (e.g. DAGL-α and β, ABHD6, ABHD12, MAGL and FAAH) in the brain are targeted with high affinity. This opens the door to broadly apply this methodology to identify and characterize not only covalent irreversible inhibitors which can potentially lead to toxicity and immunogenicity of covalent inhibitor-protein complexes and selectivity issues using reactive covalent warheads, but also reversible inhibitors that target DAGL and other enzymes involved in 2-AG biosynthesis and metabolism. These reversible inhibitors have the additional intrinsic advantage to enable a better control of partial inhibition of their target enzyme. This is especially important with respect to endocannabinoid signaling in the CNS, since complete blockade of cannabinoid CB1 receptor signaling in the CNS may lead to severe side effects.41 Thus, this feature can play an important factor in the development of clinical candidates to provide therapeutic solutions for diseases, such as obesity, related metabolic disorders and neuroinflammation, in which excessive 2-AG signaling or its metabolites play an important factor.
Supplementary Material
Acknowledgements
We thank the Dutch Research Council-Chemical Sciences (ECHO-grant: 711.014.009; MvdS), Leiden University, Faculty of Science (“Profiling Programme: Endocannabinoids”; MvdS, TH, VK), and the Chinese Scholarship Council (DH) for financial support.
Footnotes
Supporting Information Available
Experimental procedures, supporting figures, compound characterization and an appendix containing detailed proteomic data. This information is available free of charge via the internet athttp://pubs.acs.org.
References
- (1).Katona I, Freund TF. Annual review of neuroscience. 2012;35:529. doi: 10.1146/annurev-neuro-062111-150420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Science. 1992;258:1946. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- (3).Sugiura T, Kondo S, Sukagawa A, Nakane S, Shinoda A, Itoh K, Yamashita A, Waku K. Biochemical and biophysical research communications. 1995;215:89. doi: 10.1006/bbrc.1995.2437. [DOI] [PubMed] [Google Scholar]
- (4).Mechoulam R, Ben-Shabat S, Hanus L, Ligumsky M, Kaminski NE, Schatz AR, Gopher A, Almog S, Martin BR, Compton DR, Pertwee RG, Griffin G, Bayewitch M, Barg J, Vogel Z. Biochemical pharmacology. 1995;50:83. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- (5).Gao Y, Vasilyev DV, Goncalves MB, Howell FV, Hobbs C, Reisenberg M, Shen R, Zhang MY, Strassle BW, Lu P, Mark L, Piesla MJ, Deng K, Kouranova EV, Ring RH, Whiteside GT, Bates B, Walsh FS, Williams G, Pangalos MN, Samad TA, Doherty P. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:2017. doi: 10.1523/JNEUROSCI.5693-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Di Marzo V. Nature reviews. Drug discovery. 2008;7:438. doi: 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- (7).Blankman JL, Cravatt BF. Pharmacol Rev. 2013;65:849. doi: 10.1124/pr.112.006387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bisogno T, Howell F, Williams G, Minassi A, Cascio MG, Ligresti A, Matias I, Schiano-Moriello A, Paul P, Williams EJ, Gangadharan U, Hobbs C, Di Marzo V, Doherty P. J Cell Biol. 2003;163:463. doi: 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Reisenberg M, Singh PK, Williams G, Doherty P. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2012;367:3264. doi: 10.1098/rstb.2011.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tanimura A, Yamazaki M, Hashimotodani Y, Uchigashima M, Kawata S, Abe M, Kita Y, Hashimoto K, Shimizu T, Watanabe M, Sakimura K, Kano M. Neuron. 2010;65:320. doi: 10.1016/j.neuron.2010.01.021. [DOI] [PubMed] [Google Scholar]
- (11).Appiah KK, Blat Y, Robertson BJ, Pearce BC, Pedicord DL, Gentles RG, Yu XC, Mseeh F, Nguyen N, Swaffield JC, Harden DG, Westphal RS, Banks MN, O'Connell JC. Journal of biomolecular screening. 2014;19:595. doi: 10.1177/1087057113511111. [DOI] [PubMed] [Google Scholar]
- (12).Janssen FJ, Deng H, Baggelaar MP, Allara M, van der Wel T, den Dulk H, Ligresti A, van Esbroeck ACM, McGuire R, Di Marzo V, Overkleeft HS, van der Stelt M. J Med Chem. 2014;57:6610. doi: 10.1021/jm500681z. [DOI] [PubMed] [Google Scholar]
- (13).Bisogno T, Mahadevan A, Coccurello R, Chang JW, Allara M, Chen Y, Giacovazzo G, Lichtman A, Cravatt B, Moles A, Di Marzo V. British journal of pharmacology. 2013;169:784. doi: 10.1111/bph.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ortar G, Bisogno T, Ligresti A, Morera E, Nalli M, Di Marzo V. J Med Chem. 2008;51:6970. doi: 10.1021/jm800978m. [DOI] [PubMed] [Google Scholar]
- (15).Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF. Nature chemical biology. 2012;8:999. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Niphakis MJ, Cravatt BF. Annual review of biochemistry. 2014;83:341. doi: 10.1146/annurev-biochem-060713-035708. [DOI] [PubMed] [Google Scholar]
- (17).Heal WP, Dang TH, Tate EW. Chemical Society reviews. 2011;40:246. doi: 10.1039/c0cs00004c. [DOI] [PubMed] [Google Scholar]
- (18).Liu Y, Patricelli MP, Cravatt BF. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14694. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hoover HS, Blankman JL, Niessen S, Cravatt BF. Bioorg Med Chem Lett. 2008;18:5838. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Yang PY, Liu K, Ngai MH, Lear MJ, Wenk MR, Yao SQ. Journal of the American Chemical Society. 2010;132:656. doi: 10.1021/ja907716f. [DOI] [PubMed] [Google Scholar]
- (21).Baggelaar MP, Janssen FJ, van Esbroeck AC, den Dulk H, Allara M, Hoogendoorn S, McGuire R, Florea BI, Meeuwenoord N, van den Elst H, van der Marel GA, Brouwer J, Di Marzo V, Overkleeft HS, van der Stelt M. Angew Chem Int Ed Engl. 2013;52:12081. doi: 10.1002/anie.201306295. [DOI] [PubMed] [Google Scholar]
- (22).Fowler CJ. Fundamental & clinical pharmacology. 2006;20:549. doi: 10.1111/j.1472-8206.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- (23).Boger DL, Sato H, Lerner AE, Hedrick MP, Fecik RA, Miyauchi H, Wilkie GD, Austin BJ, Patricelli MP, Cravatt BF. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5044. doi: 10.1073/pnas.97.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Pedicord DL, Flynn MJ, Fanslau C, Miranda M, Hunihan L, Robertson BJ, Pearce BC, Yu XC, Westphal RS, Blat Y. Biochemical and biophysical research communications. 2011;411:809. doi: 10.1016/j.bbrc.2011.07.037. [DOI] [PubMed] [Google Scholar]
- (25).van der Wel T, Janssen FJ, Baggelaar MP, Deng H, den Dulk H, Overkleeft HS, van der Stelt M. Journal of lipid research. 2015;56:927. doi: 10.1194/jlr.D056390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Mileni M, Garfunkle J, DeMartino JK, Cravatt BF, Boger DL, Stevens RC. Journal of the American Chemical Society. 2009;131:10497. doi: 10.1021/ja902694n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20941. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Jung KM, Astarita G, Thongkham D, Piomelli D. Mol Pharmacol. 2011;80:60. doi: 10.1124/mol.110.070458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ohno-Shosaku T, Maejima T, Kano M. Neuron. 2001;29:729. doi: 10.1016/s0896-6273(01)00247-1. [DOI] [PubMed] [Google Scholar]
- (30).Wilson RI, Nicoll RA. Nature. 2001;410:588. doi: 10.1038/35069076. [DOI] [PubMed] [Google Scholar]
- (31).Hashimotodani Y, Ohno-Shosaku T, Kano M. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:1211. doi: 10.1523/JNEUROSCI.4159-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hashimotodani Y, Ohno-Shosaku T, Maejima T, Fukami K, Kano M. Neuropharmacology. 2008;54:58. doi: 10.1016/j.neuropharm.2007.06.002. [DOI] [PubMed] [Google Scholar]
- (33).Hashimotodani Y, Ohno-Shosaku T, Tanimura A, Kita Y, Sano Y, Shimizu T, Di Marzo V, Kano M. The Journal of physiology. 2013;591:4765. doi: 10.1113/jphysiol.2013.254474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Edwards DA, Zhang LH, Alger BE. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:8142. doi: 10.1073/pnas.0803558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhang LH, Wang MN, Bisogno T, Di Marzo V, Alger BE. Plos One. 2011;6 doi: 10.1371/journal.pone.0016305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Szabo B, Urbanski MJ, Bisogno T, Di Marzo V, Mendiguren A, Baer WU, Freiman I. J Physiol-London. 2006;577:263. doi: 10.1113/jphysiol.2006.119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Edwards DA, Kim J, Alger BE. J Neurophysiol. 2006;95:67. doi: 10.1152/jn.00813.2005. [DOI] [PubMed] [Google Scholar]
- (38).Min R, Testa-Silva G, Heistek TS, Canto CB, Lodder JC, Bisogno T, Di Marzo V, Brussaard AB, Burnashev N, Mansvelder HD. Journal of Neuroscience. 2010;30:2710. doi: 10.1523/JNEUROSCI.BC-3622-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Min R, Di Marzo V, Mansvelder HD. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2010;16:608. doi: 10.1177/1073858410373281. [DOI] [PubMed] [Google Scholar]
- (40).Alger BE, Kim J. Trends Neurosci. 2011;34:304. doi: 10.1016/j.tins.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Kunos G, Tam J. British journal of pharmacology. 2011;163:1423. doi: 10.1111/j.1476-5381.2011.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.